Abstract
Microbial communities are spatially organized in both the environment and the human body. Although patterns exhibited by these communities are described by microbial biogeography this discipline has previously only considered large-scale, global patterns. By contrast, the fine-scale positioning of a pathogen within an infection site can greatly alter its virulence potential. In this Review, we highlight the importance of considering spatial positioning in the study of polymicrobial infections and discuss targeting biogeography as a therapeutic strategy.
Rather than being ‘bags of enzymes’, microorganisms are social creatures that build complex communities such as biofilms. Similar to most cities, biofilms are multicultural and well engineered, or — in the parlance of microbiologists — polymicrobial and spatially organized. Traditionally, biofilms are thought of as hundreds of thousands of cells encased in a matrix and attached to a surface1, but they can also contain as few as tens of cells simply arranged as a small cluster or aggregate. Relevant to human health, polymicrobial biofilms are prevalent throughout the human body, both during health and disease. Although historically infections have been attributed to individual pathogens2, poly-microbial interactions within biofilms also hugely affect disease3. For example, some infections require colonization with multiple interacting microorganisms (for example, Porphyromonas gingivalis and commensal oral microorganisms in periodontal disease)4, whereas other infections are modulated in severity by the presence of co-infecting species (for example, Pseudomonas aeruginosa and Staphylococcus aureus in chronic wound infections)5. The timing of these interactions can also vary. As in the above examples, co-pathogens can appear in the host concurrently, or they can appear stepwise as a successional series of pathogen acquisitions (for example, bacterial pneumonia following viral infection). Moreover, in some instances a well-known ‘pathogen’ may only be a minor player in disease progression, with commensal strains having a more prominent role (for example, Streptococcus mutans in dental caries)6.
‘Synergy’ is a positive interaction term often used to describe the interactions between microbial strains or species that result in an outcome that is greater than the sum of the individual parts. In the context of infection, the primary outcome generally assessed is damage to the host. Polymicrobial infections are therefore classified as synergistic if they are more severe than infections with individual microorganisms7. Important clinical repercussions of synergy during polymicrobial infections include heightened antimicrobial resistance and a prolonged time necessary for host recovery. Although the mechanisms of synergy are often ill defined, it is not necessary that all infecting species benefit from the polymicrobial interaction for the interaction to be synergistic. In fact, one species can benefit at the expense of another as long as this results in an advantage during any of the major phases of infection: attachment, growth, host immune evasion and host damage3. Furthermore, synergistic interactions should not be thought of as randomly occurring events, as simply combining any two pathogens in an infection does not always result in synergy8,9. However, synergy is not always positively selected for over time and can instead seem ‘accidental’. For instance, P. aeruginosa (an environmental bacterium that usually resides in soil and water) and S. aureus (a bacterium that usually resides in the respiratory tract and on the skin) competitively interact in wounds5, leading to chronic infections that are resistant to antimicrobial therapy10. However, the response of these bacteria to each other is unlikely to have evolved in the context of wounds and has probably evolved owing to interactions with other microorganisms that are present in their natural habitats11. By contrast, the oral cavity exemplifies a polymicrobial environment where interactions are highly evolved, as oral microorganisms usually exist only in that environment and in the presence of each other. Unfortunately, a thorough, mechanistic understanding of polymicrobial interactions in relation to disease is still lacking, an alarming fact when considering that polymicrobial infections pose a considerable burden on society12.
Biogeography is the study of the distribution of species through space and time. Traditionally, this discipline examines the global distribution of plants and animals, a classic example being the Wallace Line, the boundary that separates Asian and Australian species13. The ecological and evolutionary forces that govern biogeographic patterns have been differentiated as selection, drift, diversification and dispersal14,15 (BOX 1). Microbial spatial patterns have also inspired significant interest and research, not only in spatial patterns occurring over continents and oceans16 but also over the landscape of a single human17–19 to the scale of a single infection site. The most basic evidence for this ‘biogeography of infection’ is the existence of endemic species, such as Helicobacter pylori, which is found only in the stomach. Further evidence, provided by the Human Microbiome Project, is that the same body sites in different individuals are often more similar in community composition than different sites in the same individual17, which indicates that microorganisms are not randomly distributed throughout the body. The processes that generate these and other spatial patterns of microbial distribution can be understood using the same framework as the one developed in traditional biogeography (BOX 1).
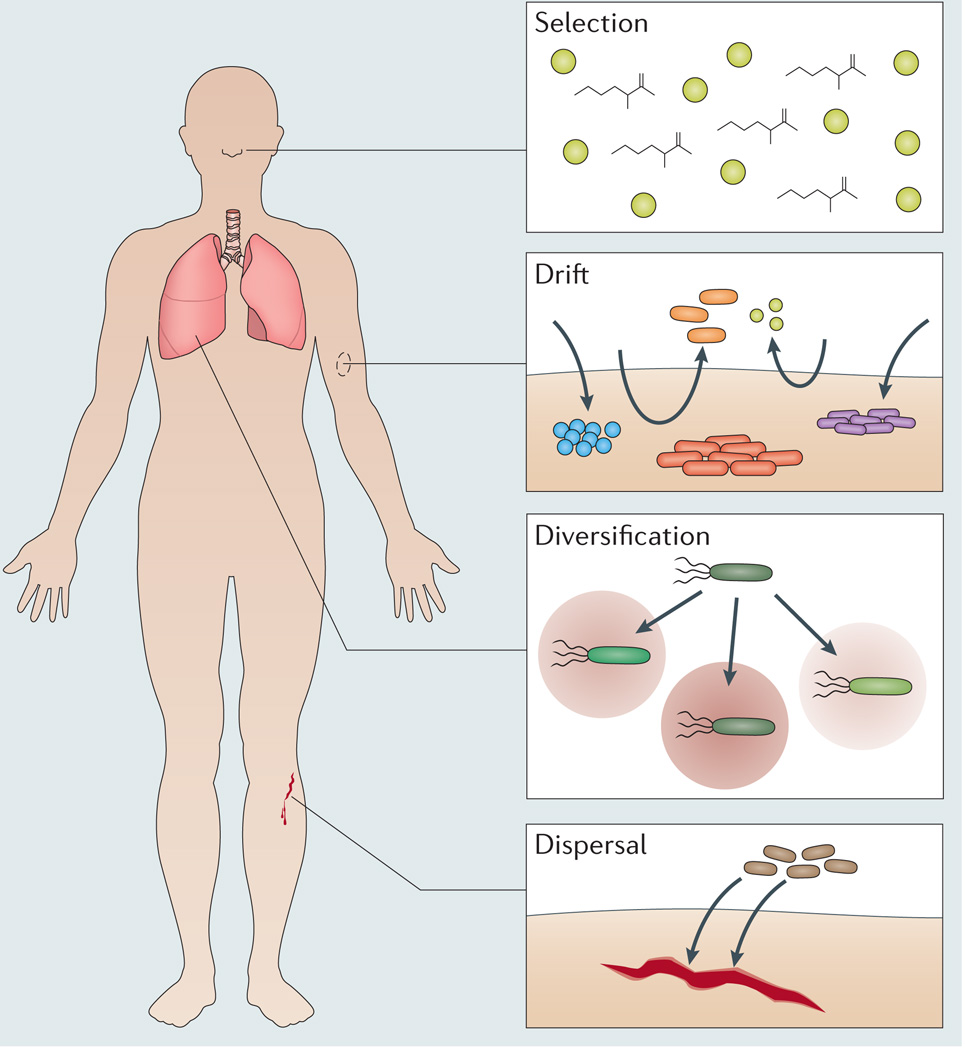
Box 1 | Forces regulating biogeography.
In traditional biogeography, four forces are described that primarily give rise to spatial patterns14. These forces also function in the context of human infections (see the figure).
Selection
As microorganisms vary markedly in the stresses that they can tolerate, one of the strongest forces governing microbial spatial patterns is environmental selection. In the host, different environments exert selective pressures that influence biogeography, as seen, for example, in the fact that the gut and oral microbiomes are more similar between different people than they are within the same person17. This suggests that these habitats exert unique but conserved selective pressures on the local microbial communities. As all microorganisms within the host must acquire nutrients to sustain growth, a major selective force determining the biogeography of host-associated microorganisms is nutrient availability. One example of this can be seen in staphylococci that differentially colonize human surfaces. This bacterial genus is prevalent on human skin119, but in contrast to many other staphylococci, Staphylococcus aureus also persistently colonizes one-third of human nasal cavities120. Supporting a nutritional basis for this pattern, S. aureus, but not skin-restricted staphylococci, can sustain growth in a synthetic nasal medium121. These experiments have revealed that the acquisition of a specific nutrient, methionine, is a key trait required for S. aureus to colonize the nose (see the figure).
Drift
Ecological drift describes changes in strain and species abundance that are due to chance. Currently, it is unknown to what extent drift, independent of selection, affects microbial community composition122,123. The enormous variability in the human microbiome124 provides strong evidence that drift, with selection, affects within-host biogeography. However, rather than drift or natural selection being the predominant force, it is more likely that their relative influence depends on the body site and time of sampling (see the figure). For example, spatial variation in drift occurs across skin regions. The forearm, an open site, is transiently colonized, whereas the inside of the ear, an occluded site, is stable over time125. Temporal variation is exemplified by the microbiomes of newborns. Babies delivered by caesarean-section have skin-like microbiomes, whereas vaginally delivered babies have vagina-like microbiomes126, but over time both microbiomes converge.
Diversification
Diversification is the divergence in phenotype among organisms that occurs through the addition, loss, or modification of traits by genetic adaptation (for example, due to horizontal gene transfer (HGT)). A notorious site of within-host diversification is the airway of patients with cystic fibrosis, where the sinuses and lungs within the same patient can harbour distinct sub-lineages of Pseudomonas aeruginosa88 (see the figure). Key to these and other diversification events is spatial structure, as it creates heterogeneity in resources and selective pressures. For example, the specific nutrients present in a specific infection site can preferentially direct evolution towards virulence. This is seen for S. aureus, which can infect many tissues, including magnesium-rich bones and kidneys, where during infection this nutrient accelerates the evolution of bacteraemia and drug resistance127. Furthermore, diversification though HGT can lead to an expansion in the biogeography of a pathogen. For example, S. aureus primarily colonizes the nose, but after acquiring a single gene from Staphylococcus epidermidis, a skin commensal, S. aureus became able to withstand the harsh conditions associated with residing on skin128.
Dispersal
Perhaps the strongest evidence for a ‘biogeography of infection’ is that many infections stem from microorganisms that migrate between host compartments. Globally, microorganisms are dispersed by water, wind, and other long-range forces, and a common debate in traditional biogeography is the scale over which this actually takes place. Dispersal has been argued as both completely unlimited (summarized in the statement “Everything is everywhere, but, the environment selects”)129,130 and severely limited123,131. It has also been proposed that microorganisms exhibit a range of dispersal capacities132. This is probably the scenario in host environments, in which dispersal is dynamic and regulated by the status of host barriers, such as the skin, gut or blood-brain barrier. These barriers maintain body sites that are sterile (representing limited dispersal), but when compromised, can no longer prevent microorganisms from entering infection sites (representing unlimited dispersal). An example of unlimited dispersal-mediated infection, in which microorganisms of many varieties can passively enter a breached infection site, is represented by open wounds, such as following a traumatic skin injury that compromises barrier function. These infections accumulate microorganisms from the skin, gut, oral cavity and environmental reservoirs (see the figure)133, leading to ‘unusual’ polymicrobial interactions that probably occur in few other places in nature, such as interactions between S. aureus (a bacterium that usually resides in the respiratory tract and on the skin) and P. aeruginosa (a bacterium that usually resides in soil and water).
Our understanding of polymicrobial infections has benefited not only from asking the questions ‘who is there?’ (which can be addressed using metagenomics) and ‘what are they doing?’ (which can be addressed using metatranscriptomics) but also ‘who is next to whom?’ (that is, ‘what is the biogeography?’). Because of their spatial arrangement, two species found in the same infection site may not directly interact. Therefore, it is beneficial to preserve native spatial organization when studying infectious communities. Although this may be a difficult task, efforts to do so are worthwhile as gaining insight from a disturbed or homogenized ecosystem is considered “similar to asking a plant ecologist to make sense out of a giant heap of plants harvested from an entire landscape” (REF. 20).
The characterization and manipulation of microbial spatial arrangements have provided insight not only into pathogenesis but also, more broadly, into bio diversity21, community stability22 and evolution23,24, further highlighting the importance of understanding the biogeography of microbial communities. In this Review, we focus on individual infection sites and examine, at the cellular level, how these communities are spatially organized and how this organization affects virulence. Although biogeography is traditionally discussed at the macroscale, we feel that, from the perspective of a microorganism, even an isolated infection site is a landscape, with microscopic features equally as diverse as those throughout the human body. These microenvironments encompass attachment sites, the physicochemical environment, nutrients and the host immune system, and these factors can vary in how well they support or suppress microbial growth, ultimately leading to complex microbial spatial patterns, or what we describe as microbiogeography. In addition to reviewing the importance of these factors during infection, we discuss the specific role of polymicrobial interactions in determining spatial organization, and highlight potential therapeutic interventions that can be derived from studying microbiogeography. Our goal is to highlight the impact of spatial organization on virulence and disease progression (FIG. 1).
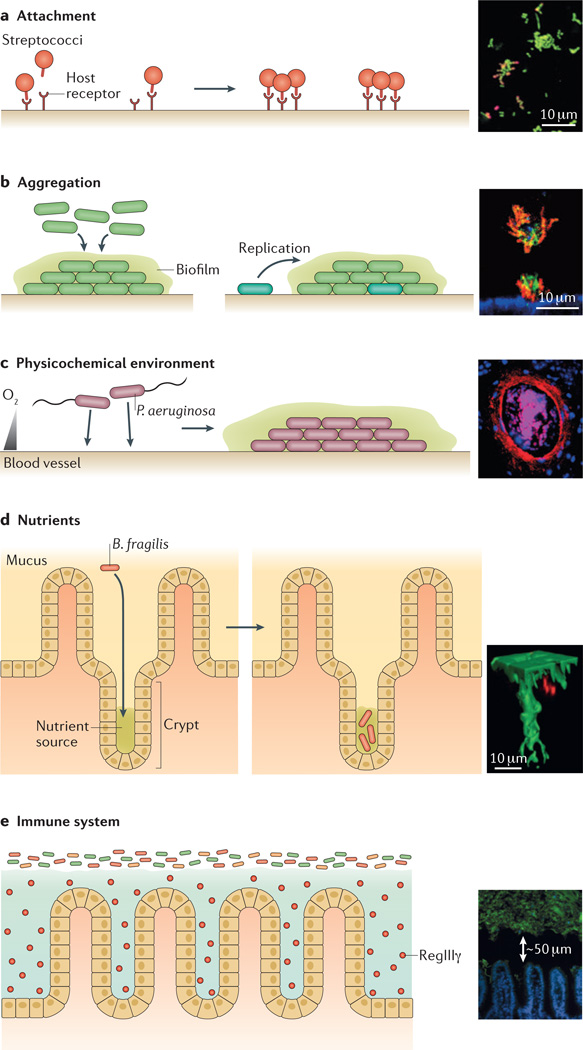
Figure 1. Factors that influence microbiogeography.
a | Attachment. Binding to specific host receptors can enable microorganisms to attach to surfaces. Therefore, the spatial distribution of such receptors may have an important role in determining the initial organization of microbial communities. For example, the insert shows streptococci (labelled in red; nonspecific nucleic acid stain labelled in green), which are initially sparse in human dental plaque communities, suggesting that the host receptors that streptococci adhere to are also sparse26. b | Aggregation. Following attachment, microorganisms usually grow and organize into aggregates. Aggregation can involve collective behaviours (left panel) or can arise from single-cell founding events followed by clonal growth (right panel, in which the cyan shaded cell is the founding cell). For example, the insert shows Pseudomonas aeruginosa (labelled in red) forming microcolonies on the surface of epithelial cells (labelled in blue), in a process that is regulated by collective behaviour and depends on the expression of a type III secretion system (T3SS)36. The aggregate exhibits biofilm-like characteristics, such as staining positive for the matrix exopolysaccharide Psl (labelled in green). c | The physicochemical environment. Gradients, such as pH gradients and oxygen gradients, can influence the distribution of bacteria within communities. For example, the insert shows P. aeruginosa (labelled in red) concentrated around a vein (host cells are labelled in pink and blue) in a cross-section of a murine burn wound, in a process that may be regulated by oxygen availability30. Scale bar not included. d | Nutrients. The ability to access and utilize certain nutrients in specific locations can also influence microbiogeography. For example, the insert shows Bacteroides fragilis (labelled in red), which carries a specific polysaccharide utilization locus that mediates its localization deep within an intestinal crypt (labelled in green)68. e | The immune system. Immune molecules, such as the lectin RegIIIγ, influence the distribution of microbial communities. For example, the insert shows how secretion of the lectin RegIIIγ by epithelial cells in the small intestine (labelled in blue) leads to the establishment of a 50-µm-wide gap between the intestinal surface and the microbiota (labelled in green)69. The insert in panel a is adapted from J. Bacteriol., 2003, 185, 3400–3409, doi:10.1128/JB.185.11.3400-3409.2003 and amended with permission from American Society for Microbiology. The insert in panel b is adapted from REF. 36. The insert in panel c is adapted from Infect. Immun., 2007, 75, 3715–3721, doi:10.1128/IAI.00586-07 and amended with permission from American Society for Microbiology. The insert in panel d is adapted from REF. 68, Nature Publishing Group. The insert in panel e is adapted from Vaishnava, S. et al. The antibacterial lectin RegIIIγ promotes the spatial segregation of microbiota and host in the intestine. Science 334, 255–258 (2011). Reprinted with permission from AAAS.
Factors that influence microbiogeography
Multiple factors influence the spatial patterns of microbial communities during infection, including the ability of microorganisms to attach and aggregate, the physicochemical environment and nutrient levels surrounding the communities, and the host immune response.
Attachment
The first requirement for successful colonization of the host is attachment. Although microorganisms can attach nonspecifically to abiotic surfaces — a behaviour that is important in the context of implanted medical devices (for example, catheters or stents)25 — in the context of host tissue, attachment often occurs through highly specific interactions between microbial cell surface structures (adhesins) and host receptors. The spatial distribution of such receptors may therefore have an important role in determining the initial organization of microbial communities on host surfaces.
The oral cavity is an environment that illustrates how attachment can influence colonization. Early colonizers of dental plaque, such as oral streptococci, can adhere to many of the receptors displayed on the acquired enamel pellicle that coats the surface of teeth. Despite this capability, spatial mapping of pioneer species on human teeth has shown that these initial communities are strikingly sparse, observed mostly as solitary cells and dense microcolonies26 (FIG. 1a). This suggests that the receptors that oral streptococci bind to are themselves sparse, implicating a direct role for the biogeography of host receptors in structuring surface-attached communities. Alternatively, these receptors could be abundant on the surface of teeth, and the observed biogeographic pattern could in fact arise by the gradual transfer of streptococci from neighbouring sites. Further insight into the factors that regulate the microbiogeography at these sites may be gained from spatially mapping the receptors themselves, similarly to what has been done for the distribution of host receptors in gut mucus27.
An important advantage of attachment in both the oral cavity as well as other infection sites is that it affords protection from flow and washout. For example, uropathogenic strains of Escherichia coli (UPEC) can successfully colonize the kidney, despite the presence of high flow rates. To achieve this, UPEC expresses multiple types of highly adhesive fimbriae; these surface appendages enable UPEC to firmly adhere to the walls of filtration tubules as well as to other UPEC cells, causing these populations to span and obstruct the lumen of the tubules. Ultimately, this spatial organization blocks urine flow, heightening the severity of kidney infection. As kidney tubules are narrow (their width is <50 µm), bacterial attachment patterns at even very small spatial scales can easily block them, triggering the onset of symptomatic infection28.
Aggregation
Once attached to the host, microorganisms generally grow and organize themselves into small (5–200 µm wide) clusters (FIG. 1b). These structures, also known as aggregates, have been termed in vivo biofilms29, and they directly contrast to the large (sub-millimetre to centimetre scale), mushroom-shaped structures often observed in biofilm studies in vitro. Many, if not most, infections are colonized with aggregates, including wounds30,31, abscesses32 and the cystic fibrosis lung33, although exceptions have been noted, such as the thick biofilms often associated with middle ear infections34. Similarly to in vitro biofilms35, the formation of in vivo biofilms is tightly regulated, with the genetic determinants for many species beginning to be examined36–38. An important insight from these studies has been that the determinants for in vivo biofilm formation are not always the same as those identified in vitro, and vice versa. Biofilm studies in P. aeruginosa demonstrate this principle. P. aeruginosa requires quorum sensing to form mushroom-like structures in flow chambers39 but not in murine burn wounds30. Similarly, P. aeruginosa requires a type III secretion system to form aggregates in murine lung infections but not on abiotic surfaces36 (FIG. 1b).
Aggregates can also be formed from single-cell founding events followed by clonal growth40, rather than through a collective microbial behaviour. In these events, a single cell replicates until it forms a multicellular aggregate; this contrasts with the active recruitment of microorganisms into an aggregate, which can occur independently of cell division (FIG. 1b). Computer simulations that model clonal growth from single cells have shown that spatial structure is an emergent property of basic cell division and crowding. Although these models have not yet perfectly reproduced the shape and size of aggregates found in vivo, they have given insight into the fundamental parameters that govern cellular organization in structured communities. For example, simulation studies have revealed that nutrient limitation fosters the spatial segregation of cell lineages41 (see nutrients section below).
As with in vitro biofilms, pathogenic microorganisms that are in aggregates have enhanced virulencerelated phenotypes, including accelerated growth42, increased stress resistance43,44, immune evasion38,45–47 and transmission37,48. Therefore, it is important for the host to physically disperse these populations to successfully eliminate them. Host factors that can disrupt this microbiogeography include two major components of mucus, lactoferrin49 and mucin50. These molecules promote motility in P. aeruginosa and thereby prevent it from aggregating. However, in immunocompromised patients with cystic fibrosis, P. aeruginosa thrives as aggregates embedded in mucus layers lining the lung. Furthermore, in these patients, P. aeruginosa aggregates are thought to release toxic molecules that stave off immune cells. One such toxin, rhamnolipid, forms a protective shield around P. aeruginosa aggregates, evidenced as a sharp, impenetrable interface against host neutrophils51. Although neutrophils cannot invade these populations, aggregate growth rates in the lungs of patients with cystic fibrosis negatively correlate with the density of the surrounding neutrophils because these host cells restrict the amount of oxygen available to the pathogen. As a result, in the respiratory tract, P. aeruginosa grows at higher rates in the respiratory zone (which is high in oxygen) than in the conducting zone (which is low in oxygen)46. Therefore, local interactions between aggregates and neutrophils affect the macroscale biogeography in the lungs of patients with cystic fibrosis.
The physicochemical environment
Gradients, such as pH gradients and oxygen gradients, provide landmarks for microbial spatial organization. For example, in the stomach, H. pylori penetrates deep into the mucus layer, where it precisely localizes to a region that is 25 µm above the epithelial surface52. Rather than relying on absolute pH or other cues of location within the mucus, such as urea or ammonia, H. pylori achieves its specific orientation in the stomach by sensing a pH gradient that guides it away from the lumen (approximately pH 3) and towards the epithelium (approximately pH 5.5)52. This mechanism enables H. pylori to colonize a precise region where it achieves high local cell densities that probably contribute to the very high rates of recombination among H. pylori strains53 and promote chronic bacterial persistence in one of most extreme, acidic environments in the human body.
Oxygen gradients also impart spatial organization to microbial communities. For example, in the gut, oxygen that diffuses out from the epithelium creates an adjacent oxic zone in which strict anaerobes are depleted but facultative anaerobes are enriched54. Oxygen therefore provides a useful cue for proximity to the epithelial layer, and this information can be utilized by pathogens such as Shigella flexneri. This intracellular pathogen secretes effectors for epithelial invasion when it senses precise oxygen levels in vivo (within 70 µm of the epithelial surface)55. At a smaller scale, gradients of oxygen can develop even within individual bacterial aggregates, owing to the consumption of oxygen by bacteria at the surface of the aggregate56. This raises the possibility of multispecies organizations in which strict anaerobes can persist in the centre of aggregates, away from the source of oxygen57, similar in concept to that which has been described in the gut54.
Despite being exposed to air, skin wounds are also highly heterogeneous in oxygen levels. In chronic, surgical wounds, this results from poor vasculature, which creates a low-oxygen environment that enables the persistence of many strict anaerobes58. However, oxygen is not completely absent, as aerobes such as P. aeruginosa also commonly thrive in these infections. P. aeruginosa is a motile bacterium that can move to the periphery of the wound31, a spatial niche where it can potentially gain greater access to oxygen. P. aeruginosa is also highly migratory in acute, burn wounds. In these infections, P. aeruginosa forms dense aggregates around arteries and veins, a spatial organization known as perivascular cuffing30 (FIG. 1c). A potential regulator of this organization is the oxygen that may leak into the infection site from the oxygen-rich blood in the vasculature. This oxygen may accumulate locally around arteries and veins, providing a chemoattractant for P. aeruginosa motility. Supporting this notion, aerotaxis and motility are required for full fitness of P. aeruginosa in burn wounds59, although many other blood-derived cues could also act as chemo-attractants in these infections. Furthermore, cuffing immediately precedes the transit of P. aeruginosa from the wound bed into the bloodstream, leading to sepsis and potentially death, highlighting the importance of these structures in pathogenesis. However, visualization of P. aeruginosa in human wounds is still needed to extend the importance of these spatial organizations to P. aeruginosa infections in humans. Nonetheless, the described murine-based studies demonstrate that microbial responses to physicochemical cues during infections can result in a spatial organization that ultimately promotes virulence.
Nutrients
As mentioned above, computer models have revealed that nutrient limitation fosters the spatial segregation of cell lineages41. For example, agent-based modelling of microbial growth in biofilms has shown that, although different strains of bacteria often intermix during growth when nutrients are plentiful, in nutrient-depleted conditions strains segregate along the growing front. This segregation occurs because as nutrient availability is reduced, the number of dividing cells is also reduced, increasing the potential for stochastic separation of lineages into distinct sectors. This formation of separate genotypic sectors has important consequences for social interactions in microorganisms (BOX 2) as it enables strains that produce secretions to avoid exploitation by non-producing cheat strains41. Therefore, as microorganisms in infection sites are often starved for nutrients, nutrient-derived spatial segregation may be a powerful source of genotype patterning in vivo.
Box 2 | Spatial structure and social evolution.
Microorganisms secrete an array of costly molecules that can confer benefits to any neighbouring cells that are suitably equipped to profit. Because secreted molecules impose costs on producer cells and potentially return benefits to neighbouring cells, they have become models for the study of bacterial cooperation and the fate of producers versus non-producers. When producers and non-producers are sufficiently well mixed (for example, in a shaken flask), the cost of secreted factor investment is paid only by the producer lineage, whereas rewards are shared among both lineages, leading to an enrichment of non-producers134,135. However, if the population is structured, the benefits of cooperation will fall preferentially on producer cells, enabling them to outcompete non-producers134,135. More recent attention has focused on the physical, behavioural and demographic forces that drive bacterial population structuring, and ultimately enable the maintenance of cooperative ‘public goods’ traits. Important factors that have been identified as drivers of lineage segregation and cooperation include nutrient levels23,41, clustered dispersal136, horizontal gene transfer137, adhesion138 and mechanisms of heterologous lineage repulsion or ‘policing’ (REF. 139).
Although many of the factors affecting bacterial population structure and the evolution of cooperation are under bacterial control (that is, they are driven by bacterial traits), others will be affected by intrinsic elements at the infection site. For example, Bacillus thuringiensis kills its insect host by transiting the midgut, a behaviour that is dependent on secreted toxins but cannot be exploited by cheats in vivo because well-separated aggregates are prevalent during early infection and restrict the sharing of virulence factors24. By contrast, mouse burn wounds infected with Pseudomonas aeruginosa are socially exploitable140, perhaps because these infections require motility59, which may homogenize the bacterial population. It has also been suggested that hosts could adjust the levels of particular nutrients to modify the composition and structure of the microbiota at certain sites61. How this variation in both disease site characteristics and bacterial traits combines to determine spatial structuring and the balance of cooperation and conflict across infections remains to be explored.
Furthermore, nutrients in the host are highly heterogeneous, as seen in the gut where host secretions can be produced in patches, forming ‘hotspots’ along the gut epithelium60, and gradients, which decrease in richness as distance from the epithelium increases61. Because of this nutrient richness closer to the epithelium, this location is the preferred niche for many enteric pathogens; when present at this location, these pathogens upregulate virulence factors that damage the host and release nutrients, ultimately enhancing bacterial growth62–64. This nutrient heterogeneity can give rise to spatial organization, which can be driven by motility40 and chemotaxis64. For example, motility is required for Salmonella enterica subsp. enterica serovar Typhimurium to infect the inflamed gut. In this environment, the host epithelium generates alternative electron acceptors that enable S. Typhimurium to outgrow the native gut microbiota65,66. Respiration using these electron acceptors acts as a signal for motility in S. Typhimurium, a behaviour known as energy taxis. As a result, S. Typhimurium undergoes chemotaxis towards, and closely associates with, the gut epithelial surface, which is the source of the electron acceptors64. Therefore, host-derived nutrients give rise to a spatial organization that underlies inflammatory pathogenesis64.
Spatial structure can also occur through local growth enrichment67, independent of motility, as non-motile bacteria can acquire specific biogeographies that are linked to nutrition. For example, in the colon, Bacteroides fragilis localizes to the intestinal crypt spaces between villi (FIG. 1d). The colonization of this spatial niche requires the expression of a specific polysaccharide utilization locus68. This locus, termed ccf (for commensal colonization factors), shows homology to the starch utilization system (Sus) family of proteins, suggesting that the uptake, and use of, glycans may regulate the ability of B. fragilis to colonize this niche. Interestingly, Vibrio cholerae also localizes to intestinal crypt spaces, but despite being motile, it does not require motility or chemotaxis for this spatial organization. This suggests that a motility-independent mechanism, perhaps similar to that used by B. fragilis to colonize this region, also regulates the ability of V. cholerae to penetrate intestinal crypt spaces62.
The immune system
The immune system also contributes to microbiogeography, most notably at epithelial surfaces where the host is at greater risk of direct contact with the microbiota. Therefore, physical separation at epithelial surfaces is crucial for the host to avoid persistent inflammation. In the intestine, the immune system establishes a narrow microorganism-free layer directly above the epithelium that is only 50 µm wide69,70 (FIG. 1e). To maintain this barrier, the small and large intestine use mechanisms that are distinct and well suited to the physiological roles of these organs71. The mucus layer in the small intestine is highly permeable to promote nutrient absorption, rendering it susceptible to the overlying microbiota. To limit the growth of this microbial population, epithelial cells in the small intestine secrete many antimicrobial factors into the lumen, but remarkably, a single antimicrobial, the lectin RegIIIγ, is responsible for local exclusion of bacteria from the epithelial cell surface69. By contrast, a physical rather than a chemical barrier is established at the mucosal surface in the large intestine. The mucus lining of the large intestine is organized into two layers. The outer layer is loosely attached, whereas the inner layer is highly stratified and dense, containing a much higher concentration of mucin 2 (MUC2), a major component of mucus. As a result, the inner layer is normally impenetrable to the microbiota in the large intestine, whereas pathogens that infect the colon can penetrate this layer by expressing MUC2-degrading enzymes72. However, some commensal bacteria, such as B. fragilis, can closely associate with the epithelial surface without triggering an overt immune response. To achieve this spatial localization, B. fragilis signals to the host through Toll-like receptor 2 (TLR2), a receptor that is normally associated with microbial clearance but is probably manipulated by many commensals (as well as some pathogens, such as S. aureus)73 to promote immune tolerance. TLR2 signalling by B. fragilis depends on poly saccharide A (PSA), as a B. fragilis mutant that lacks the ability to produce this polysaccharide de-localizes entirely to the lumen and is not found in tight association with the epithelial surface74.
The importance of the mucus layer in the large intestine to host health is emphasized by the fact that genetically deficient hosts (such as mice that lack MUC2) are more susceptible to infection by enteric pathogens. One such pathogen, Citrobacter rodentium, can penetrate the mucosal barrier even in MUC2-expressing mice, after which it forms largely clonal aggregates at the epithelial surface. However, in MUC2-deficient mice, this spatial organization is altered, which leads to C. rodentium forming mixed-species aggregates with the commensal flora, leading to increased epithelial invasion and reduced host survival75. MUC2-deficient mice also show increased incidence of microorganism-induced colitis and colon cancer70,76. In humans, most cancers of the ascending colon are associated with biofilms77. These cancer-linked biofilms are both highly invasive, with bacteria penetrating into intestinal crypts, and polymicrobial. Interestingly, when comparing the composition of paired communities from cancerous mucosa with normal mucosa in the same patient, it was found that they are highly overlapping, indicating that it is not the enrichment of particular pathogenic species but rather the spatial organization of the community into a biofilm that can intimately associ ate with the epithelium that may be responsible for carcinogenesis77. However, it is important to note that so far these studies are correlative. It is also possible that changes that occur, independently of the microbiota, in environments in which tumours develop are conducive to both tumorigenesis and the re-organization of the microbiota into biofilms. Future studies in animal models, in which the spatial organization of the colon microbiota can be directly manipulated, may establish biofilms as aetiological agents of colon cancer.
Polymicrobial infections
Similarly to living in a city, where an inhabitant of an infection chooses to live depends not only on the real estate (the host) but also the neighbours (polymicrobial interactions). As a result of such interactions between neighbouring microorganisms, multispecies communities can develop highly intricate spatial organizations, such as layers78, interdigitations26,79,80 and even ‘corn cobs’ (REF. 81). Moreover, the spatial structure that develops from these interactions can in turn affect the interactions that generate it, namely by tuning the spatial proximity of the interacting microorganisms as lineages expand. Mutualistic microbial lineages that grow towards each other will therefore enhance their metabolic interactions and preserve their mutualistic relationship, whereas competitive lineages that grow apart will segregate in space and attenuate their interaction. In broad terms, community organization can be classified as either spatially segregated or mixed (FIG. 2). In terms of infection, both classes of spatial structure and their associated interactions can influence virulence. Below, we describe four prominent examples of polymicrobial interactions that lead to emergent spatial structure. We categorize these interactions as ‘physical’ and ‘chemical’ (FIG. 2). Physical interactions are mediated by cell–cell contact or components of the biofilm matrix. Chemical interactions are mediated by diffusible molecules such as excreted metabolic by-products.
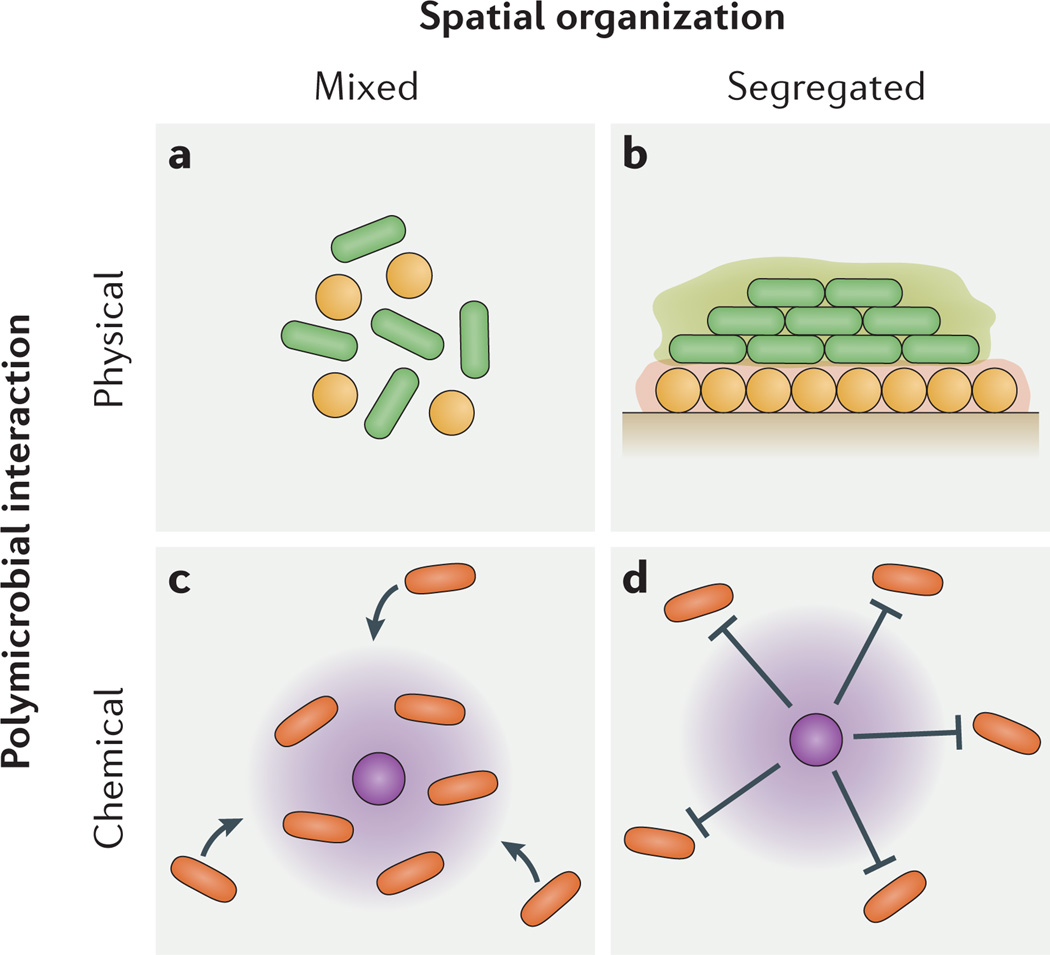
Figure 2. Polymicrobial interacions contribute to microbiogeography.
Chemical and physical polymicrobial interactions generate spatially mixed and segregated community patterns during infections. a | Co-aggregation, or intercellular binding, can cause spatial mixing. b | Excess production of biofilm polysaccharide (green and orange halos) can cause spatial segregation. c | Production of a beneficial metabolite (purple halo) can cause spatial mixing. d | Production of a harmful metabolite (purple halo) can cause spatial segregation.
Co-aggregation
Intercellular binding between distinct bacterial taxa, known as co-aggregation, is especially prevalent in the oral cavity. Most, if not all, oral bacterial species have at least one co-aggregation partner82, and these interactions are highly predictive of ‘nearest neighbours’ in human dental plaque83 (FIG. 2a). For example, streptococci co-aggregate with Veillonella spp. and Actinomyces spp. in vitro, and these species also form mixed, interdigitated aggregates on the surface of teeth26,79. As a result of these close associations, metabolic benefits are gained. Veillonella atypica cannot grow alone in saliva but can grow if co-aggregated with streptococci79,84, and Streptococcus gordonii cannot grow alone in low-arginine media but can grow if co-aggregated with Actinomyces naeslundii85. Importantly, co-aggregation is not always indicative of mutualism. For example, an extremely small (200–300 nm) TM7 phylotype (TM7x) only grows when attached to Actinomyces odontolyticus, from which it derives amino acids, but TM7x kills A. odontolyticus when starved. However, this interaction is not strictly parasitic, as TM7x strongly suppresses host immune signalling, which may benefit the growth of A. odontolyticus86. As enrichment of the TM7 group is associated with oral infections, the obligate co-aggregation between TM7x and A. odontolyticus suggests that, in addition to promoting metabolite exchange, co-aggregation can have immunosuppressive functions that affect virulence86.
Biofilm remodelling
Another physical structuring interaction that regulates microbiogeography is biofilm remodelling. Biofilms are held together by extracellular matrix components such as polysaccharides, and therefore biofilm remodelling refers to the production or breakdown of these components. In contrast to coaggregation, biofilm remodelling often acts to increase the separation between community members (FIG. 2b). For example, when growing on an agar surface, Pseudomonas fluorescens regularly mutates de novo into mucoid variants that overproduce exopolysaccharide. These variants push themselves to the top of the biofilm, where they gain greater access to oxygen, and this repositioning occurs at the expense of ‘smothered’ competitors87. Furthermore, direct disturbance to the structure of biofilms during competition experiments showed that remodelling of the biofilm structure to alter its position and reach the growing edge is key to the success of the P. fluorescens mucoid variant. Analysis of the genetic basis of this phenotype in 565 independently evolved mucoid variants showed that all variants had mutations in a single locus. This notable consistency in both phenotypic and genotypic parallel evolution highlights how traits involved in biofilm remodelling may be under very strong selection and show rapid evolution in short timescales.
Similarly, P. aeruginosa variants that overproduce the polysaccharide alginate can be observed in the lungs of patients with cystic fibrosis, in which the infecting community is also highly spatially organized, both at the macroscale as regionally isolated populations88 and at the microscale as aggregates33. Many evolutionary pressures are likely to select for mucoid variants of P. aeruginosa in the lungs of these patients, including interspecies competition. Supporting this idea, mucoidy confers a fitness advantage to P. aeruginosa when in competition with S. aureus89. Furthermore, the other major P. aeruginosa polysaccharides, Pel and Psl, have distinct structural roles in organizing multispecies biofilms with S. aureus. In these biofilms, a juxtaposed structure, in which P. aeruginosa and S. aureus are in close proximity, requires Pel, whereas a layered structure, in which P. aeruginosa forms ‘caps’ on top of S. aureus, requires Psl90. These ‘caps’ are reminiscent of the smothering phenotype exhibited by P. fluorescens mucoid variants (described above) and, incidentally, the polysaccharide mediating this organization, Psl, is strongly selected for in the lungs of patients with cystic fibrosis91.
Furthermore, polymicrobial interactions that modulate the architecture of P. aeruginosa biofilms are not limited to competition with S. aureus. Burkholderia cenocepacia is a soil bacterium that, like S. aureus, is often co-isolated with P. aeruginosa from the lungs of patients with cystic fibrosis92, in which B. cenocepacia and P. aeruginosa are thought to compete93. In mixed-species biofilms, P. aeruginosa responds to a fatty acid secreted by B. ceno-cepacia by spatially reorganizing as filaments94,95. Similar structural changes may occur in the lungs of patients with cystic fibrosis, potentially dictating the interactions between these two species. In a murine lung co-infection model, P. aeruginosa was able to outcompete B. cenocepa-cia, with a major consequence being greater inflammation in the host93. However, in-depth spatial analyses are still required to attribute biofilm remodelling to competitive interactions in these communities. Nonetheless, it is becoming evident that the re-positioning of community members in polymicrobial biofilms can influence both community interactions and disease progression.
Local growth inhibition
‘Chemical’ interactions through interspecies signals or metabolites also affect spatial structure. Interspecies signals such as autoinducer 2 (REF. 96) can spatially regulate biofilm formation97 and dispersion98, and can also stimulate other virulence properties, such as antibiotic resistance94,99 and per-sistence96. As these signals are often restricted to very short-range (<10 µm) effects100, they have the potential to generate fine-scale spatial structure. However, signalling is restricted to only those species with an appropriate receptor, suggesting that non-discriminatory metabolic interactions may be a more widespread force that generates spatial structure. For example, metabolic waste products can act as broad-spectrum toxins, affecting bacterial growth near the producer (FIG. 2d). Many streptococci exemplify this behaviour as they generate abundant lactate and hydrogen peroxide as waste products, molecules that cause acid stress and oxidative stress, respectively. As these by-products are most concentrated in the immediate vicinity of the producer, they are most effective at eliminating local competitors, which has the potential to regulate fine-scale segregation patterns. For example, hydrogen peroxide produced by S. gordonii reaches only micromolar concentrations in bulk solution, but can reach millimolar concentrations when measured only 100 µm above a S. gordonii biofilm101. Similarly, S. mutans, a bacterial species that frequently contributes to the formation of tooth cavities6, forms isolated ‘pockets’ of strong acid along the substratum of a mixed-species biofilm, corresponding to a loss in community diversity. Ironically, creation of these pockets is stimulated by the surrounding oral bacterial species, at their own demise102. As this acid stress occurs only locally (in these ‘pockets’), S. mutans does not fully displace the community. Rather, a relatively diverse microbiota is associated with cavities, suggesting a polymicrobial origin of disease6. Furthermore, ex vivo human dental biofilms rapidly develop steep pH gradients (in which the pH varies from 7.1 to 4.4) when exposed to sucrose (a carbon source that S. mutans preferentially converts to acid)102,103, further highlighting that inhibitory chemical gradients mediate polymicrobial interactions in the human microbiome.
A highly segregated spatial arrangement also manifests itself in human chronic wound infections, in which P. aeruginosa inhabits much greater depths (50–60 µm from the wound surface) than S. aureus (only 20–30 µm from the wound surface)104. In murine wound infection models, these species also rarely intermix, instead inhabiting discrete, largely clonal aggregates (M.W., unpublished observations). A probable source of this spatial segregation is the growth inhibition of S. aureus by P. aeruginosa as, even at low cell densities, the doubling time of S. aureus cells positioned near P. aeruginosa (<6 µm apart) is shorter than the doubling time of S. aureus cells positioned further away (>30 µm apart) on glass surfaces105. A specific toxin that potentially mediates the spatial segregation between these species is pyocyanin. P. aeruginosa increases the production of pyocyanin when it senses cell wall fragments shed by S. aureus5, suggesting that P. aeruginosa kills S. aureus when these species are in close proximity. Despite this, S. aureus persists in wounds that contain P. aeruginosa, possibly because the wound environment is highly viscous and restrictive of cell migration. After spatial segregation is established, this viscosity may act to prevent mixing and therefore any further interspecies killing. Supporting this notion, P. aeruginosa quickly eliminated S. aureus in well-shaken laboratory media but not in a gelatinous in vitro wound model106. Many other experimental systems also sustain higher biodiversity when viscosity is imposed21,22,107.
Local growth promotion
Whereas growth suppression causes spatial segregation, growth promotion causes spatial mixing. This can occur either by metabolic cross-feeding, whereby one species consumes the by-products of another, or it can occur by antimicrobial cross-protection, whereby one species shields another from a stress. Often, these interactions result in the clustering of the benefit receiver around the benefit giver108–110 (FIG. 2c), although this spatial interaction can also be reversed, when the giver surrounds and thereby shields the receiver instead111. Single-cell studies have shown that these interactions can generally occur across only very short distances. For example, an aggregate of S. aureus can be protected from the antibiotic ampicillin when closely surrounded by a shell of ampicillin-resistant P. aeruginosa but not when the same P. aeruginosa population is present at low density111. Importantly, the level of mixing that results from these interactions depends on the strength of the interactions themselves; species that are strongly interdependent mix more than species that are weakly interdependent, and even more so than species that conflict78,80. This concept is exemplified by the ‘food for detoxification’ interaction between the oral bacteria Aggregatibacter actinomycetem comitans and S. gordonii. S. gordonii produces lactate, which A. actinomycetemcomitans cross-feeds on, and in exchange A. actinomycetemcomitans detoxifies peroxide, one of the major by-products of S. gordonii metabolism. This metabolic synergy results in a mutualistic relationship between both species during an abscess co-infection in which both bacteria reach higher burdens when they are together than when they are apart. In these abscesses, A. actinomycetemcomitans strongly colocalizes around S. gordonii, promoting cross-feeding while also maintaining a >4 µm gap from S. gordonii to avoid growth inhibition by peroxide. A. actinomycetemcomitans acquires this spacing by sensing peroxide, immediately after which it activates an enzyme, known as dispersin B, which dissolves its biofilm matrix. Inactivation of this enzyme disrupts the spacing, placing A. actinomycetemcomitans directly next to S. gordonii, disturbing co-infection synergy32. Similar fine-scale spatial patterns are also seen for other synergistic oral pathogens, such as Fusobacterium nucleatum and Tannerella forsythia112,113. These studies firmly link microbial positioning to community virulence and also demonstrate the basic role of growth promotion by secreted metabolic products in generating community spatial structure.
Targeting biogeography as a therapeutic strategy
So far in this Review we have highlighted the importance of biogeography in microbial virulence, raising the possibility of manipulating the spatial organization of bacterial pathogens as a viable therapeutic strategy (FIG. 3). Below we present four, independent strategies focused on manipulating biogeography to dampen pathogenesis: dispersing pathogen biofilms; strengthening host epithelial barriers; depleting commensal microbiota that enhance pathogen virulence; and altering factors that guide pathogens to spatial patterns linked to virulence. An additional therapeutic approach is to analyse the structure of complex communities to discern (or ‘reverse-engineer’) the poly-microbial interactions within, such as whether they are mutual or competitive, as a diagnostic for disease severity or potential (BOX 3).
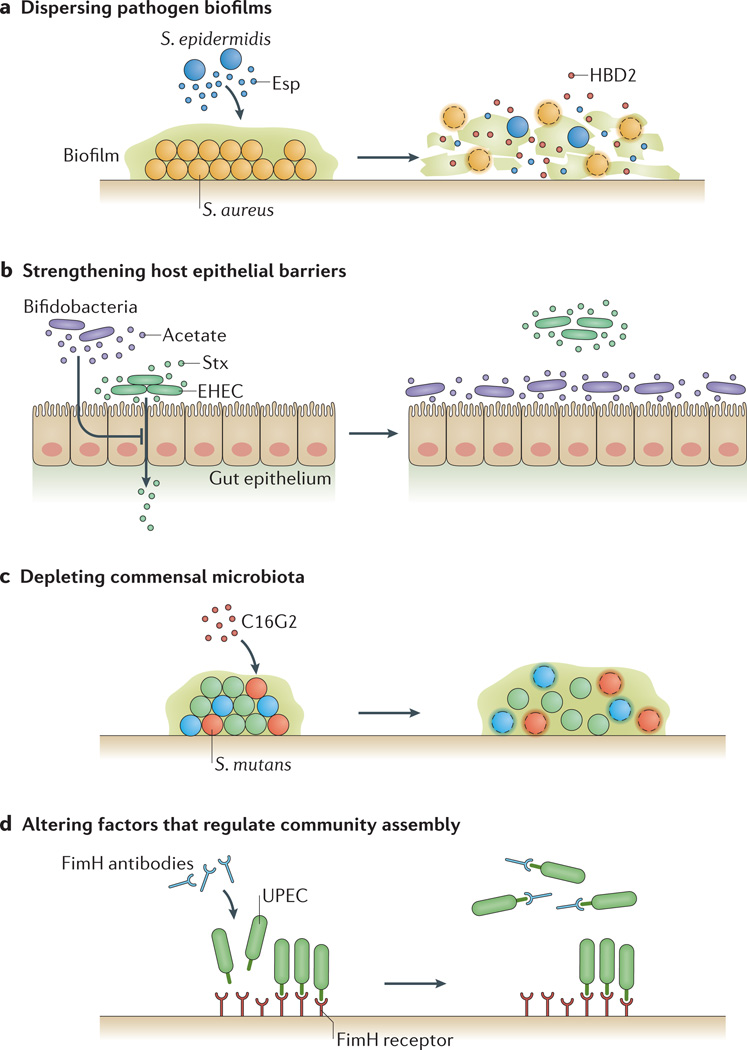
Figure 3. Targeting biogeography as a therapeutic strategy.
a | Dispersing pathogen biofilms. Spatially organizing into biofilms can make microorganisms resistant to challenges, such as host immune responses, so dispersing biofilms can provide a potential therapy against infections. For example, Staphylococcus epidermidis can destroy biofilms of Staphylococcus aureus through the production of the serine protease Esp. Furthermore, Esp enhances the susceptibility of S. aureus to human β-defensin 2 (HBD2), an antimicrobial peptide released during inflammation of the nasal cavity114. b | Strengthening host epithelial barriers. Manipulating biogeography can also be used to prevent pathogens from crossing host barriers. For example, Shiga toxin (Stx) produced by enterohaemorrhagic Escherichia coli (EHEC) can cross the gut epithelium, but the introduction of commensal bifidobacteria can protect against fatal EHEC infection. Protective bifidobacteria can consume fructose and produce acetate, which induces an anti-inflammatory response in epithelial cells in the colon that lowers their susceptibility to Stx115. c | Depleting commensal microbiota. Some commensal microorganisms can exacerbate infection by promoting pathogen virulence. Therefore, in situations in which targeting the pathogen directly has proven ineffective, depleting these commensals could represent an alternative therapeutic strategy. The feasibility of this approach is demonstrated by the synthetic antimicrobial peptide C16G2, which selectively kills Streptococcus mutans (in red), a commensal known to be involved in the progression of dental caries. Treating oral biofilms with C16G2 not only depletes communities of S. mutans (red cocci) but also other community members that directly interact with S. mutans116. d | Altering factors that regulate community assembly. Masking microbial attachment sites, or reversing or eliminating the molecular gradients that give rise to virulence-associated spatial organizations, can eliminate microbiogeography. For example, a vaccine designed against the FimH adhesin, used by uropathogenic strains of E. coli (UPEC) to adhere to the bladder epithelium, is highly effective at limiting attachment and colonization of this pathogen in bladder infection models117.
Box 3 | Reverse-engineering polymicrobial interactions from spatial structure.
The spatial structure of infections caused by mixed bacterial species is an emergent consequence of the nature of the metabolic interactions among the infecting microorganisms. In model microbial systems, the effects of different metabolic interactions on spatial structure can be elucidated using experimental manipulations to directly observe their effects on spatial structure and the ultimate consequences for disease outcomes. However, for most polymicrobial infections the nature of the interactions among species and their consequences for spatial structure are not known. For example, it is still unclear whether the spatial structure of natural infections is a consequence of competition or mutualism among constitutive species. Similarly, how the spatial structure varies among different infections is also largely unknown. We suggest that methods that are typically applied to analyse the biogeography of macroorganisms can also be applied to elucidate the nature of interactions in natural polymicrobial infections.
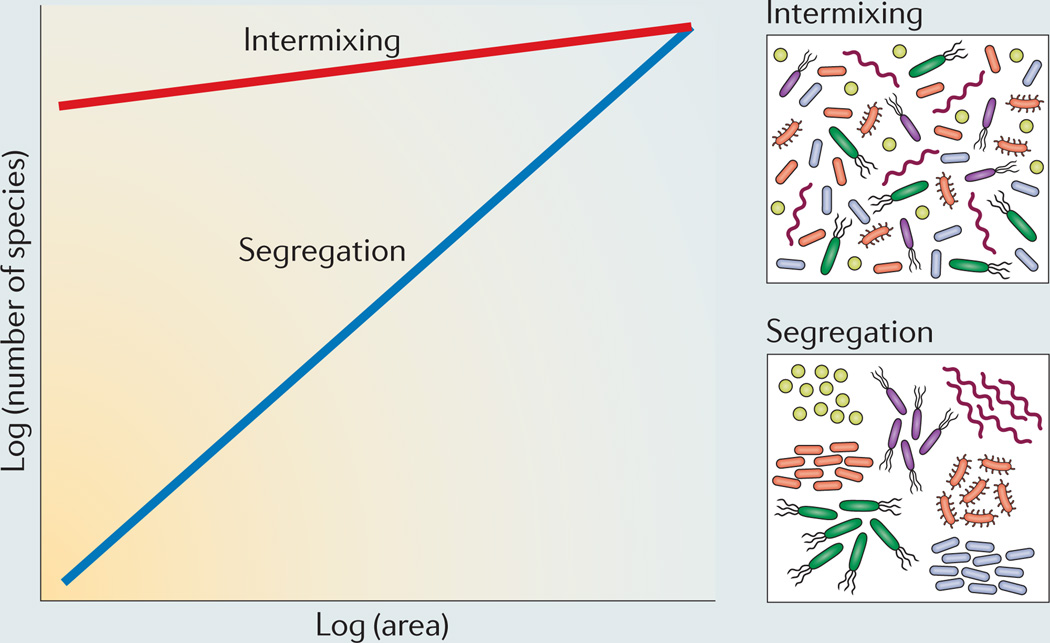
One of the most common ways to study the distribution of macroorganisms is using the ‘species-area curve’141,142 (see the figure). This curve describes the accumulation of species as larger areas within a habitat are sampled, typically modelled as a log-log regression of the number of species present in a sample on the size of the area from which that sample was taken. The slope of this species-area curve indicates the nature of microbial spatial organization. Strong competition among species is expected to lead to species segregating in space, whereas mutualism and inter-reliance are expected to lead to the spatial mixing of species80. If species show strong segregation owing to competition, then sampling larger areas during an infection will lead to the accumulation of more species (see the figure, blue slope). Conversely, if species show intermixing owing to mutualism, then there will be little accumulation of species as larger areas of an infection site are sampled (see the figure, red slope).
Although the species-area curve is obviously a great simplification of the complex distributions of biodiversity, it has become a cornerstone of biogeographic studies, playing an important part in current debates regarding the roles of both local processes (such as competition, predation and population dynamics) and regional processes (such as speciation, extinction and colonization) in driving the spatial distributions of biodiversity143–145. Species-area curves have also begun to be applied to microbial biogeography, giving insights into the drivers of spatial patterns of microbial diversity in the environment146–148, although they have not been applied at the fine spatial scales over which bacteria typically interact. Of note, when applied to this scale, this tool would need to be adapted for the three-dimensional structures that microorganisms are often found in (for example, biofilms) as species-area curves were originally developed for two-dimensional biodiversity studies (for example, the positioning of plant species across a field).
Dispersing pathogen biofilms
The biofilm lifestyle confers resistance to components of the host immune system. Therefore, probiotics that perturb this spatial organization may be therapeutically beneficial114. This was found to be the case for strains of Staphylococcus epidermidis that destroy S. aureus biofilms114 (FIG. 3a). In an epidemiological study, a negative correlation was discerned between the presence of S. aureus in the nasal cavity and strains of S. epidermidis that destroy S. aureus biofilms. The biofilm-destroying factor, identified as the serine protease Esp, disrupts even pre-formed S. aureus biofilms, including those formed by methicillin-resistant S. aureus (MRSA) and vancomycin-intermediate resistant S. aureus (VISA). Furthermore, Esp enhances the susceptibility of these strains to human β-defensin 2 (HBD2; also known as DEFB4A), an antimicrobial peptide released during inflammation of the nasal cavity. Although HBD2 has low bactericidal activity against Gram-positive bacteria such as S. aureus, in combination with Esp it shows enhanced killing of S. aureus biofilms. S. epidermidis may therefore act in concert with the innate immune system to perturb S. aureus biogeography and subsequently eliminate S. aureus from the nasal cavity114.
Strengthening host epithelial barriers
Disease often results when pathogens cross host barriers, indicating that inhibiting this process could act as another biogeography-oriented therapeutic strategy. The gut epithelium is a host barrier that, when crossed by Shiga toxin (Stx) produced by enterohaemorrhagic E. coli (EHEC) O157:H7, causes potentially fatal infections. Certain species of commensal bifidobacteria in the gut protect against fatal EHEC infection in mice. Compared with non-protective species, probiotic bifidobacteria reduce serum levels of Stx without affecting EHEC viability or virulence gene expression. These species encode ATP binding cassette (ABC)-type transporters, which are not present in non-protective bifidobacteria, that enable fructose utilization and conversion to acetate. Acetate induces an anti-inflammatory response in colonic epithelial cells, reducing their susceptibility to Stx (FIG. 3b). Mice that were fed acetylated starch were also protected from fatal EHEC infection, further supporting the role of a probiotic metabolite — acetate — in reducing pathogen virulence115. Therefore, probiotics can assist the host in defending against pathogens by simply increasing barrier function and confining pathogens to an avirulent biogeography.
Depleting commensal microbiota
Another potential biogeography-focused therapeutic strategy is to specifically deplete commensal microbiota that, although not directly harming the hosts themselves, exacerbate an ongoing infection by promoting pathogen virulence. Synthetic antimicrobial peptides make this approach feasible as they can be designed to target specific bacteria. One such peptide, C16G2 (REF. 116), selectively kills S. mutans (FIG. 3c). Despite its very narrow-spectrum activity, C16G2 was found to have a very large community-wide impact on a saliva-derived oral bacterial community, in which many of the species that were no longer detected in the C16G2-treated community were metabolically or physically dependent on S. mutans. Therefore, C16G2 acts as a proof of concept that the targeting of an individual species can have an effect that spreads to the entire community and, in the case of targeting S. mutans, reduces the community to an avirulent state116.
Targeting factors that regulate community assembly
Finally, it may be of therapeutic value to manipulate factors that modulate microbial spatial patterning. These unique therapeutic opportunities could include masking microbial attachment sites, or reversing or eliminating the molecular gradients that give rise to virulence-associated spatial organizations (FIG. 3d). For example, a vaccine designed against the FimH adhesin produced by uropathogenic strains of E. coli is highly effective at limiting attachment and colonization of this pathogen in murine bladder models117. Although not yet applied therapeutically, altering molecular gradients has been demonstrated experimentally to disrupt the biogeography of pathogens, notably of H. pylori in mammalian stomach models, whereby eliminating the pH gradient across the mucus layer abolishes its normal spatial orientation52.
All of these examples highlight the more general role of biogeographic thinking in discovering new targets for therapeutic interventions. The spatial distributions of pathogens and commensals at disease sites contain vast amounts of information on the progression of infection. Studying these patterns to understand not only where pathogens are located and what other species they are associated with, but also how these patterns develop, offers the prospect of identifying the key biogeographic events during infections that can be targeted for therapeutic intervention.
Conclusions and future directions
As described in this Review, microbial communities can be highly spatially organized throughout the human body and within sites of infection. This ‘biogeography’ arises owing to both host–microorganism and microorganism–microorganism interactions, many of which have been shown to directly or indirectly affect virulence. Although the infections discussed here cover a wide range of organisms and niches within the host, one message is clear, and that is that spatial structure is key to virulence. In future work, studies of polymicrobial infections should continue to focus on manipulating the structure of microbial communities to explore its effects on virulence118. Technologies available for creating and visualizing small bacterial aggregates will enable us to understand both mono-culture and co-culture interactions at relevant scales in the host. These studies would blend well with the consistent advancement in high-throughput genomic techniques, enabling transcriptomics and spatial organization of infections to be observed in parallel, opening many new avenues of research. Overall, there is a lot to be gained from studying the biogeography of infection, whether that is from a molecular, evolutionary, or clinical viewpoint. To stop the progression of — or eradicate — an infection, we must first understand how and why microorganisms assemble and persist within microbial communities. Therefore, studying the spatial organization of any community both in vivo and in natural environments will contribute to halting and eliminating infections.
Acknowledgments
The authors thank members of the Whiteley laboratory for critical discussion of this manuscript. This work was supported by a US National Institutes of Health (NIH) Grant 1R01DE020100 (to M.W.) and a Human Frontier Science Program (HFSP) grant HFSP RGP0011/2014 (to S.P.B. and M.W.). M.W. is a Burroughs Wellcome Investigator in the Pathogenesis of Infectious Disease program.
Glossary
- Biofilms
Surface-attached microbial communities that are encased in a matrix (for example, of polysaccharides, proteins, and/or DNA) and are often polymicrobial as well as highly resistant to antibiotic therapy and the host immune system.
- Polymicrobial
Diverse in species and/or strain content.
- Aggregate
A population of very few to several cells arranged in a cluster (otherwise known as a microcolony or in vivo biofilm) with many of the same properties that are observed in much larger, traditional biofilms studied in vitro.
- Synergy
A positive interaction term meaning ‘greater than the sum of parts’. Here we use synergy to refer to microbial interactions that shape host health (disease synergy: two or more species in combination cause more severe infection than either could when acting alone) or microbial growth (growth synergy: each species grows better in combination than when alone). Note that growth synergy often, but not always, implies disease synergy.
- Metagenomics
Sequence-based analysis of the DNA recovered from microbial communities. These studies describe the genetic repertoire of a microbial community.
- Metatranscriptomics
Sequence-based analysis of the mRNA recovered from microbial communities. These studies provide information regarding gene expression in a microbial community.
- Microbiogeography
Although ‘biogeography’. refers to the distribution of species through space and time, ‘microbiogeography,’. as defined in this Review, is the spatial organization of pathogen and commensal microbial populations at the scale of single infections.
- Pellicle
The permanent layer of proteins that coat oral surfaces and provide binding sites for early colonizers of dental plaque.
- Pioneer species
The first species to colonize previously disrupted ecosystems.
- Fimbriae
Hair-like bacterial appendages (also known as pili) that mediate surface attachment.
- Cystic fibrosis
A human genetic disorder in which a defect in a transmembrane ion channel causes the accumulation of mucus in the lungs, acting as a rich substrate for microbial growth.
- Quorum sensing
Density-dependent cell–cell communication where a constitutively produced signal, once it accumulates to a threshold concentration, can trigger microbial group behaviours.
- Type III secretion system
A needle-like bacterial apparatus that delivers effector proteins into host cells.
- Lactoferrin
A bactericidal host protein that sequesters iron.
- Mucin
A class of gel-forming proteins that give mucus its viscous property.
- Neutrophils
Host immune cells that unleash a mixture of redox-active molecules, in a process known as respiratory burst, to kill microorganisms.
- Aerotaxis
Chemotaxis in response to an oxygen gradient.
- Cheat
In social evolution, community members that exploit, but do not contribute to, the production of ‘public goods’.
- Respiration
A metabolic growth process characterized by the reduction of an electron acceptor (such as oxygen or nitrate).
- Intestinal crypts
The narrow spaces that lie between villi (multicellular host structures that assist in nutrient absorption) in the small and large intestine that, in a non-diseased state, are very low in microbial presence.
- RegIIIγ
A host antimicrobial lectin (polysaccharide-binding protein) that targets Gram-positive bacteria.
- Mucin 2
(MUC2). A primary mucin (gel-forming glycoprotein) in the mucus layers of the small and large intestine.
- Co-aggregation
Intercellular binding between genetically distinct cells, often mediated by the recognition of a polysaccharide on the target cell by a cognate surface protein on the partner cell.
- Mucoid
A phenotypic variant that overproduces exopolysaccharide (for example, alginate produced by Pseudomonas aeruginosa).
- Autoinducer 2
A signalling molecule that is synthesized and sensed by many bacterial species.
- Pyocyanin
A broad-spectrum toxin produced by Pseudomonas aeruginosa that is upregulated in response to cell-wall fragments shed by Staphylococcus aureus, underlying synergistic virulence of these species in wound infections.
- Cross-feeding
The consumption of a waste product of one microorganism by another microorganism that can utilize the waste product as a nutrient source.
- Cross-protection
The shielding of one microorganism by another microorganism from an external stress.
- Probiotics
Microorganisms that promote host health.
Footnotes
Competing interests statement
The authors declare no competing interests.
References
- 1.Hall-Stoodley L, Costerton JW, Stoodley P. Bacterial biofilms: from the natural environment to infectious diseases. Nat. Rev. Microbiol. 2004;2:95–108. doi: 10.1038/nrmicro821. [DOI] [PubMed] [Google Scholar]
- 2.Kaufmann SH, Schaible UE. 100th anniversary of Robert Koch’s Nobel Prize for the discovery of the tubercle bacillus. Trends Microbiol. 2005;13:469–475. doi: 10.1016/j.tim.2005.08.003. [DOI] [PubMed] [Google Scholar]
- 3.Smith H. The role of microbial interactions in infectious disease. Phil. Trans. R. Soc. Lond. B. Biol. Sci. 1982;297:551–561. doi: 10.1098/rstb.1982.0060. [DOI] [PubMed] [Google Scholar]
- 4.Hajishengallis G, et al. Low-abundance biofilm species orchestrates inflammatory periodontal disease through the commensal microbiota and complement. Cell Host Microbe. 2011;10:497–506. doi: 10.1016/j.chom.2011.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Korgaonkar A, Trivedi U, Rumbaugh KP, Whiteley M. Community surveillance enhances Pseudomonas aeruginosa virulence during polymicrobial infection. Proc. Natl Acad. Sci. USA. 2013;110:1059–1064. doi: 10.1073/pnas.1214550110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Simon-Soro A, Mira A. Solving the etiology of dental caries. Trends Microbiol. 2015;23:76–82. doi: 10.1016/j.tim.2014.10.010. [DOI] [PubMed] [Google Scholar]
- 7.Murray JL, Connell JL, Stacy A, Turner KH, Whiteley M. Mechanisms of synergy in polymicrobial infections. J. Microbiol. 2014;52:188–199. doi: 10.1007/s12275-014-4067-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sibley CD, et al. Discerning the complexity of community interactions using a Drosophila model of polymicrobial infections. PLoS Pathog. 2008;4:e1000184. doi: 10.1371/journal.ppat.1000184. This study defined the major categories of polymicrobial virulence, including additive and synergistic, by screening P. aeruginosa with 40 respiratory tract isolates in a Drosophila co-infection model.
- 9.Onderdonk AB, Bartlett JG, Louie T, Sullivan-Seigler N, Gorbach SL. Microbial synergy in experimental intra-abdominal abscess. Infect. Immun. 1976;13:22–26. doi: 10.1128/iai.13.1.22-26.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Trivedi U, et al. Prevalence of multiple antibiotic resistant infections in diabetic versus nondiabetic wounds. J. Pathog. 2014;2014:173053. doi: 10.1155/2014/173053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hogan DA, Kolter R. Pseudomonas–Candida interactions: an ecological role for virulence factors. Science. 2002;296:2229–2232. doi: 10.1126/science.1070784. [DOI] [PubMed] [Google Scholar]
- 12.Griffiths EC, Pedersen AB, Fenton A, Petchey OL. The nature and consequences of coinfection in humans. J. Infect. 2011;63:200–206. doi: 10.1016/j.jinf.2011.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Whitmore TC, editor. Wallace’s Line and Plate Tectonics. Oxford: Clarendon Press; Oxford University Press; 1981. [Google Scholar]
- 14.Vellend M. Conceptual synthesis in community ecology. Q. Rev. Biol. 2010;85:183–206. doi: 10.1086/652373. [DOI] [PubMed] [Google Scholar]
- 15.Nemergut DR, et al. Patterns and processes of microbial community assembly. Microbiol. Mol. Biol. Rev. 2013;77:342–356. doi: 10.1128/MMBR.00051-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hanson CA, Fuhrman JA, Horner-Devine MC, Martiny JB. Beyond biogeographic patterns: processes shaping the microbial landscape. Nat. Rev. Microbiol. 2012;10:497–506. doi: 10.1038/nrmicro2795. [DOI] [PubMed] [Google Scholar]
- 17.Costello EK, et al. Bacterial community variation in human body habitats across space and time. Science. 2009;326:1694–1697. doi: 10.1126/science.1177486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stearns JC, et al. Bacterial biogeography of the human digestive tract. Sci. Rep. 2011;1:170. doi: 10.1038/srep00170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oh J, et al. Biogeography and individuality shape function in the human skin metagenome. Nature. 2014;514:59–64. doi: 10.1038/nature13786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vos M, Wolf AB, Jennings SJ, Kowalchuk GA. Micro-scale determinants of bacterial diversity in soil. FEMS Microbiol. Rev. 2013;37:936–954. doi: 10.1111/1574-6976.12023. [DOI] [PubMed] [Google Scholar]
- 21.Kerr B, Riley MA, Feldman MW, Bohannan BJ. Local dispersal promotes biodiversity in a real-life game of rock-paper-scissors. Nature. 2002;418:171–174. doi: 10.1038/nature00823. [DOI] [PubMed] [Google Scholar]
- 22.Kim HJ, Boedicker JQ, Choi JW, Ismagilov RF. Defined spatial structure stabilizes a synthetic multispecies bacterial community. Proc. Natl Acad. Sci. USA. 2008;105:18188–18193. doi: 10.1073/pnas.0807935105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Drescher K, Nadell CD, Stone HA, Wingreen NS, Bassler BL. Solutions to the public goods dilemma in bacterial biofilms. Curr. Biol. 2014;24:50–55. doi: 10.1016/j.cub.2013.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou L, Slamti L, Nielsen-LeRoux C, Lereclus D, Raymond B. The social biology of quorum sensing in a naturalistic host pathogen system. Curr. Biol. 2014;24:2417–2422. doi: 10.1016/j.cub.2014.08.049. [DOI] [PubMed] [Google Scholar]
- 25.Harding JL, Reynolds MM. Combating medical device fouling. Trends Biotechnol. 2014;32:140–146. doi: 10.1016/j.tibtech.2013.12.004. [DOI] [PubMed] [Google Scholar]
- 26.Palmer RJ, Jr, Gordon SM, Cisar JO, Kolenbrander PE. Coaggregation-mediated interactions of streptococci and actinomyces detected in initial human dental plaque. J. Bacteriol. 2003;185:3400–3409. doi: 10.1128/JB.185.11.3400-3409.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gunning AP, et al. Mining the “glycocode” —exploring the spatial distribution of glycans in gastrointestinal mucin using force spectroscopy. FASEB J. 2013;27:2342–2354. doi: 10.1096/fj.12-221416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Melican K, et al. Uropathogenic Escherichia coli P and type 1 fimbriae act in synergy in a living host to facilitate renal colonization leading to nephron obstruction. PLoS Pathog. 2011;7:e1001298. doi: 10.1371/journal.ppat.1001298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bjarnsholt T, et al. The in vivo biofilm. Trends Microbiol. 2013;21:466–474. doi: 10.1016/j.tim.2013.06.002. [DOI] [PubMed] [Google Scholar]
- 30.Schaber JA, et al. Pseudomonas aeruginosa forms biofilms in acute infection independent of cell-to-cell signaling. Infect. Immun. 2007;75:3715–3721. doi: 10.1128/IAI.00586-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dalton T, et al. An in vivo polymicrobial biofilm wound infection model to study interspecies interactions. PLoS ONE. 2011;6:e27317. doi: 10.1371/journal.pone.0027317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stacy A, et al. Bacterial fight-and-flight responses enhance virulence in a polymicrobial infection. Proc. Natl Acad. Sci. USA. 2014;111:7819–7824. doi: 10.1073/pnas.1400586111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Singh PK, et al. Quorum-sensing signals indicate that cystic fibrosis lungs are infected with bacterial biofilms. Nature. 2000;407:762–764. doi: 10.1038/35037627. [DOI] [PubMed] [Google Scholar]
- 34.Nguyen CT, et al. Noninvasive in vivo optical detection of biofilm in the human middle ear. Proc. Natl Acad. Sci. USA. 2012;109:9529–9534. doi: 10.1073/pnas.1201592109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Monds RD, O’Toole GA. The developmental model of microbial biofilms: ten years of a paradigm up for review. Trends Microbiol. 2009;17:73–87. doi: 10.1016/j.tim.2008.11.001. [DOI] [PubMed] [Google Scholar]
- 36. Tran CS, et al. Translocon is required for biofilm formation at the epithelial barrier. PLoS Pathog. 2014;10:e1004479. doi: 10.1371/journal.ppat.1004479. This study identified a determinant for P. aeruginosa biofilm formation in vivo that is dispensable in vitro, illustrating that microorganisms should be studied in natural and infectious contexts to gain better insight into the mechanisms of spatial organization.
- 37.Travier L, et al. ActA promotes Listeria monocytogenes aggregation, intestinal colonization and carriage. PLoS Pathog. 2013;9:e1003131. doi: 10.1371/journal.ppat.1003131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Blanchette-Cain K, et al. Streptococcus pneumoniae biofilm formation is strain dependent, multifactorial, and associated with reduced invasiveness and immunoreactivity during colonization. mBio. 2013;4:e00745–e00713. doi: 10.1128/mBio.00745-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Davies DG, et al. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science. 1998;280:295–298. doi: 10.1126/science.280.5361.295. [DOI] [PubMed] [Google Scholar]
- 40.Millet YA, et al. Insights into Vibrio cholerae intestinal colonization from monitoring fluorescently labeled bacteria. PLoS Pathog. 2014;10:e1004405. doi: 10.1371/journal.ppat.1004405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nadell CD, Foster KR, Xavier JB. Emergence of spatial structure in cell groups and the evolution of cooperation. PLoS Comput. Biol. 2010;6:e1000716. doi: 10.1371/journal.pcbi.1000716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jemielita M, et al. Spatial and temporal features of the growth of a bacterial species colonizing the zebrafish gut. mBio. 2014;5:e01751–e01714. doi: 10.1128/mBio.01751-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Monier JM, Lindow SE. Differential survival of solitary and aggregated bacterial cells promotes aggregate formation on leaf surfaces. Proc. Natl Acad. Sci. USA. 2003;100:15977–15982. doi: 10.1073/pnas.2436560100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Connell JL, et al. Probing prokaryotic social behaviors with bacterial “lobster traps”. mBio. 2010;1:e00202–e00210. doi: 10.1128/mBio.00202-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guggenberger C, Wolz C, Morrissey JA, Heesemann J. Tw o distinct coagulase-dependent barriers protect Staphylococcus aureus from neutrophils in a three dimensional in vitro infection model. PLoS Pathog. 2012;8:e1002434. doi: 10.1371/journal.ppat.1002434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kragh KN, et al. Polymorphonuclear leukocytes restrict growth of Pseudomonas aeruginosa in the lungs of cystic fibrosis patients. Infect. Immun. 2014;82:4477–4486. doi: 10.1128/IAI.01969-14. This study illustrated how microbiogeography (the local suppression of microbial growth by neutrophils) can determine macrobiogeography (in which aggregates of P. aeruginosa preferentially colonize the respiratory tract).
- 47. Davis KM, Mohammadi S, Isberg RR. Community behavior and spatial regulation within a bacterial microcolony in deep tissue sites serves to protect against host attack. Cell Host Microbe. 2015;17:21–31. doi: 10.1016/j.chom.2014.11.008. This study examined how microcolonies of Yersinia pseudotuberculosis in the spleen repel attacking neutrophils, illustrating how bacterial aggregates can adeptly resist the immune system.
- 48.Faruque SM, et al. Transmissibility of cholera: in vivo-formed biofilms and their relationship to infectivity and persistence in the environment. Proc. Natl Acad. Sci. USA. 2006;103:6350–6355. doi: 10.1073/pnas.0601277103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Singh PK, Parsek MR, Greenberg EP, Welsh MJ. A component of innate immunity prevents bacterial biofilm development. Nature. 2002;417:552–555. doi: 10.1038/417552a. [DOI] [PubMed] [Google Scholar]
- 50.Caldara M, et al. Mucin biopolymers prevent bacterial aggregation by retaining cells in the free-swimming state. Curr. Biol. 2012;22:2325–2330. doi: 10.1016/j.cub.2012.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Alhede M, et al. Pseudomonas aeruginosa recognizes and responds aggressively to the presence of polymorphonuclear leukocytes. Microbiology. 2009;155:3500–3508. doi: 10.1099/mic.0.031443-0. [DOI] [PubMed] [Google Scholar]
- 52.Schreiber S, et al. The spatial orientation of Helicobacter pylori in the gastric mucus. Proc. Natl Acad. Sci. USA. 2004;101:5024–5029. doi: 10.1073/pnas.0308386101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Falush D, et al. Recombination and mutation during long-term gastric colonization by Helicobacter pylori: estimates of clock rates, recombination size, and minimal age. Proc. Natl Acad. Sci. USA. 2001;98:15056–15061. doi: 10.1073/pnas.251396098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yasuda K, et al. Biogeography of the intestinal mucosal and lumenal microbiome in the rhesus macaque. Cell Host Microbe. 2015;17:385–391. doi: 10.1016/j.chom.2015.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Marteyn B, et al. Modulation of Shigella virulence in response to available oxygen in vivo . Nature. 2010;465:355–358. doi: 10.1038/nature08970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wessel AK, et al. Oxygen limitation within a bacterial aggregate. mBio. 2014;5:e00992. doi: 10.1128/mBio.00992-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bradshaw DJ, Marsh PD, Watson GK, Allison C. Role of Fusobacterium nucleatum and coaggregation in anaerobe survival in planktonic and biofilm oral microbial communities during aeration. Infect. Immun. 1998;66:4729–4732. doi: 10.1128/iai.66.10.4729-4732.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bowler PG, Duerden BI, Armstrong DG. Wound microbiology and associated approaches to wound management. Clin. Microbiol. Rev. 2001;14:244–269. doi: 10.1128/CMR.14.2.244-269.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Turner KH, Everett J, Trivedi U, Rumbaugh KP, Whiteley M. Requirements for Pseudomonas aeruginosa acute burn and chronic surgical wound infection. PLoS Genet. 2014;10:e1004518. doi: 10.1371/journal.pgen.1004518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Berry D, et al. Host-compound foraging by intestinal microbiota revealed by single-cell stable isotope probing. Proc. Natl Acad. Sci. USA. 2013;110:4720–4725. doi: 10.1073/pnas.1219247110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schluter J, Foster KR. The evolution of mutualism in gut microbiota via host epithelial selection. PLoS Biol. 2012;10:e1001424. doi: 10.1371/journal.pbio.1001424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nielsen AT, et al. A bistable switch and anatomical site control Vibrio cholerae virulence gene expression in the intestine. PLoS Pathog. 2010;6:e1001102. doi: 10.1371/journal.ppat.1001102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kamada N, et al. Regulated virulence controls the ability of a pathogen to compete with the gut microbiota. Science. 2012;336:1325–1329. doi: 10.1126/science.1222195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rivera-Chavez F, et al. Salmonella uses energy taxis to benefit from intestinal inflammation. PLoS Pathog. 2013;9:e1003267. doi: 10.1371/journal.ppat.1003267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Thiennimitr P, et al. Intestinal inflammation allows Salmonella to use ethanolamine to compete with the microbiota. Proc. Natl Acad. Sci. USA. 2011;108:17480–17485. doi: 10.1073/pnas.1107857108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Winter SE, et al. Gut inflammation provides a respiratory electron acceptor for Salmonella . Nature. 2010;467:426–429. doi: 10.1038/nature09415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Leveau JH, Lindow SE. Appetite of an epiphyte: quantitative monitoring of bacterial sugar consumption in the phyllosphere. Proc. Natl Acad. Sci. USA. 2001;98:3446–3453. doi: 10.1073/pnas.061629598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Lee SM, et al. Bacterial colonization factors control specificity and stability of the gut microbiota. Nature. 2013;501:426–429. doi: 10.1038/nature12447. This study showed how microbiogeography (targeting of B. fragilis to the intestinal crypts) can contribute to stable colonization of the host.
- 69.Vaishnava S, et al. The antibacterial lectin RegIIIγ promotes the spatial segregation of microbiota and host in the intestine. Science. 2011;334:255–258. doi: 10.1126/science.1209791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Johansson ME, et al. The inner of the two Muc2 mucin-dependent mucus layers in colon is devoid of bacteria. Proc. Natl Acad. Sci. USA. 2008;105:15064–15069. doi: 10.1073/pnas.0803124105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Johansson ME, Hansson GC. Microbiology. keeping bacteria at a distance. Science. 2011;334:182–183. doi: 10.1126/science.1213909. [DOI] [PubMed] [Google Scholar]
- 72.Lidell ME, Moncada DM, Chadee K, Hansson GC. Entamoeba histolytica cysteine proteases cleave the MUC2 mucin in its C-terminal domain and dissolve the protective colonic mucus gel. Proc. Natl Acad. Sci. USA. 2006;103:9298–9303. doi: 10.1073/pnas.0600623103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chau TA, et al. Toll-like receptor 2 ligands on the staphylococcal cell wall downregulate superantigen-induced T cell activation and prevent toxic shock syndrome. Nat. Med. 2009;15:641–648. doi: 10.1038/nm.1965. [DOI] [PubMed] [Google Scholar]
- 74.Round JL, et al. The Toll-Like receptor 2 pathway establishes colonization by a commensal of the human microbiota. Science. 2011;332:974–977. doi: 10.1126/science.1206095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bergstrom KS, et al. Muc2 protects against lethal infectious colitis by disassociating pathogenic and commensal bacteria from the colonic mucosa. PLoS Pathog. 2010;6:e1000902. doi: 10.1371/journal.ppat.1000902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Velcich A, et al. Colorectal cancer in mice genetically deficient in the mucin Muc2. Science. 2002;295:1726–1729. doi: 10.1126/science.1069094. [DOI] [PubMed] [Google Scholar]
- 77.Dejea CM, et al. Microbiota organization is a distinct feature of proximal colorectal cancers. Proc. Natl Acad. Sci. USA. 2014;111:18321–18326. doi: 10.1073/pnas.1406199111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Momeni B, Brileya KA, Fields MW, Shou W. Strong inter-population cooperation leads to partner intermixing in microbial communities. eLIFE. 2013;2:e00230. doi: 10.7554/eLife.00230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chalmers NI, Palmer RJ, Jr, Cisar JO, Kolenbrander PE. Characterization of a Streptococcus sp.-Veillonella sp. community micromanipulated from dental plaque. J. Bacteriol. 2008;190:8145–8154. doi: 10.1128/JB.00983-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Estrela S, Brown SP. Metabolic and demographic feedbacks shape the emergent spatial structure and function of microbial communities. PLoS Comput. Biol. 2013;9:e1003398. doi: 10.1371/journal.pcbi.1003398. This study used individual-based modelling to show that the strength of metabolic dependency determines the level of mixing in microbial communities.
- 81.Zijnge V, et al. Oral biofilm architecture on natural teeth. PLoS ONE. 2010;5:e9321. doi: 10.1371/journal.pone.0009321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kolenbrander PE, Palmer RJ, Jr, Periasamy S, Jakubovics NS. Oral multispecies biofilm development and the key role of cell-cell distance. Nat. Rev. Microbiol. 2010;8:471–480. doi: 10.1038/nrmicro2381. [DOI] [PubMed] [Google Scholar]
- 83. Valm AM, et al. Systems-level analysis of microbial community organization through combinatorial labeling and spectral imaging. Proc. Natl Acad. Sci. USA. 2011;108:4152–4157. doi: 10.1073/pnas.1101134108. This study presents a ‘nearest neighbour’ network, in which frequencies of intercellular associations are described, for 15 taxonomic groups in human dental plaque.
- 84.Egland PG, Palmer RJ, Jr, Kolenbrander PE. Interspecies communication in Streptococcus gordonii-Veillonella atypica biofilms: signaling in flow conditions requires juxtaposition. Proc. Natl Acad. Sci. USA. 2004;101:16917–16922. doi: 10.1073/pnas.0407457101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Jakubovics NS, Gill SR, Iobst SE, Vickerman MM, Kolenbrander PE. Regulation of gene expression in a mixed-genus community: stabilized arginine biosynthesis in Streptococcus gordonii by coaggregation with Actinomyces naeslundii . J. Bacteriol. 2008;190:3646–3657. doi: 10.1128/JB.00088-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.He X, et al. Cultivation of a human-associated TM7 phylotype reveals a reduced genome and epibiotic parasitic lifestyle. Proc. Natl Acad. Sci. USA. 2015;112:244–249. doi: 10.1073/pnas.1419038112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Kim W, Racimo F, Schluter J, Levy SB, Foster KR. Importance of positioning for microbial evolution. Proc. Natl Acad. Sci. USA. 2014;111:E1639–E1647. doi: 10.1073/pnas.1323632111. This study showed how inter-strain competition can select for novel positioning strategies in biofilms, such as hyper-secretion to push above neighbours.
- 88.Markussen T, et al. Environmental heterogeneity drives within-host diversification and evolution of Pseudomonas aeruginosa . mBio. 2014;5:e01592–e01514. doi: 10.1128/mBio.01592-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yang L, et al. Pattern differentiation in co-culture biofilms formed by Staphylococcus aureus and Pseudomonas aeruginosa . FEMS Immunol. Med. Microbiol. 2011;62:339–347. doi: 10.1111/j.1574-695X.2011.00820.x. [DOI] [PubMed] [Google Scholar]
- 90.Chew SC, et al. Dynamic remodeling of microbial biofilms by functionally distinct exopolysaccharides. mBio. 2014;5:e01536–e01514. doi: 10.1128/mBio.01536-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Huse HK, et al. Pseudomonas aeruginosa enhances production of a non-alginate exopolysaccharide during long-term colonization of the cystic fibrosis lung. PLoS ONE. 2013;8:e82621. doi: 10.1371/journal.pone.0082621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Eberl L, Tummler B. Pseudomonas aeruginosa and Burkholderia cepacia in cystic fibrosis: genome evolution, interactions and adaptation. Int. J. Med. Microbiol. 2004;294:123–131. doi: 10.1016/j.ijmm.2004.06.022. [DOI] [PubMed] [Google Scholar]
- 93.Bragonzi A, et al. Modelling co-infection of the cystic fibrosis lung by Pseudomonas aeruginosa and Burkholderia cenocepacia reveals influences on biofilm formation and host response. PLoS ONE. 2012;7:e52330. doi: 10.1371/journal.pone.0052330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ryan RP, et al. Interspecies signalling via the Stenotrophomonas maltophilia diffusible signal factor influences biofilm formation and polymyxin tolerance in Pseudomonas aeruginosa . Mol. Microbiol. 2008;68:75–86. doi: 10.1111/j.1365-2958.2008.06132.x. [DOI] [PubMed] [Google Scholar]
- 95.Twomey KB, et al. Bacterial cis-2-unsaturated fatty acids found in the cystic fibrosis airway modulate virulence and persistence of Pseudomonas aeruginosa . ISME J. 2012;6:939–950. doi: 10.1038/ismej.2011.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Armbruster CE, et al. Indirect pathogenicity of Haemophilus influenzae and Moraxella catarrhalis in polymicrobial otitis media occurs via interspecies quorum signaling. mBio. 2014;1:e00102–e00110. doi: 10.1128/mBio.00102-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Park S, et al. Motion to form a quorum. Science. 2003;301:188. doi: 10.1126/science.1079805. [DOI] [PubMed] [Google Scholar]
- 98.Davies DG, Marques CN. A fatty acid messenger is responsible for inducing dispersion in microbial biofilms. J. Bacteriol. 2009;191:1393–1403. doi: 10.1128/JB.01214-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Vega NM, Allison KR, Samuels AN, Klempner MS, Collins JJ. Salmonella typhimurium intercepts Escherichia coli signaling to enhance antibiotic tolerance. Proc. Natl Acad. Sci. USA. 2013;110:14420–14425. doi: 10.1073/pnas.1308085110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Connell JL, Kim J, Shear JB, Bard AJ, Whiteley M. Real-time monitoring of quorum sensing in 3D–printed bacterial aggregates using scanning electrochemical microscopy. Proc. Natl Acad. Sci. USA. 2014;111:18255–18260. doi: 10.1073/pnas.1421211111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Liu X, et al. Real-time mapping of a hydrogen peroxide concentration profile across a polymicrobial bacterial biofilm using scanning electrochemical microscopy. Proc. Natl Acad. Sci. USA. 2011;108:2668–2673. doi: 10.1073/pnas.1018391108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Xiao J, et al. The exopolysaccharide matrix modulates the interaction between 3D architecture and virulence of a mixed-species oral biofilm. PLoS Pathog. 2012;8:e1002623. doi: 10.1371/journal.ppat.1002623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.von Ohle C, et al. Real-time microsensor measurement of local metabolic activities in ex vivo dental biofilms exposed to sucrose and treated with chlorhexidine. Appl. Environ. Microbiol. 2010;76:2326–2334. doi: 10.1128/AEM.02090-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Fazli M, et al. Nonrandom distribution of Pseudomonas aeruginosa and Staphylococcus aureus in chronic wounds. J. Clin. Microbiol. 2009;47:4084–4089. doi: 10.1128/JCM.01395-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hutchison JB, et al. Single-cell control of initial spatial structure in biofilm development using laser trapping. Langmuir. 2014;30:4522–4530. doi: 10.1021/la500128y. [DOI] [PubMed] [Google Scholar]
- 106.DeLeon S, et al. Synergistic interactions of Pseudomonas aeruginosa and Staphylococcus aureus in an in vitro wound model. Infect. Immun. 2014;82:4718–4728. doi: 10.1128/IAI.02198-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kirkup BC, Riley MA. Antibiotic-mediated antagonism leads to a bacterial game of rock-paper-scissors in vivo . Nature. 2004;428:412–414. doi: 10.1038/nature02429. [DOI] [PubMed] [Google Scholar]
- 108.Cowan SE, Gilbert E, Liepmann D, Keasling JD. Commensal interactions in a dual-species biofilm exposed to mixed organic compounds. Appl. Environ. Microbiol. 2000;66:4481–4485. doi: 10.1128/aem.66.10.4481-4485.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Nielsen AT, Tolker-Nielsen T, Barken KB, Molin S. Role of commensal relationships on the spatial structure of a surface-attached microbial consortium. Environ. Microbiol. 2000;2:59–68. doi: 10.1046/j.1462-2920.2000.00084.x. [DOI] [PubMed] [Google Scholar]
- 110.Leriche V, Briandet R, Carpentier B. Ecology of mixed biofilms subjected daily to a chlorinated alkaline solution: spatial distribution of bacterial species suggests a protective effect of one species to another. Environ. Microbiol. 2003;5:64–71. doi: 10.1046/j.1462-2920.2003.00394.x. [DOI] [PubMed] [Google Scholar]
- 111. Connell JL, Ritschdorff ET, Whiteley M, Shear JB. 3D printing of microscopic bacterial communities. Proc. Natl Acad. Sci. USA. 2013;110:18380–18385. doi: 10.1073/pnas.1309729110. This study illustrated how the spatial organization of multispecies communities can determine important virulence properties, such as antibiotic resistance.
- 112.Schillinger C, et al. Co-localized or randomly distributed? Pair cross correlation of in vivo grown subgingival biofilm bacteria quantified by digital image analysis. PLoS ONE. 2012;7:e37583. doi: 10.1371/journal.pone.0037583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Settem RP, El-Hassan AT, Honma K, Stafford GP, Sharma A. Fusobacterium nucleatum and Tannerella forsythia induce synergistic alveolar bone loss in a mouse periodontitis model. Infect. Immun. 2012;80:2436–2443. doi: 10.1128/IAI.06276-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Iwase T, et al. Staphylococcus epidermidis Espinhibits Staphylococcus aureus biofilm formation and nasal colonization. Nature. 2010;465:346–349. doi: 10.1038/nature09074. [DOI] [PubMed] [Google Scholar]
- 115.Fukuda S, et al. Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nature. 2011;469:543–547. doi: 10.1038/nature09646. [DOI] [PubMed] [Google Scholar]
- 116.Guo L, et al. Precision-guided antimicrobial peptide as a targeted modulator of human microbial ecology. Proc. Natl Acad. Sci. USA. 2015;112:7569–7574. doi: 10.1073/pnas.1506207112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Langermann S, et al. Prevention of mucosal Escherichia coli infection by FimH-adhesin-based systemic vaccination. Science. 1997;276:607–611. doi: 10.1126/science.276.5312.607. [DOI] [PubMed] [Google Scholar]
- 118.Wessel AK, Hmelo L, Parsek MR, Whiteley M. Going local: technologies for exploring bacterial microenvironments. Nat. Rev. Microbiol. 2013;11:337–348. doi: 10.1038/nrmicro3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Leung MH, Wilkins D, Lee PK. Insights into the pan-microbiome: skin microbial communities of Chinese individuals differ from other racial groups. Sci. Rep. 2015;5:11845. doi: 10.1038/srep11845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Gould JC, Mc KE. The carriage of Staphylococcus pyogenes var. aureus in the human nose. J. Hyg. (Lond.) 1954;52:304–310. doi: 10.1017/s0022172400027509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Krismer B, et al. Nutrient limitation governs Staphylococcus aureus metabolism and niche adaptation in the human nose. PLoS Pathog. 2014;10:e1003862. doi: 10.1371/journal.ppat.1003862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Jeraldo P, et al. Quantification of the relative roles of niche and neutral processes in structuring gastrointestinal microbiomes. Proc. Natl Acad. Sci. USA. 2012;109:9692–9698. doi: 10.1073/pnas.1206721109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Rosindell J, Hubbell SP, Etienne RS. The unified neutral theory of biodiversity and biogeography at age ten. Trends Ecol. Evol. 2011;26:340–348. doi: 10.1016/j.tree.2011.03.024. [DOI] [PubMed] [Google Scholar]
- 124.Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Grice EA, et al. Topographical and temporal diversity of the human skin microbiome. Science. 2009;324:1190–1192. doi: 10.1126/science.1171700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Dominguez-Bello MG, et al. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc. Natl Acad. Sci. USA. 2010;107:11971–11975. doi: 10.1073/pnas.1002601107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Koch G, et al. Evolution of resistance to a last-resort antibiotic in Staphylococcus aureus via bacterial competition. Cell. 2014;158:1060–1071. doi: 10.1016/j.cell.2014.06.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Planet PJ, et al. Emergence of the epidemic methicillin-resistant Staphylococcus aureus strain USA300 coincides with horizontal transfer of the arginine catabolic mobile element and speG-mediated adaptations for survival on skin. mBio. 2013;4:e00889–e00813. doi: 10.1128/mBio.00889-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.de Wit R, Bouvier T. ‘Everything is everywhere, but, the environment selects’; what did Baas Becking and Beijerinck really say? Environ. Microbiol. 2006;8:755–758. doi: 10.1111/j.1462-2920.2006.01017.x. [DOI] [PubMed] [Google Scholar]
- 130.Finlay BJ. Global dispersal of free-living microbial eukaryote species. Science. 2002;296:1061–1063. doi: 10.1126/science.1070710. [DOI] [PubMed] [Google Scholar]
- 131.Whitaker RJ, Grogan DW, Taylor JW. Geographic barriers isolate endemic populations of hyperthermophilic archaea. Science. 2003;301:976–978. doi: 10.1126/science.1086909. [DOI] [PubMed] [Google Scholar]
- 132.Martiny JB, et al. Microbial biogeography: putting microorganisms on the map. Nat. Rev. Microbiol. 2006;4:102–112. doi: 10.1038/nrmicro1341. [DOI] [PubMed] [Google Scholar]
- 133.Dowd SE, et al. Survey of bacterial diversity in chronic wounds using pyrosequencing, DGGE, and full ribosome shotgun sequencing. BMC Microbiol. 2008;8:43. doi: 10.1186/1471-2180-8-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Griffin AS, West SA, Buckling A. Cooperation and competition in pathogenic bacteria. Nature. 2004;430:1024–1027. doi: 10.1038/nature02744. [DOI] [PubMed] [Google Scholar]
- 135.Diggle SP, Griffin AS, Campbell GS, West SA. Cooperation and conflict in quorum-sensing bacterial populations. Nature. 2007;450:411–414. doi: 10.1038/nature06279. [DOI] [PubMed] [Google Scholar]
- 136.Kummerli R, Gardner A, West SA, Griffin AS. Limited dispersal, budding dispersal, and cooperation: an experimental study. Evolution. 2009;63:939–949. doi: 10.1111/j.1558-5646.2008.00548.x. [DOI] [PubMed] [Google Scholar]
- 137.Dimitriu T, et al. Genetic information transfer promotes cooperation in bacteria. Proc. Natl Acad. Sci. USA. 2014;111:11103–11108. doi: 10.1073/pnas.1406840111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Schluter J, Nadell CD, Bassler BL, Foster KR. Adhesion as a weapon in microbial competition. ISME J. 2015;9:139–149. doi: 10.1038/ismej.2014.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Wang M, Schaefer AL, Dandekar AA, Greenberg EP. Quorum sensing and policing of Pseudomonas aeruginosa social cheaters. Proc. Natl Acad. Sci. USA. 2015;112:2187–2191. doi: 10.1073/pnas.1500704112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Rumbaugh KP, et al. Quorum sensing and the social evolution of bacterial virulence. Curr. Biol. 2009;19:341–345. doi: 10.1016/j.cub.2009.01.050. [DOI] [PubMed] [Google Scholar]
- 141.Arrhenius O. Species and area. J. Ecol. 1921;9:95–99. [Google Scholar]
- 142.Gleason H. On the relation between species and area. Ecology. 1922;3:158–162. [Google Scholar]
- 143.Cornell HLJH. Species interactions, local and regional processes, and limits to the richness of ecological communities: a theoretical perspective. J. Anim. Ecol. 1992;61:1–12. [Google Scholar]
- 144.Lawton J. Are there general laws in ecology? Oikos. 1999;84:177–192. [Google Scholar]
- 145.Hillebrand HBT. Regional and local impact on species diversity - from pattern to processes. Oecologia. 2002;132:479–491. doi: 10.1007/s00442-002-0988-3. [DOI] [PubMed] [Google Scholar]
- 146.Horner-Devine MC, Lage M, Hughes JB, Bohannan BJ. A taxa-area relationship for bacteria. Nature. 2004;432:750–753. doi: 10.1038/nature03073. [DOI] [PubMed] [Google Scholar]
- 147.Bell T, et al. Larger islands house more bacterial taxa. Science. 2005;308:1884. doi: 10.1126/science.1111318. [DOI] [PubMed] [Google Scholar]
- 148.Ranjard L, et al. Turnover of soil bacterial diversity driven by wide-scale environmental heterogeneity. Nat. Commun. 2013;4:1434. doi: 10.1038/ncomms2431. [DOI] [PubMed] [Google Scholar]