Abstract
The Lim domain-binding proteins are key co-factor proteins that assemble with LIM domains of the LMO/LIM-HD family to form functional complexes that regulate cell proliferation and differentiation. Using conditional mutagenesis and comparative phenotypic analysis, we analyze the function of Ldb1 and Ldb2 in mouse retinal development, and demonstrate overlapping and specific functions of both proteins. Ldb1 interacts with Lhx2 in the embryonic retina and both Ldb1 and Ldb2 play a key role in maintaining the pool of retinal progenitor cells. This is accomplished by controlling the expression of the Vsx2 and Rax, and components of the Notch and Hedgehog signaling pathways. Furthermore, the Ldb1/Ldb2-mediated complex is essential for generation of early-born photoreceptors through the regulation of Rax and Crx. Finally, we demonstrate functional redundancy between Ldb1 and Ldb2. Ldb1 can fully compensate the loss of Ldb2 during all phases of retinal development, whereas Ldb2 alone is sufficient to sustain activity of Lhx2 in both early- and late-stage RPCs and in Müller glia. By contrast, loss of Ldb1 disrupts activity of the LIM domain factors in neuronal precursors. An intricate regulatory network exists that is mediated by Ldb1 and Ldb2, and promotes RPC proliferation and multipotency; it also controls specification of mammalian retina cells.
KEY WORDS: Isl1, Ldb1, Lhx2, Retinogenesis
Summary: Ldb1 and Ldb2 have both overlapping and specific functions in mouse retinal development, specifically in maintaining the pool of retinal progenitors and for the generation of early-born photoreceptors.
INTRODUCTION
LIM domain proteins regulate cell proliferation and cell fate in many regions of the CNS. These proteins function within complexes that assemble together with LIM domain-binding (Ldb) proteins (Matthews et al., 2008). The composition of the Ldb-LIM complex is crucial for determining the stage-specific and cell type-specific gene expression profile in multiple developmental contexts (Matthews and Visvader, 2003; Bhati et al., 2008; Love et al., 2014). In this study, we seek to provide insight into the role of Ldb proteins in the control of mouse retinal development.
The retina is a highly specialized extension of the CNS that comprises six major classes of neurons as well as Müller glia cells, organized in three layers: a photoreceptor (PR) layer consisting of cone and rod PR cells; an inner nuclear layer (INL) comprising horizontal, bipolar and amacrine interneurons and Müller glia; and a ganglion cell layer (GCL) consisting of ganglion cells, which extend axons to the brain.
The development of the retina, which is initiated during early stages of neurulation, begins with the expression of a combination of proteins known as eye-field transcription factors (Zuber et al., 2003; Zuber, 2010). Among these are the homeodomain transcription factor Rax, the homeodomain and paired domain protein Pax6, and the LIM-homeodomain protein Lhx2 (Zuber, 2010). These factors are expressed in the lateral protrusions from the ventral forebrain, which give rise to the optic vesicle (OV) that becomes the optic cup (OC) (Oliver et al., 1995; Mathers et al., 1997; Porter et al., 1997; Jean et al., 1999). The neuroepithelium of the OC gives rise to several ocular cell types: the most distal regions form the ciliary body (CB) and iris; the outer layer of the OC differentiates into the retinal pigmented epithelium; and the inner layer of the OC contains retinal progenitor cells (RPCs) that will differentiate into the various retinal cell types (Shaham et al., 2012).
The temporal order of the processes involved in retinal histogenesis is conserved among vertebrate species. Retinal ganglion cells, cone PR cells, horizontal and amacrine cells are born first, followed by bipolar neurons and Müller glial cells, whereas the rod PR cells are generated throughout retinogenesis (Marquardt, 2003). The generation of the correct numbers of the various retinal cell types is dependent on the availability of a continuous supply of progenitor cells. Therefore, in parallel to cell cycle exit and differentiation, the RPCs continue to proliferate well into the postnatal period.
The retina is known to harbor several proteins from the LIM domain family – a family that includes proteins with a LIM homeodomain (LIM-HD/Lhx) in addition to proteins consisting only of a LIM domain (LMO). Lhx2 is a LIM-HD family member that is expressed in proliferating RPCs (Balasubramanian et al., 2014), and is required for the morphogenesis and patterning of the OV and OC, as well as for maintaining the pool of RPCs (Porter et al., 1997; Yun et al., 2009; Hagglund et al., 2011; Gordon et al., 2013; Roy et al., 2013). Lhx2 expression is also maintained in mature Müller glia, where it controls stress responses (de Melo et al., 2012).
Unlike proliferating RPCs, post-mitotic neuronal precursors express multiple different types of LIM-HD family proteins. The LIM-HD protein Isl1 was shown to be crucial for the generation of ganglion cells (Wu et al., 2015). Isl1 is also expressed in precursors of bipolar and cholinergic amacrine cells, and Isl1 loss results in a reduction in bipolar cell number and loss of cholinergic amacrine cells (Elshatory et al., 2007; Mu et al., 2008; Pan et al., 2008). The LIM-class homeobox gene Lim1 (Lhx1) is required for the laminar positioning of horizontal cells (Poche et al., 2007). Finally, the LMO transcription factor Lmo4 has an important role in the generation of GABAergic amacrine and OFF-bipolar cells (Duquette et al., 2010).
The different types of LIM proteins often function in complexes, together with other transcription factors (Bach et al., 1997; Lee and Pfaff, 2003). The distinct composition of LIM proteins within a complex is termed the ‘LIM code’, as each unique combination dictates the expression of a specific set of target genes (Shirasaki and Pfaff, 2002). As noted above, Ldb proteins are key co-factor proteins that assemble with the LIM domains of the LMO/LIM-HD family and stabilize these protein complexes. They are also essential for transcriptional regulation (Matthews et al., 2008).
In mice, there are two Ldb proteins: Ldb1 and Ldb2. Ldb1 plays key roles during development and can compensate for most of the activities of Ldb2 (Mukhopadhyay et al., 2003; Narkis et al., 2012; Leone et al., 2016). Ldb2-null mice are fertile and phenotypically normal, whereas Ldb1-null embryos die at early stages of neurogenesis (E9.5-10.0), owing to multiple developmental aberrations, including the truncation of head structures (Mukhopadhyay et al., 2003). Ldb1 has been shown to fulfill several important functions: it has an essential role in the development of the CNS and is crucial for specification and maintenance of hematopoietic stem cells (Li et al., 2011). Moreover, Ldb1, together with Isl1, is a key early regulator of mammalian limb and cardiac development (Narkis et al., 2012; Caputo et al., 2015).
Notably, there are currently no reports on the function of Ldb1 and Ldb2 in mammalian retinal development, or on the manner in which they compensate for the activity of one another during this process. Herein, through comparative analysis of conditional mutations of both Ldb1 and Ldb2 (Ldb1/Ldb2), or of Ldb1, Ldb2 or Lhx2 alone in RPCs, we determine the role of Ldb1 and unravel the selective compensatory potential of Ldb2 during retinal development in mammals.
RESULTS
Ldb1 is expressed in retinal progenitors and is maintained in INL and GCL cell types
We characterized Ldb1 expression in the developing and differentiated retina as it is considered to play fundamental roles and to compensate for Ldb2 loss in various developmental processes (Mukhopadhyay et al., 2003; Narkis et al., 2012). Indeed, Ldb1 seemed to be sufficient to support retinal development, as we did not detect any alterations in OC morphology, differentiation to retinal cell types and lamination in the Ldb1loxP/loxP;Ldb2−/− embryos (Fig. 1A-E) when compared with wild-type control, testing several antibodies to cell-specific markers at indicated stages (Fig. S1). Moreover, despite Ldb2 loss, the transcript levels of Ldb1 in embryonic eye and adult retina remain unchanged (mean fold change of 1.04, P=0.76; and 1.16, P=0.37, respectively), excluding compensatory transcriptional upregulation of Ldb1 following loss of Ldb2 (Fig. S1E).
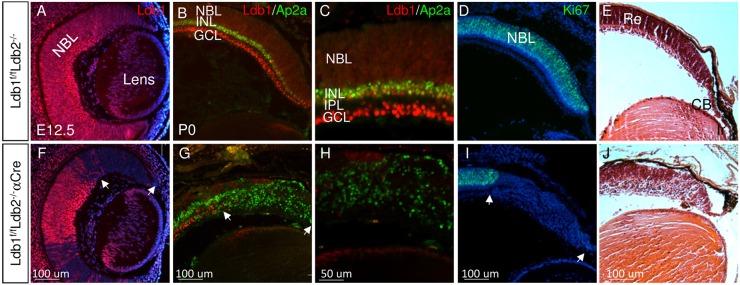
Fig. 1.
Ldb1 and Ldb2 are required for maintaining proliferation and multipotency of RPCs. Gene expression and tissue morphology were monitored in control (A-E) and in Ldb1loxP/loxP;Ldb2−/−;α-Cre (F-J) eyes, determined by immunofluorescence analyses (A-D,F-I) and Hematoxylin and Eosin staining (E,J) for Ldb1 (red in A-C,F-H), Ap2a (green in B,C,G,H) and Ki67 (green in D,I) at E12.5 (A,F) and P0 (B-E,G-J). Counterstaining was with DAPI (blue, A,D,F,I). C,H are higher magnifications of staining for Ldb1 and Ap2a. White arrows in F,G,I mark the mutation area. CB, ciliary body; GCL, ganglion cell layer; INL, inner nuclear layer; IPL, inner plexiform layer; Ir, iris; NBL, neuroblastic layer; Re, retina. Scale bars: 100 μm in A,B,D-G,I,J; 50 μm in C,H.
During early stages of retinogenesis [embryonic day (E) 12.5] Ldb1 protein was detected in the RPCs and ganglion cell precursors (Fig. 1A, Fig. S1A). At late stages of retinogenesis, postnatal day (P)1, the expression of Ldb1 was maintained in the RPCs and in the ganglion cell layer, and was also detected in INL cells where it partially overlapped with the expression of the amacrine and horizontal cell marker Ap2a (Fig. 1B,C; Fig. S1B) (Bassett et al., 2012). At P14, when retinal differentiation was complete, expression of Ldb1 was not detected in the PR cells but was maintained in the INL and GCL cell types (Fig. S1), including ganglion cells co-labeled with the transcription factor Pou4f2 (Fig. S1F) (Gan et al., 1996), horizontal cells co-labeled with the calcium-binding protein calbindin (Fig. S1G) (Wassle et al., 1998), amacrine cells co-labeled with the transcription factors Pax6 and Isl1 (Fig. S1H,I,I′) (Alexiades and Cepko, 1997), and Müller glia co-labeled with Lhx2 and glutamine synthase (GS) (de Melo et al., 2012) (Fig. S1J,K). Low expression of Ldb1 was detected in bipolar cells co-labeled with Vsx2 (Rowan and Cepko, 2004) and in Isl1-positive bipolar cells in the apical/outer INL (Elshatory et al., 2007) (Fig. S1L,I, and enlargement in I′,I″). These observations indicate that, during retinogenesis, Ldb1 is expressed in RPCs and post-mitotic precursors in the GCL and INL. In the mature retina, Ldb1 expression is maintained in the INL and GCL cells, but is not detectable in PR cells.
Ldb1 and Ldb2 are required for maintaining RPC proliferation, and are necessary for the generation of most retinal cell types
In order to study the roles of Ldb1 in retinal development, we used the α-Cre transgenic mouse line to induce a mutation of Ldb1 in Ldb1loxP/loxP;Ldb2−/−;α-Cre mice, in the distal RPCs, and in progenitors of iris and CB from E10.5 onwards (Marquardt et al., 2001). The conditional mutation of Ldb1 was induced on a background of Ldb2 deficiency to preclude any possible compensation for Ldb1 loss by Ldb2 (Tzchori et al., 2009; Narkis et al., 2012). Ldb1loxP/loxP;Ldb2−/− embryos exhibited a normal retinal phenotype, and thus were used as controls (Fig. 1, Fig. S1).
At E12.5, immunofluorescence analysis indicated a loss of Ldb1 protein expression in the distal OC of Ldb1loxP/loxP;Ldb2−/−;α-Cre embryos (Fig. 1F). Although we did not detect altered morphology in the Ldb1loxP/loxP;Ldb2−/−;α-Cre OC at E12.5, by E18.5 the NBL was reduced, while the relative fraction of postmitotic precursor cells was elevated when compared with control, based on number of Ki67- and VC1.1-expressing cells (Fig. S2A-H). At P1, in the control retina, the separation between the GCL and INL is evident with the establishment of the inner plexiform layer (IPL). By contrast, in the Ldb1 and Ldb2 (Ldb1/Ldb2)--deficient retina, most cells expressed the pan-amacrine/horizontal precursor marker Ap2a (Tfap2a) (Fig. 1G,H), while no proliferating RPCs were observed [Ki67+ (Mki67); Fig. 1I].
In addition, histological analysis indicated that the non-neuronal anterior structures of the eye, i.e. the CB and iris (Fig. 1E), fail to form in the Ldb1/Ldb2-deficient OC (Fig. 1J). These findings reveal that Ldb1 plays essential roles in the maintenance of the retinal progenitor pool, in the generation and differentiation of most retinal cell types, and in the development of the CB and iris.
Loss of function of Ldb1/Ldb2 results in depletion of the progenitor pool due to premature cell cycle exit and onset of differentiation
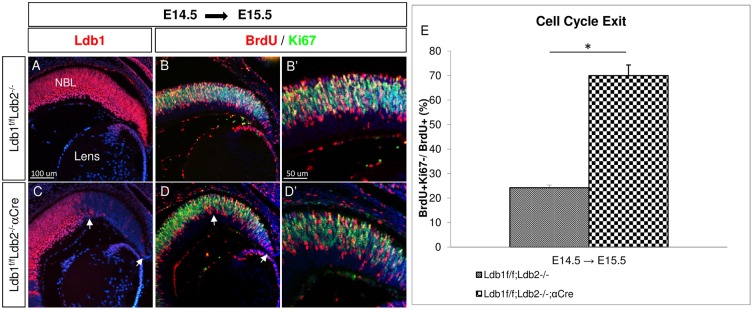
Depletion of RPCs could reflect premature neurogenesis in the Ldb1loxP/loxP;Ldb2−/−;α-Cre. We therefore used BrdU pulse-chase analysis to compare the cell cycle exit index between control and Ldb1/Ldb2 mutant cells (Fig. 2A-D). BrdU, which labels cells in S-phase, was injected into pregnant dams at E14.5. Embryos were sacrificed at E15.5, and the proportion of BrdU+/Ki67− from total BrdU+ cells was calculated in order to determine the cell cycle exit index (Fig. 2E). In the Ldb1/Ldb2 mutant retina, the proportion of cells exiting the cell cycle at E14.5 (70%) was significantly higher than that in the controls (24%) (P<0.001; Fig. 2E). In agreement with this observation, p57kip2 (Cdkn1c), which was detected in only few cells in the control retina, was detected in many Ldb1/Ldb2-deficient cells (Fig. S2I-P).
Fig. 2.
The loss of Ldb1 results in premature cell cycle exit. (A-D′) BrdU was administered at E14.5 and was followed by analysis at E15.5 in control OCs (A-B′) and in Ldb1loxP/loxP;Ldb2−/−;α-Cre OCs (C-D′). Immunofluorescence analyses of Ldb1 (A,C, red), BrdU and Ki67 (red and green; B,B′,D,D′). The Ldb1/Ldb2 mutated area is marked with white arrows. (E) The percentage of BrdU+/Ki67− from total BrdU+ cells was calculated in the control and Ldb1loxP/loxP;Ldb2−/−;α-Cre distal retina at E15.5, representing cell cycle exit. Data are mean±s.d., n=3, *P<0.001 calculated using a two-tailed t-test. NBL, neuroblastic layer. Scale bars: in A, 100 μm in A,B,C,D; in B′, 50 μm in B′,D′.
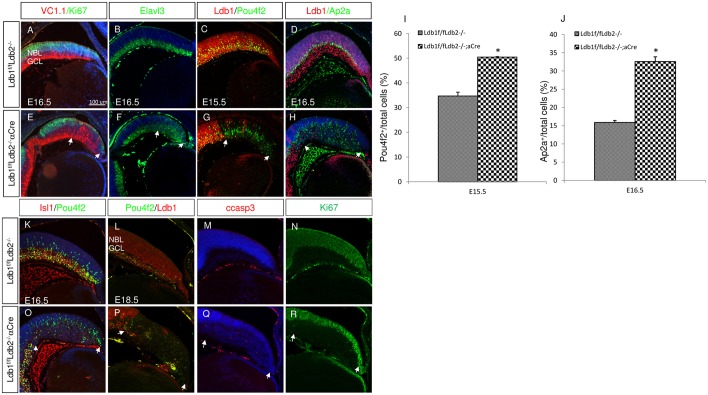
The high rate of cell cycle exit in the Ldb1/Ldb2-deficient RPCs resulted in depletion of Ki67 at E16.5, and occurred in conjunction with accumulation of ganglion and amacrine precursors, reflected in the expression levels of Vc1.1, Elavl3, Pou4f2 and Ap2a (Fig. 3A-H). Quantitative analyses revealed that the number of cells expressing Pou4f2 at E15.5 was significantly higher in the Ldb1/Ldb2-deficient OC (50.4%) than in the control peripheral OC (34.7%; P<0.002, Fig. 3I). Likewise, at E16.5 the number of cells expressing Ap2a was significantly higher in the mutant peripheral OC (32.6% in the Ldb1loxP/loxP;Ldb2−/−;α-Cre OC when compared with 15.8% in controls; P<0.001, Fig. 3J). In sum, the loss of Ldb1/Ldb2 from RPCs resulted in depletion of the progenitor pool due to premature cell cycle exit, followed by generation of the early-born precursors of the ganglion and amacrine/horizontal cell lineages.
Fig. 3.
Ldb proteins are required first for preventing premature differentiation into ganglion and amacrine lineages and later for ganglion cell survival. (A-H) In control (A-D) and Ldb1loxP/loxP;Ldb2−/−;α-Cre (E-H) retinas, immunofluorescence analyses show the expression of Vc1.1 and Ki67 (red and green; A,E), Elavl3 (B,F), Ldb1 and Pou4f2 (red and green; C,G), and Ldb1 and Ap2a (red and green; D,H). Arrows indicate mutated area. (I) The percentage of Pou4f2+ cells at E15.5 in the control retina (34.7±1.55) and in Ldb1/Ldb2 mutant retina (50.4±0.21). Data are mean±s.d., n=3, *P<0.002 calculated using a two-tailed t-test. (J) The percentage of Ap2a+ cells at E16.5 in the control retina (15.8±0.56%) and in Ldb1/Ldb2 mutant retina (32.6±1.3%). Data are mean±s.d., n=3, *P<0.001 calculated using a two-tailed t-test. (K-R) In control (K-N) and Ldb1loxP/loxP;Ldb2−/−;α-Cre (O-R) retinas, immunofluorescence analyses show the expression of Isl1 and Pou4f2 at E16.5 (red and green, K,O), Pou4f2 and Ldb1 (green and red, L,P), cCasp3 (red, M,Q) and Ki67 (green, N,R) at E18.5. GCL, ganglion cell layer; NBL, neuroblastic layer. Scale bar: 100 μm.
Ldb proteins are required for the differentiation and survival of ganglion cells, possibly by maintaining Isl1 protein levels in GCL precursors
Although Pou4f2 expression at E15.5 and E16.5 (Fig. 3C,G,I,K,O) indicated that more ganglion cells are generated in the Ldb1/Ldb2-deficient retina when compared with age-matched controls, at E18.5 when most RPCs are depleted based on reduced expression of Ki67 (Fig. 3N,R), the number of cells expressing Pou4f2 diminished in the Ldb1/Ldb2-deficient retina (Fig. 3L,P). This reduction occurred in conjunction with an elevation of the number of apoptotic cells based on detection of cleaved caspase 3 (cCasp3) (Fig. 3M,Q). This phenotype of GCL cell death is reminiscent of the phenotype resulting from an RPC-specific mutation in Isl1, a LIM-HD protein, which is co-expressed and has shared targets with Pou4f2 in GCL precursors (Mu et al., 2008; Pan et al., 2008). Indeed at E16.5 we detected lower expression of Isl1 protein in the Ldb1/Ldb2 mutant retina when compared with controls (Fig. 3K,O, red; Isl1 protein reduction in Ldb1 mutant INL was quantified at E15.5, Fig. S5). These observations suggest that the maintenance of Isl1 protein in the developing retina is dependent on Ldb1/Ldb2 activity. The loss of Isl1 could at least partly explain the eventual reduction in ganglion cell precursors in the Ldb1/Ldb2-deficient retina.
Ldb1 and Ldb2 are required for maintaining the RPC pool through regulation of multiple transcription factors and signaling pathways
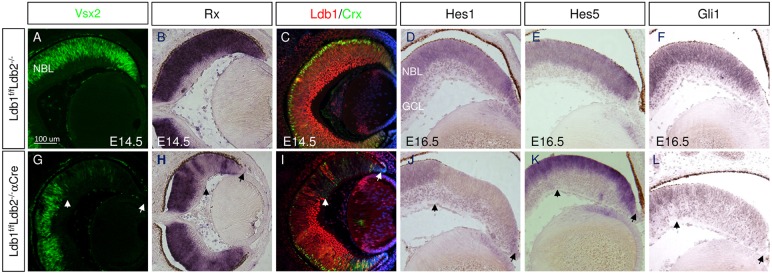
Vsx2 and Rax are transcription factors expressed in early RPCs and are required for their normal proliferation (Burmeister et al., 1996; Mathers et al., 1997; Andreazzoli et al., 1999; Casarosa et al., 2003). Reduced expression of Vsx2 protein and Rax mRNA were detected in Ldb1/Ldb2 mutants (Fig. 4A,B,G,H). Rax is also a key determinant of the specification and differentiation of PR cells, and it is required for the expression of the PR precursor transcription factor Crx, which is necessary for PR differentiation (Chen et al., 1997; Freund et al., 1997; Furukawa et al., 1997; Mathers et al., 1997; Nishida et al., 2003; Muranishi et al., 2011). Corresponding with Rax mRNA reduction, Crx protein was also reduced in the Ldb1/Ldb2-deficient OC (Fig. 4C,I), suggesting a role for Ldb1/2 proteins in the specification of PR cells during early stages of retinal neurogenesis. The reduction of Vsx2 and Rax expression is likely to contribute to the lower rates of proliferation of RPCs, although it does not explain the abrupt cell cycle exit and premature differentiation observed following loss of Ldb1/Ldb2 activity (Figs 2 and 3) (Green et al., 2003).
Fig. 4.
Loss of Ldb1/2 alters the expression pattern of Notch and Hedgehog pathway genes, and impairs specification of PR cells. Control (A-F) and Ldb1loxP/loxP;Ldb2−/−;α-Cre (G-L) retinas from E14.5 (A-C,G-I) and E16.5 (D-F,J-L) eyes labeled with antibodies to Vsx2 (A,G), and Ldb1 and Crx (red and green; C,I). The expression of Rx (B,H), Hes1 (D,J), Hes5 (E,K) and Gli1 (F,L) were analyzed by in situ hybridization. Arrows indicate mutated area. GCL, ganglion cell layer; NBL, neuroblastic layer. Scale bar: 100 μm.
Premature differentiation was previously reported to occur upon loss of Notch signaling factors (Jadhav et al., 2006a; Yaron et al., 2006), and the Notch-target gene Hes1 was recently reported to be downstream of the Ldb co-factor Lhx2 in cortical and late RPCs (Chou and O'Leary, 2013; Gordon et al., 2013; de Melo et al., 2016). Therefore, we sought to determine whether the Ldb1/Ldb2 complex regulates Notch signaling components during retinogenesis.
To this end, we characterized the expression of Hes1 and Hes5 in control and Ldb1/Ldb2-deficient E16.5 RPCs (Fig. 4D,E,J,K). Interestingly, Hes1 expression was reduced in all mutant cells, whereas Hes5 expression was maintained or possibly elevated in the residual RPCs (Fig. 4J,K). Hes1 has been found to be a target of Hedgehog protein signaling in the retina (Wall et al., 2009). Indeed, we detected a reduction of the Hedgehog target gene Gli1 (Goodrich and Scott, 1998) in the Ldb1/Ldb2-deficient RPCs compared with controls (Fig. 4F,L). Taken together, these findings suggest a role for Ldb1/Ldb2 in maintaining the RPC progenitor pool through regulating expression of multiple factors required for RPC proliferation, including Rax and Vsx2, as well as Notch and Hedgehog pathway genes.
Ldb1 and Lhx2 proteins interact during retinal development, and Ldb1 is associated with the Lhx2-bound regions of Vsx2 and Rax genes
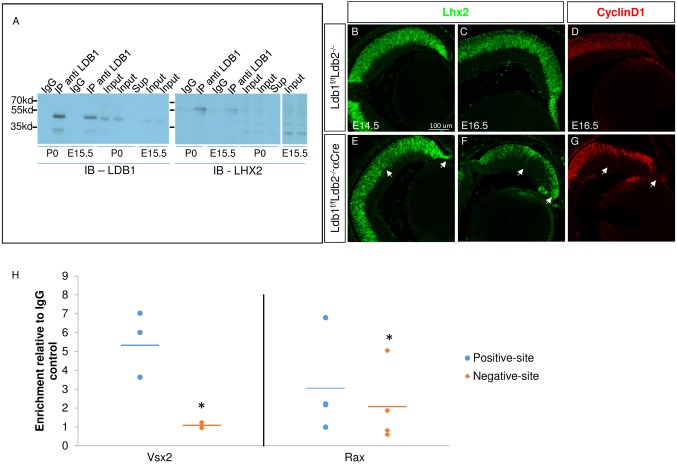
Ldb factors are known to be essential to the function and stability of LIM domain proteins. Lhx2 is the LIM-HD protein co-expressed with Ldb1 in RPCs in early and late stages of retinogenesis, and is required for maintaining the RPCs (Gordon et al., 2013; Roy et al., 2013; Balasubramanian et al., 2014). To examine the physical interaction between Ldb1 and Lhx2, we conducted co-immunoprecipitation with Ldb1 antibodies, with IgG used as control, from wild-type E15.5 eyes and P0 retinas (Fig. 5A). Lhx2 was co-precipitated with Ldb1, but not with control IgG antibodies, demonstrating the association of Ldb1 and Lhx2 at both ages. This result provides the biochemical evidence for the in vivo interaction of Lhx2 and Ldb1. Importantly, we demonstrate that this interaction occurs in the developing retina, implying shared functions for Lhx2 and Ldb1 during retina development.
Fig. 5.
Ldb1 interacts with Lhx2 in the eye, is bound to the same regulatory regions as Lhx2 in Vsx2 and Rax genes, but is dispensable for Lhx2 stability. (A) Endogenous Ldb1 was immunoprecipitated (IP) from a lysate prepared from E15.5 eyes and P0 retinas (IP anti LDB1). As a negative control, parallel lysate was incubated with normal rabbit IgG control (IgG). The precipitated complex, whole cell lysates (Input) and supernatant (Sup; P0) were subjected to western analysis for detection of endogenous Lhx2 (IB-LHX2) and Ldb1 (IB-LDB1). (B-G) In control retinas (B-D) and in Ldb1loxP/loxP;Ldb2−/−;α-Cre retinas (E-G), antibody labeling was used to detect Lhx2 (B,C,E,F) and Ccnd1 (cyclin D1, D,G) during retinogenesis. The arrows mark the peripheral retina where α-Cre transgenes are active. Scale bar: 100 μm. (H) Ldb1 ChIP was performed on retinal tissue collected at P0. Scatter plot represents fold enrichment for the immunoprecipitated fractions relative to the isotype controls at regulatory sites of Vsx2 and Rax genes that were found to be bound by Lhx2 at positive sites (blue dots; de Melo et al., 2016) and at negative sites (orange diamonds). The horizontal lines indicate the mean fold enrichment of Vsx2 (n=3, *P=0.02) and Rax (n=4, *P=0.037) calculated using a one-tailed paired-t-test.
Given the observed association between Ldb1 and Lhx2 proteins in RPC we expected that Ldb1/Ldb2 deficiency would lead to compromised stability and/or activity of Lhx2. Interestingly, the expression of Ldb1 does not seem to be necessary for maintaining the stability of Lhx2; vice versa, Ldb1 levels were maintained in Lhx2-deficient OCs (Fig. 5, Fig. S3). Nevertheless, in support of functional dependency between these proteins, the phenotype of the Ldb1loxP/loxP;Ldb2−/−;α-Cre RPCs mimicked main features of the phenotype of Lhx2-deficient RPCs (Lhx2loxP/loxP;α-Cre) (Gordon et al., 2013; Roy et al., 2013) (Fig. S3), including early reduction of Vsx2 levels (E13.5), depletion of RPCs, which exit mitosis and express markers of ganglion and amacrine precursors (Pou4f2, Vc1.1), reduction in the levels of Gli1 and Notch pathway genes, Rax and the photoreceptor precursor transcription factor Crx (Fig. S3). We also noted ectopic misexpression of the hypothalamic gene Lhx5 in the Ldb1/Ldb2 mutant at E16.5, similar to ectopic expression reported for Lhx2 mutants (Fig. S3) (Roy et al., 2013; de Melo et al., 2016). The observed interaction between Lhx2 and Ldb1, along with the phenotypic similarity between the Ldb1/Ldb2- and Lhx2-deficient RPCs, supports the notion that Ldb proteins are required for Lhx2 function in RPCs.
However, we also noted differences between the phenotypes of Ldb1loxP/loxP;Ldb2−/−;α-Cre and Lhx2loxP/loxP;α-Cre mice, suggesting that Lhx2 also has Ldb-independent roles. In particular, we noted that amacrine cells generated in the Ldb1/Ldb2 mutants expressed Ap2a, whereas in the Lhx2-deficient retina Ap2a expression was substantially reduced (Fig. 3, Fig. S3), suggesting that Lhx2 also contributes to the maturation of amacrine precursors.
Having shown a physical interaction between Ldb1 and Lhx2 in P0 retinas and a role for Ldb1/Ldb2 in maintaining the RPC progenitor pool through regulation of the expression of factors required for RPC proliferation, we further investigated [using chromatin immunoprecipitation (ChIP)-qPCR on P0 retina] whether Ldb1 directly binds candidate cis-regulatory sequences targeted by Lhx2 at this stage (de Melo et al., 2016). Ldb1 was found to be highly enriched in the regulatory regions of Vsx2, compared with IgG controls and with regions located downstream of these sites (termed ‘negative sites’). Significant enrichment of Ldb1 was also observed in the regulatory regions of Rax, although at lower affinity than that observed for Vsx2 (Fig. 5H). Taken together, Ldb1 and Lhx2 seem to maintain the RPC pool by positive transcriptional regulation of both Vsx2 and Rax.
Ldb2 is sufficient to sustain the RPC pool and Müller glia differentiation, but not for the differentiation of a subset of retinal neurons
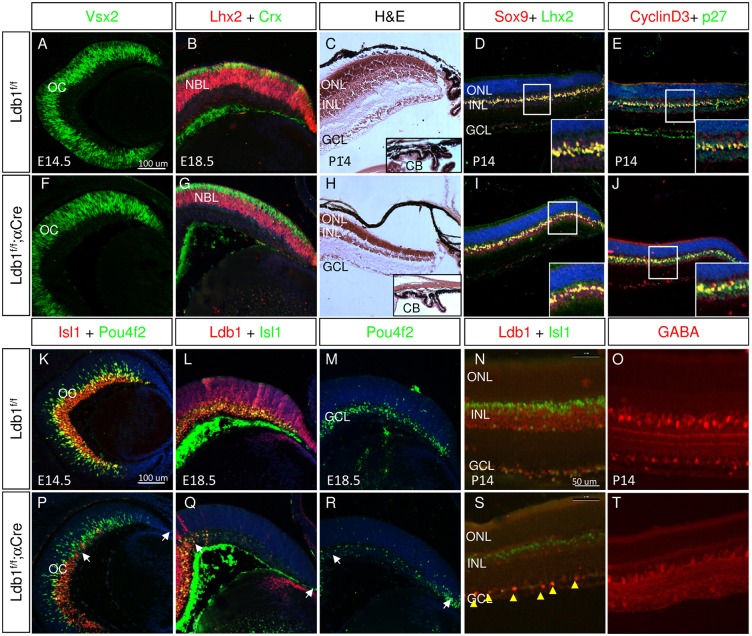
Genetic and molecular studies conducted on Ldb1 and Ldb2 have revealed functional redundancy between the two factors (Tzchori et al., 2009; Narkis et al., 2012). To determine directly the potential for Ldb2 to compensate for Ldb1 deficiency during retinal development, we analyzed the retinal phenotype of embryos deficient in Ldb1 in which Ldb2 remained intact (Ldb1loxP/loxP;α-Cre). Ldb2 transcript levels remain unchanged (mean fold change of 0.88, P=0.62) in the embryonic Ldb1 mutant retina (Fig. S4). However, the phenotype of the Ldb1loxP/loxP;α-Cre retina was strikingly different from that of the Ldb1loxP/loxP;Ldb2−/−;α-Cre retina, and further analysis led us to determine that Ldb2 is sufficient for maintaining the pool of retinal progenitors and for generation of Müller glia, both of which are known to require Lhx2 (Gordon et al., 2013; de Melo et al., 2016). This was already evident at E14.5, as in the Ldb1loxP/loxP;α-Cre retina the expression level of Vsx2 was similar to that of the control retina (Fig. 6A,F), and at E18.5 the progenitor pool was not depleted and Lhx2 expression was retained in the NBL (Fig. 6B,G). Furthermore, in the Ldb1loxP/loxP;α-Cre retina, PR precursors expressing Crx were detected in the prospective ONL at E18.5 (Fig. 6B,G) at levels similar to control based on QPCR (fold change of 1.19, P=0.47, Fig. S4). The activity of Ldb2 also eventually enabled the formation of a laminated retinal tissue, and was also sufficient for the development of auxiliary structures of the CB and iris (Fig. 6C,H), neither of which develop normally in Ldb1/Ldb2-deficient mutants (Fig. 1). Interestingly, Lhx2 activity was also preserved in the Ldb1loxP/loxP;α-Cre retina at postnatal stages. In these mice, Müller glia were generated, expression of the Müller glia factors Lhx2, Sox9, p27kip1 and Ccnd3 was maintained and properly localized in the INL, and upregulation of GFAP expression was not detected (Fig. 6D,E,I,J, data not shown). These stand in sharp contrast to in the phenotype detected following loss of Lhx2 in late-stage RPCs or in committed Müller glia precursors (de Melo et al., 2016). Taken together, these results reveal that Ldb2 alone, in the presence of Lhx2, is sufficient to fulfill the functions of the Ldb1/Ldb2 complex in maintaining the progenitor pool, generating PR cells, driving differentiation of Müller glia and contributing to the development of the auxiliary structures of the CB and iris.
Fig. 6.
Ldb1 is not essential for maintaining the retinal progenitors but is required for activities of LIM domain proteins in retinal precursors. Control (A-E,K-O) and Ldb1loxP/loxP;α-Cre (F-J,P-T) retinas were analyzed by antibody labeling, at the indicated developmental stages, for detection of Vsx2 (A,F), Lhx2 and Crx (red and green; B,G), Sox9 and Lhx2 (red and green; D,I), cyclin D3 and p27 (red and green; E,J), Isl1 and Pou4f2 (red and green; K,P), Ldb1 and Isl1 (red and green; L,Q,N,S, yellow arrowheads indicate a few cells that escape Cre activity based on maintained expression of Ldb1), Pou4f2 (M,R), and GABA (red; O,T). Hematoxylin and Eosin staining (C,H). CB, ciliary body; GCL, ganglion cell layer; INL, inner nuclear layer; NBL, neuroblastic layer; OC, optic cup; ONL, outer nuclear layer. White arrows indicate mutated areas. Scale bars: in A and K, 100 μm for A-M,P-R; in N, 50 μm in N,O,S,T.
By contrast, Ldb2 did not fully compensate for those functions of Ldb1 that are mediated by other LIM domain proteins, such as Isl1 and Lmo4. Specifically, the phenotype of the Ldb1loxP/loxP;α-Cre retina mimicked the phenotype associated with the combined loss of Isl1 and Lmo4, which is characterized by loss of GCLs, and a reduction in the number of GABAergic amacrine cells and subsets of bipolar cells (Elshatory et al., 2007; Duquette et al., 2010).
We observed a reduction in Isl1 levels in the Ldb1loxP/loxP;α-Cre retina relative to controls (E14.5, E18.5, Fig. 6K,L,P,Q). This was further corroborated by fluorescence quantification of Isl1 levels in INL regions with or without Ldb1, showing a significant halving of Isl1 levels at E15.5 (Fig. S5), which was reminiscent of the Isl1 reduction detected in Ldb1loxP/loxP;Ldb2−/−;α-Cre retina (Fig. 4, Fig. S5). However, Isl1 mRNA levels were significantly elevated in the Ldb1loxP/loxP;α-Cre retina, based on QPCR analysis (1.64 mean fold change, P=0.04) (Fig. S4). Ldb1 may therefore be required to maintain stability of Isl1 protein in retina, similar to observations made in cardiac progenitors (Caputo et al., 2015).
The reduction in Isl1 protein levels in the Ldb1-deficient retina likely contributes to the depletion of GCLs from the Ldb1loxP/loxP;α-Cre retina at E18.5 (Fig. 6M,R), even though at E14.5 the pattern of GCL production in this mutant resembled that seen in control OC (Fig. 6K,P). In Ldb1-deficient retina at P14, we did not detect Isl1+ GCLs, nor did we detect Isl1 expression in the inner portion of the INL, where Isl1+ starburst amacrine cells are normally located (Fig. 6N,S). Thus Ldb2 alone is not sufficient for the generation of the Isl1+ early-born retinal neurons. At postnatal stages, we detected some Isl1-expressing cells in the outer region of the INL where bipolar cells are normally found, although their number was 40% lower than in the control [31.2±2.07 (s.d.) cells/100 µm in control and 19.4±2.4 cells/100 µm in the Ldb1loxP/loxP;α-Cre retina; P<0.01, Fig. 6N,S]. Lmo4 is an LMO protein that is essential for the generation of GABAergic amacrine cells and a subset of bipolar cells. The number of GABAergic amacrine cells in Ldb1loxP/loxP;α-Cre was reduced when compared with control (Fig. 6O,T), suggesting compromised activity of Lmo4. Lmo4 mRNA levels, however, remain unchanged in the E15.5 Ldb1loxP/loxP;α-Cre retina, implying that Lmo4 translation and/or stability or function is dependent on Ldb1 (Fig. S4).
Taken together, these findings suggest that, in the absence of Ldb1, Ldb2 – most likely in conjunction with Lhx2 – is sufficient to maintain RPCs during both early and late stages of embryogenesis, as well as to support Muller glia differentiation. However, Ldb2 alone is not sufficient to carry out all of the functions associated with the LIM domain proteins Isl1 and Lmo4, including production of ganglion cells and generating the diversity of amacrine and bipolar lineages.
DISCUSSION
Ldb1 and Ldb2 are scaffold proteins that assemble modular, tissue-specific transcriptional complexes containing LIM-homeodomain factors. Herein, through detailed comparative phenotypic analyses of mutants in both Ldb1 and Ldb2, and co-immunoprecipitation of Ldb1, we obtain a comprehensive overview of the functions of Ldb1 and Ldb2 during mammalian retinal development.
Ldb1 and Ldb2 are obligatory co-factors of Lhx2 in retinal progenitors
The activity of Lhx2 has been shown to be dependent on Ldb proteins during both wing development and neuronal differentiation in Drosophila, and during limb development in vertebrates (van Meyel et al., 1999, 2000). However, the role of Ldb factors in regulating Lhx2 function has not been directly studied in the context of CNS development. Here, we show by immunoprecipitation of endogenous Ldb1 from embryonic eye and P0 retina that Ldb1 interacts with Lhx2 in RPCs, where they are co-expressed. This, together with the striking similarity in the phenotypes seen following loss of function of Ldb1/Ldb2 or Lhx2 in RPCs, demonstrates the importance of Ldb factors in regulating Lhx2 function in the developing retina. The finding that Ldb2 alone is sufficient for maintenance of RPC proliferation and Müller glia differentiation further implies similar interaction of Ldb2 with Lhx2, although this should be directly tested once appropriate antibodies to Ldb2 are available.
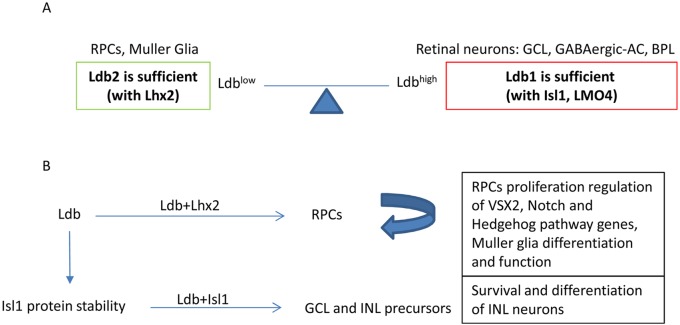
Several studies have provided insights into the roles of Lhx2 in retinogenesis. The systemic inactivation of Lhx2, or its conditional deletion at the OV stage, resulted in aberrant OC morphogenesis and elevation of genes specifically expressed in the hypothalamus and prethalamus (Yun et al., 2009; Hagglund et al., 2011; Roy et al., 2013). These phenotypes suggest a role for Lhx2 in maintaining retinal fate that is reminiscent of its roles in early stages of corticogenesis (E10.5; Mangale et al., 2008). The inactivation of either Lhx2 or Ldb1/Ldb2 following the onset of retinal neurogenesis results in abrupt cell cycle exit and onset of differentiation into early-born retinal precursors that express Pou4f2 and Vc1.1 (Gordon et al., 2013; Roy et al., 2013; this study). The striking similarity between the phenotypes of Lhx2 and of Ldb1/Ldb2 mutants suggests that Lhx2 is the main LIM domain protein functioning in early RPCs, that the functions of Ldb1/Ldb2 and Lhx2 depend on the formation of an Ldb-Lhx2 complex, and that at the OC stage this complex is primarily required for maintaining the pool of proliferating and multipotent RPCs (Fig. 7).
Fig. 7.
The proposed dose-dependent roles of Ldb complexes in retinogenesis. (A) The expression of Ldb2 alone (Ldblow) is sufficient to execute activities of the Ldb complex, which, probably through an interaction with Lhx2, is required for maintaining the RPC pool and Müller glia differentiation and function. High Ldb (Ldb1 alone; Ldbhigh) is required for maturation of ganglion and amacrine (AC) precursors, and for generation of a subset of bipolar cells (BPL). Ldb1 probably contributes to these activities by maintaining the stability and function of LIM proteins that are expressed in these precursors: Isl1 and Lmo4. (B) The levels of Ldb available for interaction with the neuronal LIM proteins (Lhx2 vs Isl1) may be involved in regulating the balance between progenitors and precursors, and eventually in generating the correct numbers of retinal cell types.
To our surprise, the inactivation of either Lhx2 or Ldb1/Ldb2 prevented PR differentiation, despite the fact that neither Ldb1 nor Lhx2 is expressed in PR precursors. This observation suggests that the Ldb-Lhx2 complex is required for the competence of early RPCs to generate PR precursors, probably by directly regulating the expression of Rax (Roy et al., 2013; de Melo et al., 2016), an RPC-expressed transcription factor that is essential for specification of PR cells and expression of Crx (Fig. 4, Fig. S3) (Nishida et al., 2003; Muranishi et al., 2011). This role of Ldb1/Ldb2 and Lhx2 in promoting generation of PR cells is restricted to early-born PR cells, as inactivation of Lhx2 at later stages of retinogenesis results in an increase in the number of PR precursors (Gordon et al., 2013), probably owing to the activity of other transcription factors and epigenetic determinants.
Phenotypic analysis, however, also implies that some of the activities of Lhx2 do not require maintained expression of Ldb proteins. This is suggested because the amacrine/horizontal precursors are generated in both mutants, as reflected in the expression of Vc1.1 and Pax6; however, in Lhx2-deficient cells, these precursors fail to express Ap2a, although they do express Ap2a in the Ldb1/Ldb2-deficient retina. This difference suggests a direct role for Lhx2 in regulating the expression of a subset of factors that are required for interneuron maturation. It is more likely, however, that the loss of Lhx2 alters the composition of the Ldb1/Ldb2 complex with other LIM domain proteins, as has been observed in the spinal cord and hematopoietic system (Song et al., 2009; Kitajima et al., 2013). The change in composition of the Ldb-LIM complex is expected to result in misexpression of genes, such as hypothalamic genes or retinal LIM domain proteins that may interfere with amacrine/horizontal differentiation.
Ldb1/Ldb2 and Lhx2 are required for maintaining the pool of retinal progenitors through regulation of RPC-enriched transcription factors, as well as Notch and Hedgehog pathway genes
Lhx2 is known to be required for maintaining the proliferation and stem cell properties of diverse lineages, including hair follicles, hematopoietic stem cells and neuronal progenitors in the telencephalon and the retina; yet, the mechanism by which Lhx2 functions in the regulation of the cell cycle remains unknown (Pinto do et al., 2002; Rhee et al., 2006; Chou and O'Leary, 2013; Gordon et al., 2013; Roy et al., 2013).
Vsx2 is known to be required for the proliferation of RPCs and for regulating the temporal onset of Hedgehog pathway activity (Burmeister et al., 1996; Green et al., 2003; Sigulinsky et al., 2008). The reduction in Vsx2 expression seen in both Lhx2 and Ldb1/Ldb2 retinal mutants occurred soon after the onset of Cre activity, suggesting that at least some of the activities of Ldb1/Ldb2 and Lhx2 in regulation of cell proliferation could be mediated by Vsx2.
In the retina, Hes1 expression is regulated by both Notch and Hedgehog signaling, and both pathways are required for proliferation of the RPCs; notably, loss of Notch1 leads to premature generation of PR cells but not GCL cells (Wang et al., 2005; Jadhav et al., 2006b; Yaron et al., 2006; Wall et al., 2009). We observed a reduction of Hes1 levels in the Ldb1/Ldb2 mutant OC, whereas Hes5 was detected in the residual NBL at E16.5 (Fig. 5). Misexpression of Hes1 prevents cell differentiation in the retina, whereas Hes1 knockout mice exhibit premature retinal neurogenesis (Tomita et al., 1996; Kageyama et al., 2008). Thus, the loss of Hes1 observed in the Ldb1/Ldb2 and Lhx2 mutant RPCs probably contributes to the premature differentiation observed following their inactivation.
The reduced expression of the Hedgehog target Gli1 observed in Lhx2 and Ldb1/Ldb2-deficient RPCs suggests that loss of Hedgehog activity may be responsible for the observed reduction in cell proliferation, as well as for the elevation in the number of GCL cells observed in the mutant retinas. Lhx2 has been reported to be required for Hedgehog signaling in the zone of polarizing activity during limb development; thus, intersection with this pathway may reflect a regulatory network that functions downstream of Lhx2 in progenitors from multiple tissues (Tzchori et al., 2009). The findings above suggest that the Ldb1/Ldb2-Lhx2 complex functions by regulating expression of Hedgehog and Notch pathway components during retinogenesis, in addition to Vsx2. The ChIP analysis for Ldb1 (Fig. 5) and for Lhx2 (de Melo et al., 2016) further supports their association with the same regulatory regions of both Rax and Vsx2, which are both important for RPC proliferation (Burmeister et al., 1996; Green et al., 2003; Muranishi et al., 2012). The RPC depletion observed in the Ldb1/Ldb2 mutants reflects the combined loss of several key regulators that dictate the timing of cell cycle exit and the proliferation rate of neuronal progenitors.
Selective roles of Ldb factors in stabilization of LIM-HD proteins could influence the balance between retinal progenitors and precursors
In Drosophila, expression of the Lhx2 homolog Apterous (Ap), is regulated by proteosomal degradation, and this proteolysis has been found to be inhibited by association of Ap with the Ldb homolog Chip (Weihe et al., 2001). Similar regulation has been observed in the vertebrate LIM-HD proteins Lhx3 and Isl1 (Ostendorff et al., 2002; Gungor et al., 2007). Deletion of the Ldb1/Ldb2 complex from RPCs enabled us to gain insight into the role of Ldb/LIM-HD complex formation in regulating the expression and stability of LIM-HD proteins during mammalian retinal development. Indeed, although Isl1 transcription was initiated (and even elevated) in the Ldb1/Ldb2- and Ldb1-deficient retinas, Isl1 protein levels gradually fell in the post-mitotic precursors compared with controls, even when only Ldb1, but not Ldb2, was deleted (Figs 3 and 6 and Fig. S5). Ldb1 was already shown to function in stabilizing the Isl1 protein in cardiac progenitors, these results suggest a similar role in the retina (Caputo et al., 2015). In contrast to the loss of Isl1 protein seen in Ldb1/Ldb2-deficient retinas, Lhx2 was maintained in the mutant cells throughout most of embryogenesis and was reduced only close to birth (E18.5, Fig. 4, not shown). This dependency of the precursor protein Isl1 on Ldb for its stability suggests a mechanism whereby the ratio between progenitor maintenance and cell cycle exit is regulated by the availability of Ldb. Lhx2 may maintain the RPC pool in embryonic retina by sequestering the Ldb and thus preventing stabilization of Isl1, a factor that is required for post-mitotic precursors (Fig. 7).
Intrinsic differences in the binding affinities of the LIM domains to Ldb proteins, and their interactions with other proteins present in developing retina, are likely to be crucial in regulating the stability of each of the multiple LIM-HD proteins that form complexes with Ldb1/Ldb2 in retinal development.
The unequal role of Ldb1 and Ldb2 in retinogenesis
Ldb1 and Ldb2 show distinct functions during retinal development. The loss of Ldb2 in the retina has no apparent retinal phenotype, suggesting that Ldb1 may be sufficient to execute the functions of Ldb in the retina. However, the loss of both Ldb1 and Ldb2 resulted in early depletion of most RPCs, while the inactivation of Ldb1 (but not of Ldb2) resulted in a phenotype that resembled a combination of the phenotypes associated with mutation of Isl1 and Lmo4, including loss of GCLs, reduction in bipolar cell number and a marked reduction in the number of GABAergic amacrine cells (Mu et al., 2008; Pan et al., 2008; Duquette et al., 2010). By contrast, Lhx2-dependent functions seemed to be preserved following loss of function of Ldb1, RPCs were maintained throughout retinogenesis, and Müller glia differentiation occurred normally.
One possible explanation for the preservation of Lhx2-dependent functions by Ldb2 could be the physical differences between Ldb1 and Ldb2, which may contribute to differences in the capacity of these proteins to selectively associate with different LIM domain proteins within functional transcriptional complexes. This notion is supported by self-association studies that reveal distinct patterns of oligomerization for Ldb1 and Ldb2, indicating that the two proteins form different transcription complexes and exert distinct biological activities (Cross et al., 2010). These studies, however, were performed in vitro with isolated self-association domains, and thus their relevance to the in vivo Ldb1 and Ldb2 transcriptional complexes remains to be explored. Moreover, in the progenitors of the hind limb bud, Ldb2 mediates Isl1 functions, rather than Lhx2 functions (Tzchori et al., 2009; Narkis et al., 2012). Thus, if there is a preferred interaction between Ldb2 and Lhx2, it is clearly context dependent.
Another possible explanation for this selective compensatory activity of Ldb2 may relate to its levels of expression (Caqueret et al., 2006), which may not be sufficient to fully compensate for Ldb1 loss at all developmental stages and in all cell types. Our experiments indicate that, in the RPCs, Ldb2 is sufficient to mediate Lhx2 activity, whereas studies in the limb bud show that Ldb2 is necessary for Isl1 activity (Tzchori et al., 2009; Narkis et al., 2012). Therefore, it is possible that only relatively low levels of Ldb-Lhx2 are required to drive expression of RPC and glial-specific genes, but that higher levels of Ldb are needed for formation of Ldb/LIM complexes required for generation of neuronal lineages. Thus, our findings suggest (Fig. 7) that Ldb expression levels in RPCs are sufficient for execution of Lhx2-dependent functions, whereas in postmitotic precursors, higher levels of Ldb are required for execution of complete neurogenic differentiation programs.
MATERIALS AND METHODS
Mouse lines
The mouse lines employed in this study are: Ldb1loxP, Ldb2−/− (Tzchori et al., 2009; Narkis et al., 2012), Lhx2 (Mangale et al., 2008) and α-Cre (Marquardt et al., 2001). All animal work was conducted according to national and international guidelines and approved by the Tel Aviv University review board.
Histology, immunofluorescence and BrdU-incorporation assays
Immunofluorescence analyses and Hematoxylin and Eosin staining was performed as described previously (Ashery-Padan et al., 2000). The antibodies are listed in Table S1. Cell cycle exit analysis was performed as described previously (Farhy et al., 2013). Slides were viewed with an Olympus BX61 fluorescent microscope, and images were analyzed using the AnalySIS. The measurements were conducted on serial sections, with well-preserved morphology, that were central according to the detection of the lens. The distal region of the OC, where α-Cre is active, was analyzed. The mutant region was defined by labeling with Ldb1 antibodies on the same section or on a section from the same series. For each eye, an average value for the presented parameter was calculated from at least three sections ∼20 µm apart. The presented ratio values are the averages from analysis of at least three eyes (n=3).
Statistical analysis
For each developmental stage and genotype, all values were averaged, and the standard deviation (s.d.) was calculated. Values obtained for control and mutant animals were compared using Student's t-test to determine statistical significance.
In situ hybridization
Hybridization was conducted overnight at 65°C with digoxigenin-labeled probes (3 µg/ml) on de-waxed paraffin sections. The slides were then treated with RNaseA, washed, blocked with 10% normal goat serum (NGS) and incubated with sheep anti-digoxigenin Fab fragments conjugated to alkaline phosphatase (1:250, Roche) in PBST with 1% NGS overnight at 4°C and incubated in BM Purple (Roche).
RNA isolation and quantitative PCR
Retinas separated from mature mouse eyes and total eyes from E15.5 mouse embryos were snap-frozen in dry ice and RNA was isolated using RNeasy kit (QIAGEN). cDNA was synthesized with the qScript cDNA Synthesis Kit (Quanta BioSciences) and quantitative PCR (QPCR) was performed using the FastStart Universal SYBR Green Master (Roche) on Applied Biosystems Viia7 real-time PCR detection system. The cycle numbers were normalized to Actin. Primer pairs are listed in Table S2.
Quantification of Isl1 and Ldb1 protein levels
Images of sections stained for Ldb1 and Isl1 were acquired by LeicaSP8 confocal microscope and subsequently were analyzed by ImageJ for their corrected total cell fluorescence (CTCF). A calculation was made as CTCF=integrated density–(area of selected cell×mean fluorescence of background readings) for regions defined as ‘Ldb1 positive’ and ‘Ldb1 negative’ based on Ldb1 fluorescent signal and equivalently for Isl1 signals in the same defined regions.
Immunoprecipitation
Eyes of E15.5 embryos and retinas of P0 mice were lysed with RIPA lysis buffer [50 mM Tris-HCl (pH 8.0), 50 mM NaCl, 1% NP-40, 0.5% Na-deoxycholate and 0.1% SDS] and protease inhibitors (Complete mini, Roche). Preclearing by incubation with protein-A agarose beads (Millipore) was followed by centrifugation and incubation of the supernatants with goat anti-Lhx2 antibody (Santa Cruz) overnight at 4°C. Then beads were added for an additional 2 h. Samples were washed four times with RIPA buffer and analyzed by western blotting using anti-Lhx2 and anti-Ldb1 antibodies for blotting. For the immunoprecipitation negative control, lysates were precipitated with normal rabbit control IgG (Santa Cruz).
Chromatin immunoprecipitation
P0 retinas were dissected in HBSS and dissociated by 30 min incubation with papain. Cross-linking, DNA shearing and immunoprecipitations were performed as previously described (Sailaja et al., 2012). Rabbit anti-Ldb1 antibody or normal rabbit IgG (Santa Cruz) were used for the immunoprecipitations. Enrichment levels were quantified by qPCR. Primers for positive regions (de Melo et al., 2016) and downstream negative regions are listed in Table S2.
Acknowledgements
We thank Drs Ryoichiro Kageyama, Takahisa Furukawa and Alex Joyner for the in situ probes. We thank Leonid Mittelman for confocal images. These studies were performed as partial requirements of K.G. for a PhD degree from Tel Aviv University.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
K.G., T.C., A.D. and R.A.-P. conceived and designed the experiments. K.G., T.C., A.D., Y.M.-L., H.N., G.N. and R.A.-P. performed the experiments. K.G., A.D., Y.M.-L., H.N., S.B. and R.A.-P. analyzed the data. T.C., L.L., P.L., J.d.M., S.B. and H.W. provided essential reagents. A.D., S.B., J.d.M. and R.A.-P. wrote the manuscript.
Funding
This work was funded by the United States - Israel Binational Science Foundation (2013016), Israel Academy of Sciences and Humanities (228/14) and by the Claire and Amédée Maratier Institute for the Study of Blindness and Visual Disorders, Tel-Aviv University. The S.B. lab is funded by the National Institutes of Health (EY02056). Deposited in PMC for release after 12 months.
Supplementary information
Supplementary information available online at http://dev.biologists.org/lookup/doi/10.1242/dev.129734.supplemental
References
- Alexiades M. R. and Cepko C. L. (1997). Subsets of retinal progenitors display temporally regulated and distinct biases in the fates of their progeny. Development 124, 1119-1131. [DOI] [PubMed] [Google Scholar]
- Andreazzoli M., Gestri G., Angeloni D., Menna E. and Barsacchi G. (1999). Role of Xrx1 in Xenopus eye and anterior brain development. Development 126, 2451-2460. [DOI] [PubMed] [Google Scholar]
- Ashery-Padan R., Marquardt T., Zhou X. and Gruss P. (2000). Pax6 activity in the lens primordium is required for lens formation and for correct placement of a single retina in the eye. Genes Dev. 14, 2701-2711. 10.1101/gad.184000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bach I., Carriere C., Ostendorff H. P., Andersen B. and Rosenfeld M. G. (1997). A family of LIM domain-associated cofactors confer transcriptional synergism between LIM and Otx homeodomain proteins. Genes Dev. 11, 1370-1380. 10.1101/gad.11.11.1370 [DOI] [PubMed] [Google Scholar]
- Balasubramanian R., Bui A., Ding Q. and Gan L. (2014). Expression of LIM-homeodomain transcription factors in the developing and mature mouse retina. Gene Expr. Patterns 14, 1-8. 10.1016/j.gep.2013.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassett E. A., Korol A., Deschamps P. A., Buettner R., Wallace V. A., Williams T. and West-Mays J. A. (2012). Overlapping expression patterns and redundant roles for AP-2 transcription factors in the developing mammalian retina. Dev. Dyn. 241, 814-829. 10.1002/dvdy.23762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhati M., Lee C., Nancarrow A. L., Lee M., Craig V. J., Bach I., Guss J. M., Mackay J. P. and Matthews J. M. (2008). Implementing the LIM code: the structural basis for cell type-specific assembly of LIM-homeodomain complexes. EMBO J. 27, 2018-2029. 10.1038/emboj.2008.123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burmeister M., Novak J., Liang M.-Y., Basu S., Ploder L., Hawes N. L., Vidgen D., Hoover F., Goldman D., Kalnins V. I. et al. (1996). Ocular retardation mouse caused by Chx10 homeobox null allele: impaired retinal progenitor proliferation and bipolar cell differentiation. Nat. Genet. 12, 376-384. 10.1038/ng0496-376 [DOI] [PubMed] [Google Scholar]
- Caputo L., Witzel H. R., Kolovos P., Cheedipudi S., Looso M., Mylona A., van I. W. F., Laugwitz K.-L., Evans S. M., Braun T. et al. (2015). The Isl1/Ldb1 Complex Orchestrates Genome-wide Chromatin Organization to Instruct Differentiation of Multipotent Cardiac Progenitors, Cell Stem Cell 17, 287-299. 10.1016/j.stem.2015.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caqueret A., Boucher F. and Michaud J. L. (2006). Laminar organization of the early developing anterior hypothalamus. Dev. Biol. 298, 95-106. 10.1016/j.ydbio.2006.06.019 [DOI] [PubMed] [Google Scholar]
- Casarosa S., Amato M. A., Andreazzoli M., Gestri G., Barsacchi G. and Cremisi F. (2003). Xrx1 controls proliferation and multipotency of retinal progenitors. Mol. Cell. Neurosci. 22, 25-36. 10.1016/S1044-7431(02)00025-8 [DOI] [PubMed] [Google Scholar]
- Chen S., Wang Q.-L., Nie Z., Sun H., Lennon G., Copeland N. G., Gilbert D. J., Jenkins N. A. and Zack D. J. (1997). Crx, a novel Otx-like paired-homeodomain protein, binds to and transactivates photoreceptor cell-specific genes. Neuron 19, 1017-1030. 10.1016/S0896-6273(00)80394-3 [DOI] [PubMed] [Google Scholar]
- Chou S. J. and O'Leary D. D. (2013). Role for Lhx2 in corticogenesis through regulation of progenitor differentiation, Mol. Cell. Neurosci. 56: 1-9. 10.1016/j.mcn.2013.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross A. J., Jeffries C. M., Trewhella J. and Matthews J. M. (2010). LIM domain binding proteins 1 and 2 have different oligomeric states. J. Mol. Biol. 399, 133-144. 10.1016/j.jmb.2010.04.006 [DOI] [PubMed] [Google Scholar]
- de Melo J., Miki K., Rattner A., Smallwood P., Zibetti C., Hirokawa K., Monuki E. S., Campochiaro P. A. and Blackshaw S. (2012). Injury-independent induction of reactive gliosis in retina by loss of function of the LIM homeodomain transcription factor Lhx2. Proc. Natl. Acad. Sci. USA 109, 4657-4662. 10.1073/pnas.1107488109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Melo J., Zibetti C., Clark B. S., Hwang W., Miranda-Angulo A. L., Qian J. and Blackshaw S. (2016). Lhx2 is an essential factor for retinal gliogenesis and notch signaling. J. Neurosci. 36, 2391-2405. 10.1523/JNEUROSCI.3145-15.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duquette P. M., Zhou X., Yap N. L., MacLaren E. J., Lu J. J., Wallace V. A. and Chen H.-H. (2010). Loss of LMO4 in the retina leads to reduction of GABAergic amacrine cells and functional deficits. PLoS ONE 5, e13232 10.1371/journal.pone.0013232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elshatory Y., Everhart D., Deng M., Xie X., Barlow R. B. and Gan L. (2007). Islet-1 controls the differentiation of retinal bipolar and cholinergic amacrine cells. J. Neurosci. 27, 12707-12720. 10.1523/JNEUROSCI.3951-07.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farhy C., Elgart M., Shapira Z., Oron-Karni V., Yaron O., Menuchin Y., Rechavi G. and Ashery-Padan R. (2013). Pax6 is required for normal cell-cycle exit and the differentiation kinetics of retinal progenitor cells. PLoS ONE 8, e76489 10.1371/journal.pone.0076489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freund C. L., Gregory-Evans C. Y., Furukawa T., Papaioannou M., Looser J., Ploder L., Bellingham J., Ng D., Herbrick J.-A. S., Duncan A. et al. (1997). Cone-rod dystrophy due to mutations in a novel photoreceptor-specific homeobox gene (CRX) essential for maintenance of the photoreceptor. Cell 91, 543-553. 10.1016/S0092-8674(00)80440-7 [DOI] [PubMed] [Google Scholar]
- Furukawa T., Morrow E. M. and Cepko C. L. (1997). Crx, a novel otx-like homeobox gene, shows photoreceptor-specific expression and regulates photoreceptor differentiation. Cell 91, 531-541. 10.1016/S0092-8674(00)80439-0 [DOI] [PubMed] [Google Scholar]
- Gan L., Xiang M., Zhou L., Wagner D. S., Klein W. H. and Nathans J. (1996). POU domain factor Brn-3b is required for the development of a large set of retinal ganglion cells. Proc. Natl. Acad. Sci. USA 93, 3920-3925. 10.1073/pnas.93.9.3920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrich L. V. and Scott M. P. (1998). Hedgehog and patched in neural development and disease. Neuron 21, 1243-1257. 10.1016/S0896-6273(00)80645-5 [DOI] [PubMed] [Google Scholar]
- Gordon P. J., Yun S., Clark A. M., Monuki E. S., Murtaugh L. C. and Levine E. M. (2013). Lhx2 balances progenitor maintenance with neurogenic output and promotes competence state progression in the developing retina. J. Neurosci. 33, 12197-12207. 10.1523/JNEUROSCI.1494-13.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green E. S., Stubbs J. L. and Levine E. M. (2003). Genetic rescue of cell number in a mouse model of microphthalmia: interactions between Chx10 and G1-phase cell cycle regulators. Development 130, 539-552. 10.1242/dev.00275 [DOI] [PubMed] [Google Scholar]
- Gungor C., Taniguchi-Ishigaki N., Ma H., Drung A., Tursun B., Ostendorff H. P., Bossenz M., Becker C. G., Becker T. and Bach I. (2007). Proteasomal selection of multiprotein complexes recruited by LIM homeodomain transcription factors. Proc. Natl. Acad. Sci. USA 104, 15000-15005. 10.1073/pnas.0703738104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagglund A. C., Dahl L. and Carlsson L. (2011). Lhx2 is required for patterning and expansion of a distinct progenitor cell population committed to eye development. PLoS ONE 6, e23387 10.1371/journal.pone.0023387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jadhav A. P., Cho S.-H. and Cepko C. L. (2006a). Notch activity permits retinal cells to progress through multiple progenitor states and acquire a stem cell property. Proc. Natl. Acad. Sci. USA 103, 18998-19003. 10.1073/pnas.0608155103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jadhav A. P., Mason H. A. and Cepko C. L. (2006b). Notch 1 inhibits photoreceptor production in the developing mammalian retina. Development 133, 913-923. 10.1242/dev.02245 [DOI] [PubMed] [Google Scholar]
- Jean D., Bernier G. and Gruss P. (1999). Six6 (Optx2) is a novel murine Six3-related homeobox gene that demarcates the presumptive pituitary/hypothalamic axis and the ventral optic stalk. Mech. Dev. 84, 31-40. 10.1016/S0925-4773(99)00068-4 [DOI] [PubMed] [Google Scholar]
- Kageyama R., Ohtsuka T. and Kobayashi T. (2008). Roles of Hes genes in neural development. Dev. Growth Differ. 50 Suppl. 1, S97-S103. 10.1111/j.1440-169X.2008.00993.x [DOI] [PubMed] [Google Scholar]
- Kitajima K., Kawaguchi M., Iacovino M., Kyba M. and Hara T. (2013). Molecular functions of the LIM-homeobox transcription factor Lhx2 in hematopoietic progenitor cells derived from mouse embryonic stem cells. Stem Cells 31, 2680-2689. 10.1002/stem.1500 [DOI] [PubMed] [Google Scholar]
- Lee S.-K. and Pfaff S. L. (2003). Synchronization of neurogenesis and motor neuron specification by direct coupling of bHLH and homeodomain transcription factors. Neuron 38, 731-745. 10.1016/S0896-6273(03)00296-4 [DOI] [PubMed] [Google Scholar]
- Leone D. P., Panagiotakos G., Heavner W. E., Joshi P., Zhao Y., Westphal H. and McConnell S. K. (2016). Compensatory actions of Ldb adaptor proteins during corticospinal motor neuron differentiation. Cereb. Cortex. 10.1093/cercor/bhw003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L. Q., Jothi R., Cui K., Lee J. Y., Cohen T., Gorivodsky M., Tzchori I., Zhao Y., Hayes S. M., Bresnick E. H. et al. (2011). Nuclear adaptor Ldb1 regulates a transcriptional program essential for the maintenance of hematopoietic stem cells. Nat. Immunol. 12, 129-136. 10.1038/ni.1978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love P. E., Warzecha C. and Li L. (2014). Ldb1 complexes: the new master regulators of erythroid gene transcription. Trends Genet. 30, 1-9. 10.1016/j.tig.2013.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangale V. S., Hirokawa K. E., Satyaki P. R. V., Gokulchandran N., Chikbire S., Subramanian L., Shetty A. S., Martynoga B., Paul J., Mai M. V. et al. (2008). Lhx2 selector activity specifies cortical identity and suppresses hippocampal organizer fate. Science 319, 304-309. 10.1126/science.1151695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquardt T. (2003). Transcriptional control of neuronal diversification in the retina. Prog. Retin. Eye Res. 22, 567-577. 10.1016/S1350-9462(03)00036-3 [DOI] [PubMed] [Google Scholar]
- Marquardt T., Ashery-Padan R., Andrejewski N., Scardigli R., Guillemot F. and Gruss P. (2001). Pax6 is required for the multipotent state of retinal progenitor cells. Cell 105, 43-55. 10.1016/S0092-8674(01)00295-1 [DOI] [PubMed] [Google Scholar]
- Mathers P. H., Grinberg A., Mahon K. A. and Jamrich M. (1997). The Rx homeobox gene is essential for vertebrate eye development. Nature 387, 603-607. 10.1038/42475 [DOI] [PubMed] [Google Scholar]
- Matthews J. M. and Visvader J. E. (2003). LIM-domain-binding protein 1: a multifunctional cofactor that interacts with diverse proteins. EMBO Rep. 4, 1132-1137. 10.1038/sj.embor.7400030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews J. M., Bhati M., Craig V. J., Deane J. E., Jeffries C., Lee C., Nancarrow A. L., Ryan D. P. and Sunde M. (2008). Competition between LIM-binding domains. Biochem. Soc. Trans. 36(Pt 6), 1393-1397. 10.1042/BST0361393 [DOI] [PubMed] [Google Scholar]
- Mu X., Fu X., Beremand P. D., Thomas T. L. and Klein W. H. (2008). Gene-regulation logic in retinal ganglion cell development: isl1 defines a critical branch distinct from but overlapping with Pou4f2. Proc. Natl. Acad. Sci. USA 105, 6942-6947. 10.1073/pnas.0802627105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay M., Teufel A., Yamashita T., Agulnick A. D., Chen L., Downs K. M., Schindler A., Grinberg A., Huang S. P., Dorward D. et al. (2003). Functional ablation of the mouse Ldb1 gene results in severe patterning defects during gastrulation. Development 130, 495-505. 10.1242/dev.00225 [DOI] [PubMed] [Google Scholar]
- Muranishi Y., Terada K., Inoue T., Katoh K., Tsujii T., Sanuki R., Kurokawa D., Aizawa S., Tamaki Y. and Furukawa T. (2011). An essential role for RAX homeoprotein and NOTCH-HES signaling in Otx2 expression in embryonic retinal photoreceptor cell fate determination. J. Neurosci. 31, 16792-16807. 10.1523/JNEUROSCI.3109-11.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muranishi Y., Terada K. and Furukawa T. (2012). An essential role for Rax in retina and neuroendocrine system development. Dev. Growth Differ. 54, 341-348. 10.1111/j.1440-169X.2012.01337.x [DOI] [PubMed] [Google Scholar]
- Narkis G., Tzchori I., Cohen T., Holtz A., Wier E. and Westphal H. (2012). Isl1 and Ldb co-regulators of transcription are essential early determinants of mouse limb development. Dev. Dyn. 241, 787-791. 10.1002/dvdy.23761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida A., Furukawa A., Koike C., Tano Y., Aizawa S., Matsuo I. and Furukawa T. (2003). Otx2 homeobox gene controls retinal photoreceptor cell fate and pineal gland development. Nat. Neurosci. 6, 1255-1263. 10.1038/nn1155 [DOI] [PubMed] [Google Scholar]
- Oliver G., Mailhos A., Wehr R., Copeland N. G., Jenkins N. A. and Gruss P. (1995). Six3, a murine homologue of the sine oculis gene, demarcates the most anterior border of the developing neural plate and is expressed during eye development. Development 121, 4045-4055. [DOI] [PubMed] [Google Scholar]
- Ostendorff H. P., Peirano R. I., Peters M. A., Schluter A., Bossenz M., Scheffner M. and Bach I. (2002). Ubiquitination-dependent cofactor exchange on LIM homeodomain transcription factors. Nature 416, 99-103. 10.1038/416099a [DOI] [PubMed] [Google Scholar]
- Pan L., Deng M., Xie X. and Gan L. (2008). ISL1 and BRN3B co-regulate the differentiation of murine retinal ganglion cells. Development 135, 1981-1990. 10.1242/dev.010751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto do Ó. P., Richter K. and Carlsson L. (2002). Hematopoietic progenitor/stem cells immortalized by Lhx2 generate functional hematopoietic cells in vivo. Blood 99: 3939-3946. 10.1182/blood.V99.11.3939 [DOI] [PubMed] [Google Scholar]
- Poche R. A., Kwan K. M., Raven M. A., Furuta Y., Reese B. E. and Behringer R. R. (2007). Lim1 is essential for the correct laminar positioning of retinal horizontal cells. J. Neurosci. 27, 14099-14107. 10.1523/JNEUROSCI.4046-07.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter F. D., Drago J., Xu Y., Cheema S. S., Wassif C., Huang S. P., Lee E., Grinberg A., Massalas J. S., Bodine D. et al. (1997). Lhx2, a LIM homeobox gene, is required for eye, forebrain, and definitive erythrocyte development. Development 124, 2935-2944. [DOI] [PubMed] [Google Scholar]
- Rhee H., Polak L. and Fuchs E. (2006). Lhx2 maintains stem cell character in hair follicles. Science 312, 1946-1949. 10.1126/science.1128004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowan S. and Cepko C. L. (2004). Genetic analysis of the homeodomain transcription factor Chx10 in the retina using a novel multifunctional BAC transgenic mouse reporter. Dev. Biol. 271, 388-402. 10.1016/j.ydbio.2004.03.039 [DOI] [PubMed] [Google Scholar]
- Roy A., de Melo J., Chaturvedi D., Thein T., Cabrera-Socorro A., Houart C., Meyer G., Blackshaw S. and Tole S. (2013). LHX2 is necessary for the maintenance of optic identity and for the progression of optic morphogenesis. J. Neurosci. 33, 6877-6884. 10.1523/JNEUROSCI.4216-12.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sailaja B. S., Cohen-Carmon D., Zimmerman G., Soreq H. and Meshorer E. (2012). Stress-induced epigenetic transcriptional memory of acetylcholinesterase by HDAC4. Proc. Natl. Acad. Sci. USA 109, E3687-E3695. 10.1073/pnas.1209990110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaham O., Menuchin Y., Farhy C. and Ashery-Padan R. (2012). Pax6: a multi-level regulator of ocular development. Prog. Retin. Eye Res. 31, 351-376. 10.1016/j.preteyeres.2012.04.002 [DOI] [PubMed] [Google Scholar]
- Shirasaki R. and Pfaff S. L. (2002). Transcriptional codes and the control of neuronal identity. Annu. Rev. Neurosci. 25, 251-281. 10.1146/annurev.neuro.25.112701.142916 [DOI] [PubMed] [Google Scholar]
- Sigulinsky C. L., Green E. S., Clark A. M. and Levine E. M. (2008). Vsx2/Chx10 ensures the correct timing and magnitude of Hedgehog signaling in the mouse retina. Dev. Biol. 317, 560-575. 10.1016/j.ydbio.2008.02.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song M.-R., Sun Y., Bryson A., Gill G. N., Evans S. M. and Pfaff S. L. (2009). Islet-to-LMO stoichiometries control the function of transcription complexes that specify motor neuron and V2a interneuron identity. Development 136, 2923-2932. 10.1242/dev.037986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomita K., Ishibashi M., Nakahara K., Ang S.-L., Nakanishi S., Guillemot F. and Kageyama R. (1996). Mammalian hairy and Enhancer of split homolog 1 regulates differentiation of retinal neurons and is essential for eye morphogenesis. Neuron 16, 723-734. 10.1016/S0896-6273(00)80093-8 [DOI] [PubMed] [Google Scholar]
- Tzchori I., Day T. F., Carolan P. J., Zhao Y., Wassif C. A., Li L., Lewandoski M., Gorivodsky M., Love P. E., Porter F. D. et al. (2009). LIM homeobox transcription factors integrate signaling events that control three-dimensional limb patterning and growth. Development 136, 1375-1385. 10.1242/dev.026476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Meyel D. J., O'Keefe D. D., Jurata L. W., Thor S., Gill G. N. and Thomas J. B. (1999). Chip and apterous physically interact to form a functional complex during Drosophila development. Mol. Cell 4: 259-265. 10.1016/S1097-2765(00)80373-1 [DOI] [PubMed] [Google Scholar]
- van Meyel D. J., O'Keefe D. D., Thor S., Jurata L. W., Gill G. N. and Thomas J. B. (2000). Chip is an essential cofactor for apterous in the regulation of axon guidance in Drosophila. Development 127, 1823-1831. [DOI] [PubMed] [Google Scholar]
- Wall D. S., Mears A. J., McNeill B., Mazerolle C., Thurig S., Wang Y., Kageyama R. and Wallace V. A. (2009). Progenitor cell proliferation in the retina is dependent on Notch-independent Sonic hedgehog/Hes1 activity. J. Cell Biol. 184, 101-112. 10.1083/jcb.200805155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Dakubo G. D., Thurig S., Mazerolle C. J. and Wallace V. A. (2005). Retinal ganglion cell-derived sonic hedgehog locally controls proliferation and the timing of RGC development in the embryonic mouse retina. Development 132, 5103-5113. 10.1242/dev.02096 [DOI] [PubMed] [Google Scholar]
- Wässle H., Peichl L., Airaksinen M. S. and Meyer M. (1998). Calcium-binding proteins in the retina of a calbindin-null mutant mouse. Cell Tissue Res. 292, 211-218. 10.1007/s004410051052 [DOI] [PubMed] [Google Scholar]
- Weihe U., Milán M. and Cohen S. M. (2001). Regulation of Apterous activity in Drosophila wing development. Development 128, 4615-4622. [DOI] [PubMed] [Google Scholar]
- Wu F., Kaczynski T. J., Sethuramanujam S., Li R., Jain V., Slaughter M. and Mu X. (2015). Two transcription factors, Pou4f2 and Isl1, are sufficient to specify the retinal ganglion cell fate. Proc. Natl. Acad. Sci. USA 112, E1559-E1568. 10.1073/pnas.1421535112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaron O., Farhy C., Marquardt T., Applebury M. and Ashery-Padan R. (2006). Notch1 functions to suppress cone-photoreceptor fate specification in the developing mouse retina. Development 133, 1367-1378. 10.1242/dev.02311 [DOI] [PubMed] [Google Scholar]
- Yun S., Saijoh Y., Hirokawa K. E., Kopinke D., Murtaugh L. C., Monuki E. S. and Levine E. M. (2009). Lhx2 links the intrinsic and extrinsic factors that control optic cup formation. Development 136, 3895-3906. 10.1242/dev.041202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuber M. E. (2010). Eye field specification in Xenopus laevis. Curr. Top. Dev. Biol. 93, 29-60. 10.1016/B978-0-12-385044-7.00002-3 [DOI] [PubMed] [Google Scholar]
- Zuber M. E., Gestri G., Viczian A. S., Barsacchi G. and Harris W. A. (2003). Specification of the vertebrate eye by a network of eye field transcription factors. Development 130, 5155-5167. 10.1242/dev.00723 [DOI] [PubMed] [Google Scholar]