Abstract
There are 3-4 million new hepatitis C virus (HCV) infections yearly. The extensive inter-genotypic sequence diversity of envelope proteins E1 and E2 of HCV and shielding of important epitopes by hypervariable region 1 (HVR1) of E2 are believed to be major hindrances to developing universally protective HCV vaccines. Using cultured viruses expressing the E1/E2 complex of isolates H77 (genotype 1a), J6 (2a), or S52 (3a), with and without HVR1, we tested HVR1-mediated neutralization occlusion in vitro against a panel of 12 well-characterized human monoclonal antibodies (HMAbs) targeting diverse E1, E2, and E1/E2 epitopes. Surprisingly, HVR1-mediated protection was greatest for S52, followed by J6 and then H77. HCV pulldown experiments showed that this phenomenon was caused by epitope shielding. Moreover, by regression analysis of HMAb binding and neutralization titer of HCV we found a strong correlation for HVR1-deleted viruses, but not for parental viruses retaining HVR1. The inter-genotype neutralization sensitivity of the parental viruses to HMAbs AR2A, AR3A, AR4A, AR5A, HC84.26, and HC33.4 varied greatly (>24- to >130-fold differences in IC50-values). However, except for AR5A, these differences decreased to less than 6.0-fold when comparing the corresponding HVR1-deleted viruses. Importantly, this simplified pattern of neutralization sensitivity in the absence of HVR1 was also demonstrated in a panel of HVR1-deleted viruses of genotypes 1a, 2a, 2b, 3a, 5a, and 6a, although for all HMAbs, except AR4A, an outlier was observed. Finally, unique amino acid residues in HCV E2 could explain these outliers in the tested cases of AR5A and HC84.26. Conclusion: HVR1 adds complexity to HCV neutralization by shielding a diverse array of unexpectedly cross-genotype-conserved E1/E2 epitopes. Thus, an HVR1-deleted antigen could be a better HCV vaccine immunogen.
Keywords: HCV, HVR1, vaccine, genotype, immune evasion
Introduction
Direct acting antivirals that can eradicate hepatitis C virus (HCV) from infected patients have become available. However, cost of treatment, frequency of occult infection, and infrastructural challenges in resource-poor parts of the world indicate that there is an urgent need for a vaccine to control the disease worldwide. There are 3-4 million new infections each year, most of whom will become chronically infected and thus be at increased risk of developing liver cirrhosis and hepatocellular carcinoma (1). HCV is an enveloped positive-strand RNA virus of the Flaviviridae family with a 9.6 kb genome consisting of 5’ and 3’ untranslated regions (UTRs) flanking an open reading frame (ORF) that encodes a single polyprotein. This polyprotein is processed into structural proteins (Core and envelope proteins E1 and E2), p7, and nonstructural proteins (NS2-NS5B) (1). HCV is a highly diverse virus, and isolates have been divided into six epidemiologically important genotypes, most containing multiple subtypes (1).
While early neutralizing antibody responses against HCV are correlated with viral clearance (2, 3), studies in chimpanzees and in human liver chimeric mice found limited cross-strain protection upon heterologous viral re-challenge (4-7). Also, HCV has been shown to persist through evolution in patients in the presence of neutralizing antibodies (8). The high genetic heterogeneity of HCV, most prominent in hypervariable region 1 (HVR1) of E2 (9), along with the high mutation rate of the virus are believed to be pivotal in escape from adaptive immunity, including anti-HCV antibodies (10).
HVR1-deleted HCV was initially proven to be viable although attenuated in chimpanzees (11). Subsequently, studies have found conserved properties of HVR1 (12) and notably a putative interaction with HCV co-receptor scavenger receptor class B, type I (13, 14). Interestingly, we and others found that HVR1 could protect HCV from neutralizing antibodies in vitro (15, 16) and we recently verified this in vivo (17). However, this was not the case for antibodies targeting non-viral epitopes such as virion-associated apolipoprotein E (18). This phenomenon was suggested to mainly involve epitopes with a role in CD81 binding during HCV entry (15), however, the extent has not been studied in detail.
High sequence variation among the viral envelope proteins combined with the large differences in neutralization sensitivity of HCV in in vitro assays (19-24) has prompted the belief that even cross-genotype neutralization-responsive epitopes are not well-conserved (25). This apparent epitope-variation could be a major obstacle for the development of an effective HCV vaccine (26, 27). One option would be to pursue a polyvalent vaccine, although cross-protection could prove difficult to achieve. Consequently, the search continues to identify novel, fully conserved, HCV envelope protein epitopes. As a part of this effort, we described two panels of HCV-specific human monoclonal antibodies (HMAbs) that contain antibodies targeting five non-overlapping epitopes namely antigenic domains A-E of E2 (22, 28, 29), and antigenic regions 1-3 (AR1-3) of E2 and AR4-5 of complexed E1/E2 (19, 20). Comparisons between the two groupings of epitopes indicates that antigenic domain A does not overlap with AR1-3, antigenic domain B partly overlaps with AR3, antigenic domain C partly overlaps with AR1-3, and antigenic domains D and E have been mapped structurally to the front layer of the E2 core domain structure within AR3 (19, 30, 31). Neutralization studies suggest that HMAbs AR3A, AR4A, AR5A, HC84.26 (anti-domain D), and HC33.4 (anti-domain E) have broad cross-reactivity although their neutralizing potential varies across diverse HCV genotypes (19, 20, 22, 29), making them interesting to study further both for immunotherapy and novel vaccine designs. These two panels of HMAbs represent the most comprehensive tool for analyzing HCV neutralization sensitivity. Although E1 appears less immunogenic, E1-specific HMAbs have also been isolated (32, 33).
We performed studies of the breadth of neutralization susceptibility of HVR1-deleted HCV using HMAbs against epitopes in antigenic domains A-E, AR1-5, and E1. Using AR1-5-specific antibodies, we performed antibody-mediated immunoprecipitation to ascertain the mechanism of increased neutralization. We showed that differences in sensitivity among HCV genotype isolates against cross-genotype reactive antibodies were primarily caused by differences in HVR1-mediated antibody shielding, rather than epitope variability, with important implications for the understanding of virus neutralization and vaccine development.
Materials and Methods
Plasmids
We used intergenotypic HCV recombinants with Core-NS2 of isolates H77, J6, or S52 and UTRs and NS3-NS5B of JFH1, which we and others developed previously (16, 21, 34, 35). Our HVR1-deleted variants of Core-NS2 HCV recombinants H77, J6, J8, S52, SA13, and HK6a were described (16). All JFH1-based recombinants are referred to by the isolate of Core-NS2, i.e. H77, J6, J8, S52, SA13, and HK6a with the subscript ΔHVR1 to signify deletion of HVR1. Coding mutations were introduced into the indicated recombinants by standard molecular cloning techniques and are referred to by their amino acid position relative to the H77 reference sequence AF009606.
Cell culture
Huh7.5 human hepatoma cells were grown in Dulbecco's modified essential medium supplemented with 10% fetal bovine serum and antibiotics. HCV transfections and infections of cells were done as described (21, 35). Infectivity titration of culture derived HCV was done as described (16, 36). Antibody 9E10 was used for immunostaining of NS5A in infected cells (34).
Antibodies for HCV neutralization
For neutralization assays we used previously published antibodies specific for HVR1 (J6.36) (37), antigenic domain A (CBH4G), B (CBH5), C (CBH7), D (HC84.26), and E (HC33.4) of E2 with control antibody R04 (22, 28), antibodies specific for antigenic region 1-5 (AR1B, AR2A, AR3A, AR4A, and AR5A) and control antibody b6 (19, 20) and antibodies against E1 (IGH520 and IGH526, Innogenetics) (32).
HCV neutralization
We plated 6×103 Huh7.5 cells/well into poly-D-lysine-coated 96-well plates (96 Well Optical Bottom Plates, Nunc). The next day, dilution series of the relevant antibody were prepared and four replicates of each dilution step were incubated for 1h at 37°C with recombinant HCV along with six or eight wells of virus only. The virus/antibody and virus only were added to the plated cells in the indicated replicates and the cells were washed after 3h incubation. Following a total infection time of 48h the cells were immunostained for the HCV NS5A protein (34), and the number of focus forming units (FFUs) were counted using an ImmunoSpot Series 5 UV Analyzer as described (36). A control antibody was included at the highest dose of the specific antibody; these data are not included in the figures, but consistently showed no effect on neutralization. Percent neutralization was calculated by relating FFU counts to mean of replicates incubated with virus only. Three- or four-parameter non-linear curve regression was used to calculate the IC50-values that were selected based on what appeared to fit the data in the most biologically relevant way (top value set to 100 and bottom value set to 0, Graphpad PRISM, v4.03).
HCV immunoprecipitation
50 μl of magnetic bead slurry (Immunoprecipitation Kit, Dynabeads® Protein G, 100.070D, Invitrogen) were incubated on a shaker with 5 μg of antibody (HMAbs targeting AR1-5 or the control antibody b6) in 50 μl for 20 min at room temperature. The beads were subsequently washed two times and incubated with the virus (106 IU HCV RNA) in 500 μl complete medium on a shaker for 1 h at room temperature. The beads were washed three times in 200 μl of washing buffer prior to RNA extraction in the lysis buffer using QIAamp MinElute Virus Vacuum kit (57714, Qiagen) along with blanks and a dilution series of known HCV RNA sample for quantitative PCR. Afterwards the beads were removed by spinning for 5 min at 1,400 × relative centrifugal force. Viral RNA was purified as per the manufacturer's instruction and eluted in 22 μl of elution buffer. 8 μl of the eluted fractions were then used in a light-cycler for RT-qPCR using primers and protocols described previously (35).
Results
A broad spectrum of neutralization epitopes within the envelope proteins E1 and E2 of HCV were protected by HVR1
We examined HCV neutralization of HMAbs targeting HCV epitopes in antigenic domains A-E, AR1-5, and E1 (19, 20, 22, 28, 32) in infectious FFU reduction assays (21). To avoid isolate-specific effects we employed previously described sequence-confirmed virus stocks of parental H77 (genotype 1a), J6 (2a), and S52 (3a) viruses as well as the HVR1-deleted variants H77ΔHVR1, J6ΔHVR1, and S52ΔHVR1 (16, 21, 34, 35). The 1a and 3a viruses with and without HVR1 had similar specific infectivity (0.00061-0.00066; Table 1), and they were only ~5-fold higher for J6 viruses (0.0027 and 0.0035; Table 1). Thus, virus particle input would be similar and should not affect the neutralization sensitivity comparisons. We initially performed neutralization assays of J6 with and without HVR1 with an HVR1-specific mouse monoclonal antibody, J6.36 (37), and found efficient dose-dependent neutralization of J6 with no effect against J6ΔHVR1 (Table 1, Figure S1).
Table 1.
Inhibitory concentration 50% for a broad panel of HCV-specific MABs against H77, J6, and S52 with and without HVR1
| Inhibitory concentration 50% (μg/ml) |
|||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Virus stock titers |
MAb Specificity |
J6.36 | CBH4G antigenic |
CBH5 antigenic |
CBH7 antigenic |
HC84.26 antigenic |
HC33.4 antigenic |
AR1B | AR2A | AR3A | AR4A | AR5A | IGH520 | IGH526 | |||
| Virus | Infectivity* | HCV RNA** | Spec. Inf.*** | HVR1 | Domain A | Domain B | Domain C | Domain D | Domain E | AR1B | AR2A | AR3A | AR4A | AR5A | E1 | E1 | |
| H77 | 4.1 | 7.3 | 0.00066 | - | >50 | >50 | >50 | 0.16 | 0.52 | >50 | 2.1 | 1.2 | 1.0 | 0.38 | >50 | >50 | |
| H77ΔHVR1 | 3.1 | 6.3 | 0.00063 | - | >50 | 9.3 | 0.68 | 0.0040 | 1.8 | 2.8 | 0.013 | 0.012 | 0.0030 | 0.0082 | 0.16 | 1.0 | |
| J6 | 5.1 | 7.6 | 0.0035 | 2.9 | >50 | 16 | >50 | 0.59 | 7.6 | >50 | >50 | 4.9 | 3.7 | 8.1 | >50 | >50 | |
| J6ΔHVR1 | 4.7 | 7.3 | 0.0027 | >50 | >50 | 0.035 | 2.4 | 0.0053 | 1.0 | >50 | 0.032 | 0.0093 | 0.0037 | 0.010 | 0.047 | 0.094 | |
| S52 | 4.4 | 7.6 | 0.00061 | - | >50 | >50 | >50 | 6.7 | 16 | >50 | >50 | >50 | 34 | >50 | >50 | >50 | |
| S52ΔHVR1 | 4.2 | 7.4 | 0.00065 | - | >50 | 0.042 | 2.3 | 0.0033 | 5.7 | >50 | 0.055 | 0.019 | 0.00094 | 2.3 | 0.11 | 0.11 | |
Data was generated by dose-response FFU reduction assays. IC50-values were calculated by four-parameter non-linear curve regression using Graphpad PRISM (v4.03).
Infectivity titer given in log10 50% tissue culture infectious dose (TCID50)/ml.
HCV RNA titer given in log10 international units (IU)/ml.
Specific infectivity given in TCID50/IU.
The antigenic domain A specific HMAb, CBH4G, had no effect against genotype 1a, 2a, and 3a viruses with and without HVR1 (Table 1, Figure S2). CBH4G was previously described to be non-neutralizing, although it cross-reacted with recombinant genotype 1 and 2 E2 proteins (28). The AR1-specific HMAb AR1B, previously shown only to react with genotype 1 (19), only neutralized the HVR1-deleted 1a virus H77ΔHVR1 (Table 1, Figure S3). However, the nine HMAbs targeting antigenic domains B-D, AR2-5 and E1 had greatly increased efficacy against all HVR1-deleted viruses compared to the parental viruses (Table 1, Figure S2-4). While this was not the case for HC33.4 (Table 1, Figure S2), targeting antigenic domain E, we found HVR1-mediated protection against two rodent-derived domain E specific monoclonal antibodies (AP33 and 3/11, data not shown) supporting recent findings that the HC33.4 epitope depended, in part, on HVR1 (38). The fact that efficient AR2A neutralization was observed for HVR1-deleted viruses of genotypes 1a, 2a and 3a was surprising as it was previously shown to only react with genotype 1 isolates (19). Thus, we found a much broader HVR1-mediated protection of HCV from neutralizing antibodies than previously described, involving every neutralization epitope in the comprehensive HMAb panels, including E1.
HVR1-mediated shielding protected HCV from binding of neutralizing antibodies
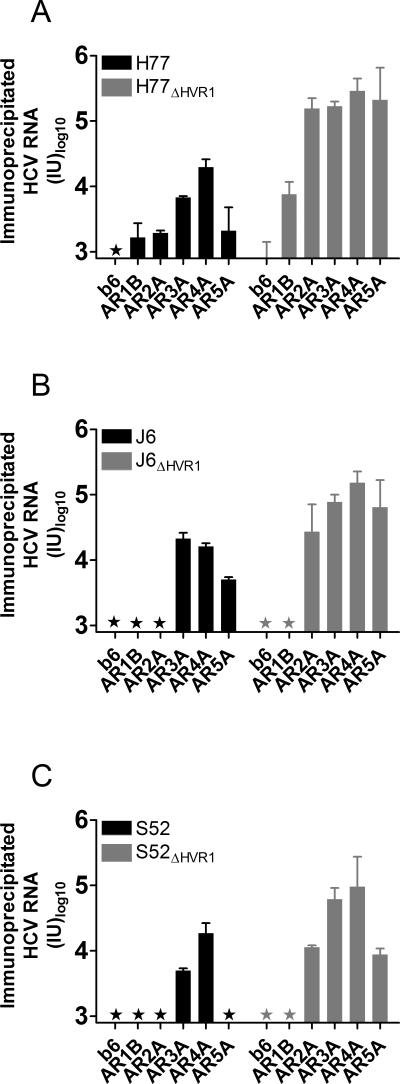
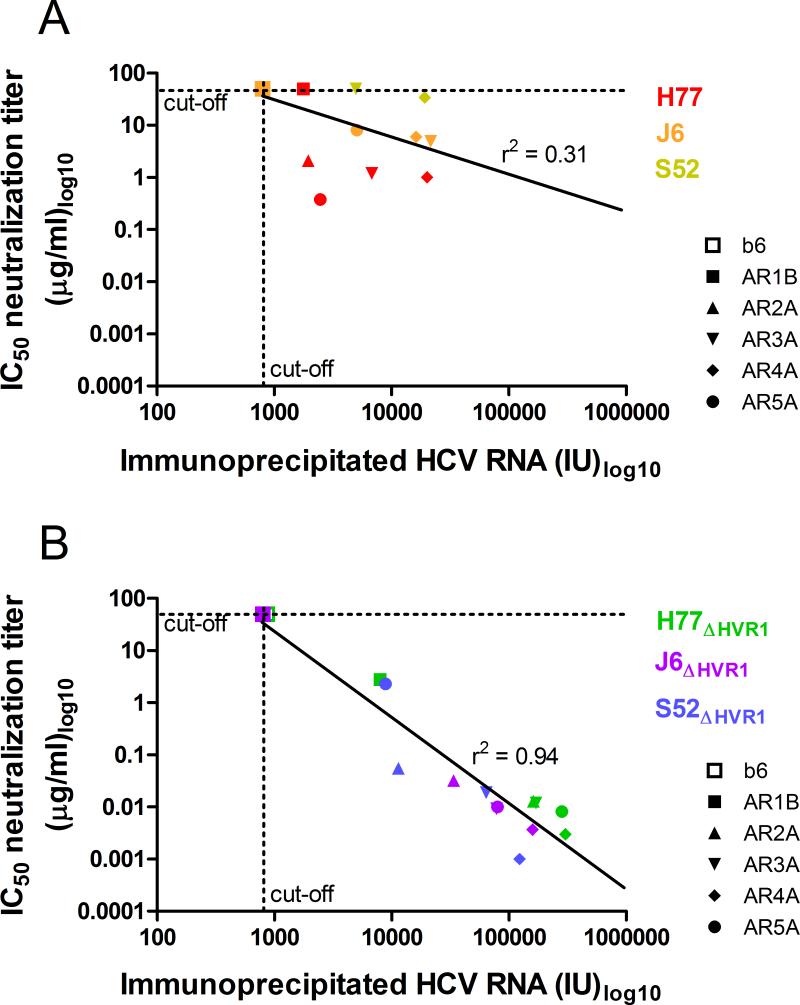
To address the mechanism of increased neutralization susceptibility of HVR1-deleted viruses we performed immunoprecipitation of H77, J6, and S52 with and without HVR1, using the AR1-5 specific antibodies. In all cases, immunoprecipitated HCV RNA either remained below the detection limit or was increased for the HVR1-deleted variants (Figure 1A-C). This indicated that the increase in neutralization susceptibility exhibited by HVR1-deleted viruses was a result of unmasking of previously inaccessible epitopes on the surface of the HCV particle. To further analyze HVR1-mediated shielding we plotted the IC50-values for a given HMAb/virus combination (data from Table 1) against the efficiency of immunoprecipitation (data from Figure 1). By performing linear regression analysis on these data points for parental viruses we found a weak correlation (r2 = 0.31) between efficiency of HCV particle immunoprecipitation and efficiency of neutralization (Figure 2A). Interestingly, we found a strong correlation (r2 = 0.94) when comparing the same two parameters for the HVR1-deleted viruses (Figure 2B). This suggested a simple relationship between antibody binding and neutralization for HVR1-deleted viruses, indicating that the target epitopes were readily accessible to the HMAbs, in the absence of HVR1-mediated shielding.
Figure 1. Immunoprecipitation of HCV strains H77 (genotype 1a), J6 (2a), and S52 (3a) with and without HVR1 using HMAbs targeting five distinct antigenic regions of E2 or E1/E2.
Magnetic beads were coated with the indicated HMAbs and incubated with 106 IU HCV RNA from infectious cell culture supernatant containing (A) H77 or H77ΔHVR1, (B) J6 or J6ΔHVR1, or (C) S52 or S52ΔHVR1. The amount of associated HCV RNA was assessed by HCV RNA extraction directly from the beads and RT-qPCR in duplicates on the eluted fractions as described in Materials and Methods. Results are shown as mean of duplicates with SD (Cut-off: 800 IU). Star, value below cut-off.
Figure 2. Strong correlation between immunoprecipitation and neutralization efficacies of HVR1-deleted viruses, but not parental viruses.
Immunoprecipitation of HCV virions and determination of IC50-values were conducted using AR1-5 specific HMAbs or control antibody b6 (as described in Table 1 and Figure 1) and plotted for (A) parental H77, J6 and, S52 or (B) the HVR1-deleted variants H77ΔHVR1, J6 ΔHVR1, and S52 ΔHVR1. The assay cut-offs were 50 μg/ml for IC50-values and 800 IU for Immunoprecipitation. Linear regression analysis was performed for both datasets (Graphpad PRISM, v4.03) and the r2 values are shown above the calculated best-fit lines.
Divergent efficiency of HVR1-mediated antibody shielding might explain differences in neutralization sensitivity between parental viruses
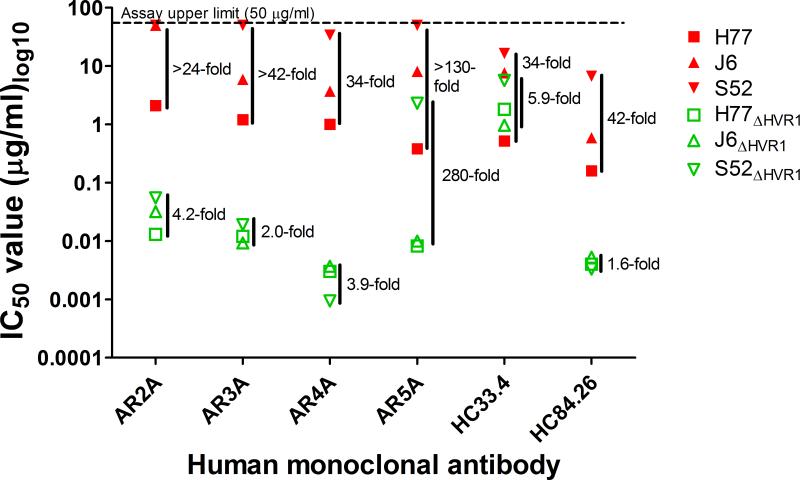
Analyzing the HMAb IC50-values for the parental viruses (Table 1) we found that only AR2A, AR3A, AR4A, AR5A, HC84.26, and HC33.4 efficiently neutralized at least one of these viruses. Plotting these we observed variation in IC50-values from >24-fold to >130-fold (Figure 3). Interestingly, comparing the IC50-values for the corresponding HVR1-deleted viruses we observed only 1.6- to 5.9-fold variation for AR2A, AR3A, AR4A, HC84.26, and HC33.4 (Figure 3). For AR5A, H77ΔHVR1 and J6ΔHVR1 were similarly sensitive (1.2-fold difference), whereas S52ΔHVR1 was about 280-fold less sensitive (Figure 3). Parental viruses were too resistant against HMAbs CBH5, CBH7, IGH520, and IGH526 to adequately compare sensitivity however variation for HVR1-deleted viruses was only ~3.5-fold for CBH7 and ~3.4-fold for IGH520. For CBH5 and IGH526 we observed about ~270-fold and ~11-fold less sensitivity for H77ΔHVR1 compared with J6ΔHVR1 and S52ΔHVR1, which were, in both cases, similar (1.2-fold differences). Taken together, this indicated that HVR1-mediated shielding, rather than the previously assumed epitope diversity, was responsible for much of the observed differences in neutralization sensitivity. Strikingly, S52 was consistently the most resistant, followed by J6 and then H77 (Figure 3), indicating that HVR1-mediated shielding varied between the isolates in an epitope-independent manner.
Figure 3. Variation in neutralization sensitivity among parental HCV strains is greatly decreased for HVR1-deleted viruses.
H77, J6, and S52 with (in red) and without HVR1 (in green) were incubated with dilution series of the indicated HMAbs starting at 50 μg/ml (upper limit of the assay) along with six replicates of virus only. 48h after inoculation of Huh7.5 cells with the virus/antibody mixes or virus only the number of FFU were visualized by HCV-specific immunostaining and the counts were normalized to the mean count of six wells of virus only.
Isotype-matched control antibody was included at the highest concentration of the specific antibody and was found to have no effect on infection. Four-parameter non-linear curve regression was used to fit the data points, top value set to 100 and bottom value set to 0 (Graphpad PRISM, v4.03). The IC50-titers of neutralization are shown in Table 1. Here the fold difference between the highest and the lowest IC50-value is given to the right of each set of values.
HCV envelope protein neutralization-epitopes for HC84.26, HC33.4, AR3A, and AR5A were highly conserved, and AR4A appeared universally conserved
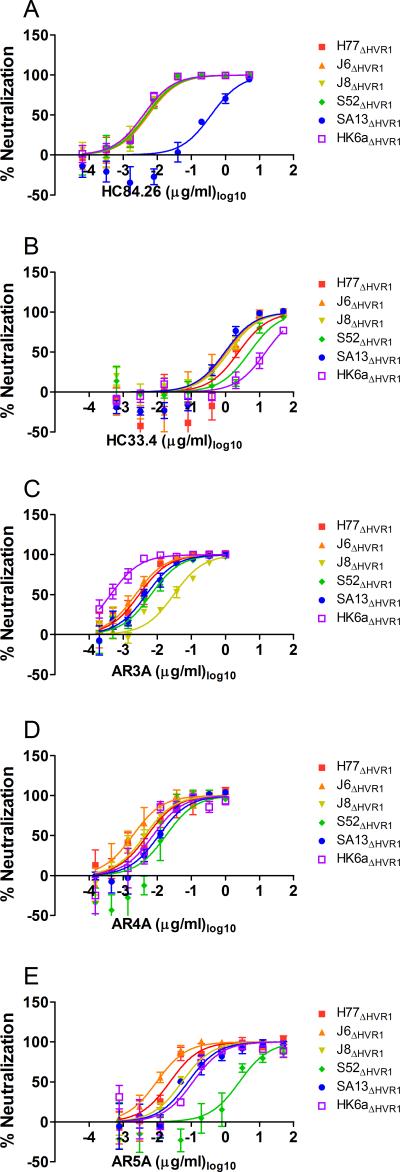
The high degree of epitope conservation led us to test neutralization sensitivity of a panel of our viable HVR1-deleted viruses (HVR1-deleted genotype 4a was previously shown to be non-viable (16)) against several HMAbs. Thus, we performed dose-response neutralization comparisons of the three previously used HVR1-deleted viruses H77 (1a), J6 (2a), and S52 (3a) along with J8 (2b), SA13 (5a), and HK6a (6a) strains without HVR1 against HC33.4, HC84.26, AR3A, AR4A, and AR5A (Figure 4). Comparing fold-differences between IC50-values among the six viruses we found 1.0-1.4 fold differences against HC84.26 (exception SA13ΔHVR1, inter-isolate difference of ~100-fold compared with all other HVR1-deleted viruses; Figure 4A), 1.1-5.4 fold differences against HC33.4 (exception HK6aΔHVR1, maximum inter-isolate difference of 16-fold compared with SA13ΔHVR1; Figure 4B), 1.3-13 fold differences against AR3A (exception HK6aΔHVR1, maximum inter-isolate difference of 71-fold compared with J8ΔHVR1; Figure 4C), 1.2-9.4 fold differences against AR4A (no exceptions, Figure 4D), and 1.5-12 fold differences against AR5A (exception S52ΔHVR1, maximum inter-isolate difference of 270-fold compared with J6ΔHVR1; Figure 4E). Thus, we confirmed the overall conservation of envelope neutralization epitopes. Furthermore, we found that the AR4A epitope was completely conserved among the tested HCV genotype isolates.
Figure 4. Neutralization epitopes in domain D and E and antigenic regions 3-5 are highly conserved across HCV genotypes.
(A-E) HVR1-deleted variants of H77 (1a), J6 (2a), J8 (2b), S52 (3a), SA13 (5a), and HK6a (6a) were incubated with dilution series of the indicated HMAbs along with eight replicates of virus only. 48h after inoculation of Huh7.5 cells with the virus/antibody mixes or virus only the number of FFU (or single spots in the cases where FFUs were too numerable to count accurately) were visualized by HCV-specific immunostaining and the counts were normalized to the mean count of virus only. Isotype-matched control antibody was included at the highest concentration of the specific antibody and was found to have no effect on infection. Three-parameter non-linear curve regression was used to fit the data points, top value set to 100 and bottom value set to 0 (Graphpad PRISM, v4.03). Error bars represent standard error of the mean.
Neutralization resistance of HVR1-deleted HCV against specific HMAbs explained by unique amino acids in the envelope proteins
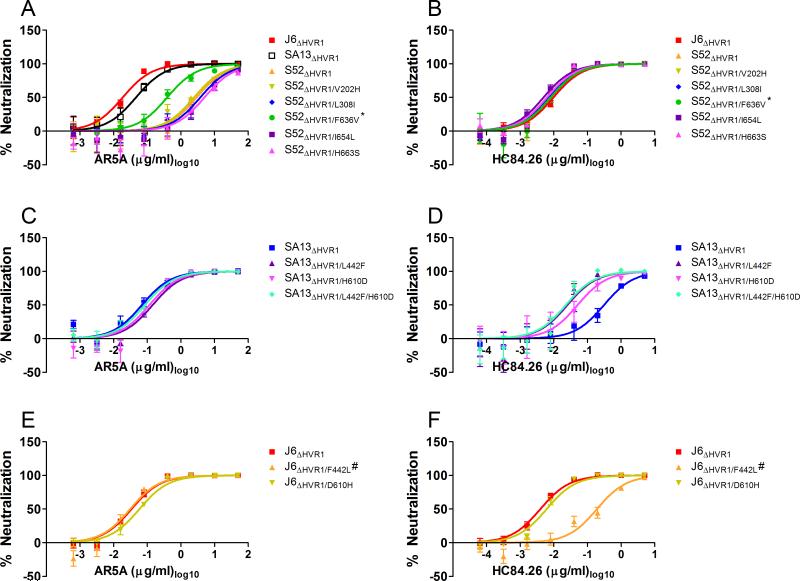
By performing a clustalW based alignment (MEGA 6) of the 6 HCV isolates used above we identified more than 50 positions with unique amino acids in E1/E2 for S52ΔHVR1 that might explain the observed resistance against AR5A. Selecting which to test was complicated by the fact that the antibody recognizes a conformational epitope on the E1/E2 heterodimer. We introduced five substitutions V202H, L308I, F636V, I654L, and H663S into S52ΔHVR1 (by choosing the most common residue at the given positions) as they were close to residues that had previously been mapped for AR5A binding by alanine scanning of recombinant E1/E2 protein (20). HCV RNA transcripts from these recombinants were transfected into Huh7.5 cells and with the exception of S52ΔHVR1/F636V the viruses spread to most culture cells as fast as the reference virus S52ΔHVR1. Virus from peak of infection was used to generate a first passage virus stock in naive Huh7.5 cells and all virus stocks had titers comparable to S52ΔHVR1. The envelope sequences of viruses recovered from these virus stocks were analyzed. Only the virus S52ΔHVR1/F636V required adaptation acquiring the mutation F291L in E1 along with N532S and D708V in E2. Interestingly, aspartic acid at position 708 was also unique to S52, suggesting a functional link between positions 636 and 708. Next, we performed dose-response neutralization comparisons against AR5A and the control antibody HC84.26. The S52ΔHVR1/F636V adapted virus showed ~5-fold decreased resistance against AR5A (Figure 5A). These effects could be reproduced in an independent assay (not shown) and importantly no change was observed for S52ΔHVR1 variants using the control antibody HC84.26 (Figure 5B).
Figure 5. Unique envelope protein residues in neutralization resistant HVR1-deleted viruses conferred their phenotype.
The indicated viruses were incubated with a 5-fold dilution series of (A, C, E) AR5A, or (B, D, F) HC84.26 starting at 50 μg/ml or 5 μg/ml, respectively, along with eight replicates of virus only. 48h after inoculation of Huh7.5 cells with the virus/antibody mixes or virus only the number of FFU were visualized by HCV-specific immunostaining and the counts were normalized to the mean count of virus only. Three-parameter non-linear curve regression was used to fit the data points, top value set to 100 and bottom value set to 0 (Graphpad PRISM, v4.03). Error bars represent standard error of the mean. *the recombinant also had the dominant substitutions F291L, N532S, and D708V. #the recombinant also had the substitution I347L.
We performed a similar analysis on the resistance of SA13ΔHVR1 against HC84.26. This analysis was simpler than the one described above as HC84.26 only targets E2 and specifically binds regions at positions 434-446 and 610-619 (31). We identified two E2 positions within these regions for which SA13 had a unique amino acid (L442 and H610), and thus possibly explaining the resistance of SA13ΔHVR1 against HC84.26. We introduced L442F, H610D, and the combination L442F/H610D into SA13ΔHVR1 as well as the corresponding mutations F442L and D610H into the HC84.26 sensitive J6ΔHVR1. With the exception of J6ΔHVR1/F442L the viruses spread to most culture cells as fast as the reference viruses. Virus from peak of infection was used to generate a first passage virus stock in naive Huh7.5 cells and all virus stocks had comparable titers. Only the virus stock for J6ΔHVR1/F442L displayed an additional substitution I347L in E1, previously described as a cell culture adaptive mutations for HVR1-deleted Jc1 (15). Next, we performed dose-response neutralization comparisons against HC84.26 and the control antibody AR5A. We observed no differences in sensitivity for the SA13ΔHVR1 and J6ΔHVR1 viruses against AR5A (Figure 5C and 5E). For SA13ΔHVR1, both L442F and H610D conferred increased sensitivity against HC84.26 (Figure 5D). These effects were confirmed in independent assays as was the fact that they did not have an additive effect on SA13ΔHVR1 sensitivity (Figure 5D). For J6ΔHVR1, we found that the single change F442L fully conferred the resistant phenotype of SA13ΔHVR1 against HC84.26 to J6ΔHVR1, whereas D610H had no effect on sensitivity (Figure 5F).
Discussion
HVR1 mediates a much broader level of neutralization shielding of HCV than previously recognized. This shielding adds complexity to the relationship between antibody binding and neutralization. We found that much of the observed differences in neutralization susceptibility among HCV isolates of various genotypes could be explained by HVR1-mediated antibody shielding rather than epitope diversity. This enabled us to identify unique amino acids important for modulating HC84.26 and AR5A sensitivity, explaining the resistant phenotypes of HVR1-deleted SA13 and S52, respectively, against these antibodies.
Epitopes in antigenic domains B-E, AR1-5, and on E1 were all effectively shielded by HVR1 for H77 (genotype 1a), J6 (2a), and S52 (3a) with and without HVR1 (Table 1). Only the HMAbs to antigenic domain B-D and AR3 (AR3A) were previously shown to block the interaction between E2 and CD81 (19, 20, 39). Therefore, HVR1 deletion not only increased exposure of the CD81 binding site (15), but also of other epitopes including the E1 IGH520/526 epitopes and epitopes in AR1-2 and AR4-5. The fact that antigenic domain A specific antibody, CBH4G, did not neutralize any of the tested viruses corroborates that this antibody is non-neutralizing (40). Our data supports that AR1 contains a high degree of sequence diversity (19). Taken together, these findings indicate that HVR1 shields most if not all known E1/E2 epitopes, which has important implications for vaccine design as it indicates that non-shielded epitopes either do not exist or are exceedingly rare. This suggests that HCV vaccine development could benefit from focusing on inducing high levels of antibodies targeting conserved epitopes with a high barrier to resistance with the goal of overcoming shielding rather than attempt to induce antibodies that circumvent it (41).
We found that the mechanism of HVR1-mediated neutralization protection was epitope shielding. Interestingly, there was a strong correlation between immunoprecipitation and neutralization of HVR1-deleted viruses, whereas only a weak correlation existed for parental viruses. We speculate that the complexity of the relationship between immunoprecipitation and neutralization for parental viruses could be because HVR1 shields neutralization epitopes present on individual E1/E2 heterodimers on the HCV virion at varying efficiencies. Thus, a single antibody might be able to bind one of the E1/E2 heterodimers, causing immunoprecipitation, but not necessarily neutralization, as other E1/E2 heterodimers on the virion, unhindered by antibody, could still mediate virus entry. This is in agreement with the observation that efficiency of immunoprecipitation versus neutralization is higher for parental viruses as compared with HVR1-deleted viruses (slopes of linear regressions in Figure 2). If such intra-virion differences in HVR1-mediated shielding of epitopes exist it would help explain the observed synergy between these HMAbs (42), as the efficient shielding of one epitope might render another epitope more accessible. Thus, the HCV virion would become increasingly susceptible to HMAbs mixtures targeting multiple exclusive epitopes.
We observed high variation in neutralization sensitivity of three parental viruses H77, J6, and S52. This variation in neutralization sensitivity among HCV strains has been described in numerous studies using both HCVcc (21, 42, 43) and HCVpp (22, 24, 44) and is commonly ascribed, in part, to variation of the envelope proteins. The envelope amino acid sequences of the isolates H77, J6, and S52 differ by 33.2-44.7%, and 31.7-43.9% upon exclusion of HVR1. However, despite the high degree of heterogeneity these pronounced differences in neutralization sensitivity decreased dramatically or disappeared when comparing HVR1-deleted viruses indicating a much greater degree of envelope protein epitope conservation than has previously been suspected based on sequence homology across highly diverse HCV genotypes. Furthermore, it indicates that HVR1-mediated epitope shielding, rather than epitope diversity, is frequently the direct cause of the differences in neutralization sensitivity observed between isolates. Our findings help explain why Tarr et al. observed no correlation between genotype and neutralization serotype (45), as sensitivity against a polyclonal serum will typically be a result of the efficiency of HVR1-mediated shielding in that particular HCV isolate, rather than epitope diversity as defined by the viral sequence. This also helps explain how sera from genotype 2 infected patients would fail to neutralize cell culture derived genotype 2 viruses (41). Interestingly, we also found that HVR1-mediated epitope shielding varied between isolates in an epitope-independent manner as we observed that parental S52 was consistently most resistant, followed by J6 and then H77 (Figure 3). This suggests that certain isolates are somehow able to depend more on HVR1-mediated shielding than others, possibly due to differences in structural flexibility of the region.
It is important to point out that while HVR1-deleted HCV is viable in vivo (11, 17) viruses without HVR1 have never been observed in patients, probably indicating an important role of the motif in the natural infection. The fact that the level of HVR1-mediated neutralizing antibody shielding varies between isolates suggests that there might be instances during infection where lower levels of antibody shielding is beneficial to the virus. We propose that lower HVR1-mediated shielding allows the virus to bypass early receptor interactions with LDLr and SR-BI, for which we and others have previously shown decreased dependency of HVR1-deleted viruses (14, 15, 18). Thus, less shielded HCV might have an entry advantage, which could be important early in the natural infection prior to the induction of HCV-specific antibodies. This subject would be of interest for future studies.
The two clearest cases of a neutralization resistant HVR1-deleted virus was AR5A against S52ΔHVR1 and HC84.26 against SA13ΔHVR1. In both cases we were able to identify, by amino acid alignments of E1 and E2, residues that were unique to the resistant viruses that we found to be partly responsible for the resistant phenotype of the virus. For HC84.26, we confirmed the effect of L442 within the epitope of the antibody, but interestingly also identified a specific role of the residue H610 within the previously described region 610-619 (31). For AR5A, which binds to a conformational epitope of the E1/E2 heterodimer, we based our mutational analysis on unique positions close to positions identified as important for binding by alanine scanning of recombinant protein (20). Although a clear shortcoming of an alanine scanning approach is that substitutions may not result in infectious virus it did allow us to narrow down our selection to five positions. Thus, we identified the novel position F636 with a role in regulating AR5A resistance, possibly in conjunction with another residue D708. Importantly, the substitutions we made at these positions did not alter the general neutralization sensitivity of the viruses, indicating that the effects were antibody-specific, probably altering the epitopes of the antibodies.
We have uncovered a novel, surprisingly high, degree of epitope conservation across genotypes by comparing neutralization sensitivity of diverse HVR1-deleted HCV. Taken together our data indicates that HVR1-mediated shielding is a powerful feature of HCV to evade antibodies against many seemingly conserved envelope epitopes. The observation that protection against HCV is often, at least partly, strain-specific during a natural infection (7) coupled with the finding that the right combination of HMAbs protect in vivo (19) indicates that an effective HCV vaccine has to present broadly conserved epitopes. Our findings, along with other studies showing that HVR1 may bind antibodies to enhance its shielding effect of conserved epitopes (38), suggest that an HVR1-deleted HCV antigen is an obvious candidate for developing an HCV vaccine, however a number of concerns exist. It is tempting to speculate that the partial success in inducing broadly reactive neutralizing antibodies in human subjects using a recombinant E1/E2 protein vaccine (43, 46) could be boosted using an HVR1-deleted recombinant protein. However, the high level of epitope exposition observed on virus particles in the absence of HVR1 has not been fully reproduced using recombinant E2 protein (47), although it has been possible to show modest increases in antigenicity (38, 48). Whether this is due to involvement of non-viral partners in HVR1-mediated neutralization protection, such as recently observed with ApoE-mediated neutralization protection of HCV (49), or because the modified recombinant antigens fail to fully emulate the native E1/E2 heterodimer remains to be determined. Alternatively, it might be feasible to develop an HVR1-deleted inactivated vaccine as we have shown that HVR1-deleted particles are more readily neutralized both ex vivo (16) and in vivo (17).
In conclusion, we found that HVR1-mediated neutralization protection was surprisingly broad involving diverse neutralization epitopes on the HCV envelope proteins. The mechanism of this protection was epitope shielding and the removal of HVR1 resulted in viruses for which neutralization was mainly determined by the affinity of the antibody/epitope interaction. The surprisingly high number of widely conserved epitopes that are exposed in the absence of HVR1 makes the HVR1-deleted antigen an interesting vaccine candidate. A high level of induction of neutralizing antibodies against several of the epitopes described here could potentially prevent chronic infection.
Supplementary Material
Acknowledgements
We are grateful to Erick Giang (The Scripps Research Institute) for technical assistance; to Steen Ladelund (Copenhagen University Hospital, Hvidovre) for statistical advice; to Bjarne Ørskov Lindhardt and Ove Andersen (Copenhagen University Hospital, Hvidovre) and Carsten Geisler (University of Copenhagen) for their support of the project; and Michael Diamond (Washington University School of Medicine, US), Arvind Patel (University of Glasgow, UK), Jean Dubuisson (Pasteur Institute, Lille, France), and Charles Rice (Rockefeller University, US) for providing reagents.
Financial support
This work was supported by a Ph.D. stipend from Faculty of Health and Medical Sciences, University of Copenhagen (RVM), an individual DFF-postdoctoral grant from the Danish Council for Independent Research, Medical Sciences (JP), and research grants from the Lundbeck Foundation (JP, JB), the Novo Nordisk Foundation (JB), the Danish Council for Independent Research, Medical Sciences (JB), and an advanced top researcher grant from the Danish Council for Independent Research (JB). JB is the 2015 recipient of the Novo Nordisk Prize. ML is funded by NIH awards AI079031, AI106005 and AI123365, and SKHF is funded by AI108024 and AI123862.
Abbreviations
- HCV
hepatitis C virus
- HVR1
hypervariable region 1
- E1
envelope protein 1
- E2
envelope protein 2
- E1/E2
envelope protein complex
- HMAbs
human monoclonal antibodies
- AR
antigenic region
- UTR
untranslated region
- ORF
open reading frame
- NS
nonstructural proteins
- FFU
focus forming units
- IU
international units
Reference List
- 1.Gottwein JM, Bukh J. Cutting the gordian knot-development and biological relevance of hepatitis C virus cell culture systems. Adv Virus Res. 2008;71:51–133. doi: 10.1016/S0065-3527(08)00002-X. [DOI] [PubMed] [Google Scholar]
- 2.Pestka JM, Zeisel MB, Blaser E, Schurmann P, Bartosch B, Cosset FL, et al. Rapid induction of virus-neutralizing antibodies and viral clearance in a single-source outbreak of hepatitis C. Proc Natl Acad Sci U S A. 2007;104(14):6025–6030. doi: 10.1073/pnas.0607026104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Osburn WO, Snider AE, Wells BL, Latanich R, Bailey JR, Thomas DL, et al. Clearance of hepatitis C infection is associated with the early appearance of broad neutralizing antibody responses. Hepatology. 2014;59(6):2140–2151. doi: 10.1002/hep.27013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bukh J, Thimme R, Meunier JC, Faulk K, Spangenberg HC, Chang KM, et al. Previously infected chimpanzees are not consistently protected against reinfection or persistent infection after reexposure to the identical hepatitis C virus strain. J Virol. 2008;82(16):8183–8195. doi: 10.1128/JVI.00142-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Prince AM, Brotman B, Lee DH, Pfahler W, Tricoche N, Andrus L, et al. Protection against chronic hepatitis C virus infection after rechallenge with homologous, but not heterologous, genotypes in a chimpanzee model. J Infect Dis. 2005;192(10):1701–1709. doi: 10.1086/496889. [DOI] [PubMed] [Google Scholar]
- 6.Meuleman P, Bukh J, Verhoye L, Farhoudi A, Vanwolleghem T, Wang RY, et al. In vivo evaluation of the cross-genotype neutralizing activity of polyclonal antibodies against hepatitis C virus. Hepatology. 2011;53(3):755–762. doi: 10.1002/hep.24171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bukh J, Engle RE, Faulk K, Wang RY, Farci P, Alter HJ, et al. Immunoglobulin with high-titer in vitro cross-neutralizing hepatitis C virus antibodies passively protects chimpanzees from homologous, but not heterologous, challenge. J Virol. 2015;89(17):9128–9132. doi: 10.1128/JVI.01194-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.von Hahn T, Yoon JC, Alter H, Rice CM, Rehermann B, Balfe P, et al. Hepatitis C virus continuously escapes from neutralizing antibody and T-cell responses during chronic infection in vivo. Gastroenterology. 2007;132(2):667–678. doi: 10.1053/j.gastro.2006.12.008. [DOI] [PubMed] [Google Scholar]
- 9.Farci P, Bukh J, Purcell RH. The quasispecies of hepatitis C virus and the host immune response. Springer Semin Immunopathol. 1997;19(1):5–26. doi: 10.1007/BF00945022. [DOI] [PubMed] [Google Scholar]
- 10.Wahid A, Dubuisson J. Virus-neutralizing antibodies to hepatitis C virus. J Viral Hepat. 2013;20(6):369–376. doi: 10.1111/jvh.12094. [DOI] [PubMed] [Google Scholar]
- 11.Forns X, Thimme R, Govindarajan S, Emerson SU, Purcell RH, Chisari FV, et al. Hepatitis C virus lacking the hypervariable region 1 of the second envelope protein is infectious and causes acute resolving or persistent infection in chimpanzees. Proc Natl Acad Sci U S A. 2000;97(24):13318–13323. doi: 10.1073/pnas.230453597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Callens N, Ciczora Y, Bartosch B, Vu-Dac N, Cosset FL, Pawlotsky JM, et al. Basic residues in hypervariable region 1 of hepatitis C virus envelope glycoprotein e2 contribute to virus entry. J Virol. 2005;79(24):15331–15341. doi: 10.1128/JVI.79.24.15331-15341.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guan M, Wang W, Liu X, Tong Y, Liu Y, Ren H, et al. Three different functional microdomains in the hepatitis C virus hypervariable region 1 (HVR1) mediate entry and immune evasion. J Biol Chem. 2012;287(42):35631–35645. doi: 10.1074/jbc.M112.382341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bankwitz D, Vieyres G, Hueging K, Bitzegeio J, Doepke M, Chhatwal P, et al. Role of hypervariable region 1 for the interplay of hepatitis C virus with entry factors and lipoproteins. J Virol. 2014;88(21):12644–12655. doi: 10.1128/JVI.01145-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bankwitz D, Steinmann E, Bitzegeio J, Ciesek S, Friesland M, Herrmann E, et al. Hepatitis C virus hypervariable region 1 modulates receptor interactions, conceals the CD81 binding site, and protects conserved neutralizing epitopes. J Virol. 2010;84(11):5751–5763. doi: 10.1128/JVI.02200-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Prentoe J, Jensen TB, Meuleman P, Serre SB, Scheel TK, Leroux-Roels G, et al. Hypervariable region 1 differentially impacts viability of hepatitis C virus strains of genotypes 1 to 6 and impairs virus neutralization. J Virol. 2011;85(5):2224–2234. doi: 10.1128/JVI.01594-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Prentoe J, Verhoye L, Moctezuma RV, Buysschaert C, Farhoudi A, Wang R, et al. HVR1-mediated antibody evasion of highly-infectious in vivo adapted hepatitis C virus in humanized mice. Gut. 2016 doi: 10.1136/gutjnl-2015-310300. (epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Prentoe J, Serre SB, Ramirez S, Nicosia A, Gottwein JM, Bukh J. Hypervariable region 1 deletion and required adaptive envelope mutations confer decreased dependency on scavenger receptor class B type I and low-density lipoprotein receptor for hepatitis C virus. J Virol. 2014;88(3):1725–1739. doi: 10.1128/JVI.02017-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Law M, Maruyama T, Lewis J, Giang E, Tarr AW, Stamataki Z, et al. Broadly neutralizing antibodies protect against hepatitis C virus quasispecies challenge. Nat Med. 2008;14(1):25–27. doi: 10.1038/nm1698. [DOI] [PubMed] [Google Scholar]
- 20.Giang E, Dorner M, Prentoe JC, Dreux M, Evans MJ, Bukh J, et al. Human broadly neutralizing antibodies to the envelope glycoprotein complex of hepatitis C virus. Proc Natl Acad Sci U S A. 2012;109(16):6205–6210. doi: 10.1073/pnas.1114927109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Scheel TK, Gottwein JM, Jensen TB, Prentoe JC, Hoegh AM, Alter HJ, et al. Development of JFH1-based cell culture systems for hepatitis C virus genotype 4a and evidence for cross-genotype neutralization. Proc Natl Acad Sci U S A. 2008;105(3):997–1002. doi: 10.1073/pnas.0711044105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Keck ZY, Xia J, Wang Y, Wang W, Krey T, Prentoe J, et al. Human monoclonal antibodies to a novel cluster of conformational epitopes on HCV E2 with resistance to neutralization escape in a genotype 2a isolate. PLoS Pathog. 2012;8(4):e1002653. doi: 10.1371/journal.ppat.1002653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Owsianka A, Tarr AW, Juttla VS, Lavillette D, Bartosch B, Cosset FL, et al. Monoclonal antibody AP33 defines a broadly neutralizing epitope on the hepatitis C virus E2 envelope glycoprotein. J Virol. 2005;79(17):11095–11104. doi: 10.1128/JVI.79.17.11095-11104.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Broering TJ, Garrity KA, Boatright NK, Sloan SE, Sandor F, Thomas WD, Jr., et al. Identification and characterization of broadly neutralizing human monoclonal antibodies directed against the E2 envelope glycoprotein of hepatitis C virus. J Virol. 2009;83(23):12473–12482. doi: 10.1128/JVI.01138-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kong L, Jackson KN, Wilson IA, Law M. Capitalizing on knowledge of hepatitis C virus neutralizing epitopes for rational vaccine design. Curr Opin Virol. 2015;11:148–157. doi: 10.1016/j.coviro.2015.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tarr AW, Khera T, Hueging K, Sheldon J, Steinmann E, Pietschmann T, et al. Genetic Diversity Underlying the Envelope Glycoproteins of Hepatitis C Virus: Structural and Functional Consequences and the Implications for Vaccine Design. Viruses. 2015;7(7):3995–4046. doi: 10.3390/v7072809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Forns X, Bukh J, Purcell RH. The challenge of developing a vaccine against hepatitis C virus. J Hepatol. 2002;37(5):684–695. doi: 10.1016/s0168-8278(02)00308-2. [DOI] [PubMed] [Google Scholar]
- 28.Hadlock KG, Lanford RE, Perkins S, Rowe J, Yang Q, Levy S, et al. Human monoclonal antibodies that inhibit binding of hepatitis C virus E2 protein to CD81 and recognize conserved conformational epitopes. J Virol. 2000;74(22):10407–10416. doi: 10.1128/jvi.74.22.10407-10416.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Keck Z, Wang W, Wang Y, Lau P, Carlsen TH, Prentoe J, et al. Cooperativity in virus neutralization by human monoclonal antibodies to two adjacent regions located at the amino terminus of hepatitis C virus E2 glycoprotein. J Virol. 2013;87(1):37–51. doi: 10.1128/JVI.01941-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kong L, Giang E, Nieusma T, Kadam RU, Cogburn KE, Hua Y, et al. Hepatitis C virus E2 envelope glycoprotein core structure. Science. 2013;342(6162):1090–1094. doi: 10.1126/science.1243876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Krey T, Meola A, Keck ZY, Damier-Piolle L, Foung SK, Rey FA. Structural basis of HCV neutralization by human monoclonal antibodies resistant to viral neutralization escape. PLoS Pathog. 2013;9(5):e1003364. doi: 10.1371/journal.ppat.1003364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meunier JC, Russell RS, Goossens V, Priem S, Walter H, Depla E, et al. Isolation and characterization of broadly neutralizing human monoclonal antibodies to the e1 glycoprotein of hepatitis C virus. J Virol. 2008;82(2):966–973. doi: 10.1128/JVI.01872-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Keck ZY, Sung VM, Perkins S, Rowe J, Paul S, Liang TJ, et al. Human monoclonal antibody to hepatitis C virus E1 glycoprotein that blocks virus attachment and viral infectivity. J Virol. 2004;78(13):7257–7263. doi: 10.1128/JVI.78.13.7257-7263.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lindenbach BD, Evans MJ, Syder AJ, Wolk B, Tellinghuisen TL, Liu CC, et al. Complete replication of hepatitis C virus in cell culture. Science. 2005;309(5734):623–626. doi: 10.1126/science.1114016. [DOI] [PubMed] [Google Scholar]
- 35.Gottwein JM, Scheel TK, Hoegh AM, Lademann JB, Eugen-Olsen J, Lisby G, et al. Robust hepatitis C genotype 3a cell culture releasing adapted intergenotypic 3a/2a (S52/JFH1) viruses. Gastroenterology. 2007;133(5):1614–1626. doi: 10.1053/j.gastro.2007.08.005. [DOI] [PubMed] [Google Scholar]
- 36.Gottwein JM, Scheel TK, Callendret B, Li YP, Eccleston HB, Engle RE, et al. Novel infectious cDNA clones of hepatitis C virus genotype 3a (strain S52) and 4a (strain ED43): genetic analyses and in vivo pathogenesis studies. J Virol. 2010;84(10):5277–5293. doi: 10.1128/JVI.02667-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sabo MC, Luca VC, Prentoe J, Hopcraft SE, Blight KJ, Yi M, et al. Neutralizing monoclonal antibodies against hepatitis C virus E2 protein bind discontinuous epitopes and inhibit infection at a postattachment step. J Virol. 2011;85(14):7005–7019. doi: 10.1128/JVI.00586-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Keck ZY, Girard-Blanc C, Wang W, Lau P, Zuiani A, Rey FA, et al. Antibody response to hypervariable region 1 interferes with broadly neutralizing antibodies to hepatitis C virus. J Virol. 2016;90(6):3112–3122. doi: 10.1128/JVI.02458-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tarr AW, Owsianka AM, Jayaraj D, Brown RJ, Hickling TP, Irving WL, et al. Determination of the human antibody response to the epitope defined by the hepatitis C virus-neutralizing monoclonal antibody AP33. J Gen Virol. 2007;88(Pt 11):2991–3001. doi: 10.1099/vir.0.83065-0. [DOI] [PubMed] [Google Scholar]
- 40.Keck ZY, Li TK, Xia J, Bartosch B, Cosset FL, Dubuisson J, et al. Analysis of a highly flexible conformational immunogenic domain a in hepatitis C virus E2. J Virol. 2005;79(21):13199–13208. doi: 10.1128/JVI.79.21.13199-13208.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pedersen J, Carlsen TH, Prentoe J, Ramirez S, Jensen TB, Forns X, et al. Neutralization resistance of hepatitis C virus can be overcome by recombinant human monoclonal antibodies. Hepatology. 2013;58(5):1587–1597. doi: 10.1002/hep.26524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Carlsen TH, Pedersen J, Prentoe JC, Giang E, Keck ZY, Mikkelsen LS, et al. Breadth of neutralization and synergy of clinically relevant human monoclonal antibodies against HCV genotypes 1a, 1b, 2a, 2b, 2c, and 3a. Hepatology. 2014;60(5):1551–1562. doi: 10.1002/hep.27298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Law JL, Chen C, Wong J, Hockman D, Santer DM, Frey SE, et al. A hepatitis C virus (HCV) vaccine comprising envelope glycoproteins gpE1/gpE2 derived from a single isolate elicits broad cross-genotype neutralizing antibodies in humans. PLoS One. 2013;8(3):e59776. doi: 10.1371/journal.pone.0059776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Meunier JC, Engle RE, Faulk K, Zhao M, Bartosch B, Alter H, et al. Evidence for cross-genotype neutralization of hepatitis C virus pseudo-particles and enhancement of infectivity by apolipoprotein C1. Proc Natl Acad Sci U S A. 2005;102(12):4560–4565. doi: 10.1073/pnas.0501275102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tarr AW, Urbanowicz RA, Hamed MR, Albecka A, McClure CP, Brown RJ, et al. Hepatitis C patient-derived glycoproteins exhibit marked differences in susceptibility to serum neutralizing antibodies: genetic subtype defines antigenic but not neutralization serotype. J Virol. 2011;85(9):4246–4257. doi: 10.1128/JVI.01332-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wong JA, Bhat R, Hockman D, Logan M, Chen C, Levin A, et al. Recombinant hepatitis C virus envelope glycoprotein vaccine elicits antibodies targeting multiple epitopes on the envelope glycoproteins associated with broad cross-neutralization. J Virol. 2014;88(24):14278–14288. doi: 10.1128/JVI.01911-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Forns X, Allander T, Rohwer-Nutter P, Bukh J. Characterization of modified hepatitis C virus E2 proteins expressed on the cell surface. Virology. 2000;274(1):75–85. doi: 10.1006/viro.2000.0419. [DOI] [PubMed] [Google Scholar]
- 48.Alhammad Y, Gu J, Boo I, Harrison D, McCaffrey K, Vietheer PT, et al. Monoclonal antibodies directed toward the hepatitis C virus glycoprotein E2 detect antigenic differences modulated by the N-terminal hypervariable region 1 (HVR1), HVR2, and intergenotypic variable region. J Virol. 2015;89(24):12245–12261. doi: 10.1128/JVI.02070-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fauvelle C, Felmlee DJ, Crouchet E, Lee J, Heydmann L, Lefevre M, et al. Apolipoprotein E Mediates Evasion From Hepatitis C Virus Neutralizing Antibodies. Gastroenterology. 2016;150(1):206–217. doi: 10.1053/j.gastro.2015.09.014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.