ABSTRACT
Recent studies have revealed that diverse cell types use mRNA localization as a means to establish polarity. Despite the prevalence of this phenomenon, much less is known regarding the mechanism by which mRNAs are localized. The Drosophila melanogaster oocyte provides a useful model for examining the process of mRNA localization. oskar (osk) mRNA is localized at the posterior of the oocyte, thus restricting the expression of Oskar protein to this site. The localization of osk mRNA is microtubule dependent and requires the plus-end-directed motor Kinesin-1. Unlike most Kinesin-1 cargoes, localization of osk mRNA requires the Kinesin heavy chain (Khc) motor subunit, but not the Kinesin light chain (Klc) adaptor. In this report, we demonstrate that a newly discovered isoform of Tropomyosin 1, referred to as Tm1C, directly interacts with Khc and functions in concert with this microtubule motor to localize osk mRNA. Apart from osk mRNA localization, several additional Khc-dependent processes in the oocyte are unaffected upon loss of Tm1C. Our results therefore suggest that the Tm1C–Khc interaction is specific for the osk localization pathway.
KEY WORDS: RNA localization, Molecular motor, Cell polarity, Actin
Highlighted Article: A previously uncharacterized isoform of Tropomyosin, referred to as Tm1C, directly interacts with Kinesin heavy chain and functions along with this microtubule motor to localize oskar mRNA in Drosophila oocytes.
INTRODUCTION
Asymmetric protein sorting underlies the establishment and maintenance of cell polarity. Recent studies have revealed that mRNA localization is a prevalent mechanism used by different cell types for restricting protein localization (Cajigas et al., 2012; Jambor et al., 2015; Lecuyer et al., 2007; Zivraj et al., 2010). Thus, understanding the mechanism of mRNA localization is crucial to gaining a deeper understanding of cell polarity establishment.
Although examples of localized mRNAs have been identified in a variety of organisms, mechanistic studies are easier to address using simpler model systems. For example, localization of ASH1 mRNA in the yeast Saccharomyces cerevisiae represents the best-understood example of a localized transcript (Gonsalvez et al., 2005). In this system, ASH1 mRNA is first identified by the RNA-binding protein She2p (Niedner et al., 2014). This complex is then coupled to the Myo4p motor through the adaptor She3p (Singer-Kruger and Jansen, 2014). In addition, the various cis-elements within ASH1 that mediate localization have also been identified (Chartrand et al., 1999).
In Drosophila melanogaster blastoderm embryos, several mRNAs are localized to the apical surface. This process is dependent on the minus-end-directed microtubule motor Dynein (Bullock, 2011). These mRNAs are thought to be identified by the RNA-binding protein Egalitarian (Egl) (Dienstbier et al., 2009). The Egl–messenger-ribonucleoprotein (mRNP) complex is then linked to Dynein through the Egl–Dynein-light-chain (Dlc, also known as Ctp) interaction or through the Egl-binding protein Bicaudal D (BicD) (Navarro et al., 2004). In mammalian cells, BicD has been shown to bind the Dynein regulator Dynactin (Hoogenraad et al., 2001). However, polarity has already been established even prior to the blastoderm stage, by events that take place in the oocyte.
The localization of oskar (osk), bicoid (bcd) and gurken (grk) mRNAs is required for establishing the polarity of the oocyte and future embryo (St Johnston, 2005). bcd and grk mRNAs localize at the anterior and dorsal-anterior regions of the oocyte, respectively (Berleth et al., 1988; Neuman-Silberberg and Schupbach, 1993). Their localization is thought to depend primarily on Dynein (MacDougall et al., 2003; Schnorrer et al., 2000). Although the exact link between these mRNAs and Dynein has not been identified, the available evidence suggests that this role is performed by Egl (Dienstbier et al., 2009; Sanghavi et al., 2016).
osk mRNA localizes at the posterior of the oocyte (Ephrussi et al., 1991; Kim-Ha et al., 1991). The primary motor responsible for this localization is the plus-end-directed microtubule motor Kinesin-1 (Brendza et al., 2000). However, recent findings suggest that Dynein is also required for efficient osk localization (Sanghavi et al., 2013). Kinesin-1 typically consists of a Kinesin heavy chain (Khc) motor subunit and a Kinesin light chain (Klc) adaptor (Hirokawa et al., 2009). Thus, most cargoes that are transported by Kinesin-1 require both Khc and Klc. However, over the past several years, numerous cargoes have been identified that are transported by Kinesin-1 in a Klc-independent manner (Hirokawa et al., 2009). In these cases, Khc typically associates with cargo using an alternative adaptor. It appears that the localization of osk might involve such an alternative adaptor. Although osk is completely delocalized in khc mutants, it remains localized in klc-null mutants (Palacios and St Johnston, 2002).
In order to better understand the mechanism by which Khc functions in osk mRNA localization, we purified the motor and identified interacting partners using mass spectrometry. Using this approach, we identified a new isoform of Tropomyosin 1, Tm1C, as an interacting partner of Khc. In this report, we demonstrate that Tm1C functions in concert with Khc to localize osk mRNA at the posterior pole. However, several additional Khc processes appear to function independently of Tm1C. Finally, we demonstrate that Tm1C expression is not restricted to the female germline. This raises the intriguing possibility that this new isoform of Tropomyosin might have functions in addition to osk localization.
RESULTS
Tm1C interacts with Kinesin heavy chain
The mechanism by which Khc localizes osk mRNA is unknown, but it is assumed that an adaptor other than Klc functions in this process. In order to identify proteins that associate with Khc, we undertook a proteomic purification strategy. Ovarian lysates were prepared from strains expressing either GFP or Khc–GFP. The tagged proteins were immunoprecipitated, and the co-precipitating proteins were eluted and identified by mass spectrometry. The entire experiment was performed twice.
Peptides corresponding to Klc were highly enriched in Khc–GFP pellets (Table 1). Thus, even though several Kinesin-1 cargoes in the oocyte are localized independent of Klc, the light chain remains abundantly associated with Khc in vivo. Excluding typical contaminants, the next most abundant protein in Khc–GFP pellets corresponded to Tropomyosin 1 (Tm1) (Table 1).
Table 1.
Candidate Khc-interacting proteins
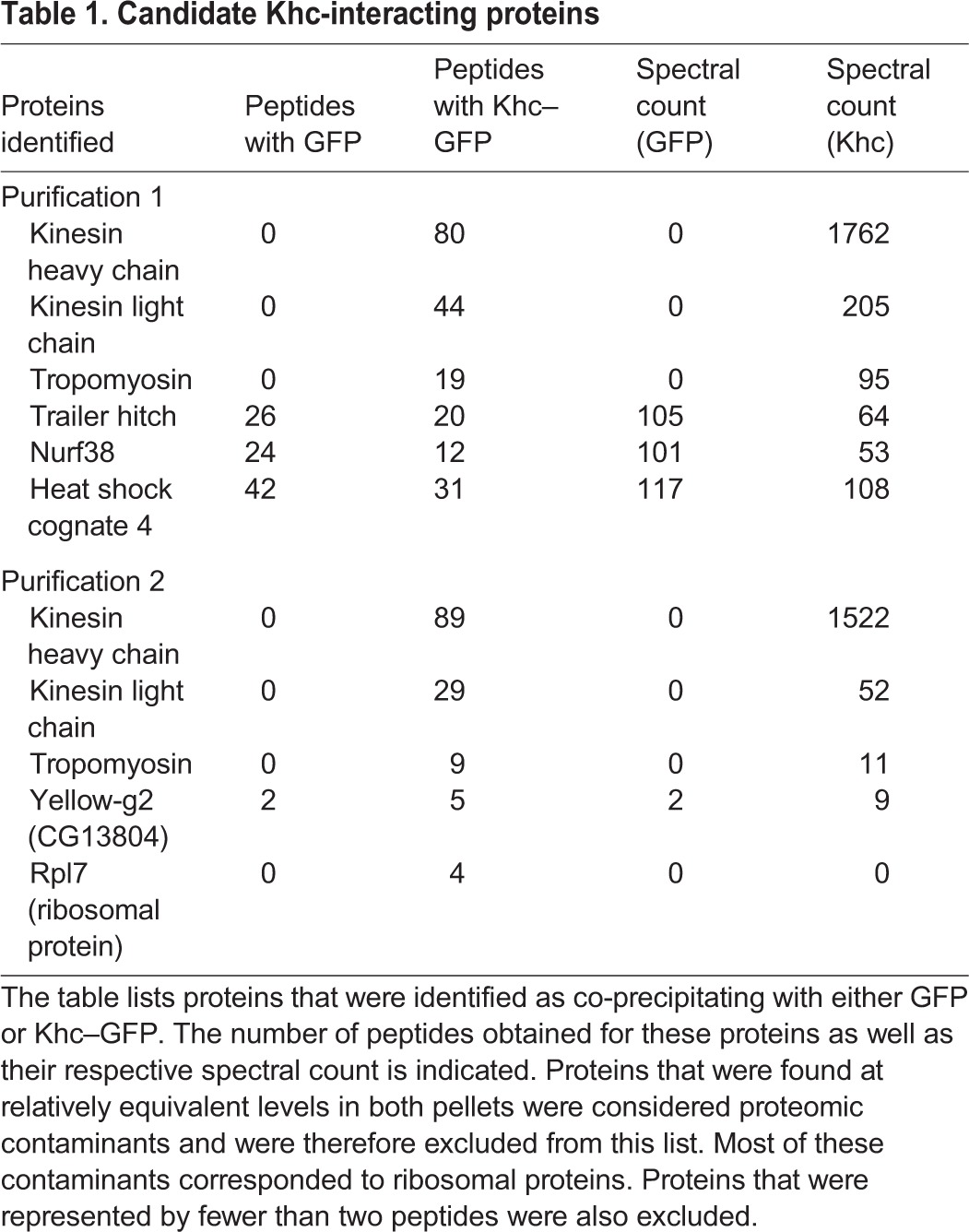
Tropomyosins are actin-binding proteins that have a well-documented role within muscle in mediating contraction (Gunning et al., 2015). However, non-muscle isoforms are also expressed and these function in a variety of actin-dependent processes (Manstein and Mulvihill, 2016). Drosophila contains two tropomyosin genes, tropomyosin 1 (Tm1) and tropomyosin 2 (Tm2). Although most publications simply refer to these genes as Tm1 and Tm2, genome annotation suggests that this family is capable of expressing numerous isoforms (Fig. S1A). In fact, a recent publication by Goins and Mullins has demonstrated that Drosophila S2 cells express two isoforms of Tm1 (Tm1A and Tm1J) and one isoform of Tm2 (Tm2A) (Goins and Mullins, 2015).
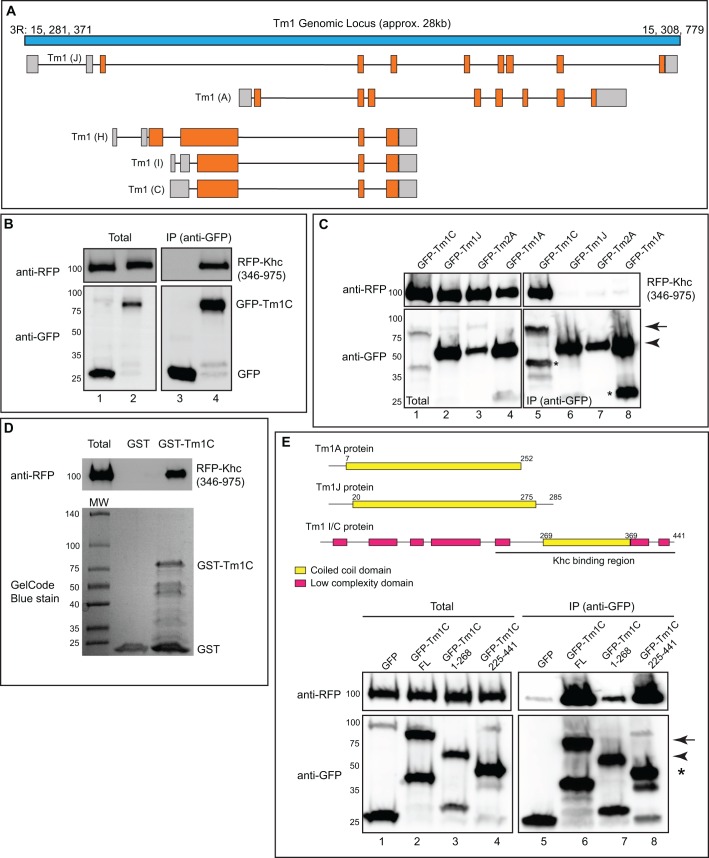
In order to determine which isoform of Tm1 associated with Khc, we analyzed peptide sequences obtained in Khc–GFP pellets. Each Tm1 peptide could be mapped to the Tm1C, Tm1H or Tm1I isoform (Fig. S1B). Tm1C and Tm1I differ in their 5′ untranslated region (UTR), but encode identical proteins (Fig. 1A). By contrast, Tm1H is predicted to contain an N-terminal extension not found in Tm1C and Tm1I (Fig. 1A). Peptides corresponding to this region were not recovered in Khc–GFP pellets. Thus, the most likely interacting partner for Khc is either Tm1C or Tm1I. Because these isoforms encode identical proteins, for the sake of simplicity, we will refer to the potential Khc-interacting isoform as Tm1C.
Fig. 1.
Tm1C is an interacting partner of Khc. (A) Schematic of the Tm1 locus. Untranslated regions are in gray and coding regions are depicted in orange. (B) Co-immunoprecipitation experiment in S2 cells between RFP–Khc and GFP (lanes 1, 3) or GFP–Tm1C (lanes 2, 4). The Khc construct did not contain the motor domain. The immunoprecipitation (IP) was performed using GFP-trap beads. The co-precipitating proteins and total fraction were analyzed by blotting using the indicated antibodies. (C) A co-immunoprecipitation experiment between RFP–Khc and GFP–Tm1C (lanes 1, 5), Tm1J (lanes 2, 6), Tm2A (lanes 3, 7) or Tm1A (lanes 4, 8). The immunoprecipitation was performed as in B and the co-precipitating proteins were detected by blotting using the indicated antibodies. The arrow indicates full-length GFP–Tm1C and the arrowhead denotes full-length GFP–Tm1J, Tm2A and Tm1A. Overexpression of GFP-tagged Tropomyosins in S2 cells often resulted in smaller faster-migrating bands (indicated by asterisks in this panel). We assume these bands represent breakdown products resulting from protein turnover. (D) A direct binding experiment using recombinant GST or GST–Tm1C produced in bacteria and RFP–Khc produced using in vitro transcription and translation. The binding reaction was run on a gel and analyzed by blotting using an anti-RFP antibody (top panel). The bottom panel is a GelCode-Blue-stained gel showing the amount of GST and GST–Tm1C used in the binding reaction. (E) The domain structure of Tm1A, Tm1J and Tm1C proteins is shown at the top of the figure. Numbers correspond to the amino acid positions for the various domains. The bottom part of the figure shows a co-immunoprecipitation experiment using RFP–Khc and either GFP (lanes 1, 5), full-length GFP–Tm1C (lanes 2, 6), an N-terminal GFP–Tm1C construct (lanes 3, 7) or a C-terminal GFP–Tm1C construct (lanes 4, 8). Immunoprecipitation was performed as in B. The arrow indicates full-length GFP–Tm1C, the arrowhead corresponds to N-terminally truncated GFP–Tm1C and the asterisk denotes C-terminally truncated GFP–Tm1C.
As a first step towards validating the Khc–Tm1C interaction, we immunoprecipitated Khc–GFP from ovarian lysates and examined the pellet using an antibody against endogenous Tm1C. The antibody was generated using peptide sequences found in both Tm1C and Tm1J, and is therefore capable of detecting both isoforms. Consistent with our proteomic results, endogenous Tm1C, but not Tm1J, co-immunoprecipitated with Khc–GFP (Fig. S1C). By using a complementary approach, we generated transgenic flies expressing GFP–Tm1C driven by the vasa promoter (Sano et al., 2002). As expected, endogenous Khc specifically co-precipitated with GFP–Tm1C (Fig. S1D).
In order to further characterize the Khc–Tm1C interaction, we expressed tagged versions of these proteins in Drosophila S2 cells and performed a co-immunoprecipitation experiment. Consistent with the ovary purification result, RFP–Khc specifically co-precipitated with GFP–Tm1C (Fig. 1B). For this experiment, a version of Khc lacking the N-terminal motor domain was used. However, a full-length RFP–Khc construct containing the motor domain is also able to bind GFP–Tm1C (data not shown). In contrast to GFP–Tm1C, RFP–Khc did not co-precipitate with GFP–Tm1A, Tm1J or Tm2A (Fig. 1C). Thus, the interaction between Khc and Tropomyosin is specific for the Tm1C isoform. We further demonstrate that purified GST–Tm1C produced in bacteria was able to interact with recombinant RFP–Khc produced using coupled in vitro transcription and translation (Fig. 1D). Thus, the interaction between Khc and Tm1C is direct.
Most tropomyosins, including those found in mammals, are relatively small, consisting of fewer than 300 amino acids. Their structure consists of α-helical coiled-coil domains suited for binding along the sides of actin filaments (Gunning et al., 2005). These features are shared by Tm1A, Tm1J and Tm2A (Fig. 1E and data not shown). By contrast, at 441 amino acids, Tm1C represents a very large tropomyosin. In addition, Tm1C contains a long N-terminal extension that is not predicted to form a coiled-coil (Fig. 1E). Because this N-terminal extension is unique to Tm1C, we predicted that this region would be involved in Khc binding. Unexpectedly, a Tm1 construct containing the C-terminal region was able to interact more strongly with Khc than the construct containing the N-terminal region (Fig. 1E). Thus, even though Khc specifically interacts with the unique Tm1C isoform, the residues that are most crucial for binding comprise the conserved coiled-coil domain.
Tm1C interacts with a C-terminal domain of Khc
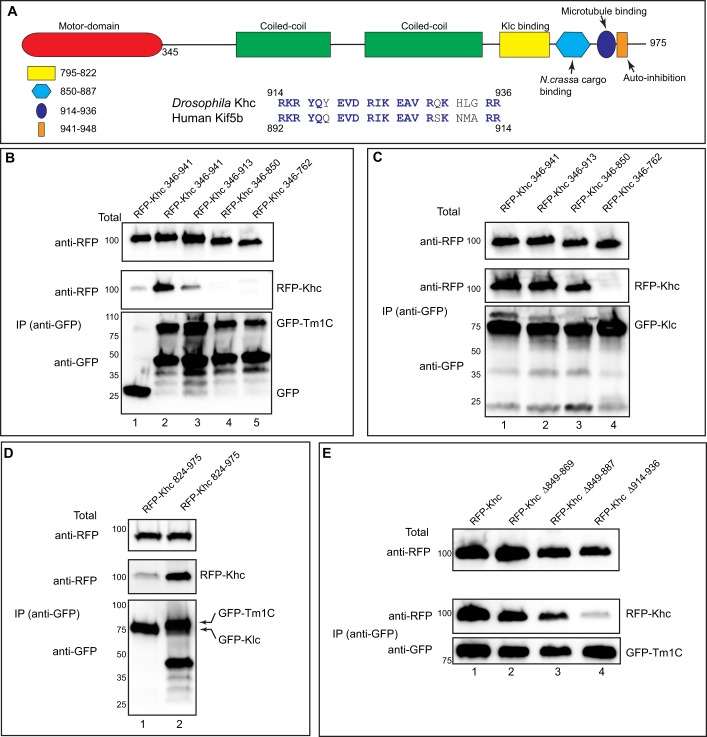
Experiments in the preceding section revealed that the motor domain of Khc (Fig. 2A, red) was not required for binding Tm1C. In addition to the motor domain, Khc also contains coiled-coil domains required for dimerization of the heavy chain (Fig. 2A, green). A smaller domain C-terminal to these is involved in binding Klc (Fig. 2A, yellow). Certain adaptors, such as the mitochondrial adaptor Milton, bind to the same region of Khc as does the light chain, thus competing for binding with Klc (Glater et al., 2006).
Fig. 2.
Tm1C binds to a C-terminal domain within Khc. (A) Domain structure of Khc. The following features are indicated: motor domain (red), coiled-coil domains required for dimerization of Khc (green), the Klc-binding site (yellow), a region important for cargo binding in N. crassa (light blue), an ATP-independent microtubule-binding region (blue) and an auto-inhibitory domain (dark blue oval). Also shown is an alignment of amino acids 914 to 936 of Khc with Kif5b, the human homolog of Khc. (B) A co-immunoprecipitation experiment using various RFP–Khc truncation constructs with full-length GFP–Tm1C. The residues included in the truncations are indicated. Lysates were prepared and the GFP-tagged proteins were immunoprecipitated. The co-precipitating proteins and the total fraction were analyzed by blotting using the indicated antibodies. (C) The same truncation constructs used in the previous panel were co-transfected with full-length GFP–Klc. The experiment was performed as in B. (D) A co-immunoprecipitation experiment using a small RFP–Khc construct and either GFP–Klc (lane 1) or Tm1C (lane 2). The immunoprecipitation was performed as described for B and analyzed using the indicated antibodies. (E) A co-immunoprecipitation experiment using GFP–Tm1C and either full-length RFP–Khc or RFP–Khc containing the indicated deletions. The co-precipitation was performed as described for B and analyzed by blotting using the indicated antibodies.
In order to determine the Tm1C-binding site within Khc, we generated several RFP-tagged C-terminal Khc truncations. These truncations were co-transfected into S2 cells along with full-length GFP–Tm1C and binding was determined using co-immunoprecipitation. A Khc construct missing the auto-inhibitory domain was able to bind to Tm1C (Fig. 2B, lane 2). However, further truncation beyond this region compromised the Khc–Tm1C interaction (Fig. 2B, lanes 3–5). The same Khc truncations were co-transfected into S2 cells along with full-length GFP–Klc, and binding was determined through co-immunoprecipitation experiments. Klc was able to interact with a subset of these truncations (Fig. 2C, lanes 1–3). However, as expected, deletion of the Klc-binding site disrupted the Khc–Klc interaction (Fig. 2C, lane 4). Consistent with these findings, a small Khc construct containing residues 824 to 975 was able to interact with Tm1C but not Klc (Fig. 2D). These results suggest that Tm1C and Klc bind to different regions of Khc. Furthermore, the binding site for Tm1C lies between residues 850 and 941.
In order to more precisely map the region of Tm1C binding, we created small deletions within full-length Khc. These internally deleted RFP–Khc constructs were transfected into S2 cells along with full-length GFP–Tm1C and binding was determined through co-immunoprecipitation experiments. Deletions spanning residues 849 to 887 had a minimal effect on Tm1C binding (Fig. 2E, lanes 1–3). This region was of interest because previous studies have shown it to function as a cargo recognition domain in Neurospora crassa (Fig. 2A, light blue) (Seiler et al., 2000). By contrast, deletion of amino acids 914 to 936 resulted in almost complete loss of binding (Fig. 2A, dark blue oval; Fig. 2E, lane 4). Thus, a region within Khc that has been shown to bind microtubules in vitro (Andrews et al., 1993; Hackney and Stock, 2000) is the primary binding site for Tm1C.
The alignment in Fig. 2A illustrates the high degree of conservation between Drosophila Khc and its mammalian homolog Kif5b within the Tm1C-binding region. Interestingly, in vivo truncation studies have identified this region of Khc as being required for osk localization (Williams et al., 2014). However, a potential adaptor or cargo that binds this region of Khc has thus far not been identified.
Localization of Tm1C
As noted in the preceding section, we generated an antibody against Tm1C. Although our antibody was capable of detecting Tm1C on western blots, we were unable to detect specific signal using immunofluorescence, thus limiting its use for localization studies. We therefore made use of a strain expressing GFP–Tm1C in which expression of the fusion protein was driven by vasa regulatory elements (Sano et al., 2002).
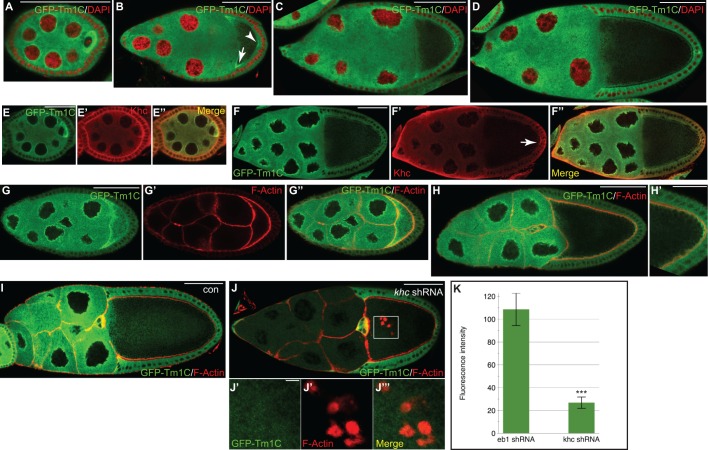
In early stage egg chambers (stages 2 through 5), GFP–Tm1C was highly enriched in the oocyte (Fig. 3A). GFP–Tm1C also displayed perinuclear enrichment in an area known as the nuage (Fig. 3A, Fig. S2A; the nuage is an electron-dense region that surrounds nurse cell nuclei). In stage 8 egg chambers, GFP–Tm1C could be detected at the anterior corners of the oocyte as well as at the posterior (Fig. 3B, arrow and arrowhead, respectively). A similar localization was observed in stage 9 egg chambers (Fig. 3C). At stage 10, the nuage localization of GFP–Tm1C persisted and, within the oocyte, GFP–Tm1C localized around the cortex (Fig. 3D).
Fig. 3.
Localization of GFP–Tm1C in the female germline. (A–D) Localization of GFP–Tm1C at various stages of egg chamber maturation. DAPI staining of nuclei is indicated in red. The arrow indicates enrichment of GFP–Tm1C at the anterior of stage 7 oocytes and the arrowhead indicates posterior enrichment. (E,F) Ovaries from the same strain as in A–D were fixed and processed using an antibody against GFP (green, E,F) and Khc (red, E′,F′). A merged image is also shown (E″,F″). The arrow in F′ indicates posterior enrichment of Khc. (G,H) Egg chambers from the same strain were processed using an antibody against GFP and were counterstained with TRITC–phalloidin to visualize F-actin. The GFP–Tm1 signal is depicted in G. The actin signal is depicted in G′ and the merge signal is shown in G″,H and H′. H′ is a magnified view of the posterior pole. (I–J) Egg chambers from strains expressing GFP–Tm1 and either a control shRNA against eb1 (I) or an shRNA against khc (J) were fixed and processed for immunofluorescence using an antibody against GFP. The samples were also counterstained with TRITC– phalloidin. J′–J‴ represent individual and merged images from the boxed region in J. (K) The intensity of the GFP–Tm1 signal in nurse cells of control (eb1 shRNA) or khc-depleted egg chambers was quantified. The results are mean±s.d. (n=25 egg chambers for each genotype). ***P<0.0001 (unpaired t-test). Scale bars: 50 μm (A–D,F,G,H,I,J); 25 μm (E,H′); 5 μm (J′–J‴).
In order to validate the localization pattern of GFP–Tm1C, a strain expressing unfused GFP in the germline was processed in a similar manner. Although unfused GFP was not specifically enriched in the oocyte and did not localize to the nuage, it could be detected around the oocyte cortex (Fig. S2B and data not shown). Based on this result, we are not able to conclude whether the cortical localization of GFP–Tm1C is a specific property of Tm1C, or whether it is an artifact of the GFP–Tm1C fusion protein.
Consistent with the protein–protein interaction result, a substantial degree of colocalization was observed between GFP–Tm1C and Khc (Fig. 3E,F). However, a few differences are worth noting. The oocyte enrichment and nuage localization was more evident for GFP–Tm1C in comparison to Khc (Fig. 3E). Furthermore, in stage 10 egg chambers, the posterior enrichment was more apparent for Khc in comparison to GFP–Tm1C (Fig. 3F′, arrow).
Within egg chambers, filamentous actin (F-actin) is highly enriched around the cortex and within ring canals. Given that Tropomyosins are actin-binding proteins, it was surprising that minimal colocalization was observed between GFP–Tm1C and F-actin (Fig. 3G,H). In order to test the dependence of GFP–Tm1C localization on F-actin, we treated egg chambers with Latrunculin A, a drug that destabilizes F-actin. Although F-actin staining was virtually eliminated by Latrunculin A treatment, the localization of GFP–Tm1C appeared to be unaffected (Fig. S2C–F).
We next determined whether the localization of GFP–Tm1C was sensitive to the presence of Khc. In order to deplete Khc, we expressed a short hairpin RNA (shRNA) against khc using a germline driver (Sanghavi et al., 2016). As expected, expression of this shRNA resulted in substantial depletion of Khc (Fig. S3A). Unexpectedly, the germline level of GFP–Tm1C was also greatly reduced in these flies. This result is specific to Khc depletion. GFP–Tm1C levels were unaffected in strains expressing a control shRNA (Fig. 3I–K). Similar results were obtained when examining ovarian lysates from these strains by western blotting (Fig. S3B). It should be noted, however, that because these lysates were prepared using whole ovaries, western analysis underestimates the true level of depletion in stage 10 egg chambers. Khc is not depleted in follicle cells or in early stage egg chambers, both of which are present in whole ovary lysates.
As shown using khc-null mutants (Januschke et al., 2002; Mische et al., 2007), depletion of Khc results in formation of numerous actin foci within the oocyte (Fig. 3J). Despite using a high-gain setting, we were not able to detect colocalization between the residual GFP–Tm1C and these foci (Fig. 3J). Thus, the localization and expression level of GFP–Tm1C in egg chambers correlates to a greater degree with the presence of Khc than with F-actin.
Tm1C depletion results in osk delocalization
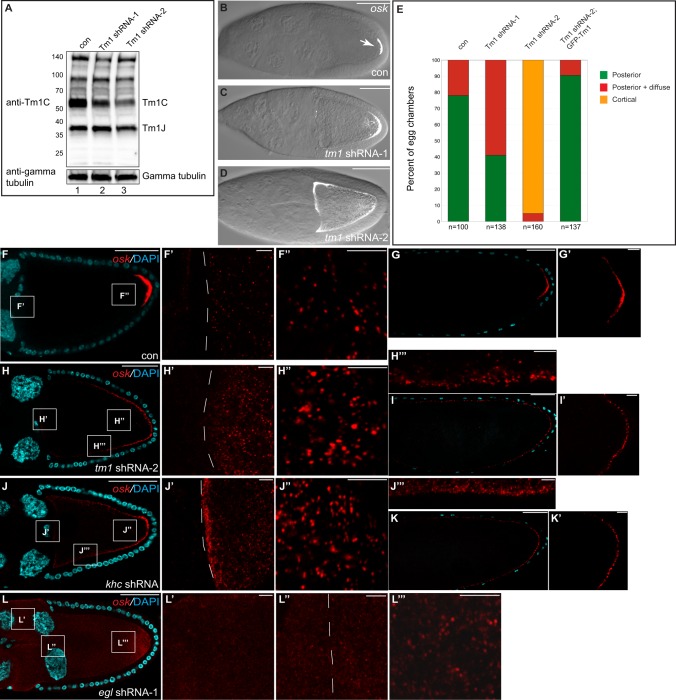
In order to determine whether Tm1C is required for osk localization, we designed shRNA constructs that were specific for this isoform. The shRNAs were expressed using a previously characterized germline driver (Sanghavi et al., 2016). Ovarian lysates were prepared from these flies and examined by blotting using our anti-Tm1 antibody. Although Tm1 shRNA-1 and Tm1 shRNA-2 were capable of depleting Tm1C, Tm1 shRNA-2 consistently displayed greater depletion (Fig. 4A). In contrast with Tm1C, neither shRNA depleted Tm1J.
Fig. 4.
Depletion of Tm1C results in osk mRNA delocalization. (A) Ovarian lysates from flies expressing a control shRNA (lane 1), Tm1 shRNA-1 (lane 2) or Tm1 shRNA-2 (lane 3) were analyzed by blotting using an antibody against Tm1C (top panel). This antibody also detects Tm1J. The blot was subsequently stripped and probed with an antibody against γ-tubulin (bottom panel). (B–D) Ovaries from these strains were dissected and processed for single-molecule fluorescence in situ hybridization (smFISH) using oligonucleotide probes against osk mRNA. The osk signal (white) is superimposed over a bright-field image. (E) The localization pattern of osk mRNA was quantified in strains expressing control shRNA, Tm1 shRNA-1 or Tm1 shRNA-2, and a strain co-expressing Tm1 shRNA-2 and the GFP–Tm1C transgene. The number of egg chambers scored is indicated. (F,G) Ovaries from females expressing a control shRNA were processed for smFISH using probes against osk (red). The samples were counterstained with DAPI (cyan). F′ and F″ are enlarged images of boxes in F. A stage 14 egg chamber is shown in G, the posterior of which is enlarged in G′. (H,I) Ovaries from females expressing Tm1 shRNA-2 were processed as above. H′,H″ and H‴ are enlarged images of boxes in H. Panel I is a stage 14 egg chamber and I′ shows an enlargement of the posterior pole. (J,K) Similar layout to the above panels using flies expressing an shRNA against khc. (L) Ovaries from females expressing egl shRNA-1 were processed as above. L′,L″ and L‴ are enlarged images of boxes in L. The dashed line in F′,H′,J′ and L″ indicate the border between nurse cells and the oocyte. Scale bars: 50 μm (B–D,F–L); 5 μm (F′,F″,G′,H′–H‴,I′,J′–J‴,K′,L′–L‴).
We next examined the localization of osk. osk mRNA was visualized using single-molecule fluorescent in situ hybridization (smFISH) (Little et al., 2015, 2013). In this approach, the target mRNA is examined using 48 unique DNA oligonucleotide probes that are directly conjugated to a fluorochrome. smFISH enables detection of mRNA with a much higher sensitivity and specificity in comparison to traditional methods. As expected, osk mRNA was highly enriched at the posterior pole in egg chambers expressing a control shRNA (Fig. 4B,E–G). As a control for specificity, we examined egg chambers using smFISH probes against Gapdh1 and Gapdh2. Higher levels of Gapdh1 and Gapdh2 mRNA were observed in nurse cells in comparison to the oocyte, and no enrichment was detected at the posterior pole (Fig. S3C).
osk mRNA was still enriched at the posterior in egg chambers expressing the less strong shRNA, Tm1 shRNA-1 (Fig. 4C,E). However, a substantial fraction of egg chambers also contained delocalized signal close to the posterior pole (Fig. 4C,E). By contrast, osk mRNA was almost completely delocalized around the cortex in egg chambers expressing the more potent Tm1 shRNA-2 (Fig. 4D,E). A similar localization was observed for the osk mRNP component Staufen in Tm1C-depleted egg chambers (Fig. S3D–F) (Zimyanin et al., 2008).
Translation of osk mRNA is regulated by several factors to ensure that only localized transcripts are translated (Kugler and Lasko, 2009). Thus, delocalization of osk mRNA often results in greatly reduced expression of Osk protein. As expected, Osk protein could not be readily detected in Tm1C-depleted egg chambers (Fig. S3G,H). Osk protein is required for recruiting pole plasm components such as Vasa to the posterior pole. Consistent with reduced Osk protein expression, Vasa was no longer enriched at the posterior pole in Tm1C-depleted oocytes (Fig. S3G′,H′).
To demonstrate the specificity of this phenotype, the GFP–Tm1C construct used in the preceding section was expressed in the background of Tm1 shRNA-2. The Tm1C coding sequence in this construct was mutated such that it was no longer recognized by the shRNA, yet still encoded wild-type protein. Thus, these flies will be depleted of endogenous Tm1C, but will express transgenic GFP–Tm1C. osk mRNA and pole plasm components were correctly localized to the posterior pole in these flies (Fig. 4E; Fig. S3I–K). We therefore conclude that these phenotypes are specific to Tm1C depletion.
A recent analysis of osk using smFISH revealed that the mRNA undergoes assembly into large particles (Little et al., 2015). This occurs in a step-wise fashion. Upon entry into the oocyte, osk particles coalesce and increase in size (Little et al., 2015). Subsequently, during transit to the posterior, osk mRNA oligomerizes further to generate larger particles (Little et al., 2015). We obtained similar results using egg chambers expressing a control shRNA (Fig. 4F′,F″). This phenotype was specific to osk and was not observed for Gapdh1 and Gapdh2 mRNA (Fig. S3C). High-resolution imaging of stage 10 egg chambers expressing Tm1 shRNA-2 revealed that osk retained the ability to coalesce into large particles upon entry into the oocyte (Fig. 4H,H′). In addition, large osk particles could also be visualized close to the posterior and lateral cortex (Fig. 4H″,H‴). In fact, these particles were easier to observe in Tm1C-depleted egg chambers (Fig. 4F versus 4H). This result is expected because, in control egg chambers, the majority of osk particles are anchored at the posterior pole (Little et al., 2015). By contrast, in Tm1C-depleted oocytes, the bulk of osk was present in delocalized particles. In late stage egg chambers, osk particles could be detected around the cortex of Tm1C-depleted oocytes, but were restricted to the posterior in controls (Fig. 4G,I). Thus, Tm1C is required for localization of osk mRNA at the oocyte posterior, but is dispensable for assembly of osk into oligomeric particles.
We next examined the localization of osk in Khc-depleted egg chambers. Loss of Khc has been shown to result in osk mRNA delocalization (Brendza et al., 2000). However, these mutants have not been examined using smFISH. Consistent with Tm1C functioning in concert with Khc in the osk localization pathway, depletion of Khc resulted in mislocalization of osk mRNA around the oocyte cortex (Fig. 4J,K). As with Tm1C depletion, oligomerization of osk was not affected upon loss of Khc (Fig. 4J′,J″,J‴).
A different phenotype was observed in Egl-depleted egg chambers. Egl is thought to function as an adaptor for Dynein (Navarro et al., 2004) and has been shown to link Dynein to localized transcripts in the embryo (Dienstbier et al., 2009). In Egl-depleted egg chambers, osk mRNA was diffusely distributed (Fig. 4L). A substantial accumulation of osk was detected in nurse cells (Fig. 4L′). This is consistent with the role of Dynein in transporting osk mRNA from nurse cells into the oocyte (Clark et al., 2007; Mische et al., 2007). As a consequence, the oligomerization of osk into larger particles upon oocyte entry was less obvious in Egl-depleted egg chambers (Fig. 4L″). Furthermore, osk particles in the posterior region of Egl-depleted egg chambers were smaller than those observed in controls (Fig. 4L‴). Thus, in contrast to Tm1C and Khc, Egl appears to be required for efficient transport of osk into the oocyte and for the formation of oligomeric particles.
Tm1C is not required for all Kinesin processes
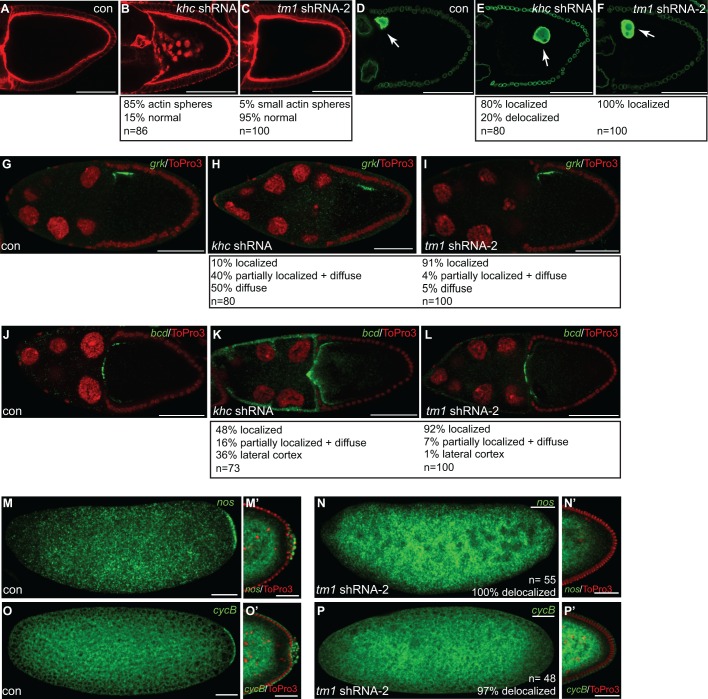
Apart from osk mislocalization, khc-null mutants are also associated with delocalization of the oocyte nucleus, aberrant formation of actin spheres, and delocalization of gurken (grk) and bicoid (bcd) mRNAs (Brendza et al., 2002; Duncan and Warrior, 2002; Januschke et al., 2002). shRNA-mediated depletion of Khc produced a similar range of phenotypes (Fig. 5A–L). In contrast, the oocyte nucleus was correctly localized in Tm1C-depleted egg chambers (Fig. 5F). Additionally, actin spheres were not observed, and grk and bcd mRNAs remained correctly localized (Fig. 5C,I,L). Thus, Tm1C appears to be required for only a subset of Khc-dependent functions.
Fig. 5.
Several Khc-dependent processes are unaffected upon Tm1C depletion. (A–C) Ovaries were fixed from flies expressing either a control shRNA (A), khc shRNA (B) or Tm1 shRNA-2 (C) and were counterstained with TRITC–phalloidin to visualize F-actin. (D–F) Ovaries were dissected from the same strains used in A–C. The egg chambers were fixed and processed using an antibody against Lamin DmO (green). Arrows indicates the oocyte nucleus. (G–I) Ovaries from flies expressing a control shRNA (G), khc shRNA (H) or Tm1 shRNA-2 (I) were processed for in situ hybridization using probes against grk mRNA (green). The egg chambers were counterstained with ToPro3 (red). (J–L) The same strains were fixed and processed for in situ hybridization using probes against bcd mRNA (green). The egg chambers were counterstained with ToPro3 (red). (M,N) Embryos were collected and fixed from mothers expressing control shRNA (M) or Tm1 shRNA-2 (N). The embryos were processed for in situ hybridization using probes against nanos mRNA (nos, green). M′ and N′ represent blastoderm stage egg chambers from the indicated strains. The posterior region of these embryos is shown. (O–P) Embryos from these same strains were fixed and processed for in situ hybridization using probes against Cyclin B mRNA (cycB, green). As with the above panel, O′ and P′ represent blastoderm stage egg chambers from the indicated strains. Quantification of phenotypes and the number of egg chambers or embryos scored are indicated. Scale bars: 50 μm (A–P); 25 μm (M′,N′,O′,P′).
Several mRNAs including nanos (nos) and Cyclin B (CycB) are localized at the posterior of late stage egg chambers and embryos (Dalby and Glover, 1992; Gavis and Lehmann, 1992). The localization of these mRNAs requires Osk protein. Consistent with the reduced expression of Osk in Tm1C-depleted egg chambers, early stage embryos from these mothers contained delocalized nos and CycB mRNA (Fig. 5M–P). As expected, given the defect in pole plasm formation, blastoderm-stage embryos from these mothers failed to form primordial germ cells (Fig. 5N′,P′).
Generation of a Tm1C-null mutant
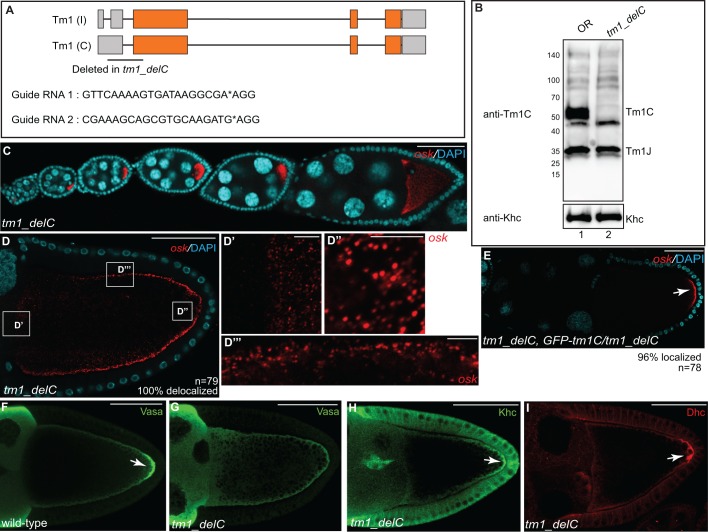
In the preceding section, we examined the role of Tm1C in osk mRNA localization using shRNA-mediated depletion. In order to independently validate this finding, and to determine whether Tm1C has roles outside of the germline, we sought to create an isoform-specific null. In order to create this null, we chose to delete a small region between the 5′UTR and the first exon of the Tm1C and Tm1I isoforms (Fig. 6A). This removes the initiating ATG codon, changes the reading frame, and results in several premature stop codons. We used a recently described CRISPR/Cas9 approach to delete this region (Gratz et al., 2014). In this strategy, two independent guide RNAs are designed to cleave at desired loci, and the intervening sequences are replaced using homology-directed repair (Gratz et al., 2014). We refer to this mutant as tm1_delC.
Fig. 6.
Generation of a Tm1C-null mutant. (A) Schematic showing the region deleted in Tm1C-null mutants. We refer to this strain as tm1_delC. Gray boxes represent untranslated regions and orange boxes indicate coding regions. The sequences of the guide RNAs used to generate this deletion are also indicated. (B) Ovarian lysates were prepared from wild-type (lane 1) or the tm1_delC mutant (lane 2). The lysates were analyzed by blotting using the anti-Tm1C antibody. The blot was subsequently stripped and probed with an anti-Khc antibody (bottom panel). (C,D) Ovaries from tm1_delC mutants were processed for smFISH using probes against osk (red) and were counterstained with DAPI (cyan). D′,D″ and D‴ are enlarged images of boxes in D. (E) Ovaries from tm1_delC mutants expressing the GFP–Tm1C transgene were processed for smFISH using probes against osk (red) and were counterstained with DAPI (cyan). The arrow indicates localized osk mRNA. (F,G) Ovaries from wild-type (F) or tm1_delC mutants (G) were fixed and processed for immunofluorescence using an antibody against Vasa. The arrow indicates localization of Vasa to the pole plasm in wild-type egg chambers. (H,I) Ovaries from tm1_delC mutants were processed for immunofluorescence using antibodies against Khc (H) or Dhc (I). Arrows indicate posterior-localized Khc and Dhc in Tm1C-null mutants. Scale bars: 50 μm (C–I); 5 μm (D′–D‴).
tm1_delC homozygotes are semi-viable. Approximately 20% of mutants die during pupation. Ovarian lysates were prepared from homozygous and wild-type adult females and analyzed by western blotting. Confirming the status of this mutant as a true null, the band corresponding to Tm1C was absent in mutant lysates (Fig. 6B). However, the levels of Tm1J and Khc were unaffected (Fig. 6B).
We next examined the localization of osk mRNA in tm1_delC females (Fig. 6C,D). As with shRNA-mediated depletion, complete loss of Tm1C resulted in delocalization of osk mRNA around the oocyte cortex (Fig. 6D). However, transport of osk into the oocyte and oligomerization into large particles was not affected (Fig. 6D). Furthermore, expression of GFP–Tm1C in the tm1_delC background completely restored posterior osk mRNA localization (Fig. 6E).
Consistent with Tm1C depletion, grk mRNA was correctly localized in tm1_delC mutants (Fig. S4A). Furthermore, the oocyte nucleus was correctly positioned and aberrant actin spheres were not observed (Fig. S4B). Finally, although the pole plasm component Vasa was delocalized in tm1_delC mutants, (Fig. 6F,G), Khc and the Dynein heavy chain (Dhc, also known as Dhc64c) remained enriched at the posterior pole (Fig. 6H,I). Thus, in the absence of Tm1C, Khc retains the ability to transport Dhc to the oocyte posterior.
The partial lethality associated with tm1_delC prompted us to examine whether this unique Tropomyosin isoform was expressed outside of the female germline. Consistent with this notion, Tm1C could be detected in lysates prepared from male flies (Fig. S4C). These findings are supported by previously published RNA-seq results (Graveley et al., 2011; Fig. S4D). However, osk expression is restricted to the female germline (Fig. S4D). This raises the intriguing possibility that Tm1C might function along with Khc to facilitate cargo transport in additional tissues. Further studies are needed to fully examine the organismal roles of Tm1C.
DISCUSSION
Localization of osk mRNA at the posterior of Drosophila oocytes requires Khc, but is independent of the canonical Kinesin-1 adaptor, Klc (Brendza et al., 2000; Palacios and St Johnston, 2002). In this report, we demonstrate that a new isoform of Tropomyosin, referred to as Tm1C, directly interacts with Khc and functions in concert with the motor to localize osk mRNA.
Tropomyosins are actin-binding proteins that are crucial for muscle contraction (Gunning et al., 2005). However, non-muscle isoforms are also expressed, and these isoforms contribute to a variety of actin-dependent processes (Gunning et al., 2005). Mammalian genomes contain four tropomyosin genes and encode more than 40 different isoforms (Wang and Coluccio, 2010). By contrast, the Drosophila genome contains two tropomyosin genes, Tm1 and Tm2. Genome annotation suggests that both genes are capable of producing multiple alternatively spliced isoforms. However, not much is known regarding the expression profile of the various tropomyosin isoforms in non-muscle tissues. A recent study by Goins and Mullins revealed that three tropomyosin isoforms are expressed in Drosophila S2 cells: Tm1A, Tm1J and Tm2A (Goins and Mullins, 2015).
The Tropomyosin isoform that interacts with Khc is Tm1C or Tm1I. These isoforms encode the same protein but differ in their untranslated regions. For simplicity, we refer to this Khc-interacting isoform as Tm1C. Although Kinesin has been shown to interact with a Myosin motor (Huang et al., 1999), this is the first report of a Tropomyosin that is capable of directly binding a microtubule motor. In comparison to canonical Tropomyosins, Tm1C has a unique domain organization. Most tropomyosins, including the high-molecular-mass isoforms detected in mammals, are composed of coiled-coil domains that enable binding along the sides of actin filaments (Gunning et al., 2015). Interestingly, the high-molecular-mass isoforms in mammalian cells are still less than 300 amino acids long. By contrast, Tm1C encodes a 441-amino-acid protein. In addition to being much larger than classical Tropomyosins, Tm1C also contains a long N-terminal extension that is devoid of coiled-coil motifs (Fig. 1E).
How might Tm1C function in osk localization? One possibility is that Tm1C might be required for organization of the microtubule cytoskeleton. Thus, a defect in this process could indirectly result in osk localization. Our results suggest that this scenario is unlikely. The distribution of α-tubulin, a core component of the microtubule polymer, was the same in control and Tm1C-null egg chambers (Fig. S4E,F). In addition, no difference was detected in the distribution of microtubule minus-ends, and Kin:βgal, a reporter of microtubule plus-ends (Clark et al., 1994), remained correctly localized at the posterior pole in Tm1C-null mutants (Fig. S4G–J). Finally, cargoes such as bcd and grk mRNA, whose localization requires a correctly polarized cytoskeleton, were correctly sorted in egg chambers lacking Tm1C (Fig. 5I,L; Fig. S4A).
We propose a model whereby Tm1C directly or indirectly links Khc to the osk mRNP. This is supported by the observation that loss of either Tm1C or Khc produces identical osk delocalization phenotypes (Figs 4 and 6). In both instances, osk mRNA localized around the oocyte cortex. In addition, we demonstrate that Tm1C binds to a small domain in the C-terminal tail of Khc (Fig. 2). Consistent with the adaptor model, Williams et al. have shown that osk mRNPs are delocalized around the cortex in mutants expressing a truncated version of Khc that is lacking this domain (Williams et al., 2014).
A key prediction of this hypothesis is that in the absence of Tm1C, Khc no longer associates with the osk mRNP. Although we were able to detect osk mRNA in Staufen immunoprecipitates, the same technique proved to be unsuccessful in detecting osk in Khc immunoprecipitates (data not shown). Thus, although there is undisputed evidence that Khc transports osk mRNA (Zimyanin et al., 2008), there have been no published reports demonstrating a physical interaction between Khc and osk. A likely possibility is that motor–cargo complexes are labile, and therefore not retained during biochemical purification. Consistent with this notion, although Khc is thought to transport Dynein to the posterior pole (Brendza et al., 2002; Duncan and Warrior, 2002; Januschke et al., 2002), we failed to detect any peptides from Dynein in Khc immunoprecipitates (Table 1 and data not shown). Given this limitation in detecting the Khc–osk interaction in wild-type egg chambers, it has not been possible to prove a lack of association in Tm1C-null egg chambers.
Several years back, Erdélyi et al. and Tetzlaff et al., demonstrated a requirement for cytoplasmic Tropomyosin in the localization of osk mRNA (Erdelyi et al., 1995; Tetzlaff et al., 1996). In these papers, the gene was referred to as tmII (Erdelyi et al., 1995; Tetzlaff et al., 1996). However, current genome annotation suggest that this P-element resides within the Tm1 gene. Furthermore, the position of the P-element transposons used in these publications suggests that the isoform most likely affected in these mutants is Tm1A. The mechanism by which loss or reduction of Tm1A could contribute to osk mRNA delocalization is unknown. However, it is unlikely to function through Kinesin because, unlike Tm1C, Khc does not appear to associate with Tm1A (Fig. 1C). In addition, osk accumulates at the anterior margin of these previous described tropomyosin mutants and not along the oocyte cortex (Erdelyi et al., 1995; Zimyanin et al., 2008). If indeed Tm1A also participates in the localization of osk mRNA, further studies will be required to reveal its mechanism of action.
Although tm1_delC adult females can be obtained, the mutant is semi-lethal. Approximately 20% of homozygotes die during pupal stages. By contrast, complete loss of osk is not associated with lethality. This raises the possibility that Tm1C has additional functions and that loss of these functions contributes to partial lethality. Consistent with this notion, Tm1C could be detected in whole male flies (Fig. S4C). In addition, RNA-seq results suggest that, unlike osk, this unique isoform of Tropomyosin is expressed at several developmental stages (Fig. S4D; Graveley et al., 2011). Future studies will seek to define whether Tm1C functions in concert with Khc to transport additional cargoes or whether Tm1C has roles that are independent of Kinesin-1.
While our manuscript was in revision, Cho et al. independently identified a role for Tm1C in border cell migration (Cho et al., 2016). Furthermore, these authors demonstrate that Tm1C shares some properties with intermediate filaments (Cho et al., 2016). Under in vitro conditions, Tm1C was able to form filamentous structures. In addition, when ovaries were incubated with a PIPES pH 6.8 buffer, mCherry–Tm1C formed filaments in vivo that colocalized with microtubules (Cho et al., 2016). We obtained similar results using our GFP–Tm1C strain (Fig. S2G). Interestingly, a similar localization pattern was also observed for Khc using these same fixation conditions (Fig. S2H,I). However, Khc maintained this filament or microtubule localization pattern in the null tm1_delC mutants (Fig. S4K,L). Thus, the role of Tm1C in the osk localization pathway does not involve recruitment of Khc to microtubules. As stated previously, we favor a model whereby Tm1C links Khc to osk mRNPs, thus facilitating posterior transport of this mRNA.
MATERIALS AND METHODS
Fly stocks
The following stocks were used: Oregon-R-C (used as wild-type; Bloomington stock center; #5); GFP-Stau (Zimyanin et al., 2008); eb1 shRNA (Bloomington stock center; #36680, donor TRiP); egl shRNA-1 (Bloomington stock center; #43550, donor TRiP); khc shRNA (Bloomington stock center; #35409, donor TRiP), and Kin:βgal (Clark et al., 1994). shRNA expression was driven using P{w[+mC]=matalpha4-GAL-VP16}V37 (Bloomington stock center; #7063, donor Andrea Brand).
GFP–Tm1C was expressed by cloning the Tm1C coding sequence into a vasa-GFP expression vector (Sano et al., 2002). The Tm1C coding sequence with silent mutations at the shRNA site was synthesized by Genewiz.
Tm1 shRNA flies were generated by cloning the following sequences into the NheI and EcoR1 sites of the Valium 22 vector (Ni et al., 2011): Tm1 shRNA-1, 5′-GCCGACGACGATGACAACCAA-3′; Tm1 shRNA-2, 5′-AAGGTCAGAGAAATCGGACAA-3′. Transgenic flies containing these constructs were generated by BestGene Inc (inserted at the attP40 site on chromosome 2). The null tm1_delC mutants were created by injecting a mix containing two guide RNA plasmids along with a donor vector for homology-directed repair (Gratz et al., 2014). Complete details regarding the cloning strategy will be provided upon request.
Antibodies and imaging reagents
Unless specifically stated, the indicated dilutions are for immunofluorescence: mouse anti-Dhc (Developmental Studies Hybridoma bank; 1:150; donor J. Scholey); rabbit anti-Khc (cat. no AKIN01-A, Cytoskeleton, Inc.; 1:150, western blotting, 1:1500); chicken anti-Oskar (Sanghavi et al., 2016; 1:50); FITC-conjugated mouse anti-α-tubulin (cat. no F2168-.2ML, Sigma; 1:100); mouse anti-β-galactosidase (cat. no Z3781, Promega; 1:1000); mouse anti-γ-tubulin (cat. no T5326-100UL, Sigma; 1:100); rat anti-GFP (cat. no 04404-84, Nacalai USA, Inc.; 1:600); mouse anti-LaminDmO (Developmental Studies Hybridoma bank; clone ADL84.12; 1:200; donor P.A. Fisher); mouse anti-GFP (cat. no 632381, Clontech; western blotting, 1:4000); rat anti-RFP (cat. no 5f8-100, Chromotek; western blotting, 1:1000); rabbit anti-Vas (1:1000 from Paul Lasko, Department of Biology, McGill University, Canada); Tm1C (custom-made as described below; western blotting, 1:1500); goat anti-rabbit-IgG conjugated to Alexa Fluor 594 and 488 (Life Technologies, 1:400 and 1:200, respectively); goat anti-mouse-IgG conjugated to Alexa 594 and 488 (Life Technologies, 1:400 and 1:200); goat anti-chicken-IgG conjugated to 594 (Life Technologies, 1:400). Anti-Tm1C antibody was generated by injecting the following peptides into rabbits: N-DKSEKSDRKKKSSGKKERSKRSNP-C and N-KEARFLAEEADKKYDEVQLK-C. The injection and purification of peptide-specific antibodies was performed by Pacific immunology (Ramona, CA). TRITC-conjugated phalloidin (Sigma Aldrich) was used to visualize F-actin and ToPro3 (Life Technologies) was used to visualize DNA.
DNA constructs
For binding studies in S2 cells, cDNA for Tm1C and klc was cloned into the pAGW gateway vector; khc constructs were cloned into the pARW vector (obtained from the Drosophila Genomics Resource Center). Internal deletions in khc were made using the Q5 site-directed mutagenesis kit (NEB, Ipswich, MA). Transfections were performed using Effectene (Qiagen). For the direct binding experiment, Tm1C was cloned into pDEST15 and the khc cDNA was cloned into the pSP64 vector (Promega). Tm1C guide RNAs were cloned into the pU6-3 Bbsi plasmid and Tm1C homology arms were cloned into the pHd-DsRed-attP plasmid from the laboratory of Kate O'Connor-Giles (Cell and Molecular Biology, University of Wisconsin, Madison). For purification of GFP and Khc–GFP from ovarian lysates, the coding sequence of either GFP or Khc–GFP was cloned into the pUASp-attB-K10 vector (Koch et al., 2009).
Protein–protein interaction
In order to purify Khc-interacting partners, ovaries were dissected from flies expressing either GFP or Khc–GFP. The ovaries were homogenized in lysis buffer (50 mM Tris-HCl pH 7.5, 50 mM NaCl, 0.2 mM EDTA, 0.05% NP40 and Halt protease inhibitor cocktail, Pierce). The lysates were cleared by centrifugation at 10,000 g at 4°C for 5 min. 1000 μg of either GFP- or Khc–GFP-expressing lysate was added to GFP-trap beads (Chromotek). The binding was performed at 4°C for 1 h. Next, the beads were washed twice with NP40-containing wash buffer (50 mM Tris-HCl pH 7.5, 200 mM NaCl, 0.2 mM EDTA, 0.05% NP40). Subsequently, the beads were washed twice with the same buffer but lacking NP40. Next, the co-precipitating proteins were eluted using a 0.2 M Glycine pH 2.5 solution. After elution, the pH was neutralized with Tris-HCl pH 8.0. The samples were digested with trypsin and analyzed by tandem mass spectrometry. The trypsin digestion and mass spectrometry analysis was performed at the Mass Spectrometry and Proteomics Resource Laboratory at Harvard University.
For detecting protein–protein interaction using S2 cells, lysates were prepared using the indicated transfected cells. The cells were lysed using RIPA buffer (50 mM Tris-HCl pH 7.5, 150 nM NaCl, 1% NP40, 1 mM EDTA). The lysates were cleared by centrifugation as described above and added to GFP-trap beads (Chromotek). After binding and wash steps, the co-precipitating proteins were eluted in Laemmli buffer, run on a gel, and analyzed by western blotting.
For the direct binding studies, GST and GST–Tm1C were expressed in BL21 DE3 pLysS cells (Life Technologies). The proteins were induced using 0.5 mM IPTG for 6 h at room temperature. Lysates were prepared using a French press. The induced proteins were purified by incubation with glutathione–Sepharose beads (Pierce). GelCode Blue (Pierce) was used to visualize these proteins in a gel. The RFP-tagged Khc construct was expressed using the TnT SP6 Coupled Wheat Germ Extract System (Promega).
Immunofluorescence and in situ hybridization
Ovaries were processed for immunofluorescence as previously described (Sanghavi et al., 2016). In situ hybridization was used to detect bcd, grk, nos and CycB mRNA and was performed using a published procedure (Sanghavi et al., 2013). smFISH was used for detection of osk. Quasar570-conjugated Stellaris DNA oligonucleotide probes against osk and Gapdh1 and Gapdh2 were obtained from LGC Biosearch Technologies (Petaluma, CA). Ovaries were fixed in 4% formaldehyde (diluted in PBS) for 20 min. Subsequently, the ovaries were washed with several changes of PBS. Next, methanol was added and the ovaries were stored at −20°C for 1 h. The ovaries were then washed with a 7:3 methanol:PBST (PBS plus 0.1% Triton X-100) solution for 10 min at room temperature. Next, 3:7 methanol:PBST was used. The ovaries were then washed 4× with PBST. PBST was removed and the samples were incubated in pre-hybridization buffer (4× SSC, 35% deionized formamide, 0.1% Tween-20) for 10 min. The prehyrbidization solution was removed and the probe diluted to 100 nM in hybridization buffer [10% dextran sulfate, 0.1 mg/ml salmon sperm single-stranded (ss)DNA, 100 µl vanadyl ribonucleoside (NEB Biolabs, Ipswich, MA), 20 μg/ml RNAse-free BSA, 4× SSC, 0.1% Tween-20 and 35% deionized formamide] was added. The ovaries were incubated with probe overnight at 37°C. The next day, the probe was removed and the ovaries were washed twice in prehybridization buffer for 30 min each. The samples were then stained with DAPI, washed in PBST, and mounted onto slides using Aqua Poly mount (Polysciences, Inc., Warrington, PA). In order to detect filaments of GFP–Tm1C and Khc, ovaries expressing the transgene were processed as previously described (Cho et al., 2016).
Microscopy
Images were captured on a Zeiss LSM 780 upright confocal microscope and were prepared for presentation using Fiji, Adobe Photoshop and Adobe Illustrator.
Quantification
Localization phenotypes were quantified by scoring oocytes of the indicated genotype from three independent experiments. The level of GFP–Tm1 in stage 10 egg chambers (Fig. 3K) was quantified using the Zen software (Zeiss). The level of fluorescence intensity in a 50-μm-square area in nurse cells of control and Khc-depleted egg chambers was determined. The quantification was done on 25 different egg chambers using a single confocal slice. An unpaired t-test was performed using standard deviation, mean and n value.
Acknowledgements
We would like to thank Caryn Navarro, Bill Saxton and Daniel St Johnston for providing antibodies and fly strains. We are extremely grateful to Kate O'Connor-Giles and Fiona Ukken for their advice and assistance with CRISPR. We are thankful to Liz Gavis for advising us on the protocol for smFISH. We thank the Bloomington Stock Center, Developmental Studies Hybridoma Bank, and the Drosophila Genomics Resource Center for providing fly strains, antibodies and DNA constructs that were crucial in completing this work.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization, R.V.-K., D.P.B. and G.B.G.; Methodology, R.V.-K., D.P.B., G.L. and G.B.G.; Investigation, R.V.-K., D.P.B., G.L. and G.B.G.; Writing, D.P.B. and G.B.G.; Funding acquisition, G.B.G. Supervision, G.B.G.
Funding
This work was supported by a grant from the National Institutes of Health to G.B.G. (R01GM100088-01). Deposited in PMC for release after 12 months.
Supplementary information
Supplementary information available online at http://jcs.biologists.org/lookup/doi/10.1242/jcs.194332.supplemental
References
- Andrews S. B., Gallant P. E., Leapman R. D., Schnapp B. J. and Reese T. S. (1993). Single kinesin molecules crossbridge microtubules in vitro. Proc. Natl. Acad. Sci. USA 90, 6503-6507. 10.1073/pnas.90.14.6503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berleth T., Burri M., Thoma G., Bopp D., Richstein S., Frigerio G., Noll M. and Nusslein-Volhard C. (1988). The role of localization of bicoid RNA in organizing the anterior pattern of the Drosophila embryo. EMBO J. 7, 1749-1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brendza R. P., Serbus L. R., Duffy J. B. and Saxton W. M. (2000). A function for kinesin I in the posterior transport of oskar mRNA and Staufen protein. Science 289, 2120-2122. 10.1126/science.289.5487.2120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brendza R. P., Serbus L. R., Saxton W. M. and Duffy J. B. (2002). Posterior localization of dynein and dorsal-ventral axis formation depend on kinesin in Drosophila oocytes. Curr. Biol. 12, 1541-1545. 10.1016/S0960-9822(02)01108-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullock S. L. (2011). Messengers, motors and mysteries: sorting of eukaryotic mRNAs by cytoskeletal transport. Biochem. Soc. Trans. 39, 1161-1165. 10.1042/BST0391161 [DOI] [PubMed] [Google Scholar]
- Cajigas I. J., Tushev G., Will T. J., tom Dieck S., Fuerst N. and Schuman E. M. (2012). The local transcriptome in the synaptic neuropil revealed by deep sequencing and high-resolution imaging. Neuron 74, 453-466. 10.1016/j.neuron.2012.02.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chartrand P., Meng X.-H., Singer R. H. and Long R. M. (1999). Structural elements required for the localization of ASH1 mRNA and of a green fluorescent protein reporter particle in vivo. Curr. Biol. 9, 333-336. 10.1016/S0960-9822(99)80144-4 [DOI] [PubMed] [Google Scholar]
- Cho A., Kato M., Whitwam T., Kim J. H. and Montell D. J. (2016). An atypical tropomyosin in Drosophila with intermediate filament-like properties. Cell Rep. 16, 928-938. 10.1016/j.celrep.2016.06.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark I., Giniger E., Ruohola-Baker H., Jan L. Y. and Jan Y. N. (1994). Transient posterior localization of a kinesin fusion protein reflects anteroposterior polarity of the Drosophila oocyte. Curr. Biol. 4, 289-300. 10.1016/S0960-9822(00)00068-3 [DOI] [PubMed] [Google Scholar]
- Clark A., Meignin C. and Davis I. (2007). A Dynein-dependent shortcut rapidly delivers axis determination transcripts into the Drosophila oocyte. Development 134, 1955-1965. 10.1242/dev.02832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalby B. and Glover D. M. (1992). 3′ non-translated sequences in Drosophila cyclin B transcripts direct posterior pole accumulation late in oogenesis and peri-nuclear association in syncytial embryos. Development 115, 989-997. [DOI] [PubMed] [Google Scholar]
- Dienstbier M., Boehl F., Li X. and Bullock S. L. (2009). Egalitarian is a selective RNA-binding protein linking mRNA localization signals to the dynein motor. Genes Dev. 23, 1546-1558. 10.1101/gad.531009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan J. E. and Warrior R. (2002). The cytoplasmic dynein and kinesin motors have interdependent roles in patterning the Drosophila oocyte. Curr. Biol. 12, 1982-1991. 10.1016/S0960-9822(02)01303-9 [DOI] [PubMed] [Google Scholar]
- Ephrussi A., Dickinson L. K. and Lehmann R. (1991). Oskar organizes the germ plasm and directs localization of the posterior determinant nanos. Cell 66, 37-50. 10.1016/0092-8674(91)90137-N [DOI] [PubMed] [Google Scholar]
- Erdelyi M., Michon A. M., Guichet A., Glotzer J. B. and Ephrussi A. (1995). Requirement for Drosophila cytoplasmic tropomyosin in oskar mRNA localization. Nature 377, 524-527. 10.1038/377524a0 [DOI] [PubMed] [Google Scholar]
- Gavis E. R. and Lehmann R. (1992). Localization of nanos RNA controls embryonic polarity. Cell 71, 301-313. 10.1016/0092-8674(92)90358-J [DOI] [PubMed] [Google Scholar]
- Glater E. E., Megeath L. J., Stowers R. S. and Schwarz T. L. (2006). Axonal transport of mitochondria requires milton to recruit kinesin heavy chain and is light chain independent. J. Cell Biol. 173, 545-557. 10.1083/jcb.200601067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goins L. M. and Mullins R. D. (2015). A novel tropomyosin isoform functions at the mitotic spindle and Golgi in Drosophila. Mol. Biol. Cell 26, 2491-2504. 10.1091/mbc.E14-12-1619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonsalvez G. B., Urbinati C. R. and Long R. M. (2005). RNA localization in yeast: moving towards a mechanism. Biol. Cell 97, 75-86. 10.1042/BC20040066 [DOI] [PubMed] [Google Scholar]
- Gratz S. J., Ukken F. P., Rubinstein C. D., Thiede G., Donohue L. K., Cummings A. M. and O'Connor-Giles K. M. (2014). Highly specific and efficient CRISPR/Cas9-catalyzed homology-directed repair in Drosophila. Genetics 196, 961-971. 10.1534/genetics.113.160713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graveley B. R., Brooks A. N., Carlson J. W., Duff M. O., Landolin J. M., Yang L., Artieri C. G., van Baren M. J., Boley N., Booth B. W. et al. (2011). The developmental transcriptome of Drosophila melanogaster. Nature 471, 473-479. 10.1038/nature09715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunning P. W., Schevzov G., Kee A. J. and Hardeman E. C. (2005). Tropomyosin isoforms: divining rods for actin cytoskeleton function. Trends Cell Biol. 15, 333-341. 10.1016/j.tcb.2005.04.007 [DOI] [PubMed] [Google Scholar]
- Gunning P. W., Hardeman E. C., Lappalainen P. and Mulvihill D. P. (2015). Tropomyosin - master regulator of actin filament function in the cytoskeleton. J. Cell Sci. 128, 2965-2974. 10.1242/jcs.172502 [DOI] [PubMed] [Google Scholar]
- Hackney D. D. and Stock M. F. (2000). Kinesin's IAK tail domain inhibits initial microtubule-stimulated ADP release. Nat. Cell Biol. 2, 257-260. 10.1038/35010525 [DOI] [PubMed] [Google Scholar]
- Hirokawa N., Noda Y., Tanaka Y. and Niwa S. (2009). Kinesin superfamily motor proteins and intracellular transport. Nat. Rev. Mol. Cell Biol. 10, 682-696. 10.1038/nrm2774 [DOI] [PubMed] [Google Scholar]
- Hoogenraad C. C., Akhmanova A., Howell S. A., Dortland B. R., De Zeeuw C. I., Willemsen R., Visser P., Grosveld F. and Galjart N. (2001). Mammalian Golgi-associated Bicaudal-D2 functions in the dynein-dynactin pathway by interacting with these complexes. EMBO J. 20, 4041-4054. 10.1093/emboj/20.15.4041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J.-D., Brady S. T., Richards B. W., Stenoien D., Resau J. H., Copeland N. G. and Jenkins N. A. (1999). Direct interaction of microtubule- and actin-based transport motors. Nature 397, 267-270. 10.1038/16722 [DOI] [PubMed] [Google Scholar]
- Jambor H., Surendranath V., Kalinka A. T., Mejstrik P., Saalfeld S. and Tomancak P. (2015). Systematic imaging reveals features and changing localization of mRNAs in Drosophila development. Elife 4, e05003 10.7554/eLife.05003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Januschke J., Gervais L., Dass S., Kaltschmidt J. A., Lopez-Schier H., St Johnston D., Brand A. H., Roth S. and Guichet A. (2002). Polar transport in the Drosophila oocyte requires Dynein and Kinesin I cooperation. Curr. Biol. 12, 1971-1981. 10.1016/S0960-9822(02)01302-7 [DOI] [PubMed] [Google Scholar]
- Kim-Ha J., Smith J. L. and Macdonald P. M. (1991). oskar mRNA is localized to the posterior pole of the Drosophila oocyte. Cell 66, 23-35. 10.1016/0092-8674(91)90136-M [DOI] [PubMed] [Google Scholar]
- Koch R., Ledermann R., Urwyler O., Heller M. and Suter B. (2009). Systematic functional analysis of Bicaudal-D serine phosphorylation and intragenic suppression of a female sterile allele of BicD. PLoS ONE 4, e4552 10.1371/journal.pone.0004552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kugler J.-M. and Lasko P. (2009). Localization, anchoring and translational control of oskar, gurken, bicoid and nanos mRNA during Drosophila oogenesis. Fly 3, 15-28. 10.4161/fly.3.1.7751 [DOI] [PubMed] [Google Scholar]
- Lecuyer E., Yoshida H., Parthasarathy N., Alm C., Babak T., Cerovina T., Hughes T. R., Tomancak P. and Krause H. M. (2007). Global analysis of mRNA localization reveals a prominent role in organizing cellular architecture and function. Cell 131, 174-187. 10.1016/j.cell.2007.08.003 [DOI] [PubMed] [Google Scholar]
- Little S. C., Tikhonov M. and Gregor T. (2013). Precise developmental gene expression arises from globally stochastic transcriptional activity. Cell 154, 789-800. 10.1016/j.cell.2013.07.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little S. C., Sinsimer K. S., Lee J. J., Wieschaus E. F. and Gavis E. R. (2015). Independent and coordinate trafficking of single Drosophila germ plasm mRNAs. Nat. Cell Biol. 17, 558-568. 10.1038/ncb3143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDougall N., Clark A., MacDougall E. and Davis I. (2003). Drosophila gurken (TGFalpha) mRNA localizes as particles that move within the oocyte in two dynein-dependent steps. Dev. Cell 4, 307-319. 10.1016/S1534-5807(03)00058-3 [DOI] [PubMed] [Google Scholar]
- Manstein D. J. and Mulvihill D. P. (2016). Tropomyosin-mediated Regulation of Cytoplasmic Myosins. Traffic. 17, 872-877. 10.1111/tra.12399 [DOI] [PubMed] [Google Scholar]
- Mische S., Li M., Serr M. and Hays T. S. (2007). Direct observation of regulated ribonucleoprotein transport across the nurse cell/oocyte boundary. Mol. Biol. Cell 18, 2254-2263. 10.1091/mbc.E06-10-0959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro C., Puthalakath H., Adams J. M., Strasser A. and Lehmann R. (2004). Egalitarian binds dynein light chain to establish oocyte polarity and maintain oocyte fate. Nat. Cell Biol. 6, 427-435. 10.1038/ncb1122 [DOI] [PubMed] [Google Scholar]
- Neuman-Silberberg F. S. and Schupbach T. (1993). The Drosophila dorsoventral patterning gene gurken produces a dorsally localized RNA and encodes a TGF alpha-like protein. Cell 75, 165-174. 10.1016/S0092-8674(05)80093-5 [DOI] [PubMed] [Google Scholar]
- Ni J.-Q., Zhou R., Czech B., Liu L.-P., Holderbaum L., Yang-Zhou D., Shim H.-S., Tao R., Handler D., Karpowicz P. et al. (2011). A genome-scale shRNA resource for transgenic RNAi in Drosophila. Nat. Methods 8, 405-407. 10.1038/nmeth.1592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niedner A., Edelmann F. T. and Niessing D. (2014). Of social molecules: the interactive assembly of ASH1 mRNA-transport complexes in yeast. RNA Biol. 11, 998-1009. 10.4161/rna.29946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palacios I. M. and St Johnston D. (2002). Kinesin light chain-independent function of the Kinesin heavy chain in cytoplasmic streaming and posterior localisation in the Drosophila oocyte. Development 129, 5473-5485. 10.1242/dev.00119 [DOI] [PubMed] [Google Scholar]
- Sanghavi P., Laxani S., Li X., Bullock S. L. and Gonsalvez G. B. (2013). Dynein associates with oskar mRNPs and is required for their efficient net plus-end localization in Drosophila oocytes. PLoS ONE 8, e80605 10.1371/journal.pone.0080605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanghavi P., Liu G., Veeranan-Karmegam R., Navarro C. and Gonsalvez G. B. (2016). Multiple Roles for Egalitarian in Polarization of the Drosophila Egg Chamber. Genetics 203, 415-432. 10.1534/genetics.115.184622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sano H., Nakamura A. and Kobayashi S. (2002). Identification of a transcriptional regulatory region for germline-specific expression of vasa gene in Drosophila melanogaster. Mech. Dev. 112, 129-139. 10.1016/S0925-4773(01)00654-2 [DOI] [PubMed] [Google Scholar]
- Schnorrer F., Bohmann K. and Nusslein-Volhard C. (2000). The molecular motor dynein is involved in targeting swallow and bicoid RNA to the anterior pole of Drosophila oocytes. Nat. Cell Biol. 2, 185-190. 10.1038/35008601 [DOI] [PubMed] [Google Scholar]
- Seiler S., Kirchner J., Horn C., Kallipolitou A., Woehlke G. and Schliwa M. (2000). Cargo binding and regulatory sites in the tail of fungal conventional kinesin. Nat. Cell Biol. 2, 333-338. 10.1038/35014022 [DOI] [PubMed] [Google Scholar]
- Singer-Kruger B. and Jansen R.-P. (2014). Here, there, everywhere. mRNA localization in budding yeast. RNA Biol. 11, 1031-1039. 10.4161/rna.29945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- St Johnston D. (2005). Moving messages: the intracellular localization of mRNAs. Nat. Rev. Mol. Cell Biol. 6, 363-375. 10.1038/nrm1643 [DOI] [PubMed] [Google Scholar]
- Tetzlaff M. T., Jackle H. and Pankratz M. J. (1996). Lack of Drosophila cytoskeletal tropomyosin affects head morphogenesis and the accumulation of oskar mRNA required for germ cell formation. EMBO J. 15, 1247-1254. [PMC free article] [PubMed] [Google Scholar]
- Wang C.-L. A. and Coluccio L. M. (2010). New insights into the regulation of the actin cytoskeleton by tropomyosin. Int. Rev. Cell Mol. Biol. 281, 91-128. 10.1016/S1937-6448(10)81003-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams L. S., Ganguly S., Loiseau P., Ng B. F. and Palacios I. M. (2014). The auto-inhibitory domain and ATP-independent microtubule-binding region of Kinesin heavy chain are major functional domains for transport in the Drosophila germline. Development 141, 176-186. 10.1242/dev.097592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimyanin V. L., Belaya K., Pecreaux J., Gilchrist M. J., Clark A., Davis I. and St Johnston D. (2008). In vivo imaging of oskar mRNA transport reveals the mechanism of posterior localization. Cell 134, 843-853. 10.1016/j.cell.2008.06.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zivraj K. H., Tung Y. C. L., Piper M., Gumy L., Fawcett J. W., Yeo G. S. H. and Holt C. E. (2010). Subcellular profiling reveals distinct and developmentally regulated repertoire of growth cone mRNAs. J. Neurosci. 30, 15464-15478. 10.1523/JNEUROSCI.1800-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]