Abstract
Background & objectives:
Dengue fever (DF) is associated with significant morbidity and mortality in the tropical and sub-tropical regions of the world. Since there are no effective antiviral drugs for treatment, clinicians often rely on the accurate diagnosis of dengue fever to begin supportive therapy at early stages of the illness. The objective of this study was to develop an in-house dengue virus serotype 2 (DENV-2) non-structural protein- 5 (NS5) based indirect ELISA.
Methods:
DENV-2 was raised in Vero cells and the viral proteins were separated and subsequently the NS5 protein was eluted. Serum samples from primary and secondary dengue fever patients; and acute and convalescent samples from Japanese encephalitis (JE) and West Nile virus (WNV) cases were used to validate the ELISA.
Results:
The assay was found to be 100 per cent specific in detecting DENV-2 specific antibodies from patient's serum. However, in terms of sensitivity, the assay could detect IgM antibodies only from 90 per cent of the primary dengue samples. The IgM/IgG ratio of the primary and secondary samples was 7.24 and 0.64, respectively.
Interpretation & conclusions:
The results indicate that the DENV-2 NS5 ELISA is dengue group specific and can be used to differentiate dengue infection from other circulating Flavivirus infections. This NS5 ELISA can also be used to distinguish between primary and secondary dengue fever on the basis of IgM/IgG ratios. Further studies with larger sample sizes and different DENV serotypes are required to validate the ELISA.
Keywords: Dengue fever, DENV-2, diagnosis, ELISA, infection, proteins, serology
Dengue fever (DF) is a mosquito-borne viral disease which causes over 50-100 million infections every year in the tropical and subtropical regions of the world. Dengue virus (DENV), the aetiological agent of this disease, belongs to the genus Flavivirus and family Flaviviridae and comprises four antigenically distinct serotypes (DENV1-4). Individuals infected by any one serotype will possess life-long immunity against re-infection by the virus, but only partial protection against infection by another dengue virus serotype. Such infections with a different serotype often lead to severe clinical manifestations such as dengue haemorrhagic fever (DHF) and/or dengue shock syndrome (DSS) which are often fatal in nature1. A reliable method to differentiate primary and secondary dengue fever would enable clinicians to predict the severity of the outcome of dengue fever in vulnerable populations2. Given the severe morbidity and mortality associated with dengue fever, there is a need for a reliable and cost-effective method to diagnose dengue fever.
Primary infection with any dengue virus serotype is characterized by a rise in virus specific IgM antibodies, 4-5 days after the appearance of symptoms. While these are detectable in serum only for 30-90 days, the IgG antibodies persist for a long time, often for life3. During a secondary infection however, the level of dengue virus specific IgM antibodies in serum is generally low and in some cases patients fail to show detectable amounts of these antibodies4. But IgG levels rise rapidly to much higher levels than that observed during the primary dengue infection and often remain at these levels for at least 30-40 days3. During primary infections, the specific IgG antibody response is initially of low avidity which gradually increases thereafter. On the contrary, secondary infections are characterized by an initial production of high-avidity antibodies. Due to this anomaly in the detection of dengue virus specific antibodies, the combined use of IgM and IgG has been proposed as an effective strategy for the serological differentiation of primary and secondary infections5. Dengue virus specific IgG avidity ELISAs work on the basis of differences in IgM and IgG levels to distinguish between primary and secondary dengue fever6.
Serological diagnosis of dengue fever mainly involves detection of DENV specific IgM or IgG antibodies present in patient's serum. A wide range of commercial diagnostic kits with varying sensitivity and specificity are available for the same7. Most of these are based on the use of either envelope/membrane (E/M) antigens or non-structural protein 1 (NS1) antigens8. E/M-specific IgG antibodies have been shown to cross-react with other Flaviviruses during secondary infections9. In case of NS1 serotype-specific antibody ELISA, the infecting DENV serotype is often not correctly identified when convalescent-serum samples from secondary infections are used10. In this study the DENV-2 non-structural protein 5 (NS5) was used to explore its diagnostic potential as an alternative to the currently used NS1 and E/M specific ELISAs. DENV-2 has been known to be frequently associated with outbreaks of dengue fever in various parts of India11. Certain structural differences in DENV-2 virions have been shown to have direct correlation with the increased virulence and pathogenicity associated with infections with this serotype12.
The DENV NS5 is highly immunogenic8 and it has been argued that antibodies directed against this protein are present more during secondary infections when compared to primary cases of dengue fever13. The NS5 protein of West Nile virus (WNV) has been shown to differentiate West Nile from dengue fever and St. Louis encephalitis cases14. We undertook this study with an aim to develop an in-house indirect ELISA based on detection of DENV-2 NS5 protein.
Material & Methods
The study was carried out at the Arbovirus research laboratory, Department of Virology, King Institute of Preventive Medicine and Research, Chennai, India, during the period 2011-2014. The study protocol was approved by the institutional ethics committee.
Case definitions: Dengue fever cases were identified using WHO case definitions15. For sample collection, dengue infection was defined as the isolation of DENV from serum or plasma samples or the detection of specific IgM antibodies against DENV as opposed to anti-JE (Japanese encephalitis) IgM antibodies. Differentiation between primary and secondary dengue fever was done by comparing the IgM/IgG values as described by Shu et al16.
Both JE and West Nile cases were screened on the basis of WHO case definitions17. JE positive cases were identified by the presence of JE virus-specific IgM antibody in a single sample of cerebrospinal fluid or serum, as detected by a JE virus specific IgM-capture ELISA18. WNV infections were confirmed by the capture of WNV specific IgM antibodies in CSF or serum by ELISA19.
Serum samples: Four panels of human serum samples were used in this study.
-
(i)
A total of 780 serum samples were tested, of which 40 collected from dengue fever patients, 20 each from primary and secondary case, were included in the study. Primary dengue fever cases were characterized by a negative IgG test (NIV Dengue IgG capture ELISA, Pune, India) for serum samples collected during acute phase at least four days after disease onset, followed by seroconversion in the convalescent-phase serum sample. Secondary cases were defined by a positive IgG test for samples obtained within four days of disease onset (acute phase) followed by a laboratory-confirmed diagnosis of acute dengue fever (NIV Dengue IgM Capture ELISA, Pune).
-
(ii)
In the case of JE, paired serum samples were collected from 20 patients (out of 660 screened) at an interval of 3-12 days after the onset of illness. Acute samples from all 20 patients were confirmed using an NIV JE IgM capture ELISA (Pune, India).
-
(iii)
Of the 2559 serum samples screened during 2011-2014, 20 paired serum samples were collected from West Nile cases, but at an interval of 4-15 days after the onset of illness. The acute samples were confirmed by PanBio WNV IgM capture ELISA (USA).
-
(iv)
Serum samples were collected from 20 healthy controls (laboratory and technical staff of the Institute) and tested negative for all the above viruses by their respective ELISA kits. All the samples were stored in aliquots at -70°C until use.
Virus and cells: The DENV-2 New Guinea C strain was obtained from the repository of National Institute of Virology (NIV, Pune, India). The virus was raised in five different batches of Vero (African green monkey kidney) cells which were cultured simultaneously in 75 cm2 tissue culture flasks. The cells were maintained in Dulbecco's modified eagles medium (DMEM, Sigma, USA) with 10 per cent heat inactivated foetal bovine serum (FBS, Thermo Fisher Scientific, USA) and 100 units/ml of penicillin and 100 μg/ml streptomycin at 37°C. The medium was aspirated from all flasks, cell monolayer washed three times with sterile phosphate buffer saline (PBS) and then inoculated with 0.5 ml of DENV-2 virus at a multiplicity of infection of 0.1. The flasks were left for 90 min at 37°C with constant agitation for adherence. Virus adsorption was allowed to proceed for 90 min at 37°C with constant agitation. Then 15 ml of DMEM supplemented with 2 per cent heat inactivated foetal bovine serum (FBS, 100 units penicillin-streptomycin per ml) was added to all the flasks and the cells incubated under standard conditions (37°C, 5% CO2). The cells were observed for cytopathic effects up to three days post infection. When the cells showed 80-90 per cent cytopathic effect, the tissue culture supernatant along with the cell monolayer was freeze-thawed consecutively three to four times, aspirated and centrifuged at 14,000 g for 30 min to remove cell debris and the supernatant were taken. The supernatants from all the flasks were pooled together and the final virus titre was determined by plaque assay.
Titration of DENV-2 by plaque assay: Vero cells were suspended in DMEM with 10 per cent heat inactivated FBS and 100 units/ml penicillin, streptomycin and seeded in a 6 cell plate (Nunc, USA) at 1 × 106 cells/well, placed at 37°C with 5 per cent CO2 for 12 h. Various dilutions of the virus were made and allowed to adsorb on the monolayer of cells for 90 min. The inoculum was removed and the cells were overlaid with two per cent agarose supplemented with DMEM containing 2 per cent FBS. After five days of incubation, the cells were stained with crystal violet, washed and number of plaques were counted.
Precipitation and concentration of viral proteins: Precipitation of the virus was carried out using poly ethylene glycol (PEG) 6000 (Sigma-Aldrich, USA) as per protocol20, but with a few modifications. Briefly, the purified virus was mixed with 6 per cent PEG 6000 in 0.5 M NaCl at the ratio of 2:1 (supernatant-PEG). The mixture was incubated overnight at 4°C with gentle agitation. The precipitate was collected, re-suspended in sodium chloride Tris-EDTA (STE) buffer (0.1M NaCl, 10mMTris, 1mM EDTA, pH 7.6) and stored at -80°C until further use.
Separation of DENV-2 proteins: The viral proteins were separated using 10 per cent (sodium dodecyl sulphate-polyacrylamide gel electrophoresis) SDS-PAGE21 with a few alterations. The virus stock without β-mercaptoethanol, was heated on a dry block at 90°C for two min before loading in the gel. A diluted suspension of Vero cells (430 μg/ml) treated in the same manner served as cell control. Molecular weight marker (Fisher Scientific-SM0431, USA) and cell control were added in the first two wells and virus suspensions were loaded in the remaining lanes. After electrophoresis, the first three lanes containing the molecular weight marker, cell control and virus proteins, respectively were cut from the gel and stained with Coomassie brilliant blue R-250 for comparison and preliminary identification of the separated viral proteins. Based on a previous study22, the viral protein band at approximately 105 kDa position was identified as the NS5 protein of dengue virus. The region corresponding to this protein in the remaining portion of the unstained gel was cut and the NS5 protein was eluted with a Nanosep Centrifugal Device with Omega membrane MWCO 300 kDa (Pall Nanosep®, USA) as per the manufacturer's instructions. The total protein content in the eluted product was estimated using a nanodrop-1000 spectrophotometer, USA.
Identification of the separated DENV-2 NS5 protein by indirect immunofluorescence assay (IFA): This method was developed in our laboratory. Briefly, 100 μl of the eluted viral protein was smeared over the surface of a clean glass slide, air dried, fixed with acetone and incubated with 100 μl of 1:1000 diluted DENV-2 NS5 monoclonal antibodies (MAb, Thermo Scientific, Pierce antibodies, product code- MA5-17296, USA). The slides were washed and subsequently incubated with a goat anti-mouse IgG antibody labelled with fluorescein isothiocyanate (Molecular Probes, Eugene, Oregon, USA). The slides were washed twice with PBS, air dried, mounted with glycerol and viewed under a fluorescent microscope (Nikon Eclipse, E200, USA).
Dengue NS5 specific IgM/IgG indirect ELISA: Three 96 well microwell plates (F96 MaxisorpNunc-Immuno Plate) were coated with the DENV-2 NS5 proteins23. The optimum concentration of coating antigen was 20 μg/ml. The samples from primary and secondary dengue fever patients were added in duplicate to separate columns of the plates. The acute and convalescent-phase samples of patients with JE and West Nile infections were also added in separate columns in the plate. Serum sample of healthy controls was used as negative control. The plates were incubated at room temperature for 1 h. After incubation, the contents of the wells were aspirated, washed and blot dried before adding the secondary antibodies. 100 μl of the HRP-conjugated mouse anti-human IgM, Thermo Scientific, Pierce antibodies, and mouse anti-human IgG secondary antibodies labelled with HRP, (Thermo Scientific, USA), both diluted 1:1000 with blocking buffer. The plates were incubated for 1 h, washed and dried and 100 μl tetramethylbenzidine (BD Pharmingen™ TMB Substrate Reagent Set, USA) substrate solution was added. Finally, the plate was incubated in dark for 20 min for colour development, after which 100 μl of 1N Sulphuric acid (H2SO4) was added to stop the reaction. The intensity of colour development was measured using an ELISA plate reader at 450 nm.
Data analysis: Positivity was determined by comparing the test samples with the OD450 values of the healthy controls. The positive and negative samples were defined as having test sample absorbance/negative (healthy) control ratio of ≥2.0 and <2.0, respectively.
Results & Discussion
Clear and distinct plaques were observed in all wells except the cell control. Based on the number of plaques counted in the well inoculated with 10-10 dilution of virus, the virus titre was estimated as 1.8x1011 plaque forming units per ml (Fig. 1). Distinct protein bands were visible in lane 1 corresponding to the Vero cell proteins. Three major bands were observed in lane 2 corresponding to the DENV proteins. The first band was seen at a position of 105 kDa, the second band at 67 and the third at 45 kDa (Fig. 2).
Fig. 1.
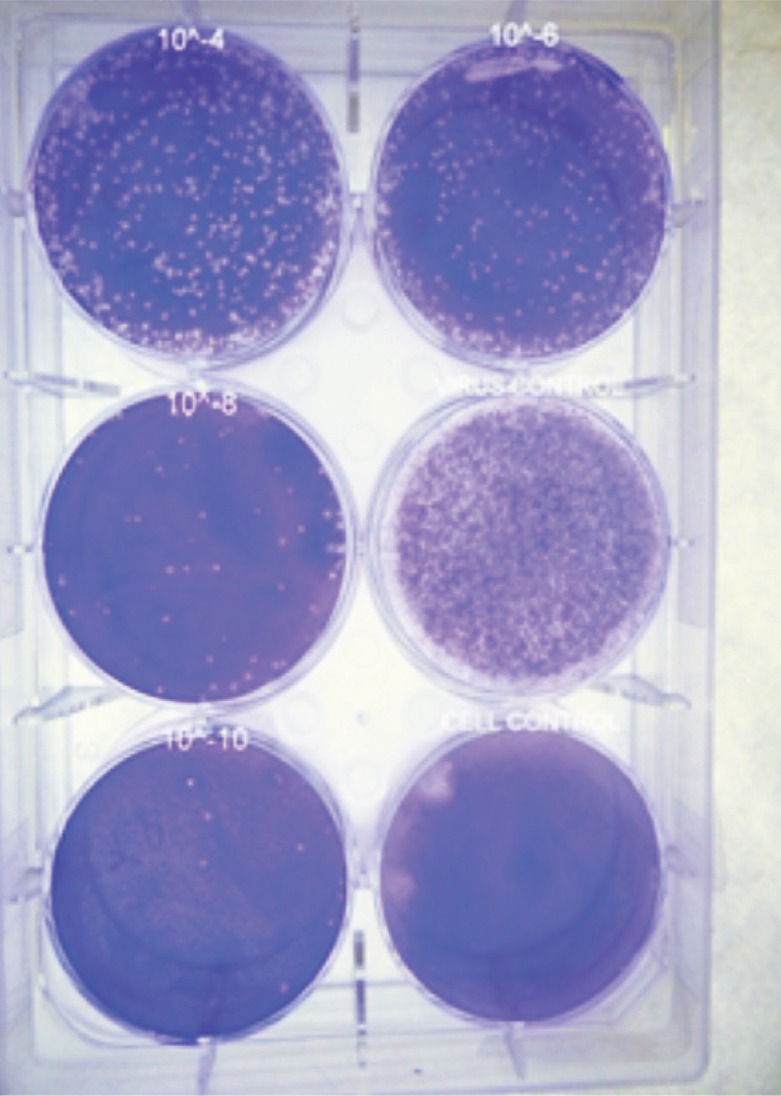
Six-well tissue culture plate showing DENV- 2 plaques. Plaques were observed in all the wells except in cell control well.
Fig. 2.
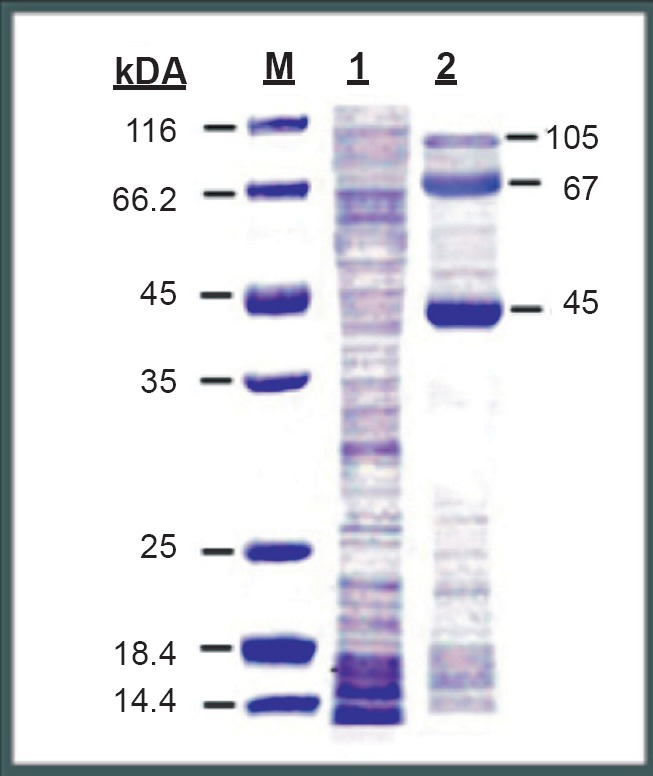
SDS-PAGE gel stained with coommassie brilliant bue-R250. Lane-M shows the protein molecular weight marker bands, Lane-1 Vero cell proteins (cell control) and Lane-2 DENV-2 proteins.
Identification of the separated DENV-2 NS5 protein by immunofluorescence assay: The DENV-2 NS5 MAbs were able to detect the presence of virus specific NS5 protein present in the eluted product. This was evidenced by the green fluorescence (Fig. 3A). No immunofluorescence was observed in the uninfected Vero cells (Fig. 3B).
Fig. 3.
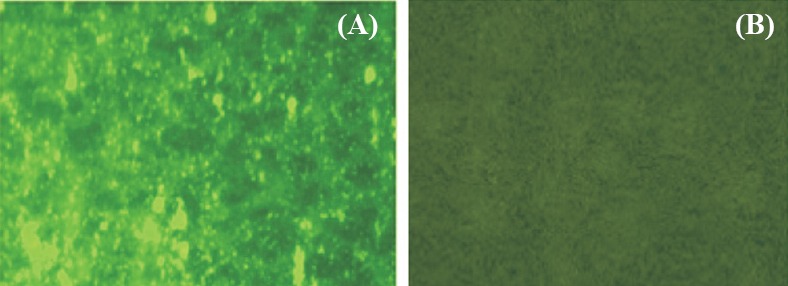
Immunofluorescence images showing (A) green fluorescence showing the presence of dengue virus NS5 protein, and (B) absence of fluorescence signals in uninfected Vero cells.
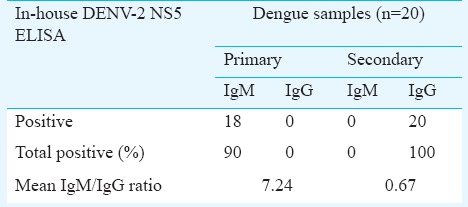
Sensitivity and specificity of DENV-2 NS5 ELISA: Of the 20 samples of primary cases of dengue fever tested, the virus specific IgM antibodies were seen in 18 samples (OD450≥2.0). The remaining two samples had OD450 values slightly lower than 2.0 (1.87 and 1.92, respectively) and were considered negative. All the 20 samples tested negative for IgG antibodies against DENV. The mean IgM/IgG ratio of all samples was found to be 7.24. Among the 20 samples from secondary dengue fever cases, none was positive for virus specific IgM antibodies, and all 20 samples gave positive results for virus specific IgG antibodies. The IgM/IgG ratio of all samples was 0.67 (Table). All the acute as well as convalescent serum samples from patients with JE and West Nile virus infections were negative for virus specific IgM and IgG antibodies. The 20 healthy samples were negative for antibodies against all the three diseases. The sensitivity and specificity of the IgM specific DENV-2 NS5 ELISA were calculated as 90 and 100 per cent, respectively. For the IgG ELISA, the test was found to be 100 per cent sensitive and 100 per cent specific for dengue antibodies.
Table.
In-house DENV-2 NS5 ELISA for primary and secondary dengue samples
A DENV-2 NS5 based indirect ELISA was developed as an alternative to the currently used NS1 E/M based ELISA diagnostic kits for the specific diagnosis of dengue infections. The DENV-2 proteins separated on polyacrylamide gel showed the presence of three distinct protein bands. Based on data from previous studies24,25, the proteins at positions 105, 67 and 45 kDa corresponded to the molecular weights of NS5, NS3 and NS1 proteins of DENV. The results of the immunofluorescence assay confirmed that the separated DENV-2 protein at 105 kDa position was viral NS5 protein. The sensitivity of the NS5 ELISA was evidenced by the results with primary and secondary dengue fever samples. The in-house DENV-2 NS5 ELISA differentiated the two groups of serum samples by detecting high levels of IgM antibodies and no IgG antibodies in all the primary cases of dengue fever. The high cut-off value of 2.0 for differentiating dengue positive and negative cases was adopted from previous work9. High levels of IgG antibodies were detected in all 20 cases of secondary dengue infection and IgM antibodies were detected in none.
This study had some major limitations. One major drawback was that only the specificity of a single dengue virus serotype could be tested. Another limitation was the small sample size. Only 20 samples from each category were tested.
In conclusion, the results indicated that the DENV-2 NS5 ELISA was dengue group specific. It may be used to differentiate dengue from other circulating Flavivirus infections. More importantly, the NS5 ELISA can also be used to distinguish between primary and secondary dengue infections on the basis of IgM/IgG ratios. Further studies with larger sample sizes and different DENV serotypes are required to validate this technique and also to check whether the DENV NS5 protein can be used to detect antibodies against all the four DENV serotypes.
Acknowledgment
The authors acknowledge the Director, Institute of Child Health and Hospital for children and Dean, Madras Medical College, and Stanley Medical College and Hospital, Chennai, for providing samples, and thank all the staff of the Arbovirus Research Laboratory at King Institute of Preventive Medicine and Research for providing technical assistance and reagents and serum samples.
Footnotes
Conflicts of Interest: None.
References
- 1.Stephenson JR. Understanding dengue pathogenesis: implications for vaccine design. Bull World Health Organ. 2005;83:308–14. [PMC free article] [PubMed] [Google Scholar]
- 2.Cordeiro MT, Neto UB, Nogueira RM, Marques ET. Reliable classifier to differentiate primary and secondary acute dengue infection based on IgG ELISA. PLoS One. 2009;4:e4945. doi: 10.1371/journal.pone.0004945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kirkpatrick BD, Durbin AP, Pierce KK, Carmolli MP, Tibery CM, Grier PL, et al. Robust and balanced immune responses to all 4 dengue virus serotypes following administration of a single dose of a live attenuated tetravalent dengue vaccine to healthy, flavivirus-naive adults. J Infect Dis. 2015;212:1–9. doi: 10.1093/infdis/jiv082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chokephaibulkit K, Perng GC. Challenges for the formulation of a universal vaccine against dengue. Exp Biol Med. 2013;238:566–78. doi: 10.1177/1535370212473703. [DOI] [PubMed] [Google Scholar]
- 5.Namekara M, Ellisa EM, O’Connella M, Elmc J, Gurarya A, Park S Y, et al. Evaluation of a new commercially available immunoglobulin M capture enzyme-linked immunosorbent assay for diagnosis of dengue virus infection. J Clin Microbiol. 2013;51:3102–6. doi: 10.1128/JCM.00351-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ueda VA, Fernandes S, Araújo ES, Tateno AF, Oliveira OMNPF, Oliveira RDR, et al. Use of an immunoglobulin G avidity test to discriminate between primary and secondary dengue virus infections. J Clin Microbiol. 2004;42:1782–4. doi: 10.1128/JCM.42.4.1782-1784.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vazqueza S, Hafnerb G, Ruiza D, Calzadaa N, Guzman MG. Evaluation of immunoglobulin M and G capture enzyme-linked immunosorbent assay Panbio kits for diagnostic dengue infections. J Clin Virol. 2007;39:194–8. doi: 10.1016/j.jcv.2007.04.003. [DOI] [PubMed] [Google Scholar]
- 8.Shu PY, Huang JH. Current advances in dengue diagnosis. Clin Diagn Lab Immunol. 2004;11:642–50. doi: 10.1128/CDLI.11.4.642-650.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shu PY, Chen LK, Chang SF, Yueh YY, Chow L, Chien LJ, et al. Comparison of capture immunoglobulin M (IgM) and IgG enzyme-linked immunosorbent assay (ELISA) and nonstructural protein NS1 serotype-specific IgG ELISA for differentiation of primary and secondary dengue virus infections. Clin Vaccine Immunol. 2003;10:622–30. doi: 10.1128/CDLI.10.4.622-630.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kao CL, King CC, Chao DY, Wu HL, Chang GJ. Laboratory diagnosis of dengue virus infection: current and future perspectives in clinical diagnosis and public health. J Microbiol Immunol Infect. 2005;38:5–16. [PubMed] [Google Scholar]
- 11.Gupta N, Srivastava S, Jain A, Umesh C. Chaturvedi. Dengue in India. Indian J Med Res. 2012;136:373–90. [PMC free article] [PubMed] [Google Scholar]
- 12.Bäck AT, Lundkvist A. Dengue viruses - an overview. Infect Ecol Epidemiol. 2013;3:10. doi: 10.3402/iee.v3i0.19839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rothman AL. Immunity to dengue virus: a tale of original antigenic sin and tropical cytokine storms. Nat Rev Immunol. 2011;11:532–43. doi: 10.1038/nri3014. [DOI] [PubMed] [Google Scholar]
- 14.Wong SJ, Boyle RH, Demarest VL, Woodmansee AN, Kramer LD, Li H, et al. Immunoassay targeting nonstructural protein 5 to differentiate West Nile virus infection from dengue and St. Louis encephalitis virus infections and from flavivirus vaccination. J Clin Microbiol. 2003;41:4217–23. doi: 10.1128/JCM.41.9.4217-4223.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.World Health Organization (WHO). Dengue: Guidelines for diagnosis, treatment, prevention and control. Geneva, Switzerland: WHO; 2009. [PubMed] [Google Scholar]
- 16.Shu PY, Chen LK, Chang SF, Yueh YY, Chow L, Chien LJ, et al. Comparison of capture immunoglobulin M (IgM) and IgG enzyme-linked immunosorbent assay (ELISA) and nonstructural protein NS1 serotype-specific IgG ELISA for differentiation of primary and secondary dengue virus infections. Clin Diagn Lab Immunol. 2003;10:622–30. doi: 10.1128/CDLI.10.4.622-630.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Centers for Disease Control and Prevention. Case definitions for infectious conditions under public health surveillance. MMWR Morb Mortal Wkly Rep. 1997;46:1–55. [PubMed] [Google Scholar]
- 18.WHO-recommended standards for surveillance of selected vaccine-preventable diseases WHO/V&B/03.01. [accessed on September 5, 2016]. Available from: http://www.path.org/files/WHO_surveillance_standards_JE.pdf .
- 19.West Nile Virus in the United States: Guidelines for Surveillance, Prevention, and Control. [accessed on September 5, 2016]. Available from: http://www.cdc.gov/westnile/resources/pdfs/wnvguidelines.pdf .
- 20.Putnak JR, Eckels K, Dubois DR. Dengue virus protection. United States patent US6254873 B1. 2001 Jul 3; [Google Scholar]
- 21.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–5. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 22.Bussetta C, Choi KH. Dengue virus nonstructural protein 5 (NS5) adopts multiple conformations in solution. Biochemistry. 2012;51:5921–31. doi: 10.1021/bi300406n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Palani G, Padmanabhan P, Ramesh K, Asadullah KS, Sambasivam M, Arunagiri K, et al. Development and evaluation of flavi-immunoglobulin M capture enzyme-linked immunosorbent assay. Indian J Pathol Microbiol. 2013;56:269–71. doi: 10.4103/0377-4929.120391. [DOI] [PubMed] [Google Scholar]
- 24.Lin SW, Chuang YC, Lin YS, Lei HY, Liu HS, Yeh TM. Dengue virus nonstructural protein NS1 binds to prothrombin/thrombin and inhibits prothrombin activation. J Infect. 2012;64:325–34. doi: 10.1016/j.jinf.2011.11.023. [DOI] [PubMed] [Google Scholar]
- 25.Perera R, Kuhn RJ. Structural proteomics of dengue virus. Curr Opin Microbiol. 2008;11:369–77. doi: 10.1016/j.mib.2008.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]


