Abstract
Direct acting antivirals (DAAs) have led to a high cure rate in treated patients with chronic hepatitis C virus (HCV) infection but this still leaves a large number of treatment failures secondary to the emergence of resistance-associated variants (RAVs). To increase the barrier to resistance, a complementary strategy is to employ neutralizing human monoclonal antibodies (HMAbs) to prevent acute infection. However, earlier efforts with the selected antibodies led to RAVs in animal and clinical studies. Therefore, we identified a HMAb that is less likely to elicit RAVs for affinity maturation to increase potency and, more importantly, breadth of protection. Selected matured antibodies show improved affinity and neutralization against a panel of diverse HCV isolates. Structural and modeling studies reveal that the affinity matured HMAb mediates virus neutralization in part by inducing conformational change to the targeted epitope and that the maturated light chain is responsible for the improved affinity and breadth of protection. A matured HMAb protected humanized mice when challenged with an infectious HCV human serum inoculum for a prolonged period. However, a single mouse experienced breakthrough infection after 63 days when the serum HMAb concentration dropped by several logs; sequence analysis revealed no viral escape mutation.
Conclusions
The findings suggest that a single broadly neutralizing antibody can prevent acute HCV infection without inducing RAVs and may complement DAAs to reduce the emergence of RAVs.
Keywords: hepatitis C virus, human antibody, resistant-associated variants and protective immunity
Advances in treatment of chronic hepatitis C virus (HCV) infection with direct-acting antivirals (DAAs) have led to high cure rates of treated patients. Although high rates of sustained virologic response of approximately 95% have been achieved, a large number of patients will still fail treatment, even with drug combination regiments. A high percentage of these treatment failures are due to the emergence of resistance-associated variants (RAVs) (1). Chronic HCV infection consequently will continue as a leading cause of cirrhosis and hepatocellular carcinoma, which are major indications for liver transplantation. The emergence of RAVs is due to a high mutation rate driven by an error-prone viral RNA-dependent polymerase and a high viral replication rate that are further increased during reinfection after liver transplantation in immunosuppressed HCV infected recipients (2). To decrease the likelihood of RAVs, complementary treatment efforts are underway to develop host-targeting agents that interfere with cellular factors involved in the viral life cycle. These include viral entry, translation, replication and assembly inhibitors, and biological response modifiers (3). Because these approaches target host proteins, a potential concern is interference with the normal functions of these proteins that can lead to host toxicity.
Another approach is to employ neutralizing antibodies to HCV as immunotherapeutics. Polyclonal and human monoclonal antibodies (HMAbs) to HCV E2 have been shown to prevent infection in a human liver-chimeric mouse model (4, 5) and in chimpanzees (6). In a clinical trial to prevent reinfection in HCV infected liver transplant recipients with a HMAb, designated as MBL-HCV1, treatment was well-tolerated and viral rebound was significantly delayed in treated patients (7). However, RAVs emerged having mutations at key residues that form part of the epitope of this therapeutic antibody. Nonetheless, this study supports a therapeutic role for neutralizing HMAbs to HCV to increase the barrier to viral resistance.
We therefore selected a neutralizing HMAb to HCV that is less likely to induce RAVs for affinity maturation in an effort to increase neutralization potency and breadth of protection. HC84.26, an IgG1 HMAb, is a member of a panel of antibodies directed at a cluster of overlapping conformational epitopes on HCV E2, designated as antigenic domain D (8). These antibodies neutralize infectious cell culture derived genotype 1–6 HCV isolates (HCVcc). When a 2a HCVcc isolate is grown under increasing concentrations of HC84.26, complete viral elimination occurs at a critical antibody concentration. Affinity maturation was undertaken by a yeast display approach to isolate affinity-matured HC84.26 clones that have improved binding and neutralization activities against HCV isolates that were neutralized poorly by wild type (wt) HC84.26. Structural studies reveal that the conformation of a synthetic peptide encompassing aa434–446 on E2 bound to an affinity-matured clone is similar to the conformation of these residues in the native E2 core protein (9, 10). When tested in the human-liver chimeric mouse model to prevent acute HCV infection, a single injection of an affinity-matured HMAb protected the majority of mice over a prolonged period. A single mouse showed breakthrough infection at a timepoint when the serum antibody concentration was reduced by several logs. Sequence analysis of the breakthrough infection revealed no viral escape mutation. Our findings suggest that a broadly neutralizing antibody can prevent acute HCV infection and complement DAAs to reduce the emergence of RAVs.
Experimental Procedures
Cells, viruses and reagents
HEK-293T cells were obtained from the ATCC. Huh7.5 cells (generously provided by Dr. C. Rice, Rockefeller University) were grown in Dulbecco's modified minimal essential medium (Invitrogen, Carlsbad, CA), supplemented with 10% fetal calf serum (Sigma-Aldrich Co., St. Louis, MO) and 2 mM glutamine. HMAbs CBH-4G, HC-84.26 and HC33.1 against HCV E2 glycoprotein were produced as described (8). Yeast Saccharomyces cerevisiae strain EBY-100 (GAL1-AGA1:URA3 ura3-52 trp1 leu2Δ1 his3Δ200 pep4::HIS2 prb1Δ1.6R can1 GAL) (Invitrogen, Carlsbad, CA) was maintained in YPD broth (Difco). The yeast display vector pYD2 was kindly provided by Dr. J. D. Marks (UCSF). HCV envelope expression constructs for pseudoparticle production included strains HCV-J (genotype 1b), J6 (2a), UKN3A1.28 (3a) and UKN4.21.16 (4) and have been described previously (11, 12).
Primary human hepatocytes
Primary human hepatocytes (PHH) were isolated and cultured from different donors as described (12) or provided by Kalycell (Plobsheim, France). Human material including liver tissue from patients undergoing surgical resection for isolation of human hepatocytes was obtained with informed consent from all patients. The protocol was approved by the Ethics Committee of the University of Strasbourg Hospitals (CPP 10–17).
Construction, characterization, and expression of various HC84.26 yeast display libraries introducing mutations
Affinity maturation was undertaken by sequential light chain shuffle, random and site-directed mutagenesis as described in Supplementary Methods.
Binding affinity and specificity measurement of selected clones
Affinity of single-chain Fv fragment (scFv) clones during screening was approximated by flow cytometry and binding kinetics were measured by surface plasmon resonance (SPR) (BIAcore 3000, Pharmacia Biosensor). Antibody specificity was evaluated in a Galanthus nivalis agglutinin (GNA)-captured E1E2 enzyme-linked immunosorbent assay (ELISA) as described (8).
In vivo protection study
Twelve Alb-uPA/SCID mice with high engraftment of human hepatocytes were allocated to the two study groups (13, 14). Six mice received the negative control R04 antibody and six received HC84.26.5D. Baseline blood draws were obtained on Day minus (-) 6 for measurement of serum human alpha-1 antitrypsin (hAAT) levels and allocation to the study groups such that the group average hAAT values were equivalent. Animals were administered their respective antibodies on Day -1 by intraperitoneal injection of a 250 mg/kg dose. On Day 0 the animals received a single intravenous challenge dose of 105 international unit (IU) gt1b HCV. Blood samples were taken weekly from day 7 to 70 for measurement of hAAT and HCV titers.
IgG half-life measurement
A modification of the standard ELISA was used to measure serum HC84.26.5D levels in treated mice (8). This required the establishment of a standard curve for this antibody, as outlined in S. Fig. 2. The serum concentrations for HC84.26.5D at different serum dilutions were back-calculated based on the standard curve and the half-life determinations were determined using a nonlinear regression curve fit (Graphpad Prism, Graphpad software, Inc., CA).
HCV-pseudotype retroviral particle (HCVpp) production and neutralization
HCVpp expressing genotype 1 HCV E1E2 glycoproteins were produced and tested in neutralization assay as described in Huh7.5 cells and in PHH (8, 15). All assays against wt and mutant H77C HCVpp were performed three times in triplicates and antibody concentrations resulting in 50% neutralization (IC50) were calculated by nonlinear regression analysis (Graphpad Prism). For neutralization against a panel of genotype 1a and 1b HCVpp in Huh7.5 cells, each antibody was tested twice in triplicates; means and standard deviations were calculated. For neutralization of HCVpp in PHH, each antibody was tested in three independent experiments performed in three biological replicates using PHH from three different donors. Means were calculated.
Clonal sequence analysis
HCV E1E2 sequence analysis of the infectious inoculum and serum from control and treated mice was performed as previously described (8). Ten individual clones from each sample containing an insert of the expected size were sequenced in both sense and antisense strands (ElimBiopharm, Hayward, CA).
Protein production and purification
The HC84.26.5D antibody was expressed as a scFv by in vitro folding from inclusion bodies produced in Escherichia coli (Supplementary Methods).
Crystallization and structure determination
The crystallization and structure determination of the HC84.26.5D–E2434–446 complex are described in Supplementary Methods. Coordinates and structure factors have been deposited in the Protein Data Bank under accession code 4Z0X.
Computational mutagenesis
In silico mutagenesis of HC84.26.5D–E2434–446 was carried out as described in Supplementary Methods.
Modeling of the HC84.26.5D–E2 core complex
The E2 core structure (9) was modeled into the HC84.26.5D–E2434–446 complex as described in Supplementary Methods.
Results
Affinity maturation of HC84.26
HC84.26 is against an epitope on E2 that includes residues at 441, 442 and 443, and 616 (8). While residues L441, W443 and W616 are conserved among all HCV isolates, F442 is only 85% conserved with the remaining 14% having either F442I or F442L mutations (16). Yet, this mutation did not occurred when a 2a HCVcc was co-cultured with HC84.26 and infectious virions were eliminated at a critical antibody concentration (8). Structural studies provide a possible explanation (16). The three residues located at 441–443 form a hydrophobic protrusion that serves as the core binding site for HC84.26. When F442I or F442L mutations is present, their interactions with the paratope formed by the heavy chain complementarity determining regions (CDRs) lead to a decrease in binding energy of the complex that can be compensated by increasing the antibody concentration. Thus, variants with 442 mutations are eliminated at higher HC84.26 concentrations. These findings suggest that binding affinity to this E2 segment is a determinant of neutralization by HC84.26 and that further affinity maturation to improve the breadth of neutralization might be feasible.
For affinity maturation, the first step was to optimize the pairing of the heavy chain variable gene regions (VH) of HC84.26 with different light chain variable gene regions (VL) for binding to a mutant H77C 1a F442I E2 recombinant protein. HC84.26 wild-type VH was cloned into a yeast vector pYD2 library that contained VL (8). The constructed HC84.26 VH/VL library was screened for scFv displayed clones that bound to F442I E2 (see Supplementary Method). A number of these clones were selected for further maturation by random mutagenesis of VH and VL fragment genes. Additional rounds of selection led to a scFv clone designated as HC84.26.5 with greater binding than HC84.26 to F442I E2 by FACS analysis (data not shown). Targeted mutagenesis on VH CDR1 and CDR2 of HC84.26.5 resulted in two scFv clones, designated as HC84.26.5D and HC84.26.5G having additional increase in binding to F442I E2. Combining the observed mutations in these two clones into a new clone, HC84.26.5DG, however, showed no further improvements by flow analysis.
After conversion of these HC84.26 affinity-matured scFv clones to full-length IgG1, they were expressed and purified, and their binding affinities to wt E2, F442I and F442L E2 mutants were measured by SPR in BIAcore (Table 1). Purified recombinant secreted E2 was first captured onto a pre-coated sensor chip with a non-neutralizing HMAb to a conformational epitope on E2. Compared to HC84.26 against wt E2, only modest improvements in Kon, Koff and KD were observed with the affinity-matured clones that ranged between 2.1–4.4 folds. Against F442I E2, Kon and Koff were dramatically improved among all matured clones with similar Kon and Koff improvements for HC84.26.5D, HC84.26.5G and HC84.26.5DG. Total improvements in KD ranged between 25–34 fold differences for these three affinity-matured clones. Against F442L E2, more moderate Kon, Koff and KD improvements were observed. KD improvements ranged between 3.2–6.3 folds. We next assessed whether improved affinities lead to improved neutralization (Table 2). Against 1a H77C HCVpp, the IC50 values were in the same range for HC84.26, HC84.26.5D, HC84.26.5G and HC84.26.5DG. Against F442I HCVpp that was not neutralized by HC84.26, IC50 values for the three matured clones ranged between 0.37–1.95 µg/ml, with HC84.26.5D having the lowest IC50 that also has the most improved KD. Against F442L HCVpp that was neutralized by HC84.26 at 15 µg/ml, the most improved clone was again HC84.26.5D with a two log reduction in IC50 value of 0.14 µg/ml. Taken together, affinity maturation of HC84.26 leads to increased breadth of protection and the increased neutralization potencies are greater than the improved antibody binding affinities.
Table 1.
Binding kinetics of HC84.26 affinity mature clones against 1a H77C E2 and variants
| Kon (M−1s−1) | Koff (s−1) | KD (M) | KD wt / KD Matured | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1aE2 | F442I E2 | F442L E2 | 1aE2 | F442I E2 | F442L E2 | 1aE2 | F442I E2 | F442L E2 | 1aE2 | F442I E2 | F442L E2 | |
| HC84.26 | 1.3E+04 | 3.2E+03 | 5.6E+03 | 2.1E−04 | 6.3E−03 | 1.6E−03 | 1.6E−08 | 2.0E−06 | 2.8E−07 | 1.0 | 1.0 | 1.0 |
| HC84.26.5 | 3.2E+04 | 1.3E+04 | 1.2E+04 | 2.5E−04 | 1.7E−03 | 1.1E−03 | 7.7E−09 | 1.3E−07 | 8.9E−08 | 2.1 | 15.7 | 3.2 |
| HC84.26.5D | 2.0E+04 | 1.1E+04 | 9.7E+03 | 1.3E−04 | 6.5E−04 | 4.4E−04 | 6.4E−09 | 5.8E−08 | 4.5E−08 | 2.5 | 34.0 | 6.3 |
| HC84.26.5G | 2.3E+04 | 1.3E+04 | 1.1E+04 | 8.5E−05 | 9.7E−04 | 6.7E−04 | 3.6E−09 | 7.7E−08 | 5.9E−08 | 4.4 | 26.0 | 4.8 |
| HC84.26.5DG | 2.7E+04 | 1.1E+04 | 1.0E+04 | 1.7E−04 | 8.7E−04 | 5.1E−04 | 6.3E−09 | 8.0E−08 | 4.9E−08 | 2.5 | 25.0 | 5.8 |
Table 2.
Neutralization against 1a H77Cpp and variants*
| 1a E2 | F442I E2 | F442L E2 | |
|---|---|---|---|
| HC84.26 wt | 0.06±0.01 | >50 | 15±0.05 |
| HC84.26.5D | 0.06±0.02 | 0.37±0.03 | 0.14±0.01 |
| HC84.26.5G | 0.09±0.02 | 1.95±0.02 | 0.76±0.03 |
| HC84.26.5DG | 0.05±0.01 | 1.25±0.02 | 0.31±0.02 |
Concentration (µg/ml) resulting in 50% neutralization (IC50)
HC84.26.5D prevents acute HCV infection in humanized mice
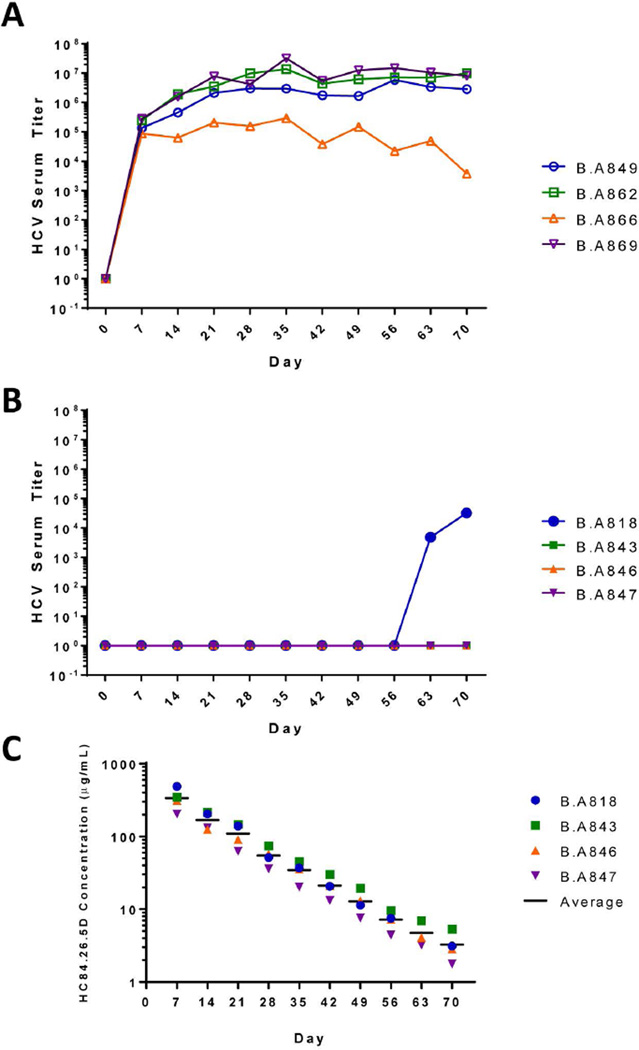
In vivo protection of acute HCV infection was tested in the human liver chimeric alb-uPA/SCID mouse model (13, 14) over a 72 day study. The mice having evidence of high engraftment of human liver cells, as determined by >400 µg/ml serum hAAT levels, were separated into two groups. The test group received HC84.26.5D antibody and the control group received R04, an isotype-matched IgG1 HMAb to human cytomegalovirus (8). Antibodies were administrated just once on Day -1 at 250 mg/kg by intraperitoneal injection based in part on earlier studies with other antibodies (4). One day later, Day 0, each mouse in both groups was challenged by intravenous injection of 105 IU of genotype 1b HCV infected human serum. Two animals in each group did not survive these procedures. No further treatment was given to the animals. The mice were followed at weekly intervals for 10 weeks after challenge and monitored for hAAT and HCV RNA titers. The serum hAAT values remained high for all mice in both groups, which confirmed the viability of the human hepatocyte grafts for the duration of the study (S. Figs. 1A; 1B). The four control mice, treated with R04, showed a progressive increase in HCV RNA to peak titers on Day 35 with approximately 105–107 RNA IU/ml, which were mainly maintained to the end of the study, except for one mouse, B.A866, that exhibited a slow decline to 103 RNA IU/ml from Day 35 to Day 70 (Fig. 1A). In contrast, three of four HC84.26.5D treated mice were completely protected without any evidence of HCV RNA for the duration of the study (Fig. 1B). The fourth mouse, designated as B.A818, had no detectable HCV RNA for 56 days. On Day 63, however, breakthrough infection occurred and serum RNA was detected at 4.8×103 RNA IU/ml. Viremia increased one week later to 3.2×104 RNA IU/ml on Day 70, when the study was terminated. To determine whether breakthrough infection in B.A818 was due to a more rapid decrease in serum HC84.26.5D level, IgG half-life measurements were determined in all treated mice. This required the optimization of the GNA-captured HCV E2 ELISA for measuring HC84.26.5D antibody concentration in mouse sera (S. Fig. 2). Of the three mice in which breakthrough infection was not observed, B.A843, B.A846 and B.A847, HC84.26.5D half-life calculated respectively at 10.4, 6.5 and 8.9 days (Fig. 1C). The IgG half-life in B.A818, while lower than the other mice at 6.4 days, was not statistically different, as determined by one-way analysis of variance (p=0.819). Breakthrough infection occurred when serum HC84.26.5D level dropped from 488 µg/ml on Day 7 to approximately 3 µg/ml on day 63. It is probable that prior to Day 63, the HCV RNA level in B.A818 was suppressed below the sensitivity of the quantitative PCR assay at 3×102 IU/ml.
Figure 1. HC84.26.5D protects humanized mice against HCV.
Human liver-chimeric mice received either (A) a control HMAb, R04, or (B) HC84.26.5D at 250 mg/kg by intraperitoneal injection 24 hours prior to challenge with genotype 1b infected human serum (105 IU of HCV RNA) by intravenous injections. Viral load measurements were determined by realtime PCR at weekly intervals on an ABI model 7300 system using TaqMan chemistry (14). (C) Serum human IgG half-life measurements in mice treated with HC84.26.5D. Each timepoint was measured by ELISA in triplicates for HMAb in each mouse serum sample tested and the concentration was back-calculated based on a standard curve (S. Fig. 2).
To assess whether breakthrough infection was associated with an escape mutation, we sequenced the E2 and E1 regions of the virions that were isolated on Day 70 from the sera of B.A818, two control mice, B.A862 and B.A869, and the 1b HCV challenge inoculum (S. Tables 1A and 1B). Of ten sequenced isolates per serum sample, random mutations were observed in all samples. In the mouse with breakthrough infection, B.A818, or the control mice, B.A862 and B.A869, no mutation having a higher frequency than 1/10 on E2 was uniquely observed when compared to the infectious inoculum. The only mutation that was observed with a higher frequency, F522S, occurred in 4/10 isolates in the infectious inoculum. This mutation was also observed in both control mice at frequencies of 3–4/10 but not in B.A818. There was no mutation at the site of known variability, F442, in the HC84.26 epitope, nor as expected at residues 441, 443 and 616. Mutations unique to B.A818 included two positions near the epitope, T435A and V515A, on E2 (S. Table 1A; Fig. 4B), although their low frequency and location away from the critical epitope residues suggest limited, if any, effects on viral escape. This is supported by the ability of HC84.26 to bind to T435A E2 mutant by 117% (8) and to V515A E2 mutant by 83% (data not shown) when compared to wt E2. On E1, mutations mainly at a frequency of 1/10 were randomly observed in all samples (S. Table 1B). The frequency of these mutations was no greater in the breakthrough mouse (B.A818) than the two control mice, which is in agreement with the HC84.26 epitope restricted to E2. These findings support our contention that no viral escape mutation was associated with the breakthrough infection. Breakthrough probably resulted from decreasing concentration of HC84.26.5D.
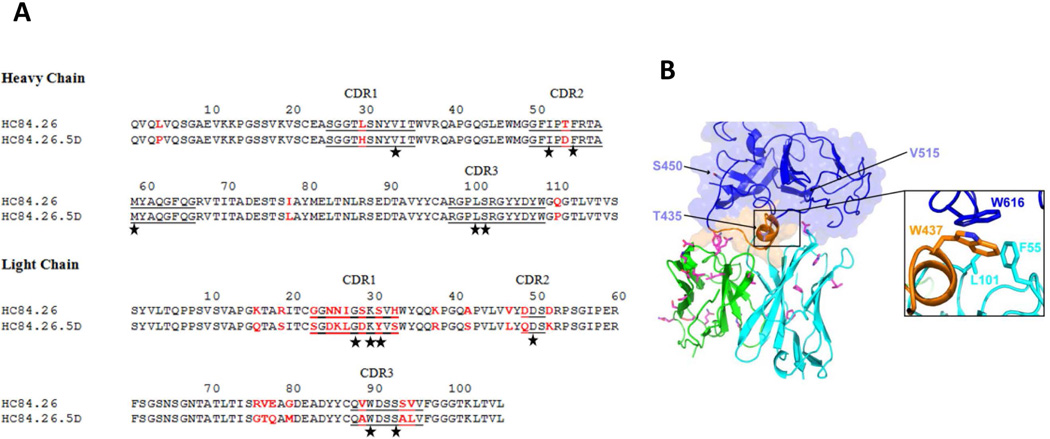
Figure 4. Affinity-matured residues in the HC84.26.5D-E2434–446 interface and model of HC84.26.5D with E2 core protein.
(A) Amino acid sequences of the VH and VL regions of wild-type HC84.26 and affinity-matured HC84.26.5D were aligned. Bars show the position of CDRs. Amino acid differences between the two antibodies are highlighted in red. Contacting residues in the HC84.26.5D–E2434–446 structure are marked with black stars. (B) Model of HC84.26.5D in complex with E2 core glycoprotein. VH and VL domains are cyan and green, respectively. Mutant antibody residues from affinity maturation are drawn in stick format, with carbon atoms in magenta, nitrogen atoms in blue, and oxygen atoms in red. Three sites that were mutated in the HC84.26.5D-treated mouse with breakthrough infection (B.A818), and not inoculum or control mice, are shown as light blue sticks on the E2 core model and labeled. Inset shows putative antibody VH hydrophobic contacts with E2 residue W616 based on the modeled complex (E2 residue W437 also shown for reference).
Structure of HC84.26.5D bound to E2434–446 peptide
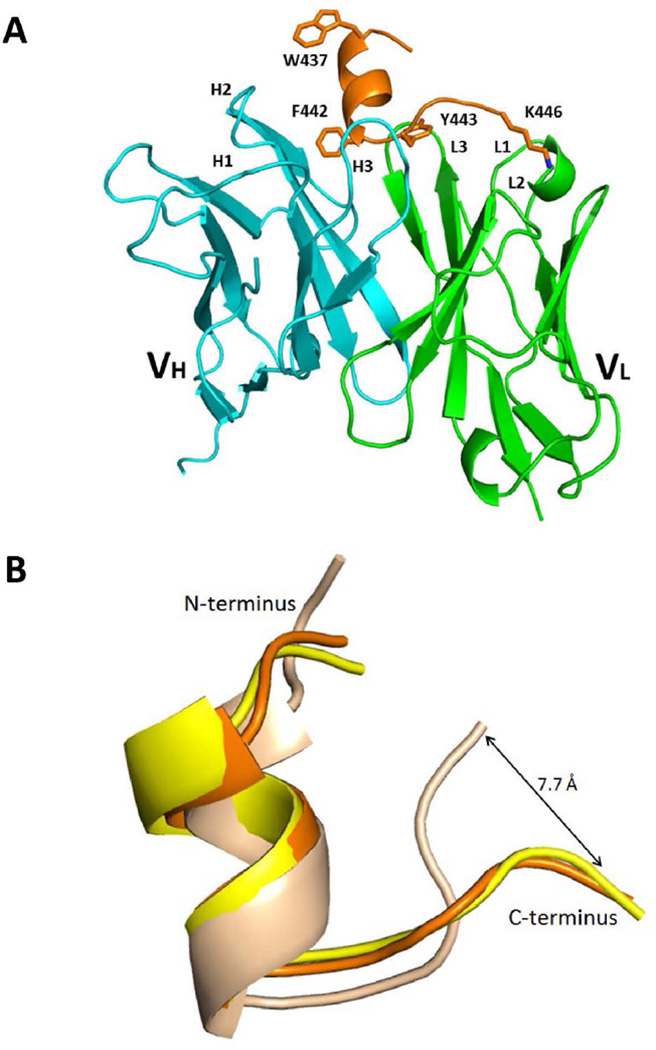
Because of the change in reactivity patterns of affinity-matured clones, we assessed whether the epitope of HC84.26.5D is structurally different than the epitope of HC84.26 and the related antigenic domain D HMAbs HC84.1 and HC84.27, which use the same VH as HC84.26 but different VL (16). Thus, we determined the structure of HC84.26.5D, expressed as a scFv, bound to a 13-mer peptide spanning residues 434–446 of the E2 glycoprotein (E2434–446) to 2.0 Å resolution (S. Table 2; Fig. 2A). The E2434–446 peptide represents a portion of the domain D epitope of E2 recognized by HC84.26, which is conformational in nature (8). Electron density difference maps revealed well-defined density for the E2434–446 peptide in which the peptide adopts an α-helical turn spanning aa437–442, while the C-terminus exhibits an extended conformation comprising aa443–446 (Fig. 2A). The conformation of E2434–446 bound to HC84.26.5D is very similar to that of this same peptide bound to the related HMAbs HC84.1 and HC84.27 (16). It is also similar to the conformation of these residues in the E2 core protein (9), except for a significant deviation in C-terminal residues 443–446 that leads to a displacement of 7.7 Å in the Cα position of K446 (Fig. 2B). In the HC84.26.5D–E2434–446 complex, K446 is firmly anchored to the antibody through a salt bridge and three hydrogen bonds (Fig. 3A), making it likely that HC84.26.5D would form similar interactions with K446 in the E2 core protein. Thus, binding of HC84.26.5D to the E2 core may induce conformational changes in residues 443–446 of the native protein. The HC84.26.5D–E2434–446 complex buries a total solvent-accessible area of 1522 Å2 with 7 peptide residues contacting 12 antibody residues (S. Table 3). The N-terminal α-helix of E2434–446 makes extensive hydrophobic contacts with VH CDR1 and CDR2, mainly through L441 and F442 (Fig. 3B), which are critical binding residues for antigenic domain D HMAbs (8).
Figure 2. Structure of the HC84.26.5D–E2434–446 complex.
(A) Ribbon diagram of the HC84.26.5D–E2434–446 complex (side view). VL, green; VH, cyan; E2434–446, orange. VLCDR loops are labeled L1–L3; VHCDR loops are labeled H1–H3. The side chains of epitope residues Trp437, Phe442, Tyr443, and Lys446 are drawn in stick representation. (B) Conformation of the E2434–446 epitope bound to HC84.26.5D and in unbound E2 core protein. The E2434–446 epitope from the complex with HC84.26.5D (orange) was superposed onto E2434–446 from the complex with HC84.1 (yellow) (Protein Data Bank accession code 4JZN) (16), and onto E2434–446 from the structure of the E2 core glycoprotein (wheat) (4MWF) (9). The E2434–446 epitopes were superposed through residues 437–442. The N- and C-termini are labeled. The epitopes are in the same overall orientation as in (A).
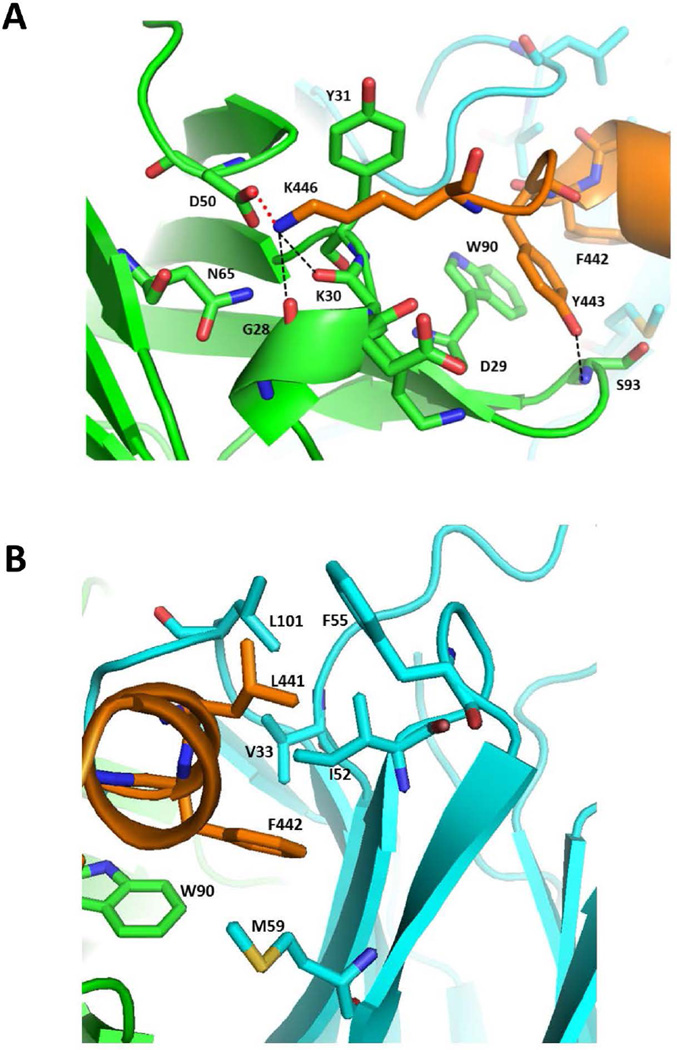
Figure 3. The HC84.26.5D–E2434–446 binding interface.
(A) Close-up view of the interactions between Lys446 of E2434–446 (orange) and and the VL (green) and VH (cyan) domains of HC84.26.5D. The side chains of contacting residues are shown in stick format with carbon atoms in orange (E2434–446) or green (VL); nitrogen atoms in blue, and oxygen atoms in red. Hydrogen bonds are drawn as dotted black lines. The salt bridge linking Lys446 to VLAsp50 is represented as a solid red line. (B) Interactions between the α-helix of E2434–446 (orange) and the VL (green) and VH (cyan) domains of HC84.26AM. The side chains of contacting residues are drawn in stick format with carbon atoms in orange (E2434–446), green (VL), or cyan (VH); nitrogen atoms in blue, oxygen atoms in red, and sulfur atom in yellow.
Regarding the C-terminal segment of E2434–446, Y443 forms stacking interactions with VL CDR3Trp90, as well as a main-chain hydrogen bond with VL CDR3Ser93 (S. Table 3; Fig. 3A). Of particular relevance to the present study is K446, whose mutation to Glu or Asn in HCV escape variants abolished (Glu) or substantially reduced (Asn) neutralization by HC84.26.5D (see below, Table 3). In the HC84.26.5D–E2434–446 structure, the positively charged side chain of K446 lies in an electronegative depression on the VL domain, between CDR1 and CDR2, where it forms a salt bridge with VL CDR2Asp50 (Fig. 3A). The predicted energetic impact of the K446E and K446N mutations on HC84.26.5D binding was assessed by modeling these substitutions and evaluating their ΔΔGs using an energy-based scoring function (17), starting with the HC84.26.5D–E2434–446 crystal structure. Reflecting the importance of the K446 positively charged side chain’s interactions with the HC84.26.5D VL domain (Fig. 3A), both the K446E (charge reversal) and K446N (charge neutralization) mutations led to >10-fold predicted affinity decreases (S. Table 4), which could explain the capacity of HCV variants bearing these substitutions to escape neutralization by HC84.26.5D (Table 3).
Table 3.
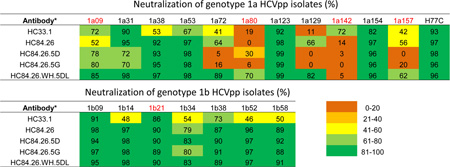
Neutralization of genotype 1a HCVpp isolates (%)
Each antibody tested at 20 µg/ml.
(Red) HCV variants with a mutation at F442 and/or at K446: 1a80 and 1a142 have F442I; 1a09 has F442L; 1a142, K446E; 1a157, K446N; and 1b21, K446H.
To investigate possible mechanisms for improved E2 affinity and HCV neutralization by HC84.26.5D, we identified residues that were mutated during affinity maturation from the parental HC84.26 HMAb (Fig. 4A). Nearly all these mutations (21 of 26) are located in VL. Surprisingly, only one mutated residue (VL CDR1Tyr31) interacts directly with the E2434–446 peptide in the HC84.26.5D–E2434–446 complex (S. Table 3). Furthermore, all four HC84.26.5D residues that contact F442 (VL CDR3Trp90, VH CDR1Val33, VH CDR2Ile52 and VH CDR2Met59) (Fig. 3B) are strictly conserved in HC84.26. Therefore, mutations in these residues cannot explain the higher affinity of HC84.26.5D than HC84.26 for E2 variants F442I and F442L, or for wt E2 (Table 1). Instead, improved binding could have resulted from contacts with the native E2 protein outside the portion of the epitope represented by the E2434–446 peptide.
To determine the putative engagement mode of E2 by HC84.26.5D and closely related HMAbs, we modeled the E2 core–HC84.26.5D complex by superposing epitope residues onto corresponding residues in the E2 core structure, followed by constrained local minimization of the HMAb–epitope region with respect to the remainder of E2 core (Fig. 4B). We identified several E2 residues outside aa434–446 that are contacted by HC84.26.5D in the model, including W616 (Fig. 4B), for which mutation to alanine had been shown to abolish binding of some domain D HMAbs (8). Several mutated HC84.26.5D residues are proximal to E2 in the modeled complex (Fig. 4B); the majority of these are in VL, suggesting that improved E2 affinity and neutralization results from that chain.
Neutralization profiles against a diverse panel of genotype 1 HCV isolates
While HC84.26 neutralized representative genotype 1–6 HCV isolates (8), the limited number in each genotype do not represent the diversity of HCV envelope polymorphisms. To gain a precise breadth of protection analysis, neutralization by HC84.26 and the affinity mature clones was tested against a panel of genotype 1 HCVpp isolates that represents 94% of envelope polymorphisms present at greater than 5% frequency in a reference panel of genotype 1 HCV isolates (15). Each HMAb, HC84.26, HC84.26.5D and HC84.26.5G, and a control HC33.1 at 20 µg/ml was tested (Table 3). HC33.1 is a HMAb directed against aa412–423 on E2 (18). Neutralizing activity <40% was viewed as no activity; 41–60% as modest; 61–80% as moderate; and >80% as strong. HC84.26 did not neutralize 1a80 and 1a142, and neutralized modestly 1a09 and 1a157 HCV isolates. Both 1a80 and 1a142 have F442I, and 1a09 has F442L mutations, which account for the lost or modest activities by HC84.26. The drop in activity against 1a157 however is not due to a mutation at 442 but is likely due to a K446N mutation. This is supported by the observation that a K446A mutation significantly reduces binding by HC84.26 by 60% (8), as well as in silico mutagenesis using the structure of the HC84.26.5D–E2434–446 complex noted above. Surprisingly, both HC84.26.5D and HC84.26.5G performed worse than HC84.26. Aside from 1a09, no improvements or decreased activities were observed against variants with mutations at either F442 or K446. In addition, neutralization against two isolates, 1a72 and 1a129, was lost by the affinity mature clones, although they were neutralized by HC84.26. This raised the question whether the specificity of the two affinity-matured clones was altered by the random mutagenesis step in VH.
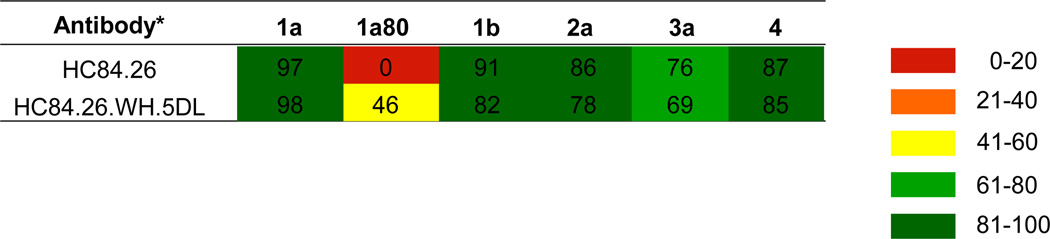
To address this issue and based on the likelihood that improved affinity shown by HC84.26.5D can be attributed to the mutations in VL, we paired VH from HC84.26 and VL from HC84.26.5D to construct a third affinity matured clone, designated as HC84.26.WH.5DL. As shown in Table 3, the entire panel of genotype 1 HCVpp was neutralized moderately to strongly by HC84.26.WH.5DL except for the 1a142 isolate. Notably, the 1a72 and 1a129 isolates that were not neutralized by HC84.26.5D and HC84.26.5G were neutralized by HC84.26.WH.5DL. This was anticipated since HC84.26 neutralized these isolates and HC84.26.WH.5DL has the wt VH. Furthermore, one of the isolates with a F442I mutation, 1a80, became sensitive to HC84.26.WH.5DL. However, the 1a142 isolate also containing a F442I mutation remained resistant to HC84.26.WH.5DL. This is likely due to 1a142 having a second mutation of K446E, which is predicted to reduce affinity of HC84.26.5D, with identical VH and putative K446 interacting residues, by >10-fold (Table S4; see Discussion). The control HMAb, HC33.1, neutralized this panel of genotype 1 isolates with a similar breadth of protection as HC84.26, albeit with lower levels of potencies against a number of isolates (Table 3). Two isolates, 1a80 and 1a129, not neutralized by HC33.1 are neutralized by HC84.26.WH.5DL. The single isolate not neutralized by HC84.26.WH.5DL is neutralized by HC33.1. While >90% of the panel is neutralized by HC84.26.WH.5DL alone, a second antibody, e.g. HC33.1, is required to achieve 100% breadth of protection coverage. To confirm breadth of neutralization, a representative panel of different genotype HCVpp was tested in primary human hepatocytes isolated from three different donors (Table 4). Potent neutralization was observed against the different HCVpp with both antibodies, except against the 1a80 isolate (as tested in Table 3). No neutralization with HC84.26 was observed, while neutralization was detected with HC84.26.WH.5DL. This pattern is similar to that observed in Huh7.5 cells (Table 3), albeit at a lower level of neutralization with the affinity matured antibody.
Table 4.
Neutralization of different genotype HCVpp isolates in primary human hepatocytes (%)
Each antibody tested at 20 µg/ml.
Discussion
Despite the advances achieved in DAAs in treating chronic hepatitis C, the emerging importance of RAVs associated with treatment failures will require alternative therapeutic approaches. This is especially likely in the transplant setting that involves a HCV-negative liver to a HCV-positive recipient when the error-prone replication rate is even higher (2). Neutralizing antibodies provide a complementary approach to DAAs in preventing or treating these patients. However, the key issue for therapeutic applications of a broadly neutralizing antibody to HCV is whether immune pressure from the selected antibody will drive viral evolution and the emergence of RAVs. We selected an antibody that is less likely to induce RAVs for affinity maturation in an effort to increase neutralization potency and breadth of protection. This has been achieved with HC84.26, an IgG1 HMAb that in earlier studies showed broad neutralization against genotypes 1–6 HCVcc. In addition, complete elimination of virus occurs at a critical concentration when this antibody is co-cultured with infectious virions (8). Affinity maturation by a yeast display approach led to the isolation of a matured HC84.26 clone, HC84.26.5D that neutralized an E2 F442I mutant HCVpp that was not neutralized by the parental antibody. When tested in the human-liver chimeric mouse model to prevent acute HCV infection, HC84.26.5D protected all four mice over a prolonged period of 56 days with a single treatment. However, one of the mice showed breakthrough infection on day 63 when the serum antibody concentration was reduced by ~100-fold. Sequence analysis of the breakthrough infection revealed no viral escape mutation. These studies demonstrate that HC84.26.5D is protective in vivo against acute HCV infection. One caveat is that the in vivo protection study was performed with a clinical genotype 1b infectious inoculum having F442 and the F442I mutation is less frequent in 1b relative to 1a (1.5% vs. 7.5% in a reference panel from Genbank). The implication is that escape with a 1b isolate will be less likely. Additional in vivo protection studies with other isolates with F442I/L mutations will be required to confirm our in vitro findings of HC84.26.5D neutralizing F442I/L variants.
Structural studies revealed that the conformation of a synthetic peptide encompassing aa434–446 on E2 bound to HC84.26.5D is similar to the conformation of these residues in the native E2 core protein, except for a significant deviation that suggests that binding of HC84.26.5D may induce conformational changes in residues 443–446 of the native protein (Fig. 2B). This apparent E2 flexibility is consistent with the highly dynamic nature of the E2 glycoprotein (9), and mobility in this region has been noted by others based on structural studies (19). The shared bound epitope conformation for HC84.26.5D and two other HC84-class HMAb structures determined separately (16) has implications for vaccine design, as stabilization of E2 with this bound epitope conformation could preferentially elicit this class of broadly neutralizing HMAbs.
Against a panel of HCV isolates representative of the polymorphism commonly observed among genotype 1 (15), it was surprising that HC84.26.5D did not neutralize some isolates that were neutralized by HC84.26 (Table 3). The pairing of VH from HC84.26 and VL from HC84.26.5D resulted in an affinity-matured clone designated as HC84.26.WH.5DL that neutralized the majority of the genotype 1 isolate panel, except for one isolate. This breadth of protection demonstrated that the heavy chain of HC84.26.WH.5DL has implications in neutralization breadth. Modeling studies with HC84.26.5D suggested that mutations in VL are most likely responsible for improved affinity (Fig. 4B), which is retained in HC84.26.WH.5DL. A lesson for vaccine design to prevent HCV infection is that a single antibody, even when affinity-matured, is not able to provide 100% protection against the diversity of HCV isolates (Table 3). At a minimum, a second broadly neutralizing antibody to a non-competing epitope is required and more likely a third antibody will be necessary since HC84.26 and HC33.1 failed to neutralize one of the genotype 1 isolates, 1a80. Only the affinity-matured HC84.26.WH.5DL neutralized this isolate.
In conclusion, our findings suggest that a broadly neutralizing antibody with the characteristics of HC84.26-derived affinity-matured clones, such as HC84.26.WH.5DL, can provide great breadth of protection and complement DAAs to reduce the emergence of RAV. In addition to the liver transplant setting, the availability of an effective means to prevent acute HCV infection in an immunosuppressed HCV-negative recipient receiving an HCV-positive donor kidney could change the high risk of acquiring HCV infection. Although there is a survival advantage to receiving an HCV-positive kidney compared with waiting on the deceased-donor waitlist, the current recommendation is to transplant HCV-positive kidneys from deceased donors to only HCV-positive recipients (20). Clinical development of HC84.26.WH.5DL could reduce the morbidity and mortality associated with these organ transplantations.
Supplementary Material
Acknowledgments
We thank Wenyan Wang for technical assistance and Dr. Rajen Koshy for helpful discussion, Professor Patrick Pessaux (Strasbourg University Hospitals) for providing liver resections for hepatocyte isolation, Kalycell (Plobsheim, France) for providing additional batches of primary hepatocytes, Dr. T. J. Liang (NIDDK), Dr. R. Purcell and Dr. Bukh (NIAID), Dr. Jonathan Ball (University of Nottingham) for providing envelope expression constructs, Dr. Mirjam Zeisel (Inserm U1110, University of Strasbourg) for helpful discussions and Laura Heydmann and Charlotte Bach for excellent technical assistance.
This work was support in part by National Institute of Allergy and Infectious Diseases/NIH grants R41-AI108024 (SKHF and FWH), U19-AI123862 (SKHF and TFB), (R21-AI126582 (SKHF, BGP and RAM), Contract No. HHSN2722010000016I to KMT Hepatech (NMK), and MPower (RAM and TRF), EU FP7 Hepamab (TFB) and Laboratory of Excellence HepSys (TFB).
Abbreviations
- DAAs
Direct acting antivirals
- HCV
hepatitis C virus
- RAVs
resistance-associated variants
- HMAbs
human monoclonal antibodies
- wt
wild-type
- scFv
single-chain Fv fragment
- SPR
surface plasmon resonance
- GNA
Galanthus nivalis agglutinin
- ELISA
enzyme-linked immunosorbent assay
- hAAT
human alpha-1 antitrypsin
- IU
international unit
- HCVpp
HCV-pseudotype retroviral particles
- IC50
antibody concentration resulting in 50% neutralization
- HCVcc
infectious cell cultured HCV
- CDRs
complementarity determining regions
- VH
heavy chain variable gene regions
- VL
light chain variable gene regions
- FACS
fluorescence-activated cell sorting
Footnotes
Conflicts of Interest Statement: The authors have declared that no conflict of interest exists.
References
- 1.Sarrazin C. The importance of resistance to direct antiviral drugs in HCV infection in clinical practice. J Hepatol. 2016;64:486–504. doi: 10.1016/j.jhep.2015.09.011. [DOI] [PubMed] [Google Scholar]
- 2.Fafi-Kremer S, Fofana I, Soulier E, Carolla P, Meuleman P, Leroux-Roels G, Patel AH, et al. Viral entry and escape from antibody-mediated neutralization influence hepatitis C virus reinfection in liver transplantation. J Exp Med. 2010;207:2019–2031. doi: 10.1084/jem.20090766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zeisel MB, Crouchet E, Baumert TF, Schuster C. Host-Targeting Agents to Prevent and Cure Hepatitis C Virus Infection. Viruses. 2015;7:5659–5685. doi: 10.3390/v7112898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Law M, Maruyama T, Lewis J, Giang E, Tarr AW, Stamataki Z, Gastaminza P, et al. Broadly neutralizing antibodies protect against hepatitis C virus quasispecies challenge. Nat Med. 2008;14:25–27. doi: 10.1038/nm1698. [DOI] [PubMed] [Google Scholar]
- 5.Meuleman P, Bukh J, Verhoye L, Farhoudi A, Vanwolleghem T, Wang RY, Desombere I, et al. In vivo evaluation of the cross-genotype neutralizing activity of polyclonal antibodies against hepatitis C virus. Hepatology. 2011;53:755–762. doi: 10.1002/hep.24171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morin TJ, Broering TJ, Leav BA, Blair BM, Rowley KJ, Boucher EN, Wang Y, et al. Human Monoclonal Antibody HCV1 Effectively Prevents and Treats HCV Infection in Chimpanzees. PLoS Pathog. 2012;8:e1002895. doi: 10.1371/journal.ppat.1002895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chung RT, Gordon FD, Curry MP, Schiano TD, Emre S, Corey K, Markmann JF, et al. Human monoclonal antibody MBL-HCV1 delays HCV viral rebound following liver transplantation: a randomized controlled study. Am J Transplant. 2013;13:1047–1054. doi: 10.1111/ajt.12083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Keck ZY, Xia J, Wang Y, Wang W, Krey T, Prentoe J, Carlsen T, et al. Human monoclonal antibodies to a novel cluster of conformational epitopes on HCV E2 with resistance to neutralization escape in a genotype 2a isolate. PLoS Pathog. 2012;8:e1002653. doi: 10.1371/journal.ppat.1002653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kong L, Giang E, Nieusma T, Kadam RU, Cogburn KE, Hua Y, Dai X, et al. Hepatitis C virus E2 envelope glycoprotein core structure. Science. 2013;342:1090–1094. doi: 10.1126/science.1243876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Khan AG, Whidby J, Miller MT, Scarborough H, Zatorski AV, Cygan A, Price AA, et al. Structure of the core ectodomain of the hepatitis C virus envelope glycoprotein 2. Nature. 2014;509:381–384. doi: 10.1038/nature13117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fofana I, Krieger SE, Grunert F, Glauben S, Xiao F, Fafi-Kremer S, Soulier E, et al. Monoclonal anti-claudin 1 antibodies prevent hepatitis C virus infection of primary human hepatocytes. Gastroenterology. 2010;139:953–964. doi: 10.1053/j.gastro.2010.05.073. [DOI] [PubMed] [Google Scholar]
- 12.Krieger SE, Zeisel MB, Davis C, Thumann C, Harris HJ, Schnober EK, Mee C, et al. Inhibition of hepatitis C virus infection by anti-claudin-1 antibodies is mediated by neutralization of E2-CD81-claudin-1 associations. Hepatology. 2010;51:1144–1157. doi: 10.1002/hep.23445. [DOI] [PubMed] [Google Scholar]
- 13.Mercer DF, Schiller DE, Elliott JF, Douglas DN, Hao C, Rinfret A, Addison WR, et al. Hepatitis C virus replication in mice with chimeric human livers. Nat Med. 2001;7:927–933. doi: 10.1038/90968. [DOI] [PubMed] [Google Scholar]
- 14.Kneteman NM, Asthana S, Lewis J, Dibben C, Douglas D, Nourbakhsh M, Tyrrell LJ, et al. Impact of calcineurin inhibitors with or without interferon on hepatitis C virus titers in a chimeric mouse model of hepatitis C virus infection. Liver Transpl. 2012;18:38–44. doi: 10.1002/lt.22400. [DOI] [PubMed] [Google Scholar]
- 15.Bailey JR, Wasilewski LN, Snider AE, El-Diwany R, Osburn WO, Keck Z, Foung SK, et al. Naturally selected hepatitis C virus polymorphisms confer broad neutralizing antibody resistance. J Clin Invest. 2015;125:437–447. doi: 10.1172/JCI78794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krey T, Meola A, Keck ZY, Damier-Piolle L, Foung SK, Rey FA. Structural basis of HCV neutralization by human monoclonal antibodies resistant to viral neutralization escape. PLoS Pathog. 2013;9:e1003364. doi: 10.1371/journal.ppat.1003364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pierce BG, Hellman LM, Hossain M, Singh NK, Vander Kooi CW, Weng Z, Baker BM. Computational design of the affinity and specificity of a therapeutic T cell receptor. PLoS Comput Biol. 2014;10:e1003478. doi: 10.1371/journal.pcbi.1003478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Keck Z, Wang W, Wang Y, Lau P, Carlsen TH, Prentoe J, Xia J, et al. Cooperativity in virus neutralization by human monoclonal antibodies to two adjacent regions located at the amino terminus of hepatitis C virus E2 glycoprotein. J Virol. 2013;87:37–51. doi: 10.1128/JVI.01941-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deng L, Ma L, Virata-Theimer ML, Zhong L, Yan H, Zhao Z, Struble E, et al. Discrete conformations of epitope II on the hepatitis C virus E2 protein for antibody-mediated neutralization and nonneutralization. Proc Natl Acad Sci U S A. 2014;111:10690–10695. doi: 10.1073/pnas.1411317111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kidney Disease: Improving Global Outcomes (KDIGO) KDIGO clinical practice guidelines for the prevention, diagnosis, evaluation, and treatment of hepatitis C in chronic kidney disease. Kidney Int. 2008;73:S1–S99. doi: 10.1038/ki.2008.81. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.