ABSTRACT
We used transcriptomics to compare instinctive and learned, reward-based honey bee behaviors with similar spatio-temporal components: mating flights by males (drones) and time-trained foraging flights by females (workers), respectively. Genome-wide gene expression profiling via RNA sequencing was performed on the mushroom bodies, a region of the brain known for multi-modal sensory integration and responsive to various types of reward. Differentially expressed genes (DEGs) associated with the onset of mating (623 genes) were enriched for the gene ontology (GO) categories of Transcription, Unfolded Protein Binding, Post-embryonic Development, and Neuron Differentiation. DEGs associated with the onset of foraging (473) were enriched for Lipid Transport, Regulation of Programmed Cell Death, and Actin Cytoskeleton Organization. These results demonstrate that there are fundamental molecular differences between similar instinctive and learned behaviors. In addition, there were 166 genes with strong similarities in expression across the two behaviors – a statistically significant overlap in gene expression, also seen in Weighted Gene Co-Expression Network Analysis. This finding indicates that similar instinctive and learned behaviors also share common molecular architecture. This common set of DEGs was enriched for Regulation of RNA Metabolic Process, Transcription Factor Activity, and Response to Ecdysone. These findings provide a starting point for better understanding the relationship between instincts and learned behaviors. In addition, because bees collect food for their colony rather than for themselves, these results also support the idea that altruistic behavior relies, in part, on elements of brain reward systems associated with selfish behavior.
KEY WORDS: Brain, Gene expression, Mushroom bodies, Neuroethology, Social insects
Highlighted Article: Brain gene expression analysis sheds new light on the relationship between instincts and learned behaviors and provides new insights into how the brain's reward system influences social behavior.
INTRODUCTION
Behaviors arise from both inborn instincts and learning. The molecular basis of several innate behavioral processes has been extensively analyzed, including monarch butterfly migration (Zhan et al., 2014), rodent mating (Shelley et al., 2006), rodent aggression (Takahashi and Miczek, 2014), bird song learning (Balakrishnan et al., 2014) and honey bee behavioral maturation (Zayed and Robinson, 2012). Similarly, much is known about the molecular basis of learned behaviors (Kandel, 2001). However, little is known about the degree to which related instincts and learned behaviors rely upon similar molecular mechanisms (Isosaka et al., 2015).
We used honey bees (Apis mellifera Linnaeus 1758) to address this issue because they have a large behavioral repertoire of both innate and learned behaviors. We focused on a pair of behaviors with similar spatiotemporal components that are performed on an innate or learned basis: mating and foraging.
When (male) drone honey bees reach sexual maturity at approximately 1 week of adult age, they begin to make daily mating flights and leave the hive to search for virgin queens at specific locations in the environment (Winston, 1987). The timing of these mating flights is both innate and species-specific within the genus Apis (Rinderer et al., 1993). Altering temperature and photoperiod can shift the time of flight (Oxley et al., 2010), suggesting that the drones are using an internal clock that is entrained to the day/night cycle.
After worker (female) honey bees have matured from working in the hive at approximately 2 to 3 weeks of adult age, they begin to forage for nectar and pollen. While the motivation of workers to undertake foraging flights also is innate, workers learn the location of floral resources and the time of their availability. This allows them to forage efficiently in the face of temporally and spatially variable food sources (Van Nest and Moore, 2012). As with drone mating flights, worker foraging flights also are under the control of an internal clock (Renner, 1957). Transcriptomic analyses revealed that different spatiotemporal foraging memories are associated with distinct patterns of brain gene expression, including genes that regulate circadian rhythms (Naeger et al., 2011). We used these two spatiotemporal flight behaviors in drone and worker honey bees to compare the patterns of gene expression in the brain associated with instinctive and learned behaviors.
A second motivation for selecting these two behaviors was to learn more about the molecular basis of reward-based behavior. Both mating and foraging involve the pursuit of natural stimuli that are generally considered to be rewarding (Young and Wang, 2004). Our recent study showed that brain transcriptomic responses to different types of food rewards in honey bees involve a mixture of similar and different molecular pathways (McNeill et al., 2016). We were interested in extending these analyses to other types of rewards to continue to explore the idea that brain responses to stimuli involved in social rewards involve subcomponents of a more general reward system, which has been shown in both insects (McNeill et al., 2016) and mammals (Cromwell and Schultz, 2003; Lardeux et al., 2009).
The mushroom bodies (MB), a region in the dorsal protocerebrum of the brain involved in multi-modal sensory processing, learning and memory (Zars, 2000), was selected for the present study, as it is involved in both instinctive (Sen Sarma et al., 2009; Lutz and Robinson, 2013; McNeill et al., 2016) and learned behavior. In addition, the MB also has recently been shown to show strong transcriptional responses to various types of food rewards (McNeill et al., 2016).
MATERIALS AND METHODS
Our objective was to obtain transcriptomic profiles of bees embarking on reward-seeking flights in response to learned and innate drives. We trained workers to fly to an artificial feeder during the same window of time in the afternoon when drones were making mating flights. We collected both workers and drones in the afternoon as they exited the hive to forage and seek mates, respectively, and in the morning when they were inactive. Collections were designed to capture the transcriptomic signatures associated with the anticipatory states of foraging and mating, without the confounding effects of flight itself.
Behavioral manipulations and bee collections
Three replicates of the experiment were performed in the summer of 2013 at the University of Illinois Bee Research Facility, Urbana, Illinois, USA. Each of the three colonies used in these experiments was headed by a naturally mated queen (unrelated to each other), contained 8000–10,000 workers and several hundred drones, and was maintained according to standard methods.
At the start of each replicate, the colony was moved into a large (20×6×3 m), outdoor, screened enclosure to control the time of food availability (Naeger et al., 2011). Every afternoon at 14:00–17:00 h CST a feeder containing 50% (w/v) sucrose solution was placed in the enclosure at the opposite end of the enclosure from the hive, coinciding with the time that mating flights begin for drones at this locality. After allowing 1 week for the colony to adjust to the new environment and the foraging workers to train themselves to the time and location of food availability, workers at the feeder were marked with a system of paint dots (Testor's PLA) on the thorax and abdomen. A different color of paint was used each day in order to track individual behavior, and only individuals exhibiting at least 3 days of consistent behavior were used for analysis. We identified bees that were anticipating food availability on their own, rather than those that might have been recruited to the feeder by successful foragers (von Frisch, 1967). This was accomplished by marking foragers at the sucrose feeder, both for several minutes before and 3 min after the feeder was made available. Drones were marked in a similar way at 14:00–15:00 h upon exiting the hive, also over several days of observation.
Paint-marked foragers and drones (N=ca. 100) were collected into liquid nitrogen at the hive entrance as they exited the hive, prior to the onset of flight, in order to prevent any effects of flight on brain gene expression. We were interested in capturing the transcriptomic signatures associated with the anticipatory states of foraging and mating, without the confounding effects of flight itself. The next morning at 09:00 h the hive was opened and remaining marked bees were collected into liquid nitrogen, to capture the transcriptomic signature of the same groups of foragers and drones, this time in inactive states.
Brain dissection and mushroom body sample preparation
Heads were removed while frozen and incubated for 16 h in RNAlater ICE (Ambion). Brains were dissected to obtain samples containing the MB and surrounding nuclei (Fig. S1), following an established protocol (Sen Sarma et al., 2009; Lutz and Robinson, 2013; McNeill et al., 2016). RNA was extracted using Picopure RNA isolation kits (Applied Biosystems) and poly-adenylated RNA was enriched using Oligo(dT)25 Dynabeads (Thermo Fisher Scientific). RNA was fragmented and converted to cDNA (as per instructions in the NEXTflex Directional RNA-Seq Kit, Bioo Scientific Corporation). Average fragment sizes were estimated with a BioAnalyzer High Sensitivity DNA assay (Agilent Technologies). cDNA libraries were labeled with NEXTflex RNA-Seq Barcodes for multiplexing during sequencing.
Final cDNA library concentrations were estimated using a Qubit (Thermo Fisher Scientific), and then eight barcoded libraries were pooled for sequencing, preserving information to allow data to be resolved at the individual bee level. The total concentration of all adapter-ligated fragments within each assembled library pools was confirmed using KAPA Library Quantification kits (Kapa Biosystems). The pooled libraries were sequenced on a HiSeq 2000 (Illumina) at the W. M. Keck Center for Comparative and Functional Genomics, University of Illinois at Urbana-Champaign. A total of 48 individual samples were sequenced, 12 individuals for each behavioral state, either just prior to flight or inactive.
RNA sequencing and analysis
Reads were trimmed to remove adapter and low-quality sequence information using Trimmomatic (v0.30; Bolger et al., 2014), and mapped to the v4.5 build of the honey bee genome (Elsik et al., 2014) using TopHat (v2.0.8; Trapnell et al., 2009). We counted reads mapping to genes (Official Gene Set 3.2) using HTseq (v0.54; Anders et al., 2015). Over 2.6×109 paired-end reads were generated; after trimming for quality and rare reads, there were on average ca. 1.8×107 pairs of reads per sample mapping to the honey bee official gene models (Assembly 4.5; Elsik et al., 2014). This translates to an average of 64% of the reads aligning to the honey bee genome, comparable to what was found in McNeill et al. (2016). Genes without at least one count per million in five individual bees were removed, leaving 11,242 genes for analysis. In addition, two drones, both from the collection of inactive drones, were obvious outliers in principal component analysis (PCA) and hierarchical clustering analyses (data not shown), and were removed from the analysis (Fig. S2).
RNA sequencing (RNA-seq) data were normalized for differences in the abundance of read counts mapped to genes between samples using the trimmed mean of M-values (TMM) normalization method in limma's voom normalization+weights function (EdgeR v3.4.2, Bioconductor; Robinson et al., 2010). Variance in gene expression was estimated using common, trended and tagwise dispersion models sequentially (McCarthy et al., 2012), and a prior degrees of freedom value was chosen that best fit all genes to the tagwise dispersion model, similar to a previously described method (McCarthy et al., 2012). Differences between groups were analyzed using a limma robust model with colony as a fixed effect, sex and time of collection as the main effects, and the interaction between sex and time. Post hoc contrasts were used to analyze the sexes independently.
The log2-transformed, TMM-normalized expression values for genes for all individuals were also used for Weighted Gene Co-Expression Network Analysis (WGCNA v1.34; Zhang and Horvath, 2005; Langfelder and Horvath, 2008) running in R (Linux, v3.0.0). WGCNA was performed to identify genes that have similar patterns of expression to each other. This unsupervised analysis finds genes that are highly correlated with each other in expression and assigns them to modules. Unsigned modules containing at least 30 co-expressed genes were formed using deepSplit=3; similarly co-expressed modules (Pearson correlation coefficients >0.9) were merged. A dendrogram was created in R using eigengene values for the genes in each module. Linear regression analyses were performed in R; the adjusted R2 values were then used to create a similarity matrix heat map and a cut-off false discovery rate (FDR) of <0.05 was used to infer significance. Gene ontology (GO) term enrichment analysis was performed using the FlyBase identification number representing the best BLAST hit for each honey bee gene and the DAVID Bioinformatics Resources Functional Annotation tool (Huang et al., 2009). GO terms returned by DAVID with a modified Fisher's exact P-value of <0.05 were considered significantly enriched.
Comparisons with other honey bee neurogenomic experiments
To determine whether the results from this study show similarities to other previously published studies on related topics, representation factor (RF) analysis was performed (Alaux et al., 2009). This involves testing for significant overlaps in lists of differentially expressed genes (DEGs) using an exact hypergeometric test.
RESULTS
Mushroom body transcriptomic signature of sex differences
Using a significance threshold of FDR<0.05, and considering both the active and inactive individuals for both behaviors, we identified 5680 DEGs between the sexes in the MB. Of the 2922 genes that were higher in drones than workers, 116 had a log2 fold change greater than 2, as did 48 of the 2758 genes higher in workers, indicating a mix of genes with high and low magnitude expression differences as is typical in behavioral genomics. PCA revealed that this sex difference, represented by PC1, accounted for the largest fraction (20.2%) of the variance in MB gene expression in the entire study (Fig. 1). GO enrichment analyses revealed 639 significantly enriched categories (Table S1), including those associated with Alternative Splicing, Nucleotide Binding, Cytoskeleton Organization, Neuron Differentiation, Embryonic Development, and Learning and Memory. Genes annotated as being involved in insect sex determination showed a diversity of expression patterns. For example, sex lethal and doublesex did not show differential expression, complimentary sex determiner and feminizer were higher in drones, and transformer-2 and fruitless were higher in workers. Because sex differences are such a strong contributor to the variance, they could potentially mask some DEGs in the following experimental variables of interest.
Fig. 1.
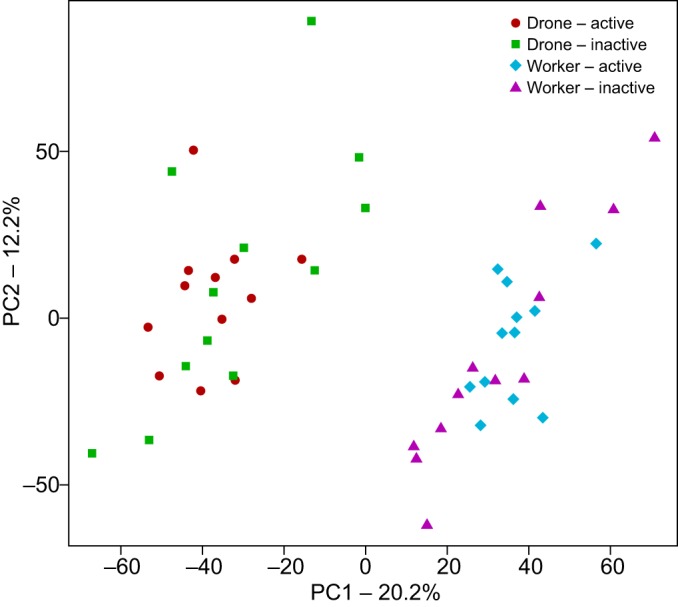
Principal component analysis demonstrates large sex differences in mushroom body gene expression, regardless of activity state. Active drones and (time-trained) workers (N=ca. 100 each) were collected just as they left the hive in the afternoon, to prevent any effects of flight on brain gene expression. Inactive workers and drones were collected the following morning in the hive.
Mushroom body transcriptomic signature unique to instinctive spatiotemporal flight behavior in drones
Because bees were captured as they exited the hive in the afternoon just prior to taking flight in the afternoon or when inactive in the morning, our analyses of differences between these two time points should reflect, in part, the motivational state to perform the behavior, rather than the consequences of performing it. Comparing these two groups of drones, there were 623 DEGs associated with the onset of drone mating behavior that did not differ in expression between active and inactive workers. Of particular note are transcription factors including the genes egr and Jun-related antigen, and the hormone receptors Hr38 and Hr51. egr showed a dramatic increase in expression as drones undertook mating flights (log2 fold change=2.7; FDR=1.91×10−8). Significantly enriched GO categories include Transcription, Unfolded Protein Binding, Post-embryonic Development, and Neuron Differentiation (Table S1).
Mushroom body transcriptomic signature unique to learned spatiotemporal flight behavior in workers
Comparing time-trained foragers exiting the hive to forage in the afternoon or when inactive in the morning, there were 473 DEGs associated with the onset of foraging that did not differ in expression between active and inactive drones. These genes include Neprilysin2, Adenosine receptor, Turtle and Arrestin2. There also were many unannotated genes on this list, including 26 of the 35 genes with the strongest expression differences (lowest FDR values). GO analysis revealed significantly enriched categories including Lipid Transport, Regulation of Programmed Cell Death, and Actin Cytoskeleton Organization (Table S1). Notably absent from this list of enriched categories was Learning and Memory, even though it was significantly enriched in the list of genes that differed between the sexes, as stated above.
Mushroom body transcriptomic signature common to both instinctive and learned spatiotemporal flight behaviors
Comparing the two active states with the two inactive states in both behaviors made it possible to identify a set of genes that showed common MB expression differences for both mating and foraging behaviors (Fig. 2). There were 166 genes that showed this combined pattern, a number that is highly significantly enriched compared with what would be expected by chance from the two separate analyses (RF=3.7, P<2.11×10−54). Remarkably, 162 of the 166 genes were concordant in the direction of change, further establishing that this set of genes shows similar transcriptional change across the two behaviors. In addition, the transcriptomic signatures for bees beginning to express either type of behavior appear to have less variation than those of bees collected while inactive; individuals of both sexes cluster closer together along the PC2 axis in the flight group as compared with the inactive group (Fig. 1). These genes could be associated with reward motivation, anticipation of flight behavior in general or circadian changes that are associated with both behaviors. However, if all of these genes were associated with circadian changes only, then we might have expected more similar levels of variation between active and inactive bees. These results indicate that the motivational states associated with the onset of mating and foraging behavior are reflected in a strong combined neurogenomic signature in the MB.
Fig. 2.

Similarities and differences in mushroom body gene expression for similar instinctive and learned behaviors: mating (drone) and time-trained foraging (workers). Venn diagram indicates the numbers of differentially expressed genes for each context.
Notable upregulated genes showing similar patterns of expression for both mating and foraging behavior included Dopamine N-acetyltransferase, the transcription factors CrebA, Diminutive and Ecdysone receptor (GB48059), and the circadian clock genes Vrille and Clockwork Orange. Downregulated genes included the transcription factor Kruppel homolog 1, the histone methyltransferase G9a and the circadian clock gene Cryptochrome2/(6-4)-photolyase. Among the GO categories that were enriched for this set of genes were Regulation of RNA Metabolic Process, Transcription Factor Activity, and Response to Ecdysone (Table S1). Despite the presence of three genes annotated as involved in circadian rhythms as part of the set of 166 genes, GO categories such as Circadian Rhythms and Rhythmic Behavior were not significantly enriched (P>0.10).
WGCNA results
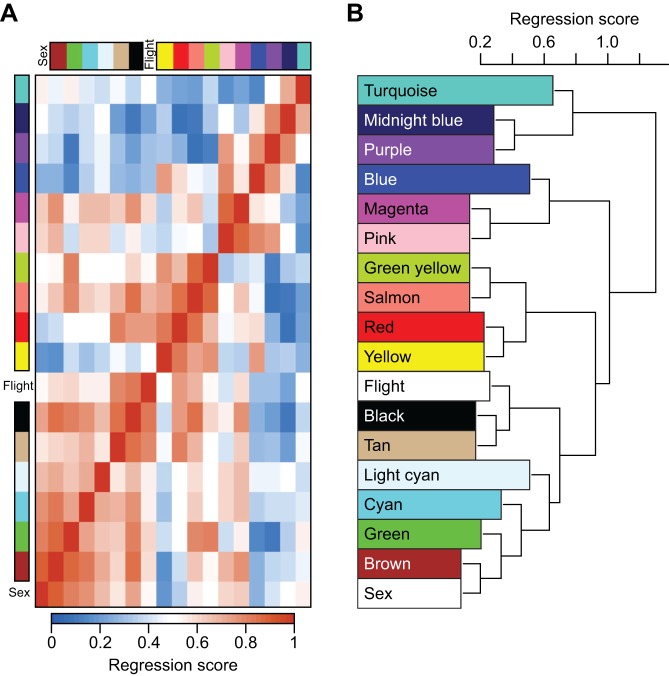
WGCNA also revealed elements of unique and common MB transcriptomic signatures for instinctive and learned spatiotemporal flight behaviors. Hierarchical clustering of the eigengene values of the modules of genes that are highly correlated with each other in expression revealed clusters of modules that correspond with the four patterns of gene expression described above (Fig. 3). There were six modules significantly correlated (FDR<0.05) with sex differences, including the second and third largest modules (Table 1). Five modules were significantly correlated with drone instinctive flight behavior, and seven modules were significantly correlated with worker learned flight behavior. Three of the modules were significantly associated with both instinctive and learned behavior, and again represent a common core of genes associated with spatiotemporal reward-related flight in general: midnight blue, red and black (Table S2) (module names are randomly assigned by the WGCNA program).
Fig. 3.
Modules from weighted gene co-expression network analysis (WGCNA) comparing similar instinctive and learned behaviors: mating (drone) and time-trained foraging (workers). (A) Heat map; (B) dendrogram. Values are based on linear regression coefficient of determination, using module eigengene values from the WGCNA analysis and the main effects of sex and flight (either at onset of flight or inactive) as binary classifying variables. Higher regression scores mean more similar patterns in expression between modules or more predictive power of the main effects on module expression in the heat map (more red) and dendogram (closer clustering).
Table 1.
Modules formed by weighted gene co-expression network analysis (WGCNA) comparing similar instinctive and learned behaviors: mating (drone) and time-trained foraging (workers)
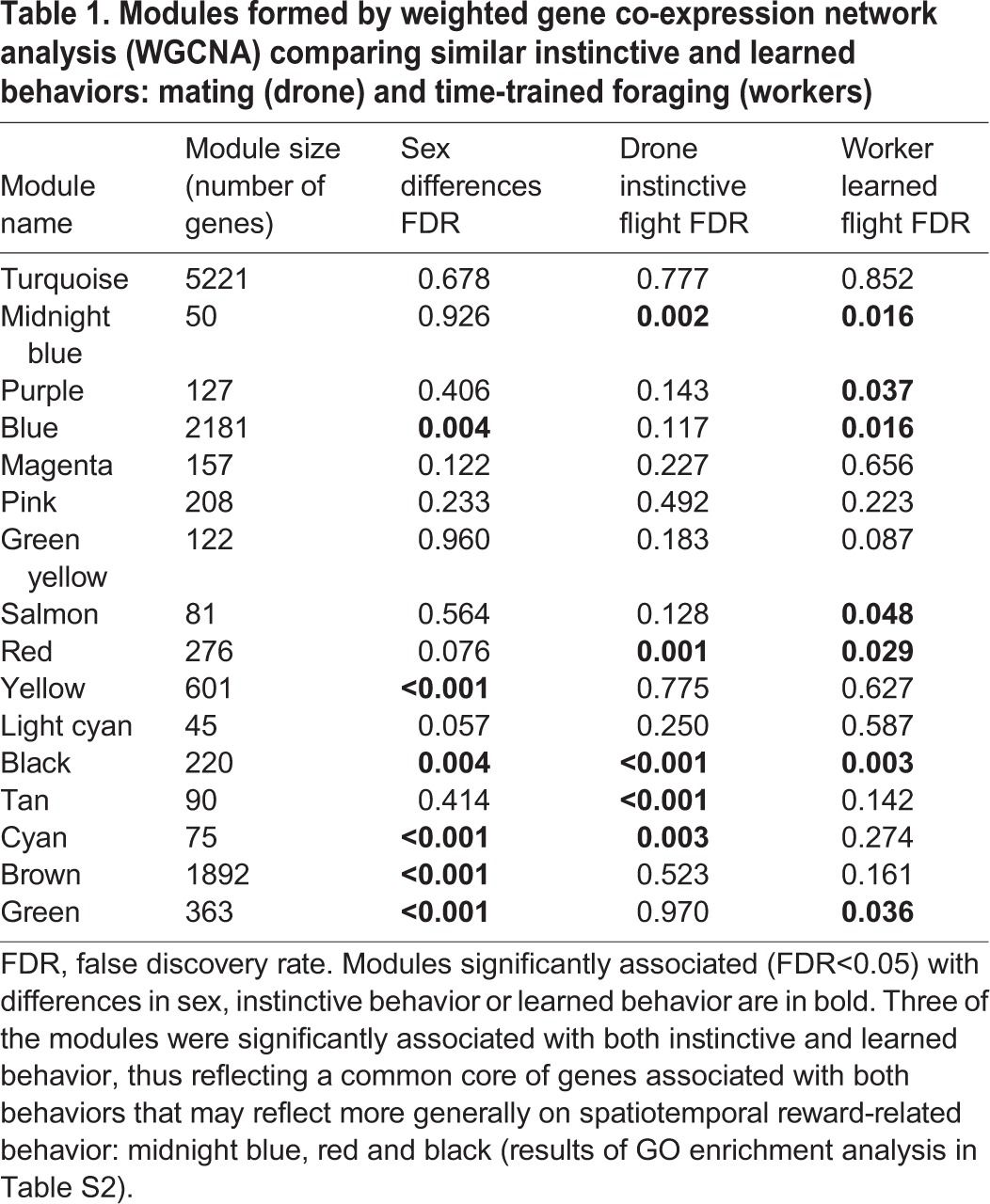
The module midnight blue was the smallest of all modules, with only 50 genes, but nevertheless was enriched for a few GO categories including Regulation of Histone Acetylation and Regulation of Histone Modification (Table S2). The module red was significantly enriched for many GO categories associated with transcriptional regulation. It included the transcription factors Hr38, Hr51, egr, Kayak and Ecdysone receptor, as well as the canonical clock genes Period, Vrille and Clockwork Orange. The black module also was enriched for a GO category associated with transcriptional regulation, Sequence-specific DNA Binding. It contained the transcription factors Creb A, Deadpan, Jun-related antigen and Trithorax, as well as the neurotransmitter-associated genes Dopa decarboxylase and Dopamine N Acetyltransferase. Many of the individual genes highlighted here also were mentioned above in the accounts of the DEG lists.
Comparisons with other honey bee neurogenomic experiments
We also compared our RNA-seq results with findings from three previously published transcriptomic studies to develop additional insights into the gene expression patterns detected here. Naeger et al. (2011) conducted a transcriptomic study of time-trained workers with the same behavioral methods as those used here, but using microarrays rather than RNA-seq, and whole brain rather than MB. There was a significant overlap of DEGs in that study and the worker DEGs in the present study (P=0.010, hypergeometric test). There also was a significant enrichment of genes shared between time-trained workers in Naeger et al. (2011) and the instinctive flight behavior of drones in the present study (P=0.049), again suggesting that there is a consistent core set of genes associated with reward-related spatiotemporal flight motivation across both learned and instinctive contexts.
Another whole-brain microarray analysis (Zayed et al., 2012) examined similarities and differences in behavioral maturation in drones and workers, related to the age at onset of mating flights and foraging behavior, respectively. There was a highly significant overlap of genes that showed sex differences in expression in that study and the present study (P=1.24×10−23). By contrast, there was no enriched overlap between genes associated with drone behavioral maturation in Zayed et al. (2012) and the immediate activation of drone mating flights in the present study (P=0.763) or between worker behavioral maturation (Zayed et al., 2012) and the immediate activation of time-trained worker foraging flights in the present study (P=0.761). Perhaps this reflects differences in the regulation of behavioral states that have longer or shorter time scales. This speculation is consistent with findings from queen honey bees: Manfredini et al. (2015) reported strong differences in whole-brain transcriptomic profiles when comparing queens returning from successful mating flights and inactive virgin queens collected inside their hives. Queen–drone mating flight transcriptomic analyses have not been conducted; with only one queen per colony, it would not be possible to compare large numbers of individuals from the same colonies, as we did with workers and drones.
McNeill et al. (2016) used RNA-seq to determine whether honey bee MB responses to food type (pollen or nectar) and food value involve different subsets of genes generally responsive to food. They found both differences and similarities in MB transcriptomic responses to these different components of food reward. An informal comparison of the enriched GO categories from their study and the present study revealed overlap for categories associated with protein folding, protein localization, extracellular matrix and cell adhesion.
DISCUSSION
Gene expression in the mushroom bodies that is associated with mating and foraging behavior in honey bees shows an intriguing mixture of similarities and differences. The similarities suggest that reward-related instinctive and learned behaviors share common molecular architecture. Because worker bees collect food for their colony rather than for themselves, these results also support the idea that social evolution has relied on elements of reward processing that function at the level of the individual in order to build a social reward system.
Overall, there were more differences than similarities in the mushroom body gene expression signatures associated with the spatiotemporal flight behavior of drones and workers. This is despite the fact that the same brain region was used, and the samples were collected at the same times of day from related individuals in the same colonies. The biggest differences were attributable to sex and were probably not directly related to the two behaviors. In addition, there were strong similarities between the sex differences in gene expression reported here and in Zayed et al. (2012), suggesting that sex differences in brain gene expression are a robust feature of behavioral regulation, in honey bees as well as in other species (e.g. Trabzuni et al., 2013). Honey bees have haplodiploid sex determination, with haploid males developing from unfertilized eggs and diploid females developing from fertilized eggs. Even though endoreduplication can occur in drones to restore diploidy within some tissues such as flight muscle (Aron et al., 2005), this process does not occur in the brain, leaving open the question of how gene dosage compensation can occur on such a mass scale when essentially half the genome is differentially expressed. These results highlight how the genome continues to orchestrate sex differences after development.
Though sex differences account for the majority of the differentially expressed genes in our experiment and may mask some positive results for our experimental variables of interest, our experimental design nonetheless enabled us to distinguish gene expression differences related to sex from those related to activity and behavior. Drone mating flights, which are initiated by an instinctive timing mechanism, were associated with differences in expression for several transcription factors including steroid nuclear receptors Hr38 and Hr51. Although sex determination in insects is largely genetic, there is evidence that ecdysteroids act like vertebrate steroids in sex-specific ways to affect behavior (De Loof, 2006). Additionally, Hr51 is known to function in axon guidance and extension in the MB of Drosophila melanogaster (Lin et al., 2009).
Another transcription factor, egr, showed a strong increase in drone MB expression at the initiation of a mating flight. egr is involved in neuroplasticity in vertebrates (Knapska and Kaczmarek, 2004) and is upregulated in the MB of workers following an orientation flight (Lutz and Robinson, 2013). During orientation flights, bees learn the location of their hive relative to prominent landmarks and the position of the sun. Tracking bees during orientation flights by harmonic radar has revealed how bees concentrate on different parts of their environment during different orientation flights (Capaldi et al., 2000). Drones take orientation flights as well, but in our study they were collected as they were leaving the hive and thus did not have the opportunity to learn their surroundings. Thus, rather than changing expression in response to a learning event, egr expression changes preceded the flight behavior. This finding suggests that egr may also be involved with priming the brain for navigational learning. This further underscores the dynamic interrelationships between instinct and learning; these two forms of behavior are often intertwined in complex ways. Many instincts are further shaped by learning, and many forms of learning act on innate neural circuits (Isosaka et al., 2015).
The genes associated specifically with time-trained foraging behavior in workers showed strong overlap with the genes found in a previous study of the same behavior (Naeger et al., 2011). Given that the previous study was performed with microarrays on whole brains in a different locality and the present study was performed with RNA-seq on the MB (a region that accounts for almost half the volume of the entire bee brain), this result indicates that there is a robust transcriptomic signature associated with time-trained foragers, regardless of when and where the training and flight occur.
The transcriptomic signature of time-trained foraging behavior in the MB involves several elements related to neural plasticity. This is evident by considering some of the DEGs and the enriched GO categories that characterize the entire DEG list. One of the clearest hints is the enrichment for cytoskeleton terms, mainly those associated with actin. Actin is involved in the remodeling of dendritic spines, an important process for neuroplasticity (Dillon and Goda, 2005). These GO terms are noticeably absent in describing the gene list associated with the onset of drone mating flights. The enriched GO terms for protein catabolism and localization also suggest that post-translational processes are involved in learned spatiotemporal flight. Other enriched terms are more opaque, such as Lipid Transport and Regulation of Programmed Cell Death, and may reflect limitations in current knowledge and annotations. Genes showing consistent expression differences associated with time-trained behavior in workers include Neprilysin2, an endopeptidase responsible for clearing proteins such as Amyloidβ from the brain (Hafez et al., 2011); Turtle, a cell adhesion gene necessary for axon guidance (Cameron et al., 2013) and flight behavior (Bodily et al., 2001); Arrestin2, a gene necessary for terminating neurotransmitter signals; and Adenosine receptor, which is targeted by caffeine, a drug shown to increase associative learning in bees and vertebrates (Wright et al., 2013). These are prime targets for future functional analyses.
As stated above, there also was a common set of differentially expressed genes that were associated with both mating and foraging behavior. Several heat shock proteins and GO categories such as Unfolded Protein Binding were significantly associated with the onset of flight behavior in both workers and drones. This result was obtained from both differential gene expression analysis and WGCNA. These changes might reflect preparation for encountering the less predictable and more stressful environment outside the hive, or might be related to the finding that heat shock proteins have also been implicated as important for circadian behavior in Drosophila (Hung et al., 2009).
The significant enrichment of GO categories such as Post-embryonic Development and Neuron Differentiation in association with the onset of flight behavior in both workers and drones implies that genes traditionally known to be involved in development also play important functions in adults. This suggests that the genes involved with building the molecular and physiological underpinnings of behaviors during development are also involved in their maintenance and expression in adulthood. These findings may also be related to the relationships between development and behavior that underlie the relationships between the development of innate neural circuits and adult learning.
The common set of genes that were differentially expressed similarly in association with the onset of flight behavior in both workers and drones also included a variety of transcription factors. This includes CrebA, well known as a regulator of neural plasticity in both vertebrates and invertebrates (Sakamoto et al., 2011), which has also been implicated as a key regulator of behavioral plasticity in the honey bee (Chandrasekaran et al., 2011). WGCNA analysis also revealed modules common to both behaviors that were enriched for transcription factors, including genes related to dopamine neurotransmission. Dopa decarboxylase and Dopamine N Acetyltransferase are two genes that control the level of dopamine in the brain that were upregulated in both the instinctive and learned contexts. Dopamine levels are higher in forager brains compared with hive worker brains (Wagener-Hulme et al., 1999), and higher in mature drone brains compared with younger drone brains (Harano et al., 2008). Dopamine is associated with motivation, in vertebrates (reviewed in Salamone and Correa, 2012) and Drosophila (Kume et al., 2011; Van Swinderen and Andretic, 2011), and the experiments in the present study in essence compared unmotivated bees with those motivated to fly.
It also is possible that some elements of the common patterns of MB gene expression between drones and workers reflect the fact that time differences were necessarily confounded with the comparisons of active and inactive states in both cases. In support of this possibility, a few of the canonical clock gene transcription factors known to regulate circadian rhythms (reviewed by Allada and Chung, 2010) were differentially expressed similarly in both the instinctive and learned behavioral contexts, including Vrille, Clockwork Orange and Cryptochrome2/(6-4)-photolyase. It is possible that this reflects the entrainment of bees to the day/night cycle, allowing for consistent timekeeping based on internal states rather than acute cues from the environment.
To our knowledge, this is only one of two studies that analyze the similarities and differences between instinctive and learned behavior at the molecular level. Isosaka et al. (2015) reported that the same serotonergic cells in the amygdala control both innate and learned fear behavior in mice, but artificial inactivation of these cells upregulated the innate freezing response and downregulated the learned freezing response. Our more general transcriptomic analysis complements this in-depth analysis of a single molecular pathway, and together they show that there are common molecular substrates for both instinctive and learned behaviors. Future challenges include identifying which genes are causative in their relationship with spatiotemporal flight behavior, and further elucidating the relationships between instincts and learned behaviors at the molecular and circuit levels.
Finally, our results provide new insights into how the brain's reward system influences social behavior. Because worker bees collect food for their colony rather than for themselves, this behavior can be interpreted as altruistic, and performed for the good of the colony. McNeill et al. (2016) performed a transcriptomic dissection of this behavior and showed that the MB responses to differences in food type or food value include a subset of the molecular pathways involved in the bee's response to a food reward that is ingested for personal sustenance. These results support the idea that altruistic behavior in bees relies, in part, on elements of brain reward systems associated with behavior that provides individual benefits, which also has been shown in mammals (Cromwell and Schultz, 2003; Lardeux et al., 2009). Our finding of commonalities in the MB transcriptomic signatures of worker foraging and drone mating – another behavior that provides individual benefits – provides additional support for this idea. Moreover, there was some overlap of enriched GO categories from McNeill et al. (2016) and our study, providing additional evidence for these connections. These findings suggest that social evolution has relied on elements of reward processing, involving both innate and learned behaviors, which function at the level of the individual in order to build a social reward system. Further transcriptomic studies of matched instinctive and learned behaviors thus hold the potential to illuminate the molecular architecture of behavior as well as the evolution of altruistic social systems.
Acknowledgements
We thank D. C. Nye for assistance with the bees; A. C. Ahmed for RNA extractions; M. S. McNeill for assistance with GO and WGCNA analyses; K. M. Kapheim and J. Drnvich (High Performance for Biological Computing Center, Carver Biotechnology Center) for assistance with statistical analyses; E. Hadley for assistance with figures; and A. B. Barron, K. M. Kapheim and M. S. McNeill for comments that improved the manuscript.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
N.L.N. and G.E.R. conceived the project, designed the study, and wrote the paper; N.L.N. performed the experiments.
Funding
This work was supported by a National Institutes of Health Director's Pioneer Award (DP1 OD006416 to G.E.R.); an Achievement Rewards for College Scientists Foundation Fellowship; and a Dissertation Completion Fellowship from the Graduate College, University of Illinois at Urbana-Champaign (to N.L.N.). Deposited in PMC for release after 12 months.
Data availability
The RNA-seq data sets have been deposited at https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE85433
Supplementary information
Supplementary information available online at http://jeb.biologists.org/lookup/doi/10.1242/jeb.144311.supplemental
References
- Alaux C., Sinha S., Hasadsri L., Hunt G. J., Guzmán-Novoa E., DeGrandi-Hoffman G., Uribe-Rubio J. L., Southey B. R., Rodriguez-Zas S. and Robinson G. E. (2009). Honey bee aggression supports a link between gene regulation and behavioral evolution. Proc. Natl. Acad. Sci. USA 106, 15400-15405. 10.1073/pnas.0907043106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allada R. and Chung B. Y. (2010). Circadian organization of behavior and physiology in Drosophila. Annu. Rev. Physiol. 72, 605-624. 10.1146/annurev-physiol-021909-135815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S., Pyl P. T. and Huber W. (2015). HTSeq – A Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166-169. 10.1093/bioinformatics/btu638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aron S., de Menten L., Van Bockstaele D. R., Blank S. M. and Roisin Y. (2005). When hymenopteran males reinvented diploidy. Curr. Biol. 15, 824-827. 10.1016/j.cub.2005.03.017 [DOI] [PubMed] [Google Scholar]
- Balakrishnan C. N., Mukai M., Gonser R. A., Wingfield J. C., London S. E., Tuttle E. M. and Clayton D. F. (2014). Brain transcriptome sequencing and assembly of three songbird model systems for the study of social behavior. Peer J. 2, e396 10.7717/peerj.396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodily K. D., Morrison C. M., Renden R. B. and Broadie K. (2001). A novel member of the Ig superfamily, turtle, is a CNS-specific protein required for coordinated motor control. J. Neurosci. 21, 3113-3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger A. M., Lohse M. and Usadel B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114-2120. 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron S., Chang W.-T., Chen Y., Zhou Y., Taran S. and Rao Y. (2013). Visual circuit assembly requires fine tuning of the novel Ig transmembrane protein borderless. J. Neurosci. 33, 17413-17421. 10.1523/JNEUROSCI.1878-13.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capaldi E. A., Smith A. D., Osborne J. L., Fahrbach S. E., Farris S. M., Reynolds D. R., Edwards A. S., Martin A., Robinson G. E., Poppy G. M. et al. (2000). Ontogeny of orientation flight in the honeybee revealed by harmonic radar. Nature 403, 537-540. 10.1038/35000564 [DOI] [PubMed] [Google Scholar]
- Chandrasekaran S., Ament S. A., Eddy J. A., Rodriguez-Zas S. L., Schatz B. R., Price N. D. and Robinson G. E. (2011). Behavior-specific changes in transcriptional modules lead to distinct and predictable neurogenomic states. Proc. Natl. Acad. Sci. USA 108, 18020-18025. 10.1073/pnas.1114093108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cromwell H. C. and Schultz W. (2003). Effects of expectations for different reward magnitudes on neuronal activity in primate striatum. J. Neurophyiol. 89, 2823-2838. 10.1152/jn.01014.2002 [DOI] [PubMed] [Google Scholar]
- De Loof A. (2006). Ecdysteroids: the overlooked sex steroids of insects? Males: the black box. Insect Sci. 13, 325-338. 10.1111/j.1744-7917.2006.00101.x [DOI] [Google Scholar]
- Dillon C. and Goda Y. (2005). The actin cytoskeleton: integrating form and function at the synapse. Annu. Rev. Neurosci. 28, 25-55. 10.1146/annurev.neuro.28.061604.135757 [DOI] [PubMed] [Google Scholar]
- Elsik C. G., Worley K. C., Bennett A. K., Beye M., Camara F., Childers C. P., de Graaf D. C., Debyser G., Deng J., Devreese B. et al. , Honey Bee Genome Sequencing Consortium (2014). Finding the missing honey bee genes: lessons learned from a genome upgrade. BMC Genomics 15, 86 10.1186/1471-2164-15-86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafez D., Huang J. Y., Huynh A. M., Valtierra S., Rockenstein E., Bruno A. M., Lu B., DesGroseillers L., Masliah E. and Marr R. A. (2011). Neprilysin-2 is an important β-amyloid degrading enzyme. Am. J. Pathol. 178, 306-312. 10.1016/j.ajpath.2010.11.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harano K.-i., Sasaki K., Nagao T. and Sasaki M. (2008). Influence of age and juvenile hormone on brain dopamine level in male honeybee (Apis mellifera): association with reproductive maturation. J. Insect Physiol. 54, 848-853. 10.1016/j.jinsphys.2008.03.003 [DOI] [PubMed] [Google Scholar]
- Huang D. W., Sherman B. T. and Lempicki R. A. (2009). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44-57. 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- Hung H.-C., Kay S. A. and Weber F. (2009). HSP90, a capacitor of behavioral variation. J. Biol. Rhythms 24, 183-192. 10.1177/0748730409333171 [DOI] [PubMed] [Google Scholar]
- Isosaka I., Matsuo T., Yamaguchi T., Funabiki K., Nakanishi S., Kobayakawa R. and Kobayakawa K. (2015). Htr2a-expressing cells in the central amygdala control the hierarchy between innate and learned fear. Cell 163, 1153-1164. 10.1016/j.cell.2015.10.047 [DOI] [PubMed] [Google Scholar]
- Kandel E. R. (2001). The molecular biology of memory storage: a dialogue between genes and synapses. Science 294, 1030-1038. 10.1126/science.1067020 [DOI] [PubMed] [Google Scholar]
- Knapska E. and Kaczmarek L. (2004). A gene for neuronal plasticity in the mammalian brain: Zif268/Egr-1/NGFI-A/Krox-24/TIS8/ZENK? Prog. Neurobiol. 74, 183-211. 10.1016/j.pneurobio.2004.05.007 [DOI] [PubMed] [Google Scholar]
- Kume K., Kume S., Park S. K., Hirsh J. and Jackson F. R. (2011). Dopamine is a regulator of arousal in the fruit fly. J. Neurosci. 25, 7377-7384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P. and Horvath S. (2008). WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9, 559 10.1186/1471-2105-9-559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lardeux S., Pernaud R., Paleressompoulle D. and Baunez C. (2009). Beyond the reward pathway: coding reward magnitude and error in the rat subthalamic nucleus. J. Neurophysiol. 102, 2526-2537. 10.1152/jn.91009.2008 [DOI] [PubMed] [Google Scholar]
- Lin S., Huang Y. and Lee T. (2009). Nuclear receptor Unfulfilled regulates axonal guidance and cell identity of Drosophila mushroom body neurons. PLoS ONE 4, e8392 10.1371/journal.pone.0008392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz C. C. and Robinson G. E. (2013). Activity-dependent gene expression in honey bee mushroom bodies in response to orientation flight. J. Exp. Biol. 216, 2031-2038. 10.1242/jeb.084905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manfredini F., Brown M. J. F., Vergoz V. and Oldroyd B. P. (2015). RNA-sequencing elucidates the regulation of behavioural transitions associated with the mating process in honey bee queens. BMC Genomics 16, 563 10.1186/s12864-015-1750-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy D. J., Chen Y. and Smyth G. K. (2012). Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 40, 4288-4297. 10.1093/nar/gks042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNeill M. S., Kapheim K. M., Brockmann A., McGill T. A. W. and Robinson G. E. (2016). Brain regions and molecular pathways responding to food reward type and value in honey bees. Genes Brain Behav. 15, 305-317. 10.1111/gbb.12275 [DOI] [PubMed] [Google Scholar]
- Naeger N. L., Van Nest B. N., Johnson J. N., Boyd S. D., Southey B., Rodriguez-Zas S. D., Moore D. and Robinson G. E. (2011). Neurogenomic signatures of spatiotemporal memories in time-trained forager honey bees. J. Exp. Biol. 214, 979-987. 10.1242/jeb.053421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oxley P. R., Hinhumpatch P., Gloag R. and Oldroyd B. P. (2010). Genetic evaluation of a novel system for controlled mating of the honeybee, Apis mellifera. J. Hered. 101, 334-338. 10.1093/jhered/esp112 [DOI] [PubMed] [Google Scholar]
- Renner M. (1957). Neue Versuche uber den Zeitsinn der Honigbiene. Z. Vergl. Physiol. 40, 85-118. 10.1007/BF00298152 [DOI] [Google Scholar]
- Rinderer T. E., Oldroyd B. P., Wongsiri S., Sylvester H. A., de Guzman L., Potichot S., Sheppard W. S. and Buchmann S. L. (1993). Time of drone flight in four honey bee species in south-eastern Thailand. J. Apicult. Res. 32, 27-33. 10.1080/00218839.1993.11101284 [DOI] [Google Scholar]
- Robinson M. D., McCarthy D. J. and Smyth G. K. (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139-140. 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto K., Karelina K. and Obrietan K. (2011). CREB: a multifaceted regulator of neuronal plasticity and protection. J. Neurochem. 116, 1-9. 10.1111/j.1471-4159.2010.07080.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salamone J. D. and Correa M. (2012). The mysterious motivational functions of mesolimbic dopamine. Neuron 76, 470-485. 10.1016/j.neuron.2012.10.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen Sarma M., Rodriguez-Zas S. L., Hong F., Zhong S. and Robinson G. E. (2009). Transcriptomic profiling of central nervous system regions in three species of honey bee during dance communication behavior. PLoS ONE 4, e6408 10.1371/journal.pone.0006408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelley D. N., Choleris E., Kavaliers M. and Pfaff D. W. (2006). Mechanisms underlying sexual and affiliative behaviors of mice: relation to generalized CNS arousal. Soc. Cogn. Affect Neurosci. 1, 260-270. 10.1093/scan/nsl032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi A. and Miczek K. A. (2014). Neurogenetics of aggressive behavior: studies in rodents . Curr. Top. Behav. Neurosci. 17, 3-44. 10.1007/7854_2013_263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trabzuni D., Ramasamy A., Imran S., Walker R., Smith C., Weale M. E., Hardy J. and Ryten M. and North American Brain Expression Consortium. (2013). Widespread sex differences in gene expression and splicing in the adult human brain. Nat. Commun. 4, 2771 10.1038/ncomms3771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Pachter L. and Salzberg S. L. (2009). TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105-1111. 10.1093/bioinformatics/btp120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Nest B. N. and Moore D. (2012). Energetically optimal foraging strategy is emergent property of time-keeping behavior in honey bees. Behav. Ecol. 23, 649-658. 10.1093/beheco/ars010 [DOI] [Google Scholar]
- Van Swinderen B. and Andretic R. (2011). Dopamine in Drosophila: setting arousal thresholds in a miniature brain. Proc. R. Soc. B Biol. Sci. 278, 906-913. 10.1098/rspb.2010.2564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Frisch K. (1967). The Dance Language and Orientation of Bees. Cambridge, MA: Belknap Press of Harvard University Press. [Google Scholar]
- Wagener-Hulme C., Kuehn J. C., Schulz D. J. and Robinson G. E. (1999). Biogenic amines and division of labor in honey bee colonies. J. Comp. Physiol. A Sens. Neural Behav. Physiol. 184, 471-479. 10.1007/s003590050347 [DOI] [PubMed] [Google Scholar]
- Winston M. L. (1987). The Biology of the Honey Bee. Cambridge, MA: Harvard University Press. [Google Scholar]
- Wright G. A., Baker D. D., Palmer M. J., Stabler D., Mustard J. A., Power E. F., Borland A. M. and Stevenson P. C. (2013). Caffeine in floral nectar enhances a pollinator's memory of reward. Science 339, 1202-1204. 10.1126/science.1228806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young L. J. and Wang Z. (2004). The neurobiology of pair bonding. Nat. Neurosci. 7, 1048-1054. 10.1038/nn1327 [DOI] [PubMed] [Google Scholar]
- Zars T. (2000). Behavioral functions of the insect mushroom bodies. Curr. Opin. Neurobiol. 10, 790-795. 10.1016/S0959-4388(00)00147-1 [DOI] [PubMed] [Google Scholar]
- Zayed A. and Robinson G. E. (2012). Understanding the relationship between brain gene expression and social behavior: lessons from the honey bee. Annu. Rev. Genet. 46, 591-615. 10.1146/annurev-genet-110711-155517 [DOI] [PubMed] [Google Scholar]
- Zayed A., Naeger N. L., Rodriguez-Zas S. L. and Robinson G. E. (2012). Common and novel transcriptional routes to behavioral maturation in worker and male honey bees. Genes Brain Behav. 11, 253-261. 10.1111/j.1601-183X.2011.00750.x [DOI] [PubMed] [Google Scholar]
- Zhan S., Zhang W., Niitepõld K., Hsu J., Haeger J. F., Zalucki M. P., Altizer S., de Roode J. C., Reppert S. M. and Kronforst M. R. (2014). The genetics of monarch butterfly migration and warning colouration. Nature 514, 317-321. 10.1038/nature13812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B. and Horvath S. (2005). A general framework for weighted gene co-expression network analysis. Stat. Appl. Genet. Mol. Biol. 4, article 17 10.2202/1544-6115.1128 [DOI] [PubMed] [Google Scholar]