SUMMARY
Bacillus subtilis is one of the best-studied organisms. Due to the broad knowledge and annotation and the well-developed genetic system, this bacterium is an excellent starting point for genome minimization with the aim of constructing a minimal cell. We have analyzed the genome of B. subtilis and selected all genes that are required to allow life in complex medium at 37°C. This selection is based on the known information on essential genes and functions as well as on gene and protein expression data and gene conservation. The list presented here includes 523 and 119 genes coding for proteins and RNAs, respectively. These proteins and RNAs are required for the basic functions of life in information processing (replication and chromosome maintenance, transcription, translation, protein folding, and secretion), metabolism, cell division, and the integrity of the minimal cell. The completeness of the selected metabolic pathways, reactions, and enzymes was verified by the development of a model of metabolism of the minimal cell. A comparison of the MiniBacillus genome to the recently reported designed minimal genome of Mycoplasma mycoides JCVI-syn3.0 indicates excellent agreement in the information-processing pathways, whereas each species has a metabolism that reflects specific evolution and adaptation. The blueprint of MiniBacillus presented here serves as the starting point for a successive reduction of the B. subtilis genome.
INTRODUCTION
Three technological revolutions dramatically changed our view of biology. The genomic revolution gives us access to genome sequences of any organism of interest at low cost. The analytical revolution, especially with respect to mass spectrometry, allows us not only to detect the presence and the fluxes of any molecule in the cell but also to study its precise concentration under any desired condition. Last but not least, the informatics revolution paves the way for the evaluation of the tremendous data sets and for the generation of meaningful models and predictions of cellular behavior. However, even knowledge of all components of a cell and of their precise concentrations does not give us a complete understanding of a living cell. For this, we have to consider all the functional interactions between different biological molecules and the dynamics of both the molecules and their interactions.
The complexity of all naturally existing organisms still precludes a deep understanding of the functions of all components of a cell and their interactions. Even small organisms such as bacteria are too complex to understand all processes in their cells. This is even the case for bacteria with naturally minimal genomes, as found in the genus Mycoplasma. These bacteria may have as few as 482 protein-coding genes and are still capable of independent life in the absence of any host cells. However, the functions of many Mycoplasma genes are so far unknown, and not the full gene set is essential, indicating that we are far from a full picture of these bacteria despite tremendous efforts in their analysis (1–8).
These limitations in our understanding of natural organisms call for a reduction of complexity: the creation of cells with a defined set of genes. Such cells can be obtained by applying either bottom-up or top-down approaches. The former approach has so far been pursued with the chemical synthesis of a bacterial genome and its application to create a semiartificial cell (9, 10). In this case, a known genome was reduced and transplanted into a closely related host cell. With only 473 protein-coding genes, the recently achieved Mycoplasma mycoides JCVI-syn3.0 minigenome is so far the smallest semiartificially designed organism. Importantly, about one-third of the proteins in this minimal cell are of unknown function (10). Moreover, there have been attempts to create so-called protocells, which are lipid vesicles that contain genetic material and/or enzymes (11–14). Even though protocells do not allow recapitulation of the evolutionary emergence of life, they are well-suited systems to study the physical and biochemical properties of basic cellular processes such as self-reproduction, permeability, enzymatic replication, and Darwinian evolution (15–17). Reduction of complexity can also be achieved by a top-down approach that starts with existing bacteria and aims at consecutively reducing their complexity. This approach is, of course, very time-consuming; on the other hand, it allows advancing from step to step. Moreover, this iterative process of genome reduction allows the immediate discovery of possible problems and, thus, finding appropriate solutions. Genome reduction is a common theme in synthetic biology, not only for pure scientific curiosity but also from an industrial point of view to create workhorses for biotechnology. Ongoing projects of genome reduction have been reported for several intensively studied bacteria such as Bacillus subtilis, Corynebacterium glutamicum, Escherichia coli, Pseudomonas putida, and Streptomyces avermitilis (18–27; for a review, see reference 28) as well as for yeast (29). All these projects are still far from the final goal, the minimal cell.
With the progress of genome reduction, it is necessary to define the set of genes that should be part of the final minimal genome. It is obvious that such a set of genes is determined by several factors, including the intended lifestyle of the final minimal cell, but also by the general biology of the organism that is to be reduced. Conceivably, a eukaryotic yeast cell will still contain a nucleus even at a late genome reduction stage. Similarly, the bacteria mentioned above differ strongly in their cellular organizations. For example, M. mycoides does not possess a cell wall, while the cell wall is differently structured yet essential in B. subtilis, C. glutamicum, and E. coli. In this work, we aim at defining the set of genes that is required for the life of a minimal cell based on B. subtilis. For several reasons, this bacterium is particularly well suited for genome minimization approaches. First, B. subtilis is one of the most intensively studied organisms, with extensive genome annotation and excellent knowledge of the major cellular processes. Second, the elaborated genetic system for B. subtilis makes all kinds of genetic manipulations very easy (see below). Finally, B. subtilis is one of the major organisms in biotechnology, suggesting that genome-reduced strains may also serve as a chassis for novel applications.
CONSIDERATIONS FOR THE DEFINITION OF THE GENE SET FOR A MINIMAL CELL
Several independent sets of information serve as the basis to define which genes are required for a viable minimal cell. First of all, this is the set of essential genes. These genes were identified for B. subtilis in 2003 (30). In addition, large dispensable regions of the chromosome have been studied, resulting in the identification of novel essential and coessential genes (31). A recent reevaluation of the essential genes of B. subtilis revealed that several metabolic genes involved in glycolysis and the tricarboxylic acid cycle originally listed as essential could be removed from the list. With the exception of the ylaN gene, all other genes of unknown function could also be removed from the list (32, 33). Moreover, recent studies indicated that the ycgG and yfkN genes as well as the rny gene, encoding RNase Y, are also dispensable (28, 34; our unpublished data). The current list of 251 essential protein-coding genes is available in the SubtiWiki database (http://subtiwiki.uni-goettingen.de/wiki/index.php/Essential_genes) (33).
The essential genes are by definition only those genes that cannot be deleted as single genes under defined optimal growth conditions (for B. subtilis, lysogeny broth [LB] with glucose at 37°C). Moreover, a recent knockdown study of essential B. subtilis genes showed that the encoded proteins are also very important for outgrowth from stationary phase, adding another level of relevance (35). However, many genes are redundant, and cellular functions can be achieved in completely different ways. The former is the case for DNA polymerase I (PolA) and its paralog YpcP or the diadenylate cyclase CdaA and one of the paralogs DisA and CdaS (36, 37). Moreover, the same function may even be fulfilled by unrelated proteins, as observed for the membrane anchors for the Z-ring protein for cell division, FtsZ. In E. coli, the essential FtsA protein serves as a membrane anchor for FtsZ. Why FtsA is nonessential in B. subtilis has been enigmatic for a long time. Only the discovery of the unrelated alternative membrane anchor SepF provided the answer (38). Finally, alternative pathways may lead to the same results. This is obvious in the acquisition of cellular building blocks such as amino acids and nucleotides. These metabolites can be either synthesized in the cell or taken up from the medium. In any case, none of the involved genes would be classified as being essential. In all these cases, a decision has to be made regarding which of the possible alternatives will be included in the minimal gene set. Accordingly, the gene complement of a minimal organism has to be designed according to essential functions.
If B. subtilis possesses multiple genes for the same function, one of them has to be selected. The criteria for the selection applied in this study are as follows. (i) The final number of genes should be as small as possible. Therefore, it seems reasonable to include transporters rather than biosynthetic genes for the acquisition of building blocks whenever possible. (ii) In some functional categories, such as cell division, the deletion of a gene may have only a mild effect; however, combination with the deletion of a second, sometimes functionally unrelated gene may be lethal (see “Cell Division,” below, for details). Therefore, such synthetic lethalities have to be considered. (iii) Prior decisions will have an impact on later selections. This is the case, for example, for cell wall-biosynthetic proteins (see below). (iv) Both gene expression and protein levels have been extensively studied in B. subtilis, and all these data are accessible in the SubtiWiki database (33, 39–41). In case of doubt, the most strongly expressed protein has been chosen. (v) Finally, conservation of genes served as a criterion. More strongly conserved genes were preferred over less conserved genes. In this respect, gene conservation and essentiality in genome-reduced Mycoplasma and other mollicutes species and the inclusion of genes in the genome of M. mycoides JCVI-syn3.0 had a high priority (8, 10, 42).
In many cases, it is not known whether a gene is truly required in the context of a minimal cell. In particular, this is the case for functions involved in RNA modification. In these cases, expression levels and gene conservation were valuable clues for deciding whether a gene should be included in the minimal gene list or not. Based on the list of the most abundant proteins (see http://subtiwiki.uni-goettingen.de/wiki/index.php/Most_abundant_proteins) (33, 43), we have decided whether there is a good reason to keep the corresponding genes or not. As an example, highly abundant enzymes required for amino acid biosynthesis were selected for deletion, whereas RNA chaperones were added. Similarly, genes conserved both in all mollicutes and in B. subtilis were regarded as being highly relevant for a minimal organism based on B. subtilis.
THE GENETIC COMPLEMENT OF A MINIMAL CELL
Based on the considerations explained above, we have selected 523 and 119 protein- and RNA-coding genes, respectively, as being important for a minimal organism that is capable of growing in LB medium supplemented with glucose at 37°C. Moreover, the growth and physiology of the minimal cell should be comparable to those of B. subtilis wild-type cells. B. subtilis has a generation time of about 20 min, whereas natural minimal organisms like M. mycoides and Mycoplasma pneumoniae divide in about 1 and 30 h, respectively. The minimal organism M. mycoides JCVI-syn3.0 has a generation time of 180 min (10, 44). This slow growth of Mycoplasma cells is another reason to rely on B. subtilis as the basis for a minimal cell. Most likely, a reduction of the growth rate has to be expected; however, the set of genes suggested in this study should allow generation times of <1 h. As a consequence, the number of genes included in the list is much larger than that of essential genes in B. subtilis 168. Moreover, not all essential genes are required for a minimal cell since several essential genes fulfill protective functions that are dispensable if, e.g., prophages have been deleted (32).
The genes of this minimal set satisfy all essential functions of the cell, such as information processing (DNA replication, transcription, and translation), metabolic pathways (metabolism of building blocks and cofactors and acquisition of ions, etc.), as well as cell division and integrity. Interestingly, there is a very good match between the genes in the MiniBacillus minimal gene set and those of M. mycoides JCVI-syn3.0 as far as information processing is concerned. In contrast, the two lists show only little overlap of genes required for metabolism, cell division, and protective functions. An overview of the set of genes required for MiniBacillus is provided in Table 1. A model of the metabolism of the minimal cell is outlined in Fig. 1, and details that include all metabolic pathways, reactions, and enzymes are provided in Fig. 2 to 11. Detailed information on each individual gene can be found in Tables 2 and 3 and Table S1 in the supplemental material. Table 2 also shows whether the components proposed to be important in the frame of a minimal B. subtilis genome are also present in the recently published minimal strain M. mycoides JCVI-syn3.0.
TABLE 1.
Overview of the genetic complement of a minimal B. subtilis cell
| Function | No. of proteins (no. of essential proteins)a | No. of RNA genes (no. of essential genes)a | Figure(s) |
|---|---|---|---|
| Information processing | 197 (125) | 119 (2) | |
| DNA replication | 18 (15) | ||
| Chromosome maintenance | 13 (9) | ||
| Transcription | 8 (5) | ||
| RNA folding and degradation | 6 (1) | ||
| Aminoacyl-tRNA synthetases | 24 (23) | 4 | |
| Ribosomal proteins | 53 (35) | ||
| rRNA and tRNA | 116 (0) | ||
| rRNA/tRNA maturation and modification | 31 (13) | 1 (1) | |
| Ribosome maturation and assembly | 9 (6) | ||
| Translation factors | 11 (9) | ||
| Translation/other | 5 (2) | 1 (0) | |
| Protein secretion | 12 (5) | 1 (1) | 2 |
| Proteolysis, protein quality control, chaperones | 7 (2) | ||
| Metabolism | 218 (59) | ||
| Central carbon metabolism | 26 (4) | 3 | |
| Respiration/energy | 16 (2) | 3 | |
| Amino acids | 30 (1)b | 3 | |
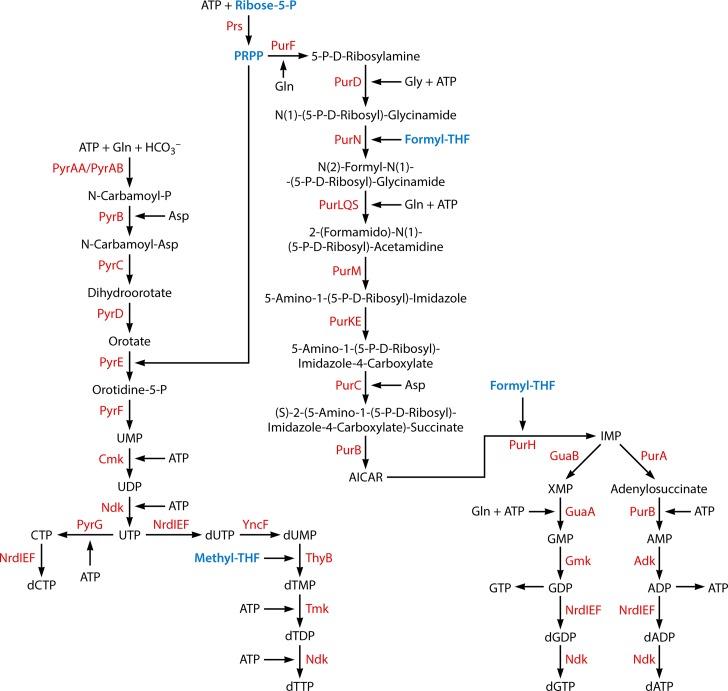
| Nucleotides/phosphate | 36 (11) | 2, 5 | |
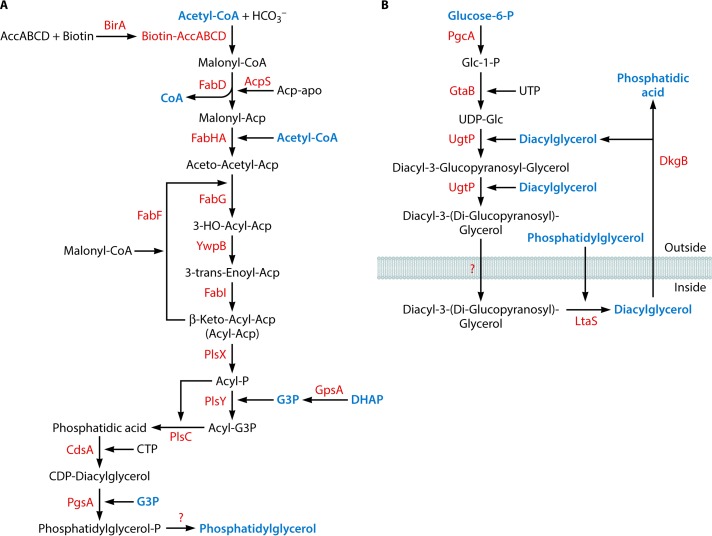
| Lipids | 19 (17) | 6 | |
| Cofactors | 62 (14)b | ||
| General components of ECF transporters | 3 (0)b | 4, 7 | |
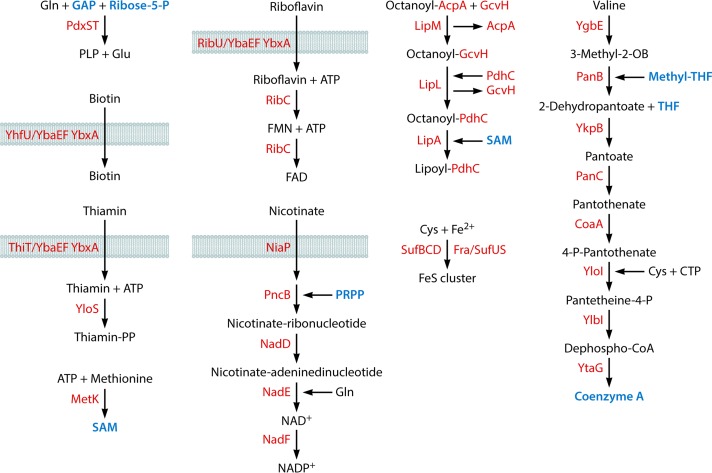
| NAD | 5 (4) | 7 | |
| FAD | 2 (1)b | 7 | |
| Pyridoxal phosphate | 2 (0) | 7 | |
| Biotin | 1 (0)b | 7 | |
| Thiamine | 2 (0)b | 7 | |
| Lipoate | 4 (1) | 7 | |
| Coenzyme A | 9 (1) | 7 | |
| S-Adenosylmethionine | 1 (1) | 7 | |
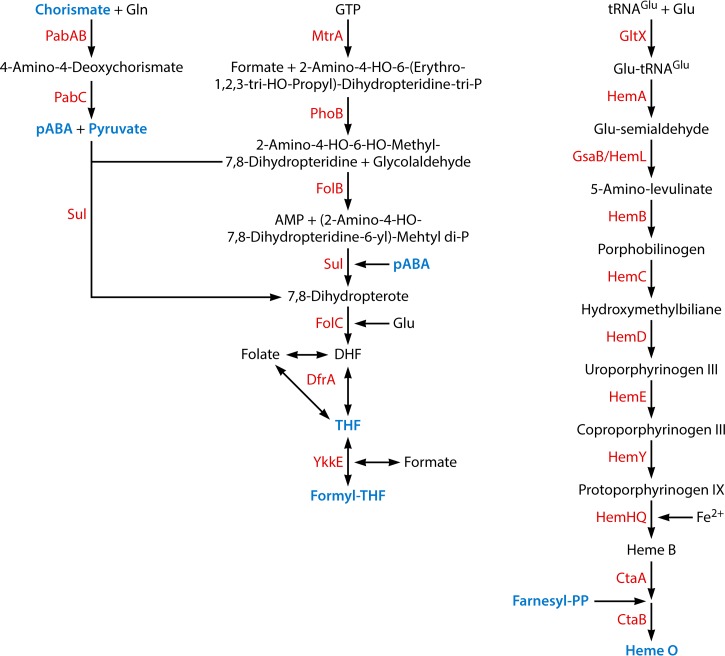
| Folate | 13 (1) | 8 | |
| Heme | 12 (0) | 8 | |
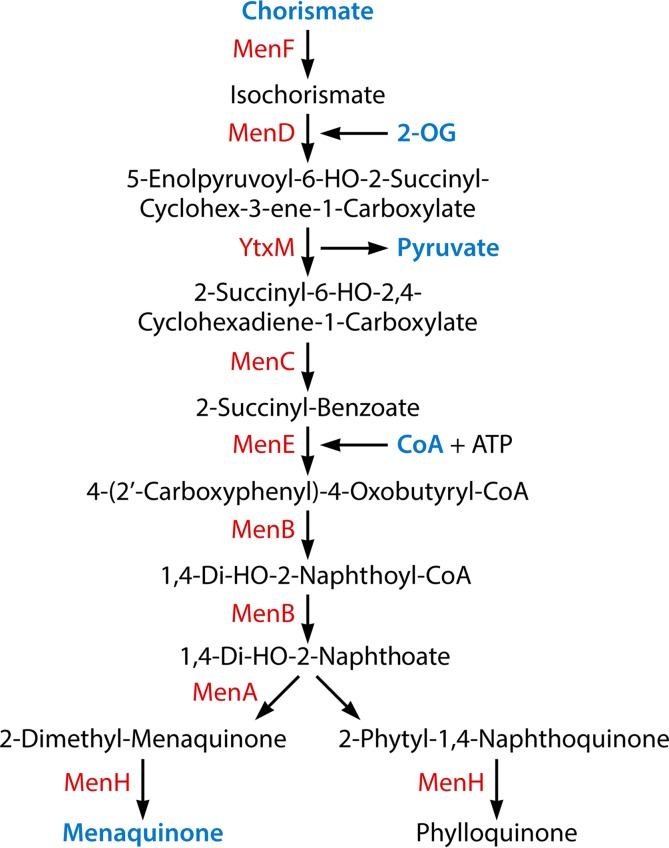
| Menaquinone | 8 (5) | 9 | |
| Metals/iron-sulfur clusters | 29 (10) | 2, 7 | |
| Cell division | 81 (52) | ||
| Cell wall synthesis | 55 (41) | ||
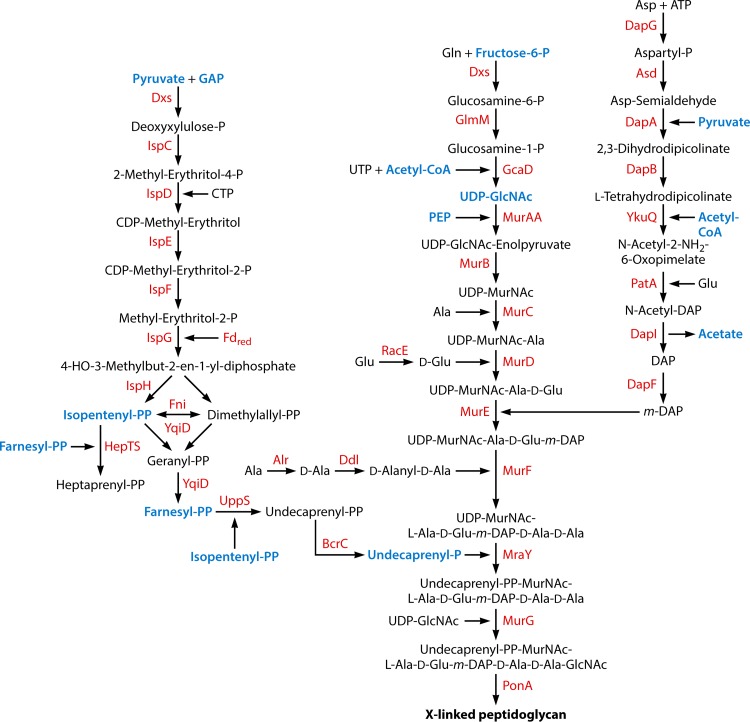
| Amino acid precursor | 11 (10) | 10 | |
| Undecaprenyl phosphate | 13 (10) | 10 | |
| Lipid II biosynthesis | 12 (11) | 10 | |
| Peptidoglycan polymerization | 5 (1) | 10 | |
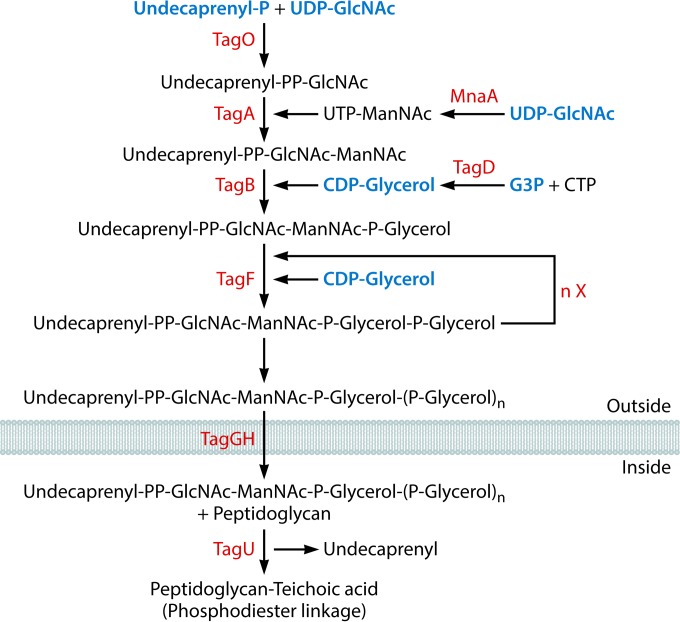
| Wall teichoic acid | 9 (8) | 11 | |
| Lipoteichoic acid | 5 (1) | 6 | |
| Coordination | 22 (9) | ||
| Signaling | 4 (2) | 2 | |
| Integrity of the cell | 16 (5) | ||
| Protection | 8 (4) | 2 | |
| Repair/genome integrity | 8 (1) | ||
| Other/unknown | 11 (2) | ||
| Total minimal genome | 523 (243) | 119 (2) |
The numbers of proteins and RNAs required for each function are listed. Numbers in parentheses indicate the numbers of proteins and RNAs that are essential in the context of B. subtilis 168.
Tryptophan, riboflavin, biotin, and thiamine are transported by transporters of the ECF (energy-coupling factor) family. The three general components are shared among all these transporters. They are listed separately with the cofactors.
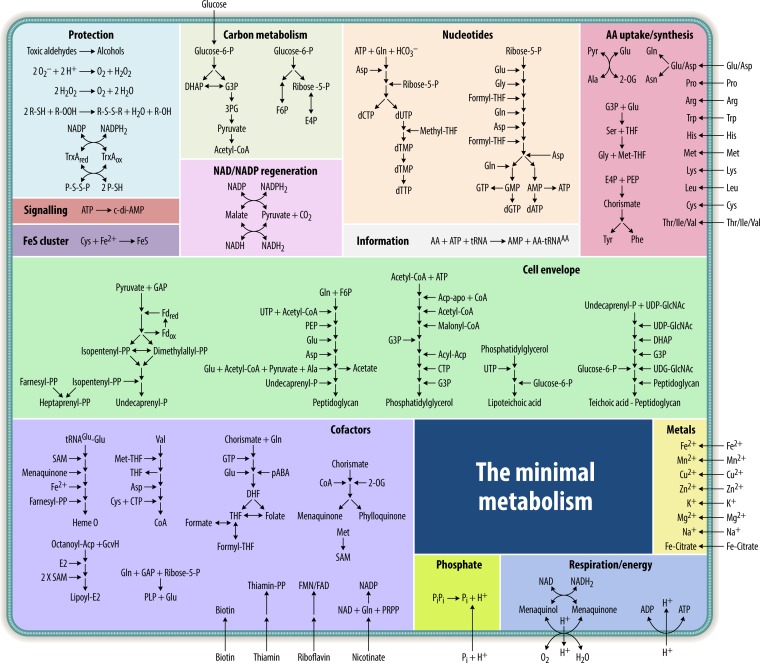
FIG 1.
Outline of the metabolic model of the minimal cell. The model gives an overview of the metabolic pathways of the intended minimal organism. Functionally related pathways are grouped in boxes. Details on all reactions and enzymes are provided in Fig. 2 to 11. DHAP, dihydroxyacetone phosphate; G3P, glycerol-3-phosphate; AA, amino acid; THF, tetrahydrofolate; 2-OG, 2-oxoglutarate; GAP, glyceraldehyde-3-phosphate; PEP, phosphoenolpyruvate; PLP, pyridoxal phosphate; DHF, 7,8-dihydrofolate; pABA, 4-aminobenzoate; FMN, flavin mononucleotide; PRPP, phosphoribosyl pyrophosphate; E4P, erythrose-4-phosphate; 3PG, 3-phosphoglycerate; PP, pyrophosphate; F6P, fructose-6-phosphate.
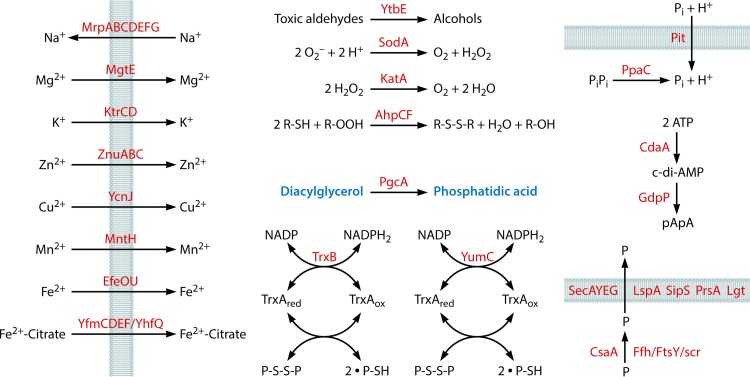
FIG 2.
Miscellaneous pathways. The model shows the uptake of metal ions and inorganic phosphate (Pi) and reactions for protective functions, for the generation of phosphatidic acid, and for the synthesis and degradation of c-di-AMP. Finally, protein secretion is included. The metabolic intermediates diacylglycerol (Fig. 6) and phosphatidic acid (Fig. 6) that occur in other pathways are labeled in blue. P, protein.
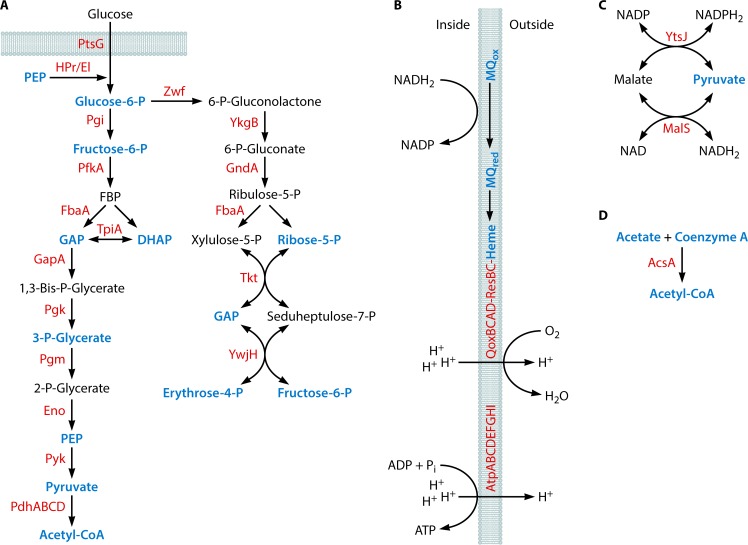
FIG 3.
Central carbon metabolism and energy conservation. (A) Glycolytic and pentose phosphate pathways. (B) The respiratory chain and ATPase. (C) The transhydrogenase cycle for balancing NADPH2. (D) Recycling of acetate derived from cell wall metabolism (Fig. 10). The following metabolic intermediates that occur in other pathways are labeled in blue: phosphoenolpyruvate (PEP) (Fig. 4 and 10), glucose-6-phosphate (Glucose-6-P) (Fig. 6), fructose-6-phosphate (Fig. 10), glyceraldehyde-3-phosphate (GAP) (Fig. 7 and 10), dihydroxyacetone phosphate (DHAP) (Fig. 6), 3-P-Glycerate (Fig. 4), pyruvate (Fig. 4 and 8 to 10), acetyl-CoA (Fig. 6 and 10), ribose-5-phosphate (Fig. 5 and 7), erythrose-4-phosphate (Fig. 4), menaquinone/menaquinol (MQ) (Fig. 9), heme (Fig. 8), acetate (Fig. 10), and coenzyme A (Fig. 6 and 10). FBP, fructose 1,6-bisphosphate.
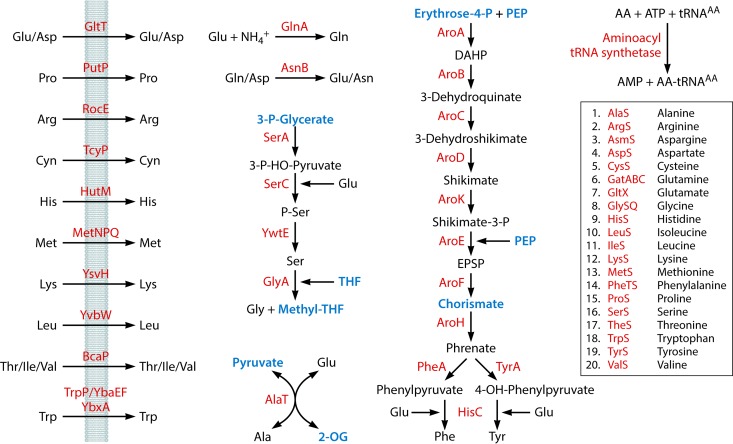
FIG 4.
Acquisition of amino acids and charging of tRNAs. The following metabolic intermediates that occur in other pathways are labeled in blue: 3-P-Glycerate (Fig. 3), tetrahydrofolate (THF) (Fig. 7 and 8), methyltetrahydrofolate (Methyl-THF) (Fig. 5 and 7), pyruvate (Fig. 3 and 8 to 10), 2-oxoglutarate (2-OG) (Fig. 9), erythrose-4-phosphate (Fig. 3), phosphoenolpyruvate (PEP) (Fig. 3 and 10), and chorismate (Fig. 8 and 9). DAHP, 3-deoxy-d-arabino-hept-2-ulosonate 7-phosphate; EPSP, 5-O-(1-carboxyvinyl)-3-phosphoshikimate; AA, amino acid.
FIG 5.
Acquisition of nucleotides. The following metabolic intermediates that occur in other pathways are labeled in blue: methyltetrahydrofolate (Methyl-THF) (Fig. 4 and 7), ribose-5-phosphate (Fig. 3 and 7), phosphoribosyl pyrophosphate (PRPP) (Fig. 7), and formyltetrahydrofolate (Formyl-THF) (Fig. 8). AICAR, 5-aminoimidazole-4-carboxamide ribonucleotide.
FIG 6.
(A) Biosynthesis of lipids. The enzyme required for the conversion of phosphatidylglycerol phosphate to phosphatidylglycerol is unknown. (B) Biosynthesis of lipoteichoic acids. The enzyme required for the export of diacyl-3-(diglucopyranosyl)-glycerol is unknown. The following metabolic intermediates that occur in other pathways are labeled in blue: acetyl-CoA (Fig. 3 and 10), CoA (Fig. 3, 7, and 9), glycerol-3-phosphate (G3P) (Fig. 11), dihydroxyacetone phosphate (DHAP) (Fig. 3), phosphatidylglycerol (this figure), glucose-6-phosphate (Fig. 3), diacylglycerol (Fig. 2), and phosphatidic acid (Fig. 2).
FIG 7.
Acquisition of cofactors and biosynthesis of iron-sulfur clusters. The following metabolic intermediates that occur in other pathways are labeled in blue: glyceraldehyde-3-phosphate (GAP) (Fig. 3 and 10), ribose-5-phosphate (Fig. 3 and 5), S-adenosylmethionine (SAM) (this figure), phosphoribosyl pyrophosphate (PRPP) (Fig. 5), methyltetrahydrofolate (Methyl-THF) (Fig. 4 and 5), tetrahydrofolate (THF) (Fig. 4 and 8), and coenzyme A (CoA) (Fig. 3, 6, and 9). PLP, pyridoxal phosphate; FMN, flavin mononucleotide; FeS, iron-sulfur cluster; 3-Methyl-2-OB, 3-methyl-2-oxobutanoate.
FIG 8.
Acquisition of cofactors. The following met1abolic intermediates that occur in other pathways are labeled in blue: chorismate (Fig. 4 and 9), 4-aminobenzoate (pABA) (this figure), pyruvate (Fig. 3, 4, 9, and 10), tetrahydrofolate (THF) (Fig. 4 and 7), formyltetrahydrofolate (Formyl-THF) (Fig. 5), farnesyl pyrophosphate (farnesyl-PP) (Fig. 10), and heme O (Fig. 3). DHF, 7,8-dihydrofolate.
FIG 9.
Acquisition of cofactors. The following metabolic intermediates that occur in other pathways are labeled in blue: chorismate (Fig. 4 and 8), 2-oxoglutarate (2-OG) (Fig. 4), pyruvate (Fig. 3, 4, 8, and 10), coenzyme A (CoA) (Fig. 3, 6, and 7), and menaquinone (Fig. 3).
FIG 10.
Biosynthesis of the cell wall. The following metabolic intermediates that occur in other pathways are labeled in blue: pyruvate (Fig. 3, 4, 8, and 9), glyceraldehyde-3-phosphate (GAP) (Fig. 3 and 7), isopentenyl pyrophosphate (isopentenyl-PP) (this figure), farnesyl pyrophosphate (farnesyl-PP) (Fig. 8), undecaprenyl phosphate (Fig. 11), fructose-6-phosphate (Fig. 3), acetyl-CoA (Fig. 3 and 6), UDP-N-acetylglucosamine (UDP-GlcNAc) (Fig. 11), phosphoenol pyrophosphate (PEP) (Fig. 3 and 4), and acetate (Fig. 3). UDP-MurNAc, UDP-N-acetylmuramic acid; DAP, diaminopimelate.
FIG 11.
Biosynthesis of wall teichoic acids. The following metabolic intermediates that occur in other pathways are labeled in blue: undecaprenyl phosphate (Fig. 10), UDP N-acetylglucosamine (UDP-GlcNAc) (Fig. 10), CDP-glycerol (this figure), and glycerol-3-phosphate (G3P) (Fig. 6). UDP-ManNAc, UDP-N-acetylmannosamine.
TABLE 2.
The complete gene set of MiniBacilluse
| Gene | BSU no.a | Essentialb | Syn3.0c | EC no. | PDB accession no. | Organismd | Function(s) |
|---|---|---|---|---|---|---|---|
| Information | |||||||
| DNA replication | |||||||
| dnaA | BSU00010 | Yes | Yes | 2Z4R | Thermotoga maritima | Replication initiation protein | |
| dnaB | BSU28990 | Yes | Initiation of chromosome replication | ||||
| dnaC | BSU40440 | Yes | Yes | 3.6.4.12 | 2VYE | Geobacillus kaustophilus | Replicative DNA helicase |
| dnaD | BSU22350 | Yes | 2V79 | B. subtilis | Initiation of chromosome replication | ||
| dnaE | BSU29230 | Yes | Yes | 2.7.7.7 | 3E0D | Thermus aquaticus | DNA polymerase III (alpha subunit) |
| dnaG | BSU25210 | Yes | Yes | 2.7.7.- | 4E2K | S. aureus | DNA primase |
| dnaI | BSU28980 | Yes | Yes | 4M4W | B. subtilis | Primosome component (helicase loader) | |
| dnaN | BSU00020 | Yes | Yes | 2.7.7.7 | 4TR6 | B. subtilis | DNA polymerase III (beta subunit), beta clamp |
| dnaX | BSU00190 | Yes | Yes | 2.7.7.7 | 1JR3 | E. coli | DNA polymerase III (gamma and tau subunits) |
| holA | BSU25560 | Yes | Yes | 2.7.7.7 | 3ZH9 | B. subtilis | DNA polymerase III, delta subunit |
| holB | BSU00310 | Yes | 2.7.7.7 | 1NJG | E. coli | DNA polymerase III (delta subunit) | |
| ligA | BSU06620 | Yes | Yes | 6.5.1.2 | 2OWO | E. coli | DNA ligase (NAD dependent) |
| priA | BSU15710 | Yes | 3.6.4.- | 4NL4 | Klebsiella pneumoniae | Primosomal replication factor Y | |
| polC | BSU16580 | Yes | Yes | 2.7.7.7 | 3F2B | G. kaustophilus | DNA polymerase III (alpha subunit) |
| rtp | BSU18490 | No | 1BM9 | B. subtilis | Replication terminator protein | ||
| ssbA | BSU40900 | Yes | Yes | 3VDY | B. subtilis | Single-strand DNA-binding protein | |
| yabA | BSU00330 | No | 5DOL | B. subtilis | Inhibitor of DnaA oligomerization | ||
| polA | BSU29090 | No | Yes | 2.7.7.7 | 1BGX | T. aquaticus | DNA polymerase I |
| Chromosome maintenance | |||||||
| scpA | BSU23220 | Yes | 3ZGX | B. subtilis | DNA segregation/condensation protein | ||
| scpB | BSU23210 | Yes | Yes | 3W6J | Geobacillus stearothermophilus | DNA segregation/condensation protein | |
| smc | BSU15940 | Yes | Yes | 3ZGX | B. subtilis | SMC protein | |
| parE | BSU18090 | Yes | Yes | 5.99.1.- | 4I3H | Streptococcus pneumoniae | Subunit of DNA topoisomerase IV |
| parC | BSU18100 | Yes | Yes | 5.99.1.- | 2INR | S. aureus | Subunit of DNA topoisomerase IV |
| spoIIIE | BSU16800 | No | 2IUT | Pseudomonas aeruginosa | ATP-dependent DNA translocase | ||
| sftA | BSU29805 | No | 2IUT | P. aeruginosa | DNA translocase | ||
| codV | BSU16140 | No | 1A0P | E. coli | Site-specific integrase/recombinase | ||
| ripX | BSU23510 | No | 1A0P | E. coli | Site-specific integrase/recombinase | ||
| gyrB | BSU00060 | Yes | Yes | 5.99.1.3 | 4I3H | S. pneumoniae | DNA gyrase (subunit B) |
| gyrA | BSU00070 | Yes | Yes | 5.99.1.3 | 4DDQ | B. subtilis | DNA gyrase (subunit A) |
| topA | BSU16120 | Yes | Yes | 5.99.1.2 | 4RUL | E. coli | DNA topoisomerase I |
| hbs | BSU22790 | Yes | 1HUE | G. stearothermophilus | Nonspecific DNA-binding protein HBsu | ||
| Transcription | |||||||
| rpoA | BSU01430 | Yes | Yes | 2.7.7.6 | 3IYD | E. coli | RNA polymerase alpha subunit |
| rpoB | BSU01070 | Yes | Yes | 2.7.7.6 | 3IYD | E. coli | RNA polymerase beta subunit |
| rpoC | BSU01080 | Yes | Yes | 2.7.7.6 | 3IYD | E. coli | RNA polymerase beta′ subunit |
| sigA | BSU25200 | Yes | Yes | 3IYD | E. coli | RNA polymerase sigma factor SigA | |
| rpoE | BSU37160 | No | 2.7.7.6 | 2KRC | B. subtilis | RNA polymerase delta subunit | |
| helD | BSU33450 | No | 3.6.4.12 | DNA 3′–5′ helicase IV | |||
| greA | BSU27320 | No | Yes | 1GRJ | E. coli | Transcription elongation factor | |
| nusA | BSU16600 | Yes | Yes | 1HH2 | T. maritima | Transcription termination factor | |
| RNA folding and degradation | |||||||
| cspD | BSU21930 | No | 1C9O | Bacillus caldolyticus | Cold shock protein | ||
| cspB | BSU09100 | No | 2ES2 | B. subtilis | Major cold shock protein | ||
| rny | BSU16960 | No | Yes | 3.1.4.16 | RNase Y | ||
| rnjA | BSU14530 | Yes | Yes | 3ZQ4 | B. subtilis | RNase J1 | |
| pnpA | BSU16690 | No | 2.7.7.8 | 3CDI | E. coli | Polynucleotide phosphorylase | |
| nrnA | BSU29250 | No | Yes | 3.1.3.7 | 3DEV | Staphylococcus haemolyticus | Oligoribonuclease (nano-RNase) |
| Aminoacyl tRNA synthetases | |||||||
| alaS | BSU27410 | Yes | Yes | 6.1.1.7 | 3HTZ | E. coli | Alanine-tRNA synthetase |
| argS | BSU37330 | Yes | Yes | 6.1.1.19 | 3FNR | Campylobacter jejuni | Arginyl-tRNA synthetase, universally conserved protein |
| asnS | BSU22360 | Yes | Yes | 6.1.1.22 | 1X54 | Pyrococcus horikoshii | Asparagyl-tRNA synthetase |
| aspS | BSU27550 | Yes | Yes | 6.1.1.12 | 1EQR | E. coli | Aspartyl-tRNA synthetase |
| cysS | BSU00940 | Yes | Yes | 6.1.1.16 | 3TQO | Coxiella burnetii | Cysteine-tRNA synthetase |
| gatC | BSU06670 | Yes | 6.3.5.7 | 2DF4 | S. aureus | Production of glutamyl-tRNAGln | |
| gatA | BSU06680 | Yes | Yes | 6.3.5.7 | 2DF4 | S. aureus | Production of glutamyl-tRNAGln |
| gatB | BSU06690 | Yes | Yes | 6.3.5.7 | 2DF4 | S. aureus | Production of glutamyl-tRNAGln |
| gltX | BSU00920 | Yes | Yes | 6.1.1.17 | 2O5R | T. maritima | Glutamyl-tRNA synthetase, universally conserved protein |
| glyS | BSU25260 | Yes | 6.1.1.14 | Glycyl-tRNA synthetase (beta subunit) | |||
| glyQ | BSU25270 | Yes | 6.1.1.14 | 1J5W | T. maritima | Glycyl-tRNA synthetase (alpha subunit) | |
| hisS | BSU27560 | Yes | Yes | 6.1.1.21 | 1QE0 | S. aureus | Histidyl-tRNA synthetase |
| ileS | BSU15430 | Yes | Yes | 6.1.1.5 | 1QU2 | S. aureus | Isoleucyl-tRNA synthetase |
| leuS | BSU30320 | Yes | Yes | 6.1.1.4 | 1OBH | Thermus thermophilus | Leucyl-tRNA synthetase |
| lysS | BSU00820 | Yes | Yes | 6.1.1.6 | 3E9H | G. stearothermophilus | Lysyl-tRNA synthetase |
| metS | BSU00380 | Yes | Yes | 6.1.1.10 | 4QRD | S. aureus | Methionyl-tRNA synthetase |
| pheT | BSU28630 | Yes | Yes | 6.1.1.20 | 2RHS | S. haemolyticus | Phenylalanyl-tRNA synthetase (beta subunit) |
| pheS | BSU28640 | Yes | Yes | 6.1.1.20 | 2RHQ | S. haemolyticus | Phenylalanyl-tRNA synthetase (alpha subunit), universally conserved protein |
| proS | BSU16570 | Yes | 6.1.1.15 | 2J3L | Enterococcus faecalis | Prolyl-tRNA synthetase | |
| serS | BSU00130 | Yes | Yes | 6.1.1.11 | 2DQ3 | Aquifex aeolicus | Seryl-tRNA synthetase |
| thrS | BSU28950 | No | Yes | 6.1.1.3 | 1NYQ | S. aureus | Threonyl-tRNA synthetase (major) |
| trpS | BSU11420 | Yes | Yes | 6.1.1.2 | 3PRH | B. subtilis | Tryptophanyl-tRNA synthetase |
| tyrS | BSU29670 | Yes | Yes | 6.1.1.1 | 2TS1 | G. stearothermophilus | Tyrosyl-tRNA synthetase (major) |
| valS | BSU28090 | Yes | Yes | 6.1.1.9 | 1GAX | T. thermophilus | Valyl-tRNA synthetase |
| Ribosomal proteins | |||||||
| rplA | BSU01030 | No | Yes | 3J9W | B. subtilis | Ribosomal protein L1 | |
| rplB | BSU01190 | Yes | Yes | 3J9W | B. subtilis | Ribosomal protein L2 | |
| rplC | BSU01160 | Yes | Yes | 3J9W | B. subtilis | Ribosomal protein L3 | |
| rplD | BSU01170 | Yes | Yes | 3J9W | B. subtilis | Ribosomal protein L4 | |
| rplE | BSU01280 | Yes | Yes | 3J9W | B. subtilis | Ribosomal protein L5 | |
| rplF | BSU01310 | Yes | Yes | 3J9W | B. subtilis | Ribosomal protein L6 | |
| rplI | BSU40500 | No | Yes | 3J9W | B. subtilis | Ribosomal protein L9 | |
| rplJ | BSU01040 | Yes | Yes | 3J9W | B. subtilis | Ribosomal protein L10 | |
| rplK | BSU01020 | No | Yes | 3J9W | B. subtilis | Ribosomal protein L11 | |
| rplL | BSU01050 | Yes | Yes | 3J9W | B. subtilis | Ribosomal protein L12 | |
| rplM | BSU01490 | Yes | Yes | 3J9W | B. subtilis | Ribosomal protein L13 | |
| rplN | BSU01260 | Yes | Yes | 3J9W | B. subtilis | Ribosomal protein L14 | |
| rplO | BSU01350 | No | Yes | 3J9W | B. subtilis | Ribosomal protein L15 | |
| rplP | BSU01230 | Yes | Yes | 3J9W | B. subtilis | Ribosomal protein L16 | |
| rplQ | BSU01440 | Yes | Yes | 3J9W | B. subtilis | Ribosomal protein L17 | |
| rplR | BSU01320 | Yes | Yes | 3J9W | B. subtilis | Ribosomal protein L18 | |
| rplS | BSU16040 | Yes | Yes | 3J9W | B. subtilis | Ribosomal protein L19 | |
| rplT | BSU28850 | Yes | Yes | 3J9W | B. subtilis | Ribosomal protein L20 | |
| rplU | BSU27960 | Yes | Yes | 3J9W | B. subtilis | Ribosomal protein L21 | |
| rplV | BSU01210 | No | Yes | 3J9W | B. subtilis | Ribosomal protein L22 | |
| rplW | BSU01180 | No | Yes | 3J9W | B. subtilis | Ribosomal protein L23 | |
| rplX | BSU01270 | Yes | Yes | 3J9W | B. subtilis | Ribosomal protein L24 | |
| rpmA | BSU27940 | Yes | Yes | 3J9W | B. subtilis | Ribosomal protein L27 | |
| rpmB | BSU15820 | No | 3J9W | B. subtilis | Ribosomal protein L28 | ||
| rpmC | BSU01240 | No | 3J9W | B. subtilis | Ribosomal protein L29 | ||
| rpmD | BSU01340 | Yes | 3J9W | B. subtilis | Ribosomal protein L30 | ||
| rpmE | BSU37070 | No | 3J9W | B. subtilis | Ribosomal protein L31 | ||
| rpmF | BSU15080 | No | 3J9W | B. subtilis | Ribosomal protein L32 | ||
| rpmGA | BSU24900 | No | 3J9W | B. subtilis | Ribosomal protein L33a | ||
| rpmGB | BSU00990 | No | 3J9W | B. subtilis | Ribosomal protein L33b | ||
| rpmH | BSU41060 | No | 3J9W | B. subtilis | Ribosomal protein L34 | ||
| rpmI | BSU28860 | No | 3J9W | B. subtilis | Ribosomal protein L35 | ||
| rpmJ | BSU01400 | No | Yes | 3J9W | B. subtilis | Ribosomal protein L36 | |
| rpsB | BSU16490 | Yes | Yes | 3J9W | B. subtilis | Ribosomal protein S2 | |
| rpsC | BSU01220 | Yes | Yes | 3J9W | B. subtilis | Ribosomal protein S3 | |
| rpsD | BSU29660 | Yes | Yes | 3J9W | B. subtilis | Ribosomal protein S4 | |
| rpsE | BSU01330 | Yes | Yes | 3J9W | B. subtilis | Ribosomal protein S5 | |
| rpsF | BSU40910 | No | Yes | 3J9W | B. subtilis | Ribosomal protein S6 | |
| rpsG | BSU01110 | Yes | Yes | 3J9W | B. subtilis | Ribosomal protein S7 | |
| rpsH | BSU01300 | Yes | Yes | 3J9W | B. subtilis | Ribosomal protein S8 | |
| rpsI | BSU01500 | Yes | Yes | 3J9W | B. subtilis | Ribosomal protein S9 | |
| rpsJ | BSU01150 | Yes | Yes | 3J9W | B. subtilis | Ribosomal protein S10 | |
| rpsK | BSU01420 | Yes | Yes | 3J9W | B. subtilis | Ribosomal protein S11 | |
| rpsL | BSU01100 | Yes | Yes | 3J9W | B. subtilis | Ribosomal protein S12 | |
| rpsM | BSU01410 | Yes | Yes | 3J9W | B. subtilis | Ribosomal protein S13 | |
| rpsN | BSU01290 | Yes | Yes | 3J9W | B. subtilis | Ribosomal protein S14 | |
| rpsO | BSU16680 | Yes | Yes | 3J9W | B. subtilis | Ribosomal protein S15 | |
| rpsP | BSU15990 | Yes | Yes | 3J9W | B. subtilis | Ribosomal protein S16 | |
| rpsQ | BSU01250 | Yes | Yes | 3J9W | B. subtilis | Ribosomal protein S17 | |
| rpsR | BSU40890 | Yes | Yes | 3J9W | B. subtilis | Ribosomal protein S18 | |
| rpsS | BSU01200 | Yes | Yes | 3J9W | B. subtilis | Ribosomal protein S19 | |
| rpsT | BSU25550 | No | 3J9W | B. subtilis | Ribosomal protein S20 | ||
| rpsU | BSU25410 | No | 3J9W | B. subtilis | Ribosomal protein S21 | ||
| rRNA/tRNA maturation and modification | |||||||
| rnpA | BSU41050 | Yes | Yes | 3.1.26.5 | 3Q1R | T. maritima | Protein component of RNase P |
| rnpB | BSU_misc_RNA_35 | Yes | 3Q1R | T. maritima | RNA component of RNase P | ||
| rnz | BSU23840 | Yes | 3.1.26.11 | 4GCW | B. subtilis | RNase Z | |
| rph | BSU28370 | No | 2.7.7.56 | 1OYP | B. subtilis | RNase PH | |
| rbfA | BSU16650 | No | 1JOS | Haemophilus influenzae | Ribosome-binding factor A | ||
| rimM | BSU16020 | No | 3H9N | H. influenzae | 16S rRNA-processing protein, RNase | ||
| cca | BSU22450 | Yes | 2.7.7.25 | 1MIY | G. stearothermophilus | tRNA nucleotidyltransferase | |
| fmt | BSU15730 | Yes | Yes | 2.1.2.9 | 4IQF | Bacillus anthracis | Methionyl-tRNA formyltransferase |
| folD | BSU24310 | Yes | Yes | 1.5.1.5 | 1B0A | E. coli | Methylenetetrahydrofolate dehydrogenase |
| rlmCD | BSU06730 | No | 2.1.1.190 | 2BH2 | E. coli | rRNA methyltransferase | |
| ysgA | BSU28650 | Yes | Yes | 2.1.1.- | 4X3M | T. thermophilus | Similar to rRNA methylase |
| mraW | BSU15140 | No | Yes | 2.1.1.199 | 1WG8 | T. thermophilus | SAM-dependent methyltransferase |
| cspR | BSU08930 | Yes | 2.1.1.- | 4PZK | B. anthracis | Similar to tRNA(Um34/Cm34) methyltransferase | |
| trmD | BSU16030 | Yes | Yes | 2.1.1.31 | 3KY7 | S. aureus | tRNA methyltransferase |
| trmU | BSU27500 | Yes | Yes | 2.8.1.- | 2HMA | S. pneumoniae | tRNA(5-methylaminomethyl-2-thiouridylate) methyltransferase |
| yrvO | BSU27510 | Yes | 2.8.1.7 | 1P3W | E. coli | Cysteine desulfurase | |
| yacO | BSU00960 | No | Yes | 2.1.1.- | 1GZ0 | E. coli | Putative 23S rRNA methyltransferase |
| ksgA | BSU00420 | No | Yes | 2.1.1.- | 3FUU | T. thermophilus | rRNA adenine dimethyltransferase |
| rluB | BSU23160 | No | Yes | 5.4.99.22 | 4LAB | E. coli | Pseudouridine synthase |
| ypuI | BSU23200 | No | rRNA pseudouridine 2633 synthase | ||||
| tilS | BSU00670 | Yes | Yes | 6.3.4.19 | 3A2K | G. kaustophilus | tRNAIle lysidine synthetase |
| tsaB | BSU05920 | Yes | 2A6A | T. maritima | Threonyl carbamoyl adenosine (t6A) modification of tRNAs that pair with ANN codons in mRNA | ||
| tsaD | BSU05940 | Yes | Yes | 2.3.1.234 | 3ZET | Salmonella enterica serovar Typhimurium | Threonyl carbamoyl adenosine (t6A) modification of tRNAs that pair with ANN codons in mRNA, universally conserved protein |
| tsaC | BSU36950 | No | 2.7.7.87 | 3AJE | Sulfolobus tokodaii | l-Threonyl carbamoyl AMP synthase, biosynthesis of the hypermodified base threonyl carbamoyl adenosine [t(6)A] | |
| gidA | BSU41010 | No | Yes | 3CP2 | E. coli | tRNA uridine 5-carboxymethyl-aminomethyl modification enzyme | |
| thdF | BSU41020 | No | Yes | 1XZP | T. maritima | GTP-binding protein, putative tRNA modification GTPase | |
| truA | BSU01480 | No | Yes | 5.4.99.12 | 1VS3 | T. thermophilus | Pseudouridylate synthase I, universally conserved protein |
| tsaE | BSU05910 | No | Yes | 1HTW | H. influenzae | P-loop ATPase | |
| trmFO | BSU16130 | No | Yes | 2.1.1.74 | 3G5Q | T. thermophilus | tRNA:m(5)U-54 methyltransferase |
| miaA | BSU17330 | No | 2.5.1.8 | 2QGN | Bacillus halodurans | tRNA isopentenylpyrophosphate transferase | |
| yaaJ | BSU00180 | No | 2B3J | S. aureus | tRNA-specific adenosine deaminase | ||
| ylyB | BSU15460 | No | Yes | 5.4.99.23 | 1V9F | E. coli | Similar to pseudouridylate synthase |
| Ribosome maturation/assembly | |||||||
| ydiD | BSU05930 | No | 2.3.1.128 | 2CNM | S. enterica | Similar to ribosomal protein alanine N-acetyltransferase | |
| ylxS | BSU16590 | No | 1IB8 | S. pneumoniae | Similar to 30S ribosomal subunit maturation protein | ||
| prp | BSU27950 | No | 4PEO | S. aureus | Maturation of L27 | ||
| engA | BSU22840 | Yes | 2HJG | B. subtilis | GTPase, ribosome 50S subunit assembly | ||
| era | BSU25290 | Yes | Yes | 3R9W | A. aeolicus | GTP-binding protein | |
| obg | BSU27920 | Yes | Yes | 3.6.5.- | 1LNZ | B. subtilis | GTP-binding protein |
| rbgA | BSU16050 | Yes | Yes | 1PUJ | B. subtilis | Assembly of the 50S subunit of the ribosome | |
| yqeH | BSU25670 | Yes | 3H2Y | B. anthracis | Assembly/stability of the 30S subunit of the ribosome, assembly of the 70S ribosome | ||
| ysxC | BSU28190 | Yes | Yes | 1SVI | B. subtilis | Assembly of the 50S subunit of the ribosome | |
| Translation factors | |||||||
| efp | BSU24450 | No | Yes | 1YBY | Clostridium thermocellum | Elongation factor P | |
| frr | BSU16520 | Yes | Yes | 4GFQ | B. anthracis | Ribosome recycling factor | |
| fusA | BSU01120 | Yes | Yes | 2XEX | S. aureus | Elongation factor G | |
| infA | BSU01390 | Yes | Yes | 4QL5 | S. pneumoniae | Translation initiation factor IF-1 | |
| infB | BSU16630 | Yes | Yes | 1ZO1 | E. coli | Translation initiation factor IF-2 | |
| infC | BSU28870 | Yes | Yes | 1TIG | G. stearothermophilus | Translation initiation factor IF-3 | |
| prfA | BSU37010 | Yes | Yes | 1ZBT | Streptococcus mutans | Peptide chain release factor 1 | |
| prfB | BSU35290 | Yes | 1MI6 | E. coli | Peptide chain release factor 2 | ||
| tsf | BSU16500 | Yes | Yes | 1EFU | E. coli | Elongation factor Ts | |
| tufA | BSU01130 | Yes | Yes | 3.6.5.3 | 4R71 | E. coli | Elongation factor Tu |
| lepA | BSU25510 | No | Yes | 4QJT | T. thermophilus | Elongation factor 4 | |
| Translation/others | |||||||
| map | BSU01380 | Yes | Yes | 3.4.11.18 | 1O0X | T. maritima | Methionine aminopeptidase |
| ywkE | BSU37000 | No | Yes | 2.1.1.297 | 2B3T | E. coli | Similar to N5-glutamine methyltransferase that modifies peptide release factors |
| ybxF | BSU01090 | No | 3V7E | B. subtilis | Similar to ribosomal protein L7 family | ||
| spoVC | BSU00530 | Yes | Yes | 3.1.1.29 | 4QT4 | S. pyogenes | Putative peptidyl-tRNA hydrolase |
| ssrA | BSU_MISC_RNA_55 | No | 1P6V | A. aeolicus | tmRNA | ||
| smpB | BSU33600 | No | Yes | 1P6V | A. aeolicus | tmRNA-binding protein | |
| Protein secretion | |||||||
| scr | BSU_misc_RNA_2 | Yes | 4UE5 | B. subtilis | Signal recognition particle RNA | ||
| ffh | BSU15980 | Yes | Yes | 4UE5 | B. subtilis | Signal recognition particle component | |
| ftsY | BSU15950 | No | Yes | 2XXA | E. coli | Signal recognition particle | |
| yidC2 | BSU23890 | No | 3WO6 | B. halodurans | Sec-independent membrane protein translocase | ||
| secA | BSU35300 | Yes | Yes | 3DL8 | B. subtilis | Preprotein translocase subunit (ATPase) | |
| secE | BSU01000 | Yes | 3DL8 | B. subtilis | Preprotein translocase subunit | ||
| secY | BSU01360 | Yes | Yes | 3DL8 | B. subtilis | Preprotein translocase subunit, universally conserved protein | |
| secG | BSU33630 | No | 3DL8 | B. subtilis | Preprotein translocase subunit | ||
| sipS | BSU23310 | No | 4NV4 | B. anthracis | Signal peptidase I | ||
| prsA | BSU09950 | Yes | 5.2.1.8 | 4WO7 | B. subtilis | Protein secretion (posttranslocation molecular chaperone) | |
| csaA | BSU19040 | No | 2NZH | B. subtilis | Molecular chaperone involved in protein secretion | ||
| lgt | BSU34990 | No | 2.4.99.- | 5AZB | E. coli | Prolipoprotein diacylglyceryl transferase | |
| lspA | BSU15450 | No | 5DIR | P. aeruginosa | Signal peptidase II | ||
| Proteolysis/quality control/chaperones | |||||||
| htrB | BSU33000 | No | 3QO6 | Arabidopsis thaliana | Serine protease | ||
| groES | BSU06020 | Yes | 1WE3 | T. thermophilus | Chaperonin, universally conserved protein | ||
| groEL | BSU06030 | Yes | 1WE3 | T. thermophilus | Chaperonin | ||
| dnaJ | BSU25460 | No | Yes | 3LZ8 | K. pneumoniae | Activation of DnaK | |
| dnaK | BSU25470 | No | Yes | 2V7Y | G. kaustophilus | Molecular chaperone | |
| grpE | BSU25480 | No | 4ANI | G. kaustophilus | Activation of DnaK | ||
| tig | BSU28230 | No | 2MLX | E. coli | Trigger factor (prolyl isomerase) | ||
| Metabolism | |||||||
| Central carbon metabolism | |||||||
| Glycolysis | |||||||
| ptsG | BSU13890 | No | Yes | 2.7.1.69 | PTS glucose permease, EIICBA(Glc) | ||
| ptsH | BSU13900 | No | Yes | 2.7.11.- | 2FEP | B. subtilis | HPr, general component of the PTS |
| ptsI | BSU13910 | No | Yes | 2.7.3.9 | 2WQD | S. aureus | Enzyme I, general component of the PTS |
| pgi | BSU31350 | No | Yes | 5.3.1.9 | 3IFS | B. anthracis | Glucose-6-phosphate isomerase |
| pfkA | BSU29190 | No | Yes | 2.7.1.11 | 4A3S | B. subtilis | Phosphofructokinase |
| fbaA | BSU37120 | No | 4.1.2.13 | 4TO8 | S. aureus | Fructose 1,6-bisphosphate aldolase | |
| tpi | BSU33920 | No | Yes | 5.3.1.1 | 2BTM | G. stearothermophilus | Triose phosphate isomerase |
| gapA | BSU33940 | Yes | Yes | 1.2.1.12 | 1GD1 | G. stearothermophilus | Glyceraldehyde-3-phosphate dehydrogenase |
| pgk | BSU33930 | No | Yes | 2.7.2.3 | 1PHP | G. stearothermophilus | Phosphoglycerate kinase, universally conserved protein |
| pgm | BSU33910 | Yes | Yes | 5.4.2.1 | 1EJJ | G. stearothermophilus | Phosphoglycerate mutase |
| eno | BSU33900 | Yes | Yes | 4.2.1.11 | 4A3R | B. subtilis | Enolase, universally conserved protein |
| pyk | BSU29180 | No | Yes | 2.7.1.40 | 2E28 | G. stearothermophilus | Pyruvate kinase |
| pdhA | BSU14580 | Yes | Yes | 1.2.4.1 | 3DUF | G. stearothermophilus | Pyruvate dehydrogenase (E1 alpha subunit) |
| pdhB | BSU14590 | No | Yes | 1.2.4.1 | 3DUF | G. stearothermophilus | Pyruvate dehydrogenase (E1 beta subunit) |
| pdhC | BSU14600 | No | Yes | 2.3.1.12 | 3DUF | G. stearothermophilus | Pyruvate dehydrogenase (dihydrolipoamide acetyltransferase E2 subunit) |
| pdhD | BSU14610 | No | Yes | 1.8.1.4 | 1EBD | G. stearothermophilus | Dihydrolipoamide dehydrogenase E3 subunit of both pyruvate and 2-oxoglutarate dehydrogenase complexes |
| Transhydrogenation cycle | |||||||
| ytsJ | BSU29220 | No | 1.1.1.38 | 2A9F | S. pyogenes | Malic enzyme | |
| malS | BSU29880 | No | 1.1.1.38 | 1LLQ | Ascaris suum | Malate dehydrogenase (decarboxylating) | |
| Pentose phosphate pathway | |||||||
| ykgB | BSU13010 | No | 3.1.1.31 | 3HFQ | Lactobacillus plantarum | 6-Phosphogluconolactonase | |
| rpe | BSU15790 | No | Yes | 5.1.3.1 | 1TQJ | Synechocystis sp. | Ribulose 5-phosphate 3-epimerase |
| tkt | BSU17890 | No | Yes | 2.2.1.1 | 3HYL | B. anthracis | Transketolase |
| zwf | BSU23850 | No | 1.1.1.49 | 1DPG | Leuconostoc mesenteroides | Glucose-6-phosphate dehydrogenase | |
| gndA | BSU23860 | No | 1.1.1.44 | 2W8Z | G. stearothermophilus | NADP-dependent phosphogluconate dehydrogenase | |
| ywlF | BSU36920 | No | Yes | 3HE8 | C. thermocellum | Ribose-5-phosphate isomerase | |
| ywjH | BSU37110 | No | 2.2.1.2 | 3R8R | B. subtilis | Transaldolase | |
| Recycling of acetate | |||||||
| acsA | BSU29680 | No | 6.2.1.1 | 2P2F | S. enterica | Acetyl-CoA synthetase | |
| Respiration/energy | |||||||
| ndh | BSU12290 | No | 4NWZ | Caldalkalibacillus thermarum | NADH dehydrogenase | ||
| Cytochrome aa3 | |||||||
| qoxD | BSU38140 | No | Cytochrome aa3 quinol oxidase (subunit IV) | ||||
| qoxC | BSU38150 | No | 1FFT | E. coli | Cytochrome aa3 quinol oxidase (subunit III) | ||
| qoxB | BSU38160 | No | 1FFT | E. coli | Cytochrome aa3 quinol oxidase (subunit I) | ||
| qoxA | BSU38170 | No | 1FFT | E. coli | Cytochrome aa3 quinol oxidase (subunit II) | ||
| Cytochrome maturation | |||||||
| resC | BSU23130 | Yes | Part of heme translocase, required for cytochrome c synthesis | ||||
| resB | BSU23140 | Yes | Part of heme translocase, required for cytochrome c synthesis | ||||
| ATPase | |||||||
| atpC | BSU36800 | No | 2E5Y | Bacillus sp. | ATP synthase, F1 (subunit epsilon) | ||
| atpD | BSU36810 | No | Yes | 3.6.3.14 | 1SKY | Bacillus sp. | ATP synthase, F1 (subunit beta) |
| atpG | BSU36820 | No | Yes | 4XD7 | G. kaustophilus | ATP synthase, F1 (subunit gamma) | |
| atpA | BSU36830 | No | Yes | 3.6.3.14 | 1SKY | Bacillus sp. | ATP synthase, F1 (subunit alpha) |
| atpH | BSU36840 | No | ATP synthase, F1 (subunit delta) | ||||
| atpF | BSU36850 | No | Yes | ATP synthase, Fo (subunit b) | |||
| atpE | BSU36860 | No | 1WU0 | Bacillus sp. | ATP synthase, Fo (subunit c) | ||
| atpB | BSU36870 | No | 1C17 | E. coli | ATP synthase, Fo (subunit a) | ||
| atpI | BSU36880 | No | ATP synthase (subunit i) | ||||
| Amino acids | |||||||
| Asp, Glu | |||||||
| gltT | BSU10220 | No | 3V8F | P. horikoshii | Major H+/Na+-glutamate symport protein | ||
| Arg | |||||||
| rocE | BSU40330 | No | 3LRB | E. coli | Amino acid permease | ||
| Pro | |||||||
| putP | BSU03220 | No | 2XQ2 | Vibrio parahaemolyticus | High-affinity proline permease | ||
| Trp | |||||||
| trpP | BSU10010 | No | S protein of tryptophan ECF transporter | ||||
| Met | |||||||
| metQ | BSU32730 | No | 4GOT | B. subtilis | Methionine ABC transporter (binding lipoprotein) | ||
| metP | BSU32740 | No | 3DHW | E. coli | Methionine ABC transporter, permease | ||
| metN | BSU32750 | No | 3DHW | E. coli | Methionine ABC transporter (ATP-binding protein) | ||
| His | |||||||
| hutM | BSU39390 | No | Histidine permease | ||||
| Cys | |||||||
| tcyP | BSU09130 | No | Yes | 3KBC | P. horikoshii | Cystine transporter | |
| Gly | |||||||
| glyA | BSU36900 | Yes | Yes | 2.1.2.1 | 1KKJ | G. stearothermophilus | Serine hydroxymethyltransferase |
| Ile, Val, Thr | |||||||
| bcaP | BSU09460 | No | Branched-chain amino acid transporter | ||||
| Lys | |||||||
| yvsH | BSU33330 | No | 3LRB | E. coli | Putative lysine transporter | ||
| Chorismate for aromatic amino acids, menaquinone, and folate | |||||||
| aroA | BSU29750 | No | 2.5.1.54 | 3NVT | L. monocytogenes | 3-Deoxy-d-arabino-heptulosonate 7-phosphate synthase/chorismate mutase isozyme 3 | |
| aroB | BSU22700 | No | 4.2.3.4 | 3CLH | Helicobacter pylori | 3-Dehydroquinate synthase | |
| aroC | BSU23080 | No | 4.2.1.10 | 1QFE | S. enterica serovar Typhi | 3-Dehydroquinate dehydratase | |
| aroD | BSU25660 | No | 1.1.1.25 | 2EGG | G. kaustophilus | Shikimate dehydrogenase | |
| aroE | BSU22600 | No | 2.5.1.19 | 3RMT | B. halodurans | 3-Phosphoshikimate 1-carboxyvinyltransferase | |
| aroF | BSU22710 | No | 4.2.3.5 | 1Q1L | A. aeolicus | Chorismate synthase | |
| aroK | BSU03150 | No | 2.7.1.71 | 2PT5 | A. aeolicus | Shikimate kinase | |
| Phe, Tyr | |||||||
| pheA | BSU27900 | No | 4.2.1.51 | 4LUB | S. mutans | Prephenate dehydratase | |
| hisC | BSU22620 | No | 2.6.1.9 | 3FFH | Listeria innocua | Histidinol-phosphate aminotransferase/tyrosine and phenylalanine aminotransferase | |
| aroH | BSU22690 | No | 1COM | B. subtilis | Chorismate mutase (isozymes 1 and 2) | ||
| tyrA | BSU22610 | No | 1.3.1.12 | 3DZB | Streptococcus thermophilus | Prephenate dehydrogenase | |
| Asn | |||||||
| asnB | BSU30540 | No | 6.3.5.4 | 1CT9 | E. coli | Asparagine synthase (glutamine hydrolyzing) | |
| Ala, Ser | |||||||
| alaT | BSU31400 | No | 2.6.1.- | 1DJU | P. horikoshii | Alanine aminotransferase | |
| serA | BSU23070 | No | 1.1.1.95 | 1YGY | Mycobacterium tuberculosis | Phosphoglycerate dehydrogenase | |
| serC | BSU10020 | No | 2.6.1.52 | 1W23 | Bacillus alcalophilus | 3-Phosphoserine aminotransferase | |
| ywtE | BSU35850 | No | 1NRW | B. subtilis | Putative phosphatase | ||
| Leu | |||||||
| yvbW | BSU34010 | No | 3GI9 | Methanocaldococcus jannaschii | Putative leucine permease | ||
| Gln | |||||||
| glnA | BSU17460 | No | 6.3.1.2 | 4S0R | B. subtilis | Glutamine synthetase | |
| Nucleotides/phosphate | |||||||
| PRPP | |||||||
| prs | BSU00510 | Yes | Yes | 2.7.6.1 | 1DKR | B. subtilis | Phosphoribosylpyrophosphate synthetase, universally conserved protein |
| Pyrimidine biosynthesis | |||||||
| pyrAA | BSU15510 | No | 6.3.5.5 | 1JDB | E. coli | Carbamoyl-phosphate synthetase (glutaminase subunit) | |
| pyrAB | BSU15520 | No | 6.3.5.5 | 1JDB | E. coli | Carbamoyl-phosphate synthetase (catalytic subunit) | |
| pyrB | BSU15490 | No | 2.1.3.2 | 3R7D | B. subtilis | Aspartate carbamoyltransferase | |
| pyrC | BSU15500 | No | 3.5.2.3 | 3MPG | B. anthracis | Dihydro-orotase | |
| pyrD | BSU15540 | No | 1.3.3.1 | 1EP1 | Lactococcus lactis | Dihydro-orotic acid dehydrogenase (catalytic subunit) | |
| pyrE | BSU15560 | No | 2.4.2.10 | 3M3H | B. anthracis | Orotate phosphoribosyltransferase | |
| pyrF | BSU15550 | No | 4.1.1.23 | 1DBT | B. subtilis | Orotidine 5′-phosphate decarboxylase | |
| cmk | BSU22890 | Yes | Yes | 2.7.4.14 | 1Q3T | S. pneumoniae | Cytidylate kinase (CMP, dCMP) |
| pyrG | BSU37150 | Yes | Yes | 6.3.4.2 | 1S1M | E. coli | CTP synthase (NH3, glutamine) |
| yncF | BSU17660 | No | 2XCD | B. subtilis | dUTPase | ||
| thyB | BSU21820 | No | 2.1.1.45 | 3IX6 | Brucella melitensis | Thymidylate synthase B | |
| tmk | BSU00280 | Yes | Yes | 2.7.4.9 | 2CCJ | S. aureus | Thymidylate kinase |
| Purine biosynthesis | |||||||
| purF | BSU06490 | No | 2.4.2.14 | 1GPH | B. subtilis | Glutamine phosphoribosyldiphosphate amidotransferase | |
| purD | BSU06530 | No | 6.3.4.13 | 2XD4 | B. subtilis | Phosphoribosylglycinamide synthetase | |
| purN | BSU06510 | No | 2.1.2.2 | 3AV3 | G. kaustophilus | Phosphoribosylglycinamide formyltransferase | |
| purS | BSU06460 | No | 1TWJ | B. subtilis | Phosphoribosylformylglycinamidine synthase | ||
| purQ | BSU06470 | No | 6.3.5.3 | 3D54 | T. maritima | Phosphoribosylformylglycinamidine synthase | |
| purL | BSU06480 | No | 6.3.5.3 | 3VIU | T. thermophilus | Phosphoribosylformylglycinamidine synthase | |
| purM | BSU06500 | No | 6.3.3.1 | 2BTU | B. anthracis | Phosphoribosylaminoimidazole synthetase | |
| purE | BSU06420 | No | 4.1.1.21 | 1XMP | B. anthracis | Phosphoribosylaminoimidazole carboxylase (ATP dependent) | |
| purK | BSU06430 | No | 4.1.1.21 | 4DLK | B. anthracis | Phosphoribosylaminoimidazole carboxylase (ATP dependent) | |
| purC | BSU06450 | No | 6.3.2.6 | 2YWV | G. kaustophilus | Phosphoribosylaminoimidazole succinocarboxamide synthase | |
| purB | BSU06440 | No | 4.3.2.2 | 1F1O | B. subtilis | Adenylsuccinate lyase | |
| purH | BSU06520 | No | 2.1.2.3 | 3ZZM | M. tuberculosis | Phosphoribosylaminoimidazole carboxamide formyltransferase | |
| guaB | BSU00090 | Yes | 1.1.1.205 | 3TSB | B. anthracis | IMP dehydrogenase | |
| guaA | BSU06360 | No | 6.3.5.2 | 1GPM | E. coli | GMP synthase (glutamine hydrolyzing) | |
| gmk | BSU15680 | Yes | Yes | 2.7.4.8 | 3TAU | L. monocytogenes | Guanylate kinase (GMP:dATP, dGMP:ATP) |
| purA | BSU40420 | No | 6.3.4.4 | 4M0G | B. anthracis | Adenylosuccinate synthetase | |
| adk | BSU01370 | Yes | Yes | 2.7.4.3 | 1P3J | B. subtilis | Adenylate kinase |
| Pyrimidine/purine biosynthesis | |||||||
| nrdE | BSU17380 | Yes | Yes | 1PEM | S. enterica | Ribonucleoside diphosphate reductase (major subunit) | |
| nrdF | BSU17390 | Yes | Yes | 4DR0 | B. subtilis | Ribonucleoside diphosphate reductase (major subunit) | |
| nrdI | BSU17370 | Yes | 1RLJ | B. subtilis | Ribonucleoside diphosphate reductase | ||
| ndk | BSU22730 | No | 2.7.4.6 | 2VU5 | B. anthracis | Nucleoside diphosphate kinase | |
| hprT | BSU00680 | Yes | Yes | 2.4.2.8 | 3H83 | B. anthracis | Hypoxanthine phosphoribosyltransferase |
| Phosphate | |||||||
| pit | BSU12840 | No | Low-affinity inorganic phosphate transporter | ||||
| Lipids | |||||||
| Malonyl-CoA synthesis | |||||||
| accC | BSU24340 | Yes | 6.3.4.14 | 2VPQ | S. aureus | Acetyl-CoA carboxylase (biotin carboxylase subunit) | |
| accB | BSU24350 | Yes | 6.4.1.2 | 4HR7 | E. coli | Acetyl-CoA carboxylase (biotin carboxyl carrier subunit) | |
| accA | BSU29200 | Yes | 6.4.1.2 | 2F9I | S. aureus | Acetyl-CoA carboxylase (alpha subunit) | |
| accD | BSU29210 | Yes | 6.4.1.2 | 2F9I | S. aureus | Acetyl-CoA carboxylase (beta subunit) | |
| birA | BSU22440 | Yes | 6.3.4.15 | 3RIR | S. aureus | Biotin protein ligase | |
| Acyl carrier | |||||||
| acpS | BSU04620 | Yes | 2.7.8.7 | 1F80 | B. subtilis | Acyl carrier protein synthase, 4′-phosphopantetheine transferase | |
| acpA | BSU15920 | Yes | 1F80 | B. subtilis | Acyl carrier protein | ||
| Aceto-acyl-Acp synthesis | |||||||
| fabD | BSU15900 | Yes | 2.3.1.39 | 3QAT | Bartonella henselae | Malonyl-CoA—acyl carrier protein transacylase | |
| fabHA | BSU11330 | No | 2.3.1.180 | 1ZOW | S. aureus | Beta-ketoacyl–acyl carrier protein synthase III | |
| β-Ketoacyl-Acp chain elongation | |||||||
| fabG | BSU15910 | Yes | 1.1.1.100 | 2UVD | B. anthracis | Beta-ketoacyl–acyl carrier protein reductase | |
| fabF | BSU11340 | Yes | 2.3.1.179 | 4LS5 | B. subtilis | Beta-ketoacyl–acyl carrier protein synthase II | |
| fabI | BSU11720 | No | 1.3.1.9 | 3OIF | B. subtilis | Enoyl-acyl carrier protein reductase | |
| ywpB | BSU36370 | Yes | 1U1Z | P. aeruginosa | β-Hydroxyacyl (acyl carrier protein) dehydratase | ||
| Phosphatidic acid synthesis | |||||||
| plsC | BSU09540 | Yes | 2.3.1.51 | Acyl-ACP:1-acylglycerolphosphate acyltransferase | |||
| plsX | BSU15890 | Yes | Yes | 1VI1 | B. subtilis | Acyl-ACP:phosphate acyltransferase | |
| plsY | BSU18070 | Yes | Yes | Acylphosphate:glycerol-phosphate acyltransferase | |||
| gpsA | BSU22830 | Yes | 1.1.1.94 | 1Z82 | T. maritima | Glycerol-3-phosphate dehydrogenase (NAD) | |
| Phosphatidylglycerol phosphate synthesis | |||||||
| cdsA | BSU16540 | Yes | Yes | 2.7.7.41 | 4Q2G | T. maritima | Phosphatidate cytidylyltransferase |
| pgsA | BSU16920 | Yes | Yes | 2.7.8.5 | Phosphatidylglycerophosphate synthase | ||
| Cofactors | |||||||
| ECF transporter (general component) for riboflavin, biotin, thiamine, tryptophan | |||||||
| ybxA | BSU01450 | No | Yes | 4HUQ | Lactobacillus brevis | ATP-binding A1 component of ECF transporters | |
| ybaE | BSU01460 | No | Yes | 4HUQ | L. brevis | ATP-binding A2 component of ECF transporters | |
| ybaF | BSU01470 | No | Yes | 4HUQ | L. brevis | Transmembrane T component of ECF transporters | |
| NAD | |||||||
| nadD | BSU25640 | Yes | 2.7.7.18 | 1KAM | B. subtilis | Nicotinamide-nucleotide adenylyltransferase | |
| nadE | BSU03130 | Yes | Yes | 6.3.1.5 | 1NSY | B. subtilis | NH3-dependent NAD+ synthetase |
| nadF | BSU11610 | Yes | Yes | 2.7.1.23 | 2I1W | L. monocytogenes | NAD kinase |
| niaP | BSU02950 | No | 4J05 | Piriformospora indica | Nicotinate transporter | ||
| pncB | BSU31750 | Yes | 2.4.2.11 | 2F7F | E. faecalis | Putative nicotinate phosphoribosyltransferase | |
| Riboflavin/FAD | |||||||
| ribC | BSU16670 | Yes | 2.7.1.26 | 3OP1 | S. pneumoniae | Riboflavin kinase/FAD synthase | |
| ribU | BSU23050 | No | 3P5N | S. aureus | Riboflavin ECF transporter, S protein | ||
| Pyridoxal phosphate | |||||||
| pdxS | BSU00110 | No | 2NV2 | B. subtilis | Pyridoxal-5′-phosphate synthase (synthase domain) | ||
| pdxT | BSU00120 | No | 2NV2 | B. subtilis | Pyridoxal-5′-phosphate synthase (glutaminase domain) | ||
| Biotin | |||||||
| yhfU | BSU10370 | No | 4DVE | L. lactis | S protein of biotin ECF transporter | ||
| Thiamine, TPP | |||||||
| yloS | BSU15800 | No | 2.7.6.2 | 3LM8 | B. subtilis | Thiamine pyrophosphokinase | |
| thiT | BSU30990 | No | 4MES | L. lactis | S protein of thiamine ECF transporter | ||
| Lipoate | |||||||
| gcvH | BSU32800 | No | 3IFT | M. tuberculosis | Glycine cleavage system protein H, 2-oxo acid dehydrogenase | ||
| lipM | BSU24530 | No | 3A7A | E. coli | Octanoyltransferase | ||
| lipL | BSU37640 | No | 2P5I | B. halodurans | GcvH:E2 amidotransferase | ||
| lipA | BSU32330 | Yes | 2.8.1.8 | 4U0O | T. elongatus | Lipoic acid synthase | |
| CoA | |||||||
| ykpB | BSU14440 | No | 3HN2 | Geobacter metallireducens | Putative ketopantoate reductase | ||
| panD | BSU22410 | No | 4.1.1.11 | 2C45 | M. tuberculosis | Aspartate 1-decarboxylase | |
| panC | BSU22420 | No | 6.3.2.1 | 2X3F | S. aureus | Pantothenate synthase | |
| panB | BSU22430 | No | 2.1.2.11 | 1M3U | E. coli | 3-Methyl-2-oxobutanoate hydroxymethyltransferase | |
| ybgE | BSU02390 | No | 2.6.1.42 | 3HT5 | M. tuberculosis | Branched-chain amino acid aminotransferase | |
| coaA | BSU23760 | No | 2.7.1.33 | 4F7W | K. pneumoniae | Probable pantothenate kinase | |
| yloI | BSU15700 | No | 4.1.1.36 | 1U7U | E. coli | Coenzyme A biosynthesis bifunctional protein CoaBC | |
| ylbI | BSU15020 | No | 2.7.7.3 | 1O6B | B. subtilis | Pantetheine-phosphate adenylyltransferase | |
| ytaG | BSU29060 | Yes | 2.7.1.24 | 4TTP | Legionella pneumophila | Dephospho-CoA kinase | |
| SAM | |||||||
| metK | BSU30550 | Yes | Yes | 2.5.1.6 | 1FUG | E. coli | S-Adenosylmethionine synthetase |
| Folate | |||||||
| folE | BSU22780 | No | 3.5.4.16 | 4UQF | L. monocytogenes | GTP cyclohydrolase I | |
| phoB | BSU05740 | No | 3.1.3.1 | 3A52 | Shewanella sp. | Alkaline phosphatase A | |
| folB | BSU00780 | No | 4.1.2.25 | 1RRI | S. aureus | Dihydroneopterin aldolase | |
| folK | BSU00790 | No | 2.7.6.3 | 4CYU | S. aureus | 2-Amino-4-hydroxy-6-hydroxymethyl-dihydropteridine diphosphokinase | |
| sul | BSU00770 | No | 2.5.1.15 | 1TWS | B. anthracis | Dihydropteroate synthase | |
| folC | BSU28080 | No | Yes | 6.3.2.17 | 1O5Z | T. maritima | Folyl-polyglutamate synthetase |
| dfrA | BSU21810 | Yes | 1.5.1.3 | 1ZDR | G. stearothermophilus | Dihydrofolate reductase | |
| pabB | BSU00740 | No | 2.6.1.85 | 5CWA | M. tuberculosis | p-Aminobenzoate synthase (subunit A) | |
| pabA | BSU00750 | No | 2.6.1.85 | 1I1Q | S. enterica serovar Typhimurium | p-Aminobenzoate synthase (subunit B)/anthranilate synthase (subunit II) | |
| pabC | BSU00760 | No | 4.1.3.38 | 4WHX | Burkholderia pseudomallei | Aminodeoxychorismate lyase | |
| gsaB | BSU08710 | No | 5.4.3.8 | 3L44 | B. anthracis | Formate dehydrogenase | |
| ykkE | BSU13110 | No | 3.5.1.10 | 3W7B | T. thermophilus | Formyltetrahydrofolate deformylase | |
| yoaE | BSU18570 | No | Formate dehydrogenase | ||||
| Heme biosynthesis | |||||||
| hemE | BSU10120 | No | 4.1.1.37 | 2INF | B. subtilis | Glutamate-1-semialdehyde aminotransferase | |
| hemH | BSU10130 | No | 1C1H | B. subtilis | Uroporphyrinogen decarboxylase (uroporphyrinogen III) | ||
| hemY | BSU10140 | No | 1.3.3.4 | 3I6D | B. subtilis | Ferrochelatase | |
| ctaA | BSU14870 | No | Protoporphyrinogen IX oxidase | ||||
| ctaB | BSU14880 | No | Heme A synthase | ||||
| hemL | BSU28120 | No | 5.4.3.8 | 3BS8 | B. subtilis | Heme O synthase (major enzyme) | |
| hemB | BSU28130 | No | 4.2.1.24 | 1W5Q | P. aeruginosa | Glutamate-1-semialdehyde aminotransferase | |
| hemD | BSU28140 | No | 4.2.1.75 | Porphobilinogen synthase | |||
| hemC | BSU28150 | No | 2.5.1.61 | 4MLQ | Bacillus megaterium | Uroporphyrinogen III synthase | |
| hemX | BSU28160 | No | Hydroxymethylbilane synthase | ||||
| hemA | BSU28170 | No | 1.2.1.70 | 4N7R | A. thaliana | Glutamyl-tRNA reductase | |
| hemQ | BSU37670 | No | 1T0T | G. stearothermophilus | Heme-binding protein | ||
| Menaquinone | |||||||
| menA | BSU38490 | Yes | Probable 1,4-dihydroxy-2-naphthoate octaprenyltransferase | ||||
| menH | BSU22750 | No | 4OBW | Saccharomyces cerevisiae | Menaquinone biosynthesis methyltransferase | ||
| menC | BSU30780 | Yes | 4.2.1.113 | 1WUE | E. faecalis | O-Succinylbenzoate-CoA synthase | |
| menE | BSU30790 | Yes | 6.2.1.26 | 5BUQ | B. subtilis | O-Succinylbenzoate-CoA ligase | |
| menB | BSU30800 | Yes | 4.1.3.36 | 2IEX | G. kaustophilus | Naphthoate synthase | |
| menD | BSU30820 | Yes | 2.2.1.9 | 2X7J | B. subtilis | 2-Succinyl-6-hydroxy-2,4-cyclohexadiene-1-carboxylate synthase/2-oxoglutarate decarboxylase | |
| ytxM | BSU30810 | No | 2XMZ | S. aureus | Similar to prolyl aminopeptidase | ||
| menF | BSU30830 | No | 5.4.4.2 | 3HWO | E. coli | Menaquinone-specific isochorismate synthase | |
| Metals and iron-sulfur clusters | |||||||
| Sodium export | |||||||
| mrpA | BSU31600 | Yes | 4HE8 | T. thermophilus | Na+/H+ antiporter subunit | ||
| mrpB | BSU31610 | Yes | Na+/H+ antiporter subunit | ||||
| mrpC | BSU31620 | Yes | Na+/H+ antiporter subunit | ||||
| mrpD | BSU31630 | Yes | 4HE8 | T. thermophilus | Na+/H+ antiporter subunit | ||
| mrpE | BSU31640 | No | Na+/H+ antiporter subunit | ||||
| mrpF | BSU31650 | Yes | Na+/H+ antiporter subunit | ||||
| mrpG | BSU31660 | No | Na+/H+ antiporter subunit | ||||
| Potassium | |||||||
| ktrD | BSU13500 | No | Yes | 4J7C | B. subtilis | Potassium transporter KtrCD | |
| ktrC | BSU14510 | No | Yes | 4J90 | B. subtilis | Potassium transporter KtrCD | |
| Iron | |||||||
| efeO | BSU38270 | No | 3AT7 | Sphingomonas sp. | Lipoprotein, elemental iron uptake system (binding protein) | ||
| efeU | BSU38280 | No | Elemental iron uptake system (permease) | ||||
| yfmF | BSU07490 | No | 4G1U | Yersinia pestis | Iron/citrate ABC transporter (ATP-binding protein) | ||
| yfmE | BSU07500 | No | 4G1U | Y. pestis | Iron/citrate ABC transporter (permease) | ||
| yfmD | BSU07510 | No | 4G1U | Y. pestis | Iron/citrate ABC transporter (permease) | ||
| yfmC | BSU07520 | No | 3EIW | S. aureus | Iron/citrate ABC transporter (binding protein) | ||
| yhfQ | BSU10330 | No | 3EIW | S. aureus | Iron/citrate ABC transporter (solute-binding protein) | ||
| Magnesium | |||||||
| mgtE | BSU13300 | No | 2YVX | T. thermophilus | Primary magnesium transporter | ||
| mntH | BSU04360 | No | 4WGV | Staphylococcus capitis | Manganese transporter (proton symport) | ||
| Zinc | |||||||
| znuA | BSU02850 | No | 2O1E | B. subtilis | ABC transporter for zinc (binding protein) | ||
| znuC | BSU02860 | No | 4YMS | Caldanaerobacter subterraneus | ABC transporter for zinc (ATP-binding protein) | ||
| znuB | BSU02870 | No | ABC transporter for zinc (permease) | ||||
| Copper | |||||||
| ycnJ | BSU03950 | No | Copper transporter | ||||
| Fe-S cluster | |||||||
| sufB | BSU32670 | Yes | 5AWF | E. coli | Synthesis of Fe-S clusters | ||
| sufU | BSU32680 | Yes | Yes | 1XJS | B. subtilis | Iron-sulfur cluster scaffold protein | |
| sufD | BSU32700 | Yes | 5AWF | E. coli | Synthesis of Fe-S clusters | ||
| sufS | BSU32690 | Yes | Yes | 2.8.1.7 | 1T3I | Synechocystis sp. | Cysteine desulfurase |
| sufC | BSU32710 | Yes | 2D2E | T. thermophilus | ABC transporter (ATP-binding protein) | ||
| fra | BSU05750 | No | 2OC6 | B. subtilis | Frataxin-like protein | ||
| yutI | BSU32220 | No | 1XHJ | Staphylococcus epidermidis | Putative iron-sulfur scaffold protein | ||
| Cell division | |||||||
| Cell wall synthesis | |||||||
| Synthesis of d-glutamate | |||||||
| racE | BSU28390 | Yes | 5.1.1.3 | 1ZUW | B. subtilis | Glutamate racemase | |
| Synthesis of d-Ala-d-Ala | |||||||
| alr | BSU04640 | Yes | 5.1.1.1 | 3ZM5 | S. pneumoniae | Alanine racemase | |
| ddl | BSU04560 | Yes | 6.3.2.4 | 2I80 | S. aureus | d-Alanine-d-alanine ligase | |
| Synthesis of m-diaminopimelate | |||||||
| dapG | BSU16760 | No | 2.7.2.4 | 1SFT | G. stearothermophilus | Aspartokinase I (alpha and beta subunits) | |
| asd | BSU16750 | Yes | 1.2.1.11 | 2GYY | S. pneumoniae | Aspartate-semialdehyde dehydrogenase | |
| dapA | BSU16770 | Yes | 4.2.1.52 | 1XKY | B. anthracis | Dihydrodipicolinate synthase | |
| dapB | BSU22490 | Yes | 1.3.1.26 | 5EER | Corynebacterium glutamicum | Dihydrodipicolinate reductase (NADPH) | |
| ykuQ | BSU14180 | Yes | 2.3.1.89 | 3R8Y | B. anthracis | Similar to tetrahydrodipicolinate succinylase | |
| patA | BSU14000 | Yes | 1GDE | P. horikoshii | Aminotransferase | ||
| dapI | BSU14190 | Yes | 3.5.1.47 | 1YSJ | B. subtilis | N-Acetyl-diaminopimelate deacetylase | |
| dapF | BSU32170 | Yes | 5.1.1.7 | 2OTN | B. anthracis | Diaminopimelate epimerase | |
| Isoprenoid biosynthesis | |||||||
| dxs | BSU24270 | Yes | 2.2.1.7 | 2O1S | E. coli | 1-Deoxyxylulose-5-phosphate synthase | |
| ispC | BSU16550 | Yes | 1.1.1.267 | 1R0K | Zymomonas mobilis | 1-Deoxy-d-xylulose-5-phosphate reductoisomerase | |
| ispD | BSU00900 | Yes | 2.7.7.60 | 2YC3 | A. thaliana | 2-C-Methyl-d-erythritol 4-phosphate cytidylyltransferase | |
| ispE | BSU00460 | Yes | 2.7.1.148 | 3PYD | M. tuberculosis | 4-Diphosphocytidyl-2-C-methyl-d-erythritol kinase | |
| ispF | BSU00910 | Yes | 4.6.1.12 | 3GHZ | S. enterica serovar Typhimurium | 2-C-Methyl-d-erythritol-2,4-cyclodiphosphate synthase | |
| ispG | BSU25070 | Yes | 3NOY | A. aeolicus | Similar to peptidoglycan acetylation, 1-hydroxy-2-methyl-2-(E)-butenyl-4-diphosphate synthase | ||
| ispH | BSU25160 | Yes | 4N7B | Plasmodium falciparum | (E)-4-Hydroxy-3-methylbut-2-enyl diphosphate reductase | ||
| fni | BSU22870 | No | 5.3.3.2 | 1P0K | B. subtilis | Isopentenyl diphosphate isomerase | |
| Undecaprenyl phosphate biosynthesis | |||||||
| yqiD | BSU24280 | No | 2.5.1.10 | 1RTR | S. aureus | Geranyltransferase | |
| uppS | BSU16530 | Yes | 2.5.1.31 | 1F75 | Micrococcus luteus | Probable undecaprenyl pyrophosphate synthetase | |
| bcrC | BSU36530 | No | Undecaprenyl pyrophosphate phosphatase | ||||
| hepT | BSU22740 | Yes | 2.5.1.30 | 3AQB | M. luteus | Heptaprenyl diphosphate synthase component II | |
| hepS | BSU22760 | Yes | 2.5.1.30 | Heptaprenyl diphosphate synthase component I | |||
| Peptidoglycan biosynthesis | |||||||
| glmS | BSU01780 | Yes | 2.6.1.16 | 4AMV | E. coli | Glutamine:fructose-6-phosphate transaminase | |
| glmM | BSU01770 | Yes | 5.4.2.10 | 3PDK | B. anthracis | Phosphoglucosamine mutase | |
| gcaD | BSU00500 | Yes | 2.7.7.23 | 4AAW | S. pneumoniae | UDP-N-acetylglucosamine pyrophosphorylase | |
| murAA | BSU36760 | Yes | 2.5.1.7 | 3SG1 | B. anthracis | UDP-N-acetylglucosamine 1-carboxyvinyltransferase | |
| murB | BSU15230 | Yes | 1.1.1.158 | 4PYT | Unidentified | UDP-N-acetylenolpyruvoylglucosamine reductase | |
| murC | BSU29790 | Yes | 6.3.2.8 | 1GQQ | H. influenzae | UDP-N-acetylmuramoyl-l-alanine synthetase | |
| murD | BSU15200 | Yes | 6.3.2.9 | 3LK7 | Streptococcus agalactiae | UDP-N-acetylmuramoyl-l-alanyl-d-glutamate synthetase | |
| murE | BSU15180 | Yes | 6.3.2.13 | 4C13 | S. aureus | UDP-N-acetylmuramoyl-l-alanyl-d-glutamyl-meso-2,6-diaminopimelate synthetase | |
| murF | BSU04570 | Yes | 6.3.2.10 | 1GG4 | E. coli | UDP-N-acetylmuramoyl-l-alanyl-d-glutamyl-meso-2,6-diaminopimeloyl-d-alanyl-d-alanine synthetase | |
| mraY | BSU15190 | Yes | 2.7.8.13 | 4J72 | A. aeolicus | Phospho-N-acetylmuramoyl-pentapeptide transferase (meso-2,6-diaminopimelate) | |
| murG | BSU15220 | Yes | 2.4.1.227 | 1F0K | E. coli | UDP-N-acetylglucosamine-N-acetylmuramyl-(pentapeptide)pyrophosphoryl-undecaprenol N-acetylglucosamine transferase | |
| murJ | BSU30050 | No | Lipid II flippase | ||||
| Peptidoglycan polymerization | |||||||
| ponA | BSU22320 | No | 3DWK | S. aureus | Penicillin-binding protein 1A/1B | ||
| PG cross-links, cell separation | |||||||
| pbpB | BSU15160 | Yes | 1RP5 | S. pneumoniae | Penicillin-binding protein 2B | ||
| pbpA | BSU25000 | No | 3VSK | S. aureus | Penicillin-binding protein 2A | ||
| lytE | BSU09420 | No | 4XCM | T. thermophilus | Cell wall hydrolase (major autolysin) for cell elongation and separation | ||
| lytF | BSU09370 | No | 4XCM | T. thermophilus | Gamma-d-glutamate-meso-diaminopimelate muropeptidase (major autolysin) | ||
| Wall teichoic acid | |||||||
| tagO | BSU35530 | Yes | 4J72 | A. aeolicus | Undecaprenyl phosphate-GlcNAc-1-phosphate transferase | ||
| mnaA | BSU35660 | Yes | 5.1.3.14 | 4FKZ | B. subtilis | UDP-N-acetylglucosamine 2-epimerase | |
| tagA | BSU35750 | Yes | 2.4.1.187 | UDP-N-acetyl-d-mannosamine transferase | |||
| tagB | BSU35760 | Yes | 3L7I | S. epidermidis | Putative CDP-glycerol:glycerol phosphate glycerophosphotransferase | ||
| tagD | BSU35740 | Yes | 2.7.7.39 | 1COZ | B. subtilis | Glycerol-3-phosphate cytidylyltransferase | |
| tagF | BSU35720 | Yes | 2.7.8.12 | 3L7I | S. epidermidis | CDP-glycerol:polyglycerol phosphate glycerophosphotransferase | |
| tagH | BSU35700 | Yes | 3.6.3.40 | ABC transporter for teichoic acid translocation (ATP-binding protein) | |||
| tagG | BSU35710 | Yes | ABC transporter for teichoic acid translocation (permease) | ||||
| tagU | BSU35650 | No | 3OWQ | L. innocua | Phosphotransferase, attachment of anionic polymers to peptidoglycan | ||
| Lipoteichoic acid | |||||||
| dgkB | BSU06720 | Yes | 2QV7 | S. aureus | Diacylglycerol kinase | ||
| pgcA | BSU09310 | No | Yes | 5.4.2.2 | 2Z0F | T. thermophilus | Alpha-phosphoglucomutase |
| gtaB | BSU35670 | No | Yes | 2.7.7.9 | 2UX8 | Sphingomonas elodea | UTP-glucose-1-phosphate uridylyltransferase |
| ltaS | BSU07710 | No | 2W8D | B. subtilis | Lipoteichoic acid synthase | ||
| ugtP | BSU21920 | No | UDP-glucose diacylglycerol glucosyltransferase | ||||
| Coordination | |||||||
| Divisome | |||||||
| divIC | BSU00620 | Yes | Cell division initiation protein (septum formation) | ||||
| ftsL | BSU15150 | Yes | Cell division protein (septum formation) | ||||
| divIB | BSU15240 | Yes | 1YR1 | G. stearothermophilus | Cell division initiation protein (septum formation) | ||
| ftsZ | BSU15290 | Yes | 2VAM | B. subtilis | Cell division initiation protein (septum formation) | ||
| ftsW | BSU14850 | Yes | Cell division protein | ||||
| ezrA | BSU29610 | No | 4UXV | B. subtilis | Negative regulator of FtsZ ring formation | ||
| sepF | BSU15390 | No | 3ZIH | B. subtilis | Part of the divisome | ||
| gpsB | BSU22180 | No | 4UG3 | B. subtilis | Removal of PBP1 from the cell pole after completion of cell pole maturation | ||
| yvcK | BSU34760 | No | 2HZB | B. halodurans | Correct localization of PBP1, essential for growth under gluconeogenic conditions | ||
| yvcL | BSU34750 | No | Yes | 3HYI | T. maritima | Involved in Z-ring assembly | |
| Division site selection | |||||||
| divIVA | BSU15420 | No | 2WUJ | B. subtilis | Cell division initiation protein (septum placement) | ||
| minC | BSU28000 | No | 2M4I | B. subtilis | Cell division inhibitor (septum placement) | ||
| minD | BSU27990 | No | 4V03 | A. aeolicus | Cell division inhibitor (septum placement) | ||
| noc | BSU40990 | No | 1VZ0 | T. thermophilus | DNA-binding protein, spatial regulator of cell division to protect the nucleoid, coordination of chromosome segregation and cell division | ||
| minJ | BSU35220 | No | Topological determinant of cell division | ||||
| Elongasome | |||||||
| mreD | BSU28010 | Yes | Cell shape-determining protein, associated with the MreB cytoskeleton | ||||
| mreC | BSU28020 | Yes | 2J5U | L. monocytogenes | Cell shape-determining protein, associated with the MreB cytoskeleton | ||
| mreB | BSU28030 | Yes | 4CZE | Caulobacter vibrioides | Cell shape-determining protein | ||
| rodA | BSU38120 | Yes | Control of cell shape and elongation | ||||
| mreBH | BSU14470 | No | 1JCF | T. maritima | Cell shape-determining protein | ||
| rodZ | BSU16910 | Maybe | Required for cell shape determination | ||||
| Coordination of cell division and DNA replication | |||||||
| walJ | BSU40370 | No | 4P62 | P. aeruginosa | Coordination of cell division and DNA replication | ||
| Signaling | |||||||
| walK | BSU40400 | Yes | 4I5S | S. mutans | Two-component sensor kinase | ||
| walR | BSU40410 | Yes | 2ZWM | B. subtilis | Two-component response regulator | ||
| cdaA | BSU01750 | No | 2.7.7.85 | 4RV7 | L. monocytogenes | Diadenylate cyclase | |
| gdpP | BSU40510 | No | c-di-AMP-specific phosphodiesterase | ||||
| Integrity of the cell | |||||||
| Protection | |||||||
| ytbE | BSU29050 | Yes | 3B3D | B. subtilis | Putative aldo/keto reductase | ||
| katA | BSU08820 | No | 1SI8 | E. faecalis | Vegetative catalase 1 | ||
| sodA | BSU25020 | No | 2RCV | B. subtilis | Superoxide dismutase | ||
| ahpC | BSU40090 | No | 1WE0 | Amphibacillus xylanus | Alkyl hydroperoxide reductase (small subunit) | ||
| ahpF | BSU40100 | No | 4O5Q | E. coli | Alkyl hydroperoxide reductase (large subunit)/NADH dehydrogenase | ||
| trxA | BSU28500 | Yes | Yes | 2GZY | B. subtilis | Antioxidative action by facilitating the reduction of other proteins by cysteine thiol-disulfide exchange | |
| yumC | BSU32110 | Yes | 1.18.1.2 | 3LZW | B. subtilis | Ferredoxin-NAD(P)+ oxidoreductase | |
| trxB | BSU34790 | Yes | Yes | 1.8.1.9 | 4GCM | S. aureus | Thioredoxin reductase (NADPH) |
| Repair/genome integrity | |||||||
| hlpB | BSU10660 | Yes | HNH nuclease-like protein, rescues AddA recombination intermediates | ||||
| mutY | BSU08630 | No | 5DPK | G. stearothermophilus | A/G-specific adenine glycosylase | ||
| polY1 | BSU23870 | No | 2.7.7.7 | 4IRC | E. coli | Translesion synthesis DNA polymerase Y1 | |
| mutM | BSU29080 | No | 3.2.2.23 | 1L1T | G. stearothermophilus | Formamidopyrimidine-DNA glycosidase | |
| mfd | BSU00550 | No | 2EYQ | E. coli | Transcription repair-coupling factor | ||
| recD2 | BSU27480 | No | 3E1S | Deinococcus radiodurans | 5′–3′ DNA helicase replication fork progression | ||
| rnhB | BSU16060 | No | 3O3G | T. maritima | RNase HII, endoribonuclease | ||
| recA | BSU16940 | No | 1UBC | Mycobacterium smegmatis | Homologous recombination and DNA repair | ||
| Other/unknown | |||||||
| ppaC | BSU40550 | Yes | 3.6.1.1 | 1WPM | B. subtilis | Inorganic pyrophosphatase | |
| ylaN | BSU14840 | Yes | 2ODM | S. aureus | Unknown | ||
| yitI | BSU11000 | No | 2JDC | Bacillus licheniformis | Unknown | ||
| yitW | BSU11160 | No | 3LNO | B. anthracis | Unknown | ||
| yqhY | BSU24330 | No | Unknown | ||||
| ykwC | BSU13960 | No | 3WS7 | Pyrobaculum calidifontis | Putative beta-hydroxy acid dehydrogenase | ||
| ylbN | BSU15070 | No | Unknown | ||||
| ypfD | BSU22880 | No | 4Q7J | E. coli | Similar to ribosomal protein S1 | ||
| yugI | BSU31390 | No | 2K4K | B. subtilis | Similar to polyribonucleotide nucleotidyltransferase | ||
| floT | BSU31010 | No | Similar to flotillin 1, orchestration of physiological processes in lipid microdomains | ||||
| yyaF | BSU40920 | No | Yes | 1JAL | H. influenzae | GTP-binding protein/GTPase |
The BSU number is the locus tag in the context of the B. subtilis 168 genome (GenBank accession no. NC_000964).
The essentiality of a gene refers to wild-type B. subtilis 168. By definition, genes that cannot be deleted as individual genes under defined optimal growth conditions (Luria-Bertani broth with glucose at 37°C) are regarded as being essential.
Each gene of the MiniBacillus gene set was tested for the presence of a homolog in Mycoplasma mycoides JCVI-syn3.0 by using a BLASTP analysis.
The organism refers to the PDB accession number.
PRPP, phosphoribosyl pyrophosphate; TPP, thiamine PPi; SAM, S-adenosylmethionine; PTS, phosphotransferase system; PBP1, penicillin-binding protein 1.
TABLE 3.
rRNAs and tRNAs
DNA Replication and Chromosome Segregation/Maintenance
A set of 18 genes was selected as being important for DNA replication. Among these 18 genes are 15 essential genes, which are absolutely required for growth of B. subtilis. With the exception of the essential NAD-dependent DNA ligase LigA, all these essential proteins are members of one or both of the protein complexes that catalyze DNA replication, i.e., the primosome and the replisome (45, 46). In addition to these essential proteins, we have added the replication termination protein Rtp, the inhibitor of DnaA oligomerization YabA, and the DNA polymerase I PolA to the list. Rtp is required for the correct termination of DNA replication and the subsequent segregation of daughter chromosomes (47). YabA, on the other hand, controls replication initiation by inhibiting the oligomerization of the initiator protein DnaA (48). Finally, DNA polymerase I fulfills an essential function, the removal of RNA primers that initiate the synthesis of Okazaki fragments. This function can be taken over by the paralog YpcP, and one of the two proteins has to be present for viability of B. subtilis. While YpcP has only the 5′-to-3′ exonuclease domain required for the removal of RNA primers, polymerase I is also capable of filling the resulting gaps (49). This additional property as well as the genetic linkage with the mutM gene, which is necessary for DNA integrity (see below), suggested that PolA should be kept in the minimal genome. Interestingly, B. subtilis needs either PolA or YpcP, whereas the M. mycoides minimal strain JCVI-syn3.0 encodes both proteins.
After DNA replication, the daughter chromosomes have to be efficiently distributed to the daughter cells. Moreover, correct chromosome condensation is essential for all biological processes that involve DNA. Chromosome segregation and condensation are highly overlapping functions; i.e., several of the proteins mentioned below are involved in both activities (for a review, see reference 50). This functional group encompasses 13 proteins, 9 of which are essential. Seven of the corresponding essential genes, but none of the nonessential genes, are present in the genome of M. mycoides JCVI-syn3.0. After the initiation of DNA replication, the newly formed origin regions are bound and separated by the condensin protein complex. The condensin complex is composed of two subunits of the Smc protein and monomers of ScpA and ScpB (51, 52). The essential topoisomerase IV formed by ParC and ParE is required for both DNA condensation and segregation (53). For the separation and segregation of the chromosome terminus, the DNA translocases SpoIIIE and SftA and the site-specific recombinases CodV and RipX are necessary. In principle, SpoIIIE and SftA as well as CodV and RipX are paralogous proteins, and one protein might be sufficient for each function. However, as we are aiming for a well-growing strain that is not significantly compromised in its major cellular functions, and as synergistic activities of these proteins were recently shown (54–56), we decided to keep all four genes. DNA topology is maintained by the interplay of gyrase and topoisomerase I activities, encoded by the essential gyrAB and topA genes, respectively (28, 57). Moreover, the histone-like protein HBsu nonspecifically binds the DNA and is involved in DNA packaging (58).
Transcription
Eight proteins, among them five essential proteins, are required for transcription. Importantly, this activity requires the RNA polymerase, which consists of the essential core subunits (RpoA, RpoB, and RpoC) and sigma factor A (SigA) for promoter binding and recognition. Moreover, we have included the RNA polymerase-interacting protein HelD and the nonessential delta subunit (RpoE). HelD binding stimulates transcription in an RpoE-dependent manner, suggesting that these two accessory proteins are important to allow rapid growth (59, 60). Interestingly, both proteins are absent from the rather slowly growing M. mycoides strain JCVI-syn3.0. Finally, GreA and the essential NusA protein are required for transcription elongation and termination, respectively (61, 62). In contrast, the transcription termination protein Rho does not affect the growth of B. subtilis in rich medium (63) and therefore has not been included in the list.
mRNA Folding and Degradation
Once the RNA has been formed by the RNA polymerase, it has to adopt a structure that is compatible with its function. The rRNAs and tRNAs adopt complex three-dimensional structures, whereas the mRNAs have to be unstructured to allow access to the ribosomes. Bacterial cells have to adapt very rapidly to changes of external conditions. A major component of this rapid adaptation is the rapid degradation of bacterial mRNAs, which have average half-lives in the minute range. RNA degradation is accomplished by a set of RNases (64). In total, six proteins are required for the proper folding and degradation of RNA. To keep RNA molecules unstructured, the cell possesses so-called RNA chaperones. Two of these RNA chaperones, CspB and CspD, belong to the most abundant proteins in B. subtilis during growth in LB medium with glucose at 37°C (43, 65). Moreover, cspD is also one of the most highly expressed genes of B. subtilis under more than 100 different conditions (39). These data suggest that these RNA chaperones should be included in the minimal cell. In contrast, we decided to exclude all DEAD-box RNA helicases since B. subtilis has no growth defect even in the absence of all four helicases at 37°C (66). Conflicting reports are available for RNases in B. subtilis. The endoribonuclease RNase Y and the 5′-to-3′ exonuclease RNase J1 have long been considered to be essential, whereas a recent report suggests that these proteins are dispensable (34, 67–69). However, the strong expression of the corresponding genes suggests that they should be part of a minimal cell. The same is true for the 3′-to-5′ exoribonuclease PnpA, which also belongs to the most abundant proteins. Moreover, PnpA has also been implicated in DNA repair (70). Finally, we decided to include the nano-RNase NrnA, which degrades and thus recycles bi- and oligoribonucleotides (71). This enzyme is conserved in all groups of mollicutes despite their significant genome reduction (42). This strong conservation is suggestive of an important function and justifies the inclusion of this protein in the minimal cell. Moreover, RNases J1 and Y as well as the nano-RNase NrnA are also encoded by M. mycoides JCVI-syn3.0.
Translation
A large set of 133 proteins and 118 RNAs is required for translation. This includes aminoacyl-tRNA synthetases, ribosomal proteins, rRNAs, tRNAs, proteins involved in rRNA and tRNA modification and maturation, ribosome biogenesis factors, and translation factors.
Twenty-four proteins constitute the set of 20 aminoacyl-tRNA synthetases (72). With the exception of ThrS, the threonyl-tRNA synthetase, all of these enzymes are essential in B. subtilis. ThrS has a weakly expressed paralog, and one of these proteins is required for the viability of B. subtilis (36). Similarly, the enzyme for tyrosine, TyrS, has a paralog. However, this paralog is not expressed under the most common conditions, rendering TyrS essential (73). It should be noted that the aminoacyl-tRNA synthetases for glycine and phenylalanine are composed of two subunits, and the transamidosome for the formation of glutamine-tRNA is composed of the three subunits (GatA, GatB, and GatC) of the glutamyl-tRNA(Gln) amidotransferase, the glutamyl-tRNA synthetase GltX, and glutamine-specific tRNA (74). In the artificial minimal organism M. mycoides JCVI-syn3.0, there are no paralogs of the genes encoding the aminoacyl-tRNA synthetases for glycine and proline. It is tempting to speculate that these functions are fulfilled by some of the 149 unknown proteins encoded by this genome.
The B. subtilis ribosome consists of 53 proteins under standard conditions, among them 33 and 20 in the large and small subunits, respectively. Several of these proteins are nonessential; however, this has been tested only with single-gene inactivation (75). It is therefore conceivable that the simultaneous lack of multiple ribosomal proteins would have a significant impact on viability. In addition, B. subtilis encodes auxiliary ribosomal proteins that are synthesized only under stress conditions or that replace zinc-containing proteins during zinc limitation (76–78). All these proteins were excluded from the list (Table 2). The genome of the artificial organism M. mycoides JCVI-syn3.0 lacks genes for the essential protein L30 (RpmD) as well as those for 10 nonessential ribosomal proteins, mainly of the large ribosomal subunit.
B. subtilis has 10 operons for rRNA. All these operons also contain genes for tRNAs. Moreover, several tRNAs are encoded by scattered genes. It is well established that the redundancy of rRNAs and tRNAs is an important determinant of the growth rate (79); therefore, we included all 30 and 86 genes encoding rRNAs and tRNAs, respectively, in the minimal genome. However, reduction of the copy numbers of rRNA and tRNA genes remains to be explored to achieve a robust minimal translation machinery.
To be functional, rRNAs and tRNAs have to be processed and modified. In the minimal gene set, we include 31 proteins and 1 RNA for rRNA and tRNA maturation/modification, respectively. Since the functional RNAs are usually expressed as parts of large operons, proper processing is the first important step in their maturation. It should be noted that RNases Y and J1 (see above) participate in the processing and maturation of these functional RNA molecules (80, 81). Moreover, RNases P, PH, and Z are involved in rRNA and tRNA processing. Of these, RNase P (composed of a protein and the catalytically active RNA component) and RNase Z are essential (82, 83). In addition, RimM and RbfA are important for ribonucleolytic 16S rRNA maturation (84, 85). rRNAs and tRNAs are subject to multiple and highly diverse modifications, including methylation, thiouridylation, and pseudouridylation. The modification of tRNAs is one of the functions that still needs substantial research effort. The proteins important for these activities have been derived from the SubtiWiki database and from a recent study on the minimal translation apparatus (33, 42). As shown in Table 2, there is a good match with the corresponding set of genes in M. mycoides JCVI-syn3.0 (10).
Nine proteins, including six essential proteins, are involved in ribosome maturation and assembly. The three nonessential proteins process or modify ribosomal proteins. The six essential proteins are all GTPases that participate in different aspects of ribosome assembly (86, 87). Four of the genes encoding these essential GTPases are also part of the genome of M. mycoides JCVI-syn3.0.
Eleven proteins function as translation factors in different steps of translation, i.e., initiation, elongation, peptide chain release, and ribosome recycling. Of these proteins, nine are essential. Of the nonessential proteins, Efp is important for the efficient translation of proteins containing multiple consecutive proline residues, among them three essential proteins (Fmt, TopA, and ValS) (see http://subtiwiki.uni-goettingen.de/wiki/index.php/Efp-dependent_proteins) (33, 88, 89). With the exception of peptide chain release factor 2 (PrfB), but including elongation factor P (Efp), all of these proteins are encoded by the genome of M. mycoides JCVI-syn3.0.
In addition, five proteins and one RNA are important for translation. These include the essential methionine aminopeptidase, which removes the N-terminal methionine from nascent proteins (90). In addition, transfer-messenger RNA (tmRNA) and its binding protein rescue stalled ribosomes, whereas the essential peptidyl-tRNA hydrolase SpoVC recycles tRNA molecules sequestered as peptidyl-tRNA as a result of premature dissociation from the ribosome (91–93). The precise functions of YwkE and YbxF still have to be discovered. YbxF binds to turns in RNA molecules and is very strongly expressed under most conditions (33, 39, 94).
Protein Secretion
Several proteins of the minimal cell have to be targeted either to the membrane or for secretion into the medium. For this activity, the minimal cell needs a set of 12 proteins (5 of them essential) and 1 essential RNA for protein secretion (Fig. 2). The first component in cotranslational targeting of the membrane and secreted proteins is the universally conserved signal recognition particle composed of the essential RNA scr, the essential protein Ffh, and FtsY (95–99). The next step in protein translocation is performed by the chaperone CsaA and by the translocation motor SecA, which provides the energy for the export of unfolded secretory precursor proteins through the channel formed by SecYEG (100–102). Alternatively, YidC2 can translocate the preproteins through the membrane. In B. subtilis, YidC2 has a paralog, SpoIIIJ, and one of the two proteins is required for viability (36, 103, 104). We included YidC2 since this protein is also important for genetic competence (105) (see below). For secreted proteins, the signal peptide is then cleaved off by signal peptidase I (SipS) (106, 107). In the case of lipoproteins, the protein is diacylglyceryl modified by Lgt for attachment to the membrane, and signal peptide cleavage is subsequently performed by LspA (108–110). Proper folding of extracellular proteins is assisted by the posttranslocation chaperone PrsA, a lipoprotein attached to the outer surface of the cytoplasmic membrane (111, 112). B. subtilis also possesses the TAT (twin arginine translocation) and type VII protein secretion systems. However, these systems are not essential in a minimal cell (113–115). Interestingly, the reduced genome of M. mycoides JCVI-syn3.0 encodes only the signal recognition particle, the translocation motor SecA, and an incomplete Sec channel. Importantly, this minimal organism seems to lack enzymes for the export and membrane attachment of lipoproteins. It is tempting to speculate that these functions are encoded by some of the unknown genes, since lipoprotein-dependent metabolite uptake is crucial for Mycoplasma bacteria with their strongly reduced metabolism. This is the case not only for artificially genome-reduced bacteria but also for natural Mycoplasma species. In M. pneumoniae, lipoproteins account for about 10% of all encoded proteins (116, 117). Moreover, many of the 149 unknown genes in the minimal genome of M. mycoides JCVI-syn3.0 encode lipoproteins (10).
Intracellular Chaperones, Protein Quality Control, and Proteolysis
To be active, proteins have to be properly folded, and misfolded proteins need to be detected and then be refolded or degraded. These activities can be performed by a set of seven proteins, including two essential proteins. The universally conserved chaperone DnaK with its cochaperones DnaJ and GrpE, the trigger factor Tig, as well as the universally conserved essential chaperonin GroES/GroEL are important for the folding of intracellular proteins (118, 119). It should be noted that DnaK, its partners, and Tig are nonessential in B. subtilis; however, the corresponding genes are highly expressed, and they are conserved among the genome-reduced mollicutes. Of note, the nonessential DnaK/DnaJ chaperone is also present in M. mycoides JCVI-syn3.0, whereas the universally conserved essential GroES/GroEL chaperonin is not (10). In addition, viability of B. subtilis requires the presence of one intramembrane quality control protease, and we selected HtrB for this activity (120, 121).
Central Carbon Metabolism
Central carbon metabolism is at the heart of any cellular metabolism. We have selected 26 proteins (among them 4 essential proteins) for this function. Glycolysis provides the cell with precursors for further metabolic pathways, ATP for substrate-level phosphorylation, and reducing power to drive respiration (see Fig. 1 and 3 for details). Moreover, the pentose phosphate pathway generates reducing power for anabolic reactions, erythrose-4-phosphate as a precursor for chorismate synthesis (Fig. 3), and ribose-5-phosphate for the synthesis of nucleic acids. To generate acetyl coenzyme A (acetyl-CoA) for lipid biosynthesis, pyruvate dehydrogenase is required (122, 123). Moreover, the acetyl-CoA synthetase recycles the acetate derived from cell wall biosynthesis (124) (Fig. 3 and 10) (see below). Both glycolysis and the pentose phosphate pathway generate a large amount of reducing power. NAD+ can be regenerated in respiration, and the YtsJ/MalS transhydrogenation cycle is used for balancing NADPH+ levels (Fig. 3) (125). The minimal cell does not need the citric acid cycle. This pathway can be completely deleted in B. subtilis (our unpublished results). The minimal organism M. mycoides JCVI-syn3.0 encodes the enzymes for glycolysis but only a strongly reduced set (three out of seven) enzymes of the pentose phosphate pathway. Moreover, these cells lack the citric acid cycle. The absence of a full pentose phosphate pathway and of the citric acid cycle is characteristic of the reduced metabolism of Mycoplasma species and reflects their reliance on the uptake of nutrients from the environment (117). Interestingly, of the glycolytic enzymes, the classical fructose 1,6-bisphosphate aldolase is missing in M. mycoides JCVI-syn3.0. The cleavage of fructose 1,6-bisphosphate may be catalyzed by the paralogous IolJ aldolase in these bacteria. It is worth noting that the nearly complete match of the gene sets for glycolysis in the suggested MiniBacillus genome and in M. mycoides JCVI-syn3.0 is exceptional among all metabolic pathways.
Respiration/Energy
ATP production is achieved efficiently by the generation of a proton motive force in respiration and its subsequent use to drive ATP synthesis. A total of 16 proteins are involved in these processes, among them 2 essential proteins (Fig. 3). NADH2 from glycolysis is reoxidized by the respiration chain consisting of NADH dehydrogenase, menaquinone, and the terminal heme-copper cytochrome aa3 oxidase. It is important to note that B. subtilis is unable to live under aerobic conditions in the absence of both terminal quinol oxidases (126). We have selected this respiration chain because it is a minimal chain and because the selected terminal oxidase is capable to pumping protons to energize the membrane. Moreover, loss of the heme-copper cytochrome aa3 oxidase results in impaired growth (126). Next, the multisubunit ATPase uses the proton gradient to provide the cell with ATP. Interestingly, several subunits of the ATPase in M. mycoides JCVI-syn3.0 seem to be too poorly conserved with those of B. subtilis to allow detection by sequence comparison.
Amino Acids
Acquisition of amino acids is one of the major activities of any living cell. This can be achieved by either the uptake of external amino acids, the uptake and subsequent degradation of peptides, or the biosynthesis of amino acids. As biosynthesis usually involves significantly more proteins than does uptake, we included mainly transport systems for amino acids in the list (see also Fig. 1 and 4 for details). Exceptions are the biosyntheses of alanine, glycine, serine, phenylalanine, tyrosine, asparagine, and glutamine. The single glycine biosynthetic enzyme (serine hydroxymethyltransferase [GlyA]) is essential, suggesting that B. subtilis has to synthesize this amino acid. For alanine, serine, asparagine, and the aromatic amino acids, no transporter is known. For glutamine, biosynthesis requires only one enzyme (glutamine synthetase [GlnA]), and the only known glutamine transporter is composed of several subunits and is expressed only during sporulation. Among the 30 proteins that are required for amino acid acquisition in the frame of the MiniBacillus genome, only 2 (the essential GlyA protein and a cysteine transporter) are also present in M. mycoides JCVI-syn3.0, suggesting that this organism acquires amino acids in a completely different way. Likely, the latter organism relies on the transport and intracellular degradation of oligopeptides to obtain amino acids (10).
Nucleotides/Phosphate
Complex media like LB cannot meet the demand of B. subtilis for nucleotides (R. Switzer, personal communication). Therefore, the minimal cell has to carry 35 genes for nucleotide de novo synthesis. Of these genes, 11 are essential. Both purine and pyrimidine biosyntheses are initiated by the synthesis of phosphoribosylpyrophosphate from ribose-5-phosphate (catalyzed by the essential and universally conserved phosphoribosylpyrophosphate synthetase Prs). The pathways of nucleotide biosynthesis are schematically shown in Fig. 1 (for more details, see Fig. 5). The pyrimidine and purine nucleotide biosynthetic pathways converge with the reduction of ribonucleotides by the NrdEIF complex and the final formation of nucleoside triphosphates by the nucleoside diphosphate kinase Ndk (127). For one of the essential proteins implicated in nucleotide biosynthesis, HprT, the reason for its essentiality is unclear (128).
One protein is required for phosphate acquisition. Pit is a constitutively expressed low-affinity phosphate transporter (Fig. 2).
Lipids
Lipid biosynthesis is an essential function in most bacterial cells. The pathway starts with the carboxylation of acetyl-CoA derived from glycolysis (Fig. 1) and ends with the addition of a so-called head group to phosphatidic acid. Fatty acid biosynthesis is an essential process in nearly all bacteria, but this pathway is lacking in Mycoplasma species (129). Accordingly, fatty acid biosynthetic genes are absent from the minimal genome of M. mycoides JCVI-syn3.0. These organisms rely on fatty acid acquisition from their hosts or the provided complex medium (10). In total, this pathway requires 19 genes (Fig. 6). With the exception of fabHA and fabI, all of these genes are essential in B. subtilis. The two nonessential genes in this pathway have functional paralogs, and it is known that the encoded functions are essential; i.e., one of the paralogs has to be present (36). The pathway is initiated by the synthesis of malonyl-CoA from acetyl-CoA by the four-subunit enzyme complex acetyl-CoA carboxylase. It should be noted that AccB, one of the proteins of this complex, needs to be biotinylated by the essential biotin protein ligase BirA (130). After the formation of acetoacetyl-acyl carrier protein (ACP), the cycle of consecutive fatty acid elongation involves FabG, YwpB, FabI, and FabF. The formation of phospholipids is initiated by the replacement of the acyl carrier protein by a phosphate group and the subsequent addition of glycerol-3-phosphate. After the addition of the second fatty acid, the head group is attached to form phosphatidylglycerol. It should be noted that the enzyme catalyzing the final reaction of the pathway, the dephosphorylation of phosphatidylglycerol phosphate, has not yet been identified in B. subtilis. The lipids of B. subtilis 168 contain up to five different head groups (131). However, it has been established that membranes consisting of only phosphatidylglycerol do not confer any growth disadvantages (131). Therefore, we decided to keep only this head group.
Cofactors
B. subtilis needs 11 distinct cofactors, i.e., NAD, flavin adenine dinucleotide (FAD), pyridoxal phosphate, biotin, thiamine, lipoic acid, coenzyme A, S-adenosylmethionine, folate, heme, and menaquinone. For some of these molecules, their acquisition is possible by uptake and biosynthesis. For the minimal cell, we have chosen to include uptake whenever possible (nicotinate and riboflavin as precursors for NAD and FAD, respectively; biotin; and thiamine) (see Fig. 1 and 7 to 9 for details). As a result, 62 proteins are required for the acquisition of cofactors. Of these proteins, those necessary for the synthesis of NAD from nicotinate, the two subunits of the pyridoxal-5-phosphate synthase, the lipoic acid synthase, the enzyme for the final step of CoA synthesis, the S-adenosylmethionine synthetase, the dihydrofolate reductase, and the enzymes for menaquinone synthesis are essential (a total of 14 proteins). It is worth noting that MenH, an enzyme of the menachinone biosynthetic pathway, was recently shown to be dispensable (132, 133). While none of the cofactor biosynthetic pathways are present in Mycoplasma species, and thus are also lacking in M. mycoides JCVI-syn3.0, this minimal genome possesses all three general components of the so-called ECF (energy-coupling factor) transporters that are capable of transporting several cofactors (10). The substrate-specific S proteins for these transporters are generally poorly conserved and barely detectable in sequence comparisons (134, 135).
Metals and Iron-Sulfur Clusters
Many enzymes and proteins need metal ions for activity. Moreover, potassium and sodium are important for the osmotic stability of the cell. The minimal B. subtilis cell has to transport sodium, potassium, iron, magnesium, manganese, zinc, and copper ions. With the exception of iron, we have included one transporter (usually of low affinity) for each ion. For iron uptake, the minimal cell should possess the EfeUO system for elemental iron uptake and the iron-citrate ABC transporter YhfQ-YfmCDEF (136, 137). Sodium is imported by amino acid-sodium symporters. It has to be exported by the Mrp complex (138) (Fig. 2). Surprisingly, in the genome of M. mycoides JCVI-syn3.0, only a potassium transporter and two putative transporters for magnesium have been annotated (10). The identities of other metal transporters in the minimal cell thus remain unclear.
Many proteins involved in redox reactions need iron-sulfur clusters for their activity. The synthesis of these clusters from cysteine and ferrous iron and their attachment to proteins involve seven proteins, five of which are essential and are therefore included (Fig. 7) (139).
Cell Wall Biosynthesis
Cell wall biosynthesis is intimately linked to cell division. First, we briefly discuss the set of enzymes that is involved in catalytic activities for the cell wall components, the peptidoglycan, and the teichoic and lipoteichoic acids. Fifty-five proteins (43 essential proteins) are required for these pathways.
Peptidoglycan synthesis starts with the synthesis of glucosamine-6-phosphate from glutamine and fructose-6-phosphate (140). The next important intermediate is UDP-N-acetylglucosamine (UDP-GlcNAc). This compound is converted to UDP-N-acetylmuramic acid, to which the 5 amino acids of the peptide are subsequently attached. The peptide contains three unusual amino acids, d-glutamate, meso-diaminopimelic acid, and d-alanine, which have to be provided by the action of racemases (d-Glu and d-Ala) or by a biosynthetic pathway starting from aspartate. The enzyme MraY replaces the activating UDP moiety by undecaprenyl phosphate, which has to be synthesized starting from pyruvate and glyceraldehyde-3-phosphate (see Fig. 1 and 10 for details). N-Acetylglucosamine is added to give rise to a molecule called lipid II (141). Owing to the undecaprenyl phosphate moiety, this molecule can integrate into the membrane and is then flipped to the outer side by the action of a flippase, MurJ. Most of the enzymes participating in the pathways mentioned above are essential. MurJ is an exception, as B. subtilis also possesses the weakly expressed functional paralog Amj (142).
Lipid II is the functional unit of peptidoglycan polymerization driven by the penicillin-binding proteins (141). These proteins catalyze the consecutive elongation of the peptidoglycan chain as well as the introduction of cross-links between the peptides. Moreover, autolysins are necessary to introduce breaks in the molecule that serve as targets for the introduction of new material. Penicillin-binding proteins and autolysins are present with several paralogs in B. subtilis. For the minimal cell, we have selected penicillin-binding proteins 1 (PonA), 2B (PbpB), and 2A (PbpA) and the autolysins LytE and LytF (143–147). As outlined above, this selection was made according to their expression profiles and the dependence on other proteins. As an example, there is a functional paralog of LytE, CwlO (148). For the activity of CwlO, B. subtilis also needs the ABC transporter FtsEX and the small protein Mbl (149). Thus, the choice of LytE allowed a smaller number of genes.
Another essential component of the Gram-positive cell wall is teichoic acids. In B. subtilis, these molecules can be attached to either peptidoglycan or the membrane via a lipid anchor. Wall teichoic acid is polyglycerol phosphate attached to the peptidoglycan via a disaccharide anchor (150). The pathway is initiated by the synthesis of the anchor from undecaprenyl phosphate and activated N-acetylglucosamine and N-acetylmannosamine (UDP-GlcNAc and UDP-N-acetylmannosamine [UDP-ManNAc]). Glycerol is activated by the synthesis of CDP-glycerol. This compound serves as the substrate for the initial addition of glycerol to the undecaprenyl phosphate-carrying disaccharide and then for consecutive rounds of chain elongation. The polymer is then exported by the TagGH ABC transporter and is finally attached to peptidoglycan by TagU (Fig. 11) (150). Lipoteichoic acid (LTA) is polyglycerol phosphate attached to the membrane via a β-diglucosyl-diacylglycerol anchor. This anchor is synthesized from glucose-6-phosphate and diacylglycerol by the enzyme UgtP (151). This anchor has to be flipped to the outside of the membrane by an as-yet-unknown flippase. The lipoteichoic acid synthase LtaS then processively adds glycerol phosphate moieties using phosphatidylglycerol, the building block of the lipid bilayer, as the substrate. This reaction releases toxic diacylglycerol, which is recycled by the essential diacylglycerol kinase DgkB to phosphatidic acid, which is used for the synthesis of phosphatidylglycerol (see above) (Fig. 6) (32, 152, 153).
Cell Division
Twenty-two proteins are necessary for the functions of cell division and cell shape determination. Of these proteins, nine are essential. Most of these proteins are part of several large protein complexes, i.e., the divisome, which links cell division to cell wall synthesis, and the elongasome, which recruits the cell wall biosynthetic enzyme to the lateral cell wall. Accordingly, cell wall biosynthetic enzymes (see above) are part of these complexes. This is the case for MraY, MurF, MurG, and penicillin-binding proteins 1 and 2B (PonA and PbpB, respectively), which are part of the divisome and/or the elongasome (154, 155). To achieve the correct placement of the division site, DivIVA in concert with the proteins of the Min system prevent the formation of the Z ring close to nascent division sites and cell poles (156, 157). Finally, WalJ coordinates cell division and DNA replication (158). Interestingly, the essential tubulin-like cell division initiation protein FtsZ has long been regarded as being present in all organisms. However, this protein is absent in some Mycoplasma species and is nonessential in those Mycoplasma species that encode it (159). Accordingly, FtsZ is also not encoded in the genome of M. mycoides JCVI-syn3.0 (10). Moreover, FtsZ is dispensable in B. subtilis L-form cells that are capable of growing without a cell wall (160, 161). However, as these cells grow very slowly and are fragile, we deemed the FtsZ-dependent cell division pathway essential in the context of the MiniBacillus genome.
Signaling in Cell Division
As in other bacteria, most signal transduction systems of B. subtilis are dispensable. This is the case for all alternative sigma factors as well as for classical transcription repressors and activators. However, there are two notable exceptions, and they are both involved in cell wall metabolism and cell division. The WalRK two-component regulatory system in B. subtilis and related Gram-positive bacteria is the only two-component system known to be essential. This system allows the expression of genes for cell division and the synthesis of wall teichoic acid, such as lytE, mreBH, ftsZ, and the tag genes (162, 163). Moreover, cyclic di-AMP (c-di-AMP) is a unique essential second messenger. The precise reason for the essentiality of this nucleotide is unknown, but c-di-AMP has been implicated in cell division and cell wall biosynthesis (164, 165). Interestingly, c-di-AMP is not only essential but also toxic if it accumulates (166). Therefore, CdaA was selected among the three diadenylate cyclases in B. subtilis, and GdpP was selected as one of two phosphodiesterases that degrade the second messenger c-di-AMP.
Integrity of the Cell (Protection and Genome Integrity)
All the functions discussed above are essential for the minimal cell. However, a minimal organism also needs activities that are not per se essential but that help to keep and protect the integrity of the cell. This involves protection against toxic metabolites and intermediates as well as against damage caused by oxygen (Fig. 2). In addition, the genome of the minimal cell has to be sufficiently stable. For this purpose, proteins involved in DNA repair and genome integrity are important components of any minimal cell that is intended for stable reproduction. In the list of required proteins, we have included eight proteins for protection and genome integrity.
Additional Proteins of the Minimal Cell
There are 11 additional proteins that may be important for a minimal cell. Two of these proteins (PpaC and YlaN) are essential, but their precise function is not known. Moreover, based on our own experimental data and those of colleagues, YitI, YitW, and YqhY are important for viability (P. Dos Santos, personal communication; our unpublished results). Four proteins (YkwC, YlbN, YpfD, and YugI) are very strongly expressed (39) and therefore certainly of vital importance. FloT is involved in the control of membrane fluidity, and the loss of both flotillin-like proteins is known to result in severe defects in cell morphology and transformation (167). Finally, the nonessential GTPase YyaF is highly conserved in Gram-positive bacteria, even among all groups of mollicutes (42). Due to its GTPase activity, it is tempting to speculate that YyaF plays a role in ribosome assembly or in translation.
Open Questions
Even though B. subtilis is one of the best-studied and best-understood organisms, there are still gaps in our knowledge. These gaps pose serious challenges to any minimal genome project. Several of these uncertainties are mentioned above. In particular, we have identified gaps in the pathways of phospholipid biosynthesis (the final phosphatase) and lipoteichoic acid synthesis (the flippase for the membrane anchor). Concerning phosphatase, there are several uncharacterized phosphatases encoded in the B. subtilis genome, leaving room for further research.
A general uncertainty is the disposal of toxic or harmful compounds. Even though we have included eight proteins for this activity, this list may be far from being complete. Another function with significant gaps in our knowledge is RNA modification. While our list is based on current knowledge, it is possible that not all of these modifications are truly essential. Moreover, some modifications might become important if other modifications are lacking due to the absence of the responsible enzymes.
Finally, only little is known about essential noncoding RNAs and essential structural DNA regions in B. subtilis. The importance of these features has been uncovered in a recent saturating-mutagenesis study with M. pneumoniae (8).
A MODEL OF MiniBacillus METABOLISM
The selection of functions, genes, and pathways presented above is like a bet. To ensure that this set allows the cell to function, we have developed a model of its metabolism. This model is schematically depicted in Fig. 1, and details are provided in Fig. 2 to 11. With this compilation of metabolic pathways, it becomes obvious that all metabolites that are needed as precursors are produced from simple building blocks. These molecules in turn can be obtained from the medium. On the other hand, all compounds that are produced in the pathways and that are not needed in other reactions are disposed of by specific reactions and pathways.
MiniBacillus AS A STARTING POINT TO DISCOVER AND STUDY NOVEL ANTIMICROBIAL DRUG TARGETS
B. subtilis is closely related to many important pathogens, including Staphylococcus aureus, Listeria monocytogenes, and Streptococcus pyogenes. The genetic complement of MiniBacillus likely represents a set of highly important genes for all of these organisms, suggesting that this set is an excellent starting point to identify promising novel potential antimicrobial drug targets. Indeed, the conserved essential diadenylate cyclase was recently proposed to be a potential drug target, screening systems have been set up, and a first drug has been discovered (168–170).
For drug targets, knowledge of the protein structure is highly beneficial to facilitate in silico screening of virtual compound libraries. Of all 523 proteins that are encoded by our proposed minimal genome, 471 (90.1%) have a known structure. This figure illustrates the huge progress made by structural biologists, especially in the framework of large structural genomic projects (171). The unique combination of high structural coverage and excellent prior knowledge of the protein components of MiniBacillus makes the list of proteins presented in Table 2 a good starting point to identify novel potential targets. Moreover, the remaining 10% of the proteins are an important challenge for structural biologists.
Knowing all the structures of the proteins and even protein complexes of MiniBacillus not only is important in the context of the search for novel drugs but also is by itself a major scientific goal. With knowledge of the structure of all the components of a minimal cell, we can start making a real image of what is going on in the cell and thus become much closer to the answer of the old question, “What is life?”
EXPERIMENTAL APPROACH TO THE CONSTRUCTION OF MiniBacillus
To obtain a minimal cell, MiniBacillus, with the described set of genes, the top-down approach seems to be the most appropriate. Such an approach requires efficient systems for markerless gene deletions, which in turn indicates a need for a highly efficient genetic system. B. subtilis is known for its natural competence. This system is under the control of a central transcription factor, ComK. In order to be able to perform consecutive deletions, the genes involved in genetic competence have to be kept until the very end of the successive genome reduction process. Therefore, another set of 54 genes has to be considered (see Table S1 in the supplemental material). Optimally, expression of competence genes would occur not just in 10% of the population as in the B. subtilis wild-type strain but in all cells of a population. For this purpose, a supercompetent B. subtilis strain was recently engineered (172). In this strain, the transcription factor ComK and ComS, a protein which prevents the proteolytic degradation of ComK, are expressed under the control of a strong mannitol-inducible promoter.
Several systems have been proposed for the construction of markerless deletions. The Cre-Lox system is very efficient, and it has been adapted to B. subtilis (173) but has the disadvantage of leaving scars behind. Such scars would accumulate if dozens of deletions were performed. Alternative systems rely on the integration of genes, which become toxic under defined conditions with a deletion cassette. Counterselection then allows the rapid and specific loss of the cassette, i.e., the markerless deletion of the desired regions. Several counterselection systems have been implemented for B. subtilis. The most popular systems rely on the mazF RNase; the upp gene, which is counterselected in the presence of 5-fluorouracil; and the mannose transporter ManP, which generates toxic mannose-6-phosphate (19). Using the latter system, a significant reduction of the B. subtilis genome was recently achieved (19, 174, 175).
In B. subtilis, many important genes that are part of the minimal gene set listed in Table 2 are scattered on the chromosome. Therefore, defragmentation of such regions will facilitate the effective progress of a genome reduction program. For this defragmentation, desired fragments can be assembled and introduced into the chromosome by using, e.g., Gibson assembly or the ligase cycling reaction in combination with the markerless deletion methods mentioned above (176, 177).
FINAL REMARKS
There is not a single solution for the question of a minimal genome, but there are many possible answers. To fully understand the functionality of living cells, minimal organisms based on different model organisms are required. Thus, the construction of M. mycoides JCVI-syn3.0 marks an important breakthrough, but with its large number of unknown genes, it cannot provide the full picture (10). It will be essential to continue genome reduction projects with very different bacteria, yeasts, and even archaea to be able to finally draw a conclusion about the essence of life.
(For more information, see http://www.minibacillus.org/.)
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to many friends and colleagues from the Bacillus scientific community who provided information on minimal gene sets required for specific functions. We give our special thanks to Valerie de Crécy-Lagard, Patricia dos Santos, Juan Alonso, Richard Daniel, Jan Maarten van Dijl, David Dubnau, Stephan Gruber, Sven Halbedel, Colin Harwood, John Helmann, Laurent Janniere, Oscar Kuipers, Mohamed Marahiel, Lyle Simmons, and Robert Switzer. We acknowledge Sarah Wilcken and Raphael Michna for the help with bioinformatic issues.
D.R.R. was supported by the Göttingen Graduate School for Neurosciences, Biophysics, and Molecular Biosciences (GGNB) (DFG grant GCS226/2). J.S. was supported by the Deutsche Forschungsgemeinschaft through grant SPP1879.
Biographies

Daniel R. Reuß studied microbiology and biochemistry at the Georg August University Göttingen, Germany. During his studies, he did an internship at DSM Nutritional Products in Basel, Switzerland, where he worked on vitamin B6 production in Bacillus subtilis. After his graduation in 2012, he worked with Prof. Ken-ichi Yoshida at Kobe University, Japan, and focused on work with the inositol pathway in B. subtilis. Afterwards, he returned to Göttingen, where he is now doing his Ph.D. working on the B. subtilis genome reduction project.

Fabian M. Commichau studied biology at the RWTH, Aachen, Germany, and obtained a Ph.D. in microbiology at the University of Göttingen, Germany. After postdoctoral work on protein-protein interactions at the University of Göttingen and at the University of Basel, Switzerland, he stayed for 2 years in industry, R&D, at DSM Nutritional Products Ltd., Kaiseraugst, Switzerland. In 2011, he returned to the University of Göttingen, where he became a group leader at the Department of General Microbiology at the Institute for Microbiology and Genetics and obtained his habilitation. In 2016, his interests are amino acid and vitamin metabolism in Gram-positive bacteria, bacterial evolution and molecular mechanisms underlying genome instability, second-messenger signaling in the human-pathogenic bacterium Listeria monocytogenes, and minimal genomes.

Jan Gundlach studied biology at the University of Göttingen, Germany. During his studies, he was working on protein-protein interactions and second-messenger signaling in Bacillus subtilis. He was also part of a group of students that participated in the iGEM competition in 2013, which was organized and held at MIT, Boston, MA. Since 2014, he has been a Ph.D. candidate at the University of Göttingen, Germany. In 2016, his interests are second-messenger signaling and ion homeostasis in Gram-positive bacteria.

Bingyao Zhu finished her bachelor's study in biology at China Agricultural University, China. Later, she pursued a master's degree in microbiology and biochemistry at the University of Göttingen, Germany. She was member of a group of students that participated in the iGEM competition in 2013, which was organized and held at MIT, Boston, MA. After obtaining her master's degree, she became a Ph.D. candidate at the Department of General Microbiology, University of Göttingen. She is currently working on the further development of SubtiWiki, an integrated annotation database for Bacillus subtilis, as her Ph.D. project. She is interested in developing new tools to collect and organize data, in order to find inner connections.

Jörg Stülke studied biology at Ernst Moritz Arndt University, Greifswald, Germany, and obtained a Ph.D. in microbiology at the University of Greifswald. After postdoctoral work on carbon catabolite repression in Bacillus subtilis at the University of Lund, Sweden, and the Institut Pasteur, Paris, France, he became a group leader at the Chair of Microbiology at the University of Erlangen, Germany. He obtained his habilitation on the control of carbon metabolism in Bacillus subtilis in 2000. In 2003, he was appointed Full Professor for Microbiology at the Georg August University Göttingen, Germany. He studies RNA-mediated control of gene expression by RNA switches and RNA degradation. Moreover, he is interested in heterogeneous gene expression and in the discovery of the function of the essential second messenger cyclic di-AMP. In the framework of the minimal genome project, he is developing tools for genome annotation and involved in the construction and analysis of genome-reduced strains of B. subtilis.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MMBR.00029-16.
REFERENCES
- 1.Glass JI, Assad-Garcia N, Alperovich N, Yooseph S, Lewis MR, Maruf M, Hutchison CA III, Smith HO, Venter JC. 2006. Essential genes of a minimal bacterium. Proc Natl Acad Sci U S A 103:425–430. doi: 10.1073/pnas.0510013103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Güell M, van Noort V, Yus E, Chen WH, Leigh-Bell J, Michalodimitrakis K, Yamada T, Arumugam M, Doerks T, Kühner S, Rode M, Suyama M, Schmidt S, Gavin AC, Bork P, Serrano L. 2009. Transcriptome complexity in a genome-reduced bacterium. Science 326:1268–1271. doi: 10.1126/science.1176951. [DOI] [PubMed] [Google Scholar]
- 3.Yus E, Maier T, Michalodimitrakis K, van Noort V, Yamada T, Chen WH, Wodke JA, Güell M, Martínez S, Bourgeois R, Kühner S, Raineri E, Letunic I, Kalinina OV, Rode M, Herrmann R, Gutiérrez-Gallego R, Russell RB, Gavin AC, Bork P, Serrano L. 2009. Impact of genome reduction on bacterial metabolism and its regulation. Science 326:1263–1268. doi: 10.1126/science.1177263. [DOI] [PubMed] [Google Scholar]
- 4.Kühner S, van Noort V, Betts MJ, Leo-Macias A, Batisse C, Rode M, Yamada T, Maier T, Bader S, Beltran-Alvarez P, Castaño-Diez D, Chen WH, Devos D, Güell M, Norambuena T, Racke I, Rybin V, Schmidt A, Yus E, Aebersold R, Herrmann R, Böttcher B, Frangakis AS, Russell RB, Serrano L, Bork P, Gavin AC. 2009. Proteome organization in a genome-reduced bacterium. Science 326:1235–1240. doi: 10.1126/science.1176343. [DOI] [PubMed] [Google Scholar]
- 5.Maier T, Schmidt A, Güell M, Kühner S, Gavin AC, Aebersold R, Serrano L. 2011. Quantification of mRNA and protein and integration with protein turnover in a bacterium. Mol Syst Biol 7:511. doi: 10.1038/msb.2011.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schmidl SR, Gronau K, Pietack N, Hecker M, Becher D, Stülke J. 2010. The phosphoproteome of the minimal bacterium Mycoplasma pneumoniae: analysis of the complete known Ser/Thr kinome suggests the existence of novel kinases. Mol Cell Proteomics 9:1228–1242. doi: 10.1074/mcp.M900267-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van Noort V, Seebacher J, Bader S, Mohammed S, Vonkova I, Betts MJ, Kühner S, Kumar R, Maier T, O'Flaherty M, Rybin V, Schmeisky A, Yus E, Stülke J, Serrano L, Russell RB, Heck AJ, Bork P, Gavin AC. 2012. Cross-talk between phosphorylation and lysine acetylation in a genome-reduced bacterium. Mol Syst Biol 8:571. doi: 10.1038/msb.2012.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lluch-Senar M, Delgado J, Chen WH, Lloréns-Rico V, O'Reilly FJ, Wodke JA, Unal EB, Yus E, Martínez S, Nichols RJ, Ferrar T, Vivancos A, Schmeisky A, Stülke J, van Noort V, Gavin AC, Bork P, Serrano L. 2015. Defining a minimal cell: essentiality of small ORFs and ncRNAs in a genome-reduced bacterium. Mol Syst Biol 11:780. doi: 10.15252/msb.20145558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gibson DG, Glass JI, Lartigue C, Noskov VN, Chuang RY, Algire MA, Benders GA, Montague MG, Ma L, Moodie MM, Merryman C, Vashee S, Krishnakumar R, Assad-Garcia N, Andrews-Pfannkoch C, Denisova EA, Young L, Qi ZQ, Segall-Shapiro TH, Calvey CH, Parmar PP, Hutchison CA III, Smith HO, Venter JC. 2010. Creation of a bacterial cell controlled by a chemically synthesized genome. Science 329:52–56. doi: 10.1126/science.1190719. [DOI] [PubMed] [Google Scholar]
- 10.Hutchison CA III, Chuang RY, Noskov VN, Assad-Garcia N, Deerinck TJ, Ellisman MH, Gill J, Kannan K, Karas BJ, Ma L, Pelletier JF, Qi ZQ, Richter RA, Strychalski EA, Sun L, Suzuki Y, Tsvetanova B, Wise KS, Smith HO, Glass JI, Merryman C, Gibson DG, Venter JC. 2016. Design and synthesis of a minimal bacterial genome. Science 351:aad6253. doi: 10.1126/science.aad6253. [DOI] [PubMed] [Google Scholar]
- 11.Xavier JC, Patil KR, Rocha I. 2014. Systems biology perspectives on minimal and simpler cells. Microbiol Mol Biol Rev 78:487–509. doi: 10.1128/MMBR.00050-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blain JC, Szostak JW. 2014. Progress toward synthetic cells. Annu Rev Biochem 83:615–640. doi: 10.1146/annurev-biochem-080411-124036. [DOI] [PubMed] [Google Scholar]
- 13.Kretschmer S, Schwille P. 2014. Toward spatially regulated division of protocells: insights into the E. coli Min system from in vitro studies. Life (Basel) 4:915–928. doi: 10.3390/life4040915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schwille P. 2015. Jump-starting life? Fundamental aspects of synthetic biology. J Cell Biol 210:687–690. doi: 10.1083/jcb.201506125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oberholzer T, Wick R, Luisi PL, Biebricher CK. 1995. Enzymatic RNA replication in self-reproducing vesicles: an approach to a minimal cell. Biochem Biophys Res Commun 207:250–257. doi: 10.1006/bbrc.1995.1180. [DOI] [PubMed] [Google Scholar]
- 16.Szostak JW, Bartel DP, Luisi PL. 2001. Synthesizing life. Nature 409:387–390. doi: 10.1038/35053176. [DOI] [PubMed] [Google Scholar]
- 17.Chen IA, Roberts RW, Szostak JW. 2004. The emergence of competition between model protocells. Science 305:1474–1476. doi: 10.1126/science.1100757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Morimoto T, Kadoya R, Endo K, Tohata M, Sawada K, Liu S, Ozawa T, Kodama T, Kakeshita H, Kageyama Y, Manabe K, Kanaya S, Ara K, Ozaki K, Ogasawara N. 2008. Enhanced recombinant protein productivity by genome reduction in Bacillus subtilis. DNA Res 15:73–81. doi: 10.1093/dnares/dsn002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wenzel M, Altenbuchner J. 2015. Development of a markerless gene deletion system for Bacillus subtilis based on the mannose phosphoenolpyruvate-dependent phosphotransferase system. Microbiology 161:1942–1949. doi: 10.1099/mic.0.000150. [DOI] [PubMed] [Google Scholar]
- 20.Westers H, Dorenbos R, van Dijl JM, Kabel J, Flanagan T, Devine KM, Jude F, Seror SJ, Beekman AC, Darmon E, Eschevins C, de Jong A, Bron S, Kuipers OP, Albertini AM, Antelmann H, Hecker M, Zamboni N, Sauer U, Bruand C, Ehrlich DS, Alonso JC, Salas M, Quax WJ. 2003. Genome engineering reveals large dispensable regions in Bacillus subtilis. Mol Biol Evol 20:2076–2090. doi: 10.1093/molbev/msg219. [DOI] [PubMed] [Google Scholar]
- 21.Hirokawa Y, Kawano H, Tanaka-Masuda K, Nakamura N, Nakagawa A, Ito M, Mori H, Oshima T, Ogasawara N. 2013. Genetic manipulation restored the growth fitness of reduced-genome Escherichia coli. J Biosci Bioeng 116:52–58. doi: 10.1016/j.jbiosc.2013.01.010. [DOI] [PubMed] [Google Scholar]
- 22.Pósfai G, Plunkett G III, Fehér T, Frisch D, Keil GM, Umenhoffer K, Kolisnychenko V, Stahl B, Sharma SS, de Arruda M, Burland V, Harcum SW, Blattner FR. 2006. Emergent properties of reduced-genome Escherichia coli. Science 312:1044–1046. doi: 10.1126/science.1126439. [DOI] [PubMed] [Google Scholar]
- 23.Kato J, Hashimoto M. 2008. Construction of long chromosomal deletion mutants of Escherichia coli and minimization of the genome. Methods Mol Biol 416:279–293. doi: 10.1007/978-1-59745-321-9_18. [DOI] [PubMed] [Google Scholar]
- 24.Krishnakumar R, Grose C, Haft DH, Zaveri J, Alperovich N, Gibson DG, Merryman C, Glass JI. 2014. Simultaneous non-contiguous deletions using large synthetic DNA and site-specific recombinases. Nucleic Acids Res 42:e111. doi: 10.1093/nar/gku509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Unthan S, Baumgart M, Radek A, Herbst M, Siebert D, Brühl N, Bartsch A, Bott M, Wiechert W, Marin K, Hans S, Krämer R, Seibold G, Frunzke J, Kalinowski J, Rückert C, Wendisch VF, Noack S. 2015. Chassis organism from Corynebacterium glutamicum—a top down approach to identify and delete irrelevant gene clusters. Biotechnol J 10:290–301. doi: 10.1002/biot.201400041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leprince A, de Lorenzo V, Völler P, van Passel MWJ, Martins dos Santos VAP. 2012. Random and cyclical deletion of large DNA segments in the genome of Pseudomonas putida. Environ Microbiol 14:1444–1453. doi: 10.1111/j.1462-2920.2012.02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Komatsu M, Uchiyama T, Omura S, Cane DE, Ikeda H. 2010. Genome-minimized Streptomyces host for the heterologous expression of secondary metabolism. Proc Natl Acad Sci U S A 107:2646–2651. doi: 10.1073/pnas.0914833107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Juhas M, Reuß DR, Zhu B, Commichau FM. 2014. Bacillus subtilis and Escherichia coli essential genes and minimal cell factories after one decade of genome engineering. Microbiology 160:2341–2351. doi: 10.1099/mic.0.079376-0. [DOI] [PubMed] [Google Scholar]
- 29.Sasaki M, Kumagai H, Takegawa K, Tohda H. 2013. Characterization of genome-reduced fission yeast strains. Nucleic Acids Res 41:5382–5399. doi: 10.1093/nar/gkt233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kobayashi K, Ehrlich SD, Albertini A, Amati G, Andersen KK, Arnaud M, Asai K, Ashikaga S, Aymerich S, Bessieres P, Boland F, Brignell SC, Bron S, Bunai K, Chapuis J, Christiansen LC, Danchin A, Débarbouille M, Dervyn E, Deuerling E, Devine K, Devine SK, Dreesen O, Errington J, Fillinger S, Foster SJ, Fujita Y, Galizzi A, Gardan R, Eschevins C, Fukushima T, Haga K, Harwood CR, Hecker M, Hosoya D, Hullo MF, Kakeshita H, Karamata D, Kasahara Y, Kawamura F, Koga K, Koski P, Kuwana R, Imamura D, Ishimaru M, Ishikawa S, Ishio I, Le Coq D, Masson A, Mauël C, et al. 2003. Essential Bacillus subtilis genes. Proc Natl Acad Sci U S A 100:4678–4683. doi: 10.1073/pnas.0730515100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tanaka K, Henry CS, Zinner JF, Jolivet E, Cohoon MP, Xia F, Bidnenko V, Ehrlich SD, Stevens RL, Noirot P. 2013. Building the repertoire of dispensable chromosome regions in Bacillus subtilis entails major refinement of cognate large-scale metabolic model. Nucleic Acids Res 41:687–699. doi: 10.1093/nar/gks963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Commichau FM, Pietack N, Stülke J. 2013. Essential genes in Bacillus subtilis: a re-evaluation after ten years. Mol Biosyst 9:1068–1075. doi: 10.1039/c3mb25595f. [DOI] [PubMed] [Google Scholar]
- 33.Michna RH, Zhu B, Mäder U, Stülke J. 2016. SubtiWiki 2.0—an integrated database for the model organism Bacillus subtilis. Nucleic Acids Res 44:D654–D662. doi: 10.1093/nar/gkv1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Figaro S, Durand S, Gilet L, Cayet N, Sachse M, Condon C. 2013. Bacillus subtilis mutants with knockouts of the genes encoding ribonucleases RNase Y and RNase J1 are viable, with major defects in cell morphology, sporulation, and competence. J Bacteriol 195:2340–2348. doi: 10.1128/JB.00164-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peters JM, Colavin A, Shi H, Czarny TL, Larson MH, Wong S, Hawkins JS, Lu CH, Koo BM, Marta E, Shiver AL, Whitehead EH, Weissman JS, Brown ED, Qi LS, Huang KC, Gross CA. 2016. A comprehensive, CRISPR-based functional analysis of essential genes in bacteria. Cell 165:1493–1506. doi: 10.1016/j.cell.2016.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thomaides HB, Davison EJ, Burston L, Johnson H, Brown DR, Hunt AC, Errington J, Czaplewski L. 2007. Essential bacterial functions encoded by gene pairs. J Bacteriol 189:591–602. doi: 10.1128/JB.01381-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mehne FMP, Gunka K, Eilers H, Herzberg C, Kaever V, Stülke J. 2013. Cyclic-di-AMP homeostasis in Bacillus subtilis: both lack and high-level accumulation of the nucleotide are detrimental for cell growth. J Biol Chem 288:2004–2017. doi: 10.1074/jbc.M112.395491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Duman R, Ishikawa S, Celik I, Strahl H, Ogasawara N, Troc P, Löwe J, Hamoen LW. 2013. Structural and genetic analyses reveal the protein SepF as a new membrane anchor for the Z ring. Proc Natl Acad Sci U S A 110:E4601–E4610. doi: 10.1073/pnas.1313978110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nicolas P, Mäder U, Dervyn E, Rochat T, Leduc A, Pigeonneau N, Bidnenko E, Marchadier E, Hoebeke M, Aymerich S, Becher D, Bisicchia P, Botella E, Delumeau O, Doherty G, Denham EL, Fogg MJ, Fromion V, Goelzer A, Hansen A, Härtig E, Harwood CR, Homuth G, Jarmer H, Jules M, Klipp E, Le Chat L, Lecointe F, Lewis P, Liebermeister W, March A, Mars RA, Nannapaneni P, Noone D, Pohl S, Rinn B, Rügheimer F, Sappa PK, Samson F, Schaffer M, Schwikowski B, Steil L, Stülke J, Wiegert T, Devine KM, Wilkinson AJ, van Dijl JM, Hecker M, Völker U, Bessières P, Noirot P. 2012. The condition-dependent whole-transcriptome reveals high-level regulatory architecture in bacteria. Science 335:1103–1106. doi: 10.1126/science.1206848. [DOI] [PubMed] [Google Scholar]
- 40.Maaβ S, Wachlin G, Bernhardt J, Eymann C, Fromion V, Riedel K, Becher D, Hecker M. 2014. Highly precise quantification of protein molecules per cell during stress and starvation responses in Bacillus subtilis. Mol Cell Proteomics 13:2260–2276. doi: 10.1074/mcp.M113.035741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Muntel J, Fromion V, Goelzer A, Maaβ S, Mäder U, Büttner K, Hecker M, Becher D. 2014. Comprehensive absolute quantification of the cytosolic proteome of Bacillus subtilis by data independent, parallel fragmentation in liquid chromatography/mass spectrometry (LC/MS(E)). Mol Cell Proteomics 13:1008–1019. doi: 10.1074/mcp.M113.032631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Grosjean H, Breton M, Sirand-Pugnet P, Tardy F, Thiaucourt F, Citti C, Barré A, Yoshizawa S, Fourmy D, de Crécy-Lagard V, Blanchard A. 2014. Predicting the minimal translation apparatus: lessons from the reductive evolution of mollicutes. PLoS Genet 10:e1004363. doi: 10.1371/journal.pgen.1004363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Eymann C, Dreisbach A, Albrecht D, Bernhardt J, Becher D, Gentner S, Tam LT, Büttner K, Buurman G, Scharf C, Venz S, Völker U, Hecker M. 2004. A comprehensive proteome map of growing Bacillus subtilis cells. Proteomics 4:2849–2876. doi: 10.1002/pmic.200400907. [DOI] [PubMed] [Google Scholar]
- 44.Halbedel S, Hames C, Stülke J. 2004. In vivo activity of enzymatic and regulatory components of the phosphoenolpyruvate:sugar phosphotransferase system in Mycoplasma pneumoniae. J Bacteriol 186:7936–7943. doi: 10.1128/JB.186.23.7936-7943.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sanders GM, Dallmann HG, McHenry CS. 2010. Reconstitution of the B. subtilis replisome with 13 proteins including two distinct replicases. Mol Cell 37:273–281. doi: 10.1016/j.molcel.2009.12.025. [DOI] [PubMed] [Google Scholar]
- 46.Briggs GS, Smits WK, Soultanas P. 2012. Chromosomal replication initiation machinery of low-G+C-content Firmicutes. J Bacteriol 194:5162–5170. doi: 10.1128/JB.00865-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Duggin IG, Matthews JM, Dixon NE, Wake RG, Mackay JP. 2005. A complex mechanism determines polarity of DNA replication fork arrest by the replication terminator complex of Bacillus subtilis. J Biol Chem 280:13105–13113. doi: 10.1074/jbc.M414187200. [DOI] [PubMed] [Google Scholar]
- 48.Merrikh H, Grossman AD. 2011. Control of the replication initiator protein DnaA by an anti-cooperativity factor. Mol Microbiol 82:434–446. doi: 10.1111/j.1365-2958.2011.07821.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fukushima S, Itaya M, Kato H, Ogasawara N, Yoshikawa H. 2007. Reassessment of the in vivo functions of DNA polymerase I and RNase H in bacterial cell growth. J Bacteriol 189:8575–8583. doi: 10.1128/JB.00653-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang X, Montero Llopis P, Rudner DZ. 2013. Organization and segregation of bacterial chromosomes. Nat Rev Genet 14:191–203. doi: 10.1038/nrg3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang X, Tang OW, Riley EP, Rudner DZ. 2014. The SMC condensing complex is required for origin segregation in Bacillus subtilis. Curr Biol 24:287–292. doi: 10.1016/j.cub.2013.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Soh YM, Bürmann F, Shin HC, Oda T, Jin KS, Toseland CP, Kim C, Lee H, Kim SJ, Kong MS, Durand-Diebold ML, Kim YG, Kim HM, Lee NK, Sato M, Oh BH, Gruber S. 2015. Molecular basis for SMC rod formation and its dissociation upon DNA binding. Mol Cell 57:290–303. doi: 10.1016/j.molcel.2014.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tadesse S, Mascarenhas J, Kösters B, Hasilik A, Graumann PL. 2005. Genetic interaction of the SMC complex with topoisomerase IV in Bacillus subtilis. Microbiology 151:3729–3737. doi: 10.1099/mic.0.28234-0. [DOI] [PubMed] [Google Scholar]
- 54.Biller SJ, Burkholder WF. 2009. The Bacillus subtilis SftA (YtpS) and SpoIIIE DNA translocases play distinct roles in growing cells to ensure faithful chromosome partitioning. Mol Microbiol 74:790–809. doi: 10.1111/j.1365-2958.2009.06893.x. [DOI] [PubMed] [Google Scholar]
- 55.Sciochetti SA, Piggot PJ, Sherratt DJ, Blakely G. 1999. The ripX locus of Bacillus subtilis encodes a site-specific recombinase involved in proper chromosome partitioning. J Bacteriol 181:6053–6062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kaimer C, Schenk K, Graumann PL. 2011. Two DNA translocases synergistically affect chromosome dimer resolution in Bacillus subtilis. J Bacteriol 193:1334–1340. doi: 10.1128/JB.00918-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Levine C, Hiasa H, Marians KJ. 1998. DNA gyrase and topoisomerase IV: biochemical activities, physiological roles during chromosome replication, and drug sensitivities. Biochim Biophys Acta 1400:29–43. doi: 10.1016/S0167-4781(98)00126-2. [DOI] [PubMed] [Google Scholar]
- 58.Fernandez S, Rojo F, Alonso JC. 1997. The Bacillus subtilis chromatin-associated protein Hbsu is involved in DNA repair and recombination. Mol Microbiol 23:1169–1179. doi: 10.1046/j.1365-2958.1997.3061670.x. [DOI] [PubMed] [Google Scholar]
- 59.Wiedermannová J, Sudzinová P, Koval T, Rabatinová A, Šanderova H, Ramaniuk O, Rittich Š, Dohnálek J, Fu Z, Halada P, Lewis P, Krásny L. 2014. Characterization of HelD, an interacting partner of RNA polymerase from Bacillus subtilis. Nucleic Acids Res 42:5151–5163. doi: 10.1093/nar/gku113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Prajapati RK, Sengupta S, Rudra P, Mukhopadhyay J. 2016. Bacillus subtilis δ factor functions as a transcriptional regulator by facilitating the open complex formation. J Biol Chem 291:1064–1075. doi: 10.1074/jbc.M115.686170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kusuya Y, Kurokawa K, Ishikawa S, Ogasawara N, Oshima T. 2011. Transcription factor GreA contributes to resolving promoter-proximal pausing of RNA polymerase in Bacillus subtilis cells. J Bacteriol 193:3090–3099. doi: 10.1128/JB.00086-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mondal S, Yakhnin AV, Sebastian A, Albert I, Babitzke P. 2016. NusA-dependent transcription termination prevents misregulation of global gene expression. Nat Microbiol 1:15007. doi: 10.1038/nmicrobiol.2015.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Quirk PG, Dunkey EA, Lee P, Krulwich TA. 1993. Identification of a putative Bacillus subtilis rho gene. J Bacteriol 175:647–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lehnik-Habrink M, Lewis RJ, Mäder U, Stülke J. 2012. RNA degradation in Bacillus subtilis: an interplay of essential endo- and exoribonucleases. Mol Microbiol 84:1005–1017. doi: 10.1111/j.1365-2958.2012.08072.x. [DOI] [PubMed] [Google Scholar]
- 65.Graumann PL, Wendrich TM, Weber MH, Schröder K, Marahiel MA. 1997. A family of cold shock proteins in Bacillus subtilis is essential for cellular growth and for efficient protein synthesis at optimal and low temperatures. Mol Microbiol 25:741–756. doi: 10.1046/j.1365-2958.1997.5121878.x. [DOI] [PubMed] [Google Scholar]
- 66.Lehnik-Habrink M, Rempeters L, Kovács Á, Wrede C, Baierlein C, Krebber H, Kuipers OP, Stülke J. 2013. The DEAD-box RNA helicases in Bacillus subtilis have multiple functions and act independent from each other. J Bacteriol 195:534–544. doi: 10.1128/JB.01475-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hunt A, Rawlins JP, Thomaides HB, Errington J. 2006. Functional analysis of 11 putative essential genes in Bacillus subtilis. Microbiology 152:2895–2907. doi: 10.1099/mic.0.29152-0. [DOI] [PubMed] [Google Scholar]
- 68.Commichau FM, Rothe FM, Herzberg C, Wagner E, Hellwig D, Lehnik-Habrink M, Hammer E, Völker U, Stülke J. 2009. Novel activities of glycolytic enzymes in Bacillus subtilis: interactions with essential proteins involved in mRNA processing. Mol Cell Proteomics 8:1350–1360. doi: 10.1074/mcp.M800546-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lehnik-Habrink M, Newman J, Rothe FM, Solovyova AS, Rodrigues C, Herzberg C, Commichau FM, Lewis RJ, Stülke J. 2011. RNase Y in Bacillus subtilis: a natively disordered protein that is the functional equivalent of RNase E from Escherichia coli. J Bacteriol 193:5431–5441. doi: 10.1128/JB.05500-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cardenas PP, Carrasco B, Sanchez H, Deikus G, Bechhofer DH, Alonso JC. 2009. Bacillus subtilis polynucleotide phosphorylase 3′-to-5′ DNase activity is involved in DNA repair. Nucleic Acids Res 37:4157–4169. doi: 10.1093/nar/gkp314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nelersa CM, Schmier BJ, Malhotra A. 2011. Purification and crystallization of Bacillus subtilis NrnA, a novel enzyme involved in nanoRNA degradation. Acta Crystallogr Sect F Struct Biol Cryst Commun 67:1235–1238. doi: 10.1107/S1744309111026509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pelchat M, Lapointe J. 1999. Aminoacyl-tRNA synthetase genes of Bacillus subtilis: organization and regulation. Biochem Cell Biol 77:343–347. doi: 10.1139/o99-040. [DOI] [PubMed] [Google Scholar]
- 73.Williams-Wagner RN, Grundy FJ, Raina M, Ibba M, Henkin TM. 2015. The Bacillus subtilis tyrZ gene encodes a highly selective tyrosyl-tRNA synthetase and is regulated by a MarR regulator and T box riboswitch. J Bacteriol 197:1624–1631. doi: 10.1128/JB.00008-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Curnow AW, Hong KW, Yuan R, Kim SI, Martins O, Winkler W, Henkin TM, Söll D. 1997. Glu-tRNAGln amidotransferase: a novel heterotrimeric enzyme required for correct decoding of glutamine codons during translation. Proc Natl Acad Sci U S A 94:11819–11826. doi: 10.1073/pnas.94.22.11819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Akanuma G, Nanamiya H, Natori Y, Yano K, Suzuki S, Omata S, Ishizuka M, Sekine Y, Kawamura F. 2012. Inactivation of ribosomal protein genes in Bacillus subtilis reveals importance of each ribosomal protein for cell proliferation and cell differentiation. J Bacteriol 194:6282–6291. doi: 10.1128/JB.01544-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Natori Y, Nanamiya H, Akanuma G, Kosono S, Kudo T, Ochi K, Kawamura F. 2007. A fail-safe system for the ribosome under zinc-limiting conditions in Bacillus subtilis. Mol Microbiol 63:294–307. doi: 10.1111/j.1365-2958.2006.05513.x. [DOI] [PubMed] [Google Scholar]
- 77.Gabriel SE, Helmann JD. 2009. Contributions of Zur-controlled ribosomal proteins to growth under zinc starvation conditions. J Bacteriol 191:6116–6122. doi: 10.1128/JB.00802-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Schmalisch M, Langbein I, Stülke J. 2002. The general stress protein Ctc of Bacillus subtilis is a ribosomal protein. J Mol Microbiol Biotechnol 4:495–501. [PubMed] [Google Scholar]
- 79.Yano K, Wada T, Suzuki S, Tagami K, Matsumoto T, Shiwa Y, Ishige T, Kawaguchi Y, Masuda K, Akanuma G, Nanamiya H, Niki H, Yoshikawa H, Kawamura F. 2013. Multiple rRNA operons are essential for cell growth and sporulation as well as outgrowth in Bacillus subtilis. Microbiology 159:2225–2236. doi: 10.1099/mic.0.067025-0. [DOI] [PubMed] [Google Scholar]
- 80.Britton RA, Wen T, Schaefer L, Pellegrini O, Uicker WC, Mathy N, Tobin C, Daou R, Szyk J, Condon C. 2007. Maturation of the 5′ end of Bacillus subtilis 16S rRNA by the essential ribonuclease YkqC/RNase J1. Mol Microbiol 63:127–138. doi: 10.1111/j.1365-2958.2006.05499.x. [DOI] [PubMed] [Google Scholar]
- 81.Gilet L, DiChiara JM, Figaro S, Bechhofer DH, Condon C. 2015. Small stable RNA maturation and turnover in Bacillus subtilis. Mol Microbiol 95:270–282. doi: 10.1111/mmi.12863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wen T, Oussenko IA, Pellegrini O, Bechhofer DH, Condon C. 2005. Ribonuclease PH plays a major role in the exonucleolytic maturation of CCA-containing tRNA precursors in Bacillus subtilis. Nucleic Acids Res 33:3636–3643. doi: 10.1093/nar/gki675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hartmann RK, Gössringer M, Späth B, Fischer S, Marchfelder A. 2009. The making of tRNAs and more—RNase P and tRNase Z. Prog Mol Biol Transl Sci 85:319–368. doi: 10.1016/S0079-6603(08)00808-8. [DOI] [PubMed] [Google Scholar]
- 84.Bylund GO, Wipemo LC, Lundberg LA, Wikström PM. 1998. RimM and RbfA are essential for efficient processing of 16S rRNA in Escherichia coli. J Bacteriol 180:73–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lövgren JM, Bylund GO, Srivastava MK, Lundberg LA, Persson OP, Wingsle G, Wikström PM. 2004. The PRC-barrel domain of the ribosome maturation protein RimM mediates binding to ribosomal protein S19 in the 30S ribosomal subunits. RNA 10:1798–1812. doi: 10.1261/rna.7720204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Schaefer L, Uicker WC, Wicker-Planquart C, Foucher AE, Jault JM, Britton RA. 2006. Multiple GTPases participate in the assembly of the large ribosomal subunit in Bacillus subtilis. J Bacteriol 188:8252–8258. doi: 10.1128/JB.01213-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Britton RA. 2009. Role of GTPases in ribosome assembly. Annu Rev Microbiol 63:155–176. doi: 10.1146/annurev.micro.091208.073225. [DOI] [PubMed] [Google Scholar]
- 88.Doerfel LK, Wohlgemuth I, Kothe C, Peske F, Urlaub H, Rodnina MV. 2013. EF-P is essential for rapid synthesis of proteins containing consecutive proline residues. Science 339:85–88. doi: 10.1126/science.1229017. [DOI] [PubMed] [Google Scholar]
- 89.Ude S, Lassak J, Starosta AL, Kraxenberger T, Wilson DN, Jung K. 2013. Translation elongation factor EF-P alleviates ribosome stalling at polyproline stretches. Science 339:82–85. doi: 10.1126/science.1228985. [DOI] [PubMed] [Google Scholar]
- 90.You C, Lu H, Sekowska A, Fang G, Wang Y, Gilles AM, Danchin A. 2005. The two authentic methionine aminopeptidase genes are differentially expressed in Bacillus subtilis. BMC Microbiol 5:57. doi: 10.1186/1471-2180-5-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Karzai AW, Roche ED, Sauer RT. 2000. The SsrA-SmpB system for protein tagging, directed degradation and ribosome rescue. Nat Struct Biol 7:449–455. doi: 10.1038/75843. [DOI] [PubMed] [Google Scholar]
- 92.Wiegert T, Schumann W. 2001. SsrA-mediated tagging in Bacillus subtilis. J Bacteriol 183:3885–3889. doi: 10.1128/JB.183.13.3885-3889.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Menez J, Buckingham RH, de Zamaroczy M, Campelli CK. 2002. Peptidyl-tRNA hydrolase in Bacillus subtilis, encoded by SpoVC, is essential to vegetative growth, whereas the homologous enzyme in Saccharomyces cerevisiae is dispensable. Mol Microbiol 45:123–129. doi: 10.1046/j.1365-2958.2002.02992.x. [DOI] [PubMed] [Google Scholar]
- 94.Baird NJ, Zhang J, Hamma T, Ferré-D'Amaré AR. 2012. YbxF and YlxQ are bacterial homologs of L7Ae and bind K-turns but not K-loops. RNA 18:759–770. doi: 10.1261/rna.031518.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Beckert B, Kedrov A, Sohmen D, Kempf G, Wild K, Sinning I, Stahlberg H, Wilson DN, Beckmann R. 2015. Translational arrest by a prokaryotic signal recognition particle is mediated by RNA interactions. Nat Struct Mol Biol 22:767–773. doi: 10.1038/nsmb.3086. [DOI] [PubMed] [Google Scholar]
- 96.Von Loeffelholz O, Knoops K, Ariosa A, Zhang X, Karuppasamy M, Huard K, Schoehn G, Berger I, Shan SO, Schaffitzel C. 2013. Structural basis of signal sequence surveillance and selection by the SRP-FtsY complex. Nat Struct Mol Biol 20:604–610. doi: 10.1038/nsmb.2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Nakamura K, Yahagi S, Yamazaki T, Yamane K. 1999. Bacillus subtilis histone-like protein, HBsu, is an integral component of a SRP-like particle that can bind the Alu domain of small cytoplasmic RNA. J Biol Chem 274:13569–13576. doi: 10.1074/jbc.274.19.13569. [DOI] [PubMed] [Google Scholar]
- 98.Nakamura K, Nishiguchi M, Honda K, Yamane K. 1994. The Bacillus subtilis SRP54 homologue, Ffh, has an intrinsic GTPase activity and forms a ribonucleoprotein complex with small cytoplasmic RNA in vivo. Biochem Biophys Res Commun 199:1394–1399. doi: 10.1006/bbrc.1994.1385. [DOI] [PubMed] [Google Scholar]
- 99.Zanen G, Antelmann H, Meima R, Jongbloed JD, Kolkman M, Hecker M, van Dijl JM, Quax WJ. 2006. Proteomic dissection of potential signal recognition particle dependence in protein secretion by Bacillus subtilis. Proteomics 6:3636–3648. doi: 10.1002/pmic.200500560. [DOI] [PubMed] [Google Scholar]
- 100.Müller JP, Bron S, Venema G, van Dijl JM. 2000. Chaperone-like activities of the CsaA protein of Bacillus subtilis. Microbiology 146:77–88. doi: 10.1099/00221287-146-1-77. [DOI] [PubMed] [Google Scholar]
- 101.Zimmer J, Nam Y, Rapoport TA. 2008. Structure of a complex of the ATPase SecA and the protein-translocation channel. Nature 455:936–943. doi: 10.1038/nature07335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Van der Wolk J, Klose M, Breukink E, Demel RA, de Kruiff B, Freudl R, Driessen AJ. 1993. Characterization of a Bacillus subtilis SecA mutant protein deficient in translocation ATPase and release from the membrane. Mol Microbiol 8:31–42. doi: 10.1111/j.1365-2958.1993.tb01200.x. [DOI] [PubMed] [Google Scholar]
- 103.Saller MJ, Fusetti F, Driessen AJ. 2009. Bacillus subtilis SpoIIIJ and YqjG function in membrane protein biogenesis. J Bacteriol 191:6749–6757. doi: 10.1128/JB.00853-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Tjalsma H, Bron S, van Dijl JM. 2003. Complementary impact of paralogous Oxa1-like proteins of Bacillus subtilis on post-translocational stages in protein secretion. J Biol Chem 278:15622–15632. doi: 10.1074/jbc.M301205200. [DOI] [PubMed] [Google Scholar]
- 105.Saller MJ, Otto A, Berrelkamp-Lahpor GA, Becher D, Hecker M, Driessen A. 2011. Bacillus subtilis YqjG is required for genetic competence development. Proteomics 11:270–282. doi: 10.1002/pmic.201000435. [DOI] [PubMed] [Google Scholar]
- 106.Tjalsma H, Bolhuis A, van Roosmalen ML, Wiegert T, Schumann W, Broekhuizen CP, Quax WJ, Venema G, Bron S, van Dijl JM. 1998. Functional analysis of the secretory precursor processing machinery of Bacillus subtilis: identification of a eubacterial homolog of archeal and eukaryotic signal peptidases. Genes Dev 12:2318–2331. doi: 10.1101/gad.12.15.2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.van Dijl JM, de Jong A, Vehmaanperä J, Venema G, Bron S. 1992. Signal peptidase I of Bacillus subtilis: patterns of conserved amino acids in prokaryotic and eukaryotic type I signal peptidases. EMBO J 11:2819–2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Tjalsma H, Kontinen VP, Prágai Z, Wu H, Meima R, Venema G, Bron S, Sarvas M, van Dijl JM. 1999. The role of lipoprotein processing by signal peptidase II in the Gram-positive eubacterium Bacillus subtilis. Signal peptidase II is required for the efficient secretion of alpha-amylase, a non-lipoprotein. J Biol Chem 274:1698–1707. [DOI] [PubMed] [Google Scholar]
- 109.Okuda S, Tokuda H. 2011. Lipoprotein sorting in bacteria. Annu Rev Microbiol 65:239–259. doi: 10.1146/annurev-micro-090110-102859. [DOI] [PubMed] [Google Scholar]
- 110.Leskelä S, Wahlstrüm E, Kontinen VP, Sarvas M. 1999. Lipid modification of prelipoproteins is dispensable for growth but essential for efficient protein secretion in Bacillus subtilis: characterization of the Lgt gene. Mol Microbiol 31:1075–1085. doi: 10.1046/j.1365-2958.1999.01247.x. [DOI] [PubMed] [Google Scholar]
- 111.Kontinen VP, Sarvas M. 1993. The PrsA lipoprotein is essential for protein secretion in Bacillus subtilis and sets a limit for high-level secretion. Mol Microbiol 8:727–737. doi: 10.1111/j.1365-2958.1993.tb01616.x. [DOI] [PubMed] [Google Scholar]
- 112.Hyyryläinen HL, Marciniak BC, Dahncke K, Pietiäinen M, Courtin P, Vitikainen M, Seppala R, Otto A, Becher D, Chapot-Chartier MP, Kuipers OP, Kontinen VP. 2010. Penicillin-binding protein folding is dependent on the PrsA peptidyl-prolyl cis-trans isomerase in Bacillus subtilis. Mol Microbiol 77:108–127. doi: 10.1111/j.1365-2958.2010.07188.x. [DOI] [PubMed] [Google Scholar]
- 113.Tjalsma H, Antelmann H, Jongbloed JD, Braun PG, Darmon E, Dorenbos R, Dubois JY, Westers H, Zanen G, Quax WJ, Kuipers OP, Bron S, Hecker M, van Dijl JM. 2004. Proteomics of protein secretion by Bacillus subtilis: separating the “secrets” of the secretome. Microbiol Mol Biol Rev 68:207–233. doi: 10.1128/MMBR.68.2.207-233.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Jongbloed JD, Grieger U, Antelmann H, Hecker M, Nijland R, Bron S, van Dijl JM. 2004. Two minimal Tat translocases in Bacillus. Mol Microbiol 54:1319–1325. doi: 10.1111/j.1365-2958.2004.04341.x. [DOI] [PubMed] [Google Scholar]
- 115.Baptista C, Barreto HC, São-José C. 2013. High levels of DegU-P activate an ESAT-6-like secretion system in Bacillus subtilis. PLoS One 8:e67840. doi: 10.1371/journal.pone.0067840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Hallamaa KM, Browning GF, Tang SL. 2006. Lipoprotein multigene families in Mycoplasma pneumoniae. J Bacteriol 188:5393–5399. doi: 10.1128/JB.01819-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Halbedel S, Hames C, Stülke J. 2007. Regulation of carbon metabolism in the mollicutes and its relation to virulence. J Mol Microbiol Biotechnol 12:147–154. [DOI] [PubMed] [Google Scholar]
- 118.Lund PA. 2001. Microbial molecular chaperones. Adv Microb Physiol 44:93–140. doi: 10.1016/S0065-2911(01)44012-4. [DOI] [PubMed] [Google Scholar]
- 119.Deuerling E, Bukau B. 2004. Chaperone-assisted folding of newly synthesized proteins in the cytosol. Crit Rev Biochem Mol Biol 39:261–277. doi: 10.1080/10409230490892496. [DOI] [PubMed] [Google Scholar]
- 120.Dalbey RE, Wang P, van Dijl JM. 2012. Membrane proteases in the bacterial protein secretion and quality control pathway. Microbiol Mol Biol Rev 76:311–330. doi: 10.1128/MMBR.05019-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Pohl S, Bhavsar G, Hulme J, Bloor AE, Misirli G, Leckenby MW, Radford DS, Smith W, Wipat A, Williamson ED, Harwood CR, Cranenburgh RM. 2013. Proteomic analysis of Bacillus subtilis strains engineered for improved production of heterologous proteins. Proteomics 13:3298–3308. doi: 10.1002/pmic.201300183. [DOI] [PubMed] [Google Scholar]
- 122.Stülke J, Hillen W. 2000. Regulation of carbon catabolism in Bacillus species. Annu Rev Microbiol 54:849–880. doi: 10.1146/annurev.micro.54.1.849. [DOI] [PubMed] [Google Scholar]
- 123.Buescher JM, Liebermeister W, Jules M, Uhr M, Muntel J, Botella E, Hessling B, Kleijn RJ, Le Chat L, Lecointe F, Mäder U, Nicolas P, Piersma S, Rügheimer F, Becher D, Bessieres P, Bidnenko E, Denham EL, Dervyn E, Devine KM, Doherty G, Drulhe S, Felicori L, Fogg MJ, Goelzer A, Hansen A, Harwood CR, Hecker M, Hubner S, Hultschig C, Jarmer H, Klipp E, Leduc A, Lewis P, Molina F, Noirot P, Peres S, Pigeonneau N, Pohl S, Rasmussen S, Rinn B, Schaffer M, Schnidder J, Schwikowski B, Van Dijl JM, Veiga P, Walsh S, Wilkinson AJ, Stelling J, Aymerich S, Sauer U. 2012. Global network reorganization during dynamic adaptations of Bacillus subtilis metabolism. Science 335:1099–1103. doi: 10.1126/science.1206871. [DOI] [PubMed] [Google Scholar]
- 124.Starai VJ, Escalante-Semerena JC. 2004. Acetyl-coenzyme A synthetase (AMP forming). Cell Mol Life Sci 61:2020–2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Rühl M, Le Coq D, Aymerich S, Sauer U. 2012. 13C flux analysis reveals NADPH-balancing transhydrogenation cycles in stationary phase of nitrogen starving Bacillus subtilis. J Biol Chem 287:27959–27970. doi: 10.1074/jbc.M112.366492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Winstedt L, von Wachenfeldt C. 2000. Terminal oxidases of Bacillus subtilis strain 168: one quinol oxidase, cytochrome aa3 or cytochrome bd, is required for aerobic growth. J Bacteriol 182:6557–6564. doi: 10.1128/JB.182.23.6557-6564.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Härtig E, Hartmann A, Schätzle M, Albertini AM, Jahn D. 2006. The Bacillus subtilis nrdEF genes, encoding a class Ib ribonucleotide reductase, are essential for aerobic and anaerobic growth. Appl Environ Microbiol 72:5260–5265. doi: 10.1128/AEM.00599-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Endo T, Uratani B, Freese E. 1983. Purine salvage pathways of Bacillus subtilis and effect of guanine on growth of GMP reductase mutants. J Bacteriol 155:169–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Rodionov DA, Mironov AA, Gelfand MS. 2002. Conservation of the biotin regulon and the BirA regulatory signal in eubacteria and archaea. Genome Res 12:1507–1516. doi: 10.1101/gr.314502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Cronan JE Jr, Waldrop GL. 2002. Multi-subunit acetyl-CoA carboxylases. Prog Lipid Res 41:407–435. doi: 10.1016/S0163-7827(02)00007-3. [DOI] [PubMed] [Google Scholar]
- 131.Salzberg LI, Helmann JD. 2008. Phenotypic and transcriptomic characterization of Bacillus subtilis mutants with grossly altered membrane composition. J Bacteriol 190:7797–7807. doi: 10.1128/JB.00720-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Block KF, Hammond MC, Breaker RR. 2010. Evidence for widespread gene control function by the ydaO riboswitch candidate. J Bacteriol 192:3983–3989. doi: 10.1128/JB.00450-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Meeske AJ, Rodrigues CD, Brady J, Lim HC, Bernhardt TG, Rudner DZ. 2016. High-throughput genetic screens identify a large and diverse collection of new sporulation genes in Bacillus subtilis. PLoS Biol 14:e1002341. doi: 10.1371/journal.pbio.1002341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Eitinger T, Rodionov DA, Grote M, Schneider E. 2011. Canonical and ECF-type ATP-binding cassette importers in prokaryotes: diversity in modular organization and cellular functions. FEMS Microbiol Rev 35:3–67. doi: 10.1111/j.1574-6976.2010.00230.x. [DOI] [PubMed] [Google Scholar]
- 135.Slotboom DJ. 2014. Structural and mechanistic insights into prokaryotic energy-coupling factor transporters. Nat Rev Microbiol 12:79–87. doi: 10.1038/nrmicro3175. [DOI] [PubMed] [Google Scholar]
- 136.Ollinger J, Song KB, Antelmann H, Hecker M, Helmann JD. 2006. Role of the Fur regulon in iron transport in Bacillus subtilis. J Bacteriol 188:3664–3673. doi: 10.1128/JB.188.10.3664-3673.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Miethke M, Monteferrante CG, Marahiel MA, van Dijl JM. 2013. The Bacillus subtilis EfeUOB transporter is essential for high-affinity acquisition of ferrous and ferric iron. Biochim Biophys Acta 1833:2267–2278. doi: 10.1016/j.bbamcr.2013.05.027. [DOI] [PubMed] [Google Scholar]
- 138.Kajiyama Y, Otagiri M, Sekiguchi J, Kosono S, Kudo T. 2007. Complex formation by the mrpABCDEFG gene products, which constitute a principal Na+/H+ antiporter in Bacillus subtilis. J Bacteriol 189:7511–7514. doi: 10.1128/JB.00968-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Dos Santos P. 2014. Fe-S assembly in Gram-positive bacteria, p 347–366. In Rouault TA. (ed), Iron-sulfur clusters in chemistry and biology. De Gruyter, Berlin, Germany. [Google Scholar]
- 140.Mouilleron S, Badet-Denizot MA, Badet B, Golinelli-Pimpaneau B. 2011. Dynamics of glucosamine-6-phosphate synthase catalysis. Arch Biochem Biophys 505:1–12. doi: 10.1016/j.abb.2010.08.008. [DOI] [PubMed] [Google Scholar]
- 141.Scheffers DJ, Tol MB. 2015. LipidII: just another brick in the wall? PLoS Pathog 11:e1005213. doi: 10.1371/journal.ppat.1005213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Meeske AJ, Sham LT, Kimsey H, Koo BM, Gross CA, Bernhardt TG, Rudner DZ. 2015. MurJ and a novel lipid II flippase are required for cell wall biogenesis in Bacillus subtilis. Proc Natl Acad Sci U S A 112:6437–6442. doi: 10.1073/pnas.1504967112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Kawai Y, Daniel RA, Errington J. 2009. Regulation of cell wall morphogenesis in Bacillus subtilis by recruitment of PBP1 to the MreB helix. Mol Microbiol 71:1131–1144. doi: 10.1111/j.1365-2958.2009.06601.x. [DOI] [PubMed] [Google Scholar]
- 144.Daniel RA, Harry EJ, Errington J. 2000. Role of penicillin-binding protein PBP 2B in assembly and functioning of the division machinery in Bacillus subtilis. Mol Microbiol 35:299–311. doi: 10.1046/j.1365-2958.2000.01724.x. [DOI] [PubMed] [Google Scholar]
- 145.Wei Y, Havasy T, McPherson DC, Popham DL. 2003. Rod shape determination by the Bacillus subtilis class B penicillin-binding proteins encoded by pbpA and pbpH. J Bacteriol 185:4717–4726. doi: 10.1128/JB.185.16.4717-4726.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Vollmer W. 2012. Bacterial growth does require peptidoglycan hydrolases. Mol Microbiol 86:1031–1035. doi: 10.1111/mmi.12059. [DOI] [PubMed] [Google Scholar]
- 147.Ohnishi R, Ishikawa S, Sekiguchi J. 1999. Peptidoglycan hydrolase LytF plays a role in cell separation with CwlF during vegetative growth of Bacillus subtilis. J Bacteriol 181:3178–3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Hashimoto M, Ooiwa S, Sekiguchi J. 2012. Synthetic lethality of the lytE cwlO genotype in Bacillus subtilis is caused by lack of d,l-endopeptidase activity at the lateral cell wall. J Bacteriol 194:796–803. doi: 10.1128/JB.05569-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Dominguez-Cuevas P, Porcelli I, Daniel RA, Errington J. 2013. Differentiated roles for MreB-actin isologues and autolytic enzymes in Bacillus subtilis morphogenesis. Mol Microbiol 89:1084–1098. doi: 10.1111/mmi.12335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Brown S, Santa Maria JP, Walker S. 2013. Wall teichoic acids of gram-positive bacteria. Annu Rev Microbiol 67:313–336. doi: 10.1146/annurev-micro-092412-155620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Matsuoka S, Chiba M, Tanimura Y, Hashimoto M, Hara H, Matsumoto K. 2011. Abnormal morphology of Bacillus subtilis ugtP mutant cells lacking glucolipids. Genes Genet Syst 86:295–304. doi: 10.1266/ggs.86.295. [DOI] [PubMed] [Google Scholar]
- 152.Matsuoka S, Hashimoto M, Kamiya Y, Miyazawa T, Ishikawa K, Hara H, Matsumoto K. 2011. The Bacillus subtilis essential gene dgkB is dispensable in mutants with defective lipoteichoic acid biosynthesis. Genes Genet Syst 86:365–376. doi: 10.1266/ggs.86.365. [DOI] [PubMed] [Google Scholar]
- 153.Percy MG, Gründling A. 2014. Lipoteichoic acid synthesis and function in gram-positive bacteria. Annu Rev Microbiol 68:81–100. doi: 10.1146/annurev-micro-091213-112949. [DOI] [PubMed] [Google Scholar]
- 154.Robson SA, Michie KA, Mackay JP, Harry E, King GF. 2002. The Bacillus subtilis cell division proteins FtsL and DivIC are intrinsically unstable and do not interact with one another in the absence of other septasomal components. Mol Microbiol 44:663–674. doi: 10.1046/j.1365-2958.2002.02920.x. [DOI] [PubMed] [Google Scholar]
- 155.Szwedziak P, Löwe J. 2013. Do the divisome and elongasome share a common evolutionary past? Curr Opin Microbiol 16:745–751. doi: 10.1016/j.mib.2013.09.003. [DOI] [PubMed] [Google Scholar]
- 156.Edwards DH, Errington J. 1997. The Bacillus subtilis DivIVA protein targets to the division septum and controls the site specificity of cell division. Mol Microbiol 24:905–915. doi: 10.1046/j.1365-2958.1997.3811764.x. [DOI] [PubMed] [Google Scholar]
- 157.Bramkamp M, van Baarle S. 2009. Division site selection in rod-shaped bacteria. Curr Opin Microbiol 12:683–688. doi: 10.1016/j.mib.2009.10.002. [DOI] [PubMed] [Google Scholar]
- 158.Biller SJ, Wayne KJ, Winkler ME, Burkholder WF. 2011. The putative hydrolase YycJ (WalJ) affects the coordination of cell division with DNA replication in Bacillus subtilis and may play a conserved role in cell wall metabolism. J Bacteriol 193:896–908. doi: 10.1128/JB.00594-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Lluch-Senar M, Querol E, Piñol J. 2010. Cell division in a minimal bacterium in the absence of ftsZ. Mol Microbiol 78:278–289. doi: 10.1111/j.1365-2958.2010.07306.x. [DOI] [PubMed] [Google Scholar]
- 160.Leaver M, Dominguez-Cuevas P, Coxhead JM, Daniel RA, Errington J. 2009. Life without a wall or division machine in Bacillus subtilis. Nature 457:849–853. doi: 10.1038/nature07742. [DOI] [PubMed] [Google Scholar]
- 161.Mercier R, Kawai Y, Errington J. 2014. General principles for the formation and proliferation of a wall-free (L-form) state in bacteria. eLife 3:e04629. doi: 10.7554/eLife.04629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Bisicchia P, Noone D, Lioliou E, Howell A, Quigley S, Jensen T, Jarmer H, Devine KM. 2007. The essential YycFG two-component system controls cell wall metabolism in Bacillus subtilis. Mol Microbiol 65:180–200. doi: 10.1111/j.1365-2958.2007.05782.x. [DOI] [PubMed] [Google Scholar]
- 163.Dubrac S, Bisicchia P, Devine KM, Msadek T. 2008. A matter of life and death: cell wall homeostasis and the WalKR (YycGF) essential signal transduction pathway. Mol Microbiol 70:1307–1322. doi: 10.1111/j.1365-2958.2008.06483.x. [DOI] [PubMed] [Google Scholar]
- 164.Luo Y, Helmann JD. 2012. Analysis of the role of Bacillus subtilis σM in β-lactam resistance reveals an essential role for c-di-AMP in peptidoglycan synthesis. Mol Microbiol 83:623–639. doi: 10.1111/j.1365-2958.2011.07953.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Commichau FM, Dickmanns A, Gundlach J, Ficner R, Stülke J. 2015. A jack of all trades: the multiple roles of the unique essential second messenger cyclic di-AMP. Mol Microbiol 97:189–204. doi: 10.1111/mmi.13026. [DOI] [PubMed] [Google Scholar]
- 166.Gundlach J, Mehne FM, Herzberg C, Kampf J, Valerius O, Kaever V, Stülke J. 2015. An essential poison: synthesis and degradation of cyclic di-AMP in Bacillus subtilis. J Bacteriol 197:3265–3274. doi: 10.1128/JB.00564-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Dempwolff F, Wischhusen HM, Specht M, Graumann PL. 2012. The deletion of bacterial dynamin and flotillin genes results in pleiotropic effects on cell division, cell growth, and in cell shape maintenance. BMC Microbiol 12:298. doi: 10.1186/1471-2180-12-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Rosenberg J, Dickmanns A, Neumann P, Gunka K, Arens J, Kaever V, Stülke J, Ficner R, Commichau FM. 2015. Structural and biochemical analysis of the essential diadenylate cyclase CdaA from Listeria monocytogenes. J Biol Chem 290:6596–6606. doi: 10.1074/jbc.M114.630418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Bai Y, Yang J, Zarella TM, Zhang Y, Metzger DW, Bai G. 2014. Cyclic di-AMP impairs potassium uptake mediated by a cyclic di-AMP binding protein in Streptococcus pneumoniae. J Bacteriol 196:614–623. doi: 10.1128/JB.01041-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Opoku-Temeng C, Sintim HO. 2016. Potent inhibition of cyclic diadenylate monophosphate cyclase by the antiparasite drug, suramin. Chem Commun (Camb) 52:3754–3757. doi: 10.1039/C5CC10446G. [DOI] [PubMed] [Google Scholar]
- 171.Stacy R, Anderson WF, Myler PJ. 2015. Structural genomics support for infectious disease drug design. ACS Infect Dis 1:127–129. doi: 10.1021/id500048p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Rahmer R, Heravi KM, Altenbuchner J. 2015. Construction of a super-competent Bacillus subtilis 168 using the pmtlA-comKS inducible cassette. Front Microbiol 6:1431. doi: 10.3389/fmicb.2015.01431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Kumpfmüller J, Kabisch J, Schweder T. 2013. An optimized technique for rapid genome modifications of Bacillus subtilis. J Microbiol Methods 95:350–352. doi: 10.1016/j.mimet.2013.10.003. [DOI] [PubMed] [Google Scholar]
- 174.Morimoto T, Ara K, Ozaki K, Ogasawara N. 2009. A new simple method to introduce marker-free deletions in the Bacillus subtilis genome. Genes Genet Syst 84:315–318. doi: 10.1266/ggs.84.315. [DOI] [PubMed] [Google Scholar]
- 175.Fabret C, Ehrlich SD, Noirot P. 2002. A new mutation delivery system for genome-scale approaches in Bacillus subtilis. Mol Microbiol 46:25–36. doi: 10.1046/j.1365-2958.2002.03140.x. [DOI] [PubMed] [Google Scholar]
- 176.Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA, Smith HO. 2009. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 177.de Kok S, Stanton LH, Slaby T, Durot M, Holmes VF, Patel KG, Platt D, Shapland EB, Serber Z, Dean J, Newman JD, Chandran SS. 2014. Rapid and reliable DNA assembly via ligase cycling reaction. ACS Synth Biol 3:97–106. doi: 10.1021/sb4001992. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.