ABSTRACT
Gene regulation by base pairing between small noncoding RNAs (sRNAs) and their mRNA targets is an important mechanism that allows bacteria to maintain homeostasis and respond to dynamic environments. In Gram-negative bacteria, sRNA pairing and regulation are mediated by several RNA-binding proteins, including the sRNA chaperone Hfq and polynucleotide phosphorylase (PNPase). PNPase and its homolog RNase PH together represent the two 3′ to 5′ phosphorolytic exoribonucleases found in Escherichia coli; however, the role of RNase PH in sRNA regulation has not yet been explored and reported. Here, we have examined in detail how PNPase and RNase PH interact to support sRNA stability, activity, and base pairing in exponential and stationary growth conditions. Our results indicate that these proteins facilitate the stability and regulatory function of the sRNAs RyhB, CyaR, and MicA during exponential growth. PNPase further appears to contribute to pairing between RyhB and its mRNA targets. During stationary growth, each sRNA responded differently to the absence or presence of PNPase and RNase PH. Finally, our results suggest that PNPase and RNase PH stabilize only Hfq-bound sRNAs. Taken together, these results confirm and extend previous findings that PNPase participates in sRNA regulation and reveal that RNase PH serves a similar, albeit more limited, role as well. These proteins may, therefore, act to protect sRNAs from spurious degradation while also facilitating regulatory pairing with their targets.
IMPORTANCE In many bacteria, Hfq-dependent base-pairing sRNAs facilitate rapid changes in gene expression that are critical for maintaining homeostasis and responding to stress and environmental changes. While a role for Hfq in this process was identified more than 2 decades ago, the identity and function of the other proteins required for Hfq-dependent regulation by sRNAs have not been resolved. Here, we demonstrate that PNPase and RNase PH, the two phosphorolytic RNases in E. coli, stabilize sRNAs against premature degradation and, in the case of PNPase, also accelerate regulation by sRNA-mRNA pairings for certain sRNAs. These findings are the first to demonstrate that RNase PH influences and supports sRNA regulation and suggest shared and distinct roles for these phosphorolytic RNases in this process.
INTRODUCTION
Bacteria rapidly alter gene expression through small noncoding RNAs (sRNAs) that act by base pairing with target mRNAs. Although classically identified as important players in bacterial stress response, sRNAs also play general roles regulating the genes involved in cell growth, nutrient acquisition, social behavior, and virulence (1, 2). sRNAs often act through translational repression, in which a given sRNA base pairs with the leader sequence of a target mRNA, obscuring the site that would otherwise form contacts with the ribosomal 30S subunit (3). This pairing can also lead to subsequent RNase E-mediated decay of the mRNA (4). In other cases, pairing with an mRNA leader sequence promotes gene expression by revealing a ribosome binding site hidden by the native mRNA secondary structure (5). sRNAs less commonly target other mRNA regions but often induce mRNA cleavage by RNase E in these cases (6, 7).
Many sRNAs act in tandem with the homohexameric Hfq chaperone to accomplish these regulatory activities. In Escherichia coli, Hfq protects such sRNAs against degradation by RNases and further facilitates pairing between these sRNAs and their mRNA targets (8). Hfq coordinates pairing by simultaneously binding both the sRNA and mRNA, thus bringing into close proximity the short complementary regions of each RNA (9, 10). Through Hfq, sRNAs are capable of pairing with multiple mRNA target sequences, and as a consequence, expression of a single sRNA can exert coordinated regulatory control over an entire set of related genes. For example, in response to iron starvation, expression of the sRNA RyhB downregulates the synthesis of more than 50 proteins involved in iron metabolism and storage (11, 12).
sRNAs and mRNAs interact with the Hfq hexamer at three distinct RNA-binding sites: the proximal face, the rim, and the distal face. RNAs with class I affinity, such as RyhB or MicA, bind the Hfq proximal and rim sites, whereas sRNAs with class II affinity, such as CyaR, interact with the Hfq proximal and distal sites (13, 14). To pair with the Hfq-bound sRNA, a target mRNA must bind to the remaining free RNA-binding site on Hfq, either to the distal face with class I sRNAs or the rim with class II sRNAs (14). Importantly, Hfq levels are limiting within the cell; sRNAs and mRNAs therefore compete for these RNA-binding sites on Hfq in a dynamic equilibrium, and regulatory outcomes are sensitive to the stoichiometry of these competing components (14–17).
Hfq is not the only protein that contributes to effective sRNA regulation. Recent studies revealed that the deletion of pnp, encoding the 3′ to 5′ phosphorolytic exoribonuclease polynucleotide phosphorylase (PNPase), paradoxically reduces the stability of several sRNAs, including CyaR and RyhB, during exponential growth (18, 19). Deletion of pnp also resulted in the decreased regulation of mRNA targets by CyaR and other sRNAs (18, 20). Notably, the activity of several sRNAs in the Δpnp mutant was restored upon further deletion of the RNase E C terminus, which is normally involved in sRNA decay (18). In vitro experiments have revealed that PNPase, Hfq, and sRNAs form a stable ternary complex (20). Together, these results suggest that PNPase helps protect sRNAs from degradation by other RNases.
PNPase is closely related to a second phosphorolytic exoribonuclease, RNase PH. These two RNases share significant core structural features (21, 22), are widely conserved throughout bacteria (23), and play important roles in processing the 3′ ends of tRNAs and other small structured RNAs (24–26). Although E. coli tolerates single deletions of either protein, a double deletion results in slower growth and cold sensitivity (27). The experiments described above that explore the impact of PNPase on sRNA stability and function were conducted in the E. coli MG1655 background. During the isolation of this common laboratory strain, MG1655 acquired an incidental frameshift mutation in the 3′ terminus of rph. The resulting rph-1 gene carries five missense codons and encodes a 228-amino-acid protein, which is a slight truncation of the native 238-amino-acid RNase PH. The rph-1 mutation abolishes poly(A) phosphorolysis activity by the encoded RNase PH enzyme (28) and, like an rph deletion, leads to slow growth and cold sensitivity in combination with Δpnp (18). Given the similarities between PNPase and RNase PH, we sought to determine what role the functional wild-type RNase PH may play in sRNA stability and function in vivo.
Here, we report that RNase PH acts in concert with PNPase during exponential growth to stabilize and promote the function of the sRNAs RyhB, CyaR, and MicA, together representing two distinct sRNA classes. We provide evidence that PNPase, but not RNase PH, significantly contributes to the decay of RyhB due to mRNA target pairing, and we demonstrate that PNPase and RNase PH regulate the stability of these three sRNAs in distinct ways during the stationary phase. Finally, we show that stabilization of MicA by RNase PH and PNPase is dependent on the Hfq-binding 3′ tail of this sRNA.
MATERIALS AND METHODS
Strains and growth conditions.
Strains and plasmids used in this study are listed in Table S1 in the supplemental material, and strain construction details are provided in the supplemental Materials and Methods. LacZ fusions were constructed in the chromosome under the control of the araBAD promoter, with the 5′ untranslated region (UTR) and the initial codons of the target gene fused in-frame to the lacZ gene as previously described (29). For strains used in MicA or CyaR sRNA stability assays, the gene encoding the sRNA was deleted from its native locus and inserted with its native +1 transcriptional start in place of ryhB at the ryhB locus by lambda Red-mediated recombination (30). All strains were grown at 37°C in Lennox lysogeny broth (LB), and cultures of strains carrying pBRplac-based plasmids were further supplemented with 100 μg/ml of ampicillin. To achieve exponential and stationary growth, overnight cultures were diluted 1:200 in fresh culture medium and incubated at 250 rpm in an orbital shaking water bath. Culture density was monitored by the optical density at 600 nm (OD600), and cultures were considered to be in exponential phase between an OD600 of 0.3 and 0.4 and in stationary phase at an OD600 of 2.0.
β-Galactosidase assays.
To measure lacZ translational fusion activity, strains carrying ompX′-′lacZ, ompC′-′lacZ, sodB′-′lacZ, and ompA′-′lacZ were transformed with the pBRplac vector control and separately with pNRD405, pMicC, pRyhB, and pMicA, respectively. Each transformed strain was then grown to exponential phase in the presence of 100 μM isopropyl-β-d-thiogalactopyranoside (IPTG) to induce sRNA expression from the plasmid and either 0.01% arabinose (ompX′-′lacZ, ompC′-′lacZ, and sodB′-′lacZ strains) or 0.1% arabinose (ompA′-′lacZ strains) to activate the expression of the reporter fusion. Samples were taken from each culture and assayed for β-galactosidase activity as described by Miller (31).
sRNA stability time courses.
Cultures were grown to exponential or stationary phase as defined above, and then each sRNA was induced from the ryhB promoter by the addition of 2,2′-dipyridyl to a final concentration of 250 μM. After 15 min of sRNA induction, a sample was collected. For standard RNA half-life experiments, all further transcription was inhibited by immediate addition of rifampin to a concentration of 250 μg/ml. For sRNA pulse expression time courses, expression from the ryhB promoter was selectively inhibited by the immediate addition of FeSO4 to a concentration of 100 μM (4). Additional samples were collected after 1, 2, 4, and 6 min.
RNA extraction.
Total RNA extractions were based on the protocol of Massé et al. (4). Samples of 700 μl were withdrawn from growing cell cultures and immediately added to 100 μl of lysis solution (320 mM sodium acetate [pH 4.6], 8% SDS, 16 mM EDTA) that was mixed with 800 μl (pH 4.5) acid-phenol–chloroform–isoamyl alcohol (IAA) (125:25:1;Ambion) and preheated to 65°C. Samples were mixed for 5 min at 65°C and centrifuged, and the supernatant was extracted once more with pH 6.7 neutral phenol-chloroform-IAA (25:24:1; Thermo Fisher Scientific). Total RNA was alcohol-precipitated from the final aqueous phase and then suspended in Tris-EDTA (TE) buffer and quantified using a NanoDrop 2000 (Thermo Fisher Scientific).
Northern blots and analysis.
Approximately 2 to 3 μg of each RNA sample was loaded on 5% or 10% Tris-borate-EDTA (TBE)–urea gels (Bio-Rad) and separated for 2 h at 80 V in 1× TBE. Samples were next transferred to a Zeta-Probe GT membrane (Bio-Rad) by the Trans-Blot SD semidry transfer apparatus (Bio-Rad) according to the manufacturer's recommendations and affixed by UV cross-linking. Membranes were incubated overnight at 42°C with 100 ng/ml of 5′ biotinylated DNA probe (see Table S2 in the supplemental material) in ULTRAhyb (Ambion) and developed by the BrightStar BioDetect kit protocol (Ambion). Blots were imaged with a ChemiDoc MP imager (Bio-Rad) and quantified using Image Lab (version 5.2.1; Bio-Rad). Blots were normalized using 5S or SsrA as the loading control. Data analysis and half-life determinations were conducted in R (version 3.2.3; R Foundation for Statistical Computing, Vienna, Austria [https://www.R-project.org]) and visualized using the ggplot2 package (version 2.1.0; http://www.ggplot2.org). The half-life of each sample was calculated from the averaged decay rates of the exponential decay curves fit to individual replicates. To visualize the decay curves of each sample, the average intensity of each replicate time course was normalized, and then an exponential decay curve was fit to the combined triplicate data. The decay curve and individual time points were each plotted as a percentage of the intensity of the decay curve at T0 (0 min after the addition of rifampin or FeSO4). Uncertainty in the difference between RNA stability and sRNA pulse expression decay probabilities was determined by the standard propagation of error rules.
RESULTS
RNase PH and PNPase support sRNA regulation.
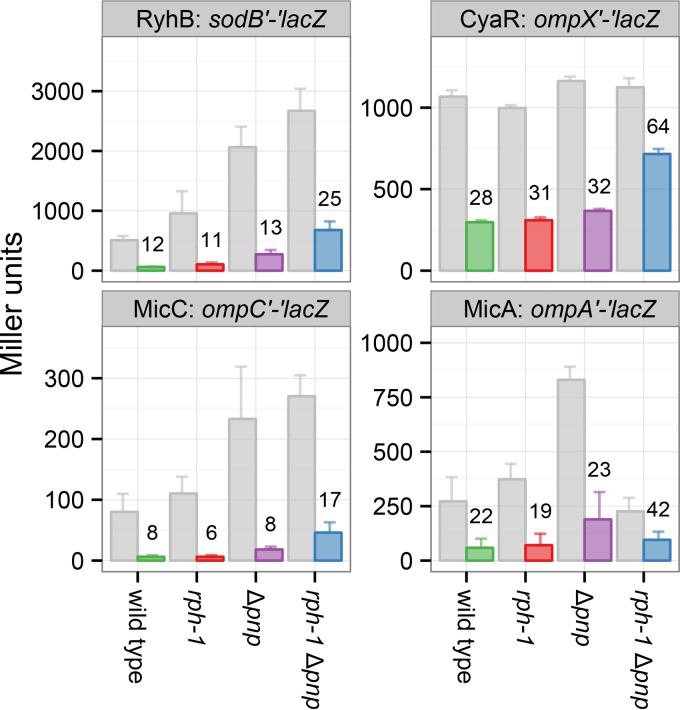
To initially examine the contribution of RNase PH to sRNA-mediated gene regulation, the activity of several sRNAs was evaluated in E. coli rph+, rph-1, rph+ Δpnp, and rph-1 Δpnp strains using β-galactosidase assays. The sRNAs RyhB, CyaR, MicC, and MicA were each expressed from a plasmid, and the strains were compared to an equivalent strain harboring the empty vector for their ability to downregulate lacZ translational reporter fusions to their target mRNAs, i.e., sodB′-′lacZ, ompX′-′lacZ, ompC′-′lacZ, and ompA′-′lacZ, respectively. The activity of each sRNA was diminished in the rph-1 Δpnp strain compared to that in the rph-1 strain (Fig. 1), which is consistent with previous findings for RyhB, CyaR, and MicC (18, 20). Interestingly, in an rph+ strain, deletion of pnp had no significant effect on the ability of RyhB, CyaR, MicC, and MicA to regulate their respective targets, suggesting substantial functional overlap between RNase PH and PNPase. These results indicate that RNase PH, like its homolog PNPase, supports the regulatory activities of these sRNAs.
FIG 1.
RNase PH and PNPase support gene regulation by sRNAs. To assess sRNA regulation in the absence of rph and/or pnp, the expression of the sodB′-′lacZ, ompX′-′lacZ, ompC′-′lacZ, and ompA′-′lacZ translational fusions was monitored by β-galactosidase assays during the exponential growth phase. Strains harbored either an empty vector or expressed the sRNAs RyhB, CyaR, MicC, or MicA, respectively, from a plasmid. The amount of β-galactosidase activity with each lacZ fusion during coexpression (colored bars) was evaluated relative to the empty plasmid vector (gray bars) in each background. Numbers above colored bars indicate the percentage of activity with sRNA expression compared to the vector control. Bars show means and 95% confidence intervals for three (CyaR, MicC, and MicA) or four (CyaR, MicC, MicA, and RyhB) replicate cultures. Each sRNA downregulates its target as long as either wild-type pnp or rph is present.
RNase PH and PNPase were not functionally identical in all aspects, however. Although no apparent change was observed in the expression of the ompX′-′lacZ fusion in the different empty vector backgrounds, expression of sodB′-′lacZ and ompC′-′lacZ with the empty vector increased with the deletion of pnp but was unaffected by rph-1 (Fig. 1). This increase may reflect the activity of other endogenous sRNAs that downregulate sodB and ompC and are dependent on PNPase but not RNase PH. Likewise, expression of ompA′-′lacZ increased with the deletion of pnp, but this increase was lost in the double mutant (rph-1 Δpnp), potentially indicating additional roles for RNase PH that are performed only in the absence of PNPase. Nonetheless, despite the variability in target expression between the different backgrounds, each of the tested sRNAs reduced target levels to similar percentages in the wild-type, Δpnp, and rph-1 strains but not in the rph-1 Δpnp double mutant.
RNase PH and PNPase promote sRNA stability.
Several previous studies have found that PNPase stabilizes unpaired sRNAs (18, 20, 32). PNPase is thus thought to support regulatory pairing between sRNAs and target mRNAs by reducing the nonproductive, premature degradation of unpaired sRNAs. We hypothesized that RNase PH may play a similar role. To test this possibility, we first examined how RNase PH affects sRNA half-life in exponentially growing cultures of the rph+, rph-1, rph+ Δpnp, and rph-1 Δpnp strains (Fig. 2). In addition to the rph-1 strains, cultures of the Δrph and Δrph Δpnp strains were also examined to ascertain whether the protein encoded by the frame-shifted rph-1 allele retains any activity toward sRNAs.
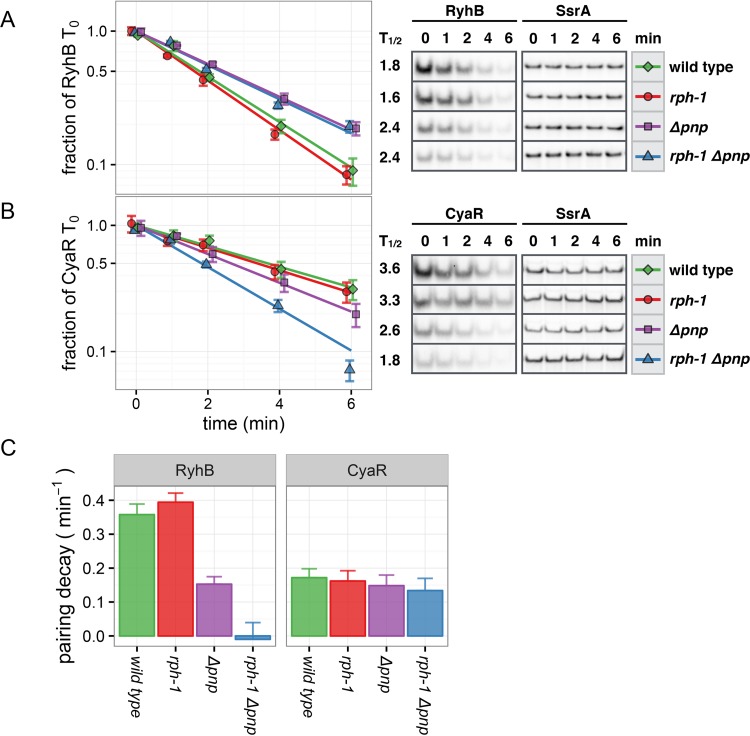
FIG 2.
RNase PH and PNPase stabilize RyhB, CyaR, and MicA. RNA stability time courses of exponentially growing cultures expressing RyhB (A), CyaR (B), or MicA (C). To measure intrinsic stability, sRNAs were induced from the ryhB promoter for 15 min in order to deplete target mRNAs that are able to pair with each sRNA prior to rifampin addition. (Left) Exponential decay curves of each sRNA in the indicated strain backgrounds. Points and error bars represent the means and standard errors of three independent cultures. Signal intensity for each sRNA band was quantified and then normalized to a 5S or SsrA loading control. Decay curves were fit to normalized replicates as described in Materials and Methods. (Right) Half-lives and representative Northern blots of each time course, presented with 5S or SsrA loading controls. Deletion of pnp decreased sRNA stability, and mutation of both pnp and rph further reduced sRNA half-life.
The sRNAs RyhB, CyaR, and MicA were individually expressed as single genomic copies under the control of the ryhB promoter. Each sRNA was induced for 15 min by the addition of dipyridyl, an iron chelator that stimulates derepression of this Fur-regulated promoter. This allowed for accumulation of the sRNA and facilitated degradation of corresponding mRNA targets prior to the addition of rifampin and the collection of time points (33). As shown in Fig. S1 in the supplemental material, downregulation of mRNA targets of CyaR and RyhB was effective in the wild-type and single-mutant strains but less effective in the double-mutant rph-1 Δpnp strain, which is consistent with the lacZ reporter fusion results (Fig. 1). After 15 min of RyhB induction, sodB levels were reduced 50% in the double mutant and 80% to 90% in other strains. For CyaR and MicA, ompX levels initially decreased rapidly and then stabilized for the remainder of the induction. This likely represents depletion of the accessible pool of ompX, the remainder recalcitrant to regulation by each sRNA. By depleting the pool of targetable transcripts, the stability of each sRNA under rifampin treatment more closely reflects the degradation of unpaired sRNAs.
Under these conditions, both RNase PH and PNPase increased sRNA stability (Fig. 2). Each sRNA was very stable in the rph+ pnp+ strain, but each was destabilized in the rph-1 Δpnp and Δrph Δpnp double mutant backgrounds. Critically, the presence of either the rph+ or pnp+ allele alone significantly increased stability, although to a greater extent with pnp+ than with rph+. We expect that the destabilization of RyhB in the double mutant backgrounds (Fig. 2A) is primarily due to the increased susceptibility of unpaired RyhB to RNases due to the minimal regulatory activity of RyhB in the rph-1 Δpnp strain (Fig. 1; see also Fig. S1 in the supplemental material). Although the stability of RyhB was unaffected by the rph-1 mutation, it was moderately reduced in the Δrph strain. However, for CyaR and MicA, mutation of rph alone, either by a frameshift mutation (rph-1) or deletion (Δrph), did not reduce sRNA stability (Fig. 2B and C), and the stability of each sRNA was similar between the rph-1 Δpnp and Δrph Δpnp strains.
To identify any differences in the expressions of proteins important to sRNA stability in these strains, the levels of RNase PH, PNPase, and Hfq were each compared between the wild-type and mutant backgrounds (see Fig. S2 in the supplemental material). Hfq protein levels were relatively consistent across all six backgrounds. PNPase levels were likewise unaffected by the rph-1 mutation but were decreased 25% by the deletion of rph. This decrease in PNPase levels may explain, in part, the modest decrease in stability that was observed between the rph-1 and Δrph strains. Expression of RNase PH was also halved in Δpnp strains. Despite this reduction, RNase PH alone still provided a substantial stabilization effect for each sRNA in the time courses. Together, these results demonstrate that RNase PH and PNPase promote the stabilization of RyhB, CyaR, and MicA.
PNPase facilitates sRNA decay as a consequence of sRNA-mRNA base pairing.
Since PNPase and RNase PH promote sRNA stability prior to base pairing (Fig. 2), we asked whether these proteins perform additional roles in sRNA-mediated regulation, such as facilitating sRNA-mRNA base pairing or the productive degradation of such duplexes. To address these possibilities, we examined whether PNPase or RNase PH impacts sRNA stability under conditions in which sRNA-mRNA pairing is allowed to occur. In these experiments, the rph+, rph-1, rph+ Δpnp, and rph-1 Δpnp strains expressing RyhB or CyaR from the Fur-regulated ryhB promoter were induced as before for 15 min with dipyridyl. However, instead of adding rifampin, FeSO4 was added in excess to selectively stop sRNA transcription from this promoter. These “sRNA pulse expression” experiments therefore reflect sRNA decay due to the degradation of unpaired sRNAs, as in the rifampin time courses, but also account for additional decay facilitated by pairing between sRNAs and nascent mRNA targets (4). Upon removal of the inducer, the levels of the mRNA targets recovered as the induced RyhB (4) or CyaR (see Fig. S3 in the supplemental material) decayed. During the RNA stability experiments, RyhB decayed more rapidly in Δpnp strains than in the parental pnp+ strains (Fig. 2A). However, when RyhB stability was examined by pulse expression time courses, turnover was more rapid in pnp+ strains than in Δpnp strains (Fig. 3A). In contrast, the relative stability of CyaR in each background was unchanged between the half-life experiments and the pulse expression time courses (compare Fig. 2B and 3B).
FIG 3.
PNPase accelerates the degradation of RyhB during pairing conditions. (A, B) sRNA pulse expression stability time courses of exponentially growing cultures expressing RyhB (A) or CyaR (B). Each sRNA was induced by dipyridyl from the ryhB promoter for 15 min, followed by the addition of excess iron to repress this Fur-regulated promoter. (Left) Exponential decay curves of each sRNA in the indicated strain backgrounds. Points and error bars represent the means and standard errors of three independent cultures. Bands were normalized to 5S or SsrA as described for Fig. 2. (Right) Half-lives and representative Northern blots of each time course, presented with SsrA loading controls. (C) Calculated decay probabilities for RyhB and CyaR due to pairing conditions. Taller bars indicate that the introduction of pairing to target mRNAs had a larger impact on RNA stability. Error bars demark standard error. Decay of RyhB due to pairing is markedly accelerated in the presence of PNPase but not RNase PH.
To quantify the additional decay observed for each sRNA due to pairing with nascent transcripts, the exponential decay constants, or probabilities, that were obtained from the exponential regressions under sRNA pulse expression conditions were compared with those obtained from the half-life experiments. By subtracting the intrinsic decay rate that was observed in the RNA half-life experiments from the total decay rate that was observed in the sRNA pulse expression time courses, we inferred the effective decay rate of RyhB and CyaR due to pairing with nascent mRNA transcripts in each background. As shown in Fig. 3C, RyhB was substantially destabilized by pairing in the presence of PNPase (rph+ or rph-1 strains), modestly destabilized by RNase PH alone (rph+ Δpnp), and unaffected in the double mutant (rph-1 Δpnp). The rph-1 mutation did not significantly impact the RyhB stability observed under pairing conditions. In contrast to RyhB, the stability of CyaR in the sRNA pulse expression experiments decreased by a similar degree in all strains (Fig. 3C). These results indicate that PNPase promotes both the intrinsic stability and decay of RyhB due to pairing but only impacts the intrinsic stability of CyaR. RNase PH has a much smaller impact on the decay of RyhB due to pairing, which indicates that it primarily supports sRNA regulation before this step, i.e., by preventing spurious degradation of Hfq-bound sRNAs prior to regulatory pairing.
RNase PH and PNPase impact sRNAs in a growth-phase-dependent manner.
Here, we have shown that PNPase and RNase PH contribute to the stability of sRNAs during exponential growth (Fig. 2). These findings are in agreement with previous studies, indicating that PNPase serves a protective role for sRNAs during exponential growth (18–20). However, PNPase was also reported to destabilize MicA during stationary phase (19, 34). We therefore asked if the protective role observed for RNase PH would be impacted by the growth phase. Since our exponential-phase data suggest that RNase PH has a greater role influencing RyhB, CyaR, and MicA stability prior to pairing, i.e., during decay in the presence of rifampin, we focused on assessing its impact during the stationary phase via standard RNA half-life experiments.
In stationary-phase conditions, PNPase and RNase PH still affected sRNA stability, although with different dynamics than those observed during exponential growth. Each sRNA was generally more stable under these conditions, even in strains harboring both rph and pnp deletions (Fig. 4). This increased stability was particularly evident with CyaR, which decayed very slowly regardless of the absence of both RNase PH and PNPase (Fig. 4B). RyhB exhibited a greater range of stability between strains, and either the mutation or deletion of rph resulted in reduced stability in the pnp+ backgrounds (Fig. 4A). The deletion of pnp resulted in the greatest reduction in RyhB stability, and the mutation of rph had no effect in the absence of pnp.
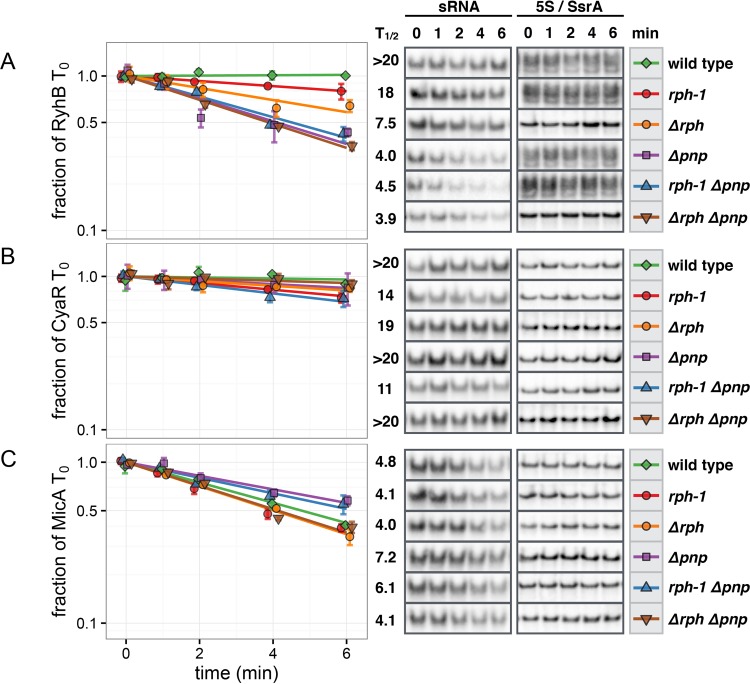
FIG 4.
Differential effects of RNase PH and PNPase on RyhB, CyaR, and MicA decay in the stationary phase. RNA stability time courses were performed as described in the legend to Fig. 2 for stationary-phase cultures expressing RyhB (A), CyaR (B), or MicA (C). (Left) Exponential decay curves of each sRNA in the indicated strain backgrounds. Points and error bars represent the means and standard errors of three independent cultures. Bands were normalized to 5S or SsrA as described for Fig. 2. (Right) Half-lives and representative Northern blots of each time course presented with 5S or SsrA loading controls. In the stationary phase, PNPase and RNase PH together stabilize RyhB. Mutation of rph and pnp does not affect the stability of CyaR. In the absence of PNPase, MicA is stabilized in strains carrying rph+ or rph-1.
Turnover of MicA was likewise altered by the shift to stationary-phase conditions and varied modestly with different combinations of rph and pnp mutations. Notably, the stability of MicA was unchanged between the wild-type (rph+ pnp+) and Δrph Δpnp strains. Nonetheless, and as reported previously in the MG1655 background (19), MicA was modestly stabilized in an rph-1 Δpnp strain compared to that in the isogenic rph-1 pnp+ strain (Fig. 4C). MicA was likewise stabilized in rph+ strains upon the deletion of pnp. Intriguingly, there also appeared to be a difference in MicA stability between the rph-1 Δpnp and Δrph Δpnp strains. However, given the modest differences in stability, additional experiments will be needed to clarify the impact of these mutations on MicA decay. Overall, the deletion of rph and/or pnp resulted in different effects on RyhB, CyaR, and MicA during stationary-phase conditions, whereas during the exponential phase, each sRNA responded similarly to the same mutations.
RNase PH and PNPase mediate the decay of MicA not bound to Hfq.
Since PNPase protects Hfq-bound sRNAs from decay but degrades unbound sRNAs (19, 20), we hypothesized that RNase PH may behave similarly. To test this hypothesis without subjecting cells to the deleterious and pleiotropic effects of an hfq deletion, we took advantage of previous work showing that Hfq does not protect a truncated MicA possessing a shortened Rho-independent terminator (35). We constructed rph+, rph-1, Δrph, rph+ Δpnp, rph-1 Δpnp, and Δrph Δpnp strains expressing MicA that was lacking the last six ribonucleotides, MicA(−6), from the chromosomal ryhB promoter. This truncation causes some transcriptional read-through past the MicA terminator (see Fig. S4 in the supplemental material), although less than that observed previously (35). This difference may be due to the promoter used to express the sRNA (ryhB versus araBAD), the downstream genetic context, or the copy number of each construct (chromosome versus plasmid). Regardless, the majority of transcripts detected on Northern blots were of the expected size for MicA and were readily discernible from read-through products (see Fig. S4). The stability of MicA(−6) was assessed in standard RNA half-life experiments in both the exponential and stationary phases as described above.
Under both conditions, the truncated MicA was dramatically destabilized in the presence of PNPase and RNase PH (Fig. 5). This loss in stability contrasts with the stabilization of wild-type MicA by PNPase and RNase PH in the exponential phase (Fig. 2C) and suggests a model in which both proteins facilitate the protection of Hfq-bound MicA and the decay of free MicA. Furthermore, MicA(−6) was much less stable in an rph+ or rph-1 strain than in a Δrph strain, suggesting that RNase PH encoded by the rph-1 allele may not be totally inert. Surprisingly, MicA(−6) was stabilized in the double deletion mutant as well as in strains harboring single deletions of pnp or rph, indicating that both PNPase and RNase PH were required for the accelerated degradation of MicA(−6). Taken together, these results support a model in which the binding of MicA by Hfq controls whether PNPase and RNase PH facilitate the protection or degradation of MicA.
FIG 5.
RNase PH destabilizes MicA lacking Hfq protection. Stability of the MicA(−6) truncation mutant was assessed by stability time courses in cultures grown to exponential (A) and stationary (B) phase as described in the legend to Fig. 2. (Left) Exponential decay curves of MicA(−6) in the indicated strain backgrounds. Points and error bars represent the means and standard errors of three independent cultures. Bands were normalized to 5S or SsrA as described for Fig. 2. (Right) Average half-lives and representative Northern blots of each time course, adjusted individually for visibility, presented with 5S or SsrA loading controls. Representative blots and decay curves correspond to the appropriately terminated transcript. In both growth phases, RNase PH and PNPase together destabilize MicA(−6).
DISCUSSION
Here, we have shown that the exoribonucleases RNase PH and PNPase impact sRNA-mediated gene regulation in E. coli by increasing the stability and function of each sRNA examined. Under exponential growth conditions, RNase PH and PNPase support the stability of the sRNAs RyhB, CyaR, and MicA and the effective regulation of their mRNA targets. However, in the stationary phase, the effect of RNase PH and PNPase on stability varies considerably among these sRNAs. PNPase further appears to assist in the pairing and/or subsequent decay of at least one sRNA. Finally, PNPase- and RNase PH-mediated stabilization of MicA, and presumably other sRNAs, is dependent upon Hfq protection. In the absence of bound Hfq, PNPase and RNase PH facilitate sRNA degradation instead.
Our results suggest that RNase PH and PNPase may serve different functions between different sRNAs and growth phases. During the exponential phase, RNase PH and PNPase function together to control the stability of RyhB, CyaR, and MicA (Fig. 2). While the stability of all three of these sRNAs significantly decreased in the absence of only PNPase (Fig. 2, compare the wild type to the Δpnp strain), the greatest defect was observed in the absence of both wild-type PNPase and RNase PH (Fig. 2, compare the Δpnp strain to the Δrph Δpnp strain or the rph-1 Δpnp strain). Furthermore, abrogation of the sRNA-mediated regulation of gene expression only occurred in the absence of both wild-type proteins (Fig. 1, compare the Δpnp strain to the rph-1 Δpnp strain; see also Fig. S1 in the supplemental material). Since rph-1 encodes a mutant RNase PH that was previously shown to lack phosphorolysis activity, these results suggest that the catalytic activity of PNPase and RNase PH may be required for sRNA stability and function during the exponential phase. One possible mechanism as to how this may occur is that PNPase and RNase PH may play a role in trimming RNase-binding sites from Hfq-bound sRNAs, which is similar to how RNase II protects certain RNAs from degradation by removing 3′ binding sites for RNase R and PNPase (36–38). Alternatively, PNPase and RNase PH may act in concert to remove RNAs that compete with these sRNAs for Hfq association. Prior work has shown that the deletion of pnp reduced MicA binding to Hfq (20). The exact mechanisms of this protection, and why it varies between sRNAs, remain to be explored in future studies.
During the stationary phase, the contributions of RNase PH and PNPase to sRNA stability differ among the three tested sRNAs, RyhB, CyaR, and MicA. For CyaR, neither PNPase nor RNase PH significantly contributed to sRNA protection (Fig. 4B). For RyhB, both proteins were necessary to achieve maximum stabilization, which is similar to the exponential phase (Fig. 4A, compare the wild type to Δpnp and Δrph strains). Likewise, PNPase clearly impacts MicA differently during the stationary phase, mediating its decay rather than promoting stability (Fig. 4C, compare the Δpnp strain to the wild type). Since PNPase degrades the MicA that is not associated with Hfq (Fig. 5), our results suggest that MicA less efficiently competes for Hfq during the stationary phase compared to the exponential phase. RNase PH may also have a role in regulating MicA stability during the stationary phase, as deletion of rph appeared to suppress the enhanced stability of MicA in a pnp deletion mutant. However, given the small differences in our decay curves, additional work will be needed to resolve the exact role of RNase PH.
Finally, the roles of PNPase and RNase PH were further differentiated by their effects on sRNA stability during the sRNA pulse expression time courses. Although RyhB decay in Δpnp strains occurred at similar rates during both RNA stability and the sRNA pulse expression time courses, RyhB turnover in pnp+ strains was much faster in the sRNA pulse expression experiments than in the RNA stability time courses (Fig. 2A and 3A). We interpret these results to mean that RyhB is susceptible to ribonucleases and is unable to either effectively pair with target mRNAs or be degraded upon pairing in the absence of PNPase. In contrast, RNase PH appears to play a much more limited role in facilitating RyhB turnover during sRNA pulse expression conditions.
Although our results suggest that PNPase and RNase PH both enhance sRNA stability and support effective regulation by sRNAs, it is apparent from our findings that PNPase performs certain functions important to sRNA-mediated gene regulation that are poorly mediated by RNase PH alone. Both proteins are phosphorolytic exoribonucleases, yet structural differences between them may be important in differentiating their roles. PNPase possesses KH and S1 domains that assist in binding RNA and guiding it to one of three active sites present in the PNPase trimer (21, 39). In contrast, RNase PH forms a simple toroid structure that is capable of binding RNA on both faces of its hexamer (22). These structural differences may have important consequences on how each protein is able to engage and interact with RNA or other proteins. These differences may only become important as different proteins and RNAs are expressed, leading to different requirements for stabilization between different sRNAs and growth conditions.
Supplementary Material
ACKNOWLEDGMENTS
We thank Susan Gottesman for her comments on the manuscript.
The content of this publication is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00624-16.
REFERENCES
- 1.Papenfort K, Vogel J. 2014. Small RNA functions in carbon metabolism and virulence of enteric pathogens. Front Cell Infect Microbiol 4:91. doi: 10.3389/fcimb.2014.00091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bobrovskyy M, Vanderpool CK, Richards GR. 2015. Small RNAs regulate primary and secondary metabolism in Gram-negative bacteria. Microbiol Spectr 3:59–94. doi: 10.1128/microbiolspec.MBP-0009-2014. [DOI] [PubMed] [Google Scholar]
- 3.Bouvier M, Sharma CM, Mika F, Nierhaus KH, Vogel J. 2008. Small RNA binding to 5′ mRNA coding region inhibits translational initiation. Mol Cell 32:827–837. doi: 10.1016/j.molcel.2008.10.027. [DOI] [PubMed] [Google Scholar]
- 4.Massé E, Escorcia FE, Gottesman S. 2003. Coupled degradation of a small regulatory RNA and its mRNA targets in Escherichia coli. Genes Dev 17:2374–2383. doi: 10.1101/gad.1127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lease RA, Cusick ME, Belfort M. 1998. Riboregulation in Escherichia coli: DsrA RNA acts by RNA:RNA interactions at multiple loci. Proc Natl Acad Sci U S A 95:12456–12461. doi: 10.1073/pnas.95.21.12456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pfeiffer V, Papenfort K, Lucchini S, Hinton JCD, Vogel J. 2009. Coding sequence targeting by MicC RNA reveals bacterial mRNA silencing downstream of translational initiation. Nat Struct Mol Biol 16:840–846. doi: 10.1038/nsmb.1631. [DOI] [PubMed] [Google Scholar]
- 7.Bandyra KJ, Said N, Pfeiffer V, Górna MW, Vogel J, Luisi BF. 2012. The seed region of a small RNA drives the controlled destruction of the target mRNA by the endoribonuclease RNase E. Mol Cell 47:943–953. doi: 10.1016/j.molcel.2012.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gottesman S, Storz G. 2011. Bacterial small RNA regulators: versatile roles and rapidly evolving variations. Cold Spring Harb Perspect Biol 3:a003798. doi: 10.1101/cshperspect.a003798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Møller T, Franch T, Højrup P, Keene DR, Bächinger HP, Brennan RG, Valentin-Hansen P. 2002. Hfq: a bacterial Sm-like protein that mediates RNA-RNA interaction. Mol Cell 9:23–30. doi: 10.1016/S1097-2765(01)00436-1. [DOI] [PubMed] [Google Scholar]
- 10.Panja S, Woodson SA. 2012. Hfq proximity and orientation controls RNA annealing. Nucleic Acids Res 40:8690–8697. doi: 10.1093/nar/gks618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Massé E, Vanderpool CK, Gottesman S. 2005. Effect of RyhB small RNA on global iron use in Escherichia coli. J Bacteriol 187:6962–6971. doi: 10.1128/JB.187.20.6962-6971.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang J, Rennie W, Liu C, Carmack CS, Prévost K, Caron M-P, Massé E, Ding Y, Wade JT. 2015. Identification of bacterial sRNA regulatory targets using ribosome profiling. Nucleic Acids Res 43:10308–10320. doi: 10.1093/nar/gkv1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang A, Schu DJ, Tjaden BC, Storz G, Gottesman S. 2013. Mutations in interaction surfaces differentially impact E. coli Hfq association with small RNAs and their mRNA targets. J Mol Biol 425:3678–3697. doi: 10.1016/j.jmb.2013.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schu DJ, Zhang A, Gottesman S, Storz G. 2015. Alternative Hfq-sRNA interaction modes dictate alternative mRNA recognition. EMBO J 34:2557–2573. doi: 10.15252/embj.201591569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moon K, Gottesman S. 2011. Competition among Hfq-binding small RNAs in Escherichia coli. Mol Microbiol 82:1545–1562. doi: 10.1111/j.1365-2958.2011.07907.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Małecka EM, Stróżecka J, Sobańska D, Olejniczak M. 2015. Structure of bacterial regulatory RNAs determines their performance in competition for the chaperone protein Hfq. Biochemistry 54:1157–1170. doi: 10.1021/bi500741d. [DOI] [PubMed] [Google Scholar]
- 17.Sagawa S, Shin J-E, Hussein R, Lim HN. 2015. Paradoxical suppression of small RNA activity at high Hfq concentrations due to random-order binding. Nucleic Acids Res 43:8502–8515. doi: 10.1093/nar/gkv777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.De Lay NR, Gottesman S. 2011. Role of polynucleotide phosphorylase in sRNA function in Escherichia coli. RNA 17:1172–1189. doi: 10.1261/rna.2531211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Andrade JM, Pobre V, Matos AM, Arraiano CM. 2012. The crucial role of PNPase in the degradation of small RNAs that are not associated with Hfq. RNA 18:844–855. doi: 10.1261/rna.029413.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bandyra KJ, Sinha D, Syrjanen J, Luisi BF, De Lay NR. 2016. The ribonuclease polynucleotide phosphorylase can interact with small regulatory RNAs in both protective and degradative modes. RNA 22:360–372. doi: 10.1261/rna.052886.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Symmons MF, Jones GH, Luisi BF. 2000. A duplicated fold is the structural basis for polynucleotide phosphorylase catalytic activity, processivity, and regulation. Structure 8:1215–1226. doi: 10.1016/S0969-2126(00)00521-9. [DOI] [PubMed] [Google Scholar]
- 22.Ishii R, Nureki O, Yokoyama S. 2003. Crystal structure of the tRNA processing enzyme RNase PH from Aquifex aeolicus. J Biol Chem 278:32397–32404. doi: 10.1074/jbc.M300639200. [DOI] [PubMed] [Google Scholar]
- 23.Condon C, Putzer H. 2002. The phylogenetic distribution of bacterial ribonucleases. Nucleic Acids Res 30:5339–5346. doi: 10.1093/nar/gkf691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kelly KO, Reuven NB, Li Z, Deutscher MP. 1992. RNase PH is essential for tRNA processing and viability in RNase-deficient Escherichia coli cells. J Biol Chem 267:16015–16018. [PubMed] [Google Scholar]
- 25.Li Z, Deutscher MP. 1994. The role of individual exoribonucleases in processing at the 3′ end of Escherichia coli tRNA precursors. J Biol Chem 269:6064–6071. [PubMed] [Google Scholar]
- 26.Basturea GN, Zundel MA, Deutscher MP. 2011. Degradation of ribosomal RNA during starvation: comparison to quality control during steady-state growth and a role for RNase PH. RNA 17:338–345. doi: 10.1261/rna.2448911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou Z, Deutscher MP. 1997. An essential function for the phosphate-dependent exoribonucleases RNase PH and polynucleotide phosphorylase. J Bacteriol 179:4391–4395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jensen KF. 1993. The Escherichia coli K-12 “wild types” W3110 and MG1655 Have an rph frameshift mutation that leads to pyrimidine starvation due to low pyrE expression levels. J Bacteriol 175:3401–3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mandin P, Gottesman S. 2009. A genetic approach for finding small RNAs regulators of genes of interest identifies RybC as regulating the DpiA/DpiB two-component system. Mol Microbiol 72:551–565. doi: 10.1111/j.1365-2958.2009.06665.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu D, Ellis HM, Lee EC, Jenkins NA, Copeland NG, Court DL. 2000. An efficient recombination system for chromosome engineering in Escherichia coli. Proc Natl Acad Sci U S A 97:5978–5983. doi: 10.1073/pnas.100127597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miller JH. 1992. A short course in bacterial genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 32.Andrade JM, Pobre V, Arraiano CM. 2013. Small RNA modules confer different stabilities and interact differently with multiple targets. PLoS One 8:e52866. doi: 10.1371/journal.pone.0052866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Massé E, Gottesman S. 2002. A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli. Proc Natl Acad Sci U S A 99:4620–4625. doi: 10.1073/pnas.032066599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Andrade JM, Arraiano CM. 2008. PNPase is a key player in the regulation of small RNAs that control the expression of outer membrane proteins. RNA 14:543–551. doi: 10.1261/rna.683308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Otaka H, Ishikawa H, Morita T, Aiba H. 2011. PolyU tail of rho-independent terminator of bacterial small RNAs is essential for Hfq action. Proc Natl Acad Sci U S A 108:13059–13064. doi: 10.1073/pnas.1107050108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mohanty BK, Kushner SR. 2003. Genomic analysis in Escherichia coli demonstrates differential roles for polynucleotide phosphorylase and RNase II in mRNA abundance and decay. Mol Microbiol 50:645–658. doi: 10.1046/j.1365-2958.2003.03724.x. [DOI] [PubMed] [Google Scholar]
- 37.Mohanty BK, Kushner SR. 2000. Polynucleotide phosphorylase, RNase II and RNase E play different roles in the in vivo modulation of polyadenylation in Escherichia coli. Mol Microbiol 36:982–994. doi: 10.1046/j.1365-2958.2000.01921.x. [DOI] [PubMed] [Google Scholar]
- 38.Hajnsdorf E, Steier O, Coscoy L, Teysset L, Régnier P. 1994. Roles of RNase E, RNase II and PNPase in the degradation of the rpsO transcripts of Escherichia coli: stabilizing function of RNase II and evidence for efficient degradation in an ams pnp rnb mutant. EMBO J 13:3368–3377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stickney LM, Hankins JS, Miao X, Mackie GA. 2005. Function of the conserved S1 and KH domains in polynucleotide phosphorylase. J Bacteriol 187:7214–7221. doi: 10.1128/JB.187.21.7214-7221.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.