Abstract
During early vertebrate embryogenesis, cell fate specification is often coupled with cell acquisition of specific adhesive, polar and/or motile behaviors. In Xenopus gastrulae, tissues fated to form different axial structures display distinct motility. The cells in the early organizer move collectively and directionally toward the animal pole and contribute to anterior mesendoderm, whereas the dorsal and the ventral-posterior trunk tissues surrounding the blastopore of mid-gastrula embryos undergo convergent extension and convergent thickening movements, respectively. While factors regulating cell lineage specification have been described in some detail, the molecular machinery that controls cell motility is not understood in depth. To gain insight into the gene battery that regulates both cell fates and motility in particular embryonic tissues, we performed RNA sequencing (RNA-seq) to investigate differentially expressed genes in the early organizer, the dorsal and the ventral marginal zone of Xenopus gastrulae. We uncovered many known signaling and transcription factors that have been reported to play roles in embryonic patterning during gastrulation. We also identified many uncharacterized genes as well as genes that encoded extracellular matrix (ECM) proteins or potential regulators of actin cytoskeleton. Co-expression of a selected subset of the differentially expressed genes with activin in animal caps revealed that they had distinct ability to block activin-induced animal cap elongation. Most of these factors did not interfere with mesodermal induction by activin, but an ECM protein, EFEMP2, inhibited activin signaling and acted downstream of the activated type I receptor. By focusing on a secreted protein kinase PKDCC1, we showed with overexpression and knockdown experiments that PKDCC1 regulated gastrulation movements as well as anterior neural patterning during early Xenopus development. Overall, our studies identify many differentially expressed signaling and cytoskeleton regulators in different embryonic regions of Xenopus gastrulae and imply their functions in regulating cell fates and/or behaviors during gastrulation.
Keywords: RNA-seq, organizer, dorsal and ventral marginal zone, convergent extension, PKDCC1
Introduction
Allocation of embryonic cells to distinct germ layers is one of the earliest events in vertebrate development. Cells in each germ layer also distinguish from each other according to their locations within the embryos, as cell positions influence their exposures to different maternal and zygotic signaling molecules and transcription factors. This patterning process endows cells not only distinct fates, but also different behaviors that are intimately linked to their fates. Hence, cells fated to become anterior mesoderm and endoderm migrate long distances to reach the head region, whereas cells that contribute to the trunk structures undertake polarized cell intercalation to alter the morphology of the tissues. Coordination of cell fate specification and cell movements in different embryonic regions is critical for proper vertebrate development.
In the frog Xenopus laevis, anterior mesendoderm is first manifested at the morphological level by the appearance of a small pigmented line in the vegetal region of early gastrula embryos. Cells surrounding this dorsal lip, the organizer, have three basic properties: they self-differentiate into the head mesoderm and the anterior endoderm; they migrate collectively as a sheet toward the animal, the future anterior, region; and they emit signaling molecules to induce adjacent tissues to adopt dorsal cell fates (Winklbauer, 1990; Winklbauer and Nagel, 1991; Winklbauer et al., 1996; Vodicka and Gerhart, 1995; Harland and Gerhard, 1997; De Robertis et al., 2001; Heasman, 2006). Cell trailing behind the organizer in involuting gastrula embryos do not spread efficiently on extracellular matrix (ECM) for migration. Instead, these cells actively modify cell-cell contact for directional cell intercalation, resulting in tissue convergence toward the midline and simultaneous extension along the anterior-posterior axis (convergent extension, or CE, cell movements), resulting in elongation of the trunk tissues (Shih and Keller, 1992a, 1992b; Smith and Howard, 1992; Symes et a., 1994; Vodicka and Gerhart, 1995; Keller and Shook, 2004, 2008). Cells located opposite to the organizer will contribute to the ventral and posterior structures. These cells also intercalate among themselves, but preferentially do so to produce multiple cell layers instead of following planar cell intercalation. This results in tissue thickening (convergent thickening, or CT, movements) at the tail end of the embryos (Wilson and Keller, 1991; Keller and Shook, 2008; Keller et al., 2008). Thus, gastrulating Xenopus embryos display region-specific cell behaviors corresponding to the distinct differentiation paths of these cells.
The molecular signatures of specific tissues in Xenopus gastrulae have been explored for almost three decades, and the functional relevance of tissue-specific molecules in embryonic patterning has also been scrutinized. These studies employ a variety of approaches, including differential gene expression analysis, functional expression library screening, individual candidate gene interrogation, and transcriptional target investigation for effectors of growth factor signals and nuclear proteins. Several prominent signaling pathways have emerged as crucial regulators of the formation and the inducing activity of the organizer (reviewed in Harland and Gerhard, 1997; Heasman, 2006). For example, maternal Wnt and zygotic nodal signals are shown to cooperate to induce the organizer, whereas the ventralizing and the caudalizing activities of bone morphogenetic proteins (BMPs) and zygotic Wnts are modulated by organizer-secreted BMP and Wnt antagonists. Fibroblast growth factors (FGFs) also participate in early embryonic induction and patterning. A plethora of transcription factors act downstream of these signals to control germ layer specification and dorsal-ventral patterning. Although these studies greatly advance our knowledge on cell fate determination, relatively limited molecules have been uncovered that can control cell movements without affecting cell fates. The most investigated signaling is the planar cell polarity (PCP) pathway, which regulates mediolateral cell intercalation during CE (reviewed in Wallingford et al., 2002; Roszko et al., 2009; Skoglund and Keller, 2010; Wallingford, 2012). Different tyrosine kinase signals have also been shown to affect gastrulation morphogenesis (Ataliotis et al., 1995; Conlon and Smith, 1999; Nutt et al., 2001; Nagel et al., 2004; Sivak et al., 2005; Nie and Chang, 2007a, 2007b; Damm and Winklbauer, 2011; Park et al., 2011; Evren et al., 2014). The PCP and the tyrosine kinase pathways may converge to control the activities of Rho family small GTPases to influence cell behaviors (Habas et al., 2001, 2003; Choi and Han, 2002; Pendo-Mendez et al., 2003; Tahinci and Symes, 2003; Ren et al., 2006). However, it is unclear whether other signals are involved, what common and divergent signal transducers are employed, and how factors involved in embryonic patterning may influence the expression and/or the function of the molecules involved in cell movements.
To fully understand molecular integration of cell fate and cell motility, it is imperative that we survey all the expressed genes in embryonic tissues with distinct fates and behaviors and investigate the function of the genes that show differential expression patterns. Several previous studies have been conducted to use non-biased approaches, such as differential library screening or microarray-based strategies, to address roles of differential gene functions in embryonic patterning (Sasai et al., 1994; Altmann et al., 2001; Munoz-Sanjuan et al., 2002; Smith and Harland, 1992; Wessely et al., 2004; Baldessari et al., 2005; Peiffer et al., 2005; Taverner et al., 2005; Hufton et al., 2006). Though productive, these studies tend to favor the discovery of genes with abundant expression due to technical limitations. Recent advances in genomic level analysis of gene expression promise to overcome the previous limitations and open the door for whole transcriptome study of each tissue without any bias. In this work, we employed the RNA sequencing (RNA-seq)-based strategy to analyze differentially expressed genes in the organizer, the dorsal trunk tissue, and the ventral-posterior region. We uncovered many known patterning molecules as well as scores of uncharacterized genes that might potentially regulate embryonic patterning and/or morphogenesis. Using activin-induced animal cap assay, we showed that several uncharacterized, differentially expressed genes had distinct ability to modulate elongation of the animal caps without affecting mesodermal cell fates. However, one ECM protein, EFEMP2, inhibited mesodermal induction by activin. Using both gain- and loss-of-function assays, we further demonstrated that the secreted protein kinase, PKDCC1, regulated gastrulation movements and anterior neural patterning during early Xenopus development.
Results
RNA-sequencing analysis of differentially expressed genes in the organizer, the dorsal trunk, and the ventral-posterior tissues
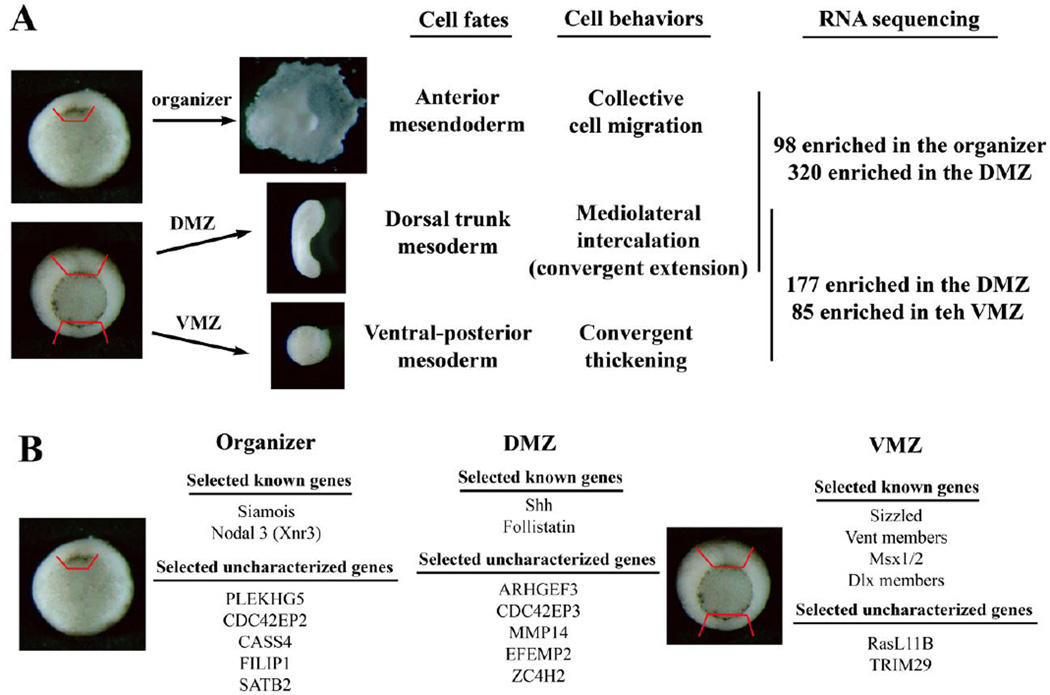
To understand how cells in different embryonic regions acquire distinct cell fates and migratory behaviors, we performed an RNA sequencing experiment using tissues dissected from three regions of gastrulating Xenopus embryos. The early organizer, which forms anterior mesendoderm and migrates collectively on the ECM, was dissected from stage 10 embryos as the tissue surrounding, and including, the dorsal lip. The dorsal and ventral marginal zones (DMZ and VMZ) that encompass the dorsal trunk and the ventral-posterior tissues that display CE or CT behaviors, respectively, were dissected from stages 11 to 11.5 embryos from the regions above the blastopore (Fig. 1). Total RNA was extracted and subjected to sequencing on Illumina HiSeq2000 platform. Two independent dissections and RNA-seq experiments were performed, and the results were analyzed based on Xenopus laevis genome version 7.1 (Suppl. Table 1). Subsequent analysis based on newly released genome version 9.1 validated the genes we identified (Suppl. Tables 1 and 2; also see Materials and Methods).
Figure 1. Schematic representation of the RNA-seq experiment.
A) The early organizer from stage 10+ embryos and the dorsal and the ventral marginal zone (DMZ and VMZ) explants from mid-gastrula stage embryos were dissected and subjected to RNA sequencing. Cells in these regions have distinct fates and motile behaviors. Pairwise comparison of differential expressed genes was performed between the organizer and the DMZ, and the DMZ and the VMZ, samples. Genes with potential to regulate embryonic patterning and/or movements were especially scrutinized in more detail in this study. B) A list of the selected known and uncharacterized genes with differential expression patterns in different embryonic regions is shown.
Pairwise comparison of the transcriptomes as well as ANOVA-like analysis for all three sample sets revealed differentially expressed genes in each region (Suppl. Table 1 and data not shown). These included the known organizer-specific genes, such as nodal-related 3 (Xnr3) and Siamois, the dorsal trunk genes, such as sonic hedgehog (Shh) and follistatin (Fst), and the ventral genes, such as Sizzled (Szl) and Vent transcription factors (Hemmati-Brivanlou et al., 1994; Ekker et al., 1995; Lemaire et al., 1995; Smith et al., 1995; Onichtchouk et al., 1996; Salic et al., 1997). In addition, hundreds of previously uncharacterized genes were uncovered from the study. A greater number of genes showed significant enrichment in the DMZ than either the organizer or the VMZ (320 and 98 genes were expressed at higher levels in the DMZ and the organizer, respectively; and 177 and 85 genes were enriched in the DMZ and VMZ, respectively). Among the genes with higher expression in the organizer, several seemed to encode proteins involved in the endodermal development, such as GATA4, Mix1, nodal-related 6, and KLF4 (Rosa, 1989; Lemaire et al., 1998; Takahashi et al., 2000; Weber et al., 2000; Cao et al., 2012). These genes might play a general role in germ layer specification rather than dorsal-ventral (DV) patterning, though several have also been implicated in cell spreading and migration of the leading edge mesendoderm (e.g. Mix1 and GATA4, Wacker et al., 1998; Fletcher et al., 2006). Other genes in this group included many transcription factors and signaling molecules, as well as a number of genes involved in metabolism, stress responses to DNA-damage or unfolded proteins in the ER, and ion and solute transporters (Suppl. Table 1). Interestingly, several genes encoded factors that might regulate Rho family signaling or actin organization. These included PLEKHG5, a Rho guanine nucleotide exchange factor (GEF) family member, CDC42EP2, an effector protein of Cdc42, CASS4, a member of the Cas scaffolding protein family that is often involved in integrin signaling, and FILIP1, a filamin A interacting protein. The preferential expression of these genes in the organizer implied that they might regulate anterior mesendoderm cell migration on the ECM. Of the genes with significant enrichment in the DMZ, 84 of them were expressed at higher levels in the DMZ than that in both the organizer and the VMZ, whereas the rest showed differential expression over only one tissue (Suppl. Table 1 and data not shown). There was a high representation of transcription factors and signaling molecules in this group. Additionally, there were many ECM proteins and ECM receptors, such as laminin β1, different collagens (e.g. type V α1, XI α2, and XIII α1), and integrins (e.g. integrin α 2 and α3). Regulators and effectors of Rho family GTPases were also present, such as ARHGEF3 and FGD5, both are RhoGEFs, and CDC42EP3. These ECM proteins and cytoskeleton regulators might participate in modulation of directional intercalation cell behaviors in the DMZ. The genes enriched in the VMZ comprised a large number of transcription factors that had been associated with ventral cell type development, including the Vent, Msx, Dlx, AP2, Grainyhead, GATA, and p63 family members. Signaling molecules and cytokeratin genes were also found in this group, but genes that might control dynamic cytoskeleton organization were conspicuously missing when compared with those expressed in the DMZ (Suppl. Table 1). Taken together, the data suggest that region-specific transcriptomes in Xenopus gastrulae contain specific members of ECM proteins and actin regulators in addition to signaling molecules and transcription factors. These ECM and actin regulatory genes may be co-modulated with patterning factors that determine cell lineages in each region, so that cells with particular fates can adopt appropriate migratory behaviors for correct morphogenesis.
Differential expression of region-specific genes during early Xenopus development
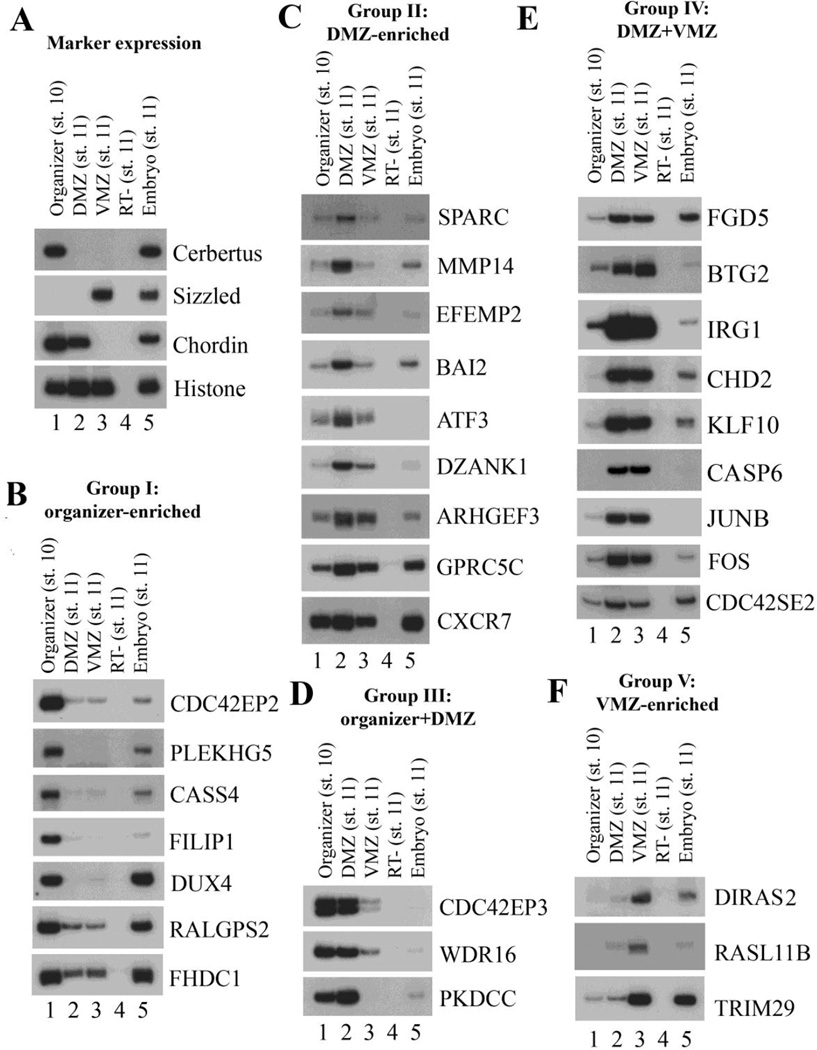
To confirm the differential expression patterns of the genes identified from our RNA-seq experiments, we dissected the organizer, the DMZ and the VMZ explants as above and performed RT-PCR to examine the expression of a subset of the genes. We focused our attention on potential regulators of embryonic patterning and movements and thus selected a group of ECM proteins, cytoskeleton regulators, signaling molecules and transcription factors. Assay for known markers expressed in the dissected regions attested the identity of these tissues (Fig. 2A). Examination of the other genes showed that they could be divided into five groups based on their RT-PCR patterns (Fig. 2 and Suppl. Fig. 1, summarized in Suppl. Table 3). Group I genes were expressed at the highest levels in the organizer and included the genes PLEKHG5, CDC42EP2, CASS4 and FILIP1. Group II genes showed higher expression in the DMZ than both the organizer and the VMZ, and included the ECM proteins SPARC, EFEMP2, the signaling molecules GPRC5C, PSKH2, the transcription factors ATF3, ZC4H2, and the RhoGEF ARHGEF3, among others. Group III genes were expressed at higher levels in both the organizer and the DMZ, whereas group IV genes showed high level expression in both the DMZ and the VMZ versus the organizer. Group V genes were enriched in the VMZ (Fig. 2, Suppl. Fig. 1, and Suppl. Table 3). In each group, some genes were probably abundantly expressed and showed strong signals, such as CDC42EP2 and GPRC5C, but some others showed low levels of expression and weak PCR signals, such as LPAR5 and PSKH2. The results indicate that RNA-seq not only uncovers abundant genes, but also identifies genes with weak, but nonetheless differential, expression profiles. The patterns of these genes in specific regions imply that they may play roles in tissue specification and/or morphogenesis in distinct embryonic regions.
Figure 2. Differential gene expression in the organizer, the DMZ, and the VMZ.
RT-PCR was performed to assay for gene expression in different embryonic regions. A) Marker analysis confirmed the expression of known region-specific genes in the dissected tissues. B) Group I genes were enriched in the organizer. C) Group II genes showed highest expression in the DMZ. D) Group III genes were expressed at high levels in both the organizer and the DMZ. E) Group IV genes were expressed at high levels in both the DMZ and the VMZ. F) Group V genes were enriched in the VMZ.
Distinct regulation of activin-induced animal cap elongation by differentially expressed genes
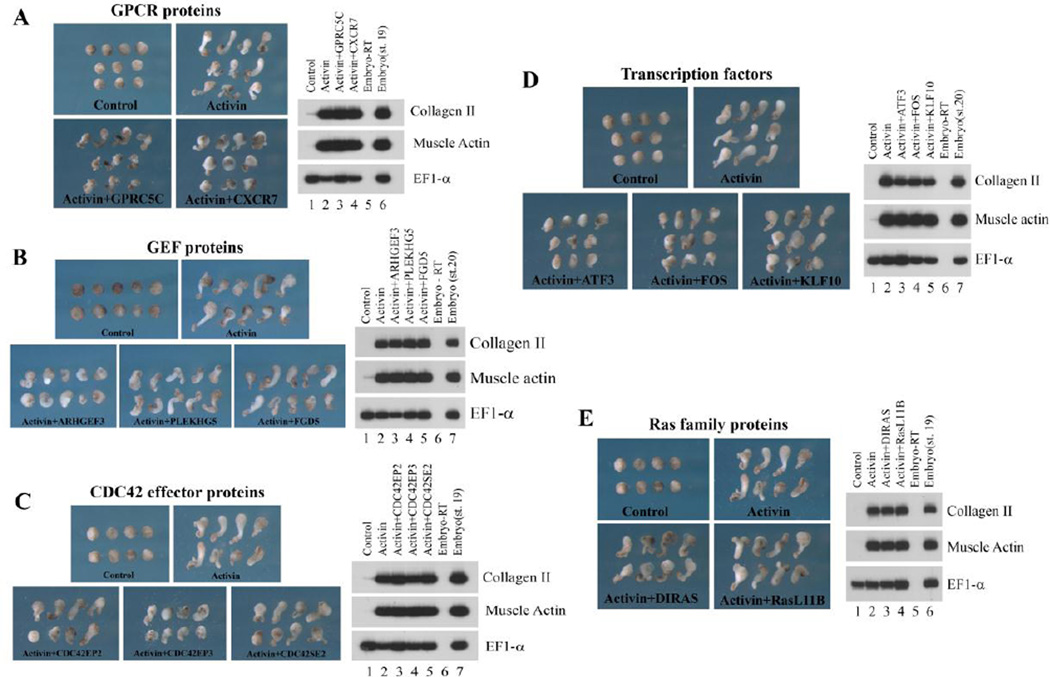
Molecules with the ability to pattern embryonic tissues during Xenopus gastrulation have been identified and described in some details for many genes, but factors that can modulate cell motility without affecting cell fates are relatively less studied. To uncover additional regulators of cell movements, we employed the animal cap assay system. Animal caps excised from the blastula ectoderm normally develop into atypical epidermis and assume a round morphology. However, when activin is included in the system, it induces dorsal mesoderm which undergoes CE movements, resulting in elongation of the animal caps (Chang, 2016). Factors that modulate CE movements can block activin-induced animal cap elongation when ectopically expressed. Using this assay system, we surveyed several molecules uncovered from our RNA-seq analysis for their ability to interfere with animal cap elongation. We chose to analyze two signaling molecules, GPRC5C and CXCR7, three RhoGEF members, ARHGEF3, PLEKHG5 and FGD5, three CDC42 effector proteins, CDC42EP2, CDC42EP3 and CDC42SE2, three transcription factors, ATF3, FOS and KLF10, and two ventrally-enriched Ras family proteins, DIRAS2 and RasL11B. Both GPRC5C and CXCR7 are seven transmembrane domain-containing G-protein coupled receptors (GPCRs) and showed higher expression in the DMZ. The regulators and the effectors of Rho family GTPases displayed differential expression in the organizer and the DMZ, whereas the three transcription factors showed higher expression in the DMZ over the organizer (Fig.2, Suppl. Fig. 1; also see in situ patterns of ARHGEF3 and CXCR7 in Hufton et al., 2006 and Mishra et al., 2013). We co-injected RNAs encoding these molecules with activin RNA into the animal region of two-cell stage embryos, dissected the animal caps at the blastula stages, cultured them to late neurula stages, observed the morphology of the caps, and examined the mesodermal marker expression in these caps (Fig. 3). In addition, we also used activin protein rather than activin RNA in these assays and obtained similar results (Suppl. Fig. 2).
Figure 3. Distinct activities of the differentially expressed genes to block activin-induced animal cap elongation.
RNAs encoding the RNA-seq clones and activin were co-injected into the animal region of 2-cell stage embryos. Animal caps were dissected at blastula stages and cultured to late neurula stages. A) Two GPRC proteins with the highest expression levels in the DMZ, GPRC5C and CXCR7, reduced activin-induced animal cap elongation without interfering with the mesodermal induction by activin. B) The DMZ-enriched GEF, ARHGEF3, efficiently blocked animal cap elongation; but the organizer-enriched GEF, PLEKHG5, or RhoGEF expressed in the DMZ and VMZ, FGD5, could not do so. C) CDC42EP3, which was expressed at high levels in both the organizer and the DMZ, blocked activin-induced animal cap elongation more efficiently than CDC42EP2, a gene enriched in the organizer. D) The transcription factors ATF3 and FOS both reduced the elongation of the animal caps, but KLF10 was ineffective in doing so. E) The ventrally-enriched Ras family proteins, DIRAS and RasL11B, did not block activin-induced animal cap elongation. The doses of RNA used: activin, 2pg; RNA-seq clones, 0.25-1ng.
As shown in Fig. 3, while none of these genes affected dorsal mesodermal induction by activin, they displayed different capacity in reducing animal cap elongation. Both the transmembrane GPCRs, GPRC5C and CXCR7, reduced the elongation of activin-induced animal caps, and GPRC5C also seemed to influence cell adhesion, as increased cell shedding was observed (Fig. 3A, Suppl. Fig. 2B, and data not shown). In contrast, among the three RhoGEF proteins, ARHGEF3 alone displayed high efficiency in blocking animal cap elongation (Fig. 3B and Suppl. Fig. 2D). Similarly, CDC42 effector proteins showed differential ability to interfere with activin-induced animal cap elongation, with CDC42EP3 as the most effective inhibitor. At higher doses, CDC42EP2 and CDC42SE2 also blocked animal cap elongation, and CDC42SE2 tended to cause dissociation of the animal caps, implying a role of this protein in regulation of cell adhesion (Fig. 3C, Suppl. Fig. 2D and data not shown). The transcription factors ATF3 and FOS also reduced animal cap elongation when ectopically expressed with activin, but KLF10 did not have much effect (Fig. 3D and Suppl. Fig. 2C). The two ventrally-enriched Ras family members could not block activin-induced animal cap elongation (Fig. 3E). Our results reveal that differentially expressed proteins belonging to the same gene family may have differential activities in modulating cell behaviors during gastrulation.
EFEMP2, an ECM protein, regulates activin signaling downstream of the activin receptors
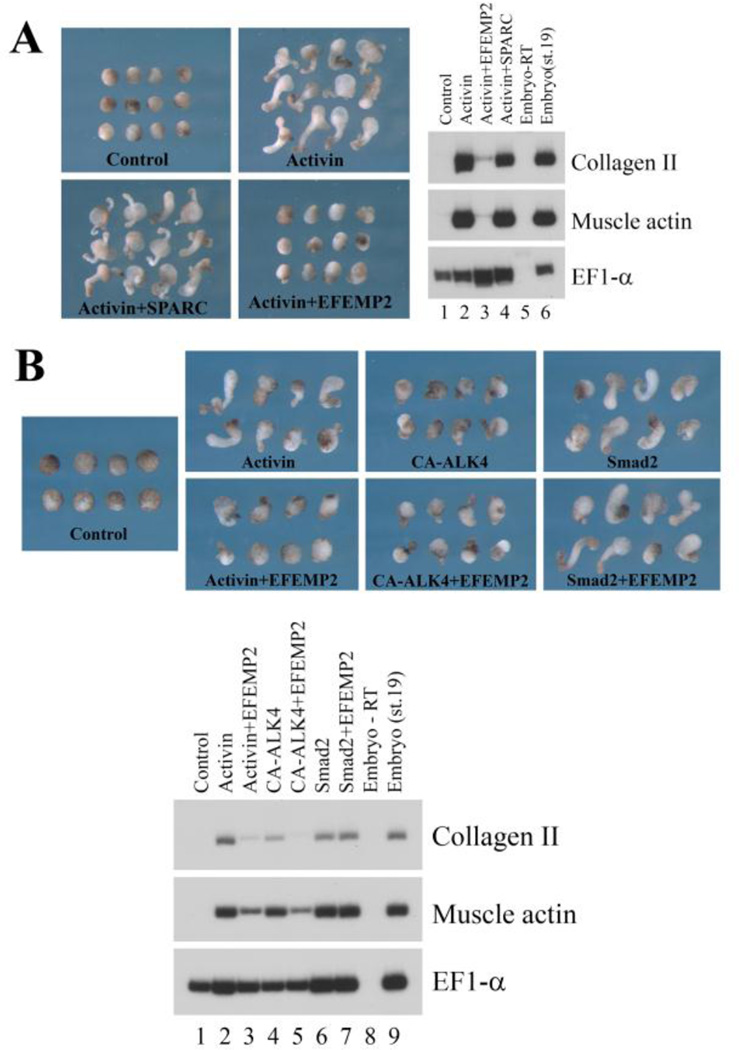
Several ECM genes were differentially expressed according to our RNA-seq analysis (Fig. 2, Suppl. Fig. 1, and Suppl. Tables 1 and 3). As ECM proteins often modulate cell adhesion and signaling to influence cell motility, we examined the effect of ectopic expression of two ECM genes, SPARC and EFEMP2, on activin-induced animal cap elongation. Co-expression of EFEMP2 with activin efficiently blocked animal cap elongation. However, unlike the other genes we assayed, EFEMP2 inhibited dorsal mesodermal induction by activin, suggesting that it regulated mesodermal cell fate to affect cell movements indirectly (Fig. 4A). To see whether EFEMP2 may block activin from interacting with its receptors to prevent downstream signaling, we co-expressed EFEMP2 with a constitutively active type I receptor CA-ALK4, which stimulated activin signaling independent of the activin ligand (Chang et al., 1997). Interestingly, EFEMP2 inhibited dorsal mesodermal induction by CA-ALK4 as well (Fig. 4B). However, co-expression of EFEMP2 with Smad2, the cytoplasmic signal transducer of activin/nodal signaling, did not interfere with mesodermal induction or animal cap elongation (Fig. 4B). The data suggest that EFEMP2 blocks activin signaling downstream of the activated receptors but upstream of Smad2. Examination of BMP-dependent ventral mesodermal induction in animal caps indicated that EFEMP2 did not inhibit BMP signaling (data not shown), demonstrating that EFEMP2 preferentially regulates the activin/nodal branch of the TGF-β signaling.
Figure 4. The ECM protein EFEMP2 inhibits activin signaling downstream of the activated receptor.
A) The ECM protein EFEMP2, but not SPARC, inhibited activin-induced mesodermal formation in animal caps. B) EFEMP2 interfered with mesodermal induction by both activin and the activated type I activin receptor ALK4 (CA-ALK4), but did not inhibit mesodermal induction by Smad2 or Smad2-induced animal cap elongation. The doses of RNA used: activin, 2pg; CA-ALK4, 1ng; Smad2, 1ng; EFEMP2, 1ng.
PKDCC1, a secreted protein kinase, regulates gastrulation and anterior neural patterning during early Xenopus development
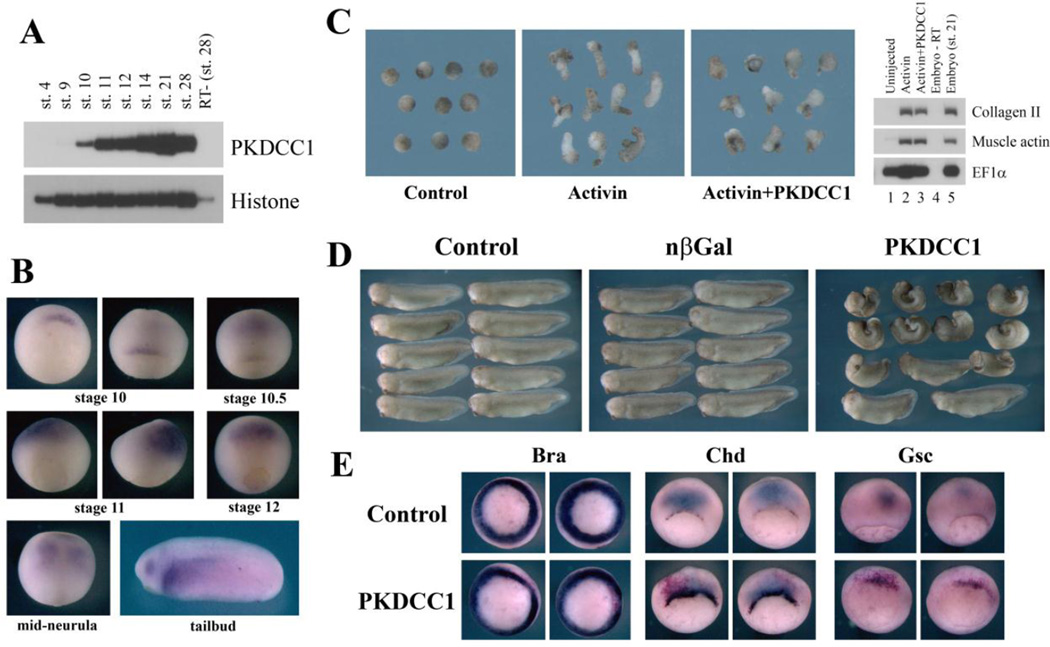
To confirm the function of differentially expressed genes in early Xenopus development, we need to perform loss-of-function studies. For this purpose, we focused our attention on one gene that we identified that had higher expression in the organizer and the DMZ over the VMZ (Fig. 2). The gene, PKDCC1 (also known as VLK, or vertebrate lonesome kinase), encoded a novel secreted protein kinase that likely exerted its function in the extracellular space (Bordoli et al., 2014). RT-PCR assay demonstrated that PKDCC1 was first expressed during early gastrulation (stage10), and its expression persisted until at least late tailbud stage (Fig. 5A). In situ hybridization revealed that PKDCC1 transcripts were localized around the organizer at stage 10, but were subsequently up-regulated in the anterior neural domain at the mid- to late gastrula stages. The anterior neural expression remained throughout the neurula stages. In tailbud embryos, PKDCC1 was detected in the eyes, the lateral plate mesoderm, and the heart primordium (Fig. 5B). Co-expression of RNAs encoding PKDCC1 and activin resulted in reduction of activin-induced animal cap elongation without defects in dorsal mesodermal cell fates (Fig. 5C). Overexpression of PKDCC1 RNA in early frog embryos led to severe gastrulation defects. The affected embryos showed shortened and bent body axis, reduced head, and split dorsal axis (Fig. 5D). These defects occurred in the absence of inhibition of the pan-mesodermal marker Brachyury (Bra, Smith et al., 1991) at the gastrula stages, though in situ hybridization indicated that the cells expressing chordin (Chd, Sasai et al., 1994) and goosecoid (Gsc, Cho et al., 1991) showed delayed involution inside the embryos (Fig. 5E). These results indicated that ectopic PKDCC1 likely interfered with gastrulation morphogenesis without affecting mesodermal cell fates.
Figure 5. PKDCC1, a dorsally enriched gene, induced gastrulation defects when ectopically expressed.
A) RT-PCR showed that PKDCC1 was first expressed during early gastrulation, and its expression persisted until at least tailbud stages. B) In situ hybridization showed that PKDCC1 was expressed in the organizer at the dorsal lip region in early gastrula embryos. Its expression was then up-regulated in the anterior neural tissues during mid-gastrula to neurula stages. At tailbud stages, PKDCC1 transcripts were seen in the eyes, the lateral plate mesoderm, and the heart primordium. C) PKDCC1 reduced activin-induced animal cap elongation without affecting mesodermal cell fates. D) Ectopic expression of PKDCC1 in early Xenopus embryos induced gastrulation defects, with the resulting embryos displaying reduced body axis, smaller head, and failure in neural tube closure. E) Overexpression of PKDCC1 did not inhibit mesodermal formation in early embryos, as indicated by normal expression of brachyury (Bra), but delayed involution and/or migration of dorsal mesodermal cells marked by chordin (Chd) and goosecoid (Gsc).
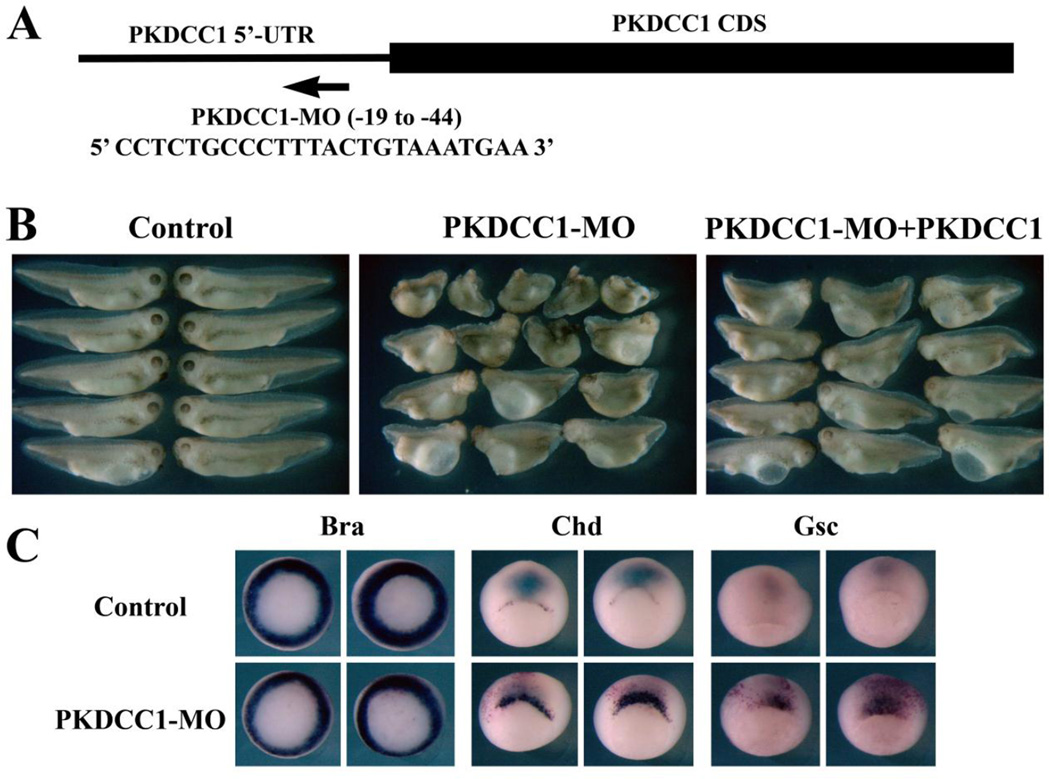
To further examine the role of PKDCC1 in early Xenopus embryogenesis, we designed a translational blocking antisense morpholino oligo (PKDCC1-MO) that hybridized to the 5’-untranslated region (5’-UTR) of the PKDCC1 transcript (Fig. 6A). Injection of PKDCC1-MO into the dorsal marginal zone region of 4-cell stage embryos resulted in dose-dependent reduction of the head structure as well as the body axis, and the defects were largely rescued by a construct that contained only the coding region, but not 5’-UTR, of PKDCC1 (Fig. 6B and data not shown). The reduction in body axis elongation was not accompanied by ventralization of the embryos, as in situ hybridization of marker expression demonstrated the presence of the somatic mesodermal marker MyoD in these embryos (not shown). In addition, at the gastrula stages, the expression levels of Bra, Chd and Gsc appeared normal, but the Chd- and Gsc-expressing cells showed delayed internalization inside the embryos and remained around the blastopore when cells expressing these markers had moved away from the blastopore in control embryos (Fig. 6C). The results demonstrated that knockdown of PKDCC1 affected Xenopus gastrulation without changing mesodermal cell fates.
Figure 6. Knockdown of PKDCC1 resulted in gastrulation defects.
A) A translational blocking antisense PKDCC1-MO was designed to hybridize to the 5’-UTR sequence of the PKDCC transcript. B) Expression of PKDCC1-MO led to embryos with reduced body axis and smaller head, which were greatly rescued by co-expression of low doses of PKDCC1 RNA that did not contain the 5’-UTR MO-target sequence. C) Knockdown of PKDCC1 resulted in delay in internalization of dorsal mesodermal cells marked by Chd and Gsc.
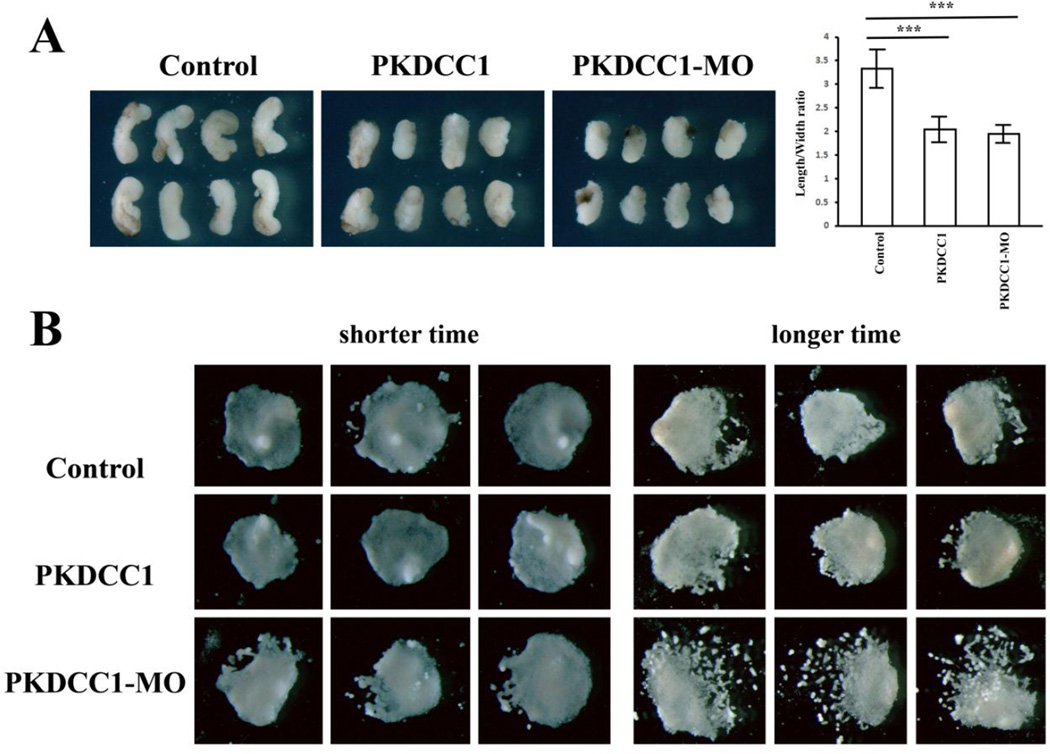
Since both gain- and loss-of-function of PKDCC1 resulted in body axis defects, we inquired whether PKDCC1 modulated gastrulation movements. We thus analyzed tissue morphogenesis in explanted dorsal mesodermal tissues in culture. Both overexpression and knockdown of PKDCC1 led to impaired CE movements, as the DMZ explants dissected from the RNA- or MO-injected embryos displayed decreased length to width ratio when cultured in vitro (Fig. 7A). We also assayed for migration of anterior mesendoderm by dissecting the organizers from the injected embryos at stage 10+ and plating them on fibronectin-coated tissue culture dishes. Examination of the explants showed that there was no discernable defects in collective cell sheet migration when the anterior mesendodermal explants were cultured for about 6 hours. However, prolonged incubation for over 8 hours resulted in dissociation of some peripheral cells from the core explants, and the effect was more pronounced in explants from the morphant embryos (Fig. 7B). The data implied that PKDCC1 might play a role in cell adhesion during collective migration of the anterior mesendoderm.
Figure 7. PKDCC1 regulates gastrulation movements.
A) Both ectopic expression and knockdown of PKDCC1 led to reduction of convergent extension of trunk mesodermal tissues derived from the DMZ. The length over width ratio of the explants decreased significantly by enhanced or reduced expression of PKDCC1. B) While altered levels of PKDCC1 did not prevent collective cell sheet migration on fibronectin, prolonged culture of the explants resulted in more pronounced cell dissociation in the absence of PKDCC1, suggesting that PKDCC1 modulated cell cohesion in the anterior mesendoderm.
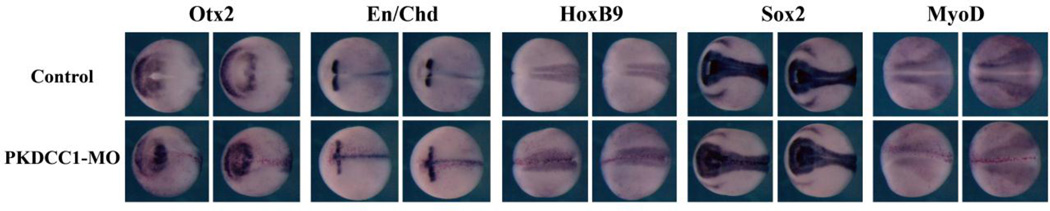
Since we observed distinct microcephaly in morphant embryos, we analyzed whether this was due to impairment of dorsal tissue formation, or it was caused by defects in anterior neural development. We thus examined dorsal mesodermal and anterior-posterior neural markers by in situ hybridization (Fig. 8). The notochord marker chordin (Sasai et al., 1994) and the paraxial mesodermal marker MyoD (Hopwood et al., 1989) were expressed normally in the morphants, and the pan-neural marker Sox2 (Mizuseki et al., 1998) was not affected either. However, the expression domain of the forebrain/midbrain marker Otx2 (Blitz and Cho, 1995; Pannese et al., 1995) was reduced, and there was a concurrent anterior shift of the midbrain marker Engrailed (En, Brivanlou and Harland, 1989) in the morphants (Fig. 8). The results suggested that anterior neural patterning, but not dorsal-ventral axis specification, was defective in PKDCC1 morphants.
Figure 8. PKDCC1 regulates anterior neural patterning.
Knockdown of PKDCC1 did not change the expression of the axial and the paraxial mesodermal markers Chd and MyoD or the neural marker Sox2, but reduced the expression domain of the forebrain marker Otx2 and shifted the midbrain marker engrailed (En) anteriorly, indicating that PKDCC1 modulated anterior-posterior neural patterning.
Discussion
Since the discovery of the organizer by Mangold and Spemann about a century ago (see a translated version of Spemann and Mangold, 2001), there has been an enduring fascination of the molecules synthesized by the organizer that can influence the surrounding tissues to adopt axial and paraxial cell fates (Harland and Gerhard, 1997; Gerhart, 2001; De Robertis, 2001). Multiple growth factor signals, including activin/nodal, BMP, Wnt, and fibroblast growth factor (FGF), have been shown to modulate organizer formation and/or activities. The downstream targets of these signals have also been investigated, including at the genomic scale (Wessely et al., 2004; Perffer et al., 2005; Hufton et al., 2006; Chiu et al., 2014; Gupta et al., 2014). However, most of the functional studies are focused on genes with the ability to induce or pattern embryonic tissues, and some large scale studies also employ techniques that are biased toward abundant genes. Organizer molecules that may control cell polarity, adhesion or motility have not been explored with the same rigor. Similarly, while it is known that temporal changes in tissues surrounding the dorsal blastopore is associated with anterior-posterior (AP) patterning of the body axis (Slack and Tannahill, 1992; Durston and Zhu, 2015), factors that may link AP cell fates and distinct motile cell behaviors are not well scrutinized. To acquire a more detailed understanding of how cells destined to differentiate into specific lineages also adopt particular motile behaviors, we performed RNA-seq analyses in this study to gain insight into distinct transcriptomes in tissues with defined cell fates and migratory patterns.
Our studies uncover a plethora of transcription factors and multitude of signaling molecules differentially enriched in the organizer, the DMZ or the VMZ. Many of these genes have been linked to cell fate determination events in previous reports. The VMZ-enriched factors are especially conspicuous in their abundance in signaling and transcription factors, with only a minor portion encoding metabolic and adhesion molecules (Suppl. Table 1). In contrast, both the organizer and the DMZ contain many distinctive potential regulators of cytoskeleton and/or cell motility, such as the ECM proteins and the actin regulators. In comparing the differentially expressed genes, we reason that those enriched in the DMZ over the organizer, but not the DMZ over the VMZ (group IV), may potentially play roles in AP patterning and/or general cell intercalation behaviors, regardless whether it is CT or CE. The observed lack of VMZ-specific enrichment of actin regulators may indicate that the type of the cell movements in the VMZ, convergent thickening, is probably the default cell behaviors in the trunk. Acquisition of new actin regulators and ECM proteins in the DMZ over the VMZ (group II) may then help to endow the DMZ cells to respond to specific dorsal or ECM signals to undergo CE. Thus, group II genes may be important for dorsal cell fates as well as CE, whereas group IV genes may be crucial for caudal cell specification and CT movements. The genes that are differentially expressed in the organizer and the DMZ (group I and II), in addition to having an effect on embryonic patterning of the head and the trunk, may also differentially influence cell-ECM and cell-cell interactions to modulate cell migration on ECM or over other cells. It is interesting to note that several genes in our dataset, such as ARHGEF3, IRG and RASL11B, overlap with those with altered expression when an ectopic organizer was induced in the ventral side by simultaneous inhibition of BMP and Wnt signals (Hufton et al., 2006). This suggests that the organizer signals can influence the expression of genes with roles in cell movements, such as ARHGEF3, to coordinate cell fate and motility.
Our initial analyses of a group of signaling, transcription and cytoskeleton regulatory factors show that members of the same gene family often have distinct functions in regulating CE in activin-induced animal caps. The difference in functionality of the genes is often associated with different expression patterns of these genes. For example, the organizer-enriched RhoGEF PLEKHG5 is not effective in blocking activin-induced animal cap elongation, but the DMZ-enriched RhoGEF ARHGEF3 interferes with CE in dorsalized animal caps efficiently. Similarly, among the Cdc42 effectors, CDC42EP3, which is expressed at high levels in both the organizer and the DMZ, displays the greatest activity in preventing activin-induced animal cap elongation, whereas CDC42EP2, which is enriched in the organizer only, has a weaker activity in this assay. The results suggest that distinct members of RhoGEF and CDC42EP families may detect and respond to different signals in the head and the trunk regions to modulate actin organization and cell behaviors. The sequences outside the conserved GEF or CDC42 binding domains are therefore crucial to determine the potency and/or the specificity of gene functions. Recognition of the sequence motif(s) that confers specific activities to particular family members is thus important in the future for us to understand how genes belonging to the same family possess unique functions in regulating different migratory cell behaviors during Xenopus gastrulation.
In addition to genes that regulate cell movements without affecting cell fates, we have also identified an ECM protein as a new regulator of the activin signaling. The gene, EFEMP2, contains multiple EGF and calcium-binding EGF domains and is also known as fibulin-4. Two other fibulin family genes have been reported to regulate TGF-β family signaling. Fibulin-1 regulates BMP signaling through direct binding to BMP-2 (Cooley et al., 2014), whereas Fibulin-3 inhibits TGF-β 1 signaling by interacting with TpRI/ALK-5 to prevent formation of a functional ligand-receptor complex (Tian et al., 2015). Fibulin-4 mutations are associated with patients with aortic aneurysms, arterial tortuosity and stenosis. TGF-β signaling is enhanced in these patients, though the underlying molecular mechanism has not been shown (Renard et al., 2010). In mice, Fibulin-4 deficiency results in increased expression of TGF-β ligands in isolated aortic smooth muscle cells, leading to enhanced TGF-β signaling (Ramnath et al., 2015). In our study, we find that EFEMP2 inhibits activin signaling downstream of the activated receptor ALK-4, suggesting that it may regulate other growth factor signals to stimulate a distinct pathway to block activation of the Smad2/3 signal transducers by activated ALK receptors. Further experiments are required to understand the detailed mechanisms underlying EFEMP2 function.
To investigate the roles of endogenous genes that show differential expression during gastrulation, we focused on PKDCC1 in this study. The protein, also known as VLK, is shown to be a novel secreted tyrosine kinase that can phosphorylate a broad range of extracellular molecules and ectodomains of transmembrane proteins. These include many ECM proteins, such as collagens, and matrix metalloproteinases (MMPs), such as MMP1 and MMP14. Phosphorylation of these proteins can potentially control their activities and impact on cell adhesion and migration (Bordoli et al., 2014). Targeted disruption of Pkdcc/Vlk gene in mice results in multiple developmental defects, such as short limb, cleft palate, and lung hypoplasia (Imuta et al., 2009; Kinoshita et al., 2009; Probst et al., 2013). In Xenopus, PKDCC1 and 2 have been reported to regulate JNK-dependent planar cell polarity pathway to control blastopore and neural tube closure (Vitorino et al., 2015). In our studies, we show that PKDCC1 regulates gastrulation movements and anterior neural patterning. PKDCC1 modulates CE of the trunk mesoderm and the sustained cohesion of the anterior mesendoderm. These phenotypes partially mimic those associated with altered expression of Arg/Abl2 (Bonacci et al., 2012), implying that PKDCC1 may influence or converge with Arg/Abl2-dependent signaling pathway. As the transcripts of PKDCC1 are expressed at high levels in the organizer and the anterior neural tissues, but not abundantly in the trunk mesoderm, it implies that the secreted protein product may diffuse a long distance to influence cell behaviors in the trunk mesoderm. PKDCC1 also regulates anterior neural patterning, with the forebrain most sensitive to the reduction of PKDCC1 levels. The AP neural patterning is controlled by multiple growth factor signals, including Wnt, FGF, BMP, and retinoid acid (Slack and Tannahill, 1992; Chang and Hemmati-Brivanlou, 1998). PKDCC1 may phosphorylate one or several extracellular components of these signals to influence the development of the anterior neural tissues. It is likely that the targets of PKDCC1 in the mesoderm and the neural tissues may differ. For example, one of the known targets of PKDCC1 is MMP14, which has been shown to modulate CE in zebrafish (Coyle et al., 2008; Williams et al., 2012). In contrast, potential regulation of the canonical Wnt signaling may contribute to the neural patterning activity of PKDCC1 (Bordoli et al., 2014). Modification of distinct substrates in different tissues will allow PKDCC1 to regulate cell movements and cell fates in two different developmental contexts.
In summary, we have uncovered in this work many differentially expressed genes in different embryonic tissues during gastrulation. These not only include factors that can control embryonic patterning, but also proteins that may potentially regulate cell adhesion, polarity and motility to influence cell movements on ECM (migration) or over other cells (intercalation). Further studies employing loss-of-function approaches are required to interrogate the endogenous roles of these genes in particular cell movements during gastrulation, and how expression and/or activities of these proteins are modulated by signals that impart cell fates. These studies promise to provide more in depth understanding on how cell fate specification and cell behaviors are coordinated during vertebrate embryogenesis.
Materials and Methods
Embryo culture and explant dissection
The Xenopus embryos were obtained and maintained using standard protocols as described (Sive et al., 2000). The early organizer was dissected from stage 10 to 10.25 embryos by removing the tissues surrounding, and including, the dorsal lip. The DMZ and the VMZ explants were dissected from stage 11 to 11.5 embryos by removing tissues from the dorsal and the ventral sides of the embryos above the blastopore, respectively. Total about 150 explants were pooled together for each sample. RNA extraction was performed using Qiagen RNeasy mini-spin columns. Two different sets of the samples were prepared independently. The second set of the samples were made at somewhat later stages, which were reflected by stronger expression of certain genes and weaker expression of the others when compared with the first set of the samples. The sample comparison was thus performed independently for each set and combined together. There were variations in calls of differentially expressed genes in the two sets, which might be due to combined effects of natural variations of gastrula embryos (Vodicka and Gerhart, 1995; Ewald et al., 2004) and variations in dissections of pooled samples. RT-PCR was thus performed to confirm and validate the differentially expressed genes, as marker expression in these samples could be easily tested.
RNA sequencing and data analyses
RNA sequencing was performed on Illumina HiSeq2000 platform with 2×50bp paired-end sequencing configuration. Briefly, the quality of the total RNA was assessed using the Agilent 2100 Bioanalyzer followed by 2 rounds of poly A+ selection and conversion to cDNA. The TruSeq library generation kit was used to construct the cDNA library as per the manufacturer’s instructions (Illumina, San Diego, CA). The cDNA library was quantitated using qPCR in a Roche LightCycler 480 with the Kapa Biosystems kit for library quantitation (Kapa Biosystems, Woburn, MA) prior to sequencing. Paired-end reads were mapped to the reference Xenopus laevis transcriptome ‘MayBall’ annotation (Chung et al., 2014), based on Xenopus laevis genome (version 7.1), using Bowtie (version 1.0.0). After the latest release of Xenopus laevis genome (version 9.1), we re-analyzed our RNA-seq data with it using BWA (version 0.7.12) against primary transcripts (based on JGI version 1.8 annotation) to quantify the expression of L and S homeologs separately (Supplemental Table 2). Differentially expressed genes were identified using edgeR packages (version 3.12.0, Robinson et al., 2010), with both classic pairwise comparison and ANOVA-like multiple comparison. However, we noticed that several genes on our original differentially expressed gene list based on 7.1 genome, such as ARHGEF3 and CXCR7, were lost in new annotation (although there are two ARHGEF3 candidates, Xelaev18003042m and Xelaev18040996m, according to sequence similarity, they were not annotated as ARHGEF3 in JGI version 1.8 annotation). Therefore, we kept using the original analysis for differential gene expression as listed in Supplemental Table 1 (only ANOVA-like multiple sample comparison was included in this table).
Animal cap assay
RNAs encoding 2pg activin and 0.25-1ng of RNA-seq clones were injected into the animal region of two-cell stage embryos. Ectodermal explants were excised from blastula stage embryos and cultured in 0.5X MMR solution until late neurula stages. The morphology of the explants was observed and photographed, and the animal caps were subjected to RNA extraction and RT-PCR as described (Chang, 2016). The primers used for RT-PCR are listed in the Suppl. Table 4. Alternatively, animal caps from the embryos injected with RNA-seq clones were dissected at blastula stages and incubated with 8–10ng/ml Activin B protein (R&D Systems) until late neurula stages before they were imaged.
Assays for dorsal mesoderm convergent extension and anterior mesendoderm migration
The DMZ explants were dissected from stage 10.5 to 11 embryos and cultured in vitro in 0.5X MMR solution. The elongation of the explants was examined and photographed at late neurula stages, and the length to width ratio was measured using the NIH ImageJ program (Bonacci et al., 2012). For anterior mesendoderm migration, the organizer region was dissected from stage 10+ embryos and plated on fibronectin-coated dishes. The explants were cultured in DFA solution at the room temperature for about 6 hours before being imaged (Nie et al., 2007a). Some of the explants were allowed for longer incubation for over 8 hours to examine the effect of altered PKDCC1 on tissue integrity and migration.
Supplementary Material
Highlights.
RNA-seq analysis of genes differentially expressed in the organizer, the dorsal marginal zone, and the ventral marginal zone of Xenopus gastrulae.
Examination of a selected subset of genes with differential expression patterns demonstrates that they display distinct activities in regulating convergent extension movements in activin-induced animal caps.
An extracellular matrix protein, EFEMP2, modulates activin signaling downstream of the activated receptor.
PKDCC1, a secreted protein kinase, regulates gastrulation movements and anterior neural patterning.
Acknowledgments
We thank National Xenopus Resource (Woods Hole, MA) for bioinformatics training in one of its workshops. We thank Xenbase (www.xenbase.org) for posting Xenopus genome assemblies and annotation of literatures to assist research in the Xenopus community. This work is funded by the NIH grant NIGMS R01GM098566 to CC, UNIST Research Fund (1.150094.01 and 1.150043.01) to TK, and NIH grant NIGMS R01GM104853 to JBW.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Altmann CR, Bell E, Sczyrba A, Pun J, Bekiranov S, Gaasterland T, Brivanlou AH. Microarray-based analysis of early development in Xenopus laevis. Dev. Biol. 2001;236:64–75. doi: 10.1006/dbio.2001.0298. [DOI] [PubMed] [Google Scholar]
- Ataliotis P, Symes K, Chou MM, Ho L, Mercola M. PDGF signalling is required for gastrulation of Xenopus laevis. Development. 1995;121:3099–3110. doi: 10.1242/dev.121.9.3099. [DOI] [PubMed] [Google Scholar]
- Baldessari D, Shin Y, Krebs O, König R, Koide T, Vinayagam A, Fenger U, Mochii M, Terasaka C, Kitayama A, Peiffer D, Ueno N, Eils R, Cho KW, Niehrs C. Global gene expression profiling and cluster analysis in Xenopus laevis. Mech. Dev. 2005;122:441–475. doi: 10.1016/j.mod.2004.11.007. [DOI] [PubMed] [Google Scholar]
- Blitz IL, Cho KW. Anterior neurectoderm is progressively induced during gastrulation: the role of the Xenopus homeobox gene orthodenticle. Development. 1995;121:993–1004. doi: 10.1242/dev.121.4.993. [DOI] [PubMed] [Google Scholar]
- Bonacci G, Fletcher J, Devani M, Dwivedi H, Keller R, Chang C. The cytoplasmic tyrosine kinase Arg regulates gastrulation via control of actin organization. Dev. Biol. 2012;364:42–55. doi: 10.1016/j.ydbio.2012.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordoli MR, Yum J, Breitkopf SB, Thon JN, Italiano JE, Jr, Xiao J, Worby C, Wong SK, Lin G, Edenius M, Keller TL, Asara JM, Dixon JE, Yeo CY, Whitman M. A secreted tyrosine kinase acts in the extracellular environment. Cell. 2014;158:1033–1044. doi: 10.1016/j.cell.2014.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brivanlou AH, Harland RM. Expression of an engrailed-related protein is induced in the anterior neural ectoderm of early Xenopus embryos. Development. 1989;106:611–617. doi: 10.1242/dev.106.3.611. [DOI] [PubMed] [Google Scholar]
- Cao Q, Zhang X, Lu L, Yang L, Gao J, Gao Y, Ma H, Cao Y. Klf4 is required for germ-layer differentiation and body axis patterning during Xenopus embryogenesis. Development. 2012;139:3950–3961. doi: 10.1242/dev.082024. [DOI] [PubMed] [Google Scholar]
- Chang C. Animal cap assay for TGF-β signaling. Methods Mol. Biol. 2016;1344:261–274. doi: 10.1007/978-1-4939-2966-5_16. [DOI] [PubMed] [Google Scholar]
- Chang C, Wilson PA, Mathews LS, Hemmati-Brivanlou A. A Xenopus type I activin receptor mediates mesodermal but not neural specification during embryogenesis. Development. 1997;124:827–837. doi: 10.1242/dev.124.4.827. [DOI] [PubMed] [Google Scholar]
- Chang C, Hemmati-Brivanlou A. Cell fate determination in embryonic ectoderm. J. Neurobiol. 1998;36:128–151. [PubMed] [Google Scholar]
- Chiu WT, Charney Le R, Blitz IL, Fish MB, Li Y, Biesinger J, Xie X, Cho KW. Genome-wide view of TGFβ/Foxh1 regulation of the early mesendoderm program. Development. 2014;141:4537–4547. doi: 10.1242/dev.107227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho KW, Blumberg B, Steinbeisser H, De Roberts EM. Molecular nature of Spemann's organizer: the role of the Xenopus homeobox gene goosecoid. Cell. 1991;67:1111–1120. doi: 10.1016/0092-8674(91)90288-a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi SC, Han JK. Xenopus Cdc42 regulates convergent extension movements during gastrulation through Wnt/Ca2+ signaling pathway. Dev. Biol. 2002;244:342–357. doi: 10.1006/dbio.2002.0602. [DOI] [PubMed] [Google Scholar]
- Chung MI, Kwon T, Tu F, Brooks ER, Gupta R, Meyer M, Baker JC, Marcotte EM, Wallingford JB. Coordinated genomic control of ciliogenesis and cell movement by RFX2. eLife. 2014;3:e01439. doi: 10.7554/eLife.01439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conlon FL, Smith JC. Interference with Brachyury function inhibits convergent extension, causes apoptosis, and reveals separate requirements in the FGF and activin signalling pathways. Dev. Biol. 1999;213:85–100. doi: 10.1006/dbio.1999.9330. [DOI] [PubMed] [Google Scholar]
- Cooley MA, Harikrishnan K, Oppel JA, Miler SF, Barth JL, Haycraft CJ, Reddy SV, Argraves WS. Fibulin-1 is required for bone formation and Bmp-2-medidated induction of Osterix. Bone. 2014;69:30–38. doi: 10.1016/j.bone.2014.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyle RC, Latimer A, Jessen JR. Membrane-type 1 matrix metalloproteinase regulates cell migration during zebrafish gastrulation: evidence for an interaction with non-canonical Wnt signaling. Exp. Cell Res. 2008;314:2150–2162. doi: 10.1016/j.yexcr.2008.03.010. [DOI] [PubMed] [Google Scholar]
- Damm EW, Winklbauer R. PDGF-A controls mesoderm cell orientation and radial intercalation during Xenopus gastrulation. Development. 2011;138:565–575. doi: 10.1242/dev.056903. [DOI] [PubMed] [Google Scholar]
- De Robertis EM, Wessely O, Oelgeschläger M, Brizuela B, Pera E, Larraín J, Abreu J, Bachiller D. Molecular mechanisms of cell-cell signaling by the Spemann-Mangold organizer. Int. J. Dev. Biol. 2001;45:189–197. [PMC free article] [PubMed] [Google Scholar]
- Durston AJ, Zhu K. A time space translation hypothesis for vertebrate axial patterning. Sem. Cell Dev. Biol. 2015;42:86–93. doi: 10.1016/j.semcdb.2015.06.001. [DOI] [PubMed] [Google Scholar]
- Ekker SC, McGrew LL, Lai CJ, Lee JJ, von Kessler DP, Moon RT, Beachy PA. Distinct expression and shared activities of members of the hedgehog gene family of Xenopus laevis. Development. 1995;121:2337–2347. doi: 10.1242/dev.121.8.2337. [DOI] [PubMed] [Google Scholar]
- Evren S, Wen JWH, Luu O, Damm EW, Nagel M, Winklbauer R. EphA4-dependent Brachyury expression is required for dorsal mesoderm involution in the Xenopus gastrula. Development. 2014;141:3649–3661. doi: 10.1242/dev.111880. [DOI] [PubMed] [Google Scholar]
- Ewald AJ, Peyrot SM, Tyszka JM, Fraser SE, Wallingford JB. Regional requirements for Dishevelled signaling during Xenopus gastrulation: separable effects on blastopore closure, mesendoderm internalization and archenteron formation. Development. 2004;131:6195–6209. doi: 10.1242/dev.01542. [DOI] [PubMed] [Google Scholar]
- Fletcher G, Jones GE, Patient R, Snape A. A role for GATA factors in Xenopus gastrulation movements. Mech. Dev. 2006;123:730–745. doi: 10.1016/j.mod.2006.07.007. [DOI] [PubMed] [Google Scholar]
- Gerhart J. Evolution of the organizer and the chordate body plan. Int. J. Dev. Biol. 2001;45:133–153. [PubMed] [Google Scholar]
- Gupta R, Wills A, Ucar D, Baker J. Developmental enhancers are marked independently of zygotic Nodal signals in Xenopus. Dev. Biol. 2014;395:38–49. doi: 10.1016/j.ydbio.2014.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habas R, Kato Y, He X. Wnt/Frizzled activation of Rho regulates vertebrate gastrulation and requires a novel Formin homology protein Daam1. Cell. 2001;107:843–854. doi: 10.1016/s0092-8674(01)00614-6. [DOI] [PubMed] [Google Scholar]
- Habas R, Dawid IB, He X. Coactivation of Rac and Rho by Wnt/Frizzled signaling is required for vertebrate gastrulation. Genes Dev. 2003;17:295–309. doi: 10.1101/gad.1022203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harland RM, Gerhart J. Formation and function of Spemann's organizer. Annu. Rev. Cell Dev. Biol. 1997;13:611–667. doi: 10.1146/annurev.cellbio.13.1.611. [DOI] [PubMed] [Google Scholar]
- Heasman J. Patterning the early Xenopus embryos. Development. 2006;133:1205–1217. doi: 10.1242/dev.02304. [DOI] [PubMed] [Google Scholar]
- Hemmati-Brivanlou A, Kelly OG, Melton DA. Follistatin, an antagonist of activin, is expressed in the Spemann organizer and displays direct neuralizing activity. Cell. 1994;77:283–295. doi: 10.1016/0092-8674(94)90320-4. [DOI] [PubMed] [Google Scholar]
- Hopwood ND, Pluck A, Gurdon JB. MyoD expression in the forming somites is an early response to mesoderm induction in Xenopus embryos. EMBO J. 1989;8:3409–3417. doi: 10.1002/j.1460-2075.1989.tb08505.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hufton AL, Vinayagam A, Suhai S, Baker JC. Genomic analysis of Xenopus organizer function. BMC Dev. Biol. 2006;6:27. doi: 10.1186/1471-213X-6-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imuta Y, Nishioka N, Kiyonari H, Sasaki H. Short limbs, cleft palate, and delayed formation of flat proliferative chondrocytes in mice with targeted disruption of a putative protein kinase gene, Pkdcc (AW548124) Dev. Dyn. 2009;238:210–222. doi: 10.1002/dvdy.21822. [DOI] [PubMed] [Google Scholar]
- Keller R, Shook D. Gastrulation in Amphibians. In: Stern CD, editor. Gastrulation: From Cells to Embryos. 2004. pp. 171–204. [Google Scholar]
- Keller R, Shook D. Dynamic determinations: patterning the cell behaviours that close the amphibian blastopore. Phil. Trans. R. Sco. B. 2008;363:1317–1332. doi: 10.1098/rstb.2007.2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller R, Shook D, Skoglund P. The forces that shape embryos: physical aspects of convergent extension by cell intercalation. Phys. Biol. 2008;5:1–24. doi: 10.1088/1478-3975/5/1/015007. [DOI] [PubMed] [Google Scholar]
- Kim GH, Han JK. JNK and ROKa function in the noncanonical Wnt/RhoA signaling pathway to regulate Xenopus convergent extension movements. Dev. Dyn. 2005;232:958–968. doi: 10.1002/dvdy.20262. [DOI] [PubMed] [Google Scholar]
- Kinoshita M, Era T, Jakt LM, Nishikawa S. The novel protein kinase Vlk is essential for stromal function of mesenchymal cells. Development. 2009;136:2069–2079. doi: 10.1242/dev.026435. [DOI] [PubMed] [Google Scholar]
- Lemaire P, Garrett N, Gurdon JB. Expression cloning of Siamois, a Xenopus homeobox gene expressed in dorsal-vegetal cells of blastulae and able to induce a complete secondary axis. Cell. 1995;81:85–94. doi: 10.1016/0092-8674(95)90373-9. [DOI] [PubMed] [Google Scholar]
- Lemaire P, Darras S, Caillol D, Kodjabachian L. A role for the vegetally expressed Xenopus gene Mix.1 in endoderm formation and in the restriction of mesoderm to the marginal zone. Development. 1998;125:2371–2380. doi: 10.1242/dev.125.13.2371. [DOI] [PubMed] [Google Scholar]
- Mishra SK, Nagata T, Furusawa K, Sasaki A, Fukui A. Expression of xSDF-1α, xCXCR4, and xCXCR7 during gastrulation in Xenopus laevis. Int. J. Dev. Biol. 2013;57:95–100. doi: 10.1387/ijdb.120130af. [DOI] [PubMed] [Google Scholar]
- Mizuseki K, Kishi M, Matsui M, Nakanishi S, Sasai Y. Xenopus Zic-related-1 and Sox-2, two factors induced by chordin, have distinct activities in the initiation of neural induction. Development. 1998;125:579–587. doi: 10.1242/dev.125.4.579. [DOI] [PubMed] [Google Scholar]
- Munoz-Sanjuan I, Bell E, Altmann CR, Vonica A, Brivanlou AH. Gene profiling during neural induction in Xenopus laevis: regulation of BMP signaling by post-transcriptional mechanisms and TAB3, a novel TAK1-binding protein. Development. 2002;129:5529–5540. doi: 10.1242/dev.00097. [DOI] [PubMed] [Google Scholar]
- Nagel M, Tahinci E, Symes K, Winklbauer R. Guidance of mesoderm cell migration in the Xenopus gastrula requires PDGF signaling. Development. 2004;131:2727–2736. doi: 10.1242/dev.01141. [DOI] [PubMed] [Google Scholar]
- Nie S, Chang C. Regulation of Xenopus gastrulation by ErbB signaling. Dev. Biol. 2007a;303:93–107. doi: 10.1016/j.ydbio.2006.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie S, Chang C. PI3K and Erk MAPK mediate ErbB signaling in Xenopus gastrulation. Mech. Dev. 2007b;124:657–667. doi: 10.1016/j.mod.2007.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nutt SL, Dingwell KS, Holt CE, Amaya E. Xenopus Sprouty2 inhibits FGF-mediated gastrulation movements but does not affect mesoderm induction and patterning. Genes Dev. 2001;15:1152–1166. doi: 10.1101/gad.191301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onichtchouk D, Gawantka V, Dosch R, Delius H, Hirschfeld K, Blumenstock C, Niehrs C. The Xvent-2 homeobox gene is part of the BMP-4 signalling pathway controlling [correction of controling] dorsoventral patterning of Xenopus mesoderm. Development. 1996;122:3045–3053. doi: 10.1242/dev.122.10.3045. [DOI] [PubMed] [Google Scholar]
- Pannese M, Polo C, Andreazzoli M, Vignali R, Kablar B, Barsacchi G, Boncinelli E. The Xenopus homologue of Otx2 is a maternal homeobox gene that demarcates and specifies anterior body regions. Development. 1995;121:707–720. doi: 10.1242/dev.121.3.707. [DOI] [PubMed] [Google Scholar]
- Park EC, Cho GS, Kim GH, Choi SC, Han JK. The involvement of Eph-Ephrin signaling in tissue separation and convergence during Xenopus gastrulation movements. Dev. Biol. 2011;350:441–450. doi: 10.1016/j.ydbio.2010.12.012. [DOI] [PubMed] [Google Scholar]
- Peiffer DA, Von Bubnoff A, Shin Y, Kitayama A, Mochii M, Ueno N, Cho KW. A Xenopus DNA microarray approach to identify novel direct BMP target genes involved in early embryonic development. Dev. Dyn. 2005;232:445–456. doi: 10.1002/dvdy.20230. [DOI] [PubMed] [Google Scholar]
- Pendo-Mendez A, Umbhauer M, Djiane A, Boucaut JC, Riou JF. Activation of Gβγ signaling downstream of Wnt-11/Xfz-7 regulates Cdc42 activity during Xenopus gastrulation. Dev. Biol. 2003;257:302–314. doi: 10.1016/s0012-1606(03)00067-8. [DOI] [PubMed] [Google Scholar]
- Probst S, Zeller R, Zuniga A. The hedgehog target Vlk genetically interacts with Gli3 to regulate chondrocyte differentiation during mouse long bone development. Differentiation. 2013;85:121–130. doi: 10.1016/j.diff.2013.03.002. [DOI] [PubMed] [Google Scholar]
- Ramnath NW, Hawinkels LJ, van Heijningen PM, Riet LT, Paauwe M, Vermeij M, Danser AH, Kanaar R, Ten Dijke P, Essers J. Fibulin-4 deficiency increases TGF-β signalling in aortic smooth muscle cells due to elevated TGF-β2 levels. Sci. Rep. 2015;5:16872. doi: 10.1038/srep16872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren R, Nagel M, Tahinci E, Winklbauer R, Symes K. Migrating anterior mesodermal cells and intercalating trunk mesodermal cells have distinct responses to Rho and Rac during Xenopus gastrulation. Dev. Dyn. 2006;235:1090–1099. doi: 10.1002/dvdy.20711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renard M, Holm T, Veith R, Callewaert BL, Adès LC, Baspinar O, Pickart A, Dasouki M, Hoyer J, Rauch A, Trapane P, Earing MG, Coucke PJ, Sakai LY, Dietz HC, De Paepe AM, Loeys BL. Altered TGFp signaling and cardiovascular manifestations in patients with autosomal recessive cutis laxa type I caused by fibulin-4 deficiency. Eur. J. Human Genet. 2010;18:895–901. doi: 10.1038/ejhg.2010.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa FM. Mix.1, a homeobox mRNA inducible by mesoderm inducers, is expressed mostly in the presumptive endodermal cells of Xenopus embryos. Cell. 1989;57:965–974. doi: 10.1016/0092-8674(89)90335-8. [DOI] [PubMed] [Google Scholar]
- Roszko I, Sawada A, Solnica-Krezel L. Regulation of convergence and extension movements during vertebrate gastrulation by the Wnt/PCP pathway. Semin. Cell Dev. Biol. 2009;20:986–997. doi: 10.1016/j.semcdb.2009.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salic AN, Kroll KL, Evans LM, Kirschner MW. Sizzled: a secreted Xwnt8 antagonist expressed in the ventral marginal zone of Xenopus embryos. Development. 1997;124:4739–4748. doi: 10.1242/dev.124.23.4739. [DOI] [PubMed] [Google Scholar]
- Sasai Y, Lu B, Steinbeisser H, Geissert D, Gont LK, De Robertis EM. Xenopus chordin : a novel dorsalizing factor activated by organizer-specific homeobox genes. Cell. 1994;79:779–790. doi: 10.1016/0092-8674(94)90068-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih J, Keller R. Cell motility driving mediolateral intercalation in explants of Xenopus laevis. Development. 1992a;116:901–914. doi: 10.1242/dev.116.4.901. [DOI] [PubMed] [Google Scholar]
- Shih J, Keller R. Patterns of cell motility in the organizer and dorsal mesoderm of Xenopus laevis. Development. 1992b;116:915–930. doi: 10.1242/dev.116.4.915. [DOI] [PubMed] [Google Scholar]
- Sivak JM, Petersen LF, Amaya E. FGF signal interpretation is directed by Sprouty and Spred proteins during mesoderm formation. Dev. Cell. 2005;8:689–701. doi: 10.1016/j.devcel.2005.02.011. [DOI] [PubMed] [Google Scholar]
- Sive HL, Grainger RM, Harland RM. Early Development of Xenopus laevis: A Laboratory Manual. New Year: Cold Spring Harbor Lab. Press, Cold Spring Harbor; 2000. [Google Scholar]
- Skoglund P, Keller R. Integration of planar cell polarity and ECM signaling in elongation of the vertebrate body plan. Curr. Opin. Cell Biol. 2010;22:589–596. doi: 10.1016/j.ceb.2010.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slack JM, Tannahill D. Mechanism of anteroposterior axis specification in vertebrates. Lessons from the amphibians. Development. 1992;114:285–302. doi: 10.1242/dev.114.2.285. [DOI] [PubMed] [Google Scholar]
- Smith JC, Howard JE. Mesoderm-inducing factors and the control of gastrulation. Development. 1992;(suppl):127–136. [PubMed] [Google Scholar]
- Smith JC, Price BMJ, Green JBA, Weigel D, Herrmann BG. Expression of a Xenopus homolog of Brachyury (T) is an immediate-early response to mesoderm induction. Cell. 1991;67:79–87. doi: 10.1016/0092-8674(91)90573-h. [DOI] [PubMed] [Google Scholar]
- Smith WC, Harland RM. Expression cloning of noggin, a new dorsalizing factor localized to the Spemann organizer in Xenopus embryos. Cell. 1992;70:829–840. doi: 10.1016/0092-8674(92)90316-5. [DOI] [PubMed] [Google Scholar]
- Smith WC, McKendry R, Ribisi S, Jr, Harland RM. A nodal-related gene defines a physical and functional domain within the Spemann organizer. Cell. 1995;82:37–46. doi: 10.1016/0092-8674(95)90050-0. [DOI] [PubMed] [Google Scholar]
- Spemann H, Mangold H. Induction of embryonic primordia by implantation of organizers from a different species. 1923. Int. J. Dev. Biol. 2001;45:13–38. [PubMed] [Google Scholar]
- Symes K, Yordan C, Mercola M. Morphological differences in Xenopus embryonic mesodermal cells are specified as an early response to distinct threshold concentrations of activin. Development. 1994;120:2339–2346. doi: 10.1242/dev.120.8.2339. [DOI] [PubMed] [Google Scholar]
- Tahinci E, Symes K. Distinct functions of Rho and Rac are required for convergent extension during Xenopus gastrulation. Dev. Biol. 2003;259:318–335. doi: 10.1016/s0012-1606(03)00206-9. [DOI] [PubMed] [Google Scholar]
- Takahashi S, Yokota C, Takano K, Tanegashima K, Onuma Y, Goto J, Asashima M. Two novel nodal-related genes initiate early inductive events in Xenopus Nieuwkoop center. Development. 2000;127:5319–5329. doi: 10.1242/dev.127.24.5319. [DOI] [PubMed] [Google Scholar]
- Taverner NV, Kofron M, Shin Y, Kabitschke C, Gilchrist MJ, Wylie C, Cho KW, Heasman J, Smith JC. Microarray-based identification of VegT targets in Xenopus. Mech. Dev. 2005;122:333–354. doi: 10.1016/j.mod.2004.10.010. [DOI] [PubMed] [Google Scholar]
- Tian H, Liu J, Chen J, Gatza ML, Blobe GC. Fibulin-3 is a novel TGF-β pathway inhibitor in the breast cancer microenvironment. Oncogene. 2015;34:5635–5647. doi: 10.1038/onc.2015.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitorino M, Silva AC, Inácio JM, Ramalho JS, Gur M, Fainsod A, Steinbeisser H, Belo JA. Xenopus Pkdcc1 and Pkdcc2 Are Two New Tyrosine Kinases Involved in the Regulation of JNK Dependent Wnt/PCP Signaling Pathway. PLoS One. 2015;10:e0135504. doi: 10.1371/journal.pone.0135504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vodicka MA, Gerhart JC. Blastomere derivation and domains of gene expression in the Spemann Organizer of Xenopus laevis. Development. 1995;121:3505–3518. doi: 10.1242/dev.121.11.3505. [DOI] [PubMed] [Google Scholar]
- Wacker S, Brodbeck A, Lemaire P, Niehrs C, Winklbauer R. Patterns and control of cell motility in the Xenopus gastrula. Development. 1998;125:1931–1942. doi: 10.1242/dev.125.10.1931. [DOI] [PubMed] [Google Scholar]
- Wallingford JB, Fraser SE, Harland RM. Convergent extension: the molecular control of polarized cell movement during embryonic development. Dev. Cell. 2002;2:695–706. doi: 10.1016/s1534-5807(02)00197-1. [DOI] [PubMed] [Google Scholar]
- Wallingford JB. Planar cell polarity and the developmental control of cell behavior in vertebrate embryos. Annu. Rev. Cell Dev. Biol. 2012;28:627–653. doi: 10.1146/annurev-cellbio-092910-154208. [DOI] [PubMed] [Google Scholar]
- Weber H, Symes CE, Walmsley ME, Rodaway AR, Patient RK. A role for GATA5 in Xenopus endoderm specification. Development. 2000;127:4345–4360. doi: 10.1242/dev.127.20.4345. [DOI] [PubMed] [Google Scholar]
- Wessely O, Kim JI, Geissert D, Tran U, De Robertis EM. Analysis of Spemann organizer formation in Xenopus embryos by cDNA macroarrays. Dev. Biol. 2004;269:552–566. doi: 10.1016/j.ydbio.2004.01.018. [DOI] [PubMed] [Google Scholar]
- Williams BB, Cantrell VA, Mundell NA, Bennett AC, Quick RE, Jessen JR. VANGL2 regulates membrane trafficking of MMP14 to control cell polarity and migration. J. Cell Sci. 2012;125:2141–2147. doi: 10.1242/jcs.097964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson P, Keller R. Cell rearrangement during gastrulation of Xenopus: direct observation of cultured explants. Development. 1991;112:289–300. doi: 10.1242/dev.112.1.289. [DOI] [PubMed] [Google Scholar]
- Winklbauer R. Mesodermal cell migration during Xenopus gastrulation. Dev. Biol. 1990;142:155–168. doi: 10.1016/0012-1606(90)90159-g. [DOI] [PubMed] [Google Scholar]
- Winklbauer R, Nagel M. Directional mesoderm cell migration in the Xenopus gastrula. Dev. Biol. 1991;148:573–589. doi: 10.1016/0012-1606(91)90275-8. [DOI] [PubMed] [Google Scholar]
- Winklbauer R, Nagel M, Selchow A, Wacker S. Mesoderm migration in the Xenopus gastrula. Int. J. Dev. Biol. 1996;40:305–311. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.