Abstract
Bacillus anthracis is a sporulating Gram-positive bacterium that is the causative agent of anthrax and a potential weapon of bioterrorism. The U.S.-licensed anthrax vaccine is made from an incompletely characterized culture supernatant of a nonencapsulated, toxigenic strain (anthrax vaccine absorbed [AVA]) whose primary protective component is thought to be protective antigen (PA). AVA is effective in protecting animals and elicits toxin-neutralizing antibodies in humans, but enthusiasm is dampened by its undefined composition, multishot regimen, recommended boosters, and potential for adverse reactions. Improving next-generation anthrax vaccines is important to safeguard citizens and the military. Here, we report that vaccination with recombinant forms of a conserved domain (near-iron transporter [NEAT]), common in Gram-positive pathogens, elicits protection in a murine model of B. anthracis infection. Protection was observed with both Freund's and alum adjuvants, given subcutaneously and intramuscularly, respectively, with a mixed composite of NEATs. Protection correlated with an antibody response against the NEAT domains and a decrease in the numbers of bacteria in major organs. Anti-NEAT antibodies promote opsonophagocytosis of bacilli by alveolar macrophages. To guide the development of inactive and safe NEAT antigens, we also report the crystal structure of one of the NEAT domains (Hal) and identify critical residues mediating its heme-binding and acquisition activity. These results indicate that we should consider NEAT proteins in the development of an improved antianthrax vaccine.
INTRODUCTION
Bacillus anthracis is a Gram-positive, encapsulated, and sporulating bacterium notable as the causative agent of anthrax. The disease is most commonly reported in wild and domestic herbivores, but because of the ability of the spores to persist in the environment and be aerosolized, B. anthracis has been regarded as one of the most serious bioterrorism agents (1, 2). The spore, which exists in a metabolically inactive form, will germinate into a highly virulent vegetative cell upon entry into a host, where it encounters a niche rich in nutrients (3). It is believed that spores are engulfed by macrophages and are transported to draining lymph nodes where they germinate into vegetative cells, which then replicate and express a series of virulence factors, including anthrax toxin and a polyglutamic acid capsule (4–12). Disease is categorized according to the route of spore exposure. These routes include spore entry through the epidermis (cutaneous anthrax), spore entry through the alveolar epithelial surface (inhalational anthrax), spore entry through the gastrointestinal epithelium (gastrointestinal anthrax), or the most recently identified form contracted through injecting drugs contaminated with spores (injectional anthrax) (13, 14). Upon anthrax infection, vegetative bacilli proliferate in the initial site of inoculation and then spread to the lymphatic tissues and disseminate to other organs and, ultimately, the bloodstream (15, 16). The precise infectious dose of B. anthracis in humans by various routes is unknown, but it is believed that inhalational anthrax can develop in susceptible hosts after exposure to a relatively small number of spores (17).
B. anthracis vaccine development efforts have included the use of attenuated strains (18), heterologous expression hosts (19, 20), capsule conjugates (21, 22), inactivated spores (23, 24), and lipid-encapsulated DNA (25). The first vaccines against anthrax were developed in the 1880s by William S. Greenfield and Louis Pasteur using live attenuated cultures of B. anthracis (26, 27). Although the vaccines were effective in livestock, the virulence of the vaccines varied, leading in 1939 to Max Sterne developing a live but attenuated vaccine from a nonencapsulated strain of B. anthracis that is the standard vaccine for livestock in the United States (18). Live attenuated vaccines have been linked with residual virulence leading to occasional animal casualties; thus, the vaccine was not regarded as safe for human use (28–30). Acellular vaccines against B. anthracis were sought. Growth of B. anthracis in chemically defined media (31, 32) and the identification of anthrax toxin and its components (33–37) led to the generation of the current licensed human anthrax vaccine, known as anthrax vaccine absorbed (AVA). AVA is a cell-free filtrate of cultures of an avirulent, nonencapsulated strain of B. anthracis. Evidence suggests that the principal component that elicits protection is protective antigen (PA), which is the cell-binding component of anthrax toxin. The mechanism of action of AVA seems to be due to anti-PA toxin neutralizing antibodies that provide protection against anthrax disease (38, 39). Historically, six immunizations over 18 months have been recommended as a part of the preexposure AVA vaccination regimen, but evidence suggests that shorter inoculation intervals can be effective at eliciting sufficient levels of protective antibodies (40–42). Injection site and systemic adverse reactions (43), the undefined nature of AVA, the potential for batch variation (44), the extended vaccination regimen with recommended boosters (45), and the instability of recombinant PA preparations (46) have spawned efforts to determine if other antigens or measures can be used to produce a next-generation vaccine (46).
In this report, we describe progress toward the development of recombinant heme transporters as potential vaccine antigens. Five such transporters have been identified in B. anthracis, each possessing at least one conserved heme-binding motif referred to as the near-iron transporter (NEAT) domain. Secreted and surface-localized NEAT-containing proteins mediate the acquisition and import of heme from host heme sources such as hemoglobin (47–51). B. anthracis encodes five proteins that contain one or more NEAT domains: IsdC, IsdX1, IsdX2, BslK, and Hal (49, 52–55). IsdX1 and IsdX2 are secreted into the culture medium and actively acquire heme from hemoglobin (49, 56). IsdC is covalently anchored to the cell wall by a sortase-mediated mechanism and can receive heme from both IsdX1 and IsdX2 (54, 57). BslK is an S-layer protein that binds heme and can also transfer heme to IsdC (52). Finally, Hal is necessary for growth on hemoglobin and heme and is also thought to be attached to the bacillus cell wall (53). Two of these NEATs, IsdX2 and IsdC, were identified as being highly antigenic by a functional genomic-serologic screen, and Hal is immunoreactive with B. anthracis antisera (58). Guinea pigs infected with the fully virulent Ames strain of B. anthracis also produce antibodies to IsdX1 and IsdX2 (49). The NEAT-containing protein iron surface determinant B (IsdB) from Staphylococcus aureus was the basis of Merck's V710 vaccine. IsdB is a cell wall-anchored protein, conserved among diverse clinical isolates of S. aureus, that harbors two NEAT motifs and functions to assist with heme iron acquisition (59). It induced rapid antibody responses in macaques and increased survival after challenge in a murine S. aureus sepsis model (60). V710 phase I trials testing the formulation of the vaccine proved successful (61, 62); however, phase III trials evaluating the administration of the vaccine preoperatively to cardiothoracic surgery patients were halted, citing a higher frequency of deaths in the vaccine recipients than in the placebo recipients (63). Considering the general importance of iron uptake to the growth and replication of bacterial pathogens and attempts to construct a vaccine from such factors to prevent staphylococcal infection, we sought to determine if recombinant NEAT proteins can protect against anthrax disease.
MATERIALS AND METHODS
Bacterial strains and cloning.
The construction of pgst-halN, pgst-isdC, pgst-isdX1, and pgst-isdX2, and pgst-bslKN was described previously and is summarized in Table 1. All strains were propagated in lysogeny broth (LB) with 100 μg/ml ampicillin (Caisson Labs, Logan, UT). Single- and double-alanine substitutions of the two aromatic residues in the heme-binding lip region of hal (Y112A, F116A, and Y112A,F116A) were generated by using QuikChange site-directed mutagenesis (Agilent, Santa Clara, CA) with the following primer pairs encoding the desired mutation (in boldface type): forward primer 5′-CCC ACT TTA GGG GCC GAT AAG GAA TTC-3′ and reverse primer 5′-GAA TTC CTT ATC GGC CCC TAA AGT GGG-3′ for HalNY112A, forward primer 5′-GGG TAC GAT AAG GAA GCC AAA ATT CAG-3′ and reverse primer 5′-CTG AAT TTT GGC TTC CTT ATC GTA CCC-3′ for HalNF116A, and forward primer 5′-CCC ACT TTA GGG GCC GAT AAG GAA GCC AAA ATT CAG-3′ and reverse primer 5′-CTG AAT TTT GGC TTC CTT ATC GGC CCC TAA AGT GGG-3′ for the HalNY112A,F116A double mutant. The HalNY112A and HalNF116A single mutants were generated according to the manufacturer's specifications using plasmid pgst-halN as the parent DNA containing the wild-type gene. This DNA template was then subjected to PCR amplification using the primer pairs mentioned above. PCR products were digested with 1 μl of DpnI (New England BioLabs, Ipswich, MA) at 10 U/μl for 2 h at 37°C. Plasmid DNA was then purified by using a QIAquick PCR purification kit (Qiagen, the Netherlands) and transformed into Escherichia coli DH5α. Mutations were verified by DNA sequencing. Single-mutation plasmid DNA was further manipulated with the HalNY112A,F116A double mutant primer set, subjected to PCR, DpnI digested, and transformed into E. coli DH5α. Once the double-alanine mutation was verified by sequencing, all of the resulting plasmids, pgst-halNY112A, pgst-halNF116A, and pgst-halNY112A,F116A, were isolated and transformed into E. coli BL21 (New England BioLabs, Ipswich, MA). All NEAT proteins were expressed and purified as described previously (49). The concentration of each protein was measured by using the Bradford assay (Bio-Rad, Hercules, CA) with bovine serum albumin (BSA; Fisher Scientific, Fair Lawn, NJ), used to generate a standard curve, and the homogeneity of the preparations was determined by using SDS-polyacrylamide gel electrophoresis (SDS-PAGE). For NEAT preparations evaluated as recombinant vaccines, all samples were applied to Pierce HighCapacity Endotoxin Removal Resin columns (Thermo Scientific, Rockford, IL) according to the manufacturer's instructions, and proteins were eluted with endotoxin-free buffer and stored at −20°C.
TABLE 1.
Bacterial strains used in this study
| Strain | Plasmid | Property | Reference(s) |
|---|---|---|---|
| E. coli BL21 | pgst-halN | Encodes the NEAT domain (amino acids 29–152) of HalN | 53 |
| E. coli BL21 | pgst-isdC | Encodes the NEAT domain (amino acids 32–212) of IsdC | 57 |
| E. coli BL21 | pgst-isdX1N | Encodes the NEAT domain (amino acids 27–152) of IsdX1 | 49 |
| E. coli BL21 | pgst-isdX2FL | Encodes the NEAT domains of IsdX2 (amino acids 30–859) | 49 |
| E. coli BL21 | pgst-bslKN | Encodes the NEAT domain (amino acids 46–165) of BslK | 52 |
| E. coli BL21 | pgst-halNY112A | Encodes HalN with an alanine substitution for tyrosine 112 | This study |
| E. coli BL21 | pgst-halNF116A | Encodes HalN with an alanine substitution for phenylalanine 116 | This study |
| E. coli BL21 | pgst-halNY112A,F116A | Encodes HalN with alanine substitutions for tyrosine 112 and phenylalanine 116 | This study |
| B. anthracis Sterne 34F2 | pUTE657 | Avirulent, attenuated, nonencapsulated strain (Specr) | 92, 93 |
Measurement of heme binding.
The Hal NEAT domains (∼10 μM for each domain, including wild-type HalN, HalNY112A, HalNF116A, and HalNY112A,F116A) were individually incubated with hemin chloride (0 to 20 μM; Sigma-Aldrich, St. Louis, MO) in phosphate-buffered saline (PBS) (pH 7.4) for 30 min at 25°C. Spectral analyses of all proteins from 280 nm to 480 nm, before and after incubation, were performed by using a DU800 spectrophotometer (Beckman Coulter, London, United Kingdom).
Measurement of heme acquisition.
Purified glutathione S-transferase (GST)-NEAT (35 μM for GST-HalN, 20 μM for HalNY112A, 20 μM for HalNF116A, and 35 μM for HalNY112A,F116A) was individually immobilized on 2 ml of glutathione-Sepharose resin, washed with 40 ml of PBS (pH 7.4), and incubated with 1 ml of bovine methemoglobin (2.5 μM; Sigma-Aldrich, St. Louis, MO) for 30 min at 25°C. After incubation, beads containing bound NEATs were centrifuged at 6,000 × g for 1 min, the supernatant (methemoglobin) was removed, and the beads were washed with 40 ml of PBS (pH 7.4). The GST-fused NEAT domains were then eluted from the beads with 0.5 ml of reduced glutathione (25 mM; Calbiochem, San Diego, CA) for 5 min, and the relative heme content was determined by comparing the intensity of the Soret band to that for a control reaction with GST-NEAT incubated with PBS only.
Reconstitution and crystallization of HalN.
The Hal NEAT (HalN) domain of the BAS0520 gene, encoding amino acids 26 to 155, was amplified out of plasmid pGEX2TK-0520_NEAT (53) with Kod polymerase (EMD), using the following primers: forward primer 5′-GCC CAT GGA GAA TAT GGC TGT ACA AA GTC CAA AAA-3′ and reverse primer 5′-CGC TCG AGT ACAA ATG TAC GCA TAT TCA CTT CAA ACT-3′. The forward and reverse primers contained the NcoI and XhoI endonuclease sites, respectively, for cloning into pET28a (Novagen) to produce a C-terminally His-tagged fusion protein. BL21-Gold(DE3) cells harboring pET28a-0520 were grown at 37°C in LB medium containing 30 μg/ml kanamycin to an optical density at 600 nm (OD600) of 0.8, and protein expression was then induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) along with the addition of 1 mM 5-aminolevulinic acid hydrochloride (heme precursor; Sigma) for 4 h, after which the cells were centrifuged at 6,000 × g and resuspended in a solution containing 50 mM Tris-Cl (pH 7.4), 350 mM NaCl, and 15 mM imidazole. Cells were lysed by sonication with the addition of 5 mg egg hen lysozyme (Sigma) and 40 μM phenylmethylsulfonyl fluoride (Sigma). The cell lysate was centrifuged at 30,000 × g for 20 min, and the supernatant was filtered by using a 1-mm glass fiber syringe filter (Pall). The supernatant was loaded onto a Ni2+-charged HisTrap column (GE Healthcare) and eluted with a linear imidazole gradient (15 to 500 mM imidazole). Fractions were analyzed by SDS-PAGE to determine HalN purity. The final purification step was performed by using an S200 10/30 Superdex column (GE Healthcare) equilibrated with a solution containing 50 mM Tris (pH 7.4) and 150 mM NaCl to yield 100% homogeneous HalN. The percentage of heme was determined by using the pyridine hemochrome and Lowry assays (Bio-Rad) (64, 65). To achieve fully heme-reconstituted HalN, a hemin solution was added to a 1:1 molar ratio of heme to protein (Sigma) (64, 65). Crystals of heme-HalN were grown at 90 mg/ml by a hanging-drop vapor diffusion method with a reservoir containing 0.2 M ammonium sulfate, 0.1 M Bis-Tris (pH 5.5), and 21% polyethylene glycol 3350 (PEG 3350) with a 1:1 (vol/vol) protein-to-reservoir drop. Crystals were harvested by using paratone oil as a cryoprotectant and frozen in liquid nitrogen. A native data set was collected at the Advanced Light Source at Berkeley National Laboratories (ALS Berkeley) on beamline 8.2.1 at a wavelength of 1.0 Å.
Structure determination of heme-HalN.
Data processing and scaling were carried out with the imosflm suite (66). Data processing statistics are summarized in Table S1 in the supplemental material. Initial models were obtained by molecular replacement in Phenix.Phaser-MR (67) using search models of B. anthracis IsdX1 (PDB accession no. 3SZ6) (56), B. anthracis IsdX2N5 (PDB accession no. 4H8P) (68), and S. aureus IsdA (PDB accession no. 2ITE) (69), followed by additional rounds of molecular replacement and autobuilding in Phenix.MR-Rosetta (67). This was followed by manual building utilizing Coot (70) with iterative cycles of refinement with Phenix.Refine (71). The final stereochemistry and geometry for each model were validated with MolProbity (72). The refinement parameters are summarized in Table S1 in the supplemental material. All molecular graphics were prepared by using PyMOL (73).
B. anthracis spore preparation.
B. anthracis Sterne 34F2 was propagated in 8 ml tryptic soy broth (TSB) and grown overnight at 250 rpm at 37°C. Cultures were then seeded into 400 ml of modified G medium containing 0.2% yeast extract, 0.0025% calcium chloride dihydrate, 0.05% dipotassium phosphate, 0.02% magnesium sulfate heptahydrate, 0.005% manganous sulfate tetrahydrate, 0.0005% zinc sulfate heptahydrate, 0.0005% cupric sulfate pentahydrate, 0.00005% ferrous sulfate heptahydrate, and 0.2% ammonium sulfate at pH 7.1. Growth was maintained for 30 h at 30°C with shaking at 200 rpm to induce sporulation, and spores were centrifuged at 6,000 × g for 10 min, washed, and resuspended in 10 ml of water. Before use, spores were heat treated at 65°C for 1 h to kill any remaining vegetative cells, followed by serial dilution of the spore preparations on LB agar to determine the number of CFU per volume of spore preparation.
B. anthracis vaccinations and infections.
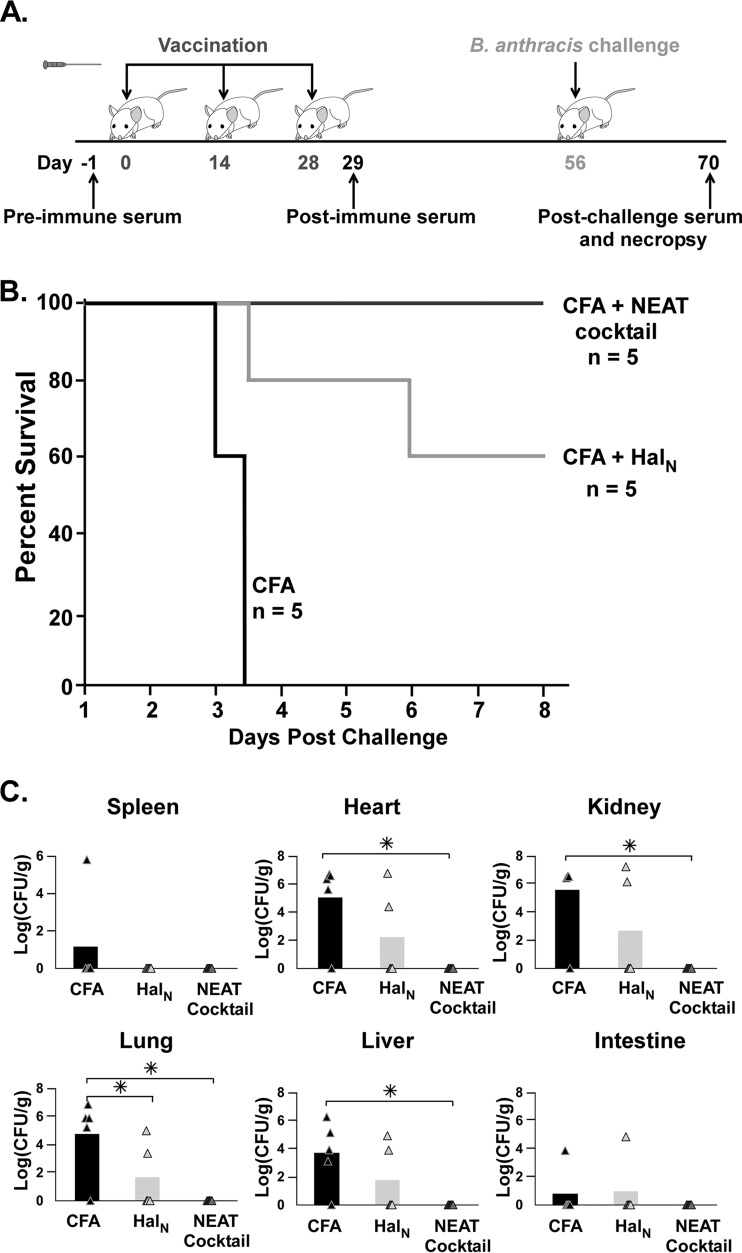
A group of 15 conventionally housed female A/J mice (Jackson Laboratory, Bar Harbor, ME) was used to determine the 50% lethal dose (LD50) of spores of B. anthracis strain Sterne 34F2. Each group, consisting of five mice, was challenged with a total volume of 200 μl containing either 1 × 103 spores, 1 × 104 spores, or 1 × 105 spores injected subcutaneously (s.c.) into the fatty tissue under the right hind leg. Prior to injection, spore preparations were heated to 65°C to kill any vegetative cells. The LD50 was determined by using the method of Reed and Muench (74). For the vaccination studies, the regimen shown in Fig. 1A was followed. Six-week-old female A/J mice were vaccinated s.c. with either adjuvant alone or complete Freund's adjuvant (CFA; Sigma-Aldrich, St. Louis, MO) or aluminum hydroxide (“alum”; Thermo Scientific, Rockford, IL) in combination with each individual (4 μg) or all (“cocktail” [4 μg of each, for 20 μg total]) recombinant NEAT domain preparations described above. After the first vaccine dose, mice given CFA were administered incomplete Freund's adjuvant for the second and third doses. After a total of three vaccinations were administered (on days 0, 14, and 28) and then 4 weeks after the third boost, the mice were challenged with either 5 or 10 times the LD50 as described above. Mice were bled a day before the first vaccination (day −1) to attain a “preimmune” serum sample, on day 29 to attain a “postvaccine” serum sample, and on day 70 or upon death to attain a “postchallenge” serum sample. Mice were monitored for morbidity by using a health index that tracks five disease characteristics (posture, activity level, coat appearance, skin turgor/tenting, and hyperpnea). Mild forms of each characteristic are scored as 0.5, while severe forms are scored as 1.0. Morbidity is achieved with a total score of 4.0, at which point the mice were euthanized and necropsied to isolate the lungs, liver, spleen, kidneys, heart, and intestine, and tissues were homogenized and plated onto LB agar to determine the number of CFU of B. anthracis per gram of tissue.
FIG 1.
Subcutaneous vaccination with NEAT domains and CFA. (A) Schematic of the vaccination regimen and collection of serum from vaccinated mice. Animals were injected subcutaneously on day 0, 14, or 28 with either the NEAT domain of Hal (HalN) (4 μg) or the NEAT domains of IsdX1, IsdX2, IsdC, BslK, and Hal (4 μg each, for 20 μg total) mixed with CFA. Serum was drawn at day −1 (preimmune), day 29 (vaccinated), and day 70 (postchallenge) and tested for anti-NEAT antibodies by ELISAs (see Materials and Methods). (B) Survival curves for mice vaccinated with adjuvant (CFA) alone, adjuvant with HalN, or adjuvant with the NEAT domain cocktail after subcutaneous challenge with 10× LD50 of B. anthracis Sterne strain spores. For CFA versus the NEAT cocktail, the P value was 0.014 as determined by a log rank (Mantel-Cox) test, and the P value was 0.017 as determined by a Wilcoxon test. For CFA versus wild-type Hal, the P value was 0.111 as determined by a log rank (Mantel-Cox) test, and the P value was 0.074 as determined by a Wilcoxon test. (C) CFU of bacilli recovered from mouse organs from the experiments described above for panels A and B (asterisks denote statistically significant values [P < 0.01], as determined by one-way analysis of variance). For CFA versus the NEAT cocktail, P values were 0.186 for spleen, 0.061 for kidney, 0.074 for heart, 0.043 for lung, 0.082 for liver, and 0.186 for gastrointestinal tract. For CFA versus wild-type Hal, P values were 0.186 for spleen, 0.009 for kidney, 0.008 for heart, 0.008 for lung, 0.012 for liver, and 0.186 for gastrointestinal tract.
Animals were handled in accordance with the Guide for the Care and Use of Laboratory Animals (75) and with approval by the Institutional Animal Care and Use Committee (protocol no. AN-5177:1) at Baylor College of Medicine.
ELISA of mouse serum samples.
Enzyme-linked immunosorbent assays (ELISAs) were performed in triplicate by using Immulon 2HB 96-well microtiter plates (Thermo Scientific, Rockford, IL) coated overnight with 0.2 μg/well of each purified NEAT domain (BslKN, HalN, IsdCN, IsdX1N, and IsdX2FL) in 1× PBS at 4°C. All reactions were carried out with a volume of 100 μl. After the initial coating, each plate was washed three times with 200 μl of 1× PBS–0.05% Tween 20 (PBST) and blotted dry by inversion on clean paper towels. Plates were then blocked with 10% nonfat dry milk overnight at 4°C. Mouse sera diluted 1:2,000 from preimmunized samples, postvaccination serum samples, and postchallenge serum samples were individually added to wells and then sequentially diluted 1:4,000, 1:8,000, and 1:16,000 in the microtiter plates and incubated for 2 h at 37°C with slow shaking at 20 rpm in a covered chamber. The plate was washed five times with PBST after each incubation, 100 μl of 1:10,000-diluted peroxidase-conjugated anti-mouse IgG (product no. A8924; Sigma-Aldrich, St. Louis, MO) was then added to each well, and the plate was incubated for 1 h at 37°C with slow shaking at 25 rpm. Plates were washed five times with PBST and then developed with the 1-Step Ultra TMB (3,3′,5′,5′-tetramethylbenzidine) ELISA substrate solution (Thermo Scientific, Rockford, IL), and the reaction was stopped with 2 M H2SO4 according to the manufacturer's specifications. Assay plate results were recorded at an OD450 with a BioTek Synergy HT plate reader (BioTek Instruments, Inc., Winooski, VT). Positive controls used included testing rabbit serum raised against individual NEAT proteins with peroxidase-conjugated anti-rabbit IgG (product no. A0545; Sigma-Aldrich, St. Louis, MO), and negative controls consisted of 1× PBS in place of serum (background absorbance).
ELISA data analysis.
Mean absorbance values at an OD450 were calculated from measurements performed in triplicate and normalized by subtraction of the average background values with standard deviation values determined by applying a propagation-of-error equation (76). Antibody titers were calculated from a mouse IgG standard by coating Immulon plates with serial dilutions of an anti-mouse IgG Fab fragment (10, 1, 0.1, and 0.01 μg/ml) in duplicate for 16 h at 4°C with periodic shaking. Wells were washed twice with 200 μl of 1× 0.05% Tween 20–PBS (wash buffer) and blocked with 2% nonfat milk and 100 μg/ml BSA overnight at 4°C. Wells were washed three times with wash buffer and incubated with 100 μl goat anti-mouse IgG-horseradish peroxidase (HRP) for 1 h at 37°C with periodic shaking. A colorimetric reaction was allowed to develop by using the 1-Step Ultra TMB ELISA substrate solution (Thermo Scientific) and 2 M H2SO4. A linear equation (y = mx + b), where y is the OD value and x is the IgG concentration in micrograms per milliliter, was generated by plotting serial dilutions of mouse IgG versus the average absorbance values and solving for x to determine IgG levels.
Assessment of the effect of anti-NEAT antibodies on B. anthracis.
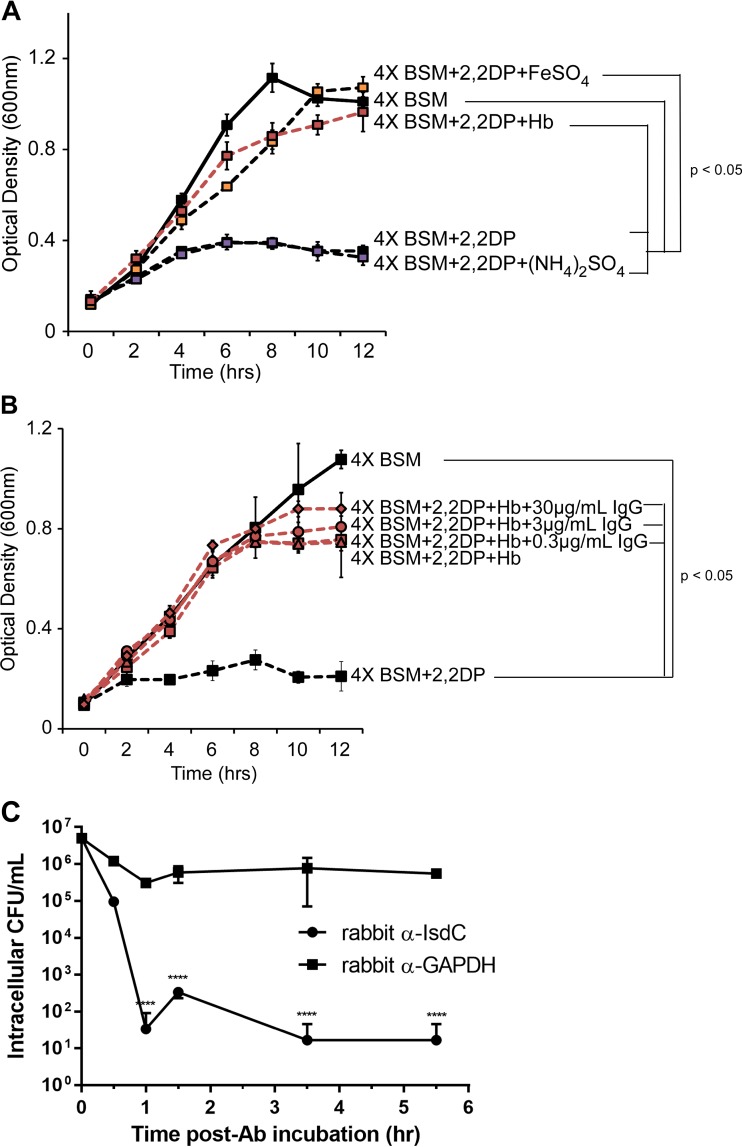
Polyclonal anti-IsdC antiserum was generated in rabbits by using standard procedures at the Baylor College of Medicine Center for Comparative Medicine. Anti-IsdC IgG was purified from serum by using an Abcam antibody purification kit (catalog no. ab102704) according to the manufacturer's instructions and used to determine the effect of anti-NEAT antibodies on B. anthracis heme uptake and opsonin-mediated killing. For heme uptake, a blood serum mimic (BSM) (and 4× BSM) was prepared, and bacillus growth in this medium was carried out as previously described (77), with 2,2-dipyridyl (500 mM; Alfa Aesar), iron sulfate (10 mM; J. T. Baker), ammonium sulfate (10 mM; BDH), and purified anti-IsdC antibody (0.3, 3, or 30 μg/ml) being added, as described in the legend to Fig. 5. Lysogeny broth (Life Technologies, Grand Island, NY) was prepared according to the manufacturer's instructions. For opsonin-mediated killing, MH-S alveolar macrophages (ATCC, Manassas, VA) were seeded at 1 day postinfection into 24-well plates at 1 × 106 cells/ml. Opsonization was initiated by adding a mixture of 30 μg/ml of anti-IsdC or anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (negative control) (catalog no. ab8425; Abcam) with B. anthracis 7702 Sterne vegetative cells at a multiplicity of infection (MOI) of 5 for 30 min at 37°C with periodic shaking. The mixture was then added to MH-S cells to begin infection, and phagocytosis was synchronized by low-speed centrifugation for 1 min at 300 × g. At each designated time point, MH-S cells were washed with 1× PBS and incubated with gentamicin (50 μg/ml) for 30 min at 37°C to eliminate extracellular bacteria. Cells were then washed with 1× PBS and lysed with 50 μl of 1% Triton X-100. Intracellular bacterial counts were determined by serial plating of the lysed samples.
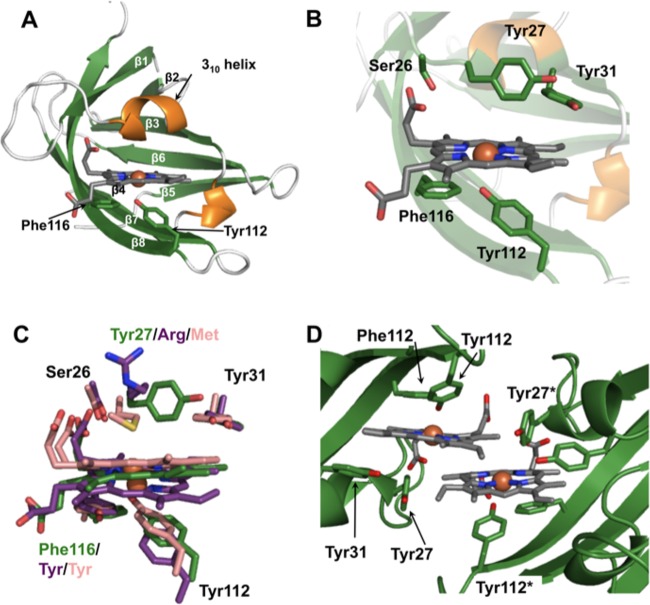
FIG 5.
Structure of heme-HalN. (A) Ribbon representation of heme-HalN, where β-strands and α-helices are shown in green and orange, respectively. Important heme-binding residues are shown in stick representation with carbon, oxygen, and nitrogen atoms in green, red, and blue, respectively, and heme is also in stick representation with carbon atoms in dark gray. (B) Closeup of the heme-binding pocket with the same color scheme as that described above for panel A. (C) Superimposition of the heme-binding sites of HalN, IsdX1, and IsdX2N5, where important heme-binding residues and heme are in stick representation, with carbon atoms of HalN in green, IsdX1 in pink, and IsdX2N5 in purple. (D) View of the dimeric interface of HalN, which is mediated predominantly by the two heme molecules. “*” denotes the second molecule in residue numbering (colored as described above for panel A).
RESULTS
Vaccination with recombinant NEAT domains can protect against a lethal dose of B. anthracis.
We initiated experimentation to determine if vaccination with recombinant NEAT proteins could protect against infection with B. anthracis. The animal model of choice for these studies was the A/J mouse, a strain commonly used to assess anthrax disease, especially toxigenic forms of B. anthracis such as those used in this study (16). The reported LD50 of B. anthracis Sterne strain 34F2 in the A/J model ranges from 103 to 105 spores (16, 30). To determine the LD50 more precisely and narrow this range, we infected three groups of mice with 102, 103, or 104 B. anthracis spores and observed survival of the mice over a 5-day period. All of the mice infected with 104 spores succumbed to infection by the fourth day, while mice challenged with 103 spores showed 80% survival by day 5, and all the mice treated with 102 spores survived (see Fig. S1 in the supplemental material). Based on these results, the LD50 was calculated to be 3.2 × 103 spores (74).
We next purified each individual NEAT domain of IsdX1, IsdC, BslK, or Hal or all five NEAT domains as a polypeptide from IsdX2 from E. coli by GST affinity chromatography. Each preparation was assessed for purity by SDS-PAGE, the protein concentration was determined by using the Bradford method, and the ability to bind heme (an indication of a functional, folded protein domain) was assessed by using assays previously developed in our laboratory (68). Lipopolysaccharide (LPS) was removed by using a series of endotoxin-binding resins, and pure preparations were mixed with complete Freund's adjuvant (CFA). The vaccination regimen for all experiments in this study is presented in Fig. 1A, with three subcutaneous (s.c.) inoculations spaced 2 weeks apart. Serum samples were collected before dose 1 and 1 day after the last dose of the vaccine. Following vaccination and on day 56, mice were challenged with 10 times the LD50 of B. anthracis strain Sterne, and mouse health was examined for 8 days postchallenge. While none of the mice receiving only CFA survived challenge, 60% of mice vaccinated with only the NEAT domain of Hal (HalN) and all of the mice vaccinated with a cocktail of every NEAT survived a lethal dose of B. anthracis (Fig. 1B). At the time of necropsy, in mice given only CFA, the spleen, heart, kidney, liver, lung, and intestine generally contained high levels of B. anthracis, whereas mice vaccinated with HalN or the NEAT cocktail showed reduced and no bacilli, respectively, in these samples (P < 0.01) (Fig. 1C).
NEAT-based protection from anthrax is associated with the production of anti-NEAT antibodies.
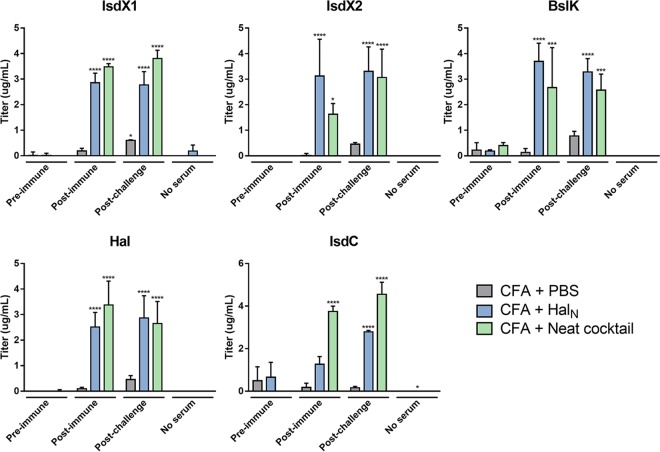
Pre- and postimmune serum samples from mice vaccinated with CFA, HalN, or the NEAT cocktail were assessed for the presence of anti-NEAT antibodies by using ELISAs with purified NEAT domains from each protein. When all the components of the ELISA mixture were added, except for serum, little to no signal was observed in any of the ELISA reactions (“no-serum” group) (Fig. 2A to E). In addition, a weak signal was observed when serum from vaccinated individuals given only CFA was tested (Fig. 2, gray bars). Furthermore, little to no signal was observed prior to vaccination (“preimmune” group) (Fig. 2). Thus, the level of background in this assay was quite low. In contrast, mice vaccinated with the NEAT cocktail yielded a strong response that was observed only if the mice were vaccinated with the NEAT domains and was not observed prior to vaccination (“postimmune” group) (Fig. 2, blue and green bars). Antibody production was observed for all five components of the NEAT cocktail with titers that ranged from 1 to 4 ng/ml. In fact, for IsdX1, IsdX2, Hal, and BslK, positive antibody was observed even when serum was diluted 10,000-fold (not shown). Of particular importance was the observation that mice vaccinated with just HalN produced antibodies that were reactive not only to HalN but also to every other NEAT domain that was analyzed (Fig. 2A to E, blue bars). Indeed, with the exception of the NEAT domain from IsdC, the titers of anti-NEAT antibodies from HalN-vaccinated mice were equivalent to those of antibodies from the cocktail-vaccinated animals (Fig. 2A to E, compare blue bars to gray bars). When considered in the context of the results from the challenge studies shown in Fig. 1, these data suggest that a 3-dose vaccination regimen using recombinant NEAT domains can substantially protect mice from a lethal dose of B. anthracis and that bacillus NEAT proteins are immunogenic, leading to the production of anti-NEAT antibodies. Furthermore, there is substantial cross-reactivity observed, in that vaccination with just a single NEAT domain (HalN) produces antibodies that also bind the NEAT domains from the other four proteins.
FIG 2.
Reactivity of sera from mice vaccinated with the NEAT domains and CFA. Preimmune, postvaccine, and postchallenge sera from mice vaccinated with either CFA, HalN, or the NEAT cocktail were evaluated for the presence of anti-NEAT antibodies by using an ELISA with purified NEAT domains from each NEAT protein: IsdX1, Hal, BslK, IsdC, and IsdX2. Asterisks represent results of Tukey's multiple-comparison test comparing CFA and Hal or CFA and the NEAT cocktail to the control (CFA plus PBS). *, P < 0.05; **, P < 0.01, ***, P < 0.001; ****, P < 0.0001.
A NEAT cocktail is efficacious when combined with alum given intramuscularly.
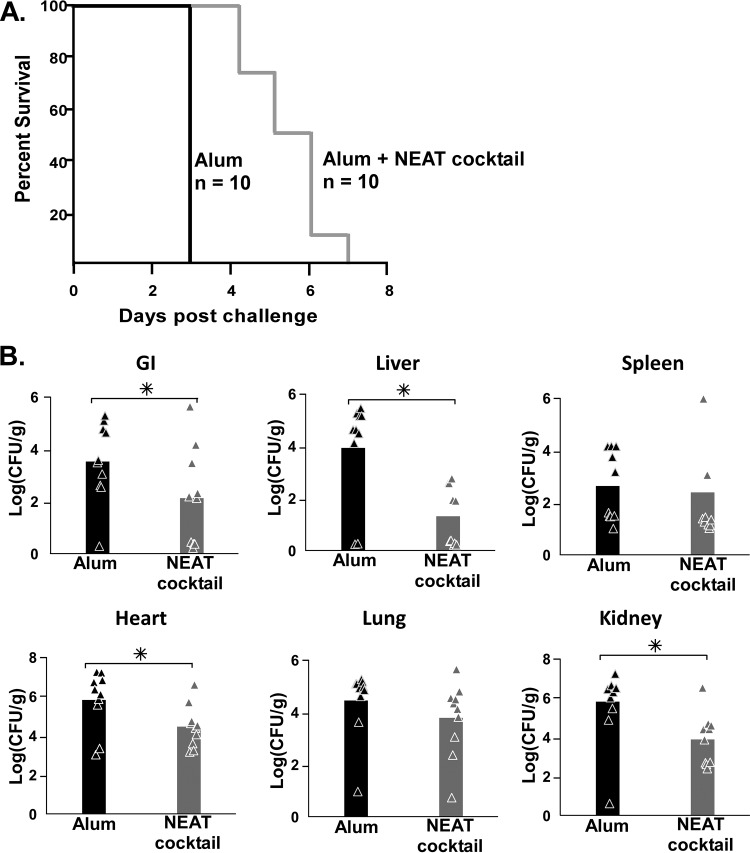
Licensed vaccines, including AVA, generally use aluminum hydroxide (alum) as the adjuvant of choice because it is safe to use in humans (78). In addition, most alum-adjuvanted vaccines administered in the United States are given intramuscularly (i.m.). It has been demonstrated that injection site reactogenicity is increased when the s.c. route is used versus the i.m. route (79). Because of this, we conducted additional vaccination studies aimed at determining the effectiveness of a NEAT domain cocktail when alum was the adjuvant and the vaccinations were administered i.m., the preferred human vaccination protocol. Each recombinant NEAT domain was purified, 4 μg of each NEAT domain was mixed together with alum, and the mixture was injected into the muscle of the right hind leg of mice (n = 10) by using the general vaccination regimen and challenge described in the legend to Fig. 1. As shown in Fig. 3A, mice receiving the NEAT cocktail survived 4 days longer on average than their alum-only counterparts (P < 0.0001). When organs were analyzed (following necropsy) for the number of CFU of bacilli, a significantly (P < 0.05) lower level of B. anthracis was observed in the heart, kidney, liver, and intestine, but not the spleen or lungs, of mice given the NEAT cocktail than in the adjuvant-only controls (Fig. 3B). When serum was analyzed for the presence of anti-NEAT antibodies, only the secreted hemophore IsdX2 yielded a detectable response upon ELISAs (see Fig. S2A and S2B in the supplemental material). No antibodies to any of the NEATs were detected in serum drawn before vaccination. Since IsdX2 harbors five nonidentical NEAT domains, while all of the other B. anthracis NEAT domain proteins have only one, these results suggest that the antibody response is dependent on the dose of the antigen, with higher titers being observed when more antigen is given. Taken as a whole, the data suggest that a NEAT protein cocktail, when used with a human-approved adjuvant and route of delivery, provides some level of protection against a lethal dose of B. anthracis, with the concomitant production of anti-IsdX2 antibodies albeit at lower titers. Partial protection and a significant reduction of the numbers of bacilli in organs indicate that even low levels of circulating anti-NEAT antibodies are sufficient to delay the development of anthrax disease.
FIG 3.
Intramuscular vaccination with NEAT domains and alum. (A) Survival curves for mice vaccinated with adjuvant alone (alum) or alum plus a NEAT cocktail (4 μg of each NEAT domain, for 20 μg total) after challenge with 5× LD50 of B. anthracis Sterne strain spores. For alum versus the NEAT cocktail, the P value was <0.0001 as determined by a log rank (Mantel-Cox) test, and the P value was <0.0001 as determined by a Wilcoxon test. (B) Comparison of the total B. anthracis CFU recovered from mouse organs (lungs, spleen, kidneys, heart, and gastrointestinal [GI] tract) treated with alum (black bars) alone versus alum with a NEAT cocktail (gray bars) after necropsy of animals on day 70. Asterisks indicate statistical significance (P < 0.01) as determined by one-way analysis of variance.
Anti-NEAT antibodies promote phagocyte-mediated killing of B. anthracis.
There are two main ways in which antibodies directed to NEAT domains may afford protection against B. anthracis. One way is through the inhibition of heme-iron import by blocking the ability of NEAT domains to bind hemoglobin, heme, or both. A second way is that the antibodies may act as an opsonin for recognition and eventual killing by phagocytic cells. We performed experiments aimed at differentiating between these possibilities by first testing if anti-NEAT antibodies prevent the growth of bacilli on heme as the only source of iron. B. anthracis strain Sterne 34F2 was grown in the presence or absence of hemoglobin (a source of heme) or FeSO4 (iron) under iron-limited conditions that are designed to mimic a blood-like environment (BSM) (77). A robust stimulation of growth was observed with the addition of iron or hemoglobin (iron in the form of heme) in the presence of an iron chelator (Fig. 4A, red, orange, and black squares). Little or no growth was observed without iron, suggesting that these conditions recapitulate an iron utilization response (Fig. 4A, purple squares and black circles). To test if antibodies that recognize NEAT proteins prevent heme-iron uptake under these conditions, we raised polyclonal antibodies to the NEAT protein considered to be the central conduit in the heme uptake cascade (IsdC) (55). Purified IgG antibodies, which recognize IsdC upon Western blotting (not shown), were next incubated with B. anthracis in BSM in the presence or absence of hemoglobin. No inhibition of bacillus growth was observed in the presence of IsdC antibodies, even at the highest concentration of IgG used (Fig. 4B).
FIG 4.
Effect of anti-NEAT antibodies on iron intake or opsonin-mediated macrophage killing. (A and B) B. anthracis was cultured in 4× BSM (solid lines) or 4× BSM with 2,2-dipyridyl (2,2DP) (500 μM) (dashed lines) in the presence of iron sulfate (150 μM) (orange boxes), ammonium sulfate (150 μM) (purple boxes), and human hemoglobin (Hb) (15 μM) (red lines) (A) or hemoglobin with increasing concentrations of purified rabbit anti-IsdC (red symbols) (B), and growth was measured by absorbance spectroscopy (OD600). Each data point represents the average of results from three independent cultures ± standard deviations. (C) Purified rabbit anti-IsdC or a purified rabbit isotype control (anti-GAPDH), each at 30 μg/ml, was preincubated with B. anthracis strain Sterne for 30 min at 37°C, washed, and added to MH-S alveolar macrophages (MOI of 5), with phagocytosis being synchronized by low-speed centrifugation. At each designated time point, MH-S cells were washed with 1× PBS, incubated with gentamicin for 30 min to eliminate extracellular bacteria, and lysed with Triton X-100. Intracellular bacterial counts were determined by serial plating of the lysed samples. The values represent the means and standard deviations of data from three independent experiments. Data were analyzed by using an unpaired t test comparing the average values at each time point (****, P < 0.00001). Ab, antibody.
To assess the effects of anti-NEAT antibodies serving as opsonins for alveolar macrophages and subsequent killing of B. anthracis once inside these cells, we cultured murine MH-S macrophages with bacilli that either were or were not preincubated with purified anti-IsdC antibodies (Fig. 4C). A decrease in the levels of bacilli of several orders of magnitude was observed for B. anthracis-incubated anti-NEAT antibody compared to the isotype-matched control group in the first 1 h of infection. In fact, a significant difference in bacterial survival under these two conditions was observed every hour for the entire 5.5-h infection. This difference was not due to an inhibition of the growth of bacilli in RPMI, the culture medium used for MH-S macrophages (not shown). Taken together, these two data sets suggest that anti-NEAT antibodies, at least those to IsdC, are not growth restrictive in iron-limiting environments for B. anthracis. Instead, such antibodies stimulate the killing of bacilli by alveolar macrophages, presumably through antibody-mediated opsonophagocytosis. By extension, these data may indicate that the protection afforded by NEAT vaccination in murine infection models may be more attributable to the phagocytosis of antibody-coated bacilli than to the inhibition of heme uptake through NEAT proteins. This may also explain why even a very low-level antibody response (alum plus i.m. inoculation) (Fig. 3) can still induce a measurable level of protection from anthrax disease.
Understanding the structural basis of NEAT activity for the design of a safer NEAT-based vaccine.
If NEAT proteins are to be evaluated or developed as potential vaccine candidates for the prevention of anthrax, it will be necessary to understand the molecular basis for their function. This knowledge will help guide the design of recombinant forms that lack heme-binding or heme extraction activity, efforts that may reduce toxicity and make vaccines safer. As mentioned above, a version of the NEAT protein IsdB (V710) was evaluated in phase I and II clinical trials as a vaccine from S. aureus, but the trials were suddenly halted due to toxicity concerns (63). Because Hal is important in heme-iron utilization in B. anthracis (53), is homologous at the amino acid level to the other four NEATs from bacilli (55), and is a component of the NEAT cocktail used here, we investigated its structure and the molecular determinants of its activity to gain a better understanding of how to develop a safer NEAT-based vaccine.
All bacilli and staphylococcal NEAT domains studied thus far have a conserved heme-binding motif, YXXXY (56, 68, 69, 80, 81). In these structures, the first tyrosine coordinates with heme-iron as the axial ligand, and the second tyrosine hydrogen bonds (H-bonds) to the axial tyrosine, stabilizing the interaction. As HalN does not contain the characteristic heme-binding motif and has a phenylalanine in the fifth position (YDKEF) (53), we determined the structure of heme-HalN to a 3.0-Å resolution to shed light on the mode of heme binding. The structure of HalN consists of an immunoglobulin-like fold, as observed in multiple NEAT domain crystal structures (56, 68, 69, 80, 81), with eight β-strands arranged in two antiparallel β-sheets that form a β-sandwich (Fig. 5A). In the structure of HalN, the predominately hydrophobic heme-binding pocket is formed by the long β-hairpin (strands β7 and β8, where β8 contains the heme binding motif) on the proximal side of the heme molecule and the 310-helix region (SXXXXY) on the distal side, which is another conserved feature of NEAT domains (51, 56, 80, 81). On the proximal side of heme, the heme-iron is coordinated by the conserved Tyr112 via its hydroxyl group, with an Fe-O distance of 2.4 Å (Fig. 5A). This Fe-O distance reported for other NEAT domains is slightly shorter (2.0 to 2.2 Å) due to the increased electronegativity of the first Fe-coordinating Tyr H-bonding to the phenolate of the second Tyr within the heme-binding motif (80). In the NEAT domain of Hal, the second Tyr within this motif is replaced with a phenylalanine (Phe116). Thus, the coordinating Tyr112 is not as electronegative as other NEAT Fe-coordinating tyrosines, resulting in a slightly longer Fe-O coordinating distance. However, Phe116 further stabilizes the heme molecule by π-stacking interactions with the pyrrole ring (Fig. 5A). Within the 310-helix region, the hydroxyl group of the conserved Ser26 H-bonds with the least-solvent-exposed heme propionate group (2.9 Å) together with the conserved Tyr31 that weakly π-stacks with the heme pyrrole ring (Fig. 5B).
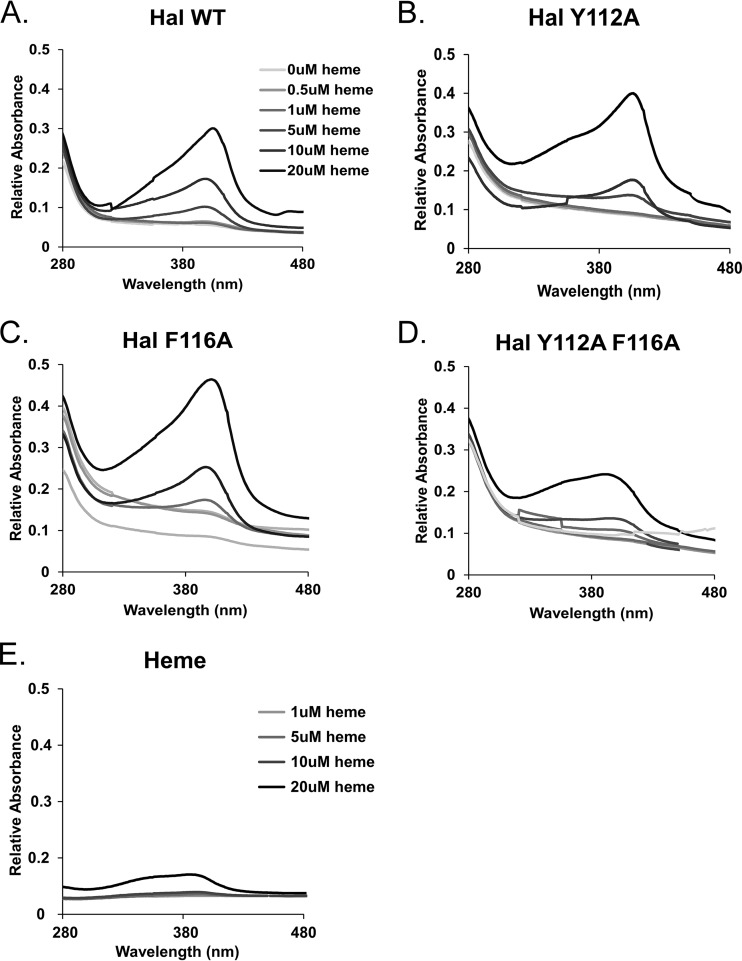
The crystal structure of the NEAT domain of Hal helped guide mutational studies to assess the effect of some of these residues on the function of Hal. This is important because if NEAT proteins are to be studied or move forward as possible anthrax vaccines, the antigens that compose this vaccine should not contain any biological activity that can introduce toxicity or adverse reactions. Because there is structural evidence that Y112 and F116 are involved in heme coordination, we used site-directed mutagenesis to individually or collectively substitute an alanine for these residues and purified their corresponding recombinant domains. The heme-binding properties of the mutant domains were next assessed by measuring the intensity of the Soret band, an optical feature of heme complexed to protein, at ∼400 nm. The titration of hemin, the oxidized form of heme, into a solution of wild-type HalN resulted in the formation of a strong Soret maximum at 402 nm in a concentration-dependent manner (Fig. 6A), which was much more intense, red shifted, and defined in shape than the hemin-only control (Fig. 6E). HalNY112A and HalNF116A, when mixed with hemin, demonstrated Soret intensities similar to those of the wild-type domain (Fig. 6B and C). However, the double mutant (HalNY112A,F116A) displayed features somewhat in between the wild-type profile and that of the hemin control, with a broad peak and lower intensity (Fig. 6D).
FIG 6.
Analysis of heme-binding properties of HalN mutants. Increasing concentrations of heme (inset) were incubated with wild-type (WT) HalN (A), HalNY112A (B), HalNF116A (C), HalNY112A,F116A (D), and heme alone (E) for 30 min, and spectral scans of the reaction mixtures were collected to assess the intensity of the Soret band, the spectral signal associated with heme binding.
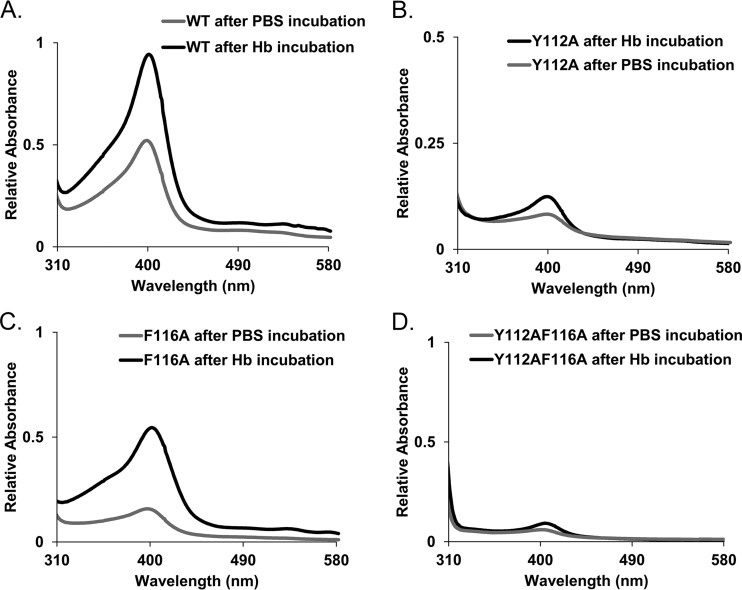
The compromised ability of the double mutant to bind hemin compelled an examination of the relative heme acquisition activity from hemoglobin. Some NEAT domains, including the two secreted hemophores IsdX1 and IsdX2 as well as Hal, can extract heme from hemoglobin at rates that are higher than the rate of thermal disassociation of heme into solution (53). To determine if these mutant NEAT domains of Hal can acquire heme from hemoglobin, we employed a simple affinity chromatography technique whereby a known amount of the NEAT domain in question is purified as a GST fusion protein and bound to glutathione-Sepharose. Hemoglobin (in this case, bovine) is next added to the reaction mixture, the beads are extensively washed, the bound NEAT is eluted with excess glutathione, and eluants are assessed for heme transfer by Soret spectroscopy. When incubated with hemoglobin and analyzed in this manner, wild-type HalN underwent an increase in the Soret band, which was suggestive of an acquisition of heme from hemoglobin (Fig. 7A). This was also observed for HalNF116A, albeit at a somewhat lower level, when analyzed under the same conditions (Fig. 7C). However, a small increase in the Soret band was observed when HalNY112A was incubated with hemoglobin (Fig. 7B), with the HalNY112A,F116A double mutant being nearly identical in heme occupancy to the control sample that was incubated with PBS only (Fig. 7D). When considered as a whole and in combination with the heme-binding data shown in Fig. 6, these results suggest that Y112 and/or F116 is sufficient to bind heme but that Y112 is important for the acquisition of heme from hemoglobin. These findings also indicate that one should consider generating inactive NEAT domain antigens as anthrax vaccine components, especially if they retain the ability to elicit protective antibodies after immunization.
FIG 7.
Analysis of the heme acquisition properties of HalN mutants. Wild-type HalN (A), HalNY112A (B), HalNF116A (C), and HalNY112A,F116A (D) were incubated with hemoglobin (black lines) or buffer alone (gray lines) for 30 min at room temperature and assessed for their ability to actively scavenge heme from hemoglobin by the intensity of the Soret band.
DISCUSSION
The key findings of this study are that (i) a 3-dose vaccination regimen using NEAT domains as antigens is efficacious against a lethal challenge of B. anthracis; (ii) protection is correlated with both the production of anti-NEAT antibodies and overall lower levels of bacilli in organs and tissues; (iii) the Hal NEAT domain alone may also be efficacious albeit at lower levels; (iv) a NEAT vaccine delays the onset of anthrax disease with human-approved alum when delivered i.m.; (v) anti-NEAT antibodies against IsdC promote killing by alveolar macrophages but do not inhibit heme-dependent growth; (vi) the structure of the Hal NEAT domain has the conserved overall NEAT immunoglobulin-like fold, with F116 and Y112 coordinating the heme-iron, while Phe116 stabilizes the heme molecule by π-stacking with the porphyrin ring; and (vii) F116 and Y112 are important for heme binding, but Tyr112 in Hal seems to be important for heme acquisition from hemoglobin. These results provide a rationale to pursue additional efforts to develop NEAT proteins as a prophylactic vaccine for anthrax disease. This development may include using either wild-type or genetically modified NEATs that retain immunogenicity but are still safe as well as combining the NEATs with other known protective antigens, such as anthrax toxin or protective antigen.
There are four main mechanisms by which antibodies against NEATs may work against a bacterial pathogen. The first mechanism is by inhibiting the binding of the NEAT to the major oxygen carrier protein in mammals, hemoglobin. Because hemoglobin coordinates heme-iron, it is a target for pathogenic bacteria. The staphylococcal NEAT protein IsdB as well as the two bacillus hemophores (IsdX1 and IsdX2) and the surface protein Hal all either stably or transiently bind hemoglobin for the purpose of releasing and acquiring its bound heme. By preventing direct access of the NEAT domain to hemoglobin, the rapid transfer of heme is inhibited, thereby decreasing the rate at which the growing pathogen can transfer heme to IsdC, the central conduit, which is thought to then relay heme and its bound iron into the cell. The second mode of inhibition is at the level of disrupting heme binding to the NEAT domain. Although the heme ligand is relatively small in comparison to the light and heavy chains of an antibody, the heme-binding site sandwiched between the 310-helix and the β-hairpin encompasses a significant amount of surface area for this small ∼15-kDa domain module. Steric hindrance blocking the entry of heme into this binding site would prevent all forms of heme scavenging, both free and direct forms of heme scavenging, thereby serving as an effective way to prevent heme-based iron uptake. Antibodies may also inhibit the transfer of heme between the NEAT domains, either from the secreted hemophores to surface NEATs (such as IsdC) or between surface NEATs themselves. Since NEAT-based heme uptake is regarded as a transfer cascade, this or a combination of all three modes of inhibition could produce a powerful mechanism to shut down this major iron uptake pathway. Torres et al. and Stranger-Jones and colleagues first reported that vaccination with staphylococcal surface proteins, one of which was IsdB (heme receptor for human hemoglobin), was effective in preventing invasive infection with human isolates of S. aureus (59, 82). Indeed, Kim and coworkers reported that antibodies against IsdB could protect mice from S. aureus when passively transferred into mice and that the mechanism of protection was linked to the ability of these antibodies to block IsdB binding to hemoglobin and, thus, heme-iron uptake and not to the ability of these antibodies to promote opsonophagocytosis (83). Another explanation is that anti-NEAT antibodies recognize surface NEAT protein and promote the recognition of bacilli by phagocytes or complement.
An antibody response is produced upon vaccination with NEAT proteins. Purified polyclonal IgG raised in rabbits against the recombinant NEAT domain of B. anthracis IsdC, which is reactive to other bacillus NEAT domains on Western blots (not shown), does not prevent heme-iron uptake from hemoglobin under iron-limiting conditions. In contrast to what is observed for antibodies to S. aureus IsdB, antibodies against IsdC did not inhibit the growth of bacilli on hemoglobin as the only iron source, thereby precluding the first three mechanisms discussed above as the cause of the negative effect on bacilli. Instead, such antibodies promoted the opsonophagocytosis of bacilli by alveolar macrophages. The doses of the NEAT antigen and the adjuvant have not yet been optimized for this formulation, and so the level of antibody needed to induce this effect is currently unknown. However, because IsdX2 is the only NEAT to induce detectable antibodies in the alum formulation (IsdX2 contains five NEATs, compared to only one each for IsdX1, IsdC, BslK, and Hal), this suggests that we have not yet reached the threshold for the upper limit of delivered antigen in this system. In addition, it is encouraging that despite the low circulating levels of antibody in both the CFA and alum studies, a strong protective response against anthrax was still observed. This may mean that a robust antianthrax response is observed with low levels of circulating anti-NEAT antibodies. The unexpected finding that such antibodies may promote phagocytosis may thus prove useful in the development of a better anthrax vaccine. The current U.S.-licensed vaccine, AVA, is believed to work by stimulating the production of antibodies that neutralize anthrax toxin. However, this leaves open the question as to whether AVA targets replicating and growing bacilli. The addition of recombinant NEATs to such a vaccine may provide a powerful one-two punch that both inhibits toxin as well as promotes the clearance of bacilli.
This raises the question as to whether capsulated bacilli would be resistant to a vaccine strategy that targets surface proteins, especially if one of the properties of the vaccine is the ability to induce the phagocytosis of opsonized bacteria. It is thought that one of the functions of the capsule of pathogenic bacteria is to protect against the host immune system, perhaps by inhibiting phagocytosis (84). The poly-γ-d-glutamic acid (PGA) capsule of B. anthracis is a poor immunogen; however, coupling PGA to a strong immunogen, including protective antigen, leads to anticapsule antibodies that promote the killing and clearance of bacilli (25, 85, 86). It currently is not understood whether antibodies to NEAT domains would bind to capsulated bacilli. Whereas IsdX1 and IsdX2 are secreted proteins, IsdC is covalently attached to the cell wall (49). BslK is an S-layer protein and may be the most surface exposed of the five NEATs (52). The surface location of Hal is unknown, but it is likely upstream of the cell wall (53). Since heme-loaded hemophores and/or hemoglobin would be expected to contact these downstream surface NEATs in the presence of capsule (otherwise, iron uptake in capsulated bacteria via heme might not occur), it may be possible for antibodies to also access these surface NEATs. Further studies will be needed to determine if the capsule represents a barrier to vaccine construction using this approach.
The suggestion that vaccines can be constructed from proteins involved in heme or iron acquisition is not unprecedented, and there already has been a major advance along these lines. IsdB also induced substantial production of IgG in rhesus macaques, suggesting that it also may do so in humans (60). In fact, the data were so convincing that Merck conducted three phase I and II clinical studies using a version of IsdB (termed V710) to determine how well it was tolerated in humans. Preliminary studies suggested that it was safe and induced an antibody response in humans (61, 62, 87). However, Merck abruptly stopped the study, citing a high rate of adverse vaccine-related events and multiorgan failure in another study (63). The details of these events have yet to be reported (88); however, in nearly all the subjects receiving V710, but not the placebo group, the prevaccination interleukin-2 (IL-2) and IL-17 levels were low, suggesting that some type of aberrant immune response contributed to an inability to resolve subsequent staphylococcal infections (89). Therefore, an understanding of the molecular properties of the NEAT domain can aid in the development of a safe anthrax vaccine.
Along these lines, our structural analysis of Hal is a step in this direction. The replacement of Y112 with an alanine indicates that HalNY112A can still bind heme but cannot accept heme from hemoglobin. This suggests that perhaps Y27 or Y30 may coordinate heme-iron in lieu of Y112, although the overall mechanism of heme acquisition from hemoglobin requires Y112. Thus, Y112 may act as an alternate ligand for heme-iron or may be required for the extraction and transfer of heme from hemoglobin, possibly at the level of the NEAT-hemoglobin interaction, as observed for R54 in IsdX1 (56). Even though HalN has a phenylalanine (F116) substitution at the second tyrosine within the highly conserved heme-binding motif (YXXXY), heme-iron is still coordinated by the first tyrosine (Y112), as observed for all other S. aureus and B. anthracis NEAT domains (56, 68, 69, 80, 81), and F116 π-stacks with the heme pyrrole ring. Interestingly, the second NEAT domain of the proposed hemophore from Listeria monocytogenes (Hbp2N2) also harbors an amino acid substitution at the second Tyr in the heme binding motif, to an alanine and not a phenylalanine, as observed for HalN. The recently solved structure of Hbp2N2 shows that unlike HalN, the conserved first Tyr of the heme-binding motif does not form an axial ligand with heme-iron (90). Instead, a tyrosine on the adjacent β-strand (β7) from the β-strand harboring the heme-binding motif (β8) coordinates its hydroxyl group to heme-iron. Furthermore, Hbp2N2 undergoes a major conformational change upon heme binding (90), which has not been observed for other NEAT domains, such as B. anthracis IsdX1 (56), S. aureus IsdA (69), and S. aureus IsdH (91). The authors of those studies suggest that this larger structural rearrangement of Hbp2N2 is a product of the lack of both tyrosine residues within the heme-binding motif, since within NEAT domains with a conserved heme-binding motif, the two tyrosine residues form a noncovalent interaction that stabilizes the β-hairpin structural element. As HalN also lacks the second tyrosine of the heme-binding motif, one may expect HalN to have the same degree of structural plasticity as Hbp2N2. This may mean that any inclusion of Hal or NEATs that are structurally similar to Hal in a vaccine may require multiple mutations at several residues in the heme-binding pocket to inactivate their heme uptake properties. Recombinant Hal containing a double substitution (Y112A and F116A), as guided by structural studies and confirmed to be inactive in functional studies, would seem to be a logical inclusion in an anthrax vaccine. Future studies will determine if vaccination with such NEATs still retains their protective effect.
Supplementary Material
ACKNOWLEDGMENTS
We thank Margaret Ellen Conner for assistance in developing a vaccine regimen and experimental advice. We also thank the Stanford Synchrotron Radiation Lightsource (SSRL) and the Advanced Light Source (ALS) at Berkeley National Laboratories for their invaluable help in data collection.
This work was supported by grants AI097167 and AI109465 (A.W.M.) and by grant AI081161 (C.W.G.) from the National Institutes of Health.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.00755-16.
REFERENCES
- 1.Nicholson WL, Munakata N, Horneck G, Melosh HJ, Setlow P. 2000. Resistance of Bacillus endospores to extreme terrestrial and extraterrestrial environments. Microbiol Mol Biol Rev 64:548–572. doi: 10.1128/MMBR.64.3.548-572.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Setlow P. 2006. Spores of Bacillus subtilis: their resistance to and killing by radiation, heat and chemicals. J Appl Microbiol 101:514–525. doi: 10.1111/j.1365-2672.2005.02736.x. [DOI] [PubMed] [Google Scholar]
- 3.Guidi-Rontani C, Weber-Levy M, Labruyere E, Mock M. 1999. Germination of Bacillus anthracis spores within alveolar macrophages. Mol Microbiol 31:9–17. doi: 10.1046/j.1365-2958.1999.01137.x. [DOI] [PubMed] [Google Scholar]
- 4.Dixon TC, Fadl AA, Koehler TM, Swanson JA, Hanna PC. 2000. Early Bacillus anthracis-macrophage interactions: intracellular survival and escape. Cell Microbiol 2:453–463. doi: 10.1046/j.1462-5822.2000.00067.x. [DOI] [PubMed] [Google Scholar]
- 5.Guidi-Rontani C, Levy M, Ohayon H, Mock M. 2001. Fate of germinated Bacillus anthracis spores in primary murine macrophages. Mol Microbiol 42:931–938. doi: 10.1046/j.1365-2958.2001.02695.x. [DOI] [PubMed] [Google Scholar]
- 6.Titball RW, Manchee RJ. 1987. Factors affecting the germination of spores of Bacillus anthracis. J Appl Bacteriol 62:269–273. doi: 10.1111/j.1365-2672.1987.tb02408.x. [DOI] [PubMed] [Google Scholar]
- 7.Smith H, Keppie HS, Stanley JI. 1953. The chemical basis of the virulence of Bacillus anthracis. I. Properties of bacteria grown in vivo and preparation of extracts. Br J Exp Pathol 34:477–485. [PMC free article] [PubMed] [Google Scholar]
- 8.Smith H, Keppie J. 1954. Observations on experimental anthrax; demonstration of a specific lethal factor produced in vivo by Bacillus anthracis. Nature 173:869–870. doi: 10.1038/173869a0. [DOI] [PubMed] [Google Scholar]
- 9.Smith H, Keppie J, Stanley JL. 1955. The chemical basis of the virulence of Bacillus anthracis. V. The specific toxin produced by Bacillus anthracis in vivo. Br J Exp Pathol 36:460–472. [PMC free article] [PubMed] [Google Scholar]
- 10.Smith H, Keppie J, Stanley JL. 1954. Observations on the cause of death in experimental anthrax. Lancet 267:474–476. [DOI] [PubMed] [Google Scholar]
- 11.Marraffini LA, Schneewind O. 2006. Targeting proteins to the cell wall of sporulating Bacillus anthracis. Mol Microbiol 62:1402–1417. doi: 10.1111/j.1365-2958.2006.05469.x. [DOI] [PubMed] [Google Scholar]
- 12.Zwartouw HT, Smith H. 1956. Polyglutamic acid from Bacillus anthracis grown in vivo: structure and aggressin activity. Biochem J 63:437–454. doi: 10.1042/bj0630437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grunow R, Verbeek L, Jacob D, Holzmann T, Birkenfeld G, Wiens D, von Eichel-Streiber L, Grass G, Reischl U. 2012. Injection anthrax—a new outbreak in heroin users. Dtsch Arztebl Int 109:843–848. doi: 10.3238/arztebl.2012.0843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Doganay M, Metan G, Alp E. 2010. A review of cutaneous anthrax and its outcome. J Infect Public Health 3:98–105. doi: 10.1016/j.jiph.2010.07.004. [DOI] [PubMed] [Google Scholar]
- 15.Glomski IJ, Piris-Gimenez A, Huerre M, Mock M, Goossens PL. 2007. Primary involvement of pharynx and Peyer's patch in inhalational and intestinal anthrax. PLoS Pathog 3:e76. doi: 10.1371/journal.ppat.0030076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Welkos SL, Keener TJ, Gibbs PH. 1986. Differences in susceptibility of inbred mice to Bacillus anthracis. Infect Immun 51:795–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Peters CJ, Hartley DM. 2002. Anthrax inhalation and lethal human infection. Lancet 359:710–711. doi: 10.1016/S0140-6736(02)07792-9. [DOI] [PubMed] [Google Scholar]
- 18.Sterne M. 1946. Avirulent anthrax vaccine. Onderstepoort J Vet Sci Anim Ind 21:41–43. [PubMed] [Google Scholar]
- 19.Baillie L, Moir A, Manchee R. 1998. The expression of the protective antigen of Bacillus anthracis in Bacillus subtilis. J Appl Microbiol 84:741–746. doi: 10.1046/j.1365-2672.1998.00405.x. [DOI] [PubMed] [Google Scholar]
- 20.Watson J, Koya V, Leppla SH, Daniell H. 2004. Expression of Bacillus anthracis protective antigen in transgenic chloroplasts of tobacco, a non-food/feed crop. Vaccine 22:4374–4384. doi: 10.1016/j.vaccine.2004.01.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chabot DJ, Joyce J, Caulfield M, Cook J, Hepler R, Wang S, Vietri NJ, Ruthel G, Shoop W, Pitt L, Leffel E, Ribot W, Friedlander AM. 2012. Efficacy of a capsule conjugate vaccine against inhalational anthrax in rabbits and monkeys. Vaccine 30:846–852. doi: 10.1016/j.vaccine.2011.12.010. [DOI] [PubMed] [Google Scholar]
- 22.Garufi G, Wang YT, Oh SY, Maier H, Missiakas DM, Schneewind O. 2012. Sortase-conjugation generates a capsule vaccine that protects guinea pigs against Bacillus anthracis. Vaccine 30:3435–3444. doi: 10.1016/j.vaccine.2012.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Duc LH, Hong HA, Atkins HS, Flick-Smith HC, Durrani Z, Rijpkema S, Titball RW, Cutting SM. 2007. Immunization against anthrax using Bacillus subtilis spores expressing the anthrax protective antigen. Vaccine 25:346–355. doi: 10.1016/j.vaccine.2006.07.037. [DOI] [PubMed] [Google Scholar]
- 24.Gauthier YP, Tournier JN, Paucod JC, Corre JP, Mock M, Goossens PL, Vidal DR. 2009. Efficacy of a vaccine based on protective antigen and killed spores against experimental inhalational anthrax. Infect Immun 77:1197–1207. doi: 10.1128/IAI.01217-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rhie GE, Roehrl MH, Mourez M, Collier RJ, Mekalanos JJ, Wang JY. 2003. A dually active anthrax vaccine that confers protection against both bacilli and toxins. Proc Natl Acad Sci U S A 100:10925–10930. doi: 10.1073/pnas.1834478100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tigertt WD. 1980. Anthrax. William Smith Greenfield, M.D., F.R.C.P., Professor Superintendent, the Brown Animal Sanatory Institution (1878-81). Concerning the priority due to him for the production of the first vaccine against anthrax. J Hyg 85:415–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pasteur L. 2002. Summary report of the experiments conducted at Pouilly-le-Fort, near Melun, on the anthrax vaccination, 1881. Yale J Biol Med 75:59–62. [PMC free article] [PubMed] [Google Scholar]
- 28.Turnbull PC, Broster MG, Carman JA, Manchee RJ, Melling J. 1986. Development of antibodies to protective antigen and lethal factor components of anthrax toxin in humans and guinea pigs and their relevance to protective immunity. Infect Immun 52:356–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Welkos SL, Friedlander AM. 1988. Comparative safety and efficacy against Bacillus anthracis of protective antigen and live vaccines in mice. Microb Pathog 5:127–139. doi: 10.1016/0882-4010(88)90015-0. [DOI] [PubMed] [Google Scholar]
- 30.Ivins BE, Welkos SL, Knudson GB, Little SF. 1990. Immunization against anthrax with aromatic compound-dependent (Aro−) mutants of Bacillus anthracis and with recombinant strains of Bacillus subtilis that produce anthrax protective antigen. Infect Immun 58:303–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wright GG, Hedberg MA, Slein JB. 1954. Studies on immunity in anthrax. III. Elaboration of protective antigen in a chemically defined, non-protein medium. J Immunol 72:263–269. [PubMed] [Google Scholar]
- 32.Wright GG, Puziss M, Neely WB. 1962. Studies on immunity in anthrax. IX. Effect of variations in cultural conditions on elaboration of protective antigen by strains of Bacillus anthracis. J Bacteriol 83:515–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Watson DW, Cromartie WJ, Bloom WL, Heckley RJ, McGhee WJ, Weissman N. 1947. Studies on infection with Bacillus anthracis; the isolation of an inflammatory factor from crude extracts of lesions of B. anthracis infection and its biological and chemical relationship to glutamyl polypeptide. J Infect Dis 80:121–136. doi: 10.1093/infdis/80.2.121. [DOI] [PubMed] [Google Scholar]
- 34.Keppie J, Smith H, Harris-Smith PW. 1955. The chemical basis of the virulence of Bacillus anthracis. III. The role of the terminal bacteraemia in death of guinea-pigs from anthrax. Br J Exp Pathol 36:315–322. [PMC free article] [PubMed] [Google Scholar]
- 35.Stanley JL, Smith H. 1961. Purification of factor I and recognition of a third factor of the anthrax toxin. J Gen Microbiol 26:49–66. [DOI] [PubMed] [Google Scholar]
- 36.Tournier JN, Quesnel-Hellmann A, Cleret A, Vidal DR. 2007. Contribution of toxins to the pathogenesis of inhalational anthrax. Cell Microbiol 9:555–565. doi: 10.1111/j.1462-5822.2006.00866.x. [DOI] [PubMed] [Google Scholar]
- 37.Moayeri M, Leppla SH. 2004. The roles of anthrax toxin in pathogenesis. Curr Opin Microbiol 7:19–24. doi: 10.1016/j.mib.2003.12.001. [DOI] [PubMed] [Google Scholar]
- 38.Little SF, Ivins BE, Fellows PF, Friedlander AM. 1997. Passive protection by polyclonal antibodies against Bacillus anthracis infection in guinea pigs. Infect Immun 65:5171–5175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Welkos S, Little S, Friedlander A, Fritz D, Fellows P. 2001. The role of antibodies to Bacillus anthracis and anthrax toxin components in inhibiting the early stages of infection by anthrax spores. Microbiology 147:1677–1685. doi: 10.1099/00221287-147-6-1677. [DOI] [PubMed] [Google Scholar]
- 40.Pittman PR, Mangiafico JA, Rossi CA, Cannon TL, Gibbs PH, Parker GW, Friedlander AM. 2000. Anthrax vaccine: increasing intervals between the first two doses enhances antibody response in humans. Vaccine 19:213–216. doi: 10.1016/S0264-410X(00)00174-2. [DOI] [PubMed] [Google Scholar]
- 41.Pittman PR, Kim-Ahn G, Pifat DY, Coonan K, Gibbs P, Little S, Pace-Templeton JG, Myers R, Parker GW, Friedlander AM. 2002. Anthrax vaccine: immunogenicity and safety of a dose-reduction, route-change comparison study in humans. Vaccine 20:1412–1420. doi: 10.1016/S0264-410X(01)00462-5. [DOI] [PubMed] [Google Scholar]
- 42.Wright JG, Plikaytis BD, Rose CE, Parker SD, Babcock J, Keitel W, El Sahly H, Poland GA, Jacobson RM, Keyserling HL, Semenova VA, Li H, Schiffer J, Dababneh H, Martin SK, Martin SW, Marano N, Messonnier NE, Quinn CP. 2014. Effect of reduced dose schedules and intramuscular injection of anthrax vaccine adsorbed on immunological response and safety profile: a randomized trial. Vaccine 32:1019–1028. doi: 10.1016/j.vaccine.2013.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sever JL, Brenner AI, Gale AD, Lyle JM, Moulton LH, Ward BJ, West DJ. 2004. Safety of anthrax vaccine: an expanded review and evaluation of adverse events reported to the Vaccine Adverse Event Reporting System (VAERS). Pharmacoepidemiol Drug Saf 13:825–840. doi: 10.1002/pds.936. [DOI] [PubMed] [Google Scholar]
- 44.Whiting GC, Rijpkema S, Adams T, Corbel MJ. 2004. Characterisation of adsorbed anthrax vaccine by two-dimensional gel electrophoresis. Vaccine 22:4245–4251. doi: 10.1016/j.vaccine.2004.04.036. [DOI] [PubMed] [Google Scholar]
- 45.Brachman PS, Gold H, Plotkin SA, Fekety FR, Werrin M, Ingraham NR. 1962. Field evaluation of a human anthrax vaccine. Am J Public Health Nations Health 52:632–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zomber G, Reuveny S, Garti N, Shafferman A, Elhanany E. 2005. Effects of spontaneous deamidation on the cytotoxic activity of the Bacillus anthracis protective antigen. J Biol Chem 280:39897–39906. doi: 10.1074/jbc.M508569200. [DOI] [PubMed] [Google Scholar]
- 47.Mazmanian SK, Skaar EP, Gasper AH, Humayun M, Gornicki P, Jelenska J, Joachimiak A, Missiakas DM, Schneewind O. 2003. Passage of heme-iron across the envelope of Staphylococcus aureus. Science 299:906–909. doi: 10.1126/science.1081147. [DOI] [PubMed] [Google Scholar]
- 48.Andrade MA, Ciccarelli FD, Perez-Iratxeta C, Bork P. 2002. NEAT: a domain duplicated in genes near the components of a putative Fe3+ siderophore transporter from Gram-positive pathogenic bacteria. Genome Biol 3:RESEARCH0047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Maresso AW, Garufi G, Schneewind O. 2008. Bacillus anthracis secretes proteins that mediate heme acquisition from hemoglobin. PLoS Pathog 4:e1000132. doi: 10.1371/journal.ppat.1000132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nobles CL, Maresso AW. 2011. The theft of host heme by Gram-positive pathogenic bacteria. Metallomics 3:788–796. doi: 10.1039/c1mt00047k. [DOI] [PubMed] [Google Scholar]
- 51.Honsa ES, Maresso AW, Highlander SK. 2014. Molecular and evolutionary analysis of near-iron transporter (NEAT) domains. PLoS One 9:e104794. doi: 10.1371/journal.pone.0104794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tarlovsky Y, Fabian M, Solomaha E, Honsa E, Olson JS, Maresso AW. 2010. A Bacillus anthracis S-layer homology protein that binds heme and mediates heme delivery to IsdC. J Bacteriol 192:3503–3511. doi: 10.1128/JB.00054-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Balderas MA, Nobles CL, Honsa ES, Alicki ER, Maresso AW. 2012. Hal is a Bacillus anthracis heme acquisition protein. J Bacteriol 194:5513–5521. doi: 10.1128/JB.00685-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Honsa ES, Fabian M, Cardenas AM, Olson JS, Maresso AW. 2011. The five near-iron transporter (NEAT) domain anthrax hemophore, IsdX2, scavenges heme from hemoglobin and transfers heme to the surface protein IsdC. J Biol Chem 286:33652–33660. doi: 10.1074/jbc.M111.241687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Honsa ES, Maresso AW. 2011. Mechanisms of iron import in anthrax. Biometals 24:533–545. doi: 10.1007/s10534-011-9413-x. [DOI] [PubMed] [Google Scholar]
- 56.Ekworomadu MT, Poor CB, Owens CP, Balderas MA, Fabian M, Olson JS, Murphy F, Bakkalbasi E, Honsa ES, He C, Goulding CW, Maresso AW. 2012. Differential function of lip residues in the mechanism and biology of an anthrax hemophore. PLoS Pathog 8:e1002559. doi: 10.1371/journal.ppat.1002559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Maresso AW, Chapa TJ, Schneewind O. 2006. Surface protein IsdC and sortase B are required for heme-iron scavenging of Bacillus anthracis. J Bacteriol 188:8145–8152. doi: 10.1128/JB.01011-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gat O, Grosfeld H, Ariel N, Inbar I, Zaide G, Broder Y, Zvi A, Chitlaru T, Altboum Z, Stein D, Cohen S, Shafferman A. 2006. Search for Bacillus anthracis potential vaccine candidates by a functional genomic-serologic screen. Infect Immun 74:3987–4001. doi: 10.1128/IAI.00174-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Torres VJ, Pishchany G, Humayun M, Schneewind O, Skaar EP. 2006. Staphylococcus aureus IsdB is a hemoglobin receptor required for heme iron utilization. J Bacteriol 188:8421–8429. doi: 10.1128/JB.01335-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kuklin NA, Clark DJ, Secore S, Cook J, Cope LD, McNeely T, Noble L, Brown MJ, Zorman JK, Wang XM, Pancari G, Fan H, Isett K, Burgess B, Bryan J, Brownlow M, George H, Meinz M, Liddell ME, Kelly R, Schultz L, Montgomery D, Onishi J, Losada M, Martin M, Ebert T, Tan CY, Schofield TL, Nagy E, Meineke A, Joyce JG, Kurtz MB, Caulfield MJ, Jansen KU, McClements W, Anderson AS. 2006. A novel Staphylococcus aureus vaccine: iron surface determinant B induces rapid antibody responses in rhesus macaques and specific increased survival in a murine S. aureus sepsis model. Infect Immun 74:2215–2223. doi: 10.1128/IAI.74.4.2215-2223.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Harro CD, Betts RF, Hartzel JS, Onorato MT, Lipka J, Smugar SS, Kartsonis NA. 2012. The immunogenicity and safety of different formulations of a novel Staphylococcus aureus vaccine (V710): results of two phase I studies. Vaccine 30:1729–1736. doi: 10.1016/j.vaccine.2011.12.045. [DOI] [PubMed] [Google Scholar]
- 62.Harro C, Betts R, Orenstein W, Kwak EJ, Greenberg HE, Onorato MT, Hartzel J, Lipka J, DiNubile MJ, Kartsonis N. 2010. Safety and immunogenicity of a novel Staphylococcus aureus vaccine: results from the first study of the vaccine dose range in humans. Clin Vaccine Immunol 17:1868–1874. doi: 10.1128/CVI.00356-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fowler VG, Allen KB, Moreira ED, Moustafa M, Isgro F, Boucher HW, Corey GR, Carmeli Y, Betts R, Hartzel JS, Chan IS, McNeely TB, Kartsonis NA, Guris D, Onorato MT, Smugar SS, DiNubile MJ, Sobanjo-ter Meulen A. 2013. Effect of an investigational vaccine for preventing Staphylococcus aureus infections after cardiothoracic surgery: a randomized trial. JAMA 309:1368–1378. doi: 10.1001/jama.2013.3010. [DOI] [PubMed] [Google Scholar]
- 64.Owens CP, Chim N, Graves AB, Harmston CA, Iniguez A, Contreras H, Liptak MD, Goulding CW. 2013. The Mycobacterium tuberculosis secreted protein Rv0203 transfers heme to membrane proteins MmpL3 and MmpL11. J Biol Chem 288:21714–21728. doi: 10.1074/jbc.M113.453076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Owens CP, Du J, Dawson JH, Goulding CW. 2012. Characterization of heme ligation properties of Rv0203, a secreted heme binding protein involved in Mycobacterium tuberculosis heme uptake. Biochemistry 51:1518–1531. doi: 10.1021/bi2018305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Battye TG, Kontogiannis L, Johnson O, Powell HR, Leslie AG. 2011. iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr D Biol Crystallogr 67:271–281. doi: 10.1107/S0907444910048675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Terwilliger TC, Dimaio F, Read RJ, Baker D, Bunkoczi G, Adams PD, Grosse-Kunstleve RW, Afonine PV, Echols N. 2012. phenix.mr_rosetta: molecular replacement and model rebuilding with Phenix and Rosetta. J Struct Funct Genomics 13:81–90. doi: 10.1007/s10969-012-9129-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Honsa ES, Owens CP, Goulding CW, Maresso AW. 2013. The near-iron transporter (NEAT) domains of the anthrax hemophore IsdX2 require a critical glutamine to extract heme from methemoglobin. J Biol Chem 288:8479–8490. doi: 10.1074/jbc.M112.430009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Grigg JC, Vermeiren CL, Heinrichs DE, Murphy ME. 2007. Haem recognition by a Staphylococcus aureus NEAT domain. Mol Microbiol 63:139–149. doi: 10.1111/j.1365-2958.2006.05502.x. [DOI] [PubMed] [Google Scholar]
- 70.Emsley P, Lohkamp B, Scott WG, Cowtan K. 2010. Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. 2010. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chen VB, Arendall WB III, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. 2010. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr 66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.DeLano WL. 2010. The PyMOL molecular graphics system, version 1.3r1. Schrödinger, LLC, New York, NY. [Google Scholar]
- 74.Reed LJ, Muench H. 1938. A simple method of estimating fifty per cent endpoints. Am J Hyg (Lond) 27:493–497. [Google Scholar]
- 75.National Research Council. 1996. Guide for the care and use of laboratory animals. National Academies Press, Washington, DC. [Google Scholar]
- 76.Ku H. 1966. Notes on the use of propagation of error formulas. J Res Natl Bur Stand Sect C Eng Instrum 70C:263–273. doi: 10.6028/jres.070C.025. [DOI] [Google Scholar]
- 77.Terwilliger A, Swick MC, Pflughoeft KJ, Pomerantsev A, Lyons CR, Koehler TM, Maresso A. 2015. Bacillus anthracis overcomes an amino acid auxotrophy by cleaving host serum proteins. J Bacteriol 197:2400–2411. doi: 10.1128/JB.00073-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Petrovsky N, Aguilar JC. 2004. Vaccine adjuvants: current state and future trends. Immunol Cell Biol 82:488–496. doi: 10.1111/j.0818-9641.2004.01272.x. [DOI] [PubMed] [Google Scholar]
- 79.Kashiwagi Y, Maeda M, Kawashima H, Nakayama T. 2014. Inflammatory responses following intramuscular and subcutaneous immunization with aluminum-adjuvanted or non-adjuvanted vaccines. Vaccine 32:3393–3401. doi: 10.1016/j.vaccine.2014.04.018. [DOI] [PubMed] [Google Scholar]
- 80.Grigg JC, Ukpabi G, Gaudin CF, Murphy ME. 2010. Structural biology of heme binding in the Staphylococcus aureus Isd system. J Inorg Biochem 104:341–348. doi: 10.1016/j.jinorgbio.2009.09.012. [DOI] [PubMed] [Google Scholar]
- 81.Gaudin CF, Grigg JC, Arrieta AL, Murphy ME. 2011. Unique heme-iron coordination by the hemoglobin receptor IsdB of Staphylococcus aureus. Biochemistry 50:5443–5452. doi: 10.1021/bi200369p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Stranger-Jones YK, Bae T, Schneewind O. 2006. Vaccine assembly from surface proteins of Staphylococcus aureus. Proc Natl Acad Sci U S A 103:16942–16947. doi: 10.1073/pnas.0606863103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kim HK, DeDent A, Cheng AG, McAdow M, Bagnoli F, Missiakas DM, Schneewind O. 2010. IsdA and IsdB antibodies protect mice against Staphylococcus aureus abscess formation and lethal challenge. Vaccine 28:6382–6392. doi: 10.1016/j.vaccine.2010.02.097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wang JY, Roehrl MH. 2005. Anthrax vaccine design: strategies to achieve comprehensive protection against spore, bacillus, and toxin. Med Immunol 4:4. doi: 10.1186/1476-9433-4-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Schneerson R, Kubler-Kielb J, Liu TY, Dai ZD, Leppla SH, Yergey A, Backlund P, Shiloach J, Majadly F, Robbins JB. 2003. Poly(gamma-D-glutamic acid) protein conjugates induce IgG antibodies in mice to the capsule of Bacillus anthracis: a potential addition to the anthrax vaccine. Proc Natl Acad Sci U S A 100:8945–8950. doi: 10.1073/pnas.1633512100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wang TT, Fellows PF, Leighton TJ, Lucas AH. 2004. Induction of opsonic antibodies to the gamma-d-glutamic acid capsule of Bacillus anthracis by immunization with a synthetic peptide-carrier protein conjugate. FEMS Immunol Med Microbiol 40:231–237. doi: 10.1016/S0928-8244(03)00366-3. [DOI] [PubMed] [Google Scholar]
- 87.Moustafa M, Aronoff GR, Chandran C, Hartzel JS, Smugar SS, Galphin CM, Mailloux LU, Brown E, Dinubile MJ, Kartsonis NA, Guris D. 2012. Phase IIa study of the immunogenicity and safety of the novel Staphylococcus aureus vaccine V710 in adults with end-stage renal disease receiving hemodialysis. Clin Vaccine Immunol 19:1509–1516. doi: 10.1128/CVI.00034-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Daum RS, Spellberg B. 2012. Progress toward a Staphylococcus aureus vaccine. Clin Infect Dis 54:560–567. doi: 10.1093/cid/cir828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.McNeely TB, Shah NA, Fridman A, Joshi A, Hartzel JS, Keshari RS, Lupu F, DiNubile MJ. 2014. Mortality among recipients of the Merck V710 Staphylococcus aureus vaccine after postoperative S. aureus infections: an analysis of possible contributing host factors. Hum Vaccin Immunother 10:3513–3516. doi: 10.4161/hv.34407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Malmirchegini GR, Sjodt M, Shnitkind S, Sawaya MR, Rosinski J, Newton SM, Klebba PE, Clubb RT. 2014. Novel mechanism of hemin capture by Hbp2, the hemoglobin-binding hemophore from Listeria monocytogenes. J Biol Chem 289:34886–34899. doi: 10.1074/jbc.M114.583013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Watanabe M, Tanaka Y, Suenaga A, Kuroda M, Yao M, Watanabe N, Arisaka F, Ohta T, Tanaka I, Tsumoto K. 2008. Structural basis for multimeric heme complexation through a specific protein-heme interaction: the case of the third neat domain of IsdH from Staphylococcus aureus. J Biol Chem 283:28649–28659. doi: 10.1074/jbc.M803383200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hammerstrom TG, Roh JH, Nikonowicz EP, Koehler TM. 2011. Bacillus anthracis virulence regulator AtxA: oligomeric state, function and CO(2)-signalling. Mol Microbiol 82:634–647. doi: 10.1111/j.1365-2958.2011.07843.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Pflughoeft KJ, Sumby P, Koehler TM. 2011. Bacillus anthracis sin locus and regulation of secreted proteases. J Bacteriol 193:631–639. doi: 10.1128/JB.01083-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.