SUMMARY
Borrelia burgdorferi possesses a sophisticated and complex chemotaxis system but how the organism utilizes this system in its natural enzootic life cycle is poorly understood. Of the three CheY chemotaxis response regulators in B. burgdorferi, we found that only deletion of cheY3 resulted in an altered motility and significantly reduced chemotaxis phenotype. Though ΔcheY3 maintained normal densities in unfed ticks, their numbers were significantly reduced in fed ticks compared to the parental or cheY3-complemented spirochetes. Importantly, mice fed upon by the ΔcheY3-infected ticks did not develop a persistent infection. Intravital confocal microscopy analyses discovered that the ΔcheY3 spirochetes were motile, but appeared unable to reverse direction and perform the characteristic backward-forward motility displayed by the parental strain. Subsequently, the ΔcheY3 became “trapped” in the skin matrix within days of inoculation, were cleared from the skin needle-inoculation site within 96 hours post-injection, and did not disseminate to distant tissues. Interestingly, although ΔcheY3 cells were cleared within 96 hours post-injection, this attenuated infection elicited significant levels of B. burgdorferi-specific IgM and IgG. Taken together, these data demonstrate that cheY3-mediated chemotaxis is crucial for motility, dissemination, and viability of the spirochete both within and between mice and ticks.
INTRODUCTION
Bacterial cells possess sophisticated signal transduction pathways that detect changes in specific parameters of their dynamic external environment, allowing them to respond appropriately to the fluctuating environment (Falke et al., 1997, Skerker et al., 2005). Chemotaxis, the cellular movement in response to chemical gradients (Wadhams et al., 2004), is one behavior that bacteria alter based on their external environment (Wolanin et al., 2002). This movement empowers bacteria to approach and remain in beneficial environments or escape from noxious ones by modulating their swimming behaviors. There are a vast array of signals, including nutrient concentrations, osmolarity, temperature, oxygen, and pH changes, which bacteria integrate together and translate into a specific response (Falke et al., 1997), either benign or pathogenic (Wadhams et al., 2004). Bacterial chemotaxis pathways are regulated by a complex two-component signal transduction system (Falke et al., 1997, Bourret et al., 2010, Hazelbauer, 2012). The system starts with the membrane-bound chemoreceptors—methyl-accepting chemotaxis proteins (MCPs)—that sense external stimuli. The MCPs are coupled (via the coupling protein, CheW) with CheA, a histidine kinase. The catalytic activity of CheA is mediated by a ligand (either attractant or repellant) bound to the MCPs. Once active, CheA utilizes ATP to autophosphorylate and then proceeds to phosphorylate the response regulator, CheY. Phosphorylated CheY (CheY-P) then binds to the flagellar switch proteins FliM and FliN, regulating the direction of rotation of the flagella (Welch et al., 1993, Toker et al., 1997, Djordjevic et al., 1998, Sarkar et al., 2010).
Borrelia burgdorferi, the causative agent of Lyme disease (Burgdorfer et al., 1982, Lane et al., 1991), is the most common arthropod-borne disease in the United States and Europe (Mead, 2015). Chemotaxis and motility genes comprise approximately 5–6% of the genome of B. burgdorferi, and we have shown that motility is required for every stage of the infectious life cycle of B. burgdorferi (Sultan et al., 2013, Motaleb et al., 2015, Sultan et al., 2015). In nature, B. burgdorferi cycles between the Ixodes tick vector and a mammalian host (Burgdorfer et al., 1982, Levine et al., 1985, Lane et al., 1991, Tsao, 2009, Brisson et al., 2012). Completion of the enzootic cycle requires that B. burgdorferi traverse through dense and complex tissues within tick and vertebrate hosts; the spirochetes must migrate from the midgut to the salivary glands within the tick to allow transmission to the next host during tick feeding (Dunham-Ems et al., 2009, Radolf et al., 2012, Sultan et al., 2013), as well as navigate through the skin matrix of a vertebrate host after deposition via the tick-bite to reach a multitude of target tissues before migrating back to a feeding tick (Ribeiro et al., 1987, Dunham-Ems et al., 2009, Pal, 2010, Radolf et al., 2012, Sultan et al., 2013). The spirochete must complete these tasks all by simultaneously detecting its current environment, determining its next optimal direction, and evading the immune systems of both hosts. Indeed, a recent global signature-tagged mutagenesis study identified that mutations in chemotaxis and motility genes were most often associated with loss in infectivity of B. burgdorferi, signifying their importance for its enzootic lifecycle (Lin et al., 2012).
While chemotaxis has been extensively studied in Escherichia coli and Salmonella enterica (Armitage, 1999, Bren et al., 2000, He et al., 2014), the chemotaxis system of B. burgdorferi is poorly understood and differs greatly from those prototypic systems. The Lyme disease spirochete is relatively long (10 to 20 μm) and thin (0.3 μm) with a distinctive flat-wave morphology, and motility is generated by rotation of the periplasmic flagella (Charon et al., 2012). Between 7 to 11 periplasmic flagella are attached near each cellular pole, causing the motility/chemotaxis behavior of B. burgdorferi and other spirochetes to be unique and complex. Tracking of B. burgdorferi swimming in vitro has described that spirochetes perform run, flex, and reverse swimming modes. Runs occur when the periplasmic flagellar motors at one pole rotate in the direction opposite that of the motors at the other pole (motors at one end rotate in clock-wise whereas motors at other end rotate counter clock-wise). The flex is a non-translational (i.e. no net motility) mode and is thought to be equivalent to the E. coli tumble. During the flex, the motors at both poles rotate in the same direction, i.e. both rotate in clock-wise (CW) or counter clock-wise (CCW). Spirochetal reversal occurs in translating (i.e. motile) cells when the motors at each end reverse their direction of rotation. For spirochetes to swim toward an attractant, the organisms must be able to coordinate the rotation of the motors at the two separate poles of the cell that are located at a considerable distance from one another (often greater than 10 μm). One of the questions related to spirochete chemotaxis is how the organisms are able to achieve this coordination (Li et al., 2002, Motaleb et al., 2005, Motaleb et al., 2011b, Charon et al., 2012, Wolgemuth, 2015). The genome of B. burgdorferi encodes multiple homologs of several chemotaxis genes (e.g. two cheA, three cheW, three cheY, two cheB, and two cheR genes), making it much more complex than E. coli or S. enterica (Fraser et al., 1997, Charon et al., 2012, Motaleb et al., 2015). In several species of pathogenic bacteria, the CheY response regulator has been shown to play an important role in chemotaxis, which is also needed for virulence (Butler et al., 2005, McGee et al., 2005, Antunez-Lamas et al., 2009, Lertsethtakarn et al., 2011). To date, only two studies have shown that chemotaxis—specifically involving the histidine kinase cheA2 and phosphatase enhancer cheD—are essential for the infectious life cycle of B. burgdorferi (Sze et al., 2012, Moon et al., 2016).
Amino acid sequence analysis indicates that B. burgdorferi CheY1, CheY2, and CheY3 share 25–37% identity with each other. Moreover, these proteins share 32%, 38%, and 25% amino acid sequence identity with E. coli CheY, respectively (Motaleb et al., 2011b). Importantly, all of the functional residues of the E. coli CheY response regulator were found to be conserved in CheY1, CheY2, and CheY3, suggesting that they all are potential chemotaxis response regulators. Previous reports also indicate that cheY3, but not cheY1 or cheY2, is important for motility and chemotaxis in vitro (Motaleb et al., 2011b). Moreover, the function of cheY3 cannot be substituted by the other cheYs in B. burgdorferi. Importantly, those cheY mutants were constructed in a high-passage, avirulent strain that cannot be evaluated in the tick vector or vertebrate hosts (Motaleb et al., 2011b). We hypothesize that cheY3 is crucial for one or more stages of the enzootic cycle. The goal of this study is to utilize a cheY3-mutated B. burgdorferi generated in a virulent genetic background to delineate the importance of CheY3 for the different host environments encountered by these bacteria. Our findings are significant in describing the deficiencies in motility and chemotaxis abilities exhibited by the ΔcheY3 strain and delineating the essential nature of this gene for every stage of the tick-mouse infection cycle. Based on our data, we propose a model indicating the importance of chemotaxis (and motility) during the enzootic life cycle of B. burgdorferi.
RESULTS
Construction and confirmation of ΔcheY3 and cheY3+ strains
B. burgdorferi genome sequence and transcriptional analyses indicated that the cheY3 gene is located on the large linear chromosome and is transcribed as a polycistronic mRNA using a σ70 promoter (Li et al., 2002). Previous reports also indicate that only cheY3 is essential for chemotaxis (Motaleb et al., 2011b). However, those studies were done in vitro under one condition, using a high-passage, non-infectious clone that cannot be evaluated in the tick-mouse infectious cycle (Motaleb et al., 2011b). To determine the importance of cheY3 in the pathogenic life cycle of B. burgdorferi, we constructed a ΔcheY3 in the low-passage B31-A3 strain using a promoterless kanamycin resistance cassette, Pl-Kan (Figure 1A) (Sultan et al., 2010). PCR data confirmed the deletion of the cheY3 gene (data not shown, see below).
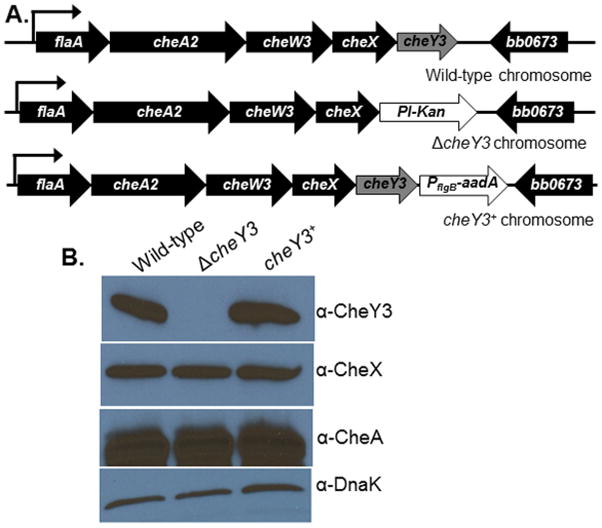
Figure 1. Construction and complementation of the ΔcheY3.
A. WT B. burgdorferi genome arrangement of flaA operon containing cheY3 (labeled as WT chromosome). The Pl-Kan (aph1) cassette was inserted after deleting the cheY3 gene by allelic exchange (ΔcheY3 chromosome). The mutant was complemented in cis by genomic reconstitution by inserting a WT copy of the cheY3 gene flanked by the PflgB-aadA cassette (cheY3+ chromosome). Arrows indicate the direction of transcription. DNAs/Plasmids are not drawn to scale. B. Immunoblot analysis of B. burgdorferi cells probed with the indicated antibodies. The CheY3 protein expression was inhibited in the mutant, but restored in the complemented cheY3+ cells as confirmed by using cell lysates from the indicated clones probed with anti-CheY3. CheY3 protein is approximately 14 kDa. The cheA2 and cheX genes are located in the same operon as the targeted cheY3, however, the expression of those gene products were not altered in the mutant or the complemented cells (see anti-CheA and anti-CheX blots). DnaK was used as a loading control.
The cheY3 gene is located at the end of its operon, thus integration of Pl-Kan in place of the cheY3 gene is unlikely to exhibit a polar effect on the expression of downstream genes. However, we complemented the ΔcheY3 in cis by genomic integration to ensure the phenotype of the mutant was solely attributed to the inactivation of the cheY3 (Figure 1A). Mutation and complementation of cheY3 were confirmed by western blotting as demonstrated by the absence or presence of CheY3 production, respectively (Figure 1B). Since cheY3 is in a polycistronic operon, we verified expression of other gene products (cheX and cheA2) in that operon. As shown in Figure 1B, expression of CheA2 and CheX proteins were not affected in ΔcheY3 bacteria. Furthermore, linear and circular endogenous plasmids of the mutant and complemented clones were verified by PCR and found that all clones retained the plasmids seen in the parental wild-type (WT) cells (data not shown).
In vitro phenotypes of ΔcheY3
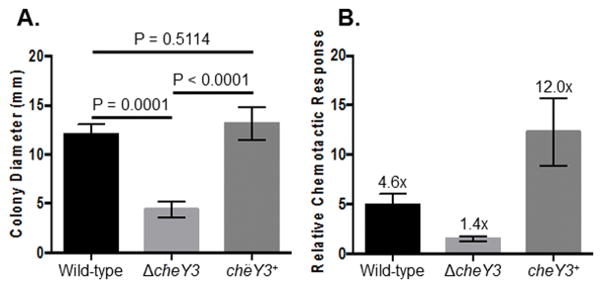
The growth, motility, and chemotaxis phenotypes of ΔcheY3 were assessed in vitro using various approaches. Dark-field microscopic analysis indicated that WT and cheY3+ cells exhibited run-flex/pause-reverse swimming patterns, whereas ΔcheY3 cells constantly ran in a single direction without reversals or switching their swimming direction. Additionally, semi-solid plating analysis showed that ΔcheY3 formed significantly smaller swarming colonies compared to WT or cheY3+ (Figure 2A). Despite the altered motility and swarming deficiencies, ΔcheY3 had no growth defect in liquid culture relative to WT or the cheY3+ (data not shown). Additionally, the chemotactic response of the ΔcheY3 was measured by using a quantitative capillary-tube chemotaxis assay. As expected, the chemotactic ability of the mutant was significantly reduced when compared to the WT or complemented cells (Figure 2B). The complemented cells exhibited a chemotactic response that was higher than the WT cells. We currently do not understand why this response was enhanced, but we speculate that the cheY3+ cells assessed for this assay were taken from a growth phase that was more chemotactic than the WT cells even though we were careful to collect all clones from the same growth phase. Together, these data are the same as reported previously, which indicate that cheY3 is important for chemotaxis as well as for governing motility in B. burgdorferi in vitro (Motaleb et al., 2011b).
Figure 2. In vitro phenotypes of the ΔcheY3.
A. ΔcheY3 colonies exhibit a significantly reduced swarming ability on semi-solid agar plates when compared to the parental WT or complemented cells (P = 0.0001). Values are mean ± SD from at least 11 individual colonies per strain. Statistical analysis was performed by using ANOVA followed by Tukey’s Multiple Comparisons test. A P<0.05 between strains is considered significant. B. The ΔcheY3 is deficient in chemotaxis as determined by the capillary tube chemotaxis assay using N-Acetylglucosamine as an attractant. A capillary tube filled with buffer without any attractant was used as a control. An increase in the number of spirochetes equal to or greater than twice that of the buffer control was considered “chemotactic”. The numbers on top of each vertical bar is the fold-increase over the buffer control. Results shown are mean ± SEM from three independent studies with three replicates per strain per assay.
ΔcheY3 spirochetes are unable to survive in mice by needle injection
To evaluate the ability of the ΔcheY3 to establish infection in a mammalian host, groups of C3H/HeN mice were needle-inoculated with WT, ΔcheY3, or cheY3+, and multiple tissues (i.e. ear, bladder, and joint tissues from each mouse) were collected at 4 weeks post-injection to determine bacterial outgrowth (Table 1). WT and complemented cells were re-isolated from one or more tissues assessed from all mice in those groups, demonstrating that these cells established an infection. Alternatively, ΔcheY3 spirochetes were not re-isolated from any mouse tissues, even when infected with 1×106 bacteria (Table 1). These results indicate that the cheY3 chemotaxis response regulator is essential for B. burgdorferi infection in a mammalian host.
Table 1.
ΔcheY3 lacks infectivity in mice via needle injectiona
| Strain | Dosage (spirochetes/mouse) | Number of Mice Infected |
|---|---|---|
| WT | 5 × 103 | 5/5 |
| ΔcheY3 | 5 × 103 | 0/5 |
| 1 × 106 | 0/5 | |
| cheY3+ | 5 × 104 | 2/2 |
C3H/HeN mice were injected intradermally using the indicated in vitro-grown spirochete clones. Mice were sacrificed four weeks post injection, and infectivity was determined by reisolation or detection of B. burgdorferi genomes by PCR from the murine ear skin, tibiotarsal joint, and bladder tissue samples. Doses shown are the actual number of spirochetes injected in each mouse.
ΔcheY3 spirochetes are unable to infect mice by Ixodes scapularis tick bite
Tick-mouse-tick infection studies were performed to determine if ΔcheY3 spirochetes are able to colonize, replicate, and transmit from infected ticks to naïve C3H/HeN mice. Since ΔcheY3 were unable to infect mice via needle inoculation, infecting ticks via feeding on infected mice was not feasible. Alternatively, we performed tick-immersion studies in order to artificially infect naïve larvae, and then the ticks were allowed to feed on naïve mice to determine the ability of the spirochetes to transmit from infected ticks to naïve mice. Additionally, the burden of spirochetes per tick was evaluated to determine the replication and survival of the spirochetes within the fed larvae 7 days post-repletion. Immunofluorescence assay using a FITC-conjugated antibody specific for B. burgdorferi demonstrated that spirochetes were intact within fed ticks (data not shown). However, while artificial immersion achieved a 90% infection rate of ticks for all strains (data not shown), the burden of ΔcheY3 spirochetes per tick was significantly less when compared to the WT or the cheY3+ cells, as determined by tick plating (Figure 3A). This result indicates that the non-chemotactic ΔcheY3 has a decreased ability to survive or multiply within fed larvae.
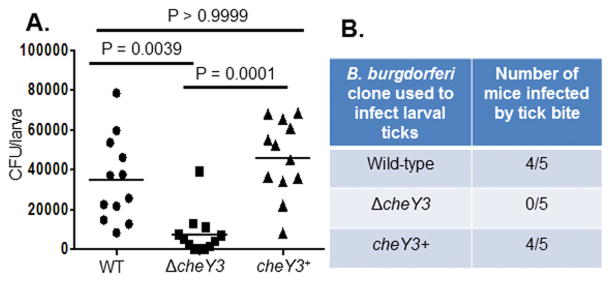
Figure 3. CheY3 is important for tick-mouse infectious cycle.
A. The burden of ΔcheY3 was significantly less in fed larval ticks compared to WT or the complemented cheY3+. Naive larval ticks were artificially infected by immersion using in vitro-grown spirochetes. Larvae were crushed individually on day 7 post-repletion, and the spirochetal density per larva was determined by plating on semi-solid growth media followed by counting viable spirochetes (CFU). The p-values were determined using Kruskal-Wallis ANOVA test followed by Dunn Multiple Comparisons test. Results shown are the spirochete burden per larva, where each dot is representative of a single tick (n = 12). The line denotes the mean of the entire group. A P<0.05 between strains is considered significant. B. CheY3 is important for establishing infection in mice by tick-bite. Naïve C3H/HeN mice were fed upon by artificially-infected larvae (~200 larvae/mouse, 3 mice per clone). Four weeks post-repletion, mice were euthanized to determine bacterial outgrowth from ear, joint, and bladder tissues.
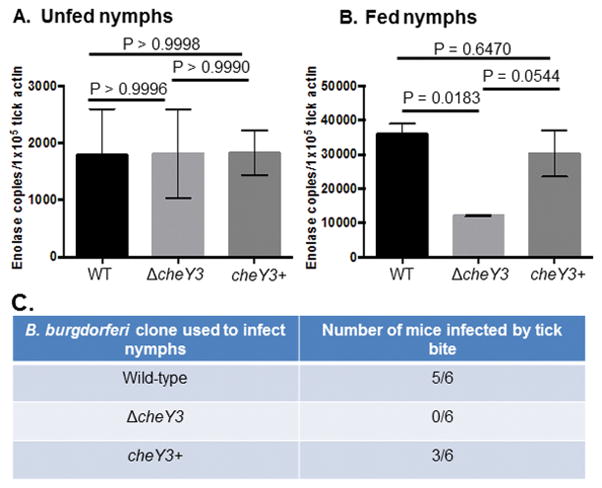
Four weeks after tick-feeding, mice were euthanized and animal tissues were cultivated to re-isolate B. burgdorferi in order to determine transmission from tick to mouse. No tissues from mice fed on by ΔcheY3-infected ticks showed bacterial outgrowth whereas 4 out of 5 mice fed upon by WT- or cheY3+-infected larvae showed regrowth (Figure 3B). These assays were also performed using encapsulated artificially immersed nymphs that produced similar results—reduced survivability of the mutants in fed but not in unfed ticks (Figures 4A, B). Moreover, we assessed the skin containing the tick-bite site, as well as ear, joint, and bladder tissues for bacterial outgrowth; PCR was also performed to detect spirochetes genomes. The artificially ΔcheY3-immersed nymphs were unable to establish an infection in any of the mice they fed upon, as the mutant bacteria were not reisolated by outgrowth in culture media or detected by PCR from those tissues, whereas five out of six mice were positive for WT bacteria and three out of six mice were positive for the cheY3+ spirochetes (Figure 4C). Together, these results indicate that chemotaxis is crucial for optimal survival of spirochetes in ticks and infection of mice by tick-bite.
Figure 4. Mice are not infected by ΔcheY3-infected nymph bite.
The burden of the mutant was significantly less in fed, but not in unfed nymphs when compared to the parental cells (A, unfed; B, fed nymphs). Naïve nymphs were artificially infected and then allowed to feed on separate naïve mice (n=3 per assay). Seven days after feeding, nymphs (fed and unfed) were processed for PCR analysis to determine spirochete-positive ticks, and subsequently qPCR to determine the number of spirochete genomes using enolase gene-specific primers (five spirochete-positive nymphs per clone). Results shown are mean ± SEM. Statistical analysis was performed using ANOVA test followed by Tukey Multiple Comparisons test. A P<0.05 between strains is considered significant. C. Naïve mice fed by the ΔcheY3-infected nymphs were not able to establish persistent infection. Fifteen infected encapsulated nymphs per mouse were allowed to feed. Four days (or 68h, data not shown) after feeding, mice were euthanized to detect B. burgdorferi DNA from tick bite site skin, ear, joint, and bladder tissues by PCR (one half of each tissue or one joint tissue from each mouse). To validate the PCR data, the other half of each of those tissues was processed for bacterial outgrowth analysis (not shown). WT and cheY3+ spirochetes were detected only in the tick-bite site skin tissues, as expected, at this early time points when B. burgdorferi cells generally do not disseminate to the distant tissues.
Intravital microscopy shows that ΔcheY3 motility behavior within skin is similar to its in vitro phenotype
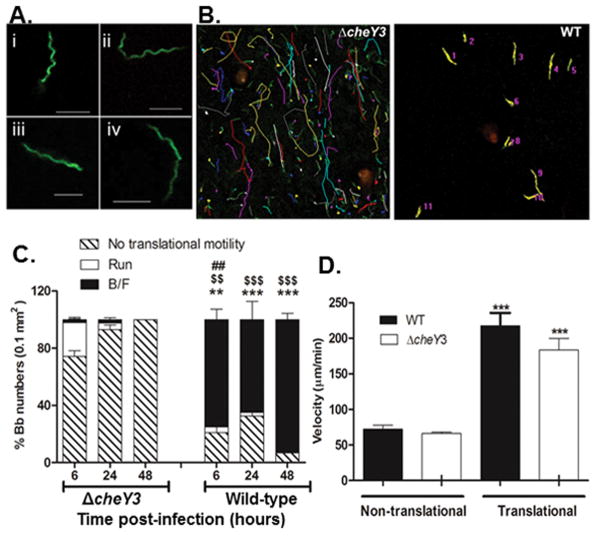
The finding that the ΔcheY3 was unable to infect mice via needle-injection or tick-bite suggested this mutant would demonstrate similar deficiencies in motility/chemotaxis in vivo as observed in vitro. To address this, intravital confocal fluorescence microscopy was used to directly observe GFP-expressing strains of WT and ΔcheY3 within the intact skin tissues of living mice. No notable differences were observed in the shapes of the WT GFP-expressing B. burgdorferi (WT-eGFP) and ΔcheY3-GFP-expressing B. burgdorferi (ΔcheY3-eGFP) spirochetes, suggesting that loss of this chemotaxis protein does not alter the morphology of B. burgdorferi within skin tissues. Assessment of time-lapse images taken at 6 hours post-injection showed that the majority of WT bacteria displayed a back-and-forth (B/F) motion (see Experimental Procedures section for a description of different motility events), with only a few showing directed runs (Figure 5B and Video 1) (Harman et al., 2012). Alternatively, while the majority of the ΔcheY3 mutants appeared non-motile, those that did display translational motility were unable to reverse their direction; even when they did stop moving, they were only able to continue movement in the same direction (Figure 5B and Video 2). Viewing the non-motile ΔcheY3 at a higher magnification showed that the majority of these mutants appeared to be trying to move forward, but could only make a minimal gain as they appeared stuck and were returned to the same spot, resulting in no net movement. This suggests that certain tissue components were hindering B. burgdorferi movement, and since the ΔcheY3 cannot reverse direction to find an unhindered route, they lose translational motility. Based on our described motility patterns (see Experimental Procedures), WT bacteria maintained the B/F motion as the major motility behavior over a period of 48 hours, whereas the majority of the ΔcheY3 bacteria were non-translational by 6 hours and almost all were non-motile at 48 hours (Figure 5C), suggesting the ΔcheY3 mutants are either stuck or become non-motile and/or cleared. Since a significant number of ΔcheY3 bacteria were non-translational by 6 hours, the velocity of bacteria that could translocate (translational, includes both run and B/F) and the ones that could not (non-translating bacteria defined above) were calculated separately. The average velocity of translating WT bacteria was 217.7 μm/min while translating ΔcheY3 bacteria was 183.4 μm/min (Figure 5D). Thus, even though ΔcheY3 were unable to reverse direction, they could still achieve velocities similar to that of the WT bacteria in skin. The velocity of bacteria at later times post-injection was not calculated, since the majority the ΔcheY3 bacteria (>90%) were non-translational after 6 hours post-inoculation. These observations suggest that the loss of cheY3 affects the motility pattern, but not the velocity potential of B. burgdorferi within murine skin.
Figure 5. Comparison of motility patterns in murine ear tissue between ΔcheY3 and WT.
A. Morphology of WT and ΔcheY3 B. burgdorferi was observed in vivo in ear skin tissue of mouse using intravital microscopy technique. 1×106 WT-eGFP or ΔcheY3-eGFP B. burgdorferi was injected intradermally into ear skin and images were collected between 2-6 hours post-injection. Both WT (i–ii) and ΔcheY3 (iii–iv) bacteria demonstrate similar characteristic flat-wave morphologies in vivo. Scale bar is 5μm. B–C. 1×106 WT-eGFP or ΔcheY3-eGFP B. burgdorferi (Bb) were injected as above and time-lapse images were collected at different times post-injection. B. Representative images of ΔcheY3 (left) and WT (right) motility path tracked at 6h post-injection using MetaMorph. Colored lines (except green) are the tracks of the bacteria. C. For both strains, the % bacteria performing run, back-and-forth (B/F) and no translational motility was calculated. The majority of WT bacteria at all times performed B/F, whereas most of the ΔcheY3 bacteria were non-translational. The translating ΔcheY3 performed mainly runs with occasional stops, rather than B/F. Results show average % motility ± SEM. ## p = 0.003 % run compared to ΔcheY3; $$ p = 0.0026, $$$ p ≤ 0.0003 % B/F compared to ΔcheY3; ** p = 0.0057, *** p ≤ 0.006 % no translational motility compared to ΔcheY3. Statistics were performed using the Mann Whitney test. n ≥ 3 mice. D. The velocity of WT and ΔcheY3 was measured at 6h post-injection using MetaMorph. Results show average ± SEM. ***p<0.0001 compared to the non-translational counterparts; Statistics were performed using the Mann Whitney test. n ≥ 30 bacteria under each bar.
ΔcheY3 bacteria fail to disseminate from the skin injection site to other target tissues and are cleared within 4 days post-injection
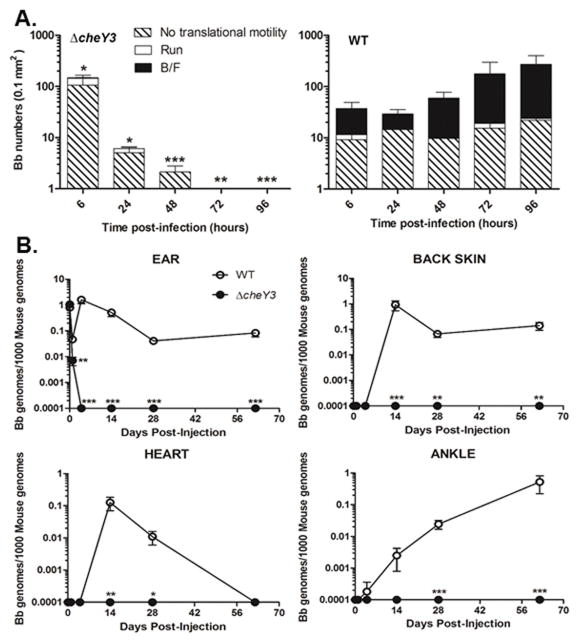
To determine the persistence of the mutant in mice, ΔcheY3-eGFP and WT-eGFP bacteria were injected intradermally into ear skin and confocal microscopy images were collected at the indicated times post-injection for manual enumeration of the spirochetes in each image. The burden of WT bacteria appeared to initially decrease between 6 hours and 24 hours post-injection, but subsequently increased dramatically between 48 hours and 96 hours post-injection (Figure 6A, right panel). Alternatively, the burden of ΔcheY3 steadily decreased after 6 hours post-injection, such that none of the mutants were visible by 96 hours post-injection, suggesting that the murine immune system cleared the ΔcheY3 from skin tissues within 96 hours post-injection. Although the same number of WT and ΔcheY3 spirochetes was injected, there appeared to be more ΔcheY3 than WT bacteria per viewing field at 6 hours post-injection (Figure 6A). We speculate that these differences are artificial due to more ΔcheY3 spirochetes becoming stuck at the injection site compared to WT spirochetes, since WT are capable of efficiently migrating through skin. These data indicate that loss of cheY3 leads to increased clearance of B. burgdorferi within murine skin tissues.
Figure 6. Dissemination and persistence of ΔcheY3 in distant target tissues.
A. Groups of C57Bl/6 mice were injected intradermally in both ears with 1×106 GFP-expressing ΔcheY3 or WT. At the indicated times, ear skin-resident bacteria were visually assessed using confocal fluorescent microscopy. Visual data was processed manually to determine motility patterns. Results shown are mean ± SD. *p = 0.04, **p < 0.0069 ***p < 0.0001 compared to the WT; Results were analyzed using an unpaired t-test; n ≥ 3 mice under each bar. B. Groups of C57Bl/6 mice were injected intradermally in both ears with 5×104 GFP-expressing ΔcheY3 or WT B. burgdorferi. Both ears, back skin, heart, and both ankles tissues were harvested at the indicated time points for DNA isolation. qPCR analysis was performed to quantify spirochete burdens. Values represent the average of B. burgdorferi genomes (flaB) per 1000 copies of mouse genome (nidogen); values of zero were assigned as 0.0001 for representation on a log scale. *p<0.05, **p<0.01, ***p<0.0001 compared to WT; Results were analyzed using the Mann Whitney test. n ≥ 5 mice from two separate experiments.
Although the ΔcheY3 appeared to be cleared relatively quickly at the inoculation site, it was still feasible that their limited mobility could allow dissemination to distant tissues via the bloodstream or other routes. To address this, B6 mice were intradermally injected with WT-eGFP or ΔcheY3-eGFP spirochetes and a number of target tissues (e.g. distant back skin, ankles, and heart) were harvested at the indicated times post-injection for bacterial enumeration by qPCR (Figure 6B). ΔcheY3 were cleared from all ear tissues (i.e. the injection site) by 96 hours post-injection, whereas WT spirochetes persisted in ear tissues (Figure 6B; top left), corroborating our intravital microscopy results (Figure 6A). Assessment of all distant tissues (i.e. back skin, heart, and ankles) indicated that WT B. burgdorferi were detected in all tested tissues by day 14–28 post-injection. However, no ΔcheY3 were detected in any of these tissues at any times post-injection, implying that these mutants did not escape the injection site but were instead cleared from the local ear tissues by the murine immune system (Figure 6B). This is consistent with the tick-mouse infection studies (Figure 3C and 4C), where ΔcheY3 spirochetes could not be re-isolated or detected from mouse tissues harvested 4–28 days after infected-ticks fed to repletion. Thus, loss of cheY3 leads to early clearance of B. burgdorferi in mice and an inability to disseminate to distant tissues.
Infection with ΔcheY3 is sufficient to elicit B. burgdorferi-specific antibodies
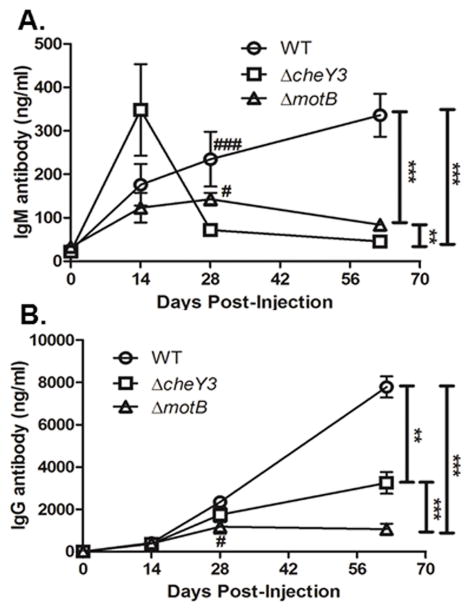
Typically during any infection, an immunocompetent vertebrate host initiates a primary antibody response that consists mainly of IgM antibodies. As this initial response starts to wane, the host then generates a stronger secondary antibody response that is primarily comprised of pathogen-specific IgG antibodies. Interestingly, B. burgdorferi infection of both mice and humans elicits a continuously increasing IgM response during an active infection together with an IgG response that is not well-maintained after the infection is cleared (Kalish et al., 2001, Hastey et al., 2012, Elsner et al., 2015). When the antisera from our current experiments were assessed by ELISA, WT bacteria elicited a strong B. burgdorferi-specific IgM response that continued to increase even after 60 days post-injection (Figure 7A), similar to those studies listed previously. Alternatively, ΔcheY3 infection elicited IgM levels similar to WT ≤28 post-injection, but these levels subsequently decrease by day 62 (Figure 7A). B. burgdorferi-specific IgG levels generated in response to the ΔcheY3 were also similar to WT ≤28 post-injection (Figure 7B); however the WT IgG levels continue to increase between days 28 and 62 post-injection, whereas antibody levels against ΔcheY3 did not increase after day 28, but was maintained for the duration of this experiment. Interestingly, the IgG levels against a non-motile ΔmotB (lacks the MotB motor stator), which we previously reported were cleared within 48–72 hours post-injection and elicited a minimal B. burgdorferi-specific antibody response, were lower than both WT and ΔcheY3 at day 62 post-injection (Sultan et al., 2015). Thus, although the IgM response is abbreviated in the ΔcheY3-infected mice, the infection is still sufficient to generate a robust B. burgdorferi-specific IgG response that persists well after bacterial clearance.
Figure 7. Infection with ΔcheY3 is sufficient to elicit B. burgdorferi-specific antibodies.
The mice used in the dissemination study (Figure 6) were sacrificed at the indicated time post-injection and sera were collected. B. burgdorferi-specific antibodies were quantified by ELISA analyses using bacterial sonicates as the capture antigen. A. Detection of IgM antibody levels in serum from WT-, ΔcheY3- and ΔmotB-infected mice. ΔmotB spirochetes was used as a control as these bacteria are reported to be non-motile, fail to disseminate from the injection site, and are cleared from the host within 48-72h after injection. **p < 0.01, ***p < 0.0001 as shown in the graph. # p < 0.05 and ### p < 0.0001 as compared to the ΔcheY3. Results were analyzed using the ANOVA test followed by the Tukey-Kramer Multiple Comparisons test. B. Detection of IgG antibody levels in serum from WT-, ΔcheY3- and ΔmotB-infected mice. **p < 0.01, ***p < 0.0001 as shown in the graph. # p < 0.05 as compared to WT. Statistical analyses were performed using ANOVA test followed by Tukey-Kramer Multiple Comparisons test.
DISCUSSION
The enzootic lifecycle of B. burgdorferi requires that it cycles between a tick vector and a vertebrate host (Burgdorfer et al., 1982, Welch et al., 1993, Toker et al., 1997). This necessitates that the spirochete sense and assess its external environment, and subsequently responds in an appropriate fashion by changing its cellular behavior and/or gene expression accordingly. The chemosensory system of B. burgdorferi would presumably aid in such sensing as well as traversing a path between the tick vector and the mammalian host.
Previous results indicate that all of the chemotaxis genes in the flaA-cheA2-cheW3-cheX-cheY3 operon are important for chemotaxis in vitro (Li et al., 2002, Motaleb et al., 2005, Motaleb et al., 2011b, Zhang et al., 2012). Additionally, previous studies have demonstrated that CheA2 is capable of both autophosphorylating and transferring that phosphoryl group to CheY3 (Motaleb et al., 2005). Therefore, it is plausible that CheA2 and CheY3 comprise a two-component system in B. burgdorferi, and that CheA2 is the cognate kinase for CheY3.
The in vivo studies with the ΔcheY3 demonstrated that burden of the ΔcheY3 in fed but not unfed ticks was significantly reduced compared to the burden of parental spirochetes (Figure 4A, B). The reason for this observed reduced burden in ticks is unknown. It is possible that since the ΔcheY3 is non-chemotactic and only able to swim in one direction it is easily identified and captured by the innate immune system of the tick or that ingested blood factors more efficiently cleared the mutant organisms (Ribeiro et al., 1990, Kern et al., 2011, Hajdusek et al., 2013). Based on our previous investigations with motility and cyclic-di-GMP mutants, we proposed that back-and-forth motility is crucial to protect the spirochetes in the fed ticks (Sultan et al., 2010, Pitzer et al., 2011, Sultan et al., 2013, Novak et al., 2014, Motaleb et al., 2015, Sultan et al., 2015). The same proposal can be applied here, as the ΔcheY3 spirochetes are unable to reverse their swimming patterns. Nevertheless, further studies are needed in order to determine the cause for the reduced burden of the ΔcheY3 in ticks.
The inability of the chemotaxis-deficient ΔcheY3 to establish an infection in mice is not completely understood, but could be due to the inability of the mutant to disseminate to target organs and/or evade the host cellular immune responses, as these mutant cells are not able to relay its CheY3-associated signals to the flagellar motor, and thus would not respond appropriately to certain chemotactic signals within their microenvironment (Table 1 and Figure 3C, 4C). We noticed that the in vivo phenotype of the mutant was partially restored in the cheY3+ cells (Figure 4C) even though we complemented the mutant by genomic reconstitution, and the CheY3 protein expression was restored to WT level (Figure 1). The reason for the partial restoration could be due to the loss of some of the endogenous B. burgdorferi plasmids in part of the population of the cheY3+ cells (Sultan et al., 2013). Nonetheless, our current data suggest that the failure of ΔcheY3 to switch its swimming behavior is causing the mutant to become trapped in the skin tissues. Skin dermis is a meshwork composed of a variety of ECM molecules, including collagen, elastin, and an extrafibrillar matrix made up of glycosaminoglycans (GAGs), proteoglycans, and glycoproteins (Leong et al., 1998, Parveen et al., 2000, Jarvelainen et al., 2009). WT B. burgdorferi appear able to detect barriers in its motility pathway and subsequently reverse or change its direction before taking an adjacent path that might be comprised of less dense tissue. Alternatively, the ΔcheY3 is unable to reverse and adjust its direction, and thus will eventually become trapped after translocating into a dense area, indicating that back-and-forth movement is essential for maneuvering around complex structures in the skin tissue (Moriarty et al., 2008, Norman et al., 2008). This is supported by the significant reduction of ΔcheY3 in skin by 24 hours and complete clearance by 96 hours post-injection (Figure 6A), suggesting that these non-translating spirochetes were quickly recognized and efficiently cleared by innate immune components.
There are currently no commercially available vaccines to protect against B. burgdorferi infection. Many of the individual purified proteins tested as vaccines do not confer complete protection (Hanson et al., 1998, Exner et al., 2000, Hagman et al., 2000, Nogueira et al., 2012, Floden et al., 2013). Passive transfer of serum from mice (or humans) injected with WT bacteria or with certain B. burgdorferi proteins seem to provide limited protection (Fikrig et al., 1994, Barthold et al., 1997, Hanson et al., 1998, Fikrig et al., 2000, Hagman et al., 2000, Floden et al., 2013, Small et al., 2014), suggesting that antibodies can provide protection if generated against critical antigens. Attenuated or killed vaccines have been successful in preventing many bacterial and viral infections (Mc, 1948, Mekalanos, 1994, Jin et al., 2015), but there are few studies screening non-infectious B. burgdorferi mutants for potential protective abilities. Immunization with killed or an aflagellar B. burgdorferi mutant conferred protection to naïve mice only if the mice were challenged early with WT bacteria (Johnson et al., 1986, Sadziene et al., 1996). But similar to passive immunization results, the protective ability of these attenuated (or killed) strains was decreased if the mice were challenged late (≥ 90 days post-immunization) (Johnson et al., 1986, Sadziene et al., 1996). One of the reasons surmised for this failure to achieve long-lasting immunity was that these mutants did not persist more than 24 hours post-injection, which is insufficient for generating an effective antibody response, particularly since it theoretically would not provide enough time for the mutants to upregulate critical virulence proteins that are expressed during vertebrate infection. Our observation that the ΔcheY3 survived ≤96 hours post-injection suggested that these mutants might persist long enough to upregulate more vertebrate host-specific virulence factors that could elicit antibodies capable of conferring protection from subsequent challenges. Our studies showed that infection with the ΔcheY3 generated similar levels of spirochete-specific IgM and IgG as WT B. burgdorferi through day 28 post-infection before reaching a significant baseline level that was maintained at least through day 62 post-infection. Notably, these levels were also significantly higher at all times tested than those generated against a non-motile ΔmotB strain that was shown to be cleared within 48–72 hours post-infection (Sultan et al., 2015). This implies that infection with the ΔcheY3 could potentially generate sufficient levels of spirochete-specific antibodies to confer protection against future challenges with fully virulent B. burgdorferi strains. If so, such studies might also allow the identification of individual antigens that can elicit protective antibodies, potentially leading to a vaccine based on recombinant proteins. Further studies are needed to demonstrate that infection with the mutant protects against WT infection.
As mentioned, B. burgdorferi contains three homologs of the cheY gene, which are located in separate operons (Motaleb et al., 2011b). However, lack of cheY3 was not compensated for by the other cheY homologs, and evidence suggests that cheY1 and cheY2 are not important for motility and chemotaxis in vitro (Motaleb et al., 2011b). So, what then is the function of these additional cheY genes? Many bacterial genomes contain multiple copies of a chemotaxis cheY gene and at least one of them is shown to be dedicated to control motility, similar to what we report in this communication with cheY3. The function of most of the additional genes is unknown (in any bacterium). There are a few instances in which multiple chemotaxis-like signaling systems appear to have very different roles in the same species and those additional cheY genes may control other cellular non-chemotactic processes (Kato et al., 1999, Szurmant et al., 2004, Whitchurch et al., 2004, Berleman et al., 2005). Consequently, it is plausible that B. burgdorferi CheY2 or CheY1 controls some non-chemotactic cellular processes such as serving as virulence determinants rather than acting as a classical chemotaxis response regulator, like CheY3 (H. Xu et al—manuscript under review; our unpublished observations).
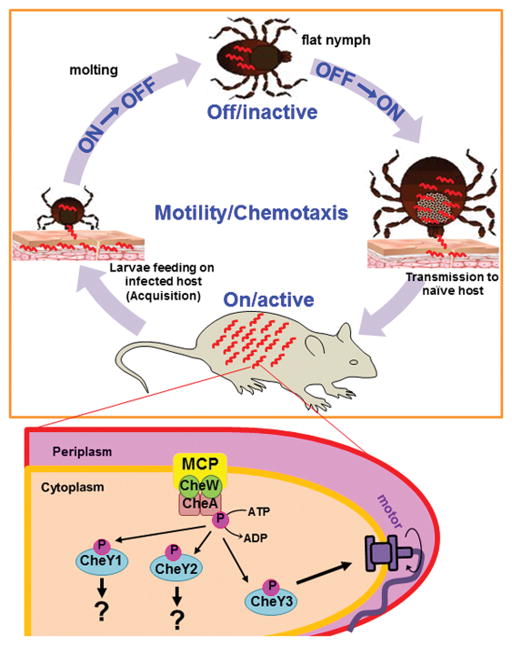
In summary, we have shown that the CheY3 response regulator is an essential component of the chemosensory system in B. burgdorferi, and that CheY3 is important for successful completion of the enzootic lifecycle and for continued perpetuation of Lyme disease. Based on our data, we propose that the chemosensory system of B. burgdorferi, and by extension motility, is critical during mammalian infection, dissemination, and possibly transmission from tick to the vertebrate host (Figure 8) (Motaleb et al., 2015). The motility/chemotaxis system is likely to be “active” or “on” during dissemination and persistent infection of the mammalian host, as well as during the tick’s blood-meal to allow navigation from the mouse into the tick and subsequent colonization of the mid-gut. However, after the nutrients from the blood-meal are depleted during the molt, B. burgdorferi must shut down flagellar rotation and chemotaxis to conserve energy, as this (motility/chemotaxis) system is unnecessary during molting (Figure 8) (Motaleb et al., 2015). Motility and chemotaxis must then be turned back “on” during nymphal feeding for subsequent transmission of the spirochetes. Although the latest time point post-injection analyzed by intravital microscopy in this study was 96 hours, we have examined WT-eGFP bacteria constantly swimming in the murine host for >2 years post-injection (R. M. Wooten and M. A. Motaleb, unpublished data). Accordingly, we propose that continuous chemotaxis/motility activities are necessary for persistent mammalian infection and are still required even after B. burgdorferi reaches its target colonization tissues in the mammal host. As outlined in Figure 8, CheY proteins are phosphorylated by CheA-P. CheY3-P then binds to the flagellar switch proteins to alter swimming behavior until it is dephosphorylated by the CheX phosphatase (Motaleb et al., 2005, Pazy et al., 2010). Based on our data, only CheY3 appears to act as a classical response regulator, as it contributes to motility and chemotaxis. Further studies are needed to not only elucidate the environmental signals that trigger the “active/on” or “inactive/off” status of the chemotaxis/motility system, but also the exact molecular components that transduce these signals within the spirochete.
Figure 8. Model of the B. burgdorferi chemotaxis/motility system during the enzootic cycle.
A simplistic chemotaxis signaling pathway of B. burgdorferi (wave-like red shapes) is depicted here and is described in the Discussion. We postulate that CheY2 or CheY1 controls some non-chemotactic cellular processes such as serving as virulence determinants rather than acting as a classical chemotaxis response regulator, like CheY3. Tick illustrations were kindly provided by the Medical Entomology and Zoonoses Ecology team at Public Health, England.
EXPERIMENTAL PROCEDURES
Ethics statement
East Carolina University and University of Toledo are both accredited by the International Association for the Assessment and Accreditation of Laboratory Animal Care. All animal procedures received Institutional Animal Care and Use Committee approvals and were in accordance with federal guidelines for the care and use of laboratory animals.
Mouse strains
All intravital microscopy experiments were performed utilizing C57BL/6 (B6; National Cancer Institute); all other experiments were completed using C3H/HeN mice (Charles River Laboratories, Raleigh, NC). These mouse strains contain equivalent susceptibilities to infection with B. burgdorferi and have been shown to contain similar bacterial numbers in most tissues during persistent infection, even though they can yield different levels of disease severity (Ma et al., 1998, Brown et al., 1999b, Weis et al., 1999).
Bacterial strains and growth conditions
Low-passage, virulent B. burgdorferi strain B31-A3 was utilized as the WT clone throughout this study (Elias et al., 2002). This clone was used to infect naïve C3H/HeN mice and then was reisolated from the mouse tissues, subcloned, and used as the parental clone for all of our subsequent studies. The genome of this strain is known to contain 12 linear and 9 circular plasmids, for a total of 21 plasmids, in addition to a 960-kbp linear chromosome (Fraser et al., 1997, Casjens et al., 2000). This strain lacks circular plasmid 9 (cp9), but still maintains its infectivity in tick-mouse infection studies (Elias et al., 2002, Jewett et al., 2009). Amplified genomic DNA from B31-A3 was used as the foundation to construct the ΔcheY3 (explained below). B. burgdorferi cells were grown in liquid Barbour-Stoenner-Kelly (BSK-II) medium, and cells were plated using plating BSK (P-BSK), which was prepared using 0.5% agarose (Motaleb et al., 2007, Stewart et al., 2008b, Sultan et al., 2013). Cells were grown at 35°C in a 2.5% CO2 incubator, as previously described (Elias et al., 2002, Smith et al., 2003, Motaleb et al., 2007). When required, culture and plating medium were supplemented with appropriate antibiotics at the following concentrations: 200 μg/ml kanamycin and 100 μg/ml streptomycin. The endogenous plasmid contents from all B. burgdorferi strains were confirmed before commencing any in vivo study involving ticks or mice.
Construction of the ΔcheY3, complement, and green-fluorescent protein (GFP) strains
The B. burgdorferi cheY3 gene was identified from the genomic sequence of B. burgdorferi and was annotated as bb0672 (441 bp) (Fraser et al., 1997). Construction of the cheY3-deletion plasmid, electroporation, and plating conditions were described previously (Sultan et al., 2010, Pitzer et al., 2011). Briefly, three pieces of DNA fragments were amplified separately by PCR: cheY3 gene plus adjacent DNA from the 5′-end (1 kb), the promoterless kanamycin resistance cassette, and the 3′-flanking DNA of cheY3 (1 kb). PCR primer sequences are not shown here but can be obtained upon request. These three pieces of fragments were gel purified and then ligated by overlapping PCR, as described in detail (Motaleb et al., 2011a). The PCR product was then ligated into the pGEM-T Easy vector (Promega, Inc.), yielding pCheY3-Pl-Kan-Easy. DNA containing cheY3-Pl-Kan was linearized by NotI restriction digestion to remove the ampicillin marker of the vector and electroporated into competent B31-A3 cells to obtain mutants (Motaleb et al., 2000, Sultan et al., 2013). Transformants were screened by PCR for proper recombination of the cheY3 inactivation cassette. The ΔcheY3 GFP-expressing strain was constructed in the same manner as the ΔcheY3, except the linearized DNA was electroporated into a B31-A3-GFP expressing strain, as described previously (Sultan et al., 2015).
In order to complement the cheY3 mutation, the cheY3 and cheX gene were amplified from genomic DNA along with adjacent flanking DNA using the primers (5′-3′): CheY3.FORWARD (TTGGATGCTGCTTCTTCGG) and CheY3.REVERSE (GCTGCTTGCATTGTTAG). The resulting PCR product was ligated into the pGEM-T Easy vector, yielding RCY3-Easy. The PflgB-aadA cassette, which confers resistance to streptomycin (Frank et al., 2003), was similarly amplified with engineered HindIII sites by PCR using the following primers (5′-3′): Flg-F (AAGCTTCCCGAGTTCAAGGAAGAT) and Strep-R (AAGCTTATTATTTGCCGACTACCTTGG). The cassette was then inserted into the unique HindIII site after the cheY3 gene. DNA containing cheX-cheY3-aadA was linearized by restriction digestion and was then electroporated into competent ΔcheY3 cells in order to obtain complemented cells. Transformants were selected with streptomycin.
Western blot analysis was used to confirm the inactivation and restoration of CheY3 in the mutant and complemented cells, respectively, and that no disruption of other gene products in the operon had occurred as a result of the genetic manipulation (see below). Linear and circular plasmid contents of all B. burgdorferi transformants were confirmed by PCR using primers described previously (Elias et al., 2002, Sultan et al., 2010, Pitzer et al., 2011).
SDS-PAGE and immunoblot analyses
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblotting with an enhanced chemiluminescent detection method (GE Health Inc.) were performed as reported previously (Motaleb et al., 2000). A Bio-Rad protein assay kit determined the concentration of protein in cell lysates. Unless otherwise noted, 5 μg of lysate protein was subjected to SDS-PAGE and immunoblotting using specific antibodies. Antibodies kindly provided by other investigators included the following: anti-DnaK by J. Benach (State University of New York [SUNY], Stony Brook, NY), and polyclonal E. coli anti-CheA by R. Silversmith (University of North Carolina, Chapel Hill, NC). CheY3 and CheX antibodies are described elsewhere (Motaleb et al., 2005, Motaleb et al., 2011b). Mouse serum collected at indicated times post-injection was utilized as the ‘primary antibody’ in western blot analysis to determine the B. burgdorferi-specific antibody response.
Dark-field microscopy and swarm plate assays
Exponentially growing B. burgdorferi clones (1–3×107 cells/ml) were imaged using a Zeiss Imager M1 dark-field microscope connected to a digital camera to determine morphology and motility. Swarming ability of individual bacterial colonies of each strain was determined by plating no more than 50 cells into a petri dish (95 mm × 15 mm) containing semi-solid P-BSK (0.35% agarose) diluted 1:10 in Dulbecco’s phosphate-buffered saline (Motaleb et al., 2000, Motaleb et al., 2007, Motaleb et al., 2011a). Plates were incubated for three weeks at which time colony diameters were measured. At least 11 colony diameters were measured for each strain in each assay.
Capillary tube chemotaxis assay
The chemotactic ability of B. burgdorferi was quantitatively determined as previously described (Motaleb et al., 2011b) with slight modifications. Briefly, WT, ΔcheY3, or cheY3 complement (cheY3+) B. burgdorferi were grown to approximately 1×108 cells/ml. Spirochetes were counted using a Petroff-Hausser chamber, and the cells needed for the assay (1×107 cells/ml) were collected by centrifuging for 15 minutes at 1,800 × g at room temperature. The cells were washed with PBS and then gently resuspended in motility buffer (136.9 mM NaCl, 8.10 mM Na2HPO4, 2.7 mM KCl, 1.47 mM KH2PO4, 2% recrystallized BSA (Sigma-Aldrich Co.), 0.1 mM EDTA, pH 7.4) to obtain a final solution of 1×107 cells/ml, 0.5% methylcellulose (400 mesh; Sigma Aldrich Inc.). This suspension (0.2 ml) was added into each 1.5 ml eppendorf tube chemotaxis chamber with a perforated lid. Capillary tubes were filled with control solution (motility buffer) or attractant solution (100 mM N-Acetylglucosamine in motility buffer, 0.22μm filter-sterilized) and sealed on one end with clay. Excess fluids on the outside of the capillary tubes were wiped off, and the capillary tubes were inserted into the appropriate eppendorf tube chemotaxis chambers (one capillary tube per chamber; 3–5 replicates per strain). Tubes were incubated at 35°C for 2 hours, at which point the tubes were carefully wiped off to remove excess liquid on the outside, and the contents was expelled into a clean eppendorf tube. The contents of each chemotaxis tube were plated individually using P-BSK to determine colony-forming units (CFU). The plates were incubated at 35°C for 2–5 weeks (until colonies appeared). Each strain was repeated three times with each repeat containing 3 replicates, and the results are expressed as the average attractant/buffer ratio. An increase in the number of spirochetes equal to or greater than twice that of the buffer control was considered significant.
Mouse infection studies using injected B. burgdorferi
Six-to-seven weeks old C3H/HeN mice were used for infection studies, as previously described (Elias et al., 2002, Stewart et al., 2008a, Sultan et al., 2013). In order to determine the infectious ability of the spirochetes, mice (n = 5 or 2 per strain) were injected subcutaneously with WT (5×103), ΔcheY3 (5×103 or 1×106), or cheY3+ cells (5×104). The number of spirochetes was determined using a Petroff-Hausser chamber and verified by colony forming units (CFUs) by plating. At four weeks post-injection, mice were euthanized and ear skin, bladder, and tibiotarsal joints were harvested and placed in BSK-II broth for up to 35 days to allow bacterial outgrowth from the animal tissues (Elias et al., 2002, Grimm et al., 2003, Stewart et al., 2008a), which is the direct determination of the ability of spirochetes to infect mice by tick bite and disseminate throughout the body. The presence of spirochetes in the growth medium was determined by dark-field microscopy (Elias et al., 2002, Grimm et al., 2003, Grimm et al., 2004, Sultan et al., 2013).
Tick-mouse studies
Transmission of spirochetes from infected ticks to naïve mice was assessed using tick-mouse infection assays (Policastro et al., 2003, Battisti et al., 2008, Stewart et al., 2008a). Naïve Ixodes scapularis larvae were purchased from Oklahoma State University. Naïve larvae were artificially inoculated by immersion in equal-density, exponential-phase (5×107 cells/ml) cultures of B. burgdorferi clones, as previously described (Policastro et al., 2003, Battisti et al., 2008, Stewart et al., 2008a). Ticks were subsequently fed to repletion on mice (3 mice per strain; ~200 larvae/mouse). After 5–7 days, fed ticks were collected once they dropped off mice. At 7 days post-repletion, a subset of ticks was individually dissected and the isolated midguts were analyzed by immunofluorescence (IFA; see below) for the presence of spirochetes (Stewart et al., 2008a). A second subset of ticks was surface-sterilized with 3% H2O2 and 70% ethanol, individually crushed in BSK-II medium, and plated in P-BSK to determine the number of viable spirochetes per tick (CFUs). Results are expressed as the spirochete burden per tick, where each dot is representative of a single tick (n = 12). A third subset of ticks was crushed individually and genomic DNA was extracted using the DNeasy blood and tissue kit, according to the manufacturer’s instructions (Qiagen Inc.). The DNA from each tick was then utilized for PCR to determine spirochete-positive ticks. Spirochete burdens in each spirochete-positive tick were determined by using quantitative real-time PCR (qPCR) using primers specific for the B. burgdorferi enolase gene, as described previously (Yang et al., 2004, Zhang et al., 2009, Pitzer et al., 2011). Copies of the B. burgdorferi enolase gene per tick were extrapolated from a standard curve generated using a known amount of plasmid DNA containing the enolase gene as the template.
To determine spirochete transmission and infection of the C3H/HeN mice, fed animals were euthanized four weeks post-repletion followed by bacterial outgrowth analysis. The presence of spirochetes in the growth medium was determined by dark-field microscopy (Sultan et al., 2010, Sultan et al., 2011).
Transmission of spirochetes to mice by encapsulated nymphs
Naïve nymphs were artificially infected by immersion, as described above. Nymphs were allowed to feed on mice using a capsulated system, as previously described (Mulay et al., 2009, Patton et al., 2011, Sultan et al., 2013). Mice were anesthetized and 15–20 nymphs were confined to capsules affixed to the shaved back of a naïve C3H/HeN mouse (n = 3 per strain per assay). The ticks were allowed to feed to repletion and then collected from the capsules. At 7 days post-repletion, spirochete burdens were determined from individually crushed ticks by qPCR, as described above. The results are expressed as mean ± SEM from at least 4 spirochete-positive tick data per clone per assay.
At 68 hours or 4 days post-repletion, mice were euthanized and the tick-bite sites were extensively washed. A section of skin comprising the tick-feeding site was excised, rinsed in 70% isopropanol, and cut into equal portions. Part of the tick-bite site skin, ear, bladder, and joint tissues were cultured separately in BSK-II medium for up to 35 days to determine bacterial outgrowth, and the other remaining tissues were processed for PCR to detect B. burgdorferi DNA using enolase gene-specific primers (Pitzer et al., 2011).
Immunofluorescence Assay
IFAs were set up as previously described (Sultan et al., 2013). Briefly, ticks were individually dissected in phosphate-buffered saline (PBS)-5 mM MgCl2 on Teflon-coated microscopic slides. Dissected tick contents were then 10-fold serially diluted to avoid quenching by hemin in the blood (Policastro et al., 2003, Sultan et al., 2010). The slides were air dried and blocked with 0.75% bovine serum albumin (BSA) in PBS-5 mM MgCl2 for 30 min. The slides were then washed twice with PBS-5 mM MgCl2, and spirochetes were detected using goat anti-B. burgdorferi antisera labeled with fluorescein isothiocyanate (1:100 dilution; Kirkegaard & Perry Laboratories, Inc.). Images were captured using a Zeiss Axio Imager M1 microscope connected to a digital camera.
Intravital microscopy
Two days prior to an experiment, the outer ear surface of the mice was depilated (Nair), rinsed immediately with H2O, and allowed to rest. Low-passage cultures of WT-eGFP and ΔcheY3-eGFP were counted to contain the desired B. burgdorferi inoculum (106 bacteria) in 10μl BSK-II medium. B6 mice were anesthetized, and the desired numbers of GFP-expressing bacteria were injected intradermally in a 10μl bolus using a 31G insulin syringe via the dorsal ear surface of the mouse. The mice were allowed to rest for 6h and then re-anesthetized for imaging at the indicated times.
Imaging was performed using an Olympus FV1000 laser confocal microscope system. For imaging, the mouse was placed on a 37°C heated imaging stage to mount the ear on a coverslip (with the template) with the dorsal surface down. For bacterial enumeration, as well as motility patterns and velocity determination of the bacteria, 2D images were collected using a 20× dry objective with a 2× optical zoom at 1 frame/1100 milliseconds for 66 seconds (60 images) at different times post-injection. At least two time-lapse images were collected for each section of the template (=10 time-lapse images per ear). For bacterial morphology studies, images were collected between 2–6 hours post-injection using a 60× water immersion objective with an additional optical zoom.
Image Analysis for intravital microscopy
For bacterial enumeration and motility assessment, the first of the 60 images in a dataset was selected and the bacteria in that image were counted and tracked manually. Motility patterns and velocity were calculated for bacteria present in the viewing field since the first frame. For motility patterns, the movement of any bacteria that remained within the viewing field for ≥ 5 seconds was manually assessed and categorized as one of three groups: (1) Run—bacteria continue to move in one direction without reversing or switching the swimming direction throughout the entire time of assessment, (2) Back/Forth (B/F)—bacteria reversed/switched their direction of motion at least once throughout the entire time of assessment, and (3) Non-translational—bacteria had no net movement, which includes both non-motile bacteria and those that could move slightly, but were unable to generate net movement (translocate). Time of assessment of the bacteria was defined as the total time the bacteria were visible in the viewing field, which ranged from 5 seconds minimum up to 60 seconds. For calculating the velocity of the bacteria, all the 60 images of the dataset were assessed using MetaMorph software (Molecular Devices), and every bacterium noted on the first image was tracked through 60 images and the average velocity of the bacterium was calculated. For tracking, one end of the spirochete was chosen and it was followed until that end is not visible for more than 1 frame. Observations for bacteria that stayed in the field of view for less than 5 frames were discarded.
Quantitative measurement of B. burgdorferi in murine skin and other target tissues
The bacterial numbers in murine tissues were quantified as previously described (Brown et al., 1999a, Morrison et al., 1999). Briefly, naïve anesthetized B6 mice were intradermally injected with 5×104 bacteria per animal via the dorsal ear surface. Multiple tissues including the entire ears, back skin, entire ankle joints, and heart were collected at various times post-injection. The tissues were appropriately processed to isolate DNA, as described (Brown et al., 1999a, Morrison et al., 1999, Sultan et al., 2015). qPCR was performed using a Light Cycler 96 (Roche Diagnostics) and Syber Green detection. Mouse DNA levels were determined by amplifying the nidogen gene and B. burgdorferi DNA was determined by amplifying the flaB gene. Copy numbers for mouse and B. burgdorferi genomes present in each sample were calculated by extrapolation to standard curves using LightCycler software (Roche Diagnostics). Normalizing B. burgdorferi genomes to 1000 mouse genomes represented the final B. burgdorferi numbers. The primers used to detect mouse nidogen were nido.F (5′-CCA GCC ACA GAA TAC CAT CC-3′) and nido.R (5′-GGA CAT ACT CTG CTG CCA TC-3′). The oligonucleotide primers used to detect B. burgdorferi flaB were flaB.F (5′-TTG CTG ATC AAG CTC AAT ATA ACC A -3′) and flaB.R (5′-TTG AGA CCC TGA AAG TGA TGC -3′).
B. burgdorferi-specific antibody detection by ELISA from intravital microscopy studies
Serum was collected at the indicated times by either retro-orbital bleeding or exsanguination, and immunoglobulin (Ig) content was assessed using previously described methods (Brown et al., 1999a). For detection of B. burgdorferi-specific antibodies, 96-well high-binding ELISA plates (Costar) were coated with B. burgdorferi sonicate or goat anti-mouse total immunoglobulin (IgG+IgM+IgA; Southern Biotech). For B. burgdorferi sonicates, the WT bacteria were grown at 33ºC and then shifted to 37ºC for the final overnight culture (Schwan et al., 2000). This temperature-shifted culture was then resuspended in PBS for sonication within a closed sterile tube using a bath sonicator (Sonifier® Cell Disruptor). The protein content of the sonicated sample was measured by Bicinchoninic acid assay (BCA; Pierce Thermo Scientific) and stored at −80ºC. The B. burgdorferi ELISA plates were made by coating appropriate wells with 5μg/ml of the sonicate (for sample and blank) or goat anti-mouse total Ig (to provide a purified Ig standard) in 0.1M carbonate-bicarbonate Buffer (pH 9.5) overnight. Serial dilutions of individual sera from uninfected or infected mice were added to the B. burgdorferi-coated plates overnight, washed to remove unbound antibodies, and bound murine Ig was detected using goat anti-mouse IgG conjugated to biotin (Southern Biotech). After incubation for 2–3 hours at room temperature, the samples were visualized by adding avidin-conjugated Horseradish Peroxidase (avidin-HRP; Vector Labs) for 30 minutes followed by adding the color solution (0.4mg/ml of O-phenylenediamine and 0.01% H2O2 in citrate buffer, pH 5) and inhibiting the reaction with 1N hydrochloride. The content was quantified by comparison to standard curves constructed using the appropriate purified mouse Ig isotype (Southern Biotech).
Statistical analyses
The significance difference between the mean values of the groups for each experiment was analyzed as follows. When comparing WT, ΔcheY3, and cheY3+ (either the cheY3+ or ΔmotB) data was checked with D’Agostino-Pearson omnibus normality test to determine if the values come from a Gaussian distribution. The data that passed normality was analyzed via a multiple-comparison analysis using a one-way analysis of variance (ANOVA), followed by a Tukey’s post hoc test; if the data did not pass normality, a multiple-comparison analysis was performed by using a Kruskal-Wallis test, followed by Dunn test. P < 0.05 is considered significant. For all intravital and detection of GFP-expressing bacteria in mice, normality was checked using Shapiro-Wilk normality test. If data was normal than the difference between WT and ΔcheY3 was assessed via an unpaired t-test, whereas non-normal data was assessed via a Mann Whitney test.
Supplementary Material
1×106 WT-eGFP B. burgdorferi were injected intradermally into mouse ear skin and time-lapse images were collected using an Olympus FV1000 laser confocal microscope system at 6 hours post-injection. 2D images were collected using a 20× dry objective with a 2X optical zoom at 1frame/1100 milliseconds for 66 seconds (60 images). The video is sped up two times the “real-time” speed.
1×106 ΔcheY3-eGFP B. burgdorferi were injected intradermally into mouse ear skin and time-lapse images were collected using an Olympus FV1000 laser confocal microscope system at 6 hours post-injection. 2D images were collected using a 20× dry objective with a 2X optical zoom at 1frame/1100 milliseconds for 66 seconds (60 images). The video is sped up two times the “real-time” speed.
Acknowledgments
We thank Dr. Ruth Silversmith for reagents and the Medical Entomology and Zoonoses Ecology team at Public Health, England for the tick’s illustrations. We also would like to acknowledge John Presloid for technical help in performing the murine infection and imaging studies. This research was supported by National Institute of Allergy and Infectious Diseases grants (1R21AI113014), National Institute of Arthritis and Musculoskeletal and Skin Diseases grant (1R01AR060834), and an American Heart Association Pre-doctoral Fellowship 14PRE20490177 (PS).
References
- Antunez-Lamas M, Cabrera-Ordonez E, Lopez-Solanilla E, Raposo R, Trelles-Salazar O, Rodriguez-Moreno A, Rodriguez-Palenzuela P. Role of motility and chemotaxis in the pathogenesis of Dickeya dadantii 3937 (ex Erwinia chrysanthemi 3937) Microbiology. 2009;155:434–442. doi: 10.1099/mic.0.022244-0. [DOI] [PubMed] [Google Scholar]
- Armitage JP. Bacterial tactic responses. Advances in microbial physiology. 1999;41:229–289. doi: 10.1016/s0065-2911(08)60168-x. [DOI] [PubMed] [Google Scholar]
- Barthold SW, Feng S, Bockenstedt LK, Fikrig E, Feen K. Protective and arthritis-resolving activity in sera of mice infected with Borrelia burgdorferi. Clinical infectious diseases: an official publication of the Infectious Diseases Society of America. 1997;25(Suppl 1):S9–17. doi: 10.1086/516166. [DOI] [PubMed] [Google Scholar]
- Battisti JM, Bono JL, Rosa PA, Schrumpf ME, Schwan TG, Policastro PF. Outer surface protein A protects Lyme disease spirochetes from acquired host immunity in the tick vector. Infection and immunity. 2008;76:5228–5237. doi: 10.1128/IAI.00410-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berleman JE, Bauer CE. Involvement of a Che-like signal transduction cascade in regulating cyst cell development in Rhodospirillum centenum. Molecular microbiology. 2005;56:1457–1466. doi: 10.1111/j.1365-2958.2005.04646.x. [DOI] [PubMed] [Google Scholar]
- Bourret RB, Silversmith RE. Two-component signal transduction. Current opinion in microbiology. 2010;13:113–115. doi: 10.1016/j.mib.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bren A, Eisenbach M. How signals are heard during bacterial chemotaxis: protein-protein interactions in sensory signal propagation. Journal of bacteriology. 2000;182:6865–6873. doi: 10.1128/jb.182.24.6865-6873.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brisson D, Drecktrah D, Eggers CH, Samuels DS. Genetics of Borrelia burgdorferi. Annual review of genetics. 2012;46:515–536. doi: 10.1146/annurev-genet-011112-112140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown CR, Reiner SL. Development of lyme arthritis in mice deficient in inducible nitric oxide synthase. The Journal of infectious diseases. 1999a;179:1573–1576. doi: 10.1086/314774. [DOI] [PubMed] [Google Scholar]
- Brown JP, Zachary JF, Teuscher C, Weis JJ, Wooten RM. Dual role of interleukin-10 in murine Lyme disease: regulation of arthritis severity and host defense. Infection and immunity. 1999b;67:5142–5150. doi: 10.1128/iai.67.10.5142-5150.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgdorfer W, Barbour AG, Hayes SF, Benach JL, Grunwaldt E, Davis JP. Lyme disease-a tick-borne spirochetosis? Science. 1982;216:1317–1319. doi: 10.1126/science.7043737. [DOI] [PubMed] [Google Scholar]
- Butler SM, Camilli A. Going against the grain: chemotaxis and infection in Vibrio cholerae. Nature reviews. Microbiology. 2005;3:611–620. doi: 10.1038/nrmicro1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casjens S, Palmer N, van Vugt R, Huang WM, Stevenson B, Rosa P, et al. A bacterial genome in flux: the twelve linear and nine circular extrachromosomal DNAs in an infectious isolate of the Lyme disease spirochete Borrelia burgdorferi. Molecular microbiology. 2000;35:490–516. doi: 10.1046/j.1365-2958.2000.01698.x. [DOI] [PubMed] [Google Scholar]
- Charon NW, Cockburn A, Li C, Liu J, Miller KA, Miller MR, et al. The unique paradigm of spirochete motility and chemotaxis. Annual review of microbiology. 2012;66:349–370. doi: 10.1146/annurev-micro-092611-150145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djordjevic S, Stock AM. Structural analysis of bacterial chemotaxis proteins: components of a dynamic signaling system. Journal of structural biology. 1998;124:189–200. doi: 10.1006/jsbi.1998.4034. [DOI] [PubMed] [Google Scholar]
- Dunham-Ems SM, Caimano MJ, Pal U, Wolgemuth CW, Eggers CH, Balic A, Radolf JD. Live imaging reveals a biphasic mode of dissemination of Borrelia burgdorferi within ticks. The Journal of clinical investigation. 2009;119:3652–3665. doi: 10.1172/JCI39401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elias AF, Stewart PE, Grimm D, Caimano MJ, Eggers CH, Tilly K, et al. Clonal polymorphism of Borrelia burgdorferi strain B31 MI: implications for mutagenesis in an infectious strain background. Infection and immunity. 2002;70:2139–2150. doi: 10.1128/IAI.70.4.2139-2150.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsner RA, Hastey CJ, Olsen KJ, Baumgarth N. Suppression of Long-Lived Humoral Immunity Following Borrelia burgdorferi Infection. PLoS pathogens. 2015;11:e1004976. doi: 10.1371/journal.ppat.1004976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Exner MM, Wu X, Blanco DR, Miller JN, Lovett MA. Protection elicited by native outer membrane protein Oms66 (p66) against host-adapted Borrelia burgdorferi: conformational nature of bactericidal epitopes. Infection and immunity. 2000;68:2647–2654. doi: 10.1128/iai.68.5.2647-2654.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falke JJ, Bass RB, Butler SL, Chervitz SA, Danielson MA. The two-component signaling pathway of bacterial chemotaxis: a molecular view of signal transduction by receptors, kinases, and adaptation enzymes. Annual review of cell and developmental biology. 1997;13:457–512. doi: 10.1146/annurev.cellbio.13.1.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fikrig E, Bockenstedt LK, Barthold SW, Chen M, Tao H, Ali-Salaam P, et al. Sera from patients with chronic Lyme disease protect mice from Lyme borreliosis. The Journal of infectious diseases. 1994;169:568–574. doi: 10.1093/infdis/169.3.568. [DOI] [PubMed] [Google Scholar]
- Fikrig E, Feng W, Barthold SW, Telford SR, 3rd, Flavell RA. Arthropod- and host-specific Borrelia burgdorferi bbk32 expression and the inhibition of spirochete transmission. Journal of immunology. 2000;164:5344–5351. doi: 10.4049/jimmunol.164.10.5344. [DOI] [PubMed] [Google Scholar]
- Floden AM, Gonzalez T, Gaultney RA, Brissette CA. Evaluation of RevA, a fibronectin-binding protein of Borrelia burgdorferi, as a potential vaccine candidate for lyme disease. Clinical and vaccine immunology: CVI. 2013;20:892–899. doi: 10.1128/CVI.00758-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank KL, Bundle SF, Kresge ME, Eggers CH, Samuels DS. aadA confers streptomycin resistance in Borrelia burgdorferi. Journal of bacteriology. 2003;185:6723–6727. doi: 10.1128/JB.185.22.6723-6727.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser CM, Casjens S, Huang WM, Sutton GG, Clayton R, Lathigra R, et al. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature. 1997;390:580–586. doi: 10.1038/37551. [DOI] [PubMed] [Google Scholar]
- Grimm D, Elias AF, Tilly K, Rosa PA. Plasmid stability during in vitro propagation of Borrelia burgdorferi assessed at a clonal level. Infection and immunity. 2003;71:3138–3145. doi: 10.1128/IAI.71.6.3138-3145.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm D, Tilly K, Byram R, Stewart PE, Krum JG, Bueschel DM, et al. Outer-surface protein C of the Lyme disease spirochete: a protein induced in ticks for infection of mammals. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:3142–3147. doi: 10.1073/pnas.0306845101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagman KE, Yang X, Wikel SK, Schoeler GB, Caimano MJ, Radolf JD, Norgard MV. Decorin-binding protein A (DbpA) of Borrelia burgdorferi is not protective when immunized mice are challenged via tick infestation and correlates with the lack of DbpA expression by B. burgdorferi in ticks. Infection and immunity. 2000;68:4759–4764. doi: 10.1128/iai.68.8.4759-4764.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajdusek O, Sima R, Ayllon N, Jalovecka M, Perner J, de la Fuente J, Kopacek P. Interaction of the tick immune system with transmitted pathogens. Frontiers in cellular and infection microbiology. 2013;3:26. doi: 10.3389/fcimb.2013.00026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson MS, Cassatt DR, Guo BP, Patel NK, McCarthy MP, Dorward DW, Hook M. Active and passive immunity against Borrelia burgdorferi decorin binding protein A (DbpA) protects against infection. Infection and immunity. 1998;66:2143–2153. doi: 10.1128/iai.66.5.2143-2153.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harman MW, Dunham-Ems SM, Caimano MJ, Belperron AA, Bockenstedt LK, Fu HC, et al. The heterogeneous motility of the Lyme disease spirochete in gelatin mimics dissemination through tissue. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:3059–3064. doi: 10.1073/pnas.1114362109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastey CJ, Elsner RA, Barthold SW, Baumgarth N. Delays and diversions mark the development of B cell responses to Borrelia burgdorferi infection. Journal of immunology. 2012;188:5612–5622. doi: 10.4049/jimmunol.1103735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazelbauer GL. Bacterial chemotaxis: the early years of molecular studies. Annual review of microbiology. 2012;66:285–303. doi: 10.1146/annurev-micro-092611-150120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He K, Bauer CE. Chemosensory signaling systems that control bacterial survival. Trends in microbiology. 2014;22:389–398. doi: 10.1016/j.tim.2014.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvelainen H, Sainio A, Koulu M, Wight TN, Penttinen R. Extracellular matrix molecules: potential targets in pharmacotherapy. Pharmacological reviews. 2009;61:198–223. doi: 10.1124/pr.109.001289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jewett MW, Lawrence KA, Bestor A, Byram R, Gherardini F, Rosa PA. GuaA and GuaB are essential for Borrelia burgdorferi survival in the tick-mouse infection cycle. Journal of bacteriology. 2009;191:6231–6241. doi: 10.1128/JB.00450-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H, Subbarao K. Live attenuated influenza vaccine. Current topics in microbiology and immunology. 2015;386:181–204. doi: 10.1007/82_2014_410. [DOI] [PubMed] [Google Scholar]
- Johnson RC, Kodner C, Russell M. Active immunization of hamsters against experimental infection with Borrelia burgdorferi. Infection and immunity. 1986;54:897–898. doi: 10.1128/iai.54.3.897-898.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalish RA, McHugh G, Granquist J, Shea B, Ruthazer R, Steere AC. Persistence of immunoglobulin M or immunoglobulin G antibody responses to Borrelia burgdorferi 10–20 years after active Lyme disease. Clinical infectious diseases: an official publication of the Infectious Diseases Society of America. 2001;33:780–785. doi: 10.1086/322669. [DOI] [PubMed] [Google Scholar]
- Kato J, Nakamura T, Kuroda A, Ohtake H. Cloning and characterization of chemotaxis genes in Pseudomonas aeruginosa. Bioscience, biotechnology, and biochemistry. 1999;63:155–161. doi: 10.1271/bbb.63.155. [DOI] [PubMed] [Google Scholar]
- Kern A, Collin E, Barthel C, Michel C, Jaulhac B, Boulanger N. Tick saliva represses innate immunity and cutaneous inflammation in a murine model of Lyme disease. Vector borne and zoonotic diseases. 2011;11:1343–1350. doi: 10.1089/vbz.2010.0197. [DOI] [PubMed] [Google Scholar]
- Lane RS, Piesman J, Burgdorfer W. Lyme borreliosis: relation of its causative agent to its vectors and hosts in North America and Europe. Annual review of entomology. 1991;36:587–609. doi: 10.1146/annurev.en.36.010191.003103. [DOI] [PubMed] [Google Scholar]
- Leong JM, Robbins D, Rosenfeld L, Lahiri B, Parveen N. Structural requirements for glycosaminoglycan recognition by the Lyme disease spirochete, Borrelia burgdorferi. Infection and immunity. 1998;66:6045–6048. doi: 10.1128/iai.66.12.6045-6048.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lertsethtakarn P, Ottemann KM, Hendrixson DR. Motility and chemotaxis in Campylobacter and Helicobacter. Annual review of microbiology. 2011;65:389–410. doi: 10.1146/annurev-micro-090110-102908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine JF, Wilson ML, Spielman A. Mice as reservoirs of the Lyme disease spirochete. The American journal of tropical medicine and hygiene. 1985;34:355–360. doi: 10.4269/ajtmh.1985.34.355. [DOI] [PubMed] [Google Scholar]
- Li C, Bakker RG, Motaleb MA, Sartakova ML, Cabello FC, Charon NW. Asymmetrical flagellar rotation in Borrelia burgdorferi nonchemotactic mutants. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:6169–6174. doi: 10.1073/pnas.092010499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin T, Gao L, Zhang C, Odeh E, Jacobs MB, Coutte L, et al. Analysis of an ordered, comprehensive STM mutant library in infectious Borrelia burgdorferi: insights into the genes required for mouse infectivity. PloS one. 2012;7:e47532. doi: 10.1371/journal.pone.0047532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Seiler KP, Eichwald EJ, Weis JH, Teuscher C, Weis JJ. Distinct characteristics of resistance to Borrelia burgdorferi-induced arthritis in C57BL/6N mice. Infection and immunity. 1998;66:161–168. doi: 10.1128/iai.66.1.161-168.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mc KB. B.C.G. vaccine in the prevention of tuberculosis. Canadian Medical Association journal. 1948;58:575–577. [PMC free article] [PubMed] [Google Scholar]
- McGee DJ, Langford ML, Watson EL, Carter JE, Chen YT, Ottemann KM. Colonization and inflammation deficiencies in Mongolian gerbils infected by Helicobacter pylori chemotaxis mutants. Infection and immunity. 2005;73:1820–1827. doi: 10.1128/IAI.73.3.1820-1827.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mead PS. Epidemiology of Lyme disease. Infectious disease clinics of North America. 2015;29:187–210. doi: 10.1016/j.idc.2015.02.010. [DOI] [PubMed] [Google Scholar]
- Mekalanos JJ. Live bacterial vaccines: environmental aspects. Current Opinion in Biotechnology. 1994;5:312–319. doi: 10.1016/0958-1669(94)90035-3. [DOI] [PubMed] [Google Scholar]
- Moon KH, Hobbs G, Motaleb MA. Borrelia burgdorferi CheD promotes various functions in chemotaxis and pathogenic life cycle of the spirochete. Infection and immunity. 2016;84(6) doi: 10.1128/IAI.01347-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriarty TJ, Norman MU, Colarusso P, Bankhead T, Kubes P, Chaconas G. Real-time high resolution 3D imaging of the lyme disease spirochete adhering to and escaping from the vasculature of a living host. PLoS pathogens. 2008;4:e1000090. doi: 10.1371/journal.ppat.1000090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison TB, Ma Y, Weis JH, Weis JJ. Rapid and sensitive quantification of Borrelia burgdorferi-infected mouse tissues by continuous fluorescent monitoring of PCR. Journal of clinical microbiology. 1999;37:987–992. doi: 10.1128/jcm.37.4.987-992.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motaleb MA, Corum L, Bono JL, Elias AF, Rosa P, Samuels DS, Charon NW. Borrelia burgdorferi periplasmic flagella have both skeletal and motility functions. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:10899–10904. doi: 10.1073/pnas.200221797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motaleb MA, Liu J, Wooten RM. Spirochetal motility and chemotaxis in the natural enzootic cycle and development of Lyme disease. Current opinion in microbiology. 2015;28:106–113. doi: 10.1016/j.mib.2015.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motaleb MA, Miller MR, Bakker RG, Li C, Charon NW. Isolation and characterization of chemotaxis mutants of the Lyme disease Spirochete Borrelia burgdorferi using allelic exchange mutagenesis, flow cytometry, and cell tracking. Methods in enzymology. 2007;422:421–437. doi: 10.1016/S0076-6879(06)22021-4. [DOI] [PubMed] [Google Scholar]
- Motaleb MA, Miller MR, Li C, Bakker RG, Goldstein SF, Silversmith RE, et al. CheX is a phosphorylated CheY phosphatase essential for Borrelia burgdorferi chemotaxis. Journal of bacteriology. 2005;187:7963–7969. doi: 10.1128/JB.187.23.7963-7969.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motaleb MA, Pitzer JE, Sultan SZ, Liu J. A novel gene inactivation system reveals altered periplasmic flagellar orientation in a Borrelia burgdorferi fliL mutant. Journal of bacteriology. 2011a;193:3324–3331. doi: 10.1128/JB.00202-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motaleb MA, Sultan SZ, Miller MR, Li C, Charon NW. CheY3 of Borrelia burgdorferi is the key response regulator essential for chemotaxis and forms a long-lived phosphorylated intermediate. Journal of bacteriology. 2011b;193:3332–3341. doi: 10.1128/JB.00362-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulay VB, Caimano MJ, Iyer R, Dunham-Ems S, Liveris D, Petzke MM, et al. Borrelia burgdorferi bba74 is expressed exclusively during tick feeding and is regulated by both arthropod- and mammalian host-specific signals. Journal of bacteriology. 2009;191:2783–2794. doi: 10.1128/JB.01802-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogueira SV, Smith AA, Qin JH, Pal U. A surface enolase participates in Borrelia burgdorferi-plasminogen interaction and contributes to pathogen survival within feeding ticks. Infection and immunity. 2012;80:82–90. doi: 10.1128/IAI.05671-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norman MU, Moriarty TJ, Dresser AR, Millen B, Kubes P, Chaconas G. Molecular mechanisms involved in vascular interactions of the Lyme disease pathogen in a living host. PLoS pathogens. 2008;4:e1000169. doi: 10.1371/journal.ppat.1000169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak EA, Sultan SZ, Motaleb MA. The cyclic-di-GMP signaling pathway in the Lyme disease spirochete, Borrelia burgdorferi. Frontiers in cellular and infection microbiology. 2014;4:56. doi: 10.3389/fcimb.2014.00056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal UaFE. Tick Interactions. In: Samuels DS, editor. Borrelia: molecular biology, host interaction, and pathogenesis. Norfolk, United Kingdom: Caister Academic Press; 2010. pp. 279–298. [Google Scholar]
- Parveen N, Leong JM. Identification of a candidate glycosaminoglycan-binding adhesin of the Lyme disease spirochete Borrelia burgdorferi. Molecular microbiology. 2000;35:1220–1234. doi: 10.1046/j.1365-2958.2000.01792.x. [DOI] [PubMed] [Google Scholar]
- Patton TG, Dietrich G, Dolan MC, Piesman J, Carroll JA, Gilmore RD., Jr Functional analysis of the Borrelia burgdorferi bba64 gene product in murine infection via tick infestation. PloS one. 2011;6:e19536. doi: 10.1371/journal.pone.0019536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazy Y, Motaleb MA, Guarnieri MT, Charon NW, Zhao R, Silversmith RE. Identical phosphatase mechanisms achieved through distinct modes of binding phosphoprotein substrate. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:1924–1929. doi: 10.1073/pnas.0911185107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitzer JE, Sultan SZ, Hayakawa Y, Hobbs G, Miller MR, Motaleb MA. Analysis of the Borrelia burgdorferi cyclic-di-GMP-binding protein PlzA reveals a role in motility and virulence. Infection and immunity. 2011;79:1815–1825. doi: 10.1128/IAI.00075-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Policastro PF, Schwan TG. Experimental infection of Ixodes scapularis larvae (Acari: Ixodidae) by immersion in low passage cultures of Borrelia burgdorferi. Journal of medical entomology. 2003;40:364–370. doi: 10.1603/0022-2585-40.3.364. [DOI] [PubMed] [Google Scholar]
- Radolf JD, Caimano MJ, Stevenson B, Hu LT. Of ticks, mice and men: understanding the dual-host lifestyle of Lyme disease spirochaetes. Nature reviews. Microbiology. 2012;10:87–99. doi: 10.1038/nrmicro2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro JM, Mather TN, Piesman J, Spielman A. Dissemination and salivary delivery of Lyme disease spirochetes in vector ticks (Acari: Ixodidae) Journal of medical entomology. 1987;24:201–205. doi: 10.1093/jmedent/24.2.201. [DOI] [PubMed] [Google Scholar]
- Ribeiro JM, Weis JJ, Telford SR. Saliva of the tick Ixodes dammini inhibits neutrophil function. Experimental parasitology. 1990;70:382–388. doi: 10.1016/0014-4894(90)90121-r. [DOI] [PubMed] [Google Scholar]
- Sadziene A, Thompson PA, Barbour AG. A flagella-less mutant of Borrelia burgdorferi as a live attenuated vaccine in the murine model of Lyme disease. The Journal of infectious diseases. 1996;173:1184–1193. doi: 10.1093/infdis/173.5.1184. [DOI] [PubMed] [Google Scholar]
- Sarkar MK, Paul K, Blair D. Chemotaxis signaling protein CheY binds to the rotor protein FliN to control the direction of flagellar rotation in Escherichia coli. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:9370–9375. doi: 10.1073/pnas.1000935107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwan TG, Piesman J. Temporal changes in outer surface proteins A and C of the lyme disease-associated spirochete, Borrelia burgdorferi, during the chain of infection in ticks and mice. Journal of clinical microbiology. 2000;38:382–388. doi: 10.1128/jcm.38.1.382-388.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skerker JM, Prasol MS, Perchuk BS, Biondi EG, Laub MT. Two-component signal transduction pathways regulating growth and cell cycle progression in a bacterium: a system-level analysis. PLoS biology. 2005;3:e334. doi: 10.1371/journal.pbio.0030334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small CM, Ajithdoss DK, Rodrigues Hoffmann A, Mwangi W, Esteve-Gassent MD. Immunization with a Borrelia burgdorferi BB0172-derived peptide protects mice against lyme disease. PloS one. 2014;9:e88245. doi: 10.1371/journal.pone.0088245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JG, Latiolais JA, Guanga GP, Citineni S, Silversmith RE, Bourret RB. Investigation of the role of electrostatic charge in activation of the Escherichia coli response regulator CheY. Journal of bacteriology. 2003;185:6385–6391. doi: 10.1128/JB.185.21.6385-6391.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart PE, Bestor A, Cullen JN, Rosa PA. A tightly regulated surface protein of Borrelia burgdorferi is not essential to the mouse-tick infectious cycle. Infection and immunity. 2008a;76:1970–1978. doi: 10.1128/IAI.00714-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart PE, Rosa PA. Transposon mutagenesis of the lyme disease agent Borrelia burgdorferi. Methods in molecular biology. 2008b;431:85–95. doi: 10.1007/978-1-60327-032-8_7. [DOI] [PubMed] [Google Scholar]
- Sultan SZ, Manne A, Stewart PE, Bestor A, Rosa PA, Charon NW, Motaleb MA. Motility is crucial for the infectious life cycle of Borrelia burgdorferi. Infection and immunity. 2013;81:2012–2021. doi: 10.1128/IAI.01228-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sultan SZ, Pitzer JE, Boquoi T, Hobbs G, Miller MR, Motaleb MA. Analysis of the HD-GYP domain cyclic dimeric GMP phosphodiesterase reveals a role in motility and the enzootic life cycle of Borrelia burgdorferi. Infection and immunity. 2011;79:3273–3283. doi: 10.1128/IAI.05153-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sultan SZ, Pitzer JE, Miller MR, Motaleb MA. Analysis of a Borrelia burgdorferi phosphodiesterase demonstrates a role for cyclic-di-guanosine monophosphate in motility and virulence. Molecular microbiology. 2010;77:128–142. doi: 10.1111/j.1365-2958.2010.07191.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sultan SZ, Sekar P, Zhao X, Manne A, Liu J, Wooten RM, Motaleb MA. Motor rotation is essential for the formation of the periplasmic flagellar ribbon, cellular morphology, and Borrelia burgdorferi persistence within Ixodes scapularis tick and murine hosts. Infection and immunity. 2015;83:1765–1777. doi: 10.1128/IAI.03097-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sze CW, Zhang K, Kariu T, Pal U, Li C. Borrelia burgdorferi needs chemotaxis to establish infection in mammals and to accomplish its enzootic cycle. Infection and immunity. 2012;80:2485–2492. doi: 10.1128/IAI.00145-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szurmant H, Ordal GW. Diversity in chemotaxis mechanisms among the bacteria and archaea. Microbiology and molecular biology reviews: MMBR. 2004;68:301–319. doi: 10.1128/MMBR.68.2.301-319.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toker AS, Macnab RM. Distinct regions of bacterial flagellar switch protein FliM interact with FliG, FliN and CheY. Journal of molecular biology. 1997;273:623–634. doi: 10.1006/jmbi.1997.1335. [DOI] [PubMed] [Google Scholar]
- Tsao JI. Reviewing molecular adaptations of Lyme borreliosis spirochetes in the context of reproductive fitness in natural transmission cycles. Veterinary research. 2009;40:36. doi: 10.1051/vetres/2009019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadhams GH, Armitage JP. Making sense of it all: bacterial chemotaxis. Nature reviews. Molecular cell biology. 2004;5:1024–1037. doi: 10.1038/nrm1524. [DOI] [PubMed] [Google Scholar]
- Weis JJ, McCracken BA, Ma Y, Fairbairn D, Roper RJ, Morrison TB, et al. Identification of quantitative trait loci governing arthritis severity and humoral responses in the murine model of Lyme disease. Journal of immunology. 1999;162:948–956. [PubMed] [Google Scholar]
- Welch M, Oosawa K, Aizawa S, Eisenbach M. Phosphorylation-dependent binding of a signal molecule to the flagellar switch of bacteria. Proceedings of the National Academy of Sciences of the United States of America. 1993;90:8787–8791. doi: 10.1073/pnas.90.19.8787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitchurch CB, Leech AJ, Young MD, Kennedy D, Sargent JL, Bertrand JJ, et al. Characterization of a complex chemosensory signal transduction system which controls twitching motility in Pseudomonas aeruginosa. Molecular microbiology. 2004;52:873–893. doi: 10.1111/j.1365-2958.2004.04026.x. [DOI] [PubMed] [Google Scholar]
- Wolanin PM, Thomason PA, Stock JB. Histidine protein kinases: key signal transducers outside the animal kingdom. Genome biology. 2002;3:REVIEWS3013. doi: 10.1186/gb-2002-3-10-reviews3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolgemuth CW. Flagellar motility of the pathogenic spirochetes. Seminars in cell & developmental biology. 2015;46:104–112. doi: 10.1016/j.semcdb.2015.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XF, Pal U, Alani SM, Fikrig E, Norgard MV. Essential role for OspA/B in the life cycle of the Lyme disease spirochete. The Journal of experimental medicine. 2004;199:641–648. doi: 10.1084/jem.20031960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Liu J, Tu Y, Xu H, Charon NW, Li C. Two CheW coupling proteins are essential in a chemosensory pathway of Borrelia burgdorferi. Molecular microbiology. 2012;85:782–794. doi: 10.1111/j.1365-2958.2012.08139.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Yang X, Kumar M, Pal U. BB0323 function is essential for Borrelia burgdorferi virulence and persistence through tick-rodent transmission cycle. The Journal of infectious diseases. 2009;200:1318–1330. doi: 10.1086/605846. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1×106 WT-eGFP B. burgdorferi were injected intradermally into mouse ear skin and time-lapse images were collected using an Olympus FV1000 laser confocal microscope system at 6 hours post-injection. 2D images were collected using a 20× dry objective with a 2X optical zoom at 1frame/1100 milliseconds for 66 seconds (60 images). The video is sped up two times the “real-time” speed.
1×106 ΔcheY3-eGFP B. burgdorferi were injected intradermally into mouse ear skin and time-lapse images were collected using an Olympus FV1000 laser confocal microscope system at 6 hours post-injection. 2D images were collected using a 20× dry objective with a 2X optical zoom at 1frame/1100 milliseconds for 66 seconds (60 images). The video is sped up two times the “real-time” speed.