ABSTRACT
Soil bacteria engage each other in competitive and cooperative ways to determine their microenvironments. In this study, we report the identification of a large number of genes required for Myxococcus xanthus to engage Bacillus subtilis in a predator-prey relationship. We generated and tested over 6,000 individual transposon insertion mutants of M. xanthus and found many new factors required to promote efficient predation, including the specialized metabolite myxoprincomide, an ATP-binding cassette (ABC) transporter permease, and a clustered regularly interspaced short palindromic repeat (CRISPR) locus encoding bacterial immunity. We also identified genes known to be involved in predation, including those required for the production of exopolysaccharides and type IV pilus (T4P)-dependent motility, as well as chemosensory and two-component systems. Furthermore, deletion of these genes confirmed their role during predation. Overall, M. xanthus predation appears to be a multifactorial process, with multiple determinants enhancing predation capacity.
IMPORTANCE Soil bacteria engage each other in complex environments and utilize multiple traits to ensure survival. Here, we report the identification of multiple traits that enable a common soil organism, Myxococcus xanthus, to prey upon and utilize nutrients from another common soil organism, Bacillus subtilis. We mutagenized the predator and carried out a screen to identify genes that were required to either enhance or diminish capacity to consume prey. We identified dozens of genes encoding factors that contribute to the overall repertoire for the predator to successfully engage its prey in the natural environment.
INTRODUCTION
The majority of biomass found in soil consists of highly diverse microbial communities which display complex networks of interspecies interactions ranging from commensalism to competition or cooperation, each of which contributes to the turnover of materials within the environment (1, 2). One important community-shaping determinant is predation. Microbial predators can range from nematodes to highly specialized endobiotic bacteria, such as Bdellovibrio bacteriovorus (3). Another ubiquitous microbial predator is Myxococcus xanthus, which secrets lytic enzymes and specialized metabolites and hunts in groups and consumes prey (4–6). M. xanthus is able to consume a diverse repertoire of microbes ranging from phage to bacterial plant pathogens and clinical isolates, and it uses the resulting nutrients to sustain growth (7–9).
M. xanthus also displays a complex life cycle that requires regulation of motility and intraspecies communication following nutrient starvation, culminating in multicellular aggregation and fruiting body formation (10). M. xanthus motility requires type IV pilus (T4P) motility and another machine involved in gliding (e.g., focal adhesion complexes and exopolysaccharide [EPS]). It has been shown that efficient predation requires the regulation of both motility systems, which are controlled, in part, by the Frz chemosensory system (7, 11, 12). Upon encountering prey, M. xanthus cells display coordinated rippling in which groups of cells move back and forth trapping prey cells to enhance predation, a phenomenon called predataxis (12).
Most bacteria tested under laboratory conditions are unable to resist M. xanthus as a predator. However, we found that Bacillus subtilis NCIB3610, an ancestral wild-type Marburg isolate (13), was able to transiently resist predation by M. xanthus; the specialized metabolite bacillaene is required for this short-term protection (14). During prolonged exposure to predation, B. subtilis produces megastructures composed of viable spores embedded within what appears to be a dense matrix of unknown composition. Our model suggests that megastructure formation provides an opportunity for B. subtilis NCIB3610 to produce spores, allowing for long-term survival of cells (7).
In general, microbial communities, such as biofilms, are known to provide protection from predation. For example, biofilm formation of Escherichia coli protects cells from predation by Caenorhabditis elegans and M. xanthus (15). Planktonic Vibrio cholerae and Pseudomonas aeruginosa cells displayed a reduction in survival against protozoan predators relative to their biofilm-associated counterparts (16, 17). Furthermore, sporulation has been shown to provide excellent protection from predation, as M. xanthus spores survive the C. elegans gut, and spores from both ancestral and laboratory strains of B. subtilis escape predation by M. xanthus (14, 18).
In other interspecies interactions, specialized metabolites have been shown to act as killing or signaling molecules (2, 19). Shank and coworkers (20) demonstrated that biofilm formation in B. subtilis is triggered in response to thiopeptide antibiotics (thiocillins), produced by a member of the same genus, Bacillus cereus (20, 21). Interestingly, they show that nonfunctional analogs of thiocillins do not alter biofilm-inducing capabilities. Thus, small molecules with antimicrobial capacity elicit complex behavioral responses in soil communities. Moreover, it is highly likely that antibiotics, such as thiocillins, are present in sublethal concentrations in soil environments, depending on their solubility or light sensitivity, and may have very localized temporal or spatial impact (2).
For M. xanthus, the specialized metabolite myxovirescin (also known as TA) is necessary for predation of proteobacteria by inhibiting lipoprotein production (9, 22, 23). Other specialized metabolites found in membrane vesicles or secreted by M. xanthus comprise part of the predation machinery. Predation by M. xanthus is, therefore, multifactorial and complex in its overall regulation (24–26). Consistent with this perspective, genome analysis indicates that M. xanthus employs 18 polyketide synthase (PKS)/nonribosomal peptide synthetase (NRPS) clusters (8.6% of the genome) to produce a large suite of specialized metabolites, some of which likely affect predation (27–29). Additionally, the M. xanthus genome encodes a large number of proteins as part of the type II and type IV secretion systems, as well as ABC transporters (30). These systems are predicted to play a role in the delivery of specialized metabolites or other elements used during predation.
In this study, we report the identification of a large number of genes required for M. xanthus predation when challenged with the B. subtilis NCIB3610 ancestral strain as a prey source. We generated and tested a transposon library of M. xanthus mutants and found many new factors required for efficient predation, including the specialized metabolite myxoprincomide, an ABC transporter permease, and the clustered regularly interspaced short palindromic repeat (CRISPR) II locus. We also identified genes known to be involved in predation, including those required for production of EPS and T4P-dependent motility, as well as chemosensory and two-component systems thought to affect predation. Deletion of many of these genes confirmed their role during predation.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
M. xanthus strains used in this study are listed in Table 1. Additionally, E. coli DH5α and B. subtilis NCIB3610 strains were used as prey strains and were grown in LB at 37°C. M. xanthus strains were cultivated in Casitone-yeast extract (CYE) medium at 32°C (31). If required, kanamycin was used at a final concentration of 50 or 100 μg/ml for M. xanthus strains.
TABLE 1.
Bacterial strains used in this study
| Bacterial strain | Genotype or characteristic | Reference or source |
|---|---|---|
| B. subtilis NCIB3610 | Ancestral strain | 13 |
| E. coli DH5α | Laboratory strain | |
| M. xanthus strains | ||
| DZ2 | Wild type | 62 |
| JK4633 | ΔdifA | This study |
| JK4634 | ΔdifE | This study |
| JK4635 | ΔhsfB | This study |
| JK4306 | Δ(MXAN_7258-MXAN_7267) (CRISPR II) | This study |
| JK4296 | Δ(MXAN_5713-MXAN_5714) (ABC transporter permease) | This study |
| JK4294 | Δ(MXAN_3778-MXAN_3779) (myxoprincomide) | This study |
Construction of M. xanthus in-frame deletion mutants.
In-frame deletion mutants were constructed as described elsewhere (32). Briefly, 1 kb of the up- and downstream regions of the gene of interest were amplified by PCR using Phusion polymerase (New England BioLabs, MA) and cloned into plasmid pBJ114. Sequencing was performed at the Nevada Genomics Center (Reno, NV). Verified plasmids were transformed into M. xanthus DZ2, and clones were selected on CYE agar plates containing kanamycin. Chromosomal insertions were verified by PCR. To obtain clean in-frame deletions, individual clones were plated onto CTT (Casitone, Tris-HCl) agar plates containing 2% galactose for counterselection. In-frame deletion mutants were identified by their ability to grow on CTT agar plates containing 2% galactose and their inability to grow on plates containing kanamycin. Additionally, the in-frame deletions were verified by PCR and sequencing of the genomic region.
Transposon mutagenesis of M. xanthus and library screen.
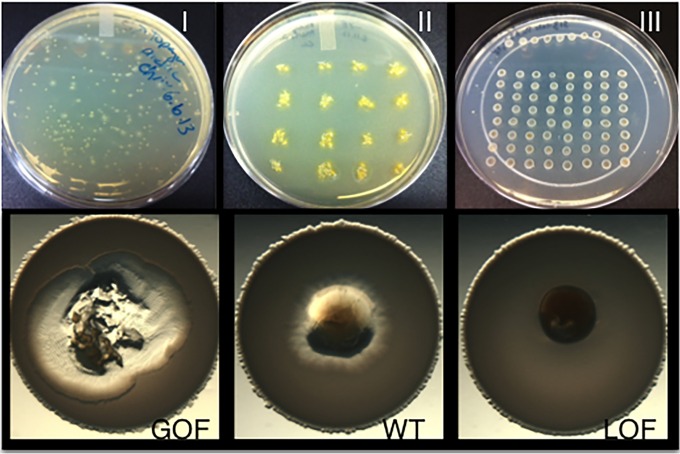
The EZ-Tn5 Transposome kit system (Epicentre) was used as a mobile genetic element to create an M. xanthus transposon library. M. xanthus DZ2 was grown to 25 Klett units (KU) in Casitone-yeast (CYE) liquid medium and spun down, and the pellet was washed two times with water. The pellet was resuspended to 1/1,000 of the initial culture volume. Fifty-microliter aliquots of cells were mixed with EZ-Tn5, according to the manufacturer, followed by electroporation at 25 mF, 400 Ω, 0.65 kV. Cells were recovered in 1 ml of CYE medium and incubated at 32°C for 4 h before plating aliquots into CYE Top agar onto CYE agar plates. Kanamycin was used for selection at a final concentration of 50 μg/ml. Incubation for 5 to 6 days at 32°C resulted in single colonies, which were transferred to new CYE-kanamycin agar plates using sterile toothpicks. Growth of kanamycin-resistant clones occurred within 2 days. Individual clones were picked onto new CYE agar plates containing kanamycin and finally onto CYE agar plates to maintain the clones for further use. To screen the transposon mutant library, individual M. xanthus clones were transferred using toothpicks into the middle of B. subtilis NCIB3610 prey spots, prepared as described below. To conduct the predation screen, we grew B. subtilis NCIB3610 to an optical density at 600 nm (OD600) of about 2.0 and washed and spotted cells onto CFL agar plates (predation/starvation medium) in 7-μl spots. A small colony of each individual M. xanthus transposon mutant was then taken from a master plate and transferred into the middle of an individual prey spot, thereby generating an inside-out predation assay. Each plate contained both a negative (loss-of-function [LOF]) control, as well as the M. xanthus parent for comparison. The LOF control used here is the mglAB mutant that was previously characterized for its loss of motility and inability to carry out predation (7). The predation plates were incubated at 32°C, and each individual prey spot was screened by microscopy each day for 5 days. Prey lysis was monitored over time up to 5 days (Fig. 1), allowing us to identify either gain-of-function (GOF) or LOF mutants.
FIG 1.
M. xanthus transposon screen outline. A library of 6,000 M. xanthus transposon mutants was generated using the EZ-Tn5 transposable element. (I) To select for single M. xanthus colonies, cells were plated into Top agar on solid CYE agar plates containing kanamycin for selection and incubated for 5 to 6 days. (II) Individual colonies were transferred onto CYE kanamycin agar plates. (III) B. subtilis NCIB3610 prey cells were spotted onto CFL agar plates (predation/starvation medium), and the M. xanthus transposon mutants were transferred into the middle, showing the inside-out assay. Screening for predation phenotypes occurred for up to 5 days. GOF and LOF strains were selected by comparing them to M. xanthus wild type (WT) and an LOF control strain (mglAB mutant), as seen in the lower part of the figure.
Rescue cloning of predation mutants.
To identify the genomic sequence into which the EZ-Tn5 transposon inserted, we isolated chromosomal DNA of the mutant strains using the DNeasy kit from Qiagen. Subsequently, we digested the chromosomal DNA using the restriction enzyme NspI, stopped the reaction by Drop dialysis, and religated the chromosomal DNA. The transposon contains the R6Kγori for rescue cloning in the pir E. coli strain and a kanamycin resistance cassette for selection (33). Kanamycin-resistant transformants were subject to plasmid isolation and sequencing performed at the Nevada Genomics Center at the University of Nevada, Reno. Obtained sequences were checked against the M. xanthus genome using the NCIB/BLAST tool. Further information about the insertion sites into the M. xanthus genome was obtained using the MiST2 database (34).
Predation assays.
M. xanthus strains were grown in CYE medium to mid-log phase. Cells were harvested and washed twice with MMC buffer (20 mM morpholinepropanesulfonic acid [MOPS] [pH 7.6], 4.0 mM MgSO4, 2 mM CaCl2). M. xanthus cells were resuspended in MMC buffer to a final concentration of 250 KU. Prey cells were grown overnight in LB medium to an OD600 of about 2. Cells were washed twice with H2O and resuspended in water to a final concentration of 1 × 1011/ml of water. Different qualitative and quantitative predation assays were performed. As seen in Fig. 2D, (wild-type) predator and prey cells were mixed in a ratio of 1:50 and spread plated onto CFL agar plates. After about 48 h of predation, megastructure formation of B. subtilis as well as the fruiting body of M. xanthus were evaluated. Another qualitative assay (inside-out assay, Fig. 2E) was performed to determine lysis of the prey (B. subtilis NCIB3610) by the predator (35). Seven microliters of prey cells was spotted onto the CFL plates. After the prey spot was dried, M. xanthus predatory cells (2 μl) were spotted into the middle of the prey spot. Predatory lysis is visible as clearing of the prey spot. Semiquantitative predation assays were performed to identify gain-of-function (GOF) and loss-of-function (LOF) mutants. Predator and prey cells were mixed in a ratio of 1:25 (Fig. 2F), 1:50 (Fig. 2G), and 1:100 (Fig. 2H). Seven microliters of predator-prey mixture was spotted onto CFL agar plates, and lysis of the prey spot was observed over time. Assay mixtures were incubated at 32°C, and pictures were taken after different times and magnifications to monitor the progression of predation. GOF can clearly be seen in Fig. 2 for both difA and difE in-frame deletion mutants in all different mixing ratios (Fig. 2F to H). LOF can clearly be seen for the hsfB, ABC transporter/permease, and myxoprincomide mutants at predator-to-prey mixing ratios of 1:25 (Fig. 2F), as prey cells that are darker remain visible compared to the wild type. LOF of the CRISPR II mutant is more visible at higher predator-to-prey cell ratios (Fig. 2G and H) and is indicated by darker prey spots and less clearing of the prey. Quantifications of prey survival and predator growth were performed as previously described (14). Briefly, prey and predator were mixed at a ratio of 50:1 and spread plated onto CFL agar plates. Additionally, both strains were plated individually on CFL agar plates. After 24 h, cells were harvested, and serial dilutions were plated onto LB agar plates to obtain B. subtilis NCIB3610 or plated into CYE Top agar onto CYE agar plates (containing 50 μg/ml kanamycin) to obtain CFU for M. xanthus strains. Assays were performed in triplicate, and the average CFU and standard deviation were calculated. Predator strains used in this study had a kanamycin resistance cassette inserted into the attB8 phage attachment site for selection.
FIG 2.
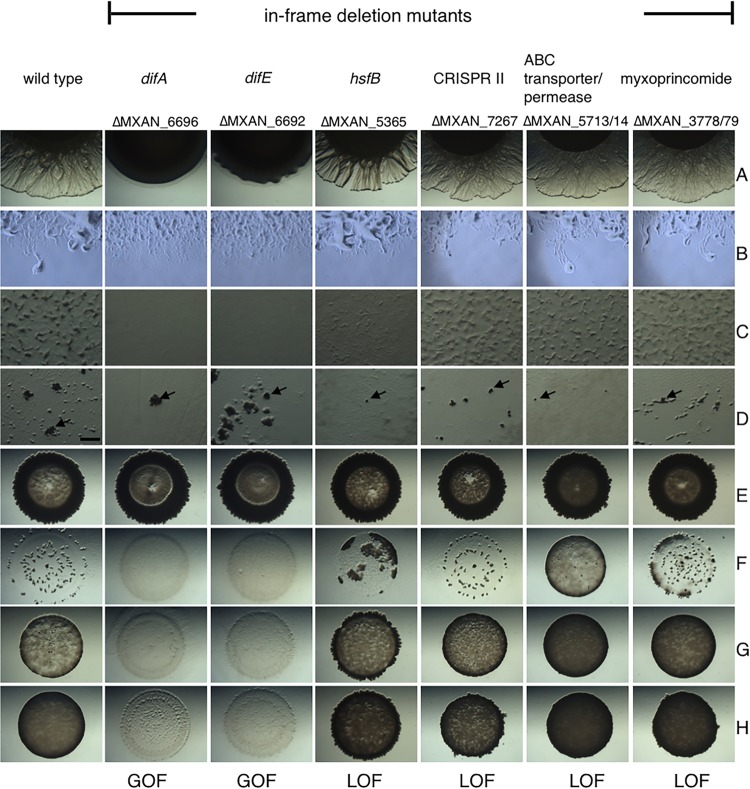
Phenotypic analysis of in-frame deletion mutants. M. xanthus transposon mutants were grown to mid-log phase, washed, and resuspended to a cell concentration of 250 KU. (A to C) Phenotypic analysis was performed to assess T4P-dependent motility on 0.5% CYE agar (A), gliding motility on 1.5% CYE agar plates (B), and fruiting body formation on CFL starvation/predation medium (C). (D and E) To investigate predatory ability, 2 μl of M. xanthus cells was spotted into the middle of B. subtilis NCIB3610 (E), or both predator and prey cells were mixed in a ratio of 1:50 and spread on plates to study megastructure formation on CFL agar plates (D). (F to H) Semiquantitative predation assays. Predator and prey were mixed in a ratio of 1:25 (F), 1:50 (G), or 1:100 (H). Pictures were taken after 24 h (B) and 48 h (A and C to H) at a magnification of ×10 (A, B, and E to H), ×30 (C and D), or ×100 (B). (D) Bar, 0.5 mm; arrows indicate individual megastructures.
Fruiting body formation and motility assays.
Fruiting body formation of M. xanthus was performed on CFL agar plates, as described before (36). Briefly, M. xanthus strains were grown to about 100 to 150 KU at 32°C in CYE medium, washed in MMC buffer, and 10 μl of a 500-KU cell suspension was spotted onto CFL agar plates and monitored for 3 days. Fruiting body formation is indicated by the formation of aggregates (as seen in Fig. 2C, wild type). To assay T4P-dependent motility as well as gliding motility, M. xanthus strains were grown as described above, and 10 μl of a 250-KU cell suspension was spotted onto 0.5% or 1.5% CYE agar plates. T4P-dependent motility is visible on soft agar plates (0.5% CYE) after about 48 h, indicated by cells swarms away from the central spot of cells (as seen in Fig. 2A, wild type is swarming, while the difA mutant is not). Focal adhesion-mediated motility is observed on hard agar surfaces as individual cells at the corner of the central spot (1.5% CYE) after 24 h (Fig. 2B, wild type).
Microscopy.
Phenotypic assays were monitored by microscopy using a Nikon SMZ10000 dissecting microscope. Images were taken using QImaging camera and QCapture software after various times.
RESULTS
Identification of M. xanthus predation mutants following a transposon mutagenesis screen.
A transposon (EZ-Tn5) mutant library of M. xanthus was constructed to screen for mutants that displayed either a gain-of-function (GOF) or a loss-of-function (LOF) phenotype during predation of B. subtilis NCIB3610 as the prey source (Fig. 1). B. subtilis NCIB3610 was chosen because it displays significant resistance to predation relative to either E. coli or laboratory strains of B. subtilis under the conditions of our assays (14).
We screened a total of approximately 6,000 M. xanthus mutants and selected 253 mutants which displayed either enhanced (1.1% GOF) or reduced (2.6% LOF) predation (Fig. 1). From those, we selected 53 mutants that displayed the most obvious predation phenotypes, 17 GOF and 36 LOF mutants, for more thorough analyses. To identify the locus into which the transposon had inserted, chromosomal DNA was isolated, digested with NspI, ligated, and transformed into E. coli pir cells capable of propagating the conditional R6Kγ origin of replication. Clones were selected on LB agar containing kanamycin, and plasmid DNA was isolated and sequenced. The results of the 53 insertions are identified in Table 2.
TABLE 2.
GOF and LOF transposon mutants
| MXAN no. | Gene | Gene product descriptiona | GOF/LOF |
|---|---|---|---|
| 0319 | 16S rRNA | 3× GOF/1× LOF | |
| 0322 | 23S rRNA | 2× GOF/1× LOF | |
| 5365 | hsfB | Response regulator sensor histidine kinase HsfB | 1× GOF |
| 6673 | Hypothetical protein, encoded in operon with GrpE; DnaK chaperone | 1× GOF | |
| 6696 | difA | Methyl-accepting protein DifA | 1× GOF |
| 0080 | Hypothetical protein, DUF1446 | 1× GOF | |
| 1112 | Hypothetical protein | 1× GOF | |
| 7234 | Hypothetical protein, DUF946 | 1× GOF | |
| 0517 | Acetyltransferase, encoded in operon with TonB-dependent receptor | 1× GOF | |
| 6679 | Hypothetical protein | 1× GOF | |
| 7509 | Inner membrane protein, 60 kDa | 1× GOF | |
| 6692 | difE | Histidine kinase DifE | 1× LOF |
| 7195 | Hypothetical protein | ||
| 7267 | Hypothetical protein, encoded in operon with CRISPR II | 1× LOF | |
| 7325 | Acetyltransferase | 1× LOF | |
| 3325 | rpsD | 30S ribosomal protein S4 | 1× LOF |
| 3779 | PKS/NRPS (myxoprincomide) | 1× LOF | |
| 0091 | HAE1 family efflux transport MFP subunit | 1× LOF | |
| 6127 | IS4 family transposase | 1× LOF | |
| 0455 | |||
| 5791 | Response regulator/GGDEF-containing protein | 1× LOF | |
| 0463 | pepP | Xaa-Pro aminopeptidase | 1× LOF |
| 3678 | Polyprenyl synthetase | 1× LOF | |
| 5220 | HhH-GPD domain-containing protein | 1× LOF | |
| 3760 | acdA | Acyl-CoA dehydrogenase | 1× LOF |
| 0180 | fis family transcriptional regulator | 1× LOF | |
| 1093 | DNA-binding response regulator | 1× LOF | |
| 5713 | ABC transporter/permease | 11× LOF | |
| 0260 | Cation transporter/universal stress protein | 1× LOF | |
| 7254 | Hypothetical protein with NAD-binding domain | 1× LOF | |
| 7143 | fis family transcriptional regulator, encoded in operon with histidine kinase | 1× LOF | |
| 3797 | Acyl-CoA dehydrogenase | 1× LOF | |
| 7466 | Hypothetical protein | 1× LOF | |
| 1449 | TonB protein | 1× LOF | |
| 4659 | Putative Flp pilus assembly protein | 1× LOF | |
| 6785 | Hypothetical protein with NaH exchange transporter domain | 1× LOF | |
| 0925 | 4-Oxalocrotonate decarboxylase | 1× LOF | |
| 7492 | Hypothetical protein | 1× LOF | |
| 6908 | pgi | Glucose-6-phosphate isomerase | 1× LOF |
| 6867 | Lipoprotein, encoded in operon with two sensor box histidine kinases | 1× LOF |
MFP, membrane fusion protein; HhH, helix-hairpin-helix; acyl-CoA, acyl coenzyme A.
All GOF transposon mutants displayed increased lysis of B. subtilis NCIB3610 prey in the inside-out assay (see Fig. S1E in the supplemental material). However, no obvious differences were visible for predation-induced megastructure formation by B. subtilis (see Fig. S1D). All LOF transposon mutants displayed reduced lysis of B. subtilis NCIB3610 (see Fig. S2E in the supplemental material) while still retaining the ability to induce B. subtilis megastructure formation (see Fig. S2D). However, it is worth noting that the LOF mutant-induced megastructures appeared to be smaller (see Fig. S2D).
Additionally, we made in-frame deletion mutants in a subset of genes identified during the transposon screen to verify that our findings were not an artifact of the transposon itself (Fig. 2). Below, we describe several transposon and in-frame deletion mutants that were expected targets (e.g., motility systems) and several that represent new loci not previously known to affect predation (e.g., specialized metabolites).
Specialized metabolite myxoprincomide is required for effective predation.
One prominent LOF transposon mutant had an insertion in locus MXAN_3779 encoding a mixed NRPS/PKS biosynthetic module, previously identified for its production of the specialized metabolite myxoprincomide (29). Myxoprincomide was discovered as a linear peptide with some unusual residues but had no known biological function (29). The transposon inserted near the 5′ end of the 42.8-kb gene. It is not yet known whether the resulting mutant produces a modified version of the polyketide.
The myxoprincomide transposon mutant displayed no defects in motility or fruiting body formation compared to the parent (see Fig. S2A to C in the supplemental material). However, B. subtilis NCIB3610 cells showed increased resistance to the myxoprincomide mutant, as indicated by less lysis of the prey spot (see Fig. S2E). Additionally, B. subtilis megastructure formation was affected such that the structures are smaller than those induced by wild-type M. xanthus (see Fig. S2D). Importantly, an in-frame deletion of the myxoprincomide synthesis cluster MXAN_3778-MXAN_3779 verified that the phenotypes observed in Fig. 2A to E were due to the loss of the intact product from the NRPS/PKS module. In addition, semiquantitative predation assays (Fig. 2F to H) clearly show reduced ability for the myxoprincomide mutant to lyse prey relative to the parent. A quantitative assay also showed that about 33% fewer prey cells were consumed by the myxoprincomide mutant than by the parent (Fig. 3A). Similarly, we observed reduced CFU for the myxoprincomide mutant in the presence of B. subtilis prey. Only about 62% survived compared to about 100% of the M. xanthus parent (Fig. 3B).
FIG 3.
Quantification of prey survival and predator growth. Predator M. xanthus wild-type or myxoprincomide mutant [Δ(MXAN_3778-MXAN_3779)] cells were mixed with B. subtilis NCIB3610 prey cells in a ratio of 1:50, and the mixture was spread plated onto CFL agar. After incubation for 24 h at 32°C, cells were harvested and serial dilutions were plated onto selective medium. Predator and prey alone were used as controls. CFU were used to calculate the percentage of prey survival and predator growth relative to the controls. (A) The myxoprincomide mutant shows a reduced ability to consume B. subtilis NCIB3610. (B) The M. xanthus wild type shows no significant growth, whereas the loss of the specialized metabolite myxoprincomide results in reduced CFU. The data represent the average of the results from 3 individual eating experiments, and the error bars show the standard deviation.
To assess expression of the myxoprincomide NRPS/PKS gene cluster, a lacZ fusion was generated with the putative promoter for MXAN_3778. The results indicated that the myxoprincomide NRPS/PKS gene cluster was constitutively expressed under the conditions of our assay, regardless of the presence of prey (see Fig. S3 in the supplemental material). Together, these data indicate that the specialized metabolite myxoprincomide is part of the predatory machinery of M. xanthus.
Hsf two-component system affects predation.
A prominent GOF phenotype was identified that resulted from a transposon insertion into the gene encoding the hybrid response regulator/histidine kinase HsfB. The transposon inserted downstream of the region encoding the response regulator domain within HsfB (see Fig. S4 in the supplemental material). For this mutant, even though predation was enhanced, we observed a reduction in both T4P-dependent motility and developmental fruiting body formation. In contrast, there was no obvious change in gliding motility, as indicated by the presence of individual cells at the colony edge (see Fig. S1 in the supplemental material). Interestingly, an in-frame deletion of hsfB displayed an LOF phenotype in the semiquantitative assays (Fig. 2F to H). However, this mutant also showed reduced motility as well as reduced fruiting body formation. Thus, the transposon insertion may have had an effect on downstream genes or resulted in dysregulation of the HsfBA two-component system.
The Hsf two-component system is encoded by an operon spanning MXAN_5365 to MXAN_5362 (see Fig. S4 in the supplemental material) and includes genes encoding HsfB, HsfA (NtrC-like response regulator), an NAD kinase, and a protein with a conserved domain of unknown function (DUF218). In previous work, we demonstrated specific phosphotransfer between HsfB and HsfA (37). In other previous work, HsfA was shown to affect the production of specialized metabolites, including DKxanthene and myxovirescin. HsfA was found to bind the putative promoter region of the gene clusters responsible for the production of each of these two molecules (38). Thus, the HsfBA system appears to be a critical regulator for the production of specialized metabolites by M. xanthus and for the regulation of predation.
Dif chemosensory system is required for predation.
The transposon insertion screen led to the identification of three mutations in the dif chemosensory system, two LOF mutants and one GOF mutant. The dif cluster encodes a chemosensory system with homologs to a prototypical chemotaxis signaling system and includes a methyl-accepting protein (MCP), DifA; a coupling protein (CheW), DifC; the response regulator DifD; the histidine kinase DifE; and a phosphatase, DifG (see Fig. S4 in the supplemental material) (39, 40). The Dif system has been well characterized and shown to regulate EPS production, T4P-based motility, and fruiting body formation in M. xanthus.
The GOF mutant resulted from an insertion into difA at bp 970, corresponding to amino acid 321 within the MCP signaling domain of the resulting protein (see Fig. S4 in the supplemental material). As an MCP, the DifA mutation could result in a truncated receptor that retains its signaling domain. This difA insertion mutant displays reduced T4P-dependent motility and is defective in fruiting body formation, while gliding motility was not affected (see Fig. S1A to C in the supplemental material). Increased lysis of the prey B. subtilis NCIB3610 was observed, while megastructure formation was not dramatically changed compared to the wild type (see Fig. S1D and E). A GOF phenotype was also obtained with an in-frame deletion mutation in difA (Fig. 2F to H). The semiquantitative predation assay clearly shows increased predation even at high prey concentrations which might otherwise enhance resistance (Fig. 2F). Additionally, the ΔdifA mutant displays no T4P-dependent motility and forms no fruiting bodies (Fig. 2A and C), as described previously.
The insertion sites for the transposons for the two LOF mutants occurred near the 5′ end of the gene encoding DifE, a CheA homolog and the central histidine kinase of the Dif chemosensory system. The transposon insertions occurred either upstream or downstream of the Hpt domain-encoding region within difE, thereby likely disrupting the production of an intact kinase (see Fig. S4 in the supplemental material). The corresponding mutants displayed reduced T4P-dependent motility and loss of fruiting body formation. However, in these cases, lysis of B. subtilis NCIB3610 was also reduced, as was induction of megastructure formation (see Fig. S2 in the supplemental material). A ΔdifE mutant, however, displayed the opposite phenotype and was a GOF mutant (Fig. 2F and G), suggesting complex regulation of predation by the Dif chemosensory system. All dif mutants described here, whether transposon insertions or in-frame deletions, displayed defects in EPS production (see Fig. S5 in the supplemental material).
An ABC transporter/permease is required for predation.
One of the most frequent transposon insertion sites (11 independent isolates) occurred in MXAN_5713, encoding an ABC transporter/permease predicted to be involved in branched-chain amino acid (BCAA) transport. BCAAs are essential to M. xanthus and function as precursors for fatty acid synthesis as well as specialized metabolite production (31, 41, 42). All 11 transposon insertion mutations in MXAN_5713 displayed an LOF phenotype (see Fig. S2 in the supplemental material). MXAN_5713 appears to be part of an operon, which includes a second ABC transporter/permease gene (MXAN_5714) and two genes located upstream encoding a putative lipoprotein (MXAN_5711) and an ABC transporter/ATP-binding protein (MXAN_5712). According to the MiST2.2 database, the M. xanthus genome contains a total of four ABC transporter/permease genes that are located in two operons, the MXAN_5711 to MXAN_5714 operon described above and the MXAN_6667 to MXAN_6659 operon (34). No transposon insertions were identified in the MXAN_6667 to MXAN_6659 cluster. Overall, these data indicate that BCAA import, possibly impacting fatty acid and specialized metabolite production, is intertwined with M. xanthus physiology to affect the efficiency of predation. An in-frame deletion mutant of the ABC transporter/permease genes MXAN_5713 and MXAN_5714 verified that loss of the transporter resulted in the observed phenotypes (Fig. 2F to H). In the semiqualitative predation assays, this in-frame deletion mutant displays the strongest LOF phenotype relative to all mutants presented in this study.
CRISPR locus affects predation.
Another LOF mutant phenotype resulted from transposon insertion into the CRISPR II locus (see Fig. S2 in the supplemental material). CRISPR/CRISPR-associated protein (Cas) systems protect prokaryotes from foreign genetic elements, like plasmids and phages, and act as prokaryotic immune systems (43). The M. xanthus CRISPR II locus contains the prototypical CRISPR/Cas genes (MXAN_7257 to MXAN_7266) of prokaryotic immunity systems (see Fig. S6 in the supplemental material). This locus also includes genes previously annotated as development genes in M. xanthus, devR (a cas7 homolog) and devS (a cas5 homolog), both of which were shown to affect sporulation levels (44, 45). The transposon mutant that we identified was inserted into MXAN_7267 (encoding a hypothetical protein) at the 5′ end of the CRISPR locus, just upstream of the newly described devI (MXAN_7266). DevI is a small protein that inhibits sporulation when overexpressed (see Fig. S6) (45). An in-frame deletion of the CRISPR II locus [Δ(MXAN_7258-MXAN_7267)] resulted in an LOF phenotype and thus verified the involvement of this locus in B. subtilis NCIB3610 predation, as seen in the semiquantitative assay (Fig. 2G and H).
DISCUSSION
Interspecies interactions can produce a variety of outcomes ranging from cooperation to competition or commensalism (1). Predation by M. xanthus influences the environment, as it provides nutrients for itself and other bacteria but also changes the composition and diversity of the soil community (8, 9, 14). M. xanthus utilizes a broad range of prey found in soil environments worldwide and is also known to produce a large suite of unique specialized metabolites thought to facilitate predation (5, 28, 46). In soil, B. subtilis would likely be a source of prey for M. xanthus. Our previous work demonstrated that the ancestral strain of B. subtilis, NCIB3610, is transiently resistant to M. xanthus predation due to its production of the specialized metabolite, bacillaene (14). Following long-term interactions, B. subtilis goes on to produce megastructures that contain B. subtilis spores embedded within a matrix, allowing cells to escape predation altogether (7). In contrast, many factors utilized by M. xanthus during predation are less well characterized. Thus, we performed a transposon mutagenesis of the predator and screened 6,000 M. xanthus mutants for their ability to consume B. subtilis NCIB3610. We identified both GOF and LOF mutants based on their capacity to consume B. subtilis cells relative to the rate of consumption by the parent M. xanthus cells.
The predicted functions of genes affecting M. xanthus predation capacity were predominantly signal transduction, motility, transport, and the production of specialized metabolites. Loss of production of the specialized metabolite myxoprincomide (either by transposon insertion or in-frame deletion) greatly reduced predation by M. xanthus. A quantitative experiment revealed that the myxoprincomide mutant survives or grows at a lower rate in the presence of prey, as only about 62% of cells were recovered after a 24-h competition experiment relative to the parent. This result may reflect an inability to lyse the NCIB3610 prey cells or be due to a killing effect from B. subtilis itself. The possibility of a killing effect should be considered because myxoprincomide was not required for the predation of sensitive strains, including E. coli or the pksL mutant of B. subtilis, which is incapable of producing bacillaene (see Fig. S7 in the supplemental material). Taken together, it is clear that specialized metabolites have an important function in interspecies interactions between M. xanthus and B. subtilis. Furthermore, the observation that myxoprincomide NRPS/PKS gene expression is constitutive strongly suggests that the production of this specialized metabolite is critical for the survival of M. xanthus.
Specialized metabolites or natural products that can act as antibiotics are known to have a variety of different functions. They can act as defensive molecules shaping microbial communities by killing or by inhibiting growth (2, 19). However, many recent publications show that the role of specialized metabolites may be more complex, as they can induce biofilm formation and act as signaling molecules (47, 48). Therefore, it was not surprising that we identified a transposon insertion mutant in the M. xanthus hsf gene cluster known to regulate specialized metabolite production (38). HsfB and HsfA form a classical two-component system (37); however, it is not known what activates this pathway or if HsfB is capable of cross talk to another response regulator. This result is of great interest, as M. xanthus is known for its large repertoire of two-component systems regulating its complex lifestyle (36–38). It is also important to point out that the hsf cluster encodes a putative GGDEF-containing protein, suggesting that local concentrations of the second messenger, cyclic di-GMP (c-di-GMP), may affect predation. Because c-di-GMP has been implicated in many processes for M. xanthus (49–51), we speculate that predation, specialized metabolite production, two-component signaling, and c-di-GMP sensing are interconnected processes. Additional experiments to investigate the metabolite profile of the hsf mutant strain during predation are under way.
M. xanthus is well known for its capacity to develop and produce spore-filled fruiting bodies. This complex process requires the production of exopolysaccharides (EPS) and coordination of two motility systems, T4P-dependent motility and gliding motility (10). These complex processes are regulated by various two-component systems (TCS) and chemosensory systems, including Frz and Dif (11, 52, 53). While we did not identify insertions in the Frz system, we identified three transposon insertion mutants in the dif cluster. Two LOF mutations mapped to the DifE (CheA) homolog, and one GOF mutant mapped to the C-terminal end of the DifA (MCP) homolog. The site of insertion in DifA is consistent with the current understanding of MCP function, suggesting that a truncated DifA receptor may produce a constitutive output; a similar mutation was isolated in M. xanthus decades ago in FrzCD (frzCDc), which produced a hyperreversing phenotype in contrast to the corresponding deletion (54, 55). Interestingly, both ΔdifA and ΔdifE mutant cells display a GOF phenotype, despite the fact that both mutants lack the ability to produce EPS. We conclude that the transposon insertion in difA led to dysregulation of the signal transduction pathway and that EPS is not required per se for predation. Additional evidence for motility as a key predation factor was the finding that disruption of a putative Flp pilus assembly protein (MXAN_4659) (Table 2) resulted in an LOF phenotype. Our results also indicate that several other putative signaling proteins are required for predation by M. xanthus, including an operon comprising a lipoprotein and two sensor histidine kinases (MXAN_6866 and MXAN_6865) (Table 2).
One critical finding was the role for the ABC transporter/permease-encoding gene cluster MXAN_5711 to MXAN_5714. The ABC transporter is most likely involved in the import of branched-chain amino acids, like leucine, isoleucine, and valine. Leucine is essential for M. xanthus growth and becomes available following the lysis of prey (31). We speculate that the transporter mutant is unable to utilize BCAAs, preventing growth, and it therefore displays a deficiency in predation. Moreover, it has been shown that BCAAs, such as leucine, serve as the precursor for fatty acid biosynthesis and also as starter molecules for polyketide synthases (41, 42). Reduced levels of available BCAAs due to disruption of the ABC transporter could result in an altered specialized metabolite or fatty acid profile and generate deficits in growth on otherwise-suitable prey. Metabolite profiling of this mutant is also under way.
Other M. xanthus functions affecting predation were identified and include a CRISPR system. The M. xanthus genome encodes three CRISPR systems, each composed of a typical arrangement of CRIPSR/Cas genes with repeat/spacer regions (44, 45, 56). The CRIPSR/Cas systems are prokaryotic immune systems that confer resistance to foreign genetic elements, such as plasmids and phages, and provide a form of acquired immunity. Because we identified an LOF mutant that mapped to one particular locus, CRIPSR II (spanning MXAN_7267 to MXAN_7258), it is possible that M. xanthus utilizes CRISPR systems during predation, since the predator is exposed to foreign DNA during prey lysis. The CRISPR II locus has been studied previously for its regulation of the development in M. xanthus and is known to contain devTRS and devI, encoding a small protein that inhibits sporulation (45, 57–59). In our hands, the LOF mutant displayed defects in predation but did not display altered motility or sporulation. It has been reported that CRISPR systems can regulate alternative functions, such as biofilm formation and swarming in Pseudomonas aeruginosa, EPS production in M. xanthus, and lipoprotein production in Francisella novicida (56, 60, 61).
In summary, our screen revealed many new genes regulating predation by M. xanthus. We also identified several functions that were expected or predicted to regulate predation. Results from the transposon insertions were independently confirmed by constructing in-frame deletion mutants. Overall, M. xanthus predation is a multifactorial process, requiring several signaling pathways, regulation of motility, branched-chain amino acid transporters, and production of specialized metabolites (Fig. 4). Myxoprincomide appears to be a key determinant to enhance the capacity of M. xanthus to prey upon B. subtilis. Further analysis is ongoing to investigate the complexity of gene regulation and specialized metabolite production during predation.
FIG 4.
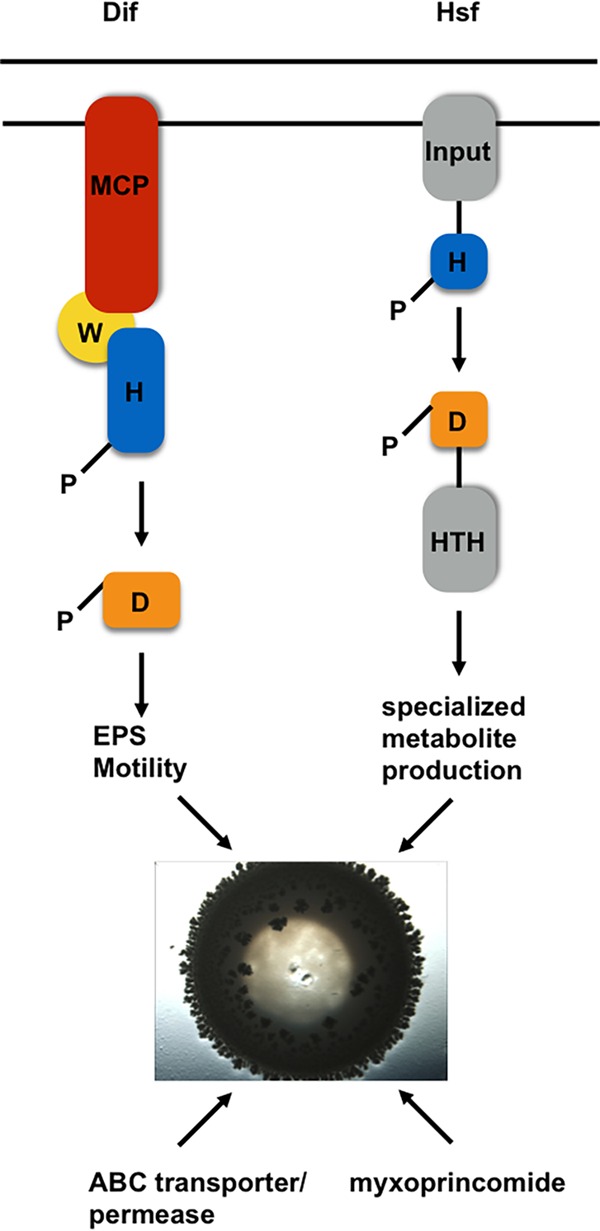
Factors and pathways involved M. xanthus predation. Our transposon mutagenesis screen identified 2 two-component systems, the Dif chemosensory system (left) and the Hsf system (right), that are involved in regulating predation. Each system utilizes a histidine kinase (blue) that becomes autophosphorylated upon activation to regulate outputs via response regulators. Upon activation, phosphoryl groups are transferred to response regulators (orange) to generate the physiological responses, such as EPS production, motility, and specialized metabolite production. Additionally, we found an ABC transporter/permease and the specialized metabolite myxoprincomide to be involved in the predation of B. subtilis NCIB3610. HTH, helix-turn-helix.
Supplementary Material
ACKNOWLEDGMENTS
Support for this work was provided by the University of Iowa, Department of Microbiology, and the Carver College of Medicine FUTUREs in Biomedicine program. Support for this work was provided in part by a grant from the NSF (MCB-1244021) to J.R.K.
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
We thank Cindy Darnell and Caroline Linke for support during this project, as well as Alfa Herrera for assistance in bioinformatic analysis of the M. xanthus CRISPR loci. DNA sequencing was performed by the Nevada Genomics Center (University of Nevada, Reno).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00575-16.
REFERENCES
- 1.Grosskopf T, Soyer OS. 2014. Synthetic microbial communities. Curr Opin Microbiol 18:72–77. doi: 10.1016/j.mib.2014.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Traxler MF, Kolter R. 2015. Natural products in soil microbe interactions and evolution. Nat Prod Rep 32:956–970. doi: 10.1039/C5NP00013K. [DOI] [PubMed] [Google Scholar]
- 3.Sockett RE. 2009. Predatory lifestyle of Bdellovibrio bacteriovorus. Annu Rev Microbiol 63:523–539. doi: 10.1146/annurev.micro.091208.073346. [DOI] [PubMed] [Google Scholar]
- 4.Majdi N, Traunspurger W. 2015. Free-living nematodes in the freshwater food web: a review. J Nematol 47:28–44. [PMC free article] [PubMed] [Google Scholar]
- 5.Berleman JE, Kirby JR. 2009. Deciphering the hunting strategy of a bacterial wolfpack. FEMS Microbiol Rev 33:942–957. doi: 10.1111/j.1574-6976.2009.00185.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pérez J, Moraleda-Munoz A, Marcos-Torres FJ, Munoz-Dorado J. 2015. Bacterial predation: 75 years and counting! Environ Microbiol 18:766–779. doi: 10.1111/1462-2920.13171. [DOI] [PubMed] [Google Scholar]
- 7.Müller S, Strack SN, Ryan SE, Kearns DB, Kirby JR. 2015. Predation by Myxococcus xanthus induces Bacillus subtilis to form spore-filled megastructures. Appl Environ Microbiol 81:203–210. doi: 10.1128/AEM.02448-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morgan AD, MacLean RC, Hillesland KL, Velicer GJ. 2010. Comparative analysis of Myxococcus predation on soil bacteria. Appl Environ Microbiol 76:6920–6927. doi: 10.1128/AEM.00414-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xiao Y, Wei X, Ebright R, Wall D. 2011. Antibiotic production by myxobacteria plays a role in predation. J Bacteriol 193:4626–4633. doi: 10.1128/JB.05052-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cao P, Dey A, Vassallo CN, Wall D. 2015. How myxobacteria cooperate. J Mol Biol 427:3709–3721. doi: 10.1016/j.jmb.2015.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mauriello EM, Mouhamar F, Nan B, Ducret A, Dai D, Zusman DR, Mignot T. 2010. Bacterial motility complexes require the actin-like protein, MreB and the Ras homologue, MglA. EMBO J 29:315–326. doi: 10.1038/emboj.2009.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Berleman JE, Scott J, Chumley T, Kirby JR. 2008. Predataxis behavior in Myxococcus xanthus. Proc Natl Acad Sci U S A 105:17127–17132. doi: 10.1073/pnas.0804387105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zeigler DR, Pragai Z, Rodriguez S, Chevreux B, Muffler A, Albert T, Bai R, Wyss M, Perkins JB. 2008. The origins of 168, W23, and other Bacillus subtilis legacy strains. J Bacteriol 190:6983–6995. doi: 10.1128/JB.00722-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Müller S, Strack SN, Hoefler BC, Straight PD, Kearns DB, Kirby JR. 2014. Bacillaene and sporulation protect Bacillus subtilis from predation by Myxococcus xanthus. Appl Environ Microbiol 80:5603–5610. doi: 10.1128/AEM.01621-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.DePas WH, Syed AK, Sifuentes M, Lee JS, Warshaw D, Saggar V, Csankovszki G, Boles BR, Chapman MR. 2014. Biofilm formation protects Escherichia coli against killing by Caenorhabditis elegans and Myxococcus xanthus. Appl Environ Microbiol 80:7079–7087. doi: 10.1128/AEM.02464-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weitere M, Bergfeld T, Rice SA, Matz C, Kjelleberg S. 2005. Grazing resistance of Pseudomonas aeruginosa biofilms depends on type of protective mechanism, developmental stage and protozoan feeding mode. Environ Microbiol 7:1593–1601. doi: 10.1111/j.1462-2920.2005.00851.x. [DOI] [PubMed] [Google Scholar]
- 17.Matz C, McDougald D, Moreno AM, Yung PY, Yildiz FH, Kjelleberg S. 2005. Biofilm formation and phenotypic variation enhance predation-driven persistence of Vibrio cholerae. Proc Natl Acad Sci U S A 102:16819–16824. doi: 10.1073/pnas.0505350102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dahl JL, Ulrich CH, Kroft TL. 2011. Role of phase variation in the resistance of Myxococcus xanthus fruiting bodies to Caenorhabditis elegans predation. J Bacteriol 193:5081–5089. doi: 10.1128/JB.05383-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hibbing ME, Fuqua C, Parsek MR, Peterson SB. 2010. Bacterial competition: surviving and thriving in the microbial jungle. Nat Rev Microbiol 8:15–25. doi: 10.1038/nrmicro2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shank EA, Klepac-Ceraj V, Collado-Torres L, Powers GE, Losick R, Kolter R. 2011. Interspecies interactions that result in Bacillus subtilis forming biofilms are mediated mainly by members of its own genus. Proc Natl Acad Sci U S A 108:E1236–E1243. doi: 10.1073/pnas.1103630108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bleich R, Watrous JD, Dorrestein PC, Bowers AA, Shank EA. 2015. Thiopeptide antibiotics stimulate biofilm formation in Bacillus subtilis. Proc Natl Acad Sci U S A 112:3086–3091. doi: 10.1073/pnas.1414272112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rosenberg E, Vaks B, Zuckerberg A. 1973. Bactericidal action of an antibiotic produced by Myxococcus xanthus. Antimicrob Agents Chemother 4:507–513. doi: 10.1128/AAC.4.5.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gerth K, Irschik H, Reichenbach H, Trowitzsch W. 1982. The myxovirescins, a family of antibiotics from Myxococcus virescens (Myxobacterales). J Antibiot (Tokyo) 35:1454–1459. doi: 10.7164/antibiotics.35.1454. [DOI] [PubMed] [Google Scholar]
- 24.Evans AG, Davey HM, Cookson A, Currinn H, Cooke-Fox G, Stanczyk PJ, Whitworth DE. 2012. Predatory activity of Myxococcus xanthus outer-membrane vesicles and properties of their hydrolase cargo. Microbiology 158:2742–2752. doi: 10.1099/mic.0.060343-0. [DOI] [PubMed] [Google Scholar]
- 25.Berleman JE, Allen S, Danielewicz MA, Remis JP, Gorur A, Cunha J, Hadi MZ, Zusman DR, Northen TR, Witkowska HE, Auer M. 2014. The lethal cargo of Myxococcus xanthus outer membrane vesicles. Front Microbiol 5:474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Remis JP, Wei D, Gorur A, Zemla M, Haraga J, Allen S, Witkowska HE, Costerton JW, Berleman JE, Auer M. 2014. Bacterial social networks: structure and composition of Myxococcus xanthus outer membrane vesicle chains. Environ Microbiol 16:598–610. doi: 10.1111/1462-2920.12187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goldman BS, Nierman WC, Kaiser D, Slater SC, Durkin AS, Eisen JA, Ronning CM, Barbazuk WB, Blanchard M, Field C, Halling C, Hinkle G, Iartchuk O, Kim HS, Mackenzie C, Madupu R, Miller N, Shvartsbeyn A, Sullivan SA, Vaudin M, Wiegand R, Kaplan HB. 2006. Evolution of sensory complexity recorded in a myxobacterial genome. Proc Natl Acad Sci U S A 103:15200–15205. doi: 10.1073/pnas.0607335103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wenzel SC, Muller R. 2009. Myxobacteria–‘microbial factories' for the production of bioactive secondary metabolites. Mol Biosyst 5:567–574. doi: 10.1039/b901287g. [DOI] [PubMed] [Google Scholar]
- 29.Cortina NS, Krug D, Plaza A, Revermann O, Muller R. 2012. Myxoprincomide: a natural product from Myxococcus xanthus discovered by comprehensive analysis of the secondary metabolome. Angew Chem Int Ed Engl 51:811–816. doi: 10.1002/anie.201106305. [DOI] [PubMed] [Google Scholar]
- 30.Konovalova A, Petters T, Sogaard-Andersen L. 2010. Extracellular biology of Myxococcus xanthus. FEMS Microbiol Rev 34:89–106. doi: 10.1111/j.1574-6976.2009.00194.x. [DOI] [PubMed] [Google Scholar]
- 31.Bretscher AP, Kaiser D. 1978. Nutrition of Myxococcus xanthus, a fruiting myxobacterium. J Bacteriol 133:763–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu SS, Kaiser D. 1996. Markerless deletions of pil genes in Myxococcus xanthus generated by counterselection with the Bacillus subtilis sacB gene. J Bacteriol 178:5817–5821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Metcalf WW, Jiang W, Wanner BL. 1994. Use of the rep technique for allele replacement to construct new Escherichia coli hosts for maintenance of R6K gamma origin plasmids at different copy numbers. Gene 138:1–7. doi: 10.1016/0378-1119(94)90776-5. [DOI] [PubMed] [Google Scholar]
- 34.Ulrich LE, Zhulin IB. 2010. The MiST2 database: a comprehensive genomics resource on microbial signal transduction. Nucleic Acids Res 38:D401–D407. doi: 10.1093/nar/gkp940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Berleman JE, Kirby JR. 2007. Multicellular development in Myxococcus xanthus is stimulated by predator-prey interactions. J Bacteriol 189:5675–5682. doi: 10.1128/JB.00544-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kirby JR, Zusman DR. 2003. Chemosensory regulation of developmental gene expression in Myxococcus xanthus. Proc Natl Acad Sci U S A 100:2008–2013. doi: 10.1073/pnas.0330944100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Willett JW, Tiwari N, Muller S, Hummels KR, Houtman JC, Fuentes EJ, Kirby JR. 2013. Specificity residues determine binding affinity for two-component signal transduction systems. mBio 4(6):e00420–13. doi: 10.1128/mBio.00420-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Volz C, Kegler C, Muller R. 2012. Enhancer binding proteins act as hetero-oligomers and link secondary metabolite production to myxococcal development, motility, and predation. Chem Biol 19:1447–1459. doi: 10.1016/j.chembiol.2012.09.010. [DOI] [PubMed] [Google Scholar]
- 39.Yang Z, Geng Y, Xu D, Kaplan HB, Shi W. 1998. A new set of chemotaxis homologues is essential for Myxococcus xanthus social motility. Mol Microbiol 30:1123–1130. doi: 10.1046/j.1365-2958.1998.01160.x. [DOI] [PubMed] [Google Scholar]
- 40.Black WP, Schubot FD, Li Z, Yang Z. 2010. Phosphorylation and dephosphorylation among Dif chemosensory proteins essential for exopolysaccharide regulation in Myxococcus xanthus. J Bacteriol 192:4267–4274. doi: 10.1128/JB.00403-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bode HB, Ring MW, Schwar G, Altmeyer MO, Kegler C, Jose IR, Singer M, Muller R. 2009. Identification of additional players in the alternative biosynthesis pathway to isovaleryl-CoA in the myxobacterium Myxococcus xanthus. Chembiochem 10:128–140. doi: 10.1002/cbic.200800219. [DOI] [PubMed] [Google Scholar]
- 42.Mahmud T, Bode HB, Silakowski B, Kroppenstedt RM, Xu M, Nordhoff S, Hofle G, Muller R. 2002. A novel biosynthetic pathway providing precursors for fatty acid biosynthesis and secondary metabolite formation in myxobacteria. J Biol Chem 277:32768–32774. doi: 10.1074/jbc.M205222200. [DOI] [PubMed] [Google Scholar]
- 43.Marraffini LA. 2015. CRISPR-Cas immunity in prokaryotes. Nature 526:55–61. doi: 10.1038/nature15386. [DOI] [PubMed] [Google Scholar]
- 44.Viswanathan P, Murphy K, Julien B, Garza AG, Kroos L. 2007. Regulation of dev, an operon that includes genes essential for Myxococcus xanthus development and CRISPR-associated genes and repeats. J Bacteriol 189:3738–3750. doi: 10.1128/JB.00187-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rajagopalan R, Wielgoss S, Lippert G, Velicer GJ, Kroos L. 2015. devI is an evolutionarily young negative regulator of Myxococcus xanthus development. J Bacteriol 197:1249–1262. doi: 10.1128/JB.02542-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Findlay BL. 2016. The chemical ecology of predatory soil bacteria. ACS Chem Biol 11:1502–1510. doi: 10.1021/acschembio.6b00176. [DOI] [PubMed] [Google Scholar]
- 47.Fajardo A, Martinez JL. 2008. Antibiotics as signals that trigger specific bacterial responses. Curr Opin Microbiol 11:161–167. doi: 10.1016/j.mib.2008.02.006. [DOI] [PubMed] [Google Scholar]
- 48.Romero VL, Manzo RH, Alovero FL. 2010. Enhanced bacterial uptake and bactericidal properties of ofloxacin loaded on bioadhesive hydrogels against Pseudomonas aeruginosa. J Chemother 22:328–334. doi: 10.1179/joc.2010.22.5.328. [DOI] [PubMed] [Google Scholar]
- 49.Hengge R. 2009. Principles of c-di-GMP signalling in bacteria. Nat Rev Microbiol 7:263–273. doi: 10.1038/nrmicro2109. [DOI] [PubMed] [Google Scholar]
- 50.Römling U, Galperin MY, Gomelsky M. 2013. Cyclic di-GMP: the first 25 years of a universal bacterial second messenger. Microbiol Mol Biol Rev 77:1–52. doi: 10.1128/MMBR.00043-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Skotnicka D, Petters T, Heering J, Hoppert M, Kaever V, Sogaard-Andersen L. 2015. Cyclic di-GMP regulates type IV pilus-dependent motility in Myxococcus xanthus. J Bacteriol 198:77–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kirby JR. 2009. Chemotaxis-like regulatory systems: unique roles in diverse bacteria. Annu Rev Microbiol 63:45–59. doi: 10.1146/annurev.micro.091208.073221. [DOI] [PubMed] [Google Scholar]
- 53.Kaimer C, Berleman JE, Zusman DR. 2012. Chemosensory signaling controls motility and subcellular polarity in Myxococcus xanthus. Curr Opin Microbiol 15:751–757. doi: 10.1016/j.mib.2012.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Blackhart BD, Zusman DR. 1985. “Frizzy” genes of Myxococcus xanthus are involved in control of frequency of reversal of gliding motility. Proc Natl Acad Sci U S A 82:8767–8770. doi: 10.1073/pnas.82.24.8767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bustamante VH, Martinez-Flores I, Vlamakis HC, Zusman DR. 2004. Analysis of the Frz signal transduction system of Myxococcus xanthus shows the importance of the conserved C-terminal region of the cytoplasmic chemoreceptor FrzCD in sensing signals. Mol Microbiol 53:1501–1513. doi: 10.1111/j.1365-2958.2004.04221.x. [DOI] [PubMed] [Google Scholar]
- 56.Wallace RA, Black WP, Yang X, Yang Z. 2014. A CRISPR with roles in Myxococcus xanthus development and exopolysaccharide production. J Bacteriol 196:4036–4043. doi: 10.1128/JB.02035-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kroos L, Kuspa A, Kaiser D. 1986. A global analysis of developmentally regulated genes in Myxococcus xanthus. Dev Biol 117:252–266. doi: 10.1016/0012-1606(86)90368-4. [DOI] [PubMed] [Google Scholar]
- 58.Kroos L, Kuspa A, Kaiser D. 1990. Defects in fruiting body development caused by Tn5 lac insertions in Myxococcus xanthus. J Bacteriol 172:484–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Thöny-Meyer L, Kaiser D. 1993. devRS, an autoregulated and essential genetic locus for fruiting body development in Myxococcus xanthus. J Bacteriol 175:7450–7462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zegans ME, Wagner JC, Cady KC, Murphy DM, Hammond JH, O'Toole GA. 2009. Interaction between bacteriophage DMS3 and host CRISPR region inhibits group behaviors of Pseudomonas aeruginosa. J Bacteriol 191:210–219. doi: 10.1128/JB.00797-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sampson TR, Napier BA, Schroeder MR, Louwen R, Zhao J, Chin CY, Ratner HK, Llewellyn AC, Jones CL, Laroui H, Merlin D, Zhou P, Endtz HP, Weiss DS. 2014. A CRISPR-Cas system enhances envelope integrity mediating antibiotic resistance and inflammasome evasion. Proc Natl Acad Sci U S A 111:11163–11168. doi: 10.1073/pnas.1323025111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Müller S, Willett JW, Bahr SM, Darnell CL, Hummels KR, Dong CK, Vlamakis HC, Kirby JR. 2013. Draft genome sequence of Myxococcus xanthus wild-type strain DZ2, a model organism for predation and development. Genome Announc 1(3):e00217–13. doi: 10.1128/genomeA.00217-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.