SUMMARY
The eukaryotic RNA exosome is an essential and conserved 3’ to 5’ exoribonuclease complex that degrades or processes nearly every class of cellular RNA. The nuclear RNA exosome includes a nine-subunit non-catalytic core that binds Rrp44 (Dis3) and Rrp6 subunits to modulate their processive and distributive 3’ to 5’ exoribonuclease activities, respectively. Here, we utilize an engineered RNA with two 3’ ends to obtain a crystal structure of an eleven-subunit nuclear exosome bound to RNA at 3.1 Å. The structure reveals an extended RNA path to Rrp6 that penetrates into the non-catalytic core, contacts between the non-catalytic core and Rrp44 that inhibit exoribonuclease activity, and features of the Rrp44 exoribonuclease site that support its ability to degrade 3’ phosphate RNA substrates. Using reconstituted exosome complexes, we show that 3’ phosphate RNA is not a substrate for Rrp6, but is readily degraded by Rrp44 in the nuclear exosome.
Graphical Abstract
eToc
Zinder et al. use an engineered 3’-3’ RNA to obtain a high-resolution structure of the yeast eleven-subunit nuclear RNA exosome with RNA bound to its exoribonucleases, Rrp6 and Rrp44. The structure reveals substrate specificities for Rrp44 and Rrp6 and features of the nine-subunit non-catalytic core that modulate their activities.
INTRODUCTION
A small fraction of most eukaryotic genomes code for proteins, but nearly all of it is transcribed (ENCODE Project Consortium, 2012). Of the non-coding RNA that is transcribed, some serves a functional purpose (e.g. tRNAs, sn(o)RNAs, rRNAs, and lncRNAs such as HOTAIR, XIST) (Cech and Steitz, 2014; Rinn and Chang, 2012) but much of it is degraded co- and post-transcriptionally by the RNA decay machinery. The RNA exosome, a conserved multi-protein complex, processes and degrades RNA in the 3’ to 5’ direction in eukaryotes (Houseley and Tollervey, 2009; Januszyk and Lima, 2014). Substrates of the nuclear RNA exosome include Cryptic Unstable Transcripts (CUTs) (Wyers et al., 2005), PROMoter uPstream Transcripts (PROMPTs) in humans (Preker et al., 2008), hypomodified tRNAs (Kadaba et al., 2004), sn(o)RNAs (Allmang et al., 1999a), and ribosomal RNAs (Mitchell et al., 1997). Mutations that result in loss of function of the RNA exosome have been identified in hematopoietic malignancies (Chapman et al., 2011) and neurodegenerative disorders (Fabre and Badens, 2014), underscoring its physiological importance.
The RNA exosome core (Exo9) includes nine subunits, a six-membered ring formed by the RNase PH-like proteins Rrp45, Rrp41, Rrp43, Rrp46, Rrp42, and Mtr3 (the PH-like ring), that is capped by a three-membered ring of S1 and K-homology (KH) domain containing proteins Rrp4, Rrp40, and Csl4 (the S1/KH ring). The structure of Exo9 from human revealed a donut-shaped architecture with a prominent central channel large enough to accommodate single-stranded RNA (Liu et al., 2006). Both yeast and human Exo9 cores are devoid of catalytic activity (Dziembowski et al., 2007; Liu et al., 2006).
In the budding yeast Saccharomyces cerevisiae, the cytoplasmic RNA exosome includes the Exo9 core and the processive exoribonuclease Rrp44 (Figure 1A) to form a ten-component complex (Exo1044). In the nucleus, Exo1044 associates with the distributive exoribonuclease Rrp6 (Figure 1A) to form an eleven-subunit complex (Exo1144/6). Protein cofactors such as Rrp47, Mpp6, and the Trf/Air/Mtr4 poladenylation (TRAMP) complex associate with the nuclear exosome to target particular RNA substrates or to alter its activities (Houseley and Tollervey, 2009; Januszyk and Lima, 2014; LaCava et al., 2005; Lubas et al., 2012; Wasmuth and Lima, 2012a; Wyers et al., 2005). All protein subunits of Exo1044 are essential for cell viability in yeast (Brouwer et al., 2000; Dziembowski et al., 2007; Mitchell et al., 1997), while deletion of Rrp6 results in temperature sensitivity, slow-growth, and RNA processing defects (Allmang et al., 1999a; 1999b; Briggs et al., 1998).
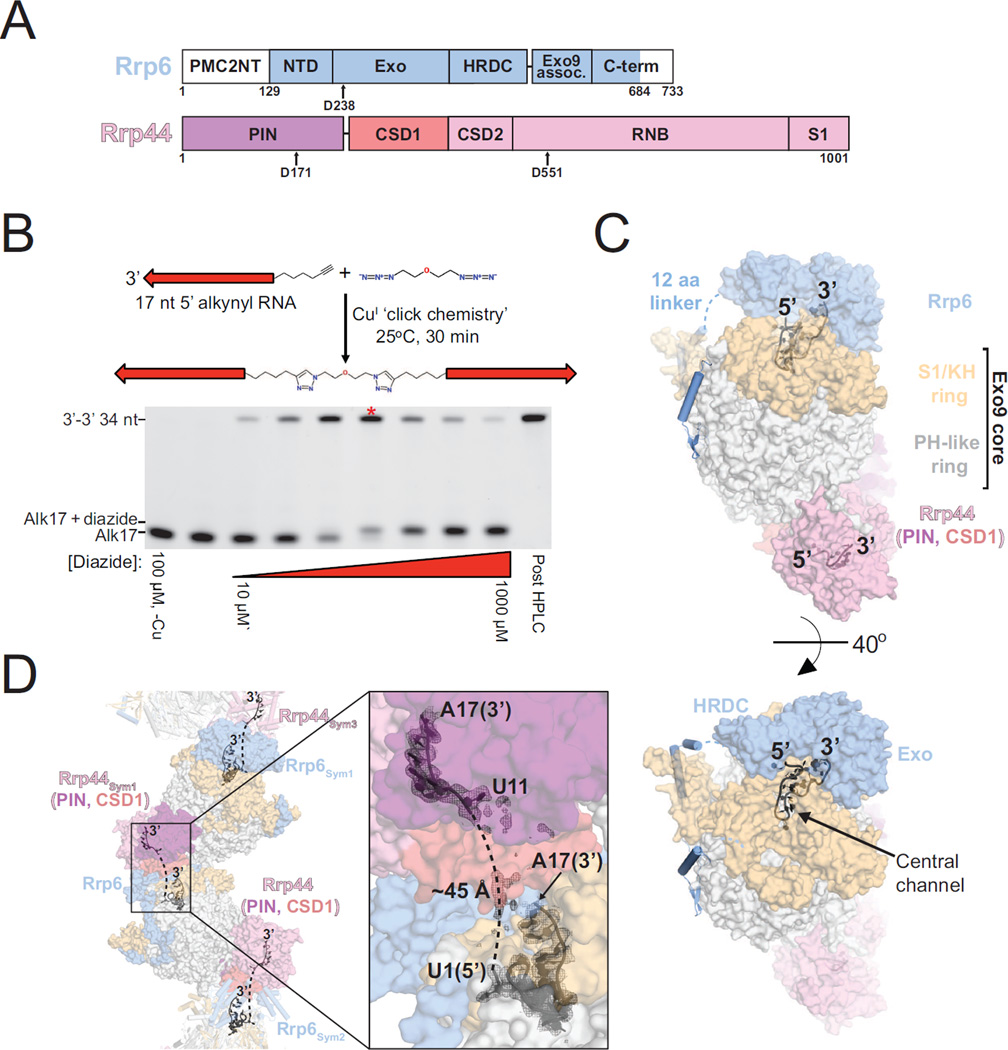
Figure 1. Architecture of Exo1144/6 Bound to 3’-3’ RNA.
(A) Domain schematics of Rrp6 and Rrp44. The shaded areas indicate the amino acid boundaries used for crystallization. Active site aspartates that were mutated to asparagine for binding assays and crystallization are indicated.
(B) Schematic for synthesis of a 34 nt RNA molecule with two 3’ ends. RNA is represented as an arrow pointing from 5’ to 3’. Gel is 15% acrylamide TBE-urea and RNA is visualized using SYBR gold stain. The lane for the reaction containing 2:1 [RNA]:[diazide] is indicated with an asterisk.
(C) Surface representation of Exo1144/6 bound to 3’-3’ 34 nt RNA. RNA is shown as a cartoon with 3’ and 5’ ends of the model indicated.
(D) RNA path in Exo1144/6/3’-3’ RNA crystals. Mesh is a simulated annealing 2Fo-Fc omit map contoured at 1.0 σ. The dashed lines indicate the most likely path connecting 5’ ends of the RNA fragments. See also Figure S1. RNB, RNase II catalytic domain; PIN, PilT N-terminal domain
Previous efforts to understand how Exo9 contributes to the RNA decay activities of the exosome revealed that the central channel was essential, that Rrp6 and Rrp44 activities are attenuated or altered when bound to the RNA exosome core, that RNA passes through the PH-like and S1/KH rings to access the Rrp44 active site, that RNA engages the S1/KH proteins to access the Rrp6 active site, and that RNA path(s) to the catalytic subunits are partially overlapping (Bonneau et al., 2009; Drazkowska et al., 2013; Makino et al., 2013b; Malet et al., 2010; Wasmuth and Lima, 2012b; Wasmuth et al., 2014).
A crystal structure of Exo1044 bound to a stem-loop RNA with a 3’ polyU30 single-stranded extension at 2.8 Å (PDB 4IFD) showed RNA passing through the Exo9 central channel and a conformation for Rrp44 that differed from that observed in the absence of RNA (Bonneau et al., 2009; Makino et al., 2013b). A similar conformation of Rrp44 was observed in a recent 2.7 Å structure of Exo1044 bound to an ∼110 amino acid fragment of the cytoplasmic RNA Exosome cofactor Ski7 and RNA (PDB 5JEA [Kowalinski et al., 2016]). Negative stain EM studies on Exo1044 at 20–25 Å resolution (Liu et al., 2014) revealed two conformations for Rrp44, one in the absence of RNA or in the presence of short RNAs or structured RNAs with short 3’ overhangs (termed the direct access or channel-independent Rrp44 conformation) and the other conformer observed in the presence of RNAs with single-stranded 3’ ends long enough to span the Exo9 central channel (termed the through channel or channel-dependent Rrp44 conformation). When Exo1044 was degrading a structured RNA that included a long single-stranded 3’ extension, sub-classification of EM data suggested that both conformations co-exist. More recent cryo-EM structures captured these same two conformations for Exo1044 in complex with Ski7, now at 4.2 Å for the channel-independent Rrp44 conformation and 5.8 Å for the channel-dependent Rrp44 conformation (Liu et al., 2016).
A structure of Exo9 bound to Rrp6 (Exo106) and 24 nt polyA RNA at 3.35 Å resolution (PDB 4OO1) revealed an RNA path to Rrp6 that partially overlapped with the channel-dependent RNA Exo1044 path (Wasmuth et al., 2014), confirming predictions based on biochemical and genetic methods (Wasmuth and Lima, 2012b). Based on mutational analysis, a similar path was posited for the human Exo106 complex (Wasmuth et al., 2014). A twinned crystal structure reported at 4.6 Å resolution (69.5% data completeness) of the nuclear exosome bound to the cofactor Rrp47 and 18 nt AU-rich RNA (PDB 5C0W) revealed its overall architecture, a speculative path for RNA bypassing Rrp6, and 12 nt of RNA bound to the channel-independent conformation of Rrp44 (Makino et al., 2015). Atomic resolution features of the nuclear exosome containing both Rrp6 and Rrp44, including the RNA path to Rrp6 and elements that stabilize the channel-independent conformation of Rrp44, remain unclear due to the nominal resolution of structures to date.
Here we report a crystal structure at 3.1 Å resolution of a nuclear Exo1144/6 complex with RNA bound to both Rrp6 and Rrp44 active sites. The structure reveals extensive RNA contacts to Rrp6 and the S1/KH subunits, Rrp44 in a channel-independent conformation bound to RNA, elements of the Exo9 core that interact with Rrp44 in the channel-independent conformation and inhibit its activities, and details within the Rrp44-RNA active site that provide a structural basis for its ability to degrade 3’ phosphorylated RNA substrates.
RESULTS
3’-3’ RNA and the nuclear RNA exosome structure
High-resolution structures of the nuclear exosome have not been achieved thus far, possibly because the Rrp44 and Rrp6 catalytic subunits are dynamic when not bound to RNA substrates. As both catalytic subunits require a 3’ end to bind RNA in their respective active sites, we employed CuI-catalyzed alkyne-azide cycloaddition, or ‘copper click chemistry,’ to synthesize an RNA with two 3’ ends that we hypothesized would enable simultaneous capture of both catalytic subunits (Figures 1B and S1). After surveying different RNA lengths, we obtained crystals that diffracted to 3.1 Å resolution of the S. cerevisiae nuclear exosome (Exo1144exo-endo-/6exo-ΔNΔC) in complex with an RNA composed of two 17 nt segments linked by 19 carbon to carbon, nitrogen, or oxygen bonds (Figures 1A, 1B, and S1). A molecular replacement solution was obtained using S. cerevisiae Exo106 (PDB 4OO1) and Rrp44 from the Rrp44-Rrp41-Rrp45 complex (PDB 2WP8) as search models (Bonneau et al., 2009; Wasmuth et al., 2014), and iterative rounds of building and refinement resulted in an Rwork/Rfree of 0.201/0.249 with good stereochemistry for a structure that includes 3731 amino acids and 24 nucleotides of RNA (Table 1).
Table 1.
Crystallographic data and refinement statistics
| Exo11Rrp44/Rrp6/RNA | |
|---|---|
| Data collection | |
| X-ray Source | APS NE-CAT 24IDC |
| Space group | P212121 |
| Cell dimensions | |
| a, b, c (Å) | 140.7, 212.3, 218.2 |
| α β γ (°) | 90.0, 90.0, 90.0 |
| Wavelength (Å) | 0.9795 |
| Resolution (Å) | 106–3.1 (3.21–3.1) |
| Rmerge | 0.111 (0.534) |
| I/σI | 14.3 (2.8) |
| CC1/2 | 0.994 (0.495) |
| Completeness (%) | 98.8 (97.0) |
| Redundancy | 3.6 (3.0) |
| Wilson B factor (Å2) | 66.2 |
| Refinement | |
| Resolution (Å) | 106–3.1 |
| No. reflections observed | 423414 |
| No. unique reflections | 106037 |
| Rwork/Rfree | 0.201/0.249 |
| No. atoms | 30252 |
| Protein | 29377 |
| RNA | 502 |
| Ligands | 92 |
| Water | 280 |
| Average B-factors | |
| Protein | 80 |
| RNA | 107 |
| Ligands | 96 |
| Water | 52 |
| R.m.s deviations | |
| Bond lengths (Å) | 0.002 |
| Bond angles (°) | 0.43 |
| Ramachandran plot | |
| % favored | 94.6 |
| % allowed | 5.2 |
| % outliers | 0.16 |
| Molprobity | |
| Clashscore/Percentile | 5.7/100th |
| MolProbity Score/Percentile | 1.68/100th |
One crystal was used. Highest resolution shell is shown in parenthesis.
The overall architecture of the complex (Figure 1C) reveals Rrp6 and Rrp44 at opposite ends of the Exo9 central channel with Rrp6 bound to RNA above the S1/KH ring of the Exo9 core and Rrp44 below the PH-like ring bound to RNA in the channel-independent Rrp44 conformation. This conformation of Rrp44 resembles that observed in previous structures (Bonneau et al., 2009; Liu et al., 2014; 2016; Makino et al., 2015), while the Rrp6 conformation resembles that observed for Exo106 bound to polyA (Wasmuth et al., 2014), a position that is rotated away from the S1/KH ring in a model of the nuclear exosome structure with Rrp6 bound to the cofactor Rrp47 and not bound to RNA (PDB 5C0W [Makino et al., 2015]). Distance constraints and electron densities suggest that the 3’-3’ RNA traverses between Rrp6 in one Exo11 complex to Rrp44 in an adjacent complex (symmetry mate) rather than traveling through the central channel of a single complex (Figure 1D).
Contacts between the Exo9 core and Rrp44 contribute to Rrp44 activities
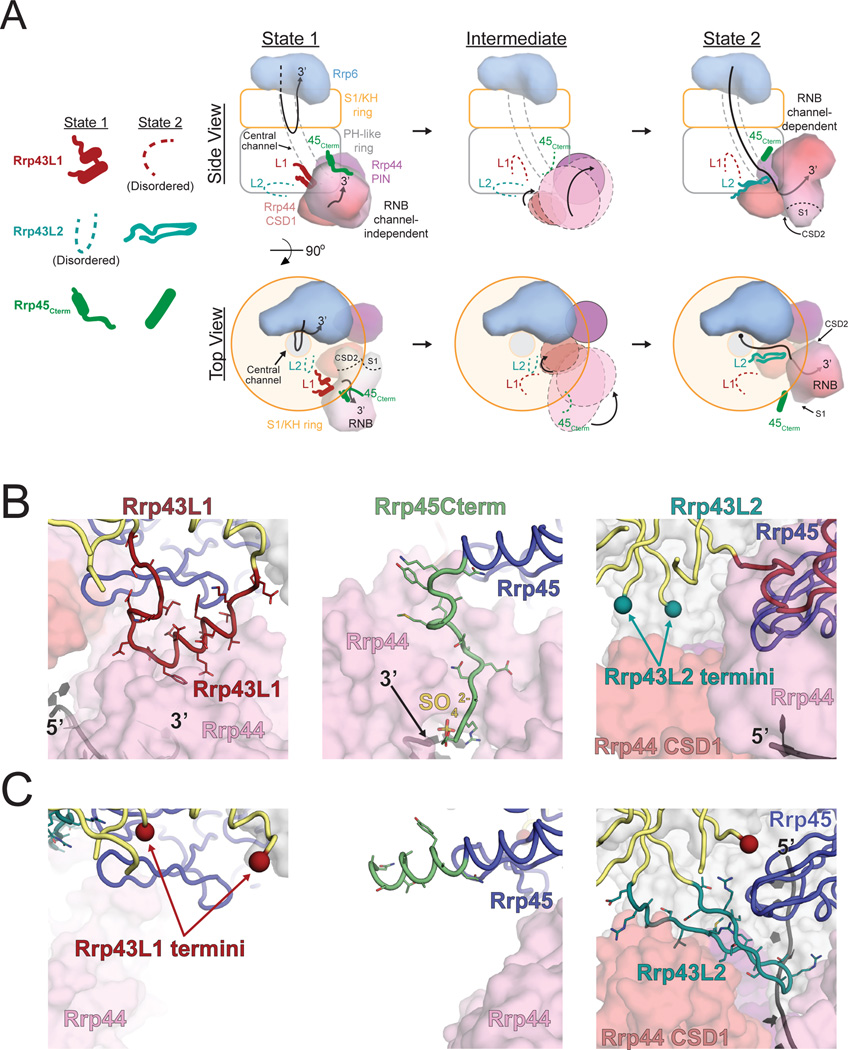
Previous structural studies observed a channel-independent conformation of Rrp44 in association with the Exo9 core (Liu et al., 2014; 2016; Makino et al., 2015), but none were determined at sufficient resolution to reveal all of the features of the Exo9 core proteins that stabilize this conformation. We now observe that Rrp43 residues 100–120 (Rrp43L1) form a loop followed by a pair of helices that contact Rrp44 near the point of RNA ingress (Figures 2A, 2B and S2). Inspection of electron density in a previous structure revealed the presence of Rrp43L1 however it was not modeled (PDB 5C0W [Makino et al., 2015]) (Figure S3). In addition, the C-terminus of Rrp45 (Rrp45Cterm) forms an extended strand that contacts Rrp44 near the 3’ end of its bound RNA in our structure as previously observed (Bonneau et al., 2009; Liu et al., 2016; Makino et al., 2015).
Figure 2. Features in the Non-Catalytic Exo9 Core Contact the RNA Channel-Independent and Channel-Dependent Conformations of Rrp44.
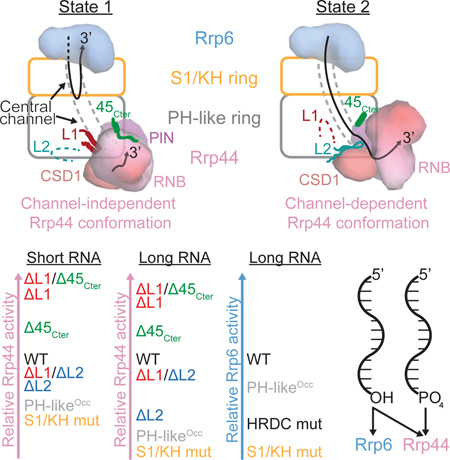
(A) Illustration of the channel-independent and channel-dependent conformations of Rrp44 along with associated features in the Exo9 core from the side and top views. The center panel depicts a theoretical transition between the two states in which the Rrp43L1, Rrp43L2 and Rrp45Cterm are disordered. Surface representations of Rrp44 were generated using the model from this work and PDB 4IFD for the channel-independent and channel-dependent conformations, respectively.
(B and C) Rrp43L1, Rrp43L2 and Rrp45Cterm features associated with the channel-independent (B) and channel-dependent (C) conformations of Rrp44 from this work and PDB 4IFD, respectively. Order/disorder boundaries of Rrp43L1 and Rrp43L2 are indicated by colored spheres. See also Figures S2 and S3.
Comparison to the Exo1044-RNA channel-dependent structure (PDB 4IFD) reveals that contacts between Rrp44 and the Rrp45Cterm and Rrp43L1 element are lost. While the Rrp43L1 element becomes disordered, the Rrp45Cterm forms a helix and does not contact Rrp44 in the channel-dependent Exo1044 (Liu et al., 2016; Makino et al., 2013b) or in the absence of Rrp44 in the Exo106 structure (Wasmuth et al., 2014) (Figures 2 and S2). Instead, different contacts are established between the Exo9 core and Rrp44 including Rrp43 residues 251–270 (Rrp43L2), which form a beta hairpin that bridges the Rrp44 cold-shock domain 1 (CSD1), catalytic domain, and RNA (Figures 2A, 2C and S2) (Liu et al., 2016; Makino et al., 2013b). By contrast, this feature of Rrp43 is disordered in our structure (Figures 2B and S2). A recent structure of Exo1144/Ski7N-term with Rrp44 bound in the through-channel conformation (PDB 5JEA) observed that Rrp43L2 was disordered (Kowalinski et al., 2016), but the authors noted that Rrp44 CSD1 is rotated relative to other RNA-through-channel structures due to crystal lattice contacts in that region, suggesting that ordering of Rrp43L2 may be dependent on rotation of Rrp44 CSD1 to that observed in the channel-dependent Rrp44 conformation. The unique contacts between Rrp44 and the Exo9 core in channel-dependent and channel-independent Rrp44 conformations suggested they might contribute differentially to the ability of Rrp44 to bind and degrade RNA.
We reconstituted exosome complexes containing deletions of Rrp43L1, Rrp43L2, and/or deletion of Rrp45Cterm (Figures 3A and 4A) to test the importance of these elements in RNA binding and degradation assays. Mutant Rrp43 and Rrp45 behaved as wild-type with respect to formation of Exo9 complexes and stable association with Rrp44. Consistent with Rrp43L1 and Rrp43L2 contacts to Rrp44 and not Rrp6, no differences in activity were observed for Rrp6 when associated with Exo9 containing the Rrp43 mutants (Figure S4C and S4D).
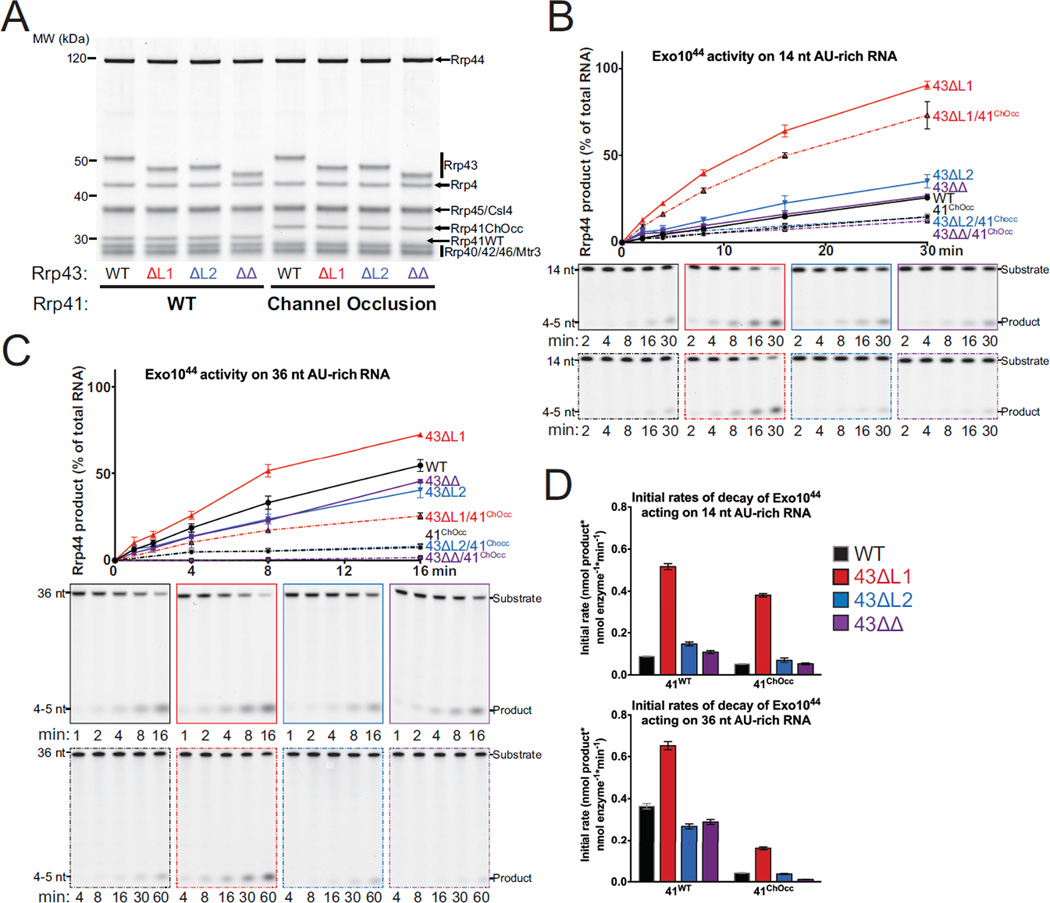
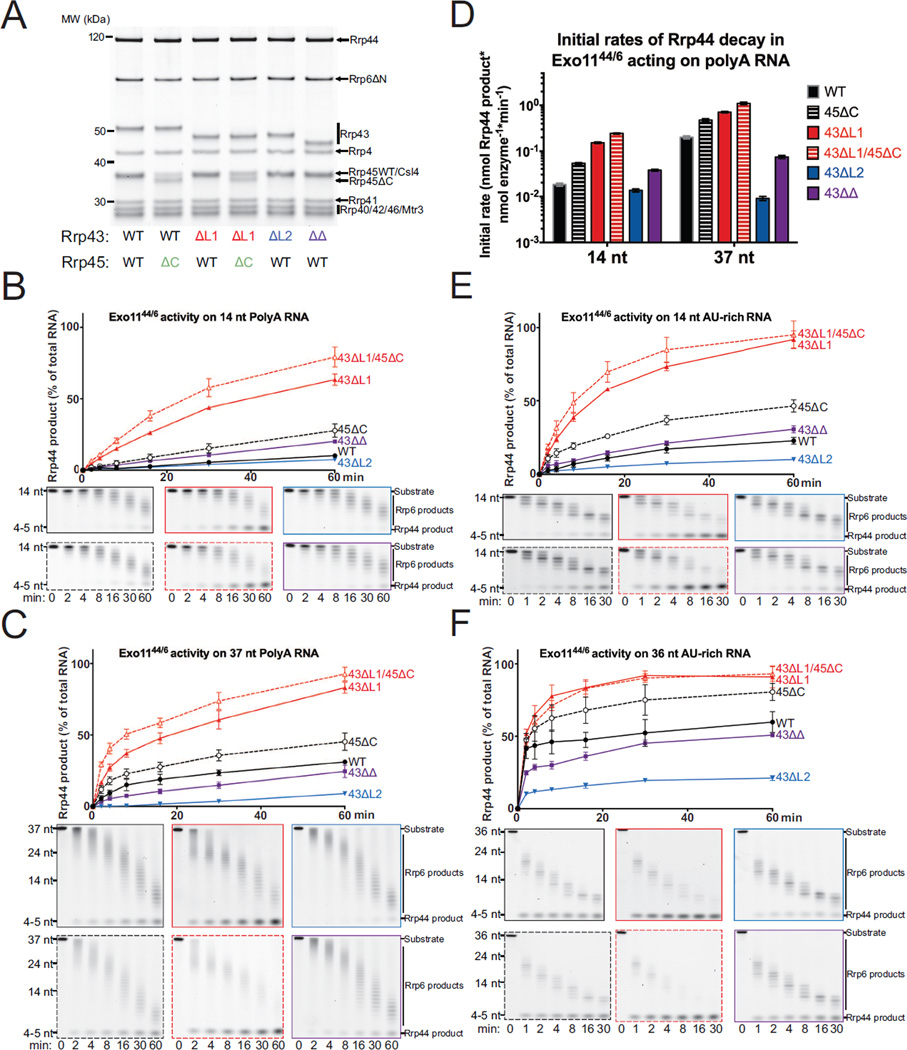
Figure 3. Exo1044 Activity on AU-rich RNAs is Enhanced by Deletion of Rrp43L1.
(A) SDS-PAGE analysis of reconstituted Exo1044 complexes containing Rrp43 mutants. ‘41ChOcc’ refers to a loop insertion mutant in Rrp41 that occludes the central channel.
(B and C) WT and mutant Exo1044 activity on 14 nt (B) and 36 nt (C) 5’ FAM AU-rich RNA substrates. The experiment was performed in triplicate and quantification of mean values is shown with error bars at ±1 standard deviation. Representative polyacrylamide TBE-urea gels are shown below.
(D) Quantification of initial rates of decay of Exo1044 complexes on 14 nt and 36 nt 5’ FAM AU-rich RNA substrates from the experiments in panels (B) and (C). Mean values of the triplicate experiments are plotted with error bars at ±1 standard deviation. See also Figure S4.
Figure 4. Deletion of Features Associated with the Channel-Independent Conformation of Rrp44 Enhance its Activity in Exo1144/6 complexes.
(A) SDS-PAGE analysis of reconstituted Exo1144/6 complexes containing mutant Rrp43 and/or Rrp45 proteins.
(B and C) WT and mutant Exo1144/6 activity on 14 nt (B) and 37 nt (C) 5’ FAM polyA RNA substrates. The experiment was performed in triplicate and quantification of mean values is shown with error bars at ±1 standard deviation. Representative polyacrylamide TBE-urea gels are shown below.
(D) Initial rates based on decay products generated by Rrp44 in Exo1144/6 complexes and free Rrp44 on short (14 nt) and long (37 nt) 5’ FAM polyA RNA substrates from the experiments in panels (B) and (C). Mean values are shown with error bars at ±1 standard deviation.
(E and F) WT and mutant Exo1144/6 activity on 14 nt (E) and 36 nt (F) AU-rich RNA substrates. The experiment was performed in triplicate and quantification of mean values is shown with error bars at ±1 standard deviation. Representative polyacrylamide TBE-urea gels are shown below. See also Figure S4.
We first assessed the impact of deleting Rrp43L1 and Rrp43L2 in the context of Exo1044 complexes in degradation of long (36/37 nt) and short (14 nt) AU-rich and polyA RNA substrates (Figures 3 and 4A). Deleting Rrp43L1 increased Rrp44 activity on short and long AU-rich RNA substrates. In contrast, deletion of Rrp43L2 had no detectable impact on degradation of short AU-rich RNA although a measurable defect was observed in degrading the long AU-rich substrate (Figure 3). Interestingly, deletion of Rrp43L2 suppressed the stimulation observed for Rrp43L1 deletion in degradation assays with short and long RNA when Rrp43L1 and Rrp43L2 deletions were combined. A similar pattern was observed for degradation of short polyA RNA for the respective Exo1044 complexes, however none of these complexes exhibited activity on 37 nt polyA RNA in comparison to Rrp44 (Figure S4A). The latter observation is consistent with previous results showing that the Rrp6 protein was required for Rrp44 to bind and degrade polyA RNA in nuclear exosome complexes (Wasmuth and Lima, 2012b).
Binding and decay assays with the AU-rich RNA substrates were also performed using Exo1044 complexes that contained a loop-insertion mutant of Rrp41 (41ChOcc) that partially occludes the central channel (Wasmuth and Lima, 2012b). We observed that binding and initial rate of decay were inhibited when the channel was occluded, supporting the conclusion that Rrp44 remains dependent on RNA passing through the central channel when bound to Exo9 cores containing the mutated Rrp43 subunit (Figures 3B, 3C, 3D and S4B). Notably, the dependence on the central channel was more pronounced for the 36 nt AU-rich RNA than the 14 nt RNA (Figures 3 and S4B), consistent with previous observations that Rrp44 conformational changes from channel-independent to the channel-dependent are promoted by incubation with long (>24 nt) single-stranded RNAs under non-degrading conditions (Liu et al., 2014).
We next assessed reconstituted wild-type and mutant nuclear Exo1144/6 complexes (Figure 4A) in degradation assays using short (14 nt) or long (37/36 nt) polyA (Figures 4B, 4C, and 4D) and AU-rich RNAs (Figures 4E and 4F). For Exo1144/6 complexes, deletion of Rrp43L1 increased Rrp44 activity for each RNA substrate tested, especially evident for the short RNA substrates. In contrast to that observed for Exo1044 complexes, deletion of Rrp43L2 resulted in a measurable loss of Rrp44 activity for the 14 nt AU-rich while deletion of Rrp43L2 had little impact on degradation of the 14 nt polyA RNA. Deletion of Rrp43L2 reduced Rrp44 activity on both 36 nt AU-rich and 37 nt polyA substrates. Consistent with results obtained using Exo1044 complexes, deletion of Rrp43L2 suppressed the stimulatory effect of removing Rrp43L1 while removal of Rrp43L1 largely suppressed the defects observed for Rrp43L2 deletion. Deletion of Rrp45Cterm tail resulted in a measurable increase in Rrp44 activity, albeit less than that observed for deletion of Rrp43L1. Deletion of both the Rrp45Cterm tail and Rrp43L1 appeared additive. The presence of Rrp6 activities in these assays could lead to artifacts in analysis of Rrp44 decay products, so we also added catalytically inactive Rrp6 (Rrp6exo-) to Exo1044 complexes in degradation assays with the polyA RNA substrates (Figure S4A). Results from these assays are qualitatively similar to those obtained for catalytically active Exo1144/6 complexes. Deletion of Rrp43L1 and Rrp45Cterm stimulated degradation, deletion of Rrp43L2 inhibited degradation, and deleting both Rrp43L1 and Rrp43L2 suppressed the effects observed for the individual deletions.
Taken together, these results suggest that Exo9 subunit contacts between the Rrp43L1, Rrp45Cterm and channel-independent Rrp44 conformation inhibit Rrp44 activity in Exo1044 and Exo1144/6, while Rrp43L2 contacts to the channel-dependent Rrp44 conformation promote activity in Exo1144/6. Longer RNA substrates presumably enter the eleven-subunit exosome via its central channel, and the greater dependence on Rrp43L2 for degrading longer RNA is consistent with Rrp43L2 contacting RNA as it exits the central channel to Rrp44 (Liu et al., 2016; Makino et al., 2013b). Rrp43L2 appears dispensable for degradation of shorter RNA substrates, but enhanced degradation of 14 nt RNA by deletion of Rrp43L1 and Rrp45Cterm was suppressed by deletion of Rrp43L2. Although other models are possible (see Discussion), we believe the simplest explanation is that Rrp44 is stabilized in the channel-independent conformation by contacts to Rrp43L1 and Rrp45Cterm, and that Rrp44 is stabilized in the channel-dependent conformation by contacts to Rrp43L2. Because deletion of Rrp43L1 enhances activity in a manner dependent on Rrp43L2, we posit that Rrp44 conformational changes are important for degrading both long and short substrates independent of whether RNA enters the exosome through the central channel (long RNA) or via direct binding to Rrp44 (short RNA). It remains unclear if this could be a regulated feature of the RNA exosome, or if it serves to inhibit Rrp44 activities when RNA is not presented through the Exo9 central channel, the presumed route for most cellular RNAs (Drazkowska et al., 2013; Han and van Hoof, 2016; Wasmuth and Lima, 2012b).
3’ phosphate RNA is a Rrp44 substrate
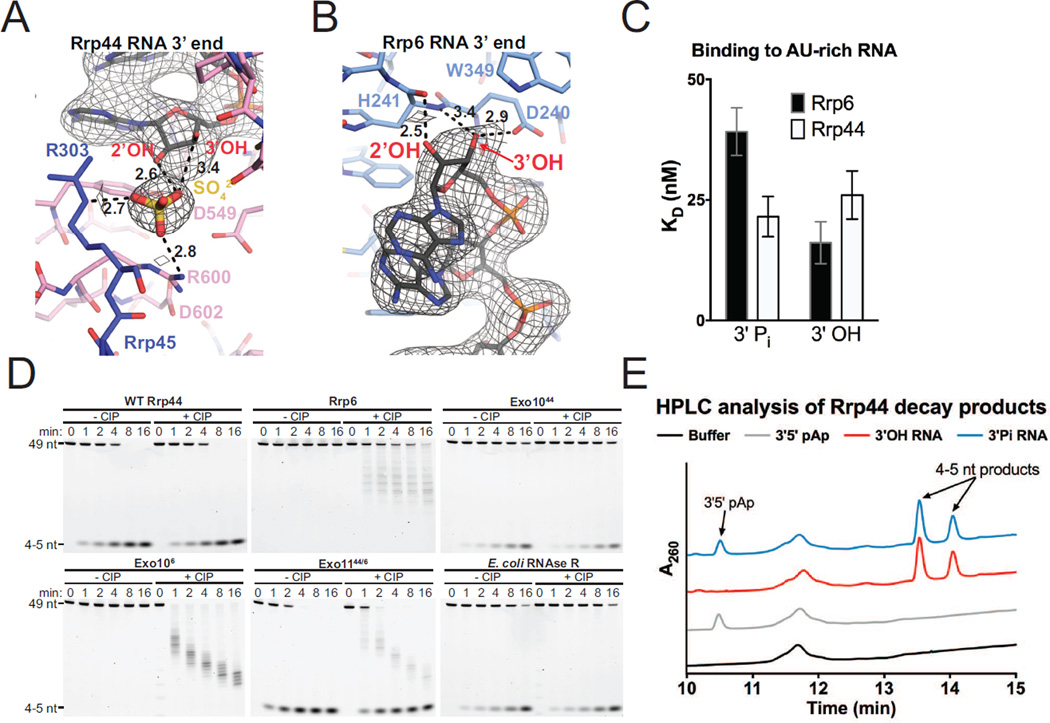
The RNA path for the last 7 nt of RNA in the Rrp44 active site in our structure resembles that previously observed in the structure of Rrp44 bound to polyA RNA (PDB 2VNU [Lorentzen et al., 2008]). A similar RNA path was modeled in the nuclear RNA exosome structure bound to an 18 nt AU-rich RNA (PDB 5C0W [Makino et al., 2015]), but its path diverges after the 7th nt and its 5’ end is modeled into densities that we assign to the Rrp43L1 loop near residue Asp105 (Figure S3). Inspection of electron densities proximal to the Rrp44 active site and 3’ end of RNA in our structure revealed a sulfate ion that was present in the crystallization medium (Figure 5A). The sulfate ion is positioned 2.6 Å or 3.4 Å from the 2’ and 3’ OH groups of the terminal base, respectively, and is situated in a positively charged pocket formed by Rrp44 Arg600 (2.8 Å) and Rrp45 Arg303 (2.7 Å).
Figure 5. 3’ Phosphate RNA is a Substrate of Rrp44 but not Rrp6.
(A and B) 3’ end of the RNA bound to Rrp44 (A) and Rrp6 (B). Mesh represents electron density from a simulated annealing 2Fo-Fc omit map contoured at 1.2 σ. Distances are shown in Ångstroms.
(C) Binding affinities of Rrp44 and Rrp6 for a 5’ FAM 3’ phosphate or 3’ OH 36 nt AU-rich RNA as measured by fluorescence anisotropy. Mean values are plotted with error bars at ±1 standard deviation of a triplicate experiment.
(D) Polyacrylamide TBE-urea gel analysis of RNA decay time-courses using a 5’ FAM 3’ phosphate AU-rich 49 nt RNA performed in the presence or absence of calf intestinal phosphatase (CIP) as indicated.
(E) Ion-pair reverse-phase HPLC analysis of Rrp44 decay products. Reactions contained Rrp44 and either 5’ FAM 3’ phosphate 49 nt AU-rich RNA, 5’ FAM 3’ OH 49 nt AU-rich RNA, 3’5’ pAp, or no substrate. Absorbance was monitored at 260 nm for detection of nucleotides and 4–5 nt RNA decay products. See also Figures S5 and S6.
The location of the sulfate ion in the Rrp44-RNA complex suggested that Rrp44 could accommodate a 3’ phosphate RNA in its active site. In contrast, inspection of the Rrp6 active site in this and previous structures of Rrp6 bound to AMP (PDB 2HBL) or Exo106 bound to polyA (PDB 4OO1) suggested that the Rrp6 active site could not accommodate a 3’ phosphate RNA because the RNA 2’ OH is 2.5 Å from the backbone carbonyl of His 241 while the 3’ OH is coordinated by side chains of Asp240 and backbone amide of His241 (Midtgaard et al., 2006; Wasmuth et al., 2014) (Figures 5B and S5A). The RNA and last nucleotide adopts a different configuration in the twinned structure of Rrp6 catalytic domain bound to polyU RNA modeled with partial occupancy (PDB 5C0Y) (Figure S5), however contacts to the RNA 3’ end in this structure would still appear to preclude accommodation of a 3’ phosphate RNA substrate (Makino et al., 2015).
Consistent with the aforementioned predictions, Rrp44 can bind and degrade a 3’ phosphorylated RNA (Figures 5C and 5D). By contrast, Rrp6 cannot degrade 3’ phosphate RNA, consistent with previous findings (Burkard and Butler, 2000; Domanski et al., 2016), despite its ability to bind it (Figures 5C and 5D). These respective activities persist when Rrp6 and Rrp44 are reconstituted in complexes as Exo1044, Exo106 or Exo1144/6 (Figure 5C). A question remained as to whether Rrp44 initiates degradation of the 3’ phosphate RNA substrate by first removing the 3’ phosphate of the terminal ribonucleotide, followed by subsequent hydrolysis and release of NMP products, or if it accommodates the 3’ phosphorylated terminal ribonucleotide in the active site, releasing 3’-5’ pNp and NMPs as products. To address this, Rrp44 decay products were analyzed by ion-pair reverse-phase HPLC. To validate our HPLC protocol, we first analyzed products of 3’ phosphate RNA decay by the Rrp44 homolog RNase R from E. coli and observed quantitative 3’-5’ pAp release, as previously observed using radiolabeled substrates and thin-layer chromatography (Vincent and Deutscher, 2006) (Figure S6C and S6D). Quantitative release of 3’-5’ pAp was also observed as a product of Rrp44 degradation for 3’ phosphorylated but not 3’ OH RNA substrates (Figures 5E, S6A, and S6B), indicating that cleavage by Rrp44 occurs at the 5’ phosphate of the terminal ribonucleotide, consistent with our structural model.
An attempt to disrupt recognition of 3’ phosphate RNA identified Rrp44 Arg600 and Asp602, as they are conserved in eukaryotic homologs (Figure S6E) and located near the sulfate ion and 3’ end of the RNA (Figure 5A). Differential binding defects were observed for the R600D/D602R mutant with 3’ OH RNA preferred over 3’ phosphate RNA (Figure S6F), however mutation of Arg600, alone or in combination with mutation of Asp602, resulted in diminished decay activities on both 3’ phosphate and 3’ OH RNA substrates. This is perhaps consistent with amino acid substitutions at these positions in RNaseR and RNaseII as both enzymes are able to degrade 3’ phosphorylated RNA substrates (Cheng and Deutscher, 2002) (Figure S6E and S6G). It is notable that Rrp44 homologs from human, including Dis3, Dis3L and DisL2, can also degrade 3’ phosphate RNA by releasing pNp, suggesting that this activity may be evolutionarily conserved (Lubas et al., 2013; Tomecki et al., 2010). Although we were unsuccessful identifying separation of function mutations to selectively diminish Rrp44 activities on 3’ OH or 3’ modified RNA substrates, future efforts along this line will be critical to determine if Rrp44 participates in the degradation of 3’ modified RNA substrates in vivo.
RNA path to Rrp6
The structure of Exo106 bound to polyA RNA (PDB 4OO1) revealed 6–8 nt of the 24 nt substrate with the RNA 3’ end anchored in the Rrp6 active site, the 5’ end extending toward the S1/KH ring, and the remaining RNA disordered (Wasmuth et al., 2014). An alternative model was proposed based on a twinned crystal structure of the isolated Rrp6 catalytic domain bound to 15 nt polyU RNA modeled with partial occupancy (PDB 5C0Y) suggesting that RNA would not contact the S1/KH ring, and that an 18 nt AU-rich RNA bypassed the Rrp6 active site altogether in a nuclear exosome complex (Makino et al., 2015) (Figure S5A).
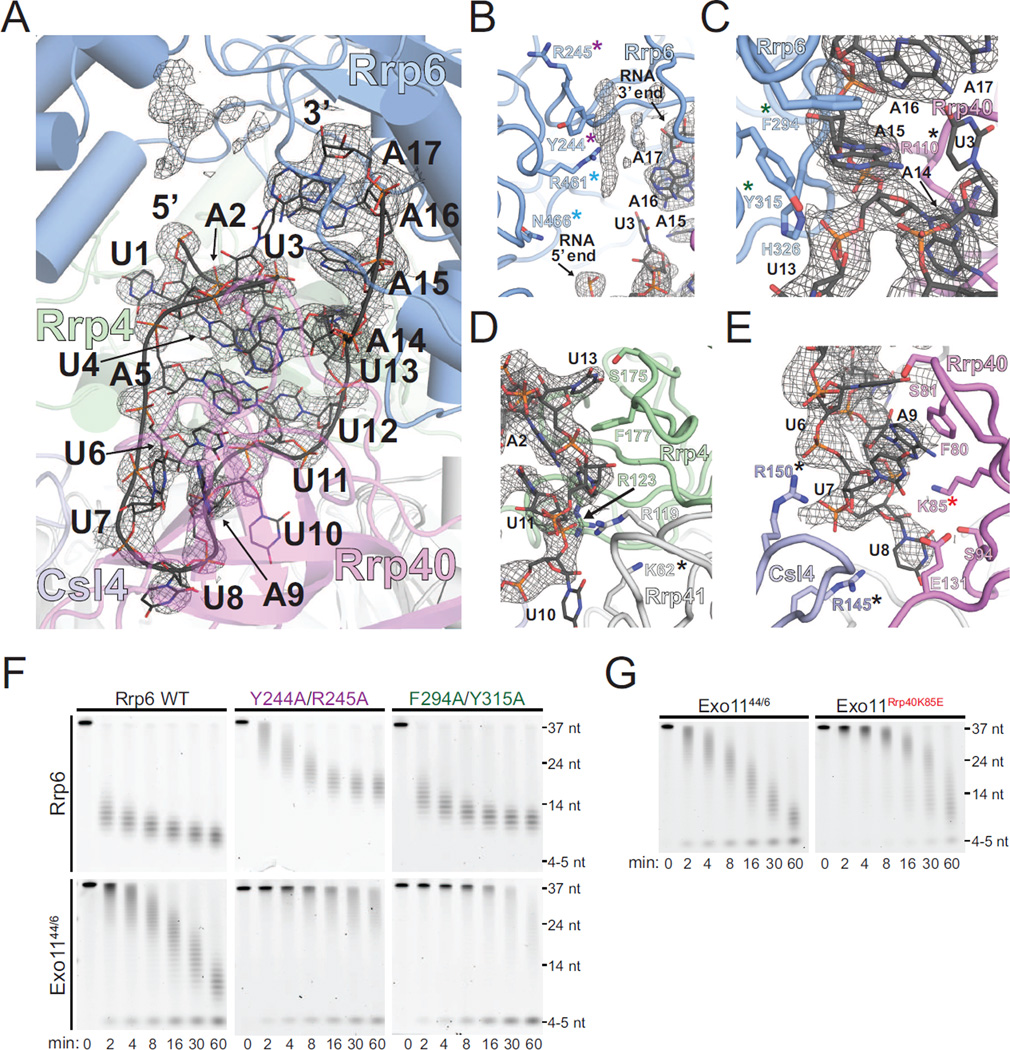
Electron densities are evident for 17 nt of RNA in our structure up to the position where the di-triazole linker connects the two segments in our symmetric 34 nt RNA substrate (Figure 6A). As observed in the Exo106 structure bound to polyA RNA (Wasmuth et al., 2014), the RNA 3’ end is anchored in the Rrp6 active site, extending toward and contacting the S1/KH ring (Figure 6A). After reaching the bottom of the S1/KH ring, the RNA is deflected and makes a U-turn back toward the Rrp6 helicase and RNase D C-terminal (HRDC) domain. Although not modeled, additional electron densities ∼20 Å from the 5’ end are observed between the HRDC and Exo domains of Rrp6 (Figures 1A, 1D, 6A and 6B), consistent with, but not identical to, a path proposed based on the twinned crystal structure of the Rrp6 catalytic domain bound to a 15 nt polyU RNA (Makino et al., 2015). Electron densities were not evident for the linker atoms, and RNA was not modeled in the HRDC/Exo domain channel due to poor definition of electron densities in this region.
Figure 6. RNA Path to Rrp6 in Exo1144/6/3’-3’RNA Structure.
(A to E) RNA contacts to Rrp6, the S1/KH proteins, and Rrp41. Mesh represents electron density from a simulated annealing 2Fo-Fc omit map contoured at 1.2 σ. Unmodeled density between the Exo and HRDC domains of Rrp6 is shown (B). Residues mutated in this and/or previous studies that contribute to the activities of the RNA exosome are highlighted with colored asterisks.
(F) Polyacrylamide TBE-urea gel analysis of decay of 5’ FAM 37 nt polyA RNA by mutants in Rrp6.
(G) Polyacrylamide TBE-urea gel analysis of decay of 5’ FAM 37 nt polyA RNA by WT Exo1144/6 and a mutant in the S1/KH protein Rrp40. See also Figure S7.
As anticipated for a non-specific exoribonuclease, protein-RNA interactions are dominated by contacts to the polyribonucleotide backbone and non-specific stacking with the nucleobases of the RNA chain. Contacts between the last 3 nt of RNA (A17-15) and Rrp6 are similar to those described for Exo106 bound to polyA RNA (Wasmuth et al., 2014), however the paths diverge at the fourth nt (Figure S5A). Instead, the nucleobase of A14 stacks on Rrp40 Arg110 while the A14 phosphate is coordinated by Rrp6 Tyr315 and His326 (Figure 6C). The U13 nucleobase is stacked on Rrp4 Phe177 and directed toward Rrp4 Ser175 (Figure 6D). The U12 and U11 nucleobases form base edge interactions with A2 and U4, respectively, with the U12 phosphate proximal to the Rrp41 Arg119 side chain and the U11 phosphate contacting Rrp4 Arg123. Density for the U10 nucleobase is not evident, but it points toward Rrp41 Lys62 (Figure 6D). The A9 nucleobase is sandwiched between Rrp40 Phe80 and the nucleobase of U7 while the A9 phosphate contacts Rrp40 Lys85 (Figure 6E). The U8 nucleobase contacts Rrp40 Ser94 above Glu131 before turning to enable contacts between the U8 phosphate and Csl4 Arg150, a side chain that is also in proximity to the U7 phosphate (Figure 6E). The U6 nucleobase is stacked between A9 and A5 and makes contacts to the Rrp40 Ser81 side chain and backbone amide (Figure 6E). The A5 nucleobase stacks between U6 and U11 with its base edge directed toward A14 and the U4 nucleobase stacks between A5 and the A2 ribose with it base edge directed toward U12. The path twists to point the U3 nucleobase toward that of A16, with the A2 nucleobase contacting Rrp4 Met137 with its edge directed toward U12. The final U1 nucleobase is sandwiched between Rrp4 Leu138 and Rrp6 Tyr430.
Previous mutational analysis reported that Rrp6 Phe294 and Tyr315 did not play a role in Rrp6 mediated decay (Makino et al., 2015), although these residues contact RNA in both the Exo106 and present structure (Wasmuth et al., 2014). Indeed, F294A/Y315A diminished Rrp6 activities when associated with Exo9 (Exo106) or in the nuclear exosome (Figures 6F, S7A, and S7C). Consistent with a path that guides RNA between the HRDC and Exo domains of Rrp6, Y244A and R245A substitution in Rrp6 reduced degradation activity in the exosome and as a free enzyme as noted previously (Makino et al., 2015) (Figures 6F and S7C). Rrp6 Arg461 and Asn466 are just below Tyr244 and Arg245, and R461V and N466D substitutions resulted in altered degradation patterns for polyA RNA in an eleven-subunit exosome complex or when combined with Rrp40 K85E (see below; Figure S7B and S7D).
RNA contacts occur deep in the S1/KH ring with Csl4 Arg145 and Arg150 side chains contacting the phosphate backbone at U7 and U8, while Rrp40 Arg110 buttresses the A14 nucleobase (Figures 6C and 6E). Consistent with these contacts, prior analysis revealed that mutation of these conserved residues diminishes both Rrp6 and Rrp44 activities (Wasmuth et al., 2014). In addition, insertions between residues 63 and 64 of Rrp41 also disrupt Rrp6 and Rrp44 activity (Wasmuth and Lima, 2012b), consistent with occluding the RNA path to Rrp6 near U10 (Figure 6D). Among several unique contacts observed in our structure are those at the deepest point of RNA ingress to the S1/KH ring between the conserved Lys85 of Rrp40 (Liu et al., 2006) and the RNA backbone phosphate of A9 (Figures 6E and S7D). Of note, Lys85 lies just below that last nucleotide modeled in the channel-dependent RNA bound exosome structure (Makino et al., 2013b). Consistent with contacts observed in our structure and predicted contacts in the channel-dependent structure, Rrp40 K85E decreased both Rrp6 and Rrp44 activities, supporting a role in coordinating RNA via overlapping but distinct paths as RNA is guided to Rrp6 or Rrp44 (Wasmuth and Lima, 2012b; Wasmuth et al., 2014) (Figures 6G and S7D).
DISCUSSION
The strategy employed here to satisfy both Rrp6 and Rrp44 exoribonuclease sites with a single RNA containing two 3’ ends enabled determination of a high-resolution structure of the nuclear RNA exosome, and it may prove useful in characterizing other RNA exosome complexes in association with additional nuclear cofactors.
Association of Rrp6 and Rrp44 with the essential non-catalytic Exo9 core attenuates or alters their activities. Previous studies suggested that attenuation is due in part to sequestration of their active sites relative to the apo enzymes by imposing a requirement that RNA must pass into the S1/KH ring to be degraded by Rrp6 or through the entire central channel to be degraded by Rrp44 (Drazkowska et al., 2013; Makino et al., 2013a; 2013b; Malet et al., 2010; Wasmuth and Lima, 2012b; Wasmuth et al., 2014). Using the high-resolution structure presented here, extensive interactions were revealed between the RNA and S1/KH ring of central channel on its way to the Rrp6 active site. While other RNA paths can be envisioned, especially for structured RNAs, mutational analysis presented here and elsewhere suggests that amino acid residues along this path contribute to both Rrp6 and Rrp44 activities, supporting the existence of overlapping yet distinct RNA paths that guide substrates to the respective catalytic subunits (Makino et al., 2015; Wasmuth and Lima, 2012b; Wasmuth et al., 2014) (Figure 6). We also resolved a feature of the Exo9 core, namely Rrp43L1, which together with Rrp45Cterm interact with the channel-independent Rrp44 conformation (Figures 2A, 2C, and S2A through S2C) and inhibit Rrp44 activity. We found that deleting these features enhanced Rrp44 activity on long and short RNAs, and that activities remained dependent on the Exo9 central channel, especially for longer RNA substrates (Figures 3D, 3C, 4, and S4B).
One alternative explanation for enhanced activity in the absence of Rrp43L1 is that Rrp43L1 is highly acidic and contacts Rrp44 near the RNA point of ingress. As such, it might create an electrostatic barrier that inhibits binding to the direct access or channel-independent Rrp44 conformation. However, this model predicts that binding and decay of RNA by the Rrp43L1 deletion mutant should not be affected by a channel occlusion nor should its activities be suppressed by deletion of Rrp43L2, which is not the case (Figures 3 and S4B). We propose a model in which Exo9 elements that bind and stabilize the channel-independent Rrp44 conformation (Rrp43L1 and Rrp45Cterm) compete with Exo9 elements that bind and stabilize the channel-dependent Rrp44 conformation (Rrp43L2). Since deletion of Rrp43L2 suppresses the activities gained by deleting Rrp43L1 on both long and short RNA substrates, we propose that conformational changes to the channel-dependent Rrp44 conformation are required for the most efficient degradation of long and short RNA substrates regardless of whether they pass through the Exo9 central channel (Figure 2A).
A recent study in S. cerevisiae showed that expressing a sole copy of Rrp44 containing mutations predicted to disrupt its channel-independent conformation was viable, though it exhibited a growth defect relative to the WT enzyme (Han and van Hoof, 2016). This strain accumulated certain RNA species such as hypomodified tRNAiMet and the rRNA 5’ externally transcribed spacer (5’ ETS). Intriguingly, expression of this mutant Rrp44 could suppress growth defects, but not certain 3’ processing defects, observed in strains harboring a channel occluding mutation in Rrp41 (Wasmuth and Lima, 2012b). Future studies comparing the biochemical activities of exosome complexes containing this mutant Rrp44 versus complexes containing the Rrp43 and Rrp45 deletions described here will be necessary to fully rationalize these observations, but they support the hypothesis that distinct Rrp44 conformations contribute differentially to the activities of the RNA exosome in vivo.
The models presented here and elsewhere provide a structural basis for the ability of Rrp44, but not Rrp6, to degrade 3’ phosphorylated RNA substrates. Although the in vivo relevance of Rrp44 activities on 3’ modified substrates remains unclear, the RNA exosome is involved in several processing steps during ribosomal RNA biogenesis, including trimming the 7S rRNA precursor to the 5.8S rRNA (Mitchell et al., 1997). In Δrrp6 strains, the 3’ end of this molecule is processed by the exosome to generate 5.8S with a ∼30 nt extension at the 3’ end (Allmang et al., 1999a; 1999b; Briggs et al., 1998), the approximate length required to span the Exo9 channel to the Rrp44 active site. This suggests a model where Rrp44 and Rrp6 cooperate to generate the mature 3’ end of 5.8S rRNA. Interestingly, a recent study identified that endoribonucleolytic cleavage of the 27SB precursor at the C2 site of internally transcribed spacer 2 (ITS2) is carried out by the Las1 complex, resulting in a 5’ OH end of the 25S fragment and a 2’, 3’-cyclic phosphate 3’ end of the 7S fragment (Gasse et al., 2015). While it is unclear if the 2’, 3’-cyclic phosphate 3’ end of the 7S precursor is acted upon by a phosphodiesterase or other enzyme prior to degradation, our structural model predicts that a 2’, 3’-cyclic phosphate 3’ end could be accommodated in the Rrp44 active site. Previous studies implicated the 5’-3’ decay pathway in degradation of 2’, 3’-cyclic phosphate tRNA introns (Wu and Hopper, 2014), but the observation that Rrp44 and related human enzymes can degrade 3’ phosphorylated RNA begs the question as to whether the RNA exosome also participates in the degradation of aberrant 3’ modified forms of RNA, or if this activity contributes to generating clean 3’OH RNA ends to promote post-transcriptional polyadenylation by TRAMP (LaCava et al., 2005; Wyers et al., 2005) or 3’ trimming by Rrp6.
EXPERIMENTAL PROCEDURES
Cloning, expression, and protein purification
Genes for the Rrp43 deletion mutants were synthesized by Life Technologies and were as follows: Rrp43ΔL1 has residues 101–119 of Rrp43 replaced with Gly-Ser-Gly-Ser-Gly-Ser, Rrp43ΔL2 has residues 254–270 deleted, and Rrp43ΔΔ contains both of these modifications. Rrp45ΔCterm (residues 1–290) has the C-terminal 14 amino acids deleted and was generated using standard PCR/restriction enzyme based cloning. Point mutants in Rrp6, Rrp40, and Rrp44 were introduced by PCR based site-directed mutagenesis. All mutants were verified by Sanger sequencing of the entire gene. Saccharomyces cerevisiae proteins were expressed as His6-Smt3 fusions in E. coli and reconstituted into complexes as described previously (Greimann and Lima, 2008) with slight modifications (see Supplemental Experimental Procedures).
3’-3’ RNA synthesis
RNA sequences can be found in Supplemental Experimental Procedures. Copper click chemistry was carried out via modification of published protocols (El-Sagheer and Brown, 2012; Paredes and Das, 2011). Click reactions (10–200 µL) contained 200 µM 5’ hexynyl oligonucleotide, 5 mM sodium ascorbate, 80 mM potassium phosphate pH 7.0 and 100 µM 1,5 diazido-3-oxapentane (Santa Cruz Biotechnology, 10 mM stock made in DMSO). Reactions were initiated with the addition 1/10 volume of 5 mM CuCl2, 5 mM Tris(3-hydroxypropltriazolemethyl) amine (Sigma-Aldrich) and incubated at 25°C for 30 minutes. Reactions were quenched by addition of EDTA to 10 mM, diluted to 500 µL with Tris-EDTA (TE) buffer and fractionated by DEAE chromatography as described in Supplemental Experimental Procedures. The concentration series for the 1,5 diazido-3-oxapentane titration shown in Figure 1B is 10, 25, 50, 100, 200, 400, and 1000 µM.
Crystallization of Exo11 in complex with 3’-3’ RNA
Exo11Rrp44 D171N/D551N, Rrp6(residues129–684)D238N at a concentration of 13–15 mg/mL (by Bradford assay) in 20 mM Tris-Cl pH 8.0, 100 mM NaCl, 0.1 mM MgCl2, 1 mM TCEP-HCl was mixed in a 1.0:0.9 protein:RNA molar ratio with 3’-3’ RNA and incubated for 2 hours on ice prior to mixing with well solution. Then, 0.8 µL of protein/RNA solution was mixed with 0.4 µL of well solution (100 mM sodium citrate pH 5.25, 7 mM NaMES pH 6.5, 6.5–7% PEG 3350, 175 mM (NH4)2SO4, 0.25-1 mM MgCl2; final pH 5.6) in 24-well hanging drop format, sealed and incubated at 4°C. Crystals appeared after 1.5 days and grew to their full size (∼350 µm in the largest dimension) after 1–2 weeks. Prior to freezing in liquid nitrogen, crystals were transferred to well solution containing 5% v/v glycerol, incubated for 1–5 min, then transferred in four steps with increasing glycerol to final well solution containing 30% v/v glycerol.
RNA binding and decay assays
RNA sequences and detailed protocols can be found in Supplemental Experimental Procedures. Briefly, for decay assays 1 nM enzyme and 10 nM 5’ 6-carboxyfluorescein (FAM) RNA substrate were incubated at 30°C in 20 mM Tris-Cl pH 8.0, 50 mM KCl, 0.5 mM MgCl2, 0.1 % v/v IGEPAL CO-630, 0.5 mM Tris(2-carboxyethyl)phosphine (TCEP)-HCl (RNA decay buffer) for the indicated times before the reaction was quenched and fractionated by urea-PAGE. RNA was visualized using a Typhoon FLA9500 instrument (GE). Binding reactions (20 µL) contained 30 nM 5’ FAM RNA and variable concentrations of catalytically inert enzymes, and were incubated at ∼22°C in the same buffer for 20 minute s prior to fluorescence anisotropy measurement using a Spectramax M5 plate reader (Molecular Devices).
Ion-pair reverse phase HPLC analysis of decay products
20 µL RNA decay reactions containing 1 µM enzyme and 10 µM 49 nt 5’ FAM AU-rich RNA (or mock substrates) were incubated for 25 minutes at 30°C in RNA decay buffer lacking IGEPAL CO-630. After incubation, reactions were quenched with the addition of EDTA to 10 mM, adjusted to 10 mM N(C4H9)4HSO4 (Sigma), and the products separated by HPLC using a Nova-Pak C18 60 Å 4 µM 3.9 x 150 mm reverse-phase column (Waters). The column was equilibrated with RP buffer A (50 mM potassium phosphate pH 7.0, 10 mM N(C4H9)4HSO4) and run at a flow rate of 1 mL/min at 40°C. Products were eluted with a linear gradient to 100% RP buffer B (RP buffer A containing 50% v/v acetonitrile) from t=5 to t=20 minutes (Figures 5E and S6A through S6D). UV absorbance was monitored at 260 nm for detection of nucleotides and small RNA decay products (Figures 5E, S6A, and S6C) and 490 nm for detection of the 5’ FAM label (Figure S6B and S6D).
Supplementary Material
Highlights.
-
-
Click chemistry produced a 3’-3’ RNA that was used to capture a nuclear RNA exosome
-
-
Contacts between the non-catalytic core and Rrp44 (Dis3) modulate its activity
-
-
Rrp44 but not Rrp6 degrades 3’ phosphate RNA in reconstituted exosomes
-
-
Extended RNA path to Rrp6 through the S1/KH ring of the non-catalytic core
Acknowledgments
Work here is based in part upon research conducted at NE-CAT beamlines (P41 GM103403, NIH NIGMS, S10 RR029205, NIH-ORIP HEI grant) which used resources of the Advanced Photon Source, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory (Contract No. DE-AC02-06CH11357). This research was supported in part by NIH/NIGMS R01GM079196 (C.D.L), R35GM118080 (C.D.L.), F31GM097910 (E.V.W.), and P30 CA008748 (NIH NCI-Cancer Center Support Grant). The content is solely the responsibility of the authors and does not represent the official views of the NIH. C.D.L is an Investigator of the Howard Hughes Medical Institute. We would also like to acknowledge members of the Lima laboratory and Malik Chaker-Margot for their suggestions and discussions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
J.C.Z. and C.D.L. designed experiments, analyzed the data, and wrote the manuscript. J.C.Z. performed all experiments and contributed to building the structural model. C.D.L. built the final structural model. E.V.W. provided reagents, designed experiments, and helped prepare the manuscript.
ACCESSION NUMBERS
Atomic coordinates and structure factors are deposited in the RCSB with accession code 5K36.
REFERENCES
- Allmang C, Kufel J, Chanfreau G, Mitchell P, Petfalski E, Tollervey D. Functions of the exosome in rRNA, snoRNA and snRNA synthesis. The EMBO Journal. 1999a;18:5399–5410. doi: 10.1093/emboj/18.19.5399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allmang C, Petfalski E, Podtelejnikov A, Mann M, Tollervey D, Mitchell P. The yeast exosome and human PM-Scl are related complexes of 3’-5’ exonucleases. Genes & Development. 1999b;13:2148–2158. doi: 10.1101/gad.13.16.2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonneau F, Basquin J, Ebert J, Lorentzen E, Conti E. The Yeast Exosome Functionsas a Macromolecular Cage to Channel RNA Substrates for Degradation. Cell. 2009;139:547–559. doi: 10.1016/j.cell.2009.08.042. [DOI] [PubMed] [Google Scholar]
- Briggs MW, Burkard KT, Butler JS. Rrp6p, the yeast homologue of the human PM-Scl 100-kDa autoantigen, is essential for efficient 5.8 S rRNA 3’ end formation. Journal of Biological Chemistry. 1998;273:13255–13263. doi: 10.1074/jbc.273.21.13255. [DOI] [PubMed] [Google Scholar]
- Brouwer R, Allmang C, Raijmakers R, van Aarssen Y, Egberts WV, Petfalski E, van Venrooij WJ, Tollervey D, Pruijn GJM. Three Novel Components of the Human Exosome. Journal of Biological Chemistry. 2000;276:6177–6184. doi: 10.1074/jbc.M007603200. [DOI] [PubMed] [Google Scholar]
- Burkard KT, Butler JS. A nuclear 3’-5’ exonuclease involved in mRNA degradation interacts with Poly(A) polymerase and the hnRNA protein Npl3p. Molecular and Cellular Biology. 2000;20:604–616. doi: 10.1128/mcb.20.2.604-616.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cech TR, Steitz JA. The Noncoding RNA Revolution— Trashing Old Rules to Forge New Ones. Cell. 2014;157:77–94. doi: 10.1016/j.cell.2014.03.008. [DOI] [PubMed] [Google Scholar]
- Chapman MA, Lawrence MS, Keats JJ, Cibulskis K, Sougnez C, Schinzel AC, Harview CL, Brunet J-P, Ahmann GJ, Adli M, et al. Initial genome sequencing and analysis of multiple myeloma. Nature. 2011;471:467–472. doi: 10.1038/nature09837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Z-F, Deutscher MP. Purification and characterization of the Escherichia coli exoribonuclease RNase R. Comparison with RNase II. J. Biol. Chem. 2002;277:21624–21629. doi: 10.1074/jbc.M202942200. [DOI] [PubMed] [Google Scholar]
- Domanski M, Upla P, Rice WJ, Molloy KR, Ketaren NE, Stokes DL, et al. Purification and analysis of endogenous human RNA exosome complexes. RNA. 2016;22:1467–1475. doi: 10.1261/rna.057760.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drazkowska K, Tomecki R, Stodus K, Kowalska K, Czarnocki-Cieciura M, Dziembowski A. The RNA exosome complex central channel controls both exonuclease and endonuclease Dis3 activities in vivo and in vitro. Nucleic Acids Res. 2013;41:3845–3858. doi: 10.1093/nar/gkt060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dziembowski A, Lorentzen E, Conti E, Séraphin B. A single subunit, Dis3, is essentially responsible for yeast exosome core activity. Nat Struct Mol Biol. 2007;14:15–22. doi: 10.1038/nsmb1184. [DOI] [PubMed] [Google Scholar]
- El-Sagheer AH, Brown T. Click Nucleic Acid Ligation: Applications in Biology and Nanotechnology. Acc. Chem. Res. 2012;45:1258–1267. doi: 10.1021/ar200321n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabre A, Badens C. Human Mendelian diseases related to abnormalities of the RNA exosome or its cofactors. Irdr. 2014;3:8–11. doi: 10.5582/irdr.3.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasse L, Flemming D, Hurt E. Coordinated Ribosomal ITS2 RNA Processing by the Las1 Complex Integrating Endonuclease, Polynucleotide Kinase, and Exonuclease Activities. Mol. Cell. 2015;60:808–815. doi: 10.1016/j.molcel.2015.10.021. [DOI] [PubMed] [Google Scholar]
- Greimann JC, Lima CD. Reconstitution of RNA exosomes from human and Saccharomyces cerevisiae cloning, expression, purification, and activity assays. Meth. Enzymol. 2008;448:185–210. doi: 10.1016/S0076-6879(08)02610-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, van Hoof A. The RNA Exosome Channeling and Direct Access Conformations Have Distinct In Vivo Functions. Cell Reports. 2016;16:3348–3358. doi: 10.1016/j.celrep.2016.08.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houseley J, Tollervey D. The Many Pathways of RNA Degradation. Cell. 2009;136:763–776. doi: 10.1016/j.cell.2009.01.019. [DOI] [PubMed] [Google Scholar]
- Januszyk K, Lima CD. The eukaryotic RNA exosome. Current Opinion in Structural Biology. 2014;24:132–140. doi: 10.1016/j.sbi.2014.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadaba S, Krueger A, Trice T, Krecic AM, Hinnebusch AG, Anderson J. Nuclear surveillance and degradation of hypomodified initiator tRNAMet in S. cerevisiae. Genes & Development. 2004;18:1227–1240. doi: 10.1101/gad.1183804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalinski E, Kögel A, Ebert J, Reichelt P, Stegmann E, Habermann B, Conti E. Structure of a Cytoplasmic 11-Subunit RNA Exosome Complex. Mol. Cell. 2016;63:125–134. doi: 10.1016/j.molcel.2016.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaCava J, Houseley J, Saveanu C, Petfalski E, Thompson E, Jacquier A, Tollervey D. RNA Degradation by the Exosome Is Promoted by a Nuclear Polyadenylation Complex. Cell. 2005;121:713–724. doi: 10.1016/j.cell.2005.04.029. [DOI] [PubMed] [Google Scholar]
- Liu J-J, Bratkowski MA, Liu X, Niu C-Y, Ke A, Wang H-W. Visualization of distinct substrate-recruitment pathways in the yeast exosome by EM. Nat Struct Mol Biol. 2014;21:95–102. doi: 10.1038/nsmb.2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J-J, Niu C-Y, Wu Y, Tan D, Wang Y, Ye M-D, Liu Y, Zhao W, Zhou K, Liu Q-S, et al. CryoEM structure of yeast cytoplasmic exosome complex. Cell Research. 2016:1–16. doi: 10.1038/cr.2016.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Greimann JC, Lima CD. Reconstitution, Activities, and Structure of the Eukaryotic RNA Exosome. Cell. 2006;127:1223–1237. doi: 10.1016/j.cell.2006.10.037. [DOI] [PubMed] [Google Scholar]
- Lorentzen E, Basquin J, Tomecki R, Dziembowski A, Conti E. Structure of the active subunit of the yeast exosome core, Rrp44: diverse modes of substrate recruitment in the RNase II nuclease family. Mol. Cell. 2008;29:717–728. doi: 10.1016/j.molcel.2008.02.018. [DOI] [PubMed] [Google Scholar]
- Lubas M, Chlebowski A, Dziembowski A, Jensen TH. Biochemistry and Function of RNA Exosomes. Enzymes. 2012;31:1–30. doi: 10.1016/B978-0-12-404740-2.00001-X. [DOI] [PubMed] [Google Scholar]
- Lubas M, Damgaard CK, Tomecki R, Cysewski D, Jensen TH, Dziembowski A. Exonuclease hDIS3L2 specifies an exosome-independent 3’-5’ degradation pathway of human cytoplasmic mRNA. The EMBO Journal. 2013;32:1855–1868. doi: 10.1038/emboj.2013.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino DL, Halbach F, Conti E. The RNA exosome and proteasome: common principles of degradation control. Nat. Rev. Mol. Cell Biol. 2013a;14:654–660. doi: 10.1038/nrm3657. [DOI] [PubMed] [Google Scholar]
- Makino DL, Baumgärtner M, Conti E. Crystal structure of an RNA-bound 11-subunit eukaryotic exosome complex. Nature. 2013b;495:70–75. doi: 10.1038/nature11870. [DOI] [PubMed] [Google Scholar]
- Makino DL, Schuch B, Stegmann E, Baumgärtner M, Basquin C, Conti E. RNA degradation paths in a 12-subunit nuclear exosome complex. Nature. 2015;524:54–58. doi: 10.1038/nature14865. [DOI] [PubMed] [Google Scholar]
- Malet H, Topf M, Clare DK, Ebert J, Bonneau F, Basquin J, Drazkowska K, Tomecki R, Dziembowski A, Conti E, et al. RNA channelling by the eukaryotic exosome. EMBO Rep. 2010;11:936–942. doi: 10.1038/embor.2010.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Midtgaard SF, Assenholt J, Jonstrup AT, Van LB, Jensen TH, Brodersen DE. Structure of the nuclear exosome component Rrp6p reveals an interplay between the active site and the HRDC domain. Proc. Natl. Acad. Sci. U.S.a. 2006;103:11898–11903. doi: 10.1073/pnas.0604731103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell P, Petfalski E, Shevchenko A, Mann M, Tollervey D. The exosome: a conserved eukaryotic RNA processing complex containing multiple 3’-5’ exoribonucleases. Cell. 1997;91:457–466. doi: 10.1016/s0092-8674(00)80432-8. [DOI] [PubMed] [Google Scholar]
- Paredes E, Das SR. Click chemistry for rapid labeling and ligation of RNA. ChemBioChem. 2011;12:125–131. doi: 10.1002/cbic.201000466. [DOI] [PubMed] [Google Scholar]
- Preker P, Nielsen J, Kammler S, Lykke-Andersen S, Christensen MS, Mapendano CK, Schierup MH, Jensen TH. RNA exosome depletion reveals transcription upstream of active human promoters. Science. 2008;322:1851–1854. doi: 10.1126/science.1164096. [DOI] [PubMed] [Google Scholar]
- Rinn JL, Chang HY. Genome Regulation by Long Noncoding RNAs. Annu. Rev. Biochem. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomecki R, Kristiansen MS, Lykke-Andersen SOR, Chlebowski A, Larsen KM, Szczesny RJ, Drazkowska K, Pastula A, Andersen JS, Stepien PP, et al. The human core exosome interacts with differentially localized processive RNases: hDIS3 and hDIS3L. The EMBO Journal. 2010;29:2342–2357. doi: 10.1038/emboj.2010.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent HA, Deutscher MP. Substrate recognition and catalysis by the exoribonuclease RNase R. J. Biol. Chem. 2006;281:29769–29775. doi: 10.1074/jbc.M606744200. [DOI] [PubMed] [Google Scholar]
- Wasmuth EV, Lima CD. Structure and Activities of the Eukaryotic RNA Exosome. Enzymes. 2012a;31:53–75. doi: 10.1016/B978-0-12-404740-2.00003-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasmuth EV, Lima CD. Exo- and endoribonucleolytic activities of yeast cytoplasmic and nuclear RNA exosomes are dependent on the noncatalytic core and central channel. Mol. Cell. 2012b;48:133–144. doi: 10.1016/j.molcel.2012.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasmuth EV, Januszyk K, Lima CD. Structure of an Rrp6-RNA exosome complex bound to poly(A) RNA. Nature. 2014;511:435–439. doi: 10.1038/nature13406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Hopper AK. Healing for destruction: tRNA intron degradation in yeast is a two-step cytoplasmic process catalyzed by tRNA ligase Rlg1 and 5’-3’ exonuclease Xrn1. Genes & Development. 2014;28:1556–1561. doi: 10.1101/gad.244673.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyers F, Rougemaille M, Badis G, Rousselle J-C, Dufour M-E, Boulay J, Régnault B, Devaux F, Namane A, Séraphin B, et al. Cryptic Pol II Transcripts Are Degraded by a Nuclear Quality Control Pathway Involving a New Poly(A) Polymerase. Cell. 2005;121:725–737. doi: 10.1016/j.cell.2005.04.030. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.