Abstract
Mitochondria are ancient organelles that are thought to have emerged from once free-living α-proto-bacteria. As such, they still possess several bacterial-like qualities, including a semi-autonomous genetic system, complete with an independent genome and a unique genetic code. The bacterial-like circular mitochondrial DNA (mtDNA) has been described to encode 37 genes, including 22 tRNAs, 2 rRNAs, and 13 mRNAs. Two additional peptides reported to originate from the mtDNA, namely humanin (Hashimoto et al., 2001; Ikone et al., 2003; Guo et al., 2003) [1–3] and MOTS-c (mitochondrial ORF of the twelve S c) (Lee et al., 2015) [4], indicate a larger mitochondrial genetic repertoire (Shokolenko and Alexeyev, 2015) [5]. These mitochondrial-derived peptides (MDPs) have profound and distinct biological activities and provide a paradigm-shifting concept of active mitochondrial-encoded signals that act at the cellular and organismal level (i.e. mitochondrial hormone) (da Cunha et al., 2015; Quiros et al., 2016) [6,7]. Considering that mitochondria are the single most important metabolic organelle, it is not surprising that these MDPs have metabolic actions. MOTS-c has been shown to target the skeletal muscle and enhance glucose metabolism. As such, MOTS-c has implications in the regulation of obesity, diabetes, exercise, and longevity, representing an entirely novel mitochondrial signaling mechanism to regulate metabolism within and between cells.
Keywords: Mitochondrial-derived peptides, MOTS-c, Muscle, Fat, Aging, Exercise, Insulin, Metabolism
1. Introduction
Mitochondria are ancient organelles that are thought to have emerged from once free-living α-proto-bacteria. As such, they still possess several bacterial-like qualities, including a semi-autonomous genetic system, complete with an independent genome and a unique genetic code. The bacterial-like circular mitochondrial DNA (mtDNA) has been described to encode 37 genes, including 22 tRNAs, 2 rRNAs, and 13 mRNAs. Two additional peptides reported to originate from the mtDNA, namely humanin [1–3] and MOTS-c (mitochondrial ORF of the twelve S c) [4], indicate a larger mitochondrial genetic repertoire [5]. These mitochondrial-derived peptides (MDPs) have profound and distinct biological activities and provide a paradigm-shifting concept of active mitochondrial-encoded signals that act at the cellular and organismal level (i.e. mitochondrial hormone) [6,7]. Considering that mitochondria are the single most important metabolic organelle, it is not surprising that these MDPs have metabolic actions. MOTS-c has been shown to target the skeletal muscle and enhance glucose metabolism. As such, MOTS-c has implications in the regulation of obesity, diabetes, exercise, and longevity, representing an entirely novel mitochondrial signaling mechanism to regulate metabolism within and between cells.
2. Mitochondrial signaling
Mitochondria regulate metabolism, growth, and apoptosis, which are vital cellular processes that have direct impact on survival. Thus, to avoid catastrophic incoordination, a sophisticated communication system between mitochondria and the cell is inevitable [7–12]. Such communication has been perceived to be predominantly, if not entirely, driven by signals encoded in the nuclear DNA (nDNA), whereas mitochondria act as functional organelles on the receiving end. However, mitochondrial communication likely co-evolved to effectively coordinate the eukaryotic system that became increasingly complex, and emerging evidence now position them as signaling units that actively regulate various biological processes [10,13]. In fact, intergenomic coordination and compatibility is evidently important. Studies in MNX (mitochon-drial–nuclear exchange) mice support the importance of mtDNA and nDNA compatibility in determining cellular and whole-body physiology, and perhaps even healthspan and lifespan [14]. The MNX mouse is a model of mtDNA polymorphism, in which the nuclear genome from one mouse strain is transferred into an enucleated recipient zygote of a different mouse strain, thus allowing each progeny to distinctly have 100% of the nDNA and mtDNA from their respective donor strains (C57BL/6J and C3H/HeN) [15]. For example, MNX mice have the nuclear genome of a C57BL/6J (C57n) or a C3H/HeN (C3Hn) mouse, and the mitochondrial gen-ome of a C3H/HeN (C3Hmt) or C57BL/6J (C57mt) mouse, respectively. Although MNX mice are generally healthy and fertile, but, depending on the mtDNA–nuclear DNA combination, exhibit different cellular bioenergetics, mitochondrial ROS generation and susceptibility to cardiac stress and atherogenic diet [15,16]. This has direct impact on human health. The United Kingdom recently approved mitochondrial replacement therapy, which would allow a woman with a mitochondrial disorder to pair her nuclear DNA with the healthy mitochondria from a donor’s egg (also referred to as three-parent babies). This has been met with concerns of mtDNA-nDNA incompatibility and dysregulation of signals encoded in the mtDNA (i.e., MDPs) [17].
Notably, some mitochondrial-derived signals are capable of acting in a non-cell-autonomous manner to regulate distal tissues. For instance, an interesting study in Caenorhabditis elegans showed that an imbalance between mitochondrial- and nuclear-encoded proteins in neurons activates the mitochondrial unfolded protein response (UPRmt) in intestinal cells, through an unidentified mi-tokine [18,19]. In addition, as discussed in detail below, peptides derived from mtDNA have hormonal actions that regulate whole-body physiology.
3. Mitochondrial-derived peptides (MDPs): a newly recognized role for an ancient organelle
3.1. mtDNA and short open reading frames (sORFs)
The mitochondrial genome was discovered approximately 50 years ago by electron microscopy and described as a bacterial-like circular DNA [20,21], and thought to encode for 13 proteins that are all components of the electron transport chain. However, emerging studies indicate that the mtDNA contains previously unknown short open reading frames (sORFs), expanding the mi-tochondrial genetic repertoire [1,4,22–24]. Notably, in various species, including humans, sORFs have been “genetic blind-spots” [23], largely due to the traditional computational cut-off of a 100 codons to avoid false-positives [25]. sORF-encoded polypeptides (SEPs) originating from the nuclear genome have significant biological activity.
3.2. Humanin and small humanin-like peptides (SHLPs)
Humanin was the first sORF to be identified in the mtDNA [1,3,26]. It is a conserved polypeptide [3] encoded as a 75-bp polycistronic sORF within the 16S rRNA, discovered in 2001 from an unbiased functional survival screen using a cDNA library created from the surviving brain fraction of an Alzheimer’s disease (AD) patient [1,27]. It was named by its discoverer with hopes that could it would recover the “humanity” of AD patients. Humanin expression is age-dependent [28,29] and detected in various tissues and in circulation in rodents and humans [1,30]. Humanin was simultaneously cloned by two other groups, who showed that it binds to the insulin-like growth factor binding protein 3 (IGFBP-3), which regulates IGF-1 bioactivity, with high affinity and specificity [2] and to the pro-apoptotic protein BAX [3]. The primary role of humanin, based on a wide range of in vitro and in vivo studies, appears to be enhancing cellular and organismal protection from various types of stress or disease states, including AD, cancer, and diabetes [26,31]. Recently, six additional peptides in the same region of mtDNA as humanin (i.e. 16S rRNA) were identified using an in silico approach, and have been named small humanin-like peptides (SHLPs) 1–6 [22]. Notably, SHLPs 2 and 3 significantly reduced staurosporine-dependent apoptosis and the generation of reactive oxygen species, while increasing mi-tochondrial respiration and ATP production in vitro [22]. SHLP2 and SHLP3 also enhanced 3T3-L1 pre-adipocyte differentiation. Intracerebral infusion of SHLP2 increased peripheral glucose uptake and suppressed hepatic glucose production, as determined by systemic hyperinsulinemic-euglycemic clamp studies, suggesting that it functions as an insulin sensitizer both peripherally and centrally [22]. Similar to HN, the levels of circulating SHLP2 declined with age [22].
3.3. The identification of MOTS-c
Following the discovery of humanin, a novel MDP named MOTS-c (Mitochondrial ORF within the Twelve S rRNA c) was recently identified [4]. MOTS-c is expressed in various tissues and in circulation (plasma) in rodents and humans, suggesting both a cell-autonomous and hormonal role [32], and co-localizes with mitochondria in the cell [4]. Notably, there were hints of additional genes in the 12S rRNA, but no specific ORFs were identified [33,34]. In a search for interferon-responsive genes in the early 80s, Maeda et al., cloned cDNAs that were induced by interferon stimulation in a myeloblast cell line [33]. Later Tsuzuki et al. reported that the majority (~80%) of such clones with strong positive responses mapped to the mitochondrial rRNA locus, but no further investigations to identify potential genes were undertaken [34]. A recent in silico search of a human 12S rRNA revealed MOTS-c, a 51 base pair sORF with a strong Kozak sequence that yields a bioactive 16 amino acid peptide (GenBank accession# KP715230) [4]. Episodic lateral gene transfer from bacteria to eu-karyotes is evident and coincides with major evolutionary transitions at the origin of mitochondria, and the entirety of the current mtDNA sequence is found scattered throughout the nDNA in a fragmented and degenerate manner, a.k.a nuclear mitochondrial sequences (NUMT) [35,36]. Using the NCBI Basic Local Alignment Search Tool (BLAST) [37], none of the NUMT sequences were fully homologous to the MOTS-c ORF and all alignments to the human expression sequence tag (EST) were identical to the mtDNA sequence [4]. This also suggests that the cDNAs that Tsukiji et al. obtained were likely mitochondrial and not nuclear [34]. Currently, targeted mutagenesis of mtDNA is not feasible, let alone the lack of sequencing sensitivity to verify the successful incorporation of such mutations in hundreds to thousands of mtDNA in a given cell [38]. However, the selective elimination of mtDNA is possible by chronic low-dose ethidium bromide treatments in HeLa cells (HeLa-ρ0) [39]. MOTS-c and mitochondrial-encoded cytochrome oxidase I and II (MT-COI/II) expression was undetectable in HeLa-ρ0 cells, whereas nuclear-encoded GAPDH levels were unaltered, supporting mitochondrial origin of MOTS-c. In addition, the selective depletion of mitochondrial RNA using actinonin [40] caused a time-dependent loss of MOTS-c and MT-COI, but not GAPDH, again, supporting mitochondrial origin of MOTS-c [4]. MOTS-c translation most likely occurs in the cytoplasm, based on the disparity between mitochondrial and nuclear genetic codes [41]; the human mitochondria use “AGA” and “AGG” as stop codons, which would be recognized as arginine using on the conventional human code. Thus, mitochondrial translation of MOTS-c would lead to tandem start and stop codons [4]. The mechanism regarding the mitochondrial export of polyadenylated MOTS-c transcripts is currently under investigation. Notably, proteomics studies have begun to provide sequence-based evidence of mi-tochondrial origin at the peptide level (unpublished data).
3.4. The conservation of MOTS-c
Mitochondria are universal among eukaryotes [42]. Mammalian mtDNA encode for two ribosomal RNA genes (12S and 16S rRNA) that are key for mitochondrial translation. There is great diversity of mtDNA sequences across the phylogenetic tree, and the mitochondrial 12S and 16S rRNA genes are widely used as molecular markers for species identification [43]. The mitochon-drial genome has a relatively higher evolutionary rate compared to the nuclear genome, likely due to a higher mutation rate and clonal propagation, which can cause sequence variations between closely related species [44–46]. However, some regions in 12S and 16S rRNA are highly conserved among species, likely resulting from a strong positive selection force [43]. The first 11 amino acid residues of MOTS-c (total 16 amino acids) are highly conserved in 14 mammalian species [4]. The ratio of divergence at non-synonymous and synonymous amino acid sites, calculated by dN/dS ratio, identified the presence and position of residues under positive or purifying selection [4]. Notably, “dwarf” sORFs that encode for peptides of ~20 amino acids are less conserved [47], which could explain the lack of MOTS-c conservation in some lower eukaryotes, including C. elegans and Drosophila melanogaster.
3.5. Cellular action of MOTS-c
3.5.1. Cell-autonomous targets of MOTS-c
Proliferating cells are in constant need of cellular building blocks for the production of nucleic acids, proteins, and lipids, and for the regulation of epigenetic and redox status [48,49]. A key cellular component to achieve this is the one-carbon metabolism comprising of the folate cycle and its tethered de novo nucleotide biosynthesis pathways and methionine cycle [48,50,51]. Recent studies show that the folate cycle is directly involved in the regulation of metabolism. The pro-longevity diabetes drug metformin [52,53], has been proposed to regulate metabolism via the folate cycle and the master metabolic regulator AMPK (AMP-activated protein kinase), which can be activated by purine intermediates [54–57].
A systematic approach combining unbiased global metabo-lomics and gene microarray identified the folate-methionine cycle and its directly tethered de novo purine biosynthesis pathway as the cellular metabolic target of MOTS-c [4]. MOTS-c inhibited the folate cycle at the level of 5Me-THF, resulting in an accumulation of AICAR [5-aminoimidazole-4-carboxamide ribonucleotide), a well-defined AMPK activator [58]. Interestingly, MOTS-c also increased cellular NAD+ levels, which are also nucleotide precursors, suggesting the possible involvement of SIRT1 in AMPK activation [59]. In fact, these pathways mediate the metabolic actions of MOTS-c [4].
3.5.2. The role of MOTS-c in cellular metabolism
MOTS-c is a mitochondrial signal that stimulates cellular glucose uptake while suppressing respiration. This is consistent with the “Crabtree effect” whereby glucose can inhibit cellular respiration and oxidative phosphorylation [60]. Notably, the glucose taken up in response to MOTS-c was routed to the anabolic pentose phosphate pathway (PPP), which provides carbon sources for the synthesis of purines, rather than being metabolized through glycolysis [4]. In addition, MOTS-c increased the levels of carnitine shuttles, which transport activated fatty acids into the mitochon-dria for β-oxidation, increased the level of a β-oxidation intermediate, and reduced intracellular levels of essential and non-essential fatty acids, suggesting enhanced lipid utilization [4]. Although β-oxidation is commonly associated with increased respiration, it can also occur under reduced respiration as shown by AICAR [61] and metformin [52].
3.6. MOTS-c as a metabolic hormone
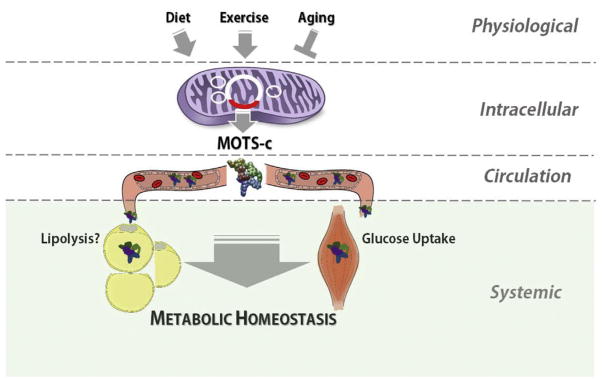
MOTS-c is detected in circulation, thus qualifying as a mi-tochondrial hormone [4,32,62,63]. Its primary target organ appears to be skeletal muscle and fat. Fasting mice for 48 h significantly reduced endogenous levels of MOTS-c in skeletal muscle and plasma [4]. In addition, acute systemic treatment of MOTS-c by intraperitoneal injections significantly reduced non-fasting glucose levels and significantly improved the response in glucose tolerance tests (GTT) [4]. The hyperinsulinemic-euglycemic clamp technique, a gold standard for measuring insulin action [64], provided evidence that MOTS-c regulates glucose homeostasis by targeting the skeletal muscle to increase clearance, but not the liver to reduce production [4]. Interestingly, MOTS-c levels in mice decline with age in skeletal muscle and in circulation concomitantly with the age-dependent development of insulin resistance [4]. Restoring MOTS-c levels by systemic injections in older mice (12 mo.) successfully reversed age-dependent skeletal muscle insulin resistance; myocytes that stably overexpress MOTS-c also exhibited increased glucose uptake [4]. In a disease model of insulin resistance in mice, established by feeding a high-fat diet (HFD), MOTS-c prevented diet-induced obesity and insulin resistance, and showed increased activation of AMPK and expression of its downstream glucose transporter in skeletal muscles [4]. Notably, MOTS-c also caused a significant reduction in HFD-induced visceral fat and hepatic steatosis, but it is unclear if this is a result of reduced adipogenesis or increased lipolysis, also observed in vitro as mentioned above (Fig. 1). The effect of MOTS-c in preventing metabolic symptoms following a HFD was not due to reduced caloric intake or increased total activity, but was associated with increased body heat production [4], indicating thermal energy dissipation as a feasible caloric outlet. This suggests a potential effect of MOTS-c on increasing the level of thermogenic brown and/or beige adipose tissue [65]. Further, these mice showed increased glucose utilization, determined by the respiratory exchange ratio (RER), likely by skeletal muscle that showed increased AMPK activation and glucose transporter expression.
Fig. 1.
A model of the systemic metabolic effects of the mitochondrial-derived peptide hormone MOTS-c. Exercise, diet (pro-longevity dietary interventions), and aging may affect the production, secretion, and activity of MOTS-c. Skeletal muscle is a major target of circulating MOTS-c, but experimental evidence also points to fat cells as a potential target. Depending on the environment, MOTS-c may have dual roles as an anabolic and a lipolytic hormone.
3.7. Implications of MOTS-c in health and disease
Mitochondria are strongly implicated in aging and age-related diseases. Given the promising beneficial effects of MOTS-c in regulating metabolic homeostasis, the therapeutic implications in obesity and diabetes are evident [23]. Future studies are required to understand the natural course of MOTS-c expression and activity in patients with diabetes, especially from a diagnostic perspective, and determine the therapeutic potential in such disease conditions. MOTS-c is also implicated in lung [66] and heart disease [67]. In addition, structural modifications, generating analogs, and conjugation for half-life extension, may be important for therapeutic development of MOTS-c [23]. Further, based on its fundamental role in regulating metabolic homeostasis, with specific effects on skeletal muscle and glucose metabolism, MOTS-c is also implicated in other areas of study, including exercise and longevity.
3.7.1. Exercise
Mitochondria are key bioenergetics sources that fuel the skeletal muscle during exercise, but they are also actively engaged in transmitting exercise-induced signals to other organs [4,32,68–71]. Muscles secrete myokines, such as irisin [72] and musclin [73], in response to exercise that mediate inter-organ communication and coordination [74]. Although the effect of exercise on regulating MOTS-c production and secretion is unknown, its beneficial effects on HFD is mirrored by MOTS-c [75]. Further, MOTS-c increases cellular levels of AICAR (an AMPK agonist) and activates AMPK [4], a well-described regulator of exercise [76–78]. MOTS-c injections in mice showed activation of skeletal muscle AMPK and increased the level of its downstream glucose transporter GLUT4 [4,79]. MOTS-c may also act as a potential mitochondrial signal that mediates an exercise-induced mitohormesis response, thereby stimulating physiological adaptation and increased tolerance to exercise [70,80,81]. Thus, it is plausible that MOTS-c may add an unprecedented mitochondrial layer to such exercise-induced signaling systems and act as a mimetic or enhancer [70,71,81–84]. The effects of exercise are broad and achieved through a complicated network of signaling factors that work seamlessly to coordinate physiological functions. Therefore, an “exercise pill” that can fully recapitulate the effects of exercise is unlikely to be developed, but exercise-mimetics could still greatly benefit those with limited mobility and improve metabolic conditions [82].
3.7.2. Longevity
Mitochondria are strongly implicated in aging, but the mechanistic details are poorly understood [85]. Mitokines [19] or MDPs [32,86], as described above, provide a paradigm-shifting concept of mitochondrial-derived endocrine factors that may regulate of aging and provide novel therapeutic and diagnostic targets for age-related diseases. For example, circulating humanin levels are (i) negatively correlated with circulating IGF-1 levels, which is seen in many long-lived animal models, and (ii) positively correlated with longevity [87]; humanin expression also declines with age in rodents and humans [29].
MOTS-c, whose levels also decline with age, has many implications in the regulation of healthspan and/or lifespan. The mitochondrial m.1382A>C polymorphism, which is found specifically in the D4b2 haplogroup that is associated with exceptional longevity in the Japanese population [88]. This polymorphism occurs within the MOTS-c ORF and causes a Lys14Gln change that may have functional alterations [89]. Further studies are required to test if MOTS-c, and its analogs, can positively extend healthspan and/or lifespan in model organisms.
There are also metabolic links between known age-modifiers and MOTS-c. NAD+ is a key metabolic coenzyme involved in redox reactions that declines with age, and restoring its levels can improve age-related disease conditions [90,91]. Further, NAD+ is a potent activator of sirtuins, which are conserved multifunctional regulators of aging and age-related diseases in various model organisms from yeast to mammals [90–92]. On that line, MOTS-c increases intracellular NAD+ levels and MOTS-c-dependent gly-colytic effects are mediated by sirtuin 1 (SIRT1) [4]. In addition, MOTS-c restricts the folate/methionine cycle, causing a reduction in methionine metabolism. In rodents, methionine restriction can increase lifespan by about 45%, decrease age-related diseases (e.g., cancer), delay lens deterioration, reduce visceral fat, and increase the major antioxidant glutathione (GSH) [93–96]. Similar observations have been made in other model organisms [97,98]. Based on these connections to known aging regulators, future studies focusing on the role of MOTS-c in modulating healthspan and lifespan may unveil novel mitochondrial-derived pro-longevity signals.
4. Conclusion
Mitochondria are multifaceted functional organelles that are increasingly being appreciated as major regulators. The survival of eukaryotic cells requires a close intergenomic coordination between mtDNA and nDNA. Communication is key to achieve such high-level coordination. Humans have two genomes, but only the nuclear genome has been thought to encode signaling factors. Thus, the neglect of the other half of our genome may hold lost opportunities for the identification of novel therapeutic and diagnostic targets for a wide range of diseases. MDPs are unprecedented candidates for such development and will allow the targeting of mitochondrial communication in health and disease. In particular, MOTS-c holds much potential as a target to treat metabolic syndromes by regulating muscle and fat physiology (Fig. 1), and perhaps even extend our healthy lifespan.
Acknowledgments
This research was supported by National Institute of Health Grants to P.C. (1R01GM090311, 1R01ES020812, 1P01AG034906) and an Ellison Medical Foundation New Scholar Award, Zumberge award, SC-CTSI grant, and Hanson Thorell Family Research Award to C.L.
References
- 1.Hashimoto Y, et al. A rescue factor abolishing neuronal cell death by a wide spectrum of familial Alzheimer’s disease genes and Abeta. Proc Natl Acad Sci USA. 2001;98(11):6336–6341. doi: 10.1073/pnas.101133498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ikonen M, et al. Interaction between the Alzheimer’s survival peptide humanin and insulin-like growth factor-binding protein 3 regulates cell survival and apoptosis. Proc Natl Acad Sci USA. 2003;100(22):13042–13047. doi: 10.1073/pnas.2135111100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guo B, et al. Humanin peptide suppresses apoptosis by interfering with Bax activation. Nature. 2003;423(6938):456–461. doi: 10.1038/nature01627. [DOI] [PubMed] [Google Scholar]
- 4.Lee C, et al. The mitochondrial-derived peptide MOTS-c promotes metabolic homeostasis and reduces obesity and insulin resistance. Cell Metab. 2015;21(3):443–454. doi: 10.1016/j.cmet.2015.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shokolenko IN, Alexeyev MF. Mitochondrial DNA: a disposable genome? Biochim Biophys Acta. 2015;1852(9):1805–1809. doi: 10.1016/j.bbadis.2015.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.da Cunha FM, Torelli NQ, Kowaltowski AJ. Mitochondrial retrograde signaling: triggers, pathways, and outcomes. Oxidative Med Cell Longev. 2015:2015. doi: 10.1155/2015/482582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Quiros PM, Mottis A, Auwerx J. Mitonuclear communication in homeostasis and stress. Nat Rev Mol Cell Biol. 2016 doi: 10.1038/nrm.2016.23. [DOI] [PubMed] [Google Scholar]
- 8.Horan MP, Gemmell NJ, Wolff JN. From evolutionary bystander to master manipulator: the emerging roles for the mitochondrial genome as a modulator of nuclear gene expression. Eur J Hum Genet. 2013;21(12):1335. doi: 10.1038/ejhg.2013.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lane N. Evolution. The costs of breathing. Science. 2011;334(6053):184–185. doi: 10.1126/science.1214012. [DOI] [PubMed] [Google Scholar]
- 10.Chandel NS. Evolution of Mitochondria as Signaling Organelles. Cell Metab. 2015 doi: 10.1016/j.cmet.2015.05.013. [DOI] [PubMed] [Google Scholar]
- 11.Pesole G, et al. The neglected genome. EMBO Rep. 2012;13(6):473–474. doi: 10.1038/embor.2012.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Finley LW, Haigis MC. The coordination of nuclear and mitochondrial communication during aging and calorie restriction. Aging Res Rev. 2009;8(3):173–188. doi: 10.1016/j.arr.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hill GE. Mitonuclear ecology. Mol Biol Evol. 2015;32(8):1917–1927. doi: 10.1093/molbev/msv104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dunham-Snary KJ, Ballinger SW. GENETICS. Mitochondrial-nuclear DNA mismatch matters. Science. 2015;349(6255):1449–1450. doi: 10.1126/science.aac5271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fetterman JL, et al. Mitochondrial genetic background modulates bioenergetics and susceptibility to acute cardiac volume overload. Biochem J. 2013;455(2):157–167. doi: 10.1042/BJ20130029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Betancourt AM, et al. Mitochondrial-nuclear genome interactions in non-alcoholic fatty liver disease in mice. Biochem J. 2014;461(2):223–232. doi: 10.1042/BJ20131433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hamilton G. The hidden risks for’three-person’babies. Nature. 2015;525(7570):444. doi: 10.1038/525444a. [DOI] [PubMed] [Google Scholar]
- 18.Durieux J, Wolff S, Dillin A. The cell-non-autonomous nature of electron transport chain-mediated longevity. Cell. 2011;144(1):79–91. doi: 10.1016/j.cell.2010.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Woo DK, Shadel GS. Mitochondrial stress signals revise an old aging theory. Cell. 2011;144(1):11–12. doi: 10.1016/j.cell.2010.12.023. [DOI] [PubMed] [Google Scholar]
- 20.Nass MM, Nass S. Intramitochondrial fibers with DNA characteristics. I. Fixation and Electron Staining Reactions. J Cell Biol. 1963;19:593–611. doi: 10.1083/jcb.19.3.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nass S, Nass MM. Intramitochondrial fibers with DNA characteristics. Ii. Enzymatic and other hydrolytic treatments. J Cell Biol. 1963;19:613–629. doi: 10.1083/jcb.19.3.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cobb LJ, et al. Naturally occurring mitochondrial-derived peptides are age-dependent regulators of apoptosis, insulin sensitivity, and inflammatory markers. Aging. 2016 doi: 10.18632/aging.100943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saghatelian A, Couso JP. Discovery and characterization of smORF-encoded bioactive polypeptides. Nat Chem Biol. 2015;11(12):909–916. doi: 10.1038/nchembio.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ma J, et al. Improved identification and analysis of small open reading frame encoded polypeptides. Anal Chem. 2016 doi: 10.1021/acs.analchem.6b00191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Andrews SJ, Rothnagel JA. Emerging evidence for functional peptides encoded by short open reading frames. Nat Rev Genet. 2014;15(3):193–204. doi: 10.1038/nrg3520. [DOI] [PubMed] [Google Scholar]
- 26.Lee C, Yen K, Cohen P. Humanin: a harbinger of mitochondrial-derived peptides? Trends Endocrinol Metab. 2013 doi: 10.1016/j.tem.2013.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hashimoto Y, et al. Mechanisms of neuroprotection by a novel rescue factor humanin from Swedish mutant amyloid precursor protein. Biochem Biophys Res Commun. 2001;283(2):460–468. doi: 10.1006/bbrc.2001.4765. [DOI] [PubMed] [Google Scholar]
- 28.Bachar AR, et al. Humanin is expressed in human vascular walls and has a cytoprotective effect against oxidized LDL-induced oxidative stress. Cardiovasc Res. 2010;88(2):360–366. doi: 10.1093/cvr/cvq191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Muzumdar RH, et al. Humanin: a novel central regulator of peripheral insulin action. PloS One. 2009;4(7):e6334. doi: 10.1371/journal.pone.0006334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Muzumdar RH, et al. Acute humanin therapy attenuates myocardial ischemia and reperfusion injury in mice. Arterioscler Thromb Vasc Biol. 2010;30(10):1940–1948. doi: 10.1161/ATVBAHA.110.205997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yen K, Lee C, Mehta H, Cohen P. The emerging role of the mitochondrial-derived peptide humanin in stress resistance. J Mol Endocrinol. 2013;50(1):R11–R19. doi: 10.1530/JME-12-0203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zarse K, Ristow M. A mitochondrially encoded hormone ameliorates obesity and insulin resistance. Cell Metab. 2015;21(3):355–356. doi: 10.1016/j.cmet.2015.02.013. [DOI] [PubMed] [Google Scholar]
- 33.Maeda S, et al. Construction and identification of bacterial plasmids containing nucleotide sequence for human leukocyte interferon. Proc Natl Acad Sci USA. 1980;77(12):7010–7013. doi: 10.1073/pnas.77.12.7010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tsuzuki T, et al. The majority of cDNA clones with strong positive signals for the interferon-induction-specific sequences resemble mitochondrial ribosomal RNA genes. Biochem Biophys Res Commun. 1983;114(2):670–676. doi: 10.1016/0006-291x(83)90833-1. [DOI] [PubMed] [Google Scholar]
- 35.Ku C, et al. Endosymbiotic origin and differential loss of eukaryotic genes. Nature. 2015;524(7566):427–432. doi: 10.1038/nature14963. [DOI] [PubMed] [Google Scholar]
- 36.Ricchetti M, Tekaia F, Dujon B. Continued colonization of the human genome by mitochondrial DNA. PLoS Biol. 2004;2(9):E273. doi: 10.1371/journal.pbio.0020273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Altschul SF, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25(17):3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kennedy SR, Salk JJ, Schmitt MW, Loeb LA. Ultra-sensitive sequencing reveals an age-related increase in somatic mitochondrial mutations that are inconsistent with oxidative damage. PLoS Genet. 2013;9(9):e1003794. doi: 10.1371/journal.pgen.1003794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hashiguchi K, Zhang-Akiyama QM. Establishment of human cell lines lacking mitochondrial DNA. Methods Mol Biol. 2009;554:383–391. doi: 10.1007/978-1-59745-521-3_23. [DOI] [PubMed] [Google Scholar]
- 40.Richter U, et al. A mitochondrial ribosomal and RNA decay pathway blocks cell proliferation. Curr Biol. 2013;23(6):535–541. doi: 10.1016/j.cub.2013.02.019. [DOI] [PubMed] [Google Scholar]
- 41.Taanman JW. The mitochondrial genome: structure, transcription, translation and replication. Biochim Biophys Acta. 1999;1410(2):103–123. doi: 10.1016/s0005-2728(98)00161-3. [DOI] [PubMed] [Google Scholar]
- 42.Embley TM, Martin W. Eukaryotic evolution, changes and challenges. Nature. 2006;440(7084):623–630. doi: 10.1038/nature04546. [DOI] [PubMed] [Google Scholar]
- 43.Yang L, et al. Species identification through mitochondrial rRNA genetic analysis. Sci Rep. 2014;4:4089. doi: 10.1038/srep04089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stewart JB, Chinnery PF. The dynamics of mitochondrial DNA heteroplasmy: implications for human health and disease. Nat Rev Genet. 2015;16(9):530–542. doi: 10.1038/nrg3966. [DOI] [PubMed] [Google Scholar]
- 45.Brown WM, George M, Jr, Wilson AC. Rapid evolution of animal mitochondrial DNA. Proc Natl Acad Sci USA. 1979;76(4):1967–1971. doi: 10.1073/pnas.76.4.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Galtier N, Enard D, Radondy Y, Bazin E, Belkhir K. Mutation hot spots in mammalian mitochondrial DNA. Genome Res. 2006;16(2):215–222. doi: 10.1101/gr.4305906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Aspden JL, et al. Extensive translation of small Open Reading Frames revealed by Poly-Ribo-Seq. eLife. 2014;3:e03528. doi: 10.7554/eLife.03528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Locasale JW. Serine, glycine and one-carbon units: cancer metabolism in full circle. Nat Rev Cancer. 2013;13(8):572–583. doi: 10.1038/nrc3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Locasale JW, Cantley LC. Metabolic flux and the regulation of mammalian cell growth. Cell Metab. 2011;14(4):443–451. doi: 10.1016/j.cmet.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tong X, Zhao F, Thompson CB. The molecular determinants of de novo nucleotide biosynthesis in cancer cells. Curr Opin Genet Dev. 2009;19(1):32–37. doi: 10.1016/j.gde.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Crider KS, Yang TP, Berry RJ, Bailey LB. Folate and DNA methylation: a review of molecular mechanisms and the evidence for folate’s role. Adv Nutr. 2012;3(1):21–38. doi: 10.3945/an.111.000992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Martin-Montalvo A, et al. Metformin improves healthspan and lifespan in mice. Nat Commun. 2013;4:2192. doi: 10.1038/ncomms3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hall SS. A Trial for the ages. Science. 2015;349(6254):1274–1278. doi: 10.1126/science.349.6254.1274. [DOI] [PubMed] [Google Scholar]
- 54.Cabreiro F, et al. Metformin retards aging in C. elegans by altering microbial folate and methionine metabolism. Cell. 2013;153(1):228–239. doi: 10.1016/j.cell.2013.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Corominas-Faja B, et al. Metabolomic fingerprint reveals that metformin impairs one-carbon metabolism in a manner similar to the antifolate class of chemotherapy drugs. Aging. 2012;4(7):480–498. doi: 10.18632/aging.100472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hardie DG, Ross FA, Hawley SA. AMPK: a nutrient and energy sensor that maintains energy homeostasis. Nat Rev Mol Cell Biol. 2012;13(4):251–262. doi: 10.1038/nrm3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li X, et al. Structural basis of AMPK regulation by adenine nucleotides and glycogen. Cell Res. 2015;25(1):50–66. doi: 10.1038/cr.2014.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hardie DG. AMP-activated protein kinase: an energy sensor that regulates all aspects of cell function. Genes Dev. 2011;25(18):1895–1908. doi: 10.1101/gad.17420111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Price NL, et al. SIRT1 is required for AMPK activation and the beneficial effects of resveratrol on mitochondrial function. Cell Metab. 2012;15(5):675–690. doi: 10.1016/j.cmet.2012.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ibsen KH. The Crabtree effect: a review. Cancer Res. 1961;21:829–841. [PubMed] [Google Scholar]
- 61.Spangenburg EE, Jackson KC, Schuh RA. AICAR inhibits oxygen consumption by intact skeletal muscle cells in culture. J Physiol Biochem. 2013 doi: 10.1007/s13105-013-0269-0. [DOI] [PubMed] [Google Scholar]
- 62.Lee MS. Effect of mitochondrial stress on systemic metabolism. Ann N Y Acad Sci. 2015;1350:61–65. doi: 10.1111/nyas.12822. [DOI] [PubMed] [Google Scholar]
- 63.Ruetenik A, Barrientos A. Dietary restriction, mitochondrial function and aging: from yeast to humans. Biochim Biophys Acta. 2015 doi: 10.1016/j.bbabio.2015.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kim JK. Hyperinsulinemic-euglycemic clamp to assess insulin sensitivity in vivo. Methods Mol Biol. 2009;560:221–238. doi: 10.1007/978-1-59745-448-3_15. [DOI] [PubMed] [Google Scholar]
- 65.Sidossis L, Kajimura S. Brown and beige fat in humans: thermogenic adipocytes that control energy and glucose homeostasis. J Clin Investig. 2015;125(2):478–486. doi: 10.1172/JCI78362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cloonan SM, Choi AM. Mitochondria in lung disease. J Clin Investig. 2016;126(3):809–820. doi: 10.1172/JCI81113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kadlec AO, Beyer AM, Ait-Aissa K, Gutterman DD. Mitochondrial signaling in the vascular endothelium: beyond reactive oxygen species. Basic Res Cardiol. 2016;111(3):26. doi: 10.1007/s00395-016-0546-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Li S, Laher I. Exercise pills: at the starting line. Trends Pharmacol Sci. 2015;36(12):906–917. doi: 10.1016/j.tips.2015.08.014. [DOI] [PubMed] [Google Scholar]
- 69.Fuku N, et al. The mitochondrial-derived peptide MOTS-c: a player in exceptional longevity? Aging Cell. 2015;14(6):921–923. doi: 10.1111/acel.12389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Merry TL, Ristow M. Mitohormesis in exercise training. Free Radic Biol Med. 2015 doi: 10.1016/j.freeradbiomed.2015.11.032. [DOI] [PubMed] [Google Scholar]
- 71.Handschin C. Caloric restriction and exercise “mimetics”: ready for prime time? Pharmacol Res. 2015;103:158–166. doi: 10.1016/j.phrs.2015.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jedrychowski MP, et al. Detection and quantitation of circulating human irisin by tandem mass spectrometry. Cell Metab. 2015;22(4):734–740. doi: 10.1016/j.cmet.2015.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Subbotina E, et al. Musclin is an activity-stimulated myokine that enhances physical endurance. Proc Natl Acad Sci USA. 2015;112(52):16042–16047. doi: 10.1073/pnas.1514250112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rai M, Demontis F. Systemic nutrient and stress signaling via myokines and myometabolites. Annu Rev Physiol. 2015 doi: 10.1146/annurev-physiol-021115-105305. [DOI] [PubMed] [Google Scholar]
- 75.Alis R, Lucia A, Blesa JR, Sanchis-Gomar F. The role of Mitochondrial Derived Peptides (MDPs) in metabolism. J Cell Physiol. 2015 doi: 10.1002/jcp.25023. [DOI] [PubMed] [Google Scholar]
- 76.Merrill GF, Kurth EJ, Hardie DG, Winder WW. AICA riboside increases AMP-activated protein kinase, fatty acid oxidation, and glucose uptake in rat muscle. Am J Physiol. 1997;273(6 Pt 1):E1107–E1112. doi: 10.1152/ajpendo.1997.273.6.E1107. [DOI] [PubMed] [Google Scholar]
- 77.Narkar VA, et al. AMPK and PPARdelta agonists are exercise mimetics. Cell. 2008;134(3):405–415. doi: 10.1016/j.cell.2008.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Fujii N, et al. Exercise induces isoform-specific increase in 5′AMP-activated protein kinase activity in human skeletal muscle. Biochem Biophys Res Commun. 2000;273(3):1150–1155. doi: 10.1006/bbrc.2000.3073. [DOI] [PubMed] [Google Scholar]
- 79.McGee SL, et al. AMP-activated protein kinase regulates GLUT4 transcription by phosphorylating histone deacetylase 5. Diabetes. 2008;57(4):860–867. doi: 10.2337/db07-0843. [DOI] [PubMed] [Google Scholar]
- 80.Yun J, Finkel T. Mitohormesis. Cell Metab. 2014;19(5):757–766. doi: 10.1016/j.cmet.2014.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Thevis M, Schanzer W. Emerging drugs affecting skeletal muscle function and mitochondrial biogenesis - Potential implications for sports drug testing programs. Rapid Commun Mass Spectrom. 2016;30(5):635–651. doi: 10.1002/rcm.7470. [DOI] [PubMed] [Google Scholar]
- 82.Hunter P. Exercise in a bottle: elucidating how exercise conveys health benefits might lead to new therapeutic options for a range of diseases from cancer to metabolic syndrome. EMBO Rep. 2016 doi: 10.15252/embr.201541835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Li S, Laher I. Exercise pills: at the starting line. Trends PharmacolSci. 2015 doi: 10.1016/j.tips.2015.08.014. [DOI] [PubMed] [Google Scholar]
- 84.Lee DE, et al. Translational machinery of mitochondrial mRNA is promoted by physical activity in Western diet-induced obese mice. Acta Physiol. 2016 doi: 10.1111/apha.12687. [DOI] [PubMed] [Google Scholar]
- 85.Bratic A, Larsson NG. The role of mitochondria in aging. J Clin Investig. 2013;123(3):951–957. doi: 10.1172/JCI64125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Long YC, Tan TM, Takao I, Tang BL. The biochemistry and cell biology of aging: metabolic regulation through mitochondrial signaling. Am J Physiol Endocrinol Metab. 2014;306(6):E581–E591. doi: 10.1152/ajpendo.00665.2013. [DOI] [PubMed] [Google Scholar]
- 87.Lee C, et al. IGF-I regulates the age-dependent signaling peptide humanin. Aging Cell. 2014;13(5):958–961. doi: 10.1111/acel.12243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Alexe G, et al. Enrichment of longevity phenotype in mtDNA haplogroups D4b2b, D4a, and D5 in the Japanese population. Hum Genet. 2007;121(3–4):347–356. doi: 10.1007/s00439-007-0330-6. [DOI] [PubMed] [Google Scholar]
- 89.Fuku N. The mitochondrial-derived peptide MOTS-c: a player in exceptional longevity? Aging Cell. 2015 doi: 10.1111/acel.12389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Imai S, Guarente L. NAD+ and sirtuins in aging and disease. Trends Cell Biol. 2014;24(8):464–471. doi: 10.1016/j.tcb.2014.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Verdin E. NAD(+) in aging, metabolism, and neurodegeneration. Science. 2015;350(6265):1208–1213. doi: 10.1126/science.aac4854. [DOI] [PubMed] [Google Scholar]
- 92.Lopez-Otin C, Blasco MA, Partridge L, Serrano M, Kroemer G. The hallmarks of aging. Cell. 2013;153(6):1194–1217. doi: 10.1016/j.cell.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zimmerman JA, Malloy V, Krajcik R, Orentreich N. Nutritional control of aging. Exp Gerontol. 2003;38(1–2):47–52. doi: 10.1016/s0531-5565(02)00149-3. [DOI] [PubMed] [Google Scholar]
- 94.Miller RA, et al. Methionine-deficient diet extends mouse lifespan, slows immune and lens aging, alters glucose, T4, IGF-I and insulin levels, and increases hepatocyte MIF levels and stress resistance. Aging Cell. 2005;4(3):119–125. doi: 10.1111/j.1474-9726.2005.00152.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Richie JP, Jr, et al. Methionine restriction increases blood glutathione and longevity in F344 rats. FASEB J. 1994;8(15):1302–1307. doi: 10.1096/fasebj.8.15.8001743. [DOI] [PubMed] [Google Scholar]
- 96.Orentreich N, Matias JR, DeFelice A, Zimmerman JA. Low methionine ingestion by rats extends life span. J Nutr. 1993;123(2):269–274. doi: 10.1093/jn/123.2.269. [DOI] [PubMed] [Google Scholar]
- 97.Lee BC, et al. Methionine restriction extends lifespan of Drosophila melanogaster under conditions of low amino-acid status. Nat Commun. 2014;5:3592. doi: 10.1038/ncomms4592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ruckenstuhl C, et al. Lifespan extension by methionine restriction requires autophagy-dependent vacuolar acidification. PLoS Genet. 2014;10(5):e1004347. doi: 10.1371/journal.pgen.1004347. [DOI] [PMC free article] [PubMed] [Google Scholar]