Abstract
Pharmacologic inhibitors of the bromodomain and extra-terminal motif (BET) protein family are in clinical trials for the treatment of hematologic malignancies, yet the functions of individual BET proteins remain largely uncharacterized. We review the molecular roles of BETs in the context of erythropoiesis. Studies in this lineage have provided valuable insights into their mechanisms of action, and helped define the individual and overlapping functions of BET protein family members BRD2, BRD3, and BRD4. These studies have important ramifications for our understanding of the molecular and physiologic roles of BET proteins, and provide a framework for elucidating some of the beneficial and adverse effects of pharmacologic inhibitors.
Introduction
BET inhibitors have great potential as treatments for cancer, inflammatory diseases, and other medical conditions. Unlocking the full potential of BET inhibitors requires an improved understanding of the physiologic functions of BETs. Here we review the molecular roles of BETs focusing on their functions in the context of erythropoiesis. We discuss how BETs associate with specific genomic locations and how this association leads to transcriptional specificity. The erythroid transcription factor GATA1 was one of the first acetylated transcription factors found to associate with BETs and remains one of the best studied. We discuss how GATA1 recruits BET family members to establish its lineage-specific gene expression program. We further consider the implications of an evolving understanding of BET mechanisms for the development of pharmacologic BET inhibitors and highlight areas where further research is needed.
BETs are chromatin interpreters
BETs play well-recognized roles in transcription [1–3]. There are four BET family members in mammalian cells. BRD2, BRD3, and BRD4 are found in all tissues, while BRDT is expressed only in the testes and in some cancers. BETs associate with chromatin in a manner requiring their bromodomains, which bind to acetylated lysine residues. This has been most closely examined in the case of BRD4. BRD4 binding correlates with histone acetylation across the genome in a number of cell types [4–6]. BET bromodomains bind histones with specific combinations of acetylated lysines and other post-translational modifications (PTMs) in vitro [7,8]. However, no histone mark or combination of marks has been found to definitively predict BET binding in vivo. The association of BETs with acetylated transcription factors accounts for at least some of their particular occupancy patterns [9–11]. Accordingly, BET association with chromatin is often maximal at transcription factor binding sites that reside in nucleosome-depleted regions [6,12]. This is particularly important as recent work has found that transcription factor binding may better predict enhancer function than histone PTMs [13]. Transcription factor binding may serve as a nucleus for BET spreading across regions of acetylated histones, perhaps through BET multimerization [14]. Another notable feature of the association of BETs with chromatin is that it remains present during mitosis when most transcription factors vacate chromatin and transcription is globally silenced [15]. Knockdown experiments have demonstrated that BETs facilitate post-mitotic gene reactivation [16–18], but whether BETs must occupy mitotic chromatin to perform this role has not been tested.
While BET proteins have conserved domains and bind to overlapping regulatory elements, they also associate with distinct regulatory complexes that may dictate their individual functions. BRD4 uses both bromodomains [19] and its unique C-terminal domain to bind and activate PTEF-b complexes [20–22]. PTEF-b subsequently phosphorylates RNA polymerase II and negative elongation factors (NELF and DSIF) to trigger productive transcriptional elongation. The concurrent recruitment of BRD4 and PTEF-b has been shown to be important for inducible gene expression programs in multiple contexts [8,23]. BRD2 and BRD3 also bind to pTEFb but do not contain any analog to BRD4’s C-terminal region, and neither has been shown to directly activate pTEFb. However, all BET proteins contain an extra-terminal (ET) domain that may promote transcription through recruitment of coactivators. The BRD4 ET domain associates with additional chromatin regulatory proteins including the nucleosome remodeling SWI/SNF complex, the histone methyltransferase NSD3, and the NuRD component CHD4 [24,25]. The ET domain is ~80% conserved between BET family members, and BRD2 and BRD3 also bind to many of the same complexes [25,26]. In addition to recruiting other factors, BRD2, BRD3, and BRD4 may have histone chaperone function and stimulate transcription through hyperacetylated nucleosomes in vitro [27,28]. Also, both BRD2 [29] and BRD4 [30] have been reported to have kinase activity. BRD4 has been characterized as an RNA Polymerase II kinase, potentially adding an intriguing additional layer of regulation.
Single cell nuclear imaging and micrococcal nuclease (MNase)-based assays have shown that BRD4 depletion leads to large-scale chromatin decondensation and increased accessibility. Furthermore, dominant-negative acting molecules encoding the double bromodomains (BDI/II) of BRD4 competitively dissociate endogenous BRD4 (and presumably other BETs from chromatin and lead to chromatin fragmentation [31] This suggests that widespread BET binding has a role in maintaining normal chromatin structure.
Work on NUT midline carcinoma (NMC) has shed further light on the interplay of BETs with chromatin architecture. NMC is an epithelial cancer that can be driven by chromosomal fusion between the NUT gene and translocation partners including BRD3 or BRD4. BRD4-NUT potentiates a positive feedback loop by recruiting p300 acetyltransferase activity, which further recruits BRD4-NUT [32–34]. Expression of a BRD4-NUT fusion protein in NMC cells expands shorter regions of acetylated chromatin into “mega-domains” that span hundreds of kilobases [35]. BRD3-NUT also binds similarly large chromatin regions. Interestingly these large acetylated domains rarely cross topologically associating domain (TAD) boundaries, suggesting that BET spreading is constrained by other levels of large-scale genomic structure.
Studies utilizing small molecule BET inhibitors have demonstrated the importance of BET proteins in diverse transcriptional programs. How inhibition of these ubiquitously expressed transcriptional regulators leads to gene-specific effects remains a critical unanswered question. Genome-wide studies examining the occupancy of BET proteins on chromatin have revealed that BRD2, BRD3, and BRD4 occupy many genes uniquely and co-occupy many others [11,36]. The degree to which BRD2, BRD3 and BRD4 control expression of the same or disparate sets of genes remains poorly understood. The molecular basis of the observed patterns of chromatin association and how these patterns relate to gene transcription are beginning to be understood in a few specific cellular contexts such as erythropoiesis. Insights from these systems are likely to be generally applicable.
BET function is required for erythropoiesis
During erythroid maturation GATA1 activates nearly all erythroid specific genes including those involved in hemoglobin synthesis. GATA1 is also a major transcriptional repressor that silences genes involved in self-renewal and the maintenance of an immature state [37–39]. GATA1 is acetylated by acetyltransferases p300/CBP on multiples lysine residues immediately adjacent to each of its two zinc fingers [40,41]. Mutations of these lysines lead to loss of chromatin occupancy in vivo, but not in vitro. This implies that the function of GATA1 acetylation is specific to chromatin [42]. Affinity purification using acetylated GATA1 peptides as bait followed by mass spectrometry identified BRD2, BRD3, and BRD4 as candidate GATA1 binding partners [9]. Evidence for the direct interaction of BRD3 with acetylated GATA1 is supported by their widespread co-occupancy on chromatin, their biochemical co-purification that is enhanced by GATA1 acetylation, the requirement of an intact acetyl-binding pocket within BDI of BRD3 for GATA1 binding, the disruption of their interaction by an acetyl-lysine mimetic, as well as structural analyses [9,43]. As BRD2 and BRD4 have similar bromodomains and also co-occupy many GATA1 bound chromatin segments, it is likely that these proteins also interact directly with GATA1. Still, the degree to which each BET occupies particular genomic GATA1 binding sites varies widely, suggesting that in vivo site selectivity is determined by additional factors.
Pharmacologic BET inhibition inhibits erythroid maturation both in erythroid cell lines and in primary mouse erythroblasts [6,9]. Interestingly, this is due to the impairment of the activating functions of GATA1 while its repressive functions are mostly left intact. This occurs through at least two mechanisms. BET inhibitors compromise the establishment of GATA1 occupancy at many sites in genome, including the beta-globin locus. Additionally, BET inhibition can suppress the transcription of many genes without significantly altering GATA1 occupancy, consistent with multiple mechanisms of BET action. The degree to which individual BET proteins carry out distinct functions remains uncertain. Regardless of the mechanism, transcriptional activation by GATA1 is a reliable predictor of sensitivity to BET inhibitors. This is consistent with the idea that BETs play major roles in inducible gene expression [23] and suggests that the major consequence of BET inhibition in this context is impaired GATA1-dependent activation. Interestingly, a small number of important genes including the transcriptional repressor Hexim1 are activated by BET inhibitor treatment in many cell types [44–46]. This effect may be driven by BRD2 inhibition as BRD2 depletion in erythroid cells activates Hexim1 transcription while BRD4 knockdown does not [6].
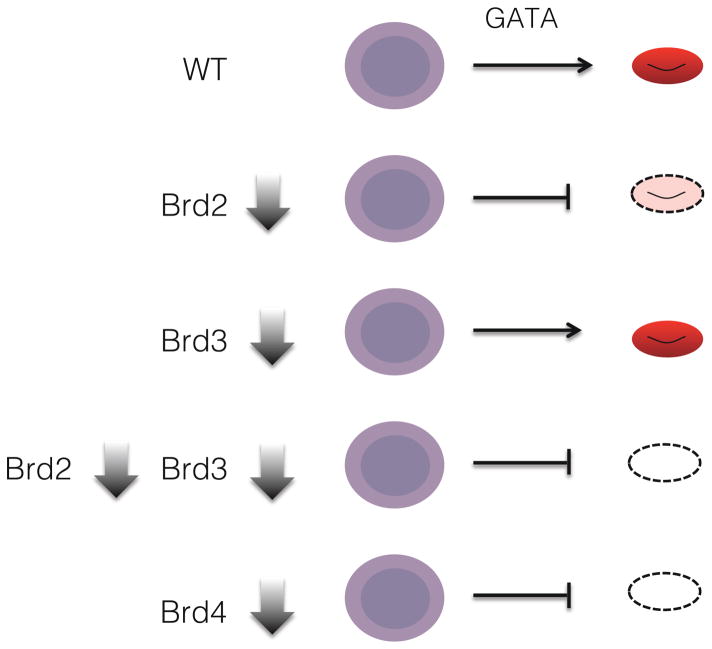
Depletion experiments demonstrate that both BRD2 and BRD4 are individually required for GATA1-driven erythroid maturation (Figure 1). Surprisingly, despite displaying the highest degree of genome-wide co-localization with GATA1, BRD3 is largely dispensable for GATA1 function. Gene knockout-rescue experiments suggest that the functions of BRD2 and BRD3 overlap extensively [6], potentially explaining the minimal phenotype associated with BRD3 loss. Consistent with this, BRD2 is expressed several times higher levels than BRD3 [47], and BRD2 is required for normal GATA1-induced erythroid maturation. The differential but overlapping requirements of BRD2 and BRD3 likely reflect functional similarities as well as their relative expression levels.
Figure 1. BET protein requirements in erythropoiesis.
BRD2 depletion partially inhibits erythroid maturation. BRD3 depletion has minimal effects by itself, but when combined with BRD2 deficiency BRD3 depletion completely abrogates erythroid maturation. BRD4 depletion also prevents erythroid maturation.
BETs are ubiquitous hematopoietic and inflammatory regulators
BETs are implicated in a rapidly growing number of hematopoietic transcriptional programs. For example, hematopoietic stem cell grafts require BRD2 to reconstitute recipient immune systems in mice [48]. Additionally BRD4 co-localizes with hematopoietic transcription factors in models of acute myeloid leukemia [12] and binds to lineage specific regulatory regions in T lymphocytes [4,49]. BRD4 also co-localizes with the transcription factor AIRE in thymic stromal cells and is required for normal expression of genes necessary for deletion of auto-reactive T cells in the thymus [50]. In addition to developmental roles, BETs are essential in the acute activation of the inflammatory response, consistent with their general importance for rapidly inducible gene expression programs. BET inhibition largely abrogates NFkB-dependent gene activation in inflammatory macrophages and cytokine-stimulated endothelial cells [51,52]. BRD4 interacts with the acetylated p65 subunit of NFkB [53], providing a potential explanation for these effects. Taken together with GATA1, the NFkB example suggests a general link between BET function and transcription factor acetylation.
BETs have also been described to play roles in metabolism, a finding likely to influence their role in many contexts including hematopoiesis and inflammation [2]. Mice with low levels of BRD2 due to disruption of the brd2 promoter have increased adiposity but less infiltration of macrophages into adipose tissue. Further, adipocytes in which BRD2 is depleted maintain insulin sensitivity in response to TNF-alpha signaling that causes insulin resistance in wild-type cells [54]. More generally, complete deletion of either BRD2 or BRD4 causes embryonic lethality in mice [54,55], and depletion of BRD4 in the hematopoietic lineage impairs the development of immune lineages, particularly T cells [56]. The increasingly recognized roles of BETs in hematopoiesis and inflammation provide insight into the pleiotropic physiologic effects of BET inhibition, including effects on the growth and maturation of various hematopoietic lineages as well as immunosuppression.
Implications for rational design of BET inhibitors
Small molecule BET inhibitors have shown promise for use as treatments for cancer and other diseases [2,3,57,58]. Many BET inhibitors chemically resemble thienodiazepines (such as JQ1, I-BET151, and OTX-015), although others have unrelated structures [59,60]. This current generation of molecules simultaneously inhibits all BET family members. A number of studies have examined the functions of BET proteins as inhibitor targets by studying the phenotypes of cells in which BET proteins are depleted individually [6,11,61–63]. While this approach has yielded significant insights, it is only able to detect non-redundant activities. Mechanistic work on BETs in erythropoiesis suggests that significant redundancy exists. While loss of BRD2 or BRD3 individually has a lesser effect on transcription, the combined depletion of these proteins reduces the expression of BET inhibitor-sensitive genes. This overlapping functionality necessitates a revised interpretation of the phenotypes elicited by currently available BET inhibitors. It suggests that the common finding in several cell types that BRD2 and BRD3 loss are individually tolerated does not allow their exclusion as functionally important targets. We speculate that this concept may hold true across cells and tissues in which the effects of BET inhibitors have been attributed to a single BET family member [62,64].
The anti-inflammatory and anti-proliferative properties of BET inhibitors will likely make them useful in diverse non-oncologic applications. BET inhibition decreases mortality in mouse models of sepsis, attenuates autoimmunity, and lessens damage from overactive inflammatory responses in the lung [49,51,65,66]. Preclinical studies have also demonstrated the potential efficacy of BET inhibition in applications that would require chronic administration such as heart failure, and type I diabetes mellitus [67,68]. Given the essential roles of BETs in development and tissue homeostasis, any pharmacologic intervention that fully inhibits BET function would be expected to be highly toxic, if not lethal, to patients [55,56]. One way in which therapeutic benefits of BET inhibitors may be achieved is through incomplete inhibition that affects only the most BET-dependent genes. Accordingly, residual BET binding in the presence of BET inhibitors has been observed in a number of systems. An additional consideration is that BET inhibition with JQ1 in mice is likely transient due to its short half-life [26], but the BET inhibitors being tested in human patients are more stable [69]. Interestingly, major side effects reported in early phase clinical trials include anemia [70] and thrombocytopenia [70,71]. Inhibition of GATA1-dependent transcription may contribute to both of these phenotypes as maturation of erythrocytes and megakaryocytes depends on GATA1. As platelets have a shorter life cycle than red blood cells, shorter treatment intervals are more likely to affect this population. Further clinical studies are needed to definitively characterize the side effects and dose-limiting toxicities of BET inhibitors.
Conclusions
BETs occupy the majority of promoters and enhancers in the genomes of mammalian cells and are required for many diverse cellular processes. BET inhibitors have demonstrated great therapeutic potential despite their ubiquitous presence and the essential functions of their targets. In-depth study of BET proteins in the context of erythroid cells has contributed to a deeper understanding of their mechanisms of action. This system allows careful teasing apart of the distinct roles of BETs in transcription factor occupancy and at subsequent steps in transcription. It has revealed that BRD2, BRD3, and BRD4 have both distinct and overlapping cellular roles. It has also presaged some of the toxicity associated with the use of BET inhibitors to treat human disease. Further study of the mechanisms of BET function in normal physiologic as well as pathologic contexts is critical to understanding the uses and limitations of the growing armamentarium of BET inhibitors.
Acknowledgments
This work was supported by National Institutes of Health grants R01-DK054937 and R56-DK065806 (G.A.B), T32-DK007780 (A.J.S.), F30-CA189553 and a Patel Family Scholar Award (S.C.H), and T32-HL007439 (M.T.W.).
Footnotes
Conflict of interest statement
The authors declare no competing financial interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Wu S-Y, Chiang C-M. The double bromodomain-containing chromatin adaptor Brd4 and transcriptional regulation. J Biol Chem. 2007;282:13141–13145. doi: 10.1074/jbc.R700001200. [DOI] [PubMed] [Google Scholar]
- 2.Belkina AC, Denis GV. BET domain co-regulators in obesity, inflammation and cancer. Nat Rev Cancer. 2012;12:465–477. doi: 10.1038/nrc3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shi J, Vakoc CR. The mechanisms behind the therapeutic activity of BET bromodomain inhibition. Mol Cell. 2014;54:728–736. doi: 10.1016/j.molcel.2014.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang W, Prakash C, Sum C, Gong Y, Li Y, Kwok JJT, et al. Bromodomain-containing protein 4 (BRD4) regulates RNA polymerase II serine 2 phosphorylation in human CD4+ T cells. J Biol Chem. 2012;287:43137–43155. doi: 10.1074/jbc.M112.413047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lovén J, Hoke HA, Lin CY, Lau A, Orlando DA, Vakoc CR, et al. Selective Inhibition of Tumor Oncogenes by Disruption of Super-Enhancers. Cell. 2013;153:320–334. doi: 10.1016/j.cell.2013.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stonestrom AJ, Hsu SC, Jahn KS, Huang P, Keller CA, Giardine BM, et al. Functions of BET proteins in erythroid gene expression. Blood Am Soc Hematology. 2015;125:2825–2834. doi: 10.1182/blood-2014-10-607309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Filippakopoulos P, Picaud S, Mangos M, Keates T, Lambert J-P, Barsyte-Lovejoy D, et al. Histone recognition and large-scale structural analysis of the human bromodomain family. Cell. 2012;149:214–231. doi: 10.1016/j.cell.2012.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zippo A, Serafini R, Rocchigiani M, Pennacchini S, Krepelova A, Oliviero S. Histone crosstalk between H3S10ph and H4K16ac generates a histone code that mediates transcription elongation. Cell. 2009;138:1122–1136. doi: 10.1016/j.cell.2009.07.031. [DOI] [PubMed] [Google Scholar]
- 9.Lamonica JM, Deng W, Kadauke S, Campbell AE, Gamsjaeger R, Wang H, et al. Bromodomain protein Brd3 associates with acetylated GATA1 to promote its chromatin occupancy at erythroid target genes. Proceedings of the National Academy of Sciences National Acad Sciences. 2011;108:E159–E168. doi: 10.1073/pnas.1102140108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu S-Y, Lee A-Y, Lai H-T, Zhang H, Chiang C-M. Phospho switch triggers Brd4 chromatin binding and activator recruitment for gene-specific targeting. Mol Cell. 2013;49:843–857. doi: 10.1016/j.molcel.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Asangani IA, Dommeti VL, Wang X, Malik R, Cieslik M, Yang R, et al. Therapeutic targeting of BET bromodomain proteins in castration-resistant prostate cancer. Nature. 2014;510:278–282. doi: 10.1038/nature13229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roe J-S, Mercan F, Rivera K, Pappin DJ, Vakoc CR. BET Bromodomain Inhibition Suppresses the Function of Hematopoietic Transcription Factors in Acute Myeloid Leukemia. Mol Cell. 2015;58:1028–1039. doi: 10.1016/j.molcel.2015.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dogan N, Wu W, Morrissey CS, Chen K-B, Stonestrom A, Long M, et al. Occupancy by key transcription factors is a more accurate predictor of enhancer activity than histone modifications or chromatin accessibility. Epigenetics Chromatin. 2015;8:16. doi: 10.1186/s13072-015-0009-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garcia-Gutierrez P, Mundi M, Garcia-Dominguez M. Association of bromodomain BET proteins with chromatin requires dimerization through the conserved motif B. J Cell Sci. 2012;125:3671–3680. doi: 10.1242/jcs.105841. [DOI] [PubMed] [Google Scholar]
- 15.Dey A, Chitsaz F, Abbasi A, Misteli T, Ozato K. The double bromodomain protein Brd4 binds to acetylated chromatin during interphase and mitosis. Proc Natl Acad Sci U S A National Acad Sciences. 2003;100:8758–8763. doi: 10.1073/pnas.1433065100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dey A, Nishiyama A, Karpova T, McNally J, Ozato K. Brd4 marks select genes on mitotic chromatin and directs postmitotic transcription. Mol Biol Cell. 2009;20:4899–4909. doi: 10.1091/mbc.E09-05-0380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang Z, He N, Zhou Q. Brd4 recruits P-TEFb to chromosomes at late mitosis to promote G1 gene expression and cell cycle progression. Mol Cell Biol American Society for Microbiology. 2008;28:967–976. doi: 10.1128/MCB.01020-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao R, Nakamura T, Fu Y, Lazar Z, Spector DL. Gene bookmarking accelerates the kinetics of post-mitotic transcriptional re-activation. Nat Cell Biol. 2011;13:1295–1304. doi: 10.1038/ncb2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schröder S, Cho S, Zeng L, Zhang Q, Kaehlcke K, Mak L, et al. Two-pronged binding with bromodomain-containing protein 4 liberates positive transcription elongation factor b from inactive ribonucleoprotein complexes. J Biol Chem. 2012;287:1090–1099. doi: 10.1074/jbc.M111.282855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jang MK, Mochizuki K, Zhou M, Jeong H-S, Brady JN, Ozato K. The Bromodomain Protein Brd4 Is a Positive Regulatory Component of P-TEFb and Stimulates RNA Polymerase II-Dependent Transcription. Mol Cell. 2005;19:523–534. doi: 10.1016/j.molcel.2005.06.027. [DOI] [PubMed] [Google Scholar]
- 21.Yang Z, Yik JHN, Chen R, He N, Jang MK, Ozato K, et al. Recruitment of P-TEFb for Stimulation of Transcriptional Elongation by the Bromodomain Protein Brd4. Mol Cell. 2005;19:535–545. doi: 10.1016/j.molcel.2005.06.029. [DOI] [PubMed] [Google Scholar]
- 22.Bisgrove DA, Mahmoudi T, Henklein P, Verdin E. Conserved P-TEFb-interacting domain of BRD4 inhibits HIV transcription. Proc Natl Acad Sci U S A National Acad Sciences. 2007;104:13690–13695. doi: 10.1073/pnas.0705053104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hargreaves DC, Horng T, Medzhitov R. Control of inducible gene expression by signal-dependent transcriptional elongation. Cell. 2009;138:129–145. doi: 10.1016/j.cell.2009.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shi J, Whyte WA, Zepeda-Mendoza CJ, Milazzo JP, Shen C, Roe J-S, et al. Role of SWI/SNF in acute leukemia maintenance and enhancer-mediated Myc regulation. Genes Dev Cold Spring Harbor Lab. 2013;27:2648–2662. doi: 10.1101/gad.232710.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rahman S, Sowa ME, Ottinger M, Smith JA, Shi Y, Harper JW, et al. The Brd4 extraterminal domain confers transcription activation independent of pTEFb by recruiting multiple proteins, including NSD3. Mol Cell Biol American Society for Microbiology. 2011;31:2641–2652. doi: 10.1128/MCB.01341-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dawson MA, Prinjha RK, Dittmann A, Giotopoulos G, Bantscheff M, Chan W-I, et al. Inhibition of BET recruitment to chromatin as an effective treatment for MLL-fusion leukaemia. Nature. 2011;478:529–533. doi: 10.1038/nature10509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.LeRoy G, Rickards B, Flint SJ. The Double Bromodomain Proteins Brd2 and Brd3 Couple Histone Acetylation to Transcription. Mol Cell. 2008;30:51–60. doi: 10.1016/j.molcel.2008.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kanno T, Kanno Y, LeRoy G, Campos E, Sun H-W, Brooks SR, et al. BRD4 assists elongation of both coding and enhancer RNAs by interacting with acetylated histones. Nat Struct Mol Biol. 2014 doi: 10.1038/nsmb.2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Denis GV, Green MR. A novel, mitogen-activated nuclear kinase is related to a Drosophila developmental regulator. Genes Dev. 1996;10:261–271. doi: 10.1101/gad.10.3.261. [DOI] [PubMed] [Google Scholar]
- 30.Devaiah BN, Lewis BA, Cherman N, Hewitt MC, Albrecht BK, Robey PG, et al. BRD4 is an atypical kinase that phosphorylates serine2 of the RNA polymerase II carboxy-terminal domain. Proc Natl Acad Sci U S A. 2012;109:6927–6932. doi: 10.1073/pnas.1120422109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang R, Li Q, Helfer CM, Jiao J, You J. Bromodomain protein Brd4 associated with acetylated chromatin is important for maintenance of higher-order chromatin structure. J Biol Chem. 2012;287:10738–10752. doi: 10.1074/jbc.M111.323493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reynoird N, Schwartz BE, Delvecchio M, Sadoul K, Meyers D, Mukherjee C, et al. Oncogenesis by sequestration of CBP/p300 in transcriptionally inactive hyperacetylated chromatin domains. EMBO J. 2010;29:2943–2952. doi: 10.1038/emboj.2010.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.French CA. Pathogenesis of NUT midline carcinoma. Annu Rev Pathol. 2012;7:247–265. doi: 10.1146/annurev-pathol-011811-132438. [DOI] [PubMed] [Google Scholar]
- 34.Wang R, You J. Mechanistic analysis of the role of bromodomain-containing protein 4 (BRD4) in BRD4-NUT oncoprotein-induced transcriptional activation. J Biol Chem. 2015;290:2744–2758. doi: 10.1074/jbc.M114.600759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alekseyenko AA, Walsh EM, Wang X, Grayson AR, Hsi PT, Kharchenko PV, et al. The oncogenic BRD4-NUT chromatin regulator drives aberrant transcription within large topological domains. Genes Dev. 2015;29:1507–1523. doi: 10.1101/gad.267583.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Anders L, Guenther MG, Qi J, Fan ZP, Marineau JJ, Rahl PB, et al. Genome-wide localization of small molecules. Nat Biotechnol. 2014;32:92–96. doi: 10.1038/nbt.2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ferreira R, Ohneda K, Yamamoto M, Philipsen S. GATA1 function, a paradigm for transcription factors in hematopoiesis. Mol Cell Biol Am Soc Microbiol. 2005;25:1215–1227. doi: 10.1128/MCB.25.4.1215-1227.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Crispino JD. GATA1 in normal and malignant hematopoiesis. Semin Cell Dev Biol. 2005;16:137–147. doi: 10.1016/j.semcdb.2004.11.002. [DOI] [PubMed] [Google Scholar]
- 39.Welch JJ, Watts JA, Vakoc CR, Yao Y, Wang H, Hardison RC, et al. Global regulation of erythroid gene expression by transcription factor GATA-1. Blood Am Soc Hematology. 2004;104:3136–3147. doi: 10.1182/blood-2004-04-1603. [DOI] [PubMed] [Google Scholar]
- 40.Hung HL, Lau J, Kim AY, Weiss MJ, Blobel GA. CREB-Binding protein acetylates hematopoietic transcription factor GATA-1 at functionally important sites. Mol Cell Biol American Society for Microbiology. 1999;19:3496–3505. doi: 10.1128/mcb.19.5.3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boyes J, Byfield P, Nakatani Y, Ogryzko V. Regulation of activity of the transcription factor GATA-1 by acetylation. Nature. 1998;396:594–598. doi: 10.1038/25166. [DOI] [PubMed] [Google Scholar]
- 42.Lamonica JM, Vakoc CR, Blobel GA. Acetylation of GATA-1 is required for chromatin occupancy. Blood. 2006;108:3736–3738. doi: 10.1182/blood-2006-07-032847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gamsjaeger R, Webb SR, Lamonica JM, Billin A, Blobel GA, Mackay JP. Structural basis and specificity of acetylated transcription factor GATA1 recognition by BET family bromodomain protein Brd3. Mol Cell Biol American Society for Microbiology. 2011;31:2632–2640. doi: 10.1128/MCB.05413-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chaidos A, Caputo V, Gouvedenou K, Liu B, Marigo I, Chaudhry MS, et al. Potent antimyeloma activity of the novel bromodomain inhibitors I-BET151 and I-BET762. Blood. 2014;123:697–705. doi: 10.1182/blood-2013-01-478420. [DOI] [PubMed] [Google Scholar]
- 45.Coudé M-M, Braun T, Berrou J, Dupont M, Bertrand S, Masse A, et al. BET inhibitor OTX015 targets BRD2 and BRD4 and decreases c-MYC in acute leukemia cells. Oncotarget. europepmc.org. 2015;6:17698–17712. doi: 10.18632/oncotarget.4131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bartholomeeusen K, Xiang Y, Fujinaga K, Peterlin BM. Bromodomain and extra-terminal (BET) bromodomain inhibition activate transcription via transient release of positive transcription elongation factor b (P-TEFb) from 7SK small nuclear ribonucleoprotein. J Biol Chem ASBMB. 2012;287:36609–36616. doi: 10.1074/jbc.M112.410746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Paralkar VR, Mishra T, Luan J, Yao Y, Kossenkov AV, Anderson SM, et al. Lineage and species-specific long noncoding RNAs during erythro-megakaryocytic development. Blood. 2014;123:1927–1937. doi: 10.1182/blood-2013-12-544494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Belkina AC, Blanton WP, Nikolajczyk BS, Denis GV. The double bromodomain protein Brd2 promotes B cell expansion and mitogenesis. J Leukoc Biol. 2014;95:451–460. doi: 10.1189/jlb.1112588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mele DA, Salmeron A, Ghosh S, Huang H-R, Bryant BM, Lora JM. BET bromodomain inhibition suppresses TH17-mediated pathology. J Exp Med. 2013;210:2181–2190. doi: 10.1084/jem.20130376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yoshida H, Bansal K, Schaefer U, Chapman T, Rioja I, Proekt I, et al. Brd4 bridges the transcriptional regulators, Aire and P-TEFb, to promote elongation of peripheral-tissue antigen transcripts in thymic stromal cells. Proc Natl Acad Sci U S A. 2015;112:E4448–57. doi: 10.1073/pnas.1512081112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nicodème E, Jeffrey KL, Schaefer U, Beinke S, Dewell S, Chung C-W, et al. Suppression of inflammation by a synthetic histone mimic. Nature. 2010;468:1119–1123. doi: 10.1038/nature09589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brown JD, Lin CY, Duan Q, Griffin G, Federation AJ, Paranal RM, et al. NF-κB directs dynamic super enhancer formation in inflammation and atherogenesis. Mol Cell. 2014;56:219–231. doi: 10.1016/j.molcel.2014.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Huang B, Yang X-D, Zhou M-M, Ozato K, Chen L-F. Brd4 coactivates transcriptional activation of NF-kappaB via specific binding to acetylated RelA. Mol Cell Biol American Society for Microbiology. 2009;29:1375–1387. doi: 10.1128/MCB.01365-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang F, Liu H, Blanton WP, Belkina A, Lebrasseur NK, Denis GV. Brd2disruption in mice causes severe obesity without Type 2 diabetes. Biochem J Portland Press Ltd. 2009;425:71–83. doi: 10.1042/BJ20090928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Houzelstein D, Bullock SL, Lynch DE, Grigorieva EF, Wilson VA, Beddington RSP. Growth and early postimplantation defects in mice deficient for the bromodomain-containing protein Brd4. Mol Cell Biol American Society for Microbiology. 2002;22:3794–3802. doi: 10.1128/MCB.22.11.3794-3802.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bolden JE, Tasdemir N, Dow LE, van Es JH, Wilkinson JE, Zhao Z, et al. Inducible in vivo silencing of Brd4 identifies potential toxicities of sustained BET protein inhibition. Cell Rep. 2014;8:1919–1929. doi: 10.1016/j.celrep.2014.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dawson MA, Kouzarides T, Huntly BJP. Targeting epigenetic readers in cancer. N Engl J Med. 2012;367:647–657. doi: 10.1056/NEJMra1112635. [DOI] [PubMed] [Google Scholar]
- 58.Filippakopoulos P, Knapp S. Targeting bromodomains: epigenetic readers of lysine acetylation. Nat Rev Drug Discov. 2014;13:337–356. doi: 10.1038/nrd4286. [DOI] [PubMed] [Google Scholar]
- 59.Hewings DS, Fedorov O, Filippakopoulos P, Martin S, Picaud S, Tumber A, et al. Optimization of 3,5-dimethylisoxazole derivatives as potent bromodomain ligands. J Med Chem. 2013;56:3217–3227. doi: 10.1021/jm301588r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Picaud S, Da Costa D, Thanasopoulou A, Filippakopoulos P, Fish PV, Philpott M, et al. PFI-1, a highly selective protein interaction inhibitor, targeting BET Bromodomains. Cancer Res. 2013;73:3336–3346. doi: 10.1158/0008-5472.CAN-12-3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tang Y, Gholamin S, Schubert S, Willardson MI, Lee A, Bandopadhayay P, et al. Epigenetic targeting of Hedgehog pathway transcriptional output through BET bromodomain inhibition. Nat Med. 2014;20:732–740. doi: 10.1038/nm.3613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zuber J, Shi J, Wang E, Rappaport AR, Herrmann H, Sison EA, et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature nature.com. 2011;478:524–528. doi: 10.1038/nature10334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shi J, Wang E, Milazzo JP, Wang Z, Kinney JB, Vakoc CR. Discovery of cancer drug targets by CRISPR-Cas9 screening of protein domains. Nat Biotechnol Nature Publishing Group. 2015;33:661–667. doi: 10.1038/nbt.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mertz JA, Conery AR, Bryant BM, Sandy P, Balasubramanian S, Mele DA, et al. Targeting MYC dependence in cancer by inhibiting BET bromodomains. Proc Natl Acad Sci U S A. 2011;108:16669–16674. doi: 10.1073/pnas.1108190108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bandukwala HS, Gagnon J, Togher S, Greenbaum JA, Lamperti ED, Parr NJ, et al. Selective inhibition of CD4+ T-cell cytokine production and autoimmunity by BET protein and c-Myc inhibitors. Proc Natl Acad Sci U S A National Acad Sciences. 2012;109:14532–14537. doi: 10.1073/pnas.1212264109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tang X, Peng R, Phillips JE, Deguzman J, Ren Y, Apparsundaram S, et al. Assessment of Brd4 inhibition in idiopathic pulmonary fibrosis lung fibroblasts and in vivo models of lung fibrosis. Am J Pathol. 2013;183:470–479. doi: 10.1016/j.ajpath.2013.04.020. [DOI] [PubMed] [Google Scholar]
- 67.Anand P, Brown JD, Lin CY, Qi J, Zhang R, Artero PC, et al. BET Bromodomains Mediate Transcriptional Pause Release in Heart Failure. Cell. 2013;154:569–582. doi: 10.1016/j.cell.2013.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fu W, Farache J, Clardy SM, Hattori K, Mander P, Lee K, et al. Epigenetic modulation of type-1 diabetes via a dual effect on pancreatic macrophages and β cells. Elife. 2014;3:e04631. doi: 10.7554/eLife.04631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Odore E, Lokiec F, Cvitkovic E, Bekradda M, Herait P, Bourdel F, et al. Phase I Population Pharmacokinetic Assessment of the Oral Bromodomain Inhibitor OTX015 in Patients with Haematologic Malignancies. Clin Pharmacokinet. 2015 doi: 10.1007/s40262-015-0327-6. [DOI] [PubMed] [Google Scholar]
- 70.Amorim S, Stathis A, Gleeson M, Iyengar S, Magarotto V, Leleu X, et al. Bromodomain inhibitor OTX015 in patients with lymphoma or multiple myeloma: a dose-escalation, open-label, pharmacokinetic, phase 1 study. Lancet Haematol Elsevier. 2016;3:e196–204. doi: 10.1016/S2352-3026(16)00021-1. [DOI] [PubMed] [Google Scholar]
- 71.Berthon C, Raffoux E, Thomas X, Vey N, Gomez-Roca C, Yee K, et al. Bromodomain inhibitor OTX015 in patients with acute leukaemia: a dose-escalation, phase 1 study. Lancet Haematol Elsevier. 2016;3:e186–95. doi: 10.1016/S2352-3026(15)00247-1. [DOI] [PubMed] [Google Scholar]