Abstract
Quality control processes are widespread and play essential roles in detecting defective molecules and removing them in order to maintain organismal fitness. Aberrant mRNA molecules, unless properly managed, pose a significant hurdle to cellular proteostasis. Often mRNAs harbor premature stop codons, possess structures that present a block to the translational machinery, or lack stop codons entirely. In eukaryotes, the three cytoplasmic mRNA-surveillance processes, nonsense-mediated decay (NMD), no-go decay (NGD) and non-stop decay (NSD), evolved to cope with these aberrant mRNAs, respectively. Non-stop mRNAs and mRNAs that inhibit translation elongation are especially problematic as they sequester valuable ribosomes from the translating ribosome pool. As a result, in addition to RNA degradation, NSD and NGD are intimately coupled to ribosome rescue in all domains of life. Furthermore, protein products produced from all three classes of defective mRNAs are more likely to malfunction. It is not surprising then that these truncated nascent protein products are subject to degradation. Over the past few years, many studies have begun to document a central role for the ribosome in initiating the RNA and protein quality control processes. The ribosome appears to be responsible for recognizing the target mRNAs, as well as for recruiting the factors required to carry out the processes of ribosome rescue and nascent protein decay.
Introduction
Cells rely on a number of template-dependent processes to maintain and decipher the genetic code. These processes are among the most accurate in biology highlighting the importance of ensuring that the sequence of protein products is a faithful interpretation of the genetic information. Even with the remarkable level of accuracy of these processes, as many as one in ten newly synthesized proteins have been estimated to contain at least one miscoded amino acid 1; most are the result of translational errors 2. Furthermore, mRNA molecules are subject to constant changes and modifications 3, 4 that could potentially have adverse consequences on proteostasis. Defective protein products are more prone to misfold and sometimes have dominant-negative effects 5, 6. As a result, it comes as no surprise that organisms evolved a number of quality control processes to detect errors in the mRNA and protein pools and subject them to rapid degradation 7. Recently, it has been appreciated that the failure to elicit these quality control processes is likely to be responsible for a number of diseases and is critical for cellular fitness 8, 9.
The biogenesis of an mRNA is complex and involves some of the most elaborate machinery in the cell, particularly in eukaryotic organisms. A mistake could result in a defective mRNA that is not true to its encoded sequence. At a minimum, transcriptional errors by the polymerase, although infrequent at a rate of about 10−5 10, can result in a nonsense mutation that can lead to the translation of truncated protein. Since many mRNAs are made from a single gene, at first glance such a mutation might seem inconsequential. However, considering that some transcripts have been estimated to produce more than a thousand copies of proteins per mRNA copy 11, the cost of a nonsense mutation is not trivial especially given the expense of protein synthesis (approximately 2000 ATP molecules per one protein molecule) 12. In addition to its cost, a truncated protein product is more likely to malfunction, adversely affecting the function of the cell 1, 6. It should be noted, however, that more often a nonsense mutation is introduced to the mRNA molecule during processing events, particularly during splicing 13. In eukaryotes, these mRNAs are subject to quality control through nonsense-mediated decay (NMD) 14.
Misprocessing of pre-mRNA often results in premature polyadenylation leading to truncated transcripts 15–18. mRNA is also subject to alterations and modifications post-synthesis with potentially-profound effects on their function 4. A key issue for maintaining the integrity of transcripts is the inherent chemical instability of the phosphodiester backbone of RNA that makes it susceptible to endonucleolytic cleavage 19, 20. Truncated transcripts, regardless of the source, often lack a stop codon causing the ribosomes to run to the end of the mRNA and stall. These mRNAs are rapidly degraded through a process termed non-stop decay (NSD) 15, 17. In addition to cleavage, RNA is also susceptible to chemical insults that modify the nucleobase interfering with the codon-anticodon interaction 3, 21. Frequently these modifications on the mRNA stall the decoding process and cause the ribosome to “not go”. Certain RNA structures such as hairpins and pseudoknots also stall the ribosome 22. No-go decay (NGD), which shares many features with NSD, is responsible for recognizing these aberrant RNAs and targeting them for degradation 23. Notably, NGD and NSD are intimately linked to the arguably more important function of ribosome rescue 24, 25. Indeed, initial studies on NGD and NSD focused on these two aspects of mRNA degradation and ribosome recycling 23. However, recent studies have also begun to address the fate of the truncated protein product 26. The defective nascent protein has been found to be the target of a quality control process that rapidly degrades it 27.
Since defective mRNAs interfere with translation, the corresponding mRNA- and protein-quality-control processes all take advantage of the ribosome for the recognition 23. In this review, we discuss our current understanding of the mechanism of this recognition process and the initial recruitment of different quality control factors to the ribosome. We also highlight some of the gaps in our understanding of the processes and potential future investigations for the field.
mRNA-surveillance
The interplay between translation and quality control
Because mRNA-surveillance and quality control of nascent proteins take advantage of changes to the translation cycle, it is worthwhile to briefly discuss the key steps of protein synthesis. The translation process is divided into four stages: initiation, elongation, termination and recycling. Initiation is the process by which the ribosome recognizes the start codon to begin protein synthesis. This process is different between eukaryotes and bacteria. Initiation is relatively complex in eukaryotes and is subject to various aspects of quality control, which are beyond the scope of this review. Instead a more detailed review of initiation can be found here 28.
The elongation phase is essentially the same in all domains of life 29, and it involves the recruitment of a ternary complex of aminoacyl-tRNA (aa-tRNA), elongation factor EFTu/eEF1A (bacteria/eukaryote, respectively) and GTP to decode the A-site codon (Fig. 1). EFTu/eEF1A is a member of translational GTPase factors that interact with a conserved region on the large subunit 30. As we shall see later, among these factors are proteins that are directly involved in initiating quality control on the ribosome. The speed of the decoding process, as it happens during the elongation phase, is sensitive to many parameters which include: 1) the identity of the A-site codon as well as its sequence context 31, 32; 2) the concentration of the corresponding aa-tRNA 22, 33; 3) the structure of the mRNA downstream 22; 4) the sequence of the nascent peptide and its interaction with the exit tunnel of the ribosome 34, 35; 5) chemical modifications to the nucleotides 3. In eukaryotes, significant decreases in the rate of peptide-bond formation, initiated by these features in the mRNA, are recognized as stalls and elicit NGD. The process is linked to ribosome disassembly and recycling of the subunits (discussed below).
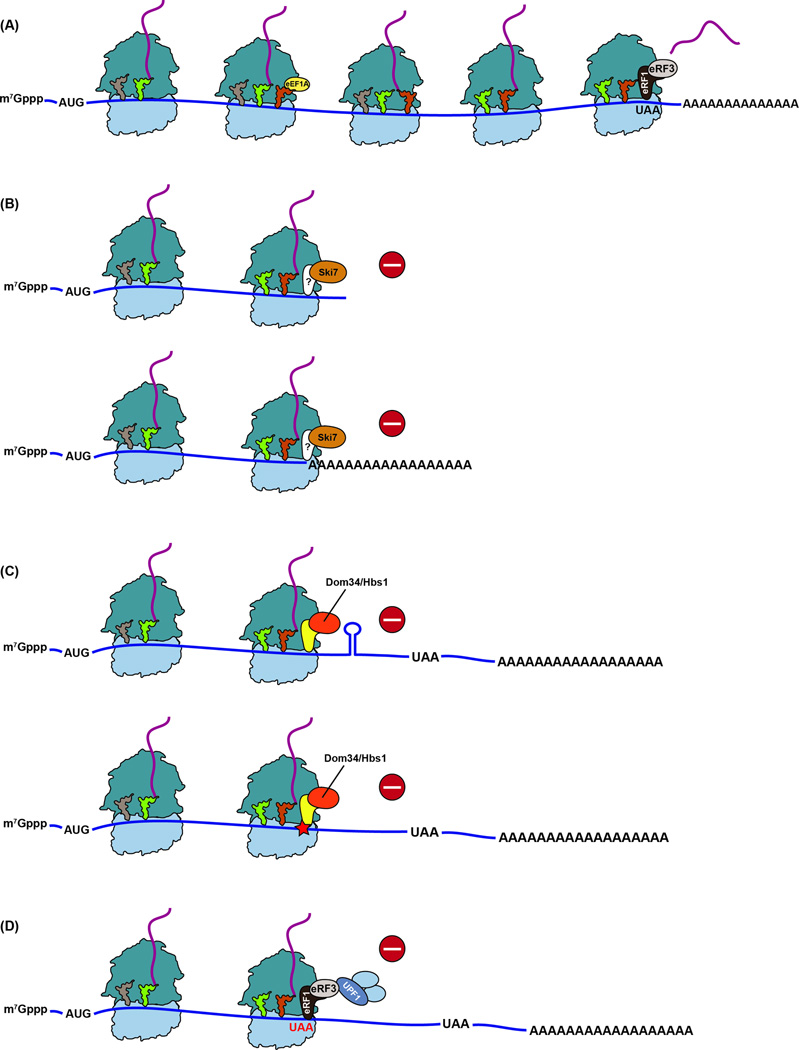
Figure 1. Translation of intact mRNAs versus aberrant mRNAs.
(A) During normal translation, a ternary complex of aa-tRNA, eEF1A (EFTu in bacteria) and GTP binds the ribosome to decode the A site codon. Following peptidyl transfer, the elongation phase continues until a stop codon arrives at the A site, where it is recognized by eRF1 release factor in a complex with eRF3-GTP. Hydrolysis of the peptidyl tRNA and dissociation of eRF3 is triggered by conformational changes to eRF1 upon GTP hydrolysis. (B) In S. cerevisiae, the GTPase Ski7 interacts with the ribosome when it is stalled at the 3’ end of a stop-codon-less mRNA (top) or when it translates a polyA tail (bottom), activating non-stop decay (NSD). A binding partner for SKI7 has not been identified (shown as ?). (C) No-go decay (NGD) is responsible for recognizing and rescuing ribosomes stalled within an mRNA, either due to stable structures that block its progression (top) or caused by damaged nucleobases or strings of rare codons (bottom - shown as a star). Dom34, together with Hbs1-GTP, binds the ribosome and recycles the stalled ribosome. The process results in an endonucleolytic cleavage event (not shown), which may precede Dom34 recruitment. (D) Premature stop codons are recognized by canonical release factors that interact with Upf1 and other factors of the nonsense mediated decay (NMD) pathway.
Although the molecular details of the termination and recycling phases differ between bacteria and eukaryotes, the processes appear to be intimately coupled to each other in both domains of life 36, 37. Peptide release is initiated when a stop codon arrives at the A site of the ribosome; this in turn is recognized by class I release factors RF1/2, eRF1 and aRF1 in bacteria, eukaryotes and archaea, respectively 38, 39. In eukaryotes, similar to aa-tRNA, eRF1 is thought to form a ternary complex with a translational GTPase eRF3 and GTP, which collectively bind the A site 40. The hydrolysis of GTP by eRF3 appears to occur after stop-codon recognition and induces a conformational change on eRF1 allowing it to engage the active site of the ribosome and promote hydrolysis of the peptidyl-tRNA 40–43. GTP hydrolysis also promotes the dissociation of eRF3, which in turn opens up a site for the ABC-type ATPase ABCE1 (Rli1 in yeast) to bind and initiate the recycling phase. Structural and biochemical studies have provided a working model for the mode of action of ABCE1/Rli1 37, 41, 44–46. Like other ABC proteins 47, the factor uses ATP binding and hydrolysis to trigger a “power stroke” to change its conformation 41. The conformational switch appears to induce ribosome dissociation through disruption of intersubunit bridges and/or further conformational changes to eRF1. Additionally in vitro experiments suggest that ABCE1/Rli1 is directly involved in peptide release since its addition to a termination reaction significantly accelerated the rate of peptide release by eRF1 37. In contrast to its ribosome dissociation activity, ABCE1/Rli1-mediated acceleration of peptide release does not require ATP. Therefore, ABCE1/Rli1 provides a link between termination and recycling with ATP hydrolysis acting as a gate between the two processes.
As a result of the union between termination and recycling, an mRNA lacking a stop codon (referred to hereafter as non-stop mRNA) leads to ribosome stalling because the subunits cannot be disassembled through canonical pathways. Similar to NGD targets, non-stop mRNAs have the potential of placing a significant burden on the cell as they remove ribosomes from the translating pool. The observation that cells evolved the specific process of NSD to rescue ribosomes suggests that non-stop mRNAs are frequent. Although non-stop mRNAs can result from multiple sources (for a review on this see 48), most likely result from premature cleavage and polyadenylation during transcription 15, 16. Non-stop mRNAs are rapidly degraded by what appears to be a combined endonucleolytic and exonucleolytic action of the cytoplasmic exosome 49. Although NSD and NGD do not appear to involve release factors for disassembly of the ribosome, they require release-factor-like proteins in addition to ABCE1/Rli1 37, 45. The details of these processes will be discussed in later sections. Due to similarities between the two pathways, particularly the sensing of a stalled ribosome, the distinction between NSD and NGD has become more ambiguous recently 23.
In contrast to NSD and NGD targets, for mRNAs harboring premature stop codons, ribosomal recycling is not a problem. Although NMD is capable of discriminating between canonical and premature stop codons, the process does not utilize specialized factors to recognize the A site and instead it continues to utilize release factors to recognize the stop codon 14. Therefore, in principle ribosome rescue ensues as it would for canonical termination. The major role for NMD is instead focused on the recognition of its target and specifying its degradation.
Non-stop decay
Similar to most mRNA-surveillance processes, NSD was first observed in yeast, for which reporter constructs lacking an in frame stop codon were shown to turn over rapidly relative to ones harboring a stop codon 15, 17. This rapid decay of non-stop mRNAs is coupled to translation as the addition of translation inhibitors such as cycloheximide has been shown to stabilize NSD targets 15. In addition to these observations, many studies documented a critical role for the ribosome in the initial recognition of the defective mRNAs 17, 19, 50, 51. In particular genetic studies identified the yeast translational GTPase Ski7 to be essential for the recognition process 17, 51. Ski7 is related to eRF3 and has been shown to interact with the ribosome 52; it is also a component of the cytoplasmic exosome in yeast, linking the process of recognition of the non-stop mRNA to its decay 17, 51, yet so far it has been found in only a handful of yeasts 53. In S. cerevisiae, Ski7 is a paralog of the NGD factor Hbs1 (discussed later); both arising from a common ancestor after a whole genome duplication 54. Indeed, in other organisms, Hbs1 appears to fulfill the function of Ski7 in NSD. Consistent with these observations, Ski7 deletion strains are complemented by introducing Hbs1 from the related yeast S. kluyveri 54. The molecular details of how Ski7 (or Hbs1) facilitates the initial recognition process on the ribosome are poorly understood, but as discussed in the next section, it is likely to resemble what occurs during NGD.
Early studies on NSD suggested that it is similar to the bacterial tmRNA rescue system, in which the process is triggered by a ribosome running to the 3’-end of the mRNA 17. This is most likely to be true for truncated transcripts. However it is most likely not the case for mRNAs lacking a stop codon that have a polyA tail, which are arguably the majority of the NSD targets 55. Although these can originate from multiple processes, most result from a premature polyadenylation event 15, 16. In contrast to truncated mRNAs, on these mRNAs the ribosome stalls as it decodes the polyA tail into poly(Lys) considerably short of reaching the end of the mRNA. Consistent with these ideas, internal polyA stretches are known to stall translation and elicit NGD as characterized by its hallmark endonucleolytic cleavage 33, 35, 55. Early studies on polyA-mediated stalling suggested that it results from extensive charge-charge interactions between the positively-charged lysine residues and the negatively-charged exit tunnel of the ribosome 34, 56. However more recent studies have argued that stalling also depends on the mRNA sequence 57, 58. In particular, poly(AAA) sequences are more likely to promote stalling than poly(AAG) ones even though both code for the same polypeptide sequence of poly(Lys). Irrespective of the mechanism of stalling, NSD and NGD appear to be closely related to each other as, for the most part, they utilize many of the same factors and in both cases the initial step in the pathway involves a stalled ribosome.
No-go decay
As described earlier, NGD is triggered by barriers that block ribosome movement on the mRNA 22. Genetic studies in yeast identified the factors Dom34 and Hbs1 to be important for the process. Interestingly, Dom34 (Pelota in mammals) and Hbs1 are homologs of the release factors eRF1 and eRF3 22, 53, respectively, highlighting the profound role for the ribosome in NGD. In contrast to eRF1, Dom34 lacks the conserved GGQ motif 59–62 required to engage the peptidyl transferase center to promote hydrolysis during termination. The factor is also missing the NIKS domain 63, which is required for recognition of the stop codons. As a result, Dom34 is incapable of carrying out the release reaction. Then, what function does Dom34 carry out on the ribosome? Biochemical reconstitution experiments by Green and colleagues provided some of the first insights into this question 46. The ternary complex of Dom34-Hbs1-GTP was shown to split the 80S ribosome into the individual subunits suggesting that these factors rescue ribosomes in vivo. Consistent with this idea, the splitting reaction is not dependent on the identity of the A-site codon 46. Furthermore, similar to normal recycling, Dom34-mediated ribosome splitting is significantly more efficient in the presence of Rli1/ABCE1 37, 45, 64. Overall the process is very similar to canonical recycling with Dom34 and Hbs1 substituting for the role of eRF1 and eRF3, respectively 41, 65. Collectively, these studies were the first to highlight the importance of ribosome recycling on defective mRNAs in eukaryotes; a process that long been appreciated in bacteria through trans-translation by tmRNA 66.
The initial observation that Dom34 is active on ribosomal complexes displaying any tested codon in the A site corroborates the essential notion that it can recycle any stalled ribosome regardless of the cause, but at the same time provided a potential conflict. In particular, how does Dom34 distinguish between an elongating ribosome and a stalled one? A solution to this would-be conundrum may be partly resolved by the kinetics of the processes 23. In vitro, ribosome splitting is slower than peptidyl transfer 46. Nevertheless, differences in rates cannot account for the overall specificity, mainly due to the necessity for efficient recycling in vivo. For instance, rate differences have to be greater than three orders of magnitude to ensure that a 1000-amino-acid-long protein is not prematurely terminated (on average). With these reduced rates of recycling, stalled ribosomes are likely to linger and sequester even more ribosomes upstream on the transcript.
The interaction between Hbs1 and the small subunit contributes significantly to the specificity of the recognition process 65. The N-terminal domain of Hbs1 binds in the mRNA entry tunnel. During elongation, the mRNA occupies this site and as a result Hbs1 cannot effectively bind, preventing premature dissociation of the ribosome. Biochemical experiments are in complete agreement with this model, for which the splitting activity of Dom34-Hbs1 is significantly accelerated for complexes having little to no mRNA downstream of the P site 37, 45, 64. Interestingly, this dependency on mRNA length is reminiscent of the processes employed by bacteria to rescue ribosomes 67, even though the molecular details are quite different.
We note that the details of the steps leading to the degradation of the mRNA is beyond the scope of this review. Relevant here, however, and to the mechanism by which the NGD targets are recognized by Dom34 and Hbs1 is the observation that the defective mRNA is endonucleolytically cleaved in the vicinity of the stall site 33, generating what essentially looks like an NSD substrate. Dom34 and Hbs1 can rescue the lagging ribosomes as they run to the end of the transcript 33, as they do not harbor mRNA in the entry tunnel. The process by which the leading ribosome is rescued is not understood, though the cleavage reaction generates an uncapped mRNA, which is rapidly degraded by the 5’-3’ exonuclease Xrn1 68–71. It is possible, as a result of its processivity, that Xrn1 may be able to displace the leading ribosome.
Nonsense-mediated decay
NMD was initially observed in yeast, for which nonsense mutations in the URA3 gene were observed to reduce the steady state levels of its transcript without affecting its rate of synthesis 72. These observations were soon extended into other eukaryotes. In particular, Maquat et al. showed that β-globin mRNA from thalassemic patients, which contains a premature stop codon, turned over much faster relative to a nonthalassemic one 73. The mechanism by which the cell recognizes NMD targets, even after almost four decades of study, is only partially understood 14. The ribosome somehow distinguishes a premature stop codon from a normal one and in addition to the ribosome and release factors, genetic studies have identified the conserved factors Upf1, Upf2 and Upf3 to be important for NMD 74–76.
One of the earlier models aimed at explaining the recognition of NMD targets relies on the presence of an exon junction complex (EJC) downstream of a premature stop codon 77–79. The EJC is a complex deposited by the splicing machinery ~24 nt upstream of exon-exon boundaries 80. During the pioneering round of translation, where mRNAs are still bound by the nuclear cap binding protein instead of eIF4E, the EJC complex is removed by the ribosome 81. As most transcripts contain a stop codon in the last exon, a typical premature stop codon has a signature of an EJC present downstream that cannot be removed by the ribosome. At a molecular level, the Upf proteins are proposed to bridge a connection between the EJC and the release factors 82, 83. This connection appears to induce posttranslational modifications to Upf1 resulting in a stimulatory signal for NMD through the recruitment of RNA degrading factors 83–86. At the same time, the connection is likely to inhibit interactions between eRF3 and polyA binding protein (PABP) 87, thought to be key for normal termination 88–90. Although appealing, the EJC model falls short of explaining all of the NMD targets; for instance in yeast robust NMD is observed on unspliced transcripts 91. There are at least two competing models that have been put forward to explain target recognition during NMD. Both rely on the fact that efficiency of decay is directly related to the length of the UTR 91. In the first model, due to the proximity between the premature stop codon and the polyA tail, interactions between eRF3 and PAB, which are critical during normal termination, are inhibited 92. In turn the Upf proteins are allowed to bind and effectively mark the RNA as an NMD target. This model has been called into question as others have argued that mRNAs lacking premature stop codons are destabilized regardless of whether they are polyadenylated or not 93. In an alternative model it has been suggested that Upf1 coats the 3’-UTR 94. Therefore for long UTRs, such as those found on NMD targets the local concentration of Upf1 is much higher, distinguishing them from non-targets. In summary, while the precise recognition of a NMD target is currently ambiguous, many in the community agree that something about the mRNA sequence downstream of a premature stop codon is responsible for initiating NMD, “the faux 3’-UTR model” 95. Future studies are needed to clarify what these signals might be. The elucidation of the signaling cascades which lead to successful recognition and degradation of aberrant transcripts are open challenges in the field of NMD.
Quality control of nascent peptides
While ribosome-based quality control processes have been typically studied in the context of the fate of the mRNA, recent studies have begun to address the fate of the nascent peptide 26. Defective mRNAs, be it NMD, NGD or NSD targets, have the potential to code for toxic protein products that are likely to misfold or malfunction 1. As a result, organisms from bacteria to man appear to have evolved at least one form of co-translational protein quality control process that target defective nascent proteins. By rapidly eliminating defective proteins on the ribosome, these processes ensure that potentially toxic protein products are not allowed to cause harm to the cell. As would be predicted, for defective mRNAs the trigger for these pathways is stalled ribosomes 27, 96, 97. But beyond this similarity, the details of co-translational protein quality control vary vastly between bacteria and eukaryotes. This is in part rationalized by differences in the mRNA-surveillance pathways.
Trans-translation in bacteria: tmRNA
In bacteria, at least one protein QC pathway is known to occur on stalled ribosomes that run to the end of an mRNA 96. These truncated mRNAs are produced through a variety of processes that include endonucleolytic cleavage, ribosome stalling, chemical insults and premature transcriptional termination. They are for the most part subject to trans-translation by tmRNA, which acts as a tRNA and an mRNA ensuring ribosomes complete the translation cycle and hence are recycled 66. Interestingly, the role of tmRNA in translation quality control was elucidated through its participation in protein quality control and ribosome rescue. In particular, although the RNA was initially discovered in the 1970s 98, its biological function was not uncovered until the 1990s 96. This discovery was made possible by the observation that heterologous expression of genes in E. coli often resulted in truncated protein products that have a defined C-terminal extension 99. This C-terminal tag was later found to be encoded by a short ORF within the tmRNA sequence, and is referred to as an ssrA (the name of the gene encoding tmRNA) tag 96. The sequence of this tag is similar to the degradation signal used by bacterial proteases suggesting that ssrA-tagged proteins are subject to proteolytic degradation 100. We now know that: 1) tmRNA, which is aminoacylated by alanine 101, binds the A site of the ribosome 102; 2) the nascent peptide is transferred to tmRNA; 103 3) translation then switches from the 3’-end of the mRNA to the ssrA-coding sequence tagging the C-terminus of the defective protein 66, 104. Similar to aa-tRNAs, tmRNA binds elongation factor EFTu 105, 106 but also requires another protein partner, SmpB 107. The molecule binds the A site in a quaternary complex with EFTu, SmpB and GTP.
An important question that emerged soon after the discovery of trans-translation is how the selectivity of the process is governed. During the elongation phase of translation, the specificity of peptidyl transfer is achieved by cognate codon-anticodon interactions between the mRNA and tRNA. These interactions are critical for initiating conformational changes in the decoding center that are pre-requisites for aa-tRNA accommodation into the peptidyl transferase center 108. However, tmRNA lacks the anticodon stem loop and as a result cannot form the same sort of interactions as aa-tRNAs 109. The first clues about the mechanism of tmRNA recognition came from in vivo studies showing that ssrA tagging occurred on ribosomes that either reach the 3’-end of the mRNA because they lack a stop codon or on stalled/paused ribosomes (resulting from rare codons or inefficient peptide release due to the peptide sequence or interactions between the nascent peptide and the exit tunnel of the ribosome) 96, 110, 111. Ribosomal complexes stalled in the middle of transcripts are converted into complexes stalled on the 3’-end of the transcript through endonucleolytic and exonucleolytic degradation of the mRNA target 111–115. Therefore, a feature that is common to all tmRNA targets is a ribosomal complex that has little to no mRNA downstream of the P site. Indeed, in vitro studies have shown that complexes with more than six nucleotides downstream of the P site are poorly recognized by tmRNA 67, 116.
Recent biochemical and structural studies have provided important clues about the molecular mechanism of the recognition process 117. Crystal structures of the tRNA-like domain of tmRNA in complex with SmpB revealed that the complex adopts a structure similar to a tRNA with the N-terminal domain of the protein substituting for the anticodon stem loop 118. Consistent with these observations, cryoEM reconstructions and chemical probing experiments showed that SmpB is likely to interact with A1492, A1493 and G530 residues of the decoding center of the ribosome 119, 120. During normal elongation, the universally conserved A1492 and A1493 residues change conformation to engage the minor groove of the codon-anticodon helix 121. The A-minor interactions are necessary to stabilize this “induced-fit” state of the ribosome 122. However, during trans-translation the decoding center is occupied by SmpB and hence A-minor interactions cannot occur. Consistent with these observations, mutating any of the decoding center residues, which is detrimental for normal decoding 123, has no effect on tmRNA activity in a reconstituted system 124. Nonetheless, a recent high-resolution crystal structure of T. thermophilus tmRNA–SmpB-EFTu complex bound to the ribosome shows that the decoding center adopts a conformation similar, with subtle differences, to that observed with normal elongation complexes 117. Therefore, although SmpB appears to induce rearrangement of A1492 and A1493, their identities are not critical to stabilize this induced state of the ribosome. Accordingly, the following steps of GTP hydrolysis and accommodation of tmRNA are essentially identical between the two processes and depend on “domain closure” of the small subunit where the head and shoulder domains of the 30S subunit rotate towards the subunit interface.
The high-resolution crystal structure of the trans-translation complex also revealed some important aspects about the selectivity of the process. In solution, the C-terminal domain of SmpB is unstructured 125, 126, but in complex with the ribosome forms a helical structure 117. The helical structure extends from the A site towards the mRNA-entry tunnel, making intimate contacts with the 16S rRNA. Earlier mutational analysis had shown that this region of SmpB and its ability to form an α-helical structure is critical for ssrA tagging 124, 127. Overall, the structure revealed that SmpB cannot bind the ribosome unless it has reached the 3’-end of the mRNA because the C-terminus of the protein occupies a site that is normally occupied by the mRNA during canonical translation. The structural clash between SmpB and the mRNA ensures that tmRNA does not bind the A site, and hence prematurely terminate protein synthesis under normal conditions. We note that these studies have addressed only the initial step of trans-translation and much more is yet to be learned about the process. For example, following the first peptidyl transfer reaction, translocation has to take place to bring the tmRNA ORF into the A site of the ribosome. The new ORF has to occupy the mRNA entry tunnel where SmpB binds initially; as a result SmpB has been predicted to change conformation to allow template switching 128. This process, by which the resume codon of tmRNA is positioned into the A site, is not understood, but appears to be dependent on key interactions between SmpB and sequence elements upstream of the ORF 129.
Eukaryotic co-translational protein quality control
In eukaryotes misfolded proteins for the most part are degraded by the ubiquitin-proteasome system 130. The process of ubiquitin conjugation involves three classes of enzymes: E1, E2 and E3. E1 and E2 are ubiquitin-activating and – conjugating enzymes, respectively. Substrate specificity is achieved by the E3 ligases, which recognize their protein substrates and polyubiquitinate them (typically K48-linked chain) targeting them for degradation. The first reports of co-translational ubiquitination came out of studies on the quality control of the cystic fibrosis transmembrane conductance regulator (CFTR) and Apolipoprotein B100 (ApoB) proteins 131, 132. CFTR is a relatively large protein and is prone to misfolding. In synchronized rabbit reticulocyte extracts, CFTR was observed to be ubiquitinated before the protein was completely synthesized suggesting that the protein is targeted for degradation on the ribosome 131. ApoB, a secretory protein, was also shown to be ubiquitinated in HepG2 cells before it is fully synthesized 132. These studies provided the first evidence that nascent proteins in eukaryotes are subject to quality control as soon as they emerge from the ribosome.
The extent to which nascent peptides are targeted for ubiquitination was initially subject to debate 133. Some initial studies suggested as much as 30% of newly synthesized proteins are subject to degradation 134; others argued that the number is much less (~6%) 135. Two recent studies, taking advantage of puromycin labeling of nascent peptides, estimated that 12–15% of newly synthesized proteins are ubiquitinated in mammalian cell culture 136, 137. Regardless of the actual number, it is evident that a significant amount of proteins are targeted for degradation before they are complete and the extent is likely to depend on the cell type and the cellular conditions. The mechanism by which the ubiquitination machinery recognizes the broadly defined folding state of the nascent proteins is not well understood, but is likely to involve the ribosome-associated chaperone machinery. In contrast, recognition of defective protein products encoded by aberrant mRNAs has been the subject of a number of recent studies and as a result is arguably better understood 26.
LTN1 targets aberrant peptides for ubiquitination
Similar to studies on mRNA-quality control processes, many of the pioneering studies on co-translational protein quality control processes came out of studies in yeast using reporter constructs. Some of these initial studies by Inada and colleagues provided the first link between the two 34, 55. In particular, protein products produced from unstable mRNAs harboring internal polyA stretches (i.e. mimicking NSD targets) were shown to be rapidly degraded by the proteasome. Genetic screens in yeast identified a set of genes that, when mutated, stabilize NSD protein products. As would be expected, most of these genes were known to affect proteasome function, but the list also included a gene encoding an uncharacterized E3 ligase RKR1/YMR247c 138. This gene is currently widely known as LTN1. At the time, the manner in which Ltn1 may recognize its substrates was not clear, especially since the only common feature of the targets is a truncated protein product. These details started to emerge soon after the discovery by Bengston and Joazeiro showing that Ltn1 is ribosome bound and targets stalled protein products for ubiquitination 27. Highlighting its participation in quality control of NSD protein products, LTN1 deletion strains are sensitive to antibiotics that promote stop-codon readthrough 27. Furthermore, mutations in the mammalian homologue Listerin have been associated with neurodegeneration 139.
The question of how Ltn1 recognizes its targets was an intriguing one. A priori, the simplest model would entail a direct association between the factor and the ribosome near the peptide exit tunnel, where it continuously monitors the status of translation speed. As soon as the ribosome stalls, the factor ubiquitinates the nascent protein. This model could easily be negated based on the relative stoichiometry of LTN1 to the ribosome; a yeast cell has an estimated 200 copies of Ltn1 relative to 200,000 ribosomes 140. A clue into Ltn1’s mode of action was made possible by the observation that inhibition of ribosome recycling by deletion of DOM34 stabilizes NGD protein products in a non-ubiquitinated form suggesting that ribosome splitting precedes ubiquitination by Ltn1 33. Further clues came out of a genome-wide screen by the Weissman group, which sought to identify factors that modulate the activity of the transcriptional regulator heat shock factor 1 (Hsf1) 141. The genetic interaction map revealed that LTN1 shares a similar interaction network with an uncharacterized factor termed RQC1. Immunoprecipitation of Rqc1 purified a complex that, in addition to Ltn1, included Tae2 (Rqc2), Cdc48 and its cofactors Ufd1 and Npl4 along with the 60S ribosomal subunit. An additional study aimed at discovering genetic interactions with Ltn1 and the Ski complex identified the same set of factors 142. Thus, Ltn1 appears to associate with a larger complex that binds the large subunit of the ribosome; this complex is now known as the ribosome quality control complex (RQC).
Role of the RQC in degrading target peptides
The next key step in understanding the mode of action of the RQC complex was assigning the role of the different factors. Initial studies showed that deletion of any of the RQC factors stabilizes protein targets 141. Furthermore, previous biochemical experiments established that ubiquitination of the target protein is fully dependent on Ltn1 and to some extent Rqc2, but not on Rqc1 and Cdc48 141, 142, suggesting that Ltn1 and Rqc2 are recruited initially to the 60S subunit. The recruitment of Cdc48 to the complex requires the presence of Rqc1. Beyond a direct role in facilitating proteasomal degradation of tRNA-linked polyubiquitinated protein targets, Cdc48 has also been suggested to play a direct physical role in the removal of the protein substrate from the 60S subunit 143. In two studies from the Hegde group, reconstitution experiments in rabbit reticulocyte extracts established a more defined view of the order of events leading up to the degradation of the protein target 144, 145. Ribosome dissociation by Pelota (the human homologue of yeast Dom34), Hbs1 and ABCE1 (the mammalian homologue of yeast Rli1) is sufficient for ubiquitination by Listerin. Subsequent binding of Listerin to the large subunit prevents its reassociation with the small subunit; cryoEM reconstruction showed that Rqc2 (or its mammalian homologue NEMF) makes contacts with the 60S subunit and the peptidyl tRNA, preventing reassociation of 60S/40S and thus allowing Ltn1/Listerin to remain bound to the large subunit 146, 147. These observations corroborate biochemical data showing that Listerin binding to the large subunit is dependent on NEMF. Its addition was sufficient to prevent the assembly of 80S ribosomes following splitting by the NGD factors. Rqc2/NEMF has an additional newly-described function that will be discussed in detail later.
Arguably much of our understanding of the molecular details of the process has come out of structural studies; in particular recent advances in cryoEM reconstructions have painted a more focused picture of the RQC complex. In yeast these structures took advantage of the fact that Ltn1 mutants lacking the RING domain are still efficiently recruited to the ribosome but are unable to ubiquitinate the protein substrate and hence trap the RQC complex on the 60S subunit 141, 142, 147. On the large subunit, Ltn1 adopts an elongated structure with the C-terminal RING domain, as expected, positioned near the exit tunnel of the ribosome 147. The extended conformation allows the N-terminal domain to interact with the sarcin-ricin loop (SRL); a conserved region on the ribosome where translation factors bind. Rqc2, in addition to its interaction with Ltn1 near the SRL, occupies the 40S-binding site explaining its function in preventing association of 40S and 60S subunits. The specificity of the factor for a rescued 60S subunit versus a free one is reconciled by the observation that Rqc2 makes intimate contacts with the peptidyl tRNA in the P site, which is the product of NGD-mediated ribosome rescue; in contrast free 60S subunits have an unoccupied P site and as a result are discriminated against. CryoEM reconstructions of the mammalian complex were in broad agreement with the yeast models highlighting a similar role for NEMF in facilitating the recognition process and recruiting Listerin to the complex 146.
C-terminal tagging of nascent peptides
Perhaps one of the most interesting discoveries to be revealed by the yeast cryoEM map is the observation of an A-site tRNA with its CCA end positioned in the peptidyl transferase center suggesting that it is likely to have participated in peptidyl transfer 147. In the absence of a 40S subunit, an A-site tRNA rapidly dissociates; however on the RQC complex, the tRNA is stabilized through interactions with Rqc2. Sequencing analysis of the RQC-associated tRNAs in the presence or absence of Rqc2 suggested that the factor might be responsible for specifically recruiting tRNAAlaAGC and tRNAThrAGT to the complex. Structural data suggests that specificity for these tRNA by Rqc2 lies in a common UUIGY motif in the anticodon loop of tRNAAlaAGC and tRNAThrAGT. The implications of these findings were clarified through the analysis of stabilized truncated NGD protein products (i.e. in the absence of Ltn1), which were of significantly higher molecular weights in the presence of Rqc2 relative to in its absence. Careful analysis of these products revealed that the C-terminus of these higher molecular weight products was extended through the enriched addition of alanine and threonine in a nonspecific sequence. These observations suggested that Rqc2 might be responsible for mRNA-template-independent addition of alanine and threonine and hence was termed “carboxy-terminal Ala and Thr extensions” (CAT tails). The exact mechanistic details of CAT addition are yet to be understood, but at least a potential role for the process has been identified. Deletion of LTN1 has been previously shown to induce a heat-shock response that is dependent on the presence of Rqc2 141. By constructing mutants that can still support clearance of the nascent peptide but not CAT extensions, the same group showed that activation of Hsf1 is dependent on the ability of Rqc2 to add nontemplated alanine and threonine amino acids 147. How CAT tails induce a heat-shock response is unclear at the moment. However, two recent studies from the Hartl and Joazeiro groups have argued that CAT extensions result in the formation of aggregated protein products, which might serve as a signal for the heat-shock response 148, 149. In addition, the CAT tails are likely required to extend nascent polypeptides, which do not harbor an appropriate lysine near the exit tunnel so that LTN1-mediated ubiquitination can proceed. Furthermore it is quite possible that the CAT tails may also fulfill some other function; for example it has been hypothesized that these extensions may be used to examine the functional integrity of the large subunit in case stalling occurred due to a nonfunctional ribosome and not due to a defective mRNA. Finally, it is intriguing that this system bears similarity to the bacterial tmRNA rescue system suggesting that C-terminal tagging may have been an ancient process to track incomplete proteins.
Conclusions
Despite many recent breakthroughs in the field of ribosome-based quality control, several outstanding questions remain. One of the major open questions related to mRNA surveillance pathways is how are the ribosome and associated factors able to differentiate between translational pausing and stalling? Particularly, how slow is slow enough to trigger NGD? The ability to distinguish between the two events is crucial for initiating mRNA and peptide degradation pathways, as well as ribosomal disassembly and recycling, but only under the right circumstances. For instance, programmed pausing is a common strategy utilized by the cell to ensure proteins are properly targeted and modified; these are not typically recognized as NGD targets. The recognition of stalled ribosomes by Dom34 is potentially determined by the kinetics of its binding in the A-site when mRNA translocation is slowed, however this has yet to be confirmed. A hallmark of NGD and NSD is an endonucleolytic cleavage upstream of the ribosome; whether this cleavage is required for recruiting Dom34 is unclear. To that end, the identity of the endonuclease and how it is recruited to the mRNA are currently unknown. Additionally, the respective molecular roles of Ski7 and its paralog Hbs1 in NSD and NGD may be overlapping, but this remains to be elucidated.
Even though NMD has been studied for decades, a unifying mechanism, which allows for the identification of every premature stop codon, remains to be elucidated. The molecular details and order of signaling events between factors at the ribosome and downstream elements need to be clarified. In addition to its role in quality control, NMD has been recently recognized to be involved in the regulation of gene expression. Its role in gene expression appears to be spatially and temporally regulated, which begs the question of how the specificity of NMD is regulated. In particular, why are certain mRNAs NMD targets in certain tissues under certain conditions, while others are in different tissues under different conditions? What about NGD, can it also be co-opted to regulate gene expression?
We have only begun to learn about the RQC process and these are exciting times as more and more details emerge. Like all quality control processes discussed here, reporter constructs have been instrumental in providing critical insights into the molecular mechanics of the process. With that said, the real cellular targets of RQC remains to be identified. Out of the 10–30% of newly synthesized proteins that are targeted for ubiquitination, what fraction is RQC’s share? Genetics and structural studies identified the factors and how they interact with the ribosome, but the mechanics of the process remain to be clarified. For instance, during CAT extensions how is translocation accomplished? Given that the addition of aa-tRNAs appears to be stochastic in nature, how does it stop? And what catalyzes the hydrolysis of the peptidyl-tRNA?
These are difficult and complex questions to address and are more than likely to require a multidisciplinary approach to tackle them. Future studies are essential not only to provide additional key insights into the mechanism of these processes but also to advance our understanding of their role in cellular fitness in health and disease.
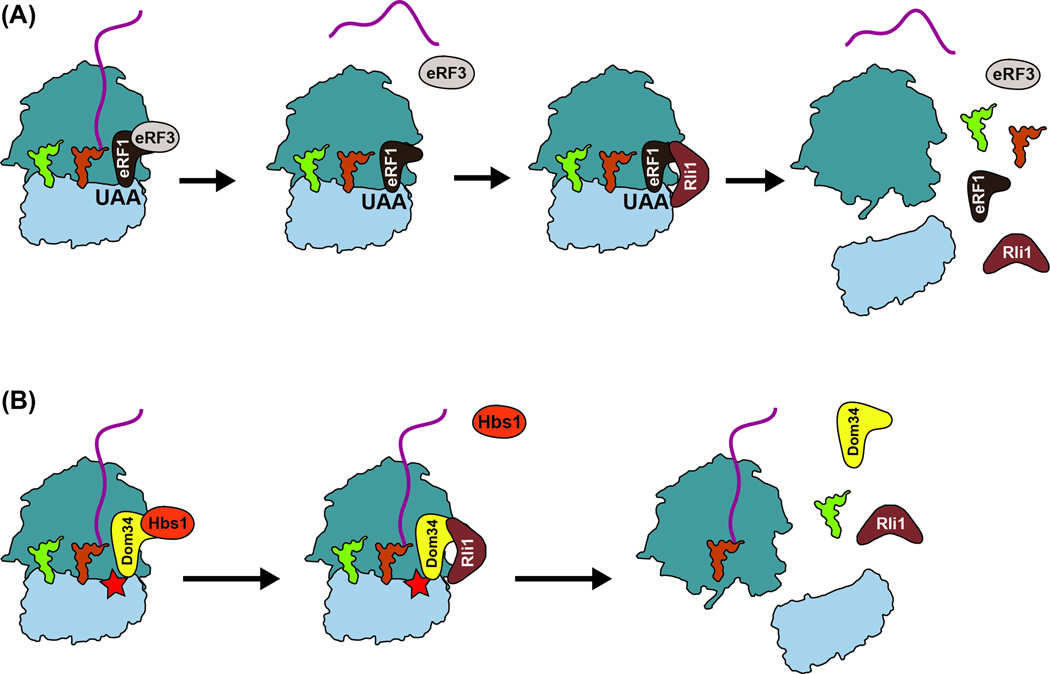
Figure 2. Ribosome recycling.
(A) During termination under normal conditions, eRF1/eRF3 release factors recognize a stop codon and bind the ribosome. GTP hydrolysis leads to conformational changes in eRF1, which mediates release of the peptide and recruitment of Rli1. Rli1 is required to promote ribosome splitting after hydrolysis of ATP. (B) During NGD, Dom34/Hbs1 recognize a stalled ribosome and bind the A site. Upon GTP hydrolysis, Hbs1 dissociates, allowing interaction with Rli1 and the subunits dissociate, but without release of the peptidyl tRNA.
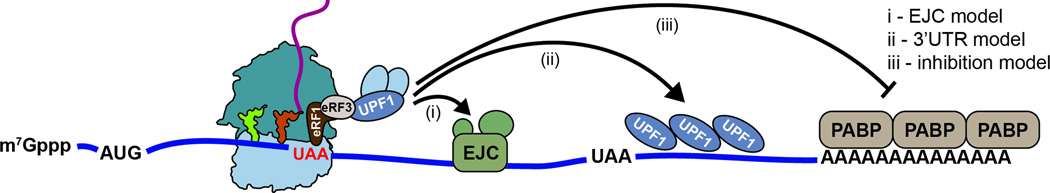
Figure 3. Models for nonsense-mediated decay.
mRNAs containing premature stop codons are recognized by the cell using several possible mechanisms. (i) the EJC model relies on interactions between an EJC located downstream of the premature stop codon and the Upf proteins that are bound to release factors on the ribosome. (ii) The 3’ UTR model suggests that Upf1 coats the UTR and the local concentration of the protein distinguishes NMD targets from other mRNAs. (iii) Interactions between eRF3 and polyA binding protein (PABP), essential during normal termination, are inhibited when the distance between the premature stop codon and polyA tail is large.
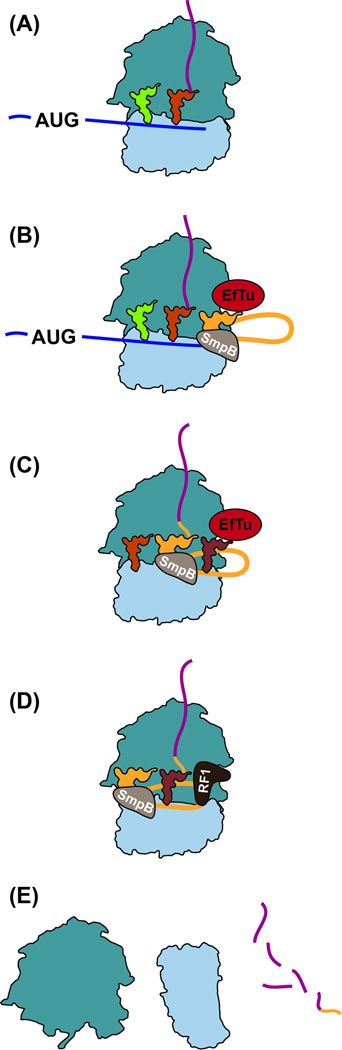
Figure 4. Ribosome rescue by trans-translation in bacteria.
(A) Ribosomes stall at the 3’ end of non-stop mRNAs or those containing rare codons. (B) A complex consisting of tmRNA, SmpB and EF-Tu, together with GTP, binds to the A site. (C) The nascent peptide is transferred to tmRNA and translation resumes on the ssrA ORF, tagging the defective protein at its 3’ end. The mRNA is released and degraded. (D) Termination occurs on the tmRNA stop codon using standard release factors. (E) Ribosomes dissociate and the tagged protein is degraded.
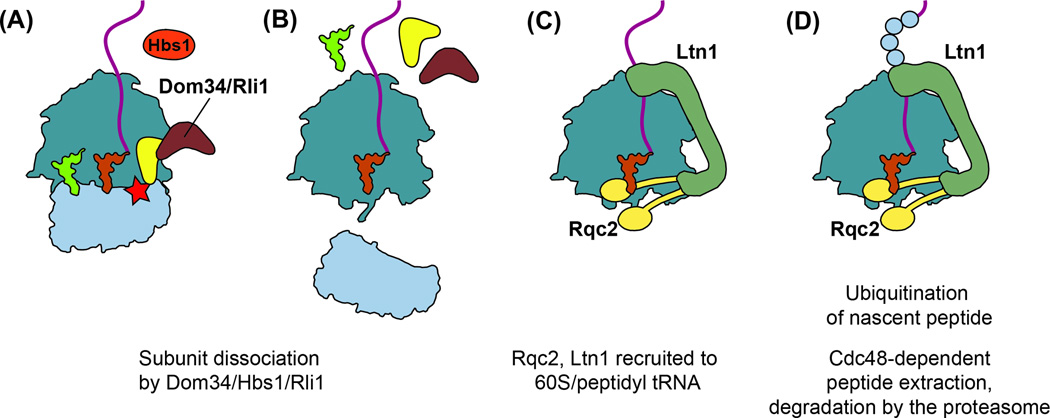
Figure 5. Co-translational protein quality control.
(A–B) Ribosomes stalled on defective mRNAs are released through the NGD pathway. (C) Rqc2 binds the 60S subunit, contacting the exposed P site tRNA and stabilizing Ltn1 binding. The C-terminal RING domain of Ltn1 contacts the exit tunnel while the N-terminus interacts with the sarcin-ricin loop on the ribosome. (D) Ltn1 ubiquitinates the nascent peptide, targeting it for degradation by the proteasome. Extraction of the peptide from the ribosome depends on Cdc48.
Acknowledgments
Research in the Zaher laboratory is supported by the National Institutes of Health (R01GM112641) and the Searle Scholars Program. We thank the members of the laboratory for comments and helpful discussions on the manuscript.
References
- 1.Wolff S, Weissman JS, Dillin A. Differential Scales of Protein Quality Control. Cell. 2014;157:52–64. doi: 10.1016/j.cell.2014.03.007. [DOI] [PubMed] [Google Scholar]
- 2.Drummond DA, Wilke CO. Mistranslation-induced protein misfolding as a dominant constraint on coding-sequence evolution. Cell. 2008;134:341–352. doi: 10.1016/j.cell.2008.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Simms CL, Hudson BH, Mosior JW, Rangwala AS, Zaher HS. An active role for the ribosome in determining the fate of oxidized mRNA. Cell Rep. 2014;9:1256–1264. doi: 10.1016/j.celrep.2014.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wurtmann EJ, Wolin SL. RNA under attack: cellular handling of RNA damage. Crit Rev Biochem Mol Biol. 2009;44:34–49. doi: 10.1080/10409230802594043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ishigame H, Mosaheb MM, Sanjabi S, Flavell RA. Truncated Form of TGF-beta RII, But Not Its Absence, Induces Memory CD8(+) T Cell Expansion and Lymphoproliferative Disorder in Mice. Journal of Immunology. 2013;190:6340–6350. doi: 10.4049/jimmunol.1300397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roman C, Cohn L, Calame K. A DOMINANT NEGATIVE FORM OF TRANSCRIPTION ACTIVATOR MTFE3 CREATED BY DIFFERENTIAL SPLICING. Science. 1991;254:94–97. doi: 10.1126/science.1840705. [DOI] [PubMed] [Google Scholar]
- 7.van Hoof A, Wagner EJ. A brief survey of mRNA surveillance. Trends in Biochemical Sciences. 2011;36:585–592. doi: 10.1016/j.tibs.2011.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ishimura R, Nagy G, Dotu I, Zhou H, Yang X-L, Schimmel P, Senju S, Nishimura Y, Chuang JH, Ackerman SL. Ribosome stalling induced by mutation of a CNS-specific tRNA causes neurodegeneration. Science. 2014;345:455–459. doi: 10.1126/science.1249749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scheper GC, van der Knaap MS, Proud CG. Translation matters: protein synthesis defects in inherited disease. Nature Reviews Genetics. 2007;8:711–723. doi: 10.1038/nrg2142. [DOI] [PubMed] [Google Scholar]
- 10.Sydow JF, Cramer P. RNA polymerase fidelity and transcriptional proofreading. Curr Opin Struct Biol. 2009;19:732–739. doi: 10.1016/j.sbi.2009.10.009. [DOI] [PubMed] [Google Scholar]
- 11.Schwanhausser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, Selbach M. Global quantification of mammalian gene expression control. Nature. 2011;473:337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- 12.Li GW, Burkhardt D, Gross C, Weissman JS. Quantifying absolute protein synthesis rates reveals principles underlying allocation of cellular resources. Cell. 2014;157:624–635. doi: 10.1016/j.cell.2014.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lewis BP, Green RE, Brenner SE. Evidence for the widespread coupling of alternative splicing and nonsense-mediated mRNA decay in humans. Proc Natl Acad Sci U S A. 2003;100:189–192. doi: 10.1073/pnas.0136770100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kervestin S, Jacobson A. NMD: a multifaceted response to premature translational termination. Nat Rev Mol Cell Biol. 2012;13:700–712. doi: 10.1038/nrm3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frischmeyer PA, van Hoof A, O’Donnell K, Guerrerio AL, Parker R, Dietz HC. An mRNA surveillance mechanism that eliminates transcripts lacking termination codons. Science. 2002;295:2258–2261. doi: 10.1126/science.1067338. [DOI] [PubMed] [Google Scholar]
- 16.Graber JH, Cantor CR, Mohr SC, Smith TF. In silico detection of control signals: mRNA 3 ‘-end-processing sequences in diverse species. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:14055–14060. doi: 10.1073/pnas.96.24.14055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van Hoof A, Frischmeyer PA, Dietz HC, Parker R. Exosome-mediated recognition and degradation of mRNAs lacking a termination codon. Science. 2002;295:2262–2264. doi: 10.1126/science.1067272. [DOI] [PubMed] [Google Scholar]
- 18.Wilson MA, Meaux S, van Hoof A. Diverse aberrancies target yeast mRNAs to cytoplasmic mRNA surveillance pathways. Biochimica et biophysica acta. 2008;1779:550–557. doi: 10.1016/j.bbagrm.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Inada T, Aiba H. Translation of aberrant mRNAs lacking a termination codon or with a shortened 30-UTR is repressed after initiation in yeast. Embo Journal. 2005;24:1584–1595. doi: 10.1038/sj.emboj.7600636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meaux S, van Hoof A. Yeast transcripts cleaved by an internal ribozyme provide new insight into the role of the cap and poly(A) tail in translation and mRNA decay. Rna-a Publication of the Rna Society. 2006;12:1323–1337. doi: 10.1261/rna.46306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gandhi R, Manzoor M, Hudak KA. Depurination of Brome mosaic virus RNA3 in vivo results in translation-dependent accelerated degradation of the viral RNA. J Biol Chem. 2008;283:32218–32228. doi: 10.1074/jbc.M803785200. [DOI] [PubMed] [Google Scholar]
- 22.Doma MK, Parker R. Endonucleolytic cleavage of eukaryotic mRNAs with stalls in translation elongation. Nature. 2006;440:561–564. doi: 10.1038/nature04530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shoemaker CJ, Green R. Translation drives mRNA quality control. Nat Struct Mol Biol. 2012;19:594–601. doi: 10.1038/nsmb.2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang CH, Wolfgang WC, Withey J, Koomey M, Friedman DI. Charged tmRNA but not tmRNA-mediated proteolysis is essential for Neisseria gonorrhoeae viability. Embo Journal. 2000;19:1098–1107. doi: 10.1093/emboj/19.5.1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Withey J, Friedman D. Analysis of the role of trans-translation in the requirement of tmRNA for lambda imm(P22) growth in Escherichia coli. Journal of Bacteriology. 1999;181:2148–2157. doi: 10.1128/jb.181.7.2148-2157.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brandman O, Hegde RS. Ribosome-associated protein quality control. Nat Struct Mol Biol. 2016;23:7–15. doi: 10.1038/nsmb.3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bengtson MH, Joazeiro CAP. Role of a ribosome-associated E3 ubiquitin ligase in protein quality control. Nature. 2010;467:470–473. doi: 10.1038/nature09371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hinnebusch AG, Lorsch JR. The mechanism of eukaryotic translation initiation: new insights and challenges. Cold Spring Harb Perspect Biol. 2012:4. doi: 10.1101/cshperspect.a011544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rodnina MV, Wintermeyer W. The ribosome as a molecular machine: the mechanism of tRNA-mRNA movement in translocation. Biochem Soc Trans. 2011;39:658–662. doi: 10.1042/BST0390658. [DOI] [PubMed] [Google Scholar]
- 30.Voorhees RM, Ramakrishnan V. Structural basis of the translational elongation cycle. Annu Rev Biochem. 2013;82:203–236. doi: 10.1146/annurev-biochem-113009-092313. [DOI] [PubMed] [Google Scholar]
- 31.Letzring DP, Dean KM, Grayhack EJ. Control of translation efficiency in yeast by codon-anticodon interactions. RNA. 2010;16:2516–2528. doi: 10.1261/rna.2411710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Letzring DP, Wolf AS, Brule CE, Grayhack EJ. Translation of CGA codon repeats in yeast involves quality control components and ribosomal protein L1. Rna. 2013;19:1208–1217. doi: 10.1261/rna.039446.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tsuboi T, Kuroha K, Kudo K, Makino S, Inoue E, Kashima I, Inada T. Dom34:hbs1 plays a general role in quality-control systems by dissociation of a stalled ribosome at the 3’ end of aberrant mRNA. Mol Cell. 2012;46:518–529. doi: 10.1016/j.molcel.2012.03.013. [DOI] [PubMed] [Google Scholar]
- 34.Dimitrova LN, Kuroha K, Tatematsu T, Inada T. Nascent Peptide-dependent Translation Arrest Leads to Not4p–mediated Protein Degradation by the Proteasome. Journal of Biological Chemistry. 2009;284:10343–10352. doi: 10.1074/jbc.M808840200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kuroha K, Akamatsu M, Dimitrova L, Ito T, Kato Y, Shirahige K, Inada T. Receptor for activated C kinase 1 stimulates nascent polypeptide-dependent translation arrest. Embo Reports. 2010;11:956–961. doi: 10.1038/embor.2010.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dever TE, Green R. The Elongation, Termination, and Recycling Phases of Translation in Eukaryotes. Cold Spring Harbor Perspectives in Biology. 2012:4. doi: 10.1101/cshperspect.a013706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shoemaker CJ, Green R. Kinetic analysis reveals the ordered coupling of translation termination and ribosome recycling in yeast. Proc Natl Acad Sci U S A. 2011;108:E1392–E1398. doi: 10.1073/pnas.1113956108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Youngman EM, McDonald ME, Green R. Peptide release on the ribosome: mechanism and implications for translational control. Annu Rev Microbiol. 2008;62:353–373. doi: 10.1146/annurev.micro.61.080706.093323. [DOI] [PubMed] [Google Scholar]
- 39.Frolova L, Le Goff X, Rasmussen HH, Cheperegin S, Drugeon G, Kress M, Arman I, Haenni AL, Celis JE, Philippe M, et al. A highly conserved eukaryotic protein family possessing properties of polypeptide chain release factor. Nature. 1994;372:701–703. doi: 10.1038/372701a0. [DOI] [PubMed] [Google Scholar]
- 40.Alkalaeva EZ, Pisarev AV, Frolova LY, Kisselev LL, Pestova TV. In vitro reconstitution of eukaryotic translation reveals cooperativity between release factors eRF1 and eRF3. Cell. 2006;125:1125–1136. doi: 10.1016/j.cell.2006.04.035. [DOI] [PubMed] [Google Scholar]
- 41.Becker T, Franckenberg S, Wickles S, Shoemaker CJ, Anger AM, Armache J-P, Sieber H, Ungewickell C, Berninghausen O, Daberkow I, et al. Structural basis of highly conserved ribosome recycling in eukaryotes and archaea. Nature. 2012;482:501–U221. doi: 10.1038/nature10829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Eyler DE, Wehner KA, Green R. Eukaryotic release factor 3 is required for multiple turnovers of peptide release catalysis by eukaryotic release factor 1. J Biol Chem. 2013;288:29530–29538. doi: 10.1074/jbc.M113.487090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brown A, Shao S, Murray J, Hegde RS, Ramakrishnan V. Structural basis for stop codon recognition in eukaryotes. Nature. 2015;524:493. doi: 10.1038/nature14896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pisarev AV, Skabkin MA, Pisareva VP, Skabkina OV, Rakotondrafara AM, Hentze MW, Hellen CUT, Pestova TV. The Role of ABCE1 in Eukaryotic Posttermination Ribosomal Recycling. Molecular Cell. 2010;37:196–210. doi: 10.1016/j.molcel.2009.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pisareva VP, Skabkin MA, Hellen CU, Pestova TV, Pisarev AV. Dissociation by Pelota, Hbs1 and ABCE1 of mammalian vacant 80S ribosomes and stalled elongation complexes. EMBO J. 2011;30:1804–1817. doi: 10.1038/emboj.2011.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shoemaker CJ, Eyler DE, Green R. Dom34:Hbs1 promotes subunit dissociation and peptidyl-tRNA drop-off to initiate no-go decay. Science. 2010;330:369–372. doi: 10.1126/science.1192430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Barthelme D, Dinkelaker S, Albers S-V, Londei P, Ermler U, Tampe R. Ribosome recycling depends on a mechanistic link between the FeS cluster domain and a conformational switch of the twin-ATPase ABCE1. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:3228–3233. doi: 10.1073/pnas.1015953108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Klauer AA, van Hoof A. Degradation of mRNAs that lack a stop codon: a decade of nonstop progress. Wiley Interdiscip Rev RNA. 2012;3:649–660. doi: 10.1002/wrna.1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schaeffer D, van Hoof A. Different nuclease requirements for exosome-mediated degradation of normal and nonstop mRNAs. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:2366–2371. doi: 10.1073/pnas.1013180108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Akimitsu N, Tanaka J, Pelletier J. Translation of nonSTOP mRNA is repressed post-initiation in mammalian cells. Embo Journal. 2007;26:2327–2338. doi: 10.1038/sj.emboj.7601679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Araki Y, Takahashi S, Kobayashi T, Kajiho H, Hoshino S, Katada T. Ski7p G protein interacts with the exosome and the Ski complex for 3 ‘-to-5 ‘ mRNA decay in yeast. Embo Journal. 2001;20:4684–4693. doi: 10.1093/emboj/20.17.4684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Benard L, Carroll K, Valle RCP, Masison DC, Wickner RB. The Ski7 antiviral protein is an EF1-alpha homolog that blocks expression of non-poly(A) mRNA in Saccharomyces cerevisiae. Journal of Virology. 1999;73:2893–2900. doi: 10.1128/jvi.73.4.2893-2900.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Atkinson GC, Baldauf SL, Hauryliuk V. Evolution of nonstop, no-go and nonsense-mediated mRNA decay and their termination factor-derived components. Bmc Evolutionary Biology. 2008:8. doi: 10.1186/1471-2148-8-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.van Hoof A. Conserved functions of yeast genes support the duplication, degeneration and complementation model for gene duplication. Genetics. 2005;171:1455–1461. doi: 10.1534/genetics.105.044057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ito-Harashima S, Kuroha K, Tatematsu T, Inada T. Translation of the poly(A) tail plays crucial roles in nonstop mRNA surveillance via translation repression and protein destabilization by proteasome in yeast. Genes & Development. 2007;21:519–524. doi: 10.1101/gad.1490207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lu J, Deutsch C. Electrostatics in the Ribosomal Tunnel Modulate Chain Elongation Rates. Journal of Molecular Biology. 2008;384:73–86. doi: 10.1016/j.jmb.2008.08.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Arthur L, Pavlovic-Djuranovic S, Smith-Koutmou K, Green R, Szczesny P, Djuranovic S. Translational control by lysine-encoding A-rich sequences. Sci Adv. 2015:1. doi: 10.1126/sciadv.1500154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Koutmou KS, Schuller AP, Brunelle JL, Radhakrishnan A, Djuranovic S, Green R. Ribosomes slide on lysine-encoding homopolymeric A stretches. Elife. 2015:4. doi: 10.7554/eLife.05534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fabre A, Charroux B, Martinez-Vinson C, Roquelaure B, Odul E, Sayar E, Smith H, Colomb V, Andre N, Hugot J-P, et al. SKIV2L Mutations Cause Syndromic Diarrhea, or Trichohepatoenteric Syndrome. American Journal of Human Genetics. 2012;90:689–692. doi: 10.1016/j.ajhg.2012.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Frolova LY, Tsivkovskii RY, Sivolobova GF, Oparina NY, Serpinsky OI, Blinov VM, Tatkov SI, Kisselev LL. Mutations in the highly conserved GGQ motif of class 1 polypeptide release factors abolish ability of human eRF1 to trigger peptidyl-tRNA hydrolysis. Rna. 1999;5:1014–1020. doi: 10.1017/s135583829999043x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Laurberg M, Asahara H, Korostelev A, Zhu J, Trakhanov S, Noller HF. Structural basis for translation termination on the 70S ribosome. Nature. 2008;454:852–857. doi: 10.1038/nature07115. [DOI] [PubMed] [Google Scholar]
- 62.Weixlbaumer A, Petry S, Dunham CM, Selmer M, Kelley AC, Ramakrishnan V. Crystal structure of the ribosome recycling factor bound to the ribosome. Nat Struct Mol Biol. 2007;14:733–737. doi: 10.1038/nsmb1282. [DOI] [PubMed] [Google Scholar]
- 63.Chavatte L, Seit-Nebi A, Dubovaya V, Favre A. The invariant uridine of stop codons contacts the conserved NIKSR loop of human eRF1 in the ribosome. Embo J. 2002;21:5302–5311. doi: 10.1093/emboj/cdf484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pisarev AV, Skabkin MA, Pisareva VP, Skabkina OV, Rakotondrafara AM, Hentze MW, Hellen CU, Pestova TV. The role of ABCE1 in eukaryotic posttermination ribosomal recycling. Mol Cell. 2010;37:196–210. doi: 10.1016/j.molcel.2009.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Becker T, Armache JP, Jarasch A, Anger AM, Villa E, Sieber H, Motaal BA, Mielke T, Berninghausen O, Beckmann R. Structure of the no-go mRNA decay complex Dom34-Hbs1 bound to a stalled 80S ribosome. Nat Struct Mol Biol. 2011;18:715–720. doi: 10.1038/nsmb.2057. [DOI] [PubMed] [Google Scholar]
- 66.Moore SD, Sauer RT. The tmRNA system for translational surveillance and ribosome rescue. Annu Rev Biochem. 2007;76:101–124. doi: 10.1146/annurev.biochem.75.103004.142733. [DOI] [PubMed] [Google Scholar]
- 67.Ivanova N, Pavlov MY, Felden B, Ehrenberg M. Ribosome rescue by tmRNA requires truncated mRNAs. Journal of Molecular Biology. 2004;338:33–41. doi: 10.1016/j.jmb.2004.02.043. [DOI] [PubMed] [Google Scholar]
- 68.Stevens A. An exoribonuclease from Saccharomyces cerevisiae: effect of modifications of 5’ end groups on the hydrolysis of substrates to 5’ mononucleotides. Biochem Biophys Res Commun. 1978;81:656–661. doi: 10.1016/0006-291x(78)91586-3. [DOI] [PubMed] [Google Scholar]
- 69.Hsu CL, Stevens A. Yeast cells lacking 5’-->3’ exoribonuclease 1 contain mRNA species that are poly(A) deficient and partially lack the 5’ cap structure. Mol Cell Biol. 1993;13:4826–4835. doi: 10.1128/mcb.13.8.4826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Stevens A, Maupin MK. A 5’----3’ exoribonuclease of Saccharomyces cerevisiae: size and novel substrate specificity. Arch Biochem Biophys. 1987;252:339–347. doi: 10.1016/0003-9861(87)90040-3. [DOI] [PubMed] [Google Scholar]
- 71.Stevens A. Purification and characterization of a Saccharomyces cerevisiae exoribonuclease which yields 5’-mononucleotides by a 5’ leads to 3’ mode of hydrolysis. J Biol Chem. 1980;255:3080–3085. [PubMed] [Google Scholar]
- 72.Losson R, Lacroute F. INTERFERENCE OF NONSENSE MUTATIONS WITH EUKARYOTIC MESSENGER-RNA STABILITY. Proceedings of the National Academy of Sciences of the United States of America. 1979;76:5134–5137. doi: 10.1073/pnas.76.10.5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Maquat LE, Kinniburgh AJ, Rachmilewitz EA, Ross J. UNSTABLE BETA-GLOBIN MESSENGER-RNA IN MESSENGER RNA-DEFICIENT BETA-O THALASSEMIA. Cell. 1981;27:543–553. doi: 10.1016/0092-8674(81)90396-2. [DOI] [PubMed] [Google Scholar]
- 74.Cui Y, Hagan KW, Zhang SA, Peltz SW. IDENTIFICATION AND CHARACTERIZATION OF GENES THAT ARE REQUIRED FOR THE ACCELERATED DEGRADATION OF MESSENGER-RNAS CONTAINING A PREMATURE TRANSLATIONAL TERMINATION CODON. Genes & Development. 1995;9:423–436. doi: 10.1101/gad.9.4.423. [DOI] [PubMed] [Google Scholar]
- 75.Leeds P, Peltz SW, Jacobson A, Culbertson MR. The product of the yeast UPF1 gene is required for rapid turnover of mRNAs containing a premature translational termination codon. Genes Dev. 1991;5:2303–2314. doi: 10.1101/gad.5.12a.2303. [DOI] [PubMed] [Google Scholar]
- 76.Leeds P, Wood JM, Lee BS, Culbertson MR. Gene products that promote mRNA turnover in Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:2165–2177. doi: 10.1128/mcb.12.5.2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Nagy E, Maquat LE. A rule for termination-codon position within intron-containing genes: when nonsense affects RNA abundance. Trends Biochem Sci. 1998;23:198–199. doi: 10.1016/s0968-0004(98)01208-0. [DOI] [PubMed] [Google Scholar]
- 78.Thermann R, Neu-Yilik G, Deters A, Frede U, Wehr K, Hagemeier C, Hentze MW, Kulozik AE. Binary specification of nonsense codons by splicing and cytoplasmic translation. EMBO J. 1998;17:3484–3494. doi: 10.1093/emboj/17.12.3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhang J, Sun X, Qian Y, LaDuca JP, Maquat LE. At least one intron is required for the nonsense-mediated decay of triosephosphate isomerase mRNA: a possible link between nuclear splicing and cytoplasmic translation. Mol Cell Biol. 1998;18:5272–5283. doi: 10.1128/mcb.18.9.5272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Le Hir H, Izaurralde E, Maquat LE, Moore MJ. The spliceosome deposits multiple proteins 20–24 nucleotides upstream of mRNA exon-exon junctions. EMBO J. 2000;19:6860–6869. doi: 10.1093/emboj/19.24.6860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ishigaki Y, Li X, Serin G, Maquat LE. Evidence for a pioneer round of mRNA translation: mRNAs subject to nonsense-mediated decay in mammalian cells are bound by CBP80 and CBP20. Cell. 2001;106:607–617. doi: 10.1016/s0092-8674(01)00475-5. [DOI] [PubMed] [Google Scholar]
- 82.Chakrabarti S, Jayachandran U, Bonneau F, Fiorini F, Basquin C, Domcke S, Le Hir H, Conti E. Molecular Mechanisms for the RNA-Dependent ATPase Activity of Upf1 and Its Regulation by Upf2. Molecular Cell. 2011;41:693–703. doi: 10.1016/j.molcel.2011.02.010. [DOI] [PubMed] [Google Scholar]
- 83.Chamieh H, Ballut L, Bonneau F, Le Hir H. NMD factors UPF2 and UPF3 bridge UPF1 to the exon junction complex and stimulate its RNA helicase activity. Nature Structural & Molecular Biology. 2008;15:85–93. doi: 10.1038/nsmb1330. [DOI] [PubMed] [Google Scholar]
- 84.Grimson A, O’Connor S, Newman CL, Anderson P. SMG-1 is a phosphatidylinositol kinase-related protein kinase required for nonsense-mediated mRNA Decay in Caenorhabditis elegans. Mol Cell Biol. 2004;24:7483–7490. doi: 10.1128/MCB.24.17.7483-7490.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Okada-Katsuhata Y, Yamashita A, Kutsuzawa K, Izumi N, Hirahara F, Ohno S. N- and C-terminal Upf1 phosphorylations create binding platforms for SMG-6 and SMG-5:SMG-7 during NMD. Nucleic Acids Res. 2012;40:1251–1266. doi: 10.1093/nar/gkr791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yamashita A, Ohnishi T, Kashima I, Taya Y, Ohno S. Human SMG-1, a novel phosphatidylinositol 3-kinase-related protein kinase, associates with components of the mRNA surveillance complex and is involved in the regulation of nonsense-mediated mRNA decay. Genes Dev. 2001;15:2215–2228. doi: 10.1101/gad.913001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ivanov PV, Gehring NH, Kunz JB, Hentze MW, Kulozik AE. Interactions between UPF1, eRFs, PABP and the exon junction complex suggest an integrated model for mammalian NMD pathways. EMBO J. 2008;27:736–747. doi: 10.1038/emboj.2008.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kashima I, Yamashita A, Izumi N, Kataoka N, Morishita R, Hoshino S, Ohno M, Dreyfuss G, Ohno S. Binding of a novel SMG-1-Upf1-eRF1-eRF3 complex (SURF) to the exon junction complex triggers Upf1 phosphorylation and nonsense-mediated mRNA decay. Genes Dev. 2006;20:355–367. doi: 10.1101/gad.1389006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wang W, Czaplinski K, Rao Y, Peltz SW. The role of Upf proteins in modulating the translation read-through of nonsense-containing transcripts. EMBO J. 2001;20:880–890. doi: 10.1093/emboj/20.4.880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kononenko AV, Mitkevich VA, Atkinson GC, Tenson T, Dubovaya VI, Frolova LY, Makarov AA, Hauryliuk V. GTP-dependent structural rearrangement of the eRF1:eRF3 complex and eRF3 sequence motifs essential for PABP binding. Nucleic Acids Res. 2010;38:548–558. doi: 10.1093/nar/gkp908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Muhlrad D, Parker R. Aberrant mRNAs with extended 3’ UTRs are substrates for rapid degradation by mRNA surveillance. RNA. 1999;5:1299–1307. doi: 10.1017/s1355838299990829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Singh G, Rebbapragada I, Lykke-Andersen J. A competition between stimulators and antagonists of Upf complex recruitment governs human nonsense-mediated mRNA decay. PLoS Biol. 2008;6:e111. doi: 10.1371/journal.pbio.0060111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Meaux S, van Hoof A, Baker KE. Nonsense-mediated mRNA decay in yeast does not require PAB1 or a Poly(A) tail. Molecular Cell. 2008;29:134–140. doi: 10.1016/j.molcel.2007.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hogg JR, Goff SP. Upf1 senses 3’UTR length to potentiate mRNA decay. Cell. 2010;143:379–389. doi: 10.1016/j.cell.2010.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Amrani N, Ganesan R, Kervestin S, Mangus DA, Ghosh S, Jacobson A. A faux 3 ‘-UTR promotes aberrant termination and triggers nonsense-mediated mRNA decay. Nature. 2004;432:112–118. doi: 10.1038/nature03060. [DOI] [PubMed] [Google Scholar]
- 96.Keiler KC, Waller PRH, Sauer RT. Role of a peptide tagging system in degradation of proteins synthesized from damaged messenger RNA. Science. 1996;271:990–993. doi: 10.1126/science.271.5251.990. [DOI] [PubMed] [Google Scholar]
- 97.Roche ED, Sauer RT. SsrA-mediated peptide tagging caused by rare codons and tRNA scarcity. Embo Journal. 1999;18:4579–4589. doi: 10.1093/emboj/18.16.4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lee SY, Bailey SC, Apirion D. SMALL STABLE RNAS FROM ESCHERICHIA-COLI - EVIDENCE FOR EXISTENCE OF NEW MOLECULES AND FOR A NEW RIBONUCLEOPROTEIN PARTICLE CONTAINING 6S–RNA. Journal of Bacteriology. 1978;133:1015–1023. doi: 10.1128/jb.133.2.1015-1023.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Tu GF, Reid GE, Zhang JG, Moritz RL, Simpson RJ. C-TERMINAL EXTENSION OF TRUNCATED RECOMBINANT PROTEINS IN ESCHERICHIA-COLI WITH A 10SA RNA DECAPEPTIDE. Journal of Biological Chemistry. 1995;270:9322–9326. doi: 10.1074/jbc.270.16.9322. [DOI] [PubMed] [Google Scholar]
- 100.Keiler KC, Sauer RT. Sequence determinants of C-terminal substrate recognition by the Tsp protease. Journal of Biological Chemistry. 1996;271:2589–2593. doi: 10.1074/jbc.271.5.2589. [DOI] [PubMed] [Google Scholar]
- 101.Komine Y, Kitabatake M, Yokogawa T, Nishikawa K, Inokuchi H. A TRANSFER-RNA-LIKE STRUCTURE IS PRESENT IN 10SA RNA, A SMALL STABLE RNA FROM ESCHERICHIA-COLI. Proceedings of the National Academy of Sciences of the United States of America. 1994;91:9223–9227. doi: 10.1073/pnas.91.20.9223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Valle M, Gillet R, Kaur S, Henne A, Ramakrishnan V, Frank J. Visualizing tmRNA entry into a stalled ribosome. Science. 2003;300:127–130. doi: 10.1126/science.1081798. [DOI] [PubMed] [Google Scholar]
- 103.Hallier M, Ivanova N, Rametti A, Pavlov M, Ehrenberg MN, Felden B. Pre-binding of small protein B to a stalled ribosome triggers trans-translation. Journal of Biological Chemistry. 2004;279:25978–25985. doi: 10.1074/jbc.M314086200. [DOI] [PubMed] [Google Scholar]
- 104.Gottesman S, Roche E, Zhou YN, Sauer RT. The ClpXP and ClpAP proteases degrade proteins with carboxy-terminal peptide tails added by the SsrA-tagging system. Genes & Development. 1998;12:1338–1347. doi: 10.1101/gad.12.9.1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Rudinger-Thirion J, Giege R, Felden B. Aminoacylated tmRNA from Escherichia coli interacts with prokaryotic elongation factor Tu. Rna-a Publication of the Rna Society. 1999;5:989–992. doi: 10.1017/s135583829999101x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Barends S, Karzai AW, Sauer RT, Wower J, Kraal B. Simultaneous and functional binding of SmpB and EF-Tu center dot GTP to the alanyl acceptor arm of tmRNA. Journal of Molecular Biology. 2001;314:9–21. doi: 10.1006/jmbi.2001.5114. [DOI] [PubMed] [Google Scholar]
- 107.Karzai AW, Susskind MM, Sauer RT. SmpB, a unique RNA-binding protein essential for the peptide-tagging activity of SsrA (tmRNA) Embo Journal. 1999;18:3793–3799. doi: 10.1093/emboj/18.13.3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Zaher HS, Green R. Fidelity at the molecular level: lessons from protein synthesis. Cell. 2009;136:746–762. doi: 10.1016/j.cell.2009.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Gutmann S, Haebel PW, Metzinger L, Sutter M, Felden B, Ban N. Crystal structure of the transfer-RNA domain of transfer-messenger RNA in complex with SmpB. Nature. 2003;424:699–703. doi: 10.1038/nature01831. [DOI] [PubMed] [Google Scholar]
- 110.Hayes CS, Sauer RT. Cleavage of the A site mRNA codon during ribosome pausing provides a mechanism for translational quality control. Molecular Cell. 2003;12:903–911. doi: 10.1016/s1097-2765(03)00385-x. [DOI] [PubMed] [Google Scholar]
- 111.Sunohara T, Jojima K, Yamamoto Y, Inada T, Aiba H. Nascent-peptide-mediated ribosome stalling at a stop codon induces mRNA cleavage resulting in nonstop mRNA that is recognized by tmRNA. Rna-a Publication of the Rna Society. 2004;10:378–386. doi: 10.1261/rna.5169404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Garza-Sanchez F, Gin JG, Hayes CS. Amino acid starvation and colicin D treatment induce A-site mRNA cleavage in Escherichia coli. Journal of Molecular Biology. 2008;378:505–519. doi: 10.1016/j.jmb.2008.02.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Garza-Sanchez F, Shoji S, Fredrick K, Hayes CS. RNase II is important for A-site mRNA cleavage during ribosome pausing. Molecular Microbiology. 2009;73:882–897. doi: 10.1111/j.1365-2958.2009.06813.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Li X, Hirano R, Tagami H, Aiba H. Protein tagging at rare codons is caused by tmRNA action at the 3 ‘ end of nonstop mRNA generated in response to ribosorne stalling. Rna-a Publication of the Rna Society. 2006;12:248–255. doi: 10.1261/rna.2212606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Li X, Yagi M, Morita T, Aiba H. Cleavage of mRNAs and role of tmRNA system under amino acid starvation in Escherichia coli. Molecular Microbiology. 2008;68:462–473. doi: 10.1111/j.1365-2958.2008.06167.x. [DOI] [PubMed] [Google Scholar]
- 116.Ivanova N, Pavlov MY, Ehrenberg M. tmRNA-induced release of messenger RNA from stalled ribosomes. Journal of Molecular Biology. 2005;350:897–905. doi: 10.1016/j.jmb.2005.05.033. [DOI] [PubMed] [Google Scholar]
- 117.Neubauer C, Gillet R, Kelley AC, Ramakrishnan V. Decoding in the Absence of a Codon by tmRNA and SmpB in the Ribosome. Science. 2012;335:1366–1369. doi: 10.1126/science.1217039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Bessho Y, Shibata R, Sekine S-i, Murayama K, Higashijima K, Hori-Takemoto C, Shirouzu M, Kuramitsu S, Yokoyama S. Structural basis for functional mimicry of long-variable-arm tRNA by transfer-messenger RNA. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:8293–8298. doi: 10.1073/pnas.0700402104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Kaur S, Gillet R, Li W, Gursky R, Frank J. Cryo-EM visualization of transfer messenger RNA with two SmpBs in a stalled ribosome. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:16484–16489. doi: 10.1073/pnas.0607438103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kurita D, Sasaki R, Muto A, Himeno H. Interaction of SmpB with ribosome from directed hydroxyl radical probing. Nucleic Acids Research. 2007;35:7248–7255. doi: 10.1093/nar/gkm677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Ogle JM, Brodersen DE, Clemons WM, Tarry MJ, Carter AP, Ramakrishnan V. Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science. 2001;292:897–902. doi: 10.1126/science.1060612. [DOI] [PubMed] [Google Scholar]
- 122.Ogle JM, Murphy FV, Tarry MJ, Ramakrishnan V. Selection of tRNA by the ribosome requires a transition from an open to a closed form. Cell. 2002;111:721–732. doi: 10.1016/s0092-8674(02)01086-3. [DOI] [PubMed] [Google Scholar]
- 123.Cochella L, Brunelle JL, Green R. Mutational analysis reveals two independent molecular requirements during transfer RNA selection on the ribosome. Nat Struct Mol Biol. 2007;14:30–36. doi: 10.1038/nsmb1183. [DOI] [PubMed] [Google Scholar]
- 124.Miller MR, Liu Z, Cazier DJ, Gebhard GM, Herron SR, Zaher HS, Green R, Buskirk AR. The role of SmpB and the ribosomal decoding center in licensing tmRNA entry into stalled ribosomes. RNA. 2011;17:1727–1736. doi: 10.1261/rna.2821711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Dong G, Nowakowski J, Hoffman DW. Structure of small protein B: the protein component of the tmRNA-SmpB system for ribosome rescue. Embo Journal. 2002;21:1845–1854. doi: 10.1093/emboj/21.7.1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Someya T, Nameki N, Hosoi H, Suzuki S, Hatanaka H, Fujii M, Terada T, Shirouzu M, Inoue Y, Shibata T, et al. Solution structure of a tmRNA-binding protein, SmpB, from Thermus thermophilus. Febs Letters. 2003;535:94–100. doi: 10.1016/s0014-5793(02)03880-2. [DOI] [PubMed] [Google Scholar]
- 127.Kurita D, Muto A, Himeno H. Role of the C-terminal tail of SmpB in the early stage of trans-translation. Rna-a Publication of the Rna Society. 2010;16:980–990. doi: 10.1261/rna.1916610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Fu J, Hashem Y, Wower I, Lei J, Liao HY, Zwieb C, Wower J, Frank J. Visualizing the transfer-messenger RNA as the ribosome resumes translation. EMBO J. 2010;29:3819–3825. doi: 10.1038/emboj.2010.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Weis F, Bron P, Giudice E, Rolland J-P, Thomas D, Felden B, Gillet R. tmRNA-SmpB: a journey to the centre of the bacterial ribosome. Embo Journal. 2010;29:3810–3818. doi: 10.1038/emboj.2010.252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Amm I, Sommer T, Wolf DH. Protein quality control and elimination of protein waste: the role of the ubiquitin-proteasome system. Biochim Biophys Acta. 2014;1843:182–196. doi: 10.1016/j.bbamcr.2013.06.031. [DOI] [PubMed] [Google Scholar]
- 131.Sato S, Ward CL, Kopito RR. Cotranslational ubiquitination of cystic fibrosis transmembrane conductance regulator in vitro. Journal of Biological Chemistry. 1998;273:7189–7192. doi: 10.1074/jbc.273.13.7189. [DOI] [PubMed] [Google Scholar]
- 132.Zhou MY, Fisher EA, Ginsberg HN. Regulated co-translational ubiquitination of apolipoprotein - B100 a new paradigm for proteasomal, degradation of a secretory protein. Journal of Biological Chemistry. 1998;273:24649–24653. doi: 10.1074/jbc.273.38.24649. [DOI] [PubMed] [Google Scholar]
- 133.Wang F, Canadeo LA, Huibregtse JM. Ubiquitination of newly synthesized proteins at the ribosome. Biochimie. 2015;114:127–133. doi: 10.1016/j.biochi.2015.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Schubert U, Anton LC, Gibbs J, Norbury CC, Yewdell JW, Bennink JR. Rapid degradation of a large fraction of newly synthesized proteins by proteasomes. Nature. 2000;404:770–774. doi: 10.1038/35008096. [DOI] [PubMed] [Google Scholar]
- 135.Vabulas RM, Hartl FU. Protein synthesis upon acute nutrient restriction relies on proteasome function. Science. 2005;310:1960–1963. doi: 10.1126/science.1121925. [DOI] [PubMed] [Google Scholar]
- 136.Duttler S, Pechmann S, Frydman J. Principles of Cotranslational Ubiquitination and Quality Control at the Ribosome. Molecular Cell. 2013;50:379–393. doi: 10.1016/j.molcel.2013.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Wang F, Durfee LA, Huibregtse JM. A Cotranslational Ubiquitination Pathway for Quality Control of Misfolded Proteins. Molecular Cell. 2013;50:368–378. doi: 10.1016/j.molcel.2013.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Wilson MA, Meaux S, van Hoof A. A genomic screen in yeast reveals novel aspects of nonstop mRNA metabolism. Genetics. 2007;177:773–784. doi: 10.1534/genetics.107.073205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Chu J, Hong NA, Masuda CA, Jenkins BV, Nelms KA, Goodnow CC, Glynne RJ, Wu H, Masliah E, Joazeiro CAP, et al. A mouse forward genetics screen identifies LISTERIN as an E3 ubiquitin ligase involved in neurodegeneration. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:2097–2103. doi: 10.1073/pnas.0812819106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Zenklusen D, Larson DR, Singer RH. Single-RNA counting reveals alternative modes of gene expression in yeast. Nat Struct Mol Biol. 2008;15:1263–1271. doi: 10.1038/nsmb.1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Brandman O, Stewart-Ornstein J, Wong D, Larson A, Williams CC, Li G-W, Zhou S, King D, Shen PS, Weibezahn J, et al. A Ribosome-Bound Quality Control Complex Triggers Degradation of Nascent Peptides and Signals Translation Stress. Cell. 2012;151:1042–1054. doi: 10.1016/j.cell.2012.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Defenouillere Q, Yao Y, Mouaikel J, Namane A, Galopier A, Decourty L, Doyen A, Malabat C, Saveanu C, Jacquier A, et al. Cdc48-associated complex bound to 60S particles is required for the clearance of aberrant translation products. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:5046–5051. doi: 10.1073/pnas.1221724110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Verma R, Oania RS, Kolawa NJ, Deshaies RJ. Cdc48/p97 promotes degradation of aberrant nascent polypeptides bound to the ribosome. Elife. 2013:2. doi: 10.7554/eLife.00308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Shao S, Hegde RS. Reconstitution of a Minimal Ribosome-Associated Ubiquitination Pathway with Purified Factors. Molecular Cell. 2014;55:880–890. doi: 10.1016/j.molcel.2014.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Shao S, von der Malsburg K, Hegde RS. Listerin-Dependent Nascent Protein Ubiquitination Relies on Ribosome Subunit Dissociation. Molecular Cell. 2013;50:637–648. doi: 10.1016/j.molcel.2013.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Shao S, Brown A, Santhanam B, Hegde RS. Structure and Assembly Pathway of the Ribosome Quality Control Complex. Molecular Cell. 2015;57:433–444. doi: 10.1016/j.molcel.2014.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Shen PS, Park J, Qin Y, Li X, Parsawar K, Larson MH, Cox J, Cheng Y, Lambowitz AM, Weissman JS, et al. Rqc2p and 60S ribosomal subunits mediate mRNA-independent elongation of nascent chains. Science. 2015;347:75–78. doi: 10.1126/science.1259724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Choe YJ, Park SH, Hassemer T, Korner R, Vincenz-Donnelly L, Hayer-Hartl M, Hartl FU. Failure of RQC machinery causes protein aggregation and proteotoxic stress. Nature. 2016;531:191–195. doi: 10.1038/nature16973. [DOI] [PubMed] [Google Scholar]
- 149.Yonashiro R, Tahara EB, Bengtson MH, Khokhrina M, Lorenz H, Chen KC, Kigoshi-Tansho Y, Savas JN, Yates JR, Kay SA, et al. The Rqc2/Tae2 subunit of the ribosome-associated quality control (RQC) complex marks ribosome-stalled nascent polypeptide chains for aggregation. Elife. 2016:5. doi: 10.7554/eLife.11794. [DOI] [PMC free article] [PubMed] [Google Scholar]