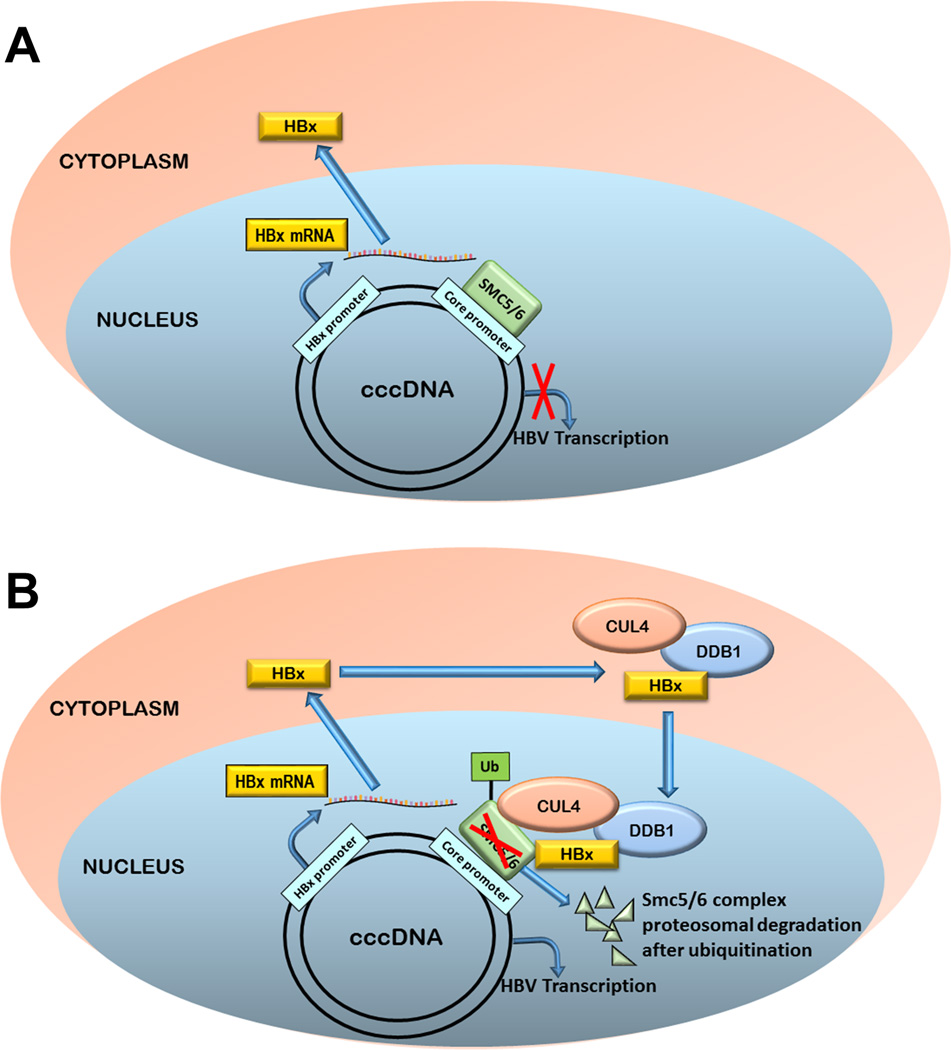
Hepatitis B virus (HBV) is a hepatotropic virus causing acute and chronic hepatitis. It consists of a partially double-stranded circular DNA genome which, upon entry into the nucleus of hepatocyte, gets converted into an episomal covalently closed circular (ccc) DNA (1). The cccDNA minichromosome serves as a template for transcription of all the four viral mRNAs by length, which are translated into seven proteins. One baffling small regulatory HBV protein is the X protein (HBx). Because of its pleiotropic functions as a viral transactivator, HBx has been a point of interest in the field for long. HBx has been reported to play an important role in regulating cccDNA function but the mechanism remains obscure. One of the best documented interactions of HBx with a host factor is with the DNA-damage binding protein 1 (DDB1) (2). However, the link between the HBx-DDB1 nexus and cccDNA regulation has yet to be established. Recently, Decorsière et al uncovered an intriguing finding showing that HBx enhances cccDNA transcription by promoting the degradation of a host factor, the structural maintenance of chromosomes (Smc)5/6 complex, through DDB1-associated ubiquitin-proteasome system (Fig.1) (3). [Decorsiere A, et al. Hepatitis B virus X protein identifies the Smc5/6 complex as a host restriction factor. Nature 2016; 531: 386-389.]
Figure 1. Schematic representations of HBx-mediated HBV cccDNA transcription via degradation of Smc5/6 complex.
(A) Shows the initial binding of SMC5/6 complex with cccDNA blocks major HBV mRNA transcription (precore mRNA, pregenomic RNA, and surface mRNAs) but not HBx mRNA transcription and protein expression. (B) Shows the initially expressed HBx binds to the DDB1-CUL4 complex in the cytoplasm. The entire HBx-DDB1-CUL4 complex is then transported to the nucleus and targets cccDNA-bound SMC5/6 complex for ubiquitin-dependent proteosomal degradation, by which relieves the transcription repression on cccDNA and allows entire viral gene expression.
In the Ubiquitin-proteasome system, the E3-Ub ligase provides substrate specificity and catalyzes ubiquitination of the lysines on the protein substrate for proteasomal degradation. Cullin-Ring E3 ligase complex 4A (CRL4) consists of CUL4 as the E3 ubiquitin ligase that has been shown to interact with HBx via the adaptor protein DDB1. CUL4-DDB1 have been known to target several viral gene products, suggesting an important involvement of the ubiquitin-proteasome system in virus maintenance within the cell. In this paper, Decorsière et al set out to identify the host target protein that is being degraded by CUL4-DDB1-HBx complex.
By fusing TAP-tagged HBx to wild-type DDB1 or mutant DDB1 (binding site for E3-ligase complex mutated and hence, no ubiquitination) and using tandem affinity purification and co-immunoprecipitation strategies, they were able to recover all the subunits of Smc5/6 complex in the presence of the mutant DDB1, suggesting that the interaction of HBx to DDB1 stabilizes the complex and allows it to interact with the target proteins, Smc5/6 complex (consisting of Smc5, Smc6 and Nse1-4 subunits). By transfecting a plasmid with luciferase reporter gene driven by HBV enhancer I, they showed increased reporter activity and reduced Smc5/6 protein levels in the presence of HBx. Also, they could pull down Smc5/6 complex with the transfected reporter plasmids or HBV episomal DNA but not with the chromosomally integrated ones and the binding was reduced in the presence of HBx. Thus, Decorsière et al concluded that Smc5/6 is a host antiviral factor that associates with cccDNA and inhibits its transcription. However, HBx can target Smc5/6 complex for degradation through ubiquitination to promote cccDNA transcription.
Smc complexes manage multiple chromosomal activities (4). Literature shows the involvement of Smc5/6 complex in many homologous recombination events including DNA repair and sister chromatid recombination; replication in normal and rDNA containing chromosome and in chromosome topology. SUMOylation by Nse2 subunit has been one of the reported mechanisms. However, researchers in this field strongly believe that there is more to the story about Smc’s physiological role and this paper adds by identifying Smc5/6 complex as a host antiviral transcriptional repressor.
One of the many merits of this paper was the use of in vitro and in vivo HBV infection systems to express HBx at its physiological level (in the context of HBV replication), thereby reducing the possibility of artifacts caused by overexpression, though showing Western blots or other direct measurements of HBx under these conditions would have completed the picture. Furthermore, the utilization of chronic hepatitis B patient liver samples to experiment the phenomena of HBx-mediated degradation of Smc5/6 would have strongly reinforced the conclusions made in the paper and made it a more plausible target for therapeutic interventions.
Decorsière et al reiterates a previous study that HBx is an immediate early gene for initial HBV infection and required to be present all the time for virus persistence (5). The conclusion by Lucifora et al that HBx is not required by the virus to enter the host cell but is essential for HBV cccDNA transcription is seconded by Decorsière et al. In fact, Lucifora et al also showed that HBx is needed for viral transcription from cccDNA but not the integrated HBV genome, which is supported to some extent by Decorsière et al while stating that HBx relieves Smc-mediated transcriptional repression exclusively on an episomal genome. However, the above statement contradicts a number of previous studies with demonstrated stimulation of host chromosome transcription by HBx. For example, Xu et al reported that the transcomplementation of HBx could enhance the X-minus HBV transgene transcription in transgenic mice (6), which argues against the episome-specific transactivation effect of HBx. This paradox awaits further investigations.
Based on the conclusions of the paper it seems that, in the absence of HBx, Smc complex specifically silences the transcription from episomal viral or plasmid DNA, but not the integrated gene. However, there is lack of proper comparisons between episomal and non-episomal DNA binding by Smc5/6 in the same cells. Furthermore, there is a need to elucidate the mechanism that makes this Smc-mediated transcriptional suppression specific to episomal DNA.
Another important question that comes around as a follow up is the mechanism behind the silencing effect of Smc5/6 complex on cccDNA transcription and what enables X mRNA still to be transcribed once Smc5/6 is already loaded on cccDNA to repress the same. One plausible explanation is that the binding of Smc complex to cccDNA minichromosome causes chromatin remodeling or epigenetic modifications and subsequently inhibits the binding of host RNA polymerase II to the viral promoters except for X promoter. Mapping the binding sites of Smc complex on cccDNA would help to answer the question.
Many avihepadnaviruses do not encode X gene. Thus, the mechanism for their cccDNA transcription regulation is the next important question that needs to be answered. Smc5/6 homologs have been predicted to exist in all living microorganisms, from bacteria to humans including birds (4). Hence, do these avian homologs resemble in function as the mammalian Smc complex and if so, then what is the protein that makes up for X’s function in avihepadnaviruses? A possible explanation would be the presence of an entirely different mechanism for the regulation or use of a viral protein other than X to target Smc complex’s inhibition. Another possibility is the presence of a conserved X-like protein in avihepadnaviruses helping in the repression of the Smc5/6 complex-induced silencing of cccDNA transcription. In line with this, earlier studies have predicted an X-like protein encoding ORF (with or without canonical start codon) in avian viruses at the position similar to that of X gene of orthohepadnaviruses (7, 8).
This paper may also excite scientists’ interest in assessing the potential involvement of Smc complex in the transcriptional regulation of other viruses with episomal DNA, such as papillomaviruses, polyomaviruses, herpesviruses and many more, especially those with circular nuclear DNA genomes. While most of these viruses have a replicating episomal DNA, HBV cccDNA minichromosome doesn’t replicate and hence this might determine the effect of Smc on transcription from different viruses. Interestingly, many DNA viruses have episomal latency, and thus the possible involvement of Smc complex in maintaining the viral latency may be another area for investigation.
In summary, the discovery reported by Decorsière and colleagues (Fig 1.) adds another new but not the final chapter to the enigmatic story of HBx. It might have unlocked another mysterious door to a better understanding of host-virus interactions and a probable futuristic therapeutic prospect with a lot more yet to unearth.
Footnotes
Conflict of Interest
Dr. Guo received grants from Arbutus and Alios. He consults for Alios and owns stock in Arbutus.
References
- 1.Guo JT, Guo H. Metabolism and function of hepatitis B virus cccDNA: Implications for the development of cccDNA-targeting antiviral therapeutics. Antiviral Res. 2015;122:91–100. doi: 10.1016/j.antiviral.2015.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Slagle BL, Bouchard MJ. Hepatitis B Virus X and Regulation of Viral Gene Expression. Cold Spring Harb Perspect Med. 2016;6:a021402. doi: 10.1101/cshperspect.a021402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Decorsiere A, Mueller H, van Breugel PC, Abdul F, Gerossier L, Beran RK, Livingston CM, et al. Hepatitis B virus X protein identifies the Smc5/6 complex as a host restriction factor. Nature. 2016;531:386–389. doi: 10.1038/nature17170. [DOI] [PubMed] [Google Scholar]
- 4.Uhlmann F. SMC complexes: from DNA to chromosomes. Nat Rev Mol Cell Biol. 2016;17:399–412. doi: 10.1038/nrm.2016.30. [DOI] [PubMed] [Google Scholar]
- 5.Lucifora J, Arzberger S, Durantel D, Belloni L, Strubin M, Levrero M, Zoulim F, et al. Hepatitis B virus X protein is essential to initiate and maintain virus replication after infection. J Hepatol. 2011;55:996–1003. doi: 10.1016/j.jhep.2011.02.015. [DOI] [PubMed] [Google Scholar]
- 6.Zhenming Xu TSBY, Wu Lanying, Madden Charles R, Tan Wenjie, Slagle Betty L, Ou Jing-hsiung. Enhancement of Hepatitis B Virus Replication by Its X Protein in Transgenic Mice Journal of Virology. 2002;76:2579–2584. doi: 10.1128/jvi.76.5.2579-2584.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang SF, Netter HJ, Hildt E, Schuster R, Schaefer S, Hsu YC, Rang A, et al. Duck hepatitis B virus expresses a regulatory HBx-like protein from a hidden open reading frame. J Virol. 2001;75:161–170. doi: 10.1128/JVI.75.1.161-170.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guo H, Mason WS, Aldrich CE, Saputelli JR, Miller DS, Jilbert AR, Newbold JE. Identification and characterization of avihepadnaviruses isolated from exotic anseriformes maintained in captivity. J Virol. 2005;79:2729–2742. doi: 10.1128/JVI.79.5.2729-2742.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]