Abstract
Autophagy, an essential eukaryotic homeostasis pathway, enables sequestration of unwanted, damaged or harmful cytoplasmic components in vesicles called autophagosomes, enabling subsequent lysosomal degradation and nutrient recycling. Autophagosome nucleation is mediated by Class III phosphatidylinositol 3-kinase complexes that include two key autophagy proteins, BECN1/Beclin 1 and ATG14/BARKOR, which form parallel heterodimers via their coiled-coil domains (CCDs). Here we present the 1.46 Å X-ray crystal structure of the anti-parallel, human BECN1 CCD homodimer, which represents BECN1 oligomerization outside the autophagosome nucleation complex. We use circular dichroism and small-angle X-ray scattering (SAXS) to show that the ATG14 CCD is significantly disordered, but becomes more helical in the BECN1:ATG14 heterodimer, although it is less well-folded than the BECN1 CCD homodimer. SAXS also indicates that the BECN1:ATG14 heterodimer is more curved than other BECN1-containing CCD dimers, which has important implications for the structure of the autophagosome nucleation complex. A model of the BECN1:ATG14 CCD heterodimer that agrees well with the SAXS data shows that BECN1 residues at the homodimer interface are also responsible for homodimerization, enabling us to identify ATG14 interface residues. Lastly, we verify the role of BECN1 and ATG14 interface residues in binding by assessing the impact of point mutations of these residues on coimmunoprecipitation of the partner, and demonstrate that these mutations abrogate starvation-induced up-regulation of autophagy, but do not impact basal autophagy. Thus, this research provides insights into structures of the BECN1 CCD homodimer and the BECN1:ATG14 CCD heterodimer, and identifies interface residues important for BECN1:ATG14 heterodimerization and for autophagy.
Keywords: Autophagy, BECN1 / Beclin 1, ATG14 / BARKOR, Intrinsically disordered region, Coiled-coil Domain
Graphical abstract
Autophagy, a catabolic cellular process conserved in all eukaryotes, plays an important role in cell proliferation, differentiation, survival and homeostasis (1–4). In cellular stress conditions such as nutrient deprivation, hypoxia, accumulation of protein aggregates and infection, double-layered membrane vesicles called autophagosomes engulf long-lived proteins, damaged organelles and pathogens (1, 5–8). The autophagosomes fuse with lysosomes to degrade and recycle the enclosed contents (1–4). Aberrant autophagy is implicated in numerous diseases (9–12).
BECN1, also called Beclin 1, was first identified as a BCL2 interacting protein (13). BECN1 is essential for autophagy (14). It is a core component of two mutually exclusive, quaternary class III phosphatidylinositol-3-kinase (PI3KC3) complexes, Complex I and Complex II, each of which comprises of PI3KC3, P150 (a ser/thr kinase that regulates PI3KC3), BECN1, and either ATG14 or UVRAG respectively, to mediate autophagosome nucleation and maturation, respectively (15–19). Several studies have demonstrated that ATG14 binding to BECN1 is important for autophagy (15–20). ATG14 competes with UVRAG to bind to BECN1, thereby dictating the function of the PI3KC3 complex (15–19). Neither BECN1 nor ATG14 have any catalytic activity, but the association of BECN1:ATG14, or BECN1:UVRAG, with the PI3KC3:P150 binary complex increases PI3KC3 catalytic activity, resulting in increased production of phosphatidylinositol-3-phopshate (PI3P) (21), a critical signal for recruitment of other autophagy proteins to the developing autophagosome and for up-regulation of autophagy.
BECN1, a 450-residue protein in humans, is highly conserved in all eukaryotes. Based on multiple structural and biophysical studies, BECN1 is a conformationally-labile protein that consists of four distinct structural domains: (i) a large, poorly-conserved, N-terminal intrinsically disordered region (IDR) that has multiple sub-domains that likely bind different partners and undergo binding-associated conformational changes (22), including a BCL2 Homology 3 Domain comprising residues 105–130 (23–26), which undergoes a dramatic disorder-to-helix conformational change upon binding to BCL2 proteins (27); (ii) a Flexible Helical Domain comprising residues 141–171, which is disordered at its N-terminus and helical at its C-terminus, but becomes fully α-helical upon binding to appropriate partners (28, 29); (iii) a Coiled-Coil Domain (CCD), comprising residues 175–265 that form an anti-parallel homodimer in the absence of other interactions (30); and (iv) a β-α repeat autophagy domain (BARAD), comprising residues 266–450 (31, 32), that is implicated in membrane association (20, 31). The BECN1 CCD enables association with many other CCD-containing proteins, including the autophagy proteins ATG14 and UVRAG.
To date, detailed structural information on ATG14 has not been obtained. The predicted ATG14 domain architecture consists of (i) an N-terminal region which includes a conserved cysteine-rich domain encompassing residues 43–58 (16) that is required for homoligomerization and ER localization (21); (ii) a CCD region encompassing residues 71–180 that is variously predicted to contain either two or three CCDs (15, 16, 18, 33); and (iii) a functionally-defined domain called the BARKOR/ATG14 autophagosome-targeting sequence (BATS) domain, comprising residues 412–492 (21, 33), that includes an IDR comprising residues 442–472 (27), and a C-terminal, amphipathic α-helix (21). The BATS domain senses membrane curvature and binds to early autophagosomal membranes in a PI3P concentration-dependent manner to recruit downstream effectors such as LC3 to mediate autophagy (21, 34). Co-immunoprecipitation (CoIP) experiments performed by different groups indicate that residues 75–95 and residues 148–178 within the larger CCD region are required for BECN1 binding (15, 16, 18, 19).
Recent 28 Å cryo-EM reconstructions of Complex I and Complex II indicate that these quaternary complexes have a similar V-shaped molecular envelope (35). This study also demonstrated that the BECN1:ATG14 or BECN1:UVRAG heterodimers constitute one arm of the V-shaped envelope, while the PI3KC3:P150 binary complex maps to the other arm. Further, the BECN1 CCD forms a parallel heterodimer with either the ATG14 or UVRAG CCDs, with the N-terminal ends of the heterodimers located at the junction of the two arms of the V. The more recent 4.4 Å structure of the VPS34:VPS15:VPS30:VPS38 complex confirms that the BECN1:UVRAG CCD heterodimer has a parallel arrangement and demonstrated that while VPS34 makes minimal interactions with the VPS30:VPS38 heterodimer, the VPS15 WD40 domain makes significant contacts with the VPS30:VPS38 CCD heterodimer (29).
However, structural details of the BECN1:ATG14 CCD interaction are still unavailable. Further, given the low number of hydrophobic residues within the ATG14 CCD, the absence of a clear heptad repeat or leucine zipper that would form a hydrophobic CCD interface, and the moderate conservation of the ATG14 CCD; the identification of ATG14 residues responsible for the interaction with the BECN1 CCD is non-trivial.
Here we present the 1.46 Å resolution X-ray crystal structure of the human BECN1 CCD homodimer. This structure confirms that BECN1 forms an anti-parallel coiled-coil homodimer in the absence of interactions with other CCD-containing binding partners. Further, we investigate the interaction of human BECN1 and ATG14 CCDs by quantifying thermodynamics of binding using isothermal titration calorimetry (ITC), and use circular dichroism (CD) spectroscopy and tandem size-exclusion chromatography – small-angle X-ray scattering (SEC-SAXS) to probe the structure of the ATG14 CCD, both as a monomer, and in complex with the BECN1 CCD. We have built a pseudo-atomic model of a parallel BECN1:ATG14 CCD heterodimer based on optimal packing interactions, and verified it against experimental SAXS data. Based on this model, we identify BECN1 and ATG14 residues at the CCD heterodimer interface. We experimentally evaluate the role of selected BECN1 and ATG14 residues by using CoIP to evaluate binding, and cellular autophagy assays to assess formation of GFP-LC3 labeled autophagosomes (36). The CoIP assays demonstrate that mutation of all BECN1 residues and many of the ATG14 residues selected, adversely impacts heterodimerization. Lastly, we show that residues important for heterodimerization are also important for the starvation-induced increase in cellular autophagy levels. Thus, we have developed an accurate pseudo-atomic model of the BECN1:ATG14 heterodimer and identified interface residues essential for heterodimerization and autophagosome nucleation.
EXPERIMENTAL PROCEDURES
Sequence Analysis
Sequences of ATG14 orthologs from five eukaryotes: Homo sapiens, Rattus norvegicus, Drosophila melanogaster, Arabidopsis thaliana, and Saccharomyces cerevisiae, were identified by BLASTP searches of Genomic RefSeq Protein databases (http://blast.ncbi.nlm.nih.gov/) for each organism. Multiple sequence alignments (Supplementary Fig. S1) of these diverse orthologs were performed with CLUSTALW (37) and displayed using Jalview (http://www.jalview.org/).
Protein Expression and Purification
The human BECN1 CCD (residues 175–265) was cloned into the pET29b expression vector between NdeI and XhoI restriction enzyme sites to enable expression of the BECN1 CCD with a C-terminal His6-tag using Kanamycin as a selection marker. The human ATG14 CCD (residues 88–178) was cloned into the pMBP-parallel-1 expression vector (38) between BamHI and NotI restriction enzyme sites to enable expression of an MBP-ATG14 CCD fusion protein using Ampicillin as a selection marker. E. coli BL21(DE3)pLysS cells were transformed with either one or both of these expression vectors. Expression of each protein individually, or co-expression of both proteins, was induced with 0.5 mM IPTG. The BECN1 CCD-His6 was expressed at 20 °C overnight. However, in order to limit degradation of the ATG14 CCD, MBP-ATG14 CCD expression and the MBP-ATG14 CCD + BECN1 CCD-His6 co-expression was performed at 37 °C for 2 hours. All proteins were first purified from cell lysate by affinity chromatography: two tandem 5 ml HisTrap columns (GE Lifesciences) were used for BECN1 CCD purification, while a 10 ml amylose column was used for MBP-ATG14 CCD purification. For the co-expressed proteins, a 10 ml amylose column was first used to bind the MBP-ATG14 CCD. The MBP-tag was removed by on-column cleavage by adding TEV protease in a 1:10 (w/w) ratio to the MBP-ATG14 CCD protein and incubating at 4 °C for 8–10 hours. Subsequently, two 5 ml tandem HisTrap columns were used to further purify the BECN1 CCD-His6 in complex with the ATG14 CCD. Finally, all proteins were purified to apparent homogeneity by SEC (Supplementary Fig. 2), using either a Superdex 200 16/60 or tandem Superdex 200 10/300 – Superdex 75 HR 10/300 columns (GE Lifesciences). Molecular weights were estimated by Kav calculations based on SEC standards (BioRad).
Crystallization, X-ray Diffraction Data Collection and Structure Solution
BECN1 CCD crystals were grown at 20 °C by hanging drop vapor diffusion from a drop consisting of 1 μl of BECN1 CCD at 5.2 mg / ml in 25 mM Tris, pH 7.5, 150 mM NaCl and 2mM β-mercaptoethanol plus 1 μl of reservoir solution comprised of 39% v/v 2-methyl-2,4-pentadiol, 9% v/v polyethylene glycol 400 and 100 mM Tris buffer, pH 7.5 suspended over a reservoir of 1 ml. Crystals were harvested and cryo-protected in the reservoir solution and then immediately flash-cooled in liquid N2. Diffraction intensities from BECN1 CCD crystals were recorded at 100 K at an X-ray wavelength of 0.9792 Å at beamline 23ID-D of GMCA@APS, Argonne National Laboratories (ANL), Argonne, IL. Images were collected at 1 second exposure per 0.5° crystal rotation per image, in a 360° sweep from a single crystal, on a MARmosaic 4×4 CCD detector (Rayonix) at a crystal-to-detector distance of 350 mm. Intensities were indexed, merged, and scaled using XDS (39). Data statistics are summarized in Table 1. The BECN1 CCD structure was solved by molecular replacement using Phaser-MR (40) with a search model extracted from the rat BECN1 CCD (residues 174–264), PDB code 3Q8T (30). The model was refined using Phenix (41) and model building performed in Coot (42) (Table 1). Coordinates of the final refined model and structure factors have been deposited in the Protein Data Bank with accession number 5HHE.
Table 1.
X-ray diffraction data collection and refinement statisticsa
| Data collection | |
|---|---|
| Wavelength (Å) | 0.97921 |
| Data range (Å) | 40.15–1.46 |
| Space group | C2 |
| Unit Cell | a=58.20 Å, b=71.58 Å, c=58.45 Å, β=112.55° |
| Unique reflections | 38596 (3766) |
| Avg. multiplicity | 7.5 (7.1) |
| Mosaicity | 0.21 |
| Completeness (%) | 99.70 (97.97) |
| CC1/2 | 0.98 (0.86) |
| 1 Rmeas | 0.11 (0.70) |
| I/σI | 15.91 (2.99) |
| Refinement | |
| Model: | |
| Chain A (number of residues) | 93 |
| Chain B (number of residues) | 93 |
| Water molecules | 255 |
| Data Range (Å) | 40.15–1.46 |
| 2Rwork (%) | 17.2 |
| 2,3Rfree (%) | 20.8 |
| Average B-values (Å2): | |
| Main Chain | 22.8 |
| Side Chain | 37.6 |
| Water | 43.0 |
| All Atoms | 32.3 |
| RMSDs from target values: | |
| Bond Lengths (Å) | 0.007 |
| Bond Angles (°) | 0.71 |
| Ramachandran outliers | 0 |
Values in parentheses pertain to the highest resolution shell.
Rmeas = Σhkl (n/n−1)1/2 Σh,i |Ihkl,i−<Ihkkl>|/ΣhklΣh,iIhkl,i.
R factor= Σh, |Fobs−|Fcalc|/Σh|Fobs|.
Test set for Rfree consisted of 5.0 % of data.
ITC
Separately purified BECN1 CCD and ATG14 CCD protein samples were loaded into separate dialysis cassettes, which were simultaneously dialyzed against 2 L of 50 mM HEPES, pH 8.0, 150 mM NaCl and 2 mM β-mercaptoethanol buffer to ensure matched buffers for the ITC experiments. ITC experiments were performed at 20 °C using a Low Volume Gold Nano ITC (TA Instruments). 400 μL of the BECN1 CCD at 0.18 mg/mL was placed in the cell and 32.4 mg/mL ATG14 CCD was titrated into the cell using 20 injections of 2.5 μL each. The data were analyzed using the NanoAnalyze Software (TA Instruments) with an independent model.
CD Spectroscopy
CD spectra were recorded at 4 °C between 195 and 250 nm using a 300 μL quartz cell with a 0.1 cm path length on a Jasco J-815 spectropolarimeter equipped with thermoelectric temperature control. MBP-ATG14 CCD, MBP and the BECN1:ATG14 CCD complex were diluted to 50 μM and dialyzed in CD buffer (10 mM potassium phosphate, pH 7.6, 100 mM ammonium sulfate) overnight. Data were analyzed using CONTIN from the CDpro program suite (43) within the Jasco Spectra Manager software.
Construction of an Atomic Model of the BECN1:ATG14 CCD complex
CCbuilder V 1.0 (44) was used to build a series of parallel heterodimer models to determine the register of the BECN1 and ATG14 helices that would result in optimal packing at the heterodimer interface. Ten different registers of ATG14 were tested, and each was used to build atomistic models of the BECN1:ATG14 CCD heterodimer using the program Protein Structure Prediction Server ((PS)2) Version 3.0, http://ps2v3.life.nctu.edu.tw) (45), which builds a model for protein complexes based on considerations of the packing density in the complex and sequence alignments with known structures. Importantly, the optimal heterodimer packing model identified by CCbuilder corresponded to the model with the highest correlation coefficient produced by (PS)2. Lastly, for comparison, another program, MODELLER (46, 47) was also used to build a coiled-coil model of the BECN1:ATG14 CCD heterodimer, based on the BECN1 CCD homodimer atomic structure and alignment of the ATG14 CCD and BECN1 CCD sequences.
SEC-SAXS Data Collection and Analysis
SEC-SAXS data were recorded at beamline 18-ID of Bio-CAT at APS, ANL, Argonne, IL on a Pilatus 1M detector at a sample-to-detector distance of 3.5 m, covering a momentum transfer range of 0.0036Å−1 < q < 0.4 Å−1. An aliquot of BECN1:ATG14 CCD complex at 5.9 mg/ml was injected onto a SEC column (Superdex 200 GL 10/300) and SAXS data were recorded by exposing the column eluate to the X-ray beam for one second with a periodicity of three seconds. SAXS signal from parts of the diffraction curve immediately preceding the BECN1:ATG14 CCD heterodimer elution peak were selected, averaged and subtracted as the buffer blank from data points within the peak corresponding to the BECN1:ATG14 CCD heterodimer. Data analysis was performed using various programs within the ATSAS program suite (48). Data were processed using PRIMUS (49) and the Rg was calculated from Guinier extrapolation. The pair distribution function P(r), calculated by Fourier inversion of the scattering intensity I(q) using AutoGNOM, was used to calculate the Rg and Dmax, and also for the reconstruction of an ab initio envelope by the application of ten cycles in DAMMIF (50). The resulting bead models were sequentially analyzed using DAMSEL, DAMSUP and DAMAVER, and then filtered using DAMMFILT (51). CRYSOL (52) was used to fit and compare theoretical scattering curves, calculated from either the BECN1 CCD homodimer crystal structure or the BECN1:ATG14 CCD heterodimer model, for comparison with the experimental SAXS curve. The BECN1 CCD homodimer crystal structure and the BECN1:ATG14 CCD heterodimer model were then fit into appropriate ab initio envelopes using the program SUPCOMB (53).
CoIP of Exogenously expressed BECN1 and ATG14 mutants
ATG14 or BECN1 mutants were created by site-directed mutagenesis (Agilent). 5×105 COS7 or MCF7 cells / ml were seeded in a 10 cm culture dish and cultured overnight in DMEM (GIBCO) with 10% FBS (GIBCO) to ~80% confluence. Lipofectamine 2000 (Invitrogen, Carlsbad, CA) was used for transfection according to the manufacturer’s instructions, using a total of 24 μg of plasmids in a 1:1 molar ratio of either Flag-tagged WT BECN1 and HA-tagged WT / mutant ATG14 to COS7 cells or HA-tagged WT ATG14 and Flag-tagged WT / mutant BECN1 to MCF7 cells. Cells were lysed in a buffer comprising 50 mM Tris HCl, pH 7.9, 150 mM NaCl, 1% Triton X-100, 1 mM DTT, protease inhibitor cocktail (1 tablet per 50 ml buffer) (Roche Applied Sciences) and 1 mM PMSF. Cell lysates were clarified by centrifugation at 14,800 × g for 10 min at 4 °C. The HA-tagged WT or mutant ATG14 supernatants were saved and subjected to immunoprecipitation using mouse monoclonal anti-Flag M2 antibodies (Sigma), whereas the Flag-tagged WT or mutant BECN1 supernatants were collected and immunoprecipitated using rabbit polyclonal anti-HA antibodies (Y-11, Santa Cruz), followed by overnight incubation with Protein G beads (Invitrogen). The samples were eluted by 2 × Laemmli buffer, analyzed by western blot using rabbit polyclonal anti-HA antibodies (Y-11, Santa Cruz) or monoclonal anti-Flag M2-HRP antibody (Sigma) detected by ECL western blotting substrate (Thermo Fisher) and imaged by a Storm Imager (GE Lifesciences).
Autophagy Assays
Autophagy levels were evaluated by monitoring cellular localization of GFP-tagged LC3 (36). 2×105 COS7 or MCF7 cells were seeded in each chamber of four-well culture slides (Millipore EZ slides) and cultured overnight in DMEM with 10% FBS until ~80% confluency. Lipofectamine 2000 was used for transfection as above, with cells in each chamber being co-transfected with 4 μg of total plasmids comprising 1.6 μg GFP-LC3; and either 1.2 μg WT BECN1 and 1.2 μg WT or mutant ATG14 expression plasmids for COS7 cells; or 1.6 μg GFP-LC3 and 1.2 μg WT ATG14 and 1.2 μg WT or mutant BECN1 expression plasmids for MCF7 cells. After transfection, the cells were cultured in either rich media (DMEM, 10% FBS, 2X amino acid mixture, GIBCO) or starvation media (Earle’s balanced salt solution, GIBCO) for 4 hours. Then the cells were fixed with 4% paraformaldehyde in PBS. GFP-LC3-positive puncta were observed using a Zeiss AxioObserver Z1 fluorescent microscope and quantified by counting a minimum of 50 cells per condition in three independent repeats using the Imaris program (Bitplane). The significance of alterations in autophagy levels was determined by a two-tailed, heteroscedastic student’s t-test, wherein p ≤ 0.05 is considered significant.
RESULTS
The Human BECN1 CCD forms an anti-parallel homodimer
We have determined the 1.46 Å X-ray crystal structure of the human BECN1 CCD, comprising residues 175–265. The crystals belonged to space group C2 with unit cell parameters of a = 58.2 Å, b = 71.58 Å, c = 58.45 Å and β = 112.55° (Table 1) and contain two BECN1 CCD monomers per asymmetric unit. The final model consists of residues 175–265 in each monomer and 255 water molecules. The two monomers in the asymmetric unit are almost identical; superimposing with an RMSD of 0.72 Å over 91 Cα atoms. Each CCD monomer in the asymmetric unit is paired with an equivalent symmetry related molecule by a crystallographic two-fold axis, to form an anti-parallel dimer (A to A and B to B).
The human and rat BECN1 CCDs (30) have similar structures, comprising of an anti-parallel, left-handed coiled coil homodimer (Fig. 1). The human and rat BECN1 CCD monomers superimpose with an average RMSD of 1.38 Å over 91 Cα atoms. For the human BECN1 CCD, the surface area buried at the interface between the two monomers is 4849 Å2, accounting for 26.4% of the total surface area of each monomer, while for the rat BECN1 CCD the surface area buried is 4969 Å2, accounting for 27% of the total surface area of each monomer. Residues that are different between the human and rat BECN1 CCDs: residues M182, I207 and T259 in human correspond to R180, V205 and M257 in rat (Fig. 1), are solvent-exposed and do not contribute to the homodimer interface. Half the residues comprising the BECN1 CCD homodimer are charged, with a predominance of acidic residues resulting in a very negatively charged surface (Fig. 2a). 40% of these charged residues are conserved.
Figure 1.
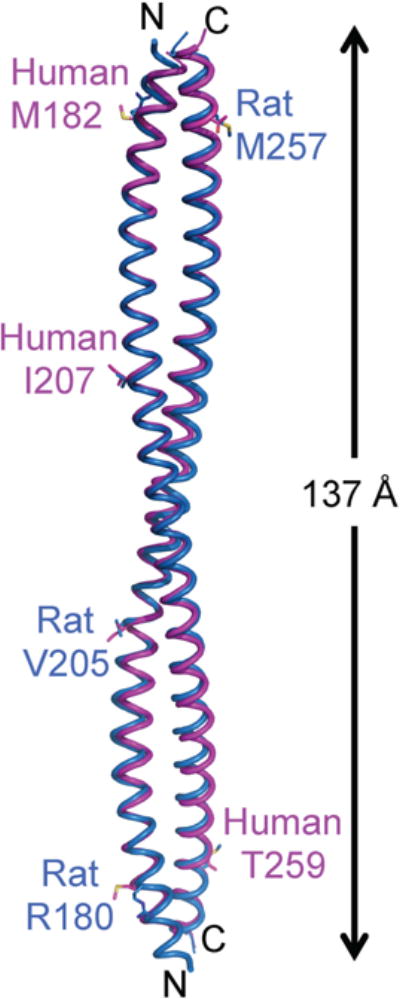
The crystal structure of the BECN1 CCD homodimer. The human BECN1 CCD homodimer and the rat BECN1 CCD (PDB ID: 3Q8T) are superimposed and displayed in magenta and marine ribbon respectively. The three residues that are different between the human and rat CCDs are rendered in stick with atoms color-coded by atom type: C, magenta (human) or marine (rat); O, red; N, blue and S, yellow. The length of the homodimer is indicated. This and all other molecular figures were prepared using the program PyMOL (79).
Figure 2.
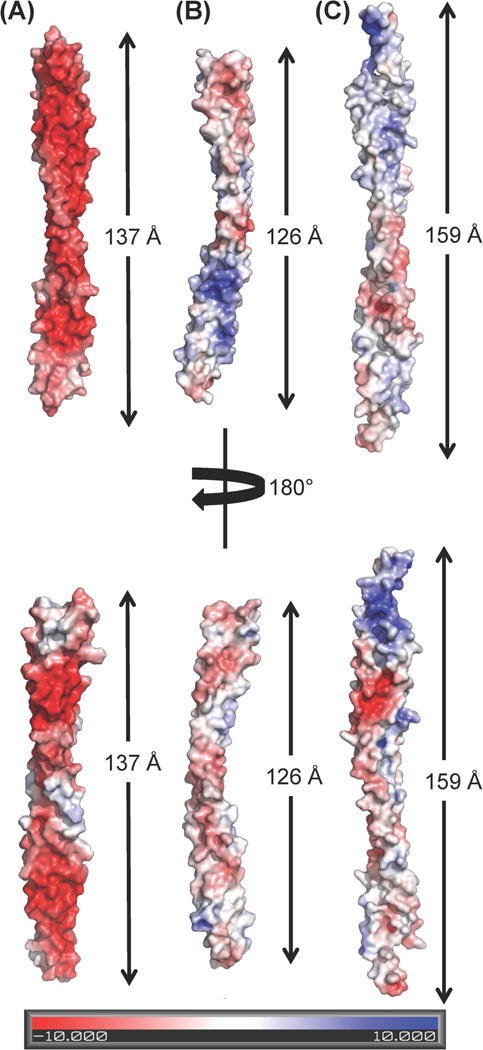
Comparison of the electrostatic potential surface of different BECN1/VPS30 CCD-containing dimers. The molecules are displayed in superimposable orientations and the length of each dimer is indicated. (A) BECN1 CCD homodimer (B) BECN1:ATG14 CCD heterodimer model (C) VPS30:VPS38 CCD heterodimer (PDB ID: 5DFZ).
The human BECN1 CCD has 26 heptad repeats (a-b-c-d-e-f-g), that stabilize the homodimer by interactions of residues at the “a” and “d” positions, similar to the rat BECN1 CCD. The 26 interacting pairs are comprised of two sets of thirteen unique pairs related by the homodimer two-fold symmetry (Fig. 3a). Six of these thirteen unique “a” and “d” positions of the heptad repeats are occupied by hydrophobic residues that form acceptable interacting pairs, including three ideal pairs (Fig. 3a, Supplementary Table 1). The remaining seven repeats bear bulky charged or polar residues resulting in non-ideal pairings at these a and d positions (Fig. 3a, Supplementary Table 1). For instance, residues at the a and d positions of the first heptad repeat of one helix, S177 and L180, pack against L264 and L261 respectively, at the d’ and a’ positions of the last heptad repeat of the partner helix, resulting in one non-ideal and one ideal pairing (Fig. 3a, Supplementary Table 1). The register of heptad interactions is maintained throughout the length of the BECN1 CCD homodimer.
Figure 3.

Comparison of the interface interactions among different BECN1/VPS30 CCD-containing dimers. Residues are shown in stick and color-coded by atom-type: O, red; N, blue; S, yellow; and C, magenta (BECN1 / VPS30), wheat (ATG14), and green (VPS38). (B) BECN1 CCD homodimer. Thirteen interaction pairs are shown, as the remaining thirteen are related by two-fold molecular and crystallographic symmetry. (B) BECN1:ATG14 CCD heterodimer. (C) VPS30:VPS38 CCD heterodimer (PDB ID: 5DFZ).
Polar interactions also play an important role in stabilizing the BECN1 CCD in both rat and humans. The BECN1 CCD homodimer contains six pairs of inter-chain polar interactions. At the interface, R205 forms a non-ideal pair with F236 of the partner helix, but in addition to the hydrophobic packing of the aliphatic part of the side chain, it also stabilizes the homodimer by forming two hydrogen bonds with Y233 and Q240 of the partner helix, two polar interface residues located a turn away on either side of F236. Further, while the aliphatic part of the interface E226 packs against the partner’s V215, the charged end makes an inter-molecular polar interaction with Q216 of the partner helix. Therefore there are three unique pairs of inter-molecule polar interactions in the BECN1 CCD homodimer that compensate for the non-ideal pairings at the interaction interface and further stabilize the CCD. Lastly, intra-molecular polar networks between charged and polar surface exposed residues formed along the whole chain, including interacting triplets of charged residues, diminish potential Coulombic repulsion between chains and stabilize CCD structure.
Despite the substantial similarity in overall structure and interface residues, the human BECN1 CCD homodimer self-associates twice as tightly (Kd = 48.3 μM) (Table 2) as the rat BECN1 CCD (Kd = 89 μM) (30). For the human homodimer, self-association is driven by entropy rather than enthalpy (Table 2). The absence of enthalpic contributions to binding is likely a result of the lower number of favorable interface interactions, relative to the total buried surface area, and results in a metastable homodimer.
Table 2.
ITC measurements of BECN1 CCD homodimerization and heterodimerization with the ATG14 CCD.
| Constructs | Kd (μM) | ΔH (kJ/mol) | ΔG (kJ/mol) | ΔS (J/K•mol) | n |
|---|---|---|---|---|---|
| Homodimer | 48.3±13.47 | 161±105 | −27.64± 0.40 | 627±356 | |
| Heterodimer | 4.44±0.08 | −27.17± 5.65 | −30.56± 0.04 | 11.361± 18.85 | 1 |
The ATG14 CCD is less helical than the BECN1 CCD
Sequence alignment of predicted ATG14 CCDs from five diverse eukaryotic organisms, ranging from human to yeast, shows that the ATG14 CCD is moderately conserved (Supplementary Fig. S1). Secondary structure prediction using JPred4 (54) indicates that the ATG14 CCD region spans residues 71–202 (Supplementary Fig. S1), similar to previously published sequence analyses. Cellular studies show that ATG14 residues 71–180 include all residues that are required for binding to the BECN1 CCD (15, 16, 18, 19). Further, biophysical and biochemical studies using purified protein fragments show that residues 88–178 are sufficient for the interaction with the BECN1 CCD (Supplementary Fig. S2) (30). Therefore, we selected ATG14 residues 88–178, which matches the length of the BECN1 CCD (residues 175–265), as the nominal ATG14 CCD for structural studies (Supplementary Fig. S1).
We utilized CD spectroscopy to analyze and compare the secondary structure content of the ATG14 CCD, BECN1 CCD homodimer and BECN1:ATG14 CCD heterodimer (Supplementary Fig. S3). Since the CCDs were expected to be highly helical, we used CONTIN (43), which has been shown to provide the most accurate estimation of α-helical content in proteins (55), to calculate the secondary structure content from the CD spectra measured from each sample. Consistent with expectations, all spectra have strong helical features comprising a positive transition at 195 nm and two negative transitions at 208 nm at 222 nm (Supplementary Fig. S3). However, there are differences in the helical and coil content estimated for each CCD sample (Table 3). The 97-residue BECN1 CCD monomer construct comprising the 91-residue BECN1 CCD (175–265) and a His6-tag is highly helical, with a helical conformation adopted by 168 residues within the 194-residue homodimer, or 84 residues within each 97-residue monomer (Table 3). However, this helicity is less than that observed in the BECN1 CCD X-ray crystal structure, wherein all observed residues form an α-helix.
Table 3.
Secondary structure content of different protein constructs estimated from CD.
| Protein | Residue number | Percentage (%) | |||
|---|---|---|---|---|---|
| Helix | Strand | Coil | Total | ||
| BECN1 CCD | 194 (dimer) | 86.8 | 0.8 | 12.4 | 100 |
| MBP-ATG14 | 487 | 56.9 | 10.8 | 32.3 | 100 |
| MBP | 371 | 57.6 | 14.1 | 28.3 | 100 |
| BECN1:ATG14 | 191 (heterodimer) | 79.6 | 0.5 | 19.9 | 100 |
The purified ATG14 CCD is unstable upon cleavage of the MBP-tag in the absence of an interacting partner; therefore CD spectra were recorded from the MBP-tagged ATG14 CCD. Analysis of this spectrum indicates that, of the 487 residues in the fusion protein (371 residues from MBP, 25 in the linker and 91 in the ATG14 CCD), 277 residues are in a helical conformation, 53 are in β-conformation and 157 are in a random coil conformation (Table 3). Analysis of a CD spectrum recorded from MBP alone indicates that of the 371 total residues, 214 residues are in helical conformation, 52 in β-conformation and 105 in random coil-conformation (Table 3). Subtraction of the secondary structure content of MBP from the MBP-ATG14 CCD enables us to deduce the secondary structure content of the ATG14 CCD when not in complex with BECN1 or other partners. Assuming the secondary structure of MBP does not change between the isolated MBP and the fusion protein, and the 25-residue linker is a random coil, we find that only 63 of the 91 residues comprising the ATG14 CCD are helical and 27 residues exist in a coil conformation. Thus, the ATG14 CCD is significantly less helical and more disordered than the BECN1 CCD.
The ATG14 CCD does not appear to form a homodimer, as no heat of self-association was detected in ITC measurements. This is not surprising as the ATG14 CCD has only 21 hydrophobic residues. Our ITC measurements indicate that the human ATG14 CCD binds to the BECN1 CCD in a 1:1 stoichiometry with a moderate affinity, that is ~10-fold tighter than BECN1 self-association under the same conditions (Table 2). Further, this heterodimerization is both enthalpically and entropically favorable, in contrast to BECN1 homodimerization, which is driven entirely by entropy (Table 2).
To further understand the structural basis of heterodimerization, we purified the BECN1:ATG14 CCD complex (Supplementary Fig. S2). We confirmed the presence of both proteins in the purified sample using SDS-PAGE (Supplementary Fig. S2) and mass spectrometry (data not shown). Analysis of CD spectra measured for the purified heterodimer indicates that 152 of the 191 residues in the BECN1:ATG14 CCD heterodimer are in a helical conformation (Table 3), sixteen residues less than that observed for the BECN1 CCD homodimer; while 38 residues are in a random coil conformation, compared to the 24 in the BECN1 CCD homodimer. Assuming the helicity of the BECN1 CCD does not change relative to that observed within a BECN1 homodimer, the ATG14 CCD has 68 residues in a helical conformation. Alternately, assuming that the helicity of both, the BECN1 and ATG14 CCDs are comparable within the heterodimer, the ATG14 CCD will have ~76 residues in helical conformation. Thus, compared to the ATG14 CCD helicity in the monomeric state, in the context of the BECN1:ATG14 CCD homodimer, the helicity of the ATG14 CCD is marginally increased by five to thirteen residues.
The BECN1:ATG14 CCD heterodimer has a partially disordered, elongated structure
We conducted extensive crystallization trials of both, the MBP-ATG14 CCD fusion protein and the ATG14:BECN1 CCD heterodimer. Although both SDS-PAGE and mass spectroscopy show that the protein samples used for crystallization contained the appropriate protein, we were unable to obtain crystals of either the fusion protein or the complex. Indeed, we find that the ATG14 CCD is very unstable even in complex with the BECN1 CCD. Careful monitoring by SDS-PAGE shows that it degrades in approximately three days even at 4 °C (data not shown). It is likely that this rapid degradation of ATG14 CCD, coupled with its inherent structural flexibility, prevented crystallization.
Therefore, in order to gain better insight into the size and shape of the BECN1 : ATG14 CCD heterodimer, we performed SAXS on samples frozen immediately after purification. To ensure that the SAXS data were collected from a homogeneous sample, SAXS was performed in tandem with size exclusion chromatography (SEC-SAXS). The SEC profile of the purified complex comprises a single peak consistent with an elongated CCD dimer and is indicative of the absence of aggregated protein (Supplemental Fig. S2). The low q-range region in the Guinier plot (Fig. 4a) is linear in the range of q × Rg < 1.3. The Radius of Gyration (Rg) estimated from the Guinier plot and the P(r) distribution function (Fig. 4b) is 34 Å and 37 Å respectively, which is comparable to the Rg of 38 Å calculated from the heterodimer model; and similar to the Rg of the elongated BECN1 CCD homodimer calculated from the SAXS data (28). The P(r) distribution was also used to calculate a Dmax of 125 Å (Fig. 4b), indicating that the heterodimer has an elongated structure, as expected for a CCD. The molecular weight derived from the Porod Volume of the heterodimer is 25.5 KD, close to its theoretical molecular weight of 23.9 KD. The most striking difference between the BECN1 CCD homodimer and the BECN1:ATG14 CCD heterodimer is observed in the Kratky plots (Fig. 4c). Consistent with the X-ray crystal structure, the Kratky plot calculated for SAXS data recorded from the BECN1 CCD homodimer indicates that this domain is well folded (28), but the Kratky plot calculated for SAXS data recorded from the BECN1:ATG14 CCD heterodimer indicates that it is partially unfolded (Fig. 4c), which is consistent with our estimation of secondary structure content from the CD spectrum of the complex. Therefore, both CD spectroscopy and SAXS indicate that the BECN1:ATG14 CCD heterodimer is unstructured relative to the BECN1 CCD homodimer.
Figure 4.
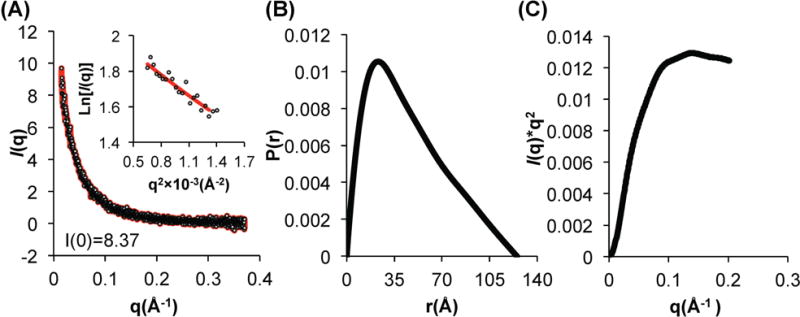
SAXS analysis of BECN1:ATG14 CCD complex. (A) Guinier plot; (B) P(r) pairwise distribution; (C) Kratky plot
BECN1 and ATG14 form a curved CCD heterodimer
The low-resolution SAXS envelope calculated for the BECN1:ATG14 CCD heterodimer has an elongated shape (Fig. 5), similar to that of the BECN1 CCD homodimer (28). However, unlike the BECN1 CCD homodimer envelope (28), the heterodimer envelope has a pronounced curvature fitting a radius of ~15 nm. In order to further investigate structural details of the BECN1:ATG14 CCD heterodimer interaction interface, we built an atomistic model of the heterodimer (Fig. 2b) as a parallel CCD, based on information from the Cryo-EM reconstruction of the human BECN1:ATG14:P150:PI3KC3 quaternary complex (35). This parallel arrangement is also analogous to the heterodimer formed by the CCDs of VPS30 and VPS38, which correlate to BECN1 and UVRAG, in the X-ray crystal structure of the yeast VPS30:VPS38:VPS15:VPS34 quaternary complex (29).
Figure 5.
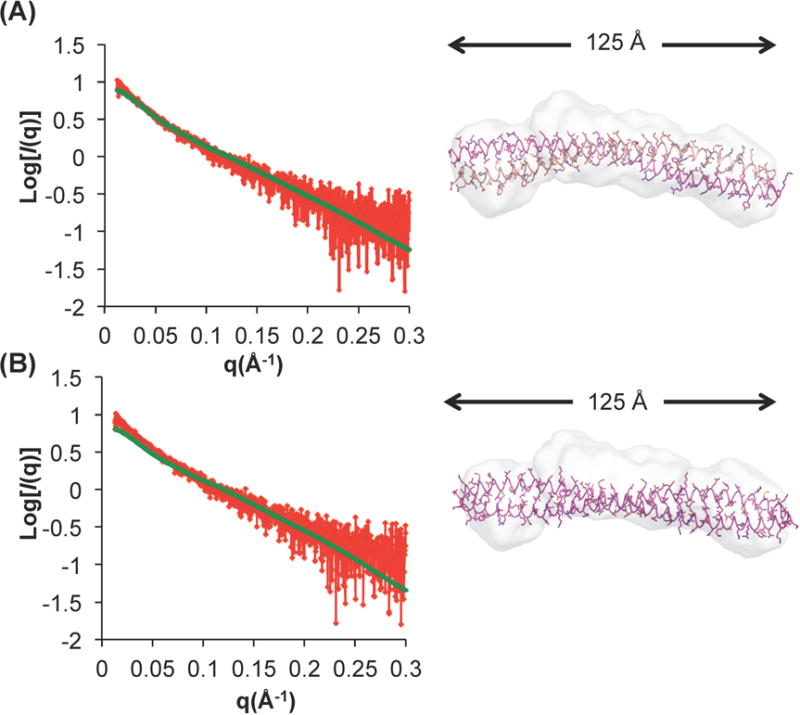
The BECN1:ATG14 heterodimer model agrees well with the experimental SAXS data. Fits of experimental SAXS data recorded from the BECN1:ATG14 CCD heterodimer to (A) the BECN1:ATG14 CCD heterodimer and (B) BECN1 CCD homodimer. The left panels show fits of the experimental scattering curve (red) measured from the BECN1:ATG14 CCD heterodimer and the theoretical scattering curve (green) calculated from the two different atomic models. The right panels show the fit of these two atomic models into the molecular envelope calculated from the SAXS data. The Dmax obtained from the P(r) function is indicated.
Strikingly, the heterodimer models obtained from the programs (PS)2 and MODELLER are both curved, with a curvature radius of 14 or 15 nm respectively (Fig. 2b), similar to the curvature radius observed for the experimental SAXS envelope (Fig. 5, right panels), but different from the BECN1 CCD homodimer, which forms a flat structure (Fig. 2a). Further, residues placed at the interface are similar in both models, therefore for subsequent analysis we primarily used the model obtained from (PS)2. The CCD heterodimer curvature results in a heterodimer CCD length of 126 Å (Fig. 2b), that agrees well with the Dmax of 125 Å calculated from the P(r) function derived from the heterodimer SAXS data, but is significantly shorter than the BECN1 CCD homodimer (Fig. 2a).
Next, we compared the experimental SAXS data recorded from the BECN1:ATG14 CCD heterodimer to the theoretical scattering curves calculated from the BECN1:ATG14 CCD heterodimer model and the BECN1 CCD homodimer structure. The experimental SAXS scattering curve fit nicely to the theoretical scattering curve calculated from the heterodimer model, with a χ2 of 1.1 (Fig. 5a, top left panel). Consistent with this, the envelope encloses the heterodimer model very well (Fig. 5a, top right panel). In contrast, the experimental SAXS data recorded for the BECN1:ATG14 CCD heterodimer does not fit well to the theoretical scattering curve calculated from the BECN1 CCD homodimer crystal structure, with a relatively higher χ2 of 2.1 (Fig. 5b, bottom left panel). Consistent with this, the BECN1 CCD homodimer crystal structure fits poorly into the molecular envelope calculated from the BECN1:ATG14 CCD heterodimer SAXS data, with both termini of the 137 Å long BECN1 CCD extending beyond the envelope and the curvature of the heterodimer envelope resulting in poor agreement along the center of the BECN1 CCD homodimer (Fig. 5b, bottom right panel). Thus, the BECN1:ATG14 CCD heterodimer model fits better than the BECN1 CCD homodimer structure to the SAXS data recorded from samples of the heterodimer.
A comparison of the BECN1:ATG14 CCD heterodimer model to the analogous CCD heterodimer structure of the BECN1:UVRAG yeast homologs VPS30:VPS38 within the VPS34:VPS30:VPS38:VPS15 complex, shows that the 159 Å long VPS30:VPS38 CCD heterodimer is less curved, with a radius of curvature of ~17 nm (Fig. 2c). Thus, the BECN1:ATG14 CCD heterodimer is more curved relative to both, the BECN1 CCD homodimer and BECN1:UVRAG CCD heterodimer (Fig. 2). As modeled, the BECN1:ATG14 CCD heterodimer (pI=7.0) has a more neutral surface (Fig. 2b) than the highly negatively charged BECN1 homodimer (pI=4.3) (Fig. 2a). In the BECN1:ATG14 CCD heterodimer, the N-terminal and central regions are negatively charged, while the C-terminal ends are positively charged (Fig. 2b). The electrostatic surface of the VPS30:VPS38 CCD heterodimer (pI=6.2) is also closer to neutral, with the N-terminal region being positively charged while the central and C-terminal parts are moderately negatively charged (Fig. 2c).
The BECN1:ATG14 CCD Heterodimer Interface
Examination of the BECN1:ATG14 CCD heterodimer model shows that the total surface area buried at the interface in the heterodimer is 4449 Å2, accounting for 25.8% of the ATG14 CCD surface area and 25.6% of the BECN1 CCD surface area. This is slightly less than the 4849 Å2 buried in the BECN1 CCD homodimer interface. In contrast, the CCD heterodimer of VPS30:VPS38 buries a total surface area of 3319 Å2, accounting for 16.8% and 15.9% of the surface area of VPS30 and VPS38, respectively, which is much less than that observed for either the BECN1:ATG14 CCD heterodimer or the BECN1 CCD homodimer.
Superposition of the BECN1:ATG14 CCD heterodimer on the BECN1 homodimer (Supplementary Fig. S4a) shows that the BECN1 CCD in the two structures aligns moderately well, with an RMSD of 2.6 Å over 91 Cα atoms. Superposition of the VPS30:VPS38 CCD heterodimer (29) (Supplementary Fig. S4b) on the BECN1 CCD homodimer is somewhat worse, with the VPS30 and BECN1 CCDs aligning with an RMSD of 3.5 for 91 Cα atoms. Superposition of the BECN1:ATG14 CCD heterodimer on equivalent regions of the VPS30:VPS38 CCD heterodimer (29) (Supplementary Fig. S4c) shows that the BECN1 CCD also aligns moderately well with equivalent VPS30 residues, with an RMSD of 2.7 Å for 91 Cα atoms; however, the partner ATG14 CCD superimposes more poorly upon the VPS38 CCD, with an RMSD of 4.1 Å for 82 Cα atoms.
In all three CCD dimer structures, the same BECN1 or equivalent VPS30 residues are pointed toward complementary residues from the partner CCD either from BECN1 or ATG14 or VPS38 (Fig. 3 and Supplementary Table S1). The BECN1:ATG14 CCD dimer is modeled as a parallel coiled-coil heterodimer, therefore, the residues at the a and d positions of heptad repeats from the BECN1 CCD interact with residues at the a′ and d′ positions of heptad repeats from the ATG14 CCD, to form 25 interacting pairs (Fig. 3b and Supplementary Table S1). A comparison of the residues at the interaction interfaces of the homodimer and heterodimer (Fig. 3a and 3b), shows that all the BECN1 residues that contribute to the homodimer interface are also involved in the heterodimer interactions with ATG14 (Supplementary Table S1), except for L264 from the last BECN1 heptad repeat. Analysis of the BECN1:ATG14 CCD heterodimer interface, shows that of the 25 interface pairs, only ten form acceptable hydrophobic interactions, including six ideal pairs (Fig. 3b and Supplementary Table 1). In the heterodimer, BECN1 residues S177 and L180 from the first heptad repeat, pack against ATG14 residues F92 and E95 (Fig. 3b and Supplementary Table S1). This register is maintained through the whole chain of interactions starting with BECN1 S177 and ATG14 F92, to BECN1 L261 and ATG14 V176.
Ten of the 25 BECN1:ATG14 interface pairs are hydrophobic (Fig 3b, Supplementary Table S1). Interestingly, most of these interface pairs are conserved amongst diverse eukaryotes, except for BECN1 A219, F236, and L261. This is particularly striking because amongst these pairs, BECN1 residues V208, L212, F236 and V250 are paired with conserved polar residues from the partner BECN1 CCD in the homodimer, and therefore make better hydrophobic interactions in the modeled heterodimer than in the BECN1 homodimer (Fig. 3, Supplementary Table S1). We find that toward the center of the heterodimer, BECN1 V208:ATG14 L123 and BECN1 L212:ATG14 I127 form two consecutive ideal hydrophobic pairings (Fig. 3b) and these residues are conserved in both BECN1 and ATG14, indicative of their importance in heterodimer formation. In addition, BECN1 V215 that was paired with a glutamate is paired with the neutral G130 from ATG14 in the heterodimer (Fig 3, Supplementary Table S1)
The invariant, charged BECN1 interface residue, E191, participates in ‘imperfect’ pairing with the highly conserved BECN1 V250 within the CCD homodimer (Fig. 3a, Supplementary Table S1). However, in the heterodimer, BECN1 V250 packs against the hydrophobic ATG14 I165, which is highly conserved amongst higher eukaryotes; while BECN1 E191 pairs with the conserved ATG14 T106, with the pairing stabilized by hydrogen bonds (Fig. 3b, Supplementary Fig. S1 and Table S1). Further, the polar BECN1 Q240, which is partnered with hydrophobic V201 in homodimer, makes better interactions as it partnered with the conserved small residue, A155, in the heterodimer.
BECN1 S177 and Q240 form non-ideal interface pairs with ATG14 F92 and A155 respectively, however, they are stabilized by inter-molecular polar interactions with two polar ATG14 interface residues, E95 and H158, located one turn away from the paired ATG14 residues. Further, the conserved BECN1 interface residue, Y233, is stabilized by an inter-chain polar interaction with the solvent-exposed, conserved ATG14 R154. Lastly, the heterodimer is also stabilized by a salt-bridge between the solvent-exposed conserved residues, BECN1 E190 and ATG14 R110. Thus, the interaction of the ATG14 CCD with the BECN1 CCD appears to involve many conserved, hydrophobic residues, as well as conserved, polar residues that are paired with conserved, polar BECN1 residues.
We used the numbering and positions in the 4.4 Å poly-alanine model of the VPS34:VPS30:VPS38:VPS15 complex, to model the residue side chains. Based on this, our analysis indicates that residues of the yeast VPS30 homolog that are equivalent to the BECN1 CCD interface residues are also pointed toward and involved in binding VPS38 (Fig. 3, Supplementary Table S1). However, residues contributed to the interface by VPS38 are mostly polar and charged (Fig. 3c and Supplementary Table S1). Amongst 19 VPS38 residues involved in heterodimer interaction, only L238, L252 and V286 are hydrophobic, but they interact with polar BECN1 D258, K272 and large side-chain F307 respectively (Fig. 3c). Thus the VPS30:VPS38 interface comprises almost entirely of non-ideal pairings. Further, the CCDs of VPS30 and VPS38 are physically shifted relative to each other, therefore VPS30 residues corresponding to four BECN1 interface residues from the N-terminal heptads, S177, L180, L184 and L187; as well as one residue from the C-terminal heptad, L264, do not interact with VPS38 residues (Fig. 3c and Supplementary Table S1). Lastly, a loop inserted in the center of the VPS38 CCD results in BECN1 residues Q286 and M289 not being paired at the interface.
Thus, overall, the VPS30:VPS38 heterodimer has very few hydrophobic interactions, with an interface that is not as extensive or well-packed as either the BECN1 CCD homodimer or BECN1:ATG14 heterodimer interfaces (Fig. 3 and Supplementary Table S1). The BECN1:ATG14 interface is slightly more extensive than that of the BECN1 homodimer with some non-ideal packing interactions involving paired hydrophobic and polar residues in the homodimer, being replaced in the heterodimer by either better hydrophobic interactions or polar pairings. These improved interactions result in a significant enthalpic contribution to BECN1: ATG14 CCD binding (Table 2). The differential packing of BECN1 homodimer and different heterodimer interfaces, likely enables competitive binding of diverse BECN1 partners in cells.
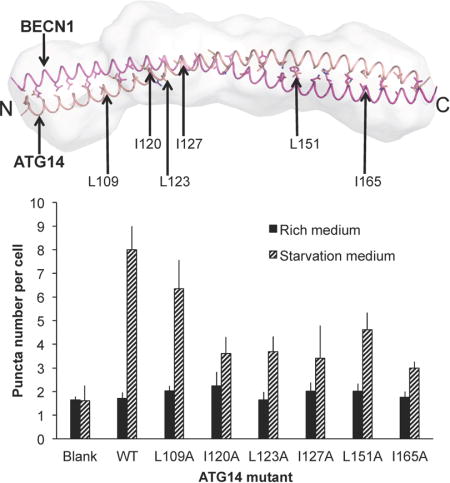
BECN1 CCD Interface Residues are Important for Binding the ATG14 CCD and are required for Starvation-Induced Autophagy
In order to assess the importance of residues at the CCD interface, we mutated selected interface residues to alanine in full-length BECN1, then assessed the impact of these mutations on CoIP with full-length ATG14 and on cellular autophagy levels. The hydrophobic BECN1 interface residues L184, L194, V208, L212, L222, Y233, F236 and V250 (Fig. 3) were selected for alanine-mutagenesis. Of these selected BECN1 residues, L194 is invariant and F236 is not conserved, while the remaining residues are conserved. Based on the heterodimer model, L194, V208, L212, and V250 form ideal hydrophobic pairings with conserved ATG14 residues. BECN1 F236 interacts with the highly conserved ATG14 L151; while L184, L222, Y233 participate in non-ideal interactions with polar or small ATG14 interface residues (Fig. 3b, Supplementary Table 1).
The cellular assays were performed using MCF7 cells, which have a defective becn1 gene and express low levels of BECN1 (14, 56–58), thereby allowing the effect of BECN1 mutations to be assayed in the absence of endogenous BECN1. Our Co-IP results show that all the BECN1 interface mutations dramatically decreased interaction with ATG14 (Fig. 6a). Amongst these, the L212A and L222A mutations completely abolish interaction with ATG14; the L194A, V208A, Y233A, F236A and V250A mutations result in barely detectable interaction with ATG14, and only the L184A mutant shows clear interaction with ATG14, although the interaction is substantially diminished relative to wild-type (WT) BECN1 (Fig. 6a). These results confirm the essential role of these BECN1 residues identified from our BECN1:ATG14 heterodimer model in mediating the interaction of full-length BECN1 and ATG14 in cells.
Figure 6.
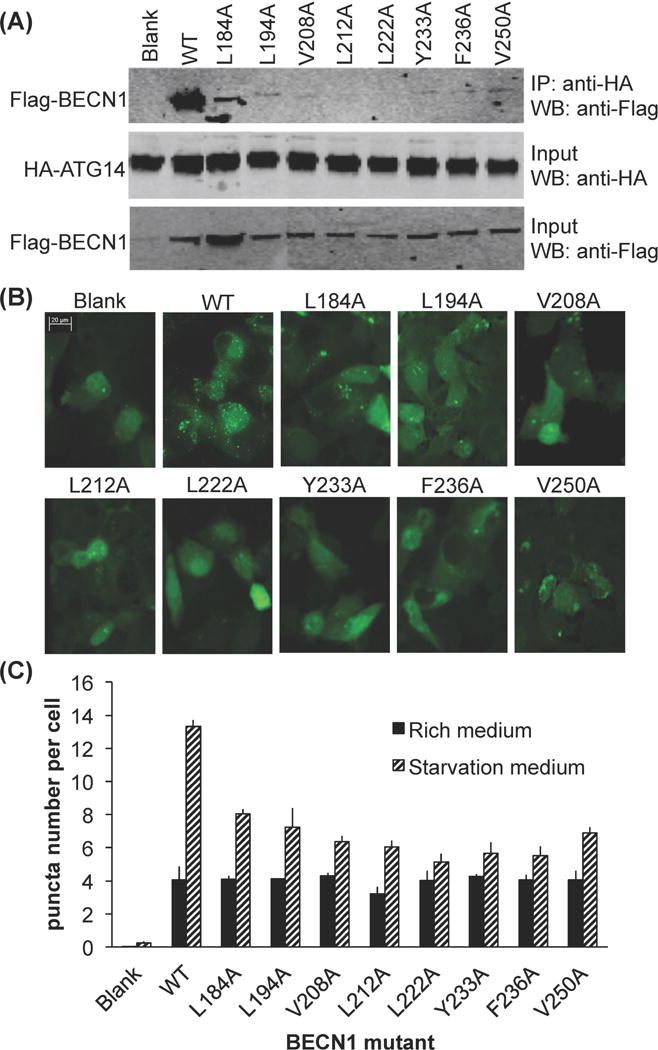
Effect of single alanine mutagenesis of conserved BECN1 interface residues. (A) Impact on ATG14 binding as assessed by CoIP of HA-ATG14 and WT or single mutant Flag-BECN1. IP with anti-HA and western blot with anti-Flag(top); Western blot with anti-HA (middle); Western blot of lysate using anti-Flag (bottom). (B) Representative images of GFP-LC3 staining in cells grown in growth media (top) and starvation media (bottom), transfected with WT ATG14 and WT or mutant BECN1 as indicated. (C) Bars represent fluorescent microscopy quantification of discrete GFP-LC3 labeled autophagic puncta per cell in GFP-positive MCF7 cells co-transfected with GFP-LC3, WT ATG14, and WT or single mutant BECN1 as indicated below the x-axis.
Earlier studies have conclusively shown that the interaction of BECN1 and ATG14 is required for autophagosome nucleation (15, 33). Therefore, here we monitored autophagy levels simply by quantifying the change in cellular localization of a GFP-tagged, transiently expressed mammalian autophagy-specific marker, LC3 (GFP-LC3) from a diffuse cytoplasmic distribution to localized punctae corresponding to autophagosomal structures (36). Due to low BECN1 expression levels, MCF7 cells do not show starvation-induced increases in autophagy unless BECN1 is ectopically expressed (14, 56–59). Transient expression of BECN1 in MCF7 cells led to a marked increase in autophagy upon starvation (p = 6.0×10−4 for starved versus nutrient-rich cells; Fig. 6b, 6c).
Basal autophagy levels in nutrient-rich media are typically lower and less consistent than in starvation conditions. Nevertheless, in nutrient rich conditions, the expression of the BECN1 CCD interface mutants results in basal autophagy levels similar to WT BECN1, (ranging between p = 0.20 to 0.98 for mutants versus WT BECN1; Fig. 6b, 6c). In contrast, a pronounced effect is seen on starvation-induced autophagy levels. Starvation triggers an increase in autophagy regardless of whether WT or mutant BECN1 is expressed (ranging between p = 4×10−5 to 0.02) for starvation versus rich conditions; Fig. 6b, 6c), except in the case of BECN1 L222A that abrogates starvation-induced autophagy (p = 0.07 for starvation versus rich conditions; Fig 6c). However, the starvation-induced increase in autophagy is markedly diminished when upon expression of each of the BECN1 mutants (ranging between p = 2×10−5 to 0.008 for mutants versus WT BECN1; Fig. 6c), with the decrease in starvation-induced autophagy caused by each mutation being somewhat co-related with the ability of the mutant to bind ATG14. This suggests that the BECN1 CCD interface residues are not essential for basal levels of autophagy in nutrient-rich conditions, but are critical for the interaction with ATG14 and for starvation-induced up-regulation of autophagy.
ATG14 CCD Residues Important for Binding the BECN1 CCD are also required for Starvation-Induced Autophagy
Based on our analysis of the BECN1:ATG14 CCD interface, hydrophobic ATG14 interface residues L109, I120, L123, I127, L151 and I165 (Fig. 2b, Supplementary Table 1) were selected for alanine-mutagenesis, to investigate their role in heterodimerization with BECN1, as well as in mediating autophagy. Each of these residues is conserved and also paired with a conserved BECN1 residue within the BECN1:ATG14 CCD heterodimer.
CoIP experiments were used to assess the impact of these mutations on the interaction between exogenously expressed full-length BECN1 and ATG14 (Fig. 7a). Our results show that compared to WT ATG14, expression of mutant ATG14 constructs decreases interaction with WT BECN1 (Fig. 7a). Expression of I120A or I165A ATG14 mutants abolishes binding to BECN1; expression of the L123A or I127A mutants substantially decreases binding; while expression of the L109A or L151A mutants only marginally decreases binding to BECN1 (Fig. 7a). Therefore, ATG14 I120 and I165 appear to be required for binding to BECN1, while L109A and L151A are less important.
Figure 7.
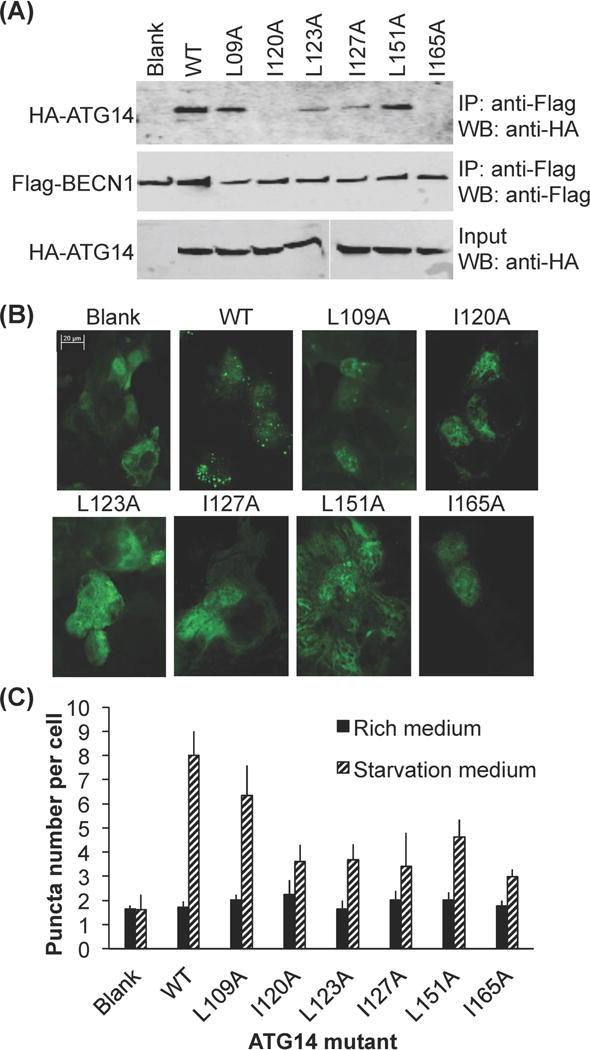
Impact of alanine mutagenesis of conserved ATG14 interface residues. (A) Impact on BECN1 binding as assessed by CoIP of Flag-BECN1 and WT and single mutant HA-ATG14. IP with anti-flag and western blot with anti-HA (top); Western blot with anti-Flag (middle); Western blot of lysate using anti-HA (bottom). (B) CoIP of Flag-BECN1 and WT or single mutant HA-ATG14. IP with anti-flag and western blot with anti-HA (top). (B) Representative images of GFP-LC3 staining in cells grown in growth media (top) and starvation media (bottom), transfected with WT BECN1 and WT or mutant ATG14 as indicated. (C) Bars represent fluorescent microscopy quantification of discrete GFP-LC3 autophagic puncta per cell in GFP-positive COS7 cells co-transfected with GFP-LC3, WT BECN1, and WT or mutant ATG14 as indicated below the x-axis.
Next we investigated the effect of these mutations on cellular autophagy, by quantifying transiently-expressed, GFP-LC3 labeled, autophagosomal puncta in mammalian cells in both nutrient-rich and starvation media. Since earlier studies have used diverse methods to establish the critical role of ATG14 and BECN1 in autophagosome nucleation, and show that in starvation conditions, ATG14 promotes autophagic flux via binding to BECN1 (15–20); the quantification of the cellular localization of the GFP-LC3 labeled, autophagosomal puncta is adequate for assessing the impact of ATG14 mutations on cellular autophagy levels (Fig. 7b and 7c). Monkey kidney COS7 cells were selected for this experiment as they have reduced endogenous expression of some autophagy-related genes including ATG14, and do not show a starvation-induced increase in autophagy unless ATG14 is ectopically expressed (Fig. 7b and 7c) (60). This allows us to assess the impact of ATG14 mutants on autophagy without background effects due to endogenous ATG14 expression.
We find that in nutrient-rich conditions, the number of autophagic puncta are comparable, regardless of whether ATG14 is exogenously expressed or not (p = 0.24 for absence vs. presence of ectopically-expressed ATG14). Indeed, in nutrient rich conditions, none of the ATG14 CCD mutants significantly impact cellular autophagy levels, as indicated by the similar numbers of autophagosome puncta per cell (p > 0.05 for mutants vs. WT ATG14; Fig. 7c). Strikingly however, starvation does not increase the number of autophagosomes (p = 0.89, for starvation vs. nutrient rich conditions in absence of ectopically-expressed ATG14), unless ATG14 is exogenously expressed (p = 0.007 for starvation vs. nutrient rich conditions when ATG14 is exogenously expressed; Fig. 7c). This suggests that ATG14 expression, and consequently the BECN1:ATG14 interaction, is not essential for basal levels of autophagosome nucleation in nutrient-rich conditions, but is critical for the starvation-induced up-regulation of autophagy.
All the ATG14 mutations tested adversely impact starvation-induced up-regulation of autophagy. Consistent with the CoIP results, up-regulation of autophagy in response to starvation is significantly diminished upon exogenous expression of either the I120A or I165A ATG14 mutant (p = 0.005 and 0.009, for mutant vs. WT ATG14 respectively, Fig 7b and 7c), for which no BECN1 binding was detected. Interestingly, there is a comparable reduction in the starvation-induced up-regulation of autophagy even when the L123A and I127A ATG14 mutants are expressed (p = 0.005 and 0.01, for mutant vs. WT ATG14 respectively, Fig 7b and 7c), which show weak binding to BECN1. Further, even the L151A ATG14 mutant that shows only a marginal decrease in binding to BECN1, shows a significant decrease in starvation-induced up-regulation of autophagy (p = 0.01, for L151A vs. WT ATG14), although the levels of starvation-induced autophagy are higher than for the preceding four mutants (Fig 7c). Only the L109A ATG14 mutant does not significantly diminish starvation-induced up-regulation of autophagy (p = 0.11 for L109A vs. WT ATG14, Fig 7b and 7c), consistent with the marginal decrease in BECN1 binding observed for this mutant.
Thus, our cellular assays validate the atomistic model, as mutations of the selected interface residues results in decreased BECN1:ATG14 interaction, and reduced autophagy levels. Further, the starvation-induced up-regulation of autophagy is correlated with the ability of the ATG14 CCD to interact with the BECN1 CCD. However, this correlation is not perfect, as the reduction in starvation-induced autophagy is more dramatic than would be expected from a response that was proportional to the reduction of interaction between ATG14 and BECN1 due to each mutation. From a biochemical mechanism perspective, this suggests that even a marginal decrease in the binding of BECN1 and ATG14 can have larger physiological effects within a cell.
DISCUSSION
BECN1 and ATG14 both contain CCDs that are essential to their role in autophagosome biogenesis and mediate a variety of interactions. However, there are many differences between the CCDs of the two proteins. The BECN1 CCD self-associates to form a stable, highly-helical, anti-parallel homodimer in the absence of other partners that interact via the CCD. In contrast, the ATG14 CCD has no detectable homodimerization, and approximately 30% of the CCD appears to be disordered in the monomeric state, and only marginally less within the BECN1:ATG14 heterodimer. It is likely that this disordered structure makes ATG14 particularly prone to proteases, preventing purification in the absence of a stabilizing tag such as MBP and complicating structural studies. Indeed, contrary to our expectations, but consistent with the observed disorder in both monomeric and heterodimeric states, the heterodimerization with BECN1 did not completely protect ATG14 from degradation, only somewhat delayed degradation. This conformational flexibility is likely an important structural and biochemical property of ATG14, perhaps facilitating interactions with diverse partners or simultaneous interaction with multiple partners as well as structural elasticity within different complexes, thereby enabling ATG14 to adopt different conformations to perform diverse functions.
The BECN1 CCD, which is well conserved amongst homologs (Figure 1), has been shown to be sufficient for self-interaction in cells (61, 62). BECN1 homo-oligomers were detected in mammalian cells via co-immunoprecipitation (CoIP) and immuno-blotting, even during starvation and rapamycin induced autophagy (62). Further, while in vitro ITC studies indicate that UVRAG, which heterodimerizes via the BECN1 CCD, disrupts the BECN1 homodimer (61); over-expression of the UVRAG CCD in cell culture diminishes, but does not abolish homo-oligomerization (61, 62). These studies suggest that the BECN1 CCD has functions as a homodimer; that extend beyond forming heterodimers within PI3KC3 complexes that mediate autophagy. BECN1 has been implicated in diverse functions besides autophagy. BECN1 contributes to endocytic trafficking in C. elegans (63) and Drosophila (64). Caspases-3, 7 and 8, cleave BECN1 at D133 or D149 to generate autophagy-inactive N- and C-terminal fragments that include the CCD, and these CCD-containing fragments translocate to the mitochondria, triggering the release of pro-apoptotic factors like cytochrome c to initiate apoptosis (65, 66). BECN1 also localizes to the nucleus where it regulates transcription of autophagosome maturation-related proteins such as LC3, lysosome associated membrane protein, and Ras-associated protein Rab-7a, by reducing Transcription Factor EB activation. The BECN1 nuclear export signal (NES) forms a complex with the chromosomal maintenance protein 1 (CRM1) to facilitate nuclear export of BECN1 (4, 56, 67–69). It is possible that the BECN1 homodimer contributes to these other functions biological functions, but this is yet to be explored.
BECN1 is well conserved and ATG14 is moderately conserved amongst eukaryotes. The CCD of each protein constitutes one of the best-conserved domains within each protein. As expected, the sequences, overall structures and interactions stabilizing the human and rat BECN1 CCD homodimer are very similar, but differ from the structure and interactions within the BECN1:ATG14 heterodimer. The pseudo-atomic, parallel BECN1:ATG14 CCD model, verified by good fits to experimental SAXS data, enables identification of potential ATG14 and BECN1 CCD residues that contribute to the BECN1:ATG14 interaction. Mutagenesis combined with cellular CoIP assays of selected BECN1 and ATG14 interface residues confirms that almost all of them are important for heterodimerization, verifying the BECN1:ATG14 CCD pseudo-atomic model presented. Further, the heterodimer model presented here is also supported by previously published studies using ITC to investigate the potential role of alanines and/or charged BECN1 residues located at the interface of the BECN1 homodimer, in binding ATG14. Interaction with the ATG14 CCD is abolished by the BECN1 single mutations: E191L and R205L; double mutations: E191L+A257L and A219L+E226L; triple mutation A219L+E226L+A257L; and the tetrad mutation E191L+A219L+E226L+A257L (30).
Strikingly, we find that exogenous expression of ATG14 in COS7 cells which have no detectable endogenous ATG14 expression, does not result in an increase in the number of autophagic puncta in nutrient-rich conditions. However, starvation-induced up-regulation of autophagy is observed only upon exogenous expression of ATG14, consistent with previous studies that indicate ATG14 is primarily required for the starvation-mediated up-regulation of autophagy (18, 19). Our studies have unambiguously identified point mutations in either BECN1 or ATG14 that abrogate heterodimerization, and shown that that the BECN1:ATG14 interaction is not essential for low, basal levels of autophagosome formation, but is critical for starvation-induced up-regulation of autophagy. Further, our results indicate that even a slight reduction of the BECN1:ATG14 interaction dramatically abrogates the increase in autophagy in response to starvation. The differential packing interaction of BECN1 in the homodimer and different heterodimers likely enables competitive binding of diverse BECN1 partners in cells. Perhaps, in cells undergoing autophagy, a slight reduction in heterodimerization affinity is sufficient to allow other BECN1 interactions to out-compete the interaction with ATG14, thereby altering the biochemical equilibrium of complex formation and preventing an up-regulation of autophagy.
The mechanism by which association of BECN1:ATG14 or BECN1:UVRAG with the PI3KC3:P150 binary complex increases PI3KC3 catalytic activity is unclear. BECN1 and ATG14 may facilitate the association of Complex I with membranes, serving as a sensor of membrane lipid composition and, in the case of ATG14, membrane curvature. Further, both BECN1 and ATG14 are implicated in additional interactions, with BECN1 specifically appearing to serve as an interaction hub for autophagy regulators (27, 66, 70, 71). Therefore, the combined role of these proteins appears to be to serve as a scaffold for the autophagosome nucleation complex, thereby up-regulating catalytic activity of PI3KC3; to sense and influence membrane curvature and lipid composition and to serve as an interaction hub for autophagy regulators.
Various studies have now shown that the CCDs of each protein are required and sufficient for the interactions of BECN1 with either ATG14 or UVRAG (15–19, 72, 73), therefore it is very important to understand the structural basis of BECN1:ATG14 (and BECN1:UVRAG) CCD heterodimerization. However, other domains from each protein likely also support these interactions. For instance, the BECN1 C-terminal BARAD packs against a BARAD at the C-terminus of UVRAG; and the FHD, a short helical region preceding the CCD in both BECN1 and UVRAG, also forms a heterodimeric coiled-coil structure in the quaternary structure complex (29). Therefore, it is possible that the ATG14 BATS domain packs against the BECN1 BARAD. Further, although unlike BEN1, ATG14 is not predicted to contain a helical region preceding the CCD, the existence of such a helical interacting region is not impossible given the known ATG14 domain architecture. Lastly, it is likely that subsequent association with other proteins within the larger PI3KC3 complex further stabilizes the heterodimer.
Our results show that the BECN1:ATG14 CCD heterodimer is shorter and more curved than the VPS30:VPS38 CCD heterodimer, which would alter the conformation of, and interactions within and outside, the quaternary PI3KC3 complexes. For instance, the different structure and curvature of the BECN1:ATG14 CCD heterodimer would result in changes in the relative positioning of membrane-binding domains within the quaternary complex (Supplementary Fig. S5). The C-terminal BECN1 BARAD and ATG14 BATS domain that are required for membrane association and for sensing membrane composition; and the PI3KC3 catalytic domain, which binds to PI-rich membranes, are located at the tips of the two arms of the V-shaped complex. The altered curvature would result in ~14 Å displacement of the BECN1 BARAD from the position occupied by the VPS30 BARAD within the quaternary complex, relative to PI3KC3 (Supplemental Fig. S5). Such changes in Complex I may dictate the preferential binding to more curved membranes. Further, the different positions, curvatures and sequences of the BECN1:ATG14 and BECN1:UVRAG heterodimers would necessitate altered positioning and interactions of the VPS15 (P150) WD50 domain within the quaternary structure. This may, in turn, alter P150:PI3KC3 interactions, resulting in stabilization of a more catalytically-active conformation of PI3KC3. As formation of the autophagosome nucleation complex increases PI3P content in the membranes, it would lead to formation of additional quaternary complexes as well as recruitment of other PI3P-binding autophagy proteins.
The CCDs of BECN1 and ATG14 have also been implicated in other, mutually exclusive interactions. The BECN1 CCD also interacts with CCDs from diverse proteins such as UVRAG/VSP38 (72, 74), TAB2 and TAB3 (75, 76). While the interaction of the BECN1 CCD with the UVRAG CCD up-regulates autophagy (15, 72–74), BECN1 CCD interaction with CCDs of either TAB2 or TAB3 down-regulates autophagy (75, 76). The ATG14 CCD also binds to the SNARE core domain of STX17, thereby stabilizing the STX17:SNAP29 binary t-SNARE complex on autophagosomes and facilitating subsequent fusion of the autophagosome and lysosome upon association of the t-SNARE and v-SNARE VAMP8 (34, 77). ATG14 binds to either STX17 or the STX17-SNAP29 binary t-SNARE ternary membrane fusion complex, but not to the STX17:SNAP29:VAMP8 complex. Further, the BECN1:ATG14 CCD and ATG14:STX17 CCD heterodimers are mutually exclusive, indicating that ATG14 plays a role in autophagy, independent of BECN1. The STX17:SNAP29:VAMP8 crystal structure (34) has a radius of curvature of 16 nm, comparable to the 15 nm radius of curvature for the BECN1:ATG14 complex. The relative curvature of these different CCDs dimers likely also affects their competitive association and function in the cell.
In summary, our studies of the BECN1 homodimer and BECN1:ATG14 heterodimer provide important structural and biochemical insights into the potential architecture, mechanism and dynamics of Complex I. We show that the BECN1:ATG14 interaction is critical for the eukaryotic cellular response to stressors such as starvation, and elucidate key structural details of this interaction. This study provides clues to the structural details of the mechanism of competitive formation of BECN1 homodimers, BECN1:ATG14 heterodimers, and perhaps BECN1:UVRAG heterodimers, in cells. It also provides clues to the conformational changes within proteins, especially in ATG14, as well as to how different BECN1 CCD interactions may affect formation and structure of different quaternary complexes that function in different stages of vesicle trafficking. Substantial additional studies will be required to elucidate details of the mechanisms by which these complexes up-regulate PI3P synthesis and influence membrane association and curvature, and how these functions are themselves regulated by additional diverse interactions. Such extensive structural, thermodynamic and cellular studies of these diverse BECN1 and ATG14 interactions are required in order to fully understand how these interactions complement and compete with each other to mediate and differentially modulate cellular autophagy levels. The information regarding the BECN1:ATG14 interaction presented here provides important insights that can be used to design future studies to ultimately elucidate the mechanism of membrane association and PI3KC3 activation. A detailed understanding of BECN1 interactions with cellular and viral BCL2 proteins recently enabled development of a potent, yet very specific, cell penetrating peptide, designed to disrupt viral BCL2-BECN1 interactions, and induce autophagy in cultured cells, without affecting normal regulation of BECN1-mediated by cellular BCL2 proteins (78). Therefore, we expect that given the importance of autophagy to human health, elucidating a detailed structure-based understanding of the various protein interactions that modulate cellular autophagy levels will greatly facilitate the future development of new therapeutics.
Supplementary Material
Table S1. Residues at the interaction interface (a, d positions of heptad repeats) in the various BECN1/VPS30-containing dimers.
Figure S1. ATG14 CCD sequence alignment. Sequence alignment of predicted CCDs from five diverse eukaryotic ATG14 proteins. Yellow, orange and red represent increasing sequence conservation, with red corresponding to invariant residues. Predicted secondary structures are shown below the sequence alignment. The predicted CCD region used in this study is boxed. Conserved residues involved in hydrophobic pairing in the heterodimer, that were selected for mutagenesis are indicated by red arrowheads.
Figure S2. SEC purification of the BECN1:ATG14 CCD heterodimer complex. SEC curves shown correspond to BECN1 CCD (green) and BECN1:ATG14 CCD (blue). Elution positions for different molecular weight markers and the molecular weight calculated from SEC are indicated. SDS-PAGE analysis of the peak fractions is shown.
Figure S3. CD spectra of different CCDs. The spectra for the MBP-ATG14 CCD fusion protein, the BECN1:ATG14 CCD complex, the BECN1 CCD, and MBP are color coded as indicated in the legend.
Figure S4. Comparison of BECN1 CCD containing dimers. All proteins are rendered in ribbon, colored as follows: BECN1, magenta; ATG14, wheat; VPS30, yellow and VPS38, green. The residues involved in interface interactions are shown in stick with atoms color-coded by atom type: O, red; N, blue; S, yellow; and C, colored according to main chain ribbon for that molecule. Superposition of (A) BECN1:ATG14 CCD complex and the BECN1 CCD homodimer. (B) VPS30:VPS38 complex and the BECN1 CCD homodimer. (C) BECN1:ATG14 CCD complex and the VPS30:VPS38 complex.
Figure S5. Displacement of the BECN1 BARAD domain in Complex I due to the curved BECN1:ATG14 quaternary structure. All proteins are shown in ribbon, colored as follows: BECN1, magenta; ATG14, wheat; VPS30, yellow; VPS38, green; VPS15, grey, and VPS34, blue. Protein domains implicated in membrane interaction, the BECN1 BARAD, VPS38 BARAD and PI3KC3 catalytic domain are labeled. Arrows indicate altered positions of equivalent BECN1/VPS30 residues in a complex containing ATG14.
Acknowledgments
The authors also acknowledge the NDSU Core Biology Facility (funded by NIH grant P30 GM103332-01) for access to tissue culture facilities and the NDSU Advanced Imaging and Microscopy Core Laboratory for access to microscopy and imaging equipment. Work performed at Bio-CAT was also supported by NIH NIGMS 9P41 GM103622. GM/CA@APS has been funded in whole or in part with Federal funds from the NCI (ACB-12002) and the National Institute of General Medical Sciences (AGM-12006). This research used resources of the APS, a U.S. DOE Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02-06CH11357.
FUNDING
This work was supported by NIH grants RO3 NS090939 (S.C.S.) and R15 GM113227 (C.L.C.); a National Science Foundation grant MCB-1413525 (S.C.S.); and NSF and North Dakota EPSCoR grant II-1355466 doctoral dissertation awards for Y.M. and M.S. (PI: S.C.S.).
ABBREVIATIONS
- ATG
autophagy-related protein
- VPS
Vacuolar protein sorting
- Beclin 1
Coiled-coil myosin-like BCL-2 interacting protein also called BECN1, VPS30 or ATG6
- ATG14
autophagy related protein 14, also called BARKOR
- BARKOR
BECN1-associated autophagy-related key regulator
- PI3KC3
class III phosphotidylinositol-3-kinase
- PI3P
phosphatidylinositol 3-phosphate
- UVRAG
UV radiation resistance associated gene protein
- CCD
coiled-coil domain
- BARAD
β-α repeat autophagy domain
- LC3
Light Chain 3
- SEC
size exclusion chromatography
- SAXS
small-angle X-ray scattering
- CD
circular dichroism spectroscopy
- CoIP
co-immunoprecipitation
- IPTG
isopropyl thio-β-D-galactoside
- TEV
tobacco etch virus
- SDS-PAGE
sodium dodecyl sulfate polyacrylamide gel electrophoresis
- MBP
maltose binding protein
- RMSD
root-mean-square deviation
- Rg
radius of gyration
- P(r)
pairwise probability distribution function
- Dmax
maximum particle distribution
- WT
wild-type
References
- 1.Klionsky DJ, Emr SD. Autophagy as a regulated pathway of cellular degradation. Science. 2000;290:1717–1721. doi: 10.1126/science.290.5497.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Levine B, Klionsky DJ. Development by self-digestion: Molecular mechanisms and biological functions of autophagy. Developmental Cell. 2004;6:463–477. doi: 10.1016/s1534-5807(04)00099-1. [DOI] [PubMed] [Google Scholar]
- 3.Xie Z, Klionsky DJ. Autophagosome formation: core machinery and adaptations. Nat Cell Biol. 2007;9 doi: 10.1038/ncb1007-1102. [DOI] [PubMed] [Google Scholar]
- 4.Mizushima N, Yoshimori T, Ohsumi Y. The role of Atg proteins in autophagosome formation. Annu Rev Cell Dev Biol. 2011;27:107–132. doi: 10.1146/annurev-cellbio-092910-154005. [DOI] [PubMed] [Google Scholar]
- 5.Mizushima N. The pleiotropic role of autophagy: from protein metabolism to bactericide. Cell Death Differ. 2005;12(Suppl 2):1535–1541. doi: 10.1038/sj.cdd.4401728. [DOI] [PubMed] [Google Scholar]
- 6.Jin S. Autophagy, mitochondrial quality control, and oncogenesis. Autophagy. 2006;2:80–84. doi: 10.4161/auto.2.2.2460. [DOI] [PubMed] [Google Scholar]
- 7.Fulda S, Gorman AM, Hori O, Samali A. Cellular stress responses: cell survival and cell death. Int J Cell Biol. 2010;2010:214074. doi: 10.1155/2010/214074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Altman BJ, Rathmell JC. Metabolic stress in autophagy and cell death pathways. Cold Spring Harb Perspect Biol. 2012;4:a008763. doi: 10.1101/cshperspect.a008763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shintani T, Klionsky DJ. Autophagy in health and disease: A double-edged sword. Science. 2004;306:990–995. doi: 10.1126/science.1099993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Levine B. Eating oneself and uninvited guests: Autophagy-related pathways in cellular defense. Cell. 2005;120:159–162. doi: 10.1016/j.cell.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 11.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mizushima N, Levine B, Cuervo A, Klionsky D. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liang XH, Kleeman LK, Jiang HH, Gordon G, Goldman JE, Berry G, Herman B, Levine B. Protection against fatal Sindbis virus encephalitis by Beclin 1, a novel Bcl-2-interacting protein. Journal of Virology. 1998;72:8586–8596. doi: 10.1128/jvi.72.11.8586-8596.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liang XH, Jackson S, Seaman M, Brown K, Kempkes B, Hibshoosh H, Levine B. Induction of autophagy and inhibition of tumorigenesis by Beclin 1. Nature. 1999;402:672–676. doi: 10.1038/45257. [DOI] [PubMed] [Google Scholar]
- 15.Itakura E, Kishi C, Inoue K, Mizushima N. Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG. Molecular biology of the cell. 2008;19:5360–5372. doi: 10.1091/mbc.E08-01-0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sun Q, Fan W, Chen K, Ding X, Chen S, Zhong Q. Identification of Barkor as a mammalian autophagy-specific factor for Beclin 1 and class III phosphatidylinositol 3-kinase. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:19211–19216. doi: 10.1073/pnas.0810452105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Itakura E, Mizushima N. Atg14 and UVRAG: mutually exclusive subunits of mammalian Beclin 1-PI3K complexes. Autophagy. 2009;5:534–536. doi: 10.4161/auto.5.4.8062. [DOI] [PubMed] [Google Scholar]
- 18.Matsunaga K, Saitoh T, Tabata K, Omori H, Satoh T, Kurotori N, Maejima I, Shirahama-Noda K, Ichimura T, Isobe T, Akira S, Noda T, Yoshimori T. Two Beclin 1-binding proteins, Atg14L and Rubicon, reciprocally regulate autophagy at different stages. Nature Cell Biology. 2009;11:385–396. doi: 10.1038/ncb1846. [DOI] [PubMed] [Google Scholar]
- 19.Zhong Y, Wang QJ, Li XT, Yan Y, Backer JM, Chait BT, Heintz N, Yue ZY. Distinct regulation of autophagic activity by Atg14L and Rubicon associated with Beclin 1-phosphatidylinositol-3-kinase complex. Nature Cell Biology. 2009;11:468–476. doi: 10.1038/ncb1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fogel AI, Dlouhy BJ, Wang CX, Ryu SW, Neutzner A, Hasson SA, Sideris DP, Abeliovich H, Youle RJ. Role of Membrane Association and Atg14-Dependent Phosphorylation in Beclin-1-Mediated Autophagy. Molecular and Cellular Biology. 2013;33:3675–3688. doi: 10.1128/MCB.00079-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fan W, Nassiri A, Zhong Q. Autophagosome targeting and membrane curvature sensing by Barkor/Atg14(L) Proceedings of the National Academy of Sciences of the United States of America. 2011;108:7769–7774. doi: 10.1073/pnas.1016472108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Glover K, Mei Y, Sinha S. Identifying Intrinsically Disordered Protein Regions Likely to Undergo Binding-Induced Helical Transitions. Biochimica et Biophysica Acta – Proteins and Proteomics. 2016 doi: 10.1016/j.bbapap.2016.05.005. In Press. http://www.sciencedirect.com/science/article/pii/S1570963916300930. [DOI] [PMC free article] [PubMed]
- 23.Oberstein A, Jeffrey PD, Shi Y. Crystal structure of the Bcl-XL-Beclin 1 peptide complex: Beclin 1 is a novel BH3 only protein. The Journal of Biological Chemistry. 2007;282:13123–13132. doi: 10.1074/jbc.M700492200. [DOI] [PubMed] [Google Scholar]
- 24.Ku B, Woo JS, Liang C, Lee KH, Hong HS, Xiaofei E, Kim KS, Jung JU, Oh BH. Structural and biochemical bases for the inhibition of autophagy and apoptosis by viral Bcl-2 of murine γ-Herpesvirus 68. PLoS Pathogens. 2008;4:e25. doi: 10.1371/journal.ppat.0040025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sinha S, Colbert CL, Becker N, Wei Y, Levine B. Molecular basis of the regulation of Beclin 1-dependent autophagy by the γ-herpesvirus 68 Bcl-2 homolog M11. Autophagy. 2008;4:989–997. doi: 10.4161/auto.6803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sinha S, Levine B. The autophagy effector Beclin 1: A novel BH3-only protein. Oncogene. 2009;27(Suppl 1):S137–S148. doi: 10.1038/onc.2009.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mei Y, Su M, Soni G, Salem S, Colbert C, Sinha S. Intrinsically disordered regions in autophagy proteins. PROTEINS: Structure, Function and Bioinformatics. 2014;82:565–578. doi: 10.1002/prot.24424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mei Y, Ramanathan A, Glover K, Christopher Stanley C, Sanishvili R, Chakravarthy S, Yang Z, Colbert CL, Sinha SC. Conformational Flexibility Enables Function of a BECN1 Region Essential for Starvation-Mediated Autophagy. Biochemistry. 2016;55:1945–1958. doi: 10.1021/acs.biochem.5b01264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rostislavleva K, Soler N, Ohashi Y, Zhang L, Pardon E, Burke JE, Masson GR, Johnson C, Steyaert J, Ktistakis NT, Williams RL. Structure and flexibility of the endosomal Vps34 complex reveals the basis of its function on membranes. Science. 2015;350:178–181. doi: 10.1126/science.aac7365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li X, He L, Che KH, Funderburk SF, Pan L, Pan N, Zhang M, Yue Z, Zhao Y. Imperfect interface of Beclin1 coiled-coil domain regulates homodimer and heterodimer formation with Atg14L and UVRAG. Nature communicaitons. 2012;3:662. doi: 10.1038/ncomms1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang W, Choi W, Hu W, Mi N, Guo Q, Ma M, Liu M, Tian Y, Lu P, Wang FL, Deng H, Liu L, Gao N, Yu L, Shi Y. Crystal structure and biochemical analyses reveal Beclin 1 as a novel membrane binding protein. Cell Research. 2012;22:473–489. doi: 10.1038/cr.2012.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Noda NN, Kobayashi T, Adachi W, Fujioka Y, Ohsumi Y, Inagaki F. Structure of the novel C-terminal domain of vacuolar protein sorting 30/autophagy-related protein 6 and its specific role in autophagy. The Journal of biological chemistry. 2012;287:16256–16266. doi: 10.1074/jbc.M112.348250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Obara K, Ohsumi Y. Atg14: a key player in orchestrating autophagy. Int J Cell Biol. 2011;2011:713435. doi: 10.1155/2011/713435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Diao JJ, Liu R, Rong YG, Zhao ML, Zhang J, Lai Y, Zhou QJ, Wilz LM, Li JX, Vivona S, Pfuetzner RA, Brunger AT, Zhong Q. ATG14 promotes membrane tethering and fusion of autophagosomes to endolysosomes. Nature. 2015;520:563–+. doi: 10.1038/nature14147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Baskaran S, Carlson LA, Stjepanovic G, Young LN, Kim DJ, Grob P, Stanley RE, Nogales E, Hurley JH. Architecture and dynamics of the autophagic phosphatidylinositol 3-kinase complex. eLife. 2014;3 doi: 10.7554/eLife.05115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, Kominami E, Ohsumi Y, Yoshimori T. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. The EMBO Journal. 2000;19:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thompson JD, Gibson TJ, Higgins DG. Multiple sequence alignment using ClustalW and ClustalX. Curr Protoc Bioinformatics. 2002 doi: 10.1002/0471250953.bi0203s00. Chapter 2: Unit 2 3. [DOI] [PubMed] [Google Scholar]
- 38.Sheffield P, Garrard S, Derewenda Z. Overcoming expression and purification problems of RhoGDI using a family of “Parallel” expression vectors. Protein Expression and Purification. 1999;15:34–39. doi: 10.1006/prep.1998.1003. [DOI] [PubMed] [Google Scholar]
- 39.Kabsch W. Integration, scaling, space-group assignment and post-refinement. Acta Crystallogr D. 2010;66:133–144. doi: 10.1107/S0907444909047374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta crystallographica. Section D, Biological crystallography. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Emsley P, Lohkamp B, Scott WG, Cowtand K. Features and development of Coot. Acta crystallographica. Section D, Biological crystallography. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Provencher SW, Glöckner J. Estimation of globular protein secondary structure from circular dichroism. Biochemistry. 1981;20:33–37. doi: 10.1021/bi00504a006. [DOI] [PubMed] [Google Scholar]
- 44.Wood CW, Bruning M, Ibarra AA, Bartlett GJ, Thomson AR, Sessions RB, Brady RL, Woolfson DN. CCBuilder: an interactive web-based tool for building, designing and assessing coiled-coil protein assemblies. Bioinformatics. 2014;30:3029–3035. doi: 10.1093/bioinformatics/btu502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huang TT, Hwang JK, Chen CH, Chu CS, Lee CW, Chen CC. (PS)2: protein structure prediction server version 3.0. Nucleic Acids Res. 2015;43:W338–342. doi: 10.1093/nar/gkv454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sali A, Blundell TL. Comparative Protein Modeling by Satisfaction of Spatial Restraints. Journal of Molecular Biology. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 47.Evans HA, Bahri PA, Vu LTT, Barnard KR. Modelling Cobalt Solvent Extraction using Aspen Custom Modeler. Comput-Aided Chem En. 2014;33:505–510. [Google Scholar]
- 48.Petoukhov MV, Franke D, Shkumatov AV, Tria G, Kikhney AG, Gajda M, Gorba C, Mertens HDT, Konarev PV, Svergun DI. New developments in the ATSAS program package for small-angle scattering data analysis. Journal of Applied Crystallography. 2012;45:342–350. doi: 10.1107/S0021889812007662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Konarev PV, Volkov VV, Sokolova AV, Koch MHJ, Svergun DI. PRIMUS: a Windows PC-based system for small-angle scattering data analysis. Journal of Applied Crystallography. 2003;36:1277–1282. [Google Scholar]
- 50.Franke D, Svergun DI. DAMMIF, a program for rapid ab-initio shape determination in small-angle scattering. Journal of Applied Crystallography. 2009;42:342–346. doi: 10.1107/S0021889809000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Volkov VV, Svergun DI. Uniqueness of ab initio shape determination in small-angle scattering. Journal of Applied Crystallography. 2003;36:860–864. doi: 10.1107/S0021889809000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Svergun D, Barberato C, Koch MHJ. CRYSOL – A program to evaluate x-ray solution scattering of biological macromolecules from atomic coordinates. Journal of Applied Crystallography. 1995;28:768–773. [Google Scholar]
- 53.Kozin MB, Svergun DI. Automated matching of high- and low-resolution structural models. Journal of Applied Crystallography. 2001;34:33–41. [Google Scholar]
- 54.Drozdetskiy A, Cole C, Procter J, Barton G. JPred4: a protein secondary structure prediction server. Nucleic Acids Research. 2015;43:W389–W394. doi: 10.1093/nar/gkv332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sreerama N, Woody RW. Estimation of protein secondary structure from circular dichroism spectra: comparison of CONTIN, SELCON, and CDSSTR methods with an expanded reference set. Analytical Biochemistry. 2000;287:252–260. doi: 10.1006/abio.2000.4880. [DOI] [PubMed] [Google Scholar]
- 56.Liang XH, Yu J, Brown K, Levine B. Beclin 1 contains a leucine-rich nuclear export signal that is required for its autophagy and tumor suppressor function. Cancer Research. 2001;61:3443–3449. [PubMed] [Google Scholar]
- 57.Furuya N, Yu F, Byfield M, Pattingre S, Levine B. The evolutionarily conserved domain of Beclin 1 is required for Vps34 binding, autophagy and tumor suppressor function. Autophagy. 2005;1:46–52. doi: 10.4161/auto.1.1.1542. [DOI] [PubMed] [Google Scholar]
- 58.Pattingre S, Tassa A, Qu X, Garuti R, Liang XH, Mizushima N, Packer M, Schneider MD, Levine B. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell. 2005;122:927–939. doi: 10.1016/j.cell.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 59.Holt JT, Thompson ME, Szabo C, Robinson-Benion C, Arteaga CL, King MC, Jensen RA. Growth retardation and tumour inhibition by BRCA1. Nature Genetics. 1996;12:298–302. doi: 10.1038/ng0396-298. [DOI] [PubMed] [Google Scholar]
- 60.Xiong XW, Tao RY, DePinho RA, Dong XC. The Autophagy-related Gene 14 (Atg14) Is Regulated by Forkhead Box O Transcription Factors and Circadian Rhythms and Plays a Critical Role in Hepatic Autophagy and Lipid Metabolism. Journal of Biological Chemistry. 2012;287:39107–39114. doi: 10.1074/jbc.M112.412569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Noble C, Dong J, Manser E, Song H. Bcl-xL and UVRAG cause a monomer-dimer switch in Beclin1. Journal of Biological Chemistry. 2008;283:26274–26282. doi: 10.1074/jbc.M804723200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Adi-Harel S, Erlich S, Schmukler E, Cohen-Kedar S, Segev O, Mizrachy L, Hirsch JA, Pinkas-Kramarski R. Beclin 1 self-association is independent of autophagy induction by amino acid deprivation and rapamycin treatment. Journal of Cellular Biochemistry. 2010;110:1262–1271. doi: 10.1002/jcb.22642. [DOI] [PubMed] [Google Scholar]
- 63.Ruck A, Attonito J, Garces KT, Nunez L, Palmisano NJ, Rubel Z, Bai Z, Nguyen KC, Sun L, Grant BD, Hall DH, Melendez A. The Atg6/Vps30/Beclin 1 ortholog BEC-1 mediates endocytic retrograde transport in addition to autophagy in C. elegans. Autophagy. 2011;7:386–400. doi: 10.4161/auto.7.4.14391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shravage BV, Hill JH, Powers CM, Wu L, Baehrecke EH. Atg6 is required for multiple vesicle trafficking pathways and hematopoiesis in Drosophila. Development. 2013;140:1321–1329. doi: 10.1242/dev.089490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wirawan E, Vande Walle L, Kersse K, Cornelis S, Claerhout S, Vanoverberghe I, Roelandt R, De Rycke R, Verspurten J, Declercq W, Agostinis P, Vanden Berghe L, Lippens S, Vandenabeele P. Caspase-mediated cleavage of Beclin-1 inactivates Beclin-1-induced autophagy and enhances apoptosis by promoting the release of proapoptotic factors from mitochondria. Cell Death Dis. 2010;1:e18. doi: 10.1038/cddis.2009.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kang R, Zeh H, Lotze M, Tang D. The Beclin 1 network regulates autophagy and apoptosis. Cell Death and Differentiation. 2011;18:571–580. doi: 10.1038/cdd.2010.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ma X, Liu H, Foyil SR, Godar RJ, Weinheimer CJ, Hill JA, Diwan A. Impaired Autophagosome Clearance Contributes to Cardiomyocyte Death in Ischemia/Reperfusion Injury. Circulation. 2012;125:3170–3181. doi: 10.1161/CIRCULATIONAHA.111.041814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang K, Chen Y, Zhang Y, Yao Q, Zhuo H. Novel CRM1 Inhibitor Prevent the Nuclear Export of Beclin-1 Blocks Autophagy in Mantle Cell Lymphoma. Blood. 2014;124 [Google Scholar]
- 69.Ma XC, Liu HY, Murphy JT, Foyil SR, Godar RJ, Abuirqeba H, Weinheimer CJ, Barger PM, Diwan A. Regulation of the transcription factor EB-PGC1alpha axis by beclin-1 controls mitochondrial quality and cardiomyocyte death under stress. Molecular and Cellular Biology. 2015;35:956–976. doi: 10.1128/MCB.01091-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.He C, Levine B. The Beclin 1 interactome. Current Opinion in Cell Biology. 2010;22:140–149. doi: 10.1016/j.ceb.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Levine B, Liu R, Dong X, Zhong Q. Beclin orthologs: integrative hubs of cell signaling, membrane trafficking, and physiology. Trends in Cell Biology. 2015;25:533–544. doi: 10.1016/j.tcb.2015.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Liang C, Feng P, Ku B, Dotan I, Canaani D, Oh BH, Jung JU. Autophagic and tumour suppressor activity of a novel Beclin1-binding protein UVRAG. Nature Cell Biology. 2006;8:688–699. doi: 10.1038/ncb1426. [DOI] [PubMed] [Google Scholar]
- 73.Liang C, Lee J-s, Inn K-S, Gack MU, Li Q, Roberts EA, Vergne I, Deretic V, Feng P, Akazawa C, Jung JU. Beclin1-binding UVRAG targets the class C Vps complex to coordinate autophagosome maturation and endocytic trafficking. Nat Cell Biol. 2008;10:776–787. doi: 10.1038/ncb1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Liang C, Feng P, Ku B, B-H O, Jung JU. UVRAG: A new player in autophagy and tumor cell growth. Autophagy. 2007;3:69–71. doi: 10.4161/auto.3437. [DOI] [PubMed] [Google Scholar]
- 75.Criollo A, Niso-Santano M, Malik SA, Michaud M, Morselli E, Marino G, Lachkar S, Arkhipenko AV, Harper F, Pierron G, Rain JC, Ninomiya-Tsuji J, Fuentes JM, Lavandero S, Galluzzi L, Maiuri MC, Kroemer G. Inhibition of autophagy by TAB2 and TAB3. Embo Journal. 2011;30:4908–4920. doi: 10.1038/emboj.2011.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Niso-Santano M, Criollo A, Malik SA, Michaud M, Morselli E, Marino G, Lachkar S, Galluzzi L, Maiuri MC, Kroemer G. Direct molecular interactions between Beclin 1 and the canonical NF kappa B activation pathway. Autophagy. 2012;8:268–270. doi: 10.4161/auto.8.2.18845. [DOI] [PubMed] [Google Scholar]
- 77.Hamasaki M, Furuta N, Matsuda A, Nezu A, Yamamoto A, Fujita N, Oomori H, Noda T, Haraguchi T, Hiraoka Y, Amano A, Yoshimori T. Autophagosomes form at ER-mitochondria contact sites. Nature. 2013;495:389–393. doi: 10.1038/nature11910. [DOI] [PubMed] [Google Scholar]
- 78.Su M, Mei Y, Sanishvili R, Levine B, Colbert CL, Sinha S. Targeting γ-herpesvirus 68 Bcl-2–mediated down-regulation of autophagy. Journal of Biological Chemistry. 2014;289:8029–8050. doi: 10.1074/jbc.M113.515361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.DeLano WL. The PyMOL Molecular Graphics System. DeLano Scientific; San Carlos, CA, USA: 2002. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Residues at the interaction interface (a, d positions of heptad repeats) in the various BECN1/VPS30-containing dimers.
Figure S1. ATG14 CCD sequence alignment. Sequence alignment of predicted CCDs from five diverse eukaryotic ATG14 proteins. Yellow, orange and red represent increasing sequence conservation, with red corresponding to invariant residues. Predicted secondary structures are shown below the sequence alignment. The predicted CCD region used in this study is boxed. Conserved residues involved in hydrophobic pairing in the heterodimer, that were selected for mutagenesis are indicated by red arrowheads.
Figure S2. SEC purification of the BECN1:ATG14 CCD heterodimer complex. SEC curves shown correspond to BECN1 CCD (green) and BECN1:ATG14 CCD (blue). Elution positions for different molecular weight markers and the molecular weight calculated from SEC are indicated. SDS-PAGE analysis of the peak fractions is shown.
Figure S3. CD spectra of different CCDs. The spectra for the MBP-ATG14 CCD fusion protein, the BECN1:ATG14 CCD complex, the BECN1 CCD, and MBP are color coded as indicated in the legend.
Figure S4. Comparison of BECN1 CCD containing dimers. All proteins are rendered in ribbon, colored as follows: BECN1, magenta; ATG14, wheat; VPS30, yellow and VPS38, green. The residues involved in interface interactions are shown in stick with atoms color-coded by atom type: O, red; N, blue; S, yellow; and C, colored according to main chain ribbon for that molecule. Superposition of (A) BECN1:ATG14 CCD complex and the BECN1 CCD homodimer. (B) VPS30:VPS38 complex and the BECN1 CCD homodimer. (C) BECN1:ATG14 CCD complex and the VPS30:VPS38 complex.
Figure S5. Displacement of the BECN1 BARAD domain in Complex I due to the curved BECN1:ATG14 quaternary structure. All proteins are shown in ribbon, colored as follows: BECN1, magenta; ATG14, wheat; VPS30, yellow; VPS38, green; VPS15, grey, and VPS34, blue. Protein domains implicated in membrane interaction, the BECN1 BARAD, VPS38 BARAD and PI3KC3 catalytic domain are labeled. Arrows indicate altered positions of equivalent BECN1/VPS30 residues in a complex containing ATG14.


