Abstract
A limited number of signaling pathways are repeatedly used to regulate a wide variety of processes during development and differentiation. The lack of tools to manipulate signaling pathways dynamically in space and time has been a major technical challenge for biologists. Optogenetic techniques, which utilize light to control protein functions in a reversible fashion, hold promise for modulating intracellular signaling networks with high spatial and temporal resolution. Applications of optogenetics in multicellular organisms, however, have not been widely reported. Here, we create an optimized bicistronic optogenetic system using Arabidopsis thaliana cryptochrome 2 (CRY2) protein and the N-terminal domain of cryptochrome-interacting basic-helix-loop-helix (CIBN). In a proof-of-principle study, we develop an optogenetic Raf kinase that allows reversible light-controlled activation of the Raf/MEK/ERK signaling cascade. In PC12 cells, this system significantly improves light-induced cell differentiation compared with co-transfection. When applied to Xenopus embryos, this system enables blue light-dependent reversible Raf activation at any desired developmental stage in specific cell lineages. Our system offers a powerful optogenetic tool suitable for manipulation of signaling pathways with high spatial and temporal resolution in a wide range of experimental settings.
KEY WORDS: Optogenetics, Xenopus, Cryptochrome, Bicistronic gene expression, Differentiation, Light-controlled protein-protein interaction
Summary: Light is used to modulate the timing of mitogen-activated protein kinase activity during PC12 cell differentiation and distinct developmental stages of Xenopus embryos.
INTRODUCTION
After several decades of intensive studies, it is well established that the majority of events during metazoan development and differentiation are regulated by cooperation of only a handful of signal transduction pathways. These signaling pathways are turned on and off precisely at the right time and in the right place to control cell proliferation, migration, cell-fate determination, differentiation and apoptosis. Effective manipulation of signaling pathways with high spatiotemporal resolution is crucial for many studies. Unfortunately, the commonly used pharmacological and genetic approaches often cannot capture the dynamic aspects of signal transduction owing to a lack of spatial and temporal control. There is an urgent need to develop new tools suitable for modulating signaling pathways at any desired period of time in any desired tissues.
The emerging optogenetic approach has enabled the development of new strategies to probe the spatial and kinetic features of signal transduction in live cells. By using a group of photoactivatable proteins, which undergo conformational changes and interact with each other upon excitation by light at specific wavelengths, optogenetics has extended its modality into interrogating complex intracellular signaling networks (Kennedy et al., 2010; Levskaya et al., 2009; Wu et al., 2009; Yazawa et al., 2009; Zhou et al., 2012). Shortly after the report of light-gated ion channel-based neuronal firing control (Boyden et al., 2005), optogenetics has been successfully used to control a variety of biological events in cell culture and single-cell organisms (reviewed by Kim and Lin, 2013; Muller et al., 2015; Schmidt and Cho, 2015; Tischer and Weiner, 2014; Toettcher et al., 2011; Tucker, 2012; Zhang and Cui, 2015; Zoltowski and Gardner, 2011). More recently, several studies have tested its application in multicellular organisms (Hallett et al., 2016).
To date, several photoactivatable proteins (Levskaya et al., 2009; Shimizu-Sato et al., 2002; Strickland et al., 2012; Wu et al., 2009; Yazawa et al., 2009) have been successfully applied in vivo. Among these is cryptochrome from Arabidopsis thaliana (CRY2) (Kennedy et al., 2010), which undergoes homo-oligomerization (Más et al., 2000) or heterodimerization with the cryptochrome-interacting basic-helix-loop-helix (CIB1) (Liu et al., 2008) in response to blue-light stimulation. It was later found that the photolyase homology region of cryptochrome 2 (CRY2PHR, abbreviated as CRY2 in this work) and the N-terminal domain of CIB1 (CIBN, 170 aa) could maintain the light-mediated interaction (Kennedy et al., 2010). Interestingly, CRY2-CIBN binding out-competes CRY2 oligomerization under the same light-activating conditions (Che et al., 2015). Both CRY2-CIBN heterodimerization (Boulina et al., 2013; Hughes et al., 2012; Idevall-Hagren et al., 2012; Kakumoto and Nakata, 2013; Kennedy et al., 2010; Konermann et al., 2013; Lee et al., 2014; Liu et al., 2012; Zhang et al., 2014) and CRY2 homo-oligomerization (Bugaj et al., 2013; Chang et al., 2014; Taslimi et al., 2014; Wend et al., 2014) have been used for optogenetic control of signal transduction. It appears that CRY2-CIBN-induced protein dimerization mimics the native interaction between the two proteins better (Zhang and Cui, 2014). So far, the CRY2 system has been used to control transcription in Drosophila (Boulina et al., 2013), zebrafish (Liu et al., 2012) and mouse cortex (Konermann et al., 2013).
Despite its advantages, the CRY2-CIBN system has a very important practical limitation – the lack of control over the ratiometric expression of CRY2- and CIBN-fusion proteins. Like any other genetically encoded heterodimerization system, when the CRY2-CIBN system is expressed in the cell, the expression level of the smaller protein is almost always higher than that of the larger one, probably owing to bias in gene delivery, gene transcription, and translation. Such biased protein expression decreases the efficiency of the system and often complicates the interpretation of light-induced phenotypes. Moreover, it is unclear whether an equal-molar expression of CIBN- and CRY2-fusion proteins leads to optimal optogenetic readout. Therefore, a system that allows ratiometric expression of CIBN- and CRY2-fusion proteins would be valuable in designing optimized optogenetic system.
The 2A peptides, which function through a ribosomal skipping mechanism, have been used for stoichiometric expression of cistrons in multicistronic constructs. Positioning the 2A peptide sequence between two cistrons prevents peptide bond formation between Gly and Pro of the consensus motif Asp-Val/Ile-Glu-X-Asn-Pro-Gly-Pro during translation, allowing the ribosome to translate the downstream cistron separately from the upstream one (Donnelly et al., 2001b). Compared with internal ribosome entry sites (IRESs), which often result in higher expression (up to threefold) of the upstream cistron compared with the downstream one (Goedhart et al., 2011; Ibrahimi et al., 2009), the 2A peptides enable much better stoichiometric expression of the cistrons. Among commonly used 2A peptides (Donnelly et al., 2001a), the porcine teschovirum-1 2A (P2A) shows the highest ribosome-skipping efficiency in mammalian cell lines, zebrafish embryos, and mouse liver (Kim et al., 2011). Here, we describe a novel P2A peptide-based bicistronic system for stoichiometric expression of CRY2- and CIBN-fusion proteins. We developed an optimal optogenetic Raf, which efficiently translocated to the plasma membrane as a consequence of light-mediated CRY2-CIBN association and elicited PC12 cell differentiation via activation of the Raf/MEK/ERK signaling cascade. Compared with the conventional co-transfection setting, this optimized bicistronic system works much more efficiently in inducing neuronal differentiation of PC12 cells. For the first time, we successfully applied this system in live Xenopus embryos, and were able to activate the Raf kinase activity in a reversible fashion at any desired time in specific cell lineages. We expect that this optimized optogenetic system can be applied to a wide range of experimental settings for manipulation of signaling pathways with high spatial and temporal resolution.
RESULTS
PC12 cells co-transfected with CRY2-mCherry-Raf1 and CIBN-GFP-CaaX show inhomogeneous light-induced cell differentiation
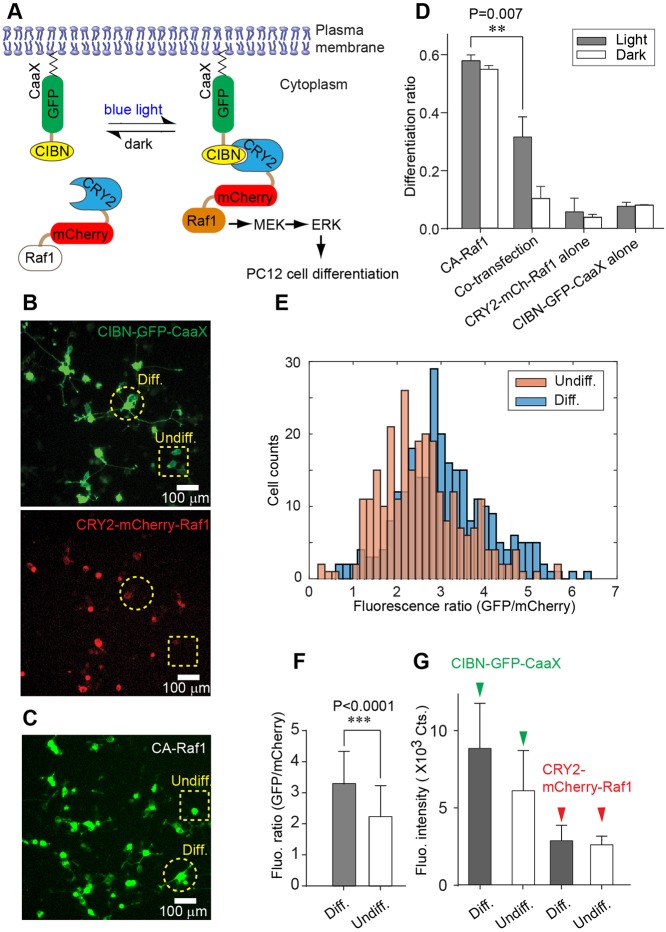
Similar to our previous report (Zhang et al., 2014), we found that blue-light stimulation induced significant neurite outgrowth in PC12 cells co-transfected with CRY2-mCherry-Raf1 and CIBN-GFP-CaaX (Fig. 1A). To assess the differentiation ratio, i.e. the number of differentiated cells over the number of transfected cells, we stimulated CRY2-mCherry-Raf1 and CIBN-GFP-CaaX co-transfected cells and cells transfected with a constitutively active Raf1 (Raf1-GFP-CaaX, referred to as CA-Raf1) with blue light for 24 h (Fig. 1B,C). The differentiation ratio of co-transfected cells is significantly lower than that of CA-Raf1-transfected cells (0.32 vs 0.54) (Fig. 1D). The suboptimal differentiation ratio in co-transfected cultures may arise from biased protein expression of CRY2- and CIBN-fusion proteins. In support of this view, a single-cell analysis in PC12 cells co-transfected with CIBN-GFP-CaaX and CRY2-mCherry-Raf1 showed that differentiated cells have a higher GFP/mCherry fluorescence intensity ratio than non-differentiated cells (Fig. 1E,F). In differentiated cells, the GFP/mCherry fluorescence ratio peaks at around 3, whereas higher or lower ratios lead to less PC12 cell differentiation (Fig. 1E, blue bars). This result (Fig. 1E) suggests that an optimal protein expression ratio of CIBN-GFP-CaaX over CRY2-mCherry-Raf1 should exist for maximum PC12 cell differentiation. As the average expression level of CRY2-mCherry-Raf1 was similar in both differentiated and non-differentiated cells (Fig. 1G, red arrowheads), the higher GFP/mCherry ratio in differentiated cells should arise from elevated CIBN-GFP-CaaX expression (Fig. 1G, green arrowheads). To find out the optimal protein expression level, we used the P2A peptide to construct bicistronic systems that allow tunable ratiometric expression of CIBN- and CRY2-fusion proteins.
Fig. 1.
Correlation between the differentiation ratio and the fluorescence intensity ratio (GFP/mCherry) in PC12 cells co-transfected with CIBN-GFP-CaaX and CRY2-mCherry-Raf1. (A) Light-induced binding between CIBN and CRY2 leads to membrane recruitment of CRY2-mCherry-Raf1, which activates the Raf/MEK/ERK signaling pathway and induces PC12 cell differentiation. (B) Representative fluorescence images of PC12 cells co-transfected with CIBN-GFP-CaaX (green) and CRY2-mCherry-Raf1 (red). A differentiated cell is marked with a dashed circle; an undifferentiated cell is marked with a dashed square. Differentiated cells are defined as those with at least one neurite of length equal to or longer than the diameter of their cell bodies. (C) A representative image of cells transfected with Raf1-GFP-CaaX, a constitutively active Raf1 (CA-Raf1). A differentiated cell is marked with a dashed circle; an undifferentiated cell is marked with a dashed square. (D) Differentiation ratios of PC12 cells transfected with CA-Raf1, co-transfected with CRY2-mCherry-Raf1 and CIBN-GFP-CaaX, singly transfected with CRY2-mCherry-Raf1, or singly transfected with CIBN-GFP-CaaX. Twenty four hours after transfection, cells were either exposed to light (0.2 mW/cm2) or incubated in the dark for another 24 h. Values represent mean±s.d. from four independent data sets. (E) Histograms of fluorescence intensity ratio (GFP/mCherry) for differentiated and undifferentiated cells in co-transfected cultures. Number of cells analyzed: differentiated cells (n=430), undifferentiated cells (n=430). (F) Quantification of GFP/mCherry ratio for the data shown in E. Values represent mean±s.d. Differentiated cells show a significantly higher GFP/mCherry ratio than undifferentiated cells. P-value was determined by a two-tailed, unpaired t-test. (G) Fluorescence intensities of GFP and mCherry in differentiated and undifferentiated cells. Values represent mean±s.d. P-values (D,F) were determined by a two-tailed, unpaired t-test.
CRY2-2A-CIBN generates stoichiometric expression of CRY2 and CIBN
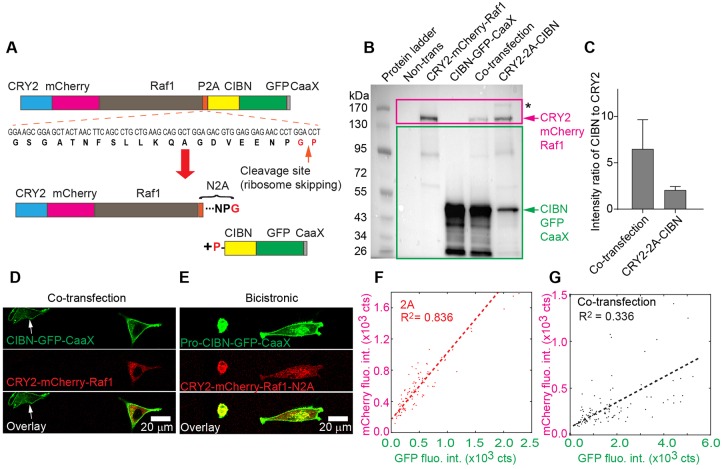
We first characterized the 2A bicistronic construct containing a CRY2-mCherry-Raf1 expression cassette, the P2A peptide and a CIBN-GFP-CaaX cassette (CRY2-mCh-Raf1-P2A-CIBN-GFP-CaaX, referred to as CRY2-2A-CIBN). Ribosomal skipping should ‘cleave’ the 2A peptide into two parts during translation, generating CRY2-mCherry-Raf1-N2A and proline-CIBN-GFP-CaaX (Fig. 2A). As a control, we transfected BHK-21 cells with an equimolar amount of CRY2-mCherry-Raf1 and CIBN-GFP-CaaX individually, or in combination. These cells expressed low levels of CRY2-mCherry-Raf1 (159 kDa), but significantly high levels of CIBN-GFP-CaaX (48 kDa) (Fig. 2B, lanes 3-5). In CRY2-2A-CIBN transfected cells, we detected both CRY2-mCherry-Raf1-N2A and proline-CIBN-GFP-CaaX proteins, confirming that this 2A-based bicistronic system did indeed undergo ribosomal skipping in mammalian cells successfully. Importantly, the ratio between CRY2-mCherry-Raf1 and CIBN-GFP-CaaX was improved significantly in CRY2-2A-CIBN-transfected cells (Fig. 2C). We also detected a faint band larger than 170 kDa, which is likely to be the residual uncleaved full-length protein (Fig. 2B, lane 6). Thus, single transfection with CRY2-2A-CIBN resulted in better stoichiometric control of CRY2- and CIBN-fusion proteins.
Fig. 2.
Characterization of the bicistronic system based on the P2A peptide. (A) Design of CRY2-2A-CIBN, a bicistronic optogenetic system. Upon ribosomal skipping, one mRNA transcript generates two proteins: CRY2-mCherry-Raf1-N2A (ending with amino acids NPG) and proline-CIBN-GFP-CaaX. (B) Western blot analysis of the CRY2-2A-CIBN system in BHK-21 cells. The blot was probed sequentially with anti-GFP (bands in the green rectangle) and anti-mCherry (bands in the pink rectangle). A composite image is shown. Lane 1: protein ladder; lane 2: non-transfection; lane 3: CRY2-mCherry-Raf1 single transfection; lane 4: CIBN-GFP-CaaX single transfection; lane 5: co-transfection of CRY2-mCherry-Raf1 and CIBN-GFP-CaaX; lane 6: CRY2-2A-CIBN single transfection. Arrows show that sizes of cleaved products from CRY2-2A-CIBN match with that of singly transfected cells. The asterisk indicates residual full-length CRY2-2A-CIBN. (C) Quantification of the intensity ratio of CIBN-GFP-CaaX to CRY2-mCherry-Raf1 on western blot. Values represent mean±s.d. (n=4). (D) Confocal fluorescence images of BHK-21 cells that were co-transfected with CRY2-mCherry-Raf1 and CIBN-GFP-CaaX. A cell expressing CIBN-GFP-CaaX, but not CRY2-mCherry-Raf1, is marked (arrow). (E) In cell cultures that were transfected with CRY2-2A-CIBN, all cells show fluorescence in both GFP and mCherry channels. (F,G) Correlation analyses between CRY2-mCherry-Raf1 and CIBN-GFP-CaaX in co-transfected and CRY2-2A-CIBN transfected cells. Fluorescence intensities between mCherry and GFP are well correlated (R2=0.836) in CRY2-2A-CIBN transfected cells (F) but are poorly correlated (R2=0.336) in co-transfected cells (G). Number of cells analyzed: CRY2-2A-CIBN (n=100), co-transfected (n=100).
To support this conclusion, we performed single-cell analyses to characterize the correlation between expression levels of CIBN-GFP-CaaX and CRY2-mCherry-Raf1, which should be proportional to the fluorescence intensities of GFP and mCherry, respectively. In co-transfected BHK-21 cell cultures, GFP and mCherry fluorescence intensities were poorly correlated. Singly transfected cells (often with CIBN-GFP-CaaX because of its smaller size) were readily detected (Fig. 2D, arrows). All cells that were transfected with CRY2-2A-CIBN, by contrast, showed fluorescence in both the GFP and mCherry channels (Fig. 2E). The variance of correlation (R2) of GFP and mCherry fluorescence intensities reveals stronger correlation in the CRY2-2A-CIBN culture (R2=0.836, n=100; Fig. 2F) than in the co-transfected culture (R2=0.336, n=100; Fig. 2G). The CIBN-GFP-CaaX fluorescence intensity spans a wide range in co-transfected cells, with a greater mean than that of CRY2-2A-CIBN-transfected cells (Fig. S1A). The mean fluorescence intensity of CRY2-mCh-Raf1, however, is lower in co-transfected cells (Fig. S1A, arrows). Such a difference in GFP and mCherry fluorescence intensities is consistent with the western blot result (Fig. 2B), suggesting that CRY2-2A-CIBN allows improved stoichiometric expression of CRY2- and CIBN-fusion proteins.
Validation of bicistronic constructs with multi-CIBN
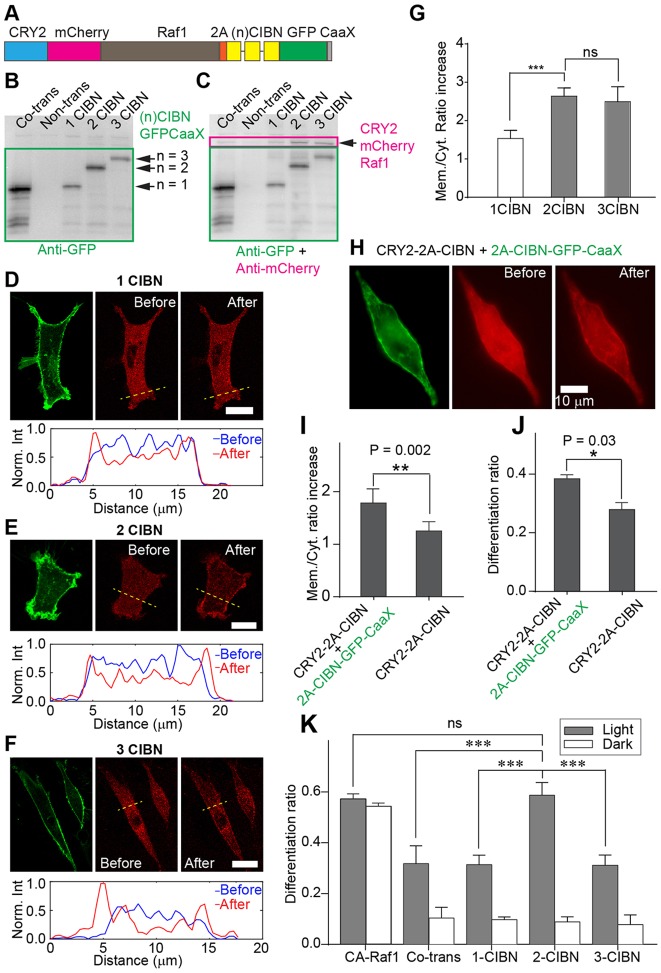
We next determined the differentiation ratio generated by the CRY2-2A-CIBN construct. Unexpectedly, the differentiation ratio showed no significant improvement compared with co-transfection. Precise control of the relative expression level of the dimerizer is crucial to successful optogenetic readout in the use of light-induced protein dimerization systems. For instance, previous work showed that successful assaying of the PIF-PhyB interaction required an appropriate expression ratio between PIF and PhyB (Toettcher et al., 2011). It is likely that the CRY2-2A-CIBN does not supply the optimal ratio between CIBN and CRY2. To determine an optimal configuration for the CRY2-CIBN system, we set out to modulate systematically the relative expression level of CIBN to CRY2 by producing a series of bicistronic constructs referred to as CRY2-2A-(n)CIBN (n=1-3). CIBN coding sequences were optimized based on the codon usage of proteins in Rattus norvegicus. GFP-CaaX was fused to the C-terminus of the last CIBN (Fig. 3A). As expected, these constructs expressed CRY2-mCherry-Raf1 and GFP-CaaX fused with the desired number of CIBNs in BHK-21 cells (Fig. 3B,C).
Fig. 3.
Characterization of bicistronic constructs with varying number of CIBNs. (A) Schematic of the CRY2-2A-(n)CIBN construct. (B) An anti-GFP western blot analysis showing the expression of (n)CIBN-GFP-CaaX in BHK-21 cells transfected with bicistronic systems. The expected sizes of (n)CIBN-GFP-CaaX proteins are 48 kDa (single-CIBN), 68 kDa (double-CIBN), and 87 kDa (triple-CIBN). The cell culture co-transfected with CRY2-mCherry-Raf1/CIBN-GFP-CaaX (lane 1) shows a band at the position of single CIBN (lane 3). (C) The same blot consequently probed with anti-mCherry without stripping. All cell cultures show a primary band that corresponds to CRY2-mCherry-Raf1 (arrow, expected size 161 kDa) in the pink rectangle. (D-F) Confocal fluorescence imaging of cells transfected with CRY2-2A-(n)CIBN. The number of CIBN is 1 (D), 2 (E) and 3 (F). In each panel, the first panel shows cleaved (n)CIBN-GFP-CaaX localized on the plasma membrane; the second panel shows a snapshot of CRY2-mCherry-Raf1 before blue-light stimulation; the third panel shows a snapshot of CRY2-mCherry-Raf1 after ten pulses of blue-light stimulation. The bottom panel shows normalized intensity profiles across the cell (marked by yellow dashed line) before and after light stimulation. Blue light-induced membrane recruitment of CRY2-mCherry-Raf1 can be observed under all three conditions but to different extents. Scale bars: 10 μm. (G) Quantification of CRY2-mCherry-Raf1 membrane translocation. The membrane/cytoplasm mCherry intensity ratios were calculated before and after light stimulation. The fold increase was then calculated by dividing the after-stimulation ratio by the before-stimulation ratio. The single-CIBN construct is less effective at inducing membrane recruitment of CRY2-mCherry-Raf1 compared with the multiple-CIBN constructs. Number of cells analyzed: 1-CIBN (n=6), 2-CIBN (n=5), 3-CIBN (n=7). Values represent mean±s.d. (H) Supplementing 2A-CIBN-GFP-CaaX to CRY2-2A-CIBN improves the membrane translocation of cytosolic CRY2-mCherry-Raf1 after blue-light stimulation. The three images are GFP fluorescence (left), and mCherry fluorescence before (middle) and after (right) ten pulses of blue-light stimulation. (I) Fold increase of the membrane/cytoplasm ratio of CRY2-mCherry-Raf1 fluorescence intensity. Supplementing additional 2A-CIBN-GFP-CaaX improves the capacity for membrane recruitment of CRY2-mCherry-Raf1. Number of cells analyzed: CRY2-2A-CIBN+2A-CIBN-GFP-CaaX (n=6), CRY2-2A-CIBN (n=7). Values represent mean±s.d. (J) Supplementing additional 2A-CIBN-GFP-CaaX increases the differentiation ratio. Values represent mean±s.d. from two independent data sets for each condition. (K) Quantification of differentiation ratios for various transfection conditions. The double-CIBN construct shows the highest differentiation ratio, double that of co-transfected constructs and reaching the limit of CA-Raf1. The single- and triple-CIBN constructs show low differentiation ratios. Number of data sets analyzed: CA-Raf1 and all light conditions (n=4), all dark conditions (n=3). Note that data for CA-Raf1 and co-transfection are the same as those in Fig. 1D. They are presented here for easier comparison between differentiation ratios from all bicistronic constructs. Values represent mean±s.d. ***P<0.001; ns, not statistically significant. P-values were determined by two-tailed, unpaired t-tests.
We then tested membrane translocation of CRY2-mCherry-Raf1 in response to blue light using confocal fluorescence microscopy (Fig. 3D-F). All 2A constructs resulted in successful membrane recruitment of cytosolic CRY2-mCherry-Raf1 upon blue-light stimulation. The extent of membrane recruitment, however, varied among different constructs. The single-CIBN 2A construct (n=1) elicited less evident membrane translocation. We could detect only a faint profile of membrane-localized CRY2-mCherry-Raf1 (Fig. 3D, right-most image). In cells transfected with double- or triple-CIBN constructs (n=2, 3), sharp profiles of membrane-localized CRY2-mCherry-Raf1 were readily observed after blue-light stimulation (Fig. 3E,F, right-most images). Statistical analysis shows that the multiple-CIBN constructs (n=2, 3) significantly increased the membrane/cytoplasm ratio of CRY2-mCherry-Raf1 in comparison with the single-CIBN construct (Fig. 3G). This result suggests that CRY2-2A-CIBN does not supply the optimal ratio between CIBN and CRY2, probably owing to a decrease in the capacity of blue light-mediated protein association. If this is the case, increasing the concentration of CIBN on the plasma membrane should improve light-mediated membrane recruitment of CRY2 fusion proteins. Indeed, membrane translocation of CRY2-mCh-Raf1 increased when we supplemented CRY2-2A-CIBN with 2A-CIBN-GFP-CaaX (Fig. 3H,I). The sequence of 2A-CIBN-GFP-CaaX is identical to that of CRY2-2A-CIBN but it lacks the CRY2-mCherry-Raf1 segment. The translational products of 2A-CIBN-GFP-CaaX are the N-terminal region of the P2A peptide and proline-CIBN-GFP-CaaX, the exact CIBN fusion protein from CRY2-2A-CIBN. Consistent with this result, light-induced PC12 cell differentiation also improved when we co-transfected CRY2-2A-CIBN with 2A-CIBN-GFP-CaaX (Fig. 3J). This result suggests that more than one copy of CIBN should be supplied for each copy of CRY2 in order to achieve optimal PC12 cell differentiation.
The double-CIBN 2A construct results in the highest differentiation ratio in PC12 cells
To determine whether the difference in membrane recruitment affects the signaling efficacy of these bicistronic constructs, we examined their activities using the PC12 cell differentiation assay. In this assay, overexpression of CA-Raf1 induced differentiation in 54% of transfected cells. Co-transfection of CRY2-mCherry-Raf1 and CIBN-GFP-CaaX worked less efficiently, with only 32% of transfected cells undergoing differentiation after a 24-h period of continuous blue-light illumination (Fig. 3K; Table S1). Consistent with the finding that the single-CIBN 2A construct was less efficient in recruiting CRY2-mCherry-Raf1 to the cell membrane upon blue-light stimulation, only about 30% of single-CIBN 2A-transfected cells underwent differentiation. Transfection of CRY2-2A-2CIBN, which allows much more efficient light-induced membrane recruitment of CRY2-mCherry-Raf1, induced differentiation very efficiently (about 60%), reaching the value generated by CA-Raf1 (Fig. 3K; Table S1). Surprisingly, CRY2-2A-3CIBN, which worked as potently as CRY2-2A-2CIBN in recruiting CRY2-mCherry-Raf1 to the cell membrane, did not further improve the differentiation ratio compared with that of the double-CIBN construct. The differentiation ratio actually dropped to approximately 30%. No significant cell differentiation occurred under dark incubation (Fig. 3K; Table S1), indicating that residual uncleaved 2A product did not cause cell differentiation. These results suggest that a CIBN:CRY2 expression ratio of 2:1 is an optimal stoichiometry for light-induced PC12 cell differentiation. Representative images of PC12 cells are shown in Fig. S2.
To explain why the 3-CIBN 2A construct is suboptimal in directing PC12 cell differentiation, we speculated that the activity of membrane-anchored Raf1 might depend on its relative position from the plasma membrane. In the case of 3-CIBN 2A, even though Raf1 can be recruited to the membrane fairly efficiently, Raf1 is likely to be located at a distance away from the plasma membrane. Because membrane association of Raf1 is required for its activation, placing Raf1 a distance away from the membrane would prevent it from being fully activated. To test this hypothesis directly, we constructed a series of membrane-anchored Raf1 by inserting one, two, or three copies of CIBN between Raf1- and GFP-CaaX. All constructs resulted in PC12 cell differentiation, although the differentiation ratio monotonically decreased as more CIBN were inserted, dropping from 60% (1-CIBN insertion) to 30% (3-CIBN insertion) (Fig. S3A). This result supports the idea that displacement of Raf1 into the cytoplasm, as may have occurred in the 3-CIBN 2A construct, reduces its capacity to induce PC12 cell differentiation. It seems likely that efficient light-induced membrane anchoring and optimal membrane association of Raf1 are both required for optogenetic activation of Raf1. Probably for this reason, the 2-CIBN configuration, which induces strong membrane association without displacing Raf1 out of the high-activity zone, is optimal for triggering PC12 cell differentiation (Fig. S3B).
Reversible activation of ERK in Xenopus embryonic tissues
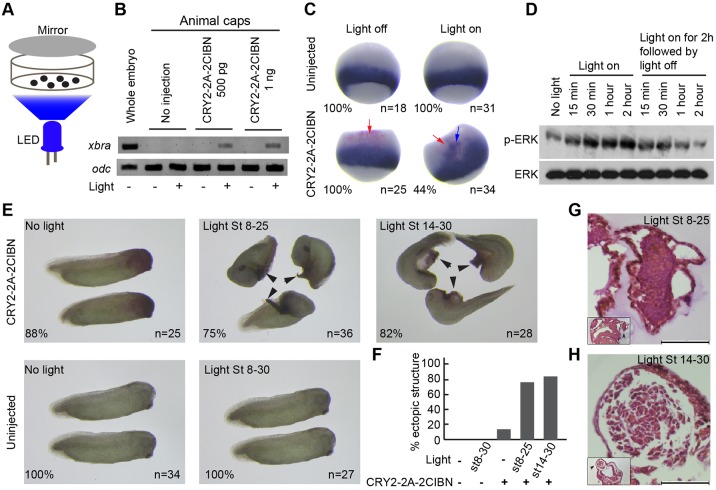
The results described above demonstrate that our bicistronic optogenetic system allows for blue light-induced activation of Raf1 in cell culture. It remains unknown if the same system would work in multicellular organisms. It is well established that Ras/Raf1/MAPK (mitogen-activated protein kinase) signaling is sufficient and necessary for vertebrate mesoderm induction. We thus applied our bicistronic optogenetic system to Xenopus embryos, a widely used model system for developmental biology. To determine whether our bicistronic optogenetic system works in Xenopus embryonic tissues, we took advantage of the Xenopus animal cap assay. Xenopus animal cap explants differentiate into atypical epidermis by default, but adapt a mesodermal cell fate in response to activation of the MAPK pathway. We injected various doses of mRNA encoding CRY2-2A-2CIBN into the animal pole of fertilized eggs and dissected animal caps at the blastula stage (stage 8). Animal caps were cultured either under normal conditions, or exposed to blue light (5 mW/cm2) from a homemade LED array (Fig. 4A). After 4 h of blue-light exposure, animal caps were harvested at the early gastrula stage (stage 11) and subjected to reverse transcription PCR (RT-PCR) for the expression of xbra, a pan-mesoderm marker that can be induced by MAPK signaling in animal caps. Indeed, blue-light exposure induced the expression of xbra in CRY2-2A-2CIBN overexpressed animal caps (Fig. 4B). As expected, no xbra expression was detected in uninjected animal caps that were exposed to blue light or in CRY2-2A-2CIBN-injected animal caps that were cultured under normal conditions. The 1- and 3-CIBN constructs did not elicit detectable xbra expression under the same condition of blue-light stimulation (Fig. S4), which is consistent with their weaker capacities to induce PC12 cell differentiation (Fig. 3K).
Fig. 4.
Application of the bicistronic optogenetic Raf1 activation system in Xenopus embryos. (A) Schematic for light-controlled Raf1 activation in Xenopus embryos or animal caps. (B) RT-PCR results showing that exposure to blue light induced the expression of xbra, a pan-mesodermal marker, in CRY2-2A-2CIBN-injected animal caps at the early gastrula stage. odc was used as loading control. (C) Expression of xbra in whole embryos at the early gastrula stage assessed by whole-mount in situ hybridization. Samples include controls (uninjected, no light treatment); embryos that were uninjected, but treated with blue light; embryos that were injected with CRY2-2A-2CIBN and cultured under normal conditions; and embryos that were injected with CRY2-2A-2CIBN and treated with blue light. Red arrows indicate the cell lineage tracer. Blue arrow indicates ectopic xbra mRNA expression. (D) Western blot analysis showing that in an animal cap assay, CRY2-2A-2CIBN induced blue light-dependent reversible phosphorylation of ERK. (E) Activation of Raf1 by treating CRY2-2A-2CIBN-injected embryos with blue light after the completion of germ layer specification is sufficient for inducing ectopic tail-like structures in the head region (arrowheads). Images show morphology of control embryos; uninjected embryos that were treated with blue light; embryos that were injected with CRY2-2A-2CIBN and cultured under normal conditions; CRY2-2A-2CIBN-injected embryos treated with blue light from stage 8 to stage 25, and CRY2-2A-2CIBN-injected embryos treated with blue light from stage 14 to stage 30. (F) Percentage of embryos with ectopic tail-like structure. (G,H) Histological analysis of the tail-like structures induced by activation of Raf1. (G) Tail-like structure induced by Raf1 in an embryo exposed to blue light from stage 8 to stage 25. (H) Tail-like structure induced by Raf1 in an embryo exposed to blue light from stage 14 to stage 30. Small inserts at the lower left corner are low-power views, which show the location of the Raf1-induced tail-like structure (arrowheads). Scale bars: 100 μm.
In parallel, we tested CRY2-2A-2CIBN in whole embryos. We injected a mixture of CRY2-2A-2CIBN mRNA (500 pg) and nuclear β-galactosidase (n-β-gal) RNA (250 pg) into the animal pole of fertilized eggs. Co-injected n-β-gal served as a lineage tracer. At the midblastula stage, embryos were cultured either under normal conditions or exposed to blue light. We harvested embryos at the gastrula stage and performed in situ hybridization to assess the expression of xbra. Our results showed that blue-light exposure induced ectopic expression of xbra in 44% of CRY2-2A-2CIBN-injected embryos. Overexpression of CRY2-2A-2CIBN alone or treatment of uninjected embryos with blue light did not induce ectopic expression of xbra (Fig. 4C). These results demonstrate that performance of the 2-CIBN optogenetic system is optimal when applied to Xenopus embryos.
We then performed an animal cap assay and studied the activation kinetics of the bicistronic system in detail. We injected mRNA encoding the CRY2-2A-2CIBN into the animal pole of fertilized eggs and dissected animal caps at the blastula stage (stage 8). After wound healing was completed, animal caps were cultured either under normal conditions, or exposed to blue light continuously for 2 h. We then turned off the blue light and cultured animal caps for an additional 2 h. Animal caps were harvested at various time points during this process and subjected to western blot analysis for phospho-ERK (p-ERK), a molecular readout for activation of Raf1. As shown in Fig. 4D, a 15-min blue-light exposure was sufficient for activating the MAPK pathway, judged by a significant increase in the level of p-ERK in CRY2-2A-2CIBN-injected animal caps. The MAPK pathway became fully activated with 30 min of light exposure. Activation of the MAPK pathway in CRY2-2A-2CIBN-injected animal caps is blue light dependent. After turning off the blue light for 15 min, the level of p-ERK was clearly decreased. The level of p-ERK further declined and returned to the baseline level after 2 h. Thus, this bicistronic system allows highly efficient light-dependent reversible activation of the MAPK pathway in embryos (Fig. 4D).
Raf1 activation induces an ectopic tail-like structure after germ layer specification in Xenopus embryos
In a previous report by Ishimura et al., overexpression of a constitutively active Raf1 induced ectopic tail-like structures in the anterior dorsal region of the embryo. It was proposed that overexpressed Raf1 might induce ectopic tail-like structures through two mechanisms. As a potent mesoderm inducer, overexpressed Raf1 can induce ectopic mesoderm during germ layer specification, which occurs during blastula and early gastrula stages, and result in the formation of ectopic tail-like structures later on. Alternatively, overexpressed Raf1 may trigger an epithelial-mesenchymal transition (EMT)-like event after germ layer specification is completed, and directly transform ectoderm to neural and mesoderm lineages, followed by proliferation and extension of these structures (Ishimura et al., 2006). Although these two mechanisms are not mutually exclusive, it was very difficult for Ishimura et al. to determine whether Raf1 could function through the second mechanism. This is because in their experiments, overexpressed Raf1 began to activate the downstream signaling cascades constitutively soon after its RNA was injected into the embryo during early cleavage stage. It was technically challenging to bypass early developmental stages and activate Raf1 specifically after germ layer specification.
To solve this puzzle and also further test the application of our new bicistronic optogenetic tool in vivo, we set out to determine whether activation of Raf1 after the completion of germ layer specification would induce ectopic tail-like structures in the anterior dorsal region of the embryo. We injected a mixture of CRY2-2A-2CIBN and n-β-gal RNAs at the 8-cell stage into one of the dorsal animal blastomeres, which gives rise to dorsal anterior tissues later on. Because Xenopus mesoderm induction occurs between the blastula stage and the mid-gastrula stage (stage 8-12), we activated Raf1 by treating injected embryos with blue light from the neurula stage to the late tailbud stage (stage 14-30). As a positive control, we also activated Raf1 from the blastula stage to the early tailbud stage (stage 8-25). As expected, activation of Raf1 from the blastula stage to the early tailbud stage induced an ectopic tail-like structure in 75% of embryos (Fig. 4E,F). Cells located in these ectopic tail-like structures are tightly associated with each other, reminiscent of somitic tissue morphologically (Fig. 4G). Intriguingly, activation of Raf1 after the completion of germ layer specification (stage 14-30) could also induce ectopic tail-like structures (82%, n=28) (Fig. 4E,F). Cells in these ectopic tail-like structures show typical fibroblast-like morphology, resembling embryonic mesenchymal cells (Fig. 4H). This observation demonstrates for the first time that activation of Raf1 after germ layer specification is sufficient for inducing ectopic tail-like structures. Furthermore, results from the histological analysis support the idea that activation of Raf1 after germ layer specification can lead to transformation of the embryonic tissues, followed by proliferation and extension of these structures. As expected, the 1- and 3-CIBN constructs showed very low activities in this assay (Fig. S5).
DISCUSSION
Several photoactivatable proteins have been applied to in vivo systems. Light-mediated control can function through photo-uncaging [e.g. light, oxygen, voltage (LOV)-based assays] or translocation (e.g. PhyB-PIF- and CRY2-CIBN-based assays). For instance, LOV has been used to control transcription (Motta-Mena et al., 2014) and cell migration in Drosophila (Wang et al., 2010) and zebrafish (Yoo et al., 2010). Successful caging of the active protein often requires trial-and-error and computationally targeted protein design. PhyB-PIF6 has been used to control protein translocation within zebrafish embryos through a global light illumination (Beyer et al., 2015). A very recent work used the PhyB-PIF6 to control protein translocation and function within zebrafish through spatially confined light illumination (Buckley et al., 2016). The PhyB-PIF6 system has the highest dynamic range and fastest reversible kinetics (reversible association and dissociation within milliseconds). One drawback is that a synthetic co-factor, phycocyanobilin or PCB, is needed for a fully functional PhyB-PIF6 system. For in vivo assays, PCB is either supplemented in buffer (Beyer et al., 2015) or injected into the body (Buckley et al., 2016). By contrast, the blue light-sensitive photoactivatable protein CRY2 from Arabidopsis thaliana requires no exogenous co-factors and displays reversible photoactivation kinetics on a time scale of several minutes. Additionally, CRY2 (498 aa) is smaller than PhyB (908 aa). Therefore, CRY2 is an attractive candidate to control protein kinase activity in live cells. A practical challenge faced by all dimerization-based optogenetic systems, however, is a lack of ratiometric control between the dimerizing proteins. For instance, it has been reported that the relative ratio between PhyB and PIF is crucial in assaying PhyB-PIF interaction (Toettcher et al., 2011). Common strategies such as co-transfection or infection with multiple viral vectors often suffer from unreliable quantitative co-expression of heterologous proteins.
In this study, we optimized the CRY2-CIBN optogenetic system using a PC12 cell differentiation assay. In the PC12 cell differentiation assay, the differentiation ratio yielded by co-transfection of CIBN-GFP-CaaX and CRY2-mCherry-Raf1 and subsequent blue light treatment was significantly lower than that induced by transfection of a CA-Raf1 (Fig. 1D). At least two factors account for this decrease in the differentiation ratio: first, bias in gene delivery (i.e. singly transfected cells) renders a subpopulation of cells that are unresponsive to light stimulation; second, in co-transfected cells, expression levels of CIBN and CRY2 fusion proteins are suboptimal. In this study, we addressed these two issues by integrating the CIBN-CRY2 dimerization system with a P2A peptide-based bicistronic system. Because the bicistronic system is contained in a single plasmid, bias in gene delivery during co-transfection of two expression plasmids is naturally resolved. In addition, the P2A-based bicistronic system allows stoichiometric co-expression of both CIBN and CRY2 fusion proteins. To achieve an optimal configuration for the CIBN-CRY2 dimerization system, we constructed bicistronic plasmids with a varying number (n=1-3) of CIBN and a single CRY2. Although all 2A constructs showed successful membrane recruitment of CRY2-mCherry-Raf1 under blue-light stimulation, they varied in recruiting capacity. The single-CIBN construct displayed weaker membrane recruitment of CRY2-mCherry-Raf1 compared with that of the multiple-CIBN constructs. Such a difference affected the capacities to induce PC12 cell differentiation under blue-light stimulation. Among these constructs, the single- and triple-CIBN constructs produced a differentiation ratio similar to that of co-transfected constructs. CRY2-2A-2CIBN, however, showed a markedly improved activity compared with co-transfected constructs. In fact, it appeared to be as potent as the CA-Raf1. We speculate that the single-CIBN construct may not generate sufficient CRY2-binding sites on the plasma membrane, which could reduce the dwelling time of Raf1 in its activation zone close to the membrane. In the case of the triple-CIBN construct, although CRY2-2A-3CIBN provides sufficient CRY2-binding sites on the cell membrane, it might undesirably position Raf1 out of its activation zone. Probably for these reasons, the CRY2-2A-CIBN and CRY2-2A-3CIBN constructs are less efficient in inducing PC12 neuronal differentiation than the CRY2-2A-2CIBN construct (Fig. 3K).
Our results demonstrate that the optimized CRY2-2A-2CIBN system is a powerful optogenetic tool for manipulating the Raf signaling cascade in vivo. In this proof-of-principle study, we applied CRY2-2A-2CIBN to Xenopus embryo. Using phosphorylation of ERK as the readout, we were able to show that CRY2-2A-2CIBN allowed reversible blue light-dependent activation of Raf1. As expected, activation of Raf1 by treating CRY2-2A-2CIBN overexpressing embryos with blue light during germ layer specification (between the blastula and gastrula stages) induced ectopic mesoderm. Intriguingly, we found that activation of Raf1 after germ layer specification could induce ectopic tail-like structures in the anterior dorsal region of Xenopus embryos. The morphology of cells in these induced structures resembled that of embryonic mesenchyme. This result provided the first direct evidence that activation of Raf1 after germ layer specification can trigger tumor-like growth of embryonic tissue. We believe this optogenetic system could be applied to many other in vivo model organisms, including zebrafish, chicken or Drosophila. As a bicistronic system, the CRY2-2A-CIBN expression cassette can be driven by a tissue-specific promoter to achieve lineage-specific expression. This makes it possible to manipulate the activity of Raf1 in any desired tissues within any desired time period.
Although combining two genes into one plasmid inevitably extends its size, the double-CIBN construct is 10.1 kb in length, which is still reasonable for cloning and transfection. If necessary, the mCherry domain can be removed from CRY2-mCherry-Raf1 to increase the cloning capacity further. The strategy of fluorophore removal will not work for the co-transfection system because one cannot tell the difference between singly and co-transfected cells. Collectively, we believe that our bicistronic optogenetic system offers the potential for light-dependent manipulation of the Raf1 signaling cascade in a manner not readily duplicated by other currently available experimental approaches. Notably, activation of kinases by membrane translocation is a common mechanism for cell signaling control. For instance, the PI3K/AKT signaling pathway can be activated by membrane targeting of the AKT domain (Kohn et al., 1996). Therefore, we expect that our strategy could be generalized to control other kinase activity in single cells or multicellular organisms. As the CRY2-CIBN dimerization platform has been increasingly used in optogenetic applications, we expect that our results will shed light on the design and optimization of such a system.
MATERIALS AND METHODS
Reagents
Phusion DNA polymerase master mix was purchased from NEB (M0530S). DreamTaq PCR Master Mix (2×) (K1071), Turbofect (FERR0531) and protease/phosphatase inhibitor cocktail (88669) were from Fisher Scientific. In-Fusion HD Cloning Plus kit was from Clontech (638909). DMEM (11995-065), F12K (21127-022) medium, and horse serum (26050088) were purchased from Life Technologies. Fetal bovine serum (12303C-500ML) and RIPA buffer (R0278) were from Sigma-Aldrich. Penicillin-streptomycin solution (30-002-CI) was from Corning. Precast protein gels (456-1024) and ECL substrate (170-5060) were from Bio-Rad. Micro cover glasses (48393 230) were from VWR. Polydimethylsiloxane (PDMS) was from Ellsworth Adhesives (184 SIL ELAST KIT 0.5KG). The sequences of oligonucleotides (IDTDNA) used in this study are summarized in Table S2. Antibodies used in this studies include anti-GFP (SICGEN, AB0040-20, 1:1000), anti-mCherry (SICGEN, AB0066-200, 1:1000), anti p-ERK (Santa Cruz, SC-23759-R, 1:1000) and anti-pan ERK (Cell Signaling, #9102, 1:1000) antibodies, and HRP-conjugated rabbit anti-goat IgG (Life Technologies, 611620, 1:1000). The sequences of oligonucleotides and double-stranded DNA used are listed in Tables S2 and S3. See the supplementary Materials and Methods for further details of plasmid construction.
Cell culture, transfection, and western blot analysis
BHK-21 cells were used for characterizing the P2A system by fluorescence microscopy and western blot. PC12 cells were used for in vitro differentiation assays. Details of cell culture, transfection, and western blot analysis can be found in the supplementary Materials and Methods.
Xenopus embryos and manipulations
Xenopus embryos were obtained and injected as described (Sive et al., 2000). Animal cap assay, RT-PCR, western blot and in situ hybridization were performed as described (Rorick et al., 2007). Primers used for RT-PCR was: odc (5′-GCCATTGTGAAGACTCTCTCCATTC-3′; 5′-TTCGGGTGCTTCCTTGCCAC-3′) and xbra (5′-GCTGGAAGTATGTGAATGGAG-3′; 5′-TTAAGTGCTGTAATCTCTTCA-3′). Histological analysis was performed as described (Sive et al., 2000).
Live cell imaging
For the light-induced membrane recruitment assay, BHK-21 cells were co-transfected with CIB1-GFP-CaaX and CRY2-mCherry-Raf1 or CRY2-2A-(n)CIBN plasmids (n=1-3). Fluorescence imaging of the transfected cells was carried out using a confocal microscope (Zeiss LSM 700). GFP fluorescence was excited by a 488-nm laser beam; mCherry fluorescence was excited by a 555-nm laser beam. Excitation beams were focused via a 40× oil objective (Plan-Neofluar NA 1.30). Ten pulsed 488-nm and 555-nm excitation light (2-s interval) were applied for each membrane recruitment experiment. CRY2-CIBN binding induced by 488-nm light was monitored by membrane recruitment of CRY2-mCherry-Raf1 to the CIBN-GFP-CaaX-anchored plasma membrane. The powers after the objective for 488-nm and 555-nm laser beam are approximately 40 µW and 75 µW, respectively. Alternatively, an epi-illumination fluorescence microscope (Leica DMI8) equipped with a 100× objective (HCX PL FLUOTAR 100×/1.30 oil) and a light-emitting diode illuminator (SOLA SE II 365) as the light source was used for the CRY2-mCherry-Raf1 membrane translocation assay.
Neurite outgrowth of PC12 cells were imaged using an epi-illumination fluorescence microscope (Leica DMI8) equipped with 10× (PLAN 10×/0.25) and 40× (HCXPL FL L 40×/0.6) objectives. Fluorescence from GFP was detected using the GFP filter cube (Leica, excitation filter 472/30, dichroic mirror 495, and emission filter 520/35); fluorescence from mCherry was detected using the Texas Red filter cube (Leica, excitation filter 560/40, dichroic mirror 595, and emission filter 645/75).
Long-term light stimulation in the neurite outgrowth assay
PC12 cells were plated and transfected in a 12-well tissue culture plate. After 24 h recovery in high-serum F12K medium [15% horse serum+2.5% fetal bovine serum (FBS)], the cell culture was exchanged to a low-serum medium (1.5% horse serum+0.25% FBS) to minimize the base-level ERK activation induced by serum. For light stimulation, the 12-well plate was placed on the top of the LED device with each well aligned above each LED. By adjusting the DC voltage and the resistors, the light intensity was set at 0.2 mW/cm2 at the cell level. Both the LED device and the cell culture plate were placed into a CO2 incubator (Fig. S6). Neurite outgrowth was quantified after 24 h of light stimulation. See the supplementary Materials and Methods for further details of construction of the programmable LED device.
Image analysis
The ROI Manager plugin from Fiji was used to analyze the correlation of fluorescence intensities for GFP and mCherry in transfected BHK-21 cells. Manual selection of the cell profile was used to enclose each cellular area. The average fluorescence intensities were read out from the plugin. A snapshot of analysis is shown in Fig. S7. The Cell Counter plugin from Fiji was used to analyze the percentage of differentiated PC12 cells within all transfected cells. A snapshot of each field of view was acquired in three channels (mCherry, GFP and phase contrast). Differentiated cells were defined as those with at least one neurite of length equal to or longer than the diameter of their cell bodies.
Statistical analysis
P-values were determined by performing two-tailed, unpaired t-test using GraphPad Prism software.
Acknowledgements
We thank Dr X. L. Nan in the Oregon Health and Science University for providing the BHK-21 cell line. We thank Dr T. Meyer in Stanford University for providing the PC12 cell line. We thank Dr S McMasters from the cell media facility of UIUC for providing DH5α competent cells and for providing comments on our experiments.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: V.V.K., J.S.K., J.Y., K.Z.; Methodology: J.Y., K.Z.; Formal analysis and investigation: V.V.K., J.S.K., W.M., A.J.T., H.M.A., P.M., D.B.P., N.R., E.E.C., J.Y., K.Z.; Writing - original draft preparation: K.Z., J.Y.; Writing - review and editing: K.Z., J.Y., V.V.K., J.S.K.; Funding acquisition: K.Z., J.Y.; Supervision: J.Y., K.Z.
Funding
This work was supported by the University of Illinois at Urbana-Champaign (UIUC) (K.Z.); and the National Institutes of Health [R01GM111816 to J.Y.]. Deposited in PMC for release after 12 months.
Supplementary information
Supplementary information available online at http://dev.biologists.org/lookup/doi/10.1242/dev.140889.supplemental
References
- Beyer H. M., Juillot S., Herbst K., Samodelov S. L., Müller K., Schamel W. W., Römer W., Schäfer E., Nagy F., Strähle U. et al. (2015). Red light-regulated reversible nuclear localization of proteins in mammalian cells and zebrafish. ACS Synth. Biol. 4, 951-958. 10.1021/acssynbio.5b00004 [DOI] [PubMed] [Google Scholar]
- Boulina M., Samarajeewa H., Baker J. D., Kim M. D. and Chiba A. (2013). Live imaging of multicolor-labeled cells in Drosophila. Development 140, 1605-1613. 10.1242/dev.088930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyden E. S., Zhang F., Bamberg E., Nagel G. and Deisseroth K. (2005). Millisecond-timescale, genetically targeted optical control of neural activity. Nat. Neurosci. 8, 1263-1268. 10.1038/nn1525 [DOI] [PubMed] [Google Scholar]
- Buckley C. E., Moore R. E., Reade A., Goldberg A. R., Weiner O. D. and Clarke J. D. (2016). Reversible optogenetic control of subcellular protein localization in a live vertebrate embryo. Dev. Cell 36, 117-126. 10.1016/j.devcel.2015.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugaj L. J., Choksi A. T., Mesuda C. K., Kane R. S. and Schaffer D. V. (2013). Optogenetic protein clustering and signaling activation in mammalian cells. Nat. Methods 10, 249-252. 10.1038/nmeth.2360 [DOI] [PubMed] [Google Scholar]
- Chang K. Y., Woo D., Jung H., Lee S., Kim S., Won J., Kyung T., Park H., Kim N., Yang H. W. et al. (2014). Light-inducible receptor tyrosine kinases that regulate neurotrophin signalling. Nat. Commun. 5, 4057 10.1038/ncomms5057 [DOI] [PubMed] [Google Scholar]
- Che D. L., Duan L., Zhang K. and Cui B. (2015). The dual characteristics of light-induced cryptochrome 2, homo-oligomerization and heterodimerization, for optogenetic manipulation in mammalian cells. ACS Synth. Biol. 4, 1124-1135. 10.1021/acssynbio.5b00048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly M. L. L., Hughes L. E., Luke G., Mendoza H., ten Dam E., Gani D. and Ryan M. D. (2001a). The ‘cleavage’ activities of foot-and-mouth disease virus 2A site-directed mutants and naturally occurring ‘2A-like’ sequences. J. Gen. Virol. 82, 1027-1041. 10.1099/0022-1317-82-5-1027 [DOI] [PubMed] [Google Scholar]
- Donnelly M. L. L., Luke G., Mehrotra A., Li X. J., Hughes L. E., Gani D. and Ryan M. D. (2001b). Analysis of the aphthovirus 2A/2B polyprotein ‘cleavage’ mechanism indicates not a proteolytic reaction, but a novel translational effect: a putative ribosomal ‘skip’. J. Gen. Virol. 82, 1013-1025. 10.1099/0022-1317-82-5-1013 [DOI] [PubMed] [Google Scholar]
- Goedhart J., van Weeren L., Adjobo-Hermans M. J. W., Elzenaar I., Hink M. A. and Gadella T. W. J. (2011). Quantitative co-expression of proteins at the single cell level – application to a multimeric FRET sensor. PLoS ONE 6, e27321 10.1371/journal.pone.0027321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallett R. A., Zimmerman S. P., Yumerefendi H., Bear J. E. and Kuhlman B. (2016). Correlating in vitro and in vivo activities of light-inducible dimers: a cellular optogenetics guide. ACS Synth. Biol. 5, 53-64. 10.1021/acssynbio.5b00119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes R. M., Bolger S., Tapadia H. and Tucker C. L. (2012). Light-mediated control of DNA transcription in yeast. Methods 58, 385-391. 10.1016/j.ymeth.2012.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahimi A., Vande Velde G., Reumers V., Toelen J., Thiry I., Vandeputte C., Vets S., Deroose C., Bormans G., Baekelandt V. et al. (2009). Highly efficient multicistronic lentiviral vectors with peptide 2A sequences. Hum. Gene Ther. 20, 845-860. 10.1089/hum.2008.188 [DOI] [PubMed] [Google Scholar]
- Idevall-Hagren O., Dickson E. J., Hille B., Toomre D. K. and De Camilli P. (2012). Optogenetic control of phosphoinositide metabolism. Proc. Natl. Acad. Sci. USA 109, E2316-E2323. 10.1073/pnas.1211305109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishimura A., Lee H.-S., Bong Y.-S., Saucier C., Mood K., Park E. K. and Daar I. O. (2006). Oncogenic Met receptor induces ectopic structures in Xenopus embryos. Oncogene 25, 4286-4299. 10.1038/sj.onc.1209463 [DOI] [PubMed] [Google Scholar]
- Kakumoto T. and Nakata T. (2013). Optogenetic control of PIP3: PIP3 is sufficient to induce the actin-based active part of growth cones and is regulated via endocytosis. PLoS ONE 8, e70861 10.1371/journal.pone.0070861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy M. J., Hughes R. M., Peteya L. A., Schwartz J. W., Ehlers M. D. and Tucker C. L. (2010). Rapid blue-light–mediated induction of protein interactions in living cells. Nat. Methods 7, 973-975. 10.1038/nmeth.1524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim B. and Lin M. Z. (2013). Optobiology: optical control of biological processes via protein engineering. Biochem. Soc. Trans. 41, 1183-1188. 10.1042/BST20130150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. H., Lee S.-R., Li L.-H., Park H.-J., Park J.-H., Lee K. Y., Kim M.-K., Shin B. A. and Choi S.-Y. (2011). High cleavage efficiency of a 2A peptide derived from porcine teschovirus-1 in human cell lines, zebrafish and mice. PLoS ONE 6, e18556 10.1371/journal.pone.0018556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohn A. D., Takeuchi F. and Roth R. A. (1996). Akt, a pleckstrin homology domain containing kinase, is activated primarily by phosphorylation. J. Biol. Chem. 271, 21920-21926. 10.1074/jbc.271.36.21920 [DOI] [PubMed] [Google Scholar]
- Konermann S., Brigham M. D., Trevino A. E., Hsu P. D., Heidenreich M., Cong L., Platt R. J., Scott D. A., Church G. M. and Zhang F. (2013). Optical control of mammalian endogenous transcription and epigenetic states. Nature 500, 472-476. 10.1038/nature12466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S., Park H., Kyung T., Kim N. Y., Kim S., Kim J. and Heo W. D. (2014). Reversible protein inactivation by optogenetic trapping in cells. Nat. Methods 11, 633-636. 10.1038/nmeth.2940 [DOI] [PubMed] [Google Scholar]
- Levskaya A., Weiner O. D., Lim W. A. and Voigt C. A. (2009). Spatiotemporal control of cell signalling using a light-switchable protein interaction. Nature 461, 997-1001. 10.1038/nature08446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Yu X., Li K., Klejnot J., Yang H., Lisiero D. and Lin C. (2008). Photoexcited CRY2 Interacts with CIB1 to Regulate Transcription and Floral Initiation in Arabidopsis. Science 322, 1535-1539. 10.1126/science.1163927 [DOI] [PubMed] [Google Scholar]
- Liu H., Gomez G., Lin S. and Lin C. (2012). Optogenetic control of transcription in zebrafish. PLoS ONE 7, e50738 10.1371/journal.pone.0050738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Más P., Devlin P. F., Panda S. and Kay S. A. (2000). Functional interaction of phytochrome B and cryptochrome 2. Nature 408, 207-211. 10.1038/35041583 [DOI] [PubMed] [Google Scholar]
- Motta-Mena L. B., Reade A., Mallory M. J., Glantz S., Weiner O. D., Lynch K. W. and Gardner K. H. (2014). An optogenetic gene expression system with rapid activation and deactivation kinetics. Nat. Chem. Biol. 10, 196-202. 10.1038/nchembio.1430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller K., Naumann S., Weber W. and Zurbriggen M. D. (2015). Optogenetics for gene expression in mammalian cells. Biol. Chem. 396, 145-152. 10.1515/hsz-2014-0199 [DOI] [PubMed] [Google Scholar]
- Rorick A. M., Mei W. Y., Liette N. L., Phiel C., El-Hodiri H. M. and Yang J. (2007). PP2A: B56 epsilon is required for eye induction and eye field separation. Dev. Biol. 302, 477-493. 10.1016/j.ydbio.2006.10.011 [DOI] [PubMed] [Google Scholar]
- Schmidt D. and Cho Y. K. (2015). Natural photoreceptors and their application to synthetic biology. Trends Biotechnol. 33, 80-91. 10.1016/j.tibtech.2014.10.007 [DOI] [PubMed] [Google Scholar]
- Shimizu-Sato S., Huq E., Tepperman J. M. and Quail P. H. (2002). A light-switchable gene promoter system. Nat. Biotechnol. 20, 1041-1044. 10.1038/nbt734 [DOI] [PubMed] [Google Scholar]
- Sive H. L., Grainger R. M. and Harland R. M. (2000). Early Development of Xenopus laevis: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Strickland D., Lin Y., Wagner E., Hope C. M., Zayner J., Antoniou C., Sosnick T. R., Weiss E. L. and Glotzer M. (2012). TULIPs: tunable, light-controlled interacting protein tags for cell biology. Nat. Methods 9, 379-384. 10.1038/nmeth.1904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taslimi A., Vrana J. D., Chen D., Borinskaya S., Mayer B. J., Kennedy M. J. and Tucker C. L. (2014). An optimized optogenetic clustering tool for probing protein interaction and function. Nat. Commun. 5, 4925 10.1038/ncomms5925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tischer D. and Weiner O. D. (2014). Illuminating cell signalling with optogenetic tools. Nat. Rev. Mol. Cell Biol. 15, 551-558. 10.1038/nrm3837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toettcher J. E., Gong D. Q., Lim W. A. and Weiner O. D. (2011). Light control of plasma membrane recruitment using the Phy-Pif system. Method Enzymol. 497, 409-423. 10.1016/B978-0-12-385075-1.00017-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker C. L. (2012). Manipulating cellular processes using optical control of protein–protein interactions. Prog. Brain Res. 196, 95-117. 10.1016/B978-0-444-59426-6.00006-9 [DOI] [PubMed] [Google Scholar]
- Wang X., He L., Wu Y. I., Hahn K. M. and Montell D. J. (2010). Light-mediated activation reveals a key role for Rac in collective guidance of cell movement in vivo. Nat. Cell Biol. 12, 591-597. 10.1038/ncb2061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wend S., Wagner H. J., Müller K., Zurbriggen M. D., Weber W. and Radziwill G. (2014). Optogenetic control of protein kinase activity in mammalian cells. ACS Synth. Biol. 3, 280-285. 10.1021/sb400090s [DOI] [PubMed] [Google Scholar]
- Wu Y. I., Frey D., Lungu O. I., Jaehrig A., Schlichting I., Kuhlman B. and Hahn K. M. (2009). A genetically encoded photoactivatable Rac controls the motility of living cells. Nature 461, 104-108. 10.1038/nature08241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yazawa M., Sadaghiani A. M., Hsueh B. and Dolmetsch R. E. (2009). Induction of protein-protein interactions in live cells using light. Nat. Biotechnol. 27, 941-945. 10.1038/nbt.1569 [DOI] [PubMed] [Google Scholar]
- Yoo S. K., Deng Q., Cavnar P. J., Wu Y. I., Hahn K. M. and Huttenlocher A. (2010). Differential regulation of protrusion and polarity by PI(3)K during neutrophil motility in live zebrafish. Dev. Cell 18, 226-236. 10.1016/j.devcel.2009.11.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K. and Cui B. X. (2014). Lighting up FGFR signaling. Chem. Biol. 21, 806-808. 10.1016/j.chembiol.2014.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K. and Cui B. (2015). Optogenetic control of intracellular signaling pathways. Trends Biotechnol. 33, 92-100. 10.1016/j.tibtech.2014.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K., Duan L., Ong Q., Lin Z., Varman P., Sung K. and Cui B. (2014). Light-mediated kinetic control reveals the temporal effect of the Raf/MEK/ERK pathway in PC12 cell neurite outgrowth. PLoS ONE 9, e92917 10.1371/journal.pone.0092917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X. X., Chung H. K., Lam A. J. and Lin M. Z. (2012). Optical control of protein activity by fluorescent protein domains. Science 338, 810-814. 10.1126/science.1226854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoltowski B. D. and Gardner K. H. (2011). Tripping the light fantastic: blue-light photoreceptors as examples of environmentally modulated protein–protein interactions. Biochemistry 50, 4-16. 10.1021/bi101665s [DOI] [PMC free article] [PubMed] [Google Scholar]