ABSTRACT
The RNA-binding protein HuR binds to elements rich in adenylate and uridylate (AU-rich elements) in target mRNAs and stabilizes them against degradation. The complete spectrum of genes whose expression is regulated by HuR and are the basis for the broad range of cellular functions of the protein is incompletely understood. We show that HuR controls the expression of multiple components of the nuclear import machinery. Consequently, HuR is crucial for the nuclear import of cellular retinoic acid-binding protein 2 (CRABP2), which delivers RA to the nuclear retinoic acid receptor (RAR) and whose mobilization to the nucleus is mediated by a ‘classical-like’ nuclear localization signal (NLS). HuR is also required for heregulin-induced nuclear translocation of the NFκB subunit p65, which contains both classical and non-canonical NLSs. HuR thus regulates the transcriptional activities of both RAR and NFκB. The observations reveal that HuR plays a central role in regulating nuclear import of proteins.
KEY WORDS: HuR, RNA stability, Nuclear import, Retinoic acid, Cellular retinoic acid-binding protein, NFκB
Summary: We show that the RNA-binding protein HuR plays a key role in controlling expression of multiple components of the nuclear import machinery, presenting a new mechanism by which nuclear import of proteins is regulated.
INTRODUCTION
One of the best-characterized RNA-binding proteins in animals is HuR (encoded for by Elavl1), a ubiquitously-expressed member of the embryonic lethal abnormal vision (ELAV)/Hu family of RNA-binding proteins. In the nucleus, HuR is involved in various functions, including RNA splicing and nuclear export. In the extranuclear compartment, HuR is associated with polysomes and binds elements rich in adenylate and uridylate (AU-rich elements; ARE) in 3′UTR regions of target transcripts and protects them against degradation (Brennan and Steitz, 2001; Chen et al., 2002; Hinman and Lou, 2008; Lebedeva et al., 2011; Lopez de Silanes et al., 2004; Mukherjee et al., 2011; Myer et al., 1997). By stabilizing target mRNAs, HuR is involved in key biological processes including cell-cycle progression, apoptosis, immune function, inflammation and oncogenic activities (Abdelmohsen and Gorospe, 2010; Ghosh et al., 2009; Gupta et al., 2012; Hinman and Lou, 2008; Lebedeva et al., 2011).
We recently discovered that transcript stabilization by HuR is aided by cellular retinoic acid-binding protein 2 (CRABP2) (Vreeland et al., 2014b). The best-understood function of CRABP2 is its ability to shuttle retinoic acid (RA) from the cytosol to the nucleus, where it is directly delivered to the nuclear retinoic acid receptor (RAR) and thereby promotes transcriptional activity (Budhu and Noy, 2002; Donato et al., 2007; Dong et al., 1999; Manor et al., 2003; Schug et al., 2007; Sessler and Noy, 2005). Surprisingly, we found that, in the absence of RA, CRABP2 binds to HuR and dramatically increases its RNA-binding affinity. Consequently, CRABP2 enhances the stability of various HuR target transcripts and upregulates their levels (Vreeland et al., 2014b). Binding of RA triggers dissociation of CRABP2 from HuR and activates a nuclear localization signal (NLS), allowing it to mobilize to the nucleus where it delivers RA to RAR. Our observations showed that the well-established anti-carcinogenic activity of CRABP2 is mediated both by its ability to activate RAR, leading to induction of anti-proliferative RAR target genes (Budhu et al., 2001; Donato and Noy, 2005; Donato et al., 2007; Manor et al., 2003), and by cooperation with HuR, resulting in stabilization of anti-oncogenic HuR target mRNAs (Vreeland et al., 2014a,b). Interestingly, we found that reducing the expression of HuR not only abolished the ability of CRABP2 to upregulate HuR target transcripts but also inhibited the transcriptional activity of RAR (Vreeland et al., 2014a). The additional observation that reducing the expression of HuR blocked nuclear translocation of CRABP2 suggested that HuR is involved in some aspects of nuclear import (Vreeland et al., 2014a).
Proteins that localize to the nucleus contain nuclear targeting sequences that link them to carrier proteins of the β-karyopherin (importin-β) superfamily. Linkage can be accomplished by direct binding to an importin-β or through mediation by adaptor proteins such as importin-α (karyopherin-α). The best-characterized nuclear targeting signal is the classical NLS recognized by an importin-α that, in turn, links the NLS-containing protein to importin-β1 (encoded for by Kpnb1) (Gorlich et al., 1995). Classical NLSs are usually identifiable in the primary sequence of a protein as a series of basic residues (Chook and Blobel, 2001; Hodel et al., 2001; Kalderon et al., 1984; Robbins et al., 1991). We previously showed that CRABP2 contains an unusual variation of a classical NLS. Instead of existing in the primary sequence, this NLS is assembled in the three-dimensional structure of the protein in response to RA binding. Consequently, holo-CRABP2 is recognized by importin-α1 (encoded for by Kpna2), enabling its ligand-induced nuclear translocation (Sessler and Noy, 2005). Other importin-β proteins, such as transportin 1/importin-β2 (encoded for by Tnpo1), directly bind cargoes that contain a broad range of ‘non-classical’ NLSs characterized by a C-terminal sequence comprised of Arg separated from a Pro-Tyr (PY) motif by two to five residues (Christie et al., 2015). An additional key component of the nuclear import machinery is the small GTPase Ran. The activation state of Ran is determined by Ran guanine nucleotide exchange factor (RanGEF), which loads the protein with GTP, and Ran GTPase-activating protein (RanGAP), which facilitates the hydrolysis of Ran-bound GTP to GDP (Bischoff et al., 1994; Bischoff and Ponstingl, 1991; Gorlich et al., 1996). GTP hydrolysis by Ran is also stimulated by the Ran-binding protein RanBP1 (Bischoff et al., 1995). Nuclear RanGEF and cytosolic RanGAP/RanBP1 thus form a RanGDP/GTP gradient that allows cargo-loaded importins to release their cargo in the nucleus and to dissociate from Ran in the cytosol (reviewed in Christie et al., 2015).
Here, we show that HuR regulates the expression of several components of the nuclear import machinery and thus controls the nuclear localization of both CRABP2, which contains a classical NLS, and the NFκB subunit p65, which contains both a classical NLS and a non-canonical NLS. We also show that, although importin-β1 and RanBP1 are necessary for nuclear import of CRABP2, they are dispensable for the cross-cytosol trafficking that allows the protein to accumulate at the nuclear membrane in response to RA.
RESULTS
HuR mediates the endoplasmic reticulum localization of apo-CRABP2
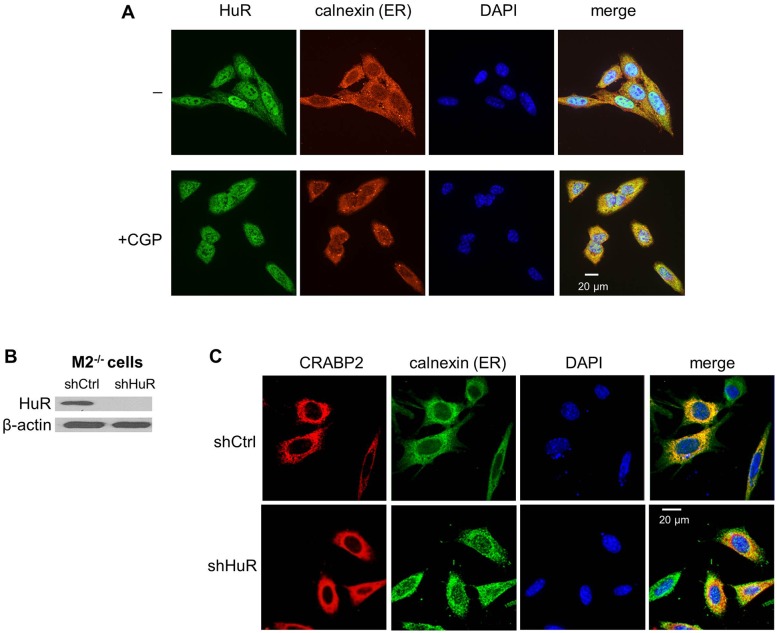
HuR shuttles between the nucleus and the cytosol but is predominantly nuclear in non-dividing cells (Fig. 1A). The cyto–nuclear shuttling of this protein is regulated by cyclin-dependent kinase 1 (Cdk1), and inhibition of the kinase results in retention of HuR in the cytosol (Kim et al., 2008). Indeed, a larger fraction of HuR was found in the extranuclear compartment of cells treated with the Cdk1 inhibitor CGP-74514A than in control cells (Fig. 2A). Cytosolic HuR extensively co-localized with the endoplasmic reticulum (ER) marker calnexin, both in the absence and presence of CGP-74514A (Fig. 1A), demonstrating that extranuclear HuR is associated with the ER.
Fig. 1.
HuR mediates the association of CRABP2 with endoplasmic reticulum. (A) M2−/− cells were untreated (−) or treated with CGP74514A (2 µM, 2 h) (+CGP). The ER marker calnexin and HuR were visualized by immunostaining. Cells were counterstained with 4′,6-diamidino-2-phenylindole (DAPI) to visualize nuclei. (B) Immunoblot demonstrating expression of HuR in M2−/− cells stably expressing shRNAs targeting luciferase (shCtrl) or HuR (shHuR). (C) M2−/− cells stably expressing shCtrl or shHuR were transiently transfected with a vector encoding Flag–CRABP2. Flag–CRABP2 (red) and ER marker calnexin (green) were immunostained and visualized using confocal microscopy. DAPI was used to visualize nuclei.
Fig. 2.
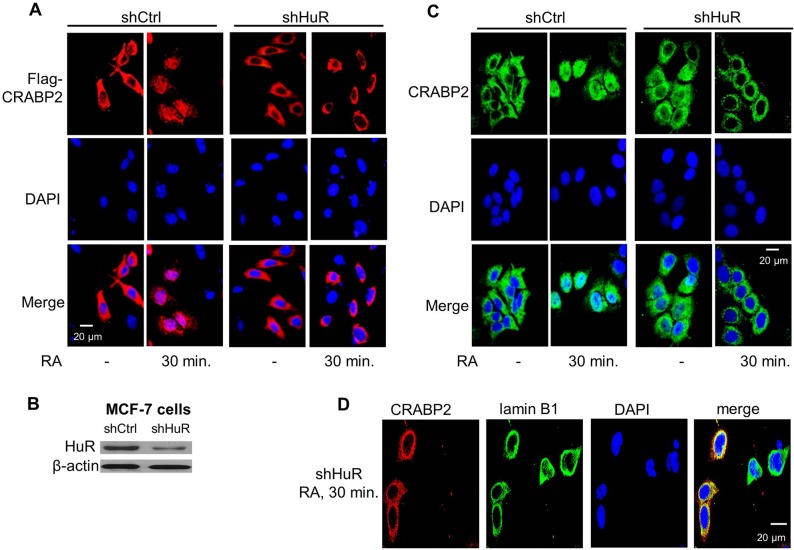
HuR is required for RA-induced nuclear translocation of CRABP2. (A) M2−/− cells stably expressing shCtrl or shHuR were cultured in delipidated medium and transfected with vector encoding Flag–CRABP2. Flag–CRABP2 was immunostained in untreated cells and in cells treated with RA for 30 min, and visualized using confocal microscopy. DAPI was used to visualize nuclei. (B) Immunoblot demonstrating expression of HuR in MCF-7 cells stably expressing shCtrl or shHuR. (C) MCF-7 cells expressing shCtrl or shHuR were cultured in delipidated medium and treated with vehicle or RA for 30 min. Endogenous CRABP2 was immunostained and visualized using confocal microscopy. DAPI was used to visualize nuclei. (D) M2−/− cells stably expressing shHuR were transfected with a vector encoding Flag–CRABP2 and treated with vehicle or RA. Flag–CRABP2 (red) and nuclear membrane marker lamin B (green) were immunostained and visualized using confocal microscopy. DAPI was used to visualize nuclei.
We previously reported that, in the absence of RA, CRABP2 localizes at the ER (Majumdar et al., 2011). Considering that CRABP2 is highly soluble, this localization is probably mediated by protein–protein interactions and not by interactions with the ER membrane. Together, the observations that HuR is associated with the ER and can bind CRABP2 (Vreeland et al., 2014b) suggest that the ER localization of CRABP2 is mediated by HuR. To examine this possibility, a cell line derived from tumors that arose in a CRABP2-null MMTV-neu mouse model of breast cancer (M2−/− cells) (Schug et al., 2008) was used. M2−/− lines that stably express short hairpin RNA (shRNA) targeting luciferase (shCtrl) or shRNA targeting HuR (shHuR) were generated (Fig. 1B). Cells were depleted of retinoids by culturing in charcoal-treated medium and were transfected with a vector encoding Flag-tagged CRABP2. Parental cells and cells with reduced expression of HuR were co-immunostained for Flag–CRABP2 and the ER marker calnexin (Fig. 1C). As expected, CRABP2 co-localized with calnexin in parental cells, reflecting its ER association. Notably, the co-localization was markedly reduced in cells with decreased expression of HuR (Fig. 1C), suggesting that CRABP2 is associated with the ER through interaction with HuR.
HuR is required for RA-induced nuclear import of CRABP2
Upon binding RA, CRABP2 dissociates from the ER and mobilizes to the nucleus, a process that is complete within 30 min of RA treatment (Majumdar et al., 2011; Sessler and Noy, 2005) (Fig. 2A). Strikingly, in M2−/− cells with stable reduced expression of HuR, RA induced a discernible shift in the subcellular localization of CRABP2, but this shift did not culminate in nuclear import. Instead, CRABP2 accumulated around the nucleus and did not enter this compartment (Fig. 2A). Similar responses were observed in M2−/− cells in which HuR expression was transiently reduced using two different shRNAs (Fig. S1), confirming that the effect reflects a reduction in HuR expression and not off-target effects of the shRNA. MCF-7 mammary carcinoma cells, which express a high level of CRABP2, were used to examine the involvement of HuR in the nuclear import of endogenous protein. MCF-7 cell lines that stably expressed either shCtrl or shHuR were generated (Fig. 2B). Cells were treated with RA, immunostained for CRABP2, and imaged. Endogenous CRABP2 in parental MCF-7 cells underwent nuclear translocation in response to RA, which was similar to its behavior in M2−/− cells that ectopically overexpress CRABP2. By contrast, in MCF-7 cells with reduced expression of HuR, treatment with RA resulted in accumulation of CRABP2 around the nucleus but, again, failed to affect complete nuclear import (Fig. 2C). Fluorescence microscopy showed that, in the presence of RA, CRABP2 displays extensive co-localization with the nuclear membrane marker lamin B1 (Fig. 2D). Hence, HuR is not necessary for mobilization of holo-CRABP2 to the nuclear membrane but is crucial for enabling translocation of the protein across this membrane into the nucleus.
HuR controls the nuclear import of CRABP2 in part by regulating the expression of importin-β1
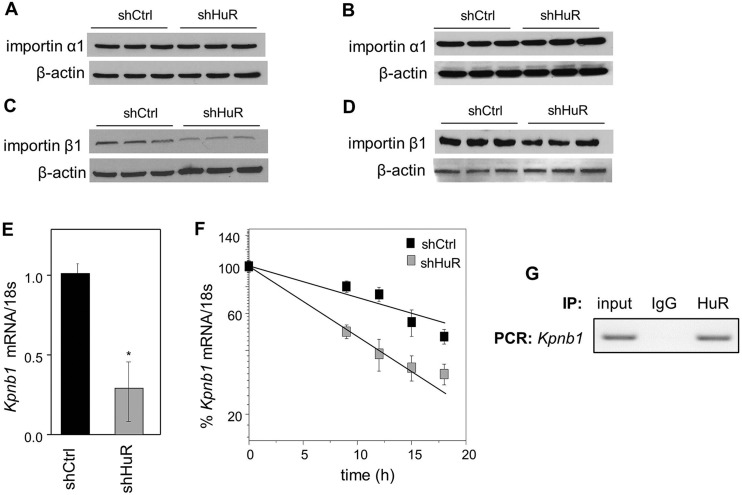
Binding of RA to CRABP2 induces structural rearrangements that activate an NLS recognized by importin-α1 (Sessler and Noy, 2005). The possibility that HuR regulates the nuclear import of CRABP2 by controlling importin-α1 expression was therefore examined. However, data showed that decreasing the expression of HuR did not affect the expression of importin-α1 in either M2−/− (Fig. 3A) or MCF-7 cells (Fig. 3B). Nuclear import of importin-α1 and its cargo is known to rely on an importin-β such as importin-β1 (Chook and Blobel, 2001; Gorlich et al., 1995; Hodel et al., 2001; Kalderon et al., 1984; Robbins et al., 1991). Examination of importin-β1 (encoded for by the Kpnb1 gene) expression revealed that decreasing the expression of HuR reduced the levels of both importin-β1 protein (Fig. 3C,D; Fig. S2A,B) and Kpnb1 mRNA (Fig. 3E). The possibility that HuR regulates importin-β1 expression by stabilizing its mRNA was then evaluated. M2−/− cells expressing shCtrl or shHuR were treated with transcription inhibitor actinomycin D (2.5 µg/ml), and the rate of degradation of Kpnb1 mRNA was monitored. The half-life of Kpnb1 mRNA was found to be 16.5±0.75 h and 10.2±1.12 h in shCtrl- and shHuR-expressing cells, respectively (Fig. 3F), indicating that this transcript is indeed stabilized by HuR. The observation that downregulation of HuR did not affect the stability of Gapdh mRNA (Fig. S2C) attested to the specificity of the response. Moreover, ribonucleoprotein immunoprecipitation (RIP) assays showed that Kpnb1 mRNA co-precipitates with HuR protein (Fig. 3G). The observations thus demonstrate that HuR modulates the level of importin-β1 by binding and stabilizing its RNA.
Fig. 3.
HuR stabilizes mRNA for importin-β1 (Kpnb1) but not for importin-α1. (A,B) Immunoblots of importin-α1 in M2−/− (A) or MCF-7 (B) cells stably expressing shCtrl or shHuR. (C,D) Immunoblots of importin-β1 in M2−/− (C) or MCF-7 (D) cells stably expressing shCtrl or shHuR. (E) Levels of mRNA for importin-β1 (Kpnb1) in M2−/− cells stably expressing shCtrl or shHuR, as measured by Q-PCR and normalized to 18s (mean±s.d.; n=3). *P<0.05, paired Student's t-test. (F) M2−/− cells stably expressing shCtrl or shHuR were treated with actinomycin D (2.5 µg/ml). Levels of Kpnb1 mRNA at various time points following treatment were measured by Q-PCR. Data (mean±s.e.m.; n=3) were normalized to corresponding values at time zero. (G) HuR was immunoprecipitated from M2−/− cells. Kpnb1 mRNA that co-precipitated with HuR was detected by semi-quantitative PCR.
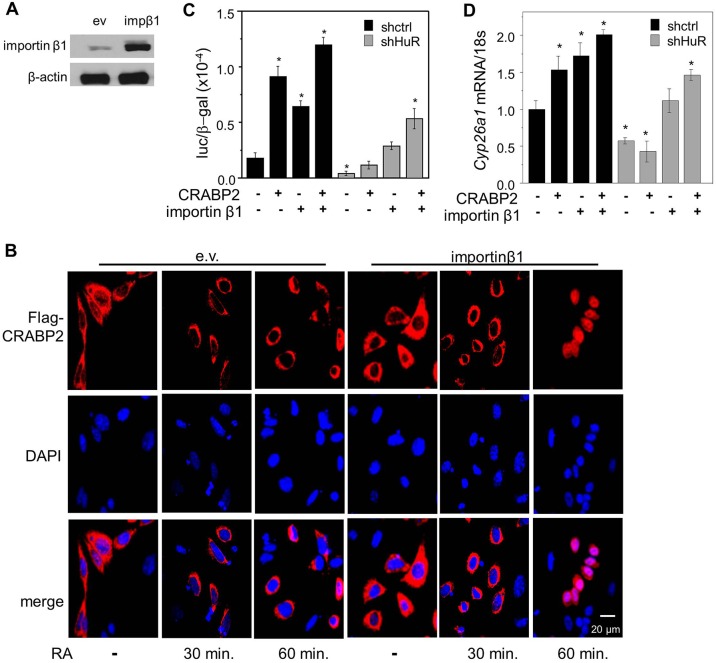
The importin-β1 inhibitor importazole (Soderholm et al., 2011) was used to evaluate whether HuR controls the nuclear import of CRABP2 by governing the expression of importin-β1. M2−/− cells ectopically expressing Flag–CRABP2, were treated with vehicle or importazole (30 µM, 1 h) prior to treatment with RA. Similar to the effect of downregulation of HuR, importazole efficiently blocked the RA-induced nuclear translocation of CRABP2 (Fig. 4). The effect of restoring the level of importin-β1 in shHuR-expressing cells was then examined. Ectopic overexpression of importin-β1 in these cells (Fig. 5A) enabled RA-induced nuclear translocation of CRABP2. However, completion of import was delayed from 30 min in parental cells (Fig. 2A) to 60 min (Fig. 5B). Hence, ectopically expressed importin-β1 rescued the nuclear import of CRABP2 but did not fully restore the kinetics of the process.
Fig. 4.
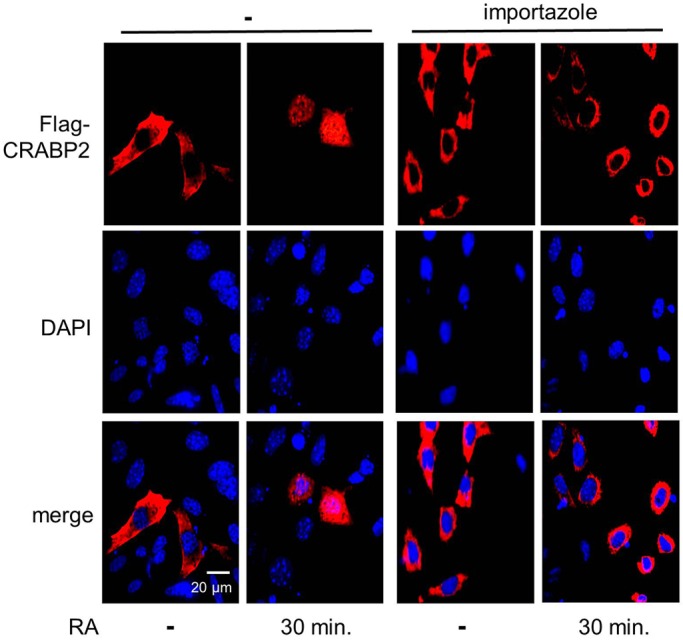
The importin-β inhibitor importazole blocks nuclear import of CRABP2. M2−/− cells were transfected with a vector encoding Flag–CRABP2. Cells were pretreated with importazole (30 µM) for 1 h prior to treatment with RA. Flag–CRABP2 (red) was immunostained and visualized using confocal microscopy. DAPI was used to visualize nuclei.
Fig. 5.
Importin-β1 partially rescues the nuclear import of holo-CRABP2 in cells with reduced expression of HuR. (A) Immunoblot demonstrating expression of importin-β1 in M2−/− cells stably expressing shHuR transfected with an empty vector (ev) or vector encoding importin-β1 (impβ1). (B) Cells were transfected with a vector encoding Flag–CRABP2 and treated with vehicle or RA for the denoted times. Flag–CRABP2 (red) was immunostained and visualized using confocal microscopy. DAPI was used to visualize nuclei. (C) Transactivation assays in denoted cells transfected with an empty vector or vector encoding CRABP2 or importin-β1 or both. Protein levels were assessed by immunoblot (Fig. S3A). Cells were treated with RA and the luciferase activity measured and normalized to β-galactosidase. (D) Expression of the RAR target gene Cyp26a1 in denoted cells transfected with empty vector, vector encoding CRABP2 or importin-β1 or both. Cells were treated with RA for 4 h and CYP26a expression measured by Q-PCR. Graphs show mean±s.e.m. (n=3). *P<0.05, paired Student's t-test, compared with cells expressing shCtrl and transfected with empty vector.
In the nucleus, CRABP2 delivers RA to the nuclear receptor RAR, thereby promoting its transcriptional activity. Transcriptional activation assays were carried out to examine whether downregulation of HuR interferes with the activation of RAR and whether importin-β1 can rescue this inhibition. M2−/− cells stably expressing shCtrl or shHuR were transfected with empty vector (ev) or vector encoding CRABP2 in the presence or absence of co-transfection of importin-β1 (Fig. S3A). Cells were also transfected with a luciferase reporter driven by an RAR response element (RARE) and a vector encoding β-galactosidase, which served as transfection control. Cells were treated with RA (1 μM, overnight) and luciferase activity measured and normalized to the activity of β-galactosidase (Fig. 5C; Fig. S3B). Ectopic expression of CRABP2 or importin-β1 alone enhanced the activation of RAR, and expression of both further promoted transactivation in parental cells. The transcriptional activity of RAR was significantly lower in shHuR-expressing cells and was partially restored upon ectopic expression of importin-β1 and CRABP2. The effects of the direct endogenous RAR target gene Cyp26a1 on the expression level were then examined. Cells were treated with RA (4 h) and then Cyp26a1 mRNA was measured by quantitative real-time PCR (Q-PCR) (Fig. 5D). Similar to the effects observed in transactivation assays, ectopic expression of CRABP2 or importin-β1 alone increased Cyp26a1 expression, and expression of both proteins further increased Cyp26a1 expression. Cyp26a1 expression was significantly lower in cells with reduced expression of HuR and, in contrast to control cells, ectopic expression of CRABP2 alone had no effect on Cyp26a1 mRNA in these cells. Overexpression of importin-β1 partially restored Cyp26a1 expression levels and recovered the effect of CRABP2. Taken together, the data indicate that importin-β1 is necessary for the nuclear import of CRABP2 and for transcriptional activation by RAR, and support the notion that HuR regulates these processes by controlling the stability of Kpnb1 mRNA and thus the level of expression of importin-β1 protein.
HuR regulates the expression of multiple components of the nuclear import machinery
The observation that importin-β1 only partially rescued the nuclear import of CRABP2 suggests that additional factors involved in nuclear import are regulated by HuR.
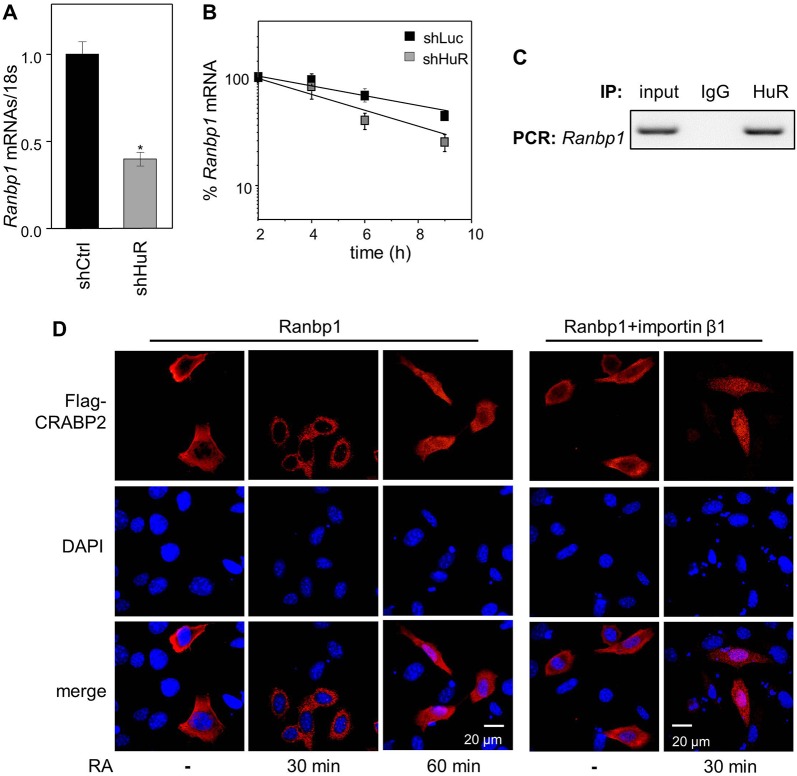
Decreasing HuR levels decreased the level of mRNA for Ranbp1 (Fig. 6A). The half-life of Ranbp1 mRNA was found to be 8.25±1.16 h and 4.88±1.10 h in cells expressing shCtrl and shHuR, respectively (Fig. 6B). Furthermore, RIP assays demonstrated that Ranbp1 mRNA co-precipitates with HuR protein (Fig. 6C). Hence, HuR regulates expression of RanBP1 by binding and stabilizing its mRNA. The ability of Ranbp1 to rescue the impaired nuclear import of CRABP2 brought about by downregulation of HuR was then examined. Ectopic expression of RANBP1 enabled RA-induced nuclear translocation of CRABP2, although completion of the process was delayed from 30 min following treatment with RA in parental cells (Fig. 2A) to 60 min (Fig. 6D). Interestingly, coexpression of RANBP1 together with importin-β1 restored the time course, allowing CRABP2 to complete its nuclear translocation within 30 min of RA treatment (Fig. 6D).
Fig. 6.
Stabilization of Ranbp1 mRNA by HuR contributes to its ability to control the nuclear import of CRABP2. (A) Levels of Ranbp1 mRNA in M2−/− cells stably expressing shCtrl or shHuR, as measured by Q-PCR and normalized to 18s. Levels of mRNAs in shHuR-expressing cells normalized to shCtrl-expressing cells are shown (mean±s.d.; n=3). *P<0.05, paired Student's t-test. (B) Cells were treated with actinomycin D (2.5 µg/ml) and then levels of Ranbp1 mRNA at various time points were measured by Q-PCR. Data (mean±s.e.m.; n=3) were normalized to corresponding values at time zero. (C) HuR was immunoprecipitated from M2−/− cells. Ranbp1 mRNA that co-precipitated with HuR was detected by semi-quantitative PCR. (D) shHuR-expressing M2−/− cells were transfected with a vector encoding Flag–CRABP2 in concert with a vector encoding RanBP1 or RanBP1 and importin-β1. Cells were treated with RA and Flag–CRABP2 (red), immunostained and visualized using confocal fluorescence microscopy. DAPI was used to visualize nuclei.
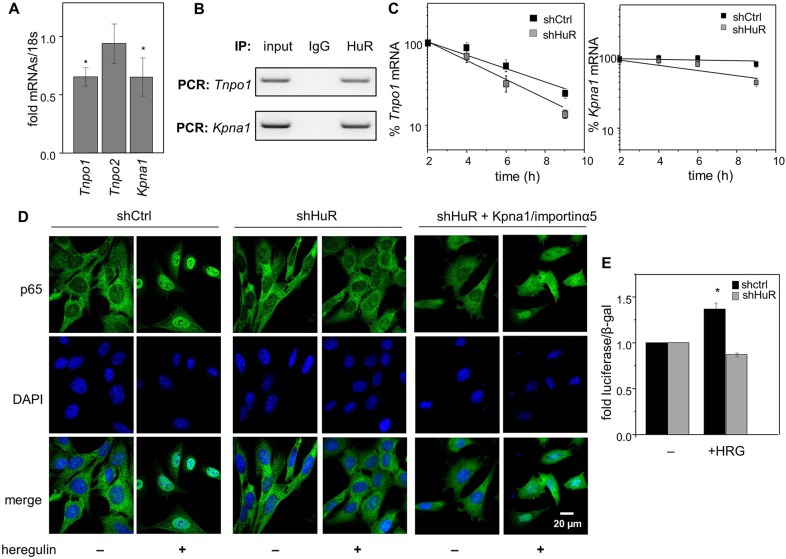
CRABP2 contains a ‘classical-like’ NLS and is mobilized to the nucleus by the importin-α1/importin-β1 pathway. However, nuclear import of proteins can also be directly mediated by an importin-β (Lee et al., 2006; Palmeri and Malim, 1999). For example, the importin-β transportin 1 (encoded for by Tnpo1) can directly bind and mobilize to nucleus cargo proteins containing non-canonical NLS (Fridell et al., 1997). Moreover, some nuclear proteins contain both classical and non-canonical NLSs. One such protein is the NFкB subunit p65, which contains a classical NLS recognized by importin-α5 (encoded for by Kpna1) (Fagerlund et al., 2008) and a non-classical NLS recognized by transportin 1 (Liang et al., 2013). Downregulation of HuR decreased the levels of both Kpna1 and Tnpo1 mRNAs (Fig. 7A), and RIP assays showed that both transcripts can be co-precipitated with HuR (Fig. 7B). The half-life of Tnpo1 mRNA decreased from 6.6±0.51 to 4.16±0.26 h in response to downregulation of HuR (Fig. 7C), whereas the half-life of Kpna1 mRNA was 25.67±4.51 and 9.63±1.89 h in parental and shHuR-expressing cells, respectively. Hence, HuR regulates the expression of both genes by stabilizing their transcripts.
Fig. 7.
Stabilization of Tnpo1 and Kpna1 mRNAs by HuR contributes to its ability to control the nuclear import of p65. (A) Levels of mRNA for Tnpo1/importin-β1, Tnpo2 and Kpna1/importin-α5 in M2−/− cells stably expressing shCtrl or shHuR, as measured by Q-PCR and normalized to 18s. Levels of mRNAs in shHuR-expressing cells normalized to cells expressing shCtrl are shown (mean±s.d.; n=3). *P<0.05, paired Student's t-test, compared to cells expressing shCtrl. (B) HuR was immunoprecipitated from M2−/− cells. Tnpo1 and Kpna1 mRNAs that co-precipitated with HuR were detected by semi-quantitative PCR. (C) Cells were treated with actinomycin D (2.5 µg/ml) and levels of Tnpo1 (left) and Kpna1 (right) mRNAs at various time points were measured by Q-PCR. Data were normalized to corresponding values at time zero (mean±s.e.m.; n=3). (D) M2−/− cells stably expressing shCtrl or shHuR and shHuR-expressing cells transfected with a vector encoding importin-α5 were treated with vehicle or heregulin-β1 (0.1 µg/ml) for 30 min. p65 was immunostained and visualized. DAPI was used to visualize nuclei. (E) Transactivation assays in denoted cells transfected with a vector harboring a luciferase reporter driven by 800 bp of the proximal promoter of FABP5. Cells were serum starved and treated with vehicle or heregulin-β1 (HRG; 50 ng/ml, overnight). Luciferase activity was measured and normalized to β-galactosidase (means±s.d.; n=3). *P<0.05, paired Student's t-test, compared to vehicle-treated cells.
NFкB subunit p65 translocates to the nucleus after activation by various signals, including the growth factor heregulin (Kannan-Thulasiraman et al., 2010). We showed that heregulin (0.1 µg/ml, 30 min) indeed induced nuclear translocation of p65 (Fig. 7D). Heregulin-induced nuclear translocation of p65 was completely abolished in cells with decreased expression of HuR, and overexpression of importin-α5 partially restored the import (Fig. 7D). In the nucleus, p65 regulates the transcription of various proteins, including the fatty acid-binding protein FABP5 (Kannan-Thulasiraman et al., 2010). The involvement of HuR in regulating the transcriptional activity of p65 was examined by transactivation assays utilizing a luciferase reporter driven by an 800-bp sequence of the proximal promoter of FABP5. The data showed that although heregulin-β1 activated the reporter in parental cells, decreasing the expression of HuR completely abolished the response (Fig. 7E).
DISCUSSION
The ability of HuR to stabilize target transcripts allows this RNA-binding protein to control biological processes ranging from cell growth and death to inflammatory responses (Abdelmohsen and Gorospe, 2010; Ghosh et al., 2009; Hinman and Lou, 2008). However, the complete spectrum of genes whose expression is regulated by HuR and are thus the basis for the broad range of cellular functions of this protein remains incompletely understood. We show here that HuR plays a central role in regulating nuclear protein import. The data reveal that HuR controls the expression of multiple components of the nuclear import machinery including RANBP1, importin-β1/KPNB1, transportin 1 and importin-α5/KPNA1. In agreement, it was previously reported that HuR binds to mRNA for both KPNB1 and TNPO1 (Mukherjee et al., 2011). At least in the cases of KPNA1, KPNB1 and TNPO1, HuR functions in this capacity by enhancing the stability of their transcripts. Consequently, downregulation of HuR severely impaired the nuclear import of CRABP2, a protein that harbors a ‘classical-like’ NLS (Fig. 2), and that of NFκB subunit p65, which reaches the nucleus using both classical and non-canonical NLSs (Fig. 7). In both cases, ectopic supplementation of factors that are lost in the absence of HuR either partially or almost completely ‘rescued’ nuclear translocation. The control of nuclear import by HuR must have important consequences for nuclear functions beyond the regulation of nuclear translocation of CRABP2, as addressed here. Indeed, decreasing the expression of HuR not only interfered with the ability of CRABP2 to promote the transcriptional activity of its cognate nuclear receptor RAR (Fig. 5C,D) but also significantly depressed the activity of the receptor in the absence of CRABP2 (Fig. 5C,D). Because HuR appears to be crucial for nuclear import mediated by different classes of NLSs, its depletion must interfere with the nuclear localization of many proteins, including those necessary for transcriptional activity in general. The role of HuR in regulating the nuclear import of such proteins (e.g. transcription factors, transcriptional co-regulators and components of the general transcription machinery) remains to be explored.
We previously reported that, in the absence of RA, CRABP2 is not uniformly distributed in the cytosol but, instead, localizes at the ER (Majumdar et al., 2011). We show here that this localization is disrupted in cells with reduced levels of HuR (Fig. 1B), indicating that CRABP2 associates with the ER through its interactions with HuR. Notably, although the nuclear translocation of CRABP2 was completely blocked in cells with reduced expression of HuR, RA induced a discernible shift in the protein's subcellular localization. RA binding not only triggered dissociation of CRABP2 from the ER, reflecting dissociation of the CRABP2–HuR complex (Fig. 1B), but also induced accumulation of the protein at the nuclear membrane (Fig. 2D). The observation that modulation of HuR expression did not affect the level of importin-α1 (Fig. 3A,B) raises the possibility that HuR mediates the movement of its cargo towards the nucleus, even in the absence of a partner importin-β. How CRABP2 travels to the nuclear surface under conditions that do not allow it to translocate into the nucleus remains to be clarified.
MATERIALS AND METHODS
Cells
We generated the M2−/− cell line from tumors that arose in MMTV-neu/CRABP2-null mice (Schug et al., 2008). MCF7 cells were recently purchased from ATCC. Cell lines were recently tested for contamination. Cells were maintained in Dulbecco's modified Eagle's medium (DMEM) containing 4.5 g/l glucose, 4.5 g/l L-glutamine, 10% fetal bovine serum (FBS; Life Technologies), 100 IU/ml penicillin and 100 µg/ml streptomycin. Cell transfections were carried out using PolyFect (Qiagen).
Reagents
Retinoic acid (RA) was purchased from Calbiochem. Actinomycin D was from Sigma-Aldrich. Antibodies against HuR (sc-5261; 1:1000 WB), β-actin (sc-57778; 1:1000 WB), karyopherin-β1 (importin-β1) (sc-1863; 1:1000 WB) and lamin B (sc-6217; 1:300 IF) were from Santa Cruz Biotechnology. Antibody against KPNA2 (importin-α1) (AB6036; 1:1000 WB) was from Abcam. Antibody against CRABP2 (1:1000 WB) was a gift from Cecile Rochette-Egly (IGBMC, Strasbourg, France).
Lentiviral shRNA production
pLKO.1 vectors harboring shRNAs for HuR (Elavl1, TRCN0000112085; Elavl1 TRCN0000112087; Elavl1 TRCN0000112088; Mouse) (ELAVL1 TRCN0000017275; Human) were from Open Biosystems; pLKO.1 vector harboring luciferase shRNA (SHC007) or non-targeting shRNA (SHC002) was from Sigma-Aldrich. Using pCMV packaging vector and pMD2.G envelope vector, lentiviruses were produced in HEK293T cells. Target cells were infected using standard protocols and selected with 5 µg/ml and 1 µg/ml puromycin for M2−/− cells and MCF-7 cells, respectively.
Real-time quantitative PCR
Q-PCR was performed using a StepOnePlus real-time PCR system with the following TaqMan probes: Elavl1, Mm00516012_m1; Kpna1, Mm00434700_m1; Kpnb1, Mm00434318_m1; Tnpo1, Mm00839059_g1; Tnpo2, Mm00520392_m1; Ranbp1, Mm00650862_m1; Cyp26a1, Mm00514486_m1; and 18s rRNA (4352930E, Applied Biosystems). Levels of mRNAs were normalized to 18s rRNA using the threshold cycle (ΔΔCT) method (Applied Biosystems technical bulletin no. 2).
Immunoblotting
Cells were lysed in RIPA buffer containing 150 mM NaCl, 10 mM Tris pH 7.2, 0.1% SDS, 1% Triton X-100, 1% deoxycholate and Halt protease inhibitor cocktail (Thermo Scientific). Protein concentration was determined using the Bradford protein assay. Cell lysates (50 µg protein) were resolved by SDS-PAGE and immunoblotted using appropriate antibodies.
Transactivation assays
Some 5×104 cells were plated in six-well plates in DMEM supplemented with 10% charcoal-treated FBS. Cells were transfected with vectors harboring a luciferase reporter driven by a RAR response element (RARE-Luc) and co-transfected with an expression vector for β-galactosidase, serving as transfection efficiency control. Cells were also co-transfected with pSG5 vector encoding CRABP2 or pCMV-Sport6 encoding importin-β1. At 24 h post-transfection, cells were treated with 1 μM RA or vehicle overnight. Luciferase activity was measured and corrected for transfection efficiency by the activity of β-galactosidase, as previously described (Budhu et al., 2001).
Confocal fluorescence microscopy
Cells, cultured in DMEM containing 10% charcoal-treated FBS, were transfected with pCMV-3Tag-1 vector encoding Flag–CRABP2. Cells were treated with RA or heregulin-β1 as described in the text, fixed with 4% paraformaldehyde in phosphate-buffered saline (PBS), and permeabilized with 0.2% Triton X-100. Endogenous CRABP2 in MCF-7 cells and Flag-tagged CRABP2 in M2−/− cells were visualized by immunostaining. Antibodies: CRABP2 (Santa Cruz sc-10065; 1:300 IF), Flag (Sigma-Aldrich F1804; 1:250 IF), calnexin (Sigma-Aldrich C4731; 1:300 IF), Alexa Fluor®-conjugated secondary antibodies (Invitrogen A11008, A11059, A11005; 1:1000 IF). Myc-DDK-pCMV-Entry vector encoding mKPNA1 and Myc-His A-pCDNA3.1(+) vector encoding mRanBP1 were used. Cells were imaged using a PerkinElmer UltraVox spinning disk confocal microscope. Analysis of images was carried out using Fiji ImageJ and Office PowerPoint.
Ribonucleoprotein immunoprecipitation
Ribonucleoprotein immunoprecipitations were performed as previously described (Keene and Tenenbaum, 2002; Vreeland et al., 2014b). Semi-quantitative PCR was performed using the following primers: Kpnb1: forward TGGCAAACCCAGGAAACAGT, reverse GCCCAACGTCTGCAAAACAT; Kpna1: forward CGAGCGTCTCCCTCTTCGTA, reverse CCTGGTGCCATCTCCATGTT; Ranbp1: forward GCAGATGAGTCCAACCACGA, reverse GCTTGACATCTCCAGTGCCT; Tnpo1: forward GCTCGTCCCTTACCTTGCTT, reverse TGTGGCAACAGACGAGAGAC.
Statistical analysis
Statistical significance of difference was analyzed by a paired Student's t-test. Analyses were performed using SPSS 16.0 software.
Acknowledgements
We thank Cecile Rochette-Egly (IGBMC, Strasbourg) for providing CRABP2 antibodies. The work utilized a confocal microscope that was acquired with National Institutes of Health shared instrument grant (SIG) [grant number 1S10OD019972-01].
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
W.Z. designed and carried out experiments and contributed to the writing of the manuscript, A.C.V. designed and carried out experiments and contributed to the writing, N.N. conceived and supervised the project, designed experiments and wrote the manuscript.
Funding
This work was supported by National Institutes of Health [grant numbers DK060684 and CA166955 to N.N.]. Deposited in PMC for release after 12 months.
Supplementary information
Supplementary information available online at http://jcs.biologists.org/lookup/doi/10.1242/jcs.192096.supplemental
References
- Abdelmohsen K. and Gorospe M. (2010). Posttranscriptional regulation of cancer traits by HuR. Wiley Interdiscip. Rev. RNA 1, 214-229. 10.1002/wrna.4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischoff F. R. and Ponstingl H. (1991). Catalysis of guanine nucleotide exchange on Ran by the mitotic regulator RCC1. Nature 354, 80-82. 10.1038/354080a0 [DOI] [PubMed] [Google Scholar]
- Bischoff F. R., Klebe C., Kretschmer J., Wittinghofer A. and Ponstingl H. (1994). RanGAP1 induces GTPase activity of nuclear Ras-related Ran. Proc. Natl. Acad. Sci. USA 91, 2587-2591. 10.1073/pnas.91.7.2587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischoff F. R., Krebber H., Smirnova E., Dong W. and Ponstingl H. (1995). Co-activation of RanGTPase and inhibition of GTP dissociation by Ran-GTP binding protein RanBP1. EMBO J. 14, 705-715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan C. M. and Steitz J. A. (2001). HuR and mRNA stability. Cell. Mol. Life Sci. 58, 266-277. 10.1007/PL00000854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budhu A. S. and Noy N. (2002). Direct channeling of retinoic acid between cellular retinoic acid-binding protein II and retinoic acid receptor sensitizes mammary carcinoma cells to retinoic acid-induced growth arrest. Mol. Cell. Biol. 22, 2632-2641. 10.1128/MCB.22.8.2632-2641.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budhu A., Gillilan R. and Noy N. (2001). Localization of the RAR interaction domain of cellular retinoic acid binding protein-II. J. Mol. Biol. 305, 939-949. 10.1006/jmbi.2000.4340 [DOI] [PubMed] [Google Scholar]
- Chen C.-Y. A., Xu N. and Shyu A.-B. (2002). Highly selective actions of HuR in antagonizing AU-rich element-mediated mRNA destabilization. Mol. Cell. Biol. 22, 7268-7278. 10.1128/MCB.22.20.7268-7278.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chook Y. M. and Blobel G. (2001). Karyopherins and nuclear import. Curr. Opin. Struct. Biol. 11, 703-715. 10.1016/S0959-440X(01)00264-0 [DOI] [PubMed] [Google Scholar]
- Christie M., Chang C. W., Rona G., Smith K. M., Stewart A. G., Takeda A. A., Fontes M. R., Stewart M., Vertessy B. G., Forwood J. K.. et al. (2015). Structural biology and regulation of protein import into the nucleus. J. Mol. Biol. 428, 2060-2090. 10.1016/j.jmb.2015.10.023 [DOI] [PubMed] [Google Scholar]
- Donato L. J. and Noy N. (2005). Suppression of mammary carcinoma growth by retinoic acid: proapoptotic genes are targets for retinoic acid receptor and cellular retinoic acid-binding protein II signaling. Cancer Res. 65, 8193-8199. 10.1158/0008-5472.CAN-05-1177 [DOI] [PubMed] [Google Scholar]
- Donato L. J., Suh J. H. and Noy N. (2007). Suppression of mammary carcinoma cell growth by retinoic acid: the cell cycle control gene Btg2 is a direct target for retinoic acid receptor signaling. Cancer Res. 67, 609-615. 10.1158/0008-5472.CAN-06-0989 [DOI] [PubMed] [Google Scholar]
- Dong D., Ruuska S. E., Levinthal D. J. and Noy N. (1999). Distinct roles for cellular retinoic acid-binding proteins I and II in regulating signaling by retinoic acid. J. Biol. Chem. 274, 23695-23698. 10.1074/jbc.274.34.23695 [DOI] [PubMed] [Google Scholar]
- Fagerlund R., Melen K., Cao X. and Julkunen I. (2008). NF-kappaB p52, RelB and c-Rel are transported into the nucleus via a subset of importin alpha molecules. Cell. Signal. 20, 1442-1451. 10.1016/j.cellsig.2008.03.012 [DOI] [PubMed] [Google Scholar]
- Fridell R. A., Truant R., Thorne L., Benson R. E. and Cullen B. R. (1997). Nuclear import of hnRNP A1 is mediated by a novel cellular cofactor related to karyopherin-beta. J. Cell Sci. 110, 1325-1331. [DOI] [PubMed] [Google Scholar]
- Ghosh M., Aguila H. L., Michaud J., Ai Y., Wu M.-T., Hemmes A., Ristimaki A., Guo C., Furneaux H. and Hla T. (2009). Essential role of the RNA-binding protein HuR in progenitor cell survival in mice. J. Clin. Invest. 119, 3530-3543. 10.1172/JCI38263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorlich D., Kostka S., Kraft R., Dingwall C., Laskey R. A., Hartmann E. and Prehn S. (1995). Two different subunits of importin cooperate to recognize nuclear localization signals and bind them to the nuclear envelope. Curr. Biol. 5, 383-392. 10.1016/S0960-9822(95)00079-0 [DOI] [PubMed] [Google Scholar]
- Gorlich D., Pante N., Kutay U., Aebi U. and Bischoff F. R. (1996). Identification of different roles for RanGDP and RanGTP in nuclear protein import. EMBO J. 15, 5584-5594. [PMC free article] [PubMed] [Google Scholar]
- Gupta S., Pramanik D., Mukherjee R., Campbell N. R., Elumalai S., de Wilde R. F., Hong S.-M., Goggins M. G., De Jesus-Acosta A., Laheru D.. et al. (2012). Molecular determinants of retinoic acid sensitivity in pancreatic cancer. Clin. Cancer Res. 18, 280-289. 10.1158/1078-0432.CCR-11-2165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinman M. N. and Lou H. (2008). Diverse molecular functions of Hu proteins. Cell. Mol. Life Sci. 65, 3168-3181. 10.1007/s00018-008-8252-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodel M. R., Corbett A. H. and Hodel A. E. (2001). Dissection of a nuclear localization signal. J. Biol. Chem. 276, 1317-1325. 10.1074/jbc.M008522200 [DOI] [PubMed] [Google Scholar]
- Kalderon D., Roberts B. L., Richardson W. D. and Smith A. E. (1984). A short amino acid sequence able to specify nuclear location. Cell 39, 499-509. 10.1016/0092-8674(84)90457-4 [DOI] [PubMed] [Google Scholar]
- Kannan-Thulasiraman P., Seachrist D. D., Mahabeleshwar G. H., Jain M. K. and Noy N. (2010). Fatty acid-binding protein 5 and PPARbeta/delta are critical mediators of epidermal growth factor receptor-induced carcinoma cell growth. J. Biol. Chem. 285, 19106-19115. 10.1074/jbc.M109.099770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keene J. D. and Tenenbaum S. A. (2002). Eukaryotic mRNPs may represent posttranscriptional operons. Mol. Cell 9, 1161-1167. 10.1016/S1097-2765(02)00559-2 [DOI] [PubMed] [Google Scholar]
- Kim H. H., Abdelmohsen K., Lal A., Pullmann R. Jr, Yang X., Galban S., Srikantan S., Martindale J. L., Blethrow J., Shokat K. M. et al. (2008). Nuclear HuR accumulation through phosphorylation by Cdk1. Genes Dev. 22, 1804-1815. 10.1101/gad.1645808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebedeva S., Jens M., Theil K., Schwanhausser B., Selbach M., Landthaler M. and Rajewsky N. (2011). Transcriptome-wide analysis of regulatory interactions of the RNA-binding protein HuR. Mol. Cell 43, 340-352. 10.1016/j.molcel.2011.06.008 [DOI] [PubMed] [Google Scholar]
- Lee B. J., Cansizoglu A. E., Suel K. E., Louis T. H., Zhang Z. and Chook Y. M. (2006). Rules for nuclear localization sequence recognition by karyopherin beta 2. Cell 126, 543-558. 10.1016/j.cell.2006.05.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang P., Zhang H., Wang G., Li S., Cong S., Luo Y. and Zhang B. (2013). KPNB1, XPO7 and IPO8 mediate the translocation of NF-kappaB/p65 into the nucleus. Traffic 14, 1132-1143. 10.1111/tra.12097 [DOI] [PubMed] [Google Scholar]
- Lopez de Silanes I., Zhan M., Lal A., Yang X. and Gorospe M. (2004). Identification of a target RNA motif for RNA-binding protein HuR. Proc. Natl. Acad. Sci. USA 101, 2987-2992. 10.1073/pnas.0306453101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majumdar A., Petrescu A. D., Xiong Y. and Noy N. (2011). Nuclear translocation of cellular retinoic acid-binding protein II is regulated by retinoic acid-controlled SUMOylation. J. Biol. Chem. 286, 42749-42757. 10.1074/jbc.M111.293464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manor D., Shmidt E. N., Budhu A., Flesken-Nikitin A., Zgola M., Page R., Nikitin A. Y. and Noy N. (2003). Mammary carcinoma suppression by cellular retinoic acid binding protein-II. Cancer Res. 63, 4426-4433. [PubMed] [Google Scholar]
- Mukherjee N., Corcoran D. L., Nusbaum J. D., Reid D. W., Georgiev S., Hafner M., Ascano M. Jr, Tuschl T., Ohler U. and Keene J. D. (2011). Integrative regulatory mapping indicates that the RNA-binding protein HuR couples pre-mRNA processing and mRNA stability. Mol. Cell 43, 327-339. 10.1016/j.molcel.2011.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myer V. E., Fan X. C. and Steitz J. A. (1997). Identification of HuR as a protein implicated in AUUUA-mediated mRNA decay. EMBO J. 16, 2130-2139. 10.1093/emboj/16.8.2130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmeri D. and Malim M. H. (1999). Importin beta can mediate the nuclear import of an arginine-rich nuclear localization signal in the absence of importin alpha. Mol. Cell. Biol. 19, 1218-1225. 10.1128/MCB.19.2.1218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins J., Dilwortht S. M., Laskey R. A. and Dingwall C. (1991). Two interdependent basic domains in nucleoplasmin nuclear targeting sequence: identification of a class of bipartite nuclear targeting sequence. Cell 64, 615-623. 10.1016/0092-8674(91)90245-T [DOI] [PubMed] [Google Scholar]
- Schug T. T., Berry D. C., Shaw N. S., Travis S. N. and Noy N. (2007). Opposing effects of retinoic acid on cell growth result from alternate activation of two different nuclear receptors. Cell 129, 723-733. 10.1016/j.cell.2007.02.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schug T. T., Berry D. C., Toshkov I. A., Cheng L., Nikitin A. Y. and Noy N. (2008). Overcoming retinoic acid-resistance of mammary carcinomas by diverting retinoic acid from PPARbeta/delta to RAR. Proc. Natl. Acad. Sci. USA 105, 7546-7551. 10.1073/pnas.0709981105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessler R. J. and Noy N. (2005). A ligand-activated nuclear localization signal in cellular retinoic acid binding protein-II. Mol. Cell 18, 343-353. 10.1016/j.molcel.2005.03.026 [DOI] [PubMed] [Google Scholar]
- Soderholm J. F., Bird S. L., Kalab P., Sampathkumar Y., Hasegawa K., Uehara-Bingen M., Weis K. and Heald R. (2011). Importazole, a small molecule inhibitor of the transport receptor importin-beta. ACS Chem. Biol. 6, 700-708. 10.1021/cb2000296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vreeland A. C., Levi L., Zhang W., Berry D. C. and Noy N. (2014a). Cellular retinoic acid-binding protein 2 inhibits tumor growth by two distinct mechanisms. J. Biol. Chem. 289, 34065-34073. 10.1074/jbc.M114.604041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vreeland A. C., Yu S., Levi L., de Barros Rossetto D. and Noy N. (2014b). Transcript stabilization by the RNA-binding protein HuR is regulated by cellular retinoic acid-binding protein 2. Mol. Cell. Biol. 34, 2135-2146. 10.1128/MCB.00281-14 [DOI] [PMC free article] [PubMed] [Google Scholar]