Abstract
The genotoxin colibactin is a secondary metabolite produced by a variety of pathogenic enterobacteria. Its biosynthesis requires the enzymatic activity of the phosphopantetheinyl transferase (PPTase) ClbA. We previously showed that ClbA can also contribute to the production of siderophores. Because the biosynthesis of siderophores is regulated by iron availability, we hypothesized that iron could also modulate the production of colibactin through the transcriptional regulation of clbA. This study revealed an increased transcription of clbA under iron-limiting conditions and a decrease of clbA expression in iron-rich media. We demonstrate that clbA transcription is regulated by both the ferric uptake regulator (Fur) and the small regulatory noncoding RNA RyhB. We evidenced that the regulation of the transcription of clbA by Fur and RyhB leads to the regulation of colibactin production. This work highlights the complex mechanism of regulation of an important virulence factor by the two major regulators of bacterial iron homeostasis, making iron a key environmental factor contributing to bacterial virulence and carcinogenesis.
INTRODUCTION
In the human body, most of the iron is intracellular or sequestered by proteins, which makes it unavailable to invading microbes. This process, i.e., nutritional immunity, is a critical host defense strategy against bacterial pathogens that perceive the scarcity of iron within the vertebrate host as a means to sense that they are in their host (1). The global response to iron deprivation in bacteria relies on the ferric uptake regulator (Fur) protein and the small regulatory noncoding RNA (sRNA) RyhB (2–4). In the classical Fur regulation paradigm, Fur binds ferrous ions and the dimeric Fe2+-Fur complex (holo-Fur) recognizes target sequences upstream of iron-regulated genes and represses their transcription. However, nowadays numerous reports support four modes of Fur regulation, i.e., apo- and holo-Fur activation and repression, establishing a significant deviation from the classical model of Fur regulation (5). In bacteria, archaea, and eukaryotes, small riboregulators have been shown to mediate posttranscriptional mechanisms of gene regulation (6). In many cases, sRNAs form base pairs with and sequester mRNA ribosome-binding sites, resulting in translational repression and accelerated transcript decay. In contrast, a growing number of examples of translational activation and mRNA stabilization by sRNAs have now been documented. A given sRNA often employs a conserved region to interact with and regulate both repressed and activated targets (6, 7).
To counteract iron deprivation, bacterial pathogens synthesize small iron-scavenging molecules, i.e., siderophores, that are crucial for their survival and play a significant role in virulence (8–10). Siderophores are nonribosomal peptides (NRP) or polyketide (PK)-NRP hybrids (11). A prerequisite for the synthesis of all NRPs, PKs, and PK-NRP hybrids is the posttranslational attachment of P-Pant arms from coenzyme A (CoA) to a conserved serine residue of a carrier protein, converting inactive apo-synthases to active holo-synthases. This reaction is catalyzed by members of the type II family of phosphopantetheinyl transferases (PPTases) (12). The core genome of Escherichia coli codes for two type II PPTases, i.e., EntD and YieE (ECK3705, b3712, JW3690) (13). Whereas the role of YieE remains elusive, EntD is the PPTase involved in the biosynthesis of the siderophores enterobactin, salmochelin, and yersiniabactin.
An additional type II PPTase, i.e., ClbA, is encoded on the pks gene cluster, responsible for the synthesis of the PK-NRP hybrid colibactin in diverse pathogenic enterobacteria, including approximately 50% of the E. coli strains that belong to phylogenetic group B2 (14). Colibactin is a genotoxin that induces DNA double-strand breaks, senescence, and chromosomal abnormalities in eukaryotic cells (15–17) and was demonstrated to be a bona fide virulence factor in a mouse model of sepsis (18) and in a rat model of neonatal meningitis (19). We have recently highlighted the existence of cross talk between colibactin and siderophore biosynthesis (20). Indeed, ClbA can replace EntD and contribute to siderophore biosynthesis. Therefore, we speculated that this interplay could constitute a fitness advantage for E. coli and that iron could play a key role in this connection.
Here we show that the expression of clbA is controlled by iron bioavailability and that the production of colibactin is regulated by Fur and RyhB. Therefore, iron could constitute a key environmental factor contributing to the virulence and carcinogenicity of E. coli strains producing colibactin.
MATERIALS AND METHODS
Bacterial strains, mutagenesis, and growth conditions.
The bacterial strains used in this study are listed in Table 1. For genetic manipulations, E. coli strains were routinely grown at 37°C with shaking at 240 rpm in 3 ml of Lennox L broth (LB; Invitrogen). Ampicillin (50 μg/ml), kanamycin (50 μg/ml), or chloramphenicol (25 μg/ml) was added to the medium when required. Gene inactivations were performed using the bacteriophage lambda Red recombinase method (21) and the primers listed in Table 1. Briefly, the ryhB::cat and fur::kan alleles were PCR amplified from the chromosomal DNA of strain MG1655 ΔryhB or Bw25113 Δfur, respectively, using primers CMD1171_ST/CMD1172_ST or Fur_F/Fur_R, respectively. The purified PCR product was used to transform the strains of interest. The allelic exchanges were confirmed by PCR using the same pairs of primers. The resulting mutants where ryhB or fur was disrupted by a resistance cassette were named the ΔryhB and Δfur mutants, respectively. To generate the fur::cat mutation in strain Nissle 1917 clbA-lux, the chloramphenicol cassette was PCR amplified using primers Fur_P1/Fur_P2. The purified PCR product was used to transform E. coli Nissle 1917 clbA-lux. The transformants were tested by PCR amplification using primers Fur_F/Fur_R. For cloning of the wild-type ryhB and fur genes in cloning vectors, the ryhB and fur genes were PCR amplified using primer pairs RyhB_F_Compl/RyhB_R_Compl and Fur_F_Compl/Fur_R_Compl, respectively. The resulting PCR products were then cloned into the pSC-A vector using a StrataClone PCR cloning kit (Agilent Technologies). The presence of the appropriate insert in the resulting plasmids was checked by PCR amplification, and the plasmids were transformed into the mutant strains.
TABLE 1.
Strains, plasmids, and primers used in this study
| Strain, plasmid, or primer | Description or sequencea | Reference or source |
|---|---|---|
| E. coli strains | ||
| Nissle 1917 clbA-lux | Luciferase fusion of the gene clbA, Kanr | 22 |
| Nissle 1917 clbR-lux | Luciferase fusion of the gene clbR, Kanr | 22 |
| Nissle 1917 clbB-lux | Luciferase fusion of the gene clbB, Kanr | 22 |
| Nissle 1917 clbQ-lux | Luciferase fusion of the gene clbQ, Kanr | 22 |
| Nissle 1917 clbA-lux ΔryhB | ryhB mutant strain, Kanr Cmr | This study |
| Nissle 1917 clbA-lux ΔryhB+p-ryhB | ryhB mutant strain carrying p-ryhB, Kanr Cmr Ampr | This study |
| Nissle 1917 clbA-lux Δfur | fur mutant strain, Kanr Cmr | This study |
| Nissle 1917 clbA-lux Δfur+p-fur | fur mutant strain carrying p-fur, Kanr Cmr Ampr | This study |
| M1/5 | Colibactin producer, commensal strain | 20 |
| M1/5 ΔryhB | ryhB mutant strain, Cmr | This study |
| M1/5 ΔryhB+p-ryhB | ryhB mutant strain carrying p-ryhB, Cmr Ampr | This study |
| M1/5 Δfur | fur mutant strain, Kanr | This study |
| M1/5 Δfur+p-fur | fur mutant strain carrying p-fur, Kanr Ampr | This study |
| Nissle 1917 | Colibactin producer, probiotic strain | 16 |
| Nissle 1917 ΔclbA+pBAD33-clbA | This study | |
| Nissle 1917 ΔclbA+pBAD33-clbA | clbA mutant strain carrying pBAD33-clbA, Kanr | This study |
| Nissle 1917 ΔclbA+pBAD33-clbAΔclbR | clbA mutant strain carrying pBAD33-clbAΔclbR, Kanr | This study |
| Nissle 1917 ΔryhB | ryhB mutant strain, Cmr | This study |
| Nissle 1917 ΔryhB+p-ryhB | ryhB mutant strain carrying p-ryhB, Cmr Ampr | This study |
| Nissle 1917 Δfur | fur mutant strain, Kanr | This study |
| Nissle 1917 Δfur+p-fur | fur mutant strain carrying p-fur, Kanr Ampr | This study |
| SP15 | Colibactin producer, pathogenic strain | 35 |
| SP15 ΔryhB | ryhB mutant strain, Cmr | This study |
| SP15 ΔryhB+p-ryhB | ryhB mutant strain carrying p-ryhB, Cmr Ampr | This study |
| SP15 Δfur | fur mutant strain, Kanr | This study |
| SP15 Δfur+p-fur | fur mutant strain carrying p-fur, Kanr Ampr | This study |
| MG1655 ΔryhB | ryhB mutant strain, Cmr | Gift from C. M. Dozois |
| Bw25113 Δfur | fur mutant strain, Kanr | Gift from C. M. Dozois |
| Plasmids | ||
| p-ryhB | High-copy-no. pSC-A plasmid carrying the ryhB gene; Ampr Kanr | This study |
| p-fur | High-copy-no. pSC-A plasmid carrying the fur gene; Ampr Kanr | This study |
| pBAD33-T7-Venus | High-copy-no. pBAD33 plasmid containing the Venus gene coding for a variant of the yellow fluorescent protein of Aequorea victoria | This study |
| pBAD33-T7-clbA | High-copy-no. pBAD33-T7 plasmid carrying the Venus fusion of the clbA gene | This study |
| pBAD33-T7-clbAΔclbR | High-copy-no. pBAD33-T7 plasmid carrying the Venus fusion of the clbA gene and mutated clbR | This study |
| Primers | ||
| Fur-P1 | AAAATCCTGGAAGTTCTTCAGGAGCCGGACAACCATCACGTCAGTGCGGAGTGTAGGCTGGAGCTGCTTC | This study |
| Fur-P2 | TTCGCGGCAATCGCCTTCGGCACAGTGACCGTAAAGATAGAGACTGTGGTCATATGAATATCCTCCTTAG | This study |
| Fur_F | CGCCCTAAAGAAAGCTGGCC | This study |
| Fur_R | CCTTCGTGCGCATGTTCATC | This study |
| Fur_F_Compl | CTGTAAGCTGTGCCACGTTTT | This study |
| Fur_R _compl | CTGAGAGCTGTAACTCTCGCTTTTC | This study |
| CMD1171_ST (ryhB::cat) | TTTGGGGTAAATGTCCCTTTC | 29 |
| CMD1172_ST (ryhB::cat) | GTGCGCATAACGAACACAAG | 29 |
| RyhB_F_Compl | CCTCTCGAGAAAGCGGACGTGGTTCCTAC | This study |
| RyhB_R_Compl | GTACTCGAGTGTTTCTGCGTGGCGTATTAC | 29 |
| Venus-XbaI | GCGCTCTAGATGGTGTCTATCACTAAAGATCAAATC | This study |
| Venus-fus-as | GCGCGTCGACTTACTTGTACAGCTCGTCCATG | This study |
| T7-term-fw | AAGGGGATCCGGCTGCTAAC | This study |
| T7-term-rev | GCGCAATTGCGGATATAGTTCCTCCTTTCAG | This study |
| pBAD33-Venus-Bam-rev | GCGCGCGGATCCGGCTGCTAACAAAG | This study |
| pBAD33-Venus-fw | P-TGGTGTCTATCACTAAAGATCAAATC | This study |
| clbAp-BamHI-fw | GCGCGGATCCAACCATCACCTTATTATCGG | This study |
| clbAp-rev1-1 | TTAGATAATCTCATTCCTGTTAGC | This study |
| clbAp-DelclbR-fw2 | CCGTTATCTAAGACAAGTATTGCGCATG | This study |
| clbAp-DelclbR-rev2 | ACTTGTCTTAGATAACGGGTTTTTTTCTTTG | This study |
| fyuA-up | CAACGCGCAGGCCTTTAC | This study |
| fyuA-rev | GCGTGCTTTCGTCTTGCTG | This study |
| clbC-up | CGAGTCAAATGCGCCATCAC | This study |
| clbC-rev | GGACCGCCATACCAATAATG | This study |
| JPN41 | CAGATACACAGATACCATTC | 14 |
| JPN42 | TCAATGAGGAAGAAATAAAAC | 14 |
Restriction sites are underlined. A P in front of the sequence indicates a phosphate residue.
For the megalocytosis assay and assays for bioluminescence and fluorescence measurements, the E. coli strains were grown overnight in Dulbecco's modified Eagle's medium (DMEM) containing HEPES (DMEM-HEPES; Gibco), supplemented with antibiotics when required, at 37°C with shaking (240 rpm). The overnight cultures were then diluted 1:50 in DMEM-HEPES supplemented or not with iron (FeCl3, 100 μM; FeSO4, 100 μM), magnesium (MgSO4, 100 μM), copper (CuCl2, 100 μM), or iron chelator (8-hydroxyquinoline [8-HQ; 40 μM] or desferrioxamine [DFO; 0.2 μM]) and grown until the optical density at 600 nm (OD600) was 0.6. The ranges of concentrations of the chelator and the iron supplement were analyzed, and the highest concentrations of substances which did not alter bacterial growth were chosen.
Construction of Venus fusions.
A low-copy-number vector system was used to generate clbA promoter fusions with the Venus gene as a reporter. For this purpose, a derivative of vector pBAD33, which was devoid of the elements required for l-arabinose-dependent expression, was employed. An overview about the construction of the plasmids is given in Fig. S3 in the supplemental material.
In more detail, the translational fusion of the first 24 bp of rplL and the Venus gene, coding for a variant of the yellow fluorescent protein of Aequorea victoria, was amplified from pMBrplL-Venus using the primers Venus-XbaI and Venus-fus-as. The PCR product was digested with XbaI and SalI and cloned into pBAD33 (see step 1 in Fig. S3 in the supplemental material). The resulting plasmid was digested with EcoRV and SacI, treated with mung bean nuclease (MBN), and religated as plasmid pBAD33-Venus in order to delete the araBAD promoter and the araC gene (see step 2 in Fig. S3). Since the aim was to clone the whole clbBR intergenic region together with the clbR gene, which harbors the clbA promoter, the transcription into the opposite direction of clbA, facing clbB, had to be blocked. To achieve this, the T7 terminator sequence of pTXB1 (NEB) was amplified with primers T7-term-for and T7-term-rev, subsequently digested with BamHI and MfeI, and cloned upstream of Venus into pBAD33-Venus, resulting in pBAD33-T7-Venus (see step 3 in Fig. S3). The plasmid was verified by sequencing.
The whole pBAD33-T7-Venus sequence was amplified via PCR as a precursor for the following plasmids by using primers pBAD33-Venus-Bam-rev and pBAD33-Venus-fw and subsequently digested with BamHI (see step 4 in Fig. S3). To yield pBAD33-T7-clbA, the 648-bp DNA sequence upstream of clbA in E. coli strain M1/5 (20), comprising the putative clbA promoter, the clbR gene, as well as the whole clbBR intergenic region (clbBR in. reg.), was amplified with primers clbAp-BamHI-fw and clbAp-rev1-1, digested with BamHI, and then ligated with the digested pBAD33-T7-Venus amplicon (see step 5a in Fig. S3). To generate pBAD33-T7-clbAΔclbR, a derivative of pBAD33-T7-clbA with a deletion of clbR nucleotides 33 to 40, a two-step-PCR was carried out using E. coli M1/5 as a template (see step 5b in Fig. S3). Amplicons were generated with the primer pairs clbAp-BamHI-fw/clbAp-DelclbR-rev2 (PCR 1) and clbAp-DelclbR-fw2/clbAp-rev1-1 (PCR 2). PCR products 1 and 2 were fused using primers clbAp-BamHI-fw and clbAp-rev1-1 (PCR 3) and then digested with BamHI and ligated with PCR-amplified and BamHI-digested pBAD33-T7-Venus, resulting in pBAD33-T7-clbAΔclbR. The plasmids were verified by sequencing.
Luciferase and Venus measurements.
The promoter activities of genes clbA, clbB, clbQ, and clbR in different media were determined by time course quantification of luciferase or Venus expression. When an OD600 of 0.6 was reached, bacterial subcultures were diluted to an OD600 of 0.1 in the appropriate medium. Samples of 100 μl were then used to inoculate a black 96-well plate (Greiner Bio-One), and the bacteria were grown without shaking at 37°C in a luminometer (Tecan Infinite Pro microplate reader). For luminescence measurements, light emission (relative light units [RLU]) was recorded (6,000-ms aperture per sample) every 30 min in parallel with the OD600. For Venus fluorescence measurements, fluorescence was recorded at 535 nm (bandwidth, 25 nm) using an excitation wavelength of 485 nm (bandwidth, 20 nm) with the luminometer. The OD600 was measured in parallel at each time point. The area under the curve (AUC), which quantifies the cumulative luminescence or fluorescence, was calculated with GraphPad Prism (version 6.0) software.
Quantification of colibactin-associated genotoxic effect by megalocytosis assay.
Quantification of the colibactin-associated genotoxic effect by megalocytosis assay was performed as previously described (14). Briefly, HeLa cells were grown in DMEM (GlutaMAX; Invitrogen) supplemented with 10% (vol/vol) fetal calf serum (FCS; Eurobio) and 1% (vol/vol) nonessential amino acids (Invitrogen) at 37°C in a 5% CO2 atmosphere. HeLa cells were dispensed in a 96-well cell culture plate (5 × 103 cells/well) for 24 h and then infected at a multiplicity of infection (MOI; number of bacteria per HeLa cell at the onset of infection) of 100 with the E. coli strains, which had previously been pregrown in the appropriate medium. At 4 h postinoculation, the cells were washed 3 times with Hanks' balanced salt solution (HBSS; Gibco) and incubated in cell culture medium for 72 h with 200 mg/ml gentamicin before fixation (4% formaldehyde) and protein staining with methylene blue (1% [wt/vol] in 0.01 M Tris-HCl). The methylene blue was extracted with 0.1 N HCl. Staining was quantified by measurement of the OD660.
Electrophoretic mobility shift assay (EMSA).
PCR products were generated by using genomic DNA from E. coli strain M1/5 as the template and the primers JPN42/JPN41 (clbA, 343 bp), clbC-up/clbC-rev (clbC, 219 bp), and fyuA-up/fyuA-rev (fuyA, 648 bp) (Table 1). The PCR products were gel purified using a Wizard SV gel and PCR cleanup system (Promega). A 20-μl binding reaction mixture containing recombinant Fur protein (MyBioSource, USA), 50 μg ml−1 of poly(dI-dC), 4 μl of 5× binding buffer (50 mM Tris-HCl, pH 7.5, 200 mM KCl, 5 mM MgCl2, 0.625 mM MnCl2, 25% [vol/vol] glycerol, 2.5 mM dithiothreitol, 500 μg ml−1 bovine serum albumin), and the DNA probe (10 nM) was incubated at room temperature for 30 min, loaded onto a 4% Tris-borate polyacrylamide gel, and electrophoresed in 0.5× Tris-borate, pH 7.5, containing 0.2 mM MnCl2 at 100 V. The gels were stained with ethidium bromide.
Statistical analysis.
Statistical analyses were conducted using GraphPad Prism (version 6.0) software. The mean with the standard error of the mean (SEM) is shown in the figures, and P values were calculated using a one-way or two-way analysis of variance (ANOVA) followed by a Bonferroni posttest, unless otherwise stated. P values of less than 0.05 were considered statistically significant.
RESULTS
Iron availability modulates clbA transcription.
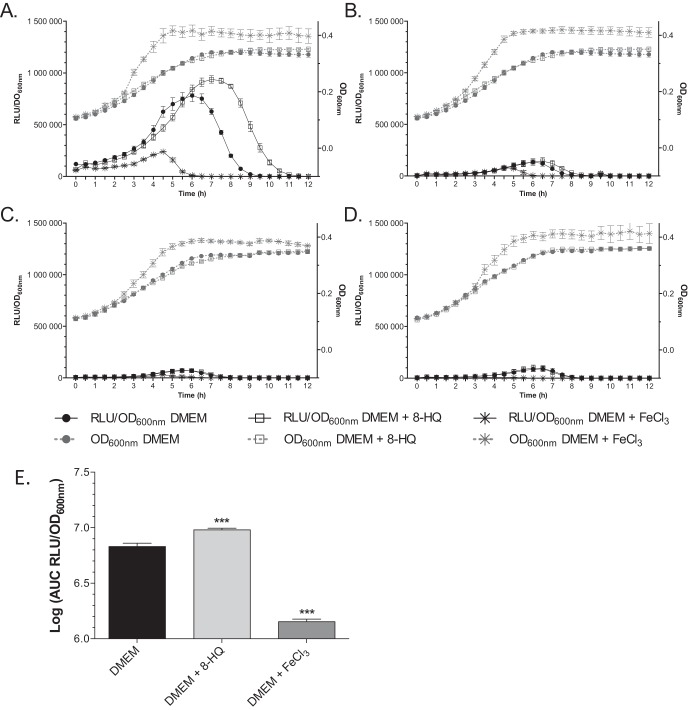
In order to test whether the promoter activities of the genes located in the pks island (see Fig. S1A in the supplemental material) were dependent on iron availability, E. coli strain Nissle 1917 harboring transcriptional luciferase fusions with the genes clbA, clbB, clbQ, and clbR, four genes involved in colibactin biosynthesis, were studied (Table 1; see also Fig. S1A and B) (22). These strains were grown in chemically defined DMEM-HEPES, DMEM-HEPES supplemented with 100 μM FeCl3, and DMEM-HEPES supplemented with an iron chelator, 8-hydroxyquinoline (8-HQ; 40 μM). The ranges of concentrations of the chelator and the iron supplement were analyzed, and the highest concentrations of substances which did not alter bacterial growth were chosen. The transcription rates of the genes clbA, clbB, clbQ, and clbR were determined as the number of OD600-standardized relative luminescence units (RLU) (Fig. 1).
FIG 1.
Iron availability modulates the transcription of clbA. Growth curves (OD600) and the RLU/OD600 of Nissle 1917 clbA-lux (A), clbB-lux (B), clbQ-lux (C), and clbR-lux (D) fusion strains grown at 37°C in DMEM-HEPES, DMEM-HEPES supplemented with 40 μM 8-HQ, or DMEM-HEPES supplemented with 100 μM FeCl3. (E) AUC, determined with GraphPad Prism (version 6.0) software, of the RLU/OD600 of clbA under the three different conditions. The given values are the mean number of RLU and SEMs from five independent experiments. Statistical analysis was performed using two-way ANOVA and the Bonferroni posttest. ***, P < 0.001 compared to the mean values obtained in DMEM-HEPES.
This analysis revealed that only clbA was significantly transcribed in DMEM-HEPES, as previously reported (22) (compare Fig. 1A to Fig. 1B to D). During the bacterial growth kinetics, a continuous increase in bioluminescence emission was observed to reach a maximal value at early stationary phase of growth, followed by a decrease of transcription (Fig. 1A).
The transcription of the genes clbB, clbQ, and clbR was not significantly modified when the culture medium was iron depleted (DMEM-HEPES supplemented with 40 μM 8-HQ) or iron replete (DMEM-HEPES supplemented with 100 μM FeCl3) (Fig. 1B, C, and D, respectively). In contrast, under iron-depleted conditions, the expression of clbA was enhanced and maintained longer than that in DMEM-HEPES (Fig. 1A). Under iron-supplemented conditions, clbA transcription was strongly repressed (Fig. 1A). Calculation of the area under the curve (AUC) of the relative number of OD600-standardized relative luminescence units (RLU/OD600) confirmed the iron-dependent differential clbA expression (Fig. 1E). Supplementation with an alternative iron chelator (desferrioxamine [DFO], 0.2 μM) or an alternative iron source (FeSO4, 100 μM) resulted in similar patterns (see Fig. S2 in the supplemental material).
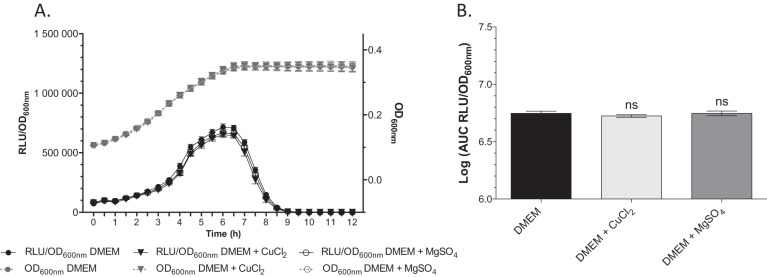
In order to test whether other divalent metals could influence clbA transcription, we monitored the expression of clbA in strain Nissle 1917 clbA-lux grown in DMEM-HEPES supplemented with copper (CuCl2, 100 μM) or magnesium (MgSO4, 100 μM) (Fig. 2). This revealed that the transcription of clbA was not altered in the presence of Mg2+ or Cu2+.
FIG 2.
The transcription of clbA is not modulated by Cu2+ or Mg2+. Growth curves (OD600) and the RLU/OD600 of E. coli Nissle 1917 clbA-lux grown at 37°C in DMEM-HEPES and DMEM-HEPES supplemented with either 100 μM MgSO4 or 100 μM CuCl2 (A) and the associated AUC (B). The given values are the mean number of RLU and SEMs from at least three independent experiments. Statistical analysis was performed by comparison of the values to the mean values obtained in DMEM-HEPES using two-way ANOVA and the Bonferroni posttest. ns, not significant.
Altogether, these results show that clbA transcription is regulated by iron availability in the medium.
The iron-dependent transcription of clbA is independent of the LuxR-like protein ClbR.
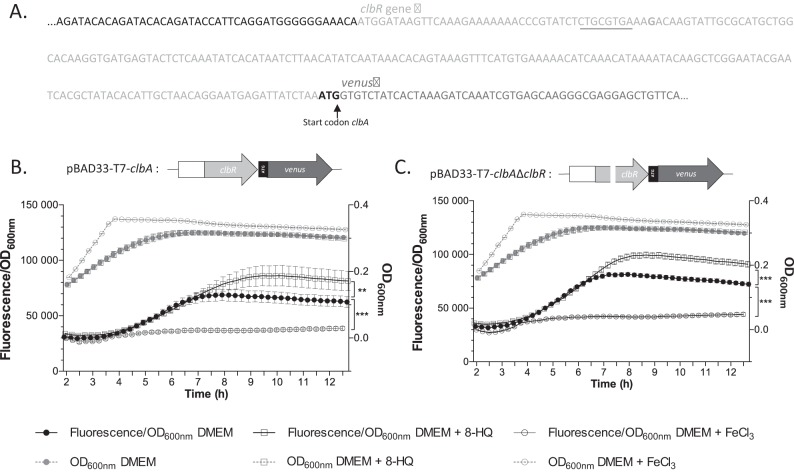
The gene clbR encodes a LuxR-like protein that exhibits a helix-turn-helix DNA-binding motif, which is suspected to be a regulator of colibactin biosynthesis (22). Moreover, cotranscription was previously demonstrated for the genes clbR and clbA in DMEM-HEPES medium with 5% fetal bovine serum (22). Our results described above showed that only clbA transcription and not clbR transcription is regulated by iron. To test whether ClbR is involved in the iron-dependent regulation of clbA, the clbA gene was transcriptionally fused with an alternative reporter gene, i.e., the Venus gene (coding for a variant of yellow fluorescent protein of Aequorea victoria) in plasmid pBAD33-T7-Venus to produce pBAD33-T7-clbA. The clbR gene was inactivated to produce plasmid pBAD33-T7-clbAΔclbR. The resulting plasmids, pBAD33-T7-clbA and pBAD33-T7-clbAΔclbR, were transformed into strain Nissle 1917 ΔclbA. Monitoring of the fluorescence of the Venus protein transcriptionally fused with the clbA gene in strain Nissle 1917 ΔclbA+pBAD33-T7-clbA confirmed the iron-dependent regulation of clbA (Fig. 3A and B). Monitoring of the fluorescence of strain Nissle 1917 ΔclbA+pBAD33-T7-clbAΔclbR under iron-supplemented and iron-chelated conditions revealed that clbA transcription was regulated by the iron concentration in the absence of functional ClbR (Fig. 3A and C). These results show that ClbR is not involved in the iron-dependent regulation of clbA.
FIG 3.
The transcription of clbA is dependent on the iron concentration in a ClbR-independent manner. (A) Sequence of the construction of the translational Venus fusion of clbA (clbA-Venus fusion). Two different constructs, one containing the wild-type clbR gene and one containing an inactive clbR gene, were generated. The underlined sequence was mutated to inactivate clbR. The wild-type and the mutated constructs were cloned into plasmid pBAD33-T7, and the plasmid was transformed into strain Nissle 1917 in which clbA was mutated. (B and C) Schematic representation of the plasmid construct obtained and results of the quantification of fluorescence of strain Nissle 1917 in which clbA was mutated and transformed with the plasmid containing the wild-type construct (B) or the mutated one (C). The fluorescence was measured in DMEM-HEPES or DMEM-HEPES supplemented with 100 μM FeCl3 or 40 μM 8-HQ. The given values are the mean fluorescence/OD600 and SEMs three independent experiments. Statistical analysis was performed by comparison of the values to the mean values obtained in DMEM-HEPES using two-way ANOVA and the Bonferroni posttest. ***, P < 0.001; **, P < 0.01.
Fur positively regulates the production of colibactin.
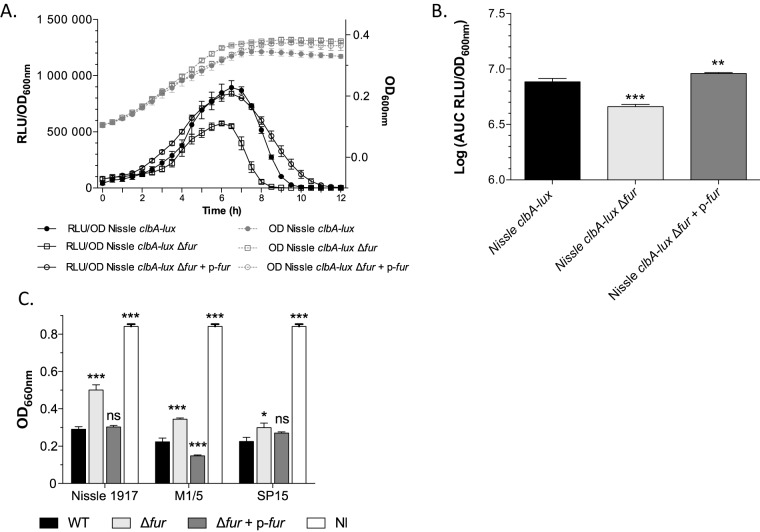
Because the global response to iron availability in bacteria relies on the ferric uptake regulator (Fur) protein, we tested whether the iron-dependent regulation of clbA transcription was mediated by Fur. We mutated the fur gene in strain Nissle 1917 clbA-lux. The complemented strain Nissle 1917 clbA-lux Δfur+p-fur, where the plasmid-borne wild-type fur gene was transformed into strain Nissle 1917 clbA-lux Δfur, was also constructed (Table 1). Monitoring of the expression of clbA in the resulting strains (Fig. 4A and B) revealed that the transcription of clbA was significantly decreased in the strain in which fur was mutated (Fig. 4A and B). Transformation of the strain in which fur was mutated with a plasmid carrying the functional wild-type fur gene (Fig. 4A and B) totally restored the expression of clbA. This indicates that the transcription of clbA is positively regulated by Fur.
FIG 4.
Fur regulates the transcription of clbA and colibactin biosynthesis. (A) Growth curves (OD600) and the RLU/OD600 of strains Nissle 1917 clbA-lux, Nissle 1917 clbA-lux Δfur, and Nissle 1917 clbA-lux Δfur+p-fur grown at 37°C in DMEM-HEPES. The given values of the number of RLU and SEMs resulted from three independent experiments. (B) AUC of the RLU/OD600 of clbA in the three different strains. (C) Quantification of colibactin production. The production of colibactin by the E. coli strains and derivatives was determined by quantification of megalocytosis as previously described (15). The multiplicity of infection was 100 bacteria per cell. Three distinct genetic contexts of E. coli were investigated: Nissle 1917 (a probiotic strain), M1/5 (a commensal strain), and SP15 (a strain isolated from a patient with neonatal meningitis). Both cells that were not infected (NI) and cells that were infected with E. coli strains that do not produce colibactin gave the same quantitative results (data not shown). The given quantification values are represented as the mean values and SEMs from three independent experiments. Statistical analysis was performed by comparison of the values to the mean values obtained for wild-type (WT) strains using two-way ANOVA and the Bonferroni posttest. ***, P < 0.001; **, P < 0.01; *, P < 0.05; ns, not significant.
Since ClbA is required for the production of colibactin, the role of Fur in the synthesis of colibactin was then investigated. Eukaryotic cells were infected with pks+ E. coli strains in which fur was mutated or not. Three different genetic contexts were analyzed: strains Nissle 1917 (a probiotic strain), M1/5 (a commensal strain), and SP15 (a pathogenic strain isolated from a patient with neonatal meningitis) (Table 1). The genotoxic effect of colibactin was monitored by quantification of megalocytosis, as previously described (14). This revealed that all the Δfur mutants displayed a genotoxicity significantly reduced compared to that of the wild-type strains (Fig. 4C). Transformation of the strains in which fur was mutated with a plasmid carrying the functional wild-type fur gene resulted in restoration of the megalocytosis phenotype (Fig. 4C). Altogether, these results evidence that Fur positively regulates colibactin production.
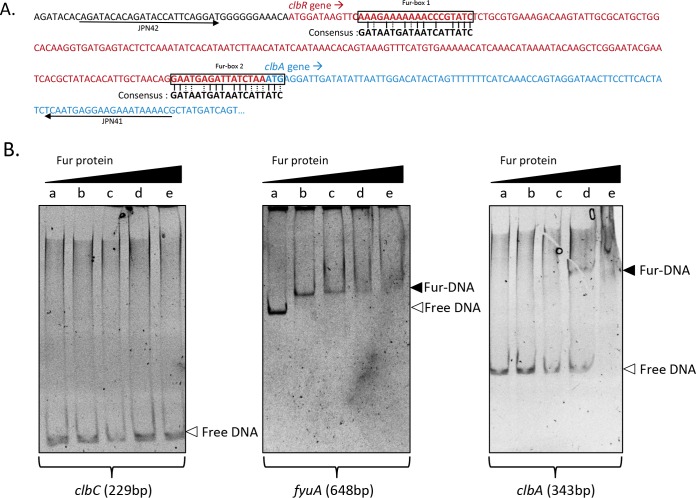
To determine whether Fur could regulate clbA directly and/or indirectly, we investigated a putative direct binding of Fur to clbA. We detected two putative Fur-binding sites in the DNA sequence of the clbA promoter (Fig. 5A). We tested whether purified Fur protein bound to its putative binding sites in the clbA promoter region (Fig. 5B). EMSA showed that migration of the 343-bp DNA fragment was retarded when it was incubated with the Fur protein. This suggests that Fur can regulate clbA expression through direct binding.
FIG 5.
Fur binds to the clbA promoter. (A) Identification and localization of the two putative Fur boxes located upstream of clbA. (B) EMSA was performed in the presence of 0 μM (lanes a), 0.25 μM (lanes b), 0.5 μM (lanes c), 1 μM (lanes d), and 4 μM (lanes e) Fur protein and 10 ng of the PCR product amplified with primers JPN41/JPN42. Three PCR products were tested: a negative control (clbC, 229 bp), a positive control (fyuA, 648 bp), and the promoter region upstream of clbA (343 bp).
RyhB negatively regulates the production of colibactin.
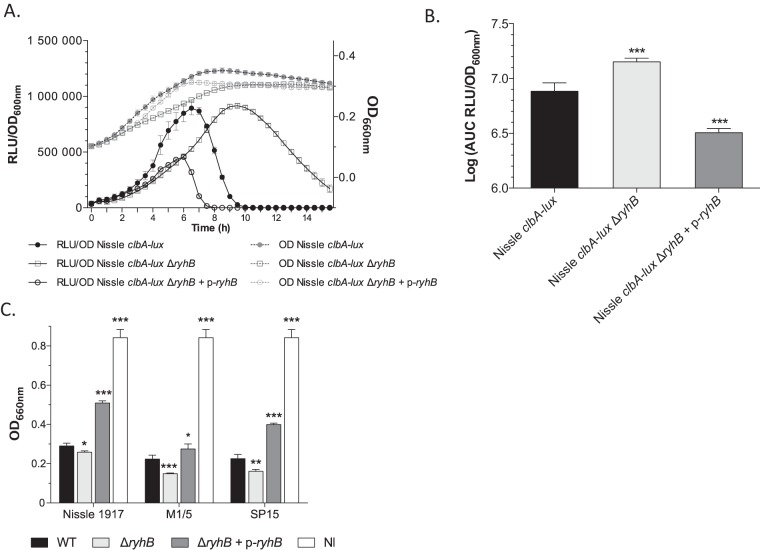
Because the second major regulator of bacterial iron metabolism is the small regulatory noncoding RNA RyhB, we investigated whether RyhB was involved in the iron-mediated regulation of clbA transcription. The gene ryhB was inactivated in strain Nissle 1917 clbA-lux, and the complemented strain was also engineered (Table 1). Monitoring of the bioluminescence in the resulting strain, Nissle 1917 clbA-lux ΔryhB, revealed that the level of transcription of clbA was enhanced and maintained longer than it was in strain Nissle 1917 clbA-lux (Fig. 6A and B). Overexpression of the wild-type ryhB gene expressed from a high-copy-number plasmid in strain Nissle 1917 clbA-lux ΔryhB resulted in the repression of clbA transcription (Nissle 1917 clbA-lux ΔryhB+p-ryhB) (Fig. 6A and B). These data evidence that RyhB plays a critical role in the transcription of clbA.
FIG 6.
RyhB regulates the transcription of clbA and colibactin biosynthesis. (A) Growth curves (OD600) and the RLU/OD600 of strains Nissle 1917 clbA-lux, Nissle 1917 clbA-lux ΔryhB, and Nissle 1917 clbA-lux ΔryhB+p-ryhB grown at 37°C in DMEM-HEPES. The given values are the mean number of RLU and SEMs from three independent experiments. (B) AUC of the RLU/OD600 of clbA in the three different strains. (C) Quantification of colibactin production. The production of colibactin by E. coli strains and its derivatives was determined by quantification of megalocytosis as previously described (14). The multiplicity of infection was 100 bacteria per cell. Three distinct genetic contexts of E. coli were investigated: Nissle 1917 (a probiotic strain), M1/5 (a commensal strain), and SP15 (a strain isolated from a patient with neonatal meningitis). Both cells that were not infected (NI) and cells that were infected with E. coli strains that do not produce colibactin gave the same quantitative results (data not shown). The given quantification values are represented as mean values and SEMs from three independent experiments. Statistical analysis was performed by comparison of the values to the mean values obtained for wild-type strains using two-way ANOVA and the Bonferroni posttest. ***, P < 0.001; **, P < 0.01; *, P < 0.05.
The role of RyhB in the synthesis of colibactin was then investigated. Eukaryotic cells were infected with pks+ E. coli strains in which ryhB was mutated or not. The genotoxic effect of colibactin, quantified by megalocytosis, revealed that all the ΔryhB mutants displayed significantly increased genotoxicity compared to the wild-type strains (Fig. 6C). Overexpression of ryhB in strains in which the ryhB gene was mutated resulted in a decrease in the level of the colibactin-associated genotoxic effect (Fig. 6C). Altogether, these results evidence that RyhB negatively regulates colibactin production under these conditions.
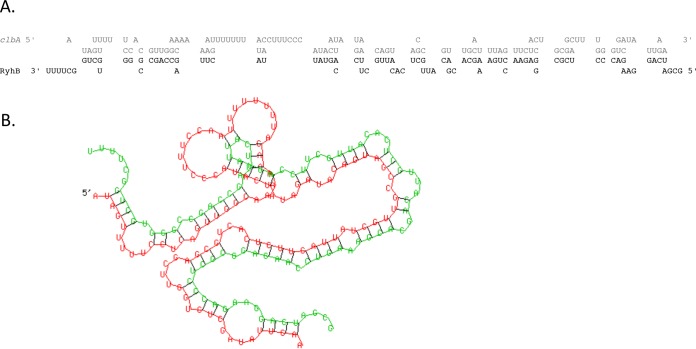
To determine whether RyhB could regulate clbA directly and/or indirectly, we investigated a putative direct pairing of RyhB to the clbA promoter. The RNA Hybrid computational program (23) predicted a region of potential pairing between RyhB and clbA (Fig. 7). The region of pairing with clbA was located 238 to 344 nucleotides downstream from the translational start (minimal free energy, −51.3 kcal/mol) (Fig. 7) in the reading frame of clbA. This suggests that ryhB could regulate clbA through direct binding.
FIG 7.
RyhB seems to interact directly with the clbA gene. The RNA Hybrid computational program predicted a region of potential interaction between RyhB and clbA. The nucleotide sequence involved in the interaction between clbA (gray) and RyhB (black) (A) and the two-dimensional structure of RyhB (green) bound to clbA (red) (B) are shown. The predicted pairing region with RyhB began at nucleotide 238 in the clbA open reading frame (minimal free energy, −51.3 kcal/mol).
DISCUSSION
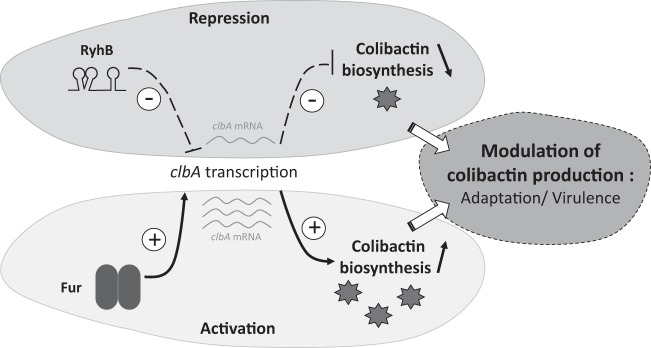
This work highlights that the transcription of clbA, the gene encoding the PPTase involved in colibactin production, is regulated by iron availability (see our model in Fig. 8). Therefore, this work provides new insights into the transcriptional regulation of clbA, as only one study on its regulation has been published previously (22). In addition to carbon source and growth phase (22), our work demonstrates that clbA is regulated by iron availability. Moreover, this regulation appears to be independent of ClbR, the LuxR-like protein predicted to regulate colibactin biosynthesis (22), although the clbR gene was demonstrated to be coexpressed with clbA under particular culture conditions (22). This suggests that clbA could be expressed from a second promoter specifically activated under specific conditions of iron availability. This iron-dependent regulation of clbA transcription could constitute a fitness advantage for the bacteria, as we previously demonstrated that ClbA can also sustain siderophore biosynthesis (20).
FIG 8.
Model for the regulation of the transcription of clbA by Fur and RyhB. The biosynthesis of the genotoxin colibactin requires the enzymatic activity of the PPTase ClbA. The transcription of clbA is dependent on both the ferric uptake regulator (Fur) and the small regulatory noncoding RNA RyhB. This makes iron a key environmental factor contributing to the regulation of colibactin production.
This work highlights that Fur is positively involved in the production of the virulence factor colibactin through the regulation of clbA. Fur is a global transcriptional regulator that controls the transcription of over 90 genes involved in iron uptake, storage, and metabolism. Only a few studies have reported the involvement of Fur in the regulation of the biosynthesis of toxins in E. coli. The stx genes, encoding Shiga toxin in enterohemorrhagic E. coli (24, 25), and hly plasmids, encoding hemolysin (26), are directly regulated by Fur. In addition to its well-known role as a repressor, Fur was characterized as a positive regulator of gene expression, such as the expression of acnA, fumA, ftnA, bfr, and sodB (27).
Our work also evidenced that RyhB regulates clbA transcription, as we observed that clbA transcription was upregulated when the ryhB gene was inactivated, demonstrating that clbA is downregulated by RyhB. RyhB is a small RNA that recruits RNase E and facilitates the degradation of mRNA targets (27, 28). Whereas only a few studies have demonstrated the impact of RyhB on virulence in pathogenic E. coli, for instance, through siderophore production (29), several studies have demonstrated that this sRNA is implicated in the virulence-associated processes of other pathogenic bacteria, such as Shigella flexneri (2) or Shigella dysenteriae (30). This study provides new evidence that RyhB is involved in the production of a virulence factor in pathogenic E. coli through the regulation of clbA transcription, which leads to the modulation of colibactin production.
Here we demonstrated that modulation of the expression of clbA via regulators of iron homeostasis leads to the modulation of colibactin production. The mutation of ryhB in a colibactin-producing E. coli strain leads to an increase in the level of colibactin production, whereas the mutation of fur results in a decrease in the level of colibactin production in three different genetic contexts of E. coli, in which the strains belonged to the B2 phylogenetic group. We confirmed that this regulation occurred in three different strains; however, the quantitative differences observed could be explained by the genetic diversity between strains. Colibactin was demonstrated to be a virulence factor, for example, in neonatal systemic infections (19), but it is also associated with the development of colorectal cancer (31–33). The fact that colibactin is regulated by the two major regulators of iron homeostasis could link iron availability in the gut and blood and E. coli-mediated carcinogenesis and systemic infections, respectively. Moreover, the main reservoir of E. coli is the intestine and, more precisely, the colon. A recent publication confirmed the emergence of the B2 group of E. coli in developed countries (34). Given the existence of a gradient of iron concentration from the lumen to the intestinal epithelial cell, it is conceivable that the fine-tuning of clbA expression allows the production of colibactin when the pathogenic E. coli isolate is located in an appropriate site in the gut. This iron-dependent regulation could be one reason for the emergence of the B2 group E. coli strains, as we know that there is cross talk between colibactin and siderophores. Integration of the regulation of virulence factors, such as siderophores and colibactin, into networks that respond to specific environmental signals, such as the local iron concentration and the balance between Fur and RyhB, could result in the accurate production of colibactin and siderophores, so that the bacteria can adapt to the competitive environment in the gut.
Further understanding of how E. coli senses environments to coordinate the expression of clbA and the mechanism of regulation of the biosynthesis of these virulence factors may uncover novel pathways for the development of potential targets against this pathogenic enterobacterium.
Supplementary Material
ACKNOWLEDGMENTS
We have no conflict of interest to declare.
We sincerely thank Charles M. Dozois for providing us with strain MG1655 ΔryhB.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.00659-16.
REFERENCES
- 1.Hood MI, Skaar EP. 2012. Nutritional immunity: transition metals at the pathogen-host interface. Nat Rev Microbiol 10:525–537. doi: 10.1038/nrmicro2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Oglesby AG, Murphy ER, Iyer VR, Payne SM. 2005. Fur regulates acid resistance in Shigella flexneri via RyhB and ydeP. Mol Microbiol 58:1354–1367. doi: 10.1111/j.1365-2958.2005.04920.x. [DOI] [PubMed] [Google Scholar]
- 3.Troxell B, Hassan HM. 2013. Transcriptional regulation by ferric uptake regulator (Fur) in pathogenic bacteria. Front Cell Infect Microbiol 3:59. doi: 10.3389/fcimb.2013.00059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Porcheron G, Dozois CM. 2015. Interplay between iron homeostasis and virulence: Fur and RyhB as major regulators of bacterial pathogenicity. Vet Microbiol 179:2–14. doi: 10.1016/j.vetmic.2015.03.024. [DOI] [PubMed] [Google Scholar]
- 5.Butcher J, Sarvan S, Brunzelle JS, Couture J-F, Stintzi A. 2012. Structure and regulon of Campylobacter jejuni ferric uptake regulator Fur define apo-Fur regulation. Proc Natl Acad Sci U S A 109:10047–10052. doi: 10.1073/pnas.1118321109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Papenfort K, Vanderpool CK. 2015. Target activation by regulatory RNAs in bacteria. FEMS Microbiol Rev 39:362–378. doi: 10.1093/femsre/fuv016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bobrovskyy M, Vanderpool CK, Richards GR. 2015. Small RNAs regulate primary and secondary metabolism in Gram-negative bacteria. Microbiol Spectr 3:MBP-0009-2014. doi: 10.1128/microbiolspec.MBP-0009-2014. [DOI] [PubMed] [Google Scholar]
- 8.Miethke M, Marahiel MA. 2007. Siderophore-based iron acquisition and pathogen control. Microbiol Mol Biol Rev 71:413–451. doi: 10.1128/MMBR.00012-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garénaux A, Caza M, Dozois CM. 2011. The ins and outs of siderophore mediated iron uptake by extra-intestinal pathogenic Escherichia coli. Vet Microbiol 153:89–98. doi: 10.1016/j.vetmic.2011.05.023. [DOI] [PubMed] [Google Scholar]
- 10.Brumbaugh AR, Smith SN, Subashchandrabose S, Himpsl SD, Hazen TH, Rasko DA, Mobley HLT. 2015. Blocking yersiniabactin import attenuates extraintestinal pathogenic Escherichia coli in cystitis and pyelonephritis and represents a novel target to prevent urinary tract infection. Infect Immun 83:1443–1450. doi: 10.1128/IAI.02904-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Walsh CT. 2008. The chemical versatility of natural-product assembly lines. Acc Chem Res 41:4–10. doi: 10.1021/ar7000414. [DOI] [PubMed] [Google Scholar]
- 12.Beld J, Sonnenschein EC, Vickery CR, Noel JP, Burkart MD. 2014. The phosphopantetheinyl transferases: catalysis of a post-translational modification crucial for life. Nat Prod Rep 31:61–108. doi: 10.1039/C3NP70054B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blattner FR, Plunkett G, Bloch CA, Perna NT, Burland V, Riley M, Collado-Vides J, Glasner JD, Rode CK, Mayhew GF, Gregor J, Davis NW, Kirkpatrick HA, Goeden MA, Rose DJ, Mau B, Shao Y. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453–1462. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 14.Putze J, Hennequin C, Nougayrède JP, Zhang W, Homburg S, Karch H, Bringer MA, Fayolle C, Carniel E, Rabsch W, Oelschlaeger TA, Oswald E, Forestier C, Hacker J, Dobrindt U. 2009. Genetic structure and distribution of the colibactin genomic island among members of the family Enterobacteriaceae. Infect Immun 77:4696–4703. doi: 10.1128/IAI.00522-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nougayrède J-P, Homburg S, Taieb F, Boury M, Brzuszkiewicz E, Gottschalk G, Buchrieser C, Hacker J, Dobrindt U, Oswald E. 2006. Escherichia coli induces DNA double-strand breaks in eukaryotic cells. Science 313:848–851. doi: 10.1126/science.1127059. [DOI] [PubMed] [Google Scholar]
- 16.Cuevas-Ramos G, Petit CR, Marcq I, Boury M, Oswald E, Nougayrède J-P. 2010. Escherichia coli induces DNA damage in vivo and triggers genomic instability in mammalian cells. Proc Natl Acad Sci U S A 107:11537–11542. doi: 10.1073/pnas.1001261107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Payros D, Secher T, Boury M, Brehin C, Ménard S, Salvador-Cartier C, Cuevas-Ramos G, Watrin C, Marcq I, Nougayrède J-P, Dubois D, Bedu A, Garnier F, Clermont O, Denamur E, Plaisancié P, Theodorou V, Fioramonti J, Olier M, Oswald E. 2014. Maternally acquired genotoxic Escherichia coli alters offspring's intestinal homeostasis. Gut Microbes 5:313–325. doi: 10.4161/gmic.28932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marcq I, Martin P, Payros D, Cuevas-Ramos G, Boury M, Watrin C, Nougayrède J-P, Olier M, Oswald E. 2014. The genotoxin colibactin exacerbates lymphopenia and decreases survival rate in mice infected with septicemic Escherichia coli. J Infect Dis 210:285–294. doi: 10.1093/infdis/jiu071. [DOI] [PubMed] [Google Scholar]
- 19.McCarthy AJ, Martin P, Cloup E, Stabler RA, Oswald E, Taylor PW. 2015. The genotoxin colibactin is a determinant of virulence in Escherichia coli K1 experimental neonatal systemic infection. Infect Immun 83:3704–3711. doi: 10.1128/IAI.00716-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martin P, Marcq I, Magistro G, Penary M, Garcie C, Payros D, Boury M, Olier M, Nougayrède J-P, Audebert M, Chalut C, Schubert S, Oswald E. 2013. Interplay between siderophores and colibactin genotoxin biosynthetic pathways in Escherichia coli. PLoS Pathog 9:e1003437. doi: 10.1371/journal.ppat.1003437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Homburg S, Oswald E, Hacker J, Dobrindt U. 2007. Expression analysis of the colibactin gene cluster coding for a novel polyketide in Escherichia coli. FEMS Microbiol Lett 275:255–262. doi: 10.1111/j.1574-6968.2007.00889.x. [DOI] [PubMed] [Google Scholar]
- 23.Krüger J, Rehmsmeier M. 2006. RNAhybrid: microRNA target prediction easy, fast and flexible. Nucleic Acids Res 34:W451–W454. doi: 10.1093/nar/gkl243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Payne SM, Wyckoff EE, Murphy ER, Oglesby AG, Boulette ML, Davies NML. 2006. Iron and pathogenesis of Shigella: iron acquisition in the intracellular environment. Biometals 19:173–180. doi: 10.1007/s10534-005-4577-x. [DOI] [PubMed] [Google Scholar]
- 25.Tobe T, Yen H, Takahashi H, Kagayama Y, Ogasawara N, Oshima T. 2014. Antisense transcription regulates the expression of the enterohemorrhagic Escherichia coli virulence regulatory gene ler in response to the intracellular iron concentration. PLoS One 9:e101582. doi: 10.1371/journal.pone.0101582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grünig HM, Rutschi D, Schoch C, Lebek G. 1987. The chromosomal fur gene regulates the extracellular haemolytic activity encoded by certain hly plasmids. Zentralbl Bakteriol Mikrobiol Hyg A 266:231–238. [DOI] [PubMed] [Google Scholar]
- 27.Massé E, Vanderpool CK, Gottesman S. 2005. Effect of RyhB small RNA on global iron use in Escherichia coli. J Bacteriol 187:6962–6971. doi: 10.1128/JB.187.20.6962-6971.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sobrero P, Valverde C. 2012. The bacterial protein Hfq: much more than a mere RNA-binding factor. Crit Rev Microbiol 38:276–299. doi: 10.3109/1040841X.2012.664540. [DOI] [PubMed] [Google Scholar]
- 29.Porcheron G, Habib R, Houle S, Caza M, Lépine F, Daigle F, Massé E, Dozois CM. 2014. The small RNA RyhB contributes to siderophore production and virulence of uropathogenic Escherichia coli. Infect Immun 82:5056–5068. doi: 10.1128/IAI.02287-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Murphy ER, Payne SM. 2007. RyhB, an iron-responsive small RNA molecule, regulates Shigella dysenteriae virulence. Infect Immun 75:3470–3477. doi: 10.1128/IAI.00112-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Arthur JC, Perez-Chanona E, Mühlbauer M, Tomkovich S, Uronis JM, Fan T-J, Campbell BJ, Abujamel T, Dogan B, Rogers AB, Rhodes JM, Stintzi A, Simpson KW, Hansen JJ, Keku TO, Fodor AA, Jobin C. 2012. Intestinal inflammation targets cancer-inducing activity of the microbiota. Science 338:120–123. doi: 10.1126/science.1224820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dalmasso G, Cougnoux A, Delmas J, Darfeuille-Michaud A, Bonnet R. 2014. The bacterial genotoxin colibactin promotes colon tumor growth by modifying the tumor microenvironment. Gut Microbes 5:675–680. doi: 10.4161/19490976.2014.969989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cougnoux A, Dalmasso G, Martinez R, Buc E, Delmas J, Gibold L, Sauvanet P, Darcha C, Déchelotte P, Bonnet M, Pezet D, Wodrich H, Darfeuille-Michaud A, Bonnet R. 2014. Bacterial genotoxin colibactin promotes colon tumour growth by inducing a senescence-associated secretory phenotype. Gut 63:1932–1942. doi: 10.1136/gutjnl-2013-305257. [DOI] [PubMed] [Google Scholar]
- 34.Massot M, Daubié A-S, Clermont O, Jauréguy F, Couffignal C, Dahbi G, Mora A, Blanco J, Branger C, Mentré F, Eddi A, Picard B, Denamur E, the COLIVILLE Group . 2016. Phylogenetic, virulence and antibiotic resistance characteristics of commensal strain populations of Escherichia coli from community subjects in the Paris area in 2010 and evolution over 30 years. Microbiology 162:642–650. doi: 10.1099/mic.0.000242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Johnson JR, Oswald E, O'Bryan TT, Kuskowski MA, Spanjaard L. 2002. Phylogenetic distribution of virulence-associated genes among Escherichia coli isolates associated with neonatal bacterial meningitis in The Netherlands. J Infect Dis 185:774–784. doi: 10.1086/339343. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.