Abstract
Hedgehog (Hh) signaling is fundamentally important for development and adult tissue homeostasis. It is well established that in vertebrates Sufu directly binds and inhibits Gli proteins, the downstream mediators of Hh signaling. However, it is unclear how the inhibitory function of Sufu towards Gli is regulated. Here we report that the Rusc family of proteins, the biological functions of which are poorly understood, form a heterotrimeric complex with Sufu and Gli. Upon Hh signaling, Rusc is displaced from this complex, followed by dissociation of Gli from Sufu. In mammalian fibroblast cells, knockdown of Rusc2 potentiates Hh signaling by accelerating signaling-induced dissociation of the Sufu-Gli protein complexes. In Xenopus embryos, knockdown of Rusc1 or overexpression of a dominant-negative Rusc enhances Hh signaling during eye development, leading to severe eye defects. Our study thus uncovers a novel regulatory mechanism controlling the response of cells to Hh signaling in vertebrates.
KEY WORDS: Hedgehog signaling, Sufu, Rusc, Gli, Xenopus, Mouse, Human
Highlighted article: Rusc proteins are identified as novel components of the vertebrate Hedgehog pathway that bind Sufu and prevent the signaling-induced dissociation of Sufu and Gli.
INTRODUCTION
The Hedgehog (Hh) signaling pathway is evolutionarily conserved and involved in a wide variety of processes during embryogenesis and adult tissue homeostasis (Jiang and Hui, 2008; Hui and Angers, 2011; Briscoe and Therond, 2013; Petrova and Joyner, 2014). One of the most important roles that Hh signaling plays during vertebrate early development is patterning of the neural tube. It is well established that a ventrally derived Sonic hedgehog (Shh) gradient counteracts dorsally derived Wnt and BMP gradients, determining fates of cells along the dorsoventral axis of the neural tube (Lupo et al., 2006; Briscoe, 2009; Briscoe and Small, 2015). In the anterior neural ectoderm, Hh signaling is essential for the formation of eye primordia. During eye development, the eye primordium is initially specified as a single morphogenetic field in the anterior neural plate. Shh, which is secreted by the prechordal plate, suppresses the expression of eye-specific genes in the midline and divides the eye field into two lateral eye primordia. Inhibition of Shh signaling impairs the eye separation process and induces cyclopia. By contrast, increased Shh signaling reduces the size of the eye (Amato et al., 2004). The proper response of cells to Shh is crucial for these developmental processes.
At the molecular level, the zinc-finger transcription factor Cubitus interruptus (Ci) and its vertebrate homologs, the Gli proteins, act at the downstream end of the pathway to mediate Hh signaling in Drosophila and vertebrates, respectively. In unstimulated cells, multiple inhibitory mechanisms act in coordination to keep Ci/Gli in check. The Hh family of proteins operates the pathway by relieving these inhibitory mechanisms, which ultimately converts Ci and Gli into transcriptional activators and induces expression of Hh target genes. Interfering with these Hh inhibitory mechanisms often has severe consequences, ranging from defective embryonic development to tumorigenesis (Huangfu and Anderson, 2006; Jia and Jiang, 2006; Jiang and Hui, 2008; Hui and Angers, 2011; Briscoe and Therond, 2013; Petrova and Joyner, 2014).
In vertebrates, one of the major Hh inhibitory mechanisms is mediated by suppressor of fused (Sufu). Sufu deficiency leads to constitutive pathway activation, resulting in severe patterning defects during development (Cooper et al., 2005; Svard et al., 2006; Min et al., 2011). Mouse embryos homozygous for the Sufu null allele die at ∼E9.5 with severely ventralized neural tubes that remain open in the anterior region (Svard et al., 2006). Knockdown of Sufu in Xenopus embryos also increases the expression of Hh target genes. As expected, Sufu-depleted Xenopus embryos develop severely reduced eyes (Min et al., 2011). In humans, inherited and sporadic mutations in SUFU have been identified in a wide variety of cancers, including medulloblastoma (Taylor et al., 2002; Brugieres et al., 2010), meningioma (Aavikko et al., 2012) and basal cell carcinoma (Pastorino et al., 2009; Kijima et al., 2012; Schulman et al., 2015). Interestingly, in contrast to Sufu in other vertebrate species, zebrafish Sufu is a weak Hh inhibitor, and knockdown of Sufu causes only a marginal increase in Hh signaling during zebrafish embryonic development (Wolff et al., 2003).
At the molecular level, Sufu directly binds Gli proteins when the Hh pathway is quiescent (Ding et al., 1999; Kogerman et al., 1999; Pearse et al., 1999; Stone et al., 1999; Zhang et al., 2013; Han et al., 2015). Sufu can inhibit Gli-dependent transcription through sequestering Gli proteins in the cytoplasm (Ding et al., 1999; Kogerman et al., 1999; Murone et al., 2000; Han et al., 2015). In the nucleus, Sufu recruits the NuRD repressor complex member p66β (Gatad2b) to the promoters of Hh target genes and suppresses Gli-dependent transcription (Lin et al., 2014). Binding of Hh ligands to their receptors triggers dissociation of the Sufu-Gli protein complexes. This relieves the inhibitory effects of Sufu on Gli proteins and allows the conversion of Gli proteins into transcriptional activators, which induce the expression of Hh target genes (Humke et al., 2010; Tukachinsky et al., 2010; Zeng et al., 2010; Lin et al., 2014). Interestingly, Sufu regulates the stability of Gli proteins as well. In the absence of Sufu, although Gli proteins become hyperactive, the total level of Gli proteins is markedly reduced (Chen et al., 2009; Jia et al., 2009; Wang et al., 2010; Liu et al., 2012). It is believed that Sufu prevents Spop-dependent proteasome degradation of Gli proteins (Wang et al., 2010). The important roles that Sufu plays in vertebrate Hh signaling are well established, but it is less clear how the inhibitory function of Sufu toward Gli proteins is regulated.
The RUN and SH3 domain-containing (Rusc) family of vertebrate proteins consists of two members. Rusc1 and Rusc2 both contain a RUN domain and a C-terminal SH3 domain. The shortest isoform of Rusc1, namely Nesca, is involved in the neurotrophin signaling pathway (MacDonald et al., 2012; Sun et al., 2012). The function of Rusc2 is not known. In this study, we report that Rusc1 and Rusc2 interact with Sufu and restrict the response of cells to Hh signaling.
RESULTS
Members of the Rusc family interact with Sufu and inhibit Hh signaling
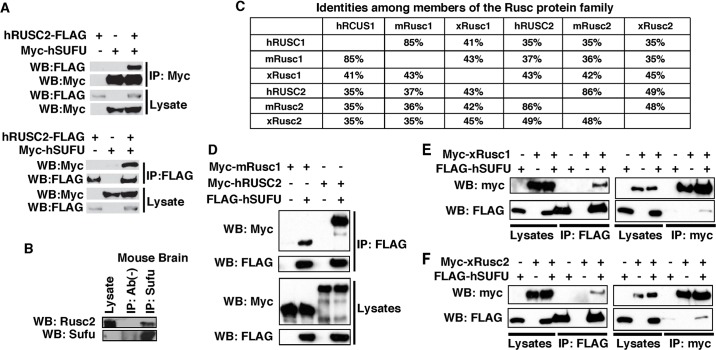
Rusc2 was identified from a yeast two-hybrid screen using full-length Sufu as bait. To verify the interaction between Sufu and Rusc2, we performed a co-immunoprecipitation (CoIP) in HEK293T cells. We were able to co-immunoprecipitate FLAG-tagged human (h) RUSC2 with myc-hSUFU (Fig. 1A, upper panel). In the reverse CoIP, myc-hSUFU co-purified with hRUSC2-FLAG (Fig. 1A, lower panel). Furthermore, we detected interaction between endogenous Rusc2 and Sufu in mouse brain (Fig. 1B). Members of the Rusc protein family are highly similar to each other (Fig. 1C). Our results reveal that like hRUSC2, mouse (m) Rusc1 interacts with hSUFU (Fig. 1D). In addition, Xenopus Rusc1 (Fig. 1E) and Rusc2 (Fig. 1F) both interacted with hSUFU.
Fig. 1.
Members of the Rusc protein family interact with Sufu. (A) Co-immunoprecipitation (CoIP) showing the interaction between hSUFU and hRUSC2. hRUSC2-FLAG and myc-hSUFU were expressed in HEK293T cells alone or in combination. CoIP was performed using an anti-myc antibody (upper panel) or an anti-FLAG antibody (lower panel). (B) CoIP showing that endogenous Sufu and Rusc2 form a complex in mouse whole brain lysate. Sufu was immunoprecipitated. (C) Identity between the Rusc proteins. Protein sequences of Rusc1 and Rusc2 from human (h), mouse (m) and Xenopus (x) were aligned using NCBI BLAST. (D) CoIP showing that mRusc1 and hRUSC2 form complexes with hSUFU. (E,F) CoIP showing that myc-xRusc1 (E) and myc-xRusc2 (F) interact with FLAG-hSUFU. IP, immunoprecipitation; WB, western blot.
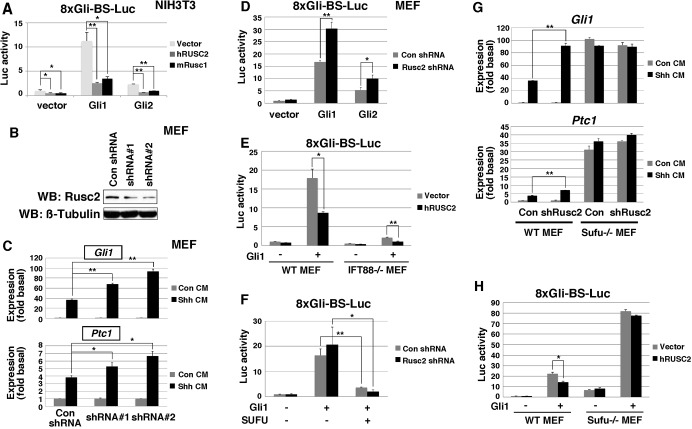
To study the functions of Rusc proteins in Hh signaling, we took advantage of an Hh-responsive luciferase reporter [8xGli-BS luciferase (Sasaki et al., 1997)]. As expected, Gli1 and Gli2 activated 8xGli-BS luciferase in mouse NIH3T3 fibroblasts. Overexpression of mRusc1 or hRUSC2 markedly reduced the activity of Gli1 and Gli2 in this assay (Fig. 2A). Interestingly, only Rusc2 is abundantly expressed in NIH3T3 cells and mouse embryonic fibroblasts (MEFs) (Fig. S1). We thus knocked down Rusc2 using two shRNAs, which target different regions of the Rusc2 mRNA (Fig. 2B). As shown in Fig. 2C, knockdown of Rusc2 in MEFs markedly enhanced Shh-induced expression of Gli1 and Ptc1 (Ptch1), two direct targets of Hh signaling. Consistently, knockdown of Rusc2 increased Gli1- and Gli2-induced 8xGli-BS luciferase activities (Fig. 2D). Similar results were obtained when the experiment was performed in NIH3T3 cells (data not shown). In addition to the shRNA knockdown experiments, we took advantage of transcription activator-like effector nuclease (TALEN) technology and generated an Rusc2 heterozygous mutant MEF cell line (Fig. S2A,B). Compared with control MEFs, Rusc2 heterozygous mutant MEFs exhibited a more robust response to overexpressed Gli1 (Fig. S2C,D) or Shh-N-conditioned medium (Fig. S2E,F). These results demonstrate that Rusc2 inhibits Hh signaling.
Fig. 2.
Rusc proteins inhibit Hh signaling. (A) Dual-luciferase assay showing that mRusc1 and hRUSC2 inhibit the activities of Gli1 and Gli2 in the 8xGli-BS luciferase reporter assay. (B) Western blot showing reduced expression of Rusc2 in MEFs infected with lentiviral shRNAs against Rusc2. (C) RT-PCR results showing that Shh-N-conditioned medium induces the expression of Gli1 and Ptc1 in MEFs. Knockdown of Rusc2 by lentiviral shRNAs enhances the activity of Shh-N-conditioned medium in this assay. The expression levels of Gli1 and Ptc1 were normalized to that of Actb. (D) Dual-luciferase assay showing that knockdown of Rusc2 in MEFs enhances the activities of Gli1 and Gli2. (E) Dual-luciferase assay showing that overexpression of hRUSC2 reduces the activity of Gli1 in wild-type MEFs and Ift88 knockout (Ift88−/−) MEFs. (F) Dual-luciferase assay showing that overexpression of hSUFU reduces the activity of Gli1 in control and Rusc2 knockdown MEFs. (G) RT-PCR results showing that knockdown of Rusc2 increases the expression of Gli1 and Ptc1 induced by Shh-N-conditioned medium in wild-type MEFs. In Sufu knockout MEFs, knockdown of Rusc2 had no effect on the expression of Gli1 and Ptc1. (H) Dual-luciferase assay showing that overexpression of Rusc2 in wild-type MEFs, but not Sufu knockout MEFs, reduces the activity of Gli1 in the 8xGli-BS luciferase reporter assay. Data are shown as mean±s.d. *P<0.05, **P<0.01.
Next, we carried out a systematic epistasis analysis. As shown in Fig. 2E, overexpression of hRUSC2 inhibited Gli1-induced 8xGli-BS luciferase in Ift88 knockout MEFs, which are deficient in primary cilia (Murcia et al., 2000). This demonstrates that Rusc2 functions independently of cilia in the Hh pathway. To define the epistatic relationship between Rusc2 and Sufu, we assayed the activity of Sufu in wild-type and Rusc2 knockdown MEFs. hSUFU reduced Gli1-induced 8xGli-BS luciferase activity in the wild-type and Rusc2 knockdown MEFs (Fig. 2F), indicating that Sufu can inhibit Hh signaling independently of Rusc2. By contrast, knockdown of Rusc2, which enhanced Shh-conditioned medium-induced expression of Gli1 and Ptc1 in wild-type MEFs, failed to do so in Sufu knockout MEFs (Fig. 2G). Consistently, overexpression of hRUSC2 reduced Gli1-induced 8xGli-BS luciferase activity in wild-type MEFs, but not in Sufu knockout MEFs (Fig. 2H). These results demonstrate that Rusc2 regulates the Hh pathway at the level of Gli. In addition, Sufu is required for the function of Rusc2 in Hh signaling.
Rusc2 inhibits signaling-induced dissociation of Sufu and Gli
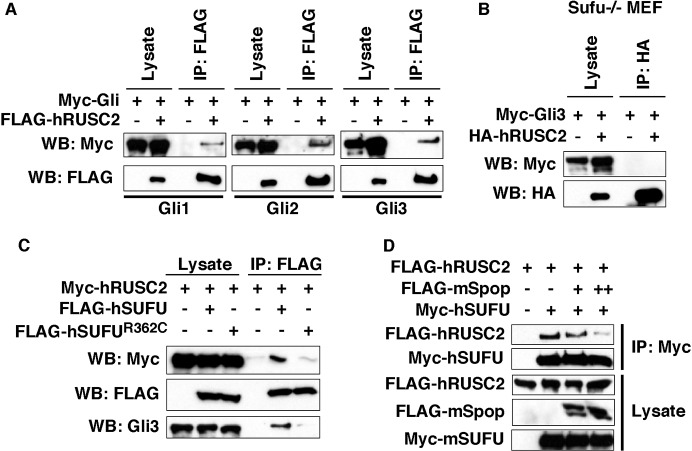
Sufu directly binds and inhibits Gli proteins. Since Rusc proteins interact with Sufu and inhibit Gli, we determined whether Rusc2 can form complexes with Gli proteins. Indeed, FLAG-hRUSC2 co-immunoprecipitated with all three Gli proteins in HEK293T cells (Fig. 3A). We found that Sufu is required for the interaction between Rusc2 and Gli proteins. In Sufu knockout MEFs, we could not detect binding between hRUSC2 and Gli3 (Fig. 3B). Interestingly, we could not detect binding between hRUSC2 and hSUFUR362C, an oncogenic form of Sufu deficient in Gli binding (Fig. 3C). Moreover, overexpression of mouse Spop, which promotes proteasome-dependent degradation of Gli proteins (Zhang et al., 2006, 2009), reduced the binding between hRUSC2 and hSUFU in a dose-dependent manner (Fig. 3D). These results suggest that Rusc2 preferentially binds Sufu that is associated with Gli proteins.
Fig. 3.
Rusc2 forms a heterotrimeric complex with Sufu and Gli proteins. (A) CoIP showing that FLAG-hRUSC2 interacts with myc-Gli1 (left), myc-Gli2 (middle) and myc-Gli3 (right). (B) CoIP showing lack of complex formation between overexpressed Rusc2 and Gli3 in Sufu knockout MEFs. (C) CoIP showing that hRUSC2 forms a complex with wild-type hSUFU, but not hSUFUR362C, which is deficient in binding Gli protein. (D) CoIP showing that overexpression of mouse Spop reduces the interaction between hSUFU and hRUSC2.
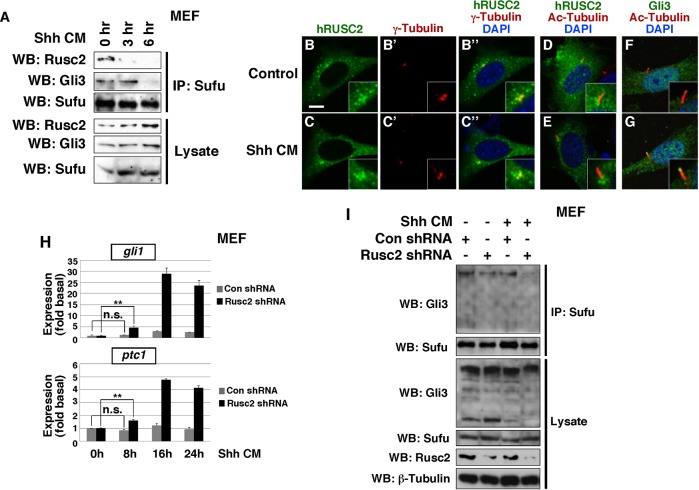
In vertebrates, Hh signaling induces translocation of the Sufu-Gli complex to the primary cilium and subsequent dissociation of the Gli-Sufu complexes. This converts Gli proteins into Gli activators that activate Hh-dependent transcription (Humke et al., 2010; Tukachinsky et al., 2010; Zeng et al., 2010; Lin et al., 2014). Since Rusc2, Sufu and Gli form a heterotrimeric complex, we investigated the effect of Hh signaling on this complex. We treated MEFs with a low dose of Shh-N-conditioned medium, which triggers the Hh pathway with slow activation kinetics. We then performed CoIP to measure the amount of endogenous Gli3 and Rusc2 that were associated with endogenous Sufu. In unstimulated MEFs, Rusc2 and Gli3 co-immunoprecipitated with Sufu. In MEFs treated with Shh-conditioned medium for 3 h, we could not detect binding between Sufu and Rusc2. By contrast, Sufu and Gli3 remained associated with each other at this time point. At 6 h post Shh-conditioned medium treatment, the Sufu-Gli3 complex was dissociated (Fig. 4A). This indicates that the Rusc2-Sufu-Gli complex is dissociated sequentially upon Hh signaling, with dissociation of Rusc2 occurring prior to the collapse of the Gli-Sufu complex.
Fig. 4.
Rusc2 inhibits signaling-induced dissociation of Sufu-Gli protein complexes. (A) CoIP showing sequential dissociation of Rusc2-Sufu-Gli3 complexes in MEFs upon Shh-conditioned medium treatment. Endogenous Sufu was immunoprecipitated. The amount of endogenous Gli3 and Rusc2 associated with Sufu was assessed by western blot. (B-G) Confocal images showing the subcellular localization of FLAG-hRUSC2 in control (upper row) and Shh-conditioned medium-stimulated (bottom row) cells. (B,C) Anti-FLAG staining for hRUSC2. (B′,C′) γ-tubulin staining. (B″,C″) Merges of B,B′ and of C,C′. (D,E) Merged images of hRUSC2 and acetylated-tubulin staining. (F,G) Merged images of Gli3 and acetylated-tubulin staining. Insets are higher magnification views of the area around cilia. Scale bar: 10 µm. (H) RT-PCR showing the expression of Gli1 and Ptc1 in unstimulated MEFs and MEFs treated with Shh-N-conditioned medium for 8, 16 and 24 h. Data are shown as mean±s.d. **P<0.01. n.s., non-significant. (I) CoIP experiments to assess the effect of Rusc2 knockdown on Shh-induced dissociation of Sufu-Gli3 protein complexes in MEFs. Endogenous Sufu and Gli3 were analyzed. The dose of Shh-N-conditioned medium used was identical to that in H.
We extended our analysis by assessing the subcellular localization of Rusc2. In unstimulated cells, hRUSC2 was mainly detected in the cytoplasm. A small amount of hRUSC2 protein overlapped with γ-tubulin, a marker for cilia basal bodies (Fig. 4B-B″). This localization pattern remained unchanged in cells treated with Shh-conditioned medium (Fig. 4C-C″). This is in stark contrast to Gli3, which is translocated to the tip of the cilium upon Shh-N-conditioned medium treatment (Fig. 4F,G). Dissociation of Sufu and Gli occurs after their ciliary translocation (Tukachinsky et al., 2010; Zeng et al., 2010). Lack of ciliary translocation of Rusc2 upon Hh signaling thus further supports the idea that Hh signaling induces the sequential dissociation of the Rusc2-Sufu-Gli complex.
In light of the above findings, we set out to determine if Rusc2 prevents signaling-induced dissociation of Sufu-Gli complexes. After titrating the dose of Shh-N-conditioned medium, we chose to treat MEFs with a low dose that was insufficient to induce the expression of Ptc1 and caused only a 3-fold increase in the expression of Gli1 at 16 h. When Rusc2 knockdown MEFs were treated with the same dose of Shh-N-conditioned medium, a significant increase in the expression of Gli1 and Ptc1 was detected 8 h later. At 16 h post treatment, we detected a robust increase in the expression of both Gli1 and Ptc1 (Fig. 4H). Under the same treatment condition, we performed Sufu CoIP to assess the effects of Rusc2 knockdown on the Sufu-Gli protein complexes. In unstimulated MEFs, knockdown of Rusc2 did not alter the interaction between Gli3 and Sufu, although a marginal reduction in the amount of Gli3 that co-immunoprecipitated with Sufu was occasionally observed. At 8 h post Shh-N-conditioned medium treatment, Gli3 and Sufu remained associated with each other in control MEFs. In Rusc2 knockdown MEFs, however, we could no longer detect binding between Gli3 and Sufu (Fig. 4I). This demonstrates that knockdown of Rusc2 accelerates the dissociation of Sufu-Gli complexes upon Hh signaling. Knockdown of Rusc2 did not alter the subcellular localization of Gli in unstimulated MEFs (Fig. S3). Taken together, we conclude that Rusc2 inhibits Hh signaling by preventing signaling-induced dissociation of the Sufu-Gli complexes.
Overexpression of Rusc2 induces cytoplasmic Gli protein aggregates
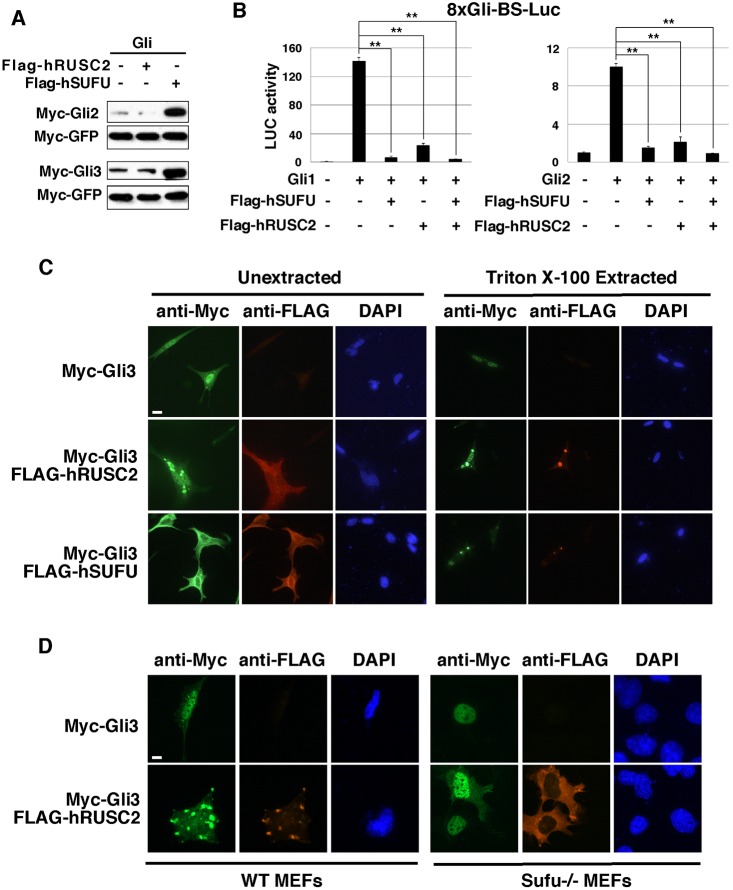
Next, we compared the activities of Sufu and Rusc2 in regulating the expression and subcellular localization of Gli proteins. We found that overexpression of hSUFU, but not hRUSC2, increased the level of Gli proteins (Fig. 5A). Both hSUFU and hRUSC2 reduced the activities of Gli1 and Gli2 in an 8xGli-BS luciferase reporter assay. However, the activity of hRUSC2 was less potent in this assay (Fig. 5B). When expressed alone in NIH3T3 cells, Gli3 was enriched in the nucleus. When Gli3 and hSUFU were co-expressed, the level of Gli3 was increased dramatically and the majority of Gli3 proteins were detected in the cytoplasm. Overexpression of hRUSC2 also decreased the amount of nuclear Gli3, albeit to a lesser extent. Strikingly, hRUSC2 overexpression induced large Gli3 protein aggregates in the cytoplasm (Fig. 5C). Similar results were obtained when Gli1 and Gli2 were co-expressed with Rusc2 (Fig. S4). Strikingly, these cytoplasmic Gli protein aggregates are resistant to extraction with Triton X-100. We found that hSUFU can also induce Triton-resistant cytoplasmic Gli3 aggregates, albeit with weaker activity (Fig. 5C, Fig. S4B).
Fig. 5.
Rusc2 induces cytosolic Gli protein aggregates. (A) Western blot showing that overexpression of hSUFU, but not hRUSC2, stabilizes Gli2 and Gli3 in NIH3T3 cells. (B) Dual-luciferase assay showing that hSUFU and hRUSC2 reduce the activity of Gli1 and Gli2. Compared with hSUFU, hRUSC2 was less potent in this assay. Data are shown as mean±s.d. **P<0.01. (C) Immunofluorescence showing the effects of hSUFU and hRUSC2 on the subcellular localization of myc-Gli3 in NIH3T3 cells. Left, cells without Triton X-100 extraction; right, cells extracted with Triton X-100 prior to fixation. (D) Immunofluorescence showing that overexpression of hRUSC2 alters the subcellular distribution of myc-Gli3 in wild-type MEFs (left), but not that in Sufu knockout MEFs (right). Scale bars: 10 µm.
We further determined whether Sufu is required for Rusc2 to induce the cytoplasmic Gli protein aggregates. In wild-type MEFs, overexpression of hRUSC2 resulted in cytoplasmic retention of Gli3 and induced cytosolic Gli3 protein aggregates. In Sufu knockout MEFs, however, hRUSC2 overexpression did not cause relocalization of Gli3 or induce Gli3 protein aggregates; instead, Gli3 protein remained in the nucleus (Fig. 5D, Fig. S4C). This indicates that Rusc2 regulates the subcellular distribution of Gli proteins in a Sufu-dependent manner. This finding further supports the idea that Rusc2 modulates the Hh pathway by regulating the interaction between Sufu and Gli.
Rusc1 inhibits Hh signaling during Xenopus embryonic development
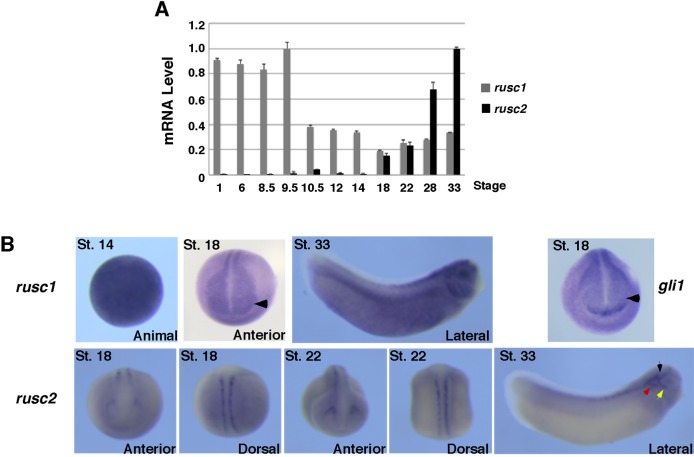
To understand the in vivo functions of Rusc proteins, we examined the expression of rusc1 and rusc2 in Xenopus embryos. rusc1 is expressed maternally and is present abundantly and ubiquitously in the embryo. Maternal rusc1 mRNA declines gradually during the gastrula and neurula stages (Fig. 6A,B). By the late neurula stage, strong expression of rusc1 was detected in the developing neural tube and eye domains (Fig. 6B). At this stage, the eye domains, which strongly express rusc1, do not express gli1, a direct target of Hh signaling (Lee et al., 1997) (Fig. 6B). This raises the possibility that Rusc1 might inhibit Hh signaling in the developing eye. As development proceeded, maternal rusc1 further declines. At the late tailbud stage, strong expression of rusc1 is observed in the dorsal neural tube, eyes and branchial arches (Fig. 6B).
Fig. 6.
Expression of rusc1 and rusc2 during Xenopus eye development. (A) RT-PCR showing the temporal expression of rusc1 and rusc2 during Xenopus development. The expression level of rusc1 and rusc2 was normalized to that of odc. Data are shown as mean±s.d. (B) Whole-mount in situ hybridization showing the spatial expression pattern of rusc1, rusc2 and gli1. St., stage. Arrowheads point to the eye domains, which express rusc1 but not gli1. Black, red and yellow arrows point to the trigeminal ganglion, middle lateral line placode, and anterodorsal lateral line placode, respectively.
In contrast to rusc1, rusc2 expression commences zygotically. We could not detected rusc2 by in situ hybridization at stage 14 (data not shown). Starting from stage 18, the expression of rusc2 can be detected in Rohon-Beard neurons, which are located along the dorsal neural tube in the trunk region. In the anterior region, rusc2 is specifically expressed in the trigeminal ganglion. At stage 33, in addition to Rohon-Beard neurons and trigeminal ganglion, rusc2 is expressed in the middle and anterodorsal lateral line placodes (Fig. 6B).
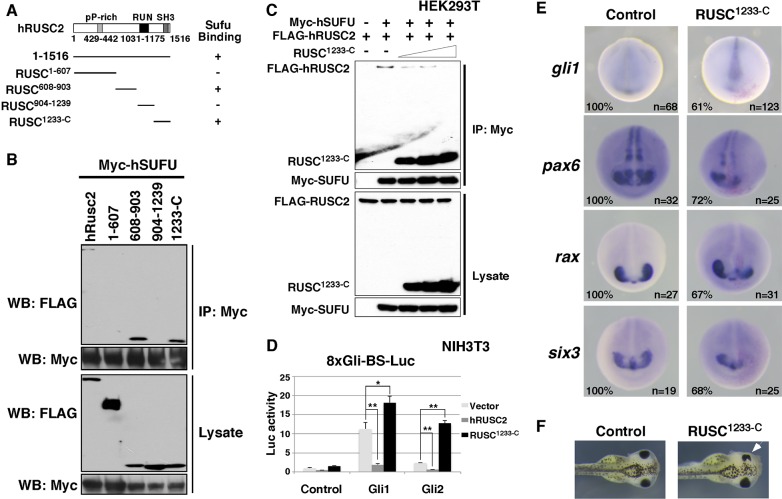
To study the functions of Rusc proteins during development, we first took a dominant-negative approach. We generated multiple hRUSC2 deletion constructs (Fig. 7A) and characterized their interaction with hSUFU in detail. Full-length hRUSC2, RUSC608-903 and RUSC1233-C interacted with hSUFU in HEK293T cells (Fig. 7A,B) and in the yeast two-hybrid system (data not shown). When RUSC1233-C was overexpressed, it interfered with complex formation between full-length hRUSC2 and hSUFU in a dose-dependent manner (Fig. 7C). We overexpressed RUSC1233-C in NIH3T3 cells and performed an 8xGli-BS luciferase reporter assay. In stark contrast to full-length hRUSC2, which inhibited Gli1 and Gli2, RUSC1233-C markedly enhanced the activities of Gli1 and Gli2 in the 8xGli-BS luciferase assay (Fig. 7D). This indicates that RUSC1233-C acts as a dominant negative.
Fig. 7.
Dominant-negative Rusc enhances Hh signaling in Xenopus embryos and impairs eye development. (A) Schematic of hRUSC2 and deletion derivatives. Whether an hRUSC2 construct interacts with hSUFU in the CoIP experiment is indicated by + or −. (B) CoIP results showing that hSUFU interacts with full-length hRUSC2, RUSC608-903 and RUSC1233-C. (C) CoIP showing that overexpression of RUSC1233-C reduces the binding between hSUFU and full-length hRUSC2. (D) Dual-luciferase assay showing that the activities of Gli1 and Gli2 are enhanced by co-overexpression of RUSC1233-C in NIH3T3 cells. Data are shown as mean±s.d. *P<0.05, **P<0.01. (E) In situ hybridization showing the expression of gli1, pax6, rax and six3 in control (left) and RUSC1233-C overexpression (right) Xenopus embryos at stage 20. At the 8-cell stage, one of the dorsal animal blastomeres was injected with a mixture of RUSC1233-C (1 ng) and n-β-gal (250 pg) encoding RNAs. (F) Overexpression of RUSC1233-C (1 ng) reduced the size of the eye (arrowhead).
It is well established that Shh separates the eye field into two distinct eye primordia by suppressing the expression of eye-specific genes in the midline. Elevated Hh signaling often reduces the expression of eye markers and decreases the size of the eye (Amato et al., 2004; Koide et al., 2006; Rorick et al., 2007; Min et al., 2011). We injected RUSC1233-C (1 ng) at the 8-cell stage into dorsal animal blastomeres of Xenopus embryos, which give rise to the neural tube and retina (Moody, 1987, 2012). To assess changes in Hh signaling, we monitored the expression of gli1, a direct target of Hh signaling (Lee et al., 1997). Indeed, overexpression of RUSC1233-C increased the expression of gli1 in cells located close to the midline in the neural ectoderm (61%, n=123; Fig. 7E). This was accompanied by a severe reduction in the expression of eye markers, including pax6 (72%, n=25), rax (67%, n=31), and six3 (68%, n=25) (Fig. 7E). At the tadpole stage, the majority of RUSC1233-C overexpression embryos (86%, n=64) exhibited reduced eyes (Fig. 7F). Thus, overexpression of a dominant-negative Rusc enhances Hh signaling and impairs Xenopus eye development.
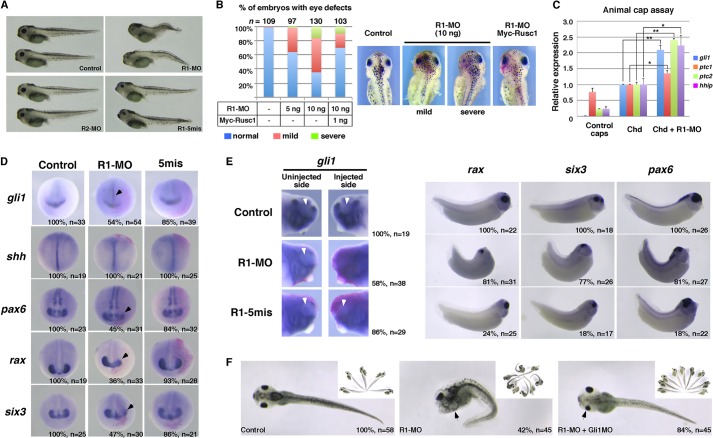
We then designed morpholinos to block the translation of rusc1 (R1-MO) and rusc2 (R2-MO) in Xenopus embryos (Fig. S5A). Injection of each morpholino (20 ng) into both dorsal blastomeres at the 4-cell stage had distinct effects on early development. We did not detect any morphological abnormalities in R2-MO-injected embryos. By contrast, injection of R1-MO induced severe defects during development. Compared with uninjected controls or embryos injected with a five-base mismatch morpholino (R1-5mis), R1-MO-injected embryos showed a shorter anterior-posterior (A/P) axis and severely reduced eyes (Fig. 8A). Histological analysis revealed that knockdown of Rusc1 did not abolish the eye completely. Retina tissues were present even in severely disrupted eyes (Fig. S5B). Both the eye and A/P axis defects induced by R1-MO were rescued by injection of myc-Rusc1 (1 ng) (Fig. 8B). To further test the specificity of Rusc1 knockdown, we designed another morpholino (R1-sb), which blocks the splicing of rusc1. Similar to phenotypes observed in R1-MO-injected embryos, R1-sb-injected embryos showed reduced eyes and shortened A/P axis (Fig. S6A-C). We thus conclude that Rusc1 is essential for normal Xenopus development.
Fig. 8.
Rusc1 inhibits Hh signaling during Xenopus eye development. (A) Whole embryo morphology of uninjected embryos and those injected with R1-MO, R1-5mis or R2-MO. Morpholinos (20 ng) were injected into both dorsal blastomeres at the 4-cell stage. (B) Overexpression of myc-xRusc1 rescued the phenotypes induced by unilateral injection of R1-MO. (Left) Summary of embryos with eye defects. (Right) Images of representative embryos. A 50% or greater reduction in eye size is considered ‘severe'; a reduction of less than 50% is considered ‘mild'. (C) RT-PCR showing the expression of gli1, ptc1, ptc2 and hhip in animal caps. Chordin (Chd, 25 pg) was injected into the animal pole of control and R1-MO (40 ng) injected embryos at the 1-cell stage. Animal caps were dissected at the late blastula stage and harvested at stage 22. Data are shown as mean±s.d. *P<0.05, **P<0.01. (D) In situ hybridization showing that unilateral injection of R1-MO (20 ng) enhances the expression of gli1, and reduces the expression of pax6, rax and six3. The expression of shh was not altered by R1-MO injection. Embryos were analyzed at stage 20. (E) In situ hybridization showing that unilateral injection of R1-MO enhances the expression of gli1 in the head region and reduces the expression of pax6, rax and six3 at stage 33. Arrowheads point to eyes on the injected side. (F) Morphology of uninjected embryos and those unilaterally injected with R1-MO alone or R1-MO together with Gli1 morpholino (Gli1 MO). Insets show further examples of the illustrated phenotype. Arrows (D,F) point to the developing eyes.
To determine if Rusc1 inhibits Hh signaling during development, we took advantage of the animal cap assay, which is an in vitro assay for studying Hh signaling in Xenopus embryonic tissues (Rorick et al., 2007; Min et al., 2011; Schwend et al., 2013). In Chordin (Chd) neuralized animal caps, injection of R1-MO caused a 2-fold increase in the expression of gli1, ptc2 and hhip at stage 22 and a modest increase in the expression of ptc1 (Fig. 8C). This demonstrates that knockdown of Rusc1 increases the expression of Hh target genes. In whole embryos, injection of R1-MO had no effect on eye-specific gene expression at stage 14 (data not shown). From the late neurula stage, we began to observe reduction in the expression of eye markers. These included pax6 (45%, n=31), rax (36%, n=33) and six3 (47%, n=30). Knockdown of Rusc1 increased the expression of gli1 in cells located close to the midline in the neural ectoderm (54%, n=54), without altering the expression of shh (Fig. 8D). The eye defect induced by Rusc1 knockdown became more pronounced by the late tailbud stage, with the expression of pax6 (81%, n=27), rax (81%, n=31) and six3 (77%, n=26) being reduced in the majority of R1-MO-injected embryos. Interestingly, the expression of pax6 in the dorsal neural tube was affected to a lesser extent, even in embryos with severely reduced eyes. We again assessed the Hh signaling activity by monitoring the expression of gli1. Although the head was generally small on the R1-MO-injected side, 58% of the injected embryos (n=38) exhibited nearly uniform expression of gli1 on the injected side. This is distinct from the uninjected side, where a ‘gli1-free' eye domain was prominent (Fig. 8E, arrowheads). Knockdown of Rusc1 by injection of R1-sb induced similar phenotypes (Fig. S6E). Since overexpression of the dominant-negative Rusc and knockdown of Rusc1 both increase gli1 expression and impair eye formation, we conclude that Rusc1, which is strongly expressed in the developing eye, inhibits Hh signaling during eye development.
To determine if the eye defects induced by Rusc1 knockdown could be attributed to elevated Hh signaling, we knocked down Gli1 in Rusc1-depleted embryos. Unilateral knockdown of Rusc1 induced eye defects in 95% of embryos [42% had severe defects and another 53% had milder eye defects (n=45)]. Interestingly, 73% of injected embryos had bodies that were bent toward the injected side, which was likely to be due to a shortened A/P axis on the injected side. Co-injection of Gli1 morpholino (Nguyen et al., 2005; Schwend et al., 2013) clearly rescued the Rusc1 knockdown phenotypes, with the majority of embryos (96%, n=45) developing a straight body axis and only 16% of embryos showing mildly affected eyes (Fig. 8F). This demonstrates that the eye development defects of Rusc1 knockdown embryos are indeed a consequence of enhanced Hh signaling.
DISCUSSION
Although the Hh pathway is evolutionarily conserved, many differences exist between vertebrate and Drosophila Hh signaling (Huangfu and Anderson, 2006; Wilson and Chuang, 2010). One major difference is Sufu, which is dispensable for Drosophila Hh signaling (Préat, 1992) but functions as a major pathway inhibitor in vertebrates (Cooper et al., 2005; Svard et al., 2006; Min et al., 2011). Sufu physically interacts with Gli proteins and regulates their stability, localization and activities (Ding et al., 1999; Kogerman et al., 1999; Murone et al., 2000; Lin et al., 2014; Han et al., 2015). Loss of Sufu elevates vertebrate Hh signaling and induces severe patterning defects during development (Wolff et al., 2003; Cooper et al., 2005; Svard et al., 2006; Min et al., 2011). In humans, oncogenic mutations in SUFU have been identified from medulloblastoma, basal cell carcinoma and other cancers (Taylor et al., 2002; Pastorino et al., 2009; Brugieres et al., 2010; Aavikko et al., 2012; Kijima et al., 2012; Schulman et al., 2015). Despite the fundamental roles played by Sufu in development and cancer, it is largely unclear how the Sufu protein itself is regulated.
Here we report that members of the vertebrate-specific Rusc protein family are novel Sufu-binding partners. Both Rusc1 and Rusc2 bind Sufu and inhibit Hh signaling. In the case of Rusc2, a domain located upstream of the RUN domain, and the C-terminal SH3 domain, are responsible for binding Sufu. During Xenopus development, it is Rusc1 that is expressed predominantly. Rusc1 is expressed maternally (i.e. from maternally inherited transcripts), and zygotic Rusc1 is strongly expressed in the developing eyes and the neural tube. Overexpression of a dominant-negative Rusc or knockdown of Rusc1 leads to increased Hh signaling, which impairs eye development. Knockdown of Rusc2, whose expression is restricted to only a few lineages, does not cause any detectable morphological defects. In contrast to Xenopus embryos, NIH3T3 and MEF cells predominantly express Rusc2. Knockdown of Rusc2 in these cells potentiates Hh signaling. These findings demonstrate that Rusc1 and Rusc2 are novel components of the vertebrate Hh pathway.
Our results reveal that Rusc2 exerts its inhibitory effect on Hh signaling through binding Sufu. As the major Gli inhibitor, Sufu forms complexes with Gli proteins and sequesters them in the cytoplasm (Ding et al., 1999; Kogerman et al., 1999; Pearse et al., 1999; Stone et al., 1999; Zhang et al., 2013; Han et al., 2015). In the nucleus, Sufu recruits p66β to the promoters of Hh target genes and represses Gli-dependent transcription (Lin et al., 2014). Hh signaling dissociates the Sufu-Gli protein complexes, converting Gli proteins into transcriptional activators, which ultimately activate the expression of Hh target genes (Humke et al., 2010; Tukachinsky et al., 2010; Zeng et al., 2010; Lin et al., 2014). Our results reveal that Rusc2, Sufu and Gli form a heterotrimeric protein complex. Upon Hh signaling, this complex is dissociated sequentially, with Rusc2 exiting first, followed by dissociation of Gli from Sufu. Although knockdown of Rusc2 is insufficient for pathway activation, it potentiates Hh signaling by accelerating signaling-induced dissociation of the Sufu-Gli complexes. It is important to note that Sufu is required for the function of Rusc2 in the Hh pathway. In the absence of Sufu, knockdown or overexpression of Rusc2 has no effect on the output of Hh signaling. These observations strongly argue that Rusc2 functions in the Hh pathway by stabilizing the Sufu-Gli complexes. In support of this hypothesis, we found that overexpression of Rusc2 decreases the amount of Gli proteins in the nucleus and induces cytosolic Gli protein aggregates, which are resistant to Triton extraction. This activity of Rusc2 is again Sufu dependent. It appears that Rusc2 inhibits Hh signaling by binding Sufu and stabilizing the Sufu-Gli complexes.
Notably, the functions of Rusc differ in several aspects from that of Sufu. Sufu deficiency results in robust pathway activation and destabilization of Gli proteins (Svard et al., 2006; Chen et al., 2009; Jia et al., 2009; Wang et al., 2010). By contrast, knockdown or overexpression of Rusc2 has no effect on the stability of Gli proteins. Knockdown of Rusc2 alone does not activate the Hh pathway. Elevated Hh signaling occurs only when cells are stimulated. In overexpression studies, Sufu sequesters Gli proteins in the cytoplasm very potently and inhibits Gli-dependent transcription. The activity of Rusc2 is weaker in these assays. Interestingly, Rusc2 is capable of inducing large cytoplasmic Gli protein aggregates. Although Sufu is required for this activity of Rusc2, Sufu itself has a weak activity in inducing these Gli protein aggregates. These findings are consistent with our hypothesis that Rusc2 stabilizes the Sufu-Gli complexes.
In Xenopus, knockdown of Rusc1 enhances Hh signaling and impairs eye development, which is reminiscent of the Sufu loss-of-function phenotypes. Nevertheless, the defects induced by Rusc1 knockdown are less severe than those observed in Sufu knockdown embryos. Sufu-deficient Xenopus embryos show robust Hh activation. Increased expression of ptc1 was detected as early as the early neurula stage (stage 15) (Min et al., 2011). In Rusc1 knockdown embryos, however, we began to detect an increase in the expression of gli1, which is very sensitive to Hh signaling, from the late neurula stage. The expression of ptc1 was increased only moderately. This suggests that knockdown of Rusc1 only causes weak Hh activation in embryos. These functional differences between Sufu and Rusc are again in agreement with the view that Rusc proteins regulate the Hh pathway by enhancing the inhibitory functions of Sufu.
Interestingly, Rusc proteins interact with kinesins (MacDonald et al., 2012) and Rab family members (Bayer et al., 2005; Fukuda et al., 2011). In vertebrates, Kif7, a member of the kinesin protein family, interacts with Gli proteins and plays an important role in Hh signaling (Tay et al., 2005; Cheung et al., 2009; Endoh-Yamagami et al., 2009; Liem et al., 2009; Law et al., 2012; Li et al., 2012; He et al., 2014). Zebrafish Kif7 potentiates the activity of Gli2 by promoting its dissociation from Sufu (Maurya et al., 2013). It is also known that Rab23, which regulates endocytic and ciliary trafficking (Evans et al., 2003; Boehlke et al., 2010), is highly expressed in the dorsal neural tube and regulates Hh signaling during neural tube patterning (Eggenschwiler et al., 2001; Li et al., 2007). Similar to Rusc proteins, Rab23 functions downstream of Smo and Ptch and inhibits Gli1 in a Sufu-dependent manner (Evans et al., 2003; Eggenschwiler et al., 2006; Chi et al., 2012). In the future, it will be of great interest to determine whether Rusc proteins physically and functionally interact with Kif7 or Rab23 in Hh signaling.
MATERIALS AND METHODS
Yeast two-hybrid screen
An adult mouse brain cDNA library (Clontech) was screened using full-length hSUFU (pGBKT7-hSUFU) as bait, according to standard protocols (Yeast Protocols Handbook, Clontech).
Plasmids
Gli1, Gli2, Gli3, hSUFU (Schwend et al., 2013) and hRUSC2 (Bayer et al., 2005) expression constructs were described previously. Mouse Rusc1 was constructed by PCR from IMAGE:6816267. Rusc1 and Rusc2 were identified in the Xenopus laevis genome using the NCBI online BLAST tool and mammalian Rusc protein sequences. Xenopus rusc1 (KX265097) and rusc2 (KX265098) were PCR cloned from Xenopus cDNA. All deletion constructs were generated by PCR and standard cloning methods. The hSUFUR362C mutant was generated by site-directed mutagenesis.
Cell lines, shRNAs, transfection and conditioned medium treatments
NIH3T3, HEK293T and MEF cells were cultured and transfected as described (Jia et al., 2009; Jin et al., 2010). Sufu−/− and Ift88−/− MEFs were provided by Dr A. Liu (Department of Biology, Pennsylvania State University). The Rusc2 heterozygous mutant MEF cell line was generated by transfection of a TALEN pair targeting the second exon of mouse Rusc2 as previously described (Mussolino et al., 2011). The targeting sequences of the Rusc2 loci are 5′-TTCTACCTGGACCTGCAGC-3′ and 5′-TGTCTTGCGAGTCCCACCA-3′, with a spacer (5′-CCTCCCCGGCTGAGTCGAGAA-3′). TALEN-transfected MEFs were selected with puromycin. A Rusc2 heterozygous mutant MEF cell line derived from TALEN-transfected single cells was then established.
Lentiviral shRNA constructs [TRCN0000252575 (targeting 5′-AGGCCATATCCATCGACATAC-3′) and TRCN0000252578 (targeting 5′-GTCCACTAGGCCGACTGATAA-3′)] were purchased from Sigma-Aldrich. Lentiviral shRNA constructs were cotransfected into HEK293T cells with the virus packaging plasmids pCMV-ΔR and VSV-G for virus preparation. Lentiviral particle-containing supernatant was collected 48 h post transfection. Infection was carried out by adding virus-containing supernatant to cell culture, followed by selection with 2 μg/ml puromycin.
Shh-N-conditioned medium was prepared from Shh-N-transfected HEK293T cells. One day after transfection, medium was replaced with DMEM containing 2% FBS, which was collected and filtered through a 0.22-μm membrane after an additional 2 days. Medium collected from non-transfected HEK293T cells served as control. To test the activity of each preparation, we treated NIH3T3 cells with Shh-N-conditioned medium and performed RT-PCR for Ptc1 and Gli1. For conditioned medium treatment, cells were starved in DMEM containing 0.5% FBS for 24 h, treated with control or Shh-N-conditioned medium, and harvested at the desired time points.
Co-immunoprecipitation, western blots, luciferase assay and immunofluorescence
Antibodies used were: anti-Rusc2 (AP12095a, Abgent, 1:500), anti-myc (5546, Sigma-Aldrich, 1:1000), anti-FLAG (F1804, Sigma-Aldrich, 1:1000), anti-HA (H9658, Sigma-Aldrich, 1:1000), anti-Sufu (sc-28847 and sc-10934, Santa Cruz, 1:200), anti-Gli3 (AF3690, R&D Systems, 1:500), anti-acetylated tubulin (T7451, Sigma-Aldrich, 1:500), anti-γ-tubulin (T6557, Sigma-Aldrich, 1:200) and anti-β-tubulin (T5293, Sigma-Aldrich, 1:1000).
Protocols for CoIP, western blot (Jin et al., 2009) and dual-luciferase reporter assay (Jin et al., 2011) were described previously. For the luciferase assay, each sample comprised three replicates. Statistical significance was determined using Student's t-test. Results are presented as mean±s.d. All experiments were performed at least three times. Immunostaining and Triton X-100 extraction experiments were carried out as previously described (Wulfkuhle et al., 1999). Prior to fixation, cells were treated with Triton X-100 extraction buffer (0.5% Triton X-100, 50 mM NaCl, 3 mM MgCl2, 30 mM sucrose, 10 mM Pipes pH 6.8) for 3 min at 4°C. After fixation, cells were stained following the standard immunostaining procedure.
RNA extraction and RT-PCR
RNA purification and reverse transcription were performed as described (Rorick et al., 2007). RT-PCR reactions were performed in triplicate using SYBR Green Master Mix (Applied Biosystems) on an Applied Biosystems 7500 real-time PCR system. Values were normalized to the control. Statistical significance was determined by Student's t-test. Results are presented as mean±s.d. Primers (5′-3′; forward and reverse) are: mouse Gli1, TCCCTGGTGGCTTTCATCAACT and GCATCATTGAACCCCGAGTAGA; mouse Ptc1, GAGGCTATGTTTAATCCTCAACTC and CTATTATCTGATCCATGTAACCTG; mouse Actb (control), AGAGGGAAATCGTGCGTGAC and CAATAGTGATGACCTGGCCGT; Xenopus gli1, AAGCTTCCTCACACTTGACC and GCTCTGCGCCATAGATAATC; Xenopus ptc1, GGACAAGAATCGCAGAGCTG and GGATGCTCAGGGAACCTTAC; Xenopus ptc2, CCAGCTCGGATCTACTGAGG and CAGTGTCTCTGGATGGAGCA; Xenopus hhip, GTTGGTGCAATGCATAGTGG and TCTTGGTTGGTGGTGTACGA; Xenopus odc (control), GCCATTGTGAAGACTCTCTCCATTC and TTCGGGTGCTTCCTTGCCAC; Xenopus rusc1, GGTCTGTTGGTTGCGATTGG and ACAGGCGGCCGATGTTACAC; Xenopus rusc2, GACCCCCTTTTCATCTCTTGC and GTGAGATCTCTTAGAAGTTGGGC.
Xenopus embryos and manipulations
Xenopus embryos were obtained as described (Sive et al., 2000). Morpholino antisense oligos (5′-3′) are: R1-MO, GGTGTCAGTCGTCAGTTACAGCCCC; R1-5mis, GcTGTCAcTCGTCAcTTACAcCCgC (lowercase letters indicate the mismatches); R1-sb, ATACAGAGAGTCACTTACCTGCCCT; R2-MO1, GCTATCCATCATCAGTGGCTTCTTC; R2-MO2, GGACATTGGTAAATCAGCAAGAGAT. Morpholino against Gli1 was described previously (Schwend et al., 2013). Microinjection, animal cap assays and in situ hybridization were performed as described (Sive et al., 2000). All procedures involving Xenopus were performed in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals and approved by the University of Illinois at Urbana-Champaign Institutional Animal Care and Use Committee.
Acknowledgements
We thank Drs A. Barnekow and A. Liu for expression constructs and cell lines; Dr J. P. Saint-Jeannet for greatly appreciated suggestions; and Drs P. Klein and I. Bagchi for reading the manuscript.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceived and designed the experiments: Z.J., H.Z. and J.Y. Performed the experiments: Z.J., T.S., J.F., Z.B., J.L., W.M. and J.Y. Analyzed the data: Z.J. and J.Y. Wrote the paper: J.Y.
Funding
T.S. was supported by a National Institutes of Health fellowship [F32EY021708]. J.Y. is supported in part by National Institutes of Health grants [R01GM093217 and R01GM111816]. Deposited in PMC for release after 12 months.
Data availability
The GenBank accessions for Xenopus rusc1 and rusc2 are KX265097 and KX265098, respectively.
Supplementary information
Supplementary information available online at http://dev.biologists.org/lookup/doi/10.1242/dev.138917.supplemental
References
- Aavikko M., Li S.-P., Saarinen S., Alhopuro P., Kaasinen E., Morgunova E., Li Y., Vesanen K., Smith M. J., Evans D. G. R. et al. (2012). Loss of SUFU function in familial multiple meningioma. Am. J. Hum. Genet. 91, 520-526. 10.1016/j.ajhg.2012.07.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amato M. A., Boy S. and Perron M. (2004). Hedgehog signaling in vertebrate eye development: a growing puzzle. Cell. Mol. Life Sci. 61, 899-910. 10.1007/s00018-003-3370-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayer M., Fischer J., Kremerskothen J., Ossendorf E., Matanis T., Konczal M., Weide T. and Barnekow A. (2005). Identification and characterization of Iporin as a novel interaction partner for rab1. BMC Cell Biol. 6, 15 10.1186/1471-2121-6-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehlke C., Bashkurov M., Buescher A., Krick T., John A.-K., Nitschke R., Walz G. and Kuehn E. W. (2010). Differential role of Rab proteins in ciliary trafficking: Rab23 regulates smoothened levels. J. Cell Sci. 123, 1460-1467. 10.1242/jcs.058883 [DOI] [PubMed] [Google Scholar]
- Briscoe J. (2009). Making a grade: Sonic Hedgehog signalling and the control of neural cell fate. EMBO J. 28, 457-465. 10.1038/emboj.2009.12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briscoe J. and Small S. (2015). Morphogen rules: design principles of gradient-mediated embryo patterning. Development 142, 3996-4009. 10.1242/dev.129452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briscoe J. and Thérond P. P. (2013). The mechanisms of Hedgehog signalling and its roles in development and disease. Nat. Rev. Mol. Cell Biol. 14, 416-429. 10.1038/nrm3598 [DOI] [PubMed] [Google Scholar]
- Brugieres L., Pierron G., Chompret A., Paillerets B. B.-d., Di Rocco F., Varlet P., Pierre-Kahn A., Caron O., Grill J. and Delattre O. (2010). Incomplete penetrance of the predisposition to medulloblastoma associated with germ-line SUFU mutations. J. Med. Genet. 47, 142-144. 10.1136/jmg.2009.067751 [DOI] [PubMed] [Google Scholar]
- Chen M.-H., Wilson C. W., Li Y.-J., Law K. K. L., Lu C.-S., Gacayan R., Zhang X., Hui C. C. and Chuang P.-T. (2009). Cilium-independent regulation of Gli protein function by Sufu in Hedgehog signaling is evolutionarily conserved. Genes Dev. 23, 1910-1928. 10.1101/gad.1794109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung H. O.-L., Zhang X., Ribeiro A., Mo R., Makino S., Puviindran V., Law K. K. L., Briscoe J. and Hui C.-c. (2009). The kinesin protein Kif7 is a critical regulator of Gli transcription factors in mammalian hedgehog signaling. Sci. Signal. 2, ra29 10.1126/scisignal.2000405 [DOI] [PubMed] [Google Scholar]
- Chi S., Xie G., Liu H., Chen K., Zhang X., Li C. and Xie J. (2012). Rab23 negatively regulates Gli1 transcriptional factor in a Su(Fu)-dependent manner. Cell. Signal. 24, 1222-1228. 10.1016/j.cellsig.2012.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper A. F., Yu K. P., Brueckner M., Brailey L. L., Johnson L., McGrath J. M. and Bale A. E. (2005). Cardiac and CNS defects in a mouse with targeted disruption of suppressor of fused. Development 132, 4407-4417. 10.1242/dev.02021 [DOI] [PubMed] [Google Scholar]
- Ding Q., Fukami S.-i., Meng X., Nishizaki Y., Zhang X., Sasaki H., Dlugosz A., Nakafuku M. and Hui C.-c (1999). Mouse suppressor of fused is a negative regulator of sonic hedgehog signaling and alters the subcellular distribution of Gli1. Curr. Biol. 9, 1119-1122. 10.1016/S0960-9822(99)80482-5 [DOI] [PubMed] [Google Scholar]
- Eggenschwiler J. T., Bulgakov O. V., Qin J., Li T. and Anderson K. V. (2006). Mouse Rab23 regulates hedgehog signaling from smoothened to Gli proteins. Dev. Biol. 290, 1-12. 10.1016/j.ydbio.2005.09.022 [DOI] [PubMed] [Google Scholar]
- Eggenschwiler J. T., Espinoza E. and Anderson K. V. (2001). Rab23 is an essential negative regulator of the mouse Sonic hedgehog signalling pathway. Nature 412, 194-198. 10.1038/35084089 [DOI] [PubMed] [Google Scholar]
- Endoh-Yamagami S., Evangelista M., Wilson D., Wen X., Theunissen J.-W., Phamluong K., Davis M., Scales S. J., Solloway M. J., de Sauvage F. J. et al. (2009). The mammalian Cos2 homolog Kif7 plays an essential role in modulating Hh signal transduction during development. Curr. Biol. 19, 1320-1326. 10.1016/j.cub.2009.06.046 [DOI] [PubMed] [Google Scholar]
- Evans T. M., Ferguson C., Wainwright B. J., Parton R. G. and Wicking C. (2003). Rab23, a negative regulator of hedgehog signaling, localizes to the plasma membrane and the endocytic pathway. Traffic 4, 869-884. 10.1046/j.1600-0854.2003.00141.x [DOI] [PubMed] [Google Scholar]
- Fukuda M., Kobayashi H., Ishibashi K. and Ohbayashi N. (2011). Genome-wide investigation of the Rab binding activity of RUN domains: development of a novel tool that specifically traps GTP-Rab35. Cell Struct. Funct. 36, 155-170. 10.1247/csf.11001 [DOI] [PubMed] [Google Scholar]
- Han Y., Shi Q. and Jiang J. (2015). Multisite interaction with Sufu regulates Ci/Gli activity through distinct mechanisms in Hh signal transduction. Proc. Natl. Acad. Sci. USA 112, 6383-6388. 10.1073/pnas.1421628112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He M., Subramanian R., Bangs F., Omelchenko T., Liem K. F. Jr, Kapoor T. M. and Anderson K. V. (2014). The kinesin-4 protein Kif7 regulates mammalian Hedgehog signalling by organizing the cilium tip compartment. Nat. Cell Biol. 16, 663-672. 10.1038/ncb2988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huangfu D. and Anderson K. V. (2006). Signaling from Smo to Ci/Gli: conservation and divergence of Hedgehog pathways from Drosophila to vertebrates. Development 133, 3-14. 10.1242/dev.02169 [DOI] [PubMed] [Google Scholar]
- Hui C.-C. and Angers S. (2011). Gli proteins in development and disease. Annu. Rev. Cell Dev. Biol. 27, 513-537. 10.1146/annurev-cellbio-092910-154048 [DOI] [PubMed] [Google Scholar]
- Humke E. W., Dorn K. V., Milenkovic L., Scott M. P. and Rohatgi R. (2010). The output of Hedgehog signaling is controlled by the dynamic association between Suppressor of Fused and the Gli proteins. Genes Dev. 24, 670-682. 10.1101/gad.1902910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia J. and Jiang J. (2006). Decoding the Hedgehog signal in animal development. Cell. Mol. Life Sci. 63, 1249-1265. 10.1007/s00018-005-5519-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia J., Kolterud Å., Zeng H., Hoover A., Teglund S., Toftgård R. and Liu A. (2009). Suppressor of Fused inhibits mammalian Hedgehog signaling in the absence of cilia. Dev. Biol. 330, 452-460. 10.1016/j.ydbio.2009.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J. and Hui C.-c. (2008). Hedgehog signaling in development and cancer. Dev. Cell 15, 801-812. 10.1016/j.devcel.2008.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Z., Shi J., Saraf A., Mei W., Zhu G.-Z., Strack S. and Yang J. (2009). The 48-kDa alternative translation isoform of PP2A:B56epsilon is required for Wnt signaling during midbrain-hindbrain boundary formation. J. Biol. Chem. 284, 7190-7200. 10.1074/jbc.M807907200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Z., Wallace L., Harper S. Q. and Yang J. (2010). PP2A:B56{epsilon}, a substrate of caspase-3, regulates p53-dependent and p53-independent apoptosis during development. J. Biol. Chem. 285, 34493-34502. 10.1074/jbc.M110.169581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Z., Mei W., Strack S., Jia J. and Yang J. (2011). The antagonistic action of B56-containing protein phosphatase 2As and casein kinase 2 controls the phosphorylation and Gli turnover function of Daz interacting protein 1. J. Biol. Chem. 286, 36171-36179. 10.1074/jbc.M111.274761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kijima C., Miyashita T., Suzuki M., Oka H. and Fujii K. (2012). Two cases of nevoid basal cell carcinoma syndrome associated with meningioma caused by a PTCH1 or SUFU germline mutation. Fam. Cancer 11, 565-570. 10.1007/s10689-012-9548-0 [DOI] [PubMed] [Google Scholar]
- Kogerman P., Grimm T., Kogerman L., Krause D., Undén A. B., Sandstedt B., Toftgård R. and Zaphiropoulos P. G. (1999). Mammalian suppressor-of-fused modulates nuclear-cytoplasmic shuttling of Gli-1. Nat. Cell Biol. 1, 312-319. 10.1038/13031 [DOI] [PubMed] [Google Scholar]
- Koide T., Hayata T. and Cho K. W. Y. (2006). Negative regulation of Hedgehog signaling by the cholesterogenic enzyme 7-dehydrocholesterol reductase. Development 133, 2395-2405. 10.1242/dev.02393 [DOI] [PubMed] [Google Scholar]
- Law K. K. L., Makino S., Mo R., Zhang X., Puviindran V. and Hui C.-c. (2012). Antagonistic and cooperative actions of Kif7 and Sufu define graded intracellular Gli activities in Hedgehog signaling. PLoS ONE 7, e50193 10.1371/journal.pone.0050193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J., Platt K. A., Censullo P. and Ruiz i Altaba A. (1997). Gli1 is a target of Sonic hedgehog that induces ventral neural tube development. Development 124, 2537-2552. [DOI] [PubMed] [Google Scholar]
- Li N., Volff J.-N. and Wizenmann A. (2007). Rab23 GTPase is expressed asymmetrically in Hensen's node and plays a role in the dorsoventral patterning of the chick neural tube. Dev. Dyn. 236, 2993-3006. 10.1002/dvdy.21331 [DOI] [PubMed] [Google Scholar]
- Li Z. J., Nieuwenhuis E., Nien W., Zhang X., Zhang J., Puviindran V., Wainwright B. J., Kim P. C. W. and Hui C.-c. (2012). Kif7 regulates Gli2 through Sufu-dependent and -independent functions during skin development and tumorigenesis. Development 139, 4152-4161. 10.1242/dev.081190 [DOI] [PubMed] [Google Scholar]
- Liem K. F. Jr., He M., Ocbina P. J. and Anderson K. V. (2009). Mouse Kif7/Costal2 is a cilia-associated protein that regulates Sonic hedgehog signaling. Proc. Natl. Acad. Sci. USA 106, 13377-13382. 10.1073/pnas.0906944106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C., Yao E., Wang K., Nozawa Y., Shimizu H., Johnson J. R., Chen J.-N., Krogan N. J. and Chuang P.-T. (2014). Regulation of Sufu activity by p66beta and Mycbp provides new insight into vertebrate Hedgehog signaling. Genes Dev. 28, 2547-2563. 10.1101/gad.249425.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Heydeck W., Zeng H. and Liu A. (2012). Dual function of suppressor of fused in Hh pathway activation and mouse spinal cord patterning. Dev. Biol. 362, 141-153. 10.1016/j.ydbio.2011.11.022 [DOI] [PubMed] [Google Scholar]
- Lupo G., Harris W. A. and Lewis K. E. (2006). Mechanisms of ventral patterning in the vertebrate nervous system. Nat. Rev. Neurosci. 7, 103-114. 10.1038/nrn1843 [DOI] [PubMed] [Google Scholar]
- MacDonald J. I. S., Dietrich A., Gamble S., Hryciw T., Grant R. I. and Meakin S. O. (2012). Nesca, a novel neuronal adapter protein, links the molecular motor kinesin with the pre-synaptic membrane protein, syntaxin-1, in hippocampal neurons. J. Neurochem. 121, 861-880. 10.1111/j.1471-4159.2012.07729.x [DOI] [PubMed] [Google Scholar]
- Maurya A. K., Ben J., Zhao Z., Lee R. T. H., Niah W., Ng A. S., Iyu A., Yu W., Elworthy S., van Eeden F. J. M. et al. (2013). Positive and negative regulation of Gli activity by Kif7 in the zebrafish embryo. PLoS Genet. 9, e1003955 10.1371/journal.pgen.1003955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min T. H., Kriebel M., Hou S. and Pera E. M. (2011). The dual regulator Sufu integrates Hedgehog and Wnt signals in the early Xenopus embryo. Dev. Biol. 358, 262-276. 10.1016/j.ydbio.2011.07.035 [DOI] [PubMed] [Google Scholar]
- Moody S. A. (1987). Fates of the blastomeres of the 16-cell stage Xenopus embryo. Dev. Biol. 119, 560-578. 10.1016/0012-1606(87)90059-5 [DOI] [PubMed] [Google Scholar]
- Moody S. A. (2012). Testing retina fate commitment in Xenopus by blastomere deletion, transplantation, and explant culture. Methods Mol. Biol. 884, 115-127. 10.1007/978-1-61779-848-1_7 [DOI] [PubMed] [Google Scholar]
- Murcia N. S., Richards W. G., Yoder B. K., Mucenski M. L., Dunlap J. R. and Woychik R. P. (2000). The Oak Ridge Polycystic Kidney (orpk) disease gene is required for left-right axis determination. Development 127, 2347-2355. [DOI] [PubMed] [Google Scholar]
- Murone M., Luoh S.-M., Stone D., Li W., Gurney A., Armanini M., Grey C., Rosenthal A. and de Sauvage F. J. (2000). Gli regulation by the opposing activities of fused and suppressor of fused. Nat. Cell Biol. 2, 310-312. 10.1038/35010610 [DOI] [PubMed] [Google Scholar]
- Mussolino C., Morbitzer R., Lutge F., Dannemann N., Lahaye T. and Cathomen T. (2011). A novel TALE nuclease scaffold enables high genome editing activity in combination with low toxicity. Nucleic Acids Res. 39, 9283-9293. 10.1093/nar/gkr597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen V., Chokas A. L., Stecca B. and Ruiz i Altaba A. (2005). Cooperative requirement of the Gli proteins in neurogenesis. Development 132, 3267-3279. 10.1242/dev.01905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastorino L., Ghiorzo P., Nasti S., Battistuzzi L., Cusano R., Marzocchi C., Garrè M. L., Clementi M. and Scarrà G. B. (2009). Identification of a SUFU germline mutation in a family with Gorlin syndrome. Am. J. Med. Genet. A 149A, 1539-1543. 10.1002/ajmg.a.32944 [DOI] [PubMed] [Google Scholar]
- Pearse R. V. II, Collier L. S., Scott M. P. and Tabin C. J. (1999). Vertebrate homologs of Drosophila suppressor of fused interact with the gli family of transcriptional regulators. Dev. Biol. 212, 323-336. 10.1006/dbio.1999.9335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrova R. and Joyner A. L. (2014). Roles for Hedgehog signaling in adult organ homeostasis and repair. Development 141, 3445-3457. 10.1242/dev.083691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Préat T. (1992). Characterization of Suppressor of fused, a complete suppressor of the fused segment polarity gene of Drosophila melanogaster. Genetics 132, 725-736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rorick A. M., Mei W., Liette N. L., Phiel C., El-Hodiri H. M. and Yang J. (2007). PP2A:B56epsilon is required for eye induction and eye field separation. Dev. Biol. 302, 477-493. 10.1016/j.ydbio.2006.10.011 [DOI] [PubMed] [Google Scholar]
- Sasaki H., Hui C., Nakafuku M. and Kondoh H. (1997). A binding site for Gli proteins is essential for HNF-3beta floor plate enhancer activity in transgenics and can respond to Shh in vitro. Development 124, 1313-1322. [DOI] [PubMed] [Google Scholar]
- Schulman J. M., Oh D. H., Sanborn J. Z., Pincus L., McCalmont T. H. and Cho R. J. (2015). Multiple hereditary infundibulocystic basal cell carcinoma syndrome associated with a germline SUFU mutation. JAMA Dermatol. 152, 323-327. 10.1001/jamadermatol.2015.4233 [DOI] [PubMed] [Google Scholar]
- Schwend T., Jin Z., Jiang K., Mitchell B. J., Jia J. and Yang J. (2013). Stabilization of speckle-type POZ protein (Spop) by Daz interacting protein 1 (Dzip1) is essential for Gli turnover and the proper output of Hedgehog signaling. J. Biol. Chem. 288, 32809-32820. 10.1074/jbc.M113.512962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sive H., Grainger R. and Harland R. (2000). Early Development of Xenopus laevis; A Laboratory Manual. Cold Spring Harbor: Cold Spring Harbor Press. [Google Scholar]
- Stone D. M., Murone M., Luoh S., Ye W., Armanini M. P., Gurney A., Phillips H., Brush J., Goddard A., de Sauvage F. J. et al. (1999). Characterization of the human suppressor of fused, a negative regulator of the zinc-finger transcription factor Gli. J. Cell Sci. 112, 4437-4448. [DOI] [PubMed] [Google Scholar]
- Sun Q., Han C., Liu L., Wang Y., Deng H., Bai L. and Jiang T. (2012). Crystal structure and functional implication of the RUN domain of human NESCA. Protein Cell 3, 609-617. 10.1007/s13238-012-2052-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svärd J., Heby-Henricson K., Persson-Lek M., Rozell B., Lauth M., Bergström Å., Ericson J., Toftgård R. and Teglund S. (2006). Genetic elimination of Suppressor of fused reveals an essential repressor function in the mammalian Hedgehog signaling pathway. Dev. Cell 10, 187-197. 10.1016/j.devcel.2005.12.013 [DOI] [PubMed] [Google Scholar]
- Tay S. Y., Ingham P. W. and Roy S. (2005). A homologue of the Drosophila kinesin-like protein Costal2 regulates Hedgehog signal transduction in the vertebrate embryo. Development 132, 625-634. 10.1242/dev.01606 [DOI] [PubMed] [Google Scholar]
- Taylor M. D., Liu L., Raffel C., Hui C.-c., Mainprize T. G., Zhang X., Agatep R., Chiappa S., Gao L., Lowrance A. et al. (2002). Mutations in SUFU predispose to medulloblastoma. Nat. Genet. 31, 306-310. 10.1038/ng916 [DOI] [PubMed] [Google Scholar]
- Tukachinsky H., Lopez L. V. and Salic A. (2010). A mechanism for vertebrate Hedgehog signaling: recruitment to cilia and dissociation of SuFu-Gli protein complexes. J. Cell Biol. 191, 415-428. 10.1083/jcb.201004108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Pan Y. and Wang B. (2010). Suppressor of fused and Spop regulate the stability, processing and function of Gli2 and Gli3 full-length activators but not their repressors. Development 137, 2001-2009. 10.1242/dev.052126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson C. W. and Chuang P.-T. (2010). Mechanism and evolution of cytosolic Hedgehog signal transduction. Development 137, 2079-2094. 10.1242/dev.045021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff C., Roy S. and Ingham P. W. (2003). Multiple muscle cell identities induced by distinct levels and timing of hedgehog activity in the zebrafish embryo. Curr. Biol. 13, 1169-1181. 10.1016/S0960-9822(03)00461-5 [DOI] [PubMed] [Google Scholar]
- Wulfkuhle J. D., Donina I. E., Stark N. H., Pope R. K., Pestonjamasp K. N., Niswonger M. L. and Luna E. J. (1999). Domain analysis of supervillin, an F-actin bundling plasma membrane protein with functional nuclear localization signals. J. Cell Sci. 112, 2125-2136. [DOI] [PubMed] [Google Scholar]
- Zeng H., Jia J. and Liu A. (2010). Coordinated translocation of mammalian Gli proteins and suppressor of fused to the primary cilium. PLoS ONE 5, e15900 10.1371/journal.pone.0015900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Zhang L., Wang B., Ou C.-Y., Chien C.-T. and Jiang J. (2006). A hedgehog-induced BTB protein modulates hedgehog signaling by degrading Ci/Gli transcription factor. Dev. Cell 10, 719-729. 10.1016/j.devcel.2006.05.004 [DOI] [PubMed] [Google Scholar]
- Zhang Q., Shi Q., Chen Y., Yue T., Li S., Wang B. and Jiang J. (2009). Multiple Ser/Thr-rich degrons mediate the degradation of Ci/Gli by the Cul3-HIB/SPOP E3 ubiquitin ligase. Proc. Natl. Acad. Sci. USA 106, 21191-21196. 10.1073/pnas.0912008106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Fu L., Qi X., Zhang Z., Xia Y., Jia J., Jiang J., Zhao Y. and Wu G. (2013). Structural insight into the mutual recognition and regulation between Suppressor of Fused and Gli/Ci. Nat. Commun. 4, 2608 10.1038/ncomms3608 [DOI] [PMC free article] [PubMed] [Google Scholar]