Abstract
Background
Chronic liver injury can result in fibrosis that may progress over years to end-stage liver disease. The most effective anti-fibrotic therapy is treatment of the underlying disease, however when not possible, interventions to reverse or slow fibrosis progression are needed.
Aim
To study the safety and tolerability of simtuzumab, a monoclonal antibody directed against lysyl oxidase-like 2 (LOXL2) enzyme, in subjects with hepatitis C virus (HCV), human immunodeficiency virus (HIV), or HCV-HIV co-infection and advanced liver disease.
Methods
Eighteen subjects with advanced liver fibrosis received simtuzumab 700mg intravenously every 2 weeks for 22 weeks. Transjugular liver biopsies were performed during screening and at the end of treatment to measure hepatic venous pressure gradient (HVPG) and to stage fibrosis.
Results
Treatment was well-tolerated with no discontinuations due to adverse events. No significant changes were seen in HVPG or liver biopsy fibrosis score after treatment. Exploratory transcriptional and protein profiling using paired pre- and post-treatment liver biopsy and serum samples suggested up-regulation of TGF-β3 and IL-10 pathways with treatment.
Conclusion
In this open-label, pilot clinical trial, simtuzumab treatment was well-tolerated in HCV- and HIV-infected subjects with advanced liver disease. Putative modulation of TGF-β3 and IL-10 pathways during simtuzumab treatment merits investigation in future trials.
Keywords: lysyl oxidases, hepatic venous pressure gradient (HVPG), magnetic resonance elastography (MRE), transforming growth factor beta-3 (TGF-β3)
Background
Liver fibrosis is the result of chronic liver injury induced by diverse etiologies, including infection with hepatitis B and C viruses (HBV, HCV), alcohol abuse, and non-alcoholic steatohepatitis (NASH). Progressive fibrosis may result in cirrhosis and manifestations of end-stage liver disease, including portal hypertension, hepatocellular dysfunction, and hepatocellular carcinoma (1). Effective therapeutic interventions for fibrotic liver disease remove or directly inhibit inciting fibrogenic stimuli. Unfortunately, therapeutic interventions to directly modulate fibrogenesis or enhance extracellular matrix degradation have met limited success in clinical trials (2, 3).
Although current treatments for chronic viral hepatitis have increased efficacy and tolerability, not all subjects with advanced liver fibrosis experience fibrosis regression, even with curative therapy (4, 5). Additionally, there are limited therapeutic options for NASH and other non-viral causes of fibrotic liver diseases, such as primary sclerosing cholangitis (6). Progression of fibrosis induced by HCV or NASH may be accelerated in immunocompromised subjects, including those with HIV-infection (7, 8). Thus, novel approaches for treatment of advanced fibrosis are urgently needed (2, 3, 9).
Hepatic fibrosis is characterized by increased production and deposition of fibril-forming collagens, particularly type-I collagen and matrix glycoconjugates, such as proteoglycans, fibronectins, and hyaluronic acid (10). Matrix synthesized and deposited in the liver undergoes dynamic turnover induced by host enzymes that modify the extracellular matrix (3). This turnover is mediated in part by the lysyl oxidase family, composed of 5 enzymes that promote cross-linking of fibrillar collagen (11, 12).
In subjects with HCV and/or NASH, expression of lysyl oxidases is confined to areas of fibrosis (12, 13). Furthermore, LOXL2 gene and protein expression are up-regulated in cancers that can arise in advanced fibrosis, including hepatocellular carcinoma (14). In contrast to LOX expression, LOXL2 is minimally expressed in normal tissues (13). Collectively, these data suggest lysyl oxidases contribute to fibrogenesis in a wide range of diseases and could be targeted therapeutically to modulate fibrosis progression.
Simtuzumab is a humanized IgG4 monoclonal antibody that specifically binds and inhibits LOXL2 function in vitro and in mouse models. Allosteric inhibition of LOXL2 using simtuzumab in mouse xenograft cancer models reduced fibroblast activation and decreased growth factors and cytokine production, including transforming growth factor-beta (TGF-β) (13). In the mouse carbon tetrachloride hepatic fibrosis model, LOXL2 was expressed in activated fibroblasts in areas of hepatic fibrosis. Furthermore, treatment with an anti-LOXL2 antibody reduced bridging fibrosis, reduced total collagen expression, and improved survival (13). In a pilot human study evaluating safety and tolerability in adults with liver fibrosis and chronic HCV and/or HIV infection, simtuzumab was safe and well-tolerated (15).
Here, to ask whether simtuzumab treatment of fibrotic subjects with HCV and/or HIV infection could impact fibrosis progression, we conducted a 6-month open-label study of simtuzumab in HCV and/or HIV-infected adults with advanced liver fibrosis and explored clinical and biologic outcomes.
Methods
Design
This phase 2a, open-label, single center exploratory study assessed the safety, tolerability and potential efficacy of simtuzumab in HCV and/or HIV infected adults (NCT01707472). Study participants received 700 mg of simtuzumab (Gilead Sciences, Inc., CA) intravenously over 30 minutes every 2 weeks for 22 weeks (12 infusions). After treatment, participants were followed every 4 weeks for an additional 12 weeks.
Study Population
Adult subjects 18 years of age or older, with HCV, HIV, or HCV/HIV infection and advanced hepatic fibrosis or cirrhosis (Ishak fibrosis score ≥3 on liver biopsy) were eligible (16). Target enrollment for this pilot study was 30 participants: 10 with HCV, 10 with HCV/HIV and 10 with HIV. Subjects with Child-Pugh class A cirrhosis and no signs or symptoms of decompensation could participate ] (17). Subjects with HBV infection, other causes of liver disease (e.g. autoimmune hepatitis or hemochromatosis), ongoing alcohol or illicit drug abuse, or contraindication to transjugular liver biopsy were excluded. A liver biopsy was required for study eligibility if not performed in the previous 6 months.
HCV infection was confirmed by positive HCV antibody and HCV RNA >2,000 IU/mL. At the time of study enrollment, no interferon-free treatments for chronic HCV infection were FDA-approved. HCV-infected subjects had a history of non-response or were unwilling or unable to receive interferon-based therapy. HIV infected subjects required an HIV viral load <40 copies/mL on stable antiretroviral therapy for over 3 months.
The study was investigator initiated, sponsored by Gilead Sciences Inc., and was approved by the National Institute of Allergy and Infectious Diseases Institutional Review Board. All participants provided written informed consent, and the study was conducted in compliance with the Declaration of Helsinki.
Safety Assessments
Safety was evaluated by review of adverse events and concomitant medications, assessment of clinical laboratory tests, vital signs, physical examinations, and ECG recordings. Treatment-emergent adverse events were graded by severity using Common Terminology Criteria for Adverse Events (version 4.03).
Transjugular Liver Biopsy with Portal Pressure Measurement
Transjugular liver biopsies were performed before and after 22 weeks of treatment with simtuzumab to determine the hepatic venous pressure gradient (HVPG) and for histologic fibrosis, inflammation and steatosis staging. Peak HVPG measurements are reported. One liver biopsy core was placed in formalin for histopathologic assessment and a second core was placed in RNAlater and stored at −80°C for gene expression analysis.
Liver Histology and Immunohistochemistry
Formalin-fixed paraffin-embedded liver biopsy sections were stained with hematoxylin and eosin, Masson’s trichrome, and for reticulin, and interpreted by a single liver pathologist (DEK). Fibrosis and inflammatory activity were scored with the modified histology activity index (Ishak) scoring system (16). Steatosis was graded on a scale of 0 to 4 based on the percentage of cells with fat.
Computer-assisted morphometry was used to quantify liver collagen content (18) and stellate cell activation was quantified by immunohistochemical staining for alpha-smooth muscle actin (19) (Center for Liver Diseases, INOVA Fairfax Hospital, Falls Church, VA).
Laboratory Assessments
CD4+ T-cell count, HIV and HCV viral loads, liver-associated enzymes (alkaline phosphatase, AST, ALT, bilirubin, albumin), non-invasive markers of liver fibrosis (APRI (20), FIB-4 (21), Forns (22) and Fibroindex (23)), and other clinical laboratory parameters, including complete blood counts, serum chemistries, amylase and lipase, were assessed at baseline and completion of therapy.
Serum LOXL2 concentrations were measured using a proprietary immunoassay (Singulex, Alameda, CA) to assess correlation with fibrosis and impact of treatment. Auto-antibodies against simtuzumab were measured in serum at baseline and week 28 using an electrochemilumenscence assay (Biologics Development Services, Tampa, FL).
Magnetic Resonance Elastography (MRE)
In participants meeting criteria for MRE (body mass index (BMI) <35 kg/m2, no contraindication to MRI), MRE was performed at baseline and the end of treatment to quantify liver elasticity, a correlate of hepatic fibrosis (24), as previously described (25). In brief, MR examination was performed on a 3.0T system (Achieva, Philips Healthcare, Best, Netherlands) using a 32 element surface coil. Mechanical stimulation was done using an investigational elastography package (Philips Research Laboratories, Hamburg, Germany). A mechanical transducer with a vibration frequency of 56 Hz was placed against the supine subjects’ side at the lowest right rib in the right–left direction. An operator blinded to subject, clinical parameters, and study time point, performed image processing to calculate shear wave speed in m/sec.
Efficacy Assessments
Exploratory efficacy end points included changes from baseline to end of treatment in hepatocellular inflammation and fibrosis as assessed by liver biopsy, HVPG, percent collagen, percent alpha-smooth muscle actin (SMA), and MRE shear wave speed.
Liver and Whole Blood Microarrays
Total RNA from 12 participants was extracted from paired pre- and post- treatment liver biopsies and whole blood. RNA was extracted from liver biopsies stored in RNAlater at −80°C as previously described (26). Whole blood collected and stored in PaxGene RNA tubes (Qiagen) at −80°C was extracted according to manufacturer’s instructions (Qiagen, Valencia, CA).
RNA quality was determined using a 2100 Bioanalyzer (Agilent Technologies) and the Agilent RNA 6000 Pico kit. The average RNA Integrity Number (RIN) value was 7.9, with a range of 5.7 to 8.8 for liver and 4.5 to 7.7 for blood. RNA was quantitated using a spectrophotometer (Eppendorf, Germany). RNA flow-throughs were stored at −80°C for small- and micro-RNA analysis.
cDNA for microarray analysis was synthesized using the Ovation Pico WTA system (Nugen Inc.) and used for hybridization on the Affymetrix GeneChip Human Gene 2.0 ST array. Gene expression results were analyzed in Partek Genomics Suite software (version 6.6) and normalized using the robust multi-array average method. Results from paired liver biopsies and whole blood samples were analyzed by paired t-test, without distinction based on underlying infection due to the small sample size. No differentially expressed transcripts were identified with multiple test corrected p-values <0.05 (27). As such, genes with an expression fold change >1.25 and unadjusted p-values <0.05 were identified by a rank order approach for analysis by Ingenuity Pathway Analysis (IPA, Qiagen), as previously described (26). Targeted validation was performed by quantitative reverse transcriptase-real time polymerase chain reaction (qRT-PCR). Microarray data were submitted to the Gene Expression Omnibus public database (GSE73634).
qRT-PCR
Total RNA isolated from liver was reverse transcribed using random primers with the High Capacity cDNA Reverse Transcriptase Kit (ThermoFischer Scientific, Waltham, MA). From 0.75–3 ng RNA was used for each Taqman gene expression assay, run as a single replicate due to limited sample availability. Gene expression was determined as cycle of threshold (Ct) based on 40 PCR cycles, using expression of GAPDH as an endogenous control. GAPDH Ct values were distributed between 23 and 27. Data from 5 of 24 samples with GAPDH Ct value greater than 27 was excluded, allowing confirmatory qRT-PCR data from only 8 of 12 subjects to be reported. Expression reactions using pre-designed Taqman assays (ThermoFischer Scientific) were run on an Applied Biosystems 7500 Real-Time PCR System, as previously described (33).
Small RNA Sequencing and Analysis
Flow-through from liver mRNA isolation was used to extract small RNAs (0 to 200 nucleotides) according to Qiagen microRNA protocol (Valencia, CA), as previously described (28).
Small RNA libraries were generated using the Illumina TruSeq Small RNA protocol following the manufacturer’s instructions. Final libraries were size selected in the range of 140bp to 340bp to collect miRNA and other noncoding RNA (ncRNA). Libraries were barcoded and sequenced on a Hiseq 2500 as 50bp single end reads. Sequencing data were deposited at the National Center for Biotechnology Information Sequence Read Archive, accession number SRP066098.
Sequencing reads were trimmed of adaptor sequence, reads less than 16 nucleotides or of low quality were filtered (FASTX-Toolkit). Reads were mapped and filtered as previously described (28). Paired t-test (unadjusted p-value) and false discovery rate corrected p-value (q-value) were computed using Partek Genomic Suite. No significant species were identified with a q-value <0.05, so species with unadjusted p-values <0.05 were considered. Only miRNA results are reported. No technical outliers were identified and all 12 subjects were included in the analysis.
Serum Biomarkers
Paired pre- and end of treatment (week 24) cryopreserved serum samples were tested by enzyme-linked immunosorbent assays (ELISA) in multiplex for tumor necrosis factor-alpha (TNF-α), interleukin (IL)-6, IL-2, IL-8, IL-10, IL-12p70, interferon gamma (IFNγ), and GM-CSF (MesoScale Discovery, Gaithersburg, MD). IL-7, osteopontin and tissue inhibitor of metalloproteinases-1 (TIMP1) were quantitated using single-plex assays, while TGF-β isoforms 1, 2 and 3 were quantitated using a triplex assay (MesoScale Discovery). Cleaved cytokeratin-18 levels were quantitated by ELISA (M30 Apoptosense, Peviva, West Chester, OH).
Statistical Methods
Clinical and safety variables were summarized with descriptive statistics. Pairwise comparisons were performed using a 2-sided Wilcoxon-Mann-Whitney test. A 2 tailed p-value of <0.05 was considered statistically significant. Microarray and miRNA statistical approaches are described above, and qRT-PCR data was analyzed with non-parametric assumptions using the Wilcoxon paired-sign rank test.
Results
Baseline Characteristics
Thirty subjects were screened and 18 enrolled prior to halting enrollment in January 2014 due to slow accrual. Twelve subjects were deemed ineligible due to: liver biopsy fibrosis (Ishak) score <3 (n=5), inadequate venous access for transjugular liver biopsy (n=2), positive illicit drug screen (n=2), Child-Pugh class >A (n=2) or cardiac arrhythmia requiring intervention (n=1). Of the 18 enrolled subjects, 6 had HCV mono-infection, 4 had HIV mono-infection with biopsy-proven NASH, and 8 had HCV/HIV co-infection. Demographic and clinical parameters are shown in Table 1.
Table 1.
Baseline demographic, clinical and laboratory parameters
| All (n=18) | HCV (n=6) | HIV/HCV (n=8) | HIV (n=4) | |
|---|---|---|---|---|
| Age, years (median, range) | 57 (45–65) | 59 (49–63) | 53 (45–61) | 55 (50–65) |
| Male sex, n (%) | 14 (78%) | 3 (50%) | 7 (87%) | 4 (100%) |
| Self-reported Race and Ethnicity, n (%) | ||||
| Black, not Hispanic/Latino | 6 (33%) | 2 (33%) | 4 (50%) | 0 (0%) |
| White, not Hispanic/Latino | 8 (44%) | 4 (67%) | 2 (25%) | 2 (50%) |
| American Indian | 1 (6%) | 0 | 1 (13%) | 0 |
| Hispanic/Latino | 3 (17%) | 0 | 1 (12%) | 2 (50%) |
| Body mass index, kg/m2 (median, range) | 30 (21–46) | 32 (22–45) | 30 (21–46) | 30 (28–32) |
| Alkaline phosphatase, U/L(median, range) | 102 (61–210) | 89 (61–143) | 114 (82–119) | 112 (75–210) |
| Aspartate aminotransferase (AST), U/L (median, range) | 63 (31–151) | 96 (31–151) | 56 (43–96) | 66 (49–116) |
| Alanine aminotransferase (ALT), U/L (median, range) | 84 (37–161) | 106 (46–144) | 58 (37–90) | 98 (67–161) |
| Total bilirubin, mg/dL (median, range) | 0.8 (0.3–2.3) | 0.7 (0.4–1.3) | 0.8 (0.3–2.3) | 0.9 (0.4–1.6) |
| Platelets, K/uL (median, range) | 152 (45–250) | 132 (75–239) | 180 (45–250) | 138 (57–166) |
| Hepatitis C genotype, n (%) | ||||
| 1a | 5 (83%) | 7 (88%) | NA | |
| 1b | 1 (17%) | 1 (12%) | NA | |
| Hepatitis C treatment experienced, n (%) | 4 (66%) | 6 (75%) | NA | |
| HIV viral load, copies/mL (median, range) | NA | <40 (<40–220) | <40 (<40–172) | |
| CD4 count, cells/mm3 (median, range) | 572 (123–2558) | 976 (410–2558) | 572 (173–1049) | 189 (123–260) |
| Hepatic venous portal gradient (HVPG), mmHg (median, range) | 9 (3–15) | 10 (4–12) | 7 (3–15) | 7 (3–15) |
| HVPG≥10mmHg, n (%) | 6 (33%) | 3 (50%) | 2 (25%) | 1 (25%) |
| Liver biopsy | ||||
| Length, mm (median, range) | 14 (5–24) | 13 (6–18) | 13 (5–18) | 15 (9–24) |
| Ishak fibrosis score (median, range) | 5 (3–6) | 6 (3–6) | 4 (3–6) | 5 (3–6) |
| Ishak inflammation score (median, range) | 8 (4–14) | 8 (4–9) | 9 (8–14) | 6 (4–9) |
| Steatosis (median, range) | Trace (0–2) | Trace (0–1) | Trace (0–2) | 2 (Trace–2) |
Most subjects were overweight (n=4, BMI 25–29.9 kg/m2) or obese (n=12, BMI≥30 kg/m2) with a median BMI of 30 kg/m2 (range 21–46). Six subjects (33%) had diabetes mellitus. Twelve subjects (67%) denied any alcohol use in the past year and 6 (33%) reported infrequent alcohol use (≤3 drinks per month).
For HIV-infected participants (n=10), the median duration of antiretroviral treatment at enrollment was 15.4 years (range 8.3–22.1 years). Antiretroviral treatment histories included past exposure to didanosine (n=5), stavudine (n=7) and zidovudine (n=7).
All 14 HCV participants had infection with genotype 1. Ten (71%) had failed prior interferon-based HCV-therapy and 4 (29%) were treatment naïve. Ten subjects (56%) had biopsy-proven cirrhosis, all Childs-Pugh class A. Baseline median HVPG was 9 mmHg (range 3–15 mmHg) and was ≥10 mmHg in 4 participants (22%). Median fibrosis stage was 5 (range 3–6), median necroinflammatory grade was 8 (range 4–14), and all subjects had minimal or no steatosis (Table 1).
Safety/Adverse Events
Simtuzumab was generally safe and well tolerated and all participants completed treatment and follow-up. Treatment-emergent adverse events are summarized in Table 2. The most common adverse events considered related to simtuzumab were fever (n=3), headache (n=3), glossitis (n=2) and asymptomatic elevations in amylase and lipase (n=5). Five participants experienced a grade 3 or 4 adverse event considered possibly related to simtuzumab. One participant experienced a serious adverse event, with right upper quadrant pain and vomiting post-liver biopsy, and subsequent E. coli bacteremia requiring parenteral antibiotic therapy. This subject made a full recovery.
Table 2.
Adverse Events
| All (n=18) | HCV (n=6) | HIV/HCV (n=8) | HIV (n=4) | |
|---|---|---|---|---|
| Any treatment-emergent adverse event, number of participants (% cohort) | 16 (89%) | 5 (83%) | 7 (88%) | 4 (75%) |
| Grade 3 and 4 treatment-related adverse events, number of participants (% cohort) | ||||
| Migraine headache | 1 (6%) | 0 | 1 (12%) | 0 |
| Elevated ALT | 1 (6%) | 0 | 0 | 1 (25%) |
| Increased lipase | 4 (22%) | 1 (17%) | 1 (12%) | 2 (50%) |
| Increased amylase | 2 (11%) | 1 (17%) | 0 | 1 (25%) |
| Discontinuation owing to adverse event | 0 | 0 | 0 | 0 |
| Serious adverse event, number of participants (% cohort) | 1* (12%) | 0 | 1* (12%) | 0 |
One participant experienced abdominal pain and vomiting following post-treatment liver biopsy and was admitted for pain control and monitoring. Course was complicated by fever and gram-negative bacteremia.
Anti-simtuzumab antibodies were not detected in serum samples drawn at baseline or week 28.
Fibrosis and HVPG
No significant change in platelet count, albumin, liver-associated enzymes, or scores on non-invasive liver fibrosis tests was seen over the course of treatment (Table 3). HCV RNA levels did not change in HCV-infected subjects. CD4+ T-cell counts did not change and HIV viral load remained <400 copies/mL in HIV-infected subjects.
Table 3.
Selected Laboratory, Clinical and Fibrosis Measures at Baseline and End of Treatment
| Baseline | End of Treatment (Week 24) | P-value* | Change from Baseline at W24 | |
|---|---|---|---|---|
| Alkaline phosphatase, U/L | 102 (61–210) | 93 (56–197) | 0.07 | −10 (−40, 35) |
| Aspartate aminotransferase (AST), U/L | 63 (31–151) | 76 (24–145) | 0.94 | −2 (−48–60) |
| Alanine aminotransferace (ALT), U/L | 84 (37–161) | 71 (30–261) | 0.62 | −6 (−53–100) |
| Gamma-glutamyl transferase (GGT), U/L | 112 (36–531) | 100 (40–612) | 0.87 | −1 (−243–222) |
| Total bilirubin, mg/dL | 0.8 (0.3–2.3) | 0.7 (0.3–2.2) | 0.96 | 0 (−0.3–0.5) |
| Albumin, g/dL | 4.2 (3.0–5.5) | 4.0 (3.0–5.3) | 0.16 | −0.1 (−1.3–0.5) |
| Platelets, K/uL | 152 (45–250) | 152 (50–268) | 0.80 | 5 (−28–51) |
| HCV viral load, IU/mL | 6.8 (4.9–7.3) | 6.5 (3.1–7.8) | 0.58 | −0.1 (−1.7–0.9) |
| HIV viral load, copies/mL | <40 (<40–220) | <40 (<40–95) | ||
| CD4+ T-cell count, cells/mm3 | 572 (123–2558) | 659 (113–1839) | 0.74 | 11 (−719–281) |
| CD4+ T-cell % | 41 (7–71) | 40 (7–68) | 0.75 | 0 (−5–10) |
| Body mass index, kg/m2 | 32 (22–47) | 32 (21–46) | 0.56 | 0.1 (−1.4–3.5) |
| Total cholesterol, fasting, mg/dL | 152 (102–235) | 153 (92–209) | 0.72 | 4 (−63–32) |
| Hepatic venous portal gradient (HVPG), mmHg | 9 (3–15) | 8 (3–20) | 0.38 | 1 (−4–5) |
| Liver biopsy** | ||||
| Ishak fibrosis score | 5 (3–6) | 6 (2–6) | 1 | 0 (−2–2) |
| Ishak inflammation score | 8 (4–14) | 8 (4–13) | 0.46 | −0.5 (−4–5) |
| Steatosis | Trace (0–2) | 1 (0–3) | 0.42 | 0 (−1–2) |
| Collagen, % | 6.3 (1.1–45.4) | 9.0 (2.8–26.5) | 0.46 | 1.6 (−34.5–21.8) |
| Smooth muscle actin, % | 5.1 (0.6–40.7) | 12.2 (1.0–23.5) | 0.21 | 3.3 (−24.8–14.4) |
| Magnetic resonance elastography, m/sec (n=12) | 2.2 (1.6–3.0) | 2.2 (1.8–3.2) | 0.01 | 0.1 (−0.1–0.3) |
| APRI | 1.4 (0.4–6.3) | 1.8 (0.3–5.2) | 0.14 | −0.3 (−1.2–1.1) |
| FIB4 | 2.6 (0.9–15.3) | 3.1 (0.8–12.5) | 0.42 | −0.1 (−2.8–1.4) |
| Forns Index | 7.6 (5.1–11.3) | 7.3 (5.8–10.9) | 0.18 | −0.1 (−1.0–1.0) |
| FibroIndex | 1.8 (1.0–3.1) | 1.7 (0.7–3.1) | 0.27 | −0.1 (−0.3–0.5) |
| Serum LOXL2 concentration (pg/mL) | 1551 (421–2739) | 1663 (612–3491) | 0.05 | 121 (−437–1645) |
Median (range) presented unless otherwise noted.
Wilcoxon matched-pairs signed rank test
One HIV-monoinfected participant is excluded from the paired analysis because of the small size of the post-treatment liver biopsy sample.
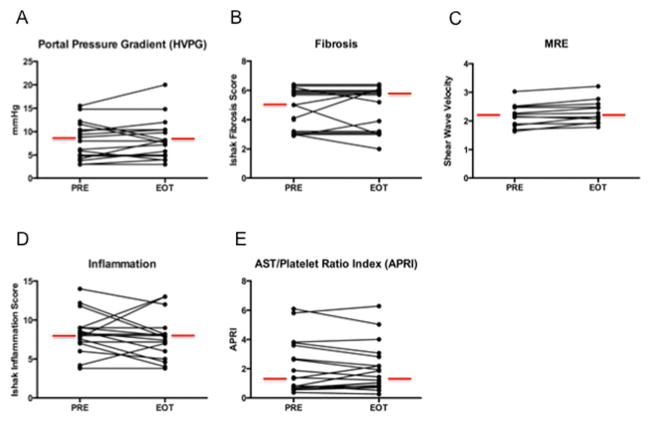
In paired liver biopsies, no significant change was seen in HVPG, fibrosis, inflammation, steatosis, or percent collagen or SMA (Figure 1). In 4 participants with baseline HVPG ≥10, HVPG was lower by ≥ 20% in 2 and unchanged in 2 others at EOT. Liver elasticity did not change in participants with baseline and end-of-treatment MRE (n=12) (Figure 1, Table 3).
Figure 1.
No significant change was seen in portal pressure gradient (HVPG) (A), fibrosis (B), MRE (C), liver inflammation (D) or AST/platelet ratio index (APRI) (E) after 22 weeks of treatment with simtuzumab. Median values are indicated with red lines. Abbreviations: PRE (pre-treatment) and EOT (end-of-treatment, week 22).
Gene Expression Analysis
We performed gene expression analysis on liver and blood samples to assess for treatment-related biologic effects. Paired pre- and post-treatment liver and whole blood samples available from 12 participants were analyzed (HIV and NASH, n=2, HCV mono-infection, n=5, HIV/HCV co-infection, n=5). Principal components analysis confirmed separation of the data by sample source (liver, blood) (data not shown). A rank order list of genes with fold change of >1.25 and unadjusted p-value <0.05, identified 549 probes from blood and 1217 probes from liver with differential expression between baseline and post-therapy.
Ingenuity Pathway Analysis of hepatic genes with differential expression suggested reduced activity of fibrosis and insulin resistance pathways (all p<0.05) and for blood, 4 pathways unrelated to fibrosis were identified (Supplementary Table 1). qRT-PCR analysis of 15 genes identified in the fibrosis pathway from liver of 8 subjects with sufficient remaining RNA revealed no significant change in expression of any individual gene (data not shown).
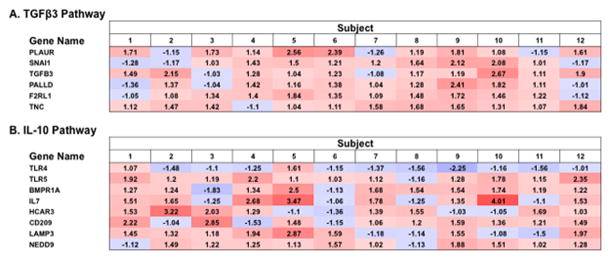
Activation analysis of hepatic gene expression suggested activation of TGF-β3 (z-score 2.4) and IL-10 pathways (z-score 2.16) with treatment (Figure 2). Examination of genes in the TGF-β3 pathway (PLAUR, TGFβ3, PALLD, F2RL1, and TNC) by qRT-PCR in 8-paired liver samples showed only F2RL1 had differential expression post-treatment (p=0.04, 1.29 fold increase), while none of 8 genes in the IL-10 pathway changed significantly (data not shown).
Figure 2.
Predicted upstream activation of TGF-β3 (A) and IL-10 (B) pathways. Shown are ratios of end of treatment (EOT, week 20) over pre-treatment (PRE) gene expression, with red representing increased post-treatment gene expression, blue representing decreased gene expression, and ratio indicated in each field. TGF-β3 and IL-10 pathways had an Ingenuity Pathway Analysis (IPA) activation Z-scores 2.4 and 2.2, respectively.
No miRNA species changed expression over treatment when results were analyzed with a multiple test corrected p-value. Considering species with an unadjusted p-value of <0.05, 19 miRNAs with differential expression over treatment were identified (Supplementary Table 2), including mir-92a (1.3 fold decrease), which is up-regulated in serum during chronic HCV infection (29), and mir-107 (1.5 fold decrease), which is down-regulated in chronic HCV and fibrosis (30).
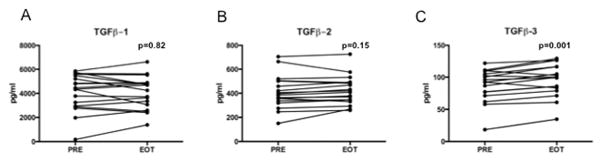
Serum cytokine analysis considering all 18 participants revealed a significant increase in TGF-β3, but not TGF-β1 or TGF-β2, over the course of therapy. No correlation was seen between TGF-β3 levels and AST, ALT or HVPG (Figure 3). No changes were observed in serum expression of IL-7, IFNγ, IL-10, IL-12 p70, IL-2, IL-8, osteopontin, or cleaved cytokeratin-18, while IL-6 concentration increased modestly (data not shown).
Figure 3.
A significant increase was seen in serum TGF-β3 (C), but not TGF-β1 (A) or TGF-β2 (B), during simtuzumab therapy (n=18). Displayed p-values are by paired t-test. Abbreviations: PRE (pre-treatment) and EOT (end-of-treatment, week 20).
Discussion
In this 6-month exploratory study we assessed the safety and tolerability of simtuzumab in HIV and/or HCV infected adults with advanced liver fibrosis, and found treatment was well-tolerated with no discontinuations due to adverse events. In contrast to preliminary findings of mild improvements in aminotransferase levels in a small dose-finding study of simtuzumab (15), however, we did not observe improvement in aminotransferases during treatment. Further, exploratory efficacy analysis identified no significant changes in liver-associated enzymes, portal pressure, liver fibrosis, liver inflammation, or liver stiffness.
While no clinical benefit was seen, our study was small and included participants with diverse viral etiologies of liver fibrosis. Absence of benefit could reflect inadequate dose or duration of treatment. Alternatively, simtuzumab inhibition of LOXL2 activity in vivo may be incomplete or compensated for by other fibrosis mediators. As is the case with many studies of antifibrotic agents, defining the optimal population, sample size, duration of treatment, and outcome measures are challenges when evaluating potential efficacy (9).
In transcriptional and protein profiling experiments, we did not identify a clear impact of treatment using stringent statistical approaches, which may relate to the small size of liver biopsies, clinical heterogeneity of the study population, absence of sufficient RNA from all subjects for validation of target transcripts, limited sample size, or lack of efficacy of the intervention. When we employed a rank order approach to gene expression analysis to analyze the data for hypothesis generation, an increase in TGF-β3 and IL-10 activity in liver and decreased expression of genes associated with progression of fibrosis and insulin resistance was suggested. While qRT-PCR studies did not validate most of the genes identified by microarray, this analysis was limited in that only 8 of 12 subjects had sufficient residual RNA.
Interestingly, the increase in predicted TGF-β3 activity suggested from transcriptional analysis was found to have a correlate in serum, as a moderate but statistically significant increase in serum TGB-β3 over the course of treatment was observed. Several reports have identified an association between TGF-β3 and attenuation or regression of fibrosis (31–33). Of note, in vitro assessment of hepatic stellate cells indicates their potential to produce TGF-β3 transcript (Don Rockey, personal communication), suggesting that modulation of TGF-β3 activity merits evaluation in future clinical trials with simtuzumab.
Limitations of our study include selection bias of subjects referred and willing to undergo transjugular liver biopsy and portal pressure measurement, underreporting of alcohol use, and sampling error inherent in biopsy-based studies.
Future studies are required to explore the long-term safety and clinical efficacy of simtuzumab and potential impact of simtuzumab treatment on TGF-β3 and IL-10 pathways. Although a Phase 2 study in idiopathic pulmonary fibrosis was recently halted due to lack of efficacy, ongoing studies of simtuzumab in NASH, primary sclerosing cholangitis, and other fibrotic conditions including myelofibrosis, will provide additional data on the safety and potential effectiveness of LOXL2 blockade in fibrotic diseases.
Supplementary Material
Key points.
Simtuzumab is an investigational monoclonal antibody against lysyl oxidase like-2 enzyme (LOXL2) being studied for the treatment of liver fibrosis.
In hepatitis C and/or HIV-infected adults with advanced liver fibrosis, simtuzumab treatment (700mg by vein every 2 weeks) for 22 weeks was well-tolerated.
No significant change was seen in portal pressure gradient or liver biopsy fibrosis after 22 weeks of simtuzumab therapy, however transcriptional and protein profiling suggested up-regulation of TGF-β3 and IL-10 pathways with treatment.
Ongoing and future studies will better define the potential role of simtuzumab in the management of liver fibrosis.
Acknowledgments
Financial support: Supported by the United States National Institutes of Health (NIH) Intramural Research Program, NIH Clinical Center, NIAID, NIDDK, and, in part, by an NIH Bench to Bedside Award funded by the Office of AIDS Research. Funded in part with federal funds from the National Cancer Institute, under contract HHSN261200800001E. The study was partially funded by a Collaborative Research and Development Agreement between NIH and Gilead Sciences.
We thank the volunteers who participated in this research study and the medical, nursing and support staff of the NIH.
List of abbreviations (in order of appearance)
- LOXL2
Lysyl oxidase-like 2
- HCV
Hepatitis C virus
- HIV
Human immunodeficiency virus
- HVPG
Hepatic venous pressure gradient
- TGF- β
Tumor growth factor beta
- IL-10
Interleukin 10
- HBV
Hepatitis B virus
- NASH
Non-alcoholic steatohepatitis
- IgG4
Immunoglobulin G4
- AST
Aspartate aminotransferase
- ALT
Alanine aminotransferase
- APRI
AST/platelet ratio index
- MRE
Magnetic resonance elastography
- SMA
Smooth muscle actin
- RNA
Ribonucleic acid
- cDNA
Circular deoxynucleic acid
- qRT-PCR
Quantitative real-time polymerase chain reaction
- miRNA
Micro RNA
Footnotes
Presented in part at The Liver Meeting (AASLD), Boston, MA, November 7–11, 2014, and Digestive Disease Week (DDW), Washington, DC, May 16–19, 2015.
Trial registration number: NCT01707472
Conflicts of interest: Dr. Meissner receives grant support from Gilead Sciences, Inc. Drs. Kanwar, Myers and Subramanian are employed by and own stock in Gilead Sciences Inc. Dr. Goodman reports grant support from Gilead Sciences, Inc., Tobira, Intercept, Galectin, Conatus, and Alexion. All other co-authors report no conflicts of interest.
References
- 1.Hernandez-Gea V, Friedman SL. Pathogenesis of liver fibrosis. Annual review of pathology. 2011;6:425–56. doi: 10.1146/annurev-pathol-011110-130246. [DOI] [PubMed] [Google Scholar]
- 2.Friedman SL, Sheppard D, Duffield JS, Violette S. Therapy for fibrotic diseases: nearing the starting line. Science translational medicine. 2013;5(167):167sr1. doi: 10.1126/scitranslmed.3004700. [DOI] [PubMed] [Google Scholar]
- 3.Rockey DC, Bell PD, Hill JA. Fibrosis--a common pathway to organ injury and failure. The New England journal of medicine. 2015;372(12):1138–49. doi: 10.1056/NEJMra1300575. [DOI] [PubMed] [Google Scholar]
- 4.Labarga P, Fernandez-Montero JV, Barreiro P, et al. Changes in liver fibrosis in HIV/HCV-coinfected patients following different outcomes with peginterferon plus ribavirin therapy. Journal of viral hepatitis. 2014;21(7):475–9. doi: 10.1111/jvh.12180. [DOI] [PubMed] [Google Scholar]
- 5.Shiffman ML, Sterling RK, Contos M, et al. Long term changes in liver histology following treatment of chronic hepatitis C virus. Annals of hepatology. 2014;13(4):340–9. [PubMed] [Google Scholar]
- 6.Younossi ZM, Reyes MJ, Mishra A, Mehta R, Henry L. Systematic review with meta-analysis: non-alcoholic steatohepatitis - a case for personalised treatment based on pathogenic targets. Alimentary pharmacology & therapeutics. 2014;39(1):3–14. doi: 10.1111/apt.12543. [DOI] [PubMed] [Google Scholar]
- 7.Morse CG, Mclaughlin M, Matthews L, et al. Nonalcoholic Steatohepatitis and Hepatic Fibrosis in HIV-1-Monoinfected Adults With Elevated Aminotransferase Levels on Antiretroviral Therapy. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America. 2015;60(10):1569–78. doi: 10.1093/cid/civ101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vodkin I, Valasek MA, Bettencourt R, Cachay E, Loomba R. Clinical, biochemical and histological differences between HIV-associated NAFLD and primary NAFLD: a case-control study. Alimentary pharmacology & therapeutics. 2015;41(4):368–78. doi: 10.1111/apt.13052. [DOI] [PubMed] [Google Scholar]
- 9.Torok NJ, Dranoff JA, Schuppan D, Friedman SL. Strategies and endpoints of antifibrotic drug trials: Summary and recommendations from the AASLD Emerging Trends Conference, Chicago, June 2014. Hepatology. 2015 doi: 10.1002/hep.27720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brenner DA, Westwick J, Breindl M. Type I collagen gene regulation and the molecular pathogenesis of cirrhosis. The American journal of physiology. 1993;264(4 Pt 1):G589–95. doi: 10.1152/ajpgi.1993.264.4.G589. [DOI] [PubMed] [Google Scholar]
- 11.Kagan HM, Li W. Lysyl oxidase: properties, specificity, and biological roles inside and outside of the cell. J Cell Biochem. 2003;88(4):660–72. doi: 10.1002/jcb.10413. [DOI] [PubMed] [Google Scholar]
- 12.Vadasz Z, Kessler O, Akiri G, et al. Abnormal deposition of collagen around hepatocytes in Wilson’s disease is associated with hepatocyte specific expression of lysyl oxidase and lysyl oxidase like protein-2. Journal of hepatology. 2005;43(3):499–507. doi: 10.1016/j.jhep.2005.02.052. [DOI] [PubMed] [Google Scholar]
- 13.Barry-Hamilton V, Spangler R, Marshall D, et al. Allosteric inhibition of lysyl oxidase-like-2 impedes the development of a pathologic microenvironment. Nat Med. 2010;16(9):1009–17. doi: 10.1038/nm.2208. [DOI] [PubMed] [Google Scholar]
- 14.Wong CC, Tse AP, Huang YP, et al. Lysyl oxidase-like 2 is critical to tumor microenvironment and metastatic niche formation in hepatocellular carcinoma. Hepatology. 2014;60(5):1645–58. doi: 10.1002/hep.27320. [DOI] [PubMed] [Google Scholar]
- 15.Talal AHF-RM, Madere J, Subramanian GM, Bornstein JD. Simtuzumab, an antifibrotic monoclonal antibody against lysyl xxidase-Like2 (LOXL2) enzyme, appears safe and well tolerated in patiens with liver disease of diverse etiology. Journal of hepatology. 2013;58(Supplement 1):S532. [Google Scholar]
- 16.Ishak K, Baptista A, Bianchi L, et al. Histological grading and staging of chronic hepatitis. Journal of hepatology. 1995;22(6):696–9. doi: 10.1016/0168-8278(95)80226-6. [DOI] [PubMed] [Google Scholar]
- 17.Cholongitas E, Papatheodoridis GV, Vangeli M, Terreni N, Patch D, Burroughs AK. Systematic review: The model for end-stage liver disease--should it replace Child-Pugh’s classification for assessing prognosis in cirrhosis? Alimentary pharmacology & therapeutics. 2005;22(11–12):1079–89. doi: 10.1111/j.1365-2036.2005.02691.x. [DOI] [PubMed] [Google Scholar]
- 18.McHutchison J, Goodman Z, Patel K, et al. Farglitazar lacks antifibrotic activity in patients with chronic hepatitis C infection. Gastroenterology. 2010;138(4):1365–73. 73 e1–2. doi: 10.1053/j.gastro.2009.12.003. [DOI] [PubMed] [Google Scholar]
- 19.Kweon YO, Goodman ZD, Dienstag JL, et al. Decreasing fibrogenesis: an immunohistochemical study of paired liver biopsies following lamivudine therapy for chronic hepatitis B. J Hepatol. 2001;35(6):749–55. doi: 10.1016/s0168-8278(01)00218-5. [DOI] [PubMed] [Google Scholar]
- 20.Wai CT, Greenson JK, Fontana RJ, et al. A simple noninvasive index can predict both significant fibrosis and cirrhosis in patients with chronic hepatitis C. Hepatology. 2003;38(2):518–26. doi: 10.1053/jhep.2003.50346. [DOI] [PubMed] [Google Scholar]
- 21.Sterling RK, Lissen E, Clumeck N, et al. Development of a simple noninvasive index to predict significant fibrosis in patients with HIV/HCV coinfection. Hepatology. 2006;43(6):1317–25. doi: 10.1002/hep.21178. [DOI] [PubMed] [Google Scholar]
- 22.Forns X, Ampurdanes S, Llovet JM, et al. Identification of chronic hepatitis C patients without hepatic fibrosis by a simple predictive model. Hepatology. 2002;36(4 Pt 1):986–92. doi: 10.1053/jhep.2002.36128. [DOI] [PubMed] [Google Scholar]
- 23.Koda M, Matunaga Y, Kawakami M, Kishimoto Y, Suou T, Murawaki Y. FibroIndex, a practical index for predicting significant fibrosis in patients with chronic hepatitis C. Hepatology. 2007;45(2):297–306. doi: 10.1002/hep.21520. [DOI] [PubMed] [Google Scholar]
- 24.Huwart L, Sempoux C, Vicaut E, et al. Magnetic resonance elastography for the noninvasive staging of liver fibrosis. Gastroenterology. 2008;135(1):32–40. doi: 10.1053/j.gastro.2008.03.076. [DOI] [PubMed] [Google Scholar]
- 25.Herzka DA, Kotys MS, Sinkus R, Pettigrew RI, Gharib AM. Magnetic resonance elastography in the liver at 3 Tesla using a second harmonic approach. Magnetic resonance in medicine. 2009;62(2):284–91. doi: 10.1002/mrm.21956. [DOI] [PubMed] [Google Scholar]
- 26.Meissner EG, Wu D, Osinusi A, et al. Endogenous intrahepatic IFNs and association with IFN-free HCV treatment outcome. The Journal of clinical investigation. 2014;124(8):3352–63. doi: 10.1172/JCI75938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society Series B (Methodological) 1995;57(1):289–300. [Google Scholar]
- 28.Meissner EG, Kohli A, Virtaneva K, et al. Achieving sustained virologic response after interferon-free hepatitis C virus treatment correlates with hepatic interferon gene expression changes independent of cirrhosis. Journal of viral hepatitis. 2016 doi: 10.1111/jvh.12510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shrivastava S, Petrone J, Steele R, Lauer GM, Di Bisceglie AM, Ray RB. Up-regulation of circulating miR-20a is correlated with hepatitis C virus-mediated liver disease progression. Hepatology. 2013;58(3):863–71. doi: 10.1002/hep.26296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sarma NJ, Tiriveedhi V, Crippin JS, Chapman WC, Mohanakumar T. Hepatitis C virus-induced changes in microRNA 107 (miRNA-107) and miRNA-449a modulate CCL2 by targeting the interleukin-6 receptor complex in hepatitis. Journal of virology. 2014;88(7):3733–43. doi: 10.1128/JVI.03060-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Deng L, Li Y, Huang JM, Zhou G, Qian W, Xu K. Effects of p-CREB-1 on transforming growth factor-beta3 auto-regulation in hepatic stellate cells. J Cell Biochem. 2011;112(4):1046–54. doi: 10.1002/jcb.23017. [DOI] [PubMed] [Google Scholar]
- 32.Xu L, Xiong S, Guo R, et al. Transforming growth factor beta3 attenuates the development of radiation-induced pulmonary fibrosis in mice by decreasing fibrocyte recruitment and regulating IFN-gamma/IL-4 balance. Immunology letters. 2014;162(1 Pt A):27–33. doi: 10.1016/j.imlet.2014.06.010. [DOI] [PubMed] [Google Scholar]
- 33.Zhang Y, Liu P, Gao X, Qian W, Xu K. rAAV2-TGF-beta(3) decreases collagen synthesis and deposition in the liver of experimental hepatic fibrosis rat. Digestive diseases and sciences. 2010;55(10):2821–30. doi: 10.1007/s10620-009-1119-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.