Abstract
DNA polymerase (pol) κ efficiently catalyzes error-free translesion DNA synthesis (TLS) opposite bulky N2-guanyl lesions induced by carcinogens such as polycyclic aromatic hydrocarbons. We investigated the biochemical effects of nine human nonsynonymous germline POLK variations on the TLS properties of pol κ, utilizing recombinant pol κ (residues 1–526) enzymes and DNA templates containing an N2-CH2(9-anthracenyl)G (N2-AnthG), 8-oxo-7,8-dihydroguanine (8-oxoG), O6-methyl(Me)G, or an abasic site. In steady-state kinetic analyses, the R246X, R298H, T473A, and R512W variants displayed 7- to 18-fold decreases in kcat/Km for dCTP insertion opposite G and N2-AnthG, with 2- to 3-fold decreases in DNA binding affinity, compared to wild-type, and further showed 5- to 190-fold decreases in kcat/Km for next-base extension from C paired with N2-AnthG. The A471V variant showed 2- to 4-fold decreases in kcat/Km for correct nucleotide insertion opposite and beyond G (or N2-AnthG) compared to wild-type. These five hypoactive variants also showed similar patterns of attenuation of TLS activity opposite 8-oxoG, O6-MeG, and abasic lesions. By contrast, the T44M variant exhibited 7- to 11-fold decreases in kcat/Km for dCTP insertion opposite N2-AnthG and O6-MeG (as well as for dATP insertion opposite an abasic site), but not opposite both G and 8-oxoG, nor beyond N2-AnthG, compared to wild-type. These results suggest that the R246X, R298H, T473A, R512W, and A471V variants cause a general catalytic impairment of pol κ opposite G and all four lesions, whereas the T44M variant induces opposite lesion-dependent catalytic impairment—i.e., only opposite O6-MeG, abasic, and bulky N2-G lesions, but not opposite G and 8-oxoG—in pol κ, which might indicate that these hypoactive pol κ variants are genetic factors in modifying individual susceptibility to genotoxic carcinogens in certain subsets of populations.
Keywords: DNA polymerase κ, genetic variation, DNA lesion, translesion DNA synthesis, carcinogen
Graphical Abstract
INTRODUCTION
A variety of endogenous and exogenous agents continuously attack cellular DNA and generate numerous DNA lesions in the cellular genome. Cells can employ elegant DNA repair systems to restore genomic integrity but these systems are not perfect, and thus unrepaired or misrepaired lesions persist to some extent in cellular DNA. These persistent DNA lesions can perturb DNA replication (i.e., cause “replication stress”) and RNA transcription in cells, which can induce mutagenesis or cell death depending on the type and extent of DNA damage, ultimately leading to carcinogenesis or other adverse health problems.1, 2 To cope with these replication obstacles and allow completion of DNA replication for cell survival, cells have evolved DNA damage tolerance mechanisms.3 Translesion DNA synthesis (TLS) is a major DNA damage tolerance mechanism whereby specialized TLS polymerases (pols) are recruited at and replicate past DNA lesions that stall replicative pols.4 TLS pols have an ability to bypass a wide variety of DNA lesions but often cause mutations, depending on the lesion type and the DNA sequence context. Indeed, every TLS pol possesses distinctive TLS properties, differing from each other in terms of accuracy (e.g., error-free to error-prone) and efficiency at particular lesions, and thus exerts anti- and pro-mutagenic effects in both a lesion- and sequence context-dependent manner during TLS processes in cells.5 For this reason and due to the low fidelity nature of TLS pols, the balanced and coordinated activities among multiple TLS pols may be critical to maintain the genomic stability in cells.
Of the at least 17 different human DNA pols, four Y-family pols (η, ι, κ, and REV1) (as well as a B-family pol ζ) are prominent in TLS as an inserter, an extender, or a scaffold, although more than half of 17 pols are involved in TLS of certain types of DNA lesions.6, 7 Defects in and dysregulation of these TLS pols can promote the genomic instability and tumorigenesis.8–13 Recent studies reveal a diverse repertoire of mutational signatures in human cancer genomes, some of which correspond well with the proposed etiologies or underlying mutational processes related to the types of DNA damage, DNA repair, and replicative transactions including TLS.1, 14, 15 In this respect, it can be speculated that the cellular capacity for faithful replication in the presence of DNA damage may be an important determinant for genome maintenance, and thus the functional and genetic alterations of TLS pols might modify overall mutation consequences (e.g., frequency and spectrum) of DNA damage induced by carcinogens in cells, potentially contributing to mutation or cancer susceptibility in the affected individuals. Our recent studies reveal that a substantial number of human natural germline genetic variants of TLS pols ι, κ, and REV1 are catalytically altered (either impaired or enhanced in TLS activity),16–18 implying their potential for individual alterations in TLS capacity and mutation risk to certain genotoxic carcinogens in genetically predisposed individuals.
Pol κ is unique in its ability to bypass bulky N2-G adducts such as N2-benzo[a]pyrene diol epoxide (BPDE)-G and N2-(1-carboxyethyl)-G, in addition to an N2-G interstrand cross-link, in a most efficient and error-free manner among TLS pols.19–22 Pol κ-deficient mouse embryonic fibroblast cells are hypersensitive to both killing and mutagenesis from benzo[a]pyrene23 and also show decreases in TLS efficiency and fidelity across an N2-BPDE-G on a shuttle vector plasmid.24 Pol κ is also required for recovery from S-phase checkpoint arrest in mouse cells following exposure to BPDE.25 In addition, pol κ functions in DNA repair processes, e.g. nucleotide excision repair after UV irradiation and strand break repair following oxidative stress, and is also required for checkpoint activation after replication fork stalling with hydroxyurea or aphidicolin in mammalian cells.26–28 Mice lacking pol κ exhibit a spontaneous mutator phenotype in various tissues and reduced survival.29 At present, no human disease linked to pol κ defect has been reported, but a haplotype and a noncoding single nucleotide polymorphism (SNP) of the POLK gene were negatively associated with lung cancer risk,30 while two noncoding POLK SNPs were positively associated with breast cancer risk,31 although specific mechanisms have not been revealed. Also, seven noncoding POLK SNPs were associated with platinum chemotherapy response, toxicity, or progression-free survival in non-small cell lung cancer patients.32 With this background, we can speculate that certain human germline POLK variations might alter the functional status of pol κ and thus modify individual risk of mutation and cancer from exposure to certain carcinogens, as well as outcomes of anticancer chemotherapy.
The human POLK gene encodes the pol κ protein, composed of 870 amino acids. To date, a total of at least 310 nonsynonymous single nucleotide variations (SNVs) in the POLK gene have been described in dbSNP (http://www.ncbi.nlm.nih.gov/projects/SNP). All of these reported nonsynonymous POLK SNVs appear to be rare in human population in that their minor allele frequencies (MAFs) are < 1% or still unknown. Rare genetic variations, which constitute a majority of the genetic variations in human population,33 have received considerable attention as a potential source of hidden disease heritability, because rare variants are predicted to be more functional than common variants (i.e. polymorphisms).34 Recent reports that novel rare variants are associated with some biomedical phenotypes in the UK10K project35, 36 also underscore the significant contribution of rare variants to human diseases and traits. Especially for the rare variants that can be missed by conventional genome-wide association studies due to the limited sample size, a biochemical approach that assesses the direct effects of genetic variations on protein function is a useful alternative method, not only to identify functional genetic variations involved in a specific gene but also to understand their biochemical mechanisms and biological consequences. Previously we reported that seven rare germline nonsynonymous POLK SNVs in the polymerase core region (residues 1–526) could at least partially impair or facilitate TLS activity of pol κ opposite a bulky N2-G adduct.17 However, the functional impacts of the other numerous POLK SNVs still remain to be explored. In the present study, we investigated the biochemical impacts of nine selected germline POLK SNVs on the catalytic activity of pol κ. We selected nine POLK SNVs based not only on their predicted effects but also on their validation statuses that are supported by multiple observations and frequency data. Thus, one nonsense SNV that causes a premature stop at codon 246 and eight missense SNVs that are located in the polymerase core domains and predicted to have deleterious effects by in silico prediction tools such as SIFT and PolyPhen-237, 38 were chosen for this study. We performed a set of experiments with full-length primer extensions, steady-state kinetics, and pol-DNA binding assays, using the recombinant catalytic core (residues 1–526) proteins for wild-type pol κ and nine variants with oligonucleotide DNA templates containing an undamaged G, a bulky N2-CH2(9-anthracenyl)G (N2-AnthG) adduct, 8-oxo-7,8-dihydroguanine (8-oxoG), O6-methyl(Me)G, or an abasic site. We found that six germline POLK SNVs can interfere with catalytic function of pol κ for DNA synthesis upon normal and/or DNA lesion templates. These findings are discussed in the context of understanding the possible mechanistic and functional consequences of impaired catalytic function with pol κ variants.
EXPERIMENTAL PROCEDURES
Materials
Protease inhibitor cocktail tablets were purchased from Roche Life Science (Indianapolis, IN). FPLC columns were obtained from GE Healthcare (Uppsala, Sweden). Unlabeled dNTPs and T4 polynucleotide kinase were purchased from New England Biolabs (Ipswich, MA). [γ-32P]ATP (specific activity 3 × 103 Ci/mmol) was obtained from PerkinElmer Life Sciences (Boston, MA). Micro Bio-Spin columns were from Bio-Rad (Hercules, CA).
Oligonucleotides
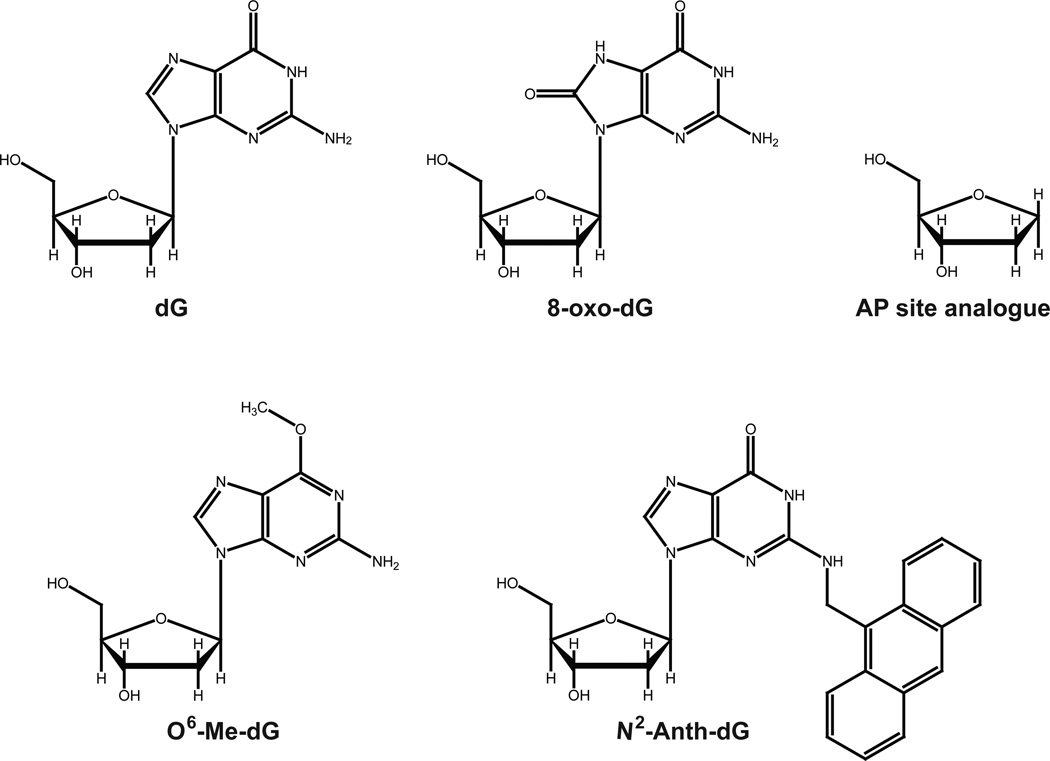
An 18-mer (5′–AGC CAG CCG CAG ACG CAG–3′), a 19-C-mer (5′–AGC CAG CCG CAG ACG CAG C–3′), an 18-FAM-mer (5′–(FAM)-AGC CAG CCG CAG ACG CAG–3′; FAM = 6-carboxyfluorescein), and a 36-mer oligonucleotides (5′–TCG GCG TCC TCX CTG CGT CTG CGG CTG GCT CGA GGC–3′; X = G, 8-oxoG, or tetrahydrofuran (abasic site analog)) containing a G, 8-oxoG, or abasic site (stable tetrahydrofuran derivative) (Figure 1) were obtained from Bioneer (Daejeon, Korea). A 36-mer (X = O6-MeG) oligonucleotides containing a O6-MeG (Figure 1) was obtained from Midland Certified Reagent Co. (Midland, TX), and a 36-N2-AnthG-mer oligonucleotide (X = N2-CH2(9-anthracenyl)G (N2-AnthG)) containing a N2-AnthG (Figure 1) was prepared as previously described.39
Figure 1. DNA adducts used in this work.
Structures of N2-CH2(9-anthracenyl)-dG (N2-Anth-dG), 8-oxo-7,8-dihydro-dG (8-oxo-dG), O6-methyl(Me)-dG, tetrahydrofuran (an abasic site analog), and normal dG are shown.
Selection of Human POLK Gene Variants to Study
We searched for germline human POLK gene variations that are highly likely to damage protein function but not yet characterized in detail from the current public databases. We searched for the natural germline POLK gene variants that are expected to be deleterious on enzymatic function of pol κ on the basis of three criteria: i) non-synonymous coding SNVs (which cause either a nonsense or a missense change), ii) SNVs located in the polymerase core domain (residues 1 to 526), and iii) SNVs predicted by SIFT and/or Polyphen-2 programs to have damaging effects on protein function,37, 38 from the public databases dbSNP (http://www.ncbi.nlm.nih.gov/projects/SNP/) and 1000 Genomes (http://browser.1000genomes.org). Thus, we selected nine SNVs that include one nonsense variation and eight missense variations and have rare minor allele frequencies (MAFs < 1%). Hence, the nonsense R246X SNV (MAF 0.02%), which is also listed as a somatic mutation found in a gastric cancer sample in the COSMIC database (http://www.sanger.ac.uk/genetics/CGP/cosmic/), was included for this study. Current information for the 9 POLK gene variations investigated in this study is summarized in Table 1, based on publically available variant databases such as dbSNP and 1000 Genomes.
Table 1.
Human POLK Gene Variations Studied
| rs IDa | nucleotide change |
amino acid change |
protein domain |
minor allele frequencyb |
predictionc | |
|---|---|---|---|---|---|---|
| SIFT | PolyPhen-2 | |||||
| rs148960463 | c.85G>A | E29K | N-clasp | 0.0006 | deleterious | possibly damaging |
| rs182817281 | c.131C>T | T44M | N-clasp | 0.0002 | deleterious | possibly damaging |
| rs150515841 | c.575T>G | F192C | Palm | 0.0001d | deleterious | probably damaging |
| rs201034973 | c.736C>T | R246X | Palm | 0.0002 | n/ae | n/a |
| rs142203892 | c.874G>A | E292K | Palm | 0.0004d | deleterious | probably damaging |
| rs151251843 | c.893G>A | R298H | Palm | 0.0004 | deleterious | probably damaging |
| rs149894654 | c.1412C>T | A471V | PAD | 0.0004 | deleterious | probably damaging |
| rs186798689 | c.1417A>G | T473A | PAD | 0.0004 | deleterious | probably damaging |
| rs142724854 | c.1534C>T | R512W | PAD | 0.0001d | deleterious | probably damaging |
Reference SNP identification number provided by dbSNP.
From 1000 Genomes project.
Possible functional effects of missense variations are predicted in silico using SIFT and PolyPhen-2 (HumVar model).
From the NHLBI ESP6500 resource described in 1000 Genomes.
Not applicable.
Construction of Expression Vectors for Catalytic Core Proteins of Nine Pol κ Variants
Nine different mutations were individually generated in the pol κ core expression vector, pBAD-wtPOLK1–52617 by PCR-based mutagenesis with nine pairs of mutagenic primers. The oligonucleotide primers for introducing the point mutation were 5'-GGA CTT AAT GAT AAT AAA GCA GGA ATG AAA GGA TTA GAT AAA GAG AAA ATT AAC A-3' for E29K, 5'-ACA AAA TTA TAA TGG AAG CCA TGA AGG GGT CCA GAT TTT ATG G-3' for T44M, 5'-GAA ATA CTT GCT GAT TAT GAT CCC AAT TGT ATG GCC ATG AGT CTT-3' for F192C, 5'-AAA CTG AGT GAG CAT GAA TGA TCC ATC TCT CCA CTA C-3' for R246X, 5'-TTT TGG AAC ATC AGC CCA GAA AGT GGT AAA GGA AAT TCG-3' for E292K, 5'-CAG GAA GTG GTA AAG GAA ATT CAT TTC AGA ATT GAG CAG AAA ACA-3' for R298H, 5'-TGA ATT TTG AAG TAA AAA CTC GTG TAT CTA CAG TTT CAT CTG TTG TTT C-3' for A471V, 5'-TGA AGT AAA AAC TCG TGC ATC TGC AGT TTC ATC TGT TGT TTC TAC-3' for T473A, 5'-GAG ATT AAG GCT TAT GGG TGT TTG GAT ATC TAG TTT TCC C-3' for R512W and its corresponding antiparallel primer for each mutation. All nine mutations were confirmed by nucleotide sequence analysis of the constructed expression vectors.
Expression and Purification of Recombinant Pol κ Proteins
The wild-type and nine different variant forms of recombinant pol κ core proteins were expressed in Escherichia coli strain TOP10 cells utilizing each of the constructed pBAD-POLK1–526 expression vectors, with arabinose induction, and the recombinant proteins were purified with a HisTrap HP column (GE Healthcare) and a Mono S column (GE Healthcare) by an ÄKTA FPLC system (GE Healthcare, Upsala, Sweden) as described previously.17 The Mono Q column (GE Healthcare) was additionally utilized for purifying the 246X variant proteins to near homogeneity. The purified proteins were stored at −80 °C in 50 mM HEPES-NaOH (pH 7.5) buffer containing 100 mM NaCl, 1 mM dithiothreitol, and 50% glycerol (v/v) after dialysis. Protein concentrations were estimated using a Bradford protein assay, and the purity of the proteins was assessed by SDS-polyacrylamide gel electrophoresis and Coomassie Brilliant Blue staining (Figure S1, Supporting Information).
DNA Polymerase Primer Extension Assays and Steady-State Kinetic Analysis
18-mer primers were 5′ end-labeled using T4 polynucleotide kinase with [γ-32P]ATP and annealed with 36-mer templates in 50 mM HEPES-NaOH (pH 7.5) buffer to make five kinds of primer-template DNA substrates: a normal 18-mer/36-G-mer and the DNA lesion-containing 18-mer/36-X-mers (X = N2-AnthG, 8-oxoG, O6-G, or abasic site). Standard DNA polymerase reactions of 8 µL were conducted in 50 mM HEPES-NaOH (pH 7.5) buffer containing 50 mM NaCl, 5 mM dithiothreitol, 100 µg mL−1 BSA (w/v), and 10% glycerol (v/v) with 100 nM DNA substrates (i.e. 5′ end-labeled 18-mer primers annealed to 36-mer templates) at 37 °C. For the next-base extension assays, 5′ end-labeled 19-C-mer primers annealed to 36-mer templates were utilized as DNA substrates. Reactions were initiated by addition of dNTPs and MgCl2 (5 mM final concentration) to preincubated pol/DNA mixtures and quenched by addition of 48 µL of a 95% formamide (v/v)/20 mM EDTA solution. For steady-state kinetic analysis, the primer-template was extended in the presence of 0.1–10 nM pol κ with increasing concentrations of individual dNTPs for 5 or 10 min, where the maximum amount of extension products was ≤20% of the total DNA substrate. Reaction products were separated by electrophoresis (16% polyacrylamide (w/v)/8 M urea gel) and visualized and quantified using a Personal Molecular Imager instrument and Quantity One software (Bio-Rad). The rate of product formation was plotted as a function of dNTP concentration and fit to the Michaelis-Menten equation using the Prism software (GraphPad, San Diego, CA) to estimate the steady-state kinetic parameters kcat and Km.
Fluorescence Polarization Assays
Two different DNA substrates containing a G or N2-AnthG were prepared for DNA binding assays. The (5´ FAM) 18-mer primer was annealed with the 36-mer template containing G or N2-AnthG for normal and lesion-containing DNA substrates. DNA substrates (2 nM) were incubated with varying concentrations of pol κ, and fluorescence polarization was measured with a Biotek Synergy NEO plate reader (Winooski, VT) using 485 and 528 nm excitation and emission filters, respectively. The polymerase-DNA binding reaction was done in the presence of 50 mM HEPES-NaOH buffer (pH 7.5) containing 10 mM potassium acetate, 10 mM KCl, 5 mM MgCl2, 0.1 mM EDTA, 2 mM β-mercaptoethanol, and 0.1 mg mL−1 BSA, as modified from a previous study.17 The fluorescence polarization data were plotted against the enzyme concentration and fit to a quadratic equation to estimate Kd,DNA using the equation: P = P0 + (Pmax −P0)((Dt + Et + Kd,DNA) − ((Dt + Et + Kd,DNA)2 – (4DtEt))1/2)/(2Dt), where P is the observed change in fluorescence polarization, P0 is the initial polarization (DNA alone), Pmax is the maximum polarization, Dt is the total DNA concentration, Et is the total enzyme concentration, and Kd,DNA is the equilibrium dissociation constant for enzyme binding to DNA.
RESULTS
Overall Work Scheme
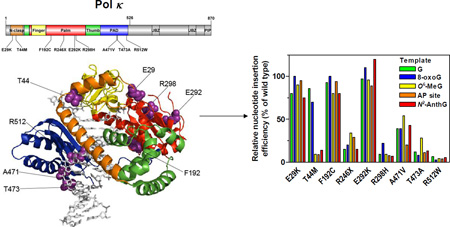
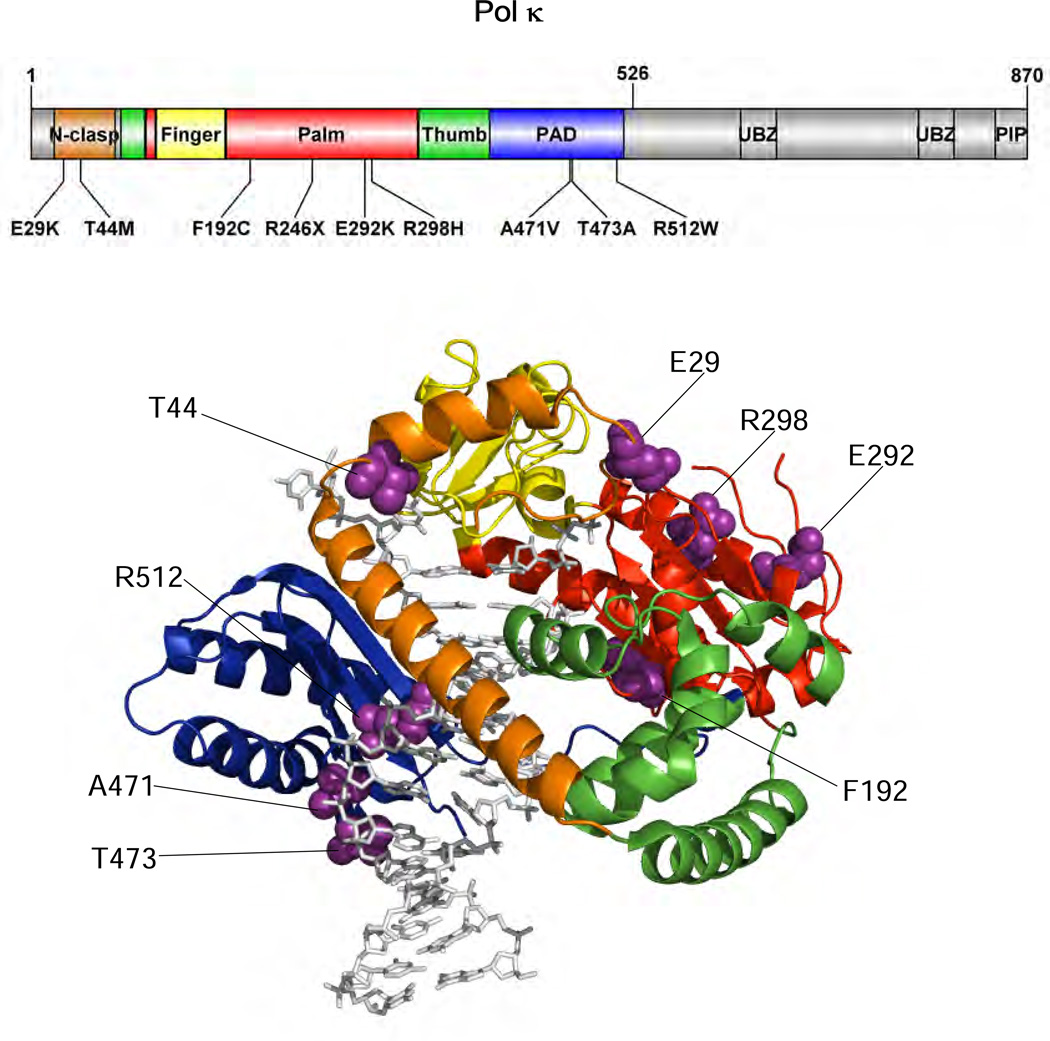
The main purpose of this study was to characterize the enzymatic properties of nine pol κ variants, which were chosen from the human germline genetic variants newly listed by the 1000 Genomes Project,33 and thus to reveal new potentially functional human germline POLK variations that impair the catalytic activity of pol κ. To achieve this, we paid particular attention to a germline POLK variation list that have been newly updated in the 1000 Genomes database since our previous work17. Among them, nine POLK SNVs were selected for functional variation candidates in this study. These candidates met at least four properties: non-synonymous coding, positioning at the catalytic core region (amino acids 1–526) of pol κ (Figure 2), validated status with multiple observations and frequency data, and putative functionality. Thus, the eight missense variations that are predicted to be damaging on protein function by prediction tools, PolyPhen-2 and SIFT,37, 38 as well as a nonsense R246X variation that causes premature protein termination, were included in this study. It is notable that the R246X variation has also been observed in a human gastric cancer sample (http://www.sanger.ac.uk/genetics/CGP/cosmic/), although its clinical relevance is unknown. The nine selected SNVs have some information available about the minor allele frequencies (MAFs) observed in human populations (Table 1), but no homozygotes have been reported for these variants yet in the current public databases. Although all of the studied POLK variations are very rare (MAF < 0.1%), they could exert influences on a small subset of people carrying these variants if functional. We performed a set of biochemical experiments, including “standing-start” primer extensions, steady-state kinetics of nucleotide incorporation opposite normal and adducted DNA templates, and DNA binding assays, using the recombinant pol κ core enzymes and DNA oligonucleotides containing a normal G, a cognate bulky N2-guanyl adduct (N2-AnthG), or each of three other common DNA lesions (8-oxoG, O6-MeG, or abasic site) at a defined site.
Figure 2. Locations of POLK genetic variations studied.
Structure of human pol κ (21–517) (PDB code, 2OH2) bound to primer/template DNA and incoming nucleotide is shown using PyMOL (http://www.pymol.org). Pol κ (21–517) is shown in cartoon ribbons, and the primer/template DNA and dNTP are shown in gray sticks. The N-clasp, finger, palm, thumb, and PAD domains are colored orange, yellow, red, green, and blue, respectively. The amino acid residues (shown in purple spheres) of genetic pol κ variants are indicated. The structural domains of pol κ are shown in the upper schematic diagram using DOG (version 2.0),60 where positions of amino acids related to nine studied variations are indicated.
Standing-Start Primer Extension Opposite G and N2-AnthG by Wild-Type Pol κ and Variants
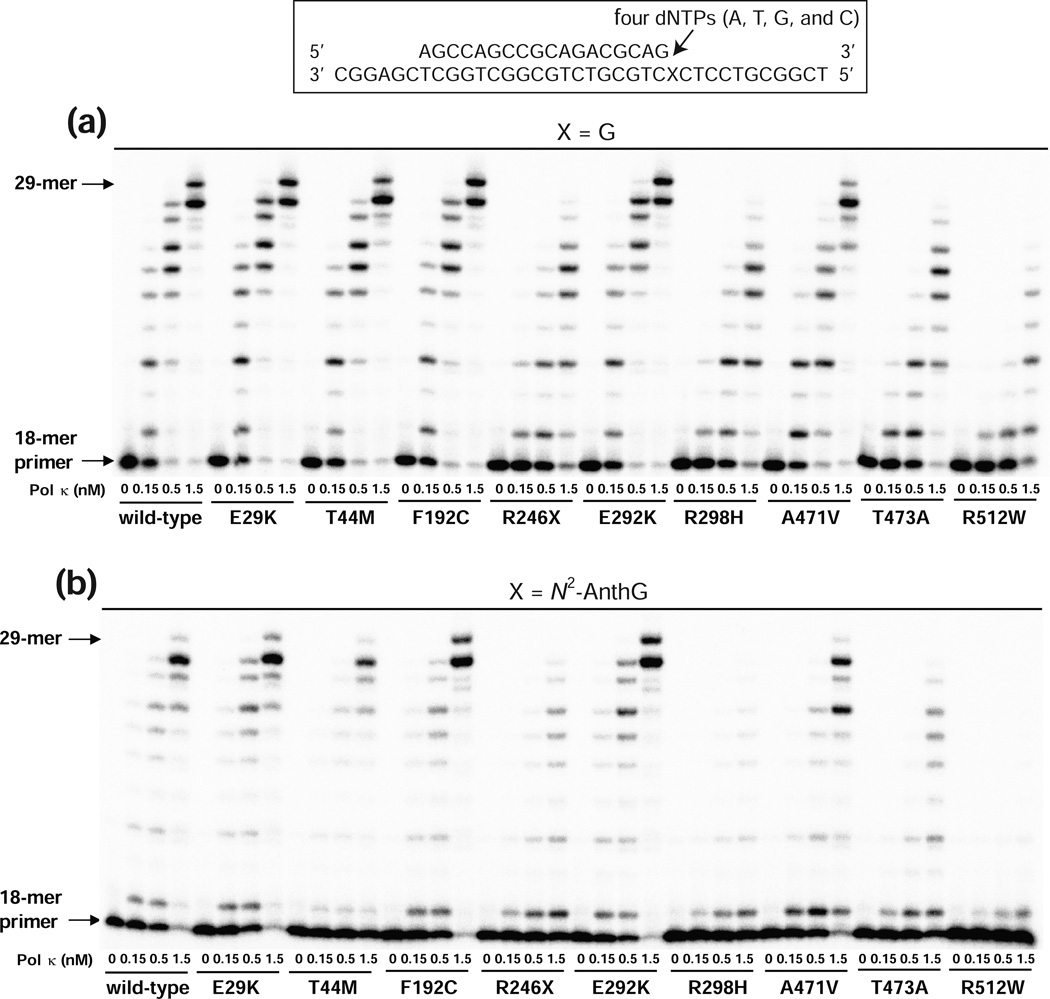
Standing-start primer extension assays were performed in order to initially observe the qualitative alterations in normal and translesion DNA synthesis activities by nine pol κ variants compared to those of the wild-type. We extended the 18-mer primers annealed to 36-mer templates containing unmodified G or N2-AnthG (Figure 1), which places the 3′-end of primers just before a template G or N2-AnthG, in the presence of all four dNTPs using the wild-type and variant pol κ core proteins (Figure 3). The 18-mer/36-mer primer-template DNAs were used as DNA substrates not only for this polymerase assay but also for DNA binding assays (see below). The N2-AnthG was utilized as a model bulky N2-G DNA adduct, because it is one of the favored TLS substrates for human pol κ.8 For polymerization at an unmodified G in the template, the wild-type pol κ readily extended the primers past G and generated extension products up to about 29-mers, and the extent of bypass synthesis was proportional to the pol concentration. The E29K, T44M, F192C, and E292K variants displayed extension activity similar to that of the wild-type enzyme. In contrast, the R246X, R298H, T473A, and R512W variants showed much lower activity than wild-type pol κ, indicating the severe attenuation of polymerase activity due to these nonsense or missense changes. Interestingly the R246X variant still retained some polymerase activity, unlike the R219X variant (which is devoid of activity),17 indicating an indispensable role of residues 219–245 of the palm domain for minimal polymerase activity. For TLS at N2-AnthG, wild-type pol κ and nine variants displayed similar patterns of primer extension as those observed with an unmodified G with the exception of the T44M variant, which showed a considerable diminution in extension activity only opposite N2-AnthG but not opposite G. The A471V variant displayed slight reductions in the extent of extension products across G and N2-AnthG compared to those with wild-type pol κ. Among the nine variants, the R512W form showed the lowest activity at both templates, generating only a trace of one- to three-base extension products across N2-AnthG even with a 1.5 nM enzyme concentration, indicating the marked diminution of its lesion bypass activity due to this substitution.
Figure 3. Extension of a 32P-labeled primer across G or N2-AnthG by human wild-type and variant pol κ (1–526) enzymes.
Two different 36-mer templates containing an unmodified G or N2-AnthG was annealed with primer (18-mer), to make primer-template DNA substrates positioning the 3′-end of primers just before a G or N2-AnthG in the template strand. Reactions were done for 15 min with increasing concentrations of pol κ (1–526) (0 – 1.5 nM) and DNA substrate (100 nM primer/template) as indicated. 32P-labeled primer was extended in the presence of all four dNTPs (25 µM each). The reaction products were analyzed by denaturing gel electrophoresis and phosphorimaging. (a) Extension across G. (b) Extension across N2-AnthG.
We also qualitatively assessed he subsequent extension activities from the N2-AnthG:C pair by nine pol κ variants compared to the wild-type, by assaying extensions of a 19-C-mer primer annealed to 36- N2-AnthG-mer template with all four dNTPs (Figure S2, Supporting Information). The patterns of primer extensions beyond the N2-AnthG:C pair by wild-type pol κ and variants were analogous to those of standing start primer extensions opposite N2-AnthG, except for the T44M variant which showed a subsequent extension activity comparable to that of the wild-type enzyme. These results indicate that none of these variations affect pol κ activity exclusively at the subsequent extension step beyond N2-AnthG, while the T44M substitution diminishes pol κ activity selectively at the insertion step opposite N2-AnthG but not the subsequent extension step.
Steady-State Kinetics of dNTP Incorporation Opposite G and N2-AnthG by Pol κ Wild-Type and Variants
To quantitatively evaluate the changes in both the efficiency and fidelity of nine pol κ variants for nucleotide incorporation opposite a unmodified G and a bulky N2-AnthG adduct, we analyzed steady-state kinetics for incorporation of single nucleotides into 18-mer/36-mer duplexes opposite each of these templates by pol κ variants in comparison to the wild-type enzyme (Tables 2 and 3). We determined the specificity constant (kcat/Km) and misinsertion frequency (f = (kcat/Km)incorrect dNTP/(kcat/Km)correct dNTP) as semi-quantitative indexes for the catalytic efficiency and fidelity of a distributive pol κ enzyme, as utilized in previous works.17, 40 For correct dCTP incorporation opposite an unmodified G, the R246X, R298H, T473A, and R512W variants showed 7- to 16-fold reductions in kcat/Km for dCTP insertion opposite G compared to that of the wild-type, while the E29K, T44M, F192C, and E292K variants showed kcat/Km values similar to that of the wild-type (Table 2). The A471V variant displayed a 3-fold decrease in kcat/Km for dCTP insertion opposite G compared to that of the wild-type. A similar trend of results was observed for dCTP incorporation opposite an N2-AnthG adduct, except for that with the T44M variant (Table 3). Interestingly, the T44M variant, which had a wild-type-like (86%) activity opposite normal G, displayed a 7-fold decrease in kcat/Km for dCTP insertion opposite N2-AnthG. The R246X, R298H, T473A, and R512W variants displayed 7- to 18-fold decreases in kcat/Km for dCTP insertion opposite N2-AnthG compared to that of wild-type pol κ, while the E29K, F192C, and E292K variants showed kcat/Km values similar to that of the wild-type (Table 3). The A471V variant displayed a 2-fold decrease in kcat/Km for dCTP insertion opposite N2-AnthG compared to that of the wild-type enzyme. These steady-state kinetic results are in good agreement with the patterns of primer extension by the variants in Figure 3. The misinsertion frequencies of the variants opposite G and N2-AnthG did not differ much from those of the wild-type but were slightly or moderately altered in some cases. Noticeably, the T44M variant showed 4- to 5-fold decreases in misinsertion frequencies of non-dCTP nucleotides opposite G but showed 4- to 6-fold increases in those of dATP and dTTP opposite N2-AnthG, while the R512W variant displayed 3- and 6-fold increases, respectively, in misinsertion frequencies of dGTP and dTTP opposite G.
Table 2.
Steady-State Kinetic Parameters for dNTP Incorporation Opposite G by Human Wild-Type Pol κ (1–526) and Variants
| pol κ(1–526) | dNTP | Km (µM) | kcat (s−1) | kcat/Km (s−1 µM−1) | finsa | relative efficiencyb |
|---|---|---|---|---|---|---|
| wild-type | A | 220 ± 50 | 0.047 ± 0.003 | 2.2 × 10−4 | 0.0024 | |
| T | 510 ± 100 | 0.17 ± 0.01 | 3.3 × 10−4 | 0.0037 | ||
| G | 71 ± 10 | 0.040 ± 0.002 | 5.6 × 10−4 | 0.0062 | ||
| C | 12 ± 1 | 1.1 ± 0.03 | 9.0 × 10−2 | 1 | 1 | |
| E29K | A | 250 ± 80 | 0.067 ± 0.007 | 2.7 × 10−4 | 0.0037 | |
| T | 590 ± 60 | 0.20 ± 0.01 | 3.4 × 10−4 | 0.0048 | ||
| G | 150 ± 30 | 0.063 ± 0.003 | 4.2 × 10−4 | 0.0058 | ||
| C | 14 ± 3 | 1.0 ± 0.1 | 7.2 × 10−2 | 1 | 0.80 | |
| T44M | A | 220 ± 3 | 0.010 ± 0.001 | 4.8 × 10−5 | 0.00062 | |
| T | 330 ± 70 | 0.026 ± 0.002 | 7.8 × 10−5 | 0.0010 | ||
| G | 55 ± 6 | 0.0053 ± 0.0001 | 1.0 × 10−4 | 0.0013 | ||
| C | 12 ± 1 | 0.93 ± 0.03 | 7.7 × 10−2 | 1 | 0.86 | |
| F192C | A | 180 ± 30 | 0.067 ± 0.003 | 3.7 × 10−4 | 0.0044 | |
| T | 470 ± 60 | 0.34 ± 0.01 | 7.2 × 10−4 | 0.0085 | ||
| G | 110 ± 10 | 0.089 ± 0.003 | 8.1 × 10−4 | 0.0096 | ||
| C | 13 ± 3 | 1.1 ± 0.1 | 8.4 × 10−2 | 1 | 0.93 | |
| R246X | A | 150 ± 2 | 0.015 ± 0.001 | 1.0 × 10−4 | 0.0075 | |
| T | 490 ± 10 | 0.052 ± 0.002 | 1.1 × 10−4 | 0.0077 | ||
| G | 92 ± 27 | 0.011 ± 0.001 | 1.2 × 10−4 | 0.0088 | ||
| C | 16 ± 3 | 0.22 ± 0.01 | 1.4 × 10−2 | 1 | 0.15 | |
| E292K | A | 150 ± 30 | 0.060 ± 0.003 | 4.0 × 10−4 | 0.0046 | |
| T | 540 ± 50 | 0.22 ± 0.01 | 4.0 × 10−4 | 0.0046 | ||
| G | 79 ± 8 | 0.050 ± 0.001 | 6.4 × 10−4 | 0.0073 | ||
| C | 13 ± 1 | 1.1 ± 0.03 | 8.7 × 10−2 | 1 | 0.97 | |
| R298H | A | 530 ± 170 | 0.026 ± 0.002 | 4.9 × 10−5 | 0.0060 | |
| T | 1400 ± 500 | 0.032 ± 0.004 | 2.3 × 10−5 | 0.0028 | ||
| G | 710 ± 190 | 0.018 ± 0.002 | 2.6 × 10−5 | 0.0031 | ||
| C | 75 ± 10 | 0.62 ± 0.02 | 8.2 × 10−3 | 1 | 0.091 | |
| A471V | A | 280 ± 70 | 0.033 ± 0.003 | 1.2 × 10−4 | 0.0034 | |
| T | 520 ± 120 | 0.19 ± 0.01 | 3.6 × 10−4 | 0.010 | ||
| G | 110 ± 20 | 0.036 ± 0.002 | 3.2 × 10−4 | 0.0091 | ||
| C | 26 ± 5 | 0.90 ± 0.06 | 3.5 × 10−2 | 1 | 0.39 | |
| T473A | A | 350 ± 90 | 0.021 ± 0.002 | 5.9× 10−5 | 0.0056 | |
| T | 920 ± 130 | 0.040 ± 0.002 | 4.4× 10−5 | 0.0041 | ||
| G | 230 ± 70 | 0.021 ± 0.002 | 9.0× 10−5 | 0.0085 | ||
| C | 48 ± 13 | 0.51 ± 0.04 | 1.1× 10−2 | 1 | 0.12 | |
| R512W | A | 550 ± 60 | 0.0086 ± 0.0003 | 1.6 × 10−5 | 0.0027 | |
| T | 410 ± 90 | 0.051 ± 0.003 | 1.3 × 10−4 | 0.022 | ||
| G | 300 ± 30 | 0.030 ± 0.001 | 1.0 × 10−4 | 0.017 | ||
| C | 80 ± 24 | 0.46 ± 0.04 | 5.8× 10−3 | 1 | 0.064 | |
Misinsertion frequency, calculated by dividing kcat/Kmfor each dNTP incorporation by the kcat/Km for dCTP incorporation opposite G.
Relative efficiency, calculated by dividing kcat/Km of each pol κ(1–526) for dCTP incorporation opposite G by kcat/Km of wild type pol κ(1–526) for dCTP incorporation opposite undamaged G.
Table 3.
Steady-State Kinetic Parameters for dNTP Incorporation Opposite N2-AnthG by Human Wild-Type Pol κ (1–526) and Variants
| pol κ(1–526) | dNTP | Km (µM) | kcat (s−1) | kcat/Km (s−1 µM−1) | finsa | relative efficiencyb |
|---|---|---|---|---|---|---|
| wild-type | A | 110 ± 10 | 0.0028 ± 0.0001 | 2.5 × 10−5 | 0.0012 | |
| T | 340 ± 40 | 0.0087 ± 0.0003 | 2.6 × 10−5 | 0.0013 | ||
| G | 85 ± 11 | 0.0024 ± 0.0001 | 2.8 × 10−5 | 0.0014 | ||
| C | 53 ± 7 | 1.0 ± 0.04 | 2.0 × 10−2 | 1 | 1 | |
| E29K | A | 200 ± 30 | 0.0013 ± 0.0001 | 6.7 × 10−6 | 0.00046 | |
| T | 350 ± 50 | 0.0055 ± 0.0002 | 1.6 × 10−5 | 0.0011 | ||
| G | 150 ± 10 | 0.0014 ± 0.00003 | 9.3 × 10−6 | 0.00064 | ||
| C | 87 ± 13 | 1.3 ± 0.3 | 1.5 × 10−2 | 1 | 0.75 | |
| T44M | A | 330 ± 80 | 0.0067 ± 0.0005 | 2.0 × 10−5 | 0.0071 | |
| T | 380 ± 80 | 0.0072 ± 0.0006 | 1.9 × 10−5 | 0.0068 | ||
| G | 100 ± 20 | 0.00064 ± 0.00006 | 6.4 × 10−6 | 0.0023 | ||
| C | 200 ± 20 | 0.55 ± 0.02 | 2.8 × 10−3 | 1 | 0.14 | |
| F192C | A | 220 ± 30 | 0.0028 ± 0.0001 | 1.3 × 10−5 | 0.00079 | |
| T | 540 ± 60 | 0.010 ± 0.0004 | 1.9 × 10−5 | 0.0012 | ||
| G | 46 ± 3 | 0.0019 ± 0.00004 | 4.1 × 10−5 | 0.0026 | ||
| C | 58 ± 16 | 0.95 ± 0.08 | 1.6 × 10−2 | 1 | 0.80 | |
| R246X | A | 130 ± 20 | 0.00074 ± 0.00003 | 5.7 × 10−6 | 0.0020 | |
| T | 670 ± 190 | 0.0023 ± 0.0002 | 3.4 × 10−6 | 0.0012 | ||
| G | 93 ± 22 | 0.00052 ± 0.00004 | 5.6 × 10−6 | 0.0020 | ||
| C | 90 ± 32 | 0.26 ± 0.03 | 2.9 × 10−3 | 1 | 0.15 | |
| E292K | A | 220 ± 40 | 0.0022 ± 0.0002 | 1.0 × 10−5 | 0.00043 | |
| T | 330 ± 80 | 0.0086 ± 0.0006 | 2.6 × 10−5 | 0.0011 | ||
| G | 76 ± 13 | 0.0015 ± 0.0001 | 2.0 × 10−5 | 0.00087 | ||
| C | 44 ± 8 | 1.0 ± 0.1 | 2.3 × 10−2 | 1 | 1.2 | |
| R298H | A | 630 ± 130 | 0.00069 ± 0.00005 | 1.1 × 10−6 | 0.00079 | |
| T | 780 ± 170 | 0.0011 ± 0.0001 | 1.4 × 10−6 | 0.0010 | ||
| G | 210 ± 30 | 0.00065 ± 0.00002 | 3.1 × 10−6 | 0.0022 | ||
| C | 400 ± 60 | 0.55 ± 0.03 | 1.4 × 10−3 | 1 | 0.070 | |
| A471V | A | 220 ± 40 | 0.0019 ± 0.0001 | 8.5 × 10−6 | 0.0010 | |
| T | 650 ± 120 | 0.0058 ± 0.0004 | 9.0 × 10−6 | 0.0011 | ||
| G | 210 ± 50 | 0.0016 ± 0.0001 | 7.8 × 10−6 | 0.00092 | ||
| C | 130 ± 20 | 1.1 ± 0.1 | 8.5 × 10−3 | 1 | 0.43 | |
| T473A | A | 410 ± 70 | 0.0014 ± 0.0001 | 3.5 × 10−6 | 0.0014 | |
| T | 510 ± 50 | 0.0015 ± 0.0001 | 3.0 × 10−6 | 0.0012 | ||
| G | 200 ± 30 | 0.00072 ± 0.00003 | 3.6 × 10−6 | 0.0014 | ||
| C | 210 ± 30 | 0.54 ± 0.03 | 2.6 × 10−3 | 1 | 0.13 | |
| R512W | A | 290 ± 60 | 0.00045 ± 0.00003 | 1.5 × 10−6 | 0.0014 | |
| T | 620 ± 80 | 0.0017 ± 0.0001 | 2.8 × 10−6 | 0.0026 | ||
| G | 220 ± 40 | 0.00023 ± 0.00001 | 1.1 × 10−6 | 0.0010 | ||
| C | 390 ± 80 | 0.42 ± 0.03 | 1.1 × 10−3 | 1 | 0.055 | |
Misinsertion frequency, calculated by dividing kcat/Kmfor each dNTP incorporation by the kcat/Km for dCTP incorporation opposite N2-AnthG.
Relative efficiency, calculated by dividing kcat/Km of each pol κ(1–526) for dCTP incorporation opposite N2-AnthG by kcat/Km of wild type pol κ(1–526) for dCTP incorporation opposite N2-AnthG.
Steady-State Kinetics of Next-Base Extension from an N2-AnthG:C Pair by Wild-Type Pol κ and Variants
To quantitatively estimate the efficiencies of the nine pol κ variants for the subsequent extension step after dCTP insertion opposite N2-AnthG, steady-state kinetic parameters were determined for insertion of the next correct nucleotide dGTP opposite template C following an adducted N2-AnthG:C base pair into a 19-C-mer primer annealed to a 36-N2-AnthG-mer by wild-type pol κ and the variants, compared to next-base extension from the normal G:C pair of a 36-G-mer/19-C-mer substrate (Table 4). The alterations in next-base extension efficiencies (i.e., kcat/Km) of each pol κ variants from the adducted N2-AnthG:C pair were similar to those from the normal G:C pair, with slight variations in some cases. For the next-base extension from the G (or N2-AnthG):C pair, the R246X, R298H, A471V, T473A, and R512W variants displayed 4- to 190-fold decreases in kcat/Km compared to that of the wild-type, while the E29K, T44M, F192C, and E292K variants showed kcat/Km values similar to that of wild-type pol κ. Interestingly the R512W variation caused a 3-fold greater decrease in kcat/Km of pol κ for next-base extension from an N2-AnthG:C pair than that from a G:C pair, indicating a more severe impairment at the subsequent extension step beyond the N2-AnthG-adducted base pair than that beyond normal base pair by this variation. However, none of the nine genetic variations caused an exclusive alteration in next-base extension efficiency only from N2-AnthG:C pair (but not from G:C pair).
Table 4.
Steady-State Kinetic Parameters for Next Base Extension from G (or N2-AnthG): C Pair Template:Primer Termini by Human Wild-Type Pol κ (1–526) and Variants
| base-pair at 3′ primer termini (template:primer) |
pol κ(1–526) | extension with dGTP (the correct nucleotide opposite next template C) | |||
|---|---|---|---|---|---|
| Km (µM) | kcat (s−1) |
kcat/Km (s−1 µM−1) |
relative extension efficiencya |
||
| G:C | wild-type | 0.66 ± 0.20 | 0.21 ± 0.02 | 3.2 × 10−1 | 1 |
| E29K | 1.4 ± 0.5 | 0.39 ± 0.04 | 2.8 × 10−1 | 0.88 | |
| T44M | 2.1 ± 0.4 | 0.70 ± 0.06 | 3.3 × 10−1 | 1.0 | |
| F192C | 1.1 ± 0.2 | 0.34 ± 0.02 | 3.1 × 10−1 | 0.97 | |
| R246X | 1.3 ± 0.2 | 0.083 ± 0.003 | 6.4 × 10−2 | 0.20 | |
| E292K | 1.1 ± 0.3 | 0.33 ± 0.03 | 3.0 × 10−1 | 0.94 | |
| R298H | 3.6 ± 0.9 | 0.24 ± 0.02 | 6.7 × 10−2 | 0.21 | |
| A471V | 3.8 ± 0.6 | 0.32 ± 0.02 | 8.4 × 10−2 | 0.26 | |
| T473A | 6.4 ± 2.0 | 0.21 ± 0.01 | 3.3 × 10−2 | 0.10 | |
| R512W | 29 ± 10 | 0.15 ± 0.02 | 5.2 × 10−3 | 0.016 | |
| N2-AnthG:C | wild-type | 3.3 ± 0.5 | 0.27 ± 0.01 | 8.2 × 10−2 | 1 |
| E29K | 3.5 ± 0.7 | 0.28 ± 0.02 | 8.0 × 10−2 | 0.98 | |
| T44M | 2.3 ± 0.6 | 0.19 ± 0.01 | 8.3 × 10−2 | 1.0 | |
| F192C | 3.3 ± 0.5 | 0.25 ± 0.01 | 7.6 × 10−2 | 0.93 | |
| R246X | 3.4 ± 0.8 | 0.031 ± 0.002 | 9.1 × 10−3 | 0.11 | |
| E292K | 2.1 ± 0.6 | 0.18 ± 0.02 | 8.6 × 10−2 | 1.0 | |
| R298H | 19 ± 2 | 0.21 ± 0.01 | 1.1 × 10−2 | 0.13 | |
| A471V | 7.4 ± 2.0 | 0.17 ± 0.01 | 2.3 × 10−2 | 0.28 | |
| T473A | 23 ± 4 | 0.14 ± 0.01 | 6.1 × 10−3 | 0.074 | |
| R512W | 200 ± 70 | 0.088 ± 0.010 | 4.4 × 10−4 | 0.0054 | |
Relative extension efficiency, calculated by dividing kcat/Km of each pol κ(1–526) for each dGTP incorporation opposite the next template C from the G (or N2-AnthG):C pair by kcat/Km of wild type pol κ(1–526) for dGTP incorporation opposite the next template C from the G (or N2-AnthG):C pair.
Binding for DNA Substrates Containing G or N2-AnthG by the Wild-Type Pol κ and Variants
We evaluated the effects of nine genetic variations on DNA substrate binding of pol κ using fluorescence polarization assays, determining the equilibrium dissociation constants (Kd,DNA) of wild-type pol κ and nine variants by fitting the fluorescence polarization values of fluorescein-labeled primer-template DNA (18-FAM-mer/36-mer) as a function of pol κ concentration to a quadratic equation (Table 5). The Kd,DNA of each pol κ enzyme for unmodified DNA was similar to that of N2-AnthG-adducted DNA, indicating that an N2-AnthG located at a primer-template junction does not substantially affect the DNA-binding affinity of wild-type pol κ and the variants. This is in a good agreement with the previous result on pol κ with DNA substrates that places the 3′-end of the primer right before a G or N2-BPDE-G.41 We also note a previous interesting report that pol κ bound 3-fold more strongly to DNAs that contains the primers extended by one to three nucleotides past N2-BPDE-G.41 Such a supportive role of pol κ in the subsequent extension steps past the bulky N2-G adduct should be considered in the future work, especially on pol κ variants impaired selectively in the subsequent extension steps past the lesion, although not applicable in this work. We found that the Kd,DNA values of the R246X and R512W variants were 2.4- to 2.9-fold higher for both N2-AnthG-adducted and unmodified DNA substrates than for wild-type pol κ, indicating slight decreases in DNA-binding affinity of pol κ by those variations. Similarly, the R298H and T473A variants showed 1.5-fold to 2-fold increases in Kd,DNA values for both DNA substrates compared to that of the wild-type. The slight reductions in the DNA binding affinity of pol κ by those four variations might partly relate to decreases in nucleotide insertion efficiencies opposite G and N2-AnthG with those variants compared to wild-type pol κ (Tables 2 and 3).
Table 5.
Binding Affinities of Human Wild-Type Pol κ (1–526) and Variants for DNA Substrates
| pol κ(1–526) | Kd (nM) | |
|---|---|---|
| 18-FAM-mer/36-G-mer | 18-FAM-mer/36-N2-AnthG-mer | |
| wild-type | 27 ± 5 | 28 ± 4 |
| E29K | 31 ± 4 | 26 ± 6 |
| T44M | 33 ± 4 | 35 ± 6 |
| F192C | 29 ± 4 | 30 ± 5 |
| R246X | 65 ± 10 | 81 ± 17 |
| E292K | 32 ± 7 | 31 ± 6 |
| R298H | 54 ± 7 | 43 ± 10 |
| A471V | 33 ± 4 | 28 ± 4 |
| T473A | 51 ± 10 | 44 ± 8 |
| R512W | 76 ± 20 | 68 ± 15 |
Steady-State Kinetics of dNTP Incorporation Opposite 8-OxoG, O6-MeG, or an Abasic Site by Wild-Type Pol κ and Variants
To assess whether six pol κ variants that were hypoactive opposite N2-AnthG have similar alterations in TLS activity opposite other DNA lesions, we performed steady-state kinetic analysis for the incorporation of single nucleotides into 18-mer/36-mer duplexes opposite three common DNA lesions—8-oxoG, O6-MeG, and an abasic site—by pol κ variants in comparison to wild-type pol κ (Tables 6 and 7). All nine pol κ variants displayed a similar trend of results opposite three other DNA lesion templates, except for the case opposite 8-oxoG with the T44M variant (Tables 6 and 7), when compared with the results opposite N2-AnthG (Table 3). Notably, the T44M variant showed a wild-type-like (70%) activity opposite 8-oxoG, whereas displaying 11-fold decreases in kcat/Km for dCTP and dATP insertion, respectively, opposite O6-MeG and abasic lesions, implying that both O6-MeG and abasic lesions but not 8-oxoG in the nascent base pair might disturb the catalysis by the T44M variant. The R246X, R298H, A471V, T473A, and R512W variants displayed 2- to 18-fold decreases in kcat/Km for dCTP insertion opposite three DNA lesions compared to that of the that of wild-type pol κ. These steady-state kinetic results are in good agreement with the patterns of standing-start primer extension in the presence of all four dNTPs by the variants (Figure S3, Supporting Information). The misinsertion frequencies or dNTP selectivity ratios of the variants opposite 8-oxoG, O6-MeG, and an abasic site were mostly similar to those of wild-type pol κ, but the hypoactive T473A and R512W variants showed 4-fold decreases in misinsertion frequencies of dATP opposite 8-oxoG.
Table 6.
Steady-State Kinetic Parameters for dNTP Incorporation Opposite 8-oxoG and O6-MeG by human wild-type κ (1–526) and variants
| template | pol κ(1–526) | dNTP | Km (µM) | kcat (s−1) | kcat/Km (s−1 µM−1) | finsa | relative efficiencyb |
|---|---|---|---|---|---|---|---|
| 8-oxoG | wild-type | A | 6.9 ± 0.7 | 0.37 ± 0.01 | 5.4 × 10−2 | 4.2 | 1 |
| C | 42 ± 7 | 0.56 ± 0.03 | 1.3 × 10−2 | 1 | |||
| E29K | A | 6.6 ± 0.8 | 0.37 ± 0.01 | 5.6 × 10−2 | 5.1 | 1.0 | |
| C | 41 ± 5 | 0.47 ± 0.02 | 1.1 × 10−2 | 1 | |||
| T44M | A | 12 ± 3 | 0.45 ± 0.03 | 3.8 × 10−2 | 3.9 | 0.70 | |
| C | 96 ± 9 | 0.94 ± 0.03 | 9.8 × 10−3 | 1 | |||
| F192C | A | 5.8 ± 0.6 | 0.31 ± 0.01 | 5.3 × 10−2 | 4.4 | 1.0 | |
| C | 48 ± 6 | 0.57 ± 0.02 | 1.2 × 10−2 | 1 | |||
| R246X | A | 9.1 ± 0.8 | 0.10 ± 0.01 | 1.1 × 10−2 | 2.6 | 0.20 | |
| C | 65 ± 12 | 0.27 ± 0.01 | 4.2 × 10−3 | 1 | |||
| E292K | A | 4.1 ± 0.8 | 0.24 ± 0.01 | 5.9 × 10−2 | 6.4 | 1.1 | |
| C | 51 ± 8 | 0.47 ± 0.02 | 9.2 × 10−3 | 1 | |||
| R298H | A | 22 ± 3 | 0.26 ± 0.01 | 1.2 × 10−2 | 8.0 | 0.22 | |
| C | 400 ± 60 | 0.59 ± 0.03 | 1.5 × 10−3 | 1 | |||
| A471V | A | 12 ± 3 | 0.25 ± 0.02 | 2.1 × 10−2 | 4.2 | 0.39 | |
| C | 110 ± 20 | 0.55 ± 0.03 | 5.0 × 10−3 | 1 | |||
| T473A | A | 50 ± 9 | 0.22 ± 0.01 | 4.4 × 10−3 | 1.1 | 0.081 | |
| C | 150 ± 20 | 0.58 ± 0.02 | 3.9 × 10−3 | 1 | |||
| R512W | A | 120 ± 10 | 0.17 ± 0.01 | 1.4 × 10−3 | 0.93 | 0.026 | |
| C | 270 ± 60 | 0.41 ± 0.03 | 1.5 × 10−3 | 1 | |||
| O6-MeG | wild-type | T | 230 ± 40 | 0.48 ± 0.02 | 2.1 × 10−3 | 0.40 | |
| C | 110 ± 30 | 0.55 ± 0.05 | 5.0 × 10−3 | 1 | 1 | ||
| E29K | T | 350 ± 40 | 0.46 ± 0.02 | 1.3 × 10−3 | 0.30 | ||
| C | 170 ± 30 | 0.76 ± 0.04 | 4.5 × 10−3 | 1 | 0.90 | ||
| T44M | T | 180 ± 40 | 0.047 ± 0.003 | 2.6 × 10−4 | 0.54 | ||
| C | 280 ± 30 | 0.13 ± 0.01 | 4.6 × 10−4 | 1 | 0.092 | ||
| F192C | T | 210 ± 30 | 0.33 ± 0.01 | 1.6 × 10−3 | 0.39 | ||
| C | 140 ± 20 | 0.56 ± 0.02 | 4.0 × 10−3 | 1 | 0.80 | ||
| R246X | T | 210 ± 30 | 0.16 ± 0.01 | 7.6 × 10−4 | 0.43 | ||
| C | 150 ± 30 | 0.26 ± 0.01 | 1.7 × 10−3 | 1 | 0.34 | ||
| E292K | T | 150 ± 40 | 0.30 ± 0.02 | 2.0 × 10−3 | 0.37 | ||
| C | 130 ± 20 | 0.62 ± 0.02 | 4.8 × 10−3 | 1 | 0.96 | ||
| R298H | T | 560 ± 110 | 0.13 ± 0.01 | 2.3 × 10−4 | 0.51 | ||
| C | 690 ± 60 | 0.31 ± 0.01 | 4.5 × 10−4 | 1 | 0.090 | ||
| A471V | T | 270 ± 70 | 0.35 ± 0.03 | 1.3 × 10−3 | 0.49 | ||
| C | 230 ± 40 | 0.61 ± 0.03 | 2.7 × 10−3 | 1 | 0.54 | ||
| T473A | T | 280 ± 50 | 0.18 ± 0.01 | 6.4 × 10−4 | 0.47 | ||
| C | 190 ± 60 | 0.26 ± 0.02 | 1.4 × 10−3 | 1 | 0.28 | ||
| R512W | T | 250 ± 60 | 0.033 ± 0.002 | 1.3 × 10−4 | 0.64 | ||
| C | 460 ± 80 | 0.096 ± 0.006 | 2.1 × 10−4 | 1 | 0.042 | ||
Misinsertion frequency, calculated by dividing kcat/Kmfor each dNTP incorporation by the kcat/Km for dCTP incorporation opposite template base.
Relative efficiency, calculated by dividing kcat/Km of each pol κ(1–526) for dNTP incorporation opposite template base by the highest kcat/Km of wild-type pol κ(1–526) for dNTP incorporation opposite template base.
Table 7.
Steady-State Kinetic Parameters for dNTP Incorporation Opposite Abasic Site by human wild-type κ (1–526) and variants
| pol κ(1–526) | dNTP | Km (µM) | kcat (s−1) | kcat/Km (s−1 µM−1) | dNTP Selectivity ratioa |
relative efficiencyb |
|---|---|---|---|---|---|---|
| wild-type | A | 220 ± 40 | 0.18 ± 0.01 | 8.2 × 10−4 | 1.2 | 1 |
| C | 140 ± 30 | 0.096 ± 0.005 | 6.9 × 10−4 | 1 | ||
| E29K | A | 180 ± 20 | 0.14 ± 0.01 | 7.8 × 10−4 | 1.5 | 0.95 |
| C | 240 ± 50 | 0.13 ± 0.01 | 5.4 × 10−4 | 1 | ||
| T44M | A | 240 ± 20 | 0.017 ± 0.001 | 7.1 × 10−5 | 0.40 | 0.087 |
| C | 63 ± 15 | 0.011 ± 0.001 | 1.7 × 10−4 | 1 | ||
| F192C | A | 130 ± 10 | 0.10 ± 0.01 | 7.7 × 10−4 | 1.2 | 0.94 |
| C | 210 ± 40 | 0.14 ± 0.01 | 6.7 × 10−4 | 1 | ||
| R246X | A | 140 ± 10 | 0.033 ± 0.001 | 2.4 × 10−4 | 1.4 | 0.29 |
| C | 250 ± 20 | 0.043 ± 0.001 | 1.7 × 10−4 | 1 | ||
| E292K | A | 130 ± 10 | 0.095 ± 0.002 | 7.3 × 10−4 | 1.6 | 0.89 |
| C | 280 ± 60 | 0.12 ± 0.01 | 4.3 × 10−4 | 1 | ||
| R298H | A | 400 ± 110 | 0.025 ± 0.002 | 6.3 × 10−5 | 0.81 | 0.077 |
| C | 320 ± 70 | 0.025 ± 0.002 | 7.8 × 10−5 | 1 | ||
| A471V | A | 350 ± 90 | 0.056 ± 0.005 | 1.6 × 10−4 | 1.2 | 0.20 |
| C | 350 ± 90 | 0.048 ± 0.005 | 1.4 × 10−4 | 1 | ||
| T473A | A | 300 ± 70 | 0.028 ± 0.002 | 9.3 × 10−5 | 0.80 | 0.11 |
| C | 360 ± 70 | 0.041 ± 0.002 | 1.1 × 10−4 | 1 | ||
| R512W | A | 260 ± 50 | 0.0083 ± 0.0005 | 3.2 × 10−5 | 0.70 | 0.039 |
| C | 190 ± 36 | 0.0085 ± 0.0005 | 4.5 × 10−5 | 1 |
dNTP selectivity ratio, calculated by dividing kcat/Km for each dNTP incorporation by the highest kcat/Km for dNTP incorporation opposite the abasic site.
Relative efficiency, calculated by dividing kcat/Km of each pol κ(1–526) for dATP incorporation opposite the abasic site by kcat/Km of wild-type pol κ(1–526) for dATP incorporation opposite the abasic site.
DISCUSSION
Pol κ is a unique polymerase involved in the error-free TLS of minor-groove DNA lesions as well as in the DNA repair synthesis, and thus it has a role in protecting the cellular genome from genotoxic stresses. Our previous report revealed that certain putatively damaging human germline variations of the POLK gene severely diminish the catalytic activity of pol κ, underlining a need for the functional characterization of other new putatively damaging variants. To date, a large number of human POLK germline variations have been newly catalogued from the 1000 Genomes and NHLBI Exome Sequencing Projects,33, 42 but their functional effects are largely unknown. In this study, we report that six germline nonsynonymous variations of the human POLK gene noticeably impair the in vitro catalytic activity of pol κ. We selected nine nonsynonymous POLK variants, i.e., eight missense and one nonsense variants, on the bases of their locations, predicted effects, and validation statuses, and then analyzed their catalytic properties. Our results clearly indicate that the R246X, R298H, T473A, and R512W variations caused severe (about two to four orders of magnitude) decreases in the overall bypass efficiency (i.e., the multiplied product of the insertion efficiency opposite a template and the next-base extension efficiency)17 of pol κ for both normal and translesion DNA synthesis across (opposite and beyond) G and N2-AnthG, as well as resulting in 2- to 3-fold decreases in the DNA-binding affinity of pol κ, while the A471V variation induces substantial (about one order of magnitude) decreases in those overall bypass efficiencies (Figure 4). Remarkably the T44M variation, which is located over the template at the n+1 position near the active site, caused a selective 7-fold decrease in TLS efficiency of pol κ only at the insertion step opposite N2-AnthG. These catalytic impairments in six pol κ variants were consistently observable opposite other common DNA lesions, except for the T44M variant that preserves TLS activity only opposite an 8-oxoG lesion.
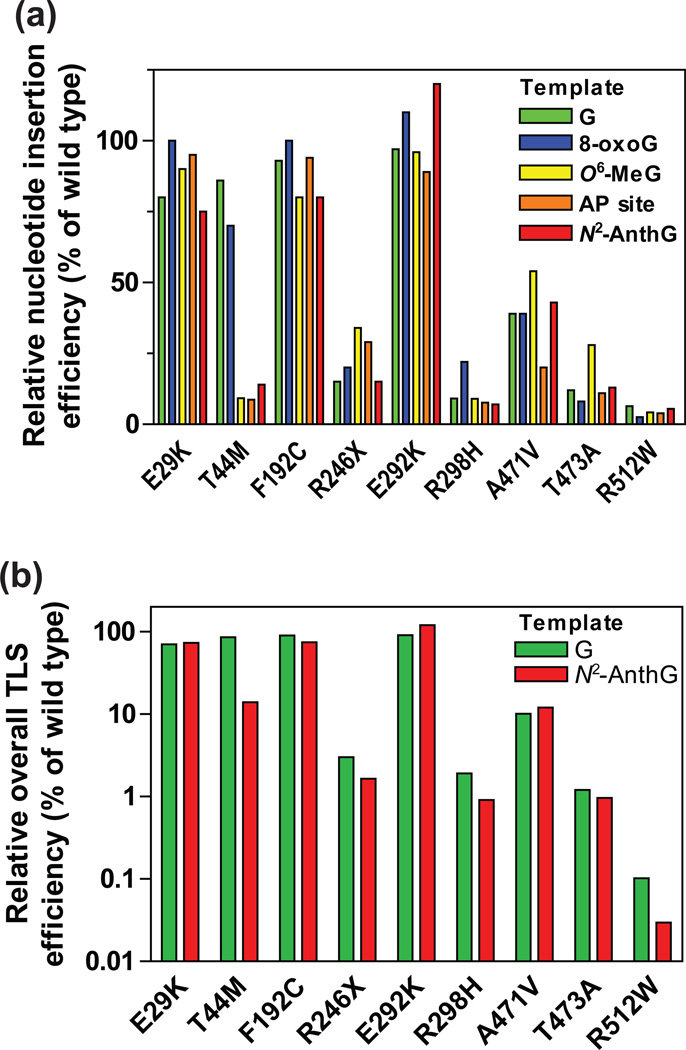
Figure 4. Relative bypass efficiencies of human variant pol κ (1–526) enzymes for G and DNA lesions compared to that of the wild-type.
(a) The relative efficiencies of nucleotide insertion opposite templates G, 8-oxoG, O6-methylG, abasic site, and N2-AnthG by variant pol κ (1–526) enzymes. The bar graph was drawn using the insertion kinetic data for insertion of the most favored dNTP opposite each template taken from the Tables 2, 3, 6, and 7. The efficiencies were normalized to that of the wild-type (set to 100%). (b) The relative efficiencies of overall TLS across (opposite and beyond) template G and N2-AnthG by variant pol κ (1–526) enzymes. The overall TLS efficiencies across G and N2-AnthG was calculated by multiplying the relative efficiency for dCTP insertion opposite G (or N2-AnthG) and the relative efficiency for the subsequent next-base extension as described previously,17 based on the insertion and extension kinetic data in Tables 2–4. The efficiencies were normalized to that of the wild-type (set to 100%).
The nine pol κ variants analyzed in this work can be classified into three types according to the alterations in their bypass efficiencies across G and four DNA lesions (Figure 4): (i) the “wild-type like” variants (E29K, F192C, and E292K), (ii) the “generally hypoactive” variants (R246X, R298H, A471V, T473A, and R512W) with diminution in both normal and TLS efficiency, (iii) the “opposite lesion-specific hypoactive” variant (T44M) with impairment only opposite N2-AnthG, O6-MeG, and abasic lesions (but not 8-oxoG and G). We observed no alterations in polymerase activity of the E29K variant opposite all four DNA lesions (Tables 3, 6, and 7), unlike the previous report on the AP site bypass with this mutant,43 the discrepancy of which might be due to the differences in the experimental conditions, e.g. expressed proteins and sequence contexts. The fidelity of nucleotide insertion opposite G and DNA lesions appeared not to be altered much in most of variants (Tables 2, 3, 6, and 7), except for several cases showing slight alterations, e.g., an increase opposite normal G but a reduction opposite N2-AnthG in the T44M variant and increases opposite 8-oxoG in the T473W and R512W variants.
The X-ray crystal structure of the catalytic core of human pol κ, in complex with primer-template DNA and incoming nucleotide, should be very useful in understanding the biochemical alterations in missense dysfunctional pol κ variants (Fig. 2). The catalytic core of pol κ has the classic polymerase domains (fingers, palm, and thumb), a PAD domain that makes major DNA contacts, and an N-clasp domain that fully encircles DNA.44, 45 It is noteworthy that the R298H variation, positioned at the palm domain, caused a severe reduction in catalytic activity of pol κ as well as a slight decrease in the DNA-binding affinity. This observed effect seems to be related to the replacement of a short side-chain residue (His) for a conserved long side-chain residue (Arg) in the αJ helix supporting a central mixed β sheet of the palm domain on the dorsal side, which might compromise its hydrogen bond interactions with nearby β sheet residues (e.g., Q332 and N330) and thus destabilize the catalytic palm domain. It is also notable that the T473A and R512W variations, which are positioned at the four-stranded β sheet of the PAD domain directly docking in the major groove of DNA, caused a strong catalytic defect with a slight decrease in DNA binding affinity. These alterations might be due to the loss of a polar or positively charged side chain at the Thr-473 or Arg-512 position that lies close to the sugar-phosphate DNA backbone, which might perturb the local conformation of the PAD domain and destabilize the interaction of pol κ with DNA. The slight activity decrease of the A471V variant seems to be due to a more bulky hydrophobic Val side chain substituted for a small Ala side chain at position 471 near the phosphodiester DNA backbone in the PAD domain. Interestingly, the T44M variant was found to have a selective catalytic impairment in nucleotide insertion only opposite N2-AnthG, O6-methylG, and abasic lesions but not a normal G and an 8-oxoG. The T44 residue is located in the N-clasp domain and its side chain hydroxyl group appears to form a hydrogen bond interaction with the first unpaired template base, which—as well as a hydrophobic pocket formed by F49, P153, F155, and I156—stabilizes this 5´ unpaired template base outside the active site cleft.45 Therefore, the loss of a hydrogen bond interaction due to the T44M substitution likely destabilizes the positioning of the 5´ unpaired template base, which might induce the positional instability of a certain bulky or distorted lesion template in the pol κ active site.
How would these dysfunctional POLK genetic variations influence cellular and organismal phenotypes? Previous work with pol κ-deficient mice and cell models,23, 24, 29, 46, 47 has suggested that pol κ protects against mutagenesis and promotes survival from exogenous and endogenous genotoxic agents such as benzo[a]pyrene and steroid derivatives by performing error-free TLS of N2-modified G lesions. In this context of TLS, it is conceivable that hypoactive pol κ variants diminish the cellular capacity for error-free TLS against cognate N2-G lesions and thus cause a compensated increase in non-C inserted mutagenic TLS events by other pols in cells. Furthermore, in view of other proposed functions of pol κ in DNA repair and replication checkpoint,26, 28 we can also speculate that hypoactive pol κ variants weaken the cellular capacity for DNA repair and checkpoint control from various types of genotoxic stress. These aforementioned deleterious effects of hypoactive pol κ variants might lead to increased incidences of mutations, genomic instability, and cancer risk from certain genotoxic stresses in the affected individuals. A large phenotypic difference would be expected between five missense variants (T44M, R298H, A471V, T473A, and R512W) and the truncated variant (R246X). The truncating R246X variant should display a POLK knockout-like phenotype irrespective of its partial activity, because it cannot be localized into the nucleus in cells due to a loss of the C-terminal NLS domain.48 By contrast, hypoactive missense variants might show a phenotype resembling but less severe than the inactivated POLK knock-in phenotype. In fact, although both the POLK knock-out cells and the inactivated POLK knock-in cells show equal hypersensitivity to benzo[a]pyrene and mitomycin C, the knock-out cells show hypersensitivity to oxidizing agents but not alkylating agents, whereas the latter cells show exactly the opposite, suggesting complex functions of inactive pol κ against different genotoxic stresses, i.e., a non-catalytic protective role against oxidative damage but a dominant negative deleterious effect on methylation damage repair.49 In this perspective, it would be desirable to carry out further investigations to delineate in vivo impacts of those dysfunctional pol κ variants at the cellular and organismal levels. It is also plausible that pol κ variants to alter protein interactions may significantly change their TLS efficiency in cells, in that the protein-protein interactions of Y-family pols have been suggested to be involved in regulation of cellular TLS processes.4, 6, 7 In this respect, it will be worth investigating whether nonsynonymous pol κ variants, particularly the variants located at C-terminal protein interaction domains (e.g., the PCNA-interacting protein (PIP) and ubiquitin-binding zinc finger (UBZ) domains), modify their interactions with PCNA and ubiquitin. Other factors such as copy number, gene expression, and epigenetic status also need to be considered, as well as genotype, in future comprehensive studies on human health impacts, in that the overexpression of pol κ also promotes genomic instability.9, 50
Among eight tested missense POLK variants that are predicted to be damaging protein function by SIFT and Polyphen-2, five variants were found to be catalytically hypoactive, which indicates a low positive predicted value (63%) of these predictions. By utilizing our present and previous data on total 31 missense Y-family pol variants among which 17 variants are catalytically impaired, we further estimated the prediction accuracies of various prediction algorithms, including MutPred (based on sequence, structural, and functional features),51 SNPs&GO (based on sequence and functional features),52 MutationTaster (based on sequence, mRNA, protein, and regulatory features),53 PolyPhen-2 (HumDIV and HumVAR models, based on sequence and structural features),37 and MetaSVM (based on nine prediction scores and allele frequencies),54 as well as five sequence homology-based methods: LRT55 SIFT,38 Mutation Assessor,56 PROVEAN,57 and FATHMM.58 The prediction accuracies of all tested algorithms were found to be relatively low (48% to 68%). Among them, the PROVEAN, SIFT, PolyPhen-2 HumVAR, and LRT methods showed prediction accuracies (68, 65, 65, and 65% respectively) somewhat higher than the other methods. Noticeably, the FATHMM and SNPs&GO methods showed high (100 and 93%, respectively) specificity but at the cost of a low (6% and 29% respectively) sensitivity, whereas the LRT, MutationTaster, and SIFT methods showed high (94%, 89%, and 89% respectively) sensitivity but at the cost of a low (29%, 15%, and 36% respectively) specificity. Only PROVEAN and Mutation Assessor methods were found to have both sensitivity and specificity ≥ 50% each, although those values were not particularly high (53% to 76%). These data emphasize the need of experimental approaches to confirm the true functional alterations in genetic variants irrespective of in silico prediction results, as well as the need for careful interpretation of in silico prediction results. In good accordance with our results, a recent report reveals a relatively low concordance between the predicted and actual effects of missense mutations in 23 immune genes and the TP53 gene.59
In conclusion, our results suggest that six germline genetic variations in the human POLK gene may impair the error-free TLS ability of pol κ across bulky N2-G DNA lesions, which might be possibly implicated as predisposing factors for mutation susceptibility to certain genotoxic agents (e.g., polycyclic aromatic hydrocarbons) in the affected individuals. Further characterization of the in vivo functional consequences of these and other as-yet unidentified POLK gene variations would contribute to a better understanding of the roles of human pol κ variants in individual’s propensity for mutations and their possible clinical implications, e.g., individual differences in cancer risks and treatment responses, with regard to specific genotoxic agents, in human populations. Our present and previous findings on Y-family pols16–18 also suggest that there likely exist a considerable number of (albeit mostly rare) dysfunctional genetic variations for Y-family pols in human population. Our growing knowledge about functional genetic variations of human TLS pols might provide some insight into understanding the complex genetic basis of cancer risks related to specific carcinogens.
Supplementary Material
Acknowledgments
Funding
This work was supported by the Basic Science Research Program through the National Research Foundation of Korea funded by the Ministry of Education (Grant NRF-2015R1D1A1A01057577) (to J.-Y.C.), Samsung Biomedical Research Institute grant, #SBRI SMX1150771 (to J.-Y.C.), the DGIST R&D Program of the Ministry of Science, ICT and Technology of Korea (20160165) (to Y.-S.L.), and the National Institutes of Health Grants R01 ES010375 and R01 ES010546 (to F.P.G.).
ABBREVIATIONS
- A
adenine
- BSA
bovine serum albumin
- C
cytosine
- dNTP
deoxynucleoside triphosphate
- FAM
6-carboxyfluorescein
- G
guanine
- MAF
minor allele frequency
- Me
methyl
- N2-Anth
N2-CH2(9-anthracenyl)
- 8-oxoG
8-oxo-7,8-dihydroguanine
- PAD
polymerase associated domain
- pol
DNA polymerase
- SDS
sodium dodecyl sulfate
- SNP
single nucleotide polymorphism (an SNV present in a population occurring at ≥1% incidence)
- SNV
single nucleotide variation
- T
thymine
- TLS
translesion synthesis
Footnotes
Supporting Information
Analysis of human pol κ (1–526) wild-type and variant proteins by SDS-polyacrylamide gel electrophoresis (Figure S1), extension of a 32P-labeled primer beyond an N2-AnthG:C pair by human wild-type and variants pol κ (1–526) enzymes (Figure S2), and the standing-start primer extensions across 8-oxoG, O6-MeG, and abasic sites by human wild-type and variant pol κ (1–526) enzymes (Figure S3). This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
REFERENCES
- 1.Zeman MK, Cimprich KA. Causes and consequences of replication stress. Nat. Cell Biol. 2014;16:2–9. doi: 10.1038/ncb2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bregeon D, Doetsch PW. Transcriptional mutagenesis: causes and involvement in tumour development. Nat. Rev. Cancer. 2011;11:218–227. doi: 10.1038/nrc3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Friedberg EC. Suffering in silence: the tolerance of DNA damage. Nat. Rev. Mol. Cell. Biol. 2005;6:943–953. doi: 10.1038/nrm1781. [DOI] [PubMed] [Google Scholar]
- 4.Sale JE. Translesion DNA synthesis and mutagenesis in eukaryotes. Cold Spring Harb. Perspect. Biol. 2013;5:a012708. doi: 10.1101/cshperspect.a012708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Choi J-Y, Eoff RE, Guengerich FP. Bypass DNA polymerases. In: Penning TM, editor. Chemical Carcinogenesis. New York, NY: Humana Press; 2011. pp. 345–373. [Google Scholar]
- 6.Lange SS, Takata K, Wood RD. DNA polymerases and cancer. Nat. Rev. Cancer. 2011;11:96–110. doi: 10.1038/nrc2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Knobel PA, Marti TM. Translesion DNA synthesis in the context of cancer research. Cancer Cell Int. 2011;11:39. doi: 10.1186/1475-2867-11-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Masutani C, Kusumoto R, Yamada A, Dohmae N, Yokoi M, Yuasa M, Araki M, Iwai S, Takio K, Hanaoka F. The XPV (xeroderma pigmentosum variant) gene encodes human DNA polymerase η. Nature. 1999;399:700–704. doi: 10.1038/21447. [DOI] [PubMed] [Google Scholar]
- 9.Bavoux C, Leopoldino AM, Bergoglio V, J OW, Ogi T, Bieth A, Judde JG, Pena SD, Poupon MF, Helleday T, Tagawa M, Machado C, Hoffmann JS, Cazaux C. Up-regulation of the error-prone DNA polymerase κ promotes pleiotropic genetic alterations and tumorigenesis. Cancer Res. 2005;65:325–330. [PubMed] [Google Scholar]
- 10.Dumstorf CA, Clark AB, Lin Q, Kissling GE, Yuan T, Kucherlapati R, McGregor WG, Kunkel TA. Participation of mouse DNA polymerase ι in strand-biased mutagenic bypass of UV photoproducts and suppression of skin cancer. Proc. Natl. Acad. Sci. U. S. A. 2006;103:18083–18088. doi: 10.1073/pnas.0605247103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ohkumo T, Kondo Y, Yokoi M, Tsukamoto T, Yamada A, Sugimoto T, Kanao R, Higashi Y, Kondoh H, Tatematsu M, Masutani C, Hanaoka F. UV-B radiation induces epithelial tumors in mice lacking DNA polymerase η and mesenchymal tumors in mice deficient for DNA polymerase ι. Mol. Cell. Biol. 2006;26:7696–7706. doi: 10.1128/MCB.01076-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wittschieben JP, Patil V, Glushets V, Robinson LJ, Kusewitt DF, Wood RD. Loss of DNA polymerase ζ enhances spontaneous tumorigenesis. Cancer Res. 2010;70:2770–2778. doi: 10.1158/0008-5472.CAN-09-4267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Iguchi M, Osanai M, Hayashi Y, Koentgen F, Lee GH. The error-prone DNA polymerase ι provides quantitative resistance to lung tumorigenesis and mutagenesis in mice. Oncogene. 2014;33:3612–3617. doi: 10.1038/onc.2013.331. [DOI] [PubMed] [Google Scholar]
- 14.Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, Bignell GR, Bolli N, Borg A, Borresen-Dale AL, Boyault S, Burkhardt B, Butler AP, Caldas C, Davies HR, Desmedt C, Eils R, Eyfjord JE, Foekens JA, Greaves M, Hosoda F, Hutter B, Ilicic T, Imbeaud S, Imielinski M, Jager N, Jones DT, Jones D, Knappskog S, Kool M, Lakhani SR, Lopez-Otin C, Martin S, Munshi NC, Nakamura H, Northcott PA, Pajic M, Papaemmanuil E, Paradiso A, Pearson JV, Puente XS, Raine K, Ramakrishna M, Richardson AL, Richter J, Rosenstiel P, Schlesner M, Schumacher TN, Span PN, Teague JW, Totoki Y, Tutt AN, Valdes-Mas R, van Buuren MM, van 't Veer L, Vincent-Salomon A, Waddell N, Yates LR, Australian Pancreatic Cancer Genome, I., Consortium, I. B. C., Consortium, I. M.-S. PedBrain I, Zucman-Rossi J, Futreal PA, McDermott U, Lichter P, Meyerson M, Grimmond SM, Siebert R, Campo E, Shibata T, Pfister SM, Campbell PJ, Stratton MR. Signatures of mutational processes in human cancer. Nature. 2013;500:415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roberts SA, Gordenin DA. Hypermutation in human cancer genomes: footprints and mechanisms. Nat. Rev. Cancer. 2014;14:786–800. doi: 10.1038/nrc3816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim J, Song I, Jo A, Shin J-H, Cho H, Eoff RL, Guengerich FP, Choi J-Y. Biochemical analysis of six genetic variants of error-prone human DNA polymerase ι involved in translesion DNA synthesis. Chem. Res. Toxicol. 2014;27:1837–1852. doi: 10.1021/tx5002755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Song I, Kim E-J, Kim I-H, Park E-M, Lee KE, Shin J-H, Guengerich FP, Choi J-Y. Biochemical characterization of eight genetic variants of human DNA polymerase κ involved in error-free bypass across bulky N2-guanyl DNA adducts. Chem. Res. Toxicol. 2014;27:919–930. doi: 10.1021/tx500072m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yeom M, Kim IH, Kim JK, Kang K, Eoff RL, Guengerich FP, Choi JY. Effects of twelve germline missense variations on DNA lesion and G-quadruplex bypass activities of human DNA polymerase REV1. Chem. Res. Toxicol. 2016;29:367–379. doi: 10.1021/acs.chemrestox.5b00513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Choi JY, Angel KC, Guengerich FP. Translesion synthesis across bulky N2-alkyl guanine DNA adducts by human DNA polymerase κ. J. Biol. Chem. 2006;281:21062–21072. doi: 10.1074/jbc.M602246200. [DOI] [PubMed] [Google Scholar]
- 20.Minko IG, Harbut MB, Kozekov ID, Kozekova A, Jakobs PM, Olson SB, Moses RE, Harris TM, Rizzo CJ, Lloyd RS. Role for DNA polymerase κ in the processing of N2-N2-guanine interstrand cross-links. J. Biol. Chem. 2008;283:17075–17082. doi: 10.1074/jbc.M801238200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rechkoblit O, Zhang Y, Guo D, Wang Z, Amin S, Krzeminsky J, Louneva N, Geacintov NE. trans-Lesion synthesis past bulky benzo[a]pyrene diol epoxide N2-dG and N6-dA lesions catalyzed by DNA bypass polymerases. J. Biol. Chem. 2002;277:30488–30494. doi: 10.1074/jbc.M201167200. [DOI] [PubMed] [Google Scholar]
- 22.Yuan B, Cao H, Jiang Y, Hong H, Wang Y. Efficient and accurate bypass of N2-(1-carboxyethyl)-2'-deoxyguanosine by DinB DNA polymerase in vitro and in vivo. Proc. Natl. Acad. Sci. U. S. A. 2008;105:8679–8684. doi: 10.1073/pnas.0711546105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ogi T, Shinkai Y, Tanaka K, Ohmori H. Pol κ protects mammalian cells against the lethal and mutagenic effects of benzo[a]pyrene. Proc. Natl. Acad. Sci. U. S. A. 2002;99:15548–15553. doi: 10.1073/pnas.222377899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Avkin S, Goldsmith M, Velasco-Miguel S, Geacintov N, Friedberg EC, Livneh Z. Quantitative analysis of translesion DNA synthesis across a benzo[a]pyrene-guanine adduct in mammalian cells: the role of DNA polymerase κ. J. Biol. Chem. 2004;279:53298–53305. doi: 10.1074/jbc.M409155200. [DOI] [PubMed] [Google Scholar]
- 25.Bi X, Slater DM, Ohmori H, Vaziri C. DNA polymerase κ is specifically required for recovery from the benzo[a]pyrene-dihydrodiol epoxide (BPDE)-induced S-phase checkpoint. J. Biol. Chem. 2005;280:22343–22355. doi: 10.1074/jbc.M501562200. [DOI] [PubMed] [Google Scholar]
- 26.Ogi T, Lehmann AR. The Y-family DNA polymerase κ (pol κ) functions in mammalian nucleotide-excision repair. Nat. Cell Biol. 2006;8:640–642. doi: 10.1038/ncb1417. [DOI] [PubMed] [Google Scholar]
- 27.Zhang X, Lv L, Chen Q, Yuan F, Zhang T, Yang Y, Zhang H, Wang Y, Jia Y, Qian L, Chen B, Zhang Y, Friedberg EC, Tang TS, Guo C. Mouse DNA polymerase κ has a functional role in the repair of DNA strand breaks. DNA Repair (Amst) 2013;12:377–388. doi: 10.1016/j.dnarep.2013.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Betous R, Pillaire MJ, Pierini L, van der Laan S, Recolin B, Ohl-Seguy E, Guo C, Niimi N, Gruz P, Nohmi T, Friedberg E, Cazaux C, Maiorano D, Hoffmann JS. DNA polymerase κ-dependent DNA synthesis at stalled replication forks is important for CHK1 activation. EMBO J. 2013;32:2172–2185. doi: 10.1038/emboj.2013.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stancel JN, McDaniel LD, Velasco S, Richardson J, Guo C, Friedberg EC. Pol κ mutant mice have a spontaneous mutator phenotype. DNA Repair (Amst) 2009;8:1355–1362. doi: 10.1016/j.dnarep.2009.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Michiels S, Danoy P, Dessen P, Bera A, Boulet T, Bouchardy C, Lathrop M, Sarasin A, Benhamou S. Polymorphism discovery in 62 DNA repair genes and haplotype associations with risks for lung and head and neck cancers. Carcinogenesis. 2007;28:1731–1739. doi: 10.1093/carcin/bgm111. [DOI] [PubMed] [Google Scholar]
- 31.Dai ZJ, Liu XH, Ma YF, Kang HF, Jin TB, Dai ZM, Guan HT, Wang M, Liu K, Dai C, Yang XW, Wang XJ. Association between single nucleotide polymorphisms in DNA polymerase κ gene and breast cancer risk in Chinese Han population: a strobe-compliant observational study. Medicine (Baltimore) 2016;95:e2466. doi: 10.1097/MD.0000000000002466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shao M, Jin B, Niu Y, Ye J, Lu D, Han B. Association of POLK polymorphisms with platinum-based chemotherapy response and severe toxicity in non-small cell lung cancer patients. Cell Biochem. Biophys. 2014;70:1227–1237. doi: 10.1007/s12013-014-0046-x. [DOI] [PubMed] [Google Scholar]
- 33.Genomes Project C, Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, Korbel JO, Marchini JL, McCarthy S, McVean GA, Abecasis GR. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhu Q, Ge D, Maia JM, Zhu M, Petrovski S, Dickson SP, Heinzen EL, Shianna KV, Goldstein DB. A genome-wide comparison of the functional properties of rare and common genetic variants in humans. Am. J. Hum. Genet. 2011;88:458–468. doi: 10.1016/j.ajhg.2011.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Consortium UK, Walter K, Min JL, Huang J, Crooks L, Memari Y, McCarthy S, Perry JR, Xu C, Futema M, Lawson D, Iotchkova V, Schiffels S, Hendricks AE, Danecek P, Li R, Floyd J, Wain LV, Barroso I, Humphries SE, Hurles ME, Zeggini E, Barrett JC, Plagnol V, Richards JB, Greenwood CM, Timpson NJ, Durbin R, Soranzo N. The UK10K project identifies rare variants in health and disease. Nature. 2015;526:82–90. doi: 10.1038/nature14962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zheng HF, Forgetta V, Hsu YH, Estrada K, Rosello-Diez A, Leo PJ, Dahia CL, Park-Min KH, Tobias JH, Kooperberg C, Kleinman A, Styrkarsdottir U, Liu CT, Uggla C, Evans DS, Nielson CM, Walter K, Pettersson-Kymmer U, McCarthy S, Eriksson J, Kwan T, Jhamai M, Trajanoska K, Memari Y, Min J, Huang J, Danecek P, Wilmot B, Li R, Chou WC, Mokry LE, Moayyeri A, Claussnitzer M, Cheng CH, Cheung W, Medina-Gomez C, Ge B, Chen SH, Choi K, Oei L, Fraser J, Kraaij R, Hibbs MA, Gregson CL, Paquette D, Hofman A, Wibom C, Tranah GJ, Marshall M, Gardiner BB, Cremin K, Auer P, Hsu L, Ring S, Tung JY, Thorleifsson G, Enneman AW, van Schoor NM, de Groot LC, van der Velde N, Melin B, Kemp JP, Christiansen C, Sayers A, Zhou Y, Calderari S, van Rooij J, Carlson C, Peters U, Berlivet S, Dostie J, Uitterlinden AG, Williams SR, Farber C, Grinberg D, LaCroix AZ, Haessler J, Chasman DI, Giulianini F, Rose LM, Ridker PM, Eisman JA, Nguyen TV, Center JR, Nogues X, Garcia-Giralt N, Launer LL, Gudnason V, Mellstrom D, Vandenput L, Amin N, van Duijn CM, Karlsson MK, Ljunggren O, Svensson O, Hallmans G, Rousseau F, Giroux S, Bussiere J, Arp PP, Koromani F, Prince RL, Lewis JR, Langdahl BL, Hermann AP, Jensen JE, Kaptoge S, Khaw KT, Reeve J, Formosa MM, Xuereb-Anastasi A, Akesson K, McGuigan FE, Garg G, Olmos JM, Zarrabeitia MT, Riancho JA, Ralston SH, Alonso N, Jiang X, Goltzman D, Pastinen T, Grundberg E, Gauguier D, Orwoll ES, Karasik D, Davey-Smith G, Consortium A, Smith AV, Siggeirsdottir K, Harris TB, Zillikens MC, van Meurs JB, Thorsteinsdottir U, Maurano MT, Timpson NJ, Soranzo N, Durbin R, Wilson SG, Ntzani EE, Brown MA, Stefansson K, Hinds DA, Spector T, Cupples LA, Ohlsson C, Greenwood CM, Consortium UK, Jackson RD, Rowe DW, Loomis CA, Evans DM, Ackert-Bicknell CL, Joyner AL, Duncan EL, Kiel DP, Rivadeneira F, Richards JB. Whole-genome sequencing identifies EN1 as a determinant of bone density and fracture. Nature. 2015;526:112–117. doi: 10.1038/nature14878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR. A method and server for predicting damaging missense mutations. Nat. Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ng PC, Henikoff S. Predicting deleterious amino acid substitutions. Genome Res. 2001;11:863–874. doi: 10.1101/gr.176601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Choi JY, Guengerich FP. Analysis of the effect of bulk at N2-alkylguanine DNA adducts on catalytic efficiency and fidelity of the processive DNA polymerases bacteriophage T7 exonuclease− and HIV-1 reverse transcriptase. J. Biol. Chem. 2004;279:19217–19229. doi: 10.1074/jbc.M313759200. [DOI] [PubMed] [Google Scholar]
- 40.Choi JY, Lim S, Kim EJ, Jo A, Guengerich FP. Translesion synthesis across abasic lesions by human B-family and Y-family DNA polymerases α, δ, η, ι, κ, and REV1. J. Mol. Biol. 2010;404:34–44. doi: 10.1016/j.jmb.2010.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Niimi N, Sassa A, Katafuchi A, Gruz P, Fujimoto H, Bonala RR, Johnson F, Ohta T, Nohmi T. The steric gate amino acid tyrosine 112 is required for efficient mismatched-primer extension by human DNA polymerase k. Biochemistry. 2009;48:4239–4246. doi: 10.1021/bi900153t. [DOI] [PubMed] [Google Scholar]
- 42.Fu W, O'Connor TD, Jun G, Kang HM, Abecasis G, Leal SM, Gabriel S, Rieder MJ, Altshuler D, Shendure J, Nickerson DA, Bamshad MJ, Project NES, Akey JM. Analysis of 6,515 exomes reveals the recent origin of most human protein-coding variants. Nature. 2013;493:216–220. doi: 10.1038/nature11690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yadav S, Mukhopadhyay S, Anbalagan M, Makridakis N. Somatic mutations in catalytic core of POLK reported in prostate cancer alter translesion DNA synthesis. Hum. Mutat. 2015;36:873–880. doi: 10.1002/humu.22820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Uljon SN, Johnson RE, Edwards TA, Prakash S, Prakash L, Aggarwal AK. Crystal structure of the catalytic core of human DNA polymerase κ. Structure. 2004;12:1395–1404. doi: 10.1016/j.str.2004.05.011. [DOI] [PubMed] [Google Scholar]
- 45.Lone S, Townson SA, Uljon SN, Johnson RE, Brahma A, Nair DT, Prakash S, Prakash L, Aggarwal AK. Human DNA polymerase κ encircles DNA: implications for mismatch extension and lesion bypass. Mol. Cell. 2007;25:601–614. doi: 10.1016/j.molcel.2007.01.018. [DOI] [PubMed] [Google Scholar]
- 46.Burr KL, Velasco-Miguel S, Duvvuri VS, McDaniel LD, Friedberg EC, Dubrova YE. Elevated mutation rates in the germline of Pol κ mutant male mice. DNA Repair (Amst) 2006;5:860–862. doi: 10.1016/j.dnarep.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 47.Singer WD, Osimiri LC, Friedberg EC. Increased dietary cholesterol promotes enhanced mutagenesis in DNA polymerase κ-deficient mice. DNA Repair (Amst) 2013;12:817–823. doi: 10.1016/j.dnarep.2013.07.010. [DOI] [PubMed] [Google Scholar]
- 48.Ogi T, Kannouche P, Lehmann AR. Localisation of human Y-family DNA polymerase κ: relationship to PCNA foci. J. Cell Sci. 2005;118:129–136. doi: 10.1242/jcs.01603. [DOI] [PubMed] [Google Scholar]
- 49.Kanemaru Y, Suzuki T, Niimi N, Gruz P, Matsumoto K, Adachi N, Honma M, Nohmi T. Catalytic and non-catalytic roles of DNA polymerase κ in the protection of human cells against genotoxic stresses. Environ. Mol. Mutagen. 2015;56:650–662. doi: 10.1002/em.21961. [DOI] [PubMed] [Google Scholar]
- 50.Jones MJ, Colnaghi L, Huang TT. Dysregulation of DNA polymerase κ recruitment to replication forks results in genomic instability. EMBO J. 2012;31:908–918. doi: 10.1038/emboj.2011.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li B, Krishnan VG, Mort ME, Xin F, Kamati KK, Cooper DN, Mooney SD, Radivojac P. Automated inference of molecular mechanisms of disease from amino acid substitutions. Bioinformatics. 2009;25:2744–2750. doi: 10.1093/bioinformatics/btp528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Calabrese R, Capriotti E, Fariselli P, Martelli PL, Casadio R. Functional annotations improve the predictive score of human disease-related mutations in proteins. Hum. Mutat. 2009;30:1237–1244. doi: 10.1002/humu.21047. [DOI] [PubMed] [Google Scholar]
- 53.Schwarz JM, Rodelsperger C, Schuelke M, Seelow D. MutationTaster evaluates disease-causing potential of sequence alterations. Nat. Methods. 2010;7:575–576. doi: 10.1038/nmeth0810-575. [DOI] [PubMed] [Google Scholar]
- 54.Dong C, Wei P, Jian X, Gibbs R, Boerwinkle E, Wang K, Liu X. Comparison and integration of deleteriousness prediction methods for nonsynonymous SNVs in whole exome sequencing studies. Hum. Mol. Genet. 2015;24:2125–2137. doi: 10.1093/hmg/ddu733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chun S, Fay JC. Identification of deleterious mutations within three human genomes. Genome Res. 2009;19:1553–1561. doi: 10.1101/gr.092619.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Reva B, Antipin Y, Sander C. Predicting the functional impact of protein mutations: application to cancer genomics. Nucleic Acids Res. 2011;39:e118. doi: 10.1093/nar/gkr407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Choi Y, Sims GE, Murphy S, Miller JR, Chan AP. Predicting the functional effect of amino acid substitutions and indels. PLoS One. 2012;7:e46688. doi: 10.1371/journal.pone.0046688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shihab HA, Gough J, Cooper DN, Day IN, Gaunt TR. Predicting the functional consequences of cancer-associated amino acid substitutions. Bioinformatics. 2013;29:1504–1510. doi: 10.1093/bioinformatics/btt182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Miosge LA, Field MA, Sontani Y, Cho V, Johnson S, Palkova A, Balakishnan B, Liang R, Zhang Y, Lyon S, Beutler B, Whittle B, Bertram EM, Enders A, Goodnow CC, Andrews TD. Comparison of predicted and actual consequences of missense mutations. Proc. Natl. Acad. Sci. U. S. A. 2015;112:E5189–E5198. doi: 10.1073/pnas.1511585112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ren J, Wen L, Gao X, Jin C, Xue Y, Yao X. DOG 1.0: illustrator of protein domain structures. Cell Res. 2009;19:271–273. doi: 10.1038/cr.2009.6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.