Abstract
Salivary agglutination is an important host defense mechanism to aggregate oral commensal bacteria as well as invading pathogens. Saliva flow and subsequent swallowing more easily clear aggregated bacteria compared to single cells. Phagocytic clearance of bacteria through polymorphonuclear neutrophil granulocytes also seems to increase to a certain extent with the size of bacterial aggregates. To determine a connection between salivary agglutination and the host innate immune response by phagocytosis, an in vitro agglutination assay was developed reproducing the average size of salivary bacterial aggregates. Using the oral commensal Streptococcus gordonii as a model organism, the effect of salivary agglutination to the phagocytic clearance through polymorphonuclear neutrophil granulocytes was investigated. Here we describe that salivary aggregates of S. gordonii are readily cleared through phagocytosis, while single bacterial cells showed a significant delay in being phagocytosed and killed. Furthermore, prior to phagocytosis the polymorphonuclear neutrophil granulocytes were able to induce a specific de-aggregation, which was dependent on serine protease activity. The herein presented data suggest that salivary agglutination of bacterial cells leads to an ideal size for recognition by polymorphonuclear neutrophil granulocytes. As a first line of defense, these phagocytic cells are able to recognize the aggregates and de-aggregate them via serine proteases to a more manageable size for efficient phagocytosis and subsequent killing in the phagolysosome. This observed mechanism not only prevents the rapid spreading of oral bacterial cells while entering the bloodstream but would also avoid degranulation of involved polymorphonuclear neutrophil granulocytes thus preventing collateral damage to nearby tissue.
Keywords: salivary agglutination, innate immunity, Streptococcus gordonii, polymorphonuclear neutrophil granulocytes
Introduction
The human oral cavity is a heavily populated habitat (Bik et al., 2010; Ahn et al., 2011; Benitez-Paez et al., 2014) where microbes come into direct contact with the host immune defense system and interface with diverse host tissues (Lang et al., 2010). Saliva, as main fluid found in the oral cavity, is the carrier of up to 109 bacterial cells per milliliter (Wade, 2013). Saliva together with the mucus flow originating from the mucosal lining of the oral cavity constitute the major system maintaining a healthy oral homeostasis (Amerongen and Veerman, 2002). Besides bacteriostatic and bactericidal functions mediated by salivary components like lactoferrin, histatins, β-defensins and lysozyme (Amerongen and Veerman, 2002; Lang et al., 2010) the salivary flow constantly reduces the abundance of planktonic bacteria by aggregation and subsequent swallowing. The major aggregating components in saliva are salivary agglutinin, the mucins MUC5B and MUC7 and the dimeric immunoglobulin sIgA (Amerongen and Veerman, 2002) which aggregate single bacterial cells into larger flocs (referred to here as salivary agglutination) for efficient clearance by swallowing.
Less well investigated is the interaction of the systemic innate immune system with bacterial cells aggregated by salivary agglutination. Events leading to this interaction are quite frequent when the oral mucosal barrier is compromised by surgical (Soldado et al., 1998) or dental procedures (Bahrani-Mougeot et al., 2008), but also by daily oral hygiene (Lucas et al., 2008) or even the ingestion of food (Diener et al., 1964). The common consequence is a transient bacteremia (Tomas et al., 2007), usually cleared efficiently by phagocytic cells of the innate immune system (Morris et al., 2007). However, inefficient clearance constitutes the initial step towards the development of deep tissue abscesses (Ulivieri et al., 2007) or infective endocarditis (Lick et al., 2005; Herzberg, 1996; Moreillon et al., 2002). Both can lead to septic shock, while the latter can also trigger acute heart failure. Polymorphonuclear neutrophil granulocytes are a major component of the innate immune defense system in the bloodstream and comprise the majority of phagocytic blood granulocytes. In general, blood circulating polymorphonuclear neutrophil granulocytes are recruited to the site of infection to respond to invading pathogens. When ingested into the phagolysosome, pathogens are usually rapidly killed by a burst of reactive oxygen and nitrogen species, cytolytic proteins and degradative enzymes (Tapper, 1996). Polymorphonuclear neutrophil granulocytes also contain different types of cytoplasmic granules with several enzymes including the serine protease elastase, gelatinase, heparanase, phospholipase (Lehrer, 2004). They also produce other toxic proteins, such as myeloperoxidase, bactericidal permeability-increasing protein, and alpha-defensin (Standish and Weiser, 2009). Contact of polymorphonuclear neutrophil granulocytes with larger bacterial aggregates resisting phagocytosis like biofilms can trigger the release of the granules in a process termed degranulation (Meyle et al., 2010) leading to an effective removal and killing of cells from the biofilm. Polymorphonuclear neutrophil granulocytes have a variety of mechanisms to clear bacteria exhibiting different degrees of aggregation.
The aim of this study was to investigate interactions between the salivary and the systemic immune system during the clearance of agglutinated cell bacteremias from the oral cavity. In the current study, Streptococcus gordonii was employed as our model species, as it and other oral streptococci make up over 80% of the species in the early oral biofilm (Rosan and Lamont, 2000). S. gordonii is also among the 10 most abundant species in dental plaque (Peterson et al., 2013). In general, S. gordonii and other Mitis group streptococci are considered commensals mainly responsible for the early colonization of tooth and oral soft tissues (Jakubovics et al., 2014). Despite their role as commensals and initiators of oral biofilm formation, some of these species, including S. gordonii, are also common etiological agents of infective endocarditis. Thus, we were interested to examine saliva-induced aggregation of S. gordonii and determine its effect upon bacterial killing by phagocytic cells of the innate immune system.
Methods
Collection of saliva and blood
To study the specific interaction between human saliva and the systemic immune system in a closed model system and to avoid problems with immune incompatibility all human body fluids used in the presented study were extracted from the same healthy mid-aged male individual.
Saliva was collected on ice one hour after regular tooth brushing procedure. The serous fraction was separated by centrifugation for 10 min at 4,500×g, filtered through a 0.22 μm pore-size polystyrene low-protein-binding filter (Corning Inc.) and stored at −80°C. Freezing-related precipitates were removed by centrifugation at 21,000×g for 10 min. immediately before use.
Freshly collected blood was taken from the right median cubital vein using Sodium Heparin Vacutainer® and Safety-Lok™ System (BD). Plasma was generated by centrifugation of fresh heparinized blood for 5 min at 2000×g. The Institutional Review Board of the University of Oklahoma Health Sciences Center approved blood (IRB # 15100) and saliva (IRB # 14327) collection protocols.
Microscopic examination of microbial flora in human saliva
To visualize the planktonic oral microbial flora 1h after toothbrushing, 1ml of fresh human saliva was incubated with LIVE/DEAD® BacLight™ Bacterial Viability Kit containing SYTO®9 and propidium iodide in a final concentration of 1.67μM (Invitrogen™) and 1μg/ml Wheat Germ Agglutinin Alexa Fluor® 350 conjugated (Invitrogen™) for 10min at 37°C. Mucous fraction obtained by 10min centrifugation at 4500×g was examined using UPlan FLN 40×/0.75 Ph2 and UPlan FLN 100×/1.30 Oil Ph3 on BX51 equipped with BX-URA2 reflected fluorescence system and X-Cite® 120W metal halide lamp (Olympus). Separate images were taken with DAPI-1160A, FITC-3540B, and TRITC-A BrightLine® fluorescence filters (Semrock), as well as U-PCD2 phase contrast condenser using DP72 CCD camera and merged with cellSens® digital imaging software Version 1.3 (Olympus).
Bacterial growth conditions
Streptococcus gordonii DL1 (Pakula and Walczak, 1963) was routinely grown in Brain Heart Infusion (BD, Difco) for 16 h at 37°C. A 1:30 dilution in fresh BHI medium was further incubated at 37°C until mid-logarithmic growth phase (A600nm = 0.4), washed twice with sterile PBS for 3 min at 2,000×g, and adjusted to A600nm = 0.5 (approx. 7×108 cfu/ml). This cell density was chosen for technical accuracy since an A600nm = 0.5 is in middle of the linear range of the spectrophotometer, and gives the best correlation between A600nm reading and cell numbers.
Determination of bacterial cell density was performed by plating a ten-fold serial dilution series of cells dispersed with a 5 s low-power sonication (VibraCell VC-50; SONICS®). An aliquot of 10 μl was inoculated on BHI agar plates and incubated at 37°C and 5% CO2 for 24h. Counting of colony forming units (cfu) was performed with SZ Trinocular Stereo microscope equipped with illuminated base (Olympus).
In vitro salivary aggregation
To investigate saliva generated bacterial aggregation, 7×108 cfu of DL1 were resuspended in 1 ml human saliva (processing of saliva was performed as described above) and incubated in a 2 ml reaction tube at 37°C on a rocking table, promoting horizontal movement from an angle of ±15° at 20 rpm (Barnstead Thermolyne Vari-Mix) which yielded an efficient mixing of the bacteria in saliva. At various time points 50 μl aliquots were sampled and saliva induced cellular aggregation was determined by comparing the counts of detectable cfu with or without 5 s low-power ultrasonic treatment (VibraCell VC-50; SONICS®).
Microscopic examination was performed directly on 10 μl aliquots using UPlan FLN 100×/1.30 Oil Ph3 on BX51 equipped with DP72 CCD camera (Olympus). Bacterial aggregate diameter was determined with UPlan FLN 40×/0.75 Ph2 and calibrated cellSens® digital imaging software Version 1.3 (Olympus) calculating the arithmetic mean of a minimum and a maximum measurement for each observed object.
In vitro blood infection
The influence of salivary aggregation on bacterial clearance in blood was investigated by a whole blood bacterial killing assay (Lancefield, 1957). A 50 ml DL1 culture from mid-logarithmic growth phase (A600nm = 0.4) was washed twice with sterile PBS and precipitated for 3 min at 2,000×g, adjusted to A600nm = 0.5 (approx. 7×108 cfu/ml) in 1 ml saliva and incubated for 30 min at 37°C on a rocking table, promoting horizontal movement from angle of ±15° at 20 rpm to form saliva induced bacterial aggregates. Washing the aggregates by gravitational pull four times with sterile PBS stopped the reaction. Where indicated, 5 s low-power ultrasonic treatments were performed to disrupt bacterial aggregates.
1 ml fresh human blood as well as freshly prepared human plasma were sealed in 2 ml borosilicate glass vials with silicone rubber caps (Wheaton) and infected with aggregated or dispersed cells (approx. 2×106 cfu/ml, MOI 1) for 3 h in a Hybaid Mini 6 Hybridization Oven (Labnet) at 37°C and 6 rpm. At hourly intervals (hourly time points were chosen for best technical reproducibility), 50 μl aliquots were added to 450 μl sterile PBS. A 5 s low-power ultrasonic treatment was performed to disrupt bacterial aggregates as well as blood cells and the sample was analyzed for bacterial cell density by serial dilution series in sterile PBS. An aliquot of 10 μl was inoculated on BHI agar plates and incubated at 37°C and 5% CO2 for 24 h. Colony counts were performed with a SZ Trinocular Stereo microscope equipped with illuminated base (Olympus).
Surface protein composition analysis
To investigate the influence of salivary aggregation and ultrasonic treatment on the protein composition of the bacterial surface during blood infection, binding experiments with saliva and plasma were performed. 50ml DL1 cultures from mid-logarithmic growth phase (A600nm = 0.4) were washed twice with sterile PBS for 3 min at 2,000×g, resuspended either in 1 ml saliva, plasma, or PBS and incubated for 30 min at 37°C on a rocking table to agitate the cells at an angle of ±15° at 20 rpm. The reaction was stopped by three washing steps with sterile PBS for 3 min at 2,000×g. When indicated, a 5 s low-power ultrasonic treatment was performed and saliva coated cells were subsequently incubated in plasma as described above. Surface bound proteins were eluted by incubating the cells in 500 μl 0.1 M glycine pH 2.0 for 15 min at room temperature. The cells were removed by centrifugation for 3 min at 6,000×g and the protein content of the supernatant was concentrated by precipitation with 50 μl 100% trichloroacetic acid in acetone for 2 h on ice. The precipitate was harvested by centrifugation at 21,000×g for 10 min at 4°C, washed twice with prechilled acetone and resuspended after air-drying in 30 μl reducing SDS-sample buffer. The acidic samples were neutralized by addition of 1 μl 1.5 M Tris-HCl pH8.8, size separated by SDS-PAGE (Laemmli, 1970), and visualized by Coomassie Brilliant Blue G-250 staining (Kang et al., 2002).
In vitro phagocytosis assay
To investigate if the observed difference in bacterial clearance in blood is caused by phagocytosis efficiency or extracellular killing, a gentamycin protection assay was performed (Medina et al., 2003). Saliva induced bacterial aggregates of DL1 were prepared as described above.
Human granulocytes were always prepared from fresh human blood on a daily basis using the Polymorphprep™ system (Axis-Shield) following the supplier’s manual. The purified cells were resuspended in HyClone® RPMI-1640 Medium + 2,05 mM L-glutamine (Thermo Scientific) containing 10% FCS (Lifetechnologies, Carlsbad, CA) and adjusted to approx. 2×106 cells/ml by trypan blue dye exclusion using an Improved Neubauer chamber (Hausser Scientific).
1 ml fresh human granulocytes in RPMI 10% FCS as well as RPMI 10% FCS without cells in 2 ml borosilicate glass vials with silicone rubber sealed caps (Wheaton) were infected with aggregated or dispersed cells (approx. 2×106 cfu/ml, MOI 1) for 3 h in a Hybaid Mini 6 Hybridization Oven (Labnet) at 37°C and 6rpm.
At hourly intervals 100 μl aliquots were taken, added to 900 μl fresh RPMI 10% FCS containing 200 μg/ml gentamycin sulfate (Calbiochem®) and incubated for 5 min in ice water to stop phagocytosis and killing processes. Extracellular bacteria were subsequently killed by incubating the samples for 1 h at 37°C in the presence of 200 μg gentamycin. Killing efficiency was tested in separate experiments (data not shown). Intracellular bacteria were collected by centrifugation after three washing steps using sterile PBS at 300×g followed by 5 s low-power ultrasonic treatment to disrupt human granulocytes and release the intracellular bacteria. 10 μl of serially diluted cells was inoculated on BHI agar plates and incubated at 37°C and 5% CO2 for 24 h. Colony counting was performed with a SZ Trinocular Stereo microscope equipped with illuminated base (Olympus).
At the same time points, 50 μl aliquots were added to 450 μl sterile PBS to determine the overall bacterial count of each infection. A 5 s low-power ultrasonic treatment was performed to disrupt bacterial aggregates as well as granulocytes and the sample was analyzed for remaining bacterial cell density by colony counting. An aliquot of 10 μl was inoculated on BHI agar plates and incubated at 37°C and 5% CO2 for 24h. Colony counting was performed with a SZ Trinocular Stereo microscope equipped with illuminated base (Olympus).
Where indicated, the specific irreversible inhibitor of serine proteases 4-(2-Aminoethyl)benzenesulfonylfluoride (AEBSF; Calbiochem) was added in a final concentration of 500 μM to the infection assay.
In vitro de-aggregation by granulocyte proteases
To determine how granulocytes disrupt saliva induced bacterial aggregates during phagocytosis, a de-aggregation assay with granule-components was performed (Standish and Weiser, 2009). Saliva induced bacterial aggregates of DL1 were prepared as described above and resuspended in 800 μl sterile PBS.
Human granulocytes were prepared as described above, resuspended in HyClone® RPMI-1640 Medium + 2,05mM L-glutamine (Thermo Scientific) without FCS and adjusted to approx. 4×107 cells/ml by trypan blue dye exclusion using an Improved Neubauer chamber (Hausser Scientific).
To degranulate the primary granules, 1 ml cell suspension in a 2 ml reaction tube was treated with 10 ng/ml PMA (Sigma Aldrich) for 30 min at 37°C and 20 rpm on a rocking table. To degranulate primary as well as azurophilic granules, the cells were first treated with 5 μg/ml cytochalasin D (Sigma Aldrich) for 5 min at 37°C and subsequently with 1 μM fMLP (Sigma Aldrich) for 30 min at 37°C and 20 rpm. The cells were removed by centrifugation at 600×g for 10 min at 20°C and the supernatant was stored over night at −80°C.
200 μl of granula-extract was added to 800 μl DL1 aggregates in PBS and incubated for 2 h at 37°C on a rocking table, promoting horizontal movement from angle of ±15° at 20 rpm. To inhibit proteolytic activity in the primary/azurophilic granula preparation, 500 μM AEBSF (Calbiochem) was added. At indicated the time points, 100 μl aliquots were added to 900 μl sterile PBS and the de-aggregation of the preformed bacterial aggregates was measured by the increase of detectable colony forming units.
To investigate the influence of each of the three proteases secreted during azurophilic granula release, a de-aggregation assay with specific protease inhibitors was performed. Where indicated, 500 μM AEBSF (Calbiochem), 100 μM Elastase Inhibitor IV (HNE IV; Calbiochem), and 200 μM Chymostatin (Sigma Aldrich) were added to the extract of primary and azurophilic granules and incubated for 5 min at 37°C prior to the addition of preformed bacterial aggregates.
Macroscopic examination was performed vertically with 1 ml samples in 2 ml reaction tubes using a Nikon D80 equipped with AT-X 100 PRO D macro lens (Tokina) in a BioDoc-It® Imaging System (UVP) with a UV to white light converter plate for proper illumination.
Statistical analysis
Statistical significance was calculated using a two-sided Student’s t test, with p values less than 0.05 considered as statistically significant. Mathematically rounded p values of the individual experiments are given in the corresponding figure legend.
RESULTS
Organizational state of bacteria in human saliva
To determine the organizational state of the bacterial flora in human saliva in vivo, a microscopic examination of freshly collected human saliva was performed (Fig. 1A). A bacteria-specific fluorescent viability stain in combination with a eukaryotic membrane stain was applied to identify bacterial as well as human cells and visualize bacterial viability (Fig. 1B). The microscopic examination indicated that nearly all bacterial cells were viable and revealed two major reservoirs of bacterial cells in human saliva. Shed buccal epithelial cells were frequently associated with high numbers of rod and spherical shaped bacterial cells in agreement with previous reports (Rudney et al., 2005) (Fig. 1C). Planktonic cells present as single cells or small clusters were detected in addition to more or less spherical aggregates with an average diameter of approximately 20μm probably caused by salivary agglutination (Fig. 1D). The red labeled components are the nuclei of shed buccal epithelial cells, suggesting that these cells are membrane compromised or possibly dead.
FIG 1.
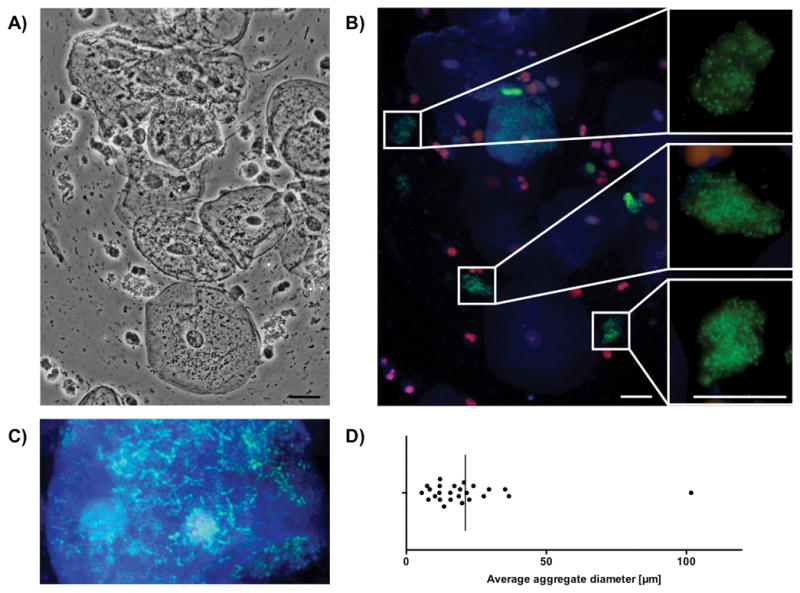
Bacterial aggregates in fresh human saliva. Morphology and viability of natural planktonic bacterial aggregates in human saliva. A) Representative phase-contrast picture. B) Merged epifluorescence picture [(blue channel = Alexa Fluor 350 WGA (staining N-acetyl-D-glucosamine and sialic acid on the cell surface of bacteria and eukaryotic cells), red channel = propidium iodide (PI) (general DNA stain to evaluate cell viability, PI can only penetrate compromised membranes), green channel = SYTO 9 (for general DNA staining)]. The close-up areas depict roughly spherical aggregates with an average diameter of approximately 20 μm. Scalebar represents a distance of 20μm. C) Rod and spherical shaped bacterial cells associated with buccal epithelial cells. D) Range of randomly selected salivary aggregates in freshly prepared saliva 1h after regular teeth brushing (n=25).
Development of an in vitro model of salivary agglutination
To investigate the biological significance of saliva-induced bacterial agglutination in a controlled environment, an in vitro model was developed using sterile filtered human serous saliva and the well described oral streptococcal strain of S. gordonii DL1. A defined number of vital bacterial cells was resuspended in the serous fraction of human saliva and incubated at 37°C with a reproducible amount of shear forces. The formation of cell aggregates was followed over time by the decrease of detectable cfu in comparison to the same sample after the aggregates were disrupted by sonication (Fig. 2A). After five minutes of incubation, a significant 13-fold reduction in cfu could be detected, that further decreased up to 45-fold after twenty minutes. The observed reduction in cfu was specific to bacterial agglutination, since no reductions in viable cell counts could be observed during the maximal incubation time of 60 min.
FIG 2.
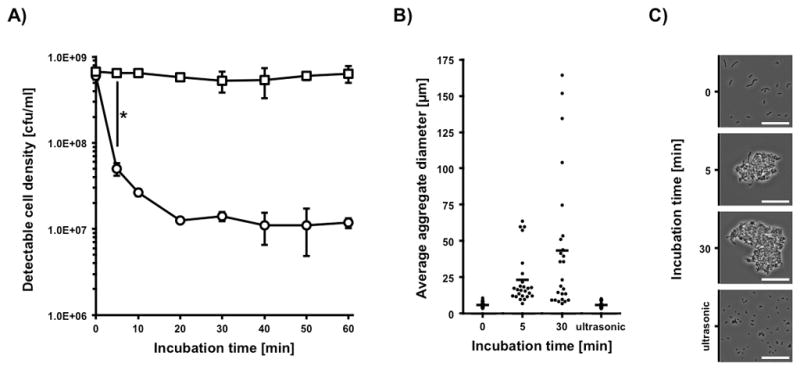
Saliva induced bacterial agglutination. Observation of saliva induced cellular agglutination kinetics and the resulting culture morphology using 5×108 cfu S. gordonii DL1. A) Decrease in detectable colony forming units. Ultrasonic treated culture aliquots are marked with squares, untreated aliquots are marked with circles. Data represent averages and standard deviations from three independent experiments (*p < 0.004). B) Distribution of aggregate diameter at different timepoints during saliva incubation and after ultrasonic treatment. Diagram shows 25 randomly chosen bacterial aggregates from one representative experiment (dots) and calculated average (bar). C) Representative microscopic phase contrast pictures of average bacterial aggregate size in saliva at different timepoints and after ultrasonic treatment. Scalebar represents a distance of 20μm.
To characterize the related aggregate morphology, microscopic observations of the samples were performed during the experiment and average aggregate sizes were determined (Fig. 2B). The in vitro incubation of bacterial cells for five minutes in saliva resulted in a population of aggregates with a diameter of 5μm to 70μm with a major group around 15μm. After thirty minutes of incubation a more heterogeneous distribution of aggregate sizes up to 160μm could be observed with an average of 45μm and several aggregates less than 25μm. When comparing the size distribution of the single species aggregates in the in vitro model to the in vivo multispecies aggregates in saliva (Fig. 1D), a better representation of the range was observed at 30 min. The 5 min time point seem to have a more even distribution, but we did not observe larger aggregates over 100 μm. Subsequent experiments were therefore performed with in vitro aggregated cells incubated in saliva for 30 min.
Microscopic examination also confirmed the initial culture exhibited typical streptococcal cell chains, whereas salivary agglutination led to the formation of more or less spherical aggregates that could be efficiently disrupted into bunches of two to ten cells by mild sonication (Fig. 2C). From these results, we concluded that these conditions could serve as a suitable in vitro approach to mimic saliva-induced bacterial agglutination for comparative studies with human blood components.
Salivary agglutination leads to accelerated clearance of bacteria in blood
To investigate the influence of salivary agglutination on bacterial survival in blood, we employed our in vitro model to compare saliva treated aggregates and dispersed bacteria. The bacterial preparations were incubated in freshly drawn human blood as well as freshly prepared human plasma as a growth control. The bacterial survival was followed over time and calculated as percentage of survival to the initial cfu (Fig. 3A). After one hour of incubation a significant 3.8-fold increase in the clearance of aggregated bacteria could be observed if compared to the non-aggregated control setup. This initial “killing-gap” in the dispersed sample resulted in a constant difference of bacterial load in comparison to the incubation with aggregated bacteria. In contrast, there was no detectable difference in viability and growth in the plasma controls between aggregated and disrupted bacterial aggregates for the duration of the experiment.
FIG 3.
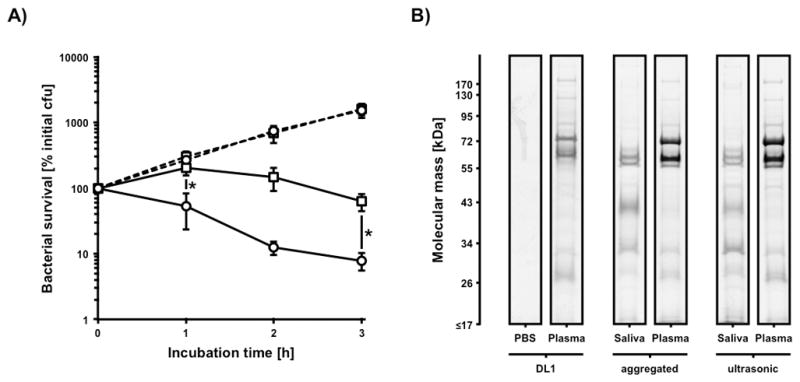
Influence of saliva induced cell agglutination on bacterial appearance in human blood. A) Survival of saliva induced aggregates (circles) and ultrasonic disrupted aggregates (squares) of 2×106 cfu S. gordonii DL1 in human plasma (dashed lines) and whole human blood (solid lines). Data represent averages and standard deviations from three independent experiments (*p < 0.04). B) Influence of low-power ultrasonic treatment on bacterial surface protein composition after incubation in PBS, human plasma, and human saliva followed by plasma incubation.
To address the possibility that sonication induced changes in bacterial surface-protein composition leading to differences in the interaction with salivary or plasma proteins, a surface protein composition analysis was performed (Fig. 3B). One-dimensional separation of acidic surface elutions showed no obvious differences in the binding behavior of saliva and plasma proteins to the bacterial surface of aggregated and dispersed bacteria. This suggests that the greater survival of dispersed cells in blood was unlikely attributable to surface protein modifications introduced through sonication.
Salivary aggregation of bacteria leads to accelerated phagocytosis by granulocytes
We were next interested to determine whether the increased killing efficiency of salivary aggregates could be attributed to differences in phagocytosis. A defined number of freshly purified human granulocytes were infected with aggregated and dispersed bacteria and we similarly observed significantly greater killing of bacterial aggregates (Fig. 4A). This result was consistent with a gentamycin protection assay, which revealed a 100-fold higher phagocytosis efficiency of saliva-aggregated bacteria compared to non-aggregated bacteria during the first hour of infection (Fig. 4B) pointing towards accelerated recognition of the bacterial cells by the neutrophils. A possible increased initial killing before the 1 hour time-point of the internalized non-aggregated cells is unlikely since non-aggregated DL1 has been shown to survive quite efficiently inside polymorphonuclear neutrophil granulocytes (Yajima et al., 2009). The bacterial aggregates were rapidly phagocytized at the beginning of the assay with a decrease of detectable intracellular bacteria after three hours of infection. In contrast, phagocytosis of the dispersed bacteria occurred slowly, following a linear course. No significant differences in viability and growth in the media controls were detected. This suggests that accelerated phagocytosis and subsequent intracellular killing caused the observed increase in the clearance efficiency of aggregated bacteria in human blood by granulocytes.
FIG 4.
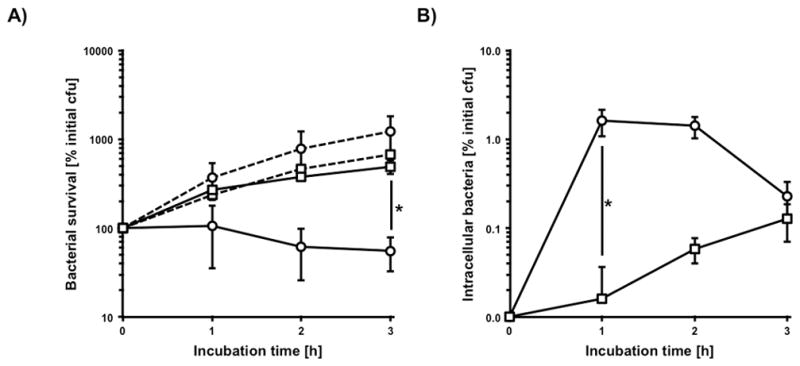
Influence of saliva induced cell agglutination on phagocytosis. A) Survival of saliva induced aggregates (circles) and ultrasonic disrupted aggregates (squares) of 2×106 cfu S. gordonii DL1 in the presence (solid lines) and absence (dashed lines) of 2×106 granulocytes in RPMI with 10% FCS. B) Corresponding data for determination of vital intracellular bacteria. Data represent averages and standard deviations from three independent experiments (*p < 0.01).
Granulocyte proteases are able to disrupt saliva-induced bacterial aggregates
To investigate how granulocytes are able to efficiently take up bacterial cells out of a compact salivary aggregate, granulocyte derived serine proteases were analyzed for their ability to disrupt bacterial aggregates. Preformed saliva-induced bacterial aggregates were treated with the content of either primary or primary + azurophilic granules and in combination with the specific, irreversible serine protease inhibitor AEBSF. Changes in aggregation were followed over time by quantifying colony forming units. In all three treatments, de-aggregation could be observed over the course of the assay (Fig. 5A). The aggregates treated with primary granule components as well as primary + azurophilic granules in combination with AEBSF both showed only a minor de-aggregation, probably caused by the applied shear forces. However, the primary + azurophilic granule components without inhibitor showed a significant 4-fold increase in detectable colony forming units after 60min of incubation (Fig. 5A). As control, we also confirmed that the different granule preparations as well as AEBSF did not affect bacterial viability and growth (data not shown).
FIG 5.
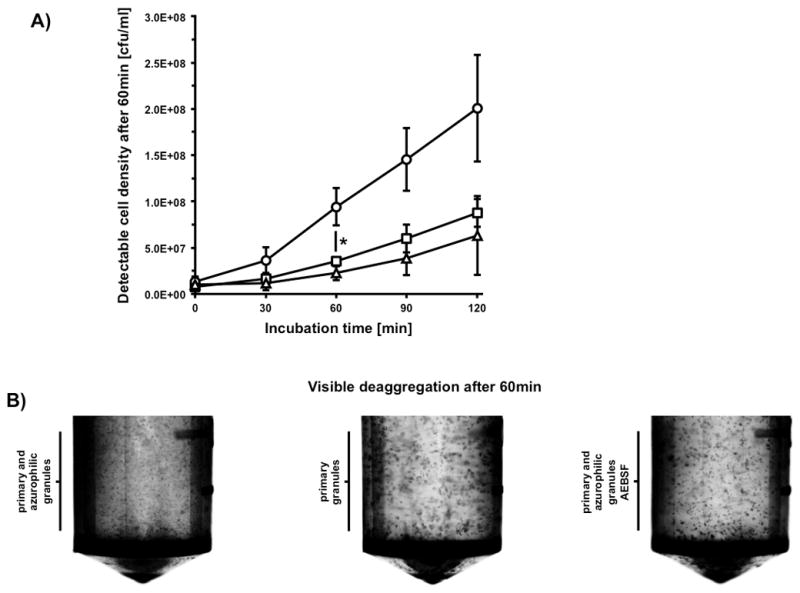
Granulocyte protease dependent deaggregation. Observation of granule-component induced deaggregation kinetics and resulting macroscopic culture morphology using preformed aggregates of 5×108 cfu S. gordonii DL1. A) Increase of detectable colony forming units of preformed bacterial aggregates during incubation with the content of primary granules (squares), primary and azurophilic granules (circles), as well as primary and azurophilic granules in combination with AEBSF (triangles). Data represent averages and standard deviations from three independent experiments (* p < 0.05). B) Representative photographic pictures of macroscopic visible bacterial aggregates in 2ml reaction-tubes.
To confirm this result, a direct macroscopic examination of the reaction was performed (Fig. 5B). After 60min of incubation, an obvious increase of the optical density and a decrease of aggregation size could be observed.
To distinguish the contribution of the different granulocyte proteases to the degradation of saliva induced bacterial aggregates, we further tested a variety of specific protease inhibitors (Fig. 6A). We detected a significant reduction in the amount of detectable colony forming units after 60min of incubation using human neutrophil-elastase inhibitor HNE IV, which was further enhanced in combination with the cathepsin g inhibitor chymostatin. Treatment with chymostatin alone was not sufficient to significantly reduce the de-aggregating effect of azurophilic granule components, which indicates neutrophil-elastase as a major factor responsible for the observed lysis of salivary aggregates. No significant effect on bacterial viability and growth could be detected using the different protease inhibitor combinations (data not shown). These observations could be confirmed independently by a macroscopic examination of the reaction tubes after 60min of incubation (Fig. 6B). A significant increase of the optical density as a result of aggregate degradation could only be seen in the positive control without any inhibitor and in the sample treated with the cathepsin g specific inhibitor chymostatin.
FIG 6.
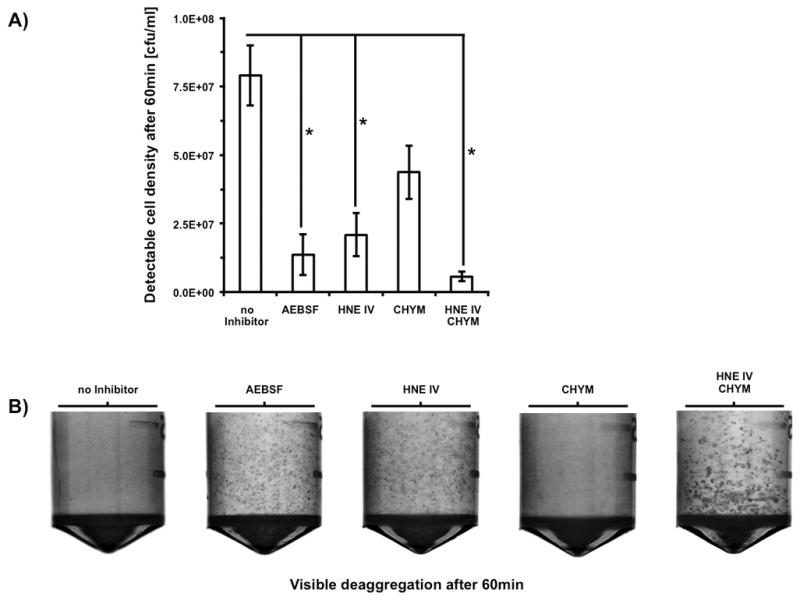
Determination of granulocyte-protease specificity. Observation of granulocyte-protease dependent deaggregation of preformed aggregates of 5×108 cfu S. gordonii DL1 in the presence of different protease-inhibitor combinations. A) Increase of detectable colony forming units of preformed bacterial aggregates after 60min of incubation. Data represent averages and standard deviations from three independent experiments (* p < 0.03). B) Representative photographic pictures of macroscopic visible bacterial aggregates in 2ml reaction-tubes.
Granulocyte protease activity is essential to take up and kill saliva aggregated bacteria
To investigate the contribution of granulocyte proteases to phagocytosis and the killing of saliva aggregated bacteria, a phagocytosis assay was performed in the presence of the AEBSF (Fig. 7A). Protease inhibitor treatment resulted in an increase in the amount of bacterial cells over the time of infection that was comparable with the plasma-control. In contrast, the samples without inhibitor exhibited a significant 9-fold decrease in bacterial load after three hours of infection. This observation could be confirmed by the determination of phagocytosis efficiency (Fig. 7B). In the presence of AEBSF only a few intracellular bacteria could be detected. In comparison, the infection without inhibitor treatment yielded a significant 500-fold increase of detectable intracellular bacteria after one hour that slowly decreased over time demonstrating the importance of granulocyte proteases during the phagocytic uptake of saliva aggregated bacteria.
FIG 7.
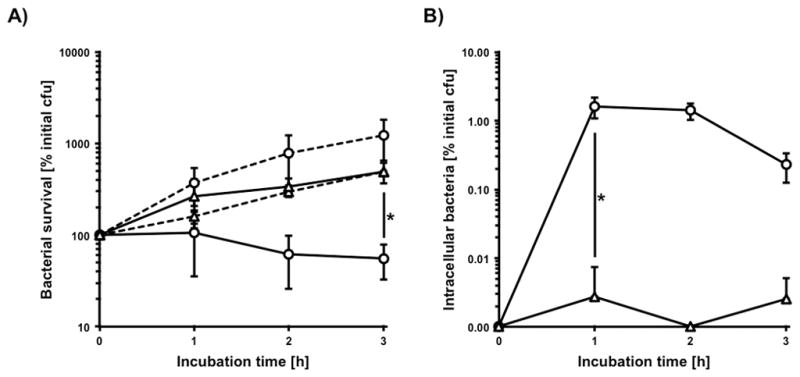
Influence of granulocyte-protease dependent deaggregation on phagocytosis and killing. A) Survival of saliva induced aggregates of 2×106 cfu S. gordonii DL1 with (triangles) and without (circles) AEBSF in the presence (solid lines) and absence (dashed lines) of 2×106 granulocytes in RPMI with 10% FCS. B) Corresponding data for determination of intracellular bacteria. Data represent averages and standard deviations from three independent experiments (* p < 0.04).
Discussion
A transient bacteremia as a consequence of a temporary breeched oral mucosal barrier is a common event induced by surgical or dental procedures, as well as daily oral hygiene or ingestion of food (Lockhart and Durack, 1999; Lockhart et al., 2008). Phagocytic cells of the innate immune system are usually very efficient in the removal of these bacterial invaders (Zhang et al., 2000). The effectiveness of this defense response, among other factors, is influenced by the size of the bacterial cell, as well as cellular aggregation state (Weiser, 2013). The size of bacteria, especially streptococci varies by the actual size of the individual cell, but can also vary as a result of incomplete separation of daughter cells leading to longer streptococcal chains (Zapun et al., 2008). For the nasopharyngal pathogen Streptococcus pneumoniae, an increase in cell-chain length was associated with increased complement deposition on the bacterial surface in vitro and promoted complement-dependent clearance during systemic infection of mice. In addition, a decrease in cell-chain length as observed with several mutants, provided a competitive advantage in vivo when compared to wild type cells. Subsequently, minimization of S. pneumoniae leads to less effective killing by opsonophagocytosis (Dalia and Weiser, 2011). Consequently microbial size seems to be proportional to the innate immune system’s ability to initiate complement deposition and clearance. It is therefore expected that host-derived mechanisms exist that promote aggregation of bacteria to prevent uncontrolled spreading of bacterial invaders and to provide an ideal recognition by phagocytic cells of the innate immune system.
The herein observed salivary agglutination bridges two important bacterial clearance mechanisms. The main route for removal of bacteria from the oral cavity through shear force dependent clearance by salivary flow and swallowing, with the gastric juice killing most of the bacterial load (Scannapieco, 1994; Zhu et al., 2006) and the salivary agglutination induced recognition and clearance of oral bacteria by polymorphonuclear neutrophil granulocytes in case the mucosal barrier is breeched and bacterial aggregates enter the blood. Published work from others (Boackle et al., 1993; Leito et al., 2011) and the herein presented data clearly support that salivary agglutination primes oral bacteria for better clearance, since single cells are more resistant to polymorphonuclear neutrophil granulocytes killing as presented in Fig. 4 and (Young Lee et al., 2006).. Overall, the ability of oral streptococci to form a biofilm as well as their successful integration into an existing biofilm structure and the salivary induced agglutination for clearance seem to be competing events. The structural biofilm components, also referred to as biofilm matrix, include polysaccharides, proteins, extracellular DNA and lipids as well as some host-components, may have potent resistance against host-defense mechanisms and shear-force depended clearance. Biofilms can protect embedded oral streptococci from being taken up by polymorphonuclear neutrophil granulocytes (among other advantages). In addition, physical and diffusion limitations might prevent the degradation of the structural components inside the biofilm. This is in contrast to salivary induced agglutination, which potentially leaves the bacteria inside the agglutinated structure seemingly protected, but our results demonstrate that the agglutinated structures can be readily digested by the polymorphonuclear neutrophil granulocytes for further clearance.
In general, it seems that microbes either try to remain as small as possible, perhaps also by avoiding biofilm development until a secure host-niche is reached and subsequently either evade the host-immune system completely by becoming an intracellular pathogen, grow into larger filamentous aggregates as described for E. coli, or most-likely develop into a biofilm (Weiser, 2013). This is further supported by investigations on the ideal particle size for phagocytosis using polymeric microspheres. Diameters between 2 to 3 μm are most effectively phagocytized (Champion et al., 2008). Since coccoid bacteria like streptococci and staphylococci are smaller than the ideal size for phagocytosis (average size between 0.5 to 1.5 μm) (Doshi and Mitragotri, 2010), remaining in the single cell state would be a perfect strategy for survival and dissemination. Effective phagocytosis of particle sizes between 2 to 3 μm is determined by the attachment to membrane ruffles found on macrophages suggesting a strong correlation between initial attachment and phagocytosis efficiency (Champion et al., 2008). Although the observed diameter of salivary induced aggregates is bigger (around 20 μm) than the ideal size for phagocytosis, the study by Doshi and Mitragotri found an increase in attachment for larger particle dimensions in the 12 μm range (Doshi and Mitragotri, 2010). Larger particle sizes or particle aggregates were not investigated in this study. However, increase in the surface area might allow additional macrophages to surround cell aggregates.
A similar strategy to avoid a certain aggregate size can also be observed with oral bacteria, which are known to colonize the tooth and mucosal surfaces as elaborate oral biofilms (Maddi and Scannapieco, 2013) and can also be found inside oral epithelial cells (Rudney et al., 2005). Furthermore, certain oral streptococci are known to secrete IgA proteases (Yokota and Oguma, 1997), which cleave specifically human sIgA1 at the hinge region to produce Fab and Fc fragments, thus preventing sIgA dependent agglutination. This protease function is of potential ecological importance in the oral cavity and it has been suggested that the protease aids in the pathogenic potential of bacteria whose systemic entry is the mucosal surface (Tyler and Cole, 1998).
Conversely, the host has developed an elaborate net of aggregating molecules, which individually target free floating single cells, but also interact with each other to form larger molecular aggregates (Scannapieco, 1994) increasing the chance to trap oral bacteria. Interestingly, while salivary agglutination mainly aids in the removal of bacteria by swallowing, it also seems to provide the systemic innate immune system with an advantage in case the mucosal barrier is breeched. In particular, the role of salivary agglutinin has been confirmed as a major mediator in this process. Salivary agglutinin is a protein complex consisting of the scavenger receptor cysteine-rich glycoprotein gp340, secretory immunoglobulin A, and an 80-kDa protein (Loimaranta et al., 2005) which interacts with streptococci and other bacteria as part of the host innate defense system at mucosal surfaces (Ericson and Rundegren, 1983; Oho et al., 1998; Prakobphol et al., 2000). The relative non-discriminate polymicrobial aggregation allows for a rigorous capture and clearance even for bacteria, which might not trigger a robust innate immune response on their own. The herein used S. gordonii DL1 interacts with salivary agglutinin through sialic acid binding adhesin Hsa and antigen I/II family adhesins SspA and SspB (Loimaranta et al., 2005). The observed salivary agglutination in our in vitro model is therefore most likely dependent upon salivary agglutinin. Besides its general aggregation ability with diverse bacterial species, salivary agglutinin is able to activate the complement system (Boackle et al., 1993; Leito et al., 2011). The activation occurs mainly through the lectin pathway involving the recognition of mannose residues on pathogens and commensal bacteria by the mannose-binding protein MBL. The classical activation through complement recognition molecule C1q only plays a minor role in the process (Leito et al., 2011). Subsequently, complement activation leads to opsonization and chemotactic attraction of innate immune cells initiating phagocytic clearance.
As shown here, salivary agglutination leads to the effective killing of S. gordonii. This was associated with a rapid intracellular accumulation of S. gordonii inside polymorphonuclear neutrophil granulocytes, indicating that the saliva driven aggregation did not interfere with phagocytosis. The incubation with components usually released by degranulating polymorphonuclear neutrophil granulocytes was associated with a visible de-aggregation that could be inhibited by the serine-protease inhibitor AEBSF, suggesting an active degradation of salivary agglutinin structures responsible for aggregation. As a consequence of AEBSF inhibition, no intracellular bacteria were detected. The herein observed events may suggest that salivary agglutination leads to an ideal size for recognition by the host defense system once bacterial aggregates enter the circulatory system. As a first line of defense, polymorphonuclear neutrophil granulocytes are able to recognize the aggregates probably by complement mediated chemotaxis and with the help of serine proteases are able to de-aggregate the cells to a more manageable size for easy phagocytosis and subsequent killing in the phagolysosome. Membrane-bound human neutrophil elastase and cathepsin G (Owen et al., 1995) seem to be responsible for the observed degradation. Although not determined here, killing via the generation of reactive oxygen species during oxidative burst is also a likely mechanisms used by polymorphonuclear neutrophil granulocytes as shown for S. aureus (Standish and Weiser, 2009), but might be ineffective againt streptococci since they are known to produce copious amounts of H2O2 themselves (Zhu and Kreth, 2012) and have potent resistant mechanisms in place (Xu et al., 2014).
A limitation of our study is the use of one blood donor. However, our results are consistent with earlier reports investigating S. gordonii polymorphonuclear neutrophil granulocyte interactions (Young Lee et al., 2006; Yajima et al., 2009) and with a report investigating the effect of bacterial size minimization on complement evasion (Dalia and Weiser, 2011).
In conclusion, the bacterial size and aggregate status are important factors for the recognition by the host defense system. The herein presented data not only support the important role for salivary agglutination in the clearance of bacteria when entering the circulatory system, it also demonstrates in general the efficient interaction of the oral salivary immune defense and the systemic immune system against invading bacterial cells. Studies investigating innate immune system-pathogen interaction should consider the actual appearance of bacteria in the host for a better understanding of mechanisms leading to eventual clearance of disseminating microbial species.
Acknowledgments
This work was supported by an NIH-NIDCR grant DE021726 to J.K. and NIH-NIDCR grants DE018893 and DE022083 to JM.
Footnotes
The authors declare no conflict of interest.
References
- Ahn J, Yang L, Paster BJ, et al. Oral microbiome profiles: 16S rRNA pyrosequencing and microarray assay comparison. PLoS One. 2011;6:e22788. doi: 10.1371/journal.pone.0022788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amerongen AV, Veerman EC. Saliva-the defender of the oral cavity. Oral Dis. 2002;8:12–22. doi: 10.1034/j.1601-0825.2002.1o816.x. [DOI] [PubMed] [Google Scholar]
- Bahrani-Mougeot FK, Paster BJ, Coleman S, Ashar J, Barbuto S, Lockhart PB. Diverse and novel oral bacterial species in blood following dental procedures. J Clin Microbiol. 2008;46:2129–2132. doi: 10.1128/JCM.02004-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benitez-Paez A, Belda-Ferre P, Simon-Soro A, Mira A. Microbiota diversity and gene expression dynamics in human oral biofilms. BMC Genomics. 2014;15:311. doi: 10.1186/1471-2164-15-311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bik EM, Long CD, Armitage GC, Loomer P, Emerson J, Mongodin EF, et al. Bacterial diversity in the oral cavity of 10 healthy individuals. ISME J. 2010;4:962–974. doi: 10.1038/ismej.2010.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boackle RJ, Connor MH, Vesely J. High molecular weight non-immunoglobulin salivary agglutinins (NIA) bind C1Q globular heads and have the potential to activate the first complement component. Mol Immunol. 1993;30:309–319. doi: 10.1016/0161-5890(93)90059-k. [DOI] [PubMed] [Google Scholar]
- Champion JA, Walker A, Mitragotri S. Role of particle size in phagocytosis of polymeric microspheres. Pharm Res. 2008;25:1815–1821. doi: 10.1007/s11095-008-9562-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalia AB, Weiser JN. Minimization of bacterial size allows for complement evasion and is overcome by the agglutinating effect of antibody. Cell Host Microbe. 2011;10:486–496. doi: 10.1016/j.chom.2011.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diener J, Schwartz SM, Shelansk M, Steinberg G. Bacteremia + Oral Sepsis with Particular Reference to Possible Reduction of Systemic Disease Originating from Oral Cavity. Journal of Periodontology. 1964;35:236. &. [Google Scholar]
- Doshi N, Mitragotri S. Macrophages recognize size and shape of their targets. PLoS One. 2010;5:e10051. doi: 10.1371/journal.pone.0010051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ericson T, Rundegren J. Characterization of a salivary agglutinin reacting with a serotype c strain of Streptococcus mutans. Eur J Biochem. 1983;133:255–261. doi: 10.1111/j.1432-1033.1983.tb07456.x. [DOI] [PubMed] [Google Scholar]
- Herzberg MC. Platelet-streptococcal interactions in endocarditis. Crit Rev Oral Biol Med. 1996;7:222–236. doi: 10.1177/10454411960070030201. [DOI] [PubMed] [Google Scholar]
- Jakubovics NS, Yassin SA, Rickard AH. Community interactions of oral streptococci. Adv Appl Microbiol. 2014;87:43–110. doi: 10.1016/B978-0-12-800261-2.00002-5. [DOI] [PubMed] [Google Scholar]
- Kang DH, Gho YS, Suh MK, Kang CH. Highly sensitive and fast protein detection with coomassie brilliant blue in sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Bulletin of the Korean Chemical Society. 2002;23:1511–1512. [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lancefield RC. Differentiation of group A streptococci with a common R antigen into three serological types, with special reference to the bactericidal test. J Exp Med. 1957;106:525–544. doi: 10.1084/jem.106.4.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang ML, Zhu L, Kreth J. Keeping the bad bacteria in check: interactions of the host immune system with oral cavity biofilms. Endodontic Topics. 2010;22:17–32. [Google Scholar]
- Lehrer RI. Primate defensins. Nat Rev Microbiol. 2004;2:727–738. doi: 10.1038/nrmicro976. [DOI] [PubMed] [Google Scholar]
- Leito JT, Ligtenberg AJ, van Houdt M, van den Berg TK, Wouters D. The bacteria binding glycoprotein salivary agglutinin (SAG/gp340) activates complement via the lectin pathway. Mol Immunol. 2011;49:185–190. doi: 10.1016/j.molimm.2011.08.010. [DOI] [PubMed] [Google Scholar]
- Lick SD, Edozie SN, Woodside KJ, Conti VR. Streptococcus viridans endocarditis from tongue piercing. J Emerg Med. 2005;29:57–59. doi: 10.1016/j.jemermed.2005.01.007. [DOI] [PubMed] [Google Scholar]
- Lockhart PB, Durack DT. Oral microflora as a cause of endocarditis and other distant site infections. Infect Dis Clin North Am. 1999;13:833–850. vi. doi: 10.1016/s0891-5520(05)70111-2. [DOI] [PubMed] [Google Scholar]
- Lockhart PB, Brennan MT, Sasser HC, Fox PC, Paster BJ, Bahrani-Mougeot FK. Bacteremia associated with toothbrushing and dental extraction. Circulation. 2008;117:3118–3125. doi: 10.1161/CIRCULATIONAHA.107.758524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loimaranta V, Jakubovics NS, Hytonen J, Finne J, Jenkinson HF, Stromberg N. Fluid- or surface-phase human salivary scavenger protein gp340 exposes different bacterial recognition properties. Infect Immun. 2005;73:2245–2252. doi: 10.1128/IAI.73.4.2245-2252.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas VS, Gafan G, Dewhurst S, Roberts GJ. Prevalence, intensity and nature of bacteraemia after toothbrushing. J Dent. 2008;36:481–487. doi: 10.1016/j.jdent.2008.03.005. [DOI] [PubMed] [Google Scholar]
- Maddi A, Scannapieco FA. Oral biofilms, oral and periodontal infections, and systemic disease. Am J Dent. 2013;26:249–254. [PubMed] [Google Scholar]
- Medina E, Goldmann O, Toppel AW, Chhatwal GS. Survival of Streptococcus pyogenes within host phagocytic cells: a pathogenic mechanism for persistence and systemic invasion. J Infect Dis. 2003;187:597–603. doi: 10.1086/373998. [DOI] [PubMed] [Google Scholar]
- Meyle E, Stroh P, Gunther F, Hoppy-Tichy T, Wagner C, Hansch GM. Destruction of bacterial biofilms by polymorphonuclear neutrophils: relative contribution of phagocytosis, DNA release, and degranulation. Int J Artif Organs. 2010;33:608–620. doi: 10.1177/039139881003300906. [DOI] [PubMed] [Google Scholar]
- Moreillon P, Que YA, Bayer AS. Pathogenesis of streptococcal and staphylococcal endocarditis. Infect Dis Clin North Am. 2002;16:297–318. doi: 10.1016/s0891-5520(01)00009-5. [DOI] [PubMed] [Google Scholar]
- Morris JA, Harrison LM, Biswas J, Telford DR. Transient bacteraemia: a possible cause of sudden life threatening events. Med Hypotheses. 2007;69:1032–1039. doi: 10.1016/j.mehy.2007.02.039. [DOI] [PubMed] [Google Scholar]
- Oho T, Yu H, Yamashita Y, Koga T. Binding of salivary glycoprotein-secretory immunoglobulin A complex to the surface protein antigen of Streptococcus mutans. Infect Immun. 1998;66:115–121. doi: 10.1128/iai.66.1.115-121.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen CA, Campbell MA, Sannes PL, Boukedes SS, Campbell EJ. Cell surface-bound elastase and cathepsin G on human neutrophils: a novel, non-oxidative mechanism by which neutrophils focus and preserve catalytic activity of serine proteinases. J Cell Biol. 1995;131:775–789. doi: 10.1083/jcb.131.3.775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pakula R, Walczak W. On the nature of competence of transformable streptococci. J Gen Microbiol. 1963;31:125–133. doi: 10.1099/00221287-31-1-125. [DOI] [PubMed] [Google Scholar]
- Peterson SN, Snesrud E, Liu J, et al. The dental plaque microbiome in health and disease. PLoS One. 2013;8:e58487. doi: 10.1371/journal.pone.0058487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakobphol A, Xu F, Hoang VM, et al. Salivary agglutinin, which binds Streptococcus mutans and Helicobacter pylori, is the lung scavenger receptor cysteine-rich protein gp-340. Journal of Biological Chemistry. 2000;275:39860–39866. doi: 10.1074/jbc.M006928200. [DOI] [PubMed] [Google Scholar]
- Rosan B, Lamont RJ. Dental plaque formation. Microbes Infect. 2000;2:1599–1607. doi: 10.1016/s1286-4579(00)01316-2. [DOI] [PubMed] [Google Scholar]
- Rudney JD, Chen R, Zhang G. Streptococci dominate the diverse flora within buccal cells. Journal of Dental Research. 2005;84:1165–1171. doi: 10.1177/154405910508401214. [DOI] [PubMed] [Google Scholar]
- Scannapieco FA. Saliva-bacterium interactions in oral microbial ecology. Crit Rev Oral Biol Med. 1994;5:203–248. doi: 10.1177/10454411940050030201. [DOI] [PubMed] [Google Scholar]
- Soldado L, Esteban F, Delgado-Rodriguez M, Solanellas J, Florez C, Martin E. Bacteraemia during tonsillectomy: a study of the factors involved and clinical implications. Clin Otolaryngol Allied Sci. 1998;23:63–66. doi: 10.1046/j.1365-2273.1998.00082.x. [DOI] [PubMed] [Google Scholar]
- Standish AJ, Weiser JN. Human neutrophils kill Streptococcus pneumoniae via serine proteases. J Immunol. 2009;183:2602–2609. doi: 10.4049/jimmunol.0900688. [DOI] [PubMed] [Google Scholar]
- Tapper H. The secretion of preformed granules by macrophages and neutrophils. J Leukoc Biol. 1996;59:613–622. doi: 10.1002/jlb.59.5.613. [DOI] [PubMed] [Google Scholar]
- Tomas I, Alvarez M, Limeres J, Potel C, Medina J, Diz P. Prevalence, duration and aetiology of bacteraemia following dental extractions. Oral Dis. 2007;13:56–62. doi: 10.1111/j.1601-0825.2006.01247.x. [DOI] [PubMed] [Google Scholar]
- Tyler BM, Cole MF. Effect of IgA1 protease on the ability of secretory IgA1 antibodies to inhibit the adherence of Streptococcus mutans. Microbiol Immunol. 1998;42:503–508. doi: 10.1111/j.1348-0421.1998.tb02316.x. [DOI] [PubMed] [Google Scholar]
- Ulivieri S, Oliveri G, Filosomi G. Brain abscess following dental procedures. Case report. Minerva Stomatol. 2007;56:303–305. [PubMed] [Google Scholar]
- Wade WG. The oral microbiome in health and disease. Pharmacol Res. 2013;69:137–143. doi: 10.1016/j.phrs.2012.11.006. [DOI] [PubMed] [Google Scholar]
- Weiser JN. The battle with the host over microbial size. Curr Opin Microbiol. 2013;16:59–62. doi: 10.1016/j.mib.2013.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Itzek A, Kreth J. Comparison of genes required for H2O2 resistance in Streptococcus gordonii and Streptococcus sanguinis. Microbiology. 2014;160:2627–2638. doi: 10.1099/mic.0.082156-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yajima A, Takahashi Y, Shimazu K, et al. Contribution of phosphoglucosamine mutase to the resistance of Streptococcus gordonii DL1 to polymorphonuclear leukocyte killing. FEMS Microbiol Lett. 2009;297:196–202. doi: 10.1111/j.1574-6968.2009.01673.x. [DOI] [PubMed] [Google Scholar]
- Yokota K, Oguma K. IgA protease produced by Streptococcus sanguis and antibody production against IgA protease in patients with Behcet’s disease. Microbiol Immunol. 1997;41:925–931. doi: 10.1111/j.1348-0421.1997.tb01951.x. [DOI] [PubMed] [Google Scholar]
- Young Lee S, Cisar JO, Bryant JL, Eckhaus MA, Sandberg AL. Resistance of Streptococcus gordonii to polymorphonuclear leukocyte killing is a potential virulence determinant of infective endocarditis. Infect Immun. 2006;74:3148–3155. doi: 10.1128/IAI.00087-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zapun A, Vernet T, Pinho MG. The different shapes of cocci. FEMS Microbiol Rev. 2008;32:345–360. doi: 10.1111/j.1574-6976.2007.00098.x. [DOI] [PubMed] [Google Scholar]
- Zhang P, Summer WR, Bagby GJ, Nelson S. Innate immunity and pulmonary host defense. Immunol Rev. 2000;173:39–51. doi: 10.1034/j.1600-065x.2000.917306.x. [DOI] [PubMed] [Google Scholar]
- Zhu H, Hart CA, Sales D, Roberts NB. Bacterial killing in gastric juice--effect of pH and pepsin on Escherichia coli and Helicobacter pylori. J Med Microbiol. 2006;55:1265–1270. doi: 10.1099/jmm.0.46611-0. [DOI] [PubMed] [Google Scholar]
- Zhu L, Kreth J. The role of hydrogen peroxide in environmental adaptation of oral microbial communities. Oxid Med Cell Longev. 2012;2012:717843. doi: 10.1155/2012/717843. [DOI] [PMC free article] [PubMed] [Google Scholar]


