Abstract
The p21-family members of Rho GTPases are important for the control of actin cytoskeleton dynamics, and are critical regulators of phagocytosis. The three-dimensional structure of phagosomes and the highly compartmentalized nature of the signaling mechanisms during phagocytosis require high-resolution imaging using ratiometric biosensors to decipher Rho GTPase activities regulating phagosome formation and function. Here we describe methods for the expression and ratiometric imaging of FRET-based Rho GTPase biosensors in macrophages during phagocytosis. As an example, we show Cdc42 activity at the phagosome over Z-serial planes. In addition, we demonstrate the usage of a new, fast, and user-friendly deconvolution package that delivers significant improvements in the attainable details of Rho GTPase activity in phagosome structures.
Keywords: Macrophages, Phagosome, Ratiometric imaging, FRET, Biosensors, Z-stack, Deconvolution
1 Introduction
Phagocytosis is a critical function of our immune system performed by phagocytes to eliminate foreign invaders such as bacteria and fungi. Phagocytosis targets particles larger than 0.5 μm and is triggered by direct ligand–receptor contacts between the particle and the phagocyte [1]. While phagocytosis can be triggered via several different receptors, one of the best-studied phagocytic receptors is the Fc gamma receptor (FcγR), which recognizes particles opsonized by IgG antibodies. FcγR-triggered phagocytosis involves a series of complex changes in cell morphology with an absolute dependence on actin reorganization [1].
Master regulators of actin dynamics are the members of the p21 Rho family of small GTPases, belonging to the Ras superfamily of small GTPases [2, 3]. These GTP-binding signaling molecules alternate between GDP-bound inactive and GTP-bound active state, in which they bind and activate wide array of downstream effector molecules to elicit a cellular response [4, 5]. The Rho GTPase family has 20 known members, and the canonical RhoA, Rac1, and Cdc42 that were first identified, are the best studied in the family [6]. Both Rac1 and Cdc42 have been shown to be required for FcγR-mediated phagocytosis [7, 8].
The traditional workhorse technique to study GTPase activation during cellular responses has been an affinity-based precipitation assay [9]. While this assay is highly useful, it provides only ensemble averages from whole cells that lack discrete resolution in space and time, which is not ideal for studying highly compartmentalized three-dimensional cellular structures, such as phagosomes. Forster resonance energy transfer (FRET)-based biosensors have become powerful tools to decipher spatial and temporal activation dynamics of Rho GTPases at high resolution on a single-cell basis, allowing researchers to gain further insights into their functional roles [10]. A previous study analyzing Rac1 and Cdc42 activity in macrophage phagosomes used bi-molecular versions of FRET biosensors, where the FRET donor and acceptor halves are on separate molecules [11]. This approach, while useful, involves cumbersome data analysis due to the non-equimolar distribution of the two separate FRET donor/acceptor components. We have overcome this issue by the development of fully genetically encoded, single-chain, FRET-based Cdc42 and Rac1 biosensors, applicable for fixed- and live-cell imaging [12, 13]. Importantly, our design maintains the C-terminal polybasic region of the Rho GTPases and allows for correct native intracellular localization and interaction with upstream regulators, including guanosine nucleotide dissociation inhibitor (GDI).
Here we detail approaches for expression of FRET-based bio-sensorsin hematopoietic cells, using a murinemonocyte/macrophage RAW 264.7 cell line as a model system and discuss important considerations relevant for successful expression of full-length biosen-sor that forms the critical basis for proper biosensor readout and data interpretation. In this chapter we describe methods for the implementation of biosensors to study phagocytosis and provide a specific example using the Cdc42 biosensor that shows Cdc42 activity at the phagosome in macrophages [12].
2 Materials
RAW/LR5 cells derived from RAW 264.7 [8].
GP2-293 packaging cell line (Clontech).
RAW/LR5 growth medium: RPMI 1640 with L-glutamine supplemented with 10 % newborn calf serum (NBCS), 1 % penicillin/streptomycin (Pen/Strep).
RAW/LR5 induction medium: RPMI 1640 with L-glutamine supplemented with 10 % fetal bovine serum (FBS), 1 % Pen/ Strep.
RAW/LR5 infection medium: RPMI 1640 with L-glutamine supplemented with 5 % NBCS, 1 % Pen/Strep.
GP2-293 growth medium: DMEM supplemented with 10 % FBS, 1 % Pen/Strep, 1 % Glutamax.
0.05% Trypsin/EDTA.
Buffer with divalent (BWD): 125 mM NaCl, 5 mM KCl, 1 mM KH2PO4, 5 mM glucose, 10 mM NaHCO3, 1 mM MgCl2, 1 mM CaCl2, and 20 mM HEPES.
Phosphate buffered saline (PBS) without calcium and magnesium.
10 mM EDTA in PBS: dilute 0.5 M EDTA (pH 8) in PBS.
3.7 % formaldehyde in BWD.
50 % glycerol in PBS.
Tris-buffered saline (TBS): 137 mM NaCl, 24.7 mM Tris- base, pH 7.4.
OptiMEM.
FuGENE HD transfection reagent (Promega).
Lipofectamine 2000 transfection reagent (Invitrogen).
Retro-X virus concentrator (Clontech).
0.01% poly-L-lysine (Sigma); dilute 1:10 in PBS for working solution.
1.5 mL microcentrifuge tubes.
15 and 50 mL polypropylene tubes.
6 and 10 cm cell culture dishes.
0.45 μm Surfactant-free Cellulose Acetate SFCA-membrane 28 mm syringe.
12-well and 24-well cell culture plates.
12 mm round glass coverslips.
3″ ×1″ × 1 mm microscope slides, plain.
8 mg/mL Polybrene aqueous stock solution.
1 mg/mL Doxycycline (Dox) aqueous stock solution.
100 mg/mL G418 (neomycin) aqueous stock solution.
100 mg/mL Zeocin aqueous stock solution.
Sheep blood alsevers.
Rabbit IgG anti-sheep red blood cell.
18 Gauge needle with syringe.
DNA constructs: VSV-g, gag/pol, Rev, Tet, Cdc42, or other biosensors [12].
3 Methods
3.1 Expression of Rho GTPase Biosensors in a Macrophage Cell Line
3.1.1 Transient Expression of Rho GTPase Biosensors in RAW/LR5 Cells
As an example, we will use our protocol for the Cdc42 biosensor [12].
(Day 1) Plate RAW/LR5 cells in a 12-well plate the day before transfection so that they will be approximately 60–80 % confluent for transfection the next day.
(Day 2) Let FuGENE HD (see Note 1) and OptiMEM come to room temperature (RT), ~10 min.
Prepare FuGENE HD transfection mix (DNA:FuGENE HD ratio 1:3): in 100 μL OptiMEM add 1 μg Cdc42 biosensor expression plasmid (see Note 2), vortex for 10 s; add 3 μL FuGENE HD and pipet to mix (do not vortex at this point). Incubate for 15 min at RT. The transfection mix can be scaled based on surface area if smaller or larger cell numbers are needed.
During incubation of the transfection mix, rinse cells once with PBS and add 500 μL of complete medium for transfection.
Add transfection mix dropwise. Swirl gently to mix.
Incubate 2–3 h (see Note 3) at 37 °C and 5 % CO2.
Transfer medium containing the transfection mix to a 15 mL tube to collect cells that may have detached during the incubation. Then lift adherent cells by adding 10 mM EDTA/PBS to the well and incubate for ~5 min at 37 °C and 5 % CO2. Tap gently to lift cells and transfer to the 15 mL tube. Rinse the well once with complete medium to collect all cells and add to the 15 mL tube. Spin cells at 300 × g for 3 min.
Aspirate the fluid and resuspend the cell pellet in 2 mL complete medium.
Set up sterile 12 mm round coverslips in 24-well plate (4 cov-erslips / 12-well transfection) (see Note 4). Add 500 μL of cell suspension per coverslip.
Let cells recover overnight at 37 °C and 5 % CO2.
(Day 3) Next day, assess transfection efficiency and perform phagocytosis assays.
3.1.2 Generation of Stable RAW/LR5 with Inducible Expression of Rho GTPase Biosensors
There are several important considerations for successful creation of an inducible system for the expression of single-chain Rho GTPase biosensors in general, as well as specifically in hematopoi-etic cells (see Notes 5 and 6). Generating stable cell lines with inducible expression of a biosensor requires the creation of a double-stable, RetroX-Tet-OFF Advanced/gene of interest (GOI) cell line, involving two consecutive retroviral transductions of the target cell line (see Notes 7 and 8). Below we outline the protocols we use routinely in our laboratory:
-
1
(Day1) Start virus production by setting up 4 × 10 cm dish per GOI (see Note 9) to plate GP2-293 packaging cells (see Note 10).
-
2
Coat 10 cm dish with 0.001 % poly-L-Lysine solution diluted in 1× PBS for at least 10 min at RT.
-
3
Plate 6×106 GP2-293 cells per coated 10 cm dish and incubate overnight at 37 °C and 5 % CO2.
-
4
(Day 2) Transfect GP2-293 cells using Lipofectamine 2000; per 10 cm dish mix (see Notes 11 and 12): in tube 1 add 4 μg pVSV-g+4 μg gag/pol+16 μg Cdc42 biosensor [12], or other carrier DNA in 500 μL OptiMEM (for lentivirus: 4 μg pVSV-g+2 μg gag/pol+2 μg Rev+2 μg Tet+14 μg carrier DNA). In tube 2 add 60 μL of Lipofectamine 2000 in 500 μL optiMEM.
-
5
Vortex tubes 1 and 2 for 10 s, spin and incubate 5 min at RT.
-
6
Combine tubes 1 and 2, vortex for 10 s, spin and incubate 20 min at RT (see Note 13).
-
7
During incubation of transfection mix, wash the cells with 1× PBS and add 4 mL DMEM containing 10 % FBS with no antibiotics.
-
8
Add 1 mL transfection mix to cells dropwise. Gently swirl to mix.
-
9
Incubate overnight at 37 °C and 5 % CO2.
-
11
(Day 3) Supplement dishes with 3 mL serum-free DMEM to adjust serum concentration to 5 %.
-
12
Transfer dishes to 32 °C since retroviruses are more stable at 32 °C (for lentivirus keep at 37 °C).
-
13
Allow cells to produce virus for 48 h.
-
14
(Day 5) Start harvesting virus by pooling supernatants from the 4 dishes into a 50 mL tube; swirl plate a little before pipetting the supernatant.
-
15
Spin 3 min at 300 × g.
-
16
Filter supernatant using 0.45 μm SFCA-membrane 28 mm syringe filter. Filter slowly, dropwise along the sides of a 50 mL tube.
-
17
Estimate the final volume of the filtrate and divide the volume by 3; that is the volume of Retro-X virus concentrator solution to add to the filtered viral titer.
-
18
Add virus concentrator straight into filtrate, pipet up and down to clean the pipet of all the virus concentrator solution. Invert gently a few times to get a homogenous solution.
-
19
Let it sit overnight at 4 °C.
-
20
Meanwhile, plate RAW/LR5 cells for infection in 6- or 10 cm dishes at approximately 20 % confluency for next day. Low confluency is recommended as infection might occur over a 3-day period, depending on GOI to be expressed.
-
21
(Day 6) Collect virus by centrifuging tubes (from step 19) at 1500×g for 45 min at 4 °C.
-
22
Resuspend the virus pellet very gently by pipetting up and down in 200 μL of RPMI without serum (or the base medium in which the target cells are normally grown) per 10 cm dish (i.e., if 4 × 10 cm dishes are pooled, add 800 μL). At first the pellet will come apart in pieces, but will become a clear solution while pipetting up and down.
-
23
Aliquot at 100–120 μL.
-
24
Store aliquots at −80 °C. If transducing cells on the same day, keep aliquots at 4 °C until use. Freeze-thawing one time reduces the viral infectivity by at least 50 %.
-
25
To transduce cells (see Note 14), start by rinsing cells once with PBS.
-
26
For transduction in 10 cm dish, add 5 mL RAW/LR5 infection medium, 5 μL of 8 mg/mL polybrene (final concentration of 8 μg/mL), and 1 aliquot of virus (for 6 cm dishes: 3 mL medium, 3 μL of polybrene stock and 1 aliquot of virus). The use of polybrene increases infectivity by 1000 times.
-
27
Incubate cells at 32 °C and 5 % CO2 (for lentivirus keep at 37 °C). Virus will have infectivity for 6–8 h.
-
28
At the end of the day, aspirate medium and repeat step 26 for overnight incubation (see Note 15). Repeat morning and evening dosing of virus for up to 3 days to maximize infection efficiency (for lentivirus, do one dose per day for 2 consecutive days maximum). For generating the stable Tet-OFF tetracycline Trans-Activator (tTA)-expressing cell line, infect for 3 days (6 doses total) to achieve optimal integration of tTA for most efficient inducible system. Check infection efficiency for biosensor expression by fluorescence microscopy. If expression is very good (meaning most cells express and they are bright enough), then there is no need to infect on the 3rd day. Usually we start seeing expression after 3–4 doses (2 days of infection). If expression is low even after a 3-day infection, then it is an indication of a problem (see Note 16).
-
29
Let cells recover from infection for one day in complete medium. To repress biosensor expression, supplement medium with 2 μg/mL Dox. It is not necessary to supplement with Dox during generation of stable tTA-expressing cell line.
-
30
Start selection. For stable tTA integration select with G418 starting at 500 μg/mL and increase by doubling the concentration, up to 2 mg/mL. For stable biosensor integration select with Zeocin starting at 250 μg/mL (in the presence of Dox) and increase by doubling the concentration, up to 1 mg/mL as cells become tolerant of the lower concentration. Selection may take up to 3 weeks. During selection cells will look stressed, however their morphology and growth will return to normal by the end of selection process.
-
31
For normal culture of double-stable RetroX Tet-OFF Advanced/ biosensor RAW/LR5 cells, maintain cells in 2 μg/mL Dox, 1 mg/mL G418 and 600 μg/mL Zeocin (see Note 17).
3.1.3 Induction of Biosensor Expression (See Note 18)
Wash cells once with PBS.
Aspirate and add 1 mL trypsin. Spread evenly and wait ~3 min. Tap side of the dish to dislodge cells. Check that the cells are detached and flowing smoothly by light microscopy or by holding the plate up against the light (see Note 19).
Add 4 mL RAW/LR5 induction medium. Pipette up and down and transfer to a 15 mL tube (see Note 20).
Rinse the plate with another 5 mL of induction medium and pool together in the 15 mL tube.
Spin 300×g for 3 min.
Rinse with 7–10 mL induction medium and spin as above.
Aspirate and resuspend cell pellet in induction medium.
Plate cells at 1:10 dilution in 10 cm dish (this is based on the 50 % confluency, see Note 17). For most efficient induction it is best to pass the cells at low confluency. One 10 cm dish will yield enough induced cells for a typical imaging session for ratiometric imaging. For western blot analysis of biosensor expression set up 4 dishes to yield enough cells for the lysate preparation.
Next day, repeat the trypsinization as above (steps 1–7). For ratiometric imaging the following day, pass at 1:3 or 1:4 onto 12 or 25 round mm coverslip. For preparing lysates for western blot analysis of biosensor expression levels over 72 h period, pass at 1:3 in 6 × 10 cm dishes for the 24 h time point, 1:3 in 3 × 10 cm dishes for 48 h time point, and 1:3 in 1 × 10 cm dishes for the 72 h time point after the 2nd trypsinization (see Note 21).
3.2 Phagocytosis Assay
While there are several ways to assay phagocytosis, here we detail synchronized phagocytosis as performed in Hanna et al. [12].
3.2.1 Opsonization of Red Blood Cells (RBCs)
This procedure typically yields approximately 1–2 × 108 IgG-coated RBCs in 1 mL.
Using sterile technique, draw some blood from sheep blood alsevers vial using an 18 gauge needle with syringe. Transfer 250 μL sheep blood into 1.5 mL microcentrifuge tube.
Pellet RBCs by spinning for 10 s at maximum speed using a bench-top mini microcentrifuge.
Wash RBCs pellet twice with 1 mL BWD. Spin as above.
Resuspend RBCs in 1 mL BWD.
In parallel, add 5 μL rabbit IgG anti-sheep RBC (see Note 22) to 9 mL BWD in 15 mL polypropylene tube and mix by inverting few times.
Add RBCs to the antibody solution and mix immediately by inverting gently few times.
Incubate for 20 min at 37 °C water bath.
Incubate for 20 min on ice.
Pellet IgG-coated RBCs by centrifuging at 1250 × g for 5 min.
Aspirate supernatant. Resuspend RBCs in 1 mL BWD and transfer to 1.5 mL microcentrifuge tube. Spin as in step 2.
Wash IgG-coated RBCs twice with 1 mL BWD. Spin as in step 2.
Resuspend in 1 mL BWD.
Store at 4 °C. Use within 7 days.
3.2.2 Synchronized Phagocytosis Assay
Cells expressing the biosensor either by transient expression or following stable induction are plated on 12 mm round cover-slips in wells of a 24-well plate and allowed to attach and spread overnight at 37 °C.
Remove coverslips from 24-well plate and chill on ice (see Note 23) by replacing medium with 35–50 μL ice-cold BWD and let sit for 5–10 min on ice.
Add 25 μL of opsonized RBCs into 500 μL of ice-cold BWD and gently tap to mix.
Replace plain BWD with 35–50 μL of ice-cold BWD containing opsonized RBCs and incubate coverslip for 15 min on ice to allow RBCs to bind to the cells.
During step 4 prewarm 500 μL BWD in 24-well plate in 37 °C water bath.
Rinse cells with ice-cold BWD on ice three times to wash away unbound RBCs.
Transfer coverslips to wells of 24-well plate containing pre-warmed BWD in water bath for 1 min, or the desired time (1 min for early events and 5 min for maximal cup formation), before fixing with 3.7 % formaldehyde in BWD.
Fix for at least 10 min. At this point, coverslips can be stored in BWD overnight at 4 °C or directly stained for imaging.
Permeabilize with 0.2 % Triton X100 in BWD for 10 min.
Stain RBCs and actin by adding TBS containing Alexa Fluor 568 anti-rabbit IgG antibody (1:400) and Alexa Fluor 680-phal-loidin (1:20) and incubate for 20–30 min at RT (see Note 24).
Rinse three times with TBS.
Mount on glass slide in 50 % glycerol in PBS.
3.3 Fixed Cell Imaging and Data Analysis
Activation of Rho GTPases at individual phagocytic events, using the Cdc42 biosensor in this example, is measured by observing the ratio of FRET emission to the donor mCerulean emission. We briefly outline the steps below:
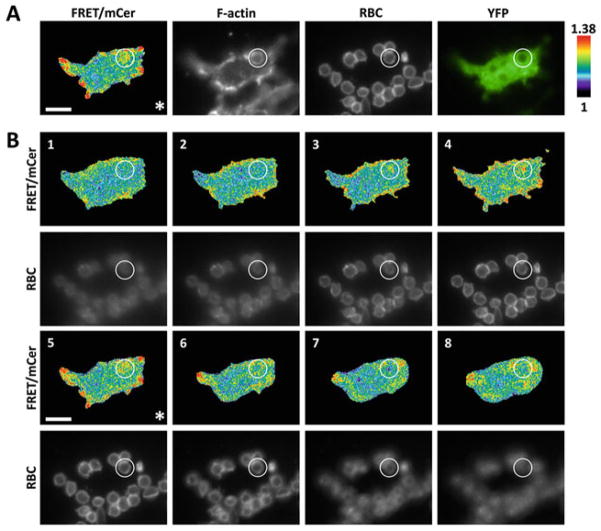
Using a 60× magnification objective lens (60× DIC N/A 1.45), acquire mCerulean (CFP), FRET, and mVenus (YFP) emission images upon excitation by mCerulean, mCerulean, and mVenus excitation wavelengths, respectively (see Note 25). As shown in our previous publication [12], Fig. 1a shows a representative ratiometric image of localized Cdc42 activity with an F-actin-rich phagocytic cup at the F-actin focal plane (see Notes 26 and 27).
The optimal focal plane for the phagocytic cup can be set by observation of co-localization of F-actin staining (phalloidin) with a bound RBC. Alternatively, confocal imaging could be used to reduce the out of focus light when obtaining a Z-stack (see Note 28).
Obtain the shading correction image set (see Note 29), and the camera noise correction image set (see Note 30) as previously described [14, 15], taking care to maintain the exposure and imaging conditions identical to the actual data acquisition. If imaging a Z-stack, obtain appropriate shading correction image sets at the correct Z-distance positions.
Process the background subtraction (see Note 31), threshold-masking (see Note 32), and ratio calculations (see Note 33) at each Z-position as previously described [14, 15].
Apply a linear pseudocolor lookup table to the ratio images and adjust the image scaling appropriately to visualize the Cdc42 activation patterns at phagocytic cups. Make sure to apply identical scaling limits to all planes if imaging a Z-series.
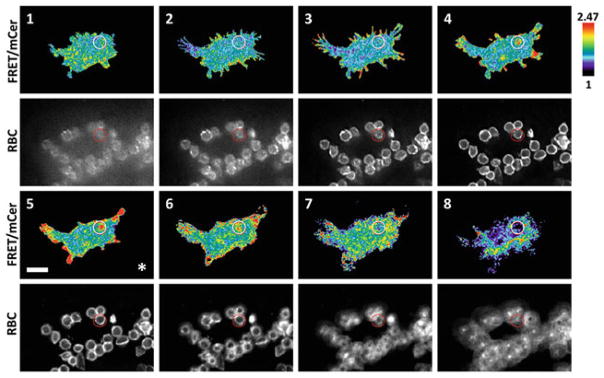
Alternatively, process the raw Z-series data set using a deconvolution algorithm to enhance the signal to noise ratio and then process the Z-series as in steps 3–5 above. This deconvolution enhances the information obtained in the ratio images. Compare Fig. 1 and the same images that have been subsequently deconvolved and shown in Fig. 2 (see Notes 34 and 35).
Fig. 1.
Cdc42 activity over a stack of Z-serial planes containing a developing phagosome. RAW/LR5 cells transiently expressing the Cdc42 biosensor were imaged (a) at optimal F-actin focal plane and (b) in Z-series at 1 μm-steps where the focal plane from A was set as the center (*). Planes 4-1 progress down towards the base of the phagocytic cup and below, while planes 6–8 move upwards from the F-actin plane. FRET/mCerulean ratio image, phalloidin staining of F-actin, red blood cell (RBC) staining and YFP for biosensor localization are shown; representative image set of n = 6 cells. Scale bar = 10 μm. Modified from Hanna et al. [12]
Fig. 2.
Cdc42 activity over serial planes of the phagosome post image deconvolution. An identical set of raw input images presented in Fig. 1 was processed for deconvolution using the Microvolution GPU-deconvolution package [28]. Image processing and ratiometric calculations were performed as in Fig. 1
Acknowledgments
This work was supported by National Institutes of Health grants T32GM007491 to VM, GM071828 to DC, and GM093121 to LH.
Footnotes
Macrophage cell lines are challenging to transfect in general. We have had the most success with FuGENE HD, giving a transfection efficiency of ~10 %. However, the transfection efficiency also varies greatly depending on the GOI being expressed. The DNA amount and DNA:FuGENE HD ratio have been carefully optimized for RAW/LR5 cells, thus the protocols described may be suboptimal for other cell types.
The Cdc42 biosensor cassette was subcloned into the pTriEX-4 vector backbone (Novagen). The pTriEX-4 vector can be used for transient overexpression of the biosensor in mammalian cells but it does not contain any mammalian antibiotic resistance gene to select for stable expression.
Overnight incubation of the cells increases transfection efficiency, while cell health is adversely affected and preactivation can also be a confounding issue. For a simple protein expression analysis an overnight incubation is tolerable. However, for downstream cellular assays we routinely opt for shorter transfection times, in the order of 2–3 h. For example, if one is interested in studying podosome function, short incubation times with the transfection mixture is critical. We found that podosomes are no longer observable if cells are incubated longer than 2–3 h with the transfection mixture.
Keep stock of 12 mm round coverslips in 100 % ethanol and flame them to sterilize.
Using viral transduction for the generation of stable cell lines of GOIs is problematic for constructs that contain tandem, repeated sequences (such as our FRET biosensors) since they are susceptible to homologous recombination from the intrinsic properties of retrovirus [16, 17]. This may result in internal deletions of the repeated sequences within the GOI. Single-chain biosensors incorporate a FRET pair of two fluorescent proteins that are highly homologous. In addition, our designs for Rac1 and Cdc42 biosensors include two tandem repeats of the p21 binding domain of PAK1 [12, 13]. The presence of two sets of repeated sequences places the biosensors at extremely high risk for homologous recombination. We recently reported a generalizable solution, in which “synonymous modification” of the DNA sequence encoding the biosensor offered a simple means to overcome this significant problem [18]. We now routinely apply synonymous modification to all of our biosensor systems.
Another important factor in stable biosensor expression is proper promoter usage for ectopic gene expression. Different promoters have variable resistance against potential promoter silencing by host cells [19]. In our second-generation Tet-OFF system we express tTA under the human elongation factor 1α (EF1α) promoter, as the traditionally used CMV promoter is subject to silencing in hematopoietic cells [20, 21].
In our laboratory we use the second-generation Tet-OFF inducible system from Clontech for stable expression of all of our biosensors. Generating stable cell lines with inducible expression of biosensor requires the creation of a double-stable, RetroX-Tet-OFF Advanced/GOI cell line, involving two consecutive infections of the target cell line. First infection generates a cell line with stable, constitutive expression of tTA. The stable tTA cell line then serves as the subsequent base cell line for a second round of infection for the stable incorporation of any GOI for inducible expression. Clontech now has available a more advanced third-generation Tet-ON 3G inducible system [22, 23], however we find that this system is not particularly suitable for FRET biosensor expression. The Tet-ON system requires the presence of Dox in medium for the induction of the GOI. Importantly, Dox possesses intrinsic fluorogenic properties [24]. Macrophages, as phagocytes, internalize and accumulate considerable amount of Dox, creating measurable background auto-fluorescence that overlaps with the spectral properties of the fluorescent protein FRET pair (i.e. mCerulean and mVenus pair) used in our Rho GTPase biosensors, which impacts the ratiometric calculations. Consequently, it is best to use a Tet-OFF system for biosensor expression that requires removal of Dox for induction of the GOI.
This procedure involves production of retrovirus that is considered a biosafety level 2 organism by the National Institute of Health and Center for Disease Control. This requires the observance of Biosafety Level 2 practices. The viral packaging system used consists of retroviral components that are on separate plasmids, and the resulting retroviruses produced are replication-incompetent, adhering to the Biosafety Level 2 requirements.
The number of plates can be scaled as needed. If using HEK293 for virus production, we recommend supplementing transfection mixture either with pCL-Eco for ecotropic or pCL-Ampho for amphotropic packaging system, depending on the target cell. Ecotropic infects rodents (but not hamster) only, while amphotropic infects rodents and human (but not hamster). Some cells have a block on retroviral infection (i.e. neurons) so those must be infected with lentivirus. VSV-g -pseudo-typed virus will infect everything including hamster cells.
It is important to achieve a single cell suspension (no clumps) of GP2-293 cells both for passaging during normal cell culture and plating for virus production. In addition, GP2-293 cells can only be kept in culture for a limited number of passages after thawing, approximately 3–4 weeks since virus production decreases as they age. One indication for discontinuing their culture is that they proliferate at higher rate. While the manufacturer does recommend plating them on gelatin-coated dishes for culture purposes we find that this is not necessary.
In our hands Lipofectamine 2000 is most optimal for highest expression by transient transfection of GP2-293 cells. In addition, we find Lipofectamine 2000 better suited for homogenous transfection of multiple-DNA plasmids in a single transfection mixture.
This protocol is for the production and transduction of retroviruses. Modifications relevant for lentivirus will be noted in the protocol. Scale amount of transfection mixture as needed and prepare a master mixture.
Transfection mixture is viable for 4–6 h.
If frozen stable tTA expressing cells are to be infected with a GOI, thaw out the cells at least a week ahead of the planned infection and keep them in G418 at 2 mg/mL. During selection for GOI, do not use G418 as all three antibiotics at these high concentrations are too toxic for the cells to tolerate.
It is not a good idea to add a new aliquot of virus to the existing virus-containing medium because too much virus could be toxic. Therefore it is best to aspirate the medium prior to applying an additional dose of virus.
There are several possible reasons for poor biosensor expression. It may be that the GP2-293 cells are too old or not handled properly during normal culture. GP2-293 cells should not be allowed to overgrow and it is important to obtain single cell suspension both during normal passage and plating for virus preparation. It may be possible that the viral aliquot is too old since frozen virus loses infectivity over time. Also, it is possible that the plasmid preparation may contain contaminants that interfere with downstream applications. We have found that it is critical to use purified water free of any bioorganic contaminants during any cloning or plasmid preparation procedures; use either commercially available molecular biology grade water or in-house purified water (such as using Millipore system) that has been certified to be free of bioorganic contaminants.
The stable inducible RAW/LR5 cells grow at somewhat slower rate than the parental counterpart. This is due to the presence of three antibiotics in the medium, and not the inducible system itself. Once the antibiotics are removed for induction, the growth rate returns to that of the parental line. Furthermore, it is not advisable to overgrow the inducible cell line; we routinely maintain them in culture at 50 % confluency.
When a stable inducible RAW/LR5 cell line is first created and then induced for biosensor expression, the cells will induce at variable expression levels, ranging from no expression to dim and moderately high expression. It is possible to sort cells by FACS to obtain a near 100 %-expressing population. Optimal expression level can be determined empirically based on the desired signal-to-noise level during FRET imaging, avoiding dominant negative effects from overexpression of the biosensor.
In our experience we find that detaching with RAW/LR5 cells with 10 mM EDTA/PBS, as used during normal culture, prevents robust induction and the cells require trypsinization instead. In addition, a complete removal of Dox from the medium is essential for a successful induction. Residual Dox could remain internally within cells or as cell-membrane associated fraction. We find that, in the case of RAW/LR5 cells, a complete removal of Dox requires two rounds of trypsinization over a 48 h period in combination with very sparse plating (<20 % confluency).
It is imperative that the serum used for induction has been checked for residual tetracycline. If there are trace amounts of tetracycline in the serum, it will likely prevent induction. We find that NBCS is suboptimal for induction, as it appears to contain trace amounts of tetracycline depending on the particular lot. We routinely use FBS that is tested to be tetracycline- free in the induction medium.
For some imaging applications, it may be that a dilution of 1:3 or 1:4 results in coverslips that are too sparse. However, for robust biosensor induction to occur the cells need to be passaged at low confluency post 2nd trypsinization. If a higher confluency is needed the cells should be trypsinized for a third time and then plated at desired confluency for next-day imaging.
It is necessary to titrate every new batch of rabbit anti-sheep RBC IgG as too little antibody will decrease the efficiency of phagocytosis. However, excess antibody will cause aggregation of the RBCs during preparation. For each new batch of antibody make small batches of IgG-coated RBCs varying the concentration of the antibody in a range from 1 to 10 μL with 5 μL being the anticipated optimal concentration. Examine the IgG-coated RBC for aggregation with light microscopy.
It is best not to place the 12 mm round coverslips directly on ice. We normally use the lid of a cell culture plate (rectangular) lined with a piece of parafilm to provide a nonslip surface.
While other fluorochromes can be utilized, these wavelengths were chosen to be compatible for imaging with CFP-YFP FRET pair of the Cdc42 biosensor. In addition, optimal filter settings are required to minimize spectral bleedthroughs.
For fixed cell imaging of macrophages we have used a single CoolsnapHQ2 camera (Roper Photometrics) attached on the bottom 100 %-throughput port of the microscope. In this configuration, excitation and emission filterwheels allow for the switching of appropriate filter sets to acquire mCerulean, FRET, and mVenus emissions. The optical specifications for this configuration are detailed in Spiering et al. [25]. Transilluminated DIC images can also be obtained.
As phagosomes are three-dimensional structures we also imaged the same phagosome over serial planes in Z-direction, where the F-actin focal plane from Fig. 1a was set as the center position of the Z-stack (Fig. 1b) [12]. For the collection of Z-stack serial planes the optimal focal plane is set as the center of the stack we have found that eight planes at 1 μm steps was sufficient.
It is important to control for the relative expression levels when imaging fixed macrophages transiently overexpressing the biosensor. It is necessary to image only those cells that have a fluorescent intensity sufficient to fill approximately 80 % of the dynamic range of a detector to maximize the signal to noise ratio of the data being captured [14, 15]. For our optical setup, this requirement translates to using excitation intensities of 0.4–1.0 mW/cm2 at the specimen plane with a camera exposure time in the range of 500–1000 ms.
Because of the three-dimensional structure of phagosomes, epifluorescence wide-field imaging, as in Fig. 1, cannot clearly resolve the overall structure of phagocytic cups due to out of focus light being included in the imaging focal plane. This problem can be exacerbated further due to the mechanics of ratiometric analysis where FRET and donor emissions could scatter differentially because of lateral chromatic aberration effects resulting in unequal levels of out of focus light being present within a ratiometric set of images at each Z-section. To address this, confocal imaging is often the first choice in attempting to resolve and to reconstruct these three-dimensional cellular structures by directly removing the out of focus light. However, the available lasers used for excitation of FRET and donor mCerulean are often suboptimal (i.e., 405 nm and/or 458 nm) in a typical laser-based confocal system. These two laser lines miss the optimal excitation peak of mCerulean excitation spectra and result in either: (a) requiring overexpression of the biosensor to compensate for weak excitation; or (b) reduced excitation of FRET and mCerulean that produces a suboptimal signal to noise ratio in the final ratio-metric data. Therefore, we recommend sourcing a 445 nm laser that will maximally excite FRET and mCerulean during imaging to allow for better final signal to noise levels in the ratiometric data set.
In a wide-field microscopy, evenness of field illumination within the imaging field of view is not always guaranteed. This is especially the case if an arc-lamp coupled to a condenser system is used for illumination (typically, the center of the field of view will be brighter than the edges). Even in cases where a light source enters the system via a fiber or a liquid light guide (i.e., lasers, monochromators, light engines etc.) coupling to a beam expander-collimator assembly, the evenness of the field illumination should also be checked and corrected, as small imperfections in alignment will ceate measurable effects in the final ratio data if uncorrected. To perform this correction, images of empty fields of view are collected at the same exposure conditions as the experimental image acquisitions, and then used to divide out any unevenness in the field brightness (typically termed “flatfield correction” or “shading correction”) [14, 15].
For a typical cooled CCD camera (sCMOS camera will be similar, albeit typically with less associated noise levels in general) used in image acquisitions, the camera “noise” is present within each frame of image set. This is a combination of the image “shot noise” associated with the actual foreground data within the image (differences in the arrival times of photons at every element contributes to this noise, thus longer exposure time averages out this effect), the number of times the data in the detector elements are “read” for digitization (“read noise”; this depends on the architecture of the data reading buffer system of the detector chip as well as the build and shielding quality of the digitization circuitry), and the time duration of exposure per frame which contributes to the production of thermal noise (“dark current noise”; thus, cooling the sensor chip is effective at reducing this noise as it is temperature dependent). The combined noise image acquired in absence of all light hitting the detector chip but using an identical exposure time as the experimental image acquisition condition produces the camera noise image containing appropriate noise levels for the particular imaging condition used for the actual experiment. The noise image is subtracted from the raw data images and the shading correction images [14, 15].
The average background intensity value is subtracted from the shading corrected FRET, mCerulean and mVenus images. The average background value is determined by drawing a small region of interest away from any foreground signal and averaging the gray values within such a region of interest. Because the shading correction and noise subtraction were applied to the data image sets, the background intensity levels should be more or less uniform everywhere within the image [14, 15].
Even after the background subtraction, regions outside of the cell are not going to be uniformly at zero pixel intensity but rather at a stochastic distribution of low pixel intensity values. If such image sets were divided against each other for ratiometric calculation, the stochastic nature of the intensity distribution will produce speckles in regions outside of the cell edge, making data interpretation difficult. To avoid this effect, a histogram of pixel intensity values is manually thresholded such that the “background” regions outside of the cell edges can be set to zero pixel intensity. This is used to produce a binary cell mask, in which regions outside of the cell is now uniformly set to zero and the region inside of the cell is set uniformly to one. This binary cell mask is then multiplied into the shade-corrected, background-subtracted data sets prior to the final ratio calculations [14, 15].
For the ratio calculations, the masked FRET image is divided by the masked donor mCerulean image. In this step, it is critical to ascertain that pixel-to-pixel match is achieved between the two images to be divided. It is not correct to assume that both images are properly aligned with each other just because a single camera is used to acquire all of the data sets. Wavelength- dependent shifts could be present in the optical setup, depending on the thickness differences and the imperfections in the mounting angles of the bandpass filters within the focused section of the microscope, field lateral chromatic dispersion effects of the objective lens, and dichoric mirror mounting imperfections, etc., which could impact pixel-to-pixel alignment of the two images to be divided. This can be a priori calibrated and corrected using multispectral beads and nonlinear coordinate transformation approaches, and using a cross-correlation-based approach to optimize the X–Y translational alignment within 1/20th subpixel accuracy [14, 26, 27].
Another approach to resolve the three-dimensionality of phagosomes, which we show an example here (Fig. 2), is to use a deconvolution technique on the wide-field epifluorescence Z-series data. Deconvolution is a mathematical operation in which the out of focus light at any Z-position is removed through inverting the effect of the point-spread function of the objective lens on the specimen. When a point-source specimen is imaged through an objective lens, the point-spread function of the lens will describe how such a point-source will spread and blur in x, y, and z directions. If this function is known, then it is possible to iteratively and quantitatively solve for the inverse function to reconstruct the original point-source specimen image, effectively removing the out of focus light. The main advantage of the deconvolution technique is that if it is used on a wide-field epifluorescence data set there will be no loss of light, unlike confocal imaging in which a physical pin-hole is used to remove out of focus light. This yields better signal to noise and reduces the level of biosensor expression needed to achieve workable fluorescence intensity levels.
The major drawback of using the deconvolution approach is the high computational load. Often, iterative calculations of a data set would take minutes to hours in order to obtain the final result, so this approach has not been amenable to live-cell imaging and analysis on-the-fly. This problem could be addressed by parallel computing in which calculations are parsed over many computers and central processing units (CPUs), but such arrangements are cost prohibitive for most ordinary researchers. This requirement has now changed through the utilization of graphical processing units (GPUs), resident on computer video cards. These newer GPU cards, which are quite affordable at only a few hundred dollars, contain thousands of parallel computing processing units to accelerate the video graphics capability of a computer. A new deconvolution software package from Microvolution LLC [28] is now taking advantage of GPUs to accelerate the deconvolution calculations by several orders of magnitudes. This package was used to deconvolve the same Z-series used in Fig. 1 to show the difference in the resulting ratiometric data (Fig. 2). The deconvolution processing using the GPU-based system from Microvolution took approximately 5 s each, and ratio calculations were performed on the deconvolved images. Immediately, we observed an improvement in the signal to noise ratio, resulting from the removal of out of focus light. This improvement was effectively in the order of a fourfold increase in the final ratio dynamic range (original normalized ratios ranged from 1.0 to 1.38; post-deconvolution range: 1.0–2.47). In addition, we now observe clear filopodial structures (Fig. 2, panels 2 and 3) in which Cdc42 is activated as expected [6, 29]. Furthermore, we observe an apparent dorsal cup closure event with associated Cdc42 activity (Fig. 2, panels 4–6), which is less clear from the ratio images prior to deconvolution (Fig. 1). We also observe additional activation “hot spots” in regions where RBCs are contacting the cell edges. These results indicate the ability of the deconvolution technique to improve the signal to noise ratio, contributing to a more precise interpretation of biosensor readouts in three dimensions. Furthermore, the speed improvement in the processing by several orders of magnitudes offered by the Microvolution system now enables an in-line processing of the Z-series, on-the-fly, as the image acquisitions are performed on the microscope. This allows integration of deconvolution processing directly in the actual image data acquisition work flow for the first time.
References
- 1.Rougerie P, Miskolci V, Cox D. Generation of membrane structures during phagocytosis and chemotaxis of macrophages: role and regulation of the actin cytoskeleton. Immunol Rev. 2013;256:222–239. doi: 10.1111/imr.12118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hall A. Rho GTPases and the actin cytoskeleton. Science. 1998;279:509–514. doi: 10.1126/science.279.5350.509. [DOI] [PubMed] [Google Scholar]
- 3.Wennerberg K, Rossman KL, Der CJ. The Ras superfamily at a glance. J Cell Sci. 2005;118:843–846. doi: 10.1242/jcs.01660. [DOI] [PubMed] [Google Scholar]
- 4.Jaffe AB, Hall A. Rho GTPases: biochemistry and biology. Annu Rev Cell Dev Biol. 2005;21:247–269. doi: 10.1146/annurev.cellbio.21.020604.150721. [DOI] [PubMed] [Google Scholar]
- 5.Bishop AL, Hall A. Rho GTPases and their effector proteins. Biochem J. 2000;348(Pt 2):241–255. [PMC free article] [PubMed] [Google Scholar]
- 6.Heasman SJ, Ridley AJ. Mammalian Rho GTPases: new insights into their functions from in vivo studies. Nat Rev Mol Cell Biol. 2008;9:690–701. doi: 10.1038/nrm2476. [DOI] [PubMed] [Google Scholar]
- 7.Caron E, Hall A. Identification of two distinct mechanisms of phagocytosis controlled by different Rho GTPases. Science. 1998;282:1717–1721. doi: 10.1126/science.282.5394.1717. [DOI] [PubMed] [Google Scholar]
- 8.Cox D, Chang P, Zhang Q, Reddy PG, Bokoch GM, Greenberg S. Requirements for both Rac1 and Cdc42 in membrane ruffling and phagocytosis in leukocytes. J Exp Med. 1997;186:1487–1494. doi: 10.1084/jem.186.9.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jennings RT, Knaus UG. Rho family and Rap GTPase activation assays. Methods Mol Biol. 2014;1124:79–88. doi: 10.1007/978-1-62703-845-4_6. [DOI] [PubMed] [Google Scholar]
- 10.Pertz O. Spatio-temporal Rho GTPase signaling—where are we now? J Cell Sci. 2010;123:1841–1850. doi: 10.1242/jcs.064345. [DOI] [PubMed] [Google Scholar]
- 11.Hoppe AD, Swanson JA. Cdc42, Rac1, and Rac2 display distinct patterns of activation during phagocytosis. Mol Biol Cell. 2004;15:3509–3519. doi: 10.1091/mbc.E03-11-0847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hanna S, Miskolci V, Cox D, Hodgson L. A new genetically encoded single-chain biosensor for Cdc42 based on FRET, useful for live-cell imaging. PLoS One. 2014;9:e96469. doi: 10.1371/journal.pone.0096469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moshfegh Y, Bravo-Cordero JJ, Miskolci V, Condeelis J, Hodgson L. A Trio-Rac1- Pak1 signalling axis drives invadopodia disassembly. Nat Cell Biol. 2014;16:574–586. doi: 10.1038/ncb2972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Spiering D, Bravo-Cordero JJ, Moshfegh Y, Miskolci V, Hodgson L. Quantitative ratiometric imaging of FRET-biosensors in living cells. Methods Cell Biol. 2013;114:593–609. doi: 10.1016/B978-0-12-407761-4.00025-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hodgson L, Shen F, Hahn K. Biosensors for characterizing the dynamics of rho family GTPases in living cells. Curr Protoc Cell Biol. 2010;Chapter 14(Unit 14):11–26. doi: 10.1002/0471143030.cb1411s46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.An W, Telesnitsky A. Effects of varying sequence similarity on the frequency of repeat deletion during reverse transcription of a human immunodeficiency virus type 1 vector. J Virol. 2002;76:7897–7902. doi: 10.1128/JVI.76.15.7897-7902.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Delviks KA, Pathak VK. Effect of distance between homologous sequences and 3' homology on the frequency of retroviral reverse transcriptase template switching. J Virol. 1999;73:7923–7932. doi: 10.1128/jvi.73.10.7923-7932.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu B, Miskolci V, Sato H, Tutucci E, Kenworthy CA, Donnelly SK, et al. Synonymous modification results in high-fidelity gene expression of repetitive protein and nucleotide sequences. Genes Dev. 2015;29:876–886. doi: 10.1101/gad.259358.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qin JY, Zhang L, Clift KL, Hulur I, Xiang AP, Ren BZ, et al. Systematic comparison of constitutive promoters and the doxycycline-inducible promoter. PLoS One. 2010;5:e10611. doi: 10.1371/journal.pone.0010611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ramezani A, Hawley TS, Hawley RG. Lentiviral vectors for enhanced gene expression in human hematopoietic cells. Mol Ther. 2000;2:458–469. doi: 10.1006/mthe.2000.0190. [DOI] [PubMed] [Google Scholar]
- 21.Salmon P, Kindler V, Ducrey O, Chapuis B, Zubler RH, Trono D. High-level trans-gene expression in human hematopoietic progenitors and differentiated blood lineages after transduction with improved lentiviral vectors. Blood. 2000;96:3392–3398. [PubMed] [Google Scholar]
- 22.Loew R, Heinz N, Hampf M, Bujard H, Gossen M. Improved Tet-responsive promoters with minimized background expression. BMC Biotechnol. 2010;10:81. doi: 10.1186/1472-6750-10-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou X, Vink M, Klaver B, Berkhout B, Das AT. Optimization of the Tet-On system for regulated gene expression through viral evolution. Gene Ther. 2006;13:1382–1390. doi: 10.1038/sj.gt.3302780. [DOI] [PubMed] [Google Scholar]
- 24.Khader H, Solodushko V, Al-Mehdi AB, Audia J, Fouty B. Overlap of doxycycline fluorescence with that of the redox-sensitive intracellular reporter roGFP. J Fluoresc. 2013;24:305–311. doi: 10.1007/s10895-013-1331-6. [DOI] [PubMed] [Google Scholar]
- 25.Spiering D, Hodgson L. Multiplex imaging of Rho family GTPase activities in living cells. Methods Mol Biol. 2012;827:215–234. doi: 10.1007/978-1-61779-442-1_15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shen F, Hodgson L, Rabinovich A, Pertz O, Hahn K, Price JH. Functional proteometrics for cell migration. Cytometry A. 2006;69:563–572. doi: 10.1002/cyto.a.20283. [DOI] [PubMed] [Google Scholar]
- 27.Danuser G. Photogrammetric calibration of a stereo light microscope. J Microsc. 1999;193:62–83. doi: 10.1046/j.1365-2818.1999.00425.x. [DOI] [PubMed] [Google Scholar]
- 28.Bruce MA, Butte MJ. Real-time GPU- based 3D deconvolution. Opt Express. 2013;21:4766–4773. doi: 10.1364/OE.21.004766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ridley AJ. Life at the leading edge. Cell. 2011;145:1012–1022. doi: 10.1016/j.cell.2011.06.010. [DOI] [PubMed] [Google Scholar]