Abstract
Recent developments in genome editing techniques have aroused substantial excitement among agricultural scientists. These techniques offer new opportunities for developing improved plant lines with addition of important traits or removal of undesirable traits. Increased adoption of genome editing has been geared by swiftly developing Clustered regularly interspaced short palindromic repeats (CRISPR). This is appearing as driving force for innovative utilization in diverse branches of plant biology. CRISPR-Cas9 mediated genome editing is being used for rapid, easy and efficient alteration of genes among diverse plant species. With approximate completion of conceptual work about CRISPR-Cas9, plant scientists are applying this genome editing tool for crop attributes enhancement. The capability of this system for performing targeted and efficient modifications in genome sequence as well as gene expression will certainly spur novel developments not only in model plants but in crop and ornamental plants as well. Additionally, due to non-involvement of foreign DNA, this technique may help alleviating regulatory issues associated with genetically modified plants. We expect that prevailing challenges in plant science like genomic region manipulation, crop specific vectors etc. will be addressed along with sustained growth of this genome editing tool. In this review, recent progress of CRISPR-Cas9 technology in plants has been summarized and discussed. We reviewed significance of CRISPR-Cas9 for specific and non-traditional aspects of plant life. It also covers strengths of this technique in comparison with other genome editing techniques, e.g., Zinc finger nucleases, Transcription activator-like effector nucleases and potential challenges in coming decades have been described.
Keywords: CRISPR, plants, genome editing, targeted modifications, novel phenotypes
Introduction
Genome editing (GE) encompasses numerous techniques of immense value for plant genome modifications. These techniques enable us to change the gene expression regulation at pre-determined sites and facilitate new insights into the plant functional genomics. GE differs from genetic engineering. So, no foreign DNA is made part of plants and they cannot be distinguished from parent plants. Genome engineering of plant cell lines or plant models has conventionally been achieved either through random mutagenesis or low-efficiency gene targeting (Hsu et al., 2014; Ma et al., 2014; Sprink et al., 2015; Wolt et al., 2016). Genome editing includes a wide variety of tools. Making the genome editing practical and reliable, techniques like Genome editing with engineered nucleases (GEEN) and programmable sequence-specific DNA nuclease etc. have granted precision to process of endogenously targeted genomic modifications. The versatile genome-editing tool CRISPR (Clustered regularly interspaced short palindromic repeats) is a comparatively precise approach to modify DNA at specific sites. CRISPR has evolved as principal technique for gene function analysis and genesis of genetic variation (Deltcheva et al., 2011; Perez-Pinera et al., 2013; Kanchiswamy et al., 2016). Particularly, success in genome modification has been noticed among species that are difficult to be modified by other techniques (Bolotin et al., 2005; Xing et al., 2014). To date, most of the studies have been conducted by using animal systems. During last few years, CRISPR-Cas9 mediated mutagenesis was performed in arabidopsis, sorghum, tobacco, proving applicability of this technique to both dicot and monocot plants (Feng et al., 2013; Li et al., 2013). Generally, CRISPR-Cas9 is highly adaptable for editing of plant genome (Charpentier and Doudna, 2013; Schaeffer and Nakata, 2015) but especially appropriate for genome editing of monocotyledons, e.g., rice due to high genomic GC content (Miao et al., 2013). With special reference to economically valuable plants, i.e., crops and ornamentals, this technique offers an extraordinary and pragmatic system to produce novel phenotypes. CRISPR together with Cas proteins form the CRISPR-Cas system (Zhou et al., 2014; Bortesi and Fischer, 2015).
The functions of CRISPR and Cas genes (CRISPR-associated) are indispensable for adaptive immunity in some bacteria and archaea. These act as facilitator in response to viral genetic material. Discovered in 1980s in Escherichia coli (Ishino et al., 1987), function of these repeats was confirmed in 2007. Till now, workers across the world have described three types of mechanisms. Type II of CRISPR is the most studied type (Bortesi and Fischer, 2015). The Types I and III system involves specific Cas endonucleases which make the pre-crRNAs (Pre-CRISPR RNA) and after attaining maturity, this crRNA assembles into Cas protein complex. This complex possesses ability to recognize and cleave nucleic bases complementary to the crRNA (Jinek et al., 2012). The CRISPR-Cas9 type II is characterized as small RNA-based immune system of archaea and bacteria (Haft et al., 2005). CRISPR-Cas9 system is featured by relative construction simplicity along with high functional efficiency in human, animal, and plant cells (Nemudryi et al., 2014). The technique allows access target recognition by using gRNAs instead of synthetic DNA-binding domains. This characteristic makes it simple in comparison with ZFNs and TALENs (Cong et al., 2013; Wang et al., 2013).
Genome editing is being adopted for economically significant plants with full trust in terms of technical viability, dogmatic acceptance and profit-making practicability (Miao et al., 2013; Bortesi and Fischer, 2015). It is noteworthy that different genetic engineering techniques can leave behind DNA alteration traces. The crop plants and ornamentals generated by means of genome editing can escape the strict statutes and regulations generally associated with GM plant development. Due to this reason many researchers believe that improvements in plant varieties through precise genome editing techniques will be highly acceptable to the public as compared to transgenic plants (Abdallah et al., 2015).
The advent of CRISPR has made it possible to rewrite host DNA by introducing some major modifications. These modifications include gene replacement, deletions, inversion, knockouts, and translocations. But more prominent are the potential prospects of this technique for producing plants with mutations linked to other disciplines of science, i.e., synthetic biology, biofuel production, disease resistance, abiotic stress tolerance, phytoremediation etc. The establishment of plants with desired gene modifications can pave the way to study complex plant biology. Unfortunately, plant science is far behind than other disciplines in application of this technology. Therefore, keeping in view the immense importance of this technique, we have summarized the prospective role of CRISPR-Cas9 for plants and related benefits. A brief comparison of CRISPR-Cas9 and other genome editing techniques has been made to justify its strengths. We attempted to sum up current progress in CRISPR-Cas9 technology especially in plant biology and potential challenges for future development.
Glimpses From History
High frequency of plant genome editing is evident in economically significant plant species due to practical feasibility and viability (Figure 1). Initially, evidences of earliest genome editing were expressed with oligonucleotide mediated mutagenesis (OMM) for herbicide resistance in rice, maize, tobacco etc. (Kochevenko and Willmitzer, 2003; Iida and Terada, 2005; Sander and Joung, 2014; Wolt et al., 2016). Gao et al. (2010) used engineered mega nuclease (EMN) for editing maize genome by using native endonuclease altered to identify and tempt very specific DSBs (Double stranded breaks) at definite locus. This resulted in disruption of gene in terms of indels (Insertions-deletions) by non-homologous end joining (NHEJ).
FIGURE 1.
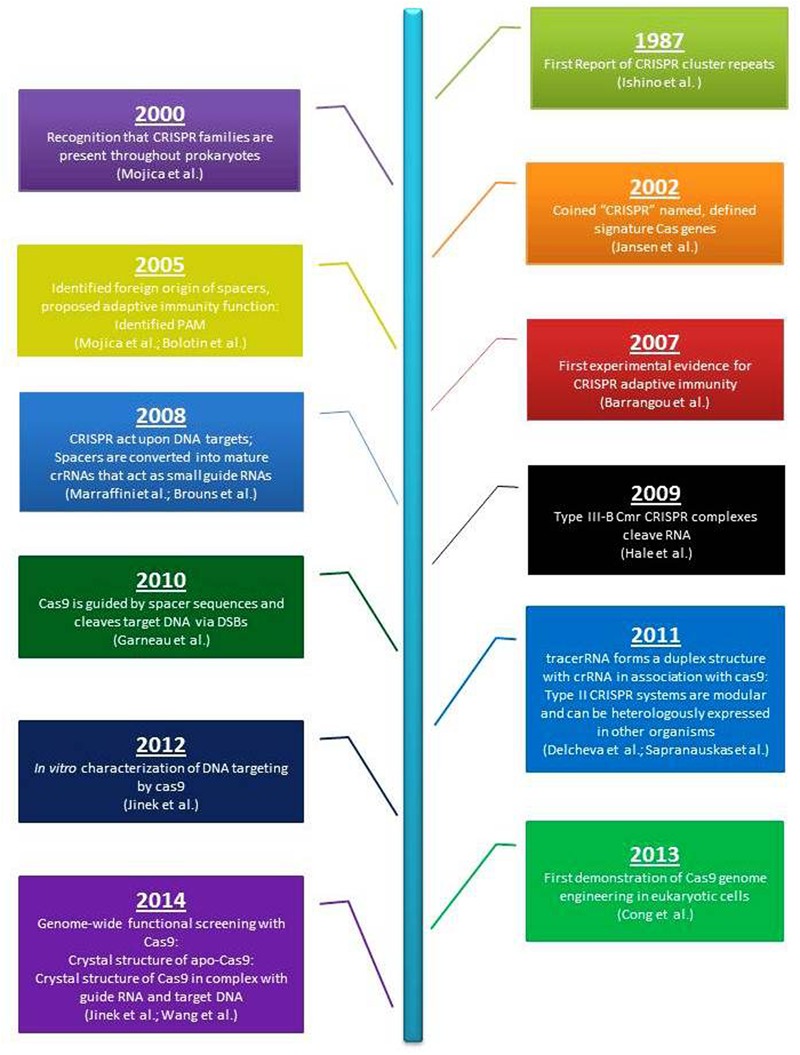
Two decades of CRISPR-Cas9 adoption and success.
In addition to this, successful target gene insertion for delivery of herbicide tolerance in cotton had been carried out through yeast endonuclease engineered EMN (D’Halluin et al., 2013). Afterward, it was observed that ZFN based site-specific trait stacking strategy produced excellent results in maize in form of new plant line possessing herbicide resistant gene.
Finally, accomplishment of CRISPR-Cas9 technique was observed for competent targeted mutagenesis in transgenic rice for improvement in growth and yield. Researchers have successfully demonstrated the production of transgenic rice having mutations in particular genes by adopting CRISPR-Cas9 technology (Miao et al., 2013; Zhang et al., 2014; Wolt et al., 2016). Xing et al. (2014) presented a toolkit for facilitating transient/stable expression of CRISPR-Cas9 in diverse plant species. The year 2016 mark the finalization of herbicide tolerant canola, e.g., Cibus 5715 approved for cultivation in Canada.
Now, several research groups have focused application of CRISPR technology on plants of significant economic worth such as rose, apple, potato, egg plant, rice (Table 1) (Wendt et al., 2013; Char et al., 2015; Sprink et al., 2015; Xiong et al., 2015; Kanchiswamy et al., 2016). Unequivocally, this technique is efficient, well-organized and flexible for editing multiplex gene. Now time is to focus on application of CRISPR-Cas9 system to other cereals with larger and complex genomes, e.g., wheat, sugar cane. Parallel with this, improvements in this technique, i.e., elimination of CRISPR-Cas9 remains after target genes mutation, will support the usage of this tools in agriculture.
Table 1.
Successful application of CRISPR-Cas9 in different plant species.
| DNA modification type | Plant | Delivery mode | Target(s) | Gene function(s) | Reference |
|---|---|---|---|---|---|
| Gene Knockout: rewriting of host DNA | A. thaliana | Stable integration | RTEL1(Regulator Of Telomere Elongation Helicase 1) | RTEL1 functions in DNA replication, DNA repair, and recombination | Schiml et al., 2014 |
| A. thaliana | Stable integration | AP1 (floral homeotic gene APETALA1), BRI1 (Brassinosteroid-insensitive2), CHLI1(Magnesium-chelatase subunit ChlI-1) | BRI1 encodes a cell surface receptor for brassinosteroids.AP1 Encodes a putative TF that acts locally to specify the identity of the floral meristem. CHLI1 plays role in chlorophyll biosynthesis. | Feng et al., 2014 | |
| A. thaliana | Stable integration | ADH1 (Alcohol dehydrogenase class-P) | ADH1 is required for survival and acclimation in hypoxic conditions, especially in roots | Fauser et al., 2014 | |
| A. thaliana N. benthamiana | Protoplasts, Agrobacterium T-DNA(Transient) | At Phytoene desaturase gene (PDS3), NbPDS3 | PDS3 is needed for primary carbon and pigment metabolism. Its activity acts as a rheostat of retrograde signaling during early chloroplast biosynthesis. | Li et al., 2013 | |
| O. sativa, S. bicolor, | Protoplasts, Agrobacterium T-DNA (Transient) | OsSWEET14, | OsSWEET14 id needed for disease resistance. | Jiang et al., 2013 | |
| O. sativa T. aestivum | Protoplasts | OsPDS,OsBADH2 (betaine aldehyde dehydrogenase-2), OsMPK2 (ortholog of tobacco SIPK), TaMLO (Mildew-resistance locus) | BADH2 plays important role in abiotic stress tolerance. OsMPK2 is activated by elicitors It negatively regulates the expression of defense-related genes. TaMLO in plant confers heritable broad-spectrum resistance to powdery mildew | Shan et al., 2013c | |
| A. thaliana, O. sativa | Stable integration | AtJAZ1 (Jasmonate ZIM-domain), (Gibberellic Acid insensitive),OsROC5 (RICE OUTMOST CELL-SPECIFIC GENE 5), OsSPP, OsYSA | AtJAZ1 is transcription repressor of jaJA)-responsive genes and major components of theJA receptor complex. AtGAI is important in attenuation of GA responses. | Feng et al., 2013 | |
| O. sativa | Stable integration | OsWEET11/13/1a/1b | OsSWEET14 id needed for disease resistance. | Zhou et al., 2014 | |
| Large deletions: | A. thaliana | Protoplasts | PDS3 | PDS3 is needed for primary carbon and pigment metabolism. | Li et al., 2013 |
| Gene replacement: | O. sativa | Protoplasts | Phytoene desaturase gene | PDS3 is needed for primary carbon and pigment metabolism. | Shan et al., 2013a |
| N. benthamiana | Protoplasts | Phytoene desaturase gene | PDS3 is needed for primary carbon and pigment metabolism. | Li et al., 2013 | |
| A. thaliana | Stable integration | ADH1 (Alcohol dehdrogenase1) | ADH1 is required for survival and acclimation in hypoxic conditions, especially in roots | Schiml et al., 2014 | |
| Controlling gene expression | N. benthamiana | Agrobacterium T-DNA (Transient) | PDS3 | PDS3 is needed for primary carbon and pigment metabolism. Its activity acts as a rheostat of retrograde signaling during early chloroplast biosynthesis. | Piatek et al., 2014 |
Mechanism of CRISPR-Cas9 Based Genome Editing
CRISPR-Cas9 system just requires three components, i.e., Cas9, tracer RNA (trRNA), CRISPR RNA (crRNA) for function. This prospective was recognized in start of this decade (Jinek et al., 2012; Hsu et al., 2014; Schaeffer and Nakata, 2015). In type II of CRISPR, attacking viral DNA or plasmids is divided into smaller pieces and integrated in CRISPR locus. The particular loci are transcribed and processed transcripts produce crRNA. These crRNAs direct effector endonuclease to target alien DNA depending upon complementarity of sequence. Cas9 produce DSBs (double-stranded breaks) at target site, which on the other hand facilitates endogenous DNA repair mechanisms leading to edited DNA (Charpentier and Doudna, 2013; Hsu et al., 2014).
Type II system, comprises of crRNA and trRNA that combine into one sgRNA (single guide RNA) (Jinek et al., 2012; Xing et al., 2014). Amazingly, the sgRNA programmed Cas9 appeared more effective in targeted gene modifications rather than individual trRNA and crRNA. Till today, genome-editing protocols have adopted three different types of Cas9 nuclease. The first Cas9 type can cut DNA site-specifically and results in the activation of DSB repair. Cellular NHEJ (Non-Homologous End Joining) mechanism is used to repair DSBs (Hsu et al., 2014; Sternberg et al., 2014). As a consequence, insertions/deletions (indels) takes place that interrupt the targeted loci (Figure 2). Otherwise, if any similarity between donor template and target locus is witnessed, the DSB may be mended by HDR pathway (homology directed repair) allowing exact substitute mutations to be prepared (Hale et al., 2009; Sternberg et al., 2014; Schaeffer and Nakata, 2015). Cong et al. (2013) introduced advanced Cas9-D10A, a mutant form having more précised nickase activity. It cuts single strand of DNA without activation of NHEJ. As an alternative, DNA repairs took place via the HDR pathway only. Hence it produces less indel mutations (Jinek et al., 2012; Cong et al., 2013). Cas9-D10A is very target specific particularly when any locus is encountered by paired Cas9 complexes for generation of contiguous DNA nicks (Ran et al., 2013). The third type is dCas9, nuclease-deficient Cas9 (Qi et al., 2013). Although mutations in the HNH domain and RuvC domain discharge cleavage activity, but do not prevent DNA binding (Gasiunas et al., 2012). Therefore, this particular variant can be utilized in sequence-specific targeting of any genome regardless of cleavage. In its place, dCas9 may be taken as a tool for either gene silencing or activation by fusion with a variety of effector domains (Maeder et al., 2013a,b). One bonus of this technique is the case of not using recombinant DNA (Figure 2). This situation can result in edited plants exempted from current GMO regulations. So we can hope for widespread application of RNA-guided genome editing in agriculture and plant biotechnology.
FIGURE 2.
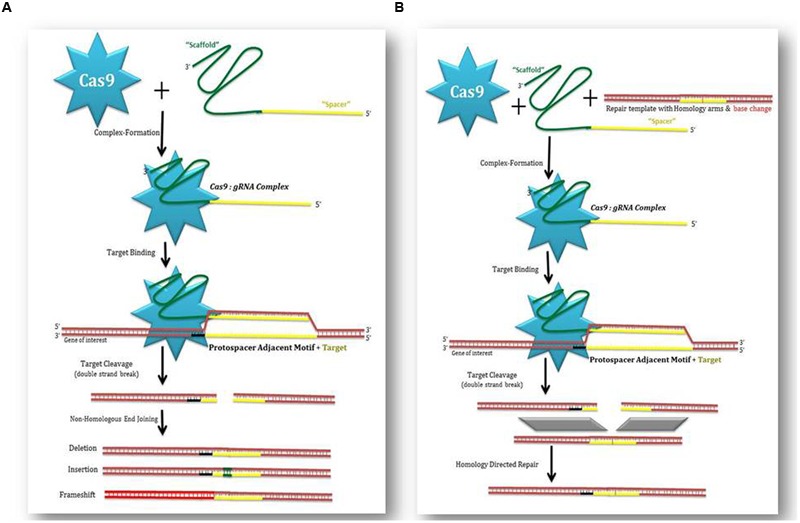
How CRISPR-Cas9 perform genome editing. Cas9 induce double stranded breaks (DSBs) at particular site. The resulting DSB is then repaired by one of these two general repair pathways, e.g., by Non-homologous end joining (NHEJ) or by Homology directed repair (HDR). (A) The NHEJ repair pathway frequently results in small nucleotide insertions or deletions (InDels) at the DSB site. This may result in gene knock out or gene insertion. (B) HDR can be used to generate precise nucleotide modifications (also called gene “edits”) ranging from a single nucleotide change to large insertions.
Why CRISPR-Cas9 is More Trustworthy than Talens and ZFNs?
For assessment of any genome editing tool, % age of achieved desired mutation known as Targeting efficiency (TE) is regarded as the most reliable attribute. The success ratio of Cas9 TE can be compared with other techniques like TALENs or ZFNs (Wendt et al., 2013; Ma et al., 2014). For example in human cells, custom-designed ZFNs and TALENs could only achieve 1–50% efficiencies (Maeder et al., 2008; Miller et al., 2011; Wolt et al., 2016). Conversely, TE of Cas9 in animals and plants, respectively, i.e., zebrafish, maize has been observed up to 70% and it ranges between 2 and 5% in case of induced pluripotent stem cells (Fu et al., 2013; Hsu et al., 2013). Later on,CRISPR-Cas9 efficiency was recorded up to 9.2% as compared to ZFN efficiency that was lower than 1% in case of pigs IGF2 (Insulin-like growth factor 2). Reports are available that broadly describe better genome targeting of single cell mouse embryo up to 78% and successful effectual germline transmission by using dual sgRNAs (Zhang et al., 2013; Zhou et al., 2014; Wolt et al., 2016). Moreover, incidence of off-target mutations is also an effective parameter for assessment of genome editor’s performance. Such mutations may be observed in sites that have dissimilarity of small number of nucleotides in comparison with original sequence till they are neighbors of Protospacer adjacent motif (PAM) sequence. The DNA sequences are used to transcribe crRNA targeting sequences known as protospacers. These consist of short sequences and found clustered in bacterial genome in form of a group called CRISPR array. The PAM sequence is absolute need of Cas9 for binding its target. Cas9 do not cleave the protospacer sequence in absence of adjacent PAM sequence. This favors the stance that Cas9 can endure mismatches up to five bases within the protospacer region (Fu et al., 2014; Sander and Joung, 2014) or one base divergence in the PAM sequence (Hsu et al., 2013). Other than facilitation activity for genome alterations, the wild-type Cas9 nuclease has capacity to be transformed into dCas9 after inactivation of catalytic domains. Furthermore, effector fusion usage can enhance the range of genome engineering modalities attainable by adopting Cas9. Normally, off-target mutations are bit difficult to detect because these require full genome sequencing to completely rule them out. So unanimous opinion is CRISPR-Cas9 facilitates plant genomes interrogation, as it enables high efficiency generation of mutants bearing multiple gene mutations (Tables 2 and 3). This effective approach endorses high specificity of wide range genome editing applications.
Table 2.
Tabular presentation of comparative attributes of plant genome editing techniques.
| CRISPR/Cas9 | Zinc Finger Nucleases (ZFNs) | Transcription factor like effector nucleases (TALENs) | Reference | |
|---|---|---|---|---|
| Mode of action | It works by inducing double-strand breaks in target DNA or single-strand DNA nicks (Cas9 nickase). | It can induce double-strand breaks in target DNA. | Induces DSBs in target DNA. | Li et al., 2013; Mao et al., 2013 |
| Off target effects | These effects can be minimized by selecting unique crRNA sequence. | These have off-target effects. | Off target effects cannot be avoided. | Hsu et al., 2013 |
| Generation of large scale libraries | YES, this is possible to generate large scale libraries. | Such generation is not possible because it requires customization of protein component for each gene. | Generation of large scale libraries is possible but technically difficult and challenging. | Cho et al., 2013; Hsu et al., 2013 |
| Protein engineering steps | It does not requires protein engineering steps, very simple to test multiple gRNA. | It requires complex to test gRNA. | TALENs need protein engineering steps to test gRNA. | Cho et al., 2013 |
| Cloning | Cloning is not necessary. | Cloning is necessary. | It requires cloning. | Cho et al., 2013 |
| gRNA production | Any number of gRNA can be produced by in vitro transcription. It keeps budget away from extra load. | Bit difficult to produce this kind of RNA. | gRNA production is bit difficult to achieve through these effector nucleases. | Cho et al., 2013 |
| Methylated DNA cleavage | It can cleave methylated DNA in human cells. This aspect is of special concern for plants as this has not been much explored | Unable to do so. | There are many question marks upon capacity of TALENs to perform methylated DNA cleavage. | Hsu et al., 2013; Ding et al., 2013 |
| Multiplexing | This is main advantage of CRISPR. Several genes can be edited at same time. Only Cas9 needed | Highly difficult to achieve this through ZFNs. | Very difficult to obtain multiplexed genes by means of TALENs. Because it needs separate dimeric proteins specific for each target | Li et al., 2013; Mao et al., 2013 |
| Structural proteins | CRISP R consists of single monomeric protein and chimeric RNA. | ZFNs work as dimeric and only protein component required. | TALENs also work as dimeric and require protein component. | Li et al., 2013; Upadhyay et al., 2013; Zhou et al., 2014 |
| Catalytic domain | It has two cleavage domains called RUVC and HNH. | ZFNs have catalytic domain of restriction endonuclease FOKI which generates a DSB. | TALENs also have FOKI catalytic domain of restriction endonuclease for DSB generation. | Jinek et al., 2012 |
| Mutation rate | Comparatively low mutation rate has been observed. | High mutation rate observed in plants. | Mutation rate is high as compared to CRISPR. | Li et al., 2013 |
| Components | crRNA, Cas9 proteins | Zn-finger domains Non-specific FokI nuclease domain | Zn-finger domains Non-specific FokI nuclease domain | Kumar and Jain, 2014 |
| Length of target sequence (bp) | 20–22 | 18–24 | 24–59 | Chen et al., 2016 |
| Target recognition efficiency | High | High | High | Kumar and Jain, 2014 |
| Level of experiment setup | Easy and very fast procedure of designing for new target site | Complicated procedure of redesigning for each new target site and need for expertise in protein engineering | Relatively easy procedure of designing for each new target site | Kumar and Jain, 2014 |
Table 3.
Technical limitations in CRISPR-Cas9 application and their effects.
| Pitfall | Reason (s) | Effects | Reference |
|---|---|---|---|
| Off-target effects | (1) Improper concentration ratio between Cas9 and sgRNA may cause off-target cleavage. (2) PAM sites may lead to undesired cleavage of DNA regions. | Unexpected mutations | Sternberg et al., 2014 |
| Cas9 codons | Insufficient Cas9 codon optimization | Inefficient translation of Cas9 proteins | Feng et al., 2013, 2014; Schiml et al., 2014 |
| Vectors | Mostly CRISPR/Cas9 systems use exogenous promoters for Cas9 and sgRNA expression. Vectors with optimal promoters should be selected. | Improper vectors can stop system proceedings. | Shan et al., 2014 |
| Gene homologs | Gene family members may complicate target sequences to be edited. | False editing of target sequence. | Song et al., 2016 |
| Epigenetic factors | DNA methylation or histone modification occurs not in regions with complex DNA compositions, such as those with repetitive sequences. | limit protein binding or RNA pairing | Song et al., 2016 |
Applications of CRISPR-Cas9 in Plant Biology and Biotechnology
The application of CRISPR-Cas9 has made it possible to rewrite host DNA by introducing some major alterations in plant genomes. Use of CRISPR-Cas9 is facilitating multiple ranges of genome engineering applications (Tables 4 and 5). Plant species with intractable genomes have now been targeted with Cas9 nuclease for introduction of various levels of genome modifications. Here, we will take into account prospective role of CRISPR-Cas9 in editing plant genome for achieving broad range goals.
Table 4.
List of promoters and gene(s) targeted through CRISPR-Cas9 system in different plants.
| Plant | sgRNA Promoter(s) | Cas9 Promoter(s) | Target (s) | Reference |
|---|---|---|---|---|
| Triticum aestivum | TaU6 | 2 × 35S | TaMLO (Wheat Mildew-resistance locus) | Shan et al., 2013b |
| TaU6 | Ub1 | TaMLO-A1 (Wheat Mildew-resistance locus1) | Wang et al., 2014 | |
| Citrus sinensis | CaMV 35S | CaMV 35S | CsPDS (Phytoene desaturase gene) | Jiang and Wang, 2014 |
| Sorghum bicolor | OsU6 | Rice Actin1 | DsRED2 (Red fluorescent protein) | Jiang et al., 2013 |
| Nicotiana benthamiana | AtU6 | 35DPPDK | NbPDS3 (Tobacco Phytoene desaturase gene) | Li et al., 2013 |
| OsU6 | 35S | GFP (Green fluorescent Protein) | Jiang et al., 2013 | |
| CaMVE35S | CaMVE 35S | Nb PDS3 (Phytoene desaturase gene) | Upadhyay et al., 2013 | |
| Marchantia polymorpha L. | MpU6-1 | CaMV 35s and MpEF1α | MpARF1 | Sugano et al., 2014 |
| Arabidopsis thaliana | AtU6 | 35DPPDK | AtPDS3, AtRACK1b (Receptor for activated C kinase 1) andAtRACK1c (Receptor for activated C kinase 1c) | Li et al., 2013 |
| AtU6-26 | 2 × 35S | BRI1 (Brassinosteroid-insensitive2), JAZ1 (Jasmonate ZIM-domain), and YFP | Feng et al., 2013 | |
| OsU6 | 35S | GFP (Green fluorescent Protein) | Jiang et al., 2013 | |
| Oryza sativa | OsU3 | 2 × 35S | OsPDS3,OsBADH2 (betaine aldehyde dehydrogenase-2), Os02g23823 and OsMPK2 (ortholog of tobacco SIPK) | Shan et al., 2013c |
| OsU6 | CaMV 35S | OsSWEET11 and OsSWEET14 (Rice bacterial blight susceptibility genes) | Jiang et al., 2013 | |
| OsU3 | Ub1 | CAO1 and LAZY1 | Miao et al., 2013 | |
| OsU6-2 | 35S | ROC5 (Rice outmost cell-specific gene 5), SPP and YSA | Feng et al., 2013 |
Table 5.
Different plasmids with their genes, vectors, and promoters used in CRISPR-Cas9 technique.
| Plasmids | Gene/Insert | Promoter | Vector type | Vector back bone | Purpose | Bacterial resistance | Reference |
|---|---|---|---|---|---|---|---|
| pK7WGF2::hCas9 | hCas9 (Syn) | 35S | Plant expression, CRISPR | pK7WGF2, | Expresses the human codon by using Cas9 with N-terminal GFP tag from the 35S promoter in the plant tissue | Spectinomycin | Nekrasov et al., 2013 |
| PHSE401 | zCas9, gRNA scaffold | 35S, AtU6-26p | Plant Expression; plant binary vector | pCambia, | CRISPR/Cas based plant genome editing and gene regulation | Kanamycin | Xing et al., 2014 |
| pHEE401E | zCas9, gRNA scaffold | U6-26p Arabidopsis U6 gene promoter, EC1.2 enhancer fused to EC1.1 promoter | Plant Expression; plant binary vector, CRISPR | pCambia | Contain gRNA scaffold for insertion of target sequence, Egg cell-specific promoter-controlled expression of 3×FLAG-NLS-zCas9-NLS | Kanamycin | Wang et al., 2015 |
| pHSN501 | zCas9D10A, gRNA scaffold (Syn) | AtU6-26p, 2 × 35Sp | CRISPR; Plant expression | pGreen-like binary vector | CRISPR/Cas based plant genome editing and gene regulation; expresses zCas9D10A | Kanamycin and Spectinomycin | Xing et al., 2014 |
| pBUN501 | gRNA scaffold (Syn), zCas9D10A (Syn) | AtU6-26p, Ubi1p, | Plant expression, CRISPR | pGreen-like binary vector | CRISPR/Cas based plant genome editing and gene regulation; | Kanamycin and Spectinomycin | Xing et al., 2014 |
| pHSN6A01 | gRNA scaffold (Syn),dCas9-VP64 (Syn) | AtU6-26p, 2 × 35Sp, | Plant expression, CRISPR | pGreen-like binary vector | expresses dCas9-VP64, gRNA scaffold for insertion of target sequence | Kanamycin and Spectinomycin | Xing et al., 2014 |
| pBUN6A11 | gRNA scaffold (Syn), dCas9-VP64 (Syn), | OsU3p, Ubi1p, | Plant expression, CRISPR | pGreen-like binary vector | expresses dCas9-VP64, gRNA scaffold for insertion of target sequence | Kanamycin and Spectinomycin | Xing et al., 2014 |
| pEGB 35s:dCas:BRD:tNos (GB1172) | dCas9:BRD | 35S | Plant expression, CRISPR | pDGB3alpha2 | Transcriptional unit of (human codon optimized) inactivated Cas9 fused to the BRD Transcriptional Repressor | Kanamycin | Vazquez-Vilar et al., 2016 |
| pEGB 35S:dCas9:Tnos (GB1191) | dCas9 | 35S | Plant Expression, CRISPR, Synthetic Biology | pDGB3alpha2 | Transcriptional unit for human codon optimized with mutated (D10A, H840A) and inactivated catalytic domains Cas9 protein plant expression driven by the 35S promoter | Kanamycin | Sarrion-Perdigones et al., 2013 |
| pJIT163-2NLSCas9 | dCas9 | 2 × 35S | Plant Expression | pJIT163 | Expression of rice codon-optimized Cas9 in plant cells | Ampicillin | Shan et al., 2013b |
| HBT-pcoCas9 | Pro Cas9 (syn) | Hybrid constitutive promoter 35SPPDK | CRISPR; Plant expression | HBT-FLAG | Transient expression of pcoCas9 gene in plant cells | Ampicillin | Li et al., 2013 |
Crispr-Cas9 and Plant Synthetic Biology
Ranging from production of primary metabolites necessary as food to secondary metabolites, plant based products are of great concern for multiple purposes. With ongoing progress in the field of plant biology in general and synthetic biology particularly, researchers are seeking to produce novel biological systems, inclusive of industrially designed plant cells and plants. One of the chief targets sets for synthetic biology is the wish for minimal plant cell, e.g., to engineer a cell devoid of non-essential components and capable of division. This desired minimal cell can then be exploited as a factory for novel biological systems. Although this minimal cell is still a dream, the prospective toolkits and strategies for generating the simplest plant cell are being operated recurrently (Baltes et al., 2015). Up till now, genome editing had been restricted to amendments in enzymatic functions within single animal or plant. Synthetic biology has already been using bacteria for engineering new absolute metabolic cycles comprising of both several enzymes and regulation of corresponding genes expression. CRISPR-Cas9 provide the most reliable and practical platform to engineer plant genome for multipurpose plant systems (Puchta and Fauser, 2014). Nitrogen fixing cereals project is classical example of goal fixed for humanity level benefits. The possibilities of genetic and metabolic engineering have been extended as a result of techniques developed for facilitating synthetic biology, especially cloning and genome editing methods (Neumann and Neumann-staubitz, 2010). At John Innes centre, different pathways have been characterized for plants capable of fixing nitrogen through bacteria (Oldroyd et al., 2011; Xie et al., 2012). The researchers are now attempting to introduce these pathways in wheat for developing ‘self-fertilizing’ cereal (Cook et al., 2014). By doing so, there will be clear cut reduction in dependency upon inorganic fertilizers because plants will be able to fix atmospheric nitrogen. At this time, there are two possible ways: either transfer the Nod factor signaling pathway to cereals or relocate the nitrogenase enzyme from nitrogen fixing bacteria into plant cells. But still different questions need to be addressed (Temme et al., 2012; Oldroyd and Dixon, 2014). A potential goal set by plant synthetic biology is C4 rice development with the help of targeted DNA insertion. Engineering rice with C4 photosynthesis pathway appears promising for increasing yield. One line of action to engineer this pathway in C3 rice is conversion of single-cell C3 cycle into a two-celled C4 cycle. The initial carbon fixation is carried out within mesophyll cells. Finally the four-carbon product is decarboxylatedforCO2 addition to RuBisCO present in bundle sheath cells. (Baltes et al., 2014). CRISPR-Cas9 has been more successfully applied to mutagenize host DNA in different plants (Li et al., 2013; Jiang et al., 2014; Zhou et al., 2014). The ability to introduce genomic amendments encourage synthetic biologists not merely remove unwanted DNA, i.e., inhibitory genes but also improve genic regulatory sequences.
Practicing plant synthetic biology needs control over nucleotide sequences in plant as well as control over expression levels of host genes. The DNA binding domain of different sequence- specific nucleases can be repurposed to help in modulation of endogenous genes expression. DNA-binding domains from ZFNs, TALENs, or dCas9 and gRNA are used to limitize repressor or activator domains in gene of interest. Uniquely, Cas9 by interfering with RNA polymerase progression can decrease gene expression (Qi et al., 2013).
The site-specific integration of DNA into plant genomes will be of special significance for plant synthetic biology research that demands the transfer of a number of genetic segments for conferring new biological function (Baltes and Voytas, 2015). CRISPR-Cas9 cannot only make traits stacking easy but also reduce variability in gene expression. Due to least targeting limitations for CRISPR-Cas9 system, almost all chromosomal positions are amenable to site-specific integration (Baltes and Voytas, 2015). First of all, transgene stacking was demonstrated in maize. After co-transformation of immature maize embryos with donor DNA and DNA encoding the ZFNs, 5% of transgenic progeny witnessed proper integration (Ainley et al., 2013). Similarly, trait stacking has successfully been done in cotton (D’Halluin et al., 2013). Other than integrating genes with the help of homologous recombination, NHEJ can be used for targeted gene insertion (Baltes et al., 2014). But this approach has not been extensively applied in plants.
However, several challenges are still to be addressed. Most importantly, successful plant system engineering and development will depend on suitable and efficient delivery systems by targeting specific tissues. There is need is to develop techniques providing command over the triplet code, therefore likely to enable us for selected amendments in DNA sequence within plant cells. We are convinced that editing genome is going to exert a material influence on the valuable scheme for plant trait improvement. It will be of supreme significance to systematically characterize the safety as well as physiological effects of Cas9 in plant synthetic biology by adopting a variety of methods.
Crispr-Cas9, a Perspective Strategy for Plant Genome Imaging
Visible genome imaging is largely executed to measure features of plant genome architecture (Sozzani et al., 2010). The intracellular organization of structural and functional elements contributes to the genomic functional output which can be dynamically enhanced or concealed. The physical genome organization has appeared specifically significant but still reckoned as obscure mechanism. Researchers used different methods like chromosome conformation capture (3C) to answer various genome related queries. 3C derived techniques, e.g., Hi-C have introduced innovative insights into genome spatial organization principles inclusive of the presence of TADs (topologically associated domains) (Chen et al., 2016). Genomic loci positioned mega bases away on same or different chromosomes could be brought closer given apt chromosomal organization, consequently mediating lengthy trans interactions (Ma et al., 2014). Conversely, there is still question mark upon manner for genome modification and in vivo modulation of their structural organization afterward (Malina et al., 2014). But, without vital methodology for DNA visualization, studying various gene interactions in different chromatin states would merely be a dream. CRISPR type II resulting from Streptococcus (Wiedenheft et al., 2012) is capable of achieving this goal (Chen et al., 2016). Griffith et al. (1999) conducted series of experiments for verification of telomere imaging efficiency and specificity by CRISPR technique. The number of telomeres identified with the help of CRISPR or specific Fluorescence in situ hybridization (FISH) with peptide nucleic acid (PNA) was similar. Thus, the matched score for cell imaging indicated similar efficiencies of FISH and CRISPR for cellular imaging. This finding declares CRISPR as an optimized toolkit for telomere visualization along with role in gene regulation enhancement. Traditional DNA labeling techniques like FISH require sample fixation and therefore, incapable to capture live course of actions. Labeling of particular DNA loci with the help of fluorescently tagged Cas9 had been introduced as a potent live-cell-imaging substitute of DNA-FISH. dCas9-EGFP and sequence-specific sgRNAs co-expression facilitate the enhancement of fluorescent signals for imaging at targeted genomic loci (Chen et al., 2013, 2016; Hsu et al., 2014). CRISPR technique endow us with vigorous repetitive element between protein-coding genes, e.g., mucin genes and telomeres. Similarly, human genome non-repetitive elements have already been visualized with the aid of multiple sgRNA (Chen et al., 2013). Not alone but together with FISH or DNA-binding proteins, this CRISPR technique recommends a matching advancement for imaging. The capacity of CRISPR to tag human cell telomeres encouraged researchers to examine whether this technique can be used for assessing telomere length. A linear correlation was noticed between PNA based FISH and CRISPR for telomere length evaluation. The superiority of CRISPR over FISH is its ability to label telomere in addition to length measurement. This has been proved by correlation between telomere count and intensity (Chen et al., 2013).
Genome functional organization mapping can be greatly aided by techniques that are helpful in directly visualizing the interactions between various genomic elements, i.e., promoters or enhancers in living cells. Therefore, multicolor imaging method would be essential for imaging and tracking numerous genomic loci (Figure 3) (Chen et al., 2016). At present, two strategies have been devised for live cell imaging by using CRISPR-Cas9. The first strategy uses fluorescent Cas9 orthologs obtained from different bacterial types. The second strategy is by means of fluorescent RNA-binding protein joined to the sgRNA. Consequently, scaffold RNA (scRNA) is formed that encodes information about the target locus and the fluorescent color (Shao et al., 2016). These approaches have been successfully used for genomic regulatory programming (Konermann et al., 2015; Zalatan et al., 2015).
FIGURE 3.
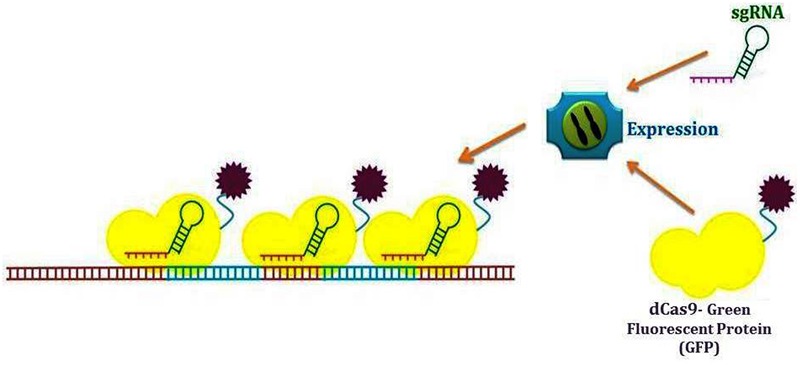
Diagrammatic illustration of live-cell DNA labeling by using CRISPR-Cas9 system.
For genome imaging, RNA-guided Cas9 system specificity can be modified by replacing a small synthetic RNA without changing the protein component. Hence, designing and production of the labeling constructs become easy and very cost-effective (Chen et al., 2016). In comparison with TALENs, CRISPR-Cas system is more flexible for target site selection. A CRISPR-Cas9 target immediately precede PAM and usually starts with G (Jinek et al., 2012), while TALENs require T at the 5 end of the target sequence (Mak et al., 2012). For co-labeling of several loci, CRISPR-Cas9 is thus an easier choice.
Another advantage of CRISPR-Cas imaging is direct measurement of spatial information by loci positions imaging (Kalhor et al., 2012; Nagano et al., 2013). Therefore, it opens up new possibilities in studying chromosome conformation. CRISPR imaging can truly state a specific chromosome number by tagging a unique sequence to that specific chromosome and can precisely detect chromosome aneuploidy and mis-segregation (Chen et al., 2016). By using CRISPR imaging system and lineage tracking, we might check the aneuploidy growth kinetics with high temporal resolution in a population of given type of cells. Blend of dCas9 variants and diverse fluorescent proteins concur to label manifold genomic sequences within single genome (Chen et al., 2013). Hence, it enables us to obtain multicolor pictures for multiplexed finding of genetic actions. Prospective engineering of CRISPR can also make possible the plant RNAs recognition other than plant genomic DNAs. Additionally, chromosome translocation and transposition can be targeted for imaging with CRISPR (Roukos et al., 2013). Imaging with the help of CRISPR offer influential strategy to comprehend the heterochromatin formation control (Grewal and Jia, 2007).
CRISPR cas9 as an emerging technique of chromatin imaging present the aim to end the gap between sequencing studies and imaging studies. Even though technical challenges lie ahead, the prospective of CRISPR imaging will assist in solving many plant genome and chromatin related mysteries through direct cell imaging. CRISPR imaging’s unparalleled flexibility and accuracy in sequence targets lead us to accept as true the best is yet to come.
Plant Epigenetic Responses and Crispr-Cas9
The dynamic events in epigenetic process determine multifaceted genome functions. Demand for dissection of plant multiple gene mutation is increasing. However, the existing methods for generating plants harboring several mutated genes involve time consumption and laborious efforts for genetic crossing of solitary-mutant plants (Xing et al., 2014; Quetier, 2015). Epigenetic alterations in DNA or histones that help in organizing chromosomes are expected to play vital roles in biological processes. Analysis of these modifications reveals their decisive value for transcriptional regulation and biological functions (Thakore et al., 2016). For example, epigenetic marks like acetylation and methylation at particular loci or histone residue can strongly effect gene expression. Responses such as histone acetylation and DNA methylation, are catalyzed by diversity of enzymes that are product of special genomic loci (Hsu et al., 2014; Song et al., 2016; Thakore et al., 2016). A multitude of enzymes can erase or produce epigenetic mark(s) on DNA. Few years back, zinc finger proteins and TAL effectors got attention and employed in many studies concerning with locus-oriented targeting of epigenetic amending enzymes (Maeder et al., 2013b; Mendenhall et al., 2013). Histone acetylation, as used in humans for transporting enzymes to specific place within genome, is of immense value and can also be used in plant epigenetics. Such epigenetic marks have particular effects (Ladford, 2016). The enzymes responsible for regulation of epigenetic state can be focused with the help of CRISPR based genome editing or used to produce genome wide perturbations in epigenetic state. This has already been observed in human embryonic stem cells (ESCs) after CRISPR-mediated observations of all DNA methyl transferases. This allows other researchers to characterize possible pluripotent cell lines with distinctive effects on the DNA methylation.
Researchers progressively need supplementary strategies for introducing epigenetic changes specifically at desired loci to test different hypotheses regarding potential implications of CRISPR-Cas technique in plant science. Epigenetic effectors are well able to cause covalent alterations to DNA and histones also (Figure 4). These can also turn on gene expression. Engineered ZFN and TALEN dependent thymine-DNA glycosylase (TDG) or 10-11 Translocation (TET) dioxygenases fusions may result in CpGs demethylation at target promoters. By this way, targeted DNA demethylation is induced, which facilitates re-activation target genes expression (Gregory et al., 2013; Chen et al., 2015; Li et al., 2015). The first observed targetable histone acetyl transferases were DBD–p300 core fusions. It is suggested that dCas9, TALENs and ZFNs fusion to p300catalytic core of histone acetyltransferase can activate gene expression from promoters as well as enhancers after depositing H3K27ac. Researchers are of the view that above mentioned fusion is predominantly important for transcriptional activation because multiplexing is not required. Additionally, distal enhancers can be activated that are unresponsive to dCas9-VP64 (Hilton et al., 2015; Thakore et al., 2016). H3K27ac augment gene expression aided by increased employment of activators in transcription along with transition of RNA Pol II to elongation from initiation. Thus enables achievement of transcriptional activation from targeted genes (Stasevich et al., 2014). Just like the affinity of the SAM complex for activation of transcription, epiCas9s (Cas9 epigenetic effectors) can also be applied for genome-wide screening to find out novel associations between chromatin states, DNA methylation and phenotypes, i.e., cellular differentiation (Hsu et al., 2014).
FIGURE 4.
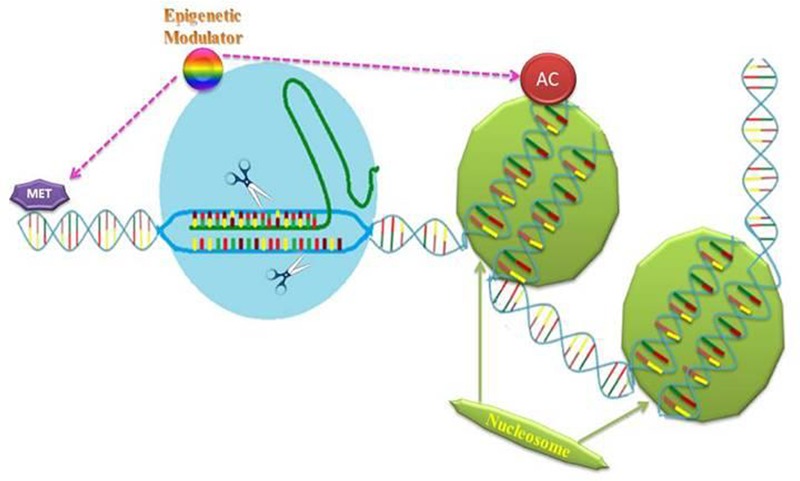
Cas-9 has ability to be coupled with epigenetic modulators, i.e., that add acetyl group (Ac) to histones or that can add methyl group (Me) to DNA. This will help researchers to find out role of precisely place modifications in effecting DNA dynamics or gene expression.
Definite epigenetic changes are adequate for influencing development of normal cells and play roles in later stages of plant development. Therefore, the enzymes for regulation of epigenetic alterations to histones or DNA can be special targets for normal plant development. CRISPR-Cas9 technology permits a catalytically inactive Cas9 to serve as targeted DNA-binding domain. When fused to epigenetic enzymes such as histone acetyl transferases (HATs) DNA methylases, or deacetylases (HDACs), the complex can simultaneously change the epigenetic state in a accurate way at a single or several specific sites. Hilton et al. (2015) presented that programmable DNA-binding proteins can be fused with p300 domain. These results support targeted acetylation as causal mechanism of trans-activation and present a strong tool for gene regulation manipulation. It is noteworthy that effector domains directly catalyzing repressive DNA methylation or histone alterations can be fused with DBDs for making epigenetic silencing proteins. Artificial ZFNs joined with DNMT3a catalyze methylation of DNA and repress transcription from endogenous promoters (Rivenbark et al., 2012; Li et al., 2015).
In the absence of capability to amend the marks at exact sites, researchers are unable to conclude whether they produce biological changes or not. As a result, for plants, system tools need extraordinary refinement for absolute results. In particular case of plant epigenetics, epigenome may be taken as the best mode for controlling activity of gene. In this way, we have to adjust plant epigenome not the plant genome itself. epiCas9 with ability to install/remove unambiguous epigenetic script at specific loci would serve as an additional stage in query of underlying effects of epigenetic amendments in determining the regulatory set-ups of genome. Obviously, the prospective for off-target doings and crosstalk between endogenous epigenetic complexes and effector domains need careful and competent characterization. One way out might be the harnessing of prokaryotic epigenetic enzymes for development of orthogonal epigenetic regulatory mechanisms that can reduce crosstalk with endogenous proteins.
Model Crop Plants
Undoubtedly, crop biologists are striving hard to engineer resistance against diseases, enhancing tolerance to low precipitation or survival under degraded rhizosphere by introducing advantageous genes taken from other varieties of similar species. But no one can deny the fact that adoption of conventional breeding to move traits may take several years and precision is still questionable. Despite the earlier met failures in establishing plant gene targeting technology (Puchta, 1999; Hsu et al., 2014), at the moment single genes are on target of CRISPR-Cas9 system rather than whole genome (Puchta and Fauser, 2013). Being RNA-guided DNA endonuclease, Cas9 can target to explicit genome sequence for making complex with the help of discretely engineered guided RNA (Wang et al., 2011; Quetier, 2015). Generally, CRISPR-Cas9 is highly adaptable for editing of plant genome (Charpentier and Doudna, 2013; Schaeffer and Nakata, 2015) but especially appropriate for genome editing of monocotyledons, e.g., rice due to high genomic GC content (Miao et al., 2013). With special reference to economically valuable plants, i.e., crops and ornamentals, this technique offers an extraordinary and pragmatic system to produce novel phenotypes.
By applying synthetic nucleases, we are able to introduce delicate changes in genome of crop plants by initiating natural repairing pathways, e.g., NHEJ can induct mutations (Puchta and Fauser, 2014). Of special concern is the point that foreign genes can be introduced through NHEJ or HR anywhere in point of interest activated by any DSB. Definitely this is the beginning. Much more is waiting ahead.
After successful demonstration as a genome editor in widely used lab plants Arabidopsis thaliana and Nicotiana benthamiana, CRISPR- Cas9 has been tested in different crops, e.g., rice, wheat, sorghum, soybeans, tomatoes, and oranges. In agriculture, CRISPR-Cas9 is presently being employed to knock-out unwanted genes from crops to promote preferable traits. For example, Chinese researchers developed wheat line resistant to powdery mildew. Genome editing may escort to a few surprising developments in agriculture. Different allergy causing proteins have been detected in peanuts (Hourihane et al., 1997; Skolnick et al., 2001). Getting rid of these proteins is not easy. But new technology may likely to offer allergy-free peanuts. CRISPR-Cas9 technique advocates important changes in plant genome within our access. Gene editing can help in overcoming a hurdle that is polyploid plants showing duplicate genome copies, i.e., Wheat. Successful editing of wheat genome in China demonstrates that CRISPR-Cas9 is definitely “multiplexed” with enormous ability to affect all gene copies or to target several genes at the same time. Certainly, any redundant natural sequence may positively be removed from plant genome by adopting this technique and designing model plants. In different chromosomes, induction of two DSBs may facilitate chromosome arms exchange (Lee et al., 2012). This supports origin of variations for survival by means of available variety of raw material (Puchta and Fauser, 2013). Genome engineering with the help of DSB is now being combined with site-specific recombinase technology in plants of economic importance especially, i.e., rice (Wang et al., 2011).
With simultaneous modification of multiple traits, the CRISPR-Cas9 system would provide highly competent method to pyramid breeding (Bortesi and Fischer, 2015). Gene knockouts mediated by NHEJ are the most direct function of CRISPR-Cas9. Negative regulators of plant disease resistance and grain development can be amended for increasing yield and granting resistance to the host plant against targeted pathogens (Song et al., 2016). Other gene editing strategies, for example regulation of gene expression and epigenetic modulation, can also be adopted for increasing agricultural benefits. Moreover, CRISPR-Cas9 offers unconventional approaches, i.e., Cas9 protein-sgRNA ribonucleoproteins, to deliver target genes into crops with no transgenic footprint. By this way we can circumvent the routine regulations on GMOs (Woo et al., 2015).
Setting up new dimensions in plant science, it tempts to speculate that chromosome engineering and plant genome construction via CRISPR-Cas9 technology is no more a dream. Improvements in nutritional values would be welcome in many crop species and some of them can be approached sensibly through genome editing (Baltes and Voytas, 2015). Extraordinarily, in few plant species, knockout mutants of homozygous nature can be created in distinct generation. Jointly with rest of sequence-specific nucleases, CRISPR-Cas9 is really a game changer skill aimed at revolutionary transformation in plant sciences.
Highly Efficient Plant Cell System
Besides application of classical methods of genetics and breeding for improvements, genome editing through Cas9 has accelerated the efforts for generating the best transgenic models and augment scientific research (Sander and Joung, 2014). Today, mutations in diseased plant populations have been focused. It is considered that CRISPR-based genome editing would be helpful in determining the underlying works of exact genetic abnormalities instead of reliance upon crop disease models. By following the same theme, technique has been applied for developing transgenic animal models few years ago (Niu et al., 2014). iPS cell disease model were engineered with definite mutations corrected or introduced with gene correction in animals (Schwank et al., 2013). The Cas9 genome editing efficiency has made it possible to modify several targets simultaneously, thus facilitates impartial genome-wide functional screens to categorize genes performing central role in development of desired phenotype. Lentivirally delivered sgRNAs directed against all genes can be used to agitate a large number of genomic elements simultaneously (Hsu et al., 2014).
For producing model plant cell systems with high efficiency, constant CRISPR-Cas9expression can be applied for mutants with super efficiency (Xing et al., 2014). These multipurpose systems can be efficiently employed for achieving objectives including production of medicinal and industrial compounds, developing resistance against abiotic as well as biotic stresses (Baltes and Voytas, 2015). Cas9 can simply be launched into the targeted cells by using transitory plasmid transfection having Cas9 and the suitable sgRNA. Like humans, Genome wide association study (GWAS) (Hsu et al., 2014) may appear useful in identifying haplotypes showing positive association with disease risk. One main aim of this technique is to design and create a cell, mean to engineer a cell of desired characteristics with no unwanted component and with ability to divide and pass triplet code ahead. Although likely plant cell is still a dream but methodologies for generating such cells have be employed. For example, CRISPR technique was used to get rid of kilobases of rice DNA that were unnecessary for plant growth. General speaking, such deletions of bases or gene knockouts will be of paramount significance for advancement. But, a strong understanding of needed or unwanted genetic components would chiefly facilitate this series.
The capability of CRISPR-Cas9 system to stack many genes is also feasible with probable application in approaches like metabolic engineering and molecular farming (Mohan, 2016). Recently it is revealed that benzylisoquinoline alkaloid (BIA) production in poppy can be modulated by modifying the particular genes expression in the BIA pathway. Transient over-expression or TRV-mediated gene silencing studies in opium poppy demonstrated that the quantity of BIA biosynthesis could be influenced in a tissue-specific mode (Hosseini et al., 2011; Desgagné-Penix and Facchini, 2012). The over-expression and the silencing of 7OMT and 4′OMT2 genes (R, S)-reticuline 7-O-methyltransferase, 3′-hydroxyl-N-methylcoclaurine 4′-O-methyltransferase) collectively proved their regulatory functions in BIA synthesis in different plant tissues. The previous strategies influenced gene expression that resulted in major reduction in gene expression but never abolished gene function (Alagoz et al., 2016). Therefore, the application CRISPR/Cas9 for knocking out such genes can help to address the challenges and increment our understanding plant cell systems.
By using this technique, we can examine the effect of single gene alternative or experiment the consequence of sole gene maneuvering on isogenic background by editing cells and then differentiating into cell of interest. Such advancements are expected to facilitate gainful, large-scale and less time consuming in vivo mutagenesis studies for avoiding perplexing off-target mutagenesis. Furthermore, Cas9 can be strap up for straight alteration of somatic tissues, precluding the requirement for embryonic exploitation and gene therapy. Although challenges to be addressed are many yet the CRISPR-Cas9 system will certainly evolve into a comprehensive strategy for biotechnology and précised crop breeding in near future. We are hopeful that with the passage of time remarkable advances would be possible in terms of genome editing in plants to produce improved plant systems with desirable traits.
Concluding Remarks and Future Perspectives
Since beginning of this decade, genome editing systems have been adopted to achieve a wide range of modifications, from subtle nucleotide alterations within host genes to the deletion of megabases in DNA (Chen et al., 2016). Together with well-defined and programmable DNA components, plant genome engineering has great potential to facilitate ambitious projects in plant biology. The availability of the CRISPR-Cas9 technology will assist the growing genomics and systems biology data to be exploited very comprehensively by accelerating discovery of genes and related traits development among plant species (Tables 5 and 6). Most of the CRISPR-Cas9 related information is currently obtained from research conducted in mammals. Apparently it is assessed that several of these findings can be universal yet it is imperative to execute analogous studies in plants to make sure that system characteristics are translatable to diverse species. This positively applies to extensive applications like orthogonal gene targeting that have yet to be experienced in plant systems (Bortesi and Fischer, 2015).
Table 6.
Specific commercial products and services available to the researchers to implement CRISPR technology.
| Commercial sources | Products and services | |||
|---|---|---|---|---|
| GeneCopoeia | Genome-wide sgRNA clones | HDR donor cloning vectors and custom HDR donor construction. | Cas9 stable cell lines. | Insertion/deletion detection system |
| Sigma–Aldrich | CRISPR Selection Too | Paired nickases | Codon-optimized Cas9 | Transfection-grade CRISPR plasmid with a guide RNA |
| Bio Labs England | Q5 Hot Start High-Fidelity 2X Master Mix, NEBuilder HiFi DNA Assembly Master Mix | Q5 Site-Directed Mutagenesis Kit (with competent cells) and Q5 Site-Directed Mutagenesis Kit (Without Competent Cells) | EnGe Cas9 Nuclease | EnGen Mutation Detection Kit |
| Integrated DNA technologies (IDT) | Human HPRT PCR Primer Mix, Mouse HPRT PCR Primer Mix, Nuclease Free Duplex Buffer | S.p. Cas9 Expression Plasmid | S.p. Cas9 Nuclease 3NLS (100, 500 μg) | CRISPR Negative Control crRNA, CRISPR Positive Control crRNA |
| DNA 2.0 | Nickase Ninja All-in-One construct expressing specific dual gRNAs | Electra Cloning Kit | ||
| Cyagen | ROSA26 large fragment knockin | |||
| ORiGene | CRISPR/Cas starter kit (HA tagging human HSP60 at C-terminus). | pCas-Guide-Nickase (D10A), pT7-Cas9-Nickase (D10A) | pCas-Guide Cloning Kit, | pCas-Guide-scramble (also available as negative control) |
| System Biosciences | Multiplex gRNA Cloning Kit Create CRISPR/Cas9 constructs with multiple gRNAs simultaneously for better genome editing | |||
| Eurofins Genomics | Cloning Oligos | Indel Detection by Amplicon Analysis | Custom Sequencing (Check the sequence of your CRISPR plasmid or genomic target sequence) | |
This table is provided only to facilitate researchers and R&D. It does not involve any commercial affiliation, marketing or financial purpose.
Future research for improving this technology will include optimization of sgRNA scaffold, which is vital for the TE due to its binding affinity for Cas9 (Jinek et al., 2014). Researchers working in polyploid crops like sugarcane, wheat need information about variation of sequence among diverse allelic forms to design precise gRNAs (Mohan, 2016). Moreover, direct engineering of these Cas9 proteins from diverse bacterial types should tender a path toward PAM independence and producing more competent Cas9 proteins. The extent of off-target mutations and differences in cleavage efficiency need to be evaluated more precisely. Another conspicuous challenge ahead is absence of high throughput screening methods to recognize transgenic plants with edited gene events. In parallel with other studies, capability of CRSIPR-Cas9 system to generate and test multiple gRNAs and availability of next-generation sequencing (NGS) technologies will grant adequate data for the comparison of this system in diversity of plant species and cell types. Keeping in view the number of researchers engaged in CRISPR-Cas9 and velocity of this technique development, additional increments in our understanding and control of the system are expected to come swiftly, promisingly guiding to the devise a new batch of genome editing tools.
Author Contributions
AN has collected research data and compiled manuscript. MA has made all figures and tables. SH has evaluated this manuscript and corrected mistakes.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- Abdallah N. A., Prakash C. S., McHughen A. G. (2015). Genome editing for crop improvement: Challenges and opportunities. GM Crops Food 6 183–205. 10.1080/21645698.2015.1129937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ainley W. M., Sastry-Dent L., Welter M. E., Murray M. G., Zeitler B., Amora R., et al. (2013). Trait stacking via targeted genome editing. Plant Biotechnol. J. 11 1126–1134. 10.1111/pbi.12107 [DOI] [PubMed] [Google Scholar]
- Alagoz Y., Gurkok T., Zhang B., Unver T. (2016). Manipulating the biosynthesis of bioactive compound alkaloids for next-generation metabolic engineering in opium poppy using CRISPR-Cas 9 genome editing technology. Sci. Rep. 6:30910 10.1038/srep30910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baltes N. J., Gil-Humanes J., Cermak T., Atkins P. A., Voytas D. F. (2014). DNA replicons for plant genome engineering. Plant Cell 26 151–163. 10.1105/tpc.113.119792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baltes N. J., Voytas D. F. (2015). Enabling plant synthetic biology through genome engineering. Trends Biotechnol. 33 120–131. 10.1016/j.tibtech.2014.11.008 [DOI] [PubMed] [Google Scholar]
- Bolotin A., Quinquis B., Sorokin A., Ehrlich S. D. (2005). Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extra chromosomal origin. Microbiology 151 2551–2561. 10.1099/mic.0.28048-0 [DOI] [PubMed] [Google Scholar]
- Bortesi L., Fischer R. (2015). The CRISPR system for plant genome editing and beyond. Biotechnol. Adv. 33 41–52. 10.1016/j.biotechadv.2014.12.006 [DOI] [PubMed] [Google Scholar]
- Char S. N., Unger-Wallace E., Frame B., Briggs S. A., Main M., Spalding M. H., et al. (2015). Heritable site-specific mutagenesis using TALENs in maize. Plant Biotechnol. J. 13 1002–1010. [DOI] [PubMed] [Google Scholar]
- Charpentier E., Doudna J. A. (2013). Rewriting a genome. Nature 495 50–51. 10.1038/495050a [DOI] [PubMed] [Google Scholar]
- Chen B., Gilbert L. A., Cimini B. A., Schnitzbauer J., Zhang W., Li G. W., et al. (2013). Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell 155 1479–1491. 10.1016/j.cell.2013.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B., Guan J., Huang B. (2015). Imaging specific genomic DNA in living cells. Annu. Rev. Biophys. 45 1–23. 10.1146/annurev-biophys-062215-010830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B., Hu J., Almeida R., Liu H., Balakrishnan S., Covill-Cooke C., et al. (2016). Expanding the CRISPR imaging toolset with Staphylococcus aureus Cas9 for simultaneous imaging of multiple genomic loci. Nucleic Acids Res. 44:e75 10.1093/nar/gkv153.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho S. W., Kim S., Kim J. M., Kim J. S. (2013). Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat. Biotechnol. 31 230–232. 10.1038/nbt.2507 [DOI] [PubMed] [Google Scholar]
- Cong L., Ran F. A., Cox D., Lin S., Barretto R., Habib N., et al. (2013). Multiplex genome engineering using CRISPR/Cas systems. Science 339 819–823. 10.1126/science.1231143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook C., Martin L., Bastow R. (2014). Opportunities in plant synthetic biology. J. Exp. Bot. 65 1921–1926. 10.1093/jxb/eru013 [DOI] [PubMed] [Google Scholar]
- Deltcheva E., Chylinski K., Sharma C. M., Gonzales K., Chao Y., Pirzada Z. A., et al. (2011). CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 471 602–607. 10.1038/nature09886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desgagné-Penix I., Facchini P. J. (2012). Systematic silencing of benzylisoquinoline alkaloid biosynthetic genes reveals the major route to papaverine in opium poppy. Plant J. 72 331–344. 10.1111/j.1365-313X.2012.05084.x [DOI] [PubMed] [Google Scholar]
- Ding Q., Regan S. N., Xia Y., Oostrom L. A., Cowan C. A., Musunuru K. (2013). Enhanced efficiency of human pluripotent stem cell genome editing through replacing TALENs with CRISPRs. Cell Stem Cell 12 393–394. 10.1016/j.stem.2013.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Halluin K., Vanderstraeten C., Van Hulle J., Rosolowska J., Van Den Brande I., Ennewaert A., et al. (2013). Targeted molecular trait stacking in cotton through targeted double-strand break induction. Plant Biotechnol. J. 11 933–941. 10.1111/pbi.12085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fauser F., Schiml S., Puchta H. (2014). Both CRISPR/Cas-based nucleases and nickases can be used efficiently for genome engineering in Arabidopsis thaliana. Plant J. 79 348–359. 10.1111/tpj.12554 [DOI] [PubMed] [Google Scholar]
- Feng Z., Mao Y., Xu N., Zhang B., Wei P., Yang D. L., et al. (2014). Multigeneration analysis reveals the inheritance, specificity and patterns of CRISPR/Cas-induced gene modifications in Arabidopsis. Proc. Natl. Acad. Sci. U S A. 111 4632–4637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Z., Zhang B., Ding W., Liu X., Yang L. Y., Wei P., et al. (2013). Efficient genome editing in plants using a CRISPR/Cas system. Cell Res. 23 1229–1232. 10.1038/cr.2013.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y., Foden J. A., Khayter C., Maeder M. L., Reyon D., Joung J. K., et al. (2013). High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat. Biotechnol. 31 822–826. 10.1038/nbt.2623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y., Sander J. D., Reyon D., Cascio V. M., Joung J. K. (2014). Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat. Biotechnol. 32 279–284. 10.1038/nbt.2808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao H., Smith J., Yang M., Jones S., Djukanovic V., Nicholson M. G., et al. (2010). Heritable targeted mutagenesis in maize using a designed endonuclease. Plant J. 61 176–187. 10.1111/j.1365-313X.2009.04041.x [DOI] [PubMed] [Google Scholar]
- Gasiunas G., Barrangou R., Horvath P., Siksnys V. (2012). Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc. Natl. Acad. Sci. U.S.A. 109 E2579–E2586. 10.1073/pnas.1208507109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory D. J., Zhang Y., Kobzik L., Fedulov A. V. (2013). Specific transcriptional enhancement of inducible nitric oxide synthase by targeted promoter demethylation. Epigenet 8 1205–1212. 10.4161/epi.26267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewal S. I., Jia S. (2007). Heterochromatin revisited. Nat. Rev. Genet. 8 35–46. 10.1038/nrg2008 [DOI] [PubMed] [Google Scholar]
- Griffith J. D., Comeau L., Rosenfield S., Stansel R. M., Bianchi A., Moss H., et al. (1999). Mammalian telomeres end in a large duplex loop. Cell 97 503–514. 10.1016/S0092-8674(00)80760-6 [DOI] [PubMed] [Google Scholar]
- Haft D. H., Selengut J., Mongodin E. F., Nelson K. E. (2005). A guild of 45 CRISPR-associated (Cas) protein families and multiple CRISPR/Cas subtypes exist in prokaryotic genomes. PLoS Comput. Biol. 1:e60 10.1371/journal.pcbi.0010060.eor [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale C. R., Zhao P., Olson S., Duff M. O., Graveley B. R., Wells L., et al. (2009). RNA-guided RNA cleavage by a CRISPR RNA-Cas protein complex. Cell 139 945–956. 10.1016/j.cell.2009.07.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilton I. B., D’Ippolito A. M., Vockley C. M., Thakore P. I., Crawford G. E., Reddy T. E., et al. (2015). Epigenome editing by a CRISPR-Cas9-based acetyltransferase activates genes from promoters and enhancers. Nat. Biotechnol. 33 510–517. 10.1038/nbt.3199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosseini B., Shahriari-Ahmadi F., Hashemi H., Marashi M. H., Mohseniazar M., Farokhzad A. (2011). Transient expression of cor gene in Papaver somniferum. Bioimpacts 1 229–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hourihane J. O., Kilburn S. A., Dean P., Warner J. O. (1997). Clinical characteristics of peanut allergy. Clin. Exp. Allergy 27 634–639. [PubMed] [Google Scholar]
- Hsu P. D., Lander E. S., Zhang F. (2014). Development and applications of CRISPRfor genome engineering. Cell 157 1262–1278. 10.1016/j.cell.2014.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu P. D., Scott D. A., Weinstein J. A., Ran F. A., Konermann S., Agarwala V., et al. (2013). DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 31 827–832. 10.1038/nbt.2647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iida S., Terada R. (2005). Modification of endogenous natural genes by gene targeting in rice and other higher plants. Plant Mol. Biol. 59 205–219. 10.1007/s11103-005-2162-x [DOI] [PubMed] [Google Scholar]
- Ishino Y., Shinagawa H., Makino K., Amemura M., Nakata A. (1987). Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli and identification of the gene product. J. Bacteriol. 169 5429–5433. 10.1128/jb.169.12.5429-5433.1987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H., Wang N. (2014). Targeted genome editing of sweet orange using Cas9/sgRNA. PLoS ONE 9:e93806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W., Zhou H., Bi H., Fromm M., Yang B., Weeks D. P. (2013). Demonstration of CRISPR/sgRNA-mediated targeted gene modification in Arabidopsis, tobacco, sorghum and rice. Nucleic Acids Res. 41:e188 10.1093/nar/gkt780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W. Z., Yang B., Weeks D. P. (2014). Efficient CRISPR-mediated gene editing in Arabidopsis thaliana and inheritance of modified genes in the T2 and T3 generations. PLoS ONE 9:e99225 10.1371/journal.pone.0099225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J. A., Charpentier E. (2012). A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337 816–821. 10.1126/science.1225829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M., Jiang F., Taylor D. W., Sternberg S. H., Kaya E., Ma E., et al. (2014). Structures of Cas9 endonucleases reveal RNA-mediated conformational activation. Science 343:1247997 10.1126/science.1247997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalhor R., Tjong H., Jayathilaka N., Alber F., Chen L. (2012). Genome architectures revealed by tethered chromosome conformation capture and population-based modeling. Nat. Biotechnol. 30 90–98. 10.1038/nbt.2057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanchiswamy C. N., Maffei M., Malnoy M., Velasco R., Kim J. S. (2016). Fine-tuning next-generation genome editing tools. Trends Biotechnol. 34 562–574. 10.1016/j.tibtech.2016.03.007 [DOI] [PubMed] [Google Scholar]
- Kochevenko A., Willmitzer L. (2003). Chimeric RNA/DNA oligonucleotide based site-specific modification of the tobacco acetolactatesyntase gene. Plant Physiol. 132 174–184. 10.1104/pp.102.016857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konermann S., Brigham M. D., Trevino A. E., Joung J., Abudayyeh O. O., Barcena C., et al. (2015). Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature 515 83–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar V., Jain M. (2014). The CRISPR–Cas system for plant genome editing: advances and opportunities. J. Exp. Bot. 66 47–57. 10.1093/jxb/eru429 [DOI] [PubMed] [Google Scholar]
- Ladford H. (2016). CRISPR: gene editing is just the beginning. Nature 531 156–159. 10.1038/531156a [DOI] [PubMed] [Google Scholar]
- Lee H. J., Kweon J., Kim E., Kim S., Kim J. S. (2012). Targeted chromosomal duplications and inversions in the human genome using zinc finger nucleases. Genome Res. 22 539–548. 10.1101/gr.129635.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J. F., Norville J. E., Aach J., McCormack M., Zhang D., Bush J., et al. (2013). Multiplex and homologous recombination-mediated genome editing in Arabidopsis and Nicotiana benthamiana using guide RNA and Cas9. Nat. Biotechnol. 31 688–691. 10.1038/nbt.2654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K., Pang J., Cheng H., Liu W. P., Di J. M., Xiao H. J., et al. (2015). Manipulation of prostate cancer metastasis by locus-specific modification of the CRMP4 promoter region using chimeric TALE DNA methyltransferase and demethylase. Oncotarget 6 10030–10044. 10.18632/oncotarget.3192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y., Zhang L., Huang X. (2014). Genome modification by CRISPR. FEBS J. 281 5186–5193. 10.1111/febs.13110 [DOI] [PubMed] [Google Scholar]
- Maeder M. L., Angstman J. F., Richardson M. E., Linder S. J., Cascio V. M., Tsai S. Q., et al. (2013a). Targeted DNA demethylation and activation of endogenous genes usingprogrammable TALE-TET1 fusion proteins. Nat. Biotechnol. 31 1137–1142. 10.1038/nbt.2726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeder M. L., Linder S. J., Cascio V. M., Fu Y., Ho Q. H., Joung J. K. (2013b). CRISPR RNA-guided activation of endogenous human genes. Nat. Methods 10 977–979. 10.1038/nmeth.2598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeder M. L., Thibodeau-Beganny S., Osiak A., Wright D. A., Anthony R. M., Eichtinger M., et al. (2008). Rapid “open-source” engineering of customized zinc-finger nucleases for highly efficient gene modification. Mol. Cell 31 294–301. 10.1016/j.molcel.2008.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mak A. N., Bradley P., Cernadas R. A., Bogdanove A. J., Stoddard B. L. (2012). The crystal structure of TAL effector PthXo1 bound to its DNA target. Science 335 716–719. 10.1126/science.1216211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malina A., Katigbak A., Cencic R., Maïga R. I., Robert F., Miura H., et al. (2014). Adapting CRISPR for functional genomics screens. Methods Enzymol. 546 193–213. 10.1016/B978-0-12-801185-0.00010-6 [DOI] [PubMed] [Google Scholar]
- Mao Y., Zhang H., Xu N., Zhang B., Gou F., Zhu J. K. (2013). Application of the CRISPR-Cas system for efficient genome engineering in plants. Mol. Plant 6 2008–2011. 10.1093/mp/sst121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendenhall E. M., Williamson K. E., Reyon D., Zou J. Y., Ram O., Joung J. K., et al. (2013). Locus-specific editing of histone modifications at endogenous enhancers. Nat. Biotechnol. 31 1133–1136. 10.1038/nbt.2701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao J., Guo D., Zhang J., Huang Q., Qin G., Zhang X., et al. (2013). Targeted mutagenesis in rice using CRISPR-Cas system. Cell Res. 23 1233–1236. 10.1038/cr.2013.123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J. C., Tan S., Qiao G., Barlow K. A., Wang J., Xia D. F., et al. (2011). A tale nuclease architecture for efficient genome editing. Nat. Biotechnol. 29 143–148. 10.1038/nbt.1755 [DOI] [PubMed] [Google Scholar]
- Mohan C. (2016). Genome editing in sugarcane: challenges ahead. Front. Plant Sci. 7:1542 10.3389/fpls.2016.01542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagano T., Lubling Y., Stevens T. J., Schoenfelder S., Yaffe E., Dean W., et al. (2013). Single-cell Hi-C reveals cell-tocell variability in chromosome structure. Nature 502 59–64. 10.1038/nature12593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nekrasov V., Staskawicz B., Weigel D., Jones J. D., Kamoun S. (2013). Targeted mutagenesis in the model plant Nicotiana benthamiana using Cas9 RNA-guided endonuclease. Nat. Biotechnol. 31 691–693. 10.1038/nbt.2655 [DOI] [PubMed] [Google Scholar]
- Neumann H., Neumann-Staubitz P. (2010). Synthetic biology approaches in drug discovery and pharmaceutical biotechnology. Appl. Microbiol. Biotechnol. 87 75–86. 10.1007/s00253-010-2578-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemudryi A. A., Valetdinova K. R., Medvedev S. P., Zakian S. M. (2014). TALEN and CRISPR/Cas genome editing systems: tools of discovery. Acta Nat. 6 19–40. [PMC free article] [PubMed] [Google Scholar]
- Niu Y., Shen B., Cui Y., Chen Y., Wang J., Wang L., et al. (2014). Generation of gene-modified cynomolgus monkey via Cas9/RNA-mediated gene targeting in one-cell embryos. Cell 156 836–843. 10.1016/j.cell.2014.01.027 [DOI] [PubMed] [Google Scholar]
- Oldroyd G. E., Murray J. D., Poole P. S., Downie J. A. (2011). The rules of engagement in the legume–rhizobial symbiosis. Ann. Rev. Gen. 45 119–144. 10.1146/annurev-genet-110410-132549 [DOI] [PubMed] [Google Scholar]
- Oldroyd G. E. D., Dixon R. (2014). Biotechnological solutions to the nitrogen problem. Curr. Opin. Biotechnol. 26 19–24. 10.1016/j.copbio.2013.08.006 [DOI] [PubMed] [Google Scholar]
- Perez-Pinera P., Kocak D. D., Vockley C. M., Adler A. F., Kabadi A. M., Polstein L. R., et al. (2013). RNA-guided gene activation by CRISPR-Cas9-based transcription factors. Nat. Methods 10 973–976. 10.1038/nmeth.2600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piatek A., Ali Z., Baazim H., Li L., Abulfaraj A., Al-Shareef S., et al. (2014). RNA-guided transcriptional regulation in planta via synthetic dCas9 –based transcription factors. Plant Biotechnol. J. 13 578–589. 10.1111/pbi.12284 [DOI] [PubMed] [Google Scholar]
- Puchta H. (1999). Double-strand break-induced recombination between ectopic homologous sequences in somatic plant cells. Genetics 152 1173–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puchta H., Fauser F. (2013). Gene targeting in plants: 25 years later. Int. J. Dev. Biol. 57 629–637. 10.1387/ijdb.130194hp [DOI] [PubMed] [Google Scholar]
- Puchta H., Fauser F. (2014). Synthetic nucleases for genome engineering in plants: prospects for a bright future. Plant J. 78 727–741. 10.1111/tpj.12338 [DOI] [PubMed] [Google Scholar]
- Qi L. S., Larson M. H., Gilbert L. A., Doudna J. A., Weissman J. S., Arkin A. P., et al. (2013). Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 152 1173–1183. 10.1016/j.cell.2013.02.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quetier F. (2015). The CRISPRtechnology:Closer to the ultimate toolkit for targeted genome editing. Plant Sci. 242 65–76. 10.1016/j.plantsci.2015.09.003 [DOI] [PubMed] [Google Scholar]
- Ran F. A., Hsu P. D., Lin C. Y., Gootenberg J. S., Konermann S., Trevino A. E., et al. (2013). Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell 154 1380–1389. 10.1016/j.cell.2013.08.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivenbark A. G., Stolzenburg S., Beltran A. S., Yuan X., Rots M. G., Strahl B. D., et al. (2012). Epigenetic reprogramming of cancer cells via targeted DNA methylation. Epigenet 7 350–360. 10.4161/epi.19507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roukos V., Voss T. C., Schmidt C. K., Lee S., Wangsa D., Misteli T. (2013). Spatial dynamics of chromosome translocations in living cells. Science 341 660–664. 10.1126/science.1237150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander J. D., Joung J. K. (2014). CRISPR-Cas systems for editing, regulating and targeting genomes. Nat. Biotechnol. 32 347–355. 10.1038/nbt.2842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarrion-Perdigones A., Vazquez-Vilar M., Palaci J., Castelijns B., Forment J., Ziarsolo P., et al. (2013). GoldenBraid 2.0: a comprehensive DNA assembly framework for plant synthetic biology. Plant Physiol. 162 1618–1631. 10.1104/pp.113.217661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaeffer S. M., Nakata P. A. (2015). CRISPR-mediated genome editing and gene replacement in plants: transitioning from lab to field. Plant Sci. 240 130–142. 10.1016/j.plantsci.2015.09.011 [DOI] [PubMed] [Google Scholar]
- Schiml S., Fauser F., Puchta H. (2014). The CRISPR/Cas system can be used as nuclease for in planta gene targeting and as paired nickases for directed mutagenesis in Arabidopsis resulting in heritable progeny. Plant J. 80 1139–1150. 10.1111/tpj.12704 [DOI] [PubMed] [Google Scholar]
- Schwank G., Koo B. K., Sasselli V., Dekkers J. F., Heo I., Demircan T., et al. (2013). Functional repair of CFTR by CRISPR in intestinal stem cell organoids of cystic fibrosis patients. Cell Stem Cell 13 653–658. 10.1016/j.stem.2013.11.002 [DOI] [PubMed] [Google Scholar]
- Shan Q., Wang Y., Chen K., Liang Z., Li J., Zhang Y., et al. (2013a). Rapid and efficient gene modification in rice and Brachypodium using TALENs. Mol. Plant 6 1365–1368. 10.1093/mp/sss162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan Q., Wang Y., Li J. (2013b). Targeted genome modification of crop plants using a CRISPR–Cas system. Nat. Biotechnol. 31 686–688. 10.1038/nbt.2650 [DOI] [PubMed] [Google Scholar]
- Shan Q., Wang Y., Li J., Zhang Y., Chen K., Liang Z., et al. (2013c). Targeted genome modification of crop plants using the CRISPR–Cassystem.Nat. Biotechnology 31 686–688. [DOI] [PubMed] [Google Scholar]
- Shan Q., Wang Y., Li J., Gao C. (2014). Genome editing in rice and wheat using CRISPR/Cas system. Nat. Protoc. 9 2395–2410. 10.1038/nprot.2014.157 [DOI] [PubMed] [Google Scholar]
- Shao S., Zhang W., Hu H., Xue B., Qin J., et al. (2016). Long-term dual-color tracking of genomic loci by modified sgRNAs of the CRISPR/Cas9 system. Nucleic Acids Res. 44 e86. 10.1093/nar/gkw066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skolnick H. S., Conover-Walker M. K., Koerner C. B., Sampson H. A., Burks W., Wood R. A. (2001). The natural history of peanut allergy. J. Allergy Clin. Immunol. 107 367–374. 10.1067/mai.2001.112129 [DOI] [PubMed] [Google Scholar]
- Song G., Jia M., Chen K., Kong X., Khattak B., Xie C., et al. (2016). CRISPR: a powerful tool for crop genome editing. Crop J. 4 75–82. 10.1016/j.cj.2015.12.002 [DOI] [Google Scholar]
- Sozzani R., Cui H., Moreno-Risueno M. A., Busch W., Van Norman J. M., Vernoux T., et al. (2010). Spatiotemporal regulation of cell-cycle genes by short root links patterning and growth. Nature 466 128–132. 10.1038/nature09143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprink T., Metje J., Hartung F. (2015). Plant genome editing by novel tools: TALEN and other sequence specific nucleases. Curr. Opin. Biotechnol. 32 47–53. 10.1016/j.copbio.2014.11.010 [DOI] [PubMed] [Google Scholar]
- Stasevich T. J., Hayashi-Takanaka Y., Sato Y., Maehara K., Ohkawa Y., Sakata-Sogawa K., et al. (2014). Regulation of RNA polymerase II activation by histone acetylation in single living cells. Nature. 516 272–275. 10.1038/nature13714 [DOI] [PubMed] [Google Scholar]
- Sternberg S. H., Redding S., Jinek M., Greene E. C., Doudna J. A. (2014). DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature. 507 62–67. 10.1038/nature13011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugano S. S., Shirakawa M., Takagi J., Matsuda Y., Shimada T., Hara-Nishimura I. (2014). CRISPR/Cas9 mediated targeted mutagenesis in the liverwort Marchantia polymorpha L. Plant Cell Physiol. 55 475–481. 10.1093/pcp/pcu014 [DOI] [PubMed] [Google Scholar]
- Temme K., Zhao D., Voigtb C. A. (2012). Refactoring the nitrogen fixation gene cluster from Klebsiellaoxytoca. Proc. Natl. Acad. Sci. U.S.A. 109 7085–7090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakore P. I., Black J. B., Hilton I. B., Gersbach C. A. (2016). Editing the epigenome: technologies for programmable transcription and epigenetic modulation. Nat. Methods 12 127–137. 10.1038/nmeth.3733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upadhyay S. K., Kumar J., Alok A., Tuli R. (2013). RNA guided genome editing for target gene mutations in wheat. G3 (Bethesda) 3 2233–2238. 10.1534/g3.113.008847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez-Vilar M., Bernabe-Orts J. M., Fernandez-Del-Carmen A., Ziarsolo P., Blanca J., Granell A. (2016). A modular toolbox for gRNA-Cas9 genome engineering in plants based on the golden braid standard. Plant Methods 12:10 10.1186/s13007-016-0101-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Yang H., Shivalila C. S., Dawlaty M. M., Cheng A. W., Zhang F., et al. (2013). One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 153 910–918. 10.1016/j.cell.2013.04.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Cheng X., Shan Q., Zhang Y., Liu J., Gao C., et al. (2014). Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nat. Biotechnol. 32 947–951. 10.3410/f.718498343.793497666 [DOI] [PubMed] [Google Scholar]
- Wang Y., Yau Y. Y., Perkins-Balding D., Thomson J. G. (2011). Recombinase technology: applications and possibilities. Plant Cell Rep 30 267–285. 10.1007/s00299-010-0938-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z. P., Xing H. L., Dong L., Zhang H. Y., Han C. Y., Wang X. C., et al. (2015). Egg cell-specific promoter-controlled CRISPR/Cas9 efficiently generates homozygous mutants for multiple target genes in Arabidopsis in a single generation. Genom. Biol. 16:144 10.1186/s13059-015-0715-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendt T., Holm P. B., Starker C. G., Christian M., Voytas D. F., Brinch-Pedersen H., et al. (2013). TAL effector nucleases induce mutations at a pre-selected location in the genome of primary barley transformants. Plant Mol. Biol. 83 279–285. 10.1007/s11103-013-0078-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedenheft B., Sternberg S. H., Doudna J. A. (2012). RNA-guided genetic silencing systems in bacteria and archaea. Nature 82 331–338. [DOI] [PubMed] [Google Scholar]
- Wolt J. D., Wang K., Yang B. (2016). The regulatory status of genome-edited crops. Plant Biotechnol. J. 14 510–518. 10.1111/pbi.12444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo J. W. J., Kim J., Kwon S. I. I., Corvalán C., Cho S. W., Kim H., et al. (2015). DNA-free genome editing in plants with preassembled CRISPR-Cas9 ribonucleoproteins. Nat. Biotechnol. 33 1162–1164. 10.1038/nbt.3389 [DOI] [PubMed] [Google Scholar]
- Xie F., Murray J. D., Kim J., Heckmann A. B., Edwards A., Oldroyd G. E., et al. (2012). Legume pectatelyase required for root infection by rhizobia. Proc. Natl. Acad. Sci. U.S.A. 109 633–638. 10.1073/pnas.1113992109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing H. L., Li D., Zhi-Ping W., Hai-Yan Z., Chun-Yan H., Bing L., et al. (2014). A CRISPR toolkit for multiplex genome editing in plants. BMC Plant Biol. 14:327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong J. S., Ding J., Li Y. (2015). Genome-editing technologies and their potential application in horticultural crop breeding. Hortic. Res 2:15019 10.1038/hortres.2015.19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalatan J. G., Lee M. E., Almeida R., Gilbert L. A., Whitehead E. H., La Russa M., et al. (2015). Engineering complex synthetic transcriptional programs with CRISPR RNA scaffolds. Cell 160 339–350. 10.1016/j.cell.2014.11.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F., Wen Y., Guo X. (2014). CRISPR for genome editing: progress, implications and challenges. Hum. Mol. Genet. 23 40–46. 10.1093/hmg/ddu125 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Zhang F., Li X., Baller J. A., Qi Y., Starker C. G., et al. (2013). Transcription activator-like effector nucleases enable efficient plant genome engineering. Plant Physiol. 161 20–27. 10.1104/pp.112.205179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H., Liu B., Weeks D. P., Spalding M. H., Yang B. (2014). Large chromosomal deletions and heritable small genetic changes induced by CRISPR in rice. Nucleic Acids Res. 42 10903–10914. 10.1093/nar/gku806 [DOI] [PMC free article] [PubMed] [Google Scholar]


