Abstract
Purpose
The oncogenic transcription factor STAT3 promotes gene transcription involved in cancer and its activation by IL-6 is found in head and neck squamous cell carcinoma. Four triazolothiadizine STAT3 pathway inhibitors were evaluated to prioritize a single compound for in vivo examination.
Methods
Metabolic stability in mouse liver microsome incubation was used to evaluate four triazolothiadizine analogues, and UPCDC-10205 was administered to mice IV as single or multiple doses to evaluate toxicity. Single dose pharmacokinetics (PK), bioavailability and metabolism were studied after IV 4 mg/kg, PO 4 mg/kg, or PO 30 mg/kg suspension in 1% carboxymethyl cellulose. Mice were euthanized between 5 min to 24 h after dosing and plasma and tissues were analyzed by LC-MS. Non-compartmental PK parameters were determined.
Results
Of the four triazolothiadizine analogues evaluated, UPCDC-10205 was metabolically most stable. The maximum soluble dose of 4 mg/kg in 10% Solutol™ was not toxic to mice after single and multiple doses. PK analysis showed extensive tissue distribution and rapid plasma clearance. Bioavailability was ~5%. A direct glucuronide conjugate was identified as the major metabolite which was recapitulated in vitro.
Conclusions
Rapid clearance of UPCDC-10205 was thought to be the result of phase II metabolism despite its favorable stability in a phase I in vitro metabolic stability assay. The direct glucuronidation explains why microsomal stability (reflective of phase I metabolism) did not translate to in vivo metabolic stability. UPCDC-10205 did not demonstrate appropriate exposure to support efficacy studies in the current formulation.
Keywords: Pharmacokinetics, small molecule inhibitor of STAT3, IL-6, LC/MS
1 INTRODUCTION
Signal transducer and activator of transcription 3 (STAT3) has been implicated in most cancer types including head and neck cancer [1–6]. Head and neck squamous cell carcinoma (HNSCC) accounts for 90% of diagnosed head and neck cancers.[2] HNSCC is the sixth most common cancer with approximately 46,000 Americans diagnosed and 8,600 Americans dying from the disease annually [7,2]. Patients diagnosed with recurrent or metastatic HNSCC currently have no curative options [8]. There has been an urgent and unmet need, especially in cases of recurrent and metastatic disease, to identify new chemotherapeutic options for HNSCC.
STAT3 has been found to be constitutively active in HNSCC cell lines and tumors [1,6]. Aberrant STAT3 activation can lead to the initiation, development and metastasis of cancers by misregulating the genes and proteins needed to overcome cancerous progression barriers [3,9,10, 5, 1]. STAT3 activation has been labeled an oncogene and can lead to the initiation, development and metastasis in cancer by regulating functions such as cellular proliferation, cell cycle progression, apoptosis, angiogenesis, immune invasion, survival, inflammation, invasion and metastasis [3,1,4,9,11,12]. The role of STAT3 as a point of convergence for multiple cancerous pathways has made it an attractive target for the development of new chemotherapeutic therapies.
Development of therapies targeting STAT3 should consider specificity. STAT3 is part of a seven member protein family that are structurally similar and act as cytosolic signal transducers and transcription factors that regulate gene transcription [1,4,13,5]. The member STAT1 acts a tumor suppressor and this function should be preserved [4,14]. A growing body of evidence has implicated IL-6 as a prominent activator of STAT3 in HNSCC [6,1]. Based on three hits identified in a high content screening (HCS) campaign that selectively inhibited IL-6-induced pSTAT3 activation over IFNγ-induced pSTAT1 activation and inhibited HNSCC tumor cell line growth in vitro [15,16], the 1,2,4-triazolo-[3,4-b] thiadiazines were selected for further chemical optimization [17]. Our goal was to identify a lead candidate for in vivo examination among four analogues through mouse liver microsomal incubations and evaluate the toxicity, pharmacokinetics and metabolism, and ultimately efficacy studies in mice.
2 MATERIALS AND METHODS
2.1 Chemicals and Solvents
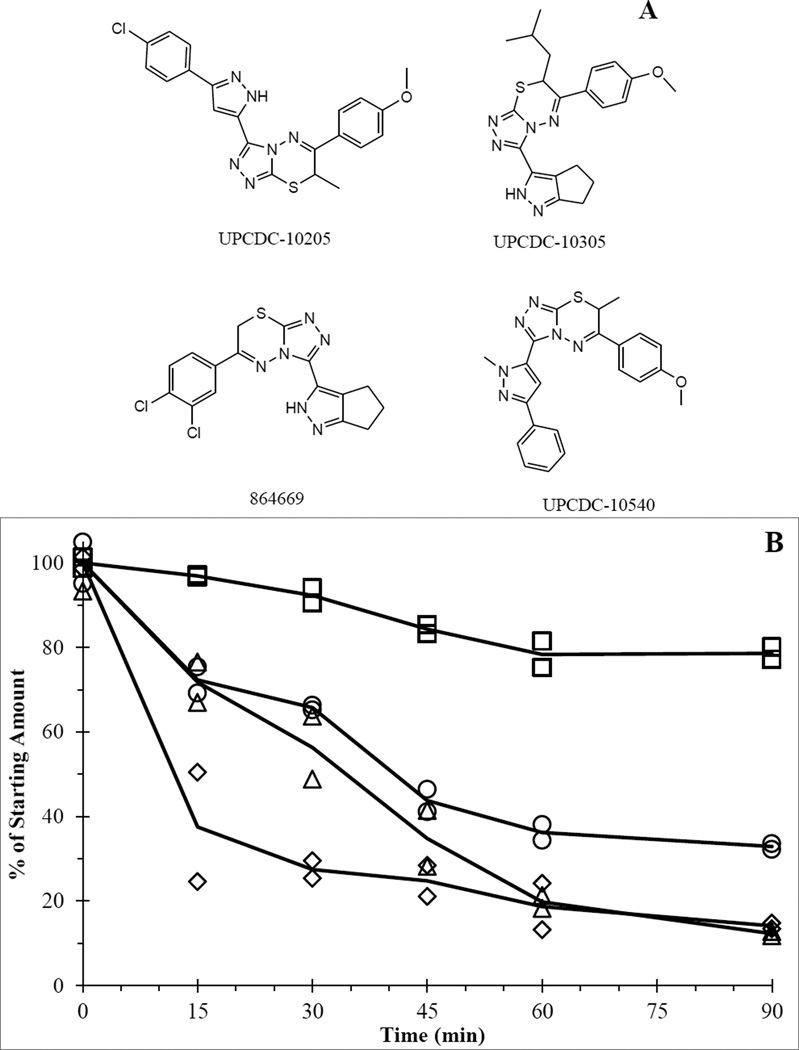
Compounds UPCDC-10205, UPCDC-10305, 864669, and UPCDC-10540 (see Figure 1A for structures) were synthesized and provided by the University of Pittsburgh Chemical Diversity Center (Pittsburgh, PA) [17]. The internal standard 2H7-UPCDC-10205 ([2H7]-3-(3-(4-chlorophenyl)-1H-pyrazole-5-yl)-6-(4-methoxyphenyl)-7-methyl-7H-[1,2,4]triazolo[3,4-b][1,3,4]thiadiazine) was custom synthesized and purchased from ALSACHIM (Illkirch-Graffenstaden, France). Water and acetonitrile (both HPLC grade), formic acid, monobasic and dibasic potassium phosphate, tris and DMSO were obtained through Fisher Scientific (Fairlawn, NJ). Bovine serum albumin (BSA), NADPH, carboxymethylcellulose, UDPGA, MgCl2, alamethicin and formic acid were purchased from Sigma-Aldrich (St. Louis, MO). Solutol™ HS15 was a gift from BASF (Florham Park, New Jersey).
Figure 1.
A) 669 series chemical structures. B) Mouse liver microsome incubation of UPCDC-10205 (□),UPCDC-10540 (○), UPCDC-10305 (Δ), and 864669 (◊).
2.2 Microsome Incubation and LC-MS Analysis
Microsomes were prepared and isolated from livers of heterozygous athymic nude-Foxn1nu/Foxn1+ female mice as previously published [18]. The microsomal pellet was stored at − 80 °C until measurement of protein concentration using a protein assay kit from BioRad (Hercules, CA) with BSA as the standard. Absorbance readings were recorded at 630 nm using an Infinite M100 Pro plate reader from Tecan (Männedorf, Switzerland). Actual incubation volumes were adapted from the published method to allow for a 200 µL incubation volume.
The 200 µL incubation contained 0.5 mg/mL microsomal protein, 1,000 ng/mL drug concentration, 1 µM NADPH, 0.1 M phosphate buffer (pH 7.4) and less than 0.1% acetontrile. The incubation was terminated with 1.0 mL acetonitrile. Sample times (point of organic reaction termination) were performed in triplicate at 0 (acetonitrile added prior to addition of microsomes), 15, 30, 45, 60 and 90 min. LMP400 (0 and 90 min incubations) was used as a positive control. Respective internal standard (10 µL of 1.0 µg/mL) of either UPCDC-10540 or UPCDC-10205 was added to samples, followed by briefly vortexing using a Vortex Genie-2 (VWR International, Radnor, PA) set at 8 and storage at −80 °C until LC-MS analysis.
An LC-MS method was developed to simultaneously quantitate the four STAT3 inhibitor analogues. Two separate MS methods (using identical LC-MS conditions) were utilized. Compound UPCDC-10540 was used as an internal standard to quantitate compounds 864669, UPCDC-10305 and UPCDC-10205. Compound UPCDC-10205 was used as an internal standard to quantitate compound UPCDC-10540.
The HPLC method utilized an Agilent 1100 autosampler and Agilent 1100 binary pump (Agilent Technologies, Palo Alto, CA) with a Synergi Hydro-RP 80A (4 µm particle size, 2 mm × 100 mm) column at ambient temperature. Mobile phase solvent A was 0.1% formic acid (v/v) in acetonitrile, and mobile phase solvent B was 0.1% formic acid (v/v) in water. The initial mobile phase was composed of 55% solvent A pumped at 0.3 mL/min for 5.0 min, changed to 99% solvent A and was held there for 1.0 min. At 6.1 min the percentage of solvent A returned to initial conditions of 55% for 4 min followed by injection of the next sample. Total run time was 10 min. A Quattro Micro mass spectrometer (Waters Corporation, Milford, MA) was used in positive-ion SRM mode (4.0 kV capillary voltage, 40V cone voltage) to monitor m/z 391.0 for 864669, m/z 409.0 for UPCDC-10305, m/z 437.0 for UPCDC-10205, and m/z 416.0 for UPCDC-10540. Retention times were as follows: 3.4 min for 864669, 4.6 min for UPCDC-10205, 4.2 min for UPCDC-10305, and 4.9 min for 540.
Calibrators (30, 100, 300, 500, and 1000 ng/mL) were prepared in a 0.1 M phosphate buffer (pH 7.4) that contained 0.5 mg/mL BSA to match the protein concentration of the microsomal incubation samples. A volume of 0.2 mL of the buffer mixture was placed into a 1.5 mL microcentrifuge tube. An internal standard solution (10 µL of 10 µg/mL) was added to each tube before being briefly vortexed. A volume of 1.0 mL of acetonitrile was then added and the samples vortexed for 1 minute. Samples were centrifuged at 14,000 × g for 4 min. The dried supernatant was resuspended in 100 µL of starting condition mobile phase. The sample injection volume was 5 µL. The regression was weighted 1/y2 and fit quadratically (y = a + b*x + c*x2). A triplicate standard curve was prepared and analyzed for accuracy (−9.5 to 6.0%) and precision (CV<10.1%) prior to sample analysis.
2.3 Mice
Specific pathogen-free heterozygous Foxn1nu/Foxn1+ mice (5–7 weeks of age) were purchased from Envigo (Indianapolis, IN, USA). Mice were allowed to acclimate to the University of Pittsburgh Animal Facility for at least 1 week before studies were initiated. To minimize exogenous infection, mice were maintained in microisolator cages and handled in accordance with the Guide for the Care and Use of Laboratory Animals (National Research Council, 2011) and on a protocol approved by the University of Pittsburgh IACUC. Ventilation and airflow in the animal facility was set to 12 changes/h. Room temperature was regulated at 72 ± 4 °F and the rooms were kept on automatic 12-h light/dark cycles. The mice received Prolab ISOPRO RMH 3000, Irradiated Lab Diet (PMI Nutrition International, Brentwood, MO) and water ad libitium. Mice were fasted overnight prior to administration of UPCDC-10205. Mice were stratified based on body weights into dose or time point groups to eliminate statistical differences in body weight.
2.4 Toxicity Studies
The vehicle 10% Solutol™ HS15 in sterile water provided the maximum solubilized concentration of UPCDC-10205 at 0.4 mg/mL and was administered to mice at a volume of 0.01 ml/g body weight. Mice were dosed IV at 4 mg/kg using this solution for toxicity studies. Other formulations did not increase the maximum soluble dose.
A single dose of UPCDC-10205 (4 mg/kg, 0.01 ml/g body weight) was administered to both male and female mice (5 per group) IV by lateral tail vein injection along with mice administered a vehicle control. After the single dose, mice were observed for a 14 day period to monitor clinical health, and body weights were recorded twice weekly. After the observation period, necropsies were performed on the mice to identify gross pathologies.
For the multiple dose toxicity study, 5 female mice per treatment group were administered UPCDC-10205 daily for 5 days IV by lateral tail vein in addition to mice administered a vehicle control. UPCDC-10205 doses used were the single dose MTD (4 mg/kg), 2/3 the single dose MTD (2.7 mg/kg) and 1/3 the single dose MTD (1.3 mg/kg). After the 5 day treatment, mice were observed for a 14 day period where clinical health was checked daily and body weights were measured twice weekly. After the observation period, mice were euthanized and necropsies performed on mice to identify gross pathologies.
2.5 Pharmacokinetics
Three female mice per treatment group per time point (5, 15, 30, 60, 120, 240, 360, 1440 min) were administered a single dose of 4.0 mg/kg UPCDC-10205 in 10% Solutol™ HS by either lateral tail vain IV injection or PO administration by oral gavage or 30 mg/kg PO administration in a suspension of 1% carboxymethyl cellulose (CMC). Control mice were treated with vehicle and euthanized at 5 and 1440 min. Mice were euthanized by CO2 inhalation and the following tissues collected: blood, liver, kidney, spleen, lung, heart, fat, skeletal muscle, and brain. Blood was collected by cardiac puncture using EDTA anticoagulated needles and syringes, transferred to microcentrifuge tubes and centrifuged at 12,000 × g for 4 min to separate plasma and red blood cells (RBCs). Urine and feces were collected over ice from mice in 1440 min groups by gang housing the 3 mice/group in metabolic cages. Tissues were weighed and all samples were flash frozen using liquid nitrogen and stored at −80 °C until analysis.
2.6 Quantitative LC-MS/MS Assay
To support the murine PK study, a sensitive LC-MS/MS assay was developed to quantitate UPCDC-10205. A stable isotope, deuterated, 2H7 -UPCDC-10205, was used as the internal standard. The HPLC method utilized an Agilent 1100 binary pump and 1200 series autosampler (Santa Clara, CA). Chromatographic separation was achieved using a Phenomenex (Torrance, CA) Synergi Hydro-RP 80A column (4 µm particle size, 2 mm × 100 mm) at ambient temperature. Mobile phase solvent A was 0.1% formic acid (v/v) in acetonitrile, and mobile phase solvent B was 0.1% formic acid (v/v) in water. The initial mobile phase was composed of 50% solvent A pumped at 0.3 mL/min for 12.0 min, changed to 95% solvent A and was held constant for 4.0 min. At 16.1 min the percentage of solvent A returned to initial conditions of 50% and allowed to equilibrate for 2 min followed by an injection of the next sample. Total run time was 18 min. The retention times were 9.6 and 9.4 min for UPCDC-10205 and 2H7 - UPCDC-10205, respectively.
A Waters Quattro Micro mass spectrometer (Milford, Massachusetts) was used in positive-ion MRM mode (4.5 kV capillary voltage, 30 V cone voltage, 450 °C desolvation temp.) to monitor m/z 437.0>304 for UPCDC-10205 and m/z 444.0>304.0 for the internal standard. The calibration curve had a range of 1 to 1000 ng/mL (1, 3, 10, 30, 100, 300, 500, 1000 ng/mL) and were prepared fresh in control mouse plasma obtained from Lampire Biological Labs Inc. (Ottsville, PA). Calibrators were prepared from serial dilutions of UPCDC-10205 in DMSO from a 1.0 mg/mL UPCDC-10205 stock solution. Quality controls (QCs) were prepared in control mouse plasma at three concentrations (2.5, 30, 800 ng/mL). Two QCs at each level were analyzed alongside each sample batch.
The sample volume was 100 µL of plasma or tissue homogenate (homogenized with 3 parts PBS (v/g)). All samples were spiked with 10 µL of 0.2 µg/mL of the 2H7-UPCDC-10205 internal standard. Extraction and protein precipitation with 500 µL of acetonitrile, vortexing for 2 min set at 8, and centrifugation was followed by evaporating supernatants at 37 °C under a gentle stream of nitrogen.
Assay performance and stability was evaluated. A triplicate standard curve weighted 1/y2 and fit linearly yielded accurate (−6.3–8.1%) and precise (CV< 8.3%) back-calculated data. The recovery of UPCDC-10205 at the QCM was 46.2% (CV 8.0%). Ion-suppression at the QCM was 8.2% (CV 6.6%). The effect of 3 consecutive −80 °C freeze/thaw cycles at QCM was acceptable with 111.5% recovery. The stability of UPCDC-10205 in plasma during sample preparation at QCM for 4 h at room temperature was 100.8%. Quantitation of UPCDC-10205 in kidney and liver tissue homogenates analyzed against a plasma calibration curve resulted in recoveries of 98.8– 105.1% with CVs ranging from 2.3 –7.4%.
The values quantitated for each of the three mice per time point were averaged to obtain a mean that served as the representative value for that time point. PK parameters (AUC0-t, AUC0-∞, T1/2, Vd, and Vss) were derived through non-compartmental analysis using PK Solutions (Summit PK, Montrose, CO)
2.7 Protein Binding
To assess the level plasma protein binding of UPCDC-10205, a rapid equilibrium dialysis (RED) method (Thermo Scientific, Pittsburgh, PA) was used. Control mouse plasma was spiked at 1,000 ng/mL using a stock prepared in DMSO (organic < 0.1%). Four replicates of 200 µL from the spiked 1000 ng/mL plasma were placed into the RED insert placed within a Teflon plate and 350 µL of PBS (pH 7.2) was aliquoted in the opposite side of the insert. The plate was placed on a shaker in an incubator at 37 °C for 4 hours. After this time, both saline and plasma portions were isolated and stored at −80 °C until analysis. Quantitation was achieved by creating standard curve in a matrix of 1:1 PBS:plasma and prepared and analyzed using the quantitation LC-MS/MS assay. Samples were diluted with either PBS or plasma to match the calibrator matrix.
2.8 Metabolite Profiling
To identify metabolites of UPCDC-10205, a series of LC-MS/MS methods were developed and utilized. Both plasma (IV 30 min 4 mg/kg) and urine (IV 0-6 hr 4 mg/kg) from mice used in the PK study were separately evaluated and compared to their respective vehicle controls to identify unique traces in treated mouse samples. Unique traces were confirmed using a selected reaction monitoring (SRM) method that monitors specified ions. Unique trace potential metabolites were postulated by comparing the unique m/z trace to the parent UPCDC-10205 m/z (437).
Sample preparation was identical to the quantification method (see 2.6 Quantitative LCMS/ MS Assay). The LC method was extended compared to the quantitative method to a run time of 75 minutes and used a gradient to spread out eluates using a Phenomenex Luna phenyl-hexyl column (3µ, 100×2.0 mm). Mobile phase conditions for mobile phase A (acetonitrile with 0.1% formic acid) increased from the initial 5% to 55% at 0.3 mL/min for 60 min. This composition was then at 55% for 5 minutes then increased to 80% and held for 5 additional minutes until returning to initial conditions for equilibration during the final 5 minutes.
Full mass (MS) scans were conducted to obtain a total scan range of 400–900 m/z. MS parameters were identical to the quantification method, with the exception of collision voltage and collision gas being turned off.
Unique chromatographic peaks detected in urine or plasma were analyzed for their m/z trace composition. These m/z values were then used in an SRM MS method and the samples reanalyzed. To gain structural information, product ion scans (PIS) were used to identify sites of metabolic alterations using varying collision voltages (10, 20, 30, 40, and 50 V). PIS fragments were compared to UPCDC-10205 fragmentation using ChemDraw (PerkinElmer, Waltham, MS).
The natural abundance of the stable isotope of 37-chlorine (37Cl) is approximately 25%, and this was used to fingerprint UPCDC-10205 metabolites. Unique chromatographic peaks with m/z traces and m/z +2 were considered derived from UPCDC-10205.
2.9 In Vitro Metabolism
Microsomes were prepared and isolated from heterozygous Foxn1nu/Foxn1+ female mice as described previously (see 2.2 Microsome Incubation and LC-MS Analysis). The microsomal pellet was stored at −80 °C until measurement of protein concentration using the Biorad assay using BSA as a standard. The incubation for glucuronide metabolism was adapted from a previously published method to allow for a 1,000 µL incubation volume [19]. The incubation mixture contained 0.1 M phosphate buffer (pH 7.4), 0.25 mg/mL microsomal protein concentration, 25 µg/mL alamethicin, 50 mM Tris-HCl, 10 mM MgCl2, and a 1,000 ng/mL drug concentration. The reaction was catalyzed with the addition of 5 µM UPDGA. At 0, 15, 30, 45, and 60 minutes, 100 µL aliquots were sampled from the reaction mixture and 500 µL of acetonitrile added to terminate the reaction. 10 µL of 1.0 µg/mL the internal standard (2H7 - UPCDC-10205) was added to each sample. Samples were processed as described previously. Samples were analyzed twice along with a standard curve produced in phosphate buffer matrix (pH 7.4) with 0.25 mg/mL BSA. The first analysis used the qualitative SRM LC-MS assay to monitor a 613 m/z (437+176) glucuronide channel and the second used the quantitative MRM LC-MS assay to quantitate UPCDC-10205 substrate depletion. An incubation mixture without UPDGA cofactor was used as a negative control.
3 RESULTS
3.1 Microsome Incubation
For all microsome incubations, compound concentrations fell within the standard calibration curve range. Concentration values were expressed as a % of the average starting concentration (0 min), seen in Figure 1B. UPCDC-10205 had the largest proportion of the starting amount remaining (79%) after 90 minutes compared to 14.1% for 864669, 12.3% for UPCDC-10305 and 32.9% for UPCDC-10540. Based on its superior metabolic stability, UPCDC-10205 was prioritized for in vivo experimentation.
3.2 Toxicity Studies of UPCDC-10205
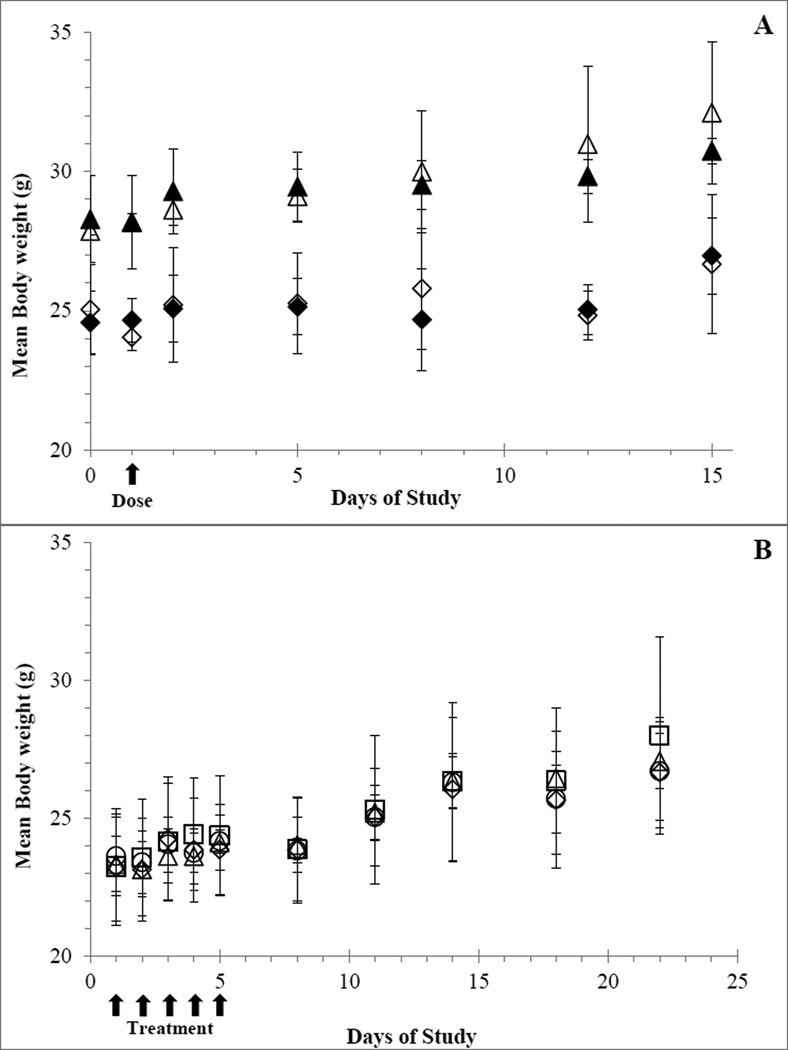
Single dose administration of the maximum soluble dose of UPCDC-10205 at 4 mg/kg resulted in mice that appeared normal and similar to vehicle dosed control mice with no apparent weight loss measured within the first 24 h after injection. During the 14 day observation period, all mice were healthy and gained weight (Figure 2A). Upon necropsy, no gross pathology was detected in any of the mice. One male had bite marks and local irritation on the skin between the scapulae where he had been bitten. The testes in this mouse were also smaller by about 30% compared to his cage mates and his spleen weight was slightly elevated, most likely due to infection. All other male mice appeared normal. The maximum solubilized dose of 4 mg/kg was determined to be the maximum tolerable dose (MTD).
Figure 2.
Average mouse body weights (N=5; Error bars represent ±1 SD). A) Single dose toxicity study before and following a single IV injection of UPCDC-10205 at 4 mg/kg to male treated (▲), male vehicle (Δ), female treated (◊), and female vehicle (♦) mice. B) Multiple dose toxicity study, before, during and following QDx5 IV administration of UPCDC-10205 to female mice at 4 mg/kg (□), 2.7 (Δ), 1.3 (○) and vehicle (◊).
Because there were no obvious differences between sexes in both clinical observations and body weights between vehicle treated and UPCDC-10205 treated mice, only female mice were used for the multiple dose toxicity study. No signs or symptoms of toxicity were detected during the five day treatment period or the 14 day observation period. All mice gained weight during the study and weights between treatment groups showed no statistical differences to the vehicle control group (Figure 2B). There were no signs of gross pathology upon necropsy.
3.3 Pharmacokinetics of UPCDC-10205
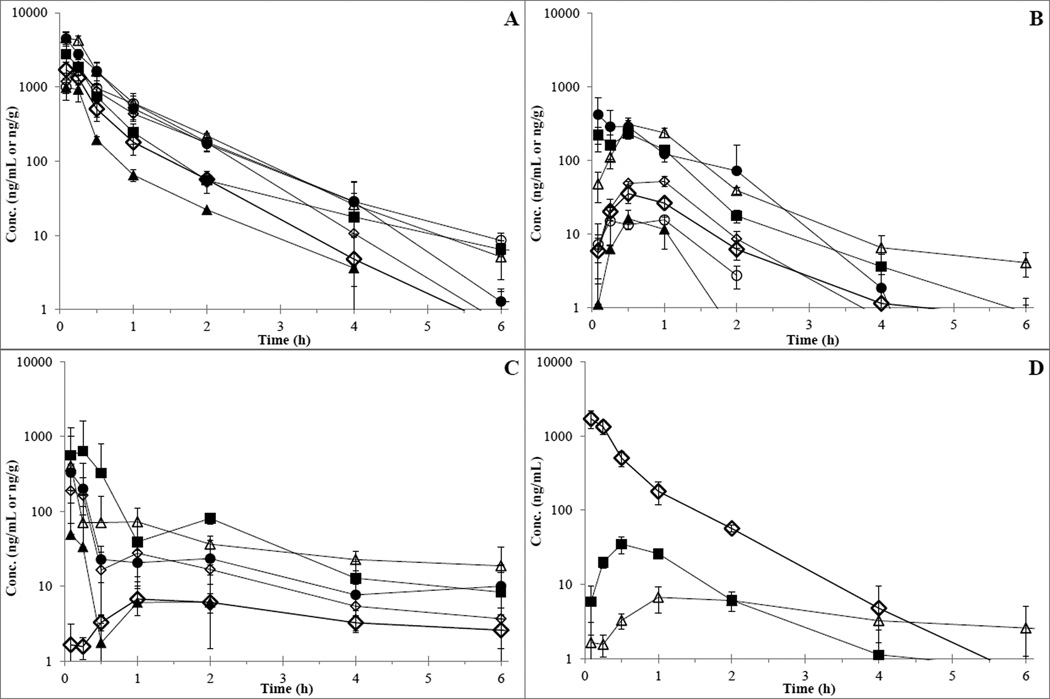
Plasma and tissue concentration versus time profiles are depicted in Figure 3 and pharmacokinetic parameters are listed in Table 1. The plasma Cmax for IV administered UPCDC-10205 was 1,707 ng/mL (± 455 ng/mL) and occurred at the first observation of 5 minutes. Plasma UPCDC-10205 levels were below the lower limit of quantitation (LLQ) by the 6 h time point. The in vivo profile appears biphasic indicating two compartmental behavior. The half-life in plasma was calculated to be 0.6 h and the AUC0-t to be 1021 ng•h/ml. AUC0-inf was 1021 ng•h/ml, clearance was 3.91 L/h/kg, Vd was 3.4 L/kg, and Vss was 2.0 L/kg. The Cmax for orally administered UPCDC-10205 at 4 mg/kg was 35.0 ng/mL and occurred at 0.5 hours after administration. Plasma UPCDC-10205 concentrations were below the LLQ after the 2 h time point. The half-life was 0.8 h and oral bioavailability (using AUC0-t) was 5.2%. The Cmax for PO administered UPCDC-10205 at 30 mg/kg in 1% CMC suspension was 6.7 ng/mL and occurred at 1.0 h after administration. The half-life was 3.4 h and the bioavailability was 0.3%. All tissues, including brain, exhibited higher levels of UPCDC-10205 than plasma indicating extensive tissue distribution.
Figure 3.
Concentration vs time results following administration of UPCDC-10205. Points represent the mean of the mice (N=3) and error bars represent ±1 SD. A) Plasma and tissues from IV 4 mg/kg UPCDC-10205. B) Plasma and tissues from PO 4 mg/kg UPCDC-10205. C) Plasma and tissues from PO 30 mg/kg UPCDC-10205 in 1% CMC; ◊=plasma, ■=liver, Δ=kidney, ●=lung, ▲=RBC, ◊=muscle, ○=brain. D) Plasma UPCDC-10205 concentration versus time from all three administration types. ◊= IV 4 mg/kg, ■= PO 4 mg/kg, Δ= PO 30 mg/kg in 1% CMC.
Table 1.
Pharmacokinetic parameters of UPCDC-10205 in mice.
| Route | mg/kg | Parameter | Plasma | Liver | Kidney | Lung | RBC | Muscle | Brain |
|---|---|---|---|---|---|---|---|---|---|
| IV | 4 | Cmax (ng/mL) | 1708 | 2779 | 4621 | 4494 | 980 | 1900 | 1643 |
| Tmax (min) | 5 | 30 | 5 | 5 | 5 | 15 | 15 | ||
| AUC0−t (ng/mL*h) | 1021 | 1472 | 3240 | 2681 | 556 | 1634 | 1731 | ||
| %F | 100 | - | - | - | - | - | - | ||
| PO | 4 | Cmax (ng/mL) | 35 | 220 | 308 | 414 | 16 | 51.2 | 15.4 |
| Tmax (min) | 30 | 5 | 30 | 4.98 | 30 | 60 | 60 | ||
| AUC0−t (ng/mL*h) | 52.1 | 300 | 428 | 630 | 23.5 | 91.4 | 30 | ||
| %F | 5.1 | - | - | - | - | - | - | ||
| CMC | 30 | Cmax (ng/mL) | 6.7 | 564 | 404 | 331 | - | 189 | 49 |
| Tmax (min) | 60 | 5 | 5 | 5 | - | 5 | 5 | ||
| AUC0−t (ng/mL*h) | 25.7 | 542 | 272 | 186 | - | 140 | 33.4 | ||
| %F | 0.3 | - | - | - | - | - | - |
Cmax and Tmax were derived from averages (N=3). UPCDC-10205 AUC0−t calculated non-compartmentally between all three administration routes and tissues. The percentage of AUC0−inf extrapolated beyond AUC0−t was less than 2.5% for IV and PO, and 32.7% for CMC. %F represents the equation of (IV plasma AUC0−t/PO plasma AUC0−t)*(IV dose/PO dose)*100.
UPCDC-10205 levels in pooled urine from the IV 4 mg/kg mice represented 0.0011% of the total amount administered for 0-6 h collection and 0.0010% for 6–24 h collection. Levels in pooled urine from the PO 4 mg/kg mice represented 0.0077% of the total dose between 0-6 h. Quantitation of UPCDC-10205 in the 6–24 h as well as all urine samples from the PO 30 mg/kg in 1% CMC had concentrations below the LLQ.
The average from the 4 replicates in the plasma portion of the RED protein binding device was 1,091 ng/mL and the value detected in the saline buffer portion was below the 1 ng/mL assay LLQ at 0.2 ng/mL. These data result in a calculated protein binding of >99.91% (using the LLQ value) or approximately 99.98% (using 0.2 ng/mL).
3.4 Metabolite Profiling of UPCDC-10205
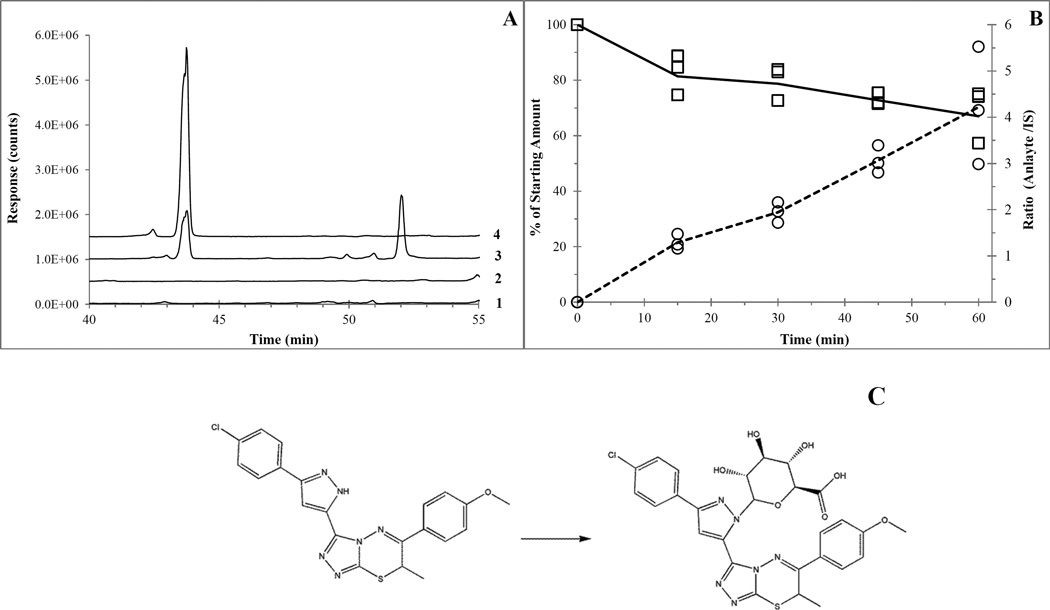
MS scans (400–900 m/z) of urine (4 mg/kg IV 0–6 hour pooled) and plasma (4 mg/kg IV 30 min) revealed a single unique peak at a retention time of 43.6 min that contained m/z traces of 437, 439, 613 and 615. 612 is the postulated molecular weight of a direct glucuronide conjugate of UPCDC-10205 (436 + 176 = 612). The smaller (1/3rd) +2 m/z trace of 615 is in line with the presence of a chlorine isotope fingerprint. The 437 and 439 m/z traces are likely due to in-source fragmentation. The SRM method (613 and 437 m/z) conducted on plasma yielded similar results, see Figure 4A. The product ion scan (PIS) of the 613 ion (20V collision voltage) in plasma revealed unique product ions of 477 and 479 m/z at the 43.2 min RT peak, revealing a similar 37Cl fingerprint. This m/z is greater than the parent (437), indicating the glucuronide is still conjugated and a separate part of the parent has been fragmented. Not enough information was available to deduce structure based on this fragmentation. Based on its structure, direct glucuronidation of UPCDC-10205 is postulated to occur on one of the nitrogens of the pyrazole group, although only a single chromatographic peak was observed and the specific regioisomer was not determined (see Figure 4C). Samples from microsomal incubations focused on glucuronidation (addition of alamethicin and appropriate cofactors) were analyzed using the qualitative SRM LC-MS assay for the glucuronide (see 2.8 Metabolite Profiling) and analyzed separately with the quantitative assay for UPCDC-10205 (see 2.6 Quantitative LC-MS/MS Assay). The rate of metabolite formation is 0.068 response ratio/min/0.25 mg protein (Figure 4B). Measured concentrations of UPCDC-10205 were expressed relative to time 0. On average, only 67% remained after 60 min (Figure 4B). One of the postulated regioisomer possibilities of the glucuronide metabolite is shown in Figure 4C.
Figure 4.
A) Chromatogram of the SRM scan (613 and 437 m/z channels) of mouse plasma from the PK study between 40 to 55 minutes. Line 1 is vehicle 437 m/z, line 2 is vehicle 613 m/z (5×106 count offset), line 3 is treated 437 m/z (1.0×107 count offset) and line 4 is treated 613 m/z (1.5×107 count offset). B) 60 minute mouse liver microsome incubation with UPCDC-10205, alamethicin and UPDGA to produce direct N-glucuronide (613 m/z). Left axis shows quantification of UPCDC-10205 (□) represented by a percent of the starting amount. Right axis shows production of metabolite (○) represented as the ratio of analyte to internal standard values. C) Proposed metabolism of UPCDC-10205 to the conjugated N-glucuronide; note that there are 2 possible regioisomers for the glucuronide, one of which is shown.
4 DISCUSSION
Our goal was to identify a lead candidate for in vivo examination. Four analogues from a series of triazolothiadiazines [17] were selected for liver microsomal screening, revealing that compound UPCDC-10205 was the most metabolically stable in the presence of phase I enzymes, with approximately 80% of the compound remaining after 90 min. UPCDC-10205 demonstrated efficacy in inhibiting pSTAT3 from IL-6 induction in a Cal33 cell line with an IC50 of 2.2 uM [17].
Neither single nor multiple dose UPCDC-10205 administrations to mice demonstrated signs of toxicity. The lack of toxicity may very well be due to the maximum soluble concentrations of UPCDC-10205, which limited the explored dose to 4 mg/kg or less. Other formulations were unable to achieve higher concentrations.
Plasma pharmacokinetics showed a biphasic profile with a terminal half-life of 0.6 h. The peak plasma concentration of UPCDC-10205 after IV dosing of 1.7 µg/mL (4.0 µM) is in the cell media concentration range reported to be active in vitro [17]. However, higher concentrations and ability to maintain concentrations for prolonged periods of time would be desirable. UPCDC-10205 displayed very high plasma protein binding of >99.9%, yet, this did not prevent extensive tissue distribution. Highly perfused tissues, such as kidney, displayed a similarly early Tmax of 5 min indicating rapid distribution, at 3 times the exposure compared to plasma. Less perfused skeletal muscle experienced approximately 50% higher concentrations at a later Tmax of 15 min. Brain exposure was similar to skeletal muscle, demonstrating the ability of UPCDC-10205 to traverse the blood brain barrier.
Clearance after IV dosing at 3.9 L/h/kg was higher than reported for mouse liver plasma flow of 2.7 L/h/kg [20]. We found no evidence for phase I metabolism, and a single direct glucuronide was the only metabolite detected. The molecular weight of UPCDC-10205 (436 a.m.u) is above the reported threshold for rodent biliary excretion (250 a.m.u.), and this route could have contributed to clearance, and this would need to be confirmed as bile was not collected in the current study. The human threshold (500–600 a.m.u.) is somewhat higher, suggesting that in humans this route of excretion may be less important [21]. Renal excretion did not appear to contribute significantly to clearance based on the amount excreted in urine (<0.1% of dose). Comparison of IV and PO pharmacokinetics revealed UPCDC-10205’s poor bioavailability at 5.2%. Increasing the amount dosed by formulating UPCDC-10205 as a suspension did not increase the plasma exposure, though it did extend the duration of absorption somewhat with a later Tmax. The major factors affecting bioavailability are absorption and presystemic metabolism (first pass effect) [22]. The first pass effect may limit bioavailability of UPCDC-10205 by metabolism mediated by liver UGTs. Additionally, biliary excretion may further decrease bioavailability.
In vivo administration of UPCDC-10205 resulted in a shorter half-life and lower exposure than anticipated based on the first liver microsome stability study targeting susceptibility to oxidative biotransformation. Identification of the direct-glucuronide metabolite as the only biotransformation product validates the prioritization of UPCDC-10205 in the original microsome incubation studies. However, although translatable, the liver microsomal incubation did not account for the direct glucuronidation of UPCDC-10205. In selecting a candidate, metabolic stability, in addition to in vitro potency, must be considered to yield efficacious leads in vivo [23,24]. In vitro metabolic stability is used as a predictor for in vivo metabolic clearance because cytochrome P450 enzymes account for the metabolism of approximately 90% of all drugs [25,26]. The 20% substrate depletion of UPCDC-10205 observed in the initial incubation did not account for the glucuronidation of UPCDC-10205, and the susceptibility of UPCDC-10205 to glucuronidation was demonstrated in the second microsome incubation, with UGTs depleting approximately 25% of UPCDC-10205 after 60 min. Although our prediction that UPCDC-10205 would be metabolically stable in vivo was not substantiated, the other analogues all contain the same pyrazole groups postulated as the site for direct glucuronidation, and have the potential to generate similar metabolites. Not only would they have been similarly susceptible to glucuronidation, but they would be expected to be more susceptible to phase I metabolism than UPCDC-10205 based on the results from the first liver microsome incubation (Figure 1).
The nitrogen of the pyrazole group is able to be targeted by UGTs for conjugation. Nglucuronidation is largely accomplished through UGT isoforms 1A4 and 2B10 and the enzyme levels of these isoforms is higher in humans than in preclinical species [27]. Glucuronidation rate differences between species are variable and further complicate extrapolation of metabolic rates to human [28]. Furthermore, both UGT2B10 and 1A4 exhibit high variability in expression levels in an analysis of human liver samples [29]. These UGT enzymes exist primarily in the liver, but UGT1A4 expression has also been detected in kidneys [30]. Glucuronidation can also occur in intestinal microsomes, which could also to contribute to decreased bioavailability [31]. Direct N-glucuronidation to pyrazole groups of small molecule drugs has been observed previously [32–35], and has prompted the synthesis of modified analogues designed to decrease susceptibility to this metabolism [35]. The addition of electronegative groups to metabolically active heterocycles such as pyrazoles may decrease glucuronidation [36]. Similar alterations to UPCDC-10205 could potentially decrease metabolic clearance, increasing bioavailability, plasma half-life, and ultimately exposure. However, there is no data on what these structural changes might do to STAT3 activity.
While UPCDC-10205 demonstrated in vitro potency, selectivity, extensive tissue distribution and metabolic stability, the current studies indicate areas of future improvements for this series. Solubility, susceptibility to glucuronidation, and plasma protein binding have been identified as potential limiting factors to achieving efficacious exposures. Investigations are currently underway to optimize this structure to generate next-generation lead candidates.
Acknowledgments
This project has been funded, in part, with Federal Funds from the National Cancer Institute, National Institutes of Health, under Contract No. HSN261200800001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government. This work was supported by the NExT-CBC Project ID #1015, S08-221 Task Order 6 ‘‘STAT3 Pathway Inhibitor HCS’’ (Grandis, PI), NCI Chemical Biology Consortium, Pittsburgh Specialized Application Center (PSAC) (Lazo and Johnston co-PIs), and University of Pittsburgh Chemical Diversity Center (Huryn, PI). The project was also supported, in part, by funds from the American Cancer Society (Grandis) and a Head and Neck Spore P50 award (Grandis, CA097190). This project was funded in the National Cancer Institute, National Institutes of Health, under Chemical Biology Consortium Contract No. HSN261200800001E. We are grateful for scientific contributions from Peter Wipf, Matthew LaPorte, Atefeh Garzan, Mary Liang, Vsevolod A. Peshkov, and Zhuzhu Wang. This project used the UPCI Animal Facility (AF) and Clinical Pharmacokinetic and Pharmacodynamic Facility (CPPF) and was supported in part by award P30CA047904.
Abbreviations
- CMC
Carboxymethyl cellulose
- LC-MS/MS
Liquid chromatography-tandem mass spectrometry
- PK
Pharmacokinetics
- UGT
UDP-glucuronosyltransferase
- HCS
High Content Screen
- AUC
Area Under the Curve
- Cmax
Maximum peak concentration
- Tmax
Time of maximum concentration
BIBLIOGRAPHY
- 1.Geiger JL, Grandis JR, Bauman JE. The STAT3 pathway as a therapeutic target in head and neck cancer: Barriers and innovations. Oral Oncol. 2016;56:84–92. doi: 10.1016/j.oraloncology.2015.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Argiris A. EGFR inhibition for recurrent or metastatic HNSCC. The Lancet Oncology. 2015;16(5):488–489. doi: 10.1016/S1470-2045(15)70178-6. [DOI] [PubMed] [Google Scholar]
- 3.Argiris A, Karamouzis MV, Raben D, Ferris RL. Head and neck cancer. Lancet (London, England) 2008;371(9625):1695–1709. doi: 10.1016/S0140-6736(08)60728-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Frank DA. STAT3 as a central mediator of neoplastic cellular transformation. Cancer letters. 2007;251(2):199–210. doi: 10.1016/j.canlet.2006.10.017. [DOI] [PubMed] [Google Scholar]
- 5.Wang X, Crowe PJ, Goldstein D, Yang JL. STAT3 inhibition, a novel approach to enhancing targeted therapy in human cancers (review) International journal of oncology. 2012;41(4):1181–1191. doi: 10.3892/ijo.2012.1568. [DOI] [PubMed] [Google Scholar]
- 6.Sriuranpong V, Park JI, Amornphimoltham P, Patel V, Nelkin BD, Gutkind JS. Epidermal growth factor receptor-independent constitutive activation of STAT3 in head and neck squamous cell carcinoma is mediated by the autocrine/paracrine stimulation of the interleukin 6/gp130 cytokine system. Cancer research. 2003;63(11):2948–2956. [PubMed] [Google Scholar]
- 7.Siegel RL, Miller KD, Jemal A, Yu H, Lee H, Herrmann A, Buettner R, Jove R, Spitzner M, Ebner R, Wolff HA, Ghadimi BM, Wienands J, Grade M. Cancer statistics, 2015. CA Cancer J Clin. 2015;65(1):5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 8.Sacco AG, Cohen EE, Waring MJ, Arrowsmith J, Leach AR, Leeson PD, Mandrell S, Owen RM, Pairaudeau G, Pennie WD, Pickett SD, Wang J, Wallace O, Weir A, Argiris A, Oh DY, Lee SH, Han SW, Kim MJ, Kim TM, Kim TY, Heo DS, Yuasa M, Yanagihara Y, Bang YJ. Current Treatment Options for Recurrent or Metastatic Head and Neck Squamous Cell Carcinoma. J Clin Oncol. 2015;33(29):3305–3313. doi: 10.1200/JCO.2015.62.0963. [DOI] [PubMed] [Google Scholar]
- 9.Sen M, Joyce S, Panahandeh M, Li C, Thomas SM, Maxwell J, Wang L, Gooding WE, Johnson DE, Grandis JR. Targeting Stat3 abrogates EGFR inhibitor resistance in cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. 2012;18(18):4986–4996. doi: 10.1158/1078-0432.CCR-12-0792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aggarwal BB, Kunnumakkara AB, Harikumar KB, Gupta SR, Tharakan ST, Koca C, Dey S, Sung B. Signal transducer and activator of transcription-3, inflammation, and cancer: how intimate is the relationship? Annals of the New York Academy of Sciences. 2009;1171:59–76. doi: 10.1111/j.1749-6632.2009.04911.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johnston PA, Grandis JR. STAT3 signaling: anticancer strategies and challenges. Mol Interv. 2011;11(1):18–26. doi: 10.1124/mi.11.1.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Niu G, Wright KL, Huang M, Song L, Haura E, Turkson J, Zhang S, Wang T, Sinibaldi D, Coppola D, Heller R, Ellis LM, Karras J, Bromberg J, Pardoll D, Jove R, Yu H. Constitutive Stat3 activity up-regulates VEGF expression and tumor angiogenesis. Oncogene. 2002;21(13):2000–2008. doi: 10.1038/sj.onc.1205260. [DOI] [PubMed] [Google Scholar]
- 13.Bromberg JF, Wrzeszczynska MH, Devgan G, Zhao Y, Pestell RG, Albanese C, Darnell JE., Jr Stat3 as an oncogene. Cell. 1999;98(3):295–303. doi: 10.1016/s0092-8674(00)81959-5. [DOI] [PubMed] [Google Scholar]
- 14.Avalle L, Pensa S, Regis G, Novelli F, Poli V. STAT1 and STAT3 in tumorigenesis: A matter of balance. Jakstat. 2012;1(2):65–72. doi: 10.4161/jkst.20045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnston PA, Sen M, Hua Y, Camarco D, Shun TY, Lazo JS, Grandis JR. High-content pSTAT3/1 imaging assays to screen for selective inhibitors of STAT3 pathway activation in head and neck cancer cell lines. Assay and drug development technologies. 2014;12(1):55–79. doi: 10.1089/adt.2013.524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnston PA, Sen M, Hua Y, Camarco DP, Shun TY, Lazo JS, Wilson GM, Resnick LO, LaPorte MG, Wipf P, Huryn DM, Grandis JR. HCS campaign to identify selective inhibitors of IL-6-induced STAT3 pathway activation in head and neck cancer cell lines. Assay and drug development technologies. 2015;13(7):356–376. doi: 10.1089/adt.2015.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.LaPorte MG, Wang Z, Colombo R, Garzan A, Peshkov VA, Liang M, Johnston PA, Schurdak ME, Sen M, Camarco DP, Hua Y, Pollock NI, Lazo JS, Grandis JR, Wipf P, Huryn DM. Optimization of pyrazole-containing 1,2,4-triazolo-[3,4-b]thiadiazines, a new class of STAT3 pathway inhibitors. Bioorganic & medicinal chemistry letters. 2016;26(15):3581–3585. doi: 10.1016/j.bmcl.2016.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hill JR. In vitro drug metabolism using liver microsomes. Current protocols in pharmacology/editorial board, SJ Enna. 2004;Chapter 7(Unit7):8. doi: 10.1002/0471141755.ph0708s23. [DOI] [PubMed] [Google Scholar]
- 19.Seo KA, Kim HJ, Jeong ES, Abdalla N, Choi CS, Kim DH, Shin JG, Safdari Y, Khalili M, Farajnia S, Asgharzadeh M, Yazdani Y, Sadeghi M, Wang SW, Sun YM, Johnston PA, Sen M, Hua Y, Camarco D, Shun TY, Lazo JS, Grandis JR. In vitro assay of six UDP-glucuronosyltransferase isoforms in human liver microsomes, using cocktails of probe substrates and liquid chromatography-tandem mass spectrometry. Drug Metab Dispos. 2014;42(11):1803–1810. doi: 10.1124/dmd.114.058818. [DOI] [PubMed] [Google Scholar]
- 20.Davies B, Morris T. Physiological parameters in laboratory animals and humans. Pharmaceutical research. 1993;10(7):1093–1095. doi: 10.1023/a:1018943613122. [DOI] [PubMed] [Google Scholar]
- 21.Ghibellini G, Leslie EM, Brouwer KL. Methods to evaluate biliary excretion of drugs in humans: an updated review. Molecular pharmaceutics. 2006;3(3):198–211. doi: 10.1021/mp060011k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pond SM, Tozer TN. First-pass elimination. Basic concepts and clinical consequences. Clinical pharmacokinetics. 1984;9(1):1–25. doi: 10.2165/00003088-198409010-00001. [DOI] [PubMed] [Google Scholar]
- 23.Gleeson MP, Hersey A, Montanari D, Overington J. Probing the links between in vitro potency, ADMET and physicochemical parameters. Nature reviews Drug discovery. 2011;10(3):197–208. doi: 10.1038/nrd3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Horst W, Doelle JSR, Marin Berovic. BIOTECHNOLOGY: Fundamentals in Biotechnology, vol XII. Encyclopedia of Life Support Systems. 2009 [Google Scholar]
- 25.Pelkonen O, Turpeinen M, Uusitalo J, Rautio A, Raunio H. Prediction of drug metabolism and interactions on the basis of in vitro investigations. Basic Clin Pharmacol Toxicol. 2005;96(3):167–175. doi: 10.1111/j.1742-7843.2005.pto960305.x. [DOI] [PubMed] [Google Scholar]
- 26.Cayen MN. Early Drug Development: Strategies and Routes to First-in-Human Trials. Hoboken, N.J: Wiley; 2010. [Google Scholar]
- 27.Kaivosaari S, Finel M, Koskinen M. N-glucuronidation of drugs and other xenobiotics by human and animal UDP-glucuronosyltransferases. Xenobiotica. 2011;41(8):652–669. doi: 10.3109/00498254.2011.563327. [DOI] [PubMed] [Google Scholar]
- 28.Chiu SH, Huskey SW. Species differences in N-glucuronidation. Drug Metab Dispos. 1998;26(9):838–847. [PubMed] [Google Scholar]
- 29.Izukawa T, Nakajima M, Fujiwara R, Yamanaka H, Fukami T, Takamiya M, Aoki Y, Ikushiro S, Sakaki T, Yokoi T. Quantitative analysis of UDP-glucuronosyltransferase (UGT) 1A and UGT2B expression levels in human livers. Drug Metab Dispos. 2009;37(8):1759–1768. doi: 10.1124/dmd.109.027227. [DOI] [PubMed] [Google Scholar]
- 30.Harbourt DE, Fallon JK, Ito S, Baba T, Ritter JK, Glish GL, Smith PC. Quantification of human uridine-diphosphate glucuronosyl transferase 1A isoforms in liver, intestine, and kidney using nanobore liquid chromatography-tandem mass spectrometry. Anal Chem. 2012;84(1):98–105. doi: 10.1021/ac201704a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fisher MB, Paine MF, Strelevitz TJ, Wrighton SA. The role of hepatic and extrahepatic UDP-glucuronosyltransferases in human drug metabolism. Drug metabolism reviews. 2001;33(3–4):273–297. doi: 10.1081/dmr-120000653. [DOI] [PubMed] [Google Scholar]
- 32.Yan Z, Caldwell GW, Gauthier D, Leo GC, Mei J, Ho CY, Jones WJ, Masucci JA, Tuman RW, Galemmo RA, Jr, Johnson DL. N-glucuronidation of the platelet-derived growth factor receptor tyrosine kinase inhibitor 6,7-(dimethoxy-2,4-dihydroindeno[1,2-C]pyrazol-3-yl)-(3-fluoro-phenyl)-amine by human UDP-glucuronosyltransferases. Drug Metab Dispos. 2006;34(5):748–755. doi: 10.1124/dmd.106.009274. [DOI] [PubMed] [Google Scholar]
- 33.Vourvahis M, Gleave M, Nedderman AN, Hyland R, Gardner I, Howard M, Kempshall S, Collins C, LaBadie R. Excretion and metabolism of lersivirine (5-{[3,5-diethyl-1-(2-hydroxyethyl)(3,5-14C2)-1H-pyrazol-4-yl]oxy}benzene-1,3-dic arbonitrile), a next-generation non-nucleoside reverse transcriptase inhibitor, after administration of [14C]Lersivirine to healthy volunteers. Drug Metab Dispos. 2010;38(5):789–800. doi: 10.1124/dmd.109.031252. [DOI] [PubMed] [Google Scholar]
- 34.Peterson EA, Boezio AA, Andrews PS, Boezio CM, Bush TL, Cheng AC, Choquette D, Coats JR, Colletti AE, Copeland KW, DuPont M, Graceffa R, Grubinska B, Kim JL, Lewis RT, Liu J, Mullady EL, Potashman MH, Romero K, Shaffer PL, Stanton MK, Stellwagen JC, Teffera Y, Yi S, Cai T, La DS. Discovery and optimization of potent and selective imidazopyridine and imidazopyridazine mTOR inhibitors. Bioorg Med Chem Lett. 2012;22(15):4967–4974. doi: 10.1016/j.bmcl.2012.06.033. [DOI] [PubMed] [Google Scholar]
- 35.Dressen D, Garofalo AW, Hawkinson J, Hom D, Jagodzinski J, Marugg JL, Neitzel ML, Pleiss MA, Szoke B, Tung JS, Wone DW, Wu J, Zhang H. Preparation and optimization of a series of 3-carboxamido-5-phenacylaminopyrazole bradykinin B1 receptor antagonists. J Med Chem. 2007;50(21):5161–5167. doi: 10.1021/jm051292n. [DOI] [PubMed] [Google Scholar]
- 36.Ye XM, Konradi AW, Smith J, Aubele DL, Garofalo AW, Marugg J, Neitzel ML, Semko CM, Sham HL, Sun M, Truong AP, Wu J, Zhang H, Goldbach E, Sauer JM, Brigham EF, Bova M, Basi GS. Discovery of a novel sulfonamide-pyrazolopiperidine series as potent and efficacious gamma-secretase inhibitors (Part II) Bioorg Med Chem Lett. 2010;20(12):3502–3506. doi: 10.1016/j.bmcl.2010.04.148. [DOI] [PubMed] [Google Scholar]