Abstract
For over half a century, the field of developmental biology has leveraged computation to explore mechanisms of developmental processes. More recently, computational approaches have been critical in the translation of high throughput data into knowledge of both developmental and stem cell biology. In the last several years, a new sub-discipline of computational stem cell biology has emerged that synthesizes the modeling of systems-level aspects of stem cells with high-throughput molecular data. In this review, we provide an overview of this new field and pay particular attention to the impact that single-cell transcriptomics is expected to have on our understanding of development and our ability to engineer cell fate.
Keywords: Computational biology, Stem cell biology, cell fate engineering, single cell transcriptomics, network biology
Computation in stem and developmental biology
Computational tools have played incisive roles in developmental biology since at least the 1950s, when Alan Turing wrote a computer program to model how morphogen concentrations might affect pattern formation in an in silico embryo [1]. From this time until the advent of OMICs (see Glossary), the role of computational tools in developmental and stem cell biology was limited largely to exploring theoretical mechanisms of morphogenesis by, for example, modeling the emergence of positional information during embryogenesis [2], and to modeling the dynamics of adult stem cell self-renewal [3]. Beginning with the large genome sequencing projects around fifteen years ago, the predominant use of computational tools in developmental and stem cell biology shifted away from modeling to the processing of large molecular data sets [4, 5].
More recently, two trends have emerged that warrant an exposition of the state-of-the-art in computational stem cell biology. First, systems biology and network biology approaches have begun to successfully synthesize large-scale molecular data with systems-level modeling of stem cell behavior and function. Second, new technologies have matured that allow single cell genome-wide molecular profiling. In this review, we concentrate on these two trends after we have briefly described the impact of OMICs and their affiliated computational techniques on stem cell biology.
OMICs in stem cell biology
The application of OMICs to stem cell biology has almost always closely followed (or coincided with) the initial description of the new technology. Here, as an introduction to the most widely-applied computational algorithms and the data on which they operate, we highlight two seminal questions in stem cell biology. We have summarized these and other common OMICs techniques and exemplary applications to stem cell biology in Table 1 and in Boxes 1 and 2.
Table 1.
A nomenclature for big data in biology: cellular measurements and applications
| Method | Cellular entity/event | Exemplary application in stem cell biology |
|---|---|---|
| Microarray profiling | DNA: SNPs | [63] |
| DNA:CNVs | [64] | |
| RNA | [65] | |
| microRNA | [66] | |
| lncRNA | [67] | |
|
| ||
| Sequencing | DNA: genetic | [8] [59] [68] [69] |
| RNA: mRNA | [70] [71] [67] | |
| RNA: microRNA | [72] | |
| RNA: lncRNA | [73] | |
|
|
||
| ChIP-Seq | Protein-DNA interaction | [74] [75] |
| ChIP-chip | Protein-DNA interaction, histone modification, DNA methylation | [6] [7] [76] |
|
| ||
| Clip-seq | RNA-binding protein | [11] [77] [78] |
| Rip-seq | Protein-binding RNA | [10] [79] |
|
| ||
| Chip-seq of histone modification | epigenetic | [80] |
| Bisulfite-seq for DNA methylation | [20] [81] | |
|
| ||
| Proteomics | Protein abundance (Mass spectrometry-based methods) | [82] [83] |
| Post translational modification(Ribo-seq/Ribosome profiling) | [9, 84] [85] | |
|
| ||
| Y2H | Protein-protein interaction | [86] |
|
| ||
| DNase hypersensitivity mapping | Gene regulatory regions | [8] [87] |
BOX 1. Common OMICs analytical tools.
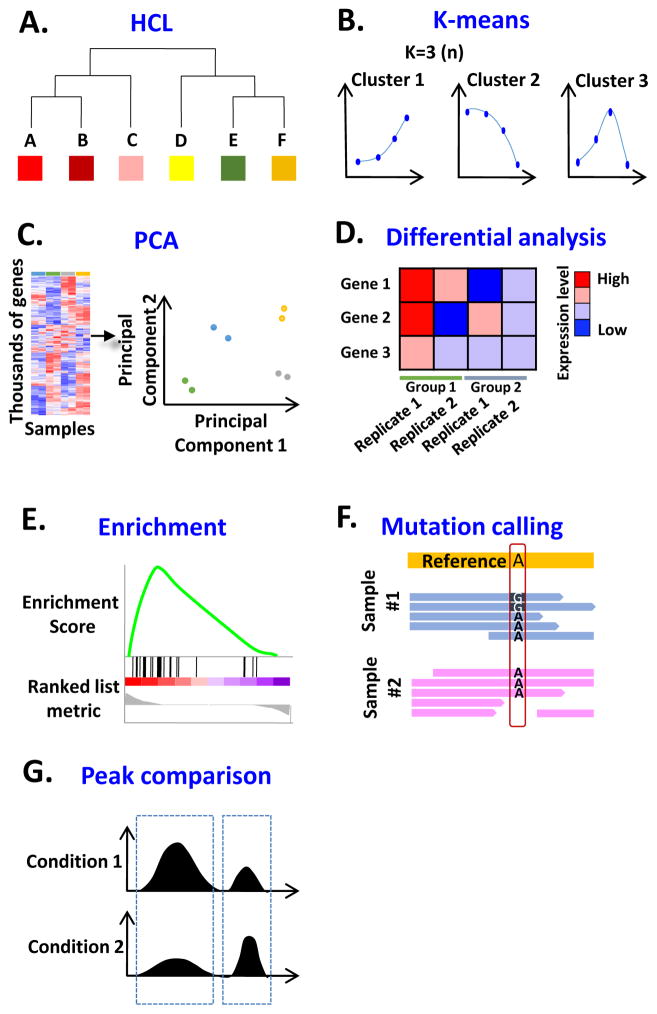
Hierarchical Clustering (HCL): Aims to build a hierarchy of clusters. It takes as input a matrix representing pairwise distances between entities, it joins the closest pairs of entities, then calculates a new distance between this merged entity and all others, and repeats until all entities have been merged (Figure IA). K-means Clustering: Aims to group data into a pre-defined number (k) of clusters by first randomly assigning entities to clusters, calculating a mean profile of each cluster, determining the inter- and intra-cluster distances, then assigning entities to the nearest cluster and re-computing the mean profiles. This process is repeated either a pre-determined number of times, or until the entities do not change their cluster membership (Figure IB). Principal Component Analysis (PCA): A dimension-reduction technique that finds axes or directions that are linear combinations of variables that maximize the total variation in the data set and are orthogonal to each other (Figure IC). Differential analysis: Aims to identify genes differentially expressed between distinct groups using approaches that account for the typically large number of statistical tests being performed (Figure ID). Enrichment analysis: Gene Set Analysis (GSA) using programs such as GSEA [88] determines whether the expression of predefined sets of genes tend to cluster towards the top or bottom of a ranked list of all genes assayed. The ranking is typically based on differential expression between two conditions (Figure IE). Mutation calling: The identification of genetic differences between a sample (e.g., from an individual’s germline or from a tumor) compared with a reference genomic sequence (Figure IF). Peak comparison: To identify genomic loci that are enriched with NGS reads that have been obtained by ChIP-seq or DNase-seq. Some peak calling tools are optimized for specific assays such as Hotspot [89] and F-Seq [90] for DNase-seq data, while some serve as generic tool for a variety of data types such as Model-based Analysis of ChIP-seq (MACS) [91, 92] and DFilter [93] (Figure IG).
Figure I.
(A) HCL clusters samples based on their similarity. A–F represent different samples. (B) K-means divides variables into a user-selected number of groups. (C) PCA reduces the number of dimensions in data. (D) Two duplicates of each condition. Gene 1 is considered differentially expressed whereas Genes 2 and 3 are not. (E) GSEA showing whether a set of genes have statistically significant difference between two conditions. (IF) A->G mutation detected by next generation sequencing (NGS). (IG) Peak comparison between two conditions.
BOX 2. Machine learning classifiers.
Support vector machines (SVMs): Separate two data classes by maximizing the margin and creating the largest distance between the separating hyperplane. (Figure IA). Naïve Bayes classifiers (NBC): A direct application of Bayes Theorem to compute the probability that a sample comes from a class with a predetermined likelihood distribution (Figure IB). Random forest (RF): Random forests are constructed by sampling with replacement from all of the cases of the training data, and also sampling a subset of possible predictor variables (most often in our context the predictor variables are genes), then generating a collection of decision trees that are collectively used to classify new data (Figure IC).
Figure I.
(A) SVMs aims to separate two groups (green and red). (B) Given expression distributions of gene x in each of RBCs and HSCs, it is possible to compute the posterior probability that a sample is either a RBC or HSC using Bayes’ theorem. For convenience, the prior probability is usually assumed to be equal across all cell types. (C) A RF classifier treats the classification of each decision tree as a vote and returns the classification with the greatest number of votes.
The pluripotency gene regulatory network
Since the isolation of embryonic stem cells (ESCs) in 1981 a question of intense focus has been, ‘What are the molecular mechanisms by which pluripotent stem cells (PSCs) maintain multi-lineage potential indefinitely?’ OMICs techniques have played a central role in answering this question and have revealed previously unanticipated complexity in the regulation of pluripotency. As a first step towards understanding the molecular basis of pluripotency, the transcriptional profile of ESCs, and its relationship to that of adult stem cells was defined. Subsequently, the predecessor to Chip-Seq, Chip-Chip, was used to reconstruct to transcriptional regulatory network of core pluripotency transcription factors in human ESCs, uncovering an auto-regulatory loop that helps to maintain the pluripotent state by buffering against transient down regulation of any single pluripotency transcription factor [6]. This network motif was found by Chip-Chip to be conserved in mouse ESCs [7]. To further define the pluripotency regulatory network at the level of genomic regulatory elements, Stamatoyannopoulos’s group paired distal DNase hypersensitivities (DHSs) with target promoters of pluripotency-specific transcription factors (TFs) such as KLF4, SOX2 and OCT4 [8].
A host of novel OMICs techniques, mainly based on next-generation sequencing, facilitated the investigation of post-transcriptional regulation such as Ribo-Seq [9], RIP-Seq [10], and CLIP-seq [11] in pluripotency. For example, the pluripotency network of mouse and human ESCs at the m6A methylome level was described using MeRIP-Seq [12], and using this method, the chromatin-associated zinc finger protein 217 (ZFP217) was shown to interact with epigenetic networks to regulate pluripotency in hESCs [13].
Taken together, these and many other genome-wide molecular profiling studies have collectively contributed to our understanding of the multilayered regulation of pluripotency, and furthermore have served as a model to understand the regulation of cell type identity for other, less-investigated lineages.
Epigenetic memory in induced pluripotent stem cells
In 2006, Yamanaka et al showed that it is possible to convert somatic cells to an ESC-like state, opening up the use of induced pluripotent stem cells (iPSCs) for disease modeling, and, in the future, for personalized regenerative medicine [14, 15]. In order for iPSC to be used in these contexts it is critical to understand in what ways iPSC and ESC are distinct, and how reprogramming itself might affect in vitro lineage bias. One hypothesis that emerged is that iPSC retain residual epigenetic marks that are transcriptionally silent in the pluripotent state but are apparent upon directed differentiation and would result in lineage bias. To explore this hypothesis, “comprehensive high-throughput arrays for relative methylation” (CHARM) [16] was used to identify differentially methylated genomic regions (DMRs) in iPSC derived from distinct starting cell types, and these DMRs were found to be enriched in promoters of TFs that specify lineages distinct from the starting cell type in mice and human iPSCs [17, 18], an effect that is moderated by extended passage [19]. A distinct technique based on targeted bisulfite sequencing revealed that residual repressive DNA-methylation at promoters of pluripotency TFs might contribute to stable, partially reprogrammed lines [20].
These are just a few of the hundreds of studies that have leveraged genome-wide profiling combined with advanced and often custom algorithms to investigate the molecular basis of pluripotency and the processes associated with reprogramming to pluripotency. Many of the algorithms used to analyze and extract biological knowledge from OMICS technologies were first developed and deployed for stem cell biology, and they have subsequently been adapted for use in many other biological contexts. However, there are pressing issues especially relevant to stem cell biology, which are beginning to be addressed with new analytical tools and single cell approaches as described below.
Computational stem cell biology
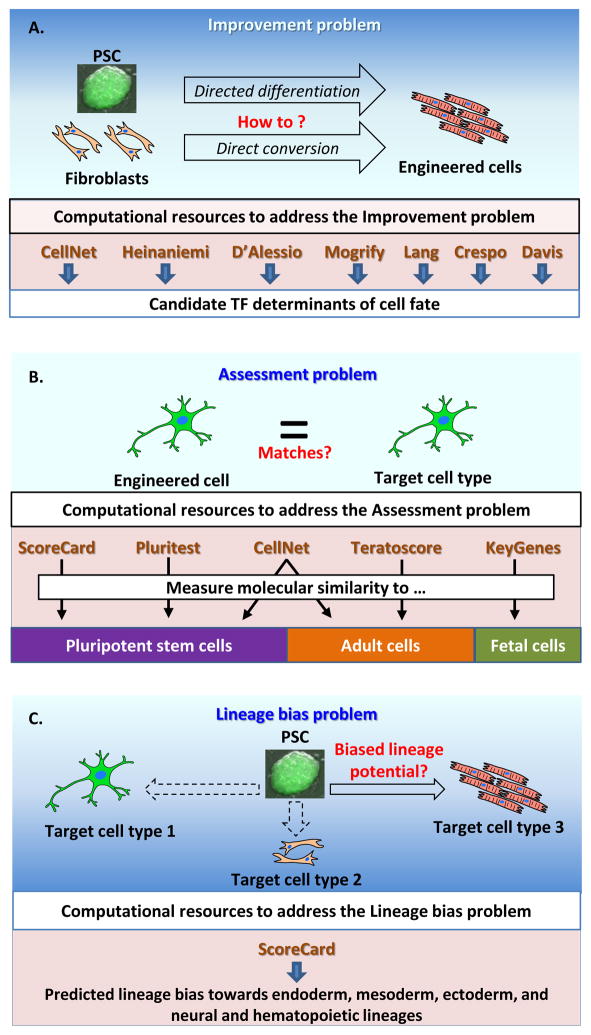
Cell fate engineering, for example the directed differentiation of PSCs or the direct conversion among somatic cell types (e.g., the conversion of fibroblasts to cardiomyocytes through the ectopic expression of Gata4, Mef2c, and Tbx5 [21]) is practiced in thousands of labs worldwide to model diseases, to explore inaccessible time points in development, to screen drugs, and to develop regenerative medicine therapies. However, there are three daunting classes of barriers that impede cell fate engineering from fulfilling its promise to broadly transform the biomedical enterprise. The first class of barriers concerns the absence of rational, proven, and hypothesis-driven systems to select conditions to guide directed differentiation or to select factors to use for direct conversion (Figure 1, Key Figure). Directed differentiation methods, inspired by our understanding of signaling cues and forces in mouse development, are limited by our inability to study highly transient, embryonic states. On the other hand, methods to select ‘master regulators’ for use in direct conversion are based on the assumption of a ‘kernel’ GRN comprised of a small number of transcription factors that auto-regulate their own expression, positively regulate the transcription of cell type associated genes, and repress alternative lineages [22]. While this strategy to identify and use the transcription factors of a kernel gene regulatory network (GRN) was successful in reprogramming back to pluripotency, the extent to which it applies to other cell types is unknown. We refer to this set of questions as the ‘Improvement problem’.
Figure 1. Computational tools to address the three major barriers to achieving cell fate engineering.
(A) The ‘Improvement problem’: how can we devise protocols to improve the fidelity of engineered cells? (B) The ‘Assessment problem’: to what extent is the engineered population equivalent to the desired cell type? (C) The ‘Lineage bias problem’: how can we quantify the in vitro lineage bias of a PSC line?
The second class of barriers concerns our limited ability to assess comprehensively the fidelity with which engineered cells resemble their in vivo counterparts (Figure 1B). We refer to this set of questions the ‘Assessment problem’. The third class of barriers concerns the long-observed variability between PSC lines in the efficiency and fidelity with which they can be guided to select lineages, which we refer to as the ‘Lineage bias problem’ [23] (Figure 1C). The molecular contributors to this variation are an area of intense scrutiny [24], and both genetic and epigenetic factors have been implicated. In this section we comprehensively review all computational tools designed to address these three major barriers (summarized in Figure 1 and Table 2).
Table 2.
Computational stem cell biology tools
| Method | Input data type | Species | Cell and tissue types | Output | Extensibility | Resource access |
|---|---|---|---|---|---|---|
| ScoreCard | Gene expression data (Nanostring and qPCR), DNA methylation data | Human | Genome-wide reference maps of DNA methylation and gene expression for 20 ES 12 iPS cell lines | Lineage scorecard summarizing cell-line specific differentiation propensities based on expression of three germ layers and hematopoietic and neural lineages. Deviation scorecard and reference corridor for DNA methylation and gene expression. | Theoretically possible to incorporate new cell types in the prediction of the lineage scorecard. | http://scorecard.computational-epigenetics.org/ |
| Pluritest | Gene expression data from Illumina HT12v3 and HT12v4 microarrays (.idat) | Human | Gene expression profiles of 233 ES 41 iPS cell lines | Predict pluripotency and deviation from pluripotency of query sample (either PSC or non PSC) | Theoretically possible to extend to global DNA methylation and RNA-Seq | http://www.pluritest.org |
| Teratoscore | Gene expression data from Affymetrix Human Genome U133 Plus 2.0 Array (.CEL file) | Human | Gene expression profiles of 12 cell lines 26 tissues and cell types | Provides a quantitative estimation of pluripotency based on germ layer contribution to teratomas. | Theoretically possible to extend to RNA-Seq, qPCR. | http://benvenisty.huji.ac.il/teratoscore.php |
| KeyGene | Gene expression profile (Affymetrix, Illumina), next-generation sequencing (NGS) data | Human | NGS data from 21 fetal organs | Predicts tissue of origin, identity of stem cell derivatives, and estimates a developmental stage. | http://www.keygenes.nl/ | |
| CellNet | Gene expression profile (Affymetrix and Illumina BeadArray microarray data) | Human, mouse | Gene expression profile of 20 cell/tissue types | Assesses similarity to 20 cell and tissue types, measures the extent to which a cell type specific GRN is established, and predicts TFs that can enable cell fate change. | Bulk and single-cell RNA-Seq | Web application: http://cellnet.hms.harvard.edu Code: https://github.com/pcahan1/CellNet |
| Mogrify | Starting and finishing cell type | Human | Gene expression data of ~ 300 cell and tissue types | Predict TFs for cell conversion/transdifferentiation | http://www.mogrify.net | |
| Heinaniemi | A cell type or a gene | Human | Gene expression data of 166 cell types and 2602 TFs | 3D epigenetic landscape, heatmap for the selected data feature (cell type or gene). Exploring and understanding the transcriptional regulation of cells | RNA-Seq | http://trel.systemsbiology.net/ |
| D’Alessio | Target cell type | Human | TFs expression for 233 tissue and cell types | List of putative fate altering TFs | Lists of candidate TFs available as a Supplementary table at Stem Cell Reports. | |
| Lang | Gene expression profile | Mouse | Gene expression profiles of 63 tissue/cell types and 1337 TFs | Identify candidate TFs for cell type conversion. | Landscape could be constructed with other genes, microRNAs, histone modification data | Online Supplementary Information |
| Crespo | Gene expression data from stable cellular phenotype and predefined GRN | Human, mouse | Published GRNs for T-helper differentiation, cell fate decisions during hematopoiesis, 24 genes in liver development | Hierarchically organized GRN, and predict core TFs for cell conversion/transdifferentiation. | ||
| Davis | Gene expression profile and chromatin profile | Human, mouse | 65 datasets (38 human and 27 mouse) | Identifies TFs for cell conversion/transdifferentiation |
Note: “Extensibility” refers to the ability to extend the tool to analyze new data types (for example extend a classifier trained on microarray data to understand RNA-Seq data), or to include new cell types not initially included in the tool. For consistency, we only include possibilities that are mentioned in the paper describing the method itself.
The central aim of ScoreCard is to predict the differentiation propensity of PSC lines. Bock et al. trained Scorecard initially on expression and DNA methylation data in ESC lines, reasoning that either inappropriate expression or DNA methylation of lineage-specific regulators could impede in vitro differentiation to certain lineages. They then documented the divergence of several iPS lines relative to ESCs in gene expression and DNA methylation at genes relevant to lineage differentiation to provide a reference table from which iPS lines can be selected for specific applications. To facilitate the prospective scoring of new ESC and iPS, Bock et al. also selected a set of 500 genes that mark each of the three germ layers as well as neural and hematopoietic lineages. They demonstrated that by monitoring the expression of these genes during undirected embryoid body (EB) differentiation it is possible to quantitatively evaluate the differentiation propensity of PSCs. For example, they confirmed that the HUES8 line was predisposed towards endoderm lineages while H1 and H9 lines exhibited the high propensity for neural lineage differentiation [25]. This platform has been subsequently improved by extending it to a more widely accessible expression technique: qPCR [26]. Importantly, no other current method attempts to predict the in vitro lineage bias of PSC lines.
Pluritest was introduced to assess the pluripotency of cells based on their gene expression profiles. In this study, the authors created a pluripotency-related gene expression database by curating publicly available gene expression data of hundreds of hESC and hiPSC lines, as well as samples representing non-pluripotent states. Then the authors used this data to create two classifiers: (1) a pluripotency probability score that distinguishes pluripotent from non-pluripotent classes based on logistic regression, and (2) a novelty score based on non-negative matrix factorization (NMF) that measures the extent of deviation from the pluripotent state. As a demonstration of these classifiers, Pluritest was applied to a neural differentiation time-course experiment and the pluripotency score remained high until three days of differentiation, after which it dropped substantially, whereas the novelty score concomitantly increased. Overall, Pluritest predicts pluripotency with both high degrees of sensitivity and specificity [27], and has proven informative in evaluating PSCs derived from diverse sources including chemically induced iPSCs and those derived from human amniotic fluid stem cells [28].
Teratoscore was designed for quantitatively assessing the differentiation potential of hPSCs in terms of gene expression pattern in teratomas. This algorithm was built on the theoretical basis that teratoma formation is one of the gold standards for evaluating hPSCs potency (the ability to differentiate to derivatives of all three germ layers), and Teratoscore also classifies whether a tumor originates from a specific tissue or from pluripotent cells [29]. The intended purpose of Teratoscore is to provide a quantitative metric in addition to the typical qualitative pathological assessment of germ layer contributions to teratomas, but it assumes and relies upon on unbiased sampling of the tumor. Future improvements to this type of tool could include adaptation to single cell molecular profiling data.
KeyGenes is a platform to evaluate tissue differentiation efficiency based on gene signatures of 21 different human fetal tissues generated by RNA-Seq or microarray at several developmental stages. KeyGenes was applied to publicly available data and new data including hPSCs differentiated to three germ lineages, tissue organoids, and human fetal and adult organs. KeyGenes was able to predict the tissue of origin of the samples and was able to identify stem cell derivatives with high accuracy. KeyGenes can also be used to assign developmental stages to differentiated hPSC derivatives [30].
CellNet was developed to assess and improve stem cell engineering paradigms using as its basis cell and tissue-specific GRNs [31]. CellNet takes as input gene expression profiles of directed differentiation or direct conversion (including reprogramming to pluripotency) and returns three outputs. First, it returns the probability that a sample is indistinguishable from each of the 20 cell and tissue types in the training data set in terms of its expression profile. Secondly, it returns an assessment of the extent to which a cell type specific GRN has been established in the query sample. Third, CellNet returns a scored list of transcription factors that are scored according to how important they are to the target cell type GRN, and how dysregulated they and their target genes are relative to the target cell type. This third output was validated by using it to identify TFs responsible for the incomplete erasure of the B-cell program in the direct conversion of pre-B cells to induced macrophages. Knocking down the expression of the two most highly scored factors, Pou2af1 and Ebf1, improved the in vitro functionality of the resulting induced macrophages [32]. CellNet is currently limited to microarray data, and its predictive ability is hampered as it is based on GRNs reconstructed largely from bulk tissue rather than homogenous populations of cells.
Heinaniemi et al. developed a novel method to identify transcriptional regulators that control lineage choice, which could be used to improve cell fate engineering efforts [33]. Their approach is based on the observation that mutually antagonist TFs frequently are also master regulators of sister lineages (e.g. the myeloid factor SPI1 inhibits the erythroid factor GATA1) [34], and this information can be used to score individual factors as contributors to specific cell types. Heinaniemi et al implemented an approach using a ‘reversal gene expression’ pattern to score 2,602 transcriptional regulators across 166 human cell types successfully recovering both known cell fate reprogramming factors and a host of new predictions.
To identify the TFs that serve as determinants of cell types, D’Alessio et al. searched for TFs characterized by a cell-type-specificity and a high expression level across a compendium of 233 human tissue and cell types [35]. The top scoring core TFs were presented as an atlas of factors that can be used as starting point to direct cell fate engineering. Importantly, the authors experimentally tested their predicted retinal progenitor cocktail of OTX2, SIX3, LHX2, PAX6, FOXD1, MITF, ZNF92, GLIS3, and SOX9 retinal pigment epithelial-like cells from fibroblasts was validated by their experimental evidence via ectopic expression of core TFs.
Rackham et al. specifically designed the Mogrify system to predict combinations of TFs that facilitate direction conversions between 173 human cell and tissues types [36]. Mogrify uses previously described regulatory and interaction networks to estimate the global expression changes that each TF might have when ectopically expressed in a specific starting cell type. By searching all TFs, it is possible to determine a set of TFs that most parsimoniously will up-regulate the target cell type expression program. Using this platform, the authors successfully predicted two novel TF transdifferentiation cocktails: dermal fibroblasts to keratinocytes and keratinocytes to microvascular endothelial cells.
Lang et al. modeled the epigenetic landscape of 63 cell and tissues types using expression data of 1,337 TFs based on a technique that has been applied previously to model neural networks [37]. By defining cell types as attractor states, the authors were able to use this model to predict key reprogramming TFs in both existing and novel reprogramming protocols.
Similar to Heinaniemi’s concept of pairs of TF with opposite function, Crespo et al. leveraged the concept of TF cross-repression in cell fate decision-making to define TFs that may play a key role in the induction of cell identity transitions. The approach of Crespo relies on predetermined GRNs, from which a regulatory hierarchy is derived that is used to find TFs with the highest putative impact on a desired fate transition, taking into consideration feedback loops that stabilize or lock in a cell fate [38]. The authors assessed this novel method in silico by three comparing their predictions to three transdifferentiation examples: from Th2 to Th1 T-helper lymphocytes, from myeloid to erythroid cells, and from fibroblasts to hepatocytes. The method was successful in finding experimentally demonstrated fate-altering TFs.
Davis et al. compared both the expression level and H3K27me3 mark of transcriptional repression of TFs that participate in cellular transdifferentiation, based on analysis of 65 published datasets (38 human, 27 mouse), to TFs that have not been proven to enable fate changes. They found transdifferentiation factors were more likely to be highly expressed in target cell types and marked by H3K27me3 in the source or starting cell types, providing another pattern by which candidate cell fate engineering transcription factors can be prioritized for experimental validation [39].
In this section, we have attempted to briefly describe all of the recently published computational approaches that use molecular profiling data to evaluate and improve cell fate engineering efforts. Because of the proliferation of methods, we are now in a position to draw some general conclusions and suggest areas where further research is needed. First, this nascent sub-field would benefit greatly from community-accepted ‘gold standards’ because they would allow for unbiased method comparison and they would enable method optimization. For example, ideally we would assess methods to predict conditions or factors that promote fate change by comparison to a gold standard consisting of experimentally verified sets of conditions/factors. Rackahm et al. evaluated Mogrify in this way but limited their analysis to positive controls (i.e. those TFs that have worked previously) and did not include negative controls [36]. Negative controls in this context would include TFs that had been tested but failed to enact a fate change, and would be highly useful in reducing the false positive rates of the algorithms.
Second, Scorecard is the only method that explicitly purports to define the lineage propensity of PSC lines. Developing new and improved methods to define lineage bias in PSC is a pressing problem for the thousands of labs that are using iPS to model diseases and it warrants more effort.
Third, most of the methods described in this section focus explicitly on transcriptomic and/or epigenetic information, but as we mentioned in the section entitled ‘The pluripotency gene regulatory network,’ the establishment and maintenance of cell identity are regulated at multiple layers. Analytical methods need to be extended to include these dimensions of cell state, including protein expression, post-translational modifications, and small RNA abundances, as they will provide a more accurate reflection of cell type identity. Finally, all of the methods above were based on data derived from tissues or bulk samples rather than single cells. This is problematic in that a primary goal of cell fate engineering is the derivation of homogenous populations of a specific cell type. Therefore, approaches such as CellNet or KeyGenes, or Teratoscore, which base their metrics of comparison upon bulk data, will be unable to achieve the level cellular resolution desired for cell fate engineering. For example, CellNet is able to classify samples as resembling ‘heart’ or ‘liver’ but does not have sufficient training data to classify ‘ventricular cardiomyocyte’ or ‘hepatocyte’. However, as discussed in the next section, new technologies have enabled single cell genome-wide molecular profiling, and thus the issue of cellular resolution should be addressable as sufficient data is generated in the near future.
Single cell OMICs
Although there has been a rapidly widening interest in using single cell profiling across a range of applications, from cancer heterogeneity [40] to the identification of new cell types [41], and a concomitant blossoming of reviews on the molecular techniques [42, 43], the computational side of appropriately handling this data has received relatively less attention. Here, we explore the types of questions that can be addressed with single cell OMICs, some of the analytical approaches that have been brought to bear thus far, and we end with a discussion of areas where analytics need to be improved to handle and take advantage of the idiosyncrasies of this data type.
Transcriptional heterogeneity in stem cells
Single cell profiling promises to provide a richer picture of the molecular basis of multi-lineage potential. The functional relevance of fluctuations in critical regulators of pluripotency has remained unclear since the documentation of NANOG variability [44]. One hypothesis is that stem cells regulate transcriptional heterogeneity in order to facilitate access to lineage differentiation upon exposure to appropriate signaling events [45, 46]. To explore this further, MacArthur et al. determined the single cell gene expression patterns of mESCs during transient Nanog down-regulation. SVM classification (see Box 2) separated these genes into pluripotent and lineage-primed classes. Genes in the latter class were up-regulated in response to Nanog loss, indicating that Nanog represses these lineage specifiers and supporting the hypothesis that regulated heterogeneity is a fundamental contributor to multi-lineage potential [47]. Similarly, Kumar et al. investigated the contribution of transcriptional heterogeneity to pluripotency by RNA-Seq of 283 single PSCs under different conditions, finding that the expression of signaling pathways and lineage specifiers was coupled to the down-regulation of pluripotency factors [48].
Population sub-structure in stem cells
Single cell OMICs is especially valuable in stem cell investigations because they can be used to further refine the sub-populations that contain stem properties. For instance, van Wolfswinkel et al. discovered that a nominally homogenous population of regenerative planarian progenitors could be further fractionated to subsets with different regenerative capacities based on single-cell transcriptional profiles [49]. Similarly, by analyzing the expression profiles of 704 single mESCs by PCA and gene set analysis (see Box 1), Kolodziejczyk et al identified three distinct sub-populations: a ground state, a primed state, and a state comprised of cells that had initiated differentiation [50].
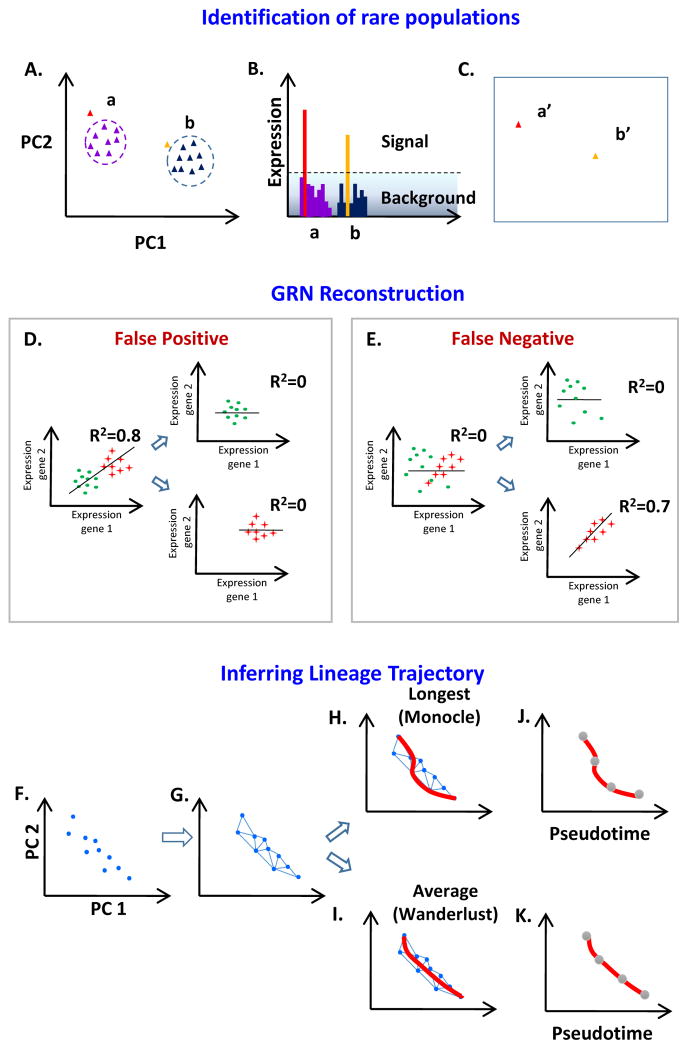
A practical concern for using single cell RNA-Seq is the total number of cells required to robustly detect rare populations. Grun et al. developed an algorithm for rare cell type identification (RaceID) based on single cell mRNA sequencing. By combining HCL and k-means clustering (see Box 1) with t-distributed stochastic neighbor embedding (t-SNE), they were able to distinguish biological and technical heterogeneity from the occurrence of rare cells, thereby identifying rare cell types within a population of intestinal cells. By applying this method to organoid-derived intestinal crypts, the authors were able to discover rare Paneth cells within the Lgr5-positive population of intestinal stem cells. This novel method will also be useful to discriminate adult stem cell types in different states such as healthy and diseased situations [51] (Figure 2A–C). As the number of single cells that can be profiled simultaneously increases, our power to detect rare sub-populations will also increase, as will our ability to define sub-states and sub-types with finer resolution.
Figure 2. Single cell molecular profiling to identify rare populations, to reconstruct gene regulatory networks, and to define lineage trajectories.
(A–C) Distinguishing expression heterogeneity from population sub-structure. (A) An example of applying dimension reduction, in this case PCA, to single cell RNA-Seq data comprised of two major cell types (a and b). The analytical task is to determine whether two (red and light blue) single cells should be considered as distinct, rare cell types or are more likely to represent statistical outliers. (B) A background model of transcriptional noise (based on the average transcript count variance across all cells) is used to weigh the relative likelihood that the outlier cells belong to one of the predetermined clusters or represent a distinct class. (C) Identification of distinct type of cells (a′ and b′). (D–E) GRN reconstruction. Reconstructing GRNs from expression profiling (either array or RNA-Seq) is based on correlations in expression between genes. Expression profiling from bulk samples can stymie this type of analysis by producing both false positives and false negatives. (D) False positives can occur when a correlation is a result of population sub-structure rather than because of a regulatory relationship. (E) Similarly population sub-structure can mask regulatory relationships present in on sub-population. (F–K) Inferring lineage trajectory. Steps common to lineage trajectory inference algorithms include: (F) reducing the dimensionality of the data (every point represents a single cell expression profile), and (G) finding a minimal spanning tree. Methods differ in how they assign a path through the minimal spanning tree to lineage progression, with Monocle using the longest path (H) and Wanderlust using an average of all possible paths (I). (J, K) The resulting path is referred to as pseudotime, and can be used to order cells in a temporal progression.
Construction of novel gene regulatory networks
Single cell analysis makes it possible to infer regulatory relationships between genes that were obscured in bulk data (Figure 2D and E). For example, Moignard et al. used single-cell gene expression analysis of 597 mouse hematopoietic stem and progenitor cells to identify a putative regulatory relationship among the transcription factors Gata2, Gfi1 and Gfi1b. The predicted repression of Gata2 by GFI1 was validated by a combination of ChIP-Seq, luciferase reporter assays, and transgenic reporter analysis, demonstrating that high-throughput single cell gene expression analysis is sufficiently powerful to identify of novel regulatory networks despite the overall low sensitivity of single cell methods [52]. Similarly, Klein et al. developed and used the inDrop RNA-Seq technology to profile 935 single mESCs, and discovered novel regulatory relationships between Nanog, Sox2 and Cyclin B [53]. More generally, the ability to sample thousands of individual cells may address several seminal issues in reconstructing gene regulatory networks, including the confounding effects of population substructure (Figure 2D–E) and the reliance on non-physiological perturbations to elicit correlated changes in gene expression.
Lineage trajectories
Examining cell states through development or in time course experiments are especially informative because they can help to reconstruct lineage hierarchies. As a proof of principle, Guo et al. determined the gene expression pattern of cell surface markers in single mouse hematopoietic cells and used it to reconstruct the differentiation hierarchy of hematopoiesis [54]. Similarly, Bendall et al. introduced a new algorithm termed Wanderlust, in which machine learning methods were used to reconstruct the sequence of cell states (or lineage trajectory) as hematopoietic stem cells (HSCs) differentiate to B-cells based on mass cytometry data [55] (Figure 2F–G, I, K).
Determining the lineage trajectory de novo will be especially useful to identify upstream regulators of developmental processes. Trapnell et al. developed the Monocle algorithm that places cells profiled with single cell RNA-Seq along a differentiation trajectory [56]. By applying this technique to the differentiation of primary human myoblasts, the authors were able to identify and validate eight novel regulators of skeletal muscle differentiation. Similarly, Shin et al. developed another 15ioinformatics workflow to approximate lineage trajectories. By applying it to single cell RNA-Seq of neurogenesis, they identified multiple novel regulators of adult neurogenesis such as the homeobox protein Dbx2, which previously had only been appreciated in embryonic neurogenesis [57].
The temporal resolution of cell state transitions during development promises to reveal how cell type specific gene regulatory networks are established. This information can be used as a foundation to reformulate algorithms to address the ‘Improvement problem’ (Figure 1A), and is likely to reveal regulatory circuits that appear only transiently during ontogeny and thus would be missed by algorithms based on compendiums of adult or even late fetal cell types. Ultimately, when sufficient data has accumulated, it will be possible to analyze the finding across lineages to devise rules that govern the establishment of cell type specific GRNs during development.
Spatially-resolved single cell genomics
The fluorescent in situ RNA sequencing (FISSEQ) described by Lee et al. quantitates hundreds to thousands of RNAs in situ in fixed cells, in tissue sections, and in whole-mount embryos. Although FISSEQ is less sensitive than single cell RNA-seq, it can still detect highly expressed, functionally relevant transcripts. Approaches such as FISSEQ will be invaluable to identify cell types in their natural environment as well as to investigate how the niche contributes to stem cell state in situ [58, 59]. Another approach to profile single cells in situ is based on adding an engineered molecule (the ‘TIVA tag’) into the cell membrane that can be photo-activated for mRNA capture. When this new technique was combined with RNA-Seq, it enabled the quantitation of mRNA in live single cells in vivo [60]. More generally, spatially resolved single cell RNA-Seq data will enable the investigation of how niche composition contributes to the maintenance of cell identity, and thus will also enhance the development of improved algorithms to address the ‘Improvement problem’.
Conclusion
The emergence of ‘computational stem cell biology’ resembles the transition from cancer biology to cancer genomics, where ‘Big Data’, especially whole genome sequences, required the recruitment and training of computationalists specifically focused on cancer biology. This trend is having significant consequences: it has transformed of our understanding of the initiation and progression of cancer and it has lead to the development of novel therapeutics. Stem cell biology now faces a similar inflection point. With the emergence of both ‘do-it-yourself’ and commercial single cell platforms, the field is facing a flood of single cell OMICs data [61, 62]. We propose that the sub-discipline of computational stem cell biology will yield transformative insights into developmental biology and will enhance our ability to engineer cell fate with fidelity and efficiency by synthesizing single cell data with predictive modeling approaches. Yet there are many open challenges (see Outstanding Questions Box). We anticipate that computational stem cell biology will produce a quantitative basis to define cell type identity, and will define the molecular logic of lineage commitment and maturation, and thereby will form the foundation to understand how genetic and epigenetic abnormalities disrupt healthy development and contribute to disease states.
Outstanding box.
What data and analysis will be needed to develop a quantitative definition of ‘cell type’?
Will gene regulatory networks and accurate molecular profiling enable quantitative models that predict population level behavior of stem cells?
Can the field produce methods to predict how a stem cell line will differentiate in vitro based on molecular data alone?
What are the limits in terms of noise and volume of data that will be needed to incorporate methods from physics to infer causality based on temporal information?
What analytical methods will prove most efficient and informative in integrating spatial information with single cell expression data?
What standards, experimental designs, and analytical methods will be required to enable cross-study comparison of tools designed to assess and improve cell fate engineering methods?
What standards, experimental designs, and analytical methods will be required to enable cross-study comparison of single cell OMICs?
What are the most appropriate experimental techniques to assess the robustness and precision of pseudotime analyses?
Trends box.
High-throughput data molecular profiling, mainly based on nucleic acid sequencing (e.g. RNA-Seq), but increasingly other modalities such as metabolomics and proteomics, has necessitated the development of sophisticated analysis algorithms.
The combination of OMICs and targeted analytics has enabled seminal observations in stem cell biology.
Computational stem cell biology has emerged as its own sub-discipline that is concerned with synthesizing the modeling of systems-level aspects of stem cells with large-scale molecular data.
Single cell genomics is poised to transform stem cell biology by identifying new cell types; by clarifying the relationship between transcriptional noise, lineage priming, and lineage potential; and by enabling a higher resolution dissection of genetic circuits underlying commitment and differentiation.
Glossary
- Chip-Chip
Chromatin immunoprecipitation (ChIP) combined with DNA microarray-based profiling to characterize the interactions of protein with genomic DNA.
- Chip-Seq
Genome-wide profiling of specific protein interactions with genomic DNA by combining ChIP with next-generation sequencing [94].
- CLIP-Seq/HITS-CLIP
Combination of UV cross-linking and immunoprecipitation with high-throughput sequencing to identify binding sites of RNA-binding proteins such as LIN28A, Argonaute (Ago), Puf5p [11, 92, 95]
- Differentially methylated genomic regions (DMRs)
typically identified on a large scale by bisulfite sequencing or CHARM.
- DNase hypersensitivity (DHS) mapping
A high-throughput sequencing method designed to infer regulatory elements including promoters, enhancers, silencers, insulators, and locus control regions by identifying genomic sequences that are accessible to DNA nucleases and thus not occluded by nucleosomes.
- ESCs
Embryonic stem cells are PSCs derived from the inner cell mass of the blastocyst.
- Hematopoietic stem cells (HSCs)
adult stem cells that are capable of reconstituting the complete hematopoietic system of immune system-ablated recipients.
- LncRNAs
Poly-adenylated, non-coding transcripts longer than 200 nucleotides initially discovered by the epigenetic profile of the corresponding genomic sequence (histone modifications reflective of transcriptionally active regions) [73].
- MeRIP-Seq
Method to define the N6-methyladenosine (m6A) post-transcriptional modification of mRNA by combining methylated RNA immunoprecipitation with high-throughput sequencing [96, 97].
- MicroRNAs
Small non-coding RNAs of 21–25 nucleotides first discovered in Caenorhabditis elegans that regulate expression by binding to complementary seed sequences and modulating translation and/or promoting mRNA degradation [98, 99].
- MS
Mass spectrometry determines the molecular signature of each peptide in a sample by ionizing and fragmenting proteins calculating their mass-to-charge ratios.
- OMICs
Techniques that generate nearly comprehensive data of a particular molecular type. For example, proteomics quantifies the proteome, functional genomics measures gene expression, and metabolomics measures concentrations of metabolic reaction products and intermediates.
- Pluripotent stem cells (PSCs)
Cells that can give rise to all of the cell types of an adult organism. Under appropriate conditions, PCSs will self-renew indefinitely and maintain their pluripotency.
- Pseudotime
A theoretical progression or timeline rather than a strictly temporal sequence of events. The intuition behind pseudotime is that there will be heterogeneity within a population in terms of how far along in a process a particular cell has traversed. Progressions include developmental (specification, differentiation, maturation), cyclical (circadian rhythm), and pathological (tumorigenesis, metastasis).
- Ribo-Seq
The translational correlate of RNA-Seq provides a read-out of active translation by preferentially sequencing ribosome-bound/protected RNA [84].
- RIP-Seq
like CLIP-Seq/HITS-CLIP, a technique meant to identify RNA that is bound by RNA-binding proteins. However, the RNA-binding sites identified are broader than in CLIP-Seq because RIP-Seq is based on RNA immunoprecipitation coupled to reverse transcription followed by high-throughput sequencing.
- Transcription factors (TFs)
proteins that bind to specific DNA sequences (transcription factor binding sites, TFBSs) to promote or inhibit recruitment and activation of the transcriptional machinery.
- Transdifferentiation
The conversion of one somatic cell type into another somatic cell type without transiting through a pluripotent cell state. Also known as direct conversion.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Turing AM. The Chemical Basis of Morphogenesis. Philosophical transactions of the Royal Society of London. Series B, Biological sciences. 1952;237:37–72. doi: 10.1098/rstb.2014.0218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lander AD. Morpheus unbound: reimagining the morphogen gradient. Cell. 2007;128:245–256. doi: 10.1016/j.cell.2007.01.004. [DOI] [PubMed] [Google Scholar]
- 3.Till JE, et al. A Stochastic Model of Stem Cell Proliferation, Based on the Growth of Spleen Colony-Forming Cells. Proceedings of the National Academy of Sciences of the United States of America. 1964;51:29–36. doi: 10.1073/pnas.51.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lander ES, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 5.Waterston RH, et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 6.Boyer LA, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Loh YH, et al. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nature genetics. 2006;38:431–440. doi: 10.1038/ng1760. [DOI] [PubMed] [Google Scholar]
- 8.Thurman RE, et al. The accessible chromatin landscape of the human genome. Nature. 2012;489:75–82. doi: 10.1038/nature11232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ingolia NT, et al. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell. 2011;147:789–802. doi: 10.1016/j.cell.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang L, et al. The THO complex regulates pluripotency gene mRNA export and controls embryonic stem cell self-renewal and somatic cell reprogramming. Cell stem cell. 2013;13:676–690. doi: 10.1016/j.stem.2013.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cho J, et al. LIN28A is a suppressor of ER-associated translation in embryonic stem cells. Cell. 2012;151:765–777. doi: 10.1016/j.cell.2012.10.019. [DOI] [PubMed] [Google Scholar]
- 12.Batista PJ, et al. m(6)A RNA modification controls cell fate transition in mammalian embryonic stem cells. Cell stem cell. 2014;15:707–719. doi: 10.1016/j.stem.2014.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aguilo F, et al. Coordination of m(6)A mRNA Methylation and Gene Transcription by ZFP217 Regulates Pluripotency and Reprogramming. Cell stem cell. 2015;17:689–704. doi: 10.1016/j.stem.2015.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 15.Takahashi K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 16.Irizarry RA, et al. Comprehensive high-throughput arrays for relative methylation (CHARM) Genome research. 2008;18:780–790. doi: 10.1101/gr.7301508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim K, et al. Epigenetic memory in induced pluripotent stem cells. Nature. 2010;467:285–290. doi: 10.1038/nature09342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim K, et al. Donor cell type can influence the epigenome and differentiation potential of human induced pluripotent stem cells. Nature biotechnology. 2011;29:1117–1119. doi: 10.1038/nbt.2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Polo JM, et al. Cell type of origin influences the molecular and functional properties of mouse induced pluripotent stem cells. Nature biotechnology. 2010;28:848–855. doi: 10.1038/nbt.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mikkelsen TS, et al. Dissecting direct reprogramming through integrative genomic analysis. Nature. 2008;454:49–55. doi: 10.1038/nature07056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qian L, et al. Reprogramming of mouse fibroblasts into cardiomyocyte-like cells in vitro. Nature protocols. 2013;8:1204–1215. doi: 10.1038/nprot.2013.067. [DOI] [PubMed] [Google Scholar]
- 22.Davidson EH, Erwin DH. Gene regulatory networks and the evolution of animal body plans. Science. 2006;311:796–800. doi: 10.1126/science.1113832. [DOI] [PubMed] [Google Scholar]
- 23.Osafune K, et al. Marked differences in differentiation propensity among human embryonic stem cell lines. Nature biotechnology. 2008;26:313–315. doi: 10.1038/nbt1383. [DOI] [PubMed] [Google Scholar]
- 24.Cahan P, Daley GQ. Origins and implications of pluripotent stem cell variability and heterogeneity. Nature reviews Molecular cell biology. 2013;14:357–368. doi: 10.1038/nrm3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bock C, et al. Reference Maps of human ES and iPS cell variation enable high-throughput characterization of pluripotent cell lines. Cell. 2011;144:439–452. doi: 10.1016/j.cell.2010.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tsankov AM, et al. A qPCR ScoreCard quantifies the differentiation potential of human pluripotent stem cells. Nature biotechnology. 2015;33:1182–1192. doi: 10.1038/nbt.3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Muller FJ, et al. A bioinformatic assay for pluripotency in human cells. Nature methods. 2011;8:315–317. doi: 10.1038/nmeth.1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Slamecka J, et al. Non-integrating episomal plasmid-based reprogramming of human amniotic fluid stem cells into induced pluripotent stem cells in chemically defined conditions. Cell Cycle. 2015 doi: 10.1080/15384101.2015.1121332. 0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Avior Y, et al. TeratoScore: Assessing the Differentiation Potential of Human Pluripotent Stem Cells by Quantitative Expression Analysis of Teratomas. Stem cell reports. 2015;4:967–974. doi: 10.1016/j.stemcr.2015.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roost MS, et al. KeyGenes, a Tool to Probe Tissue Differentiation Using a Human Fetal Transcriptional Atlas. Stem cell reports. 2015;4:1112–1124. doi: 10.1016/j.stemcr.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cahan P, et al. CellNet: network biology applied to stem cell engineering. Cell. 2014;158:903–915. doi: 10.1016/j.cell.2014.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morris SA, et al. Dissecting engineered cell types and enhancing cell fate conversion via CellNet. Cell. 2014;158:889–902. doi: 10.1016/j.cell.2014.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heinaniemi M, et al. Gene-pair expression signatures reveal lineage control. Nature methods. 2013;10:577–583. doi: 10.1038/nmeth.2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang P, et al. Negative cross-talk between hematopoietic regulators: GATA proteins repress PU.1. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:8705–8710. doi: 10.1073/pnas.96.15.8705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.D’Alessio AC, et al. A Systematic Approach to Identify Candidate Transcription Factors that Control Cell Identity. Stem cell reports. 2015 doi: 10.1016/j.stemcr.2015.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rackham OJ, et al. A predictive computational framework for direct reprogramming between human cell types. Nature genetics. 2016 doi: 10.1038/ng.3487. [DOI] [PubMed] [Google Scholar]
- 37.Lang AH, et al. Epigenetic landscapes explain partially reprogrammed cells and identify key reprogramming genes. PLoS computational biology. 2014;10:e1003734. doi: 10.1371/journal.pcbi.1003734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Crespo I, Del Sol A. A general strategy for cellular reprogramming: the importance of transcription factor cross-repression. Stem Cells. 2013;31:2127–2135. doi: 10.1002/stem.1473. [DOI] [PubMed] [Google Scholar]
- 39.Davis FP, Eddy SR. Transcription factors that convert adult cell identity are differentially polycomb repressed. PloS one. 2013;8:e63407. doi: 10.1371/journal.pone.0063407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Patel AP, et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science. 2014;344:1396–1401. doi: 10.1126/science.1254257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Treutlein B, et al. Reconstructing lineage hierarchies of the distal lung epithelium using single-cell RNA-seq. Nature. 2014;509:371–375. doi: 10.1038/nature13173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Macaulay IC, Voet T. Single cell genomics: advances and future perspectives. PLoS genetics. 2014;10:e1004126. doi: 10.1371/journal.pgen.1004126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Etzrodt M, et al. Quantitative single-cell approaches to stem cell research. Cell stem cell. 2014;15:546–558. doi: 10.1016/j.stem.2014.10.015. [DOI] [PubMed] [Google Scholar]
- 44.Chambers I, et al. Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell. 2003;113:643–655. doi: 10.1016/s0092-8674(03)00392-1. [DOI] [PubMed] [Google Scholar]
- 45.MacArthur BD, Lemischka IR. Statistical mechanics of pluripotency. Cell. 2013;154:484–489. doi: 10.1016/j.cell.2013.07.024. [DOI] [PubMed] [Google Scholar]
- 46.Graf T, Stadtfeld M. Heterogeneity of embryonic and adult stem cells. Cell stem cell. 2008;3:480–483. doi: 10.1016/j.stem.2008.10.007. [DOI] [PubMed] [Google Scholar]
- 47.MacArthur BD, et al. Nanog-dependent feedback loops regulate murine embryonic stem cell heterogeneity. Nature cell biology. 2012;14:1139–1147. doi: 10.1038/ncb2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kumar RM, et al. Deconstructing transcriptional heterogeneity in pluripotent stem cells. Nature. 2014;516:56–61. doi: 10.1038/nature13920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.van Wolfswinkel JC, et al. Single-cell analysis reveals functionally distinct classes within the planarian stem cell compartment. Cell stem cell. 2014;15:326–339. doi: 10.1016/j.stem.2014.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kolodziejczyk AA, et al. Single Cell RNA-Sequencing of Pluripotent States Unlocks Modular Transcriptional Variation. Cell stem cell. 2015;17:471–485. doi: 10.1016/j.stem.2015.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Grun D, et al. Single-cell messenger RNA sequencing reveals rare intestinal cell types. Nature. 2015;525:251–255. doi: 10.1038/nature14966. [DOI] [PubMed] [Google Scholar]
- 52.Moignard V, et al. Characterization of transcriptional networks in blood stem and progenitor cells using high-throughput single-cell gene expression analysis. Nature cell biology. 2013;15:363–372. doi: 10.1038/ncb2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Klein AM, et al. Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell. 2015;161:1187–1201. doi: 10.1016/j.cell.2015.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Guo G, et al. Mapping cellular hierarchy by single-cell analysis of the cell surface repertoire. Cell stem cell. 2013;13:492–505. doi: 10.1016/j.stem.2013.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bendall SC, et al. Single-cell trajectory detection uncovers progression and regulatory coordination in human B cell development. Cell. 2014;157:714–725. doi: 10.1016/j.cell.2014.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Trapnell C, et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nature biotechnology. 2014;32:381–386. doi: 10.1038/nbt.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shin J, et al. Single-Cell RNA-Seq with Waterfall Reveals Molecular Cascades underlying Adult Neurogenesis. Cell stem cell. 2015;17:360–372. doi: 10.1016/j.stem.2015.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lee JH, et al. Highly multiplexed subcellular RNA sequencing in situ. Science. 2014;343:1360–1363. doi: 10.1126/science.1250212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lee JH, et al. Fluorescent in situ sequencing (FISSEQ) of RNA for gene expression profiling in intact cells and tissues. Nature protocols. 2015;10:442–458. doi: 10.1038/nprot.2014.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lovatt D, et al. Transcriptome in vivo analysis (TIVA) of spatially defined single cells in live tissue. Nature methods. 2014;11:190–196. doi: 10.1038/nmeth.2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Macosko EZ, et al. Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell. 2015;161:1202–1214. doi: 10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pollen AA, et al. Low-coverage single-cell mRNA sequencing reveals cellular heterogeneity and activated signaling pathways in developing cerebral cortex. Nature biotechnology. 2014;32:1053–1058. doi: 10.1038/nbt.2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Josephson R, et al. A molecular scheme for improved characterization of human embryonic stem cell lines. BMC biology. 2006;4:28. doi: 10.1186/1741-7007-4-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Abyzov A, et al. Somatic copy number mosaicism in human skin revealed by induced pluripotent stem cells. Nature. 2012;492:438–442. doi: 10.1038/nature11629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bhattacharya B, et al. Gene expression in human embryonic stem cell lines: unique molecular signature. Blood. 2004;103:2956–2964. doi: 10.1182/blood-2003-09-3314. [DOI] [PubMed] [Google Scholar]
- 66.Laurent LC, et al. Comprehensive microRNA profiling reveals a unique human embryonic stem cell signature dominated by a single seed sequence. Stem Cells. 2008;26:1506–1516. doi: 10.1634/stemcells.2007-1081. [DOI] [PubMed] [Google Scholar]
- 67.Guttman M, et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477:295–300. doi: 10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gore A, et al. Somatic coding mutations in human induced pluripotent stem cells. Nature. 2011;471:63–67. doi: 10.1038/nature09805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Quinlan AR, et al. Genome sequencing of mouse induced pluripotent stem cells reveals retroelement stability and infrequent DNA rearrangement during reprogramming. Cell stem cell. 2011;9:366–373. doi: 10.1016/j.stem.2011.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wilhelm BT, et al. RNA-seq analysis of 2 closely related leukemia clones that differ in their self-renewal capacity. Blood. 2011;117:e27–38. doi: 10.1182/blood-2010-07-293332. [DOI] [PubMed] [Google Scholar]
- 71.Gifford CA, et al. Transcriptional and epigenetic dynamics during specification of human embryonic stem cells. Cell. 2013;153:1149–1163. doi: 10.1016/j.cell.2013.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Clancy JL, et al. Small RNA changes en route to distinct cellular states of induced pluripotency. Nature communications. 2014;5:5522. doi: 10.1038/ncomms6522. [DOI] [PubMed] [Google Scholar]
- 73.Guttman M, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mikkelsen TS, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Klauke K, et al. Polycomb Cbx family members mediate the balance between haematopoietic stem cell self-renewal and differentiation. Nature cell biology. 2013;15:353–362. doi: 10.1038/ncb2701. [DOI] [PubMed] [Google Scholar]
- 76.Mathur D, et al. Analysis of the mouse embryonic stem cell regulatory networks obtained by ChIP-chip and ChIP-PET. Genome biology. 2008;9:R126. doi: 10.1186/gb-2008-9-8-r126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kwon SC, et al. The RNA-binding protein repertoire of embryonic stem cells. Nature structural & molecular biology. 2013;20:1122–1130. doi: 10.1038/nsmb.2638. [DOI] [PubMed] [Google Scholar]
- 78.Balakrishnan I, et al. Genome-wide analysis of miRNA-mRNA interactions in marrow stromal cells. Stem Cells. 2014;32:662–673. doi: 10.1002/stem.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Van Wynsberghe PM, et al. LIN-28 co-transcriptionally binds primary let-7 to regulate miRNA maturation in Caenorhabditis elegans. Nature structural & molecular biology. 2011;18:302–308. doi: 10.1038/nsmb.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lienert F, et al. Genomic prevalence of heterochromatic H3K9me2 and transcription do not discriminate pluripotent from terminally differentiated cells. PLoS genetics. 2011;7:e1002090. doi: 10.1371/journal.pgen.1002090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lister R, et al. Hotspots of aberrant epigenomic reprogramming in human induced pluripotent stem cells. Nature. 2011;471:68–73. doi: 10.1038/nature09798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Baharvand H, et al. Proteomic signature of human embryonic stem cells. Proteomics. 2006;6:3544–3549. doi: 10.1002/pmic.200500844. [DOI] [PubMed] [Google Scholar]
- 83.Kratchmarova I, et al. Mechanism of divergent growth factor effects in mesenchymal stem cell differentiation. Science. 2005;308:1472–1477. doi: 10.1126/science.1107627. [DOI] [PubMed] [Google Scholar]
- 84.Ingolia NT, et al. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324:218–223. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Grow EJ, et al. Intrinsic retroviral reactivation in human preimplantation embryos and pluripotent cells. Nature. 2015;522:221–225. doi: 10.1038/nature14308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zheng Y, et al. Generation and characterization of yeast two-hybrid cDNA libraries derived from two distinct mouse pluripotent cell types. Molecular biotechnology. 2013;54:228–237. doi: 10.1007/s12033-012-9561-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Vierstra J, et al. Mouse regulatory DNA landscapes reveal global principles of cis-regulatory evolution. Science. 2014;346:1007–1012. doi: 10.1126/science.1246426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.John S, et al. Chromatin accessibility pre-determines glucocorticoid receptor binding patterns. Nature genetics. 2011;43:264–268. doi: 10.1038/ng.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Boyle AP, et al. F-Seq: a feature density estimator for high-throughput sequence tags. Bioinformatics. 2008;24:2537–2538. doi: 10.1093/bioinformatics/btn480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Feng J, et al. Identifying ChIP-seq enrichment using MACS. Nature protocols. 2012;7:1728–1740. doi: 10.1038/nprot.2012.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wilinski D, et al. RNA regulatory networks diversified through curvature of the PUF protein scaffold. Nature communications. 2015;6:8213. doi: 10.1038/ncomms9213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kumar V, et al. Uniform, optimal signal processing of mapped deep-sequencing data. Nature biotechnology. 2013;31:615–622. doi: 10.1038/nbt.2596. [DOI] [PubMed] [Google Scholar]
- 94.Johnson DS, et al. Genome-wide mapping of in vivo protein-DNA interactions. Science. 2007;316:1497–1502. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- 95.Leung AK, et al. Genome-wide identification of Ago2 binding sites from mouse embryonic stem cells with and without mature microRNAs. Nature structural & molecular biology. 2011;18:237–244. doi: 10.1038/nsmb.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Meyer KD, et al. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell. 2012;149:1635–1646. doi: 10.1016/j.cell.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Dominissini D, et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485:201–206. doi: 10.1038/nature11112. [DOI] [PubMed] [Google Scholar]
- 98.Lee RC, et al. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 99.Wightman B, et al. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]