Abstract
MicroRNAs (miRNAs) have been established as key regulators of various biological processes with possible involvement in the pathobiology of periodontal disease. Expanding our earlier observations of substantial differential expression of specific miRNAs between clinically healthy and periodontitis-affected gingival tissues, we used miRNA inhibitors (sponges) in loss-of-function experiments to investigate the involvement of specific miRNAs in the response of pocket epithelium-derived, telomerase-immortalized human gingival keratinocytes (TIGKs) to microbial infection. We constructed stable knockdown (KD) cell lines for five epithelium-expressed miRNAs (miR-126, miR-141, miR-155, miR-210, and miR-1246) and assessed their response to infection with periodontal pathogens using microarray analysis, quantitative PCR (qPCR), enzyme-linked immunosorbent assay (ELISA), and Western blot assay. miR-126 KD cells showed lower expression of interleukin 8 (IL-8) and CXCL1, both on the mRNA and protein levels, than did controls upon stimulation by heat-killed wild-type Porphyromonas gingivalis, live P. gingivalis protease-deficient mutant KDP128, and live Aggregatibacter actinomycetemcomitans. In contrast, infection of miR-155 KD and miR-210 KD cells with the same organisms resulted in higher IL-8 and CXCL1 mRNA and protein expression. These effects appeared to be regulated by NF-κB, as suggested by altered transcription and/or phosphorylation status of components of the NF-κB system. Reduced neutrophil-like HL-60 cell chemotactic activity was observed in response to infection of miR-126 KD cells, indicating that miR-126 plays an important role in immune responses. Our findings indicate that specific miRNAs regulate the expression of inflammatory cytokines in human gingival epithelial cells in response to microbial infection.
INTRODUCTION
MicroRNAs (miRNAs) are noncoding, single-stranded, ∼22-base RNA sequences that have been shown to play critical roles in posttranscriptional regulation, primarily through their ability to induce mRNA destabilization and/or repression of protein translation (1). MicroRNAs have been shown to play important roles in the regulation of immune responses (2–4) and have also emerged as novel biomarkers for several diseases of multifactorial etiology, including cancer (5, 6), cardiovascular disease (7–9), and Alzheimer's disease (10, 11).
Mapping of the function of miRNAs in biological systems is the focus of intense ongoing research efforts. Experimental studies have employed loss-of-function strategies, i.e., inhibition of miRNAs using intracellular delivery of either chemically synthesized miRNA inhibitors or plasmids in nonviral vectors such as cationic polymers or liposomes (12–14). These methods can provide transient inhibition of target miRNAs and generally lead to low transfection efficacy in primary cells that may not be sufficient for inducing functional consequences (15). In addition, the use of inhibitory reagents has been shown to induce cellular immune responses, suggesting that this strategy is unsuitable for mapping the role of miRNAs in immune response regulation (16). In contrast, the use of lentiviral vectors that carry specific miRNA inhibitors (sponges) has been shown to overcome these challenges (17, 18). These vectors integrate into the host genome and allow stable expression of miRNA sponges and long-term inhibition of specific miRNAs.
Limited data are available on the function of miRNAs in the gingival tissues and their role in the pathobiology of human periodontitis (19). Benakanakere et al. (20) showed increased miR-105 expression in primary human keratinocytes challenged with heat-inactivated Porphyromonas gingivalis and identified miR-105 as a modulator of Toll-like receptor 2 (TLR-2) protein translation. Nahid et al. (21) showed a persistent association of miR-146a expression in both the periodontium and spleens isolated from ApoE−/− mice infected with P. gingivalis, Treponema denticola, and Tannerella forsythia, suggesting that it may modulate the course of infection by these microorganisms. Moffatt and Lamont (22) studied the differential expression of several miRNAs between P. gingivalis-infected and uninfected primary gingival epithelial cells and showed an upregulation of miR-203 in infected cells followed by inhibition of important host signaling responses. Our group recently described the differential expression of miRNAs and their target genes in inflamed or noninflamed gingival tissue samples harvested from patients with periodontitis (23). Given that the epithelial lining of the gingival crevice/periodontal pocket constitutes the barrier that interfaces with the microbial biofilm of the dental plaque (24), we selected a number of miRNAs (miR-126, miR-141, miR-146a, miR-155, miR-210, miR-223, miR-451, miR-486, and miR-1246) earlier shown by us (23) to be strongly differentially expressed between states of gingival health and disease and studied their role as regulators of cytokine responses in gingival epithelial cells.
MATERIALS AND METHODS
Cell culture and bacteria.
Telomerase-immortalized human gingival keratinocytes (TIGKs; a generous gift from Richard Lamont, University of Louisville, KY) (25) were cultured in keratinocyte serum-free medium (K-SFM; Invitrogen, Carlsbad, CA) supplemented with 0.4 mM calcium chloride, 25 μg/ml of bovine pituitary extract, 0.2 ng/ml of epidermal growth factor, penicillin (100 U/ml), and streptomycin (100 μg/ml). HL-60 promyelocytic cells (ATCC CCL-240) were maintained in Iscove's modified Dulbecco's medium with 10% heat-inactivated fetal bovine serum (FBS) and 100 U/ml of penicillin-streptomycin at 37°C. To induce differentiation, cells were diluted (2.5 × 105 cells/ml) with 1.25% dimethyl sulfoxide (DMSO) for 4 to 5 days. Wild-type (WT) Porphyromonas gingivalis strain ATCC 33277 and the KDP128 mutant, which is deficient in the production of proteases encoded by rgpA, rgpB, and kgp (a generous gift from M. R. Benakanakere, University of Pennsylvania), were cultured in Trypticase soy broth (30 mg/ml) supplemented with yeast extract (5 mg/ml), hemin (5 μg/ml), and vitamin K1 (1 μg/ml), anaerobically, at 37°C. Overnight cultures were diluted to an optical density at 600 nm (OD600) of 1.0 (109 CFU/ml). Since the WT P. gingivalis strain has profound proteolytic capabilities, we used heat-killed WT P. gingivalis cells to avoid extensive degradation of cytokines in the culture medium. These were produced by heating at 70°C for 1 h, washing three times with sterile phosphate-buffered saline (PBS) at 6,000 × g for 10 min, and suspension in cell culture medium. Aggregatibacter actinomycetemcomitans strain ATCC 33384 was cultured in brain heart infusion broth (37 mg/ml) and grown at 37°C in 5% CO2. Overnight cultures were diluted to an OD600 of 1.0 (109 CFU/ml), followed by three washes with sterile PBS at 6,000 × g for 10 min and suspension in cell culture medium.
Confirmation of specific miRNA expression in wild-type TIGKs.
MicroRNAs were isolated using the mirVana miRNA isolation kit (Life Technologies, Grand Island, NY). miRNA-specific cDNA templates were prepared from 10 ng of total RNA using a TaqMan microRNA reverse transcription kit (Life Technologies). The constitutive expression of miR-126, miR-141, miR-146a, miR-155, miR-210, miR-223, miR-451, miR-486, and miR-1246 in TIGKs was assessed by real-time quantitative PCR (qPCR; TaqMan microRNA assay; Life Technologies). RNU6B was used as the reference gene.
MicroRNA sponge design and generation and lentivirus packaging and production.
Specific miRNA inhibitors (sponges) each containing 10 tandemly arrayed miRNA binding sites (MBS) separated by 4-nucleotide-long spacer sequences (Fig. 1C) were synthesized by Life Technologies for those miRNAs found to be constitutively expressed in wild-type TIGKs, using a central-mismatch design (26) that results in better knockdown (KD) efficiency than a perfect-match design. Sponge sequences are listed in Table S1 in the supplemental material, and their sites of binding to each target miRNA are listed in Table S2 in the supplemental material. Each sponge was cloned into the XhoI and EcoRI sites of the pLVX-AcGFP1-N1 lentiviral vectors (Clontech, Mountain View, CA). For lentivirus packing and production, Lenti-X 293T packaging cells (Clontech) were cultured to 80% confluence in 175-cm2 culture flasks in Dulbecco's modified Eagle medium (DMEM) containing 10% FBS, 100 U/ml of penicillin, and 100 μg/ml of streptomycin. Lentiviruses were produced by cotransfection of lentiviral vector plasmids (16 μg) and packing plasmids Δ8.9 (8 μg) and vesicular stomatitis virus G protein (VSV-G) (8 μg) with 96 μl of TransIT-LT1 (Mirus, Madison, WI) and 3.2 ml of Opti-MEM (Life Technologies) into 293T packaging cells according to the manufacturer's protocol. On the second day, medium was replaced with fresh medium. On the third day, the supernatant was collected and cellular debris was removed by low-speed centrifugation (300 × g for 5 min). The supernatant was filtered through a 0.45-mm low-protein-binding membrane (Pall Life Science, Port Washington, NY). To concentrate the viral particles, Lenti-X concentrator (Clontech) was used according to the manufacturer's protocol.
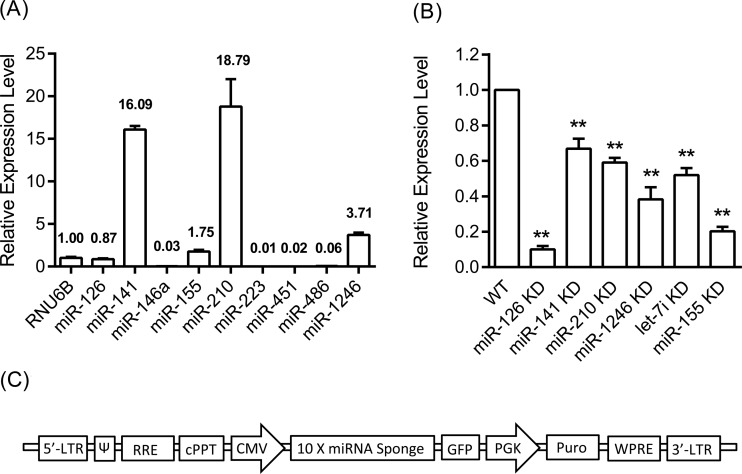
FIG 1.
(A) Constitutive expression of the selected miRNAs in wild-type (WT) TIGKs, assessed through TaqMan miRNA qPCR. Five miRNAs (miR-126, miR-141, miR-155, miR-210, and miR-1246) were expressed at levels higher than or comparable to those of the reference RNU6B gene and were therefore chosen for further analysis. Bars indicate relative expression calculated by delta CT values in relation to RNU6B reference gene expression (=1). (B) Relative expression level of miRNAs in the knockdown cells assessed through qPCR. Relative miRNA expression in each miRNA knockdown cell line was normalized to their corresponding miRNA level in the WT cells (=100%). Bars express means and standard deviations, based on triplicate experiments. Asterisks indicate statistically significant differences between WT and miRNA KD cells (**, P < 0.01). (C) Schematic representation of miRNA sponge expression cassette.
Production of miRNA KD cells and assessment of miRNA KD efficiency.
To produce miRNA KD isogenic clones, TIGKs were infected with lentiviruses containing miRNA sponges. Transduced cells (positive for green fluorescent protein [GFP+]) were single-cell sorted into 96-well plates by flow cytometry. Seven to 9 days after sorting, cell colonies were transferred to larger plates for further expansion. Mock-transduced cells were generated by transducing wild-type cells with lentiviruses that did not contain an miRNA sponge; GFP+ cells were subsequently sorted by flow cytometry. MicroRNAs were isolated using mirVana miRNA isolation kit (Life Technologies). cDNA templates were prepared from 10 ng of total RNA using a TaqMan MicroRNA reverse transcription kit (Life Technologies). The knockdown efficiency was assessed by real-time qPCR (TaqMan microRNA assay; Life Technologies). RNU6B was used as the endogenous control.
Gene expression analysis and GSEA.
Total RNA was isolated from P. gingivalis-stimulated (multiplicity of infection [MOI] of 100 for 6 h) TIKG wild-type cells and miRNA KD cells using TRIzol Plus RNA purification kit (Thermo Fisher Scientific, New York, NY). RNA quantity was measured spectrophotometrically on a NanoDrop, and RNA quality was assessed using an Agilent 2100 bioanalyzer (Agilent Technologies, Santa Clara, CA), demonstrating an RNA integrity number ≥9. One hundred nanograms of total RNA was reverse transcribed and labeled using the Illumina TotalPrep RNA amplification kit (Life Technologies). Labeled RNA was hybridized on HumanHT-12 v4 Expression BeadChips (Illumina, San Diego, CA). Gene expression profiles were analyzed with R and Bioconductor statistical frameworks. Gene set enrichment analysis (GSEA; http://software.broadinstitute.org/gsea/index.jsp) was carried out.
Screening for cytokine expression.
Wild-type TIGKs and miR-126-KD cells were stimulated with heat-killed P. gingivalis and assessed for simultaneous expression of 36 different cytokines using the human cytokine array, panel A (R&D Systems, Minneapolis, MN).
Confirmation of selected cytokine expression by ELISA.
A total of 5 × 105 WT TIGKs, mock-transduced cells, and miRNA KD cells were seeded on 12-well plates. Twenty-four hours later, 108 CFU of heat-killed WT P. gingivalis, P. gingivalis mutant KDP128, or A. actinomycetemcomitans at an MOI of 100 were added to the culture medium for 6 h. Concentrations of CXCL1 and interleukin 8 (IL-8) in the supernatants were determined using CXCL1 (R&D Systems) and IL-8 (Thermo Fisher Scientific, New York, NY) ELISA kits. Assessments were replicated in three independent experiments.
Gene expression in wild-type and miRNA KD TIGKs by qPCR.
To assess gene expression for IL-8 and CXCL1 in WT TIGKs and miRNA KD cells, 12-well plates were seeded at a density of 5 × 105 per well. Twenty-four hours later, heat-killed WT P. gingivalis, live P. gingivalis mutant KDP128, or A. actinomycetemcomitans was added to the culture medium at an MOI of 100 for 6 h. Cells were lysed with TRIzol (Life Technologies) and total RNA was isolated according to the manufacturer's protocol. Total RNA was treated with DNase I (Promega, Madison, WI), a high-capacity cDNA reverse transcription kit was used to produce cDNA, and ABsolute blue qPCR SYBR green mixes (Thermo Scientific, New York, NY) were used for qPCR (primer sequences are listed in Table S3 in the supplemental material). The qPCR experiments were performed using the ViiA 7 real-time PCR system (Life Technologies). The relative amount of each gene was calculated using the comparative threshold cycle (CT) method, using glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as an endogenous control. Assessments were replicated in three independent experiments.
Western blotting.
Whole-cell extracts were fractionated by SDS-PAGE and transferred to a polyvinylidene difluoride membrane using a semidry transfer apparatus according to the manufacturer's protocols (Bio-Rad). After incubation with 5% nonfat milk and 2% bovine serum albumin (BSA) in Tris-buffered saline with Tween 20 (TBST) for 1 h at room temperature, the membrane was washed once with TBST and incubated with antibodies against phospho-NF-κB p65 (Ser536) (Cell Signaling Technology), NF-κB P65 (Santa Cruz Biotechnology, TX), or GAPDH (Thermo Fisher Scientific, NY) at 4°C overnight. Membranes were washed three times for 10 min and incubated with a 1:3,000 dilution of horseradish peroxidase-conjugated anti-mouse or anti-rabbit antibodies for 1 h. Blots were washed with TBST three times and developed with the ECL system (Amersham Biosciences) according to the manufacturer's protocol.
Chemotaxis and myeloperoxidase activity assays.
A leukocyte chemotaxis/myeloperoxidase activity assay was carried out as described previously (27), using 24-well Transwell tissue-culture permeable inserts with 3-μm pores (Corning, NY, USA). A total of 106 induced HL-60 cells in 100 μl of Hanks' balanced salt solution (HBSS) buffer (devoid of calcium and magnesium) were added to the upper compartment, and 600 μl of supernatant collected from WT and miRNA KD cells stimulated with heat-killed WT P. gingivalis for 6 h was added to the lower compartment. After 2 h of incubation, the number of migrated HL-60 cells was quantified by assessing relative myeloperoxidase activity against a linear standard curve of known numbers of HL-60 cells. To prevent any effects of the cell culture supernatant on myeloperoxidase activity, the plates were centrifuged at 200 × g for 5 min with no brake, the supernatant was discarded, and 600 μl of PBS was added into each well to resuspend the cells. After resuspension, 50 μl of 10% Triton X-100 was added to each well and the plate was rotated at 4°C for 20 min. Thereafter, 50 μl of 1 M citrate buffer was added to each well, followed by thorough mixing. One hundred microliters from each sample was transferred to a 96-well plate, and 100 μl of fresh prepared 2,2′-azino-bis(3-ethylbenzothiazoline-6-sulfonic acid) (ABTS) solution with 0.003% H2O2 was added. The plate was allowed to develop for 10 min in the dark and was read at a wavelength of 405 nm using a microplate reader.
Statistical analysis.
Mean values and standard deviations of triplicate experiments were calculated. Differences between control and KD cells were analyzed using the Student t test. A P value of <0.05 was considered statistically significant.
RESULTS
Expression of miRNAs by TIGKs and efficacy of miRNA inhibition in miRNA KD cells.
Assessment of the constitutive expression level of the nine selected miRNAs by wild-type TIGKs using qPCR showed that five miRNAs (miR-126, miR-141, miR-155, miR-210, and miR-1246) were expressed at levels higher than or comparable to those of the reference RNU6B gene (Fig. 1A). Therefore, sponges for these particular miRNAs were designed, and corresponding miRNA KD cells were produced, as described in Materials and Methods. Assessment of miRNA expression in KD cells by qPCR showed that use of the sponges resulted in variable miRNA inhibition efficiencies, ranging from a low of 33% for miR-140 KD cells to a high of 90% for miR-126 KD cells (Fig. 1B).
Gene set enrichment analysis.
To screen for an overall effect of miRNA knockdown on gene expression, we selected miR-126 KD cells, which showed the highest miRNA inhibition efficiency, and performed whole-genome mRNA profiling, followed by gene set enrichment analysis to further analyze the gene expression data. The top 50 differentially expressed genes between WT and miR-126KD cells are listed in Table S4 in the supplemental material. Table S5 in the supplemental material lists the top 25 gene sets enriched in P. gingivalis-stimulated wild-type cells, most of which were related to immune regulation and host defense responses.
Cytokine responses in miRNA KD TIGKs.
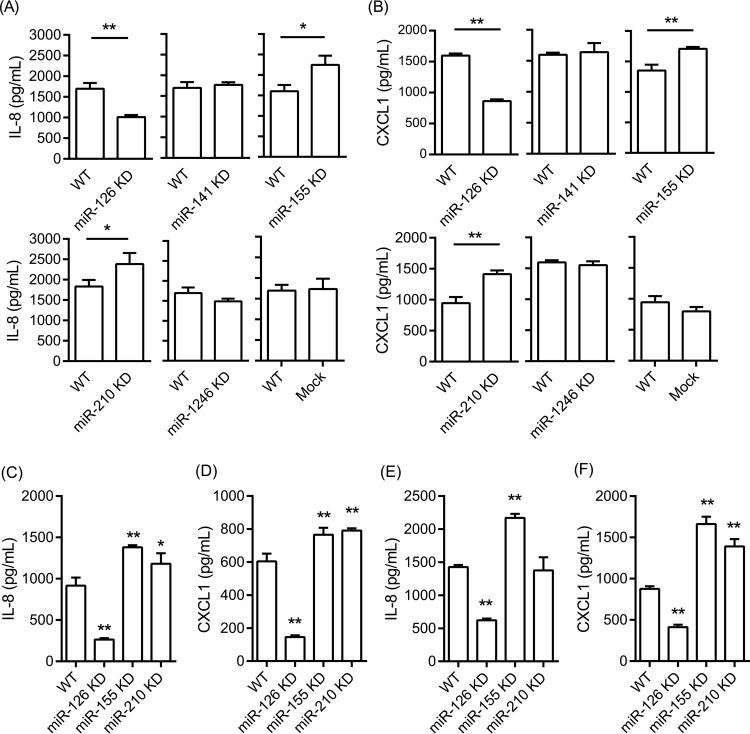
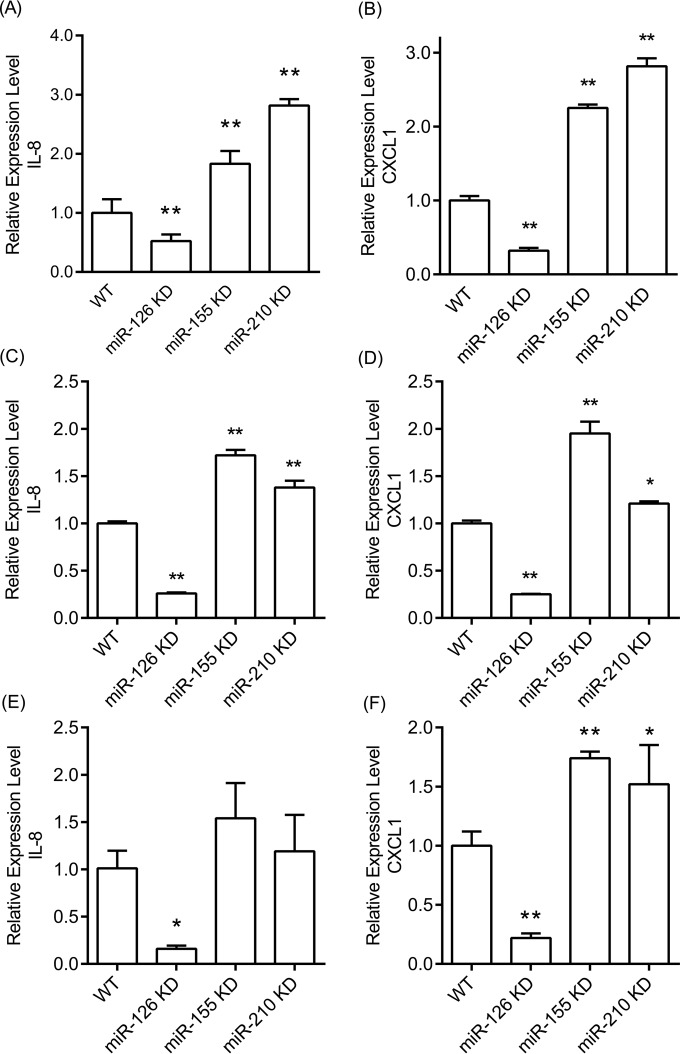
To further assess the role of miR-126 in the regulation of inflammatory responses, we stimulated wild-type and miR-126 KD TIGKs with the prominent periodontal pathogen P. gingivalis and assessed cytokine production using human cytokine arrays. Since P. gingivalis has pronounced proteolytic properties, these experiments were performed with heat-killed P. gingivalis, to avoid extensive degradation of cytokines in the culture medium. We observed that two closely related proinflammatory cytokines with potent neutrophil-attracting properties, IL-8 and CXCL1 alpha, were induced upon stimulation. In order to carry out more quantitative comparisons, we stimulated wild-type, mock-transduced, and miRNA KD cells for all 5 miRNAs with heat-killed P. gingivalis and measured IL-8 and CXCL1 by ELISA (Fig. 2A and B). We observed lower CXCL1 and IL-8 production by P. gingivalis-stimulated miR-126 KD than by WT cells. In contrast, P. gingivalis-stimulated miR-155 KD and miR-210 KD cells showed higher IL-8 and CXCL1 levels than wild-type cells, while no statistically significantly different secretion of either cytokine was noted between miR-141KD, miR-1246 KD, mock-transduced, and WT cells. A similar pattern of cytokine induction by epithelial cells was observed when we used live P. gingivalis protease-deficient mutant strain KDP128 (Fig. 2C and D). Stimulation with viable cells by a second periodontal pathogen, A. actinomycetemcomitans, which does not induce pronounced proteolysis, led to similar observations (Fig. 2E and F). We also performed qPCR experiments to examine whether the observed differences in IL-8 and CXCL1 cytokine production were reflected by corresponding differences in mRNA expression levels. The results showed concordance between protein and mRNA levels for both cytokines (Fig. 3).
FIG 2.
Expression of IL-8 (A, C, and E) and CXCL1 (B, D, and F) assessed by ELISA after stimulation with heat-killed WT P. gingivalis (A and B), live P. gingivalis mutant KDP128 (C and D), and live A. actinomycetemcomitans (E and F), at a multiplicity of infection (MOI) of 100 for 6 h. Bars represent mean values and standard deviations based on triplicate experiments. Asterisks indicate statistically significant differences between WT and miRNA KD cells (*, P < 0.05; **, P < 0.01).
FIG 3.
mRNA expression of IL-8 and CXCL1 in WT and miRNA KD TIGKs stimulated with heat-killed WT P. gingivalis (A and B), live P. gingivalis mutant KDP128 (C and D), and live A. actinomycetemcomitans (E and F) at an MOI of 100 for 6 h. Bars represent mean values and standard deviations based on triplicate experiments. Asterisks indicate statistically significant differences between WT and miRNA KD cells (*, P < 0.05; **, P < 0.01).
Regulation of the effects of miRNA knockdown by NF-κB.
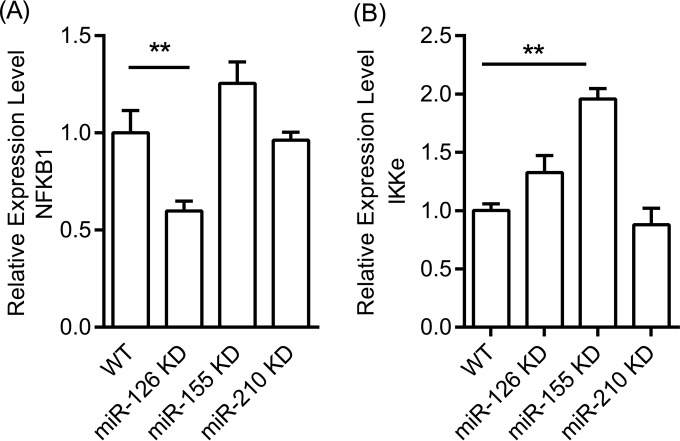
We tested the hypothesis that the observed effect on IL-8 and CXCL1 expression induced by miRNA knockdown involves interaction with components of the NF-κB family. Thus, we tested the expression of NF-κB1 and inhibitor-κB kinase ε (IKKε), as well as the phosphorylation of the p65 subunit of NF-κB in the miRNA knockdown cells.
First, we measured NF-κB1 mRNA expression in wild-type and miRNA KD cells stimulated with heat-killed P. gingivalis. As shown in Fig. 4A, the level of NF-κB1 mRNA expression was lower in miR-126 KD than in wild-type cells, indicating that miR-126 may affect upstream signaling of NF-κB and that the impaired cytokine production is due to lower NF-κB transcription. A modest (∼1.2-fold), non-statistically significant increase in NF-κB 1 mRNA was observed in miR-155 KD cells, while no effect was apparent in miR-210 KD cells.
FIG 4.
(A) Relative mRNA expression level of NFKB1 in WT TIGKs and miRNA KD cells; (B) IKKε mRNA expression level in WT and miR-155 KD cells. TIGK WT and miRNA KD cells were stimulated with heat-killed WT P. gingivalis at an MOI of 100 for 6 h. Bars represent mean values and standard deviations based on triplicate experiments. Asterisks indicate statistically significant differences between WT and miRNA KD cells (**, P < 0.01).
IKKε is a multifunctional IKB kinase protein known to regulate NF-κB transcription (28, 29) that was shown to be a potential target for miR-155 (30). Therefore, we measured IKKε mRNA expression by qPCR in wild-type and miR-155 KD cells stimulated by heat-killed P. gingivalis. These experiments (Fig. 4B) showed an approximate 2-fold increase in IKKε mRNA expression in miR-155 KD cells over that in wild-type cells, suggesting that the higher levels of IKKε in miR-155 KD cells contributed to higher levels of NF-κB and, subsequently, more pronounced cytokine production.
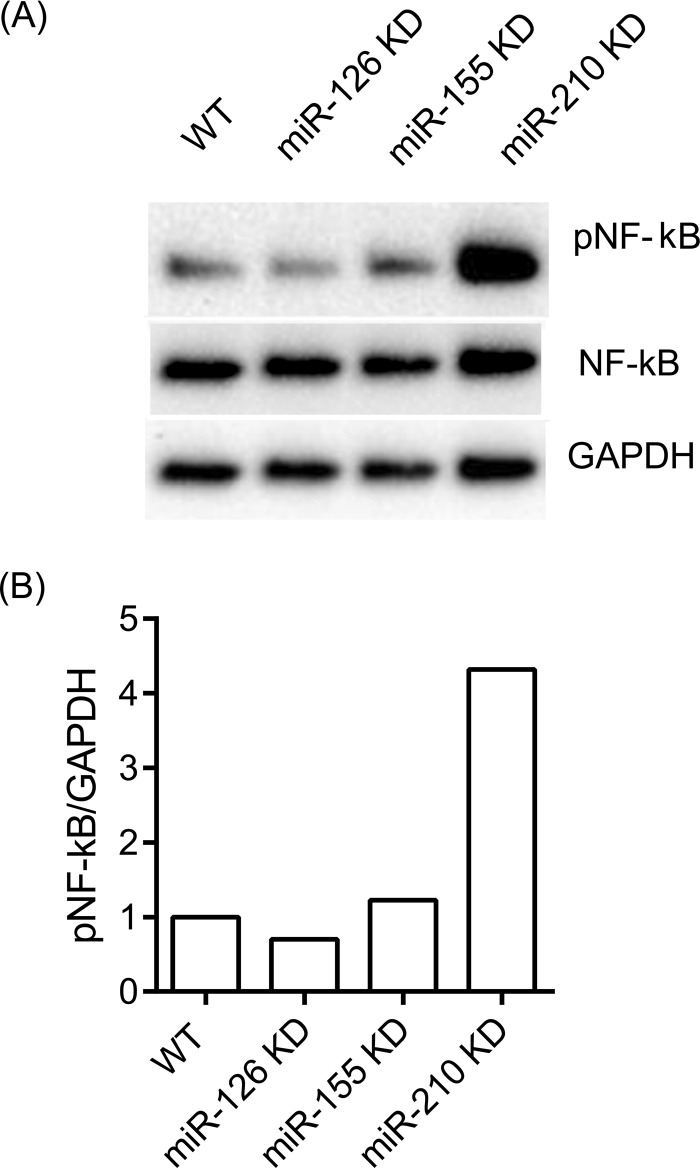
Lastly, we assessed phosphorylation of the p65 subunit of NF-κB that is directly related to its activation. Indeed, the level of phosphorylated p65 was found to be strongly increased in miR-210 KD cells after stimulation with heat-killed P. gingivalis for 30 min (Fig. 5).
FIG 5.
Levels of phosphorylation of NF-κB protein in WT and miRNA KD cells. (A) Western blot of epithelial cells stimulated with heat-killed WT P. gingivalis for 30 min; (B) ratio metric analysis (pNFKB/GAPDH) of Western blot intensity in epithelial cells.
Neutrophil-like HL-60 cell chemotactic capacity is lower in miRNA-126 KD cells.
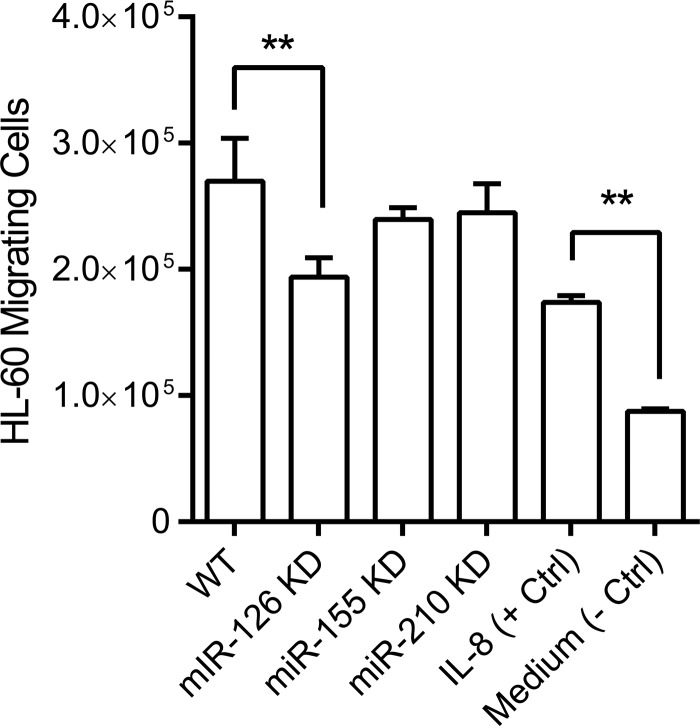
Above, we showed that the knockdown of miR-126, miR-155, and miR-210 in gingival epithelial cells resulted in a significant alteration of the production of two neutrophil-attracting cytokines, IL-8 and CXCL1, when challenged with heat-killed WT P. gingivalis, live protease-deficient P. gingivalis, or live A. actinomycetemcomitans. To assess whether these alterations would lead to functional differences in the ability of challenged epithelial cells to recruit neutrophil-like HL-60 cells, we performed chemotaxis assays to compare the abilities of the different cell lines engineered by us to attract these cells. We observed a significant reduction of the capacity of miR-126 KD cells to attract neutrophil-like HL-60 cells in comparison to controls (Fig. 6), which reflected the reduced production of the neutrophil-attracting chemokines IL-8 and CXCL1 by these cells upon infection. However, although higher levels of IL-8 and CXCL1 secretion were observed in miR-155 and miR-210 KD cells, we did not observe any significant difference in leukocyte chemotaxis by these two miRNA KD cells compared to WT cells.
FIG 6.
Neutrophil-like HL-60 cell chemotaxis toward WT and miRNA KD cells. Supernatants collected from WT and miRNA KD cells stimulated with heat-killed WT P. gingivalis were used. IL-8 (100 ng/ml) was used as a positive control, and pure cell culture medium was used as a negative control. Bars represent mean values and standard deviations based on triplicate experiments (**, P < 0.01).
DISCUSSION
miRNAs are key regulators of gene expression in mammalian cells, affecting a multitude of molecular and cellular pathways, notably those involved in the innate and adaptive immunity and the stress response. Their role in the homeostasis of the periodontal tissues or in the pathobiology of human periodontitis is largely unexplored and has only recently gained attention (19). In an earlier study by our group, we identified miRNAs that were differentially expressed in healthy and periodontitis-affected gingival tissues and examined the concomitant expression of mRNA in the same samples, making a first step toward the identification of in vivo targets of miRNAs in the setting of human periodontitis (23). In our current work, we characterized the role of specific miRNAs as regulators of inflammatory responses in gingival epithelial cells. To do so, we first studied the constitutive expression of a number of the earlier-identified differentially expressed miRNAs in human gingival keratinocytes (Fig. 1A) and documented that miR-210 and miR-141 are the most abundantly expressed, followed by miR-1246, miR-155, and miR-126, while miR-146a, miR-223, miR-451, and miR-486 were all expressed at levels that were <0.1% that of the housekeeping gene, suggesting that the observed differential expression of the latter miRNAs between healthy and diseased gingival tissues (23) was likely due to other cellular components. Of the miRNAs found to be expressed in TIGKs, miR-210 has been earlier documented in gastric epithelium (31) as well as in human skin keratinocytes (32); members of the miR-200 family (to which miR-141 belongs) are known to be involved in epithelial to mesenchymal transition during tumorigenesis (33, 34). miR-155 was found to be expressed in nasal epithelium cytology brushings in children with respiratory syncytial virus infection (35), while miR-126 was found in normal basal and luminal mammary epithelium (36). We are unaware of any literature describing epithelial expression of miR-1246.
Our next step was to proceed with establishing stable miRNA knockdown (KD) cells using miRNA sponge technology (17, 18). Consistent with the observation that highly abundant miRNAs are more likely to mediate target suppression and that miRNA/target ratio is important in miRNA-regulated gene networks (37, 38), we achieved more efficient suppression in KD cells for the less abundant miRNAs, such as miR-126 and miR-155, than for the more abundantly expressed miRNAs (Fig. 1B).
Since miR-126 KD cells were the ones in which we achieved the strongest miRNA suppression (90%), we screened for global differential gene expression between wild-type and KD cells and carried out gene set enrichment analysis, which indicated a broad dysregulation of inflammatory and immune responses in miR-126 KD cells (see Table S4 in the supplemental material). Further screening for cytokine induction in wild-type and miR-126 KD cells stimulated by the keystone periodontal species P. gingivalis showed upregulation of IL-8 and CXCL1. A quantitative assessment by ELISA comparing all 5 miRNA KD clones with wild-type TIGKs under similar bacterial stimulation showed significantly lower induction of IL-8 and CXCL1 in miR-126 KD cells and higher induction of both cytokines in miR-155 KD and miR-210 KD cells (Fig. 2). These observations were fully corroborated by qPCR findings that demonstrated that the observed differences in cytokine induction were regulated at the mRNA level (Fig. 3). Infection of miRNA KD epithelial cells with live protease-deficient mutant P. gingivalis and live A. actinomycetemcomitans showed identical patterns of cytokine expression with the exception of miR-210 KD cells, which showed higher induction of CXCL1 but not IL-8 than in control cells. Importantly, the reduced expression of chemoattractant cytokines by miR-126 KD epithelial cells appeared to be functionally relevant, as supernatants of infected KD cells showed a lower capacity to attract neutrophil-like HL-60 cells than supernatants from control cells. In addition, our results showed that the lower cytokine expression in miR-126 KD cells concurred with lower NF-κB transcription (Fig. 4A), while the higher cytokine levels in miR-155 KD cells corresponded to higher transcription levels of the inhibitor-κB kinase ε (Fig. 4B).
Our findings must be interpreted within the larger context of periodontitis pathogenesis and miRNA biology. Of the three identified miRNAs shown to affect cytokine expression in epithelial cells, miR-126 and miR-155 were found to be upregulated and miR-210 downregulated in periodontitis-affected gingival tissues (23). Thus, the decreased cytokine expression in miR-126 KD cells and the increased cytokine expression in miR-210 KD cells are in accordance with the higher mRNA expression of IL-8 (2.48-fold) and CXCL1 (3.55-fold) in periodontitis-affected versus healthy gingival tissues (39), and with the higher mRNA expression for both cytokines in gingival tissues from periodontal pockets with high versus low levels of colonization by P. gingivalis (40). In contrast, the increased cytokine expression in miR-155 KD cells, and the observed higher expression of both miR-155 (23) and IL-8 and CXCL1 (41, 42) in inflamed gingival tissues may suggest either that the effects of this particular miRNA on cytokine induction are overwhelmed by other regulatory networks acting toward an opposite direction or that the observed increased expression of both miR-155 and these particular cytokines in gingival tissues should be attributed to nonepithelial cellular sources.
All three miRNAs mentioned above have been shown to have biologic functions of potential importance for pathological conditions that involve an inflammatory component. Thus, miRNA-126 has been primarily identified as a potent suppressor of several validated pathways involved in tumorigenesis (43), but it has also been associated with inflammatory conditions such as obesity and arthritis (44). Although miRNA-126 is primarily expressed in endothelial cells and involved in angiogenesis, miR-126-deficient mice were found to have increased susceptibility to viral infection (45). There is a body of vast literature on miR-155 involvement in immunity and inflammation, with one proposed mode of action being a positive correlation between miR-155 upregulation and NF-κB activation (46). Nevertheless, this positive association may be context or cell type specific and was not corroborated by our present findings in gingival keratinocytes. Instead, our results indicate that miR-155 is an NF-κB transactivational target that is involved in a negative-feedback loop through downregulation of IKKs and possible other genes. Lastly, miR-210 is strongly linked to hypoxia pathways (47, 48) that are critically important in host defenses during the course of bacterial infections (49). Although miR-210 was reported to negatively regulate production of proinflammatory cytokines in lipopolysaccharide (LPS)-stimulated murine macrophages through NF-κB targeting (50), we did not observe any significant alteration of NF-κB1 mRNA levels in miR-210 KD cells infected with P. gingivalis for 6 h, despite the observed increased cytokine induction. However, we did observe a strong increase of phosphorylated NF-κB in miR-210 KD cells at a much earlier time point (30 min) after P. gingivalis infection (Fig. 5).
In conclusion, our data indicate that miR-126, miR-155, and miR-210 are potential regulators of inflammatory responses by gingival epithelial cells and add to the literature on the emerging role of miRNAs in the pathobiology of periodontitis and other chronic infectious and inflammatory conditions.
Supplementary Material
ACKNOWLEDGMENTS
The content of this article is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
We declare no potential conflicts of interest with respect to the authorship and/or publication of this article.
Funding Statement
This work was supported by the NIH/NIDCR (DE015649, DE021820, DE024735) and an unrestricted gift by Colgate-Palmolive, NJ, USA, to P. N. Papapanou and by a National Center for Advancing Translational Sciences Award NIH/NCATS (TR000040). M. Kebschull was supported by the German Research Foundation (DFG KFO208 TP6 & TP9), the German Society for Periodontology (DGP), and the DGZMK. Selected experiments reported in this publication were performed in the CCTI Flow Cytometry Core, supported in part by the Office of the Director, National Institutes of Health, under award S10OD020056.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.00263-16.
REFERENCES
- 1.Bartel DP. 2004. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 2.Xiao C, Rajewsky K. 2009. MicroRNA control in the immune system: basic principles. Cell 136:26–36. doi: 10.1016/j.cell.2008.12.027. [DOI] [PubMed] [Google Scholar]
- 3.Zhou R, O'Hara SP, Chen XM. 2011. MicroRNA regulation of innate immune responses in epithelial cells. Cell Mol Immunol 8:371–379. doi: 10.1038/cmi.2011.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.O'Connell RM, Rao DS, Baltimore D. 2012. microRNA regulation of inflammatory responses. Annu Rev Immunol 30:295–312. doi: 10.1146/annurev-immunol-020711-075013. [DOI] [PubMed] [Google Scholar]
- 5.Jeffrey SS. 2008. Cancer biomarker profiling with microRNAs. Nat Biotechnol 26:400–401. doi: 10.1038/nbt0408-400. [DOI] [PubMed] [Google Scholar]
- 6.Nana-Sinkam SP, Croce CM. 2013. Clinical applications for microRNAs in cancer. Clin Pharmacol Ther 93:98–104. doi: 10.1038/clpt.2012.192. [DOI] [PubMed] [Google Scholar]
- 7.Min PK, Chan SY. 22 July 2015. The biology of circulating microRNAs in cardiovascular disease. Eur J Clin Invest doi: 10.1111/eci.12475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Seeger T, Boon RA. 5 July 2015. MicroRNAs in cardiovascular ageing. J Physiol doi: 10.1113/JP270557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Andreou I, Sun X, Stone PH, Edelman ER, Feinberg MW. 2015. miRNAs in atherosclerotic plaque initiation, progression, and rupture. Trends Mol Med 21:307–318. doi: 10.1016/j.molmed.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kumar P, Dezso Z, MacKenzie C, Oestreicher J, Agoulnik S, Byrne M, Bernier F, Yanagimachi M, Aoshima K, Oda Y. 2013. Circulating miRNA biomarkers for Alzheimer's disease. PLoS One 8:e69807. doi: 10.1371/journal.pone.0069807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Femminella GD, Ferrara N, Rengo G. 2015. The emerging role of microRNAs in Alzheimer's disease. Front Physiol 6:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ebert MS, Neilson JR, Sharp PA. 2007. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat Methods 4:721–726. doi: 10.1038/nmeth1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ebert MS, Sharp PA. 2010. MicroRNA sponges: progress and possibilities. RNA 16:2043–2050. doi: 10.1261/rna.2414110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tay FC, Lim JK, Zhu H, Hin LC, Wang S. 2014. Using artificial microRNA sponges to achieve microRNA loss-of-function in cancer cells. Adv Drug Deliv Rev doi: 10.1016/j.addr.2014.05.010. [DOI] [PubMed] [Google Scholar]
- 15.Nayerossadat N, Maedeh T, Ali PA. 2012. Viral and nonviral delivery systems for gene delivery. Adv Biomed Res 1:27. doi: 10.4103/2277-9175.98152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jacobsen L, Calvin S, Lobenhofer E. 2009. Transcriptional effects of transfection: the potential for misinterpretation of gene expression data generated from transiently transfected cells. Biotechniques 47:617–624. doi: 10.2144/000113132. [DOI] [PubMed] [Google Scholar]
- 17.Gentner B, Schira G, Giustacchini A, Amendola M, Brown BD, Ponzoni M, Naldini L. 2009. Stable knockdown of microRNA in vivo by lentiviral vectors. Nat Methods 6:63–66. doi: 10.1038/nmeth.1277. [DOI] [PubMed] [Google Scholar]
- 18.Haraguchi T, Ozaki Y, Iba H. 2009. Vectors expressing efficient RNA decoys achieve the long-term suppression of specific microRNA activity in mammalian cells. Nucleic Acids Res 37:e43. doi: 10.1093/nar/gkp040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kebschull M, Papapanou PN. 2015. Mini but mighty: microRNAs in the pathobiology of periodontal disease. Periodontol 2000 69(1):201–220. doi: 10.1111/prd.12095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Benakanakere MR, Li Q, Eskan MA, Singh AV, Zhao J, Galicia JC, Stathopoulou P, Knudsen TB, Kinane DF. 2009. Modulation of TLR2 protein expression by miR-105 in human oral keratinocytes. J Biol Chem 284:23107–23115. doi: 10.1074/jbc.M109.013862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nahid MA, Rivera M, Lucas A, Chan EK, Kesavalu L. 2011. Polymicrobial infection with periodontal pathogens specifically enhances microRNA miR-146a in ApoE−/− mice during experimental periodontal disease. Infect Immun 79:1597–1605. doi: 10.1128/IAI.01062-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moffatt CE, Lamont RJ. 2011. Porphyromonas gingivalis induction of microRNA-203 expression controls suppressor of cytokine signaling 3 in gingival epithelial cells. Infect Immun 79:2632–2637. doi: 10.1128/IAI.00082-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stoecklin-Wasmer C, Guarnieri P, Celenti R, Demmer RT, Kebschull M, Papapanou PN. 2012. MicroRNAs and their target genes in gingival tissues. J Dent Res 91:934–940. doi: 10.1177/0022034512456551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lindhe J, Wennström JL, Berglundh T. 2015. The mucosa at teeth and implants, p 83–99. In Lang NP, Lindhe J (ed), Clinical periodontology and implant dentistry, 6th ed, vol 1 Wiley-Blackwell, Chichester, West Sussex, United Kingdom. [Google Scholar]
- 25.Moffatt-Jauregui CE, Robinson B, de Moya AV, Brockman RD, Roman AV, Cash MN, Culp DJ, Lamont RJ. 2013. Establishment and characterization of a telomerase immortalized human gingival epithelial cell line. J Periodontal Res 48:713–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kluiver J, Gibcus JH, Hettinga C, Adema A, Richter MK, Halsema N, Slezak-Prochazka I, Ding Y, Kroesen BJ, van den Berg A. 2012. Rapid generation of microRNA sponges for microRNA inhibition. PLoS One 7:e29275. doi: 10.1371/journal.pone.0029275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kusek ME, Pazos MA, Pirzai W, Hurley BP. 2014. In vitro coculture assay to assess pathogen induced neutrophil trans-epithelial migration. J Vis Exp 2014(83):e50823. doi: 10.3791/50823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li Q, Verma IM. 2002. NF-kappaB regulation in the immune system. Nat Rev Immunol 2:725–734. doi: 10.1038/nri910. [DOI] [PubMed] [Google Scholar]
- 29.Shen RR, Hahn WC. 2011. Emerging roles for the non-canonical IKKs in cancer. Oncogene 30:631–641. doi: 10.1038/onc.2010.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tili E, Michaille JJ, Cimino A, Costinean S, Dumitru CD, Adair B, Fabbri M, Alder H, Liu CG, Calin GA, Croce CM. 2007. Modulation of miR-155 and miR-125b levels following lipopolysaccharide/TNF-alpha stimulation and their possible roles in regulating the response to endotoxin shock. J Immunol 179:5082–5089. doi: 10.4049/jimmunol.179.8.5082. [DOI] [PubMed] [Google Scholar]
- 31.Kiga K, Mimuro H, Suzuki M, Shinozaki-Ushiku A, Kobayashi T, Sanada T, Kim M, Ogawa M, Iwasaki YW, Kayo H, Fukuda-Yuzawa Y, Yashiro M, Fukayama M, Fukao T, Sasakawa C. 2014. Epigenetic silencing of miR-210 increases the proliferation of gastric epithelium during chronic Helicobacter pylori infection. Nat Commun 5:4497. doi: 10.1038/ncomms5497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hildebrand J, Rutze M, Walz N, Gallinat S, Wenck H, Deppert W, Grundhoff A, Knott A. 2011. A comprehensive analysis of microRNA expression during human keratinocyte differentiation in vitro and in vivo. J Invest Dermatol 131:20–29. doi: 10.1038/jid.2010.268. [DOI] [PubMed] [Google Scholar]
- 33.Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, Vadas MA, Khew-Goodall Y, Goodall GJ. 2008. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol 10:593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 34.Cao Q, Lu K, Dai S, Hu Y, Fan W. 2014. Clinicopathological and prognostic implications of the miR-200 family in patients with epithelial ovarian cancer. Int J Clin Exp Pathol 7:2392–2401. [PMC free article] [PubMed] [Google Scholar]
- 35.Inchley CS, Sonerud T, Fjaerli HO, Nakstad B. 2015. Nasal mucosal microRNA expression in children with respiratory syncytial virus infection. BMC Infect Dis 15:150. doi: 10.1186/s12879-015-0878-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bockmeyer CL, Christgen M, Muller M, Fischer S, Ahrens P, Langer F, Kreipe H, Lehmann U. 2011. MicroRNA profiles of healthy basal and luminal mammary epithelial cells are distinct and reflected in different breast cancer subtypes. Breast Cancer Res Treat 130:735–745. doi: 10.1007/s10549-010-1303-3. [DOI] [PubMed] [Google Scholar]
- 37.Mullokandov G, Baccarini A, Ruzo A, Jayaprakash AD, Tung N, Israelow B, Evans MJ, Sachidanandam R, Brown BD. 2012. High-throughput assessment of microRNA activity and function using microRNA sensor and decoy libraries. Nat Methods 9:840–846. doi: 10.1038/nmeth.2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bosson AD, Zamudio JR, Sharp PA. 2014. Endogenous miRNA and target concentrations determine susceptibility to potential ceRNA competition. Mol Cell 56:347–359. doi: 10.1016/j.molcel.2014.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kebschull M, Guarnieri P, Demmer RT, Boulesteix AL, Pavlidis P, Papapanou PN. 2013. Molecular differences between chronic and aggressive periodontitis. J Dent Res 92:1081–1088. doi: 10.1177/0022034513506011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Papapanou PN, Behle JH, Kebschull M, Celenti R, Wolf DL, Handfield M, Pavlidis P, Demmer RT. 2009. Subgingival bacterial colonization profiles correlate with gingival tissue gene expression. BMC Microbiol 9:221. doi: 10.1186/1471-2180-9-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Demmer RT, Behle JH, Wolf DL, Handfield M, Kebschull M, Celenti R, Pavlidis P, Papapanou PN. 2008. Transcriptomes in healthy and diseased gingival tissues. J Periodontol 79:2112–2124. doi: 10.1902/jop.2008.080139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kebschull M, Demmer R, Behle JH, Pollreisz A, Heidemann J, Belusko PB, Celenti R, Pavlidis P, Papapanou PN. 2009. Granulocyte chemotactic protein 2 (GCP-2/CXCL6) complements interleukin-8 in periodontal disease. J Periodontal Res 44:465–471. doi: 10.1111/j.1600-0765.2008.01134.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ebrahimi F, Gopalan V, Smith RA, Lam AK. 2014. miR-126 in human cancers: clinical roles and current perspectives. Exp Mol Pathol 96:98–107. doi: 10.1016/j.yexmp.2013.12.004. [DOI] [PubMed] [Google Scholar]
- 44.Marques-Rocha JL, Samblas M, Milagro FI, Bressan J, Martinez JA, Marti A. 11 June 2015. Noncoding RNAs, cytokines, and inflammation-related diseases. FASEB J doi: 10.1096/fj.14-260323. [DOI] [PubMed] [Google Scholar]
- 45.Agudo J, Ruzo A, Tung N, Salmon H, Leboeuf M, Hashimoto D, Becker C, Garrett-Sinha LA, Baccarini A, Merad M, Brown BD. 2014. The miR-126-VEGFR2 axis controls the innate response to pathogen-associated nucleic acids. Nat Immunol 15:54–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ma X, Becker Buscaglia LE, Barker JR, Li Y. 2011. MicroRNAs in NF-kappaB signaling. J Mol Cell Biol 3:159–166. doi: 10.1093/jmcb/mjr007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kelly TJ, Souza AL, Clish CB, Puigserver P. 2011. A hypoxia-induced positive feedback loop promotes hypoxia-inducible factor 1alpha stability through miR-210 suppression of glycerol-3-phosphate dehydrogenase 1-like. Mol Cell Biol 31:2696–2706. doi: 10.1128/MCB.01242-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jiang Y, Wang HY, Cao HM, Wang CY, Zhang L, Wang H, Liu L, Li Y, Cai JH. 2014. Peripheral blood miRNAs as a biomarker for chronic cardiovascular diseases. Sci Rep 4:5026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schaffer K, Taylor CT. 2015. The impact of hypoxia on bacterial infection. FEBS J 282:2260–2266. doi: 10.1111/febs.13270. [DOI] [PubMed] [Google Scholar]
- 50.Qi J, Qiao Y, Wang P, Li S, Zhao W, Gao C. 2012. microRNA-210 negatively regulates LPS-induced production of proinflammatory cytokines by targeting NF-kappaB1 in murine macrophages. FEBS Lett 586:1201–1207. doi: 10.1016/j.febslet.2012.03.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.