Abstract
Jatropha curcas seeds are an excellent biofuel feedstock, but seed yields of Jatropha are limited by its poor flowering and fruiting ability. Thus, identifying genes controlling flowering is critical for genetic improvement of seed yield. We isolated the JcLFY, a Jatropha ortholog of Arabidopsis thaliana LEAFY (LFY), and identified JcLFY function by overexpressing it in Arabidopsis and Jatropha. JcLFY is expressed in Jatropha inflorescence buds, flower buds, and carpels, with highest expression in the early developmental stage of flower buds. JcLFY overexpression induced early flowering, solitary flowers, and terminal flowers in Arabidopsis, and also rescued the delayed flowering phenotype of lfy-15, a LFY loss-of-function Arabidopsis mutant. Microarray and qPCR analysis revealed several flower identity and flower organ development genes were upregulated in JcLFY-overexpressing Arabidopsis. JcLFY overexpression in Jatropha also induced early flowering. Significant changes in inflorescence structure, floral organs, and fruit shape occurred in JcLFY co-suppressed plants in which expression of several flower identity and floral organ development genes were changed. This suggests JcLFY is involved in regulating flower identity, floral organ patterns, and fruit shape, although JcLFY function in Jatropha floral meristem determination is not as strong as that of Arabidopsis.
Jatropha curcas (hereafter Jatropha; Euphorbiaceae) is a perennial deciduous shrub that is widespread throughout the tropics and subtropics of central America1. Jatropha is monoecious, with male and female flowers borne on the same inflorescences2. The potential for Jatropha as a biofuel crop in tropical and subtropical countries is widely recognized3,4. Jatropha has been cultivated for its unique biodiesel potential because of its high oil content, high biomass productivity, adaptability to marginal land across various agro-climatic conditions, and non-competitiveness with food production2,3,4. This high agro-industrial potential for biofuel production, including bio-jet fuel production, has motivated cultivation of Jatropha5,6. The economic value of Jatropha is derived from its seed oil; Jatropha seeds contain up to 40% oil7. Accordingly, Jatropha cultivation may alleviate future energy crises and reduce environment pollution associated with petroleum production8. However, Jatropha exhibits an overabundance of vegetative shoots and leaves and a long juvenile stage5,9. In addition, unreliable and poor flowering is an important factor that contributes to low seed productivity in Jatropha. Breeders chiefly aim to shorten the juvenile phase; increase seed yield; reduce plant height; increase the ratio of female to male flowers; and improve oil content and oil fuel properties5,7. Thus, identifying flowering genes is one of the key steps to improve seed yield and enhance industrial use of Jatropha.
Flowering is the developmental turning point from the vegetative to reproductive phase and critical to crop yields. The precise timing of flowering is crucial to reproductive fitness, as plants must ensure the energy and resources accumulated during the vegetative phase are optimally allocated to offspring10. Both environmental and endogenous cues determine the onset of reproductive development11. Identification of flowering time genes is a key step for breeding plant varieties adapted to different agricultural conditions and seasons10. These signals modulate the level and activity of flowering-time regulators that initiate the reproductive phase and induce expression of the meristem identity genes12. Flowering has been most extensively studied in Arabidopsis thaliana, in which flowering is regulated by at least five parallel pathways: photoperiod, vernalization, gibberellin (GA)-dependent, autonomous, and age-related pathways11. LEAFY (LFY), FLOWERING LOCUS T (FT), and SUPPRESSOR OF OVEREXPRESSION OF CONSTANS1 (SOC1)/AGAMOUS-LIKE 20 (AGL20) are genes that integrate signals from multiple genetic pathways13.
The LFY gene was identified by Weigel et al.14 in Arabidopsis, and its orthologous gene, FLORICAULA (FLO), was identified by Coen et al.15 in Antirrhinum majus. A single LFY ortholog is found in most land plant species, even in monocots whose evolutionary histories include several polyploidization events16. LFY is a plant-specific transcription factor that binds to the regulatory region of its target genes at a helix-turn-helix motif within a unique protein fold17. In seeds plants, LFY is the central flower meristem identity gene; it is mainly expressed in inflorescence and flower primordia14,15,18. LFY expression level is an important determinant of flower initiation19. LFY acts as a master regulator, orchestrating the floral gene network; it activates downstream genes that determine the unique identities of floral meristem (FM) tissue and floral organ primordia. LFY directly regulates flower organ identity genes including CAULIFLOWER (CAL), APETALA1 (AP1), APETALA3 (AP3), AGAMOUS (AG), SEPALLATA (SEP), and TERMINAL FLOWER1 (TFL1)20,21. Accordingly, LFY controls multiple aspects of floral morphogenesis, including phyllotaxis, organ number, organ identity, and determinacy22. LFY executes its meristem identity role by activating AP1 expression, as is supported by the recovery of the lfy mutant phenotype by overexpression of AP122. LFY is also involved in inflorescence and flower development. In maize (Zea mays), LFY homologs ZFL1 and ZFL2 are required for proper expression of B and C genes in flowers23. In contrast, the LFY homolog in rice, RFL, controls inflorescence structure; RFL is unexpressed in FMs, and rice flowers appear fertile even when RFL is silenced24. LFY homologs have an ancestral role in regulating cell division and arrangement in gymnosperms, ferns, and mosses—taxa that lack flowers17. In pea and Medicago truncatula, LFY orthologs are involved in leaf development and are required to form leaflets25,26.
LFY overexpression in Arabidopsis induces early flowering and converts all buds into flower buds while making all inflorescences occur as solitary and terminal flowers19,27. Additionally, LFY overexpression accelerates flowering in a variety of other plants, including tobacco28, rice24, Sinningia speciosa29, Brassica juncea30, and Citrus31. In Arabidopsis lfy mutants, FM development is dramatically delayed with late flowering32, flower buds are surrounded by extra bracts, and the early flowers are completely transformed into inflorescence shoots. Additionally, the first four floral organs develop into sepal-like organs and develop fewer petals and stamens. Flowers produced by plants with intermediate alleles are often male sterile, but female fertile14. In Antirrhinum, mutations in flo transform flowers into inflorescence shoots completely, and these inflorescences exhibited meristems that proliferated without producing flowers15.
Several studies have reported that LFY interacts with hormone pathways. For example, LFY controls auxin biosynthesis through repressing the auxin synthesis gene YUC4; in turn, auxin increases LFY transcription, and LFY promotes auxin signaling, leading to further increases in LFY activity33. In Arabidopsis, GA regulates LFY transcription to promote flower initiation34,35,36. LFY represses the cytokinin signaling negative regulator ARR7 through a direct interaction with its promoter20.
LFY orthologs have been cloned and characterized in several woody species such as Eucalyptus37, Monterey pine38, Kiwifruit39, and papaya40. However, few is known about the specific role of LFY in tree reproductive development. In woody species, juvenile phases can last years to decades such that trees delay flowering and fruit production for a very long time. The existence of a lengthy juvenile phase in woody species is a limiting factor for their genetic improvement, and this prevents the full domestication of many economically important woody species. Currently, only Jatropha FLOWERING LOCUS T (JcFT)41,42, and JcAP143 have been functionally analyzed in Jatropha. However, cloning and molecular characterization of JcLFY has not yet been reported. In this study, we cloned JcLFY from Jatropha and analyzed the function of JcLFY by transforming both wild-type (WT) and lfy-15 mutant Arabidopsis. We further analyzed JcLFY function in flowering induction and floral organ specification using JcLFY overexpression and co-suppression in transgenic Jatropha plants.
Results
Cloning and bioinformatic analysis of JcLFY in Jatropha
Full length JcLFY cDNA sequence was obtained from Jatropha flower bud tissue by RT-PCR and RACE. JcLFY gDNA and cDNA were amplified by long-distance PCR. The full 2302-bp length of JcLFY gDNA contains two introns and three exons, and the full 1350-bp length of JcLFY cDNA contains a 1155-bp open reading frame, encoding 384 amino acids (Supplementary Fig. S1A). Alignment of the JcLFY sequence with Ricinus communis LFY (RcLFY), Populus trichocarpa LFY (PtLFY), Arabidopsis thaliana LFY (AtLFY), Oryza sativa FLO (OsFLO), and Zea mays FLO (ZmFLO) showed amino acid sequence identities of 88%, 84%, 72%, 69%, and 66%, respectively. LFY proteins show higher similarity in the C-terminal region (Supplementary Fig. S1B).
A total of 41 LFY amino acid sequences from different species were used to construct a phylogenetic tree showing the relationship of LFY homologs (Supplementary Fig. S2). These genes can be classified into five classes, which generally differentiate lower plants from higher plants (Supplementary Fig. S2). Specifically, two genes from mosses form class I; ten genes from ferns form class II; five genes from gymnosperms form class III; fifteen genes from eudicots form class IV; and nine genes from monocots form class V (Supplementary Fig. S2). JcLFY clustered together with sequences from other eudicots plants and belongs to class IV, and JcLFY has the highest identity to RcLFY from Ricinus communis (Supplementary Fig. S2). RcLFY and JcLFY clustered in the same clade, consistent with the close evolutionary relationship between Jatropha and Ricinus communis.
Expression pattern of JcLFY in Jatropha
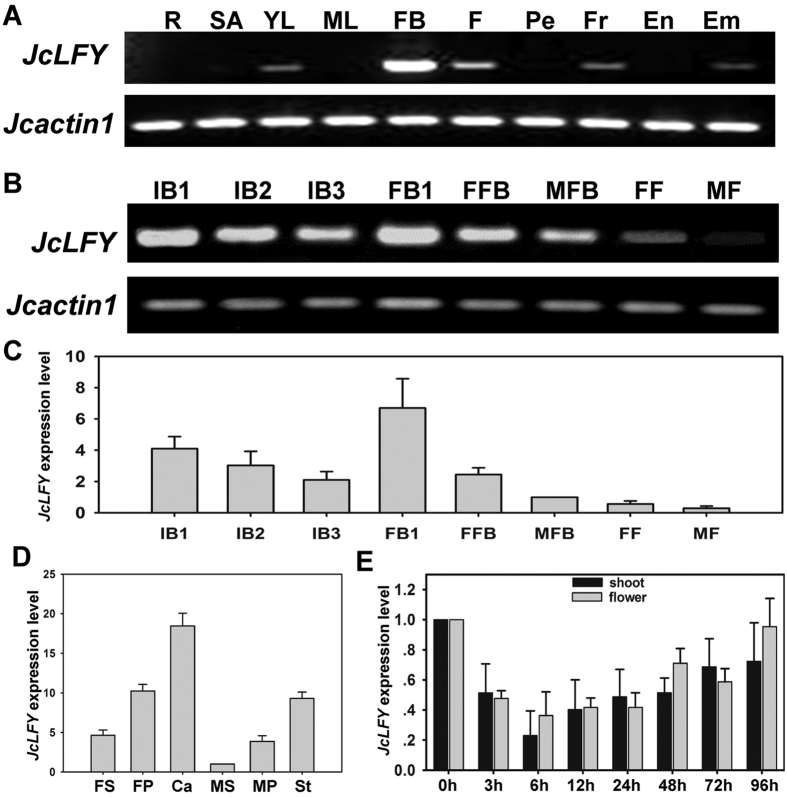
RT-PCR was used to investigate the expression of JcLFY across different organs and tissues. We first tested the expression of JcLFY in various tissues, revealing that this gene is highly expressed in flower buds and the transcript occurs in young leaves, flowers, fruits, and embryos. However, transcripts were not detected in roots, shoot apices, mature leaves, pedicels, and endosperms (Fig. 1A). To better analyze JcLFY transcripts, samples were collected from flowers at different developmental stages (Fig. 1B). JcLFY was highly expressed in inflorescence buds (IB1, IB2, and IB3), flower buds (FB), male flower buds (MFB), and female flower buds (FFB), but exhibited lower expression levels in bloomed male and female flowers (Fig. 1B). Overall, JcLFY was highly expressed during the early-stage inflorescence buds (e.g., IB1) and early-stage flower buds (e.g., FB1). During inflorescence and floral organ development, JcLFY expression levels decreased. Real-time qPCR agreed with the semi RT-PCR results of JcLFY expressions across flower development stages (Fig. 1C). We further analyzed JcLFY expression in different floral whorls, finding JcLFY expression higher in stamens and carpels than in sepals and petals (Fig. 1D). To determine whether JcLFY was induced by GA, we assessed JcLFY expression levels in shoot apex and flower bud tissues after application of 1 mM GA. Unexpectedly, JcLFY expression exhibited a slight decrease after GA application (Fig. 1E). Hence, JcLFY was not induced by GA in Jatropha, in contrast with Arabidopsis34,35 and Chrysanthemum44.
Figure 1. Analysis of JcLFY expression in different Jatropha tissues.
(A,B) Semi-quantitative RT-PCR analysis of JcLFY expression in different tissues; (C) Real-time qPCR performed on inflorescence buds and flower buds to validate semi-quantitative RT-PCR results; (D) Real-time qPCR analysis of JcLFY expression in floral organs; (E) Real-time qPCR analysis of JcLFY expression in shoot and flower bud tissues after 1 mM GA application. R, root; SA, shoot apex; YL, young leaf; ML, mature leaf; FB, flower bud; F, flower; Pe, pedicel; Fr, fruit; En, endosperm; Em, embryo; FS, female sepal; FP, female petal; Ca, carpel; MS, male sepal; MP, male petal; St, stamen; IB1, inflorescence bud stage 1 (0–5 days, inflorescence buds are visible); IB2, inflorescence bud stage 2 (1 week after IB1); IB3, inflorescence bud stage 3 (1 week after IB2); FB1, flower bud stage 1 (1 week after IB3); and FB2, flower bud stage 2 (1 week after FB1). In this stage, male flower bud (MFB) and female flower bud (FFB) are identifiable: MF, male flower (1 week after MFB) and FF, female flower (1 week after FFB). Fruits were harvested 15 days after fertilization. Endosperm and embryo tissues were harvested from mature seeds. The levels of detected amplicons were normalized using the amplified products of JcActin1. The mRNA levels in FFB (C), MS (D), and before GA application to FB (E) tissues were standardized with a value of 1. The total PCR cycles of JcActin1 in (A) and (B) are 26 and 25, respectively, while the PCR cycles of JcLFY in (A) and (B) are both 30.
JcLFY overexpression in Arabidopsis induced early flowering, solitary flowers, and terminal flowers
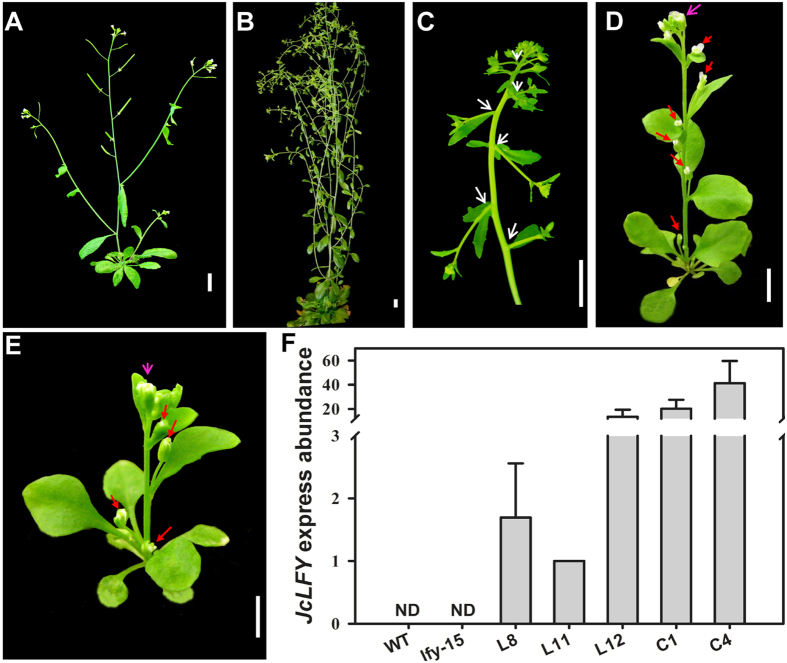
To determine whether JcLFY is involved in the regulation of flowering time, JcLFY cDNA driven by the CaMV 35S promoter was transformed into WT Arabidopsis (untransformed WT plants were controls). Transgenic plants were confirmed via qRT-PCR analysis of JcLFY expression. Forty independent T0 transgenic lines were generated with the 35S:JcLFY construct. In a majority of transgenic lines, bolting occurred notably earlier than in WT plants under both LD and SD conditions.
The phenotypes of three independent homozygous lines from the T2 generation were examined. Relative to WT plants under LD conditions, L8, L11, and L12 bolted 9–15 days earlier, produced 2–6 fewer rosette leaves, produced 2–4 fewer branches, and were 3–17.5 cm shorter (Fig. 2B–D and Table 1). Under SD conditions, L8, L11, and L12 flowered approximately 1–2 months earlier, produced 16–36 fewer rosette leaves, and were 6.1–32.7 cm shorter (Table 2). Thus, JcLFY overexpression in Arabidopsis significantly reduced the vegetative growth period.
Figure 2. Phenotypes of transgenic Arabidopsis overexpressing JcLFY.
(A) Wild-type (WT) Arabidopsis under LD conditions (30 d). (B) Transgenic Arabidopsis L8 grown under LD conditions (40 d); the white arrow indicates a branch and a solitary flower formed in a cauline leaf axil. (C,D) Transgenic Arabidopsis L11 and L12 grown under LD conditions (20 d); the red arrows indicate the solitary flowers, while the pink arrow indicates the terminal flower. (E) WT Arabidopsis grown under SD conditions (120 d). (F) Detailed image of contents of the red box in (E). (G) Transgenic plant L11 grown under SD conditions, exhibiting opposite leaves and solitary flowers formed in leaf axils; fruit development was normal. (H) Transgenic plant L12 grown under SD conditions; two flowers formed in a leaf axil and fruit development was normal. All Arabidopsis plants were derived from the Columbia ecotype. (I,J) Schematic comparison of WT and 35S:JcLFY transgenic Arabidopsis plants grown under LD and SD conditions, respectively. Arrowheads indicate shoot meristems, while circles indicate flowers. In WT plants, the primary shoot can be subdivided into a basal rosette, which contains leaves separated by short internodes, and an apical shoot with elongated internodes; the apical shoot, often referred to as an inflorescence, bears a few bracts (small stem leaves) with associated secondary shoots, as well as a potentially indeterminate number of flowers. Scale bar = 1 cm.
Table 1. Overexpression of JcLFY promotes flowering in Arabidopsis under LD conditions.
| Lines | N | Rosette leaves | Flower bud formation time/Day | Flowering time/Day | Height/cm | Branches | Cauline leaves |
|---|---|---|---|---|---|---|---|
| WT | 22 | 12.36 ± 1.56 | 32.27 ± 3.87 | 40.59 ± 3.78 | 21.82 ± 2.52 | 4.14 ± 0.83 | 4.09 ± 0.61 |
| Line8 | 17 | 10.41 ± 1.06* | 23.24 ± 2.59** | 32.59 ± 3.66** | 18.06 ± 2.97* | 4.06 ± 0.97 | 4.06 ± 1.39 |
| Line11 | 16 | 9.25 ± 1.24* | 19.43 ± 3.72** | 26.94 ± 3.60** | 16.16 ± 4.13* | 2.25 ± 1.06** | 3.38 ± 1.36 |
| Line12 | 16 | 6.19 ± 1.11** | 17.19 ± 2.83** | 25.19 ± 1.94** | 4.47 ± 1.59** | 0.00 ± 0.00** | 4.13 ± 1.15 |
WT plants and three independent JcLFY-overexpressing lines (L8, L11 and L12) grown under LD conditions (16 h light/8 h dark) were subjected to the analysis of rosette leaves, Flower bud formation time, flowering time, height, branches, and cauline leaves. N = plant number. The values are presented as the mean ± standard deviation. *Significantly different from the control at the 5% level; **significantly different from the control at the 1% level.
Table 2. Overexpression of JcLFY promotes flowering in Arabidopsis under SD conditions.
| Lines | N | Rosette leaves | Flower bud formation time/Day | Flowering time/Day | Height/cm |
|---|---|---|---|---|---|
| WT | 10 | 50.58 ± 5.14 | 126.50 ± 5.77 | 138.70 ± 14.47 | 38.88 ± 2.98 |
| Line8 | 7 | 34.28 ± 4.22** | 92.57 ± 8.62** | 118.00 ± 11.76* | 32.71 ± 7.96* |
| Line11 | 15 | 25.58 ± 3.49** | 79.56 ± 6.69** | 97.33 ± 10.13** | 27.22 ± 2.90** |
| Line12 | 12 | 14.85 ± 2.07** | 54.25 ± 3.70** | 60.33 ± 4.52** | 6.17 ± 3.29** |
WT plants and three independent JcLFY-overexpressing lines (L8, L11 and L12) grown under SD conditions (8 h light/12 h dark) were subjected to the analysis of rosette leaves, flower bud formation time, flowering time, and height. N = plant number. The values are presented as the mean ± standard deviation. *Significantly different from the control at the 5% level; **significantly different from the control at the 1% level.
In transgenic plants, the primary shoots were converted into terminal flowers (Fig. 2D). The secondary shoots produced in cauline and rosette leaf axils were converted into solitary flowers. In extreme phenotypes of transgenic plants, all branches and inflorescences were replaced by solitary flowers (Fig. 2C,D). Furthermore, transgenic plants under SD conditions exhibited opposite leaves, solitary flowers in leaf axils (Fig. 2G), and two flowers within a single leaf axil (Fig. 2H). Transgenic plants L12 exhibited the highest JcLFY expression levels (Fig. 3F) and the most severe phenotypes (Fig. 2D).
Figure 3. 35S:JcLFY recovered lfy-15 mutant Arabidopsis phenotypes.
(A) WT Arabidopsis (45 d). (B) lfy-15 mutant (70 d). (C) Inflorescence of a lfy-15 mutant with flowers transformed into inflorescences/branches (white arrows). (D) lfy-15 mutant harboring 35S:JcLFY transgenic plant C1 (25 d). (E) lfy-15 mutant harboring 35S:JcLFY transgenic plant C4 (20 d). All Arabidopsis were derived from the Columbia ecotype. The red arrows in (D) and (E) indicate solitary flowers; the pink arrows in (D) and (E) indicate terminal flowers (scale bar = 1 cm). (F) Data generated by qRT-PCR were used to quantitatively assess the expression of JcLFY transgenes. Expression profiles were normalized against AtActin2. Expression analysis was conducted using quantitative RT-PCR with RNA extracted from seedlings of T2 transgenic L8, L11, L12, and lfy-15 mutants harboring JcLFY (C1 and C4). WT and lfy-15 plants grown under the same conditions were used as controls; JcLFY expression was not detected in both WT and lfy-15 mutant. Error bars represent the standard deviation (SD) across three independent replicates.
To explore genes involved in the JcLFY mediated pathway, 2-week-old soil-grown transgenic 35S:JcLFY L8, L11, and L12 plants as well as WT Arabidopsis were grown under LD conditions for microarray analysis. In total, 1,854 genes exhibited more than 2-fold changes, 122 genes exhibited more than 10-fold changes, and 22 genes exhibited more than 50-fold changes (Supplementary Table S3, and Fig. S4A). We classified 2-fold or higher expression changes in these three lines into six groups. Overall, 48 flower-related genes were detected by microarray at this threshold (Supplementary Fig. S4); 71 phytohormone-related genes changed expression, including genes related to auxin, cytokinin, gibberellin, abscisic acid, brassinosteroids, and jasmonate; 58 stress-related genes were changed; 480 genes encoding metabolic enzymes were changed; and 474 genes of unknown function were changed (Supplementary Table S3 and Fig. S4B).
Microarray data and qRT-PCR results indicated the promotion of flowering and terminal flowers in 35S:JcLFY transgenic Arabidopsis was associated with a significant upregulation of the FM identity genes AP1, SOC1, and CAL, and the floral organ identity genes AG, AP3, SEP1, SEP2, and SEP3 (Supplementary Fig. S3). The expression levels of these genes were highest in L12. TFL1 expression was downregulated in L12 plants (Supplementary Fig. S3), which likely induced solitary terminal flowers. Thus, the early-flowering and terminal flower phenotypes induced by ectopic JcLFY expression in transgenic Arabidopsis were similar to those induced by AtLFY overexpression27. Under both LD and SD conditions, JcLFY overexpression in Arabidopsis induced early flowering.
Chromatin immuno-precipitation experiment45 showed Arabidopsis LFY has 15 direct target genes, and each target gene was upregulated more than 2-fold. However, most of these target genes were upregulated in the present study, though the fold changes were much lower (e.g., AT5G60630, AT5G49770, At5G03790, AT5G03230, and At2g44450 in Supplementary Table S2). Some of these target genes exhibited no significant change (e.g., AT4G22780 and AT5G46660), while others were downregulated (e.g., AT3G52470 and AT3G19390) Supplementary Table S2.
JcLFY overexpression in lfy-15 mutant Arabidopsis induced early flowering and complemented mutant phenotypes
To further determine whether JcLFY is an AtLFY ortholog, the 35S:JcLFY construct was transformed into Arabidopsis lfy-15 mutant plants. Ten independent T0 transgenic lines were generated and confirmed through qRT-PCR of JcLFY expression. WT plants and lfy-15 mutants under the same growth conditions were controls. Most transgenic lines bolted earlier than the WT and lfy-15 mutant plants under LD conditions.
To examine phenotypes, we selected two independent homozygous lines in the T2 generation that exhibited high JcLFY expression (Fig. 3D,E). Under LD conditions, lines C1 and C4 bolted 8–13 days earlier and produced 2–6 fewer rosette leaves than WT plants, and bolted 18–23 days earlier and produced 9–13 fewer rosette leaves than lfy-15 mutants (Fig. 3A–E; Table 3). In transgenic plants, solitary flowers appeared in the axils of rosette and cauline leaves, and terminal flowers appeared on primary shoots (Fig. 3D,E). The lfy-15 mutant flowers were converted to inflorescences, and floral organs were abnormal and less fertile (Fig. 3C)14. The transgenic mutant C1 and C4 lines rescued mutant late bolting, and the conversion of flowers to inflorescences was repressed (Fig. 3D,E).
Table 3. Overexpression of JcLFY in lfy-15 Arabidopsis plants promotes flowering under LD conditions.
| Lines | N | Rosette leaves | Flower bud formation time/Day | Flowering time/Day | Height/cm | Branches | Cauline leaves |
|---|---|---|---|---|---|---|---|
| WT | 18 | 10.50 ± 2.09 | 25.39 ± 6.00 | 37.39 ± 6.10 | 31.11 ± 8.98 | 6.78 ± 1.90 | 5.78 ± 1.26 |
| lfy-15 | 17 | 17.79 ± 3.36 | 35.18 ± 8.50* | 51.71 ± 10.35** | 32.47 ± 3.94 | 32.71 ± 7.30** | 35.18 ± 9.02** |
| C1 | 16 | 8.06 ± 1.18** | 17.19 ± 2.20** | 23.31 ± 3.54** | 7.31 ± 2.36** | 0.00 ± 0.00** | 5.62 ± 1.09 |
| C4 | 15 | 4.36 ± 1.21** | 12.46 ± 1.57** | 18.23 ± 2.09** | 5.25 ± 1.77** | 0.00 ± 0.00** | 4.79 ± 0.88 |
WT plants, the lfy-15 mutant, and two independent JcLFY-overexpressing lines (C1 and C4) grown under LD growing conditions (16 h light/8 h dark) were subjected to the analysis of rosette leaves, flower bud formation time, flowering time, height, branches, and cauline leaves. N = plant number. The values are presented as the mean ± standard deviation. *Significantly different from the control at the 5% level; **significantly different from the control at the 1% level.
Both C1 and C4 exhibited high JcLFY expression (Fig. 3F). Promotion of flowering in the 35S:JcLFY transgenic Arabidopsis mutant was associated with a significant up-regulation of FM identity genes AP1, SOC1, and SEPs (Supplementary Fig. S3). The branch reduction may have occurred through the repression of TFL1 by JcLFY (Supplementary Fig. S3I).
These results demonstrate that the constitutive expression of JcLFY complements increased branches (i.e., inflorescences) in later flowering stages and abnormal flowers in lfy-15 mutant; thus, JcLFY functions as a LFY homolog.
JcLFY overexpression in Jatropha induced early flowering
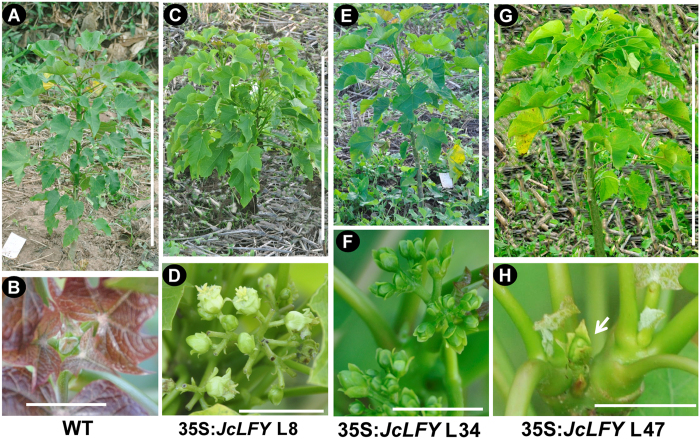
Jatropha expression profiles and Arabidopsis transgenic analyses suggested JcLFY is a floral identity gene in Jatropha. To test this hypothesis, we generated transgenic Jatropha with the 35S:JcLFY construct. Non-transgenic plants were used as a control. Fifty independent transgenic lines were confirmed via PCR. Unexpectedly, most transgenic Jatropha plants lacked an early-flowering phenotype, while some transgenic plants (approximately 10) showed slightly early flowering. When regenerated plantlets (Fig. 4) were grown in the field for 2 months, flower buds emerged from transgenic plants (Fig. 4C–H), while control plants didn’t produce flower buds (Fig. 4A,B). We chose early-flowering plants from L8, L34, and L47 for further analysis. qRT-PCR indicated JcLFY expression was higher in these plants, while JcLFY expression was much higher in the L47 sample (Supplementary Fig. S5A). We further analyzed several floral identity genes and flower organ identity genes, finding that the transcript levels of JcAP1, JcAP3, JcAG, JcSEP1, and JcSEP3 were obviously increased in transgenic flower buds; however, JcAP2 and JcTFL1c expression was reduced. The inflorescence structure and floral organ patterns were normal. Furthermore, we grew 15 transgenic plants in a greenhouse in Kunming located in a subtropical area of southwestern China (Supplementary Fig. S6). We found 2 transgenic plants produced flowers in the second spring, and 5 transgenic plants produced flowers in the third spring (Supplementary Fig. S6). However, the WT plants didn’t produce flowers up to five years after plantation in the greenhouse (Supplementary Fig. S6). The results obtained from the transgenic Jatropha plants indicate that JcLFY is involved in FM determination in Jatropha.
Figure 4.
35S:JcLFY-overexpressing transgenic Jatropha exhibited slightly early flowering (A) Wild-type (WT) Jatropha grown in the field for 2 months and still in the vegetative stage, bar = 50 cm. (B) Shoot apex of WT Jatropha in the field, no flower produced. bar = 1 cm. (C,E,G) 35S:JcLFY transgenic Jatropha (L8, L34, and L47, respectively) grown in the field for 2 months and at the anthesis stage, bar = 50 cm. (D,F,H) Inflorescence of transgenic Jatropha in the field with a single terminal flower on L47 (white arrow indicated in H). bar = 1 cm.
JcLFY co-suppression changed inflorescence structure and flower organ pattern
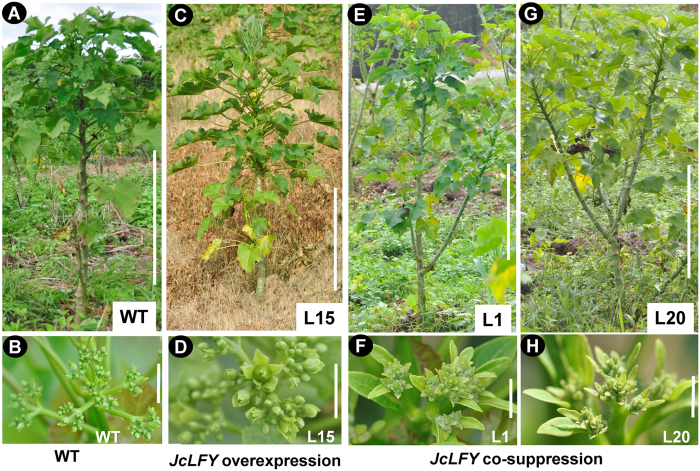
Among transgenic Jatropha generated with the 35S:JcLFY construct, we found three JcLFY co-suppressed plants. qRT-PCR results showed that JcLFY expression levels were more than 10 folds lower than WT in flower buds; L1 exhibited the lowest JcLFY expression levels (Supplementary Fig. S5A). Such co-suppression was first reported by Napoli et al.46, and it has been widely reported in many transgenic plants and animals47. These co-suppressed transgenic Jatropha plants exhibited no late-flowering phenotypes. When regenerated plantlets were grown in the field for 4 months, flower buds emerged in both co-suppressed transgenic and control plants (Fig. 5A,C,E,G).
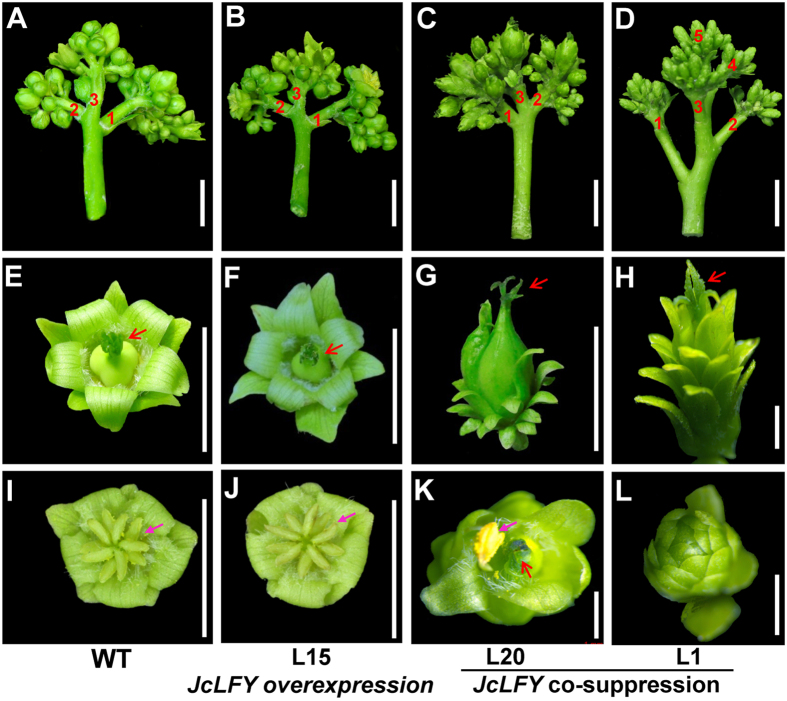
Figure 5. Co-suppression of JcLFY in Jatropha led to severe deficiencies in inflorescences and flowers.
(A) Wild type (WT) grown in the field for 4 months (at the anthesis stage), bar = 50 cm. (B) WT inflorescence in (A), bar = 1 cm. (C) 35S:JcLFY transgenic Jatropha L15 grown in the field for 4 months (at the anthesis stage); the plants exhibited normal flowering times and floral organs, bar = 50 cm. (D) Transgenic Jatropha inflorescence from L15 in (C), bar = 1 cm. (E,G) 35S:JcLFY co-suppressed Jatropha (L1 and L20) grown in the field for 4 months; plants exhibited normal flowering times, bar = 50 cm. (F,H) Inflorescences of L1 in (E) and L20 in (G), these plants exhibited abnormal inflorescences. bar = 1 cm.
Unlike many other species, Jatropha flowers are not subtended by small leaves called bracts (Fig. 5B,D). Jatropha flowers are composed of three concentric rings of organs: five sepals in the first, outermost whorl; five petals in the second whorl; and ten stamens (male flower) or one carpel (female flower) in the third whorl.
Inflorescences of JcLFY co-suppressed plants exhibited many bracts surrounding florets (Fig. 5F,H). One primary inflorescence branch was analyzed for its secondary inflorescence structure revealing there were more secondary inflorescence branches in co-suppressed plants.
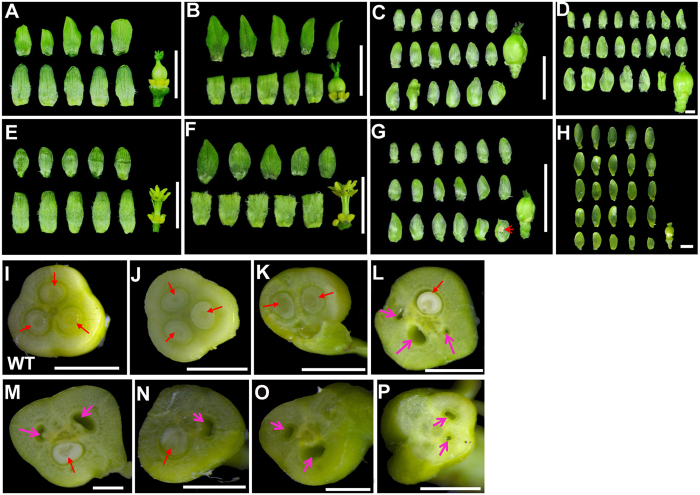
Flowers of weakly JcLFY co-suppressed plants (i.e., L20) bloomed, but all floral organs were abnormal. In male and female flowers, sepals and petals were replaced by sepal-like structures (sepaloid organs). In female flowers, stigmas were abnormal (Fig. 6G); in male flowers, only 1–2 stamens were observed, but abnormal stigmas also occurred (Fig. 6K). The first few flowers were more abnormal than later flowers; most of the early arising flowers were aborted. Such female flowers (the central flowers, specifically) have 15–20 sepaloid organs and an abnormal carpel (Fig. 7C); marginal flowers (bisexual flowers; in WT, the flower in this position is specifically male) of L20 had 15–20 sepaloid organs and a stamen fused to sepaloid organs in each flower (Fig. 7C). A cross section of the carpels of this plant revealed abnormal ovule and ovule cavity numbers, with only 0–2 observable ovules (Fig. 7K–M). However, WT and JcLFY-overexpression plants exhibited three ovules and ovule cavities per flower (Fig. 7I,J). Since well-developed stamens are rare and ovule numbers were reduced, the fertility of weakly JcLFY co-suppressed plants was likely reduced.
Figure 6. Phenotypes of co-suppressed JcLFY Jatropha flowers.
(A) A primary inflorescence branch from a wild-type (WT) plant; 3 secondary inflorescence branches (noted by numerals) were produced (bar = 1 cm, unless otherwise noted). (B) A primary inflorescence branch of JcLFY overexpression plant (L15); 3 secondary inflorescence branches (noted by numerals) were produced. (C) A primary inflorescence branch of a JcLFY co-suppressed plant (L20); 3 secondary inflorescence branches (noted by numerals) were produced. (D) A primary inflorescence branch of JcLFY co-suppressed plant (L1); 5 secondary inflorescence branches (noted by numerals) were produced. (E) Female flower (central flower) of WT plant. (F) Female flower (central flower) of JcLFY-overexpression plants (L15). (G) Female flower (central flower) of JcLFY co-suppression plant (L20). (H) Central flower of JcLFY co-suppressed plant (L1, bar = 1 mm). (I) Male flower of WT plant. (J) Male flower (marginal flower) of JcLFY overexpression plants (L15). (K) Marginal flower of JcLFY co-suppressed plant (L20) with 1 stamen and 1 stigma exhibited in the same flower (bar = 1 mm). (L) Marginal flower of a JcLFY co-suppressed plant (L1); the flower could not bloom (bar = 1 mm). Red arrows indicate stigmas; pink arrows indicate stamens.
Figure 7. Anatomy of flowers from JcLFY overexpression and co-suppressed plants.
(A) Wild-type (WT) female flower (bar = 1 cm, unless otherwise noted). (B) JcLFY overexpression female flower. (C) JcLFY co-suppression female flower (L20) with 15–20 sepaloid organs in each flower. (D) JcLFY co-suppression female flower (L1) with 20–25 sepaloid organs in each flower (bar = 1 mm). (E) WT male flower. (F) JcLFY overexpression male flower. (G) JcLFY co-suppressed marginal flower (bisexual flower; L20) with a stamen fused to a sepaloid organ (red arrow). There are 15–20 sepaloid organs in each flower. (H) JcLFY co-suppressed marginal flower (no ovule, female flower; L20). There are 22–28 sepaloid organs in each flower (bar = 1 mm). (I) WT carpel cross section with three ovules (bar = 1 mm). (J) JcLFY overexpression carpel cross section with three ovules (bar = 1 mm). (K–P) JcLFY co-suppressed carpel cross section with ovules and the ovule cavity partially or completely absent (bar = 1 mm). Red arrows indicate ovules and pink arrows indicate the ovule cavities without ovules.
Flowers of strongly JcLFY co-suppressed plants (i.e., L1) were all aborted, all flower organs were abnormal, and the flowers were smaller than those of the controls (Fig. 6D). Male and female flowers’ sepals and petals were replaced by sepaloid organs. Female flower stigmas were severely abnormal (Fig. 6H); in male flowers, stamens were unobserved (Fig. 6L). We dissected the flowers of such plants, finding the female flowers (i.e., central flowers) had 20–25 sepaloid organs and abnormal carpels (Fig. 7D); the marginal flower (in WT, this is a male flower) of L1 plants had 22–28 sepaloid organs and an abnormal carpel-like organ (Fig. 7H). Cross sections of the carpels of this plant revealed abnormal ovule and ovule cavity numbers, with only 0–1 ovules in central flowers (Fig. 7N,O) and no ovules in marginal flowers (Fig. 7P). Flowers of this plant were male and female sterile.
Comparison of these JcLFY co-suppressed plants demonstrated that stronger JcLFY co-suppression was associated with more sepaloid organs and fewer stamens and ovules. This indicates that JcLFY is important in regulating floral organ development.
Altered expression of JcLFY affected fruit and seed development
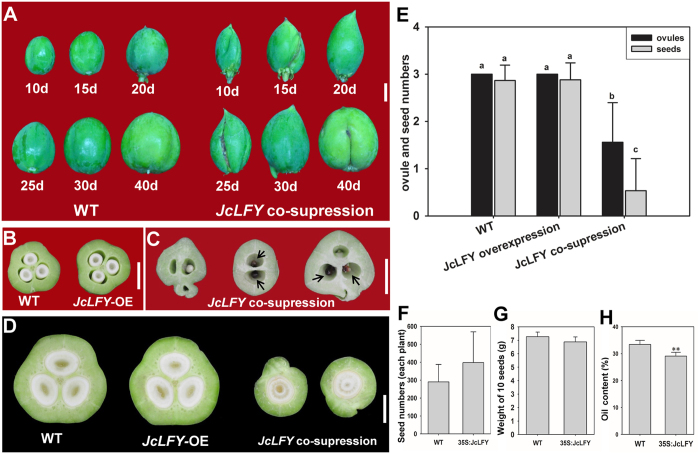
To further analyze whether JcLFY can affect fruit development, we analyzed fruit phenotypes at various developmental stage. In JcLFY co-suppressed plants, we found fruits were longer than in WT plants. Additionally, fruits were narrower, and the shapes were severely abnormal, which was maintained to maturity (Fig. 8A). Cross sections of 10-day-old fruits revealed the WT and JcLFY-overexpression plants had fruits with 3 seeds each (Fig. 8B); however, the JcLFY co-suppressed plant fruits had only 0–1 seed each, and most of the seeds aborted at an early stage (Fig. 8C). Normally, seedless-fruits are aborted at an early stage, and most developable fruits have only one seed in each fruit (Fig. 8D,E). Because of ovule abortion in co-suppressed plants, number of seeds per fruit was fewer than number of ovules per female flower (Fig. 8E).
Figure 8. Fruit and seed development in JcLFY co-suppressed and JcLFY overexpressing transgenic plants.
(A) Wild-type (WT, left) and JcLFY co-suppressed (L20, right) fruit at different development stages (10 d, 15 d, 20 d, 25 d, 30 d, and 40 d after fertilization); the fruits varied in size and shape; (B) WT (left) and JcLFY overexpression (right) fruit cross sections (15 d after fertilization). (C) Cross section of JcLFY co-suppressed fruits (10 d after fertilization). Most of the seeds were aborted (black arrows). (D) Cross section of WT, JcLFY overexpressing, and JcLFY co-suppressed fruits (40 d after fertilization). (E) Ovule and seed numbers in WT, JcLFY overexpression, and JcLFY co-suppression fruits. Each value is the mean of 30 fruits. Error bars represent standard deviations. Means with different letters are different according to Tukey’s tests (p < 0.05). Comparison of the seed number per plant (F), weight of 10 seeds (G) and seed oil content (H) between 35S:JcLFY T0 transgenic plants and WT. Fifteen plants from WT and T0 transgenic plants respectively were used for statistical analysis. The plants were 1-year old after transfer to the field. Scale bars = 1 cm. **Significantly different from the control at the 1% level.
To assess the potential of JcLFY-overexpression plants in the genetic improvement of Jatropha, we analyzed the seed number per plant, seed weight and seed oil content in T0 transgenic plants of 35S:JcLFY (Fig. 8F–H). There was no statistical difference in seed number per plant between transgenic plants and wild-types, although the average seed number per plant of 35S:JcLFY was slightly more than that of wild-type (Fig. 8F). There was also no significant change in seed weight (Fig. 8G), but oil content in seeds of 35S:JcLFY transgenic plants was significantly decreased (Fig. 8H). Because extensive variation in seed number per plant was observed among T0 transgenic plants of 35S:JcLFY (Fig. 8F), homozygous plants from the transgenic lines by self-pollination and subsequent vegetative propagation need to be used for further assessment of the overall effect of JcLFY-overexpression on oil yield of transgenic plants.
Discussion
JcLFY is an ortholog of Arabidopsis LFY
JcLFY shares high amino acid sequence similarity with other LFY proteins and most closely resembles Ricinus communis LFY. JcLFY contains conserved domains such as a proline-rich region and a leucine zipper motif in the N-terminal domain (Supplementary Fig. S1B), suggesting that JcLFY might have a similar function as LFY. We detected JcLFY transcripts in several tissues, finding the highest accumulation in flower buds (Fig. 1A). Moreover, JcLFY expression was highest in early stage inflorescence buds (IB1) and early stage flower buds (FB1) (Fig. 1B,C), implying a possible role of JcLFY in regulating Jatropha flowering.
The present study has shown that over-expressing 35S:JcLFY in Arabidopsis can inhibit vegetative growth, hence reducing branch number and heights of transgenic plants while promoting early flowering, by approximately 15 days and 2 months relative to WT plants under LD and SD conditions, respectively (Tables 1 and 2). This is quite similar to phenotypes under constitutive LFY ortholog expression in Arabidopsis27, Gloxinia29, Brassica juncea30, and Nicotiana tabacum28. Constitutive LFY expression driven by the CaMV35S promoter attenuates development of both vegetative and inflorescence phases, inducing the production of terminal flowers on Arabidopsis primary shoots27.
This study has shown that JcLFY overexpression in Arabidopsis produced terminal and solitary flowers (Fig. 2). These findings are similar to the phenotypic changes caused by constitutive LFY expression in Arabidopsis27,48. The production of terminal and solitary flowers in LFY-overexpressing plants is caused by the inhibition of TFL1 expression induced by LFY49. TFL1 expression was reduced in transgenic L12 plants (Supplementary Fig. S3I). Overexpression of JcLFY in the Arabidopsis lfy-15 mutant recovered the late flowering phenotype, rescued the abnormal flowers of lfy mutant plants, and repressed inflorescence development (Fig. 3B–E). This suggests that JcLFY acted as a functional homolog of LFY in Arabidopsis.
JcLFY expression was not induced by GA
GA has been generally found to strongly inhibit flowering in some woody perennial plants, such as citrus, rose, grape, and apple36,50,51,52. However, GA accelerates flowering in Arabidopsis and Chrysanthemum by inducing expression of the FM identity gene LFY34,35,36,44. We detected JcLFY expression levels in GA-treated shoot apex and flower bud tissues at different time points, revealing that JcLFY expression levels were not induced by GA. In contrast, JcLFY expression levels decreased between 3 and 48 h after GA application (Fig. 1E). This contrasts with Arabidopsis findings, suggesting GA may play a negative function in regulating flowering in Jatropha. Recently research revealed that although GA promoted termination of vegetative development, it inhibited flower formation in Arabidopsis36. Ghosh et al.9 found that paclobutrazol, a GA biosynthesis inhibitor, promotes flower initiation in Jatropha, further suggesting GA inhibits flower initiation in Jatropha. We propose that GA inhibition of flowering in Jatropha might occur via repression of JcLFY expression. This hypothesis need to be tested in future studies by determination of endogenous GA levels in different developmental stages of wild-type plants, and by analysis of phenotypic changes in flowering time in transgenic plants with altered GA biosynthesis or perception.
JcLFY regulated flowering time and floral organs
LFY is a FM identity gene that plays an important role in promoting flowering in Arabidopsis13,19. The JcLFY-overexpressing transgenic Arabidopsis bolted two months earlier than WT plants under SD conditions (Table 2). However, JcLFY transgenic Jatropha plants took more than seven months to produce flower buds, although two months earlier than WT plants (Fig. 4). And in a subtropical area, the 35S:JcLFY transgenic Jatropha produced flowers until two or three years after plantation when no flower was found in WT plants (Supplementary Fig. S6). Compared to the 35S:LFY transgenic citrus described by Peña et al.31, the flowering time of transgenic Jatropha was very late; in citrus, transgenic shoots flowered just five weeks after regeneration. Therefore, the JcLFY-overexpression transgenic lines used in this work showed a slightly early flowering, but weaker phenotype than plants overexpressing the Arabidopsis LFY gene in Arabidopsis27 and citrus31. The early flowering phenotype of JcLFY transgenic Jatropha was also weaker than observed in studies of transgenic Jatropha plants overexpressing the florigen gene JcFT41,42, in which flower buds initiated directly from transformed callus 7 weeks after in vitro culture47. However, the JcLFY co-suppressed transgenic Jatropha plants did not exhibit late flowering (Fig. 5). These analysis of flowering time in overexpression and co-suppression plants suggest that JcLFY may not be a key flowering promoter.
In Jatropha, FMs are derived from inflorescence meristems (IMs), but FMs execute a developmental program very different from those of IMs. Thus, there must be factors that promote the determination of FMs but not IMs. JcLFY is one of these factors because co-suppression of JcLFY delayed flower formation, leading to production of more secondary inflorescence branches (Fig. 6D). Inactivation of JcLFY induced abnormal flower development in many aspects. First, the inflorescences and flowers were subtended by bracts (Fig. 5F,H). Second, most flowers were aborted, especially in the strongly co-suppressed plants. Third, the outermost floral organs were sepal-like. These sepaloid organs substantially outnumbered sepals and petals combined in WT plants (Fig. 7A–H). Fourth, the number of stamens was reduced, and the morphology of stamens was changed (Figs 6K,L and 7G,H). Fifth, carpels occurred in every flower of the co-suppressed plants, but their morphologies were abnormal, with irregular shapes and sizes (Figs 6G,H,K and 7C,D,G,H), which may result in the abnormal fruits in JcLFY co-suppressed plants (Fig. 8). Sixth, the ovules and ovule cavities were partially or completely absent in the co-suppressed plants (Fig. 7I–P).
The flower phenotypes of JcLFY co-suppressed plants were different from those of Arabidopsis single mutants of ap1, ap2, ap3, ag, gi, or sep53,54,55. The co-suppressed plants produced more inflorescences, bracts and sepaloid organs, and exhibited reduced fertility, closely resembling Arabidopsis lfy mutants14. Our qRT-PCR analysis (Supplementary Fig. S5) suggest these phenotypes may be induced by downregulation of JcAP1, JcAP3, JcAG, JcSEP1, JcSEP2, and JcSEP3 in JcLFY co-suppressed Jatropha.
Materials and Methods
Plant materials
Wild-type Jatropha plants were grown in the Xishuangbanna Tropical Botanical Garden (XTBG; 21°54′N, 101°46′E, 580 m in altitude) of the Chinese Academy of Sciences located in Mengla County, Yunnan Province, southwestern China. Young transgenic Jatropha plants were grown in a greenhouse. Mature transgenic Jatropha were planted in a field plot within the XTBG in 2014–2015. The field plot for transgenic Jatropha plantation was isolated by other horticultural plant species and fruit trees, such as Bauhinia, papaya, and grapefruit. The roots, stems, mature leaves, inflorescence buds, flower buds, male flowers, female flowers, and fruits of Jatropha were collected during the summer. All of the tissues collected for qRT-PCR were immediately frozen in liquid N2 and stored at −80 °C until use. The Arabidopsis thaliana WT and lfy-15 mutant of the Columbia ecotype (Col-0) were obtained from The Arabidopsis Information Resource (TAIR) (http://www.arabidopsis.org/). Arabidopsis seeds were germinated on 1/2 Murashige and Skoog (MS) medium for one week. Then, the seedlings were transferred to peat soil in plant growth chambers maintained at 22 ± 2 °C under long-day (LD; 16 h light/8 h dark) or short-day (SD; 8 h light/16 h dark) conditions. Phenotype analysis was performed on homozygous (T2) Arabidopsis plants and heterozygous (T0) Jatropha plants. More than 20 plants were used for the characterization of each Arabidopsis genotype. The number of rosette leaves, branches, and time (days) between transfer to soil and the appearance of the first flower bud as well as plant heights were recorded. The aboveground tissues of 15-day-old Arabidopsis seedlings were harvested to analyze mRNA transcription levels.
Cloning full length JcLFY cDNA
Total RNA was isolated from Jatropha flower buds using the silica particle method56. cDNA synthesis was performed by using the SMARTTM cDNA Library Construction Kit (Clontech, Mountain View, CA, USA) according to the manufacturer’s instructions.
Two pairs of primers (XT97/XT98 and XT99/XT100) were designed according to the conserved LFY sequences of other woody species: Ricinus communis (gi|255540036|), Populus trichocarpa (gi|224136347|), Salix discolor (gi|29423801|), Mangifera (gi|323650468|), Dimocarpus longan (gi|144686979|), and Buddleja davidii (gi|76495760|). A 552-bp JcLFY fragment was isolated from Jatropha cDNA by nested PCR. JcLFY full length cDNA was obtained by RACE-PCR with gene-specific primers (XT130 for JcLFY 5′ RACE, XT131 for JcLFY 5′ RACE nested PCR, XT132 for JcLFY 3′ RACE, and XT133 for JcLFY 3′ RACE nested PCR), which were designed based on the 552-bp JcLFY fragment. A detailed sequence alignment was performed using BLAST against the NCBI database.
Sequence and phylogenetic analyses
Specific primers (XT134/XT135) were designed to obtain the full length cDNA and genomic DNA sequences of JcLFY. Genomic sequences were amplified from 30-ng samples of DNA, and 2 μl of cDNA was used as a template for amplifying JcLFY full length cDNA sequences. The amplified PCR products were subjected to 1% agarose gel electrophoresis. Purified PCR fragments were cloned into a T&A cloning vector (Promega, Madison, Wisconsin, USA) for sequencing. Sequence alignment was carried out using Vector NTI 11 software (Invitrogen, Carlsbad, USA). To determine the amino acid sequence, the alignment results were subjected to pairwise comparisons using DNAMAN 5.0 (Lynnon Biosoft, Quebec, Canada). A phylogenetic tree based on the protein sequences was constructed with MEGA 5.057. A neighbor-joining phylogenetic tree was generated with MEGA 5.0 using a Poisson model with gamma-distributed substitution rates and 1000 bootstrap replicates.
Vector construction and plant transformation
To characterize the function of JcLFY, the coding region of JcLFY was cloned into a derived pORE R4 vector (obtained from TAIR) driven by the cauliflower mosaic virus (CaMV) 35S promoter at the SmaI and SalI sites of the pORE R4 vector. The construct was transformed into Agrobacterium tumefaciens strain EHA 105 via the freeze-thaw method58 and transformed into WT and lfy-15 mutant Arabidopsis by the Agrobacterium tumefaciens-mediated floral dip transformation method59. Transgenic lines were selected on 1/2 MS medium containing 50 μg/ml kanamycin, and transformed seedlings were further identified by JcLFY-specific PCR analysis. Verified transgenic T2 generation seedlings were transplanted into pots and grown in different light conditions for further experiments. Transformation of Jatropha with the Agrobacterium strain EHA105 carrying the same construct was performed according to the protocol described by Fu et al.60. All transgenic plants were confirmed using genomic PCR and RT-PCR.
Semi-quantitative PCR and real-time qPCR analyses
Total RNA was isolated from different tissues of Jatropha using the silica particles method56. Ten RNA samples from different Jatropha tissues were obtained, including roots, young leaves, mature leaves, flower buds, flowers, shoot apices, pedicels, fruits, endosperms, and embryos. Total RNA was extracted from frozen Arabidopsis tissues derived from 15-day-old WT plants, lfy-15 mutant plants, WT transgenic plants (lines 8, 11, and 12, henceforth L8, L11, and L12), and lfy-15 mutant transgenic plants harboring JcLFY (plants C1 and C4) using TRIzol reagent (Biocentury Transgene, Shenzhen, China). Total RNA samples of transgenic and WT Jatropha were isolated from the flower buds using the silica particles method56. First-strand cDNA was synthesized with the PrimeScript® RT Reagent Kit with gDNA Eraser (TAKARA, Dalian, China). The cDNA templates of first-strand cDNA were diluted 5-fold with sterilized double-distilled water. qRT-PCR was performed using SYBR® Premix Ex Taq™ II (TAKARA) on a Roche 480 Real-Time PCR Detection System (Roche, Mannheim, Germany). The primers employed for qRT-PCR and semi-quantitative PCR are listed in Supplementary Table S1. qRT-PCR was conducted with three independent biological replicates and three technical replicates for each sample. The data were analyzed using the 2−ΔΔCT method described by Livak and Schmittgen61. RT-PCR was carried out as described by Brownie et al.62. The transcript levels of specific genes were normalized using Jatropha JcActin1 or Arabidopsis Actin2.
Gene array analyses
The 35S:JcLFY transgenic Arabidopsis were transplanted into soil under LD conditions. Total RNA was extracted from the aboveground tissues of 2-week-old soil-grown WT Arabidopsis and transgenic Arabidopsis using an RNeasy® Plant Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. Total RNA was quantified, and its quality was assessed using an Agilent 2100 Bioanalyzer (Santa Clara, CA, USA). Microarray analysis was performed using the Arabidopsis (V4, 4 × 44) Gene Expression Microarray, Design ID: 021169 (Agilent) containing 43,603 Arabidopsis gene probes and 1,417 Agilent control probes. A total of at least 1 μg purified RNA was required for gene array analyses.
Labeled cRNA probes were fragmented using fragmentation buffer and hybridized to the Arabidopsis arrays in the presence of the Gene Expression Hybridization buffer HI-RPM and blocking agent for 17 h at 65 °C with a 10-rpm rotation speed in a hybridization oven. After the 17 h incubation, the arrays were washed using low stringency wash buffer 1 at room temperature for 1 min followed by a high stringency wash using wash buffer 2 at 37 °C. The arrays were air-dried and scanned using a high-resolution array scanner (Agilent) with the appropriate settings for one-color gene expression arrays. The signal intensities were extracted from the scanned images with the aid of Feature Extraction software 10.7.1.1 (Agilent) and subjected to background subtraction and spatial detrending. The outliers and abnormal features were flagged, and the data were normalized using intra-array percentile shift normalization (minimum threshold of 75) and median-based inter-array normalization. GeneSpring GX (Agilent) was used to calculate intensity ratios and fold changes. All genes with a P-value below 0.05 and at least 2-fold expression changes were chosen for a Gene Ontology enrichment analysis. The gene array hybridization experiments were performed by Shanghai Biotechnology Corporation (Shanghai, China).
Analysis of flower, fruit, and seed phenotypes
One-year-old T0 transgenic plants grown in the Xishuangbanna Tropical Botanical Garden (XTBG; 21°54′N, 101°46′E, 580 m in altitude) of the Chinese Academy of Sciences were used to analyze the flower, fruit, and seed phenotypes. The seeds were harvested in autumn 2015. Flower and fruit anatomies were examined and photographed with a light Leica DM IRB anatomical lens (Leica, Heerbrugg, Switzerland) equipped with a Leica DFC425 C camera (Leica).
Additional Information
How to cite this article: Tang, M. et al. An ortholog of LEAFY in Jatropha curcas regulates flowering time and floral organ development. Sci. Rep. 6, 37306; doi: 10.1038/srep37306 (2016).
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
We thank Mr. Congcong Gao, Miss Dongyun Bao, Miss Xiulan Wang, and Mr. Zhiyu Pu for transplanting and maintaining transgenic Jatropha plants. This work was supported by funding from the National Natural Science Foundation of China (31370595 and 31300568), the Natural Science Foundation of Yunnan Province (2014FB186), and the CAS 135 Program (XTBG-T02). The authors gratefully acknowledge the Central Laboratory of the Xishuangbanna Tropical Botanical Garden for providing the research facilities.
Footnotes
Author Contributions M.T. designed the experiments, performed the experiments, analyzed the results, and wrote the manuscript. Z.-F.X. conceived and designed the study, analyzed the results, and revised the manuscript. Y.-B.T., Q.F., Y.S. and L.-J.N. performed some of the experiments or revised the manuscript. All authors reviewed and approved the manuscript.
References
- Kumar A. & Sharma S. An evaluation of multipurpose oil seed crop for industrial uses (Jatropha curcas L.): A review. Ind. Crop. Prod. 28, 1–10, doi: 10.1016/j.indcrop.2008.01.001 (2008). [DOI] [Google Scholar]
- Pandey V. C. et al. Jatropha curcas: A potential biofuel plant for sustainable environmental development. Renew. Sust. Energ. Rev. 16, 2870–2883, doi: 10.1016/j.rser.2012.02.004 (2012). [DOI] [Google Scholar]
- Akashi K. Jatropha research: A new frontier for biofuel development. Plant Biotechnol. 29, 121, doi: 10.5511/plantbiotechnology.12.0003p (2012). [DOI] [Google Scholar]
- Khalil H. P. S. A. et al. A Jatropha biomass as renewable materials for biocomposites and its applications. Renew. Sust. Energ. Rev. 22, 667–685, doi: 10.1016/j.rser.2012.12.036 (2013). [DOI] [Google Scholar]
- Ong H. C., Mahlia T., Masjuki H. & Norhasyima R. Comparison of palm oil, Jatropha curcas and Calophyllum inophyllum for biodiesel: a review. Renew. Sust. Energ. Rev. 15, 3501–3515, doi: 10.1016/j.rser.2011.05.005 (2011). [DOI] [Google Scholar]
- Pramanik K. Properties and use of Jatropha curcas oil and diesel fuel blends in compression ignition engine. Renew. Energ. 28, 239–248, doi: 10.1016/S0960-1481(02)00027-7 (2003). [DOI] [Google Scholar]
- Pan B.-Z. & Xu Z.-F. Benzyladenine treatment significantly increases the seed yield of the biofuel plant Jatropha curcas. J. Plant Growth Regul. 30, 166–174, doi: 10.1007/s00344-010-9179-3 (2011). [DOI] [Google Scholar]
- Fairless D. Biofuel: The little shrub that could - maybe. Nature 449, 652–655, doi: Doi 10.1038/449652a (2007). [DOI] [PubMed] [Google Scholar]
- Ghosh A. et al. Paclobutrazol arrests vegetative growth and unveils unexpressed yield potential of Jatropha curcas. J. Plant Growth Regul. 29, 307–315, doi: 10.1007/s00344-010-9137-0 (2010). [DOI] [Google Scholar]
- Roux F., Touzet P., Cuguen J. & Le Corre V. How to be early flowering: an evolutionary perspective. Trends Plant Sci. 11, 375–381, doi: 10.1016/j.tplants.2006.06.006 (2006). [DOI] [PubMed] [Google Scholar]
- Albani M. C. & Coupland G. Comparative analysis of flowering in annual and perennial plants in Curr. Top Dev. Biol. (ed Timmermans Marja C. P.) 323–348, doi: 10.1016/S0070-2153(10)91011-9 (Academic Press, 2010). [DOI] [PubMed] [Google Scholar]
- Amasino R. Seasonal and developmental timing of flowering. Plant J. 61, 1001–1013, doi: 10.1111/j.1365-313X.2010.04148.x (2010). [DOI] [PubMed] [Google Scholar]
- Araki T. Transition from vegetative to reproductive phase. Curr. Opin. Plant Biol. 4, 63–68, doi: 10.1016/s1369-5266(00)00137-0 (2001). [DOI] [PubMed] [Google Scholar]
- Weigel D., Alvarez J., Smyth D. R., Yanofsky M. F. & Meyerowitz E. M. LEAFY controls floral meristem identity in Arabidopsis. Cell 69, 843–859, doi: 10.1016/0092-8674(92)90295-n (1992). [DOI] [PubMed] [Google Scholar]
- Coen E. S. et al. FLORICAULA a homeotic gene required for flower development in Antirrhinum-majus. Cell 63, 1311–1322, doi: 10.1016/0092-8674(90)90426-f (1990). [DOI] [PubMed] [Google Scholar]
- Wagner D. Flower morphogenesis: timing is key. Dev. Cell 16, 621–622, doi: 10.1016/j.devcel.2009.05.005 (2009). [DOI] [PubMed] [Google Scholar]
- Moyroud E., Kusters E., Monniaux M., Koes R. & Parcy F. LEAFY blossoms. Trends Plant Sci. 15, 346–352, doi: 10.1016/j.tplants.2010.03.007 (2010). [DOI] [PubMed] [Google Scholar]
- Benlloch R., Berbel A., Serrano-Mislata A. & Madueno F. Floral initiation and inflorescence architecture: A comparative view. Ann. Bot. 100, 1609–1609, doi: 10.1093/aob/mcm293 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blazquez M. A., Soowal L. N., Lee I. & Weigel D. LEAFY expression and flower initiation in Arabidopsis. Development 124, 3835–3844 (1997). [DOI] [PubMed] [Google Scholar]
- Chahtane H. et al. A variant of LEAFY reveals its capacity to stimulate meristem development by inducing RAX1. Plant J. 74, 678–689, doi: 10.1111/tpj.12156 (2013). [DOI] [PubMed] [Google Scholar]
- Siriwardana N. S. & Lamb R. S. The poetry of reproduction: the role of LEAFY in Arabidopsis thaliana flower formation. Int. J. Dev. Biol. 56, 207–221, doi: 10.1387/ijdb.113450ns (2012). [DOI] [PubMed] [Google Scholar]
- Liljegren S. J., Gustafson-Brown C., Pinyopich A., Ditta G. S. & Yanofsky M. F. Interactions among APETALA1, LEAFY, and TERMINAL FLOWER1 specify meristem fate. Plant Cell 11, 1007–1018 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bomblies K. et al. Duplicate FLORICAULA/LEAFY homologs zfl1 and zfl2 control inflorescence architecture and flower patterning in maize. Development 130, 2385–2395, doi: 10.1242/dev.00457 (2003). [DOI] [PubMed] [Google Scholar]
- Kyozuka J., Konishi S., Nemoto K., lzawa T. & Shimamoto K. Down-regulation of RFL, the FLO/LFY homolog of rice, accompanied with panicle branch initiation. P. Natl. Acad. Sci. USA 95, 1979–1982, doi: 10.1073/pnas.95.5.1979 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H. et al. Control of compound leaf development by FLORICAULA/LEAFY ortholog SINGLE LEAFLET1 in Medicago truncatula. Plant Physiol. 146, 1759–1772, doi: 10.1104/pp.108.117044 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofer J. et al. UNIFOLIATA regulates leaf and flower morphogenesis in pea. Curr. Biol. 7, 581–587, doi: 10.1016/s0960-9822(06)00257-0 (1997). [DOI] [PubMed] [Google Scholar]
- Weigel D. & Nilsson O. A developmental switch sufficient for flower initiation in diverse plants. Nature 377, 495–500 doi: 10.1038/377495a0 (1995). [DOI] [PubMed] [Google Scholar]
- Ahearn K. P., Johnson H. A., Weigel D. & Wagner D. R. NFL1, a Nicotiana tabacum LEAFY-like gene, controls meristen initiation and floral structure. Plant Cell Physiol. 42, 1130–1139, doi: 10.1093/pcp/pce143 (2001). [DOI] [PubMed] [Google Scholar]
- Zhang M.-Z. et al. Overexpression of the cucumber LEAFY homolog CFL and hormone treatments alter flower development in gloxinia (Sinningia speciosa). Plant Mol. Biol. 67, 419–427, doi: 10.1007/s11103-008-9330-8 (2008). [DOI] [PubMed] [Google Scholar]
- Roy S. D., Saxena M. & Bhalla-Sarin N. Overexpression of AtLEAFY accelerates flowering in Brassica juncea. Crop Sci. 49, 930–936, doi: 10.2135/cropsci2008.03.0118 (2009). [DOI] [Google Scholar]
- Peña L. et al. Constitutive expression of Arabidopsis LEAFY or APETALA1 genes in citrus reduces their generation time. Nat. Biotechnol. 19, 263–267, doi: 10.1038/85719 (2001). [DOI] [PubMed] [Google Scholar]
- Huala E. & Sussex I. M. LEAFY interacts with floral homeotic genes to regulate Arabidopsis floral development. Plant Cell 4, 901–913 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W. et al. LEAFY controls auxin response pathways in floral primordium formation. Sci. Signal 6, doi: 10.1126/scisignal.6277er3 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson S., Bohlenius H., Moritz T. & Nilsson O. GA4 is the active gibberellin in the regulation of LEAFY transcription and Arabidopsis floral initiation. Plant Cell 18, 2172–2181, doi: 10.1105/tpc.106.042317 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blazquez M. A., Green R., Nilsson O., Sussman M. R. & Weigel D. Gibberellins promote flowering of Arabidopsis by activating the LEAFY promoter. Plant Cell 10, 791–800 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi N. et al. Gibberellin acts positively then negatively to control onset of flower formation in Arabidopsis. Science 344, 638–641, doi: 10.1126/science.1250498 (2014). [DOI] [PubMed] [Google Scholar]
- Southerton S. G. et al. Eucalyptus has a functional equivalent of the Arabidopsis floral meristem identity gene LEAFY. Plant Mol. Biol. 37, 897–910, doi: 10.1023/a:1006056014079 (1998). [DOI] [PubMed] [Google Scholar]
- Mellerowicz E. J., Horgan K., Walden A., Coker A. & Walter C. PRFLL - a pinus radiata homologue of FLORICAULA and LEAFY is expressed in buds containing vegetative shoot and undifferentiated male cone primordia. Planta 206, 619–629, doi: 10.1007/s004250050440 (1998). [DOI] [PubMed] [Google Scholar]
- Walton E. F., Podivinsky E. & Wu R. M. Bimodal patterns of floral gene expression over the two seasons that kiwifruit flowers develop. Physiol. Plantarum 111, 396–404, doi: 10.1034/j.1399-3054.2001.1110318.x (2001). [DOI] [PubMed] [Google Scholar]
- Yu Q. Y., Moore P. H., Albert H. H., Roader A. H. K. & Ming R. Cloning and characterization of a FLORICAULA/LEAFY ortholog, PFL, in polygamous papaya. Cell Research 15, 576–584, doi: 10.1038/sj.cr.7290327 (2005). [DOI] [PubMed] [Google Scholar]
- Li C., Luo L., Fu Q., Niu L. & Xu Z.-F. Isolation and functional characterization of JcFT, a FLOWERING LOCUS T (FT) homologous gene from the biofuel plant Jatropha curcas. BMC Plant Biol. 14, 125, doi: 10.1186/1471-2229-14-125 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J. et al. The Jatropha FT ortholog is a systemic signal regulating growth and flowering time. Biotechnol. Biofuels 7, 91, doi: 10.1186/1754-6834-7-91 (2014). [DOI] [Google Scholar]
- Tang M., Tao Y.-B. & Xu Z.-F. Ectopic expression of Jatropha curcas APETALA1 (JcAP1) caused early flowering in Arabidopsis, but not in Jatropha. PeerJ 4, e1969, doi: 10.7717/peerj.1969 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumitomo K., Li T. & Hisamatsu T. Gibberellin promotes flowering of chrysanthemum by upregulating CmFL, a chrysanthemum FLORICAULA/LEAFY homologous gene. Plant Sci. 176, 643–649, doi: 10.1016/j.plantsci.2009.02.003 (2009). [DOI] [Google Scholar]
- William D. A. et al. Genomic identification of direct target genes of LEAFY. P. Natl. Acad. Sci. USA 101, 1775–1780, doi: 10.1073/pnas.0307842100 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napoli C., Lemieux C. & Jorgensen R. Introduction of a chimeric chalcone synthase gene into petunia results in reversible co-suppression of homologous genes in trans. Plant Cell 2, 279–289, doi: 10.1105/tpc.2.4.279 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannon G. J. RNA interference. Nature 418, 244–251, doi: 10.1038/418244a (2002). [DOI] [PubMed] [Google Scholar]
- Melzer S., Kampmann G., Chandler J. & Apel K. FPF1 modulates the competence to flowering in Arabidopsis. Plant J. 18, 395–405, doi: 10.1046/j.1365-313X.1999.00461.x (1999). [DOI] [PubMed] [Google Scholar]
- Blazquez M. A., Ferrandiz C., Madueno F. & Parcy F. How floral meristems are built. Plant Mol. Biol. 60, 855–870 (2006). [DOI] [PubMed] [Google Scholar]
- Wilkie J. D., Sedgley M. & Olesen T. Regulation of floral initiation in horticultural trees. J Exp Bot 59, 3215–3228, doi: 10.1093/jxb/ern188 (2008). [DOI] [PubMed] [Google Scholar]
- Goldschmidt E. E., Tamim M. & Goren R. In Eight International Symposium on Plant Bioregulators in Fruit Production. Acta Horticulturae (eds Guardiola J. L. & Martinez J. L. G.) 201–208 (1997). [Google Scholar]
- Randoux M. et al. Gibberellins regulate the transcription of the continuous flowering regulator, RoKSN, a rose TFL1 homologue. J Exp Bot 63, 6543–6554, doi: 10.1093/jxb/ers310 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman J. L., Smyth D. R. & Meyerowitz E. M. Genetic interactions among floral homeotic genes of Arabidopsis. Development 112, 1–20 (1991). [DOI] [PubMed] [Google Scholar]
- Mandel M. A., Gustafson-Brown C., Savidge B. & Yanofskay M. F. Molecular chanracterization of the Arbidopsis floral homeotic gene APETALA1. Nature 360, 5 (1992). [DOI] [PubMed] [Google Scholar]
- Favaro R. et al. MADS-box protein complexes control carpel and ovule development in Arabidopsis. Plant Cell 15, 2603–2611, doi: 10.1105/tpc.015123 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L.-W., Sun Q.-Y., Wang Z.-Y., Sun Y.-B. & Xu Z.-F. Using silica particles to isolate total RNA from plant tissues recalcitrant to extraction in guanidine thiocyanate. Anal. Biochem. 374, 426–428, doi: 10.1016/j.ab.2007.11.030 (2008). [DOI] [PubMed] [Google Scholar]
- Tamura K. et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739, doi: 10.1093/molbev/msr121 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- An G. Binary ti vectors for plant transformation and promoter analysis. Method Enzymo. 153, 292–305 (1987). [Google Scholar]
- Clough S. J. & Bent A. F. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743 (1998). [DOI] [PubMed] [Google Scholar]
- Fu Q. et al. An efficient protocol for Agrobacterium-mediated transformation of the biofuel plant Jatropha curcas by optimizing kanamycin concentration and duration of delayed selection. Plant Biotechnol. Rep. 9, 405–416, doi: 10.1007/s11816-015-0377-0 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K. J. & Schmittgen T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2 ΔΔC T method. Methods 25, 402–408 (2001). [DOI] [PubMed] [Google Scholar]
- Brownie J. et al. The elimination of primer-dimer accumulation in PCR. Nucleic Acids Res. 25, 3235–3241, doi: 10.1093/nar/25.16.3235 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.