Abstract
Background and purpose
Contradictory data exist on the association between host interleukin-28B (IL28B) rs12979860 genotype and liver fibrosis in patients with chronic hepatitis C (CHC). This large, international, observational study (NCT01675427/MV25600) investigated relationships between IL28B rs12979860 genotype and liver fibrosis stage in CHC patients.
Methods
A total of 3003 adult, treatment-naive CHC patients were enrolled into the study. Patients made one study visit to provide a blood sample for genotyping; other data were obtained from medical records.
Results
2916 patients comprised the analysis population; the majority were enrolled in Europe (n = 2119), were Caucasian (n = 2582) and had hepatitis C virus (HCV) genotype (G)1 infection (n = 1702) (G2 = 323, G3 = 574, G4 = 260). Distribution of IL28B genotypes varied according to region of enrolment, patient ethnicity and HCV genotype. A significant association was observed between increasing number of IL28B T alleles and the prevalence of cirrhosis/transition to cirrhosis (based on biopsy or non-invasive assessments) in G1-infected patients (CC = 22.2% [111/499], CT = 27.5% [255/928], TT = 32.3% [87/269]; p = 0.0018). The association was significant in the large subgroup of European Caucasian G1 patients (n = 1245) but not in the smaller Asian (n = 25), Latin American (n = 137) or Middle Eastern (n = 289) G1 subgroups. IL28B genotype was not associated with liver fibrosis stage in patients with HCV G2, G3 or G4 infection.
Conclusion
This large, international study found that IL28B rs12979860 genotype is significantly associated with liver fibrosis stage in CHC patients with HCV G1 infection. This association was evident in European Caucasians but not in G1-infected patients from Asia, Latin America or the Middle East.
Electronic supplementary material
The online version of this article (doi:10.1186/s40064-016-3663-6) contains supplementary material, which is available to authorized users.
Keywords: Hepatitis C, Gen-C study, IL28B, Advanced fibrosis, Chronic hepatitis C
Background
Host interleukin-28B (IL28B) genotype is associated with spontaneous clearance of acute hepatitis C virus (HCV) infection, response to interferon (IFN)-based treatment, and with the development of hepatocellular carcinoma (HCC) in patients with chronic hepatitis C (CHC) (Thomas et al. 2009; Ge et al. 2009; Zhang et al. 2016). Genome-wide association studies have identified single-nucleotide polymorphisms (SNPs) in close proximity to the IL28B gene that encode for IFN lambda (IFN-λ), with strong associations with spontaneous or IFN-induced clearance of HCV (Thomas et al. 2009; Ge et al. 2009; Rauch et al. 2010; Suppiah et al. 2009; Tanaka et al. 2009). In particular, a host IL28B rs12979860 CC genotype is associated with the highest, and TT genotype the lowest, rates of response to IFN-based therapy (Thomas et al. 2009; Ge et al. 2009; Mangia et al. 2010; Asselah et al. 2012; De Nicola et al. 2012; Poordad et al. 2012; Lawitz et al. 2013; Susser et al. 2014). The T allele also increases the risk of HCC associated with HCV (Zhang et al. 2016). In addition, differences in the distribution of IL28B genotypes explain, in part, historical observations of low sustained virological response (SVR) rates in Black or Latino patients and high SVR rates in Asian patients (Muir et al. 2004; Yu et al. 2008; Rodriguez-Torres et al. 2009).
Despite the established role of host IL28B genotype as a predictor of response to therapy, the association between IL28B polymorphisms and the natural history of HCV is currently the subject of debate. CHC patients with an IL28B CC genotype had higher alanine transaminase (ALT) levels and greater hepatic necro-inflammatory activity at baseline, with worse clinical outcomes over 4 years of follow-up than patients with non-CC genotypes (Noureddin et al. 2013). However, the same study observed no difference in the rate of fibrosis progression between patients with CC and non-CC genotypes. Other studies have yielded contradictory results on the association between IL28B genotype and fibrosis progression (Asselah et al. 2012; Falleti et al. 2011; Fabris et al. 2011; Marabita et al. 2011; Di Marco et al. 2012; D’Ambrosio et al. 2014).
While the efficacy of recently introduced, IFN-free treatment regimens is not affected by host IL28B genotype, their use is limited to patients with advanced fibrosis in many jurisdictions (Lawitz et al. 2013; Jacobson et al. 2013). Therefore, predictors of fibrosis progression may provide a valuable tool for selection of patients eligible for treatment. In addition, dual peginterferon alfa/ribavirin therapy and protease inhibitor-based triple therapy continue to be used in many regions (Pawlotsky 2014). Given that advanced fibrosis is associated with lower rates of SVR to IFN-based therapies, the identification factors predictive of fibrosis stage has implications for treatment selection and optimization (Poordad et al. 2012; Lawitz et al. 2013; Hadziyannis et al. 2004; Bonnet et al. 2014; Manns et al. 2014; Jacobson et al. 2014).
The primary objective of the Gen-C study was to investigate associations between IL28B genotype and fibrosis stage in CHC patients, in addition to gathering further information on the distribution of IL28B genotypes by HCV genotype, geographic region and ethnicity. The final results from treatment-naive patients are reported here.
Methods
Patients and study design
Gen-C is a large, international, observational study of adults with CHC (clinicaltrials.gov, trial identifier NCT01675427). Patients were excluded if they had hepatitis B virus co-infection, a history of decompensated liver disease, major organ transplantation or end-stage renal disease. Only treatment-naive patients were included in the present analysis. Enrolment was at the discretion of the investigator.
The primary objective of the Gen-C study was to investigate the relationship between IL28B genotypes and liver fibrosis stage in patients with CHC.
Secondary objectives included relationships between IL28B genotypes and liver inflammation, ALT levels and patient demographics, and the distribution of IL28B genotypes by HCV genotype, geographic region and ethnicity.
Data collection
Patients who provided written, informed consent made one study visit for blood sample collection, which was analysed in a central laboratory in Germany or Italy.
A 5 ml sample of whole blood was collected in a tube containing ethylenediaminetetraacetic acid. After collection, the tube was gently inverted 10 times and was not to be centrifuged. A bar-coded label was then applied to the tube and the sample was stored at −20 °C before being packed in dry ice and shipped by courier to the central laboratory. Samples were stored at −20 °C until DNA extraction then were thawed overnight (i.e., for approximately 16 h) and rolled for approximately 30 min before opening.
Standard polymerase chain reaction technology was used to genotype SNPs near IL28B (rs12979860 [CC, TC and TT] and rs8099917 [TT, GT or GG]) and inosine triphosphate pyrophosphatase (ITPA) (rs1127354 [AA, CA or CC] and rs7270101 [CC, AC or AA]). Positive controls for IL28B rs12979860 TC and rs8099917 TG were obtained from TIB MOLBIOL GmbH, Berlin, Germany. Nucleic acid purification was performed using the MagNa Pure 96 System (Roche Diagnostics, Mannheim, Germany). Genomic DNA was extracted from a 50 µL sample by adding MagNa Pure96 to make up an eluate of 100 µL. Genotype was then determined by commercial genotyping assays (LightMix Kit rs12979860 IL28B, LightSNiP rs8099917 IL28B, LightSNiP rs7270101 ITPA, LightSNiP rs1127354 ITPA; Roche Diagnostics, GmbH) using an eluate of 5 µL.
The results of previous invasive or non-invasive fibrosis assessments were entered in the electronic Case Report Form. Investigators recorded the date, type (invasive or non-invasive) and result of the assessment. Fibrosis stage was documented categorically (Cirrhosis, Transition to cirrhosis, Advanced fibrosis-non-cirrhotic, Mild/minimal fibrosis, No fibrosis). Cirrhosis/transition to cirrhosis was defined by biopsy (Ishak 4–6, METAVIR 3–4, Batts and Ludwig 3–4, Knodell 3–4 or Scheuer 3–4) or non-invasive assessment.
Statistical analyses
Enrolment of 1500 treatment-naive and 1500 treatment-experienced patients with evaluable data was estimated to provide 80% power for a Chi square test to detect an association between liver fibrosis stage (five degrees of freedom) and IL28B genotype (three degrees of freedom) at a significance level of 0.05. The final enrolment target of 4000–6000 patients was arrived at after allowing for patient dropout and patients without evaluable data.
The analysis population was defined as all patients with blood samples and available data for IL28B SNPs. For analysis of the primary endpoint, patients were categorized as having cirrhosis/transition to cirrhosis or no cirrhosis. The relationship between cirrhosis status (cirrhosis/transition to cirrhosis vs no cirrhosis) and number of rs12979860 IL28B T alleles was investigated by Cochran-Armitage trend test. The relationship between IL28B genotype and all other fibrosis measurements (e.g. METAVIR fibrosis stage) was investigated using the Jonckheere-Terpstra trend test. The same methods were used to investigate the relationship between IL28B and other continuous, ordinal or binary variables, while the Pearson Chi square test was used to compare the association between IL28B genotypes and nominal variables.
Relationships between baseline characteristics and cirrhosis status were explored by multivariate logistic regression (MLR) analysis. Variables considered in the analysis were: alcohol consumption, ALT ratio, aspartate transaminase (AST) ratio, age, autoimmune disease, body mass index (BMI), body weight, ethnic origin, glucose intolerance/diabetes, HCV genotype, HCV RNA level, HIV–HCV co-infection, liver disease other than CHC, platelets, region of enrolment and years since HCV infection.
Results
A total of 3003 treatment-naive patients were enrolled at 213 centres across 30 countries between August 2011 (first patient) and September 2013 (last patient last visit) (Fig. 1). Overall, 87 patients were excluded, and therefore the analysis population comprised 2916 patients.
Fig. 1.
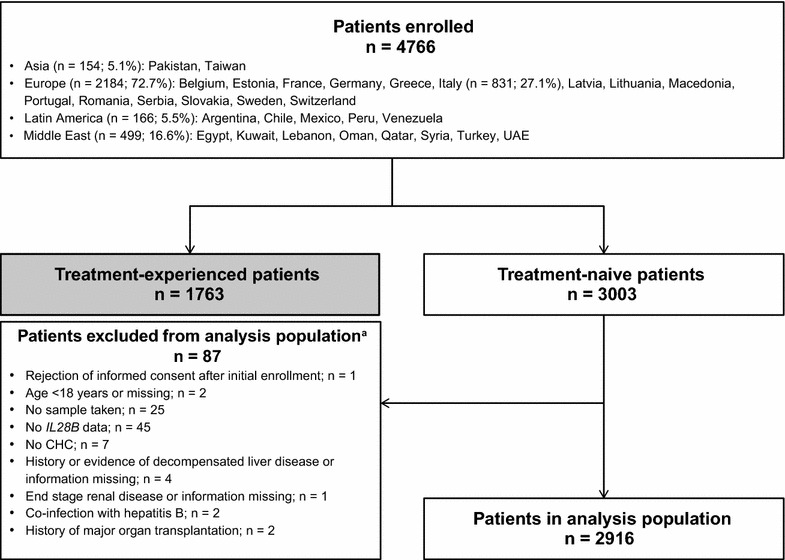
Patient flow diagram. aPatients may have been excluded for more than one reason
Among the total population of 2916 patients, a fibrosis assessment was conducted in 2902 individuals (99.5%), of whom 26.1% were diagnosed with cirrhosis/transition to cirrhosis (Table 1). The majority of patients were male, Caucasian and had HCV G1 infection, with similar characteristics across IL28B rs12979860 genotypes (Table 1). More patients with CC or CT compared with TT genotypes had a high viral load and elevated ALT levels. The distribution of HCV genotypes varied by region (Fig. 2).
Table 1.
Patient demographics and background disease characteristics
| IL28B rs12979860 genotype | ||||
|---|---|---|---|---|
| CC (n = 978) | CT (n = 1518) | TT (n = 420) | P valuee | |
| Mean age, years ± SD | 46.8 ± 13.6 | 47.3 ± 13.2 | 46.8 ± 12.6 | 0.6973e |
| G1 (n = 1702) | 47.9 ± 13.3 | 48.5 ± 13.2 | 48.1 ± 12.2 | 0.6602e |
| G2 (n = 323) | 54.7 ± 13.1 | 55.1 ± 12.6 | 51.1 ± 15.9 | 0.5182e |
| G3 (n = 574) | 39.2 ± 9.8 | 39.6 ± 10.3 | 39.0 ± 9.4 | 0.6864e |
| G4 (n = 260) | 45.5 ± 13.2 | 44.3 ± 10.2 | 46.0 ± 10.4 | 0.7789e |
| Male, n (%) | 582 (59.5) | 802 (52.8) | 224 (53.3) | 0.0049f |
| Ethnic origin, n (%) | <0.0001g | |||
| Asian | 89 (9.1) | 83 (5.5) | 9 (2.1) | |
| Black | 5 (0.5) | 24 (1.6) | 19 (4.5) | |
| Caucasian | 860 (87.9) | 1349 (88.9) | 373 (88.8) | |
| Othera | 24 (2.5) | 62 (4.1) | 19 (4.5) | |
| Region, n (%) | <0.0001g | |||
| Asia | 78 (8.0) | 66 (4.3) | 8 (1.9) | |
| Europe | 721 (73.7) | 1095 (72.1) | 303 (72.1) | |
| Middle East | 138 (14.1) | 272 (17.9) | 75 (17.9) | |
| Latin America | 41 (4.2) | 85 (5.6) | 34 (8.1) | |
| BMI, kg/m2, mean ± SD | 25.8 ± 4.4 | 26.0 ± 4.5 | 26.0 ± 4.7 | 0.2617e |
| HCV genotype, n (%) | <0.0001g | |||
| Known genotype | 957 (97.9) | 1501 (98.9) | 414 (98.6) | |
| G1b | 500 (52.2) | 930 (62.0) | 272 (65.7) | |
| G2b | 126 (13.2) | 160 (10.7) | 37 (8.9) | |
| G3b | 232 (24.2) | 277 (18.5) | 65 (15.7) | |
| G4b | 92 (9.6) | 129 (8.6) | 39 (9.4) | |
| G5/6b | 7 (0.7) | 5 (0.3) | 1 (0.2) | |
| Unknown/missing | 21 (2.1) | 17 (1.1) | 6 (1.4) | |
| Duration of HCV infection, mean years ± SD | 14.0 ± 12.3 | 14.0 ± 12.6 | 13.0 ± 12.4 | 0.1860e |
| G1 (n = 1615) | 14.8 ± 13.1 | 14.5 ± 13.0 | 12.7 ± 12.5 | 0.0536e |
| G2 (n = 303) | 16.6 ± 12.9 | 15.9 ± 13.5 | 16.6 ± 15.8 | 0.5129e |
| G3 (n = 552) | 10.9 ± 9.6 | 11.8 ± 10.7 | 11.3 ± 8.5 | 0.4443e |
| G4 (n = 249) | 13.7 ± 10.6 | 13.1 ± 10.4 | 14.8 ± 13.8 | 0.9733e |
| HIV–HCV co-infection, n (%) | 38 (3.9) | 49 (3.2) | 15 (3.6) | 0.6000f |
| Autoimmune disease, n (%) | 23 (2.4) | 46 (3.0) | 6 (1.4) | 0.6433f |
| Liver disease other than CHC, n (%) | 34 (3.5) | 35 (2.3) | 13 (3.1) | 0.3728f |
| Glucose intolerance/diabetes, n (%) | 73 (7.5) | 125 (8.2) | 33 (7.9) | 0.6614f |
| Alcohol consumption, mean units/week ± SD | 1.83 ± 8.7 | 1.10 ± 5.2 | 0.87 ± 3.5 | 0.1423e |
| Previous/current drug use/methadone or substitute therapy, n (%) | 289 (29.6) | 361 (23.8) | 96 (22.9) | 0.0013f |
| Smoking, n (%)b | 0.0070g | |||
| Never | 496 (50.9) | 887 (58.5) | 230 (54.9) | |
| Previous | 141 (14.5) | 192 (12.7) | 56 (13.4) | |
| Current | 338 (34.7) | 438 (28.9) | 133 (31.7) | |
| ITPA rs1127354, CC, n (%) | 818 (83.6) | 1338 (88.1) | 372 (88.6) | 0.0020f |
| ITPA rs7270101, AA, n (%) | 793 (81.1) | 1254 (82.6) | 342 (81.4) | 0.6563f |
| Mean ALT, IU/L ± SD | 104.1 ± 103.5 | 83.4 ± 79.2 | 72.9 ± 63.8 | <0.0001e |
| Mean ALT ratio ± SDc | 2.28 ± 2.07 | 1.88 ± 1.55 | 1.65 ± 1.31 | <0.0001e |
| G1 (n = 1671) | 2.09 ± 1.94 | 1.77 ± 1.38 | 1.63 ± 1.30 | 0.0007e |
| G2 (n = 312) | 2.42 ± 2.58 | 1.96 ± 1.96 | 1.21 ± 0.829 | 0.0061e |
| G3 (n = 565) | 2.80 ± 2.15 | 2.32 ± 1.91 | 2.05 ± 1.62 | <0.0001e |
| G4 (n = 247) | 2.00 ± 1.75 | 1.66 ± 1.11 | 1.65 ± 1.09 | 0.3885e |
| Mean HCV RNA, log10 IU/mL ± SD | 5.94 ± 0.99 | 5.83 ± 0.81 | 5.68 ± 0.83 | <0.0001e |
| HCV RNA > 800,000 IU/mL, n (%)bd | 607/959 (63.3) | 764/1498 (51.0) | 179/416 (43.0) | <0.0001f |
| G1 | 320/495 (64.6) | 493/921 (53.5) | 116/268 (43.3) | <0.0001f |
| G2 | 81/124 (65.3) | 80/157 (51.0) | 23/37 (62.2) | 0.1853 |
| G3 | 152/231 (65.8) | 131/276 (47.5) | 22/65 (33.8) | <0.0001 |
| G4 | 44/92 (47.8) | 53/128 (41.4) | 14/39 (35.9) | 0.1792 |
| Assessment of cirrhosis, n (%) | 0.0723g | |||
| Biopsy | 295 (30.2) | 419 (27.6) | 114 (27.1) | |
| Noninvasive | 646 (66.0) | 1046 (68.9) | 289 (68.8) | |
| Both | 37 (3.8) | 46 (3.0) | 12 (2.9) | |
| None | 1 (0.1) | 7 (0.5) | 5 (1.2) | |
| Fibrosis stage (biopsy or noninvasive), n (%) | ||||
| Assessment made | 976 (99.8) | 1511 (99.5) | 415 (98.8) | |
| Cirrhosis/transition to cirrhosisb | 248 (25.4) | 390 (25.8) | 119 (28.7) | 0.2702f |
| No cirrhosisb | 728 (74.6) | 1121 (74.2) | 296 (71.3) | |
| Missing | 2 (0.2) | 7 (0.5) | 5 (1.2) | |
ALT alanine transaminase, IL28B interleukin-28B, ITPA inosine triphosphate pyrophosphatase
aEthnic origin was selected from a drop-down menu in the CSR; “Other” may include patients who are Hispanic, Latino, or identified themselves as Mixed Race
bPercentages calculated for patients with available data only
cALT ratio is ALT divided by upper limit of normal range
dIncludes HCV RNA categories reported as ≤800,000 or >800,000 IU/mL
eJonckheere-Terpstra Test for a trend of a continuous variable across the three IL28B genotype categories
fCochran-Armitage Trend Test for a trend in binomial proportions across the three IL28B genotype categories
gPearson Chi square test
Fig. 2.
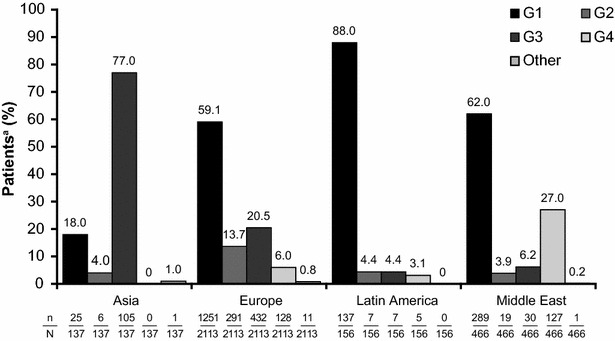
Distribution of IL28B rs12979860 genotype by HCV genotype and geographic region. The distribution of HCV genotype showed significant variation (p < 0.0001) by region of enrolment. p < 0.0001b for association between HCV genotype and region of enrolment. aPatients with multiple genotypes were assigned to disjoint groups according to the following hierarchy: G1, G4, G3, G2, other bPearson’s Chi square test
IL28B genotype and liver fibrosis stage
Among HCV G1-infected patients, the prevalence of cirrhosis/transition to cirrhosis increased with the number of rs12979860 T alleles overall (p = 0.0018) (Fig. 3a), in Caucasians (p = 0.0010) (Fig. 3b) and in European Caucasians (p = 0.0023) (Fig. 3c).
Fig. 3.
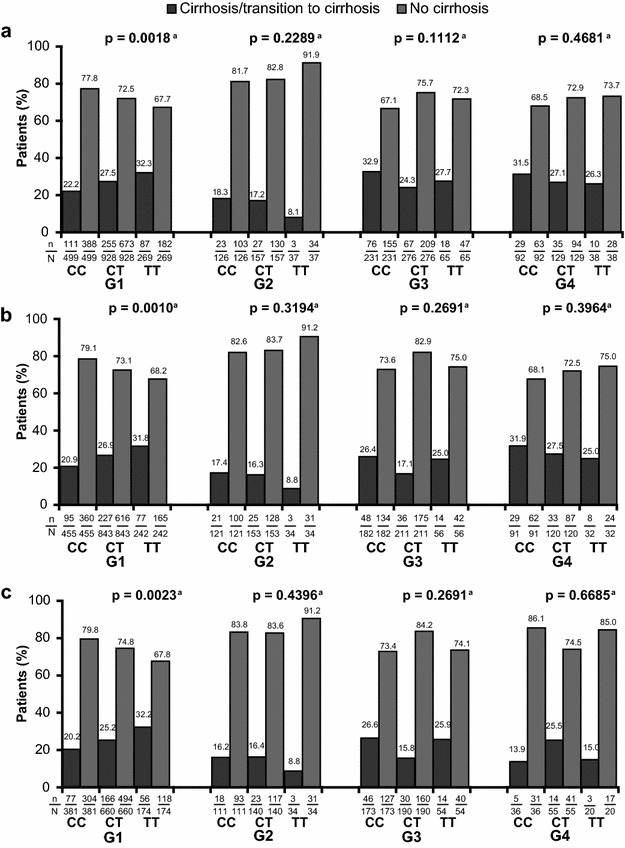
The association between IL28B rs12979860 genotype and liver fibrosis by HCV genotype overall (a), in Caucasian patients with HCV G1 infection (b), and in European Caucasian patients with HCV G1 infection (c). Significant associations were found between IL28B rs12979860 genotype and liver fibrosis in patients with HCV G1 infection (p = 0.0018) (a), in Caucasian treatment-naive patients with HCV G1 infection (p = 0.0010) (b), and in treatment-naive European Caucasian patients with HCV G1 infection (p = 0.0023) (c). aCochran-Armitage Trend Test for a trend in binomial proportions across the three IL28B genotypes
No statistically significant associations were observed for other HCV genotypes. Among HCV G1 patients enrolled at European study sites (n = 1245), an association was observed between the prevalence of cirrhosis/transition to cirrhosis and the number of rs12979860 T alleles (p = 0.0030, Table 2). For patients enrolled in Asia, Latin America or the Middle East, no significant associations were observed (Table 2; Additional file 1: Tables S1–S4). However, these results should be interpreted with caution due to low patient numbers.
Table 2.
Association between liver fibrosis and IL28B rs12979860 genotype in patients with HCV G1 infection, by region of enrolment
| Patients, n (%) | IL28B rs12979860 genotype | |||
|---|---|---|---|---|
| CC | TC | TT | p valuea | |
| Asia (n = 25) | ||||
| Cirrhosis/transition to cirrhosis | 8 (50.0) | 5 (55.6) | ND | |
| No cirrhosis | 8 (50.0) | 4 (52.3) | ND | 0.7896 |
| European (n = 1245) | ||||
| Cirrhosis/transition to cirrhosis | 78 (20.2) | 168 (24.8) | 57 (31.7) | |
| No cirrhosis | 309 (79.8) | 510 (75.2) | 123 (68.3) | 0.0030 |
| Latin America (n = 137) | ||||
| Cirrhosis/transition to cirrhosis | 9 (29.0) | 25 (33.8) | 10 (31.3) | |
| No cirrhosis | 22 (71.0) | 49 (66.2) | 22 (68.8) | 0.8547 |
| Middle East (n = 289) | ||||
| Cirrhosis/transition to cirrhosis | 16 (24.6) | 57 (34.1) | 20 (35.1) | |
| No cirrhosis | 49 (75.4) | 110 (65.9) | 37 (64.9) | 0.2022 |
ND no data available
aCochran-Armitage trend test for a trend in binomial proportions across the three IL28B genotype categories
MLR analyses
Older age, higher BMI, HCV G1 infection (vs G2), higher AST ratio, lower platelet count, enrolment at an Asian/Middle Eastern site (vs European), liver disease other than CHC and glucose intolerance/diabetes were significantly associated with an increased risk of cirrhosis/transition to cirrhosis (Fig. 4a). IL28B genotype was not associated with cirrhosis/transition to cirrhosis in the final MLR model. In contrast, when only HCV G1-infected patients were considered, rs12979860 genotype (CT or TT vs CC) was significantly associated with cirrhosis/transition to cirrhosis (Fig. 4b). When the analysis was further restricted to European HCV G1 patients, rs8099917 GG and TG genotype (vs TT) was significantly associated with cirrhosis/transition to cirrhosis (Fig. 4c).
Fig. 4.
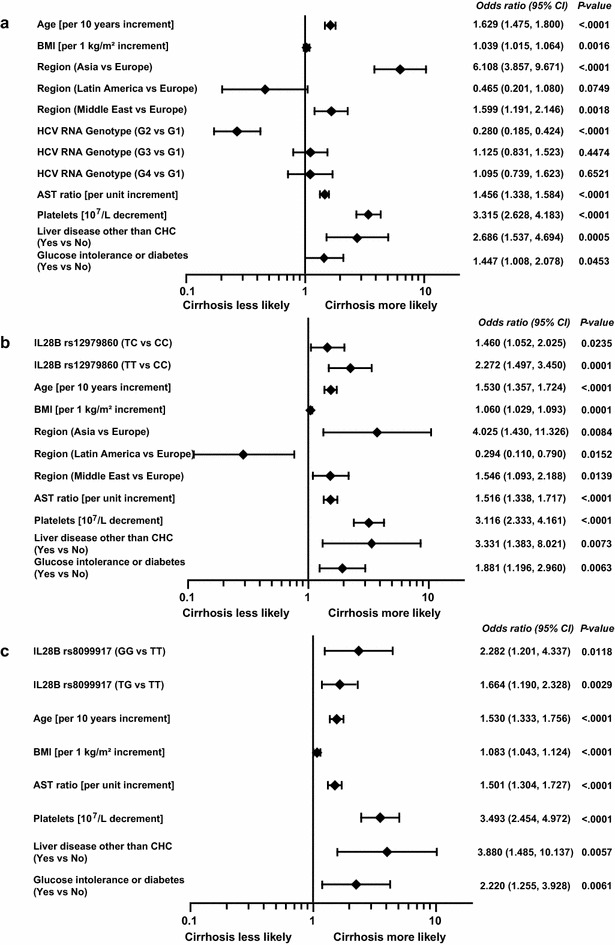
Multiple logistic regression analysis for cirrhosis or transition to cirrhosis in all patients (a), all patients with HCV G1 infection (b), and all European Caucasian patients with HCV G1 infection (c). AST aspartate transaminase; MLR multiple logistic regression
IL28B rs12979860 genotype, serum ALT levels and necro-inflammatory grade
A statistically significant decrease in mean ALT ratio was observed with an increasing number of rs12979860 T alleles for G1 (p = 0.0007), G2 (p = 0.0061) and G3 (p < 0.0001) but not G4 patients (Additional file 1: Table S5).
No association was observed between IL28B genotype and METAVIR fibrosis stage or necro-inflammatory grade (Additional file 1: Tables S6 and S7), or liver stiffness (Additional file 1: Table S8) overall or when stratified by HCV genotype. There was however significant association between IL28B genotype and AST to platelet ratio index (APRI) score and Fibrosis-4 (FIB-4) score (age, AST, platelet count and ALT) in the overall population and in the subgroup of patients with G2 and G3 infection (Additional file 1: Tables S9 and S10).
rs12979860 genotype distributions
In general, an rs12979860 CT genotype was more common than a CC or TT genotype (Fig. 5a) and there was a significant association between HCV genotype and IL28B genotype (p < 0.0001), with the CC genotype occurring more frequently in G2 and G3 patients than in G1 and G4 patients.
Fig. 5.
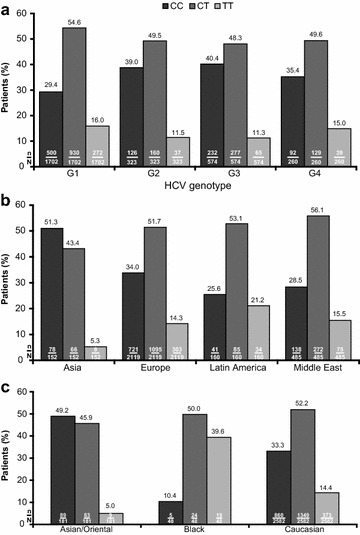
Distribution of IL28B rs12979860 genotype according to HCV genotype. The distribution of IL28B rs12979860 genotype varied significantly by HCV genotype (a), region of enrolment (b), and ethnicity (c). p < 0.0001a for the association between HCV genotype and IL28B rs12979860 genotype, p < 0.0001a for the association between geographic region and IL28B rs12979860 genotype, and p < 0.0001a for the association between ethnicity and IL28B rs12979860 genotype. aPearson’s Chi square test
rs12979860 genotype was significantly associated with region of enrolment overall (p < 0.0001, Fig. 5b) and in HCV G1 and G4 patients [p < 0.0001 and p = 0.0467, respectively (Table 3)]. rs12979860 distribution was not, however, homogenous across the countries within each region (Additional file 1: Table S8).
Table 3.
Distribution of IL28B rs12979860 genotype by region of enrolment and HCV RNA genotype
| HCV genotype | IL28B rs12979860 genotype | p valuea | ||
|---|---|---|---|---|
| CC | CT | TT | ||
| G1 (n = 1702) | ||||
| Asia (n = 25) | 16 (64.0) | 9 (36.0) | 0 (0.0) | |
| Europe (n = 1251) | 388 (31.0) | 680 (54.4) | 183 (14.6) | |
| Middle East (n = 289) | 65 (22.5) | 167 (57.8) | 57 (19.7) | |
| Latin America (n = 137) | 31 (22.6) | 74 (54.0) | 32 (23.4) | |
| Total (n = 1702) | 500 (29.4) | 930 (54.6) | 272 (16.0) | <0.0001 |
| G2 (n = 323) | ||||
| Asia (n = 6) | 4 (66.7) | 2 (33.3) | 0 (0.0) | |
| Europe (n = 291) | 111 (38.1) | 144 (49.5) | 36 (12.4) | |
| Middle East (n = 19) | 7 (36.8) | 12 (63.2) | 0 (0.0) | |
| Latin America (n = 7) | 4 (57.1) | 2 (28.6) | 1 (14.3) | |
| Total (n = 323) | 126 (39.0) | 160 (49.5) | 37 (11.5) | 0.3572 |
| G3 (n = 574) | ||||
| Asia (n = 105) | 45 (42.9) | 52 (49.5) | 8 (7.6) | |
| Europe (n = 432) | 177 (41.0) | 200 (46.3) | 55 (12.7) | |
| Middle East (n = 30) | 8 (26.7) | 20 (66.7) | 2 (6.7) | |
| Latin America (n = 7) | 2 (28.6) | 5 (71.4) | 0 (0.0) | |
| Total (n = 574) | 232 (40.4) | 277 (48.3) | 65 (11.3) | 0.2041 |
| G4 (n = 260) | ||||
| Asia (n = 0) | 0 | 0 | 0 | |
| Europe (n = 128) | 37 (28.9) | 64 (50.0) | 27 (21.1) | |
| Middle East (n = 127) | 53 (41.7) | 62 (48.8) | 12 (9.4) | |
| Latin America (n = 5) | 2 (40.0) | 3 (60.0) | 0 (0.0) | |
| Total (n = 260) | 92 (35.4) | 129 (49.6) | 39 (15.0) | 0.0467 |
aPearson’s Chi square test for differences in IL28B rs12979860 genotype by variables
rs12979860 genotype was significantly associated with patient ethnic origin (p < 0.0001); CC was the most common genotype in Asian patients, but was infrequent in Black patients (Fig. 5c).
Discussion
The primary finding of the Gen-C study was a positive association between an increasing number of rs12979860 T alleles and the prevalence of cirrhosis/transition to cirrhosis in treatment-naïve G1 patients. While the positive association between T allele and fibrosis stage observed for HCV G1 broadly supports findings from several previous studies, it should be taken into account that these studies did not differentiate between HCV genotypes, and also included relatively small numbers of patients (Falleti et al. 2011; Fabris et al. 2011; Di Marco et al. 2012).
There is also evidence that contradicts the results of the present study: analyses in G1 patients have reported that a CC genotype is associated with a higher prevalence of cirrhosis/transition to cirrhosis (Abe et al. 2010), and that there is no relationship between IL28B and fibrosis stage (D’Ambrosio et al. 2014; Bochud et al. 2012). The studies by D’Ambrosio et al. and Abe et al. measured fibrosis by biopsy; however, they included relatively small numbers of patients (D’Ambrosio et al. 2014; Abe et al. 2010). While the study by Bochud et al. (2012) included a similar number (n = 919) of Caucasian G1-infected, treatment-naive patients, the median duration of HCV infection was longer (21 vs ~14 years), and more patients had HIV–HCV co-infection (5 vs 3.5%). Two further studies found no association between rs12979860 genotype and fibrosis stage (Noureddin et al. 2013; Marabita et al. 2011).
Associations between IL28B genotype and liver fibrosis were restricted to HCV G1 infection in the present study. When the data were analysed by geographic region, the association between IL28B genotype and fibrosis status remained statistically significant in the large subgroup of European Caucasian patients with G1 infection but was not statistically significant in other populations. The lack of a relationship in the smaller subgroup of Asian, Latin American and Middle Eastern patients with G1 infection may simply be due to the smaller sample size, or may be due to factors related to ethnicity that were not measured in this analysis. Interestingly there was also a significant association between IL28B genotype and APRI score and FIB-4 score in the overall population and in patients with G2 and G3 infection, but not in those with G1 infection. Strong conclusions cannot be drawn regarding the overall association between IL28B genotype and fibrosis status, as significant associations between C allele frequency and cirrhosis/transition to cirrhosis have been reported previously (Bochud et al. 2012; Rembeck et al. 2012).
The primary immune response to HCV infection is mediated predominantly through IFN-λ cytokines and may be influenced by IL28B genotype (Thomas et al. 2012; Watanabe et al. 2013). Patients with a “favourable” rs12979860 CC genotype are more likely to experience spontaneous viral clearance and respond to IFN-based therapy (Thomas et al. 2009; Ge et al. 2009), while those with the T allele may be at increased risk of HCV-related HCC (Zhang et al. 2016). Interestingly, there is a well-established association between this “favourable” genotype for viral clearance (CC), and increased markers of intra-hepatic inflammatory response to HCV infection (Noureddin et al. 2013; D’Ambrosio et al. 2014; Abe et al. 2010; Bochud et al. 2012; Rembeck et al. 2012). The presence of significantly higher necro-inflammatory activity and serum ALT levels in patients with CC genotypes has been interpreted as an indication of an enhanced immune response in patients with CC genotypes as compared with non-CC genotypes (Noureddin et al. 2013; D’Ambrosio et al. 2014; Rembeck et al. 2012). A more vigorous immune response would be expected to lead to more rapid fibrosis progression over time in patients who do not experience spontaneous viral clearance. However, an association with liver fibrosis stage and IL28B genotype is less well established (Noureddin et al. 2013; D’Ambrosio et al. 2014; Abe et al. 2010; Bochud et al. 2012; Rembeck et al. 2012). In the current study, the CC genotype was associated with higher ALT ratios and a lower prevalence of cirrhosis/transition to cirrhosis in patients with HCV G1 infection. These results support the interpretation that the “favourable” CC genotype is associated with a more vigorous antiviral immune response but not with higher incidence of fibrosis. Unfortunately, inflammatory grade was not available for many patients, which limits the strength of this conclusion.
Recently, it has been shown that different combinations of IL28B genotype and toll-like receptor-2 (TLR-2) variants are associated with different manifestations of HCV-related liver disease and lymphoproliferative diseases (De Re et al. 2016). The IL28B CC genotype exerts a protective effect on CHC and the development of cirrhosis and HCC, while the TLR-2 del/del genotype was associated with an increased risk of HCC. In combination, the presence of one or more TLR-2 del alleles abolished the protective effect of the CC genotype. These data suggest that IL28B and TLR-2 are functionally interconnected in HCV disease-specific phenomena.
Baseline factors predictive of cirrhosis/transition to cirrhosis are similar to those reported elsewhere (Noureddin et al. 2013; Falleti et al. 2011; Bochud et al. 2012; Rueger et al. 2015; Wright et al. 2003). It should be noted that attendance at a clinic in Asia (vs Europe) was significantly associated with cirrhosis/transition to cirrhosis in G1 patients, which seems counterintuitive given that the CC genotype was more prevalent in Asian patients. However, this result may be explained by the greater proportion of Asian than European patients with cirrhosis at baseline (~53 vs ~25%).
Overall, the Gen-C study supports previously published frequency distributions of IL28B genotypes. The CC genotype was less common in G1 and G4 than G2 and G3 patients, and the proportions of CC patients in each HCV genotype agreed with previous estimates (De Nicola et al. 2012; Falleti et al. 2011; Bochud et al. 2012; Mottola et al. 2015). The geographic distribution of rs12979860 genotypes also agrees with previous reports, with the highest prevalence of the C allele in Asia, and the highest prevalence of the TT genotype in Latin America (Thomas et al. 2009; Ge et al. 2009). Finally, ethnic associations with s12979860 genotypes agree with previous reports, with the lowest prevalence of the C allele in Black patients, and the highest in Asian patients (Thomas et al. 2009; Ge et al. 2009).
Limitations include the small amount of METAVIR data collected, which tempers conclusions regarding IL28B genotype, inflammatory grade and fibrosis stage. Rapid fibrosis progression is associated with factors other than IL28B genotype, such as alcohol consumption, liver disease other than CHC, HIV–HCV co-infection and glucose intolerance, which, notwithstanding our MLR analysis, could have confounded the results.
In summary, an increasing number of rs12979860 T alleles is associated with an increased prevalence of cirrhosis/transition to cirrhosis in treatment-naïve patients with HCV G1 infection. When the G1 population was categorized geographically, this association was evident in the large subgroup of European Caucasians with HCV G1 infection but not in the smaller subgroups of G1-infected patients from Asia, Latin America or the Middle East. Thus, the presence of the T allele may be used to prioritize treatment for G1 patients who are at increased risk of progressing to cirrhosis.
Authors’ contributions
AM, GRF and FT participated in the study concept and design. VdL, FB, JB, JK, JV, and NR were involved in acquisition of data. DM, FT, GB, AM and GRF were involved in analysis and interpretation of data. AM, VdL, FB, JB, JK, JV, NR, DM and GRF were involved in drafting the manuscript. All authors read and approved the final manuscript.
Acknowledgements
This research was funded by F. Hoffmann-La Roche Ltd, Basel, Switzerland. Support for third-party writing assistance for this manuscript, furnished by Jake Burrell PhD and Blair Jarvis MSc, Health Interactions, was provided by F. Hoffmann-La Roche Ltd, Basel, Switzerland.
Competing interests
AMangia served in advisory committees or review panels for Gilead Sciences, Bristol-Myers Squibb and Achillion, as a speaker for Janssen Cilag and MSD and received research funding from Roche. V De Ledinghen served in advisory committees or review panels for Gilead Sciences, Bristol-Myers Squibb, Janssen Cilag, MSD and AbbVie. F Bailly served in advisory committees or review panels and as a speaker for AbbVie, Bristol-mYers Squibb, Merck, Gilead Sciences and Janssen. J Brahm served in advisory committees or review panels and as a speaker for Gilead Sciences, Bristol-Myers Squibb, AbbVie and Roche. D Messinger is an employee of PROMETRIS GmbH, the clinical research organization providing data management and statistical services to Roche. F Tatsch was an employee of F. Hoffmann-La Roche at the time of the study. Now an employee of AbbVie. G Bakalos is an employee of F. Hoffmann-La Roche. GR Foster has received speaker and consultancy fees from AbbVie, Bristol-Myers Squibb, Merck, Gilead, Janssen, Tekmira, Alnylam and Roche. J Keiss, J Valantinas and N Rasmann declare that they have no competing interests.
Ethics committees
The following ethics committees/institutional review boards, listed by country, considered and approved the study protocol.
Argentina: Comité de Ética en Investigacion, Mar del Plata; Comité de Docencio del Centro Oncologico Integral, Mar del Plata; Comité de Ética DIM Clinica Privada, Ramos Mejía La Matanza; Comité de Docencia e Investigación DIM Clínica Privada, Ramos Mejía La Matanza.
Belgium: Ethisch Comité Uz Gent, Universitair Ziekenhuis Gent, Gent.
Chile: Comité de Ética Cientifico Servicio de Salud Metropolitano Norte, Independencia; Comité de Ética Hospital Clínico Universidad de Chile, Independencia; Comité de Ética Cientifico Servicio de Salud Metropolitano Central, Santiago; Comité de Ética en Investigación Pontificia Universidad Católica de Chile Facultad de Medicina, Santiago.
Egypt: Ain Shams University EC, Cairo; Tanta University EC, Tanta-Gharbia; Menoufiya University National Liver Institute IRB, Shibin El Kom; Theodre Institute, Imbaba Giza; Alexandria University EC, Alexandria; Kasr Eini Internal Medecin Department, Gastroenterology and Hepatology unit EC, Cairo.
Estonia: Tallinn Medical Research Ethics Committee, Tallinn.
France: Comité de Protection des Personnes Sud-Ouest et Outre Mer III, Bordeaux.
Germany: Ethik-Kommission der Friedrich-Alexander Universität (FAU) Erlangen-Nürnberg, Erlangen; Ethik-Kommission der Universität zu Lübeck, Lübeck; Ärztekammer Nordrhein, Düsseldorf; Ethik-Kommission an der Medizinischen Fakultät der Eberhard-Karls-Universität und am Universitätsklinikum Tübingen, Tübingen; Med. Ethik-Kommission II, Mannheim; Geschäftsstelle der Ethikkommission - Universität zu Köln, Köln; Landesärztekammer Baden-Württemberg, Stuttgart; Ethik-Kommission bei der Landesärztekammer Hessen, Frankfurt; Ethik-Kommission der Otto-von-Guericke-Universität Magdeburg, Magdeburg; Ethikkommission an der Medizinischen Fakultät der Universität Rostock, Rostock; Ethik-Kommission - Medizinische Fakultät der Universität Duisburg-Essen, Essen; Ethik-Kommission an der Medizinischen Fakultät der RWTH Aachen, Aachen; Ethik-Kommission Würzburg, Würzburg; Ethik-Kommission Universitätsklinikum Frankfurt, Frankfurt.
Greece: Hippokratio Hospital Institutional Review Board, Thessaloniki; University Hospital of Alexandroupolis Institutional Review Board, Alexandroupolis; University Hospital of Ioannina Institutional Review Board, Ioannina; University Hospital of Larissa Institutional Review Board, Larissa; Hippokratio Hospital Institutional Review Board, Athens; Agioi Anargiroi Hospital of Kifissia Institutional Review Board, Kifissia; Anticancer Hospital Agios Savvas Institutional Review Board, Athens; University Hospital of Patras Institutional Review Board, Patras.
Italy: Comitato Etico Ospedale San Camillo Forlanini, Roma; Comitato Etico Azienda Policlinico Umberto I, Roma; Comitato Etico per la sperimentazione clinica dei medicinali AUSL 8 di Arezzo, Arezzo; Comitato Etico AOU. Maggiore della Carità, Novara; Comitato Etico ASL NA 3 Sud, Brusciano; Comitato Etico IRCCS S. De Bellis, Castellana Grotte; Comitato Etico AOU Policlinico P. Giaccone, Palermo; Comitato Etico AO Treviglio AO Papa Giovanni XXIII, Treviglio; Comitato Etico AOU. Cagliari, Cagliari; Comitato Etico ASL TO2, Torino; Comitato Etico AO Bianchi Melacrino Morelli, Calabria; Comitato Etico ASL BT, Andria; Ufficio Ricerche Cliniche Ospedale San Raffaele IRCCS Lombardia, Milano; Comitato Etico IRCCS AOU.San Martino – IST, Genova; Comitato Etico Provinciale di Modena, Modena; Comitato Etico IRCCS Casa Sollievo della Sofferenza, San Giovanni Rotondo; Comitato Etico AO Ospedale di Circolo Busto Arsizio, Busto Arsizio; Comitato Etico AOU. Policlinico G. Martino, Messina; Comitato Etico Ospedale Luigi Sacco Milano Area A, Milano; Comitato Etico per la Sperimentazione AOU Padova, Padova; Comitato Etico AOU S.Maria della Misericordia – CERU Udine, Udine; Comitato Etico AO San Paolo - Milano Area A, Milano; Comitato Etico Indipendente Policlinico S. Orsola-Malpighi, Bologna; Comitato Etico Fondazione IRCCS Ca’ Granda Ospedale Maggiore Policlinico – Milano Area B, Milano; Comitato Etico ASL NA 1, Napoli; Comitato Etico Scientifico AO Ospedale Niguarda Ca’ Granda – Milano Area C. Milano; Comitato Etico ASL 1 Sassari – ASL Sassari, Sassari; Comitato Etico ASL Caserta, Caserta; Sottocomitato Etico per le sperimentazioni cliniche sull’uomo AUSL 6, Livorno; Comitato Etico ARNAS Garibaldi, Catania; Comitato Etico AO A.Cardarelli, Napoli; Comitato Etico Indipendente AO. Policlinico Consorziale – CEI, Bari; Comitato Bioetico ARNAS Ospedali Civico Di Cristina Benfratelli, Palermo; Comitato Etico AOU. Careggi – CEAVCE, Firenze; Comitato di Etica ASL Salerno, Nocera Inferiore; Comitato Etico Ospedali FBF S.Giuseppe Milano e Sacra Famiglia Erba – Milano Area B, Milano; Comitato Etico Università Cattolica S. Cuore - Policlinico Gemelli, Roma; Comitato Etico Azienda Ospedaliera Sant’Andrea, Roma; Comitato Etico Provinciale di Reggio Emilia, Reggio Emilia; Comitato Etico AUSL di Piacenza, Piacenza; Comitato Etico Aziendale ASL 2 Savonese – CER S.Martino, Savona; Comitato Etico AOU. San Luigi Gonzaga – CEI S.Luigi Gonzaga, Orbassano; Comitato di Etica Università degli studi G. D’Annunzio e ASL 2 Lanciano-Vasto-Chieti – ASL 1 CH, Chieti Scalo; Comitato Etico Provinciale di Parma – CE per Parma, Parma; Comitato Etico AOU Seconda Università di Napoli, Napoli; Comitato Etico AORN. San Giuseppe Moscati, Avellino; Comitato Etico per la sperimentazione clinica Provincia di Rovigo, Rovigo; Comitato Etico Indipendente AOG Brotzu - AOU Cagliari, Cagliari; CE dell’ Azienda Ospedaliero-Universitaria AOU Ospedali Riuniti “Umberto I, Lancisi, Salesi”, Ancona; Comitato Etico IRCCS INMI L. Spallanzani, Roma.
Kuwait: Joint Committee For The Protection of Human Subjects In Research, Safat.
Latvia: Ethics Committee for Clinical Research at Pauls Stradins Clinical University Hospital Development Society, Riga.
Lebanon: American University of Beirut IRB, Hamra; Nabatieh Governmental Hospital IEC, Nabatieh; Hotel Dieu De France IEC, Ashrafieh; Haykel Hospital IEC, Tripoli; Sahel General Hospital IEC, Dahieh; Rafic Hariri University Hospital IRB, Beirut; Commission d’Ethique de la Recherche (CER), Baabda.
Lithuania: State Medicines Control Agency/Lithuanian Bioethics Committee, Vilnius.
Macedonia: Eticka Komisija, Ministerstvo za zdravstvo, Skopje.
Mexico: Comité de Investigación para estudios en Humanos, Mexico City; Comision Nacional de Investigación Científica, Piso.
Oman: College of Medicine and Health Sciences, Sultanate of Oman
Pakistan: Allied Med Ethics, Shan Hospital, Karachi; Ethics Review Committee, South City Hospital, Karachi; Institutional Review Board, Lahore General Hospital, Post Graduate Medical Institute, Lahore; Institutional Review Board, Shaikh Zayed Hospital, Lahore; Ethics Review Committee, Chaudhry Hospital, Gujranwala; Institutional Review Board/Ethics Committee, Doctors Hospital and Medical Center, Lahore; Secretariat Ethical Review Committee (ERC), Faisalabad.
Peru: Comité de Etica del Hospital Rebagliati, Lima; Comité de Etica en Investigacion del Hospital Nacional Guillermo Almenara Irigoyen, Lima;
Portugal: Comissão de Ética para a Saúde do Centro Hospitalar do Baixo Vouga, EPE, Aveiro; Comissão de Ética para a Saúde do Hospital Professor Dr. Fernando da Fonseca, EPE, Amadora; Comissão de Ética para a Saúde do Hospital de Faro, EPE, Faro; Comissão de Ética para a Saúde do Centro Hospitalar do Porto, EPE, Porto; Comissão de Ética para a Saúde do Centro Hospitalar de Lisboa Ocidental, EPE, Lisboa; Comissão de Ética para a Saúde do Hospital Garcia de Orta, EPE, Almada; Comissão de Ética para a Saúde do Centro Hospitalar Lisboa Norte, EPE, Lisboa; Comissão de Ética para a Saúde do Hospital de São João, EPE, Porto.
Qatar: Medical Research Center at Hamad Medical Corporation, Doha.
Romania: Comisia Naţională de Bioetică a Medicamentului şi a Dispozitivelor Medicale, Bucharest.
Serbia: Ethics Committee Clinical Center of Vojvodina, Novi Sad; Ethics Committee Clinical Hospital Center Zvezdara, Belgrade.
Slovakia: Eticka Komisia BSK, Bratislava; Eticka Komisia SZU, Bratislava; Eticka Komisia FNsP JA Reimanna, Presov; Eticka Komisia UNM, Martin; Eticka Komisia un Bratislava Nemocnica Akad. L. Derera, Bratislava; Eticka Komisia FNsP FDR, Banska Bystrica.
Sweden: Regionala Etikprovningsnamnden i Uppsala, Uppsala.
Switzerland: Comitato Etico Cantonale, Bellinzona; Ethikkomission Kantonsspital, St Gallen.
Syria: Ministry of Health Syria, Damascus.
Taiwan: China Medical University and Hospital Research Ethics Committee, Taichung City; Chang Gung Medical Foundation Institutional Review Board, Guishan Township, Taoyuan County; National Taiwan University Hospital Research Ethics Committee, Taipei City; Kaohsiung Medical University Chung-Ho Memorial Hospital Institutional Review Board, Kaohsiung City; Institutional Review Board, Taipei Veterans General Hospital, Taipei City
Turkey: Ankara Universtiesi Tıp Fakultesi Klinik Arastırmalar Etik Kuurlu, Ankara.
UAE: Al Qassimi Clinical Research Center, Sharjah; Medical Research Committee, Dubai Health authority, Dubai.
Venezuela: Comité de Etica Hospital Dr. Domingo Luciani, Caracas; Comité de Ética Hospital Clinico Maracaibo, Maracaibo; Comité de Ética para la Investigación Centro Medico Docente La Trinidad, Caracas; Comité de Bioetica Centro Médico La Floresta, Caracas; Comité de Bioetica para la Investigación Hospital de Clinicas Caracas, Caracas.
Informed consent in studies with human subjects
All procedures followed were in accordance with the ethical standards of the responsible committee on human experimentation (institutional and national) and with the Helsinki Declaration of 1975, as revised in 2008 and in line with the guidelines for good clinical practice set out in the International Conference on Harmonisation (ICH) Tripartite Guideline. All patients included in the study provided written, informed consent.
Abbreviations
- ALT
alanine transaminase
- AST
aspartate transaminase
- APRI
AST to platelet ratio
- CHC
chronic hepatitis C
- HCV
hepatitis C virus
- G
genotype
- IFN
interferon
- IL28B
interleukin 28B
- MLR
multiple logistic regression
- SNP
single nucleotide polymorphism
- SVR
sustained virological response
- TLR
toll-like receptor
Additional file
Additional file 1. List of investigators, and associations between IL28B genotype and liver fibrosis/inflammation in different subgroups and HCV genotypes.
Contributor Information
Alessandra Mangia, Phone: +39 0882 416375, Email: a.mangia@tin.it.
Victor De Ledinghen, Phone: +33 5 57 65 64 39, Email: victor.deledinghen@chu-bordeaux.fr.
François Bailly, Phone: +33 4 26 10 92 07, Email: francois.bailly@chu-lyon.fr.
Javier Brahm, Phone: +56 2 29788350, Email: jbrahm@hcuch.cl.
Jazeps Keiss, Phone: +371 67014595, Email: keiss.jazeps@inbox.lv.
Jonas Valantinas, Phone: +370 52365230, Email: jonas.valantinas@santa.lt.
Nele Rasmann, Phone: +372 6598659, Email: nele.rasmann@keskhaigla.ee.
Diethelm Messinger, Phone: +49 621 861 992 515, Email: Diethelm.Messinger@prometris.com.
Fernando Tatsch, Phone: +1 800 255 5162, Email: fernando.tatsch@abbvie.com.
Georgios Bakalos, Phone: +41 61 688 11 11, Email: george.bakalos@roche.com.
Graham R. Foster, Phone: +44 20 7882 7241, Email: g.r.foster@qmul.ac.uk
References
- Abe H, Ochi H, Maekawa T, Hayes CN, Tsuge M, Miki D, et al. Common variation of IL28 affects gamma-GTP levels and inflammation of the liver in chronically infected hepatitis C virus patients. J Hepatol. 2010;53:439–443. doi: 10.1016/j.jhep.2010.03.022. [DOI] [PubMed] [Google Scholar]
- Asselah T, De Muynck S, Broët P, Masliah-Planchon J, Blanluet M, Bièche I, et al. IL28B polymorphism is associated with treatment response in patients with genotype 4 chronic hepatitis C. J Hepatol. 2012;56:527–532. doi: 10.1016/j.jhep.2011.09.008. [DOI] [PubMed] [Google Scholar]
- Bochud PY, Bibert S, Kutalik Z, Patin E, Guergnon J, Nalpas B, et al. IL28B alleles associated with poor hepatitis C virus (HCV) clearance protect against inflammation and fibrosis in patients infected with non-1 HCV genotypes. Hepatology. 2012;55:384–394. doi: 10.1002/hep.24678. [DOI] [PubMed] [Google Scholar]
- Bonnet D, Guivarch M, Berard E, Combis JM, Remy AJ, Glibert A, et al. Telaprevir- and boceprevir-based tritherapies in real practice for F3-F4 pretreated hepatitis C virus patients. World J Hepatol. 2014;6:660–669. doi: 10.4254/wjh.v6.i9.660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Ambrosio R, Aghemo A, De Francesco R, Rumi MG, Galmozzi E, De Nicola S, et al. The association of IL28B genotype with the histological features of chronic hepatitis C is HCV genotype dependent. Int J Mol Sci. 2014;15:7213–7224. doi: 10.3390/ijms15057213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Nicola S, Aghemo A, Rumi MG, Galmozzi E, Valenti L, Soffredini R, et al. Interleukin 28B polymorphism predicts pegylated interferon plus ribavirin treatment outcome in chronic hepatitis C genotype 4. Hepatology. 2012;55:336–342. doi: 10.1002/hep.24683. [DOI] [PubMed] [Google Scholar]
- De Re V, De Zorzi M, Caggiari L, Lauletta G, Tornesello ML, Fognani E, et al. HCV-related liver and lymphoproliferative diseases: association with polymorphisms of IL28B and TLR2. Oncotarget. 2016 doi: 10.18632/oncotarget.9303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Marco V, Bronte F, Calvaruso V, Capra M, Borsellino Z, Maggio A, et al. IL28B polymorphisms influence stage of fibrosis and spontaneous or interferon-induced viral clearance in thalassemia patients with hepatitis C virus infection. Haematologica. 2012;97:679–686. doi: 10.3324/haematol.2011.050351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabris C, Falleti E, Cussigh A, Bitetto D, Fontanini E, Bignulin S, et al. IL-28B rs12979860 C/T allele distribution in patients with liver cirrhosis: role in the course of chronic viral hepatitis and the development of HCC. J Hepatol. 2011;54:716–722. doi: 10.1016/j.jhep.2010.07.019. [DOI] [PubMed] [Google Scholar]
- Falleti E, Bitetto D, Fabris C, Cussigh A, Fornasiere E, Cmet S, et al. Role of interleukin 28B rs12979860 C/T polymorphism on the histological outcome of chronic hepatitis C: relationship with gender and viral genotype. J Clin Immunol. 2011;31:891–899. doi: 10.1007/s10875-011-9547-1. [DOI] [PubMed] [Google Scholar]
- Ge D, Fellay J, Thompson AJ, Simon JS, Shianna KV, Urban TJ, et al. Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature. 2009;461:399–401. doi: 10.1038/nature08309. [DOI] [PubMed] [Google Scholar]
- Hadziyannis SJ, Sette H, Jr, Morgan TR, Balan V, Diago M, Marcellin P, et al. Peginterferon-alpha2a and ribavirin combination therapy in chronic hepatitis C: a randomized study of treatment duration and ribavirin dose. Ann Intern Med. 2004;140:346–355. doi: 10.7326/0003-4819-140-5-200403020-00010. [DOI] [PubMed] [Google Scholar]
- Jacobson IM, Gordon SC, Kowdley KV, Yoshida EM, Rodriguez-Torres M, Sulkowski MS, et al. Sofosbuvir for hepatitis C genotype 2 or 3 in patients without treatment options. N Engl J Med. 2013;368:1867–1877. doi: 10.1056/NEJMoa1214854. [DOI] [PubMed] [Google Scholar]
- Jacobson IM, Dore GJ, Foster GR, Fried MW, Radu M, Rafalsky VV, et al. Simeprevir with pegylated interferon alfa 2a plus ribavirin in treatment-naive patients with chronic hepatitis C virus genotype 1 infection (QUEST-1): a phase 3, randomised, double-blind, placebo-controlled trial. Lancet. 2014;384:403–413. doi: 10.1016/S0140-6736(14)60494-3. [DOI] [PubMed] [Google Scholar]
- Lawitz E, Mangia A, Wyles D, Rodriguez-Torres M, Hassanein T, Gordon SC, et al. Sofosbuvir for previously untreated chronic hepatitis C infection. N Engl J Med. 2013;368:1878–1887. doi: 10.1056/NEJMoa1214853. [DOI] [PubMed] [Google Scholar]
- Mangia A, Thompson AJ, Santoro R, Piazzolla V, Tillmann HL, Patel K, et al. An IL28B polymorphism determines treatment response of hepatitis C virus genotype 2 or 3 patients who do not achieve a rapid virologic response. Gastroenterology. 2010;139:821–827. doi: 10.1053/j.gastro.2010.05.079. [DOI] [PubMed] [Google Scholar]
- Manns M, Marcellin P, Poordad F, de Araujo ES, Buti M, Horsmans Y, et al. Simeprevir with pegylated interferon alfa 2a or 2b plus ribavirin in treatment-naive patients with chronic hepatitis C virus genotype 1 infection (QUEST-2): a randomised, double-blind, placebo-controlled phase 3 trial. Lancet. 2014;384:414–426. doi: 10.1016/S0140-6736(14)60538-9. [DOI] [PubMed] [Google Scholar]
- Marabita F, Aghemo A, De Nicola S, Rumi MG, Cheroni C, Scavelli R, et al. Genetic variation in the interleukin-28B gene is not associated with fibrosis progression in patients with chronic hepatitis C and known date of infection. Hepatology. 2011;54:1127–1134. doi: 10.1002/hep.24503. [DOI] [PubMed] [Google Scholar]
- Mottola L, Cenderello G, Piazzolla VA, Forte P, Carretta V, Mecenate F, et al. Interleukin-28B genetic variants in untreated Italian HCV-infected patients: a multicentre study. Liver Int. 2015;35:482–488. doi: 10.1111/liv.12630. [DOI] [PubMed] [Google Scholar]
- Muir AJ, Bornstein JD, Killenberg PG. Peginterferon alfa-2b and ribavirin for the treatment of chronic hepatitis C in blacks and non-Hispanic whites. N Engl J Med. 2004;350:2265–2271. doi: 10.1056/NEJMoa032502. [DOI] [PubMed] [Google Scholar]
- Noureddin M, Wright EC, Alter HJ, Clark S, Thomas E, Chen R, et al. Association of IL28B genotype with fibrosis progression and clinical outcomes in patients with chronic hepatitis C: a longitudinal analysis. Hepatology. 2013;58:1548–1557. doi: 10.1002/hep.26506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawlotsky JM. New hepatitis C therapies: the toolbox, strategies, and challenges. Gastroenterology. 2014;146:1176–1192. doi: 10.1053/j.gastro.2014.03.003. [DOI] [PubMed] [Google Scholar]
- Poordad F, Bronowicki JP, Gordon SC, Zeuzem S, Jacobson IM, Sulkowski MS, et al. Factors that predict response of patients with hepatitis C virus infection to boceprevir. Gastroenterology. 2012;143:608–618. doi: 10.1053/j.gastro.2012.05.011. [DOI] [PubMed] [Google Scholar]
- Rauch A, Kutalik Z, Descombes P, Cai T, Di Iulio J, Mueller T, et al. Genetic variation in IL28B is associated with chronic hepatitis C and treatment failure: a genome-wide association study. Gastroenterology. 2010;138:1338–1345. doi: 10.1053/j.gastro.2009.12.056. [DOI] [PubMed] [Google Scholar]
- Rembeck K, Alsiö A, Christensen PB, Färkkilä M, Langeland N, Buhl MR, et al. Impact of IL28B-related single nucleotide polymorphisms on liver histopathology in chronic hepatitis C genotype 2 and 3. PLoS ONE. 2012;7:e29370. doi: 10.1371/journal.pone.0029370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Torres M, Jeffers LJ, Sheikh MY, Rossaro L, Ankoma-Sey V, Hamzeh FM, et al. Peginterferon alfa-2a and ribavirin in Latino and non-Latino whites with hepatitis C. N Engl J Med. 2009;360:257–267. doi: 10.1056/NEJMoa0805062. [DOI] [PubMed] [Google Scholar]
- Rueger S, Bochud PY, Dufour JF, Mullhaupt B, Semela D, Heim MH, et al. Impact of common risk factors of fibrosis progression in chronic hepatitis C. Gut. 2015;64:1605–1615. doi: 10.1136/gutjnl-2014-306997. [DOI] [PubMed] [Google Scholar]
- Suppiah V, Moldovan M, Ahlenstiel G, Berg T, Weltman M, Abate ML, et al. IL28B is associated with response to chronic hepatitis C interferon-alpha and ribavirin therapy. Nat Genet. 2009;41:1100–1104. doi: 10.1038/ng.447. [DOI] [PubMed] [Google Scholar]
- Susser S, Herrmann E, Lange C, Hamdi N, Muller T, Berg T, et al. Predictive value of interferon-lambda gene polymorphisms for treatment response in chronic hepatitis C. PLoS ONE. 2014;9:e112592. doi: 10.1371/journal.pone.0112592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y, Nishida N, Sugiyama M, Kurosaki M, Matsuura K, Sakamoto N, et al. Genome-wide association of IL28B with response to pegylated interferon-alpha and ribavirin therapy for chronic hepatitis C. Nat Genet. 2009;41:1105–1109. doi: 10.1038/ng.449. [DOI] [PubMed] [Google Scholar]
- Thomas DL, Thio CL, Martin MP, Qi Y, Ge D, O’Huigin C, et al. Genetic variation in IL28B and spontaneous clearance of hepatitis C virus. Nature. 2009;461:798–801. doi: 10.1038/nature08463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas E, Gonzalez VD, Li Q, Modi AA, Chen W, Noureddin M, et al. HCV infection induces a unique hepatic innate immune response associated with robust production of type III interferons. Gastroenterology. 2012;142:978–988. doi: 10.1053/j.gastro.2011.12.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Sugauchi F, Tanaka Y, Matsuura K, Yatsuhashi H, Murakami S, et al. Hepatitis C virus kinetics by administration of pegylated interferon-alpha in human and chimeric mice carrying human hepatocytes with variants of the IL28B gene. Gut. 2013;62:1340–1346. doi: 10.1136/gutjnl-2012-302553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright M, Goldin R, Fabre A, Lloyd J, Thomas H, Trepo C, et al. Measurement and determinants of the natural history of liver fibrosis in hepatitis C virus infection: a cross sectional and longitudinal study. Gut. 2003;52:574–579. doi: 10.1136/gut.52.4.574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu ML, Dai CY, Huang JF, Chiu CF, Yang YH, Hou NJ, et al. Rapid virological response and treatment duration for chronic hepatitis C genotype 1 patients: a randomized trial. Hepatology. 2008;47:1884–1893. doi: 10.1002/hep.22319. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Zhu SL, Chen J, Li LQ. Meta-analysis of associations of interleukin-28B polymorphisms rs8099917 and rs12979860 with development of hepatitis virus-related hepatocellular carcinoma. Onco Targets Ther. 2016;9:3249–3257. doi: 10.2147/OTT.S104904. [DOI] [PMC free article] [PubMed] [Google Scholar]


