Abstract
Ecological theory suggests that pathogens are capable of regulating or limiting host population dynamics, and this relationship has been empirically established in several settings. However, although studies of childhood diseases were integral to the development of disease ecology, few studies show population limitation by a disease affecting juveniles. Here, we present empirical evidence that disease in lambs constrains population growth in bighorn sheep (Ovis canadensis) based on 45 years of population-level and 18 years of individual-level monitoring across 12 populations. While populations generally increased (lambda =1.11) prior to disease introduction, most of these same populations experienced an abrupt change in trajectory at the time of disease invasion, usually followed by stagnant-to-declining growth rates (lambda = 0.98) over the next twenty years. Disease-induced juvenile mortality imposed strong constraints on population growth that were not observed prior to disease introduction, even as adult survival returned to pre-invasion levels. Simulations suggested that models including persistent disease-induced mortality in juveniles qualitatively matched observed population trajectories, whereas models that only incorporated all-age disease events did not. We use these results to argue that pathogen persistence may pose a lasting, but under-recognized, threat to host populations, particularly in cases where clinical disease manifests primarily in juveniles.
Keywords: Pathogen persistence, childhood disease, integrated population model, bighorn sheep
Introduction
Disease-induced die-offs are often viewed as random events, after which host populations quickly return to disease-free, “healthy” dynamics. This perception rests on assumptions that host population dynamics are relatively stable, and that perturbations from disease-induced die-offs do not permanently alter host vital rates. In reality, some pathogen introductions push host populations onto trajectories headed toward new equilibria. Pathogens that generate these “phase transitions” in host dynamics are capable of regulating or limiting host population growth (Turchin et al. 2003). Pathogen-induced phase transitions shape population trajectories of several well-studied host species. Macroparasite dynamics help switch host populations into and out of cyclic dynamics by altering fecundity in adult females as opposed to mortality of offspring (Hudson et al. 1998, Albon et al. 2002); and pathogens that reduce adult survival can directionally suppress host population growth, pushing these populations into periods of sustained decline (e.g., Monello et al. 2014, McCallum et al. 2009). Yet there are few examples of juvenile diseases regulating or limiting vertebrate host populations.
To produce phase transitions, pathogens must exert lasting pressure on host vital rates. This is rarely possible for pathogens that fade out (i.e., go locally extinct) rapidly, but it can occur when pathogens persist. On-going effects of pathogen persistence are visible in systems with temporally autocorrelated disease incidence. For example, epidemics of acute, perfectly immunizing pathogens like measles and mumps in humans buffer populations against future epidemics by temporarily depleting the pool of susceptible hosts (Grenfell et al. 2002). Other diseases, such as chronic wasting disease (Miller et al. 2000) or bovine tuberculosis (Rodwell et al. 2001), gradually accumulate in host populations so that the risk of new infections increases with time since invasion (Storm et al. 2013). In both cases, current disease intensity shapes future disease intensity, and this autocorrelation keeps host population dynamics confined to pathogen-determined trajectories.
Host vital rates that drive long-term shifts in population trajectories may not be the rates most altered by pathogen introduction events. Disease invasions often reduce survival for all members of a host population, regardless of age. Over time, however, morbidity and mortality from chronic diseases may become most severe in older individuals with long infection histories. By contrast, pathogens that induce lifelong immunity in their hosts gradually become sequestered in the youngest host age classes as older individuals become immune. Detecting the effects of these latter agents in nature is particularly challenging, since juvenile survival is both hard to measure and often subject to more natural variability than that of adults (e.g., Gaillard 2001). As a consequence, although phase transitions in host population dynamics due to disease-induced changes in fecundity and adult survival are well-known, the hypothesis that sustained juvenile disease could drive phase transitions in host dynamics remains untested.
Here, we ask whether pneumonia in bighorn sheep (Ovis canadensis) induces a phase transition in host population dynamics by placing long-term constraints on juvenile survival. This disease is caused by invasion of pathogens commonly carried by domestic sheep (O. aries) and goats (Capra hircus) into naïve wild sheep populations usually accompanied by easily recognizable all-age die-offs; indeed, managers often rely on these die-offs as a proxy for pathogen presence (Besser et al. 2012). Although adult bighorns appear to acquire protective immunity following exposure to pneumonia-causing agents (Plowright et al. 2013), disease continues to impede lamb survival in the years-to-decades following disease introduction events (Cassirer et al. 2013). Yet most existing models assume that the impact of disease invasion on juvenile survival is relatively short-lived (e.g., Cahn et al. 2011, Clifford et al. 2009). We investigate whether pathogens induce long-term phase transitions in bighorn sheep vital rates.
We use timeseries data from 12 well-studied bighorn sheep populations to examine how temporal autocorrelation and demographic variation in disease intensity shape population dynamics. We tested two hypotheses. First, that disease persistence causes a phase transition in bighorn sheep population dynamics. Second, that stagnant population growth during the disease persistence phase is attributable to juvenile mortality. We employed individual-level survival and lamb rearing data to identify specific vital rates that shift when populations transition from the pre-invasion “healthy” phase to the post-invasion “disease persistence” phase. We then applied these estimates to compare the immediate costs of pathogen invasion and the long-term costs of pathogen persistence to address the hypothesis that long-term pathogen persistence could have greater population-level consequences than all-age die-off events. Finally, we simulated population trajectories under different autocorrelation and disease intensity scenarios to explore the predictive capacity of models that make varying assumptions about the disease persistence period.
Methods
Data Collection
We analyzed data collected on radio-collared ewes and their lambs in 12 bighorn sheep populations from the Hells Canyon metapopulation in Idaho, Washington, and Oregon. Bighorn sheep populations in Hells Canyon have experienced periodic pneumonia outbreaks since the first reintroduction in 1971. In 1995–96 several northern Hells Canyon populations underwent a major pneumonia-induced die-off, and disease spread through much of the metapopulation by 2002 (Fig. 1a). From 1997 to 2015 we collected demographic data from marked individuals and conducted annual population surveys (Cassirer et al. 2013). Prior to 1997, our analyses rely solely on annual population composition counts.
Figure 1. Empirical patterns of pneumonia in bighorn sheep.
(a) Disease status of twelve focal Hells Canyon bighorn populations since 1995. Open squares are years with no documented pneumonia. Grey circles are years with documented pneumonia in adults. Black circles are years with documented or suspected (Cassirer et al. 2013) pneumonia in lambs. Population-years with no data are left blank. (b) Lambda and recruitment (March lamb:ewe ratio) for every population-year in the Hells Canyon study. Colors represent different disease classes, and point size scales with population size. Dashed lines are at lambda = 1 (i.e., static population size) and recruitment = 0.2, (this equates to 20 lambs per hundred ewes, a current Western Association of Fish and Wildlife Agencies guideline for a healthy population). Boxplots show the marginal distributions of recruitment and estimated lambda for healthy years (blue) as compared to years with any documented disease (red). Lines extend to 2.5th and 97.5th quantiles. (c) Disease status classifications as a function of years since the first recorded large die-off for each Hells Canyon population Number of populations contributing data for each year following invasion are reported above the columns. (d) Recruitment (March lamb:ewe ratio) as a function of years since disease introduction in Hells Canyon. Number of populations with recruitment for each year following invasion are reported below the columns. The blue zone is the middle 50% of recruitment values for population-years with no detected disease.
Throughout the entire timeseries (1971–2015), we assigned each population to one of three disease states in each year: disease-free and healthy; disease introduction (defined as the first reported pneumonia event in a given population); and post-invasion disease persistence. Disease status for each year was based on criteria described in Cassirer et al. (2013). Healthy estimates were primarily based on data from one population with no evidence of invasion and no detection of disease through 2011, but also included data from other populations during years with no evidence of active disease.
For each year, we calculated age-specific ewe survival and productivity (measured as lamb survival to weaning). Lambs born to radio-collared ewes that survived to October 1st were classified as surviving to weaning. Recruitment was based on ground or helicopter surveys conducted in late February or March.
Throughout these analyses, we use confirmed presence of clinical pneumonia (i.e., from necropsies of animals with pneumonic lesions in their lungs) or low summer lamb survival paired with observed pneumonia signs (coughing, nasal discharge, lethargy; Cassirer and Sinclair 2007) to classify population-years as “persistently diseased” or “healthy”. This definition is explicitly independent of the presence or absence any specific causal agent. Although a large body of evidence – much of it from this system (Besser et al. 2008; Besser et al. 2011) – suggests that Mycoplasma ovipneumoniae is the primary pathogen driving pneumonia dynamics in bighorn sheep, our animal handling intensity was insufficient to confirm M. ovipneumoniae status by pathogen detection for every population-year included here. Additional details on field data collection are available in the online supplement SI: Field methods.
Empirical data analysis
We applied hierarchical generalized linear models to the dataset from 1997 onwards to assess whether recruitment and health status changed in the years following disease introduction. In each case, we modelled the response variable (lambs per 100 ewes in the first case; proportion of years classified as healthy in the second) as a function of years since invasion, with a hierarchical term capturing heterogeneity across populations (SI: Post-invasion empirical trends).
We then applied hierarchical change-point models (Toms and Lesperance 2003) to the entire timeseries of population counts to determine whether trends in female population counts changed before and after the first disease detection. Change-point models identify the location of shifts in a timeseries trajectory, and describe the slope before and after the shift. We used trajectories of female counts from nine populations that were established well before the initial disease introduction event. Population-specific timeseries ranged in length from 13 to 45 years (median = 33 years). Populations 2, 8, and 9 in Fig. 1a were excluded due to very limited data prior to disease introduction. Trajectories were aligned so that the first year of observed disease was set to zero for all populations. The model incorporated data from all nine populations simultaneously, while accounting for variation in baseline population size, growth rate before and after the change-point, and change-point's position in the time series through hierarchical effect terms (SI: Trajectory change-point models).
We used an integrated population model (“IPM”; Kery and Schaub 2012) to combine ewe annual productivity and survival with lamb counts (i.e., lamb survival to weaning) from 1997 onwards to estimate survival and population growth under healthy and persistent disease conditions. However, we could not estimate vital rates during disease introduction due to limited individual-level data collected during die-offs. The IPM simultaneously leverages individual- and population-level data to estimate age-specific survival and lamb-weaning probabilities.
The IPM incorporated data from 217 known-age ewes monitored for a total of 1555 ewe-years. We combined ewes into age classes of 1, 2–3, 4–7, 8–13, >13, following Festa-Bianchet et al. (2006), except that unlike Festa-Bianchet, we combined 2 and 3 three-year-olds due to a sparsity of two-year-old records in healthy years. Productivity data consisted of 525 lambs born to 141 known-aged ewes (Table 1). We also used ewe and lamb counts from 10 Hells Canyon populations over the 18 years from 1997 to 2014 (a total of 156 population-years). Recruitment was empirically measured as the lamb:ewe ratio in March of each year. In the IPM, it was estimated by combining individual-level summer lamb survival data with population-level counts of overwinter lamb survival, since lamb status was not determined for specific ewes after October 1st. In its entirety, the IPM estimated 24 parameters, 11 each – one over-winter lamb survival term, five age-specific ewe survival terms, and five ewe-age-specific lamb survival-to-weaning terms – for healthy and diseased years, and two hierarchical year effects associated with adult survival and reproduction (SI: Integrated Population Model). The IPM was fit using JAGS (Plummer, 2015) accessed through R (R Core Team, 2015).
Table 1.
Number of animal-years of individual survival and lamb weaning contributed by radiocollared ewes, structured by age class and population-level health status.
| Ewe age class | Annual ewe survival | Lamb survival to weaning |
|---|---|---|
| 2–3 | Pneumonia: 149 Healthy: 150 |
Pneumonia: 13 Healthy: 14 |
| 4–7 | Pneumonia: 348 Healthy : 203 |
Pneumonia: 139 Healthy: 69 |
| 8–13 | Pneumonia: 315 Healthy: 90 |
Pneumonia: 172 Healthy: 61 |
| >13 | Pneumonia: 65 Healthy: 14 |
Pneumonia: 49 Healthy: 8 |
Population projections and simulations
We used age-structured Leslie matrices to project population growth and demography separately for disease-free and persistent-disease states. The matrices were constructed so that births were pulsed (Caswell 2001). Since population counts occur prior to lambing in the spring, we used a female-only post-breeding model with a 1:1 sex ratio at birth (SI Population Projection Models). Leslie matrices were parameterized using draws from posterior vital rate distributions generated under the IPM. Population growth rate under each disease state was estimated as the dominant eigenvalue of the Leslie matrix parameterized from posterior estimates for that state (denoted λHealthy and λPersist, respectively).
We then compared recovery time -- years required to return to pre-die-off population size -- for die-offs followed by immediate return to healthy rates to recovery time for die-offs followed by a specified period of persistent disease. Populations must increase by k% to return to pre-die-off size following a die-off killing k% of the population. Under immediate recovery, this occurs when . As long as λHealthy > 1, recovery is achieved when t = yr, and
| (1) |
When disease persists, populations continue declining until persistence ends (if λPersist < 1), and begin growing again only after pathogen fade-out. In a case where pathogens persist for p years, recovery is achieved when . Years to recovery (yr for a population experiencing a die-off killing k% of the population, with persistence of p years, was calculated as
| (2) |
Finally, we simulated population growth under varying periods of disease persistence. We assumed that populations experience three disease states, healthy, disease introduction, and persistent disease, and transition through disease states according to a Markov process. In this model, populations are initially healthy, but experience disease introduction events with some probability, α. When the population is in either an introduction or a persistent disease state, another introduction event can occur with probability α, the disease can fade out (returning the population to a healthy state) with probability (1 − α) γ, or the disease can persist and shift the population into a disease persistence state with probability (1 − α) (1− γ). The expected number of years between disease introduction events is equal to 1/α; the expected number of years from disease introduction to fade out (i.e., return to healthy) is 1/γ. Transitions among disease states are summarized in the row-stochastic Markov transition matrix in (3).
| (3) |
We ran 100 replicate simulations for each scenario. All simulations started with a population of 500 ewes and we initiated the disease process after 30 years of healthy growth. For each year, we first updated the disease status using binomial distributions based on that simulation's disease model (i.e. age structured vs all-age models). Then we parameterized the Leslie matrix with values drawn from the posterior distribution generated for that disease state by the IPM. Since we had limited age-specific survival data during invasion years, we assumed that disease introduction years experienced a 30% reduction in adult survival (although many published accounts exceed this value – e.g., Coggins 1988 -- we suspect that many smaller outbreak events go undetected). We also examined simulations that did not account for age-specific disease effects (i.e., in these simulations, every “persistent disease” year was given “disease introduction” demographic rates), and simulations in which disease years occurred at random, instead of in sequence.
We do not incorporate density-dependence in these simulations, because these are small populations (median population size 75) with an estimated 6 sq km of potential habitat per sheep (IDFG unpubl. data), and we observed no evidence of density effects on vital rates prior to pathogen introduction (Fig. 2). Population size accounted for less than 7% of the variation in recruitment or population growth in healthy years. We recognize the importance of density-dependent population regulation in healthy bighorn sheep populations (Jorgenson et al. 1997) but density did not play a role in the parameter space of these data and simulations.
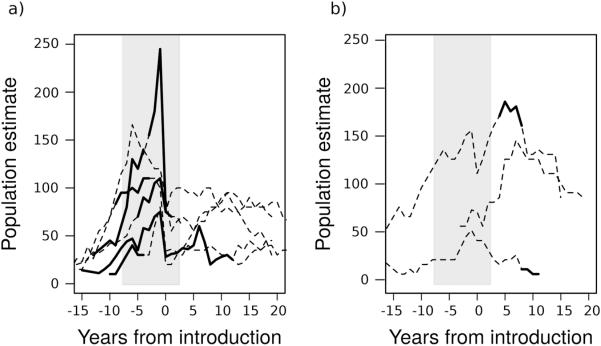
Figure 2. Change-points in complete ewe count trajectories.
Populations are aligned so that the first year of documented pneumonia is defined to be zero such that all negative years are disease-free. The grey shaded region showed the 95% posterior credible intervals on the mean change-point calculated across all populations. Solid black sections of the individual trajectories show 95% posterior credible intervals on that particular trajectory's change-point. (a) Trajectories1 for all populations whose individual change-points fell within the 95% posterior credible interval for the mean change-point. (b) Three populations' individual 95% change-point credible intervals fell entirely outside the 95% posterior credible interval for the mean change-point.
1The highest peak to trough includes removal of 72 sheep (43% of estimated mortalities) during the outbreak, but removing that trajectory had negligible effects on posterior estimates.
Results
Bighorn sheep populations did not fully recover following die-off events
Disease is known to persist in bighorn populations following pathogen invasion (e.g., Cassirer et al. 2013). Here we show that disease presence is associated with both stagnant growth and low recruitment (Fig. 1b), and that disease costs are not alleviated over time (Fig. 1c). The probability of a year being classified as “Healthy” after pathogen invasion was 0.23 and decreased by a multiplicative factor of 0.84 with each successive year-post-invasion. Healthy years actually became more rare with increasing time since pathogen introduction (95% posterior credible interval [0.54, 1.25], Fig. 1c, Table S2). Recruitment was not associated with time since disease introduction, suggesting that the magnitude of disease in lambs remained constant (lamb:ewe median = 0.32 in the year of invasion; 95% posterior credible interval on yearly change in lamb:ewe ratio = [−0.01, 0.01]; Fig. 1d, Table S1). Taken together, these results indicate that population dynamics do not return quickly to pre-die-off rates; instead, growth remains constrained as long as pathogens persist in the host populations. Full descriptions of model posteriors are included in the online supplement.
Die-offs during pathogen invasion events may eliminate weaker hosts, selecting for the most fit individuals and leading to accelerated population growth in the absence of disease transmission (as shown by Jolles et al. 2006 for TB in African buffalo). However, we saw no evidence of improved population growth after die-offs, even in healthy years (mean λ in healthy years = 1.14 before die-offs and 1.10 after die-offs). There is no evidence in this system that hosts who survive all-age die-off events have post-die-off vital rates that facilitate rapid return to pre-die-off population dynamics; instead, growth remains constrained as long as pathogens persist in the host populations.
Population trajectories changed in tandem with disease invasion
Ewe count trajectories in six of nine populations with data before and after disease introduction showed a statistically significant decline associated with the first observation of pneumonia (posterior median year of trajectory change = 2.57 years prior to reported pathogen introduction, 95% posterior credible interval from 7.31 years prior to reported introduction to 2.20 years after reported introduction; Table S4).
Populations generally grew prior to pathogen introduction (posterior median increase in numbers per year = 5.54, 95% posterior credible interval for the population change per year before the change-point, μβ1= [4.26, 7.08]), but stagnated or declined thereafter (posterior median change in count per year = −3.17, 95% posterior credible interval for the population change per year after the change-point = [−8.08, 1.59]). We detected no evidence of a second change-point corresponding to recovery in the aggregated post-invasion timeseries. Instead, a weakly-characterized second change-point occurred in several populations during the first 15 years following invasion (posterior median = 4.42, 95% posterior credible interval for μu = [1.18,15.70]; Table S5), and was associated with accelerating decreases in population growth (posterior median = −3.76, 95% posterior credible interval for the mean annual change in population size before the change-point was [2.71, 15.25], whereas after the change-point the posterior median was −3.76 with a 95% credible interval of [−11.75, 3.23]). Credible intervals for the specific change-points for three populations fell entirely outside the posterior credible interval for the mean change-point for all populations. These populations initially grew following disease introduction, but declined 6–8 years later. No populations showed long-term recovery to pre-invasion population size or growth rates (Fig. 2).
Adult survival returned to pre-die-off rates following disease introduction, but juvenile survival remained low
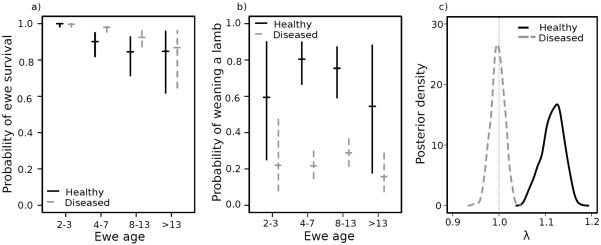
Adult female survival was not significantly different in disease-free and post-invasion disease persistence years (Fig. 3a; Table S6). In fact, we saw some weak suggestion that adult survival might be marginally better in diseased years than healthy years (Fig. 3a), though this effect was not significant. Estimated lamb weaning rates were lower in disease persistence years than in healthy years, especially for prime-aged ewes (indicated by the non-overlapping posterior credible intervals in Fig. 3b; Table S6). We did see weak evidence that disease mortality may be somewhat compensatory in lambs, since estimated overwinter lamb survival in health years approximately 12% lower than in infected years (Table S6). However, this effect was insufficient to overcome the dramatic costs disease imposed on lamb survival until weaning. Consequently, population age structure was predicted to shift toward older age classes under disease persistence (Fig. S1b).
Figure 3. Posterior estimates for vital rates and λ.
Point estimate and posterior credible intervals for (a) age-specific ewe survival and (b) probably of weaning a lamb in persistent diseased (grey) and healthy (black solid) years. (c) Posterior distribution of stochastic population growth rate in healthy (black solid) and diseased (grey dashed) years.
Disease persistence period plays a major role in determining time to population recovery
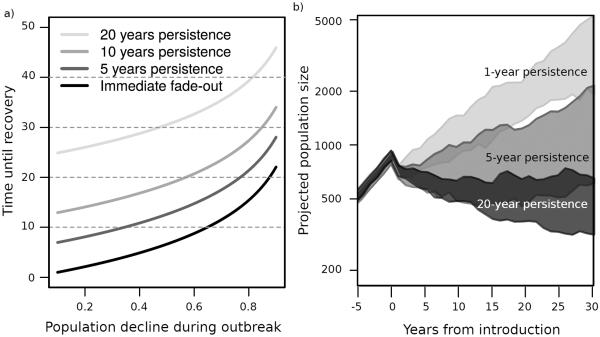
Projections based on the Leslie matrices projected population growth in disease-free conditions (posterior median = 1.11; 95% posterior credible interval = [1.05, 1.14]; Fig. 3c), but gradual decline under disease persistence (posterior median = 0.98; 95% posterior credible interval = 0.95, 1.01; Fig. 3c). As expected, the simulation studies suggested that increasing persistence sharply increased expected time to recovery. A five-year increase in disease persistence had a similar effect on time to recovery as doubling the mortality in the initial outbreak (Fig. 4a). This is reflected in the distance on the y-axis between immediate fade-out and increasing persistence time lines in Fig. 4a: the 25 year difference in projected recovery time between initial population declines of 10% vs 90% (reflected in the bottom curve in Fig. 4a) was similar to, but slightly less than the difference in projected recovery time for populations with immediate disease fade-out vs. 20-year persistence, regardless of initial die-off size (Fig. 4a). These effects remained apparent even after incorporating stochasticity (Fig. 4b). In general, population growth projections were lower when age-structure was overlooked, but this effect was less pronounced when dynamics were dominated by disease introduction (right panels of Fig. S2 and S3) as opposed to persistence (left panels of Fig. S2 and S3). Temporal sequencing of disease was not crucial to population dynamics following disease introduction: random vs. ordered disease states had relatively little effect on population growth, so long as disease impacted juveniles, and not all age-classes (Fig. S4). This was likely because the vast majority of years following invasion were diseased.
Figure 4. Simulated effects of disease persistence.
(a) The number of years required for a population to recover, i.e., regain pre-die-off sizes, over varying persistence periods. (b) Population projections based on 500 simulations under the Markov transition model in equation (3). Populations are subject to continuous healthy growth rates until year 0, and experience their first disease event between years 0 and 1. The expected invasion rates is set to one invasion per five years, and expected persistence periods of 1 (light grey), 5 (medium grey), or 20 (black) years. Polygons bound the middle 50% of the simulated trajectories. Additional simulation outputs are shown in Figs. S2 – S4.
Discussion
Pathogens constrain host vital rates, and therefore push host population trajectories toward new equilibrium dynamics including extinction (McCallum and Dobson 1995, De Castro and Bolker 2005). This may be of limited importance for transient pathogens that go locally extinct shortly after introduction, where the period of pathogen-constraint is short with healthy dynamics resuming quickly. When novel pathogens persist, however, they can induce phase transitions in host population dynamics, sometimes with serious consequences for host species conservation and management. Our analysis suggests that such a phase transition may be underway in pneumonic populations of bighorn sheep.
The majority of the bighorn sheep populations in this study did not resume pre-disease-invasion growth rates during up to twenty years following recorded disease-induced die-off events. Instead, most populations declined abruptly near the time of the first all-age disease event (Fig. 2a), and remained stagnant-to-declining thereafter. While pathogen invasion events reduced survival regardless of age (Fig. 2), we found that as the pathogen persisted, its effects differentiated across age classes. Adult survival returned to pre-invasion rates even as juvenile survival-to-weaning remained low. The projected cost of persistent disease on host population growth could rapidly exceed the cost of all-age die-offs (Fig. 4a). A steady onslaught of short-lived spillover events could produce population trajectories similar to those observed in the field (middle row of Fig. S2). However, that pattern would require years of strong recruitment between all-age events, which is inconsistent with our field observations of sustained lamb disease (Cassirer et al. 2013, Fig. 1a). The complete suite of disease dynamics was therefore best captured by simulations that accounted for long-term juvenile disease persistence; neither a disease process consisting of only die-off events, nor a process of persistent disease in all age classes could accurately capture both observed population trajectories and vital rates (Fig. 4b, Fig. S2–S4).
Assumptions about how long populations are subject to disease-structured growth rates have important consequences on the accuracy of population models, because increasing persistence period dramatically increased the expected time to population recovery in our study (Fig. 4). Relatively rapid short-term recovery does sometimes occur, even in the Hells Canyon populations. Three of our populations did not show immediate downward shifts in population trajectories following die-off events (Fig. 2b). Additionally, we regularly documented at least one year of good recruitment during the 3–5 years following disease introduction, and our study populations regularly surpassed the management agency threshold for “healthy” vital rates (greater than 0.2 lambs recruited per ewe; WAFWA WHC 2014), even in the presence of clinical disease (Fig. 1b). Yet despite this fact, and the observation that local bighorn sheep populations remain well below historic or habitat-based carrying capacity estimates, neither recruitment nor population growth returned to pre-die-off rates in the twenty years following die-off events (Fig. 1, Fig. 3).
Our models illustrate that even pathogen introduction events corresponding to very small initial die-offs could have severe long-term consequences on population growth. While our results may overestimate the cost of disease persistence following very mild initial outbreaks (since mild initial outbreaks may reflect underlying factors like low-virulence pathogen strains or strong spatial structuring that might also effect persistence dynamics), this scenario remains empirically undercharacterized, and merits further consideration.
Disease-induced mortality rates in bighorn sheep vary substantially in the field, and this is reflected in both the empirical data analysis (Fig. 2) and the simulation study (Fig. 4). Variation could be attributable to multiple processes including contact heterogeneity and social substructuring (Manlove et al. 2014), pathogen virulence, host susceptibility to pathogen establishment, and factors associated with each individual's unique mucosal immunity and carriage status. The IPM did not parse these drivers, but they are accounted for in the breadth of the posterior estimates, and pushed forward directly into the simulations. As a consequence, the simulations capture a realistic range of disease impacts on host vital rates, insofar as those rates are empirically represented within our dataset. Partitioning the variance attributable to each source of heterogeneity is critical for management, and remains an area of active inquiry. Nevertheless, even amidst heterogeneity, the basic trend remains clear: most bighorn sheep populations experience lasting reductions in juvenile survival in the aftermath of disease-induced die-off events, and this reduction may persist until the causal agent goes locally extinct.
Evidence that pathogen persistence might induce phase transitions in host vital rates is beginning to emerge from a number of other wildlife disease systems. Acutely immunizing infections produce negatively autocorrelated prevalence oscillations, which can then drive host population cycles. This has been demonstrated for macroparasites of red grouse (Hudson et al. 1998) and reindeer (Albon et al. 2002), and is increasingly evident for morbilliviruses like canine (Viana et al. 2015, Almberg et al. 2012) and phocine distemper viruses (Swinton et al. 1998), hantavirus (Luis et al. 2015), and plague (Kausrud et al. 2007). Oscillating population trajectories are usually centered above zero, so the major conservation concern is whether host populations can persist through epidemic peaks.
Invading chronic pathogens, however, can shift previously stable populations into monotonic declines toward zero. Small but sustained downward shifts in host viral rates “redden” the host demographic spectrum (e.g., Schwager et al. 2006). As a consequence, persistent chronic pathogens can severely diminish the host population's long-term growth, resilience, and viability. There are increasing indications that this may be the case for chronic wasting disease in elk at Rocky Mountain National Park (Monello et al. 2014), and simian immunodeficiency virus in chimpanzees (Keele et al. 2009), both of which appear to produce phase-transitions in adult survival. Brucellosis is projected to have a similar directional effect in African buffalo, this time through a phase-transition in fecundity (Gorsich et al. 2015). Fungal pathogens of amphibians and bats also produce directional shifts in population growth rates, although the precise demography may vary between particular host species (Langwig et al. 2012, Kilpatrick et al. 2010).
If invading chronic pathogens do not cause high mortality initial die-offs, as occurred in a third of the bighorn sheep populations in this study, they may go undetected, reducing opportunities for early control. Overlooking persistence and modeling only highly-visible disease introductions produces a dramatically inflated projection of future population growth for bighorn sheep, and this bias may also apply to other disease conservation systems. Wildlife and conservation biologists must be alert to invading and establishing pathogens -- even those with apparently negligible costs. When left unchecked, these agents have the potential to quietly but dramatically alter long-term population trajectories.
Supplementary Material
Acknowledgements
This research was supported by Morris Animal Foundation grant D13ZO-081. KRM was supported through a Penn State Academic Computing Fellowship. The RAPIDD program provided support to PJH. RKP was supported by National Institutes of Health IDeA Program grants P20GM103474 and P30GM110732, P. Thye, and Montana University System Research Initiative: 51040-MUSRI2015-03. Data were collected under the Hells Canyon Initiative with support from the wildlife agencies of Idaho, Oregon, and Washington, Federal Aid in Wildlife Restoration, U.S. Forest Service, Bureau of Land Management, the Wild Sheep Foundation National, Idaho, Oregon, and Washington chapters, Oregon Hunters Association, and Shikar-Safari Club International. Any use of trade, firm, or product names is for descriptive purposes only and does not imply endorsement by the U.S. Government.
Literature Cited
- Albon SD, Stien A, Irvine RJ, Langvatn R, Ropstad E, Halvorsen O. The role of parasites in the dynamics of a reindeer population. Proceedings of the Royal Society B: Biological Sciences. 2002;269:1625–1632. doi: 10.1098/rspb.2002.2064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almberg ES, Cross PC, Dobson AP, Smith DW, Hudson PJ. Parasite invasion following host reintroduction: a case study of Yellowstone's wolves. Philosophical transactions of the Royal Society B: Biological Sciences. 2012;367:2840–2851. doi: 10.1098/rstb.2011.0369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besser TE, Cassirer EF, Potter KA, VanderSchalie J, Fischer A, Knowles DP, Herndon DR, Rurangirwa FR, Weiser GC, Srikumaran S. Association of Mycoplasma ovipneumoniae infection with population-limiting respiratory disease in free-ranging Rocky Mountain bighorn sheep (Ovis canadensis canadensis) Journal of Clinical Microbiology. 2008;46:423–430. doi: 10.1128/JCM.01931-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besser TE, Highland MA, Baker K, Cassirer EF, Anderson NJ, Ramsey JM, et al. Causes of pneumonia epizootics among bighorn sheep, Western United States, 2008–2010. Emerging Infectious Diseases. 2012;18(3):406–414. doi: 10.3201/eid1803.111554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahn ML, Conner MM, Schmitz OJ, Stephenson TR, Wehausen JD, Johnson HE. Disease, population viability, and recovery of endangered Sierra Nevada bighorn sheep. Journal of Wildlife Management. 2011;75(8):1753–1762. [Google Scholar]
- Cassirer EF, Sinclair ARE. Dynamics of pneumonia in a bighorn sheep metapopulation. Journal of Wildlife Management. 2007;71(4):1080–1088. [Google Scholar]
- Cassirer EF, Plowright RK, Manlove KR, Cross PC, Dobson AP, Potter KA, et al. Spatio-temporal dynamics of pneumonia in bighorn sheep. Journal of Animal Ecology. 2013;82:518–528. doi: 10.1111/1365-2656.12031. [DOI] [PubMed] [Google Scholar]
- Caswell H. Matrix Population Models: Construction, Analysis, and Interpretation. 2nd Edition Sinaur Associates, Inc. Publishers; Sunderland, MA, USA: 2001. [Google Scholar]
- Clifford DL, Shumaker BA, Stephenson TR, Bleich VC, Cahn ML, Gonzales BJ, Boyce WM, Mazet JAK. Assessing disease risk at the wildlife-livestock interface: A study of Sierra Nevada bighorn sheep. Biological Conservation. 2009;149:2559–2568. [Google Scholar]
- Coggins VL. The Lostine Rocky Mountain bighorn sheep die-off and domestic sheep. Biennial Symposium of the Northern Wild Sheep and Goat Council. 1988;6:57–64. [Google Scholar]
- Daszak P, Cunningham AA, Hyatt AD. Anthropogenic environmental change and the emergence of infectious disease in wildlife. Acta Tropica. 2001;78:103–116. doi: 10.1016/s0001-706x(00)00179-0. [DOI] [PubMed] [Google Scholar]
- De Castro F, Bolker B. Mechanisms of disease-induced extinction. Ecology Letters. 2005;8:117–126. [Google Scholar]
- Festa-Bianchet M, Gaillard J, Cote S. Variable age structure and apparent density dependence in survival of adult ungulates. Journal of Animal Ecology. 2003;72(4):640–649. doi: 10.1046/j.1365-2656.2003.00735.x. [DOI] [PubMed] [Google Scholar]
- Festa-Bianchet M, Coulson T, Gaillard J, Hogg JT, Pelletier F, F. Stochastic predation events and population persistence in bighorn sheep. Proceedings of the Royal Society B: Biological Sciences. 2006;273:1537–1543. doi: 10.1098/rspb.2006.3467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaillard JM, Festa-Bianchet M, Yoccoz NG, Loison A, Toigo C. Temporal variation in fitness components and population dynamics of large herbivores. Annual Reviews in Ecology Evolution and Systematics. 2000;31:367–393. [Google Scholar]
- Gorsich EE, Ezenwa VO, Cross PC, Gengis RG, Jolles AE. Context-dependent survival, fecundity and predicted population-level consequences of brucellosis in African buffalo. Journal of Animal Ecology. 2015;84:999–1009. doi: 10.1111/1365-2656.12356. [DOI] [PubMed] [Google Scholar]
- Grenfell BT, Bjornstad ON, Finkenstadt BF. Dynamics of measles epidemics: Scaling noise, determinism, and predictability with the TSIR model. Ecological Monographs. 2002;72(2):185–202. [Google Scholar]
- Hudson PJ, Dobson AP, Newborn D. Prevention of population cycles by parasite removal. Science. 1998;282:2256–2258. doi: 10.1126/science.282.5397.2256. [DOI] [PubMed] [Google Scholar]
- Jolles A, Etienne RS, Olff H. Independent and competing disease risks: implications for host populations in variable environments. The American Naturalist. 2006;167(5):745–757. doi: 10.1086/503055. [DOI] [PubMed] [Google Scholar]
- Jorgenson JT, Festa-Bianchet M, Gaillard J, Wishart WD. Effects of age, sex, disease, and density on survival of bighorn sheep. Ecology. 1997;78(4):1019–1032. [Google Scholar]
- Kausrud KL, Viljugrein H, Frigessi A, Begon M, Davis S, S., Leirs H, H., et al. Climatically driven synchrony of gerbil populations allows large-scale plague outbreaks. Proceedings of the Royal Society B: Biological Sciences. 2007;274(1621):1963–1969. doi: 10.1098/rspb.2007.0568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keele B, Jones JH, Terio KA, Estes JD, Rudicell RS, Wilson ML, et al. Increased mortality and AIDS-like immunopathology in wild chimpanzees infected with SIVcpz. Nature. 2009;460:515–519. doi: 10.1038/nature08200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kery M, Schaub M. Bayesian Population Analysis Using WinBUGS: A hierarchical perspective. Academic Press; Waltham MA: 2012. [Google Scholar]
- Kilpatrick AM, Briggs CJ, Daszak P. The ecology and impact of chytridiomycosis: an emerging disease of amphibians. Trends In Ecology and Evolution. 2010;25(2):109–118. doi: 10.1016/j.tree.2009.07.011. [DOI] [PubMed] [Google Scholar]
- Langwig KE, Frick WF, Bried JT, Hicks AC, Kunz TH, Kilpatrick AM. Sociality, density-dependence and microclimates determine persistence of populations suffering from a novel fungal disease, white-nose syndrome. Ecology Letters. 2012;15(9):2050–2057. doi: 10.1111/j.1461-0248.2012.01829.x. [DOI] [PubMed] [Google Scholar]
- Luis A, Douglass RJ, Mills JN, Bjornstad ON. Environmental fluctuations lead to predictability in Sin Nombre hantavirus outbreaks. Ecology. 2015;96(6):1691–1701. [Google Scholar]
- Manlove KR, Cassirer EF, Cross PC, Plowright RK, Hudson PJ. Costs and benefits of group living with disease: A case study of pneumonia in bighorn lambs (Ovis canadensis) Proceedings of the Royal Society B: Biological Sciences. 2014;281:20142331. doi: 10.1098/rspb.2014.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCallum H, Dobson AP. Detecting disease and parasite threats to endangered species and ecosystems. Trends in Ecology and Evolution. 1995;10(5):190–194. doi: 10.1016/s0169-5347(00)89050-3. [DOI] [PubMed] [Google Scholar]
- McCallum H, Jones M, Hawkins C, Hamede R, Lachish S, Sinn DL, Beeton N, Lazenby B. Transmission dynamics of Tasmanian devil facial tumor disease may lead to disease-induced extinction. Ecology. 2009;90(12):3379–3392. doi: 10.1890/08-1763.1. [DOI] [PubMed] [Google Scholar]
- Miller MW, Williams ES, McCarty CW, Spraker TR, Kreeger TJ, Larsen CT, et al. Epizootiology of chronic wasting disease in free-ranging cervids in Colorado and Wyoming. Journal of Wildlife Diseases. 2000;36(4):676–690. doi: 10.7589/0090-3558-36.4.676. [DOI] [PubMed] [Google Scholar]
- Monello RJ, Powers JG, Hobbs NT, Spraker TR, Watry MK, Wild MA. Survival and population growth of a free-ranging elk population with a long history of exposure to chronic wasting disease. Journal of Wildlife Management. 2014;78(2):214–223. [Google Scholar]
- Plowright RK, Manlove KR, Cassirer EF, Cross PC, Besser TE, Hudson PJ, P.J. Use of exposure history to identify immunity to pneumonia in bighorn sheep (Ovis canadensis) PLoS One. 2013;8:e61919. doi: 10.1371/journal.pone.0061919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plummer M. Rjags: Bayesian graphical models using MCMC. R package version 4-4. 2015 https://CRAN.R-project.org/package=rjags.
- R Core Team . R Foundation for Statistical Computing. Vienna, Austria: 2015. R: A language and environment for statistical computing. https://www.R-project.org. [Google Scholar]
- Rodwell TC, Whyte IJ, Boyce WM. Evaluation of population effects of bovine tuberculosis in free-ranging African Buffalo (Syncerus caffer) Journal of Mammalogy. 2001;82(1):231–238. [Google Scholar]
- Schwager M, Johst K, Jeltsch F. Does red noise increase or decrease extinction risk? Single extreme events versus series of unfavorable conditions. The American Naturalist. 2006;167(6):879–889. doi: 10.1086/503609. [DOI] [PubMed] [Google Scholar]
- Storm DJ, Samuel MD, Rolley RE, Shelton P, Keuler NS, Richards BJ, B.J., et al. Deer density and disease prevalence influence transmission of chronic wasting disease in white-tailed deer. Ecosphere. 2013;4(1):article 10. [Google Scholar]
- Swinton J, Harwood J, Grenfell BT, Gilligan CA. Persistence thresholds for phocine distemper virus infection in harbor seal Phoca vitulina metapopulations. Journal of Animal Ecology. 1998;67(1):54–68. [Google Scholar]
- Stubben CJ, Milligan BG. Estimating and analyzing demographic models using the popbio package in R. Journal of Statistical Software. 2007;22(11) [Google Scholar]
- Toms JD, Lesperance ML. Piecewise regression: A tool for identifying ecological thresholds. Ecology. 2003;84(8):2034–2041. [Google Scholar]
- Turchin P, Wood SN, Ellner SP, Kendall BE, Murdoch WM, Fischlin A, Casas J, McCauley E, Briggs CJ. Dynamical effects of plant quality and parasitism on population cycles of larch budmoth. Ecology. 2003;84(5):1207–1214. [Google Scholar]
- Viana M, Cleaveland S, Matthiopoulos J, Halliday J, Packer C, Craft ME, et al. Dynamics of a morbillivirus at the domestic-wildlife interface: Canine distemper virus in domestic dogs and lions. Proceedings of the National Academy of Sciences, USA. 2015;112(5):1464–1469. doi: 10.1073/pnas.1411623112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WAFWA WHC . 2014 Bighorn sheep herd health monitoring recommendation. Western Association of Fish & Wildlife Agencies Wildlife Health Committee; 2015. http://www.wafwa.org/Documents%20and%20Settings/37/Site%20Documents/Working%20Groups/Wild%20Sheep/Disease/BHS%20herd%20health%20monitoring_Final%201_3_2015.pdf. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.