Abstract
Many mutations in rhodopsin gene linked to retinitis pigmentosa (RP) cause rhodopsin misfolding. Rod photoreceptor cells expressing misfolded rhodopsin eventually die. Identifying mechanisms to prevent rhodopsin misfolding or to remove irreparably misfolded rhodopsin could provide new therapeutic strategies. IRE1, ATF6, and PERK signaling pathways, collectively called the unfolded protein response (UPR), regulate the functions of endoplasmic reticulum, responsible for accurate folding of membrane proteins such as rhodopsin. We used chemical and genetic approaches to selectively activate IRE1, ATF6, or PERK signaling pathways one at a time and analyzed their effects on mutant rhodopsin linked to RP. We found that both artificial IRE1 and ATF6 signaling promoted the degradation of mutant rhodopsin with lesser effects on wild-type rhodopsin. Furthermore, IRE1 and ATF6 signaling preferentially reduced levels of aggregated rhodopsins. By contrast, PERK signaling reduced levels of wild-type and mutant rhodopsin. These studies indicate that activation of either IRE1, ATF6, or PERK prevents mutant rhodopsin from accumulating in the cells. In addition, activation of IRE1 or ATF6 can selectively remove aggregated or mutant rhodopsin from the cells and may be useful in treating RP associated with rhodopsin protein misfolding.
Keywords: Unfolded protein response, Rhodopsin, P23H, Misfolded protein, IRE1, PERK, ATF6, Retinitis pigmentosa, Retinal degeneration
83.1 Instruction
The most common form of inherited blindness is retinitis pigmentosa (RP) [1]. Over 100 distinct mutations in rhodopsin have been identified that cause RP (www.sph.uth.tmc.edu/retnet/). Rhodopsin gene encodes a G protein-coupled receptor that is folded in the endoplasmic reticulum (ER) prior to export to the rod photoreceptor outer segment [2]. Many class II mutations in rhodopsin gene that cause RP generate misfolded rhodopsin proteins that are retained within the ER [3–6]. Rod photo-receptors expressing misfolded mutant rhodopsin eventually die, leading to retinal degeneration. Studies have demonstrated that reducing misfolded rhodopsin protein levels by introducing chaperones or ribozymes specifically against mutant rhodopsin mRNA can delay retinal degeneration [7–9]. Therefore, identifying strategies to reduce misfolded rhodopsin protein in cells holds promise in preventing the death of rod photoreceptor cells.
Mammalian cells respond to ER stress by activating three distinct branches of signal transduction pathways, collectively called the unfolded protein response (UPR), to restore ER proteostasis. UPR is controlled by three ER-resident membrane proteins, inositol requiring enzyme-1 (IRE1), activating transcription factor 6 (ATF6), and protein kinase RNA-like endoplasmic reticulum kinase (PERK), which all contain ER luminal domains that monitor the quality of ER proteins and cytosolic domains that activate mechanistically-distinct signal transduction cascades [10]. In response to ER stress, IRE1, PERK, and ATF6 signaling pathways are activated and restore ER proteostasis by upregulating genes involved in ER protein folding and ER-associated protein degradation (ERAD) [11–13], and attenuating protein translation [14]. IRE1, PERK, and ATF6 signaling therefore enhance ER protein folding capabilities and also reduce protein levels by dampening translation and promoting protein degradation.
UPR regulation of ER protein-folding fidelity offers potential means to repair or remove misfolded proteins, such as class II rhodopsin mutant. In this review, we summarize how activation of each pathway, using artificial chemical or genetic means, affects the fate of mutant rhodopsin in cells.
83.2 Selective Activation of IRE1 Promotes the Degradation of Misfolded Mutant Rhodopsin in the Cells
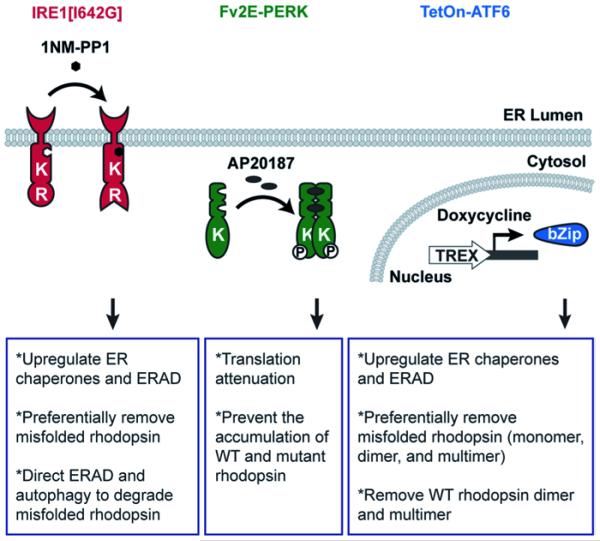
The cytosolic domain of IRE1 includes kinase and endoribonuclease (RNase) domains that control the splicing of Xbp-1 mRNA [15–17]. To selectively activate IRE1, we used a genetically altered version of human IRE1, IRE1 [I642G], in which an isoleucine at residue 642 is substituted by a glycine. In this system, the RNase function of IRE1 [I642G] can be enabled by an ATP analog, 4-amino-1-tert-butyl-3-(1’-naphthylmethyl)pyrazolo[3,4-d]pyrimidine (1NM-PP1) [18, 19], to induce the splicing of Xbp-1 mRNA (Fig. 83.1). Previously, we demonstrated that in cells stably expressing IRE1[I642G], the addition of 1NM-PP1 rapidly increased Xbp-1 mRNA splicing without activating ATF6 and PERK pathways [19–21].
Fig. 83.1.
Chemical-genetic strategies to selectively activate IRE1, PERK, and ATF6 pathways
Using this system, we found that selectively activating IRE1 pathway tremendously reduced the protein levels of all class II rhodopsin mutants tested, including P23H, T17M, Y178C, C185R, D190G, and K296E rhodopsin, whereas WT rhodopsin was minimally affected by the activation of IRE1. Furthermore, we found that the effects of IRE1 were almost exclusively targeted to the insoluble (aggregated) fractions of P23H rhodopsin, indicating that IRE1 removed mutant proteins at the monomeric state as well as when they were aggregated. Levels of mutant rhodopsin mRNAs were not changed in our system. Instead, activation of IRE1 promoted the degradation of mutant rhodopsin protein through two separate protein degradation pathways: the ubiquitin-proteasome system via ERAD and the autophagy-lysosome system [22].
83.3 Selective Activation of ATF6 Promotes the Degradation of Misfolded Mutant Rhodopsin in the Cells
In response to ER stress, ATF6 undergoes proteolysis to release its cytosolic bZIP-containing transcription factor domain (ATF6f) that migrates to the nucleus to upregulate genes involved in ER protein folding and ERAD [12, 13]. To selectively activate ATF6, we developed a system to induce the transcription of human ATF6f upon doxycycline addition (Fig. 83.1) without activating IRE1 and PERK pathways. Using this system, we showed that selectively activating ATF6 pathway significantly reduced the protein levels of class II mutant rhodopsin, such as P23H, T17M, Y178C, C185R, D190G, and K296E rhodopsin [23]. Furthermore, ATF6 reduced the protein levels of all species of P23H rhodopsin protein (monomer, dimer, and multimers). Interestingly, the protein level of WT rhodopsin was mildly reduced by ATF6, but this effect was limited to abnormal dimer and multimeric WT rhodopsin species [23]. Monomeric WT rhodopsin levels were unaffected. Our findings indicate that ATF6 signaling reduces misfolded rhodopsins as well as aggregated rhodopsin.
83.4 Selective Activation of IRE1 or ATF6 Promotes the Degradation of S334ter Mutant Rhodopsin
A non-class II mutant rhodopsin, S334ter rhodopsin lacking the last 15 amino acid residues in the C-terminus, also causes severe retinal degeneration.[24–28]. Surprisingly, selective activation of IRE1 or ATF6 signaling also decreased the protein level of S334ter rhodopsin in the cells [23]. Unlike the class II mutant rhodopsin that form multimers, S334ter is predominantly found as a monomer in cells, similar to the WT rhodopsin, suggesting that S334ter might not be misfolded. However, we found that S334ter is more sensitive to Endo H deglycosylation than WT rhodopsin [23]. Sensitivity to Endo H indicates that the protein contains high mannose N-linked glycans, a typical characteristic of proteins that have not matured beyond the ER [29]. Increase of S334ter rhodopin’s sensitivity to Endo H indicated that there might be more S334ter rhodopsin retained in the ER and therefore more susceptible to ER quality control upregulated by selective activation of IRE1 or ATF6 signaling.
83.5 Selective Activation of PERK Reduces the Accumulation of WT and P23H Rhodopsin Proteins in the Cells
PERK bears a cytosolic kinase domain that phosphorylates eukaryotic translation initiating factor 2 subunit α (eIF2α), thereby impairing ribosomal assembly on mRNAs and attenuating protein translation [14]. To selectively activate PERK, we used a genetically altered PERK protein, Fv2E-PERK, which consists of the eIF2α kinase domain of PERK fused to two modified FK506 binding domains (Fv2E) [30]. In this system, addition of a chemical dimerizer, AP20187, rapidly triggered the activation of Fv2E-PERK’s eIF2α kinase domain, leading to the activation of PERK signaling (Fig. 83.1) without activating other endogenous UPR pathways [20, 30].
When we selectively activated PERK through this system, we found great reduction in protein levels of WT or P23H rhodopsin [23]. A similar reduction was also observed in cells transfected with other proteins such as VCAM-1 or GFP upon the activation of Fv2E-PERK [23]. These findings showed that PERK signaling reduced the protein levels of mutant and WT rhodopsin and other transfected proteins, consistent with the role of PERK in global attenuation of protein translation in the cells [31].
83.6 Conclusion
Here we dissected the effects of three separate UPR signaling pathways on rhodopsins linked to RP. We found that both IRE1 and ATF6 selectively reduced levels of misfolded rhodopsins with minimal effects on wild-type rhodopsin. Furthermore, both IRE1 and ATF6 preferentially target aggregated rhodopsins compared to the normal monomeric form. Given the ability of IRE1 and ATF6 to induce genes involved in ER protein folding and ER associated degradation, the effects of IRE1 and ATF6 signaling may be to disrupt the mutant rhodopsin aggregate and direct them for degradation. By contrast, PERK signaling non-specifically reduced levels of all rhodopsins. These findings suggest that artificial IRE1 or ATF6 signaling might be beneficial in enhancing photoreceptor cell survival in certain types of RP arising from expression of misfolded rhodopsins.
Abbreviations
- ATF6
Activating transcription factor 6
- eIF2α
Eukaryotic translation initiating factor 2 subunit α
- ER
Endoplasmic reticulum
- ERAD
ER-associated degradation
- IRE1
Inositol requiring enzyme-1
- PERK
Protein kinase RNA-like endoplasmic reticulum kinase
- RP
Retinitis pigmentosa
- UPR
Unfolded protein response
- 1NM-PP1
4-amino-1-tert-butyl-3-(1’-naphthylmethyl)pyrazolo[3,4-d] pyrimidine
Contributor Information
Wei-Chieh Jerry Chiang, Email: wcchiang@ucsd.edu.
Jonathan H. Lin, Email: jlin@ucsd.edu.
References
- 1.Berson EL. Retinitis pigmentosa. The Friedenwald lecture. Invest Ophthalmol Vis Sci. 1993;34(5):1659–1676. [PubMed] [Google Scholar]
- 2.Sung CH, Chuang JZ. The cell biology of vision. J Cell Biol. 2010;190(6):953–963. doi: 10.1083/jcb.201006020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kaushal S, Khorana HG. Structure and function in rhodopsin. 7. Point mutations associated with autosomal dominant retinitis pigmentosa. Biochemistry. 1994;33(20):6121–6128. doi: 10.1021/bi00186a011. [DOI] [PubMed] [Google Scholar]
- 4.Sung CH, Schneider BG, Agarwal N, Papermaster DS, Nathans J. Functional heterogeneity of mutant rhodopsins responsible for autosomal dominant retinitis pigmentosa. Proc Natl Acad Sci USA. 1991;88(19):8840–8844. doi: 10.1073/pnas.88.19.8840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Illing ME, Rajan RS, Bence NF, Kopito RR. A rhodopsin mutant linked to autosomal dominant retinitis pigmentosa is prone to aggregate and interacts with the ubiquitin proteasome system. J Biol Chem. 2002;277(37):34150–34160. doi: 10.1074/jbc.M204955200. [DOI] [PubMed] [Google Scholar]
- 6.Saliba RS, Munro PM, Luthert PJ, Cheetham ME. The cellular fate of mutant rhodopsin: quality control, degradation and aggresome formation. J Cell Sci. 2002;115:2907–2918. doi: 10.1242/jcs.115.14.2907. Pt 14. [DOI] [PubMed] [Google Scholar]
- 7.Lewin AS, Drenser KA, Hauswirth WW, Nishikawa S, Yasumura D, Flannery JG, et al. Ribozyme rescue of photoreceptor cells in a transgenic rat model of autosomal dominant retinitis pigmentosa. Nat Med. 1998;4(8):967–971. doi: 10.1038/nm0898-967. [DOI] [PubMed] [Google Scholar]
- 8.Kosmaoglou M, Kanuga N, Aguila M, Garriga P, Cheetham ME. A dual role for EDEM1 in the processing of rod opsin. J Cell Sci. 2009;122:4465–4472. doi: 10.1242/jcs.055228. Pt 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mendes HF, Cheetham ME. Pharmacological manipulation of gain-of-function and dominant-negative mechanisms in rhodopsin retinitis pigmentosa. Hum Mol Genet. 2008;17(19):3043–3054. doi: 10.1093/hmg/ddn202. [DOI] [PubMed] [Google Scholar]
- 10.Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol. 2007;8(7):519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- 11.Lee AH, Iwakoshi NN, Glimcher LH. XBP-1 regulates a subset of endoplasmic reticulum resident chaperone genes in the unfolded protein response. Mol Cell Biol. 2003;23(21):7448–7459. doi: 10.1128/MCB.23.21.7448-7459.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haze K, Yoshida H, Yanagi H, Yura T, Mori K. Mammalian transcription factor ATF6 is synthesized as a transmembrane protein and activated by proteolysis in response to endoplasmic reticulum stress. Mol Biol Cell. 1999;10(11):3787–3799. doi: 10.1091/mbc.10.11.3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ye J, Rawson RB, Komuro R, Chen X, Dave UP, Prywes R, et al. ER stress induces cleavage of membrane-bound ATF6 by the same proteases that process SREBPs. Mol Cell. 2000;6(6):1355–1364. doi: 10.1016/s1097-2765(00)00133-7. [DOI] [PubMed] [Google Scholar]
- 14.Harding HP, Zhang Y, Ron D. Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature. 1999;397(6716):271–274. doi: 10.1038/16729. [DOI] [PubMed] [Google Scholar]
- 15.Cox JS, Shamu CE, Walter P. Transcriptional induction of genes encoding endoplasmic reticulum resident proteins requires a transmembrane protein kinase. Cell. 1993;73(6):1197–1206. doi: 10.1016/0092-8674(93)90648-a. [DOI] [PubMed] [Google Scholar]
- 16.Calfon M, Zeng H, Urano F, Till JH, Hubbard SR, Harding HP, et al. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature. 2002;415(6867):92–96. doi: 10.1038/415092a. [DOI] [PubMed] [Google Scholar]
- 17.Yoshida H, Matsui T, Yamamoto A, Okada T, Mori K. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell. 2001;107(7):881–891. doi: 10.1016/s0092-8674(01)00611-0. [DOI] [PubMed] [Google Scholar]
- 18.Bishop AC, Ubersax JA, Petsch DT, Matheos DP, Gray NS, Blethrow J, et al. A chemical switch for inhibitor-sensitive alleles of any protein kinase. Nature. 2000;407(6802):395–401. doi: 10.1038/35030148. [DOI] [PubMed] [Google Scholar]
- 19.Lin JH, Li H, Yasumura D, Cohen HR, Zhang C, Panning B, et al. IRE1 signaling affects cell fate during the unfolded protein response. Science. 2007;318(5852):944–949. doi: 10.1126/science.1146361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin JH, Li H, Zhang Y, Ron D, Walter P. Divergent effects of PERK and IRE1 signaling on cell viability. PLoS ONE. 2009;4(1):e4170. doi: 10.1371/journal.pone.0004170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hiramatsu N, Joseph VT, Lin JH. Monitoring and manipulating mammalian unfolded protein response. Meth Enzymol. 2011;491:183–198. doi: 10.1016/B978-0-12-385928-0.00011-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chiang WC, Messah C, Lin JH. IRE1 directs proteasomal and lysosomal degradation of misfolded rhodopsin. Mol Biol Cell. 2012;23(5):758–770. doi: 10.1091/mbc.E11-08-0663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chiang WC, Hiramatsu N, Messah C, Kroeger H, Lin JH. Selective activation of ATF6 and PERK endoplasmic reticulum stress signaling pathways prevent mutant rhodopsin accumulation. Invest Ophthalmol Vis Sci. 2012;53(11):7159–7166. doi: 10.1167/iovs.12-10222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shinde VM, Sizova OS, Lin JH, Lavail MM, Gorbatyuk MS. ER stress in retinal degeneration in S334ter rho rats. PLoS One. 2012;7(3):e33266. doi: 10.1371/journal.pone.0033266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dykens JA, Carroll AK, Wiley S, Covey DF, Cai ZY, Zhao L, et al. Photoreceptor preservation in the S334ter model of retinitis pigmentosa by a novel estradiol analog. Biochem Pharmacol. 2004;68(10):1971–1984. doi: 10.1016/j.bcp.2004.06.042. [DOI] [PubMed] [Google Scholar]
- 26.Lee D, Geller S, Walsh N, Valter K, Yasumura D, Matthes M, et al. Photoreceptor degeneration in Pro23His and S334ter transgenic rats. Adv Exp Med Biol. 2003;533:297–302. doi: 10.1007/978-1-4615-0067-4_36. [DOI] [PubMed] [Google Scholar]
- 27.Anderson RE, Maude MB, McClellan M, Matthes MT, Yasumura D, LaVail MM. Low docosahexaenoic acid levels in rod outer segments of rats with P23H and S334ter rhodopsin mutations. Mol Vis. 2002;8:351–358. [PubMed] [Google Scholar]
- 28.Kroeger H, Messah C, Ahern K, Gee J, Joseph V, Matthes MT, et al. Induction of endoplasmic reticulum stress genes, BiP and Chop, in genetic and environmental models of retinal degeneration. Invest Ophthalmol Vis Sci. 2012;53(12):7590–7599. doi: 10.1167/iovs.12-10221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sherblom AP, Smagula RM. High-mannose chains of mammalian glycoproteins. Methods Mol Biol. 1993;14:143–149. doi: 10.1385/0-89603-226-4:143. [DOI] [PubMed] [Google Scholar]
- 30.Lu PD, Jousse C, Marciniak SJ, Zhang Y, Novoa I, Scheuner D, et al. Cytoprotection by pre-emptive conditional phosphorylation of translation initiation factor 2. EMBO J. 2004;23(1):169–179. doi: 10.1038/sj.emboj.7600030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Harding HP, Zhang Y, Bertolotti A, Zeng H, Ron D. Perk is essential for translational regulation and cell survival during the unfolded protein response. Mol Cell. 2000;5(5):897–904. doi: 10.1016/s1097-2765(00)80330-5. [DOI] [PubMed] [Google Scholar]