Abstract
Intrauterine infection is a major detriment for maternal-child health and occurs despite local mechanisms that protect the maternal-fetal interface from a wide variety of pathogens. The bacterial pathogen Listeria monocytogenes causes spontaneous abortion, stillbirth, and preterm labor in humans and serves as a model for placental pathogenesis. Given the unique immunological environment of the maternal-fetal interface, we hypothesized that virulence determinants with placental tropism are required for infection of this tissue. We performed a genomic screen in pregnant guinea pigs that led to the identification of 201 listerial genes important for infection of the placenta but not maternal liver. Among these genes was lmrg1778 (lmo2470), here named inlP, predicted to encode a secreted protein that belongs to the internalin family. InlP is conserved in virulent L. monocytogenes strains but absent in Listeria species that are nonpathogenic for humans. The intracellular life cycle of L. monocytogenes deficient in inlP (ΔinlP) was not impaired. In guinea pigs and mice, InlP increased the placental bacterial burden by a factor of 3 log10 while having only a minor role in other maternal organs. Furthermore, the ΔinlP strain was attenuated in intracellular growth in primary human placental organ cultures and trophoblasts. InlP is a novel virulence factor for listeriosis with a strong tropism for the placenta. This virulence factor represents a tool for the development of new modalities to prevent and treat infection-related pregnancy complications.
INTRODUCTION
The immunological environment of the maternal-fetal interface is unique because protection of the fetus from pathogens has to be balanced with tolerance of the fetus by the maternal immune system (1, 2). How this is accomplished is one of the major enigmas of mammalian reproduction. Contrary to the long-standing hypothesis that the pregnant mother is immunocompromised (3), recent evidence suggests that the maternal immune system is intricately regulated during pregnancy, and the placenta is well guarded against infection (4–6). A few predominantly intracellular microbes are able to infect the placenta and cause pregnancy complications such as preterm labor, fetal damage, and death (5, 7). Given the unique immunological environment of the maternal-fetal interface and the inability of many pathogens to colonize the placenta, we hypothesized that specific virulence determinants are required for microbes to survive and replicate in this tissue.
Listeria monocytogenes is a facultative intracellular bacterial pathogen that causes spontaneous abortion, preterm labor, and stillbirth in humans and other mammals (8, 9). There are ∼1,600 human cases in the United States per year, and about one-third of these cases are pregnancy associated (10). L. monocytogenes is also extremely amenable to experimental analysis and therefore has been exploited over the past 5 decades to understand host-pathogen interactions of intracellular microbes (11, 12). L. monocytogenes can infect a wide variety of phagocytic and nonphagocytic cells. Cell wall surface proteins that belong to the internalin (Inl) family of virulence factors promote bacterial adherence and internalization into nonphagocytic host cells via binding to receptors on the host cell membrane (13). After internalization, the bacterium's intracellular life cycle is facilitated by key virulence determinants that are expressed under the control of the transcriptional regulator PrfA (14): vacuolar escape is mediated largely by listeriolysin O (LLO), with contributions from two phospholipases (PlcA and PlcB) and a metalloproteinase (Mpl). Once in the cytoplasm, ActA orchestrates actin tail formation that allows the bacteria to migrate into cell wall protrusions, which are ingested by neighboring cells, where the life cycle begins anew. Importantly, all pathogens that are able to infect the maternal-fetal interface via the hematogenous route have intracellular life cycles (5, 7). Thus, we chose to exploit L. monocytogenes for two reasons: its importance for human health and its utility as a model for intracellular pathogenesis.
The internalin family contains several important virulence factors (13). Twenty-five members of the internalin family have been identified in L. monocytogenes. All members contain leucine-rich repeats (LRRs), and most are anchored to the bacterial cell wall. The best-characterized internalins are InlA and InlB, which play a critical role in crossing the intestinal barrier (15–17). However, their role in crossing the maternal-fetal barrier has been controversial (18–20). Nine cell wall-bound internalins, in addition to InlA and InlB, have been evaluated, and some of these internalins contribute to the adhesion to and invasion of various cell types and/or virulence in the nonpregnant mouse model (21–24). Four members of the internalin family lack any known cell wall anchor domains. The prototype of this subgroup, InlC, is secreted (25) and was recently shown to modulate innate host immune responses (26). Furthermore, InlC binds to a cytosolic mammalian adaptor protein, which affects cell-to-cell spread (27). To date, the functions of the other secreted internalins are unknown.
Among all the small-animal models, the maternal-fetal interface of guinea pigs resembles that of humans most closely (28). Therefore, we performed a genomic screen in pregnant guinea pigs to identify tissue-specific L. monocytogenes virulence determinants in vivo. We used transposon site hybridization, a negative-selection screen based on transposon mutagenesis and microarray technology, which led to identification of bacterial genes that are required for survival of Mycobacterium tuberculosis in murine liver and spleen (29), chronic infection with Salmonella enterica serovar Typhimurium in mice (30), virulence of Francisella novicida (31), and host specificity of Legionella pneumophila (32).
Among the genes identified as being important for infection of the placenta was lmrg1778 (lmo2470), here named inlP, which is predicted to encode a protein of previously unknown function that belongs to the secreted internalins (13). Our data show that InlP strongly promotes placental infection while having only a minor role in infection of other maternal organs. Discovery of its mechanism of action will unravel virulence strategies required for colonization of the maternal-fetal interface.
MATERIALS AND METHODS
Ethics statement.
For human subjects, this study was conducted according to the principles expressed in the Declaration of Helsinki. The study was approved by the Institutional Review Board (IRB) at the University of California, San Francisco (UCSF), where all experiments were performed (IRB number 11-05530). All patients provided written informed consent for the collection of samples and subsequent analysis. For vertebrate animals, this study was carried out in strict accordance with recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health (33). All protocols were reviewed and approved by the Animal Care and Use Committee (IACUC) at the University of California, San Francisco (IACUC number AN079731-03A).
Bacterial strains.
L. monocytogenes strains used in this study are 10403S (erythromycin susceptible) (34) and DP-L3903 (erythromycin resistant) (35). The transposon mutant library was created as described previously (36) and contained ∼30,000 colonies. Bacterial inocula for infection of guinea pigs (18) and human organ cultures (37) were prepared as described previously.
Bacterial mutant strains generated for this study were constructed by using the temperature-sensitive plasmid pKSV7 as previously described (38). Briefly, ∼500-bp DNA fragments containing the sequences flanking the target genes were amplified. DNA fragments were designed with HindIII/KpnI (5′-CCAATTATCAGGTTTCACATAGA AAGCTTCTAC-3′/5′-GTATATTTTTCAATCTATTTATGGTACCATGAATAATAG-3′) or KpnI/EcoRI (5′-GTATAATCACAATTATGCTACTGGAGGGGTACCCTCTTAT G-3′/5′-CATTATCACGGAGCAAAAGCAGGAATTCAATTAGCGCACG-3′) restriction sites (sequences are underlined). KpnI sites were used for the ligation of the two DNA fragments, and HindIII and EcoRI sites were used for the integration of the fragments into pKSV7, followed by electroporation into 10403S cells. Bacteria were grown at 42°C (restrictive temperature) in the presence of chloramphenicol (7.5 μg/ml). This led to the generation of a merodiploid intermediate. To excise the respective wild-type (WT) gene and cure the plasmid, strains were grown at 30°C (permissive temperature) without chloramphenicol. L. monocytogenes revertants (chloramphenicol susceptible) were tested for the deletion of target genes by PCR. Complementation of deletion strains was performed as previously described, using the pPL2 site-specific integration vector (39). DNA fragments were designed with BamHI/SalI restriction sites (sequences are underlined) (5′-CCGCTCCGGATCCAAGCATCGTTAAATCAAACG-3′ and 5′-GCTGGAAGTCGACAAACTCTGAACTTCCAG-3′).
Microarray hybridizations.
Extraction of genomic DNA (gDNA), enzymatic digestion with AluI and MseI, T7 transcription, and preparation of samples for microarray hybridization were performed as described previously (30). Briefly, gDNA was purified from bacterial cultures by using the Epicentre Gram-positive DNA purification kit (Epicentre Biotechnologies, Madison, WI), replacing lysozyme with mutanolysin (5 U/μl; Sigma-Aldrich, St. Louis, MO). Each gDNA sample was divided in two and incubated separately with AluI or MseI (New England BioLabs, Ipswich, MA). Two micrograms of digested gDNA was used as the template for in vitro transcription with the AmpliScribe T7-Flash transcription kit (Epicentre Biotechnologies) according to the manufacturer's protocol, except that all reaction volumes and reagent concentrations were doubled, and the reaction was allowed to proceed for 16 h. RNA was purified with the MEGAclear kit (Ambion, Austin, TX), and 2 μg was used for a reverse transcriptase (RT) step using SuperScript III with random hexamers (Invitrogen, Grand Island, NY) as primers. Amino-allyl dUTP (Sigma-Aldrich) was incorporated during the RT reaction, followed by CyDye (Amersham Biosciences, Little Chalfont, United Kingdom) conjugation. All samples were hybridized against a common reference in which gDNA from a separate culture of the library grown overnight was used as the template for the in vitro transcription reactions. The reference was conjugated to Cy3, whereas all the samples were conjugated to Cy5. Oligonucleotides (70-mers) for the L. monocytogenes microarray (version 2) were obtained from the J. Craig Venter Institute (http://www.jcvi.org/cms/home/), and microarrays were printed at the Center for Advanced Technology at UCSF (http://cat.ucsf.edu/) (40). A total of 6,247 oligomers were spotted onto each array. Oligonucleotide sequences were based on EGDe (41) (2,843 oligonucleotides) and three food isolates, F6854 (916 oligonucleotides), H7858 (704 oligonucleotides), and F2365 (1,884 oligonucleotides) (42). Hybridizations were performed as described previously (30).
Microarray data analysis.
Microarrays were gridded by using SpotReader software (Niles Scientific) and GenePix Pro 6 (Molecular Devices) and analyzed by using Acuity 4 software (Molecular Devices). The highest-quality spots meeting extra-stringent criteria were identified. These highest-quality spots were used to calculate normalization factors such that the median Cy5/Cy3 ratio was brought to 1. These factors were then applied to the entire data set after removal of low-quality spots. Each experimental sample was zero transformed to the input/input control. To identify transposon mutants whose abundance changed in vivo, microarray data were filtered to include only genes present in all organs and where at least 40% of animals had a signal over the background. The values were then log2 transformed and median centered. Significance analysis of microarrays (SAM) was then used to determine which genes were significantly overrepresented or relatively absent in organs, with significance being a false discovery rate (FDR) of <1%. Significant genes were then exported for further analysis. Statistically overabundant or absent genes in the liver and placenta were clustered by using a Euclidean distance similarity metric and a complete linkage clustering algorithm in Cluster3 (43). Heat maps were then generated by using JavaTree (44).
Eukaryotic cells.
JEG-3 (choriocarcinoma cell line), J774 (mouse macrophage-like cell line), and MDCK (Madin-Darby canine kidney) cells were purchased from ATCC (https://www.atcc.org/) and propagated according to the instructions of ATCC. Human first-trimester placental fibroblasts and human trophoblast progenitor cells (hTPCs) were a generous gift from Susan Fisher and were propagated and differentiated as previously described (45, 46). Mixed glial cell cultures were generated from newborn mice and plated onto poly-d-lysine (Millipore)-coated plates as previously described (47). Intracellular growth curves were performed as previously described (18). Cells were plated and analyzed based on optimal growth conditions for each cell type and/or the preferences of the investigator performing the experiment: JEG-3 and J774 cells were evaluated as monolayers on glass coverslips, primary human first-trimester placental fibroblasts were analyzed as monolayers in 24-well tissue culture plates, undifferentiated hTPCs were grown on 0.5% gelatin (Sigma)-coated 6-well plates and were 80% confluent at the time of infection, and differentiated hTPCs were grown on Matrigel-coated transwell filters and formed regularly interspersed clusters of cells at the time of inoculation. Primary mixed murine glial cell cultures were grown in 24-well plates (Corning) and analyzed as monolayers. MDCK cells were seeded onto 12-mm polycarbonate transwell inserts with 0.4-μm pores (Corning) and grown for 72 h to a density of ∼1.5 × 106 cells/well to form a polarized epithelial monolayer at the time of infection. Cell-to-cell spread was determined as described previously (27). Polarized MDCK cultures were infected with L. monocytogenes wild-type or ΔinlP strain at a multiplicity of infection (MOI) of 140 bacteria per cell. At 8 h postinfection, cells were fixed in fresh 4% paraformaldehyde, rinsed in phosphate-buffered saline (PBS), blocked in 0.7% fish skin gelatin (Sigma), and permeabilized with 0.2% Triton in PBS. Primary staining was performed with rat anti-ZO-1 clone R40.76 (1:300; Santa Cruz Biotechnology) and polyclonal rabbit Listeria O antiserum (1:500; BD Biosciences). Secondary staining was done with CF405M phalloidin (1:40; Biotium, Hayward, CA), Alexa Fluor 488 goat anti-rat (1:500; Thermo Fisher Scientific), and Alexa Fluor 568 goat anti-rabbit (1:500; Thermo Fisher Scientific). Samples were mounted in Vectashield mounting medium (Vector Laboratories) and imaged with a Yokagawa CSU22 spinning-disk confocal microscope (Yokagawa Electrical Corporation, Sugarland TX). Images were analyzed by using FIJI software (48).
Vertebrate animals. (i) Guinea pigs.
For screens, pregnant female Hartley outbred guinea pigs (gestational age, 25 days) were purchased from Elm Hill Breeding Labs (Chelmsford, MA) and injected intravenously (i.v.) with 1 × 109 CFU of L. monocytogenes as previously described (18). Placenta and maternal liver were removed at 24 h postinfection (p.i.). For additional explanations regarding dose and time point, see the legend of Fig. S2 in the supplemental material. Tissues were homogenized in PBS, plated onto brain heart infusion (BHI) agar plates containing 2 μg/ml erythromycin, and incubated overnight at 37°C. Approximately 10,000 CFU from each organ were scraped off the agar plates into PBS, the bacteria were pelleted, and the pellets were resuspended in 5 ml BHI broth–40% glycerol. The initial input inoculum was also plated and collected to account for the effect that this step may have on the complexity of the library. One-milliliter aliquots were frozen at −80°C. For strain-specific i.v. infections, pregnant guinea pigs and nonpregnant female guinea pigs (aged 3 to 4 weeks) were injected i.v. with 5 × 105 CFU. Maternal liver, spleen, and placenta were processed at 24 and 72 h p.i. as described above for CFU enumeration. Placentas harvested at 24 h p.i. were symmetrically halved. One half was used for CFU enumeration, and the other half was fixed in fresh 10% neutral buffered formalin for 24 h and paraffin embedded by using standard techniques. At least two full-thickness sections that included the maternal uterus, implantation site, and placental labyrinth were stained with hematoxylin and eosin (H&E) and evaluated in a blind fashion by a pathologist (G.A.R.). Immunohistochemistry (IHC) using polyclonal rabbit Listeria antisera (BD Difco) was performed by using standard techniques. For strain-specific oral infections, nonpregnant female guinea pigs (aged 3 to 4 weeks) were orally inoculated with 1 × 108 CFU as previously described (49). Liver, spleen, mesenteric lymph nodes, and the distal 30 cm of the small intestine were processed at 7 days p.i. as described above for CFU enumeration.
(ii) Mice.
Male and female C57BL/6J mice aged 6 to 8 weeks were purchased from the Jackson Laboratory. After mating, the timing of pregnancy was evaluated by visualization of a copulation plug (embryonic day 0.5), and the mice were used 9 to 10 days thereafter (embryonic days 9.5 to 10.5). Nonpregnant and pregnant females were injected i.v. with 104 to 105 CFU and euthanized at the indicated time points (24 to 72 h p.i.). Placenta, maternal liver, and spleen were homogenized in PBS and plated onto BHI agar plates, and numbers of bacteria per organ were determined.
Human tissues.
Human placental and decidual organ cultures were prepared and infected as previously described (37, 50). For competitive-index experiments, cultures were infected with a 1:1 ratio of wild-type to specific mutant strains with differential erythromycin susceptibilities, as described previously (51). For microscopy, organ cultures were fixed and stained as previously described (37). Primary antibodies used were mouse anti-HCG-61 clone SC-51606 (1:500; Santa Cruz) and polyclonal rabbit Listeria O antiserum (1:500; BD Biosciences, San Jose, CA). Secondary antibodies used were Alexa Fluor 594 goat anti-mouse or Alexa Fluor 568 goat anti-mouse (1:500; Thermo Fisher Scientific) and Alexa-Fluor 488 goat anti-rabbit (1:500; Thermo Fisher Scientific), nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI) (Affymetrix, Santa Clara, CA), and sections were mounted in Vectashield mounting medium (Vector Laboratories, Burlingame, CA). Stitched high-power images were acquired on a Nikon Ti-E epifluorescence microscope with a DS-Ri2 camera and NIS-Elements 4.30 software (Nikon Corporation, Tokyo, Japan) or with a Keyence BZ-X700 fluorescence microscope and BZ-X Analyzer software (Keyence Corporation of America, Chicago, IL). The area and perimeter of the villous stroma were measured by using FIJI software, and L. monocytogenes bacteria were counted with the Manual Cell Counter plug-in. Brightness and contrast were adjusted with Photoshop (Adobe Systems).
Analysis of Lmrg1778 conservation in Listeria strains.
Homologs of the protein sequence of Lmrg1778 were found by using NCBI BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi) and by searching for highly aligned proteins in the Listeria genus only. Homologous sequences were aligned with CLC Main Workbench 6.7.1 (Qiagen, Alameda, CA) to generate a neighbor-joined phylogram. The Newick file of the phylogram was exported, and ETE Toolkit v3.0 was then used to visualize the phylogram (52).
Statistics.
See above for analysis of microarray data. The Mann-Whitney U test was used to compare bacterial burdens, and a log rank (Mantel-Cox) test was used to compare percentages of infected placentas.
RESULTS
Negative-selection screen for identification of virulence determinants in vivo.
We performed a negative-selection screen to identify L. monocytogenes genes that contribute to organ tropism in vivo (see Fig. S1 in the supplemental material). We used a mariner transposon mutant library (36) for i.v. inoculation of pregnant guinea pigs with 109 CFU. A high dose was chosen because placental colonization is restricted (51, 53), and we wanted to decrease the effect of the bottleneck as much as possible. At 24 h p.i., a time point when the animals still appeared healthy, we compared input pools to output pools by using microarray technology. The input pool consisted of L. monocytogenes transposon mutants grown in BHI broth. The output pools were composed of L. monocytogenes transposon mutants isolated from maternal liver (n = 4) or placenta (n = 8). Genes that were relatively absent in the output pools in comparison to the input pools were considered potentially important for infection in vivo, because bacterial mutants with transposon insertions in these genes were less fit to grow and survive in liver or placenta than in BHI medium. The numbers of genes in this category were 128 and 240 for liver and placenta, respectively (see Fig. S2 and Table S1 in the supplemental material). On the other hand, overabundant genes in the output pools may convey a fitness disadvantage for growth in BHI medium. In this category, we identified 183 genes in the liver but none in the placenta.
We focused on the genes that were relatively absent because we wanted to identify genes that convey an advantage for fitness in vivo. One hundred twenty-eight genes in the liver (4.5% of the genome) were highly reproducible across biological replicates. Among these were key virulence determinants of the core prfA regulon (54), specifically the four genes that encode the proteins that facilitate vacuolar escape: hly, plcA, plcB, and mpl. We did not identify actA, probably because a loss of its function results in a virulence defect at time points later than 24 h (35). We identified additional genes that have been shown to be important for virulence in the murine model of listeriosis, e.g., the fibronectin binding protein fbpA (55) and the enzyme responsible for d-alanine esterification of lipoteichoic and teichoic acids (dltD) (56). In summary, the negative selection of genes encoding previously identified listerial pathogenicity determinants in liver versus BHI broth validated our screen.
In comparison to maternal liver, bacterial seeding of the placenta is relatively restricted (51, 53), which leads to greater variability in the pool of transposon mutants that colonize the placenta at the time of i.v. inoculation. Thus, a relative absence or overabundance of specific genes may reflect the ratio of transposon mutant strains that seeded the placenta at the time of inoculation rather than advantages or disadvantages in bacterial fitness in the placenta. To evaluate the effect of the bottleneck in the placenta, we took advantage of the many mutants that are not required for growth in vivo. Our analysis found that there were 1,935 genes with transposon insertions shared between the liver, placenta, and BHI broth. This represents ∼60% of the coding genes in L. monocytogenes and confirms that there is a bottleneck in the placenta. However, we found that only 240 genes were significantly absent in the placenta versus BHI broth. Since passage through a bottleneck is a random process, the finding that 240 genes (8.4% of the genome) were relatively absent in eight biological replicates is most likely due to negative selection.
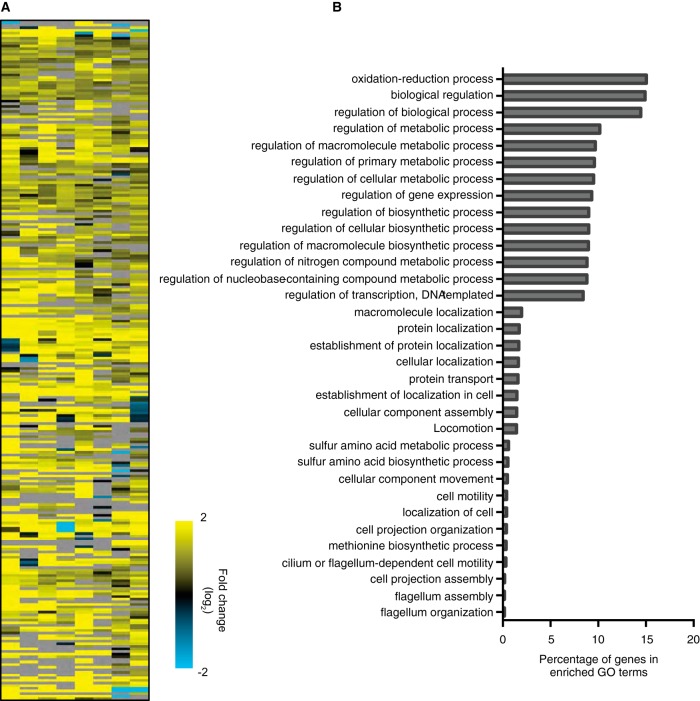
To identify virulence factors with placental tropism, we compared the genes that were relatively absent in the placenta versus BHI medium to the genes that were not necessary for survival in the liver (see Fig. S3 in the supplemental material). This group contained 201 genes, which fell into 33 broad categories based on gene ontology (Fig. 1; see also Table S1 in the supplemental material). The 3 largest categories encompassed genes with functions related to oxidation reduction and the regulation of biological processes, followed by 10 categories with genes involved in metabolic processes. A smaller percentage of relevant genes were involved in cellular motility and flagellum organization.
FIG 1.
Negative selection of L. monocytogenes mutants in vivo. Pregnant guinea pigs were injected i.v. with 109 CFU of the transposon mutant library. Bacteria surviving at 24 h p.i. in maternal liver (n = 4) and placenta (n = 8) were collected. The transposon mutants present in each sample were identified by microarray analysis. Data for the genes negatively selected in placenta only are represented. (A) Heat map of genes relatively absent in placenta versus BHI broth that are also not necessary for survival in the liver. Yellow represents a relative absence of the signal in the placenta compared with the liver, and blue represents a relative overabundance of the signal in the placenta compared with the liver. (B) Role category diagram representing the distribution of genes relatively absent in placenta, shown in panel A, according to gene ontology (GO) categories.
InlP is highly conserved in L. monocytogenes.
Among the genes specifically important for placental infection was lmrg1778 (lmo2470), here named inlP, which is predicted to encode a protein (388 amino acids [aa]) with five or six LRRs. The presence of a signal peptide but the lack of a predicted cell wall-anchoring domain places InlP in the category of secreted internalins, which has four known members (13). The prototype of this subgroup, InlC, is secreted, modifies innate host immune responses (26), and boosts cell-to-cell spread (27) via binding to cytosolic host proteins. The other three secreted internalins share little homology to InlC, and their functions for virulence in vivo were previously unknown. Phylogenetic analysis of InlP identified closely related homologs as being overrepresented in the human pathogen L. monocytogenes and absent in strains not pathogenic for humans (see Fig. S4 in the supplemental material).
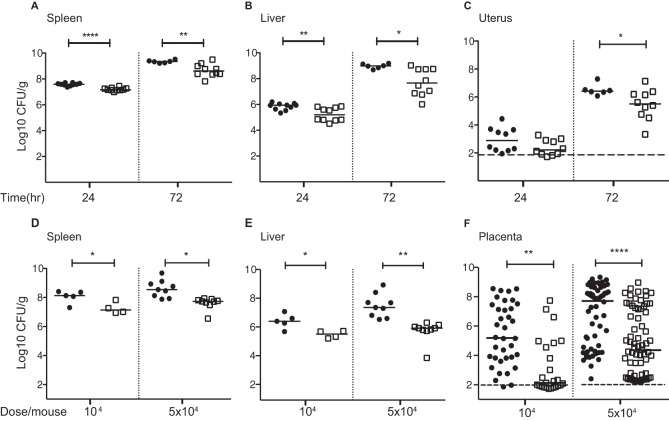
Therefore, we decided to investigate the role of InlP in the pathogenesis of listeriosis. We generated an isogenic mutant strain of L. monocytogenes strain 10403S (wild type) deficient in inlP (ΔinlP). The phenotype of the ΔinlP strain did not differ from that of the wild type in standard in vitro assays, including intracellular growth and cell-to-cell spread (see Fig. S5 in the supplemental material). Next, we infected nonpregnant mice with either the wild-type or the ΔinlP strain. Twenty-four hours after i.v. inoculation with 104 CFU, the bacterial burdens in liver and spleen did not differ in wild-type- versus ΔinlP mutant-infected animals (see Fig. S6A and S6B in the supplemental material). When the inoculum was increased to 7 × 104 CFU, the ΔinlP mutant was slightly attenuated in spleen (0.5 log10) and liver (0.4 log10) (Fig. 2A and B), a difference that was restored by isogenic complementation of the ΔinlP mutant with inlP (see Fig. S6C in the supplemental material). The attenuation of the ΔinlP mutant was more pronounced at 72 h p.i., as evidenced by 0.7 and 1.3 log virulence defects in spleen and liver, respectively (Fig. 2A and B) as well as a difference in survival (see Fig. S6D in the supplemental material). The wild-type and ΔinlP strains did not differ in their ability to colonize the nonpregnant uterus at 24 h p.i., suggesting that InlP does not affect trafficking. However, the ΔinlP strain was attenuated in virulence in the uterus by 1 log10 at 72 h p.i. (Fig. 2C). We concluded that InlP is a novel virulence factor for systemic infection in mice.
FIG 2.
InlP is a virulence determinant in mice. Shown are data for intravenous infection of nonpregnant (A to C) and pregnant (D to F) mice with the wild-type (black circles) or ΔinlP (empty squares) strain. (A to C) CFU per gram in spleen (A), liver (B), and uterus (C) 24 and 72 h p.i. with 7 × 104 CFU. (D to F) CFU per gram in spleen (D), liver (E), and (F) placenta 48 h p.i. with 104 CFU or 5 × 104 CFU. Total numbers of placentas were 36 for wild-type and 25 for ΔinlP mutant infection with an inoculum of 104 CFU and 62 for wild-type and 66 for ΔinlP mutant infection with an inoculum of 5 × 104 CFU. Mann-Whitney U test was used for statistical analysis (*, P < 0.05; **, P < 0.005; ****, P < 0.0001). Horizontal bars indicate medians, and horizontal dashed lines indicate the detection limit.
InlP strongly promotes placental infection in vivo.
To test the role of InlP during pregnancy, we inoculated pregnant mice i.v. on day 10.5 of gestation with 104 or 5 × 104 CFU of the wild-type or ΔinlP strain. At 48 h p.i., the ΔinlP mutant was attenuated in maternal spleen (1 log10) and liver (0.8 to 1.7 log10) (Fig. 2D and E). In contrast to the liver and spleen, bacterial colonization of the placenta is restricted, leading to a much higher variability in the bacterial burden among the placentas from each animal. To demonstrate this variability, we randomly picked four wild-type-infected pregnant mice and showed CFU in their placentas in individual plots (see Fig. S6E and S6F in the supplemental material). Gross examination of gravid uteri from mice infected with 1 × 104 CFU showed widespread hemorrhages in wild-type- but not ΔinlP mutant-infected fetoplacental units (see Fig. S6G in the supplemental material). Mice given ΔinlP bacteria had fewer infected placentas than did mice given the wild type, 52% (13/25) versus 94% (34/36) for infection with 1 × 104 CFU and 81% (54/66) versus 100% (62/62) for infection with 5 × 104 CFU. These differences reached statistical significance for both doses (P < 0.005). The median number of CFU per gram was reduced by 3 log10 in the placentas of animals infected with the ΔinlP strain in comparison to the wild type regardless of the infectious dose (Fig. 2F).
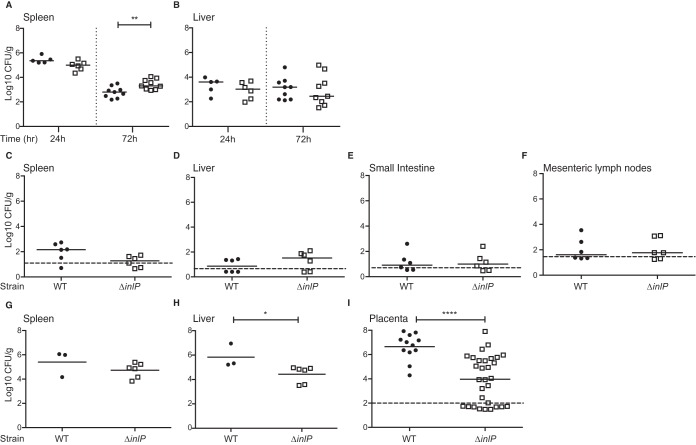
Next, we evaluated the phenotype of the ΔinlP strain in guinea pigs. First, we infected nonpregnant animals i.v. with 5 × 105 CFU or orally with 1 × 108 CFU of the wild-type or ΔinlP strain. We did not observe a difference between the wild-type and ΔinlP strains in liver, spleen, small intestine, and mesenteric lymph nodes except for the spleen 72 h after i.v. inoculation, when the bacterial burden of the ΔinlP strain was 5-fold higher than that of the wild-type strain (Fig. 3A to F). Next, we infected pregnant guinea pigs i.v. on day 32 of gestation with 5 × 105 CFU of the wild-type or ΔinlP strain. At 72 h p.i., the wild-type and ΔinlP strains were equally virulent in maternal spleen, and the ΔinlP strain was attenuated by 0.5 log10 in maternal liver (P < 0.05) (Fig. 3G and H). The percentages of infected placentas were 100% (12/12) and 67% (20/30) for the wild-type and ΔinlP strains, respectively (P < 0.01), and the median numbers of CFU per gram were 3 log10 lower in ΔinlP mutant-infected than in wild-type-infected placentas (P < 0.0001) (Fig. 3I). In conclusion, data from both rodent models suggest that InlP strongly (1,000-fold) promotes placental infection while having only minor effects on other tissues.
FIG 3.
InlP is a virulence determinant with strong placental tropism in pregnant guinea pigs. Nonpregnant (A to F) and pregnant (G to I) guinea pigs were infected i.v. (A, B, and G to I) with 5 × 105 CFU or orally (C to F) with 1 × 108 CFU of the wild-type (black circles) or ΔinlP (empty squares) strain. (A and B) CFU per gram in spleen (A) and liver (B) of nonpregnant guinea pigs 24 and 72 h after i.v. inoculation. (C to F) CFU per gram in spleen (C), liver (D), small intestine (E), and mesenteric lymph nodes (F) of nonpregnant guinea pigs 7 days after oral inoculation. (G to I) CFU per gram in spleen (G), liver (H), and placenta (I) of pregnant guinea pigs 72 h after i.v. inoculation. The total numbers of placentas are 12 for wild-type-infected and 30 for ΔinlP mutant-infected animals. Data for each group represent the results of at least 2 independent experiments. Mann-Whitney U test was used for statistical analysis (*, P < 0.05; **, P < 0.005; ****, P < 0.0001). The detection limit varies between organs and is between 2 and 100 CFU/g per organ (horizontal dashed lines). Horizontal bars indicate medians.
Histological examination of the maternal-fetal interface in guinea pigs.
During pregnancy, fetal trophoblasts invade the decidua (uterine implantation site) and remodel the maternal arteries. Consequently, maternal blood flows into the placenta, where nutrient and gas exchanges between mother and fetus occur. We and others previously demonstrated that infection of the maternal-fetal interface begins in decidua and invasive trophoblasts (37, 57). To further investigate the role of InlP, we microscopically evaluated the placenta and decidua of guinea pigs infected with the wild-type or ΔinlP strain (Fig. 4A). The median bacterial burdens in these tissues were 1.6 × 106 CFU/g (range, 1.9 × 104 to 6.3 × 107 CFU/g) and 3.9 × 104 CFU/g (range, 1 × 102 to 7.9 × 107 CFU/g) for the wild-type and ΔinlP strains, respectively (CFU data are included in Fig. 3). Sections of the maternal-fetal interface were stained with H&E or by IHC with antisera against L. monocytogenes (2 sections per placenta). Focal collections of acute inflammatory cells, predominantly neutrophils, with various degrees of necrotic debris were identified in the placenta (Fig. 4B) and within the walls of the decidual vasculature (Fig. 4D). IHC revealed large aggregates of L. monocytogenes in most of the lesions (Fig. 4C). The microscopic appearances of the lesions appeared similar for both strains, but the median numbers of inflammatory lesions per tissue (placenta and decidua) were 3 and 0 for wild-type- and ΔinlP mutant-infected animals, respectively (Fig. 4F). This difference did not reach statistical significance (P = 0.11) but was consistent with the observed difference in bacterial burdens between the two strains in the placenta, which was statistically significant (Fig. 3I). However, the most striking difference between the wild type and the ΔinlP mutant was the degree of decidual necrosis, which was scored on a scale from 1 (healthy to minimal) to 4 (severe) (Fig. 4G and H; see also Table S2 in the supplemental material). The median decidual necrosis scores were 4 and 2 for wild-type- and ΔinlP mutant-infected deciduas, respectively (P = 0.007) (Fig. 4I).
FIG 4.
Histological evaluation of the maternal-fetal interface of guinea pigs infected with wild type (n = 8) or ΔinlP mutant (n = 13) for 24 h. (A) Whole-tissue section showing the anatomy of the maternal-fetal interface, including placenta (black diamond), decidua (*), maternal arteries (MA), and umbilical cord (UC). (B) H&E staining of a representative placental lesion composed of neutrophils, mononuclear cells, and necrotic debris. (C) IHC of the lesion shown in panel B demonstrating abundant L. monocytogenes bacteria. (D and E) Inflammatory lesions (*) in walls of maternal decidual arterioles (MA labels the lumen) (D) versus unaffected arterioles (MA labels the lumen) (E). (F) Numbers of lesions per placenta and decidua for wild-type (WT) (black circles) versus ΔinlP mutant (empty squares) infection. (G and H) Representative images of decidual necrosis with a score of 1 (G) versus a score of 4 (H). The contiguous zone of coagulative necrosis is >2 mm long (*). (I) Decidual necrosis scores for wild-type (black circles) versus ΔinlP mutant (empty squares) infection. Decidual necrosis was scored as follows: 1 for normal to minimal, 2 for mild, 3 for moderate, and 4 for severe (see Table S2 in the supplemental material for a detailed description of scoring). Bars, 6 mm (A), 100 μm (B to E), and 200 μm (G and H). Mann-Whitney U test was used for statistical analysis (**, P = 0.007). Horizontal bars in panels F and I indicate medians.
Role of InlP in infection of primary human placental organ cultures.
To further investigate the role of InlP in infection of the human maternal-fetal interface, we utilized primary human organ cultures. Since infection begins in the decidua (5), we first compared wild-type and ΔinlP mutant infections in decidual organ cultures, which showed no statistically significant difference between these two strains (see Fig. S7A in the supplemental material).
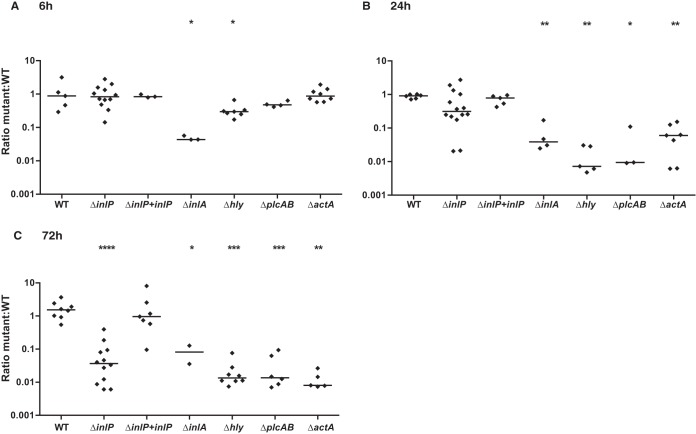
Next, we analyzed infection of placental organ cultures (37, 58). These organ cultures are derived from first-trimester placentas and vary in shape and size. Therefore, we used the competitive-index assay to determine the role of InlP in infection of primary human placental tissues. We infected placental organ cultures with the ΔinlP and wild-type strains at a 1:1 ratio, with each strain having differential susceptibilities to erythromycin (35). Equal colonization rates at 6 h p.i. suggested that InlP does not have a functional role in host cell invasion (Fig. 5A). Over time, the intracellular growth of the ΔinlP mutant decreased, resulting in a >1-log10 attenuation at 72 h p.i. (P < 0.0001) (Fig. 5C). It is possible that InlP secreted by wild-type bacteria can compensate for the lack of InlP in the ΔinlP strain in these experiments. In this case, the observed virulence defect of the ΔinlP strain in comparison to wild-type L. monocytogenes might be an underestimation. Isogenic complementation of the ΔinlP strain with inlP restored the wild-type phenotype (Fig. 5A to C).
FIG 5.
Role of virulence factors in human placenta. Primary human placental organ cultures were infected with a 1:1 ratio of WT (erythromycin resistant) and WT (erythromycin susceptible) or the following mutant strains (all erythromycin susceptible): ΔinlP, ΔinlP mutant complemented with inlP, ΔinlA, Δhly, ΔplcAB, and ΔactA. The ratio of erythromycin-resistant to erythromycin-susceptible CFU was determined at 6 h p.i. (A), 24 h p.i. (B), and 72 h p.i. (C). Mann-Whitney U test was used for statistical analysis (*, P < 0.05; **, P < 0.005; ***, P < 0.001; ****, P < 0.0001). Horizontal bars indicate medians.
For comparison, we coinfected primary human placental organ cultures with wild-type L. monocytogenes and L. monocytogenes strains deficient in known key virulence factors at a 1:1 ratio. All of the deletion mutants were significantly attenuated (Fig. 5A to C). An L. monocytogenes strain deficient in InlA was 1-log10 attenuated at 6, 24, and 72 h p.i., consistent with a defect in invasion and normal intracellular growth. The attenuation of mutants with an impaired intracellular life cycle was more pronounced at later time points: L. monocytogenes strains defective in vacuolar escape due to a lack of LLO (Δhly) or PlcAB were 3-fold attenuated at 6 h p.i., a difference that increased to 1 to 2 log10 at 24 h and 72 h p.i.; the L. monocytogenes strain deficient in ActA was equally as virulent as the wild type at 6 h p.i. and was attenuated by 1 log10 and 2 log10 at 24 and 72 h p.i., respectively. Thus, the degree of attenuation of ΔinlP mutant infection in comparison to wild-type infection of primary human placental tissue is within the range of attenuation of previously known key virulence determinants.
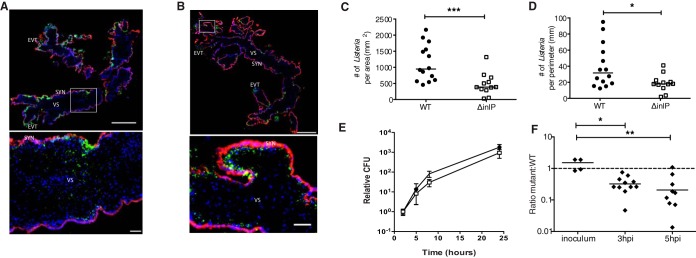
Next, we examined these placental organ cultures by immunofluorescence microscopy. As previously described, L. monocytogenes infection starts in extravillous trophoblasts (EVTs), spreads along subsyncytial trophoblasts, and reaches the villous stroma by 72 h p.i. (37). This pattern of invasion and spread was observed with the wild-type and ΔinlP strains (Fig. 6A and B). However, the intracellular growth and/or spread of the ΔinlP mutant was impaired, as demonstrated by the significantly lower numbers of ΔinlP bacteria than of wild-type bacteria per area or perimeter of villous stroma (Fig. 6C and D). In contrast, the ΔinlP mutant was not attenuated for growth and spread during infection of isolated primary human placental fibroblasts (Fig. 6E).
FIG 6.
InlP is important for virulence in human placenta. (A to D) Human placental organ cultures from 3 donors were analyzed 72 h p.i. with either wild type (WT) or ΔinlP mutant. (A) Representative image of a WT-infected placental organ culture stained for L. monocytogenes (green), βHCG (human chorionic gonadotropin) (to stain syncytiotrophoblasts) (red), and DAPI (blue). The rectangle in the top image denotes the location of the high-power image at the bottom. SYN, syncytiotrophoblast; EVT, extravillous trophoblast; VS, villous stroma. (B) Representative image of a placental organ culture infected with ΔinlP mutant and stained as described above for panel A. (C and D) Numbers of L. monocytogenes bacteria present per area (C) and perimeter (D) of villous stroma quantified by immunofluorescence microscopy for 14 WT- and 12 ΔinlP mutant-infected organ cultures. (E) Intracellular growth of the WT (black circles) and ΔinlP mutant (empty squares) in primary human placental fibroblasts as determined by a gentamicin protection assay (averages of data from two independent experiments performed in triplicate). Error bars indicate standard errors of the means. (F) Competitive index in differentiated primary human trophoblast progenitor cells infected with erythromycin-resistant WT and erythromycin-susceptible ΔinlP mutant bacteria at a 1:1 ratio. Mann-Whitney U test was used for statistical analysis of data in panels C to E, and a Kruskal-Wallis test with multiple comparisons was used for analysis of data in panel F (*, P < 0.05; **, P < 0.005; ***, P < 0.001). Horizontal bars indicate medians. The dashed line represents a ratio of 1. Bars, 500 μm (low-power images) and 50 μm (high-power insets).
Since escaping the growth restriction in EVTs is the first hurdle that L. monocytogenes has to overcome when colonizing the placenta (59), we tested whether InlP promotes intracellular growth in EVTs. For this purpose, we utilized primary human trophoblast progenitor cells (hTPCs), which can be differentiated in vitro to resemble EVTs (45). Although wild-type L. monocytogenes grows well in undifferentiated hTPCs, its growth is reduced in differentiated hTPCs (see Fig. S7B in the supplemental material), consistent with previously reported observations in human organ cultures showing that less-differentiated mononuclear trophoblasts are more permissive for bacterial growth than EVTs (59). In undifferentiated hTPCs, the ΔinlP strain behaved like the wild-type strain (see Fig. S7C in the supplemental material). In contrast, infection of differentiated hTPCs with ΔinlP and wild-type bacteria at a 1:1 ratio revealed reduced intracellular growth of the ΔinlP strain in comparison to the wild type (Fig. 6F), suggesting that InlP contributes to overcoming the EVT barrier.
Finally, we tested the role of InlP in infection of primary murine mixed glial cell cultures, which were used previously to evaluate target cells for L. monocytogenes infection of the central nervous system (60). Intracellular growth curves for mixed glial cells showed no difference between the wild-type and ΔinlP strains (see Fig. S7D in the supplemental material).
DISCUSSION
The results of this study demonstrate that InlP strongly promotes placental infection in two pregnant rodent models. The systemic defects observed in nonpregnant mice validate InlP as a novel virulence factor and suggest that InlP could be important for bacterial virulence in other tissues as well. In primary human placental organ cultures, the ΔinlP strain was significantly attenuated in competitive-index experiments against wild-type L. monocytogenes. This degree of attenuation is comparable to that of an L. monocytogenes strain deficient in LLO, one of the most attenuated strains in vivo, due to its impaired intracellular life cycle (51, 61). In summary, the highlight of this study is the discovery of InlP, a novel virulence factor for listeriosis with strong placental tropism.
Most likely, there are other genes among the 201 hits of our screen that confer bacterial fitness with a tropism for the placenta. One of the largest groups of genes was related to oxidation reduction processes and includes, e.g., mrsA and fri. MrsA (methionine S-sulfoxide reductase) repairs proteins that have been oxidatively damaged by the formation of methionine sulfoxides (62), and Fri (ferritin) protects DNA from oxidative damage (63). Interestingly, mammalian development occurs in physiological hypoxia (64), and the placenta produces many reactive oxygen species (65). Thus, placental pathogens that are able to withstand oxidative stress can adapt to the placental niche. The results of our screen also included genes that encode the highly conserved flagellar export apparatus (66). This was surprising because most L. monocytogenes strains do not produce flagella at 37°C (67). However, 2% of L. monocytogenes 10403S bacteria express flagella when grown at 37°C (68), and among 100 human clinical isolates, 20 strains had flagellar motility at 37°C (69). Although flagellin is not essential for systemic listeriosis in mice (70), perhaps it contributes to placental virulence.
InlP is conserved in L. monocytogenes. Listeria strains without homology to InlP are not associated with human disease, including L. innocua (41) and L. ivanovii (71). We did not find any other prokaryotes that contain sequences with homology to InlP, suggesting that this virulence factor is specific for the pathogenesis of listeriosis. InlP appears to be particularly important for placental infection compared to infection of liver, spleen, nonpregnant uterus, small intestine, and mesenteric lymph nodes in vivo and primary murine mixed glial cell cultures in vitro. There is typically a large amount of variability in placental infections even among all the placentas from one animal (see Fig. S6E and S6F in the supplemental material) (19, 72). The variability in placental CFU is due to the bottleneck in placental colonization, which does not seem to differ between wild-type and ΔinlP strains. However, the net bacterial burden of the ΔinlP strain in the placenta is 3 log10 lower than that in wild-type-infected placentas. The bacterial burden is a composite of bacterial replication, elimination, and trafficking. Typically, the net bacterial burden reaches stationary phase before beginning to decrease. It is likely that bacteria in highly infected placentas have reached stationary phase, and even wild-type bacteria cannot continue to grow any further due to limits in the nutrients available and/or immune responses that increase bacterial elimination.
InlP belongs to the internalin family, which includes InlA and InlB. Evaluation of the roles of InlA and InlB in placental infection in vivo has been complicated by the species specificity of the receptor interactions: both internalins bind to human and gerbil E-cadherin and Met, but InlA and InlB have a reduced affinity for murine E-cadherin (73) and guinea pig cMet (74), respectively. InlA plays no role in placental infection of guinea pigs (18), perhaps due to the lack of an interaction between InlB and guinea pig Met (75). However, in three pregnant rodent models with functional InlA- and InlB-host cell receptor interactions, the role of InlA and InlB in placental infection, if present, is small: placental infection of mice injected with L. monocytogenes expressing InlA engineered to interact with murine E-cadherin does not differ from wild-type infection (19). Infection of pregnant gerbils or transgenic mice expressing human E-cadherin with L. monocytogenes bacteria deficient in InlA, InlB, or both revealed an ∼0.5-log virulence defect in the placenta in comparison to wild type (20). We previously infected primary human placental organ cultures with ΔinlA, ΔinlB, or ΔinlAB bacteria (see Fig. 4B in reference 37), which showed significantly decreased placental invasion of the ΔinlA strain in comparison to wild type. We did not observe an effect of the ΔinlB mutant on invasion of human placental organ cultures, and the ΔinlAB double-deletion mutant did not differ significantly from the ΔinlA mutant. Thus, in this study, we compared the roles of InlA and InlP in infection of placental organ cultures in a competitive-index assay (Fig. 5).
Our results from placental cell and organ culture models do not support a significant role for InlP in host cell invasion because CFU at early time points were similar for the wild-type and ΔinlP strains, while the ΔinlA strain was attenuated 2 log10 in invasion of human trophoblasts that express E-cadherin in vitro as well as primary human placental organ cultures (Fig. 5A) (18, 76). In contrast, InlP promotes bacterial pathogenesis at stages downstream of host cell invasion, as evidenced by the reduced intracellular growth in differentiated human trophoblasts and human placental organ cultures. It is possible that InlP and InlA may have synergistic or additive effects for infection in the placenta, and further studies will be performed to evaluate this hypothesis.
What is the reason for the observed discrepancy in InlA's importance for trophoblast invasion in vitro and placental infection in vivo? In addition to internalin-mediated host cell invasion, L. monocytogenes can spread from cell to cell via actin-based motility. Interestingly, previously reported studies in mice and guinea pigs indicate that most of the bacterial burden in vivo is inside host cells (53, 77). These studies suggest that cell-to-cell spread is an important mechanism for placental infection in vivo and can compensate for a lack of InlA–E-cadherin and/or InlB-cMet interactions.
Like InlC, InlP belongs to the small subgroup of secreted internalins. Thus, potential functional roles can be inferred for InlP in comparison to InlC. Interestingly, there is evidence that InlC interacts directly with IκB kinase α (IKKα) and modifies innate host cell responses to infection (26). Thus, an intriguing possible functional role for InlP is in interference with specific host defense pathways via direct binding to host cell proteins, which in turn could lead to decreased intracellular bacterial replication or survival. InlC also binds to Tuba, a cytosolic mammalian adaptor protein, to promote bacterial cell-to-cell spread (27). We previously observed that L. monocytogenes grows to a high density in subsyncytial trophoblasts, whereas only very few bacteria reach the villous stroma, and suggested that the basal membrane represents another barrier against listerial spread into deeper layers of the placenta (37). In placental organ cultures infected with the wild-type or ΔinlP strain, the appearance of infected subsyncytial trophoblasts did not differ. Both strains heavily infected subsyncytial trophoblasts, but the number of bacteria that reached the villous stroma was lower for ΔinlP mutant infection than for wild-type infection. While it is still possible that InlP facilitates cell-to-cell spread in subsyncytial trophoblasts, we did not observe such a role for InlP in MDCK cells (see Fig. S5D and S5E in the supplemental material). Thus, we favor the hypothesis that InlP is involved in promoting bacterial spread across the basement membrane. Once L. monocytogenes has reached the villous stroma, it can grow in fetal placental trophoblasts whether InlP is present or not (Fig. 6E). Further studies to evaluate the mechanistic roles of InlP are currently in progress.
There are several possibilities for the observed organ tropism of InlP. One possibility is organ-specific expression of host protein binding partners. On the other hand, evidence is mounting that the metabolic environment of the host cell can regulate the expression and/or activity of bacterial virulence determinants (78). Thus, an alternate possibility is that InlP is regulated by host signals that are differentially expressed in maternal liver and spleen versus placenta.
Our detailed histological examination of the maternal-fetal interface of infected guinea pig placentas at 24 h p.i. demonstrated inflammatory lesions with abundant bacteria and neutrophils, consistent with data from previous studies in pregnant mice (57). The appearances of inflammatory lesions were similar in ΔinlP and wild-type infections. It is possible that evaluation by flow cytometry or at additional time points would uncover distinct differences in immune cell composition. On the other hand, the finding that the ΔinlP strain is attenuated in human placental organ cultures argues that the effects of InlP, at least in part, occur independently of the influx of maternal immune cells. The most striking histological difference between ΔinlP and wild-type infections was the degree of decidual necrosis. Tissue necrosis in vivo correlates with a widespread infiltration of neutrophils that are recruited to contain bacterial infection (79). Consistent with this, we observed bacterial foci, neutrophil infiltration, and tissue necrosis in the decidua (Fig. 4). This does not occur in an ex vivo system such as decidual organ cultures. Thus, the lack of neutrophil recruitment and tissue necrosis in decidual organ cultures (see Fig. S7A in the supplemental material) may have affected the bacterial burden. It is interesting to note that previous reports by Redline and Lu described decidual necrosis in pseudopregnant mice after intrauterine inoculation with L. monocytogenes as well (57). Furthermore, decidual senescence and necrosis have been associated with pregnancy complications, including preterm labor (80, 81), one of the clinical manifestations of pregnancy-associated listeriosis. Despite the high rates of morbidity and mortality associated with preterm birth, the underlying molecular mechanisms of preterm labor remain largely undefined (82).
In summary, we have discovered a novel virulence determinant for L. monocytogenes that is predicted to belong to the secreted internalins. Although InlP contributes to systemic virulence in mice, the phenotype is strongest in the placenta in two rodent models. InlP represents a novel tool that we can use to understand host-pathogen interactions at the maternal-fetal interface and may serve as a springboard for the development of new modalities to prevent and treat infection-related pregnancy complications such as preterm labor.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Sandi Wong and Fay Pon for technical assistance; Anita Sil, Mark Voorhies, and David Weiss for helpful discussions; Susan Fisher, Tippi Mackenzie, Shaeri Mukherjee, Adrian Erlebacher, Joanne Engel, and Stephen Gitelman for critical reading of the manuscript; and Cedric Brimacombe for useful comments. hTPCs were a generous gift from Susan Fisher. We acknowledge Mark Weinstein and the San Francisco General Hospital Pathology Department for expert assistance with immunohistochemistry.
This work was supported by National Institutes of Health grant R01AI084928 and Burroughs Wellcome Fund grant 41259 to A.I.B. G.A.R. is supported by grant F32AI108195, a Society for Pediatric Pathology Young Investigator Research grant, and the University of California Partnerships for Faculty Diversity President's Postdoctoral Fellowship. C.C. received support from grant T32AI060537. D.E.L. received support from grants T32AI060537 and F32AI120676.
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.00625-16.
REFERENCES
- 1.Erlebacher A. 2013. Immunology of the maternal-fetal interface. Annu Rev Immunol 31:387–411. doi: 10.1146/annurev-immunol-032712-100003. [DOI] [PubMed] [Google Scholar]
- 2.Zeldovich VB, Bakardjiev AI. 2012. Host defense and tolerance: unique challenges in the placenta. PLoS Pathog 8:e1002804. doi: 10.1371/journal.ppat.1002804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Medawar PB. 1953. Some immunological and endocrinological problems raised by the evolution of viviparity in vertebrates. Symp Soc Exp Biol 7:320–338. [Google Scholar]
- 4.Mor G, Cardenas I. 2010. The immune system in pregnancy: a unique complexity. Am J Reprod Immunol 63:425–433. doi: 10.1111/j.1600-0897.2010.00836.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Robbins JR, Bakardjiev AI. 2012. Pathogens and the placental fortress. Curr Opin Microbiol 15:36–43. doi: 10.1016/j.mib.2011.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jiang TT, Chaturvedi V, Ertelt JM, Kinder JM, Clark DR, Valent AM, Xin L, Way SS. 2014. Regulatory T cells: new keys for further unlocking the enigma of fetal tolerance and pregnancy complications. J Immunol 192:4949–4956. doi: 10.4049/jimmunol.1400498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vigliani MB, Bakardjiev AI. 2013. First trimester typhoid fever with vertical transmission of Salmonella Typhi, an intracellular organism. Case Rep Med 2013:973297. doi: 10.1155/2013/973297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lamont RF, Sobel J, Mazaki-Tovi S, Kusanovic JP, Vaisbuch E, Kim SK, Uldbjerg N, Romero R. 2011. Listeriosis in human pregnancy: a systematic review. J Perinat Med 39:227–236. doi: 10.1515/JPM.2011.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mylonakis E, Paliou M, Hohmann EL, Calderwood SB, Wing EJ. 2002. Listeriosis during pregnancy: a case series and review of 222 cases. Medicine (Baltimore) 81:260–269. doi: 10.1097/00005792-200207000-00002. [DOI] [PubMed] [Google Scholar]
- 10.Scallan E, Hoekstra RM, Angulo FJ, Tauxe RV, Widdowson MA, Roy SL, Jones JL, Griffin PM. 2011. Foodborne illness acquired in the United States—major pathogens. Emerg Infect Dis 17:7–15. doi: 10.3201/eid1701.P11101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cossart P, Lebreton A. 6 June 2014. A trip in the “New Microbiology” with the bacterial pathogen Listeria monocytogenes. FEBS Lett doi: 10.1016/j.febslet.2014.05.051. [DOI] [PubMed] [Google Scholar]
- 12.Witte CE, Archer KA, Rae CS, Sauer JD, Woodward JJ, Portnoy DA. 2012. Innate immune pathways triggered by Listeria monocytogenes and their role in the induction of cell-mediated immunity. Adv Immunol 113:135–156. doi: 10.1016/B978-0-12-394590-7.00002-6. [DOI] [PubMed] [Google Scholar]
- 13.Bierne H, Sabet C, Personnic N, Cossart P. 2007. Internalins: a complex family of leucine-rich repeat-containing proteins in Listeria monocytogenes. Microbes Infect 9:1156–1166. doi: 10.1016/j.micinf.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 14.Freitag NE, Port GC, Miner MD. 2009. Listeria monocytogenes—from saprophyte to intracellular pathogen. Nat Rev Microbiol 7:623–628. doi: 10.1038/nrmicro2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lecuit M, Vandormael-Pournin S, Lefort J, Huerre M, Gounon P, Dupuy C, Babinet C, Cossart P. 2001. A transgenic model for listeriosis: role of internalin in crossing the intestinal barrier. Science 292:1722–1725. doi: 10.1126/science.1059852. [DOI] [PubMed] [Google Scholar]
- 16.Pentecost M, Kumaran J, Ghosh P, Amieva MR. 2010. Listeria monocytogenes internalin B activates junctional endocytosis to accelerate intestinal invasion. PLoS Pathog 6:e1000900. doi: 10.1371/journal.ppat.1000900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pentecost M, Otto G, Theriot JA, Amieva MR. 2006. Listeria monocytogenes invades the epithelial junctions at sites of cell extrusion. PLoS Pathog 2:e3. doi: 10.1371/journal.ppat.0020003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bakardjiev AI, Stacy BA, Fisher SJ, Portnoy DA. 2004. Listeriosis in the pregnant guinea pig: a model of vertical transmission. Infect Immun 72:489–497. doi: 10.1128/IAI.72.1.489-497.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wollert T, Pasche B, Rochon M, Deppenmeier S, van den Heuvel J, Gruber AD, Heinz DW, Lengeling A, Schubert WD. 2007. Extending the host range of Listeria monocytogenes by rational protein design. Cell 129:891–902. doi: 10.1016/j.cell.2007.03.049. [DOI] [PubMed] [Google Scholar]
- 20.Disson O, Grayo S, Huillet E, Nikitas G, Langa-Vives F, Dussurget O, Ragon M, Le Monnier A, Babinet C, Cossart P, Lecuit M. 2008. Conjugated action of two species-specific invasion proteins for fetoplacental listeriosis. Nature 455:1114–1118. doi: 10.1038/nature07303. [DOI] [PubMed] [Google Scholar]
- 21.Dramsi S, Dehoux P, Lebrun M, Goossens PL, Cossart P. 1997. Identification of four new members of the internalin multigene family of Listeria monocytogenes EGD. Infect Immun 65:1615–1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Raffelsbauer D, Bubert A, Engelbrecht F, Scheinpflug J, Simm A, Hess J, Kaufmann SH, Goebel W. 1998. The gene cluster inlC2DE of Listeria monocytogenes contains additional new internalin genes and is important for virulence in mice. Mol Gen Genet 260:144–158. doi: 10.1007/s004380050880. [DOI] [PubMed] [Google Scholar]
- 23.Sabet C, Lecuit M, Cabanes D, Cossart P, Bierne H. 2005. LPXTG protein InlJ, a newly identified internalin involved in Listeria monocytogenes virulence. Infect Immun 73:6912–6922. doi: 10.1128/IAI.73.10.6912-6922.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stachowiak R, Jagielski T, Roeske K, Osinska O, Gunerka P, Wisniewski J, Bielecki J. 2015. Lmo0171, a novel internalin-like protein, determines cell morphology of Listeria monocytogenes and its ability to invade human cell lines. Curr Microbiol 70:267–274. doi: 10.1007/s00284-014-0715-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Engelbrecht F, Chun SK, Ochs C, Hess J, Lottspeich F, Goebel W, Sokolovic Z. 1996. A new PrfA-regulated gene of Listeria monocytogenes encoding a small, secreted protein which belongs to the family of internalins. Mol Microbiol 21:823–837. doi: 10.1046/j.1365-2958.1996.541414.x. [DOI] [PubMed] [Google Scholar]
- 26.Gouin E, Adib-Conquy M, Balestrino D, Nahori MA, Villiers V, Colland F, Dramsi S, Dussurget O, Cossart P. 2010. The Listeria monocytogenes InlC protein interferes with innate immune responses by targeting the IkappaB kinase subunit IKKalpha. Proc Natl Acad Sci U S A 107:17333–17338. doi: 10.1073/pnas.1007765107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rajabian T, Gavicherla B, Heisig M, Muller-Altrock S, Goebel W, Gray-Owen SD, Ireton K. 2009. The bacterial virulence factor InlC perturbs apical cell junctions and promotes cell-to-cell spread of Listeria. Nat Cell Biol 11:1212–1218. doi: 10.1038/ncb1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carter AM. 2007. Animal models of human placentation—a review. Placenta 28(Suppl A):S41–S47. doi: 10.1016/j.placenta.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 29.Sassetti CM, Rubin EJ. 2003. Genetic requirements for mycobacterial survival during infection. Proc Natl Acad Sci U S A 100:12989–12994. doi: 10.1073/pnas.2134250100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lawley TD, Chan K, Thompson LJ, Kim CC, Govoni GR, Monack DM. 2006. Genome-wide screen for Salmonella genes required for long-term systemic infection of the mouse. PLoS Pathog 2:e11. doi: 10.1371/journal.ppat.0020011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weiss DS, Brotcke A, Henry T, Margolis JJ, Chan K, Monack DM. 2007. In vivo negative selection screen identifies genes required for Francisella virulence. Proc Natl Acad Sci U S A 104:6037–6042. doi: 10.1073/pnas.0609675104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.O'Connor TJ, Adepoju Y, Boyd D, Isberg RR. 2011. Minimization of the Legionella pneumophila genome reveals chromosomal regions involved in host range expansion. Proc Natl Acad Sci U S A 108:14733–14740. doi: 10.1073/pnas.1111678108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.National Research Council. 2011. Guide for the care and use of laboratory animals, 8th ed National Academies Press, Washington, DC. [Google Scholar]
- 34.Bishop DK, Hinrichs DJ. 1987. Adoptive transfer of immunity to Listeria monocytogenes. The influence of in vitro stimulation on lymphocyte subset requirements. J Immunol 139:2005–2009. [PubMed] [Google Scholar]
- 35.Auerbuch V, Lenz LL, Portnoy DA. 2001. Development of a competitive index assay to evaluate the virulence of Listeria monocytogenes actA mutants during primary and secondary infection of mice. Infect Immun 69:5953–5957. doi: 10.1128/IAI.69.9.5953-5957.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zemansky J, Kline BC, Woodward JJ, Leber JH, Marquis H, Portnoy DA. 2009. Development of a mariner-based transposon and identification of Listeria monocytogenes determinants, including the peptidyl-prolyl isomerase PrsA2, that contribute to its hemolytic phenotype. J Bacteriol 191:3950–3964. doi: 10.1128/JB.00016-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Robbins JR, Skrzypczynska KM, Zeldovich VB, Kapidzic M, Bakardjiev AI. 2010. Placental syncytiotrophoblast constitutes a major barrier to vertical transmission of Listeria monocytogenes. PLoS Pathog 6:e1000732. doi: 10.1371/journal.ppat.1000732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Camilli A, Tilney LG, Portnoy DA. 1993. Dual roles of plcA in Listeria monocytogenes pathogenesis. Mol Microbiol 8:143–157. doi: 10.1111/j.1365-2958.1993.tb01211.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lauer P, Chow MY, Loessner MJ, Portnoy DA, Calendar R. 2002. Construction, characterization, and use of two Listeria monocytogenes site-specific phage integration vectors. J Bacteriol 184:4177–4186. doi: 10.1128/JB.184.15.4177-4186.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Verdugo RA, Medrano JF. 2006. Comparison of gene coverage of mouse oligonucleotide microarray platforms. BMC Genomics 7:58. doi: 10.1186/1471-2164-7-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Glaser P, Frangeul L, Buchrieser C, Rusniok C, Amend A, Baquero F, Berche P, Bloecker H, Brandt P, Chakraborty T, Charbit A, Chetouani F, Couve E, de Daruvar A, Dehoux P, Domann E, Dominguez-Bernal G, Duchaud E, Durant L, Dussurget O, Entian KD, Fsihi H, Garcia-del Portillo F, Garrido P, Gautier L, Goebel W, Gomez-Lopez N, Hain T, Hauf J, Jackson D, Jones LM, Kaerst U, Kreft J, Kuhn M, Kunst F, Kurapkat G, Madueno E, Maitournam A, Vicente JM, Ng E, Nedjari H, Nordsiek G, Novella S, de Pablos B, Perez-Diaz JC, Purcell R, Remmel B, Rose M, Schlueter T, Simoes N, et al. 2001. Comparative genomics of Listeria species. Science 294:849–852. [DOI] [PubMed] [Google Scholar]
- 42.Nelson KE, Fouts DE, Mongodin EF, Ravel J, DeBoy RT, Kolonay JF, Rasko DA, Angiuoli SV, Gill SR, Paulsen IT, Peterson J, White O, Nelson WC, Nierman W, Beanan MJ, Brinkac LM, Daugherty SC, Dodson RJ, Durkin AS, Madupu R, Haft DH, Selengut J, Van Aken S, Khouri H, Fedorova N, Forberger H, Tran B, Kathariou S, Wonderling LD, Uhlich GA, Bayles DO, Luchansky JB, Fraser CM. 2004. Whole genome comparisons of serotype 4b and 1/2a strains of the food-borne pathogen Listeria monocytogenes reveal new insights into the core genome components of this species. Nucleic Acids Res 32:2386–2395. doi: 10.1093/nar/gkh562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.de Hoon MJ, Imoto S, Nolan J, Miyano S. 2004. Open source clustering software. Bioinformatics 20:1453–1454. doi: 10.1093/bioinformatics/bth078. [DOI] [PubMed] [Google Scholar]
- 44.Saldanha AJ. 2004. Java Treeview—extensible visualization of microarray data. Bioinformatics 20:3246–3248. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- 45.Genbacev O, Donne M, Kapidzic M, Gormley M, Lamb J, Gilmore J, Larocque N, Goldfien G, Zdravkovic T, McMaster MT, Fisher SJ. 2011. Establishment of human trophoblast progenitor cell lines from the chorion. Stem Cells 29:1427–1436. doi: 10.1002/stem.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ilic D, Kapidzic M, Genbacev O. 2008. Isolation of human placental fibroblasts. Curr Protoc Stem Cell Biol Chapter 1:Unit 1C.6. doi: 10.1002/9780470151808.sc01c06s5. [DOI] [PubMed] [Google Scholar]
- 47.Schildge S, Bohrer C, Beck K, Schachtrup C. 19 January 2013. Isolation and culture of mouse cortical astrocytes. J Vis Exp doi: 10.3791/50079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A. 2012. Fiji: an open-source platform for biological-image analysis. Nat Methods 9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Melton-Witt JA, Rafelski SM, Portnoy DA, Bakardjiev AI. 2012. Oral infection with signature-tagged Listeria monocytogenes reveals organ-specific growth and dissemination routes in guinea pigs. Infect Immun 80:720–732. doi: 10.1128/IAI.05958-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rizzuto GA, Kapidzic M, Gormley M, Bakardjiev AI. 21 July 2016. Human placental and decidual organ cultures to study infections at the maternal-fetal interface. J Vis Exp doi: 10.3791/54237. [DOI] [PubMed] [Google Scholar]
- 51.Bakardjiev AI, Stacy BA, Portnoy DA. 2005. Growth of Listeria monocytogenes in the guinea pig placenta and role of cell-to-cell spread in fetal infection. J Infect Dis 191:1889–1897. doi: 10.1086/430090. [DOI] [PubMed] [Google Scholar]
- 52.Huerta-Cepas J, Dopazo J, Gabaldon T. 2010. ETE: a python environment for tree exploration. BMC Bioinformatics 11:24. doi: 10.1186/1471-2105-11-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bakardjiev AI, Theriot JA, Portnoy DA. 2006. Listeria monocytogenes traffics from maternal organs to the placenta and back. PLoS Pathog 2:e66. doi: 10.1371/journal.ppat.0020066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Scortti M, Monzo HJ, Lacharme-Lora L, Lewis DA, Vazquez-Boland JA. 2007. The PrfA virulence regulon. Microbes Infect 9:1196–1207. doi: 10.1016/j.micinf.2007.05.007. [DOI] [PubMed] [Google Scholar]
- 55.Dramsi S, Bourdichon F, Cabanes D, Lecuit M, Fsihi H, Cossart P. 2004. FbpA, a novel multifunctional Listeria monocytogenes virulence factor. Mol Microbiol 53:639–649. doi: 10.1111/j.1365-2958.2004.04138.x. [DOI] [PubMed] [Google Scholar]
- 56.Abachin E, Poyart C, Pellegrini E, Milohanic E, Fiedler F, Berche P, Trieu-Cuot P. 2002. Formation of d-alanyl-lipoteichoic acid is required for adhesion and virulence of Listeria monocytogenes. Mol Microbiol 43:1–14. doi: 10.1046/j.1365-2958.2002.02723.x. [DOI] [PubMed] [Google Scholar]
- 57.Redline RW, Lu CY. 1988. Specific defects in the anti-listerial immune response in discrete regions of the murine uterus and placenta account for susceptibility to infection. J Immunol 140:3947–3955. [PubMed] [Google Scholar]
- 58.Genbacev O, Schubach SA, Miller RK. 1992. Villous culture of first trimester human placenta—model to study extravillous trophoblast (EVT) differentiation. Placenta 13:439–461. doi: 10.1016/0143-4004(92)90051-T. [DOI] [PubMed] [Google Scholar]
- 59.Zeldovich VB, Robbins JR, Kapidzic M, Lauer P, Bakardjiev AI. 2011. Invasive extravillous trophoblasts restrict intracellular growth and spread of Listeria monocytogenes. PLoS Pathog 7:e1002005. doi: 10.1371/journal.ppat.1002005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dramsi S, Levi S, Triller A, Cossart P. 1998. Entry of Listeria monocytogenes into neurons occurs by cell-to-cell spread: an in vitro study. Infect Immun 66:4461–4468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Portnoy DA, Jacks PS, Hinrichs DJ. 1988. Role of hemolysin for the intracellular growth of Listeria monocytogenes. J Exp Med 167:1459–1471. doi: 10.1084/jem.167.4.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ezraty B, Aussel L, Barras F. 2005. Methionine sulfoxide reductases in prokaryotes. Biochim Biophys Acta 1703:221–229. doi: 10.1016/j.bbapap.2004.08.017. [DOI] [PubMed] [Google Scholar]
- 63.Dussurget O, Dumas E, Archambaud C, Chafsey I, Chambon C, Hebraud M, Cossart P. 2005. Listeria monocytogenes ferritin protects against multiple stresses and is required for virulence. FEMS Microbiol Lett 250:253–261. doi: 10.1016/j.femsle.2005.07.015. [DOI] [PubMed] [Google Scholar]
- 64.Maltepe E, Bakardjiev AI, Fisher SJ. 2010. The placenta: transcriptional, epigenetic, and physiological integration during development. J Clin Invest 120:1016–1025. doi: 10.1172/JCI41211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Myatt L. 2010. Review: reactive oxygen and nitrogen species and functional adaptation of the placenta. Placenta 31(Suppl):S66–S69. doi: 10.1016/j.placenta.2009.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Macnab RM. 2003. How bacteria assemble flagella. Annu Rev Microbiol 57:77–100. doi: 10.1146/annurev.micro.57.030502.090832. [DOI] [PubMed] [Google Scholar]
- 67.Peel M, Donachie W, Shaw A. 1988. Temperature-dependent expression of flagella of Listeria monocytogenes studied by electron microscopy, SDS-PAGE and Western blotting. J Gen Microbiol 134:2171–2178. [DOI] [PubMed] [Google Scholar]
- 68.Grundling A, Burrack LS, Bouwer HG, Higgins DE. 2004. Listeria monocytogenes regulates flagellar motility gene expression through MogR, a transcriptional repressor required for virulence. Proc Natl Acad Sci U S A 101:12318–12323. doi: 10.1073/pnas.0404924101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bigot A, Pagniez H, Botton E, Frehel C, Dubail I, Jacquet C, Charbit A, Raynaud C. 2005. Role of FliF and FliI of Listeria monocytogenes in flagellar assembly and pathogenicity. Infect Immun 73:5530–5539. doi: 10.1128/IAI.73.9.5530-5539.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Way SS, Thompson LJ, Lopes JE, Hajjar AM, Kollmann TR, Freitag NE, Wilson CB. 2004. Characterization of flagellin expression and its role in Listeria monocytogenes infection and immunity. Cell Microbiol 6:235–242. doi: 10.1046/j.1462-5822.2004.00360.x. [DOI] [PubMed] [Google Scholar]
- 71.Buchrieser C, Rusniok C, Garrido P, Hain T, Scortti M, Lampidis R, Karst U, Chakraborty T, Cossart P, Kreft J, Vazquez-Boland JA, Goebel W, Glaser P. 2011. Complete genome sequence of the animal pathogen Listeria ivanovii, which provides insights into host specificities and evolution of the genus Listeria. J Bacteriol 193:6787–6788. doi: 10.1128/JB.06120-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Redline RW, Lu CY. 1987. Role of local immunosuppression in murine fetoplacental listeriosis. J Clin Invest 79:1234–1241. doi: 10.1172/JCI112942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lecuit M, Dramsi S, Gottardi C, Fedor-Chaiken M, Gumbiner B, Cossart P. 1999. A single amino acid in E-cadherin responsible for host specificity towards the human pathogen Listeria monocytogenes. EMBO J 18:3956–3963. doi: 10.1093/emboj/18.14.3956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Khelef N, Lecuit M, Bierne H, Cossart P. 2006. Species specificity of the Listeria monocytogenes InlB protein. Cell Microbiol 8:457–470. doi: 10.1111/j.1462-5822.2005.00634.x. [DOI] [PubMed] [Google Scholar]
- 75.Disson O, Lecuit M. 2013. In vitro and in vivo models to study human listeriosis: mind the gap. Microbes Infect 15:971–980. doi: 10.1016/j.micinf.2013.09.012. [DOI] [PubMed] [Google Scholar]
- 76.Lecuit M, Nelson DM, Smith SD, Khun H, Huerre M, Vacher-Lavenu MC, Gordon JI, Cossart P. 2004. Targeting and crossing of the human maternofetal barrier by Listeria monocytogenes: role of internalin interaction with trophoblast E-cadherin. Proc Natl Acad Sci U S A 101:6152–6157. doi: 10.1073/pnas.0401434101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Drevets DA, Jelinek TA, Freitag NE. 2001. Listeria monocytogenes-infected phagocytes can initiate central nervous system infection in mice. Infect Immun 69:1344–1350. doi: 10.1128/IAI.69.3.1344-1350.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Reniere ML, Whiteley AT, Hamilton KL, John SM, Lauer P, Brennan RG, Portnoy DA. 2015. Glutathione activates virulence gene expression of an intracellular pathogen. Nature 517:170–173. doi: 10.1038/nature14029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Witter AR, Okunnu BM, Berg RE. 2016. The essential role of neutrophils during infection with the intracellular bacterial pathogen Listeria monocytogenes. J Immunol 197:1557–1565. doi: 10.4049/jimmunol.1600599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hirota Y, Daikoku T, Tranguch S, Xie H, Bradshaw HB, Dey SK. 2010. Uterine-specific p53 deficiency confers premature uterine senescence and promotes preterm birth in mice. J Clin Invest 120:803–815. doi: 10.1172/JCI40051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Goldenberg RL, Faye-Petersen O, Andrews WW, Goepfert AR, Cliver SP, Hauth JC. 2007. The Alabama Preterm Birth Study: diffuse decidual leukocytoclastic necrosis of the decidua basalis, a placental lesion associated with preeclampsia, indicated preterm birth and decreased fetal growth. J Matern Fetal Neonatal Med 20:391–395. doi: 10.1080/14767050701236365. [DOI] [PubMed] [Google Scholar]
- 82.Romero R, Dey SK, Fisher SJ. 2014. Preterm labor: one syndrome, many causes. Science 345:760–765. doi: 10.1126/science.1251816. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.