Abstract
Once thought to live independently, bacteria are now known to be highly social organisms. Their behavior ranges from cooperatively forming complex multispecies communities to fiercely competing for resources. Work over the past fifty years has shown that bacteria communicate through diverse mechanisms including exchanging diffusible molecules, exporting molecules in membrane vesicles, and interacting through direct cell-cell contact. These methods allow bacteria to sense and respond to other cells around them and coordinate group behavior. In this review, share the discoveries and lessons learned in the field of bacterial communication in the hope of providing insights to parasitologists and other researchers working on related questions.
Introduction
The natural world is teeming with social creatures. Male peacocks attract mates with striking ornamental feathers, honey bees perform intricate dances to direct relatives to food, lions roar in unison to intimidate competitors, and the list goes on. However, until relatively recently communication and social behavior was thought possible only in multicellular organisms. The discovery that bacteria can coordinate complex behaviors as a community via cell-cell communication has led to an exponential increase in related research. The accumulated literature makes it clear that not only are mechanisms for communication diverse and widespread among bacteria but communication is also frequently critical for their survival. In this review, we walk through the development of the field and describe the fundamental mechanics underlying bacterial communication systems and the functional roles these systems play in nature. We conclude by discussing potential therapeutics that interfere with bacterial communication and likely future directions for the field. We hope that our overview of the relatively mature field of bacterial communication will inspire new ideas about communication between other organisms such as parasites.
A brief history of bacterial communication research
Bacterial communication began to attract interest in the 1960s with studies of the symbiotic relationship between the marine bacterium Vibrio fischeri and the Hawaiian bobtail squid, Euprymna scalopes (Nealson and Hastings 1979). This bioluminescent bacterium resides in the squid’s ventral light organ and is thought to help the squid hide its shadow from predators by matching the intensity of the overlying moonlight (Figure 1-A). Researchers noted that the bacteria became bioluminescent only after reaching a high cellular density (Kempner and Hanson 1968). It was first suggested that the artificial media used to grow V. fischeri in the laboratory contained an unknown inhibitory substance that prevented bioluminescence until the inhibitor was consumed (Kempner and Hanson 1968). However, in 1970 Kenneth Nealson and colleagues demonstrated that bioluminescence was actually induced by a molecule released by the bacteria themselves. This molecule accumulates to high concentrations during growth and induces bioluminescence in a process the researchers termed autoinduction (Nealson, Platt et al. 1970) (see glossary for definitions of bolded terms). The autoinducing molecule, N-(3-oxohexanoyl) homoserine lactone, was isolated in 1981 (Eberhard, Burlingame et al. 1981), and the genes involved in the regulatory pathway were soon identified (Engebrecht, Nealson et al. 1983).
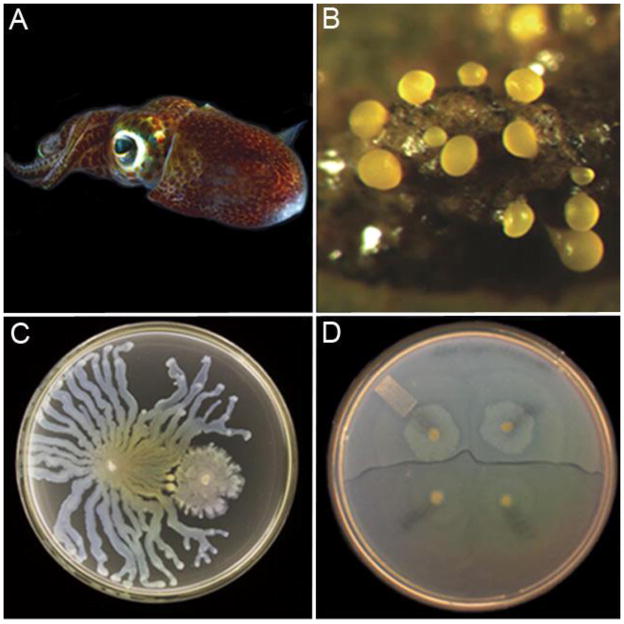
Figure 1. Examples of bacterial communication.
A) Bioluminescent Vibrio fischeri in symbiosis with the Hawaiian Bobtail Squid. V. fischeri controls its bioluminescence through Quorum Sensing (QS), a process involving the exchange of diffusible, density-dependent signal molecules. The squid uses bioluminescence as a camouflaging mechanism. (Photograph courtesy Margaret McFall-Ngai) B) Fruiting body of Myxococcus xanthus. Communities of M. xanthus use a membrane-bound, contact-dependent signal to aggregate and coordinate the formation of a structure called a fruiting body. Cells in the center differentiate into environmentally resistant, metabolically inert spores and are released to germinate at a future time when environmental conditions are more favorable. (Photograph courtesy Michiel Vos) C) Wild-type Pseudomonas aeruginosa on the left regulates genes required for swarming behavior via QS. QS-deficient mutants on the right are unable to communicate and activate QS-controlled genes including those important for swarming (Photograph by Steve Diggle and Edgar Lis-sel). D) Swarms of Proteus mirabilis use a contact-dependent system to differentiate between members of their own strain and a competing strain. Approaching swarms merge with other members of a genetically identical clonal population but distance themselves from members of a competing strain, forming a visible boundary (Photographs courtesy of Margaret McFall-Ngai, Michiel Vos, Steve Diggle and Edgar Lissel, and Karine Gibbs, respectively)
For decades after the initial study of bioluminescence in V. fischeri, autoinduction was considered a peculiarity specific to only a few closely related marine bacteria. This began to change in the 1990’s when autoinducers and genetic regulatory systems similar to those of V. fischeri’s were identified in distantly related bacteria (Gambello and Iglewski 1991, Bainton, Bycroft et al. 1992). Assays were developed to screen for homologous communication systems in other Gram-negative (Winson, Swift et al. 1998) and even the more evolutionarily distant Gram-positive bacteria (Wuster and Babu 2008). This led to the discovery of many similar systems widespread across the prokaryotic domain. These density-dependent genetic regulatory pathways mediated by diffusible molecules have been termed quorum sensing (QS) systems. Since the surprising discovery of QS, the field of bacterial communication has grown rapidly.
An evolutionary perspective on communication
In recent years, QS and other methods of bacterial communication have attracted the interest of evolutionary biologists who have long been interested in understanding the mechanisms of animal and insect communication (Maynard Smith and Harper 2003). Bacteria are seen as an ideal model system to study the principles of communication because they reproduce quickly, are often genetically tractable, and most importantly, they release a variety of cooperative secretions. These secretions, termed public goods, are extracellular products ranging from digestive enzymes to iron-scavenging molecules that can improve the fitness (ability to survive and reproduce) of neighboring bacteria, making them “social” traits (West, Diggle et al. 2007). Some researchers focus on fundamental questions about how cell-cell communication regulates these social traits, such as “why do bacteria convey honest information to benefit other cells rather than giving a false signal to their own advantage?” or “if some bacteria don’t send honest information, why should the receiver even sense it?” In relation to QS specifically, they are also interested in uncovering how bacterial communities avoid being overrun by cheaters that either don’t produce public goods or don’t respond to the autoinducer molecules of cooperative bacteria around them (Strassmann, Gilbert et al. 2011).
Evolutionary biologists have made other important contributions to the field by helping to elucidate several key theoretical concepts. For example, drawing on past work in the animal communication literature, they clarified the distinction between a signal and a cue (Maynard Smith and Harper 2003, Diggle, Gardner et al. 2007). A signal must 1) have evolved for the specific purpose of communication and 2) benefit both the recipient and the producer who have evolved specifically to sense and respond to that signal (Maynard Smith and Harper 2003). In contrast, interactions through cues benefit only the recipient, and only the recipient evolved to sense the cue. For instance, the brightly colored pattern on the venomous coral snake is a signal because the sender (the snake) and receiver (would-be predators) both benefit by the transmission of the information that the snake is venomous (would-be predators avoid a deadly bite and the snake is unharmed). In contrast, a shark smelling the blood of prey is a cue because the prey does not benefit from the shark’s receipt of the cue. In the past, microbiologists often misinterpreted cues as signals, leading to confusion. Therefore, a thorough molecular understanding of any social interaction between microbes is essential for accurately describing it. This distinction is important because it affects predictions about how organisms will behave under different circumstances, and will be referenced throughout this review (see (Diggle, Gardner et al. 2007) for a more complete discussion).
How bacteria communicate
There are several methods bacteria use to communicate with each other. These include exchanging diffusible molecules, packaging molecules into membrane vesicles, and transferring information through direct cell-cell contact with neighboring cells (Figure 2).
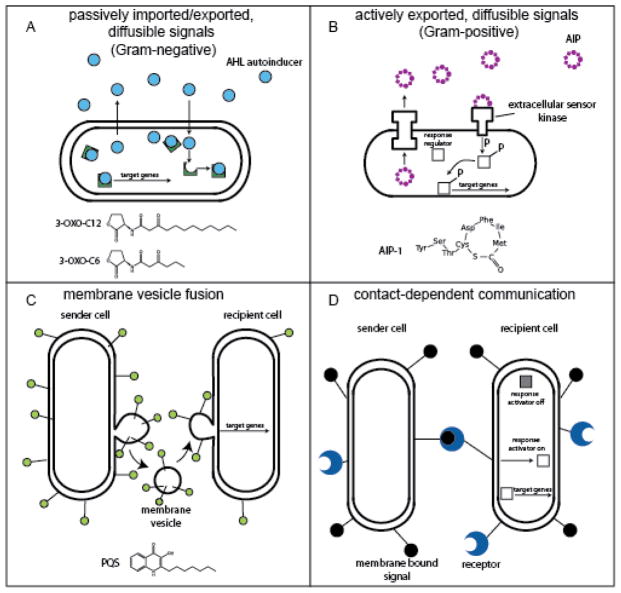
Figure 2. Methods of bacterial communication.
A) Quorum sensing (QS) in Gram-negative bacteria. Acyl-homoserine lactone (AHL) autoinducers freely diffuse out of cells and activate target gene transcription after reaching a critical cytoplasmic concentration. 3-OXO-C12 and 3-OXO-C6 are examples of two AHL autoinducers. B) QS in Gram-positive bacteria. Autoinducing-peptides (AIPs) are exported from the cell and bind extracellular receptors. After reaching a critical concentration, they also activate transcription of target genes. AIP-1 is an example of an AIP. C) Transport of molecules using outer membrane vesicles (OMVs). The outer membrane of Gram-negative bacteria containing hydrophobic signals (green lollipops) blebs off of a producing cell to fuse with a recipient cell to activate transcription of target genes. The PQS autoinducer is transported via OMVs D) Contact-dependent signal transmission. Membrane-bound signaling proteins bind to their cognate membrane-bound receptor in a neighboring cell, triggering changes in response regulators that alter transcription of target genes.
Diffusible molecules
The best understood form of bacterial communication is mediated by diffusible signal molecules known as autoinducers. This includes the QS system of V. fischeri and other bacteria. The characteristic autoinducer for QS in Gram-negative bacteria is the acyl homoserine lactone, or AHL (Figure 2-A). The specific AHLs produced by different species may vary in the length and structure of the acyl group, but all AHLs contain identical homoserine lactone moieties (Figure 2-A) (Fuqua, Parsek et al. 2001). AHLs freely diffuse out of and into cells and accumulate in direct proportion to cell density. After reaching a critical concentration, they bind to and activate their cognate cytoplasmic receptor, causing a conformational change in the receptor that allows it to bind DNA and alter the transcription of target genes (Fuqua, Parsek et al. 2001). Receptor-AHL activation also induces a positive feedback loop by increasing the transcription of AHL-synthesis genes, therefore resulting in rapid extracellular accumulation of AHL and a synchronized change in gene expression and behavior in the population.
In contrast, most Gram-positive bacteria use small peptides as QS molecules (Figure 2-B). These peptides, known as autoinducing peptides (AIPs), are different from AHLs in that they vary widely in sequence and structure and are actively exported from the cell by dedicated transporters. AIPs are first synthesized as pro-peptides and are post-translationally modified into mature AIPs during or after export from the cell (Thoendel and Horswill 2010). Gram-positive bacteria typically use two-component genetic regulatory systems that generally consist of a membrane-bound AIP-receptor and a DNA-binding response regulator. After a critical concentration of AIP-receptor binding is reached, the receptor activates the response regulator such that it can bind target DNA and alter the transcription of target genes regulated by QS (Thoendel and Horswill 2010).
There are variations on these QS systems. A notable example is Vibrio harveyi, a Gram-negative bacterium that uses an AHL autoinducer but instead uses a two-component regulatory system to control gene expression similar to Gram-positives (Bassler, Wright et al. 1993). Likewise, AHLs and AIPs are the canonical autoinducers for Gram-negative and Gram-positive bacteria, respectively, but several bacteria use non-canonical autoinducers with unique molecular structures. The Gram-negative plant pathogen, Ralstonia solanacearum, uses a gaseous volatile compound (3-hydroxypalmitic acid methyl ester, or 3-OH PAME) as its autoinducer. Use of a gaseous molecule allows R. solanacearum to communicate long distances between isolated colonies and regulate gene expression appropriate for either soil or plant environments (Flavier, Clough et al. 1997). It is very likely that many unusual diffusible molecules like 3-OH PAME have been overlooked and are yet to be discovered.
Membrane vesicles
Some molecules used for communication are not freely diffusible, but instead are packaged into membrane vesicles (Figure 2-C) (Mashburn-Warren and Whiteley 2006). Membrane vesicles are small, spherical, bi-layered containers that bleb off of the outer membrane of Gram-negative bacteria and fuse with other cells in a process reminiscent of the vesicle trafficking common in eukaryotes. Membrane vesicles can be used to transport a variety of molecules between cells, such as toxins, DNA, and signal molecules (Mashburn-Warren and Whiteley 2006). The Gram-negative opportunistic pathogen Pseudomonas aeruginosa transports an autoinducer with a unique structure (Pseudomonas quinolone signal, or PQS) inside membrane vesicles (Mashburn and Whiteley 2005). PQS can then activate the expression of a variety of virulence factors in the recipient cell. Packaging PQS inside membrane vesicles may serve to compensate for the very hydrophobic nature of PQS, to concentrate PQS before dissemination, or to protect it from extracellular degradation and thus enhance the molecule’s delivery (Mashburn and Whiteley 2005). It is speculated that other Gram-negative bacteria may transport hydrophobic signaling molecules in this fashion (Mashburn-Warren and Whiteley 2006).
Contact-dependent mechanisms
Not all communication is mediated by the exchange of small molecules. Contact-dependent cell-cell communication is used to sense adjacent cells and to coordinate spatial arrangement as a community with more precision than a diffusible molecule (figure 2-D)(Blango and Mulvey 2009). The binding of one cell’s membrane-bound signal protein to a neighboring cell’s membrane-bound receptor triggers activation of a response regulator in the recipient cell and alters transcription of target genes. A striking example is demonstrated by the Gram-negative soil bacterium Myxococcus xanthus, which uses a contact-dependent signal to spatially and temporally coordinate the formation of complex multicellular structures. When the community senses nutrient starvation, they begin to express low levels of surface protein. Surface receptor-ligand binding between cells triggers changes in gene expression and behavior resulting in organized, ripple-like gliding chains of cells, synchronized cell aggregation, and eventual formation of a large structure called a fruiting body (Figure 1-B) (Kim and Kaiser 1990). Inside the fruiting body, surface protein contact further directs centrally located cells to differentiate into dormant, environmentally resistant spores that germinate at a later time when nutrients are more available. For more information, see (Jelsbak and Sogaard-Andersen 2000).
Other methods of contact-dependent communication utilize type VI secretion systems (T6SSs). These systems, which are structurally similar to the tails of bacteriophages, are used like a syringe to inject neighboring bacteria (or even eukaryotic cells) with toxins (Pukatzki, Ma et al. 2006). T6SS can also be used to detect and transmit information from nearby hostile cells. Interestingly, P. aeruginosa is able to sense penetration of its outer membrane by another species’s T6SS and responds with its own T6SS counter-attack oriented specifically at the offending bacterium (Basler, Ho et al.). T6SSs are also thought to be used to positively influence the behavior of neighboring clonal (genetically identical) cells. Its speculated that they can enable communities to protect themselves from aggressive species, enforce cooperative behaviors in cheating cells, direct the spatial structure of a community, and defend against phage-infected cells (Russell, Peterson et al. 2014). In the next section we discuss how T6SSs are also used specifically by Proteus mirabilis to recognize and avoid competitors.
Why bacteria communicate
We have just reviewed the main mechanisms through which bacteria communicate. Now we focus on why they communicate and the information that is actually being conveyed. Ultimately, communication is used to either facilitate cooperation or competition.
Coordinating cooperative activities among clonal cells
Bacteria are most cooperative toward clonal relatives. Since clonal bacteria share identical genes, they have a collective motivation to enhance their population’s fitness by cooperating with each other (West, Griffin et al. 2006). The best studied mechanism of bacterial communication within a clonal community is QS. As discussed previously, QS involves secretion of autoinducer molecules that accumulate in proportion to population density and induce a population-wide shift in behavior once a threshold density is reached (Nealson 1977). This process regulates a diverse array of cooperative functions across many species. These include the production of public goods used to acquire nutrients such as proteases and iron-scavenging molecules; the release of antimicrobials that inhibit competing bacteria; the formation of community-wide protective coatings called biofilms; and secreted surfactants that promote motility and migration (Fig 1-C). There are many other known QS-regulated genes, but of those that are well-understood, most appear to be cooperative.
Though several fundamental assumptions about the adaptive function of QS have not been explicitly tested, recent work has empirically validated the most basic explanation of the benefit and role of QS: it regulates behaviors that are effective only at high cell density (Darch, West et al. 2012). This study demonstrated that QS-controlled public goods, such as extracellular proteases, are most beneficial for the population at the same high cell densities at which they are induced. QS regulation was shown to prevent inefficient protease production at low population densities when most molecules would diffuse away rather than benefit the community (Darch, West et al. 2012). Consistent with this evidence, independent work has also shown that the regulon of P. aeruginosa’s primary QS regulatory circuit, which directly controls up to 74 genes, is more enriched for genes encoding public goods than any other functional category (Whiteley, Lee et al. 1999). It is important to note that environmental factors independent of cell number can also influence external autoinducer levels. In particular, physical forces like diffusion and mass transfer can significantly reduce external autoinducer concentration (Redfield 2002, West, Winzer et al. 2012). If these forces are great enough, autoinduction may not occur even at high cell density (West, Winzer et al. 2012) Therefore, in natural settings both cell density and diffusion forces can impact QS (West, Winzer et al. 2012).
Many clinically relevant bacterial pathogens use QS to control expression of numerous virulence factors. QS-deficient mutants often show severe attenuation in animal infection models (Rumbaugh, Griswold et al. 1999). Evidence suggests that pathogens strictly regulate expression of metabolically expensive virulence factors to avoid wasteful production when not living within a host (e.g in the soil or water). It is also speculated that pathogens restrict virulence factor production during the initial stages of an infection when cell numbers are low in order to avoid prematurely triggering the host immune response, although there is currently little empirical evidence supporting this theory. In some pathogens, QS seems to function as a switch to adjust expression of virulence genes in accordance to the various stages of disease progression. Staphylococcus aureus uses QS to switch between genes appropriate for either the early or late stage of an infection (Mayville, Ji et al. 1999, Geisinger, Muir et al. 2009). Similarly, Vibrio cholerae actually uses QS to suppress toxin and biofilm gene expression at high cell density in order to facilitate dispersal from the host after the infection has run its course (Ng and Bassler 2009). The abundance of virulence factors controlled by QS makes it an attractive target for QS-blocking therapy, as discussed later in this review.
Coordinating cooperative activities among unrelated bacteria
While cooperative behaviors are more difficult to evolve and maintain between bacteria that are not clonal, different species occasionally form cooperative mutualistic relationships. One important type of communication used to coordinate such interactions is the proposed “universal language” mediated by autoinducer-2 (AI-2) (Vendeville, Winzer et al. 2005). AI-2 is the name for several inter-converting molecules that are synthesized by the highly conserved enzyme LuxS (Chen, Schauder et al. 2002). AI-2 was first discovered in Vibrio harveyi, which responds to AI-2 with bioluminescence. To screen for production of this molecule in other species, culture supernatants from many diverse bacteria were added to V. harveyi cultures. Any supernatants that stimulated bioluminescence in V. harveyi indicated the production of AI-2 in that species. Many genetically distant species were identified in this screen (Bassler, Greenberg et al. 1997). Furthermore, AI-2 has been shown to stimulate a variety of responses in numerous bacteria.
Since so many species make and respond to AI-2, it has been argued that AI-2 may function as an interspecies signal molecule that coordinates behavior in complex multispecies communities (Stacy, Diggle et al. 2012). The human microbiome contains several of these communities, such as those found in the mouth. For instance, the oral bacterium Streptococcus gordonii produces AI-2 which induces carbohydrate metabolism in other oral microbes as well as plaque formation by the oral community (McNab, Ford et al. 2003, Kolenbrander, Palmer et al. 2010). There are other examples of AI-2-mediated communication throughout the human body, including the nasopharynx and gut (see (Pereira, Thompson et al. 2013). Caution, however, is necessary in interpreting AI-2 interactions. LuxS, the enzyme responsible for synthesizing AI-2, is an essential component of cellular metabolism in some bacteria, and many bacteria that produce AI-2 lack a corresponding receptor (Rezzonico and Duffy 2008). For this reason, it is sometimes difficult to clearly determine whether any two species are specifically signaling to each other through AI-2 or if one species is just using the metabolic byproduct of the other as a cue (Winzer, Hardie et al. 2002, Diggle, Gardner et al. 2007, Pereira, Thompson et al. 2013). Improved techniques for studying complicated multispecies environments at the molecular level will be useful for further investigating these interactions.
Sensing competitors
Though there are notable examples of bacterial mutualisms, many microbiologists think competition is potentially a more common outcome when different species coexist (Foster and Bell 2012). Bacteria often live in diverse mixed-species environments where competition for resources is fierce, and they must be able to avoid harm from aggressive bacteria and kill or inhibit the growth of competitors. Interactions between strains of the same species tend to be even more antagonistic than interactions between two different species because different strains occupy the same ecological niche and thus must compete for identical resources (Russell, Peterson et al. 2014). It is often in a bacterium’s interest to detect potential competitors before direct conflict arises. Unlike communication within a clonal population, mixed-species communication generally relies on cues instead of signals. This is because the bacteria receiving the cue benefit from detecting the presence of other organisms, but the producer does not benefit and did not evolve its cue for that purpose.
One way in which cells can sense the presence of competitors is by detecting specific molecules that are associated with a particular species. In principle, a bacterium could tailor its response to each of the various species that it is capable of identifying. QS bacteria recognize their own autoinducers, but some Gram-negative bacteria also have QS receptors that can recognize AHLs from other species. This is possible because the general AHL structure is highly conserved among Gram-negative bacteria (Figure 2-A). Termed bacterial “cross-talk” or “eavesdropping,” this activity may provide a fitness advantage to the recipient by alerting it to the presence of a competitor (Strassmann, Gilbert et al. 2011). The soil bacterium Chromobacterium violaceum responds to Burkholderia thailandensis’s AHL by upregulating its own antibiotic production (Chandler, Heilmann et al. 2012). Therefore, the ability to sense foreign AHLs allows C. violaceum to detect and inhibit the growth of competing bacteria. However, there are difficulties with tailoring responses to particular competitors. In complex communities it is difficult for a cell to maintain the genetic machinery required to coordinate separate responses for all interacting bacteria, and further, competitors may alter or stop producing their identifiable molecules in order to go undetected.
An alternative method is to sense generic cues from competitors. This can be accomplished either by sensing a non-specific molecule released by competing bacteria or by sensing the harm itself caused by an attacking cell. An example of the first strategy is P. aeruginosa’s ability to sense the peptidoglycan cell wall fragments shed from the surface of Gram-positive bacteria. Upon detection of these fragments, P. aeruginosa responds with induction of an antimicrobial that causes non-specific lysis of surrounding bacteria (Korgaonkar, Trivedi et al. 2013). Interestingly, due to the nonspecific nature of the secreted toxins, this response was shown to not only kill Gram-positives but to also damage the host and enhance P. aeruginosa’s virulence. Use of this system also diminishes the need for specific responses tailored to individual species. Alternatively, rather than detecting a released molecule, a bacterium can sense and respond to the direct physiological injury caused by a hostile cell. In this way, a cell can react to competitors precisely when it is most urgent: while it is being harmed. The types of harm often caused by other bacteria may include DNA damage, compromised cell wall integrity, oxidative stress, or starvation. Upon detection of competitor-induced damage, many bacteria defend themselves by releasing their own antibiotics and upregulating detoxification mechanisms. By directly sensing the harm caused by competitors instead of the particular molecules that those competitors release, bacteria can accurately and robustly respond to a broad range of threats.
Lastly, bacteria can also antagonize competitors via contact-dependent processes. As described previously, contact-dependent communication is useful as a defense against hostile cells and for maintaining genetic relatedness in a community (Russell, Peterson et al. 2014). Some species use type VI secretion systems (T6SS) to inject other cells with toxins in conjunction with their own toxin-antitoxin systems that provide the host cell and clonal relatives with immunity from their own toxin. Proteus mirabilis, a causative agent of urinary tract infections, uses these two systems to recognize members of its own strain and display territorial behavior. Strains of P. mirablis appear to encode their own characteristic toxin-antitoxin system, making them immune to the T6SS toxins of their own strain but sensitive to the toxins of unrelated strains (Wenren, Sullivan et al. 2013). On agar plates, approaching swarms of P. mirabilis merge with swarms of the same strain, while a striking visible boundary forms between two competing strains (Figure 1-D)(Wenren, Sullivan et al. 2013). The ability to maintain strain homogeneity in a community is therefore useful for carving out a territory in a competitive environment (Budding, Ingham et al. 2009).
Medical applications for disrupting bacterial communication
Because communication is of vital importance to bacteria, research efforts are also being directed towards the development of novel therapeutics that interfere with bacterial communication systems. Since conserved QS systems are used by many clinically relevant pathogens to regulate expression of their virulence genes, QS is an attractive target for intervention. Importantly, since these “anti-virulence” treatments do not kill or inhibit bacterial growth but rather prevent the expression of virulence factors, there is less selective pressure for bacteria to develop resistance to them (Dong, Xu et al. 2000, Dong, Wang et al. 2001, Chen, Gao et al. 2013). One approach for treating infection is through the use of quorum-quenching enzymes. These enzymes degrade QS molecules and are naturally produced by some bacteria and plants as a likely defense against harmful invading bacteria (Fuqua, Parsek et al. 2001). Similarly, QS inhibitors are small molecules that hinder QS by acting as QS-receptor antagonists (Kalia 2013). Both of these techniques are promising because they show protection in vivo (Hentzer, Wu et al. 2003). Researchers are also experimenting with anti-QS antibodies for prophylactic therapy. Passive immunization with anti-autoinducer antibodies is appealing because their pharmaceutical properties are more consistent and better defined than other experimental compounds (Park, Jagasia et al. 2007, Kaufmann, Park et al. 2008). Lastly, vaccinations that stimulate production of anti-QS antibodies in the host are a new potential pathway for mitigating the severity of bacterial infections (Golpasha, Mousavi et al. 2015).
It may also be possible to effectively treat certain bacterial infections by disrupting mixed-species communication. Polymicrobial infections often have worsened disease severity for the host than infection by a single species. Interfering with communication between species, like the ability of P. aeruginosa to sense peptidoglycan fragments shed by S. aureus (Korgaonkar, Trivedi et al. 2013) or AI-2 exchange between bacteria in dental biofilms (Kolenbrander, Palmer et al. 2010), could substantially lessen disease severity. Furthermore, targeting a particular species involved in a cooperative relationship may be a more effective strategy than directly treating the main pathogen. For instance, non-pathogenic Escherichia coli in the gut release the metabolite indole that acts as a cue to stimulate antibiotic resistance in Salmonella typhimurium (Vega, Allison et al. 2013). Therefore, compounds designed to specifically eliminate E. coli could make S. typhimurium more susceptible to antibiotic treatment. As basic research involving bacterial communication accumulates, the field may see significant breakthroughs in treating deadly bacterial pathogens.
Future perspectives
Microbiologists have made great progress in characterizing the main features of bacterial communication, but many basic questions remain. It is not clear why certain genes and not others are under QS regulation, (Cornforth, Popat et al. 2014), why many bacteria use several communication systems simultaneously (Cornforth, Popat et al. 2014), or how chemical inputs and growth conditions influence communication systems (Duan and Surette 2007). Perhaps the most important step in understanding bacterial communication will be moving away from standard in vitro models toward more biologically relevant ones. There are several shortcomings with standard in vitro models including: growth at unrealistically high population densities, use of single-species cultures that lack interactions with other microbes and the use of artificial media that doesn’t reflect normal nutrient conditions (Duan and Surette 2007). Fortunately, new technologies are being developed that may address these shortcomings. Microfabrication techniques are being used to carefully place bacteria into realistic, small, high-density clusters to study cell-cell communication on a relevant spatial scale (Connell, Wessel et al. 2010). Researchers are also working on methods for studying complex bacterial communities in their natural arrangement and composition (Valm, Welch et al. 2011). These techniques, in combination with advancing transcriptomic approaches, may answer basic questions of how bacteria communicate in nature. Despite all that has been learned since the discovery of bacterial communication, in many ways the field still has much room to grow. As research advances, it will be exciting to see what parallels are uncovered between bacterial communication and communication between other organisms such as parasites.
Glossary
- Strain
A taxonomic subset within a species that is differentiated by distinct phenotypes
- Autoinduction
A change in gene expression triggered by extracellular accumulation of a signal molecule (autoinducer) that is produced by the host cell
- Gram negative bacteria
A large group of bacteria characterized by a thin peptidoglycan cell wall outside of their cytoplasmic membrane followed by an additional outer membrane
- Gram positive bacteria
A large group of bacteria characterized by a thick peptidoglycan cell wall outside of their cytoplasmic membrane
- Quorum sensing (QS)
A system of communication used by bacteria to regulate community gene expression through diffusible autoinducer signal molecules that accumulate in direct proportion to population density
- Public goods
Extracellular products released by individual cells that benefit the community. For example, secreted proteases and elastases release nutrients from the environment and are beneficial to other cells regardless of the source
- Fitness
The relative ability of an organism to survive and pass on its genes through reproduction
- Cheaters
Individual members of a community that benefit from the cooperative activities and public goods of other members of their community but do not perform cooperative behaviors in return
- Signaling
A specific type of communication that benefits both the producing cell and the recipient cell. Signaling molecules must have evolved for the explicit purpose of communication and they must induce a greater physiological response in the recipient than only what is necessary to detoxify or metabolize the molecule
- Cue
Information that is transferred from one organism to another, but the interaction only benefits the recipient that perceives the cue, not the producer. Depending on the context, examples of cues may include QS molecules, cellular components, metabolites and cell-cell contact
- Acyl homoserine lactone (AHL)
The typical autoinducer used by quorum sensing Gram-negative bacteria. The structure of the AHL is highly conserved with subtle structural variations used by different species
- Autoinducing peptides (AIPs)
The typical autoinducer used by quorum sensing Gram-positive bacteria. AIPs are often cyclic peptides but can vary widely in sequence and structure. AIPs usually activate a two-component regulatory system to regulate target genes
- Type IV secretion systems (T6SS)
Syringe-like protrusions on the surface of bacteria that are used to transfer materials such a toxins, DNA, or effector proteins to other cells
- Clonal
Refers to a population of cells that are genetically identical and are derived from the same parent genotype
- Biofilm
“Slime cities” or communities of bacteria surrounded by a thick, protective layer of polysaccharide. Biofilms protect bacteria from antibiotics, disinfectants, desiccation, and immune defenses
- Microbiome
The vast collection of all bacterial species that live in or on the human body
- Toxin–antitoxin systems
A genetic system whereby a cell carries both the genes to produce a toxin and the genes for detoxifying that toxin within itself, thereby providing immunity to its own poison
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Literature cited
- Bainton NJ, Bycroft BW, Chhabra SR, Stead P, Gledhill L, Hill PJ, Rees CE, Winson MK, Salmond GP, Stewart GS, et al. A general role for the lux autoinducer in bacterial cell signalling: control of antibiotic biosynthesis in Erwinia. Gene. 1992;116(1):87–91. doi: 10.1016/0378-1119(92)90633-z. [DOI] [PubMed] [Google Scholar]
- Basler M, Ho Brian T, Mekalanos John J. Tit-for-Tat: Type VI Secretion System Counterattack during Bacterial Cell-Cell Interactions. Cell. 152(4):884–894. doi: 10.1016/j.cell.2013.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassler BL, Greenberg EP, Stevens AM. Cross-species induction of luminescence in the quorum-sensing bacterium Vibrio harveyi. J Bacteriol. 1997;179(12):4043–4045. doi: 10.1128/jb.179.12.4043-4045.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassler BL, Wright M, Showalter RE, Silverman MR. Intercellular signalling in Vibrio harveyi: sequence and function of genes regulating expression of luminescence. Mol Microbiol. 1993;9(4):773–786. doi: 10.1111/j.1365-2958.1993.tb01737.x. [DOI] [PubMed] [Google Scholar]
- Blango MG, Mulvey MA. Bacterial landlines: contact-dependent signaling in bacterial populations. Curr Opin Microbiol. 2009;12(2):177–181. doi: 10.1016/j.mib.2009.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budding A, Ingham C, Bitter W, Vandenbroucke-Grauls C, Schneeberger P. The Dienes phenomenon: competition and territoriality in swarming Proteus mirabilis. J Bacteriol. 2009;191(12):3892–3900. doi: 10.1128/JB.00975-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler JR, Heilmann S, Mittler JE, Greenberg EP. Acyl-homoserine lactone-dependent eavesdropping promotes competition in a laboratory co-culture model. ISME J. 2012;6(12):2219–2228. doi: 10.1038/ismej.2012.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Gao Y, Chen X, Yu Z, Li X. Quorum Quenching Enzymes and Their Application in Degrading Signal Molecules to Block Quorum Sensing-Dependent Infection. Interntl J Mol Sci. 2013;14(9):17477. doi: 10.3390/ijms140917477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Schauder S, Potier N, Van Dorsselaer A, Pelczer I, Bassler BL, Hughson FM. Structural identification of a bacterial quorum-sensing signal containing boron. Nature. 2002;415(6871):545–549. doi: 10.1038/415545a. [DOI] [PubMed] [Google Scholar]
- Connell JL, Wessel AK, Parsek MR, Ellington AD, Whiteley M, Shear JB. Probing prokaryotic social behaviors with bacterial “lobster traps”. MBio. 2010;1(4):e00202–00210. doi: 10.1128/mBio.00202-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornforth DM, Foster KR. Competition sensing: the social side of bacterial stress responses. Nature Rev Microbiol. 2013;11(4):285–293. doi: 10.1038/nrmicro2977. [DOI] [PubMed] [Google Scholar]
- Cornforth DM, Popat R, McNally L, Gurney J, Scott-Phillips TC, Ivens A, Diggle SP, Brown SP. Combinatorial quorum sensing allows bacteria to resolve their social and physical environment. Proc Natl Acad Sci USA. 2014;111(11):4280–4284. doi: 10.1073/pnas.1319175111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darch SE, West SA, Winzer K, Diggle SP. Density-dependent fitness benefits in quorum-sensing bacterial populations. Proc Natl Acad Sci USA. 2012;109(21):8259–8263. doi: 10.1073/pnas.1118131109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diggle SP, Gardner A, West SA, Griffin AS. Evolutionary theory of bacterial quorum sensing: when is a signal not a signal? Phil Transact Roy Soc London B: Biol Sci. 2007;362(1483):1241–1249. doi: 10.1098/rstb.2007.2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong YH, Xu JL, Li XZ, Zhang LH. AiiA, an enzyme that inactivates the acylhomoserine lactone quorum-sensing signal and attenuates the virulence of Erwinia carotovora. Proc Natl Acad Sci USA. 2000;97(7):3526–3531. doi: 10.1073/pnas.060023897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong YH, Wang LH, Xu JL, Zhang HB, Zhang XF, Zhang LH. Quenching quorum-sensing-dependent bacterial infection by an N-acyl homoserine lactonase. Nature. 2001;411(6839):813–817. doi: 10.1038/35081101. [DOI] [PubMed] [Google Scholar]
- Duan K, Surette MG. Environmental regulation of Pseudomonas aeruginosa PAO1 Las and Rhl quorum-sensing systems. J Bacteriol. 2007;189(13):4827–4836. doi: 10.1128/JB.00043-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberhard A, Burlingame AL, Eberhard C, Kenyon GL, Nealson KH, Oppenheimer NJ. Structural identification of autoinducer of Photobacterium fischeri luciferase. Biochemistry. 1981;20(9):2444–2449. doi: 10.1021/bi00512a013. [DOI] [PubMed] [Google Scholar]
- Engebrecht J, Nealson K, Silverman M. Bacterial bioluminescence: isolation and genetic analysis of functions from Vibrio fischeri. Cell. 1983;32(3):773–781. doi: 10.1016/0092-8674(83)90063-6. [DOI] [PubMed] [Google Scholar]
- Flavier AB, Clough SJ, Schell MA, Denny TP. Identification of 3-hydroxypalmitic acid methyl ester as a novel autoregulator controlling virulence in Ralstonia. Mol Microbiol. 1997;26(2):251–259. doi: 10.1046/j.1365-2958.1997.5661945.x. [DOI] [PubMed] [Google Scholar]
- Foster Kevin R, Bell T. Competition, Not Cooperation, Dominates Interactions among Culturable Microbial Species. Curr Biol. 2012;22(19):1845–1850. doi: 10.1016/j.cub.2012.08.005. [DOI] [PubMed] [Google Scholar]
- Fuqua C, Parsek MR, Greenberg EP. Regulation of gene expression by cell-to-cell communication: acyl-homoserine lactone quorum sensing. Ann Rev Genet. 2001;35(1):439–468. doi: 10.1146/annurev.genet.35.102401.090913. [DOI] [PubMed] [Google Scholar]
- Gambello MJ, Iglewski BH. Cloning and characterization of the Pseudomonas aeruginosa lasR gene, a transcriptional activator of elastase expression. J Bacteriol. 1991;173(9):3000–3009. doi: 10.1128/jb.173.9.3000-3009.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisinger E, Muir TW, Novick RP. Agr receptor mutants reveal distinct modes of inhibition by staphylococcal autoinducing peptides. Proc Natl Acad Sci USA. 2009;106(4):1216–1221. doi: 10.1073/pnas.0807760106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golpasha ID, Mousavi SF, Owlia P, Siadat SD, Irani S. Immunization with 3-oxododecanoyl-L-homoserine lactone-r-PcrV conjugate enhances survival of mice against lethal burn infections caused by Pseudomonas aeruginosa. Bosn J Basic Med Sci. 2015;15(2):15–24. doi: 10.17305/bjbms.2015.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentzer M, Wu H, Andersen JB, Riedel K, Rasmussen TB, Bagge N, Kumar N, Schembri MA, Song Z, Kristoffersen P, Manefield M, Costerton JW, Molin S, Eberl L, Steinberg P, Kjelleberg S, Hoiby N, Givskov M. Attenuation of Pseudomonas aeruginosa virulence by quorum sensing inhibitors. Embo j. 2003;22(15):3803–3815. doi: 10.1093/emboj/cdg366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jelsbak L, Søgaard-Andersen L. Pattern formation: fruiting body morphogenesis in Myxococcus xanthus. Curr OpinMicrobiol. 2000;3(6):637–642. doi: 10.1016/s1369-5274(00)00153-3. [DOI] [PubMed] [Google Scholar]
- Kalia VC. Quorum sensing inhibitors: an overview. Biotechnol Advances. 2013;31(2):224–245. doi: 10.1016/j.biotechadv.2012.10.004. [DOI] [PubMed] [Google Scholar]
- Kaufmann GF, Park J, Mee JM, Ulevitch RJ, Janda KD. The quorum quenching antibody RS2-1G9 protects macrophages from the cytotoxic effects of the Pseudomonas aeruginosa quorum sensing signalling molecule N-3-oxo-dodecanoyl-homoserine lactone. Mol Immunol. 2008;45(9):2710–2714. doi: 10.1016/j.molimm.2008.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempner ES, Hanson FE. Aspects of light production by Photobacterium fischeri. J Bacteriol. 1968;95(3):975–979. doi: 10.1128/jb.95.3.975-979.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SK, Kaiser D. C-factor: a cell-cell signaling protein required for fruiting body morphogenesis of M. xanthus. Cell. 1990;61(1):19–26. doi: 10.1016/0092-8674(90)90211-v. [DOI] [PubMed] [Google Scholar]
- Kolenbrander PE, Palmer RJ, Periasamy S, Jakubovics NS. Oral multispecies biofilm development and the key role of cell–cell distance. Nature Rev Microbiol. 2010;8(7):471–480. doi: 10.1038/nrmicro2381. [DOI] [PubMed] [Google Scholar]
- Korgaonkar A, Trivedi U, Rumbaugh KP, Whiteley M. Community surveillance enhances Pseudomonas aeruginosa virulence during polymicrobial infection. Proc Natl Acad Sci USA. 2013;110(3):1059–1064. doi: 10.1073/pnas.1214550110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashburn-Warren LM, Whiteley M. Special delivery: vesicle trafficking in prokaryotes. Mol Microbiol. 2006;61(4):839–846. doi: 10.1111/j.1365-2958.2006.05272.x. [DOI] [PubMed] [Google Scholar]
- Mashburn LM, Whiteley M. Membrane vesicles traffic signals and facilitate group activities in a prokaryote. Nature. 2005;437(7057):422–425. doi: 10.1038/nature03925. [DOI] [PubMed] [Google Scholar]
- Maynard Smith J, Harper D. Animal signals. New York: Oxford University Press; 2003. [Google Scholar]
- Mayville P, Ji G, Beavis R, Yang H, Goger M, Novick RP, Muir TW. Structure-activity analysis of synthetic autoinducing thiolactone peptides from Staphylococcus aureus responsible for virulence. Proc Natl Acad Sci USA. 1999;96(4):1218–1223. doi: 10.1073/pnas.96.4.1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNab R, Ford SK, El-Sabaeny A, Barbieri B, Cook GS, Lamont RJ. LuxS-based signaling in Streptococcus gordonii: autoinducer 2 controls carbohydrate metabolism and biofilm formation with Porphyromonas gingivalis. J Bacteriol. 2003;185(1):274–284. doi: 10.1128/JB.185.1.274-284.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nealson KH. Autoinduction of bacterial luciferase. Occurrence, mechanism and significance. Arch Microbiol. 1977;112(1):73–79. doi: 10.1007/BF00446657. [DOI] [PubMed] [Google Scholar]
- Nealson KH, Hastings JW. Bacterial bioluminescence: its control and ecological significance. Microbiol Rev. 1979;43(4):496–518. doi: 10.1128/mr.43.4.496-518.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nealson KH, Platt T, Hastings JW. Cellular Control of the Synthesis and Activity of the Bacterial Luminescent System. J Bacteriol. 1970;104(1):313–322. doi: 10.1128/jb.104.1.313-322.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng W-L, Bassler BL. Bacterial quorum-sensing network architectures. Ann Rev Genet. 2009;43:197. doi: 10.1146/annurev-genet-102108-134304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J, Jagasia R, Kaufmann GF, Mathison JC, Ruiz DI, Moss JA, Meijler MM, Ulevitch RJ, Janda KD. Infection control by antibody disruption of bacterial quorum sensing signaling. Chem Biol. 2007;14(10):1119–1127. doi: 10.1016/j.chembiol.2007.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira CS, Thompson JA, Xavier KB. AI-2-mediated signalling in bacteria. FEMS Microbiol Rev. 2013;37(2):156–181. doi: 10.1111/j.1574-6976.2012.00345.x. [DOI] [PubMed] [Google Scholar]
- Pukatzki S, Ma AT, Sturtevant D, Krastins B, Sarracino D, Nelson WC, Heidelberg JF, Mekalanos JJ. Identification of a conserved bacterial protein secretion system in Vibrio cholerae using the Dictyostelium host model system. Proc Natl Acad Sci USA. 2006;103(5):1528–1533. doi: 10.1073/pnas.0510322103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redfield RJ. Is quorum sensing a side effect of diffusion sensing? Trends Microbiol. 2002;10(8):365–370. doi: 10.1016/s0966-842x(02)02400-9. [DOI] [PubMed] [Google Scholar]
- Rezzonico F, Duffy B. Lack of genomic evidence of AI-2 receptors suggests a non-quorum sensing role for luxS in most bacteria. BMC Microbiol. 2008;8(1):154. doi: 10.1186/1471-2180-8-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rumbaugh KP, Griswold JA, Iglewski BH, Hamood AN. Contribution of quorum sensing to the virulence ofpseudomonas aeruginosa in burn wound infections. Infect Immun. 1999;67(11):5854–5862. doi: 10.1128/iai.67.11.5854-5862.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell AB, Peterson SB, Mougous JD. Type VI secretion system effectors: poisons with a purpose. Nat Rev Microbiol. 2014;12(2):137–148. doi: 10.1038/nrmicro3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stacy AR, Diggle SP, Whiteley M. Rules of engagement: defining bacterial communication. Curr Opin Microbiol. 2012;15(2):155–161. doi: 10.1016/j.mib.2011.11.007. [DOI] [PubMed] [Google Scholar]
- Strassmann JE, Gilbert OM, Queller DC. Kin discrimination and cooperation in microbes. Ann Review Microbiol. 2011;65:349–367. doi: 10.1146/annurev.micro.112408.134109. [DOI] [PubMed] [Google Scholar]
- Thoendel M, Horswill AR. Biosynthesis of peptide signals in gram-positive bacteria. Adv Appl Microbiol. 2010;71:91–112. doi: 10.1016/S0065-2164(10)71004-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valm AM, Welch JLM, Rieken CW, Hasegawa Y, Sogin ML, Oldenbourg R, Dewhirst FE, Borisy GG. Systems-level analysis of microbial community organization through combinatorial labeling and spectral imaging. Proc Natl Acad Sci USA. 2011;108(10):4152–4157. doi: 10.1073/pnas.1101134108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vega NM, Allison KR, Samuels AN, Klempner MS, Collins JJ. Salmonella typhimurium intercepts Escherichia coli signaling to enhance antibiotic tolerance. Proc Natl Acad Sci USA. 2013;110(35):14420–14425. doi: 10.1073/pnas.1308085110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vendeville A, Winzer K, Heurlier K, Tang CM, Hardie KR. Making ‘sense’ of metabolism: autoinducer-2, LUXS and pathogenic bacteria. Nat Rev Micro. 2005;3(5):383–396. doi: 10.1038/nrmicro1146. [DOI] [PubMed] [Google Scholar]
- Wenren LM, Sullivan NL, Cardarelli L, Septer AN, Gibbs KA. Two independent pathways for self-recognition in Proteus mirabilis are linked by type VI-dependent export. MBio. 2013;4(4) doi: 10.1128/mBio.00374-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West SA, Diggle SP, Buckling A, Gardner A, Griffin AS. The social lives of microbes. Ann Rev Ecology, Evolution, and Systematics. 2007:53–77. [Google Scholar]
- West SA, Griffin AS, Gardner A, Diggle SP. Social evolution theory for microorganisms. Nat Rev Microbiol. 2006;4(8):597–607. doi: 10.1038/nrmicro1461. [DOI] [PubMed] [Google Scholar]
- West SA, Winzer K, Gardner A, Diggle SP. Quorum sensing and the confusion about diffusion. Trends Microbiol. 2012;20(12):586–594. doi: 10.1016/j.tim.2012.09.004. [DOI] [PubMed] [Google Scholar]
- Whiteley M, Lee KM, Greenberg EP. Identification of genes controlled by quorum sensing in Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 1999;96(24):13904–13909. doi: 10.1073/pnas.96.24.13904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winson MK, Swift S, Fish L, Throup JP, Jorgensen F, Chhabra SR, Bycroft BW, Williams P, Stewart GS. Construction and analysis of luxCDABE-based plasmid sensors for investigating N-acyl homoserine lactone-mediated quorum sensing. FEMS Microbiol Lett. 1998;163(2):185–192. doi: 10.1111/j.1574-6968.1998.tb13044.x. [DOI] [PubMed] [Google Scholar]
- Winzer K, Hardie KR, Williams P. Bacterial cell-to-cell communication: sorry, can’t talk now - gone to lunch! Curr Opin Microbiol. 2002;5(2):216–222. doi: 10.1016/s1369-5274(02)00304-1. [DOI] [PubMed] [Google Scholar]
- Wuster A, Babu MM. Conservation and evolutionary dynamics of the agr cell-to-cell communication system across firmicutes. J Bacteriol. 2008;190(2):743–746. doi: 10.1128/JB.01135-07. [DOI] [PMC free article] [PubMed] [Google Scholar]