Summary
Methods to visualize, track, measure, and perturb or activate proteins in living cells are central to biomedical efforts to characterize and understand the spatial and temporal underpinnings of life inside cells. Although fluorescent proteins have proven to be extremely useful for in vivo studies of protein function, their utility is inherently limited because their spectral and structural characteristics are interdependent. These limitations have spurred the creation of alternative approaches for the chemical labeling of proteins. We describe in this protocol the use of fluorescence resonance emission transfer (FRET)-quenched DnaE split-inteins for the site-specific labeling and concomitant fluorescence activation of proteins in living cells. We have successfully employed this approach for the site-specific in-cell labeling of the DNA binding domain (DBD) of the transcription factor YY1 using several human cell lines. Moreover, we have shown that this approach can be also used for modifying proteins in order to control their cellular localization and potentially alter their biological activity.
Keywords: split-intein, protein trans-splicing, Npu intein, protein labeling, fluroescence
1. Introduction
Understanding the roles of specific proteins in cellular processes is a fundamental goal of molecular biology [1–3]. Methods to label and visualize proteins inside living cells are extremely useful in the study of localization, movement, interactions, and microenvironments of proteins in living cells. Although fluorescent proteins have revolutionized such studies, they have numerous shortcomings, which have spurred the creation of alternative approaches to chemically label proteins in living cells. These next generation approaches combine the genetic targeting capabilities of fluorescent proteins with the diversity and environmental sensitivity of fluorescent small molecules and/or other biophysical probes. Most of the available techniques, however, provide only limited temporal resolution for labeling of biomolecules in living cells.
Ideally these approaches should be modular, thus making possible the introduction of a wide variety of fluorophores or other type of biophysical probes. The kinetics of the labeling reaction should be fast enough to provide temporal resolution to satisfy the most time-sensitive biological assays. It should also allow spatial control during the in-cell labeling process. The labeling reaction should introduce minimal modifications on the target protein in order to preserve its original structure and biological function. Finally, it should make possible the simultaneous introduction of different probes onto multiple target proteins for simultaneous tracking purposes.
One of the most promising approaches for in-cell protein labeling involves the use of intein-mediated protein trans-splicing (Fig. 1) [4]. Protein trans-splicing is a naturally occurring post-translational modification similar to protein splicing with the difference being that the intein self-processing domain is split in two fragments, called N-intein (IN) and C-intein (IC), respectively [5, 6]. These two intein fragments are inactive individually, however, they can bind each other with high specificity under appropriate conditions to form a functional protein-splicing domain. Split mini-inteins have been widely used by our group and others for the site-specific modification of proteins in vitro [7–10] and in living cells [11–13]. In-cell labeling of proteins can be easily accomplished by expressing the protein of interest fused to the IN fragment. The second half of the split intein can be chemically synthesized to contain any chemical probe at the C-extein moiety, and then introduced into the cells by using peptide transducing domains (PTD) [11, 13].
Figure 1.
A. Site-specific labeling and fluorescence activation of a protein of interest (POI) by FRET-quenched protein trans-splicing. Key to this approach is the introduction of fluorescence quencher (Q) into the IC polypeptide, which blocks the fluorescence signal of the fluorophore (F) located at the C-terminus of the IC polypeptide before protein trans-splicing happens. When protein trans-splicing occurs the fluorophore is covalently attached to the C-terminus of the POI triggering its fluorescence. The use of this approach for in-cell modification and fluorescence tagging of proteins minimizes the fluorescence background from the unreacted IC polypeptide thus facilitating the optical tracking of the labeled protein inside the cell. B. Scheme showing the approach used for in-cell labeling of a POI with a fluorophore inside a live cell using protein trans-splicing (figure modified from [13]).
Intein-mediated labeling of proteins is highly modular allowing the covalent site-specific incorporation of a myriad of biophysical probes into proteins [8–10]. The kinetics of protein splicing is also relatively fast, with a number of split-inteins having reaction times in the order of several minutes [9, 14, 15]. Moreover, the recent development of conditional protein splicing, both through chemical and photochemical means, makes possible the chemical modification of proteins in living cells with temporal and spatial control [16–19].
One of the best-characterized naturally occurring split-inteins are α-subunit DNA polymerase III (DnaE) intein [20], with many known orthologs with high sequence homology in many cyanobacteria species (Fig. 2A) [14, 21]. The DnaE split-inteins are characterized for having IN and IC fragments with ≈120 and ≈30 residues, respectively. The relatively small size of the IC fragment facilitates its chemical synthesis thus allowing the use of synthetic IC fragments bearing different biophysical probes in the C-extein segment to be used for the chemical modification of proteins through protein trans-splicing [9, 13, 17, 18].
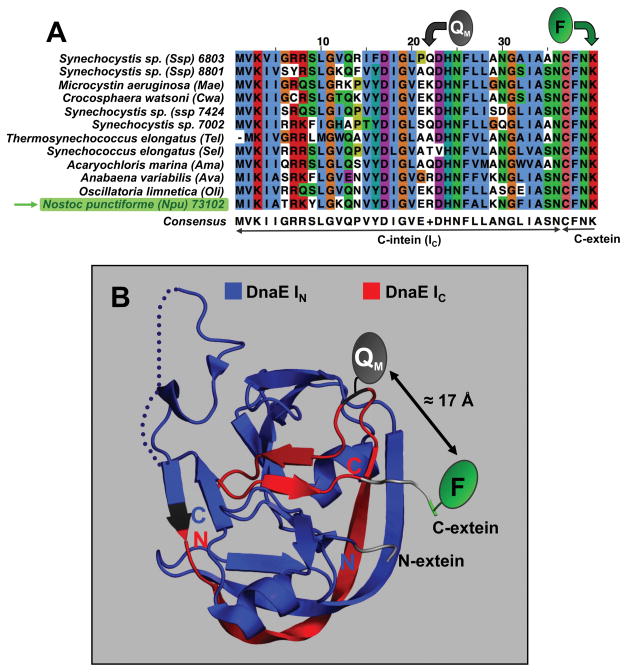
Figure 2.
Design of FRET-quenched DnaE split inteins. A. Multiple sequence alignment of the DnaE IC for different species indicating the positions used for the introduction of the quencher group in the IC polypeptide. Multiple sequence alignment was performed using T-Coffee and visualized using Jalview [26]. Molecular representations of the DnaE inteins were generated using the PyMol software package. B. Crystal structure of the Npu DnaE intein in the pre-spliced state (PDB code: 2KEQ) [27]. DnaE IC and IN are shown in red and blue respectively. The structural secondary elements are also shown. The position used to place the quencher and fluorophore groups at the IC and C-extein, as well as the distances are indicated (figure modified from [13]).
We will use in this protocol the Nostoc puntiforme PCC73102 (Npu) DnaE split-intein. This particular DnaE split-intein has one of the highest rate reported for protein trans-splicing (τ1/2 ≈ 60 s) [15] and a high splicing yield [15, 22]; and therefore is ideal for in-cell protein labeling purposes.
The use of protein trans-splicing for the site-specific labeling of proteins with fluorogenic dyes for in-cell tracking purposes requires that the labeling process must be linked to the simultaneous activation of fluorescence (Fig. 1). This can be accomplished by making use of fluorescence resonance emission transfer (FRET)-quenched DnaE split-inteins for the site-specific labeling and concomitant fluorescence activation of proteins in living cells. In this protocol we use fluorescein and dabcyl as fluorescence donor and FRET-quencher, respectively (Fig. 2), but any other combination of donor and quencher could be also used.
The fluorescein group is introduced at the C-terminus of the first four residues (Cys-Phe-Asn-Lys) of the C-extein, which are required for efficient trans-splicing (Fig. 2) [13] The dabcyl group (QM, Fig. 2) is introduced on residue 22 of the Npu DnaE IC polypeptide (peptide IC, Table 1). This position is in close proximity to the C-extein (≈17 Å, Fig. 2B) and provides an excellent FRET-quenching (>99% FRET-quenching) [13].
Table 1.
Sequence of the DnaE C-intein (IC) polypeptide used in this protocol. Standard single code letters are used for the peptides sequences. Single letter codes B and X stand for norleucine and 6-amino hexanoic acid, respectively. 5-(Iodoacetamide)-fluoresceine (IAF) is used to introduce fluorescein into specific Lys or Cys residues, respectively. Dabcyl, Cam, Cam-Fl stand for 4-dimethylaminoazobenzene-4’-carboxyl, carboxamidomethyl, and fluorescein-carboxamidomethyl, respectively. Residues in blue, magenta and yellow represent the Npu DnaE IC intein, Npu DnaE C-extein and NLS peptide, respectively. The quencher and fluorophore groups are in red and green respectively.
| Peptide Name | Compound | Sequence | Molecular Weight Found (Expecteda)/Da |
|---|---|---|---|
| Npu QM-IC-NLS-Fl | IC |
|
6449.0±0.1 (6447.6) |
Average molecular weight
In this chapter we describe the protocol for in-cell C-terminal labeling of the DNA binding domain (DBD) of the transcription factor Yin Yang 1 (YY1) in live U2OS and HeLa cells. YY1 is a ubiquitously distributed multifunctional transcription factor belonging to the GLI-Kruppel class of zinc finger proteins [23]. The protein is involved in repressing and activating a diverse number of promoters including negative regulation of p53, thus making it of particular interest [24, 25]. In the example described in this chapter the DBD of YY1 was labeled at its C-terminal with a fluorophore (fluorescein) and a nuclear localization (NLS) signal peptide to demonstrate the potential of this technique to control the localization of and biological function of a protein/protein domain. The protocol described uses humans U2OS cells but it could be easily adapted to any other mammalian cell line.
It is important to note that this approach is highly modular and can be used for in-cell labeling of proteins with other biophysical probes or peptide sequences required for activity. In addition, the use of different orthogonal split inteins should also make possible the simultaneous labeling of multiple proteins with different probes.
Before performing the labeling experiment in-cell, it is advisable to test the labeling reaction in vitro first. This protocol also provides instructions on how to evaluate labeling by protein trans-splicing in vitro.
2. Materials
All solutions were prepared using ultrapure water with a resistivity of 18 MΩ × cm at 25° C and analytical grade reagents. All reagents and solutions were stored at room temperature unless indicated otherwise.
2.1 Instruments
Water bath able to operate at 42° C and 94°C.
Table-top micro centrifuge capable of operating at 14,000 rpm.
Microbiology incubator set at 37°C.
Temperature controlled incubator Shaker.
Orbital shaker.
Polymerase chain reaction thermocycler.
Agarose gel electrophoresis unit.
Electrophoresis power pack able to operate up to 250 V.
UV-visible spectrophotometer.
Sonicator to lyse E. coli cells.
High speed centrifuge (e.g. Sorvall RC 5C Plus, Thermo Fisher scientific).
SDS-PAGE electrophoresis apparatus.
Centrifuge tubes of 0.5 mL, 1.5 mL, 15 mL and 30 mL of capacity.
5 ml Polypropylene Columns.
Class II, type A2 biosafety cabinets.
Gel and Blot Imaging System.
Fluorescence microscope.
CO2 incubator.
2.2 Cloning of YY1-IN construct
Synthetic DNA primers used to amplify genes encoding Npu DnaE IN and DBD-YY1 (20 nmol scale, HPLC purified) (Table 2).
Genomic DNA from Nostoc punctiforme strain ATCC 29133/PCC 73102 (obtained from ATCC).
DNA encoding human YY1 proteins (cDNA clone IMAGE: 5261384) (can be obtained from many sources, eg. Genscript, USA).
TE buffer: 10 mM Tris-HCl, 1 mM EDTA, pH 8.0.
Vent DNA polymerase, TaqDNA polymerase, dNTPs solution, 10X thermopol PCR buffer, 10X Taq DNA polymerase buffer.
Restriction enzymes Not I, Nde I, BamH I, Kpn I and Sal I.
PCR Purification Kit (e.g. QIAquick from QIAGEN).
Miniprep Kit (e.g. QIAprep from QIAGEN).
Gel Extraction Kit (e.g. QIAquick from QIAGEN).
Chemical competent DH5α cells.
Expression plasmid pET28a (Novagen-EMD Millipore).
Mammalian Expression vector pcDNA4/TO/myc-His (Invitrogen).
T4 DNA ligase and T4 DNA ligase buffer.
Ampicillin stock solution: 100 mg ampicillin/mL in pure H2O, sterilized by filtration. Store in 1 mL aliquots at −20° C.
Kanamycin stock solution: 25 mg kanamycin/mL in pure H2O, sterilized by filtration. Store in 1 mL aliquots at −20° C.
Chloramphenicol stock solution: 34 mg chloramphenicol/mL in EtOH. Store in 1 mL aliquots at −20° C.
LB medium: 25 g of LB broth was dissolved in 1 L of pure H2O and sterilized by autoclaving at 120° C for 30 min.
LB medium-agar: 3.3 g of LB agar was suspended in 100 mL of pure H2O and sterilized by autoclaving at 120° C for 30 min. To prepare plates, allow LB medium-agar to cool to ≈50° C, then add 0.1 mL of antibiotic stock solution.
SOC Medium: 20 g of tryptone, 5 g yeast extract, 0.5 g NaCl and 0.186 g KCl were suspended into 980 ml of pure water and sterilized by autoclaving at 120° C for 30 min. Dissolve 4.8 g MgSO4, 3.603 g dextrose in 20 mL of pure H2O and filter sterilize over a 45 μm filter and add to the autoclaved medium.
Table 2.
DNA oligonucleotides used to generate the dsDNA encoding the YY1-IN and DnaE IN.
| Primer Name | Nucleotide Sequence |
|---|---|
| p5-YY1 | 5′-AAA GAA GAT CAT ATG CCA AGA ACA ATA GCT TGC CCT C-3′ |
| p3-YY1 | 5′-C TTC GGA TCC CTG GTT GTT TTT GGC CTTA GC-3′ |
|
| |
| p5-IN | 5′-CTA GTC GAC AAG CTT TTA AGT TTG CGG AAT ATT GTT TAA G CTA TG -3′ |
| p3-IC | 5′-TTT GCG GCC GCT TAA TTC GGC AAA TTA TCA ACC CGC AT -3′ |
|
| |
| p5-YY1-IN | 5′-AGG GGT ACC ACC ATG GGC AGC AGC -3′ |
| p3-YY1-IN | 5′-GT GGT GCT CGA GTG CGG CCG CA -3′ |
2.3 Bacterial expression YY1-IN construct
Chemical competent BL21 (DE3) and Origami2 (DE3) cells (EMD Millipore).
Isopropyl-thio-β-D-galactopyranoside (IPTG): Prepare a stock solution of 1 M analytical grade IPTG in H2O and sterilize by filtration over 45 μm filter. Store at −20° C.
Lysis buffer: 0.1 mM EDTA, 25 mM sodium phosphate, 150 mM NaCl, 10% (v/v) glycerol, pH 7.4.
Phosphate buffer saline (PBS): 25 mM sodium phosphate, 150 mM NaCl, pH 7.4
100 mM phenylmethylsulphonyl fluoride (PMSF) in EtOH (better to prepare fresh before use).
4X SDS-PAGE sample buffer: 1.5 mL of 1 M Tris-HCl buffer at pH 6.8, 3 mL of 1 M DTT (dithiothreitol) in pure H2O, 0.6 g of sodium dodecyl sulfate (SDS), 30 mg of bromophenol blue, 2.4 mL of glycerol, bring final volume to 7.5 mL.
SDS-PAGE sample buffer: dilute 4 times 4×SDS-PAGE sample buffer in pure H2O and add 20% (v/v) 2-mercaptoethanol. Prepare fresh.
SDS-4–20% PAGE gels, 1X SDS running buffer.
Gel stain: Coomassie brilliant blue or Gelcode® Blue (Thermo scientific, USA) or silver stain kit.
2.4 In-vitro trans-splicing
Pure labeled DnaE IC polypeptide shown in Table 1. The synthetic peptides used in this study were generated in-house but they could be ordered from any chemical supplier specialized in providing synthetic peptides.
Trans-splicing buffer: 0.5 mM EDTA, 1 mM tris-(2-carboxyethyl)phosphine (TCEP), 50 mM NaH2PO4, 250 mM NaCl, pH 7.0 (Note 1).
2.5 In-cell trans-splicing reaction
U2OS cells, available from ATCC (ATCC® HTB-96™).
HeLa cells, available from ATCC (ATCC® CCL-2™).
Dulbecco’s modified eagle medium (DMEM) containing 10% heat inactivated FBS (Fetal Bovine Serum), 1% L-glutamine and 1% penicillin-streptomycin solution.
RIPA BUFFER: 50 mM Tris-HCl, 150 mM NaCl buffer, pH 8.0, 1% NP-40, 0.5% sodium deoxycholate, 1% Triton X-100.
TBST buffer: 50 mM Tris-HCl, 150 mM NaCl, pH 7.6.
Nonpyrogenic sterile 100 × 20 mm polystrene plates.
Nonpyrogenic sterile 35 mm glass bottom plates (e.g. MatTek).
Transfection reagent for mammalian cells (e.g. Fugene-6 from Promega or equivalent).
Complete protease cocktail (e.g. Thermo Scientific).
PVDF membrane for western blotting.
5% skim milk in TBST buffer.
Anti-His Antibody (e.g. murine IgG). Store at −20° C.
Secondary antibody (e.g. horseradish peroxidase-conjugated anti-murine IgG, Vector Lab).
ECL kit (e.g. Life Technologies).
Chariot protein delivery reagent (Active Motif).
3. Methods
3.1 Cloning of DNA encoding Npu DnaE IN into expression vector pET28a(+)
Amplify by PCR the gene containing the Npu DnaE IN (residues 770–876, UniProtKB: B2J066) using a plasmid containing the DnaE gene from Nostoc punctiforme (Strain ATCC 2913/PCC 73102) as template using primers p5-IN and p3-IN (Table 2). The 5′-primer encodes a Sal I restriction site. The 3′-primer introduces a Not I restriction site and stop codon. Carry out the PCR reaction as follows: 40 μL sterile pure H2O, 1 μL of DNA template (≈10 ng/μL), 5 μL of 10× thermopol reaction buffer, 1.0 μL of dNTP solution (10 mM each), 1 μL of p5-IN primer solution (0.2 μM), 1 μL of p3-IN r primer solution (0.2 μM), and 1 μL Vent DNA polymerase (2 units). PCR cycle conditions used: initial denaturation at 94° C for 5 min followed by 30 cycles (94° C denaturation for 30 s, annealing at 52° C for 45 s, and extension at 72° C for 60 s) and final extension at 72° C for 10 min.
Purify the PCR amplified fragment encoding Npu Dna IN using a PCR purification kit following the manufacturer instructions and quantify it by UV absorption (for a 1-cm pathlength, an optical density at 260 nm (OD260) of 1.0 equals to a concentration of 50 μg/mL solution of dsDNA).
Digest plasmid pET28a(+) (Novagen-EMD Millipore) and PCR-amplified gene encoding the DnaE IN polypeptide with restriction enzymes Sal I and Not I. Use a 0.5 mL centrifuge tube and add 5 μL of 10× restriction buffer (e.g. NEB buffer 2.1 from New England Biolabs), add enough pure sterile water to have a final volume reaction of 50 μL, add ≈10 μg of the corresponding dsDNA to be digested and finally add 1 μL (20 units) of restriction enzyme Not I. Incubate at 37° C for 3 h. Then, add 1 μL (20 units) of restriction enzyme Sal I to the same tube and incubate at 37° C for 1 h.
Purify the double digested PCR-product and pET28a plasmid by agarose (0.8% and 2% agarose gels for pET28a(+) and PCR product should be used, respectively) gel electrophoresis. Cut out the bands corresponding to the double digested DNA and purify the DNA using a gel extraction kit. Elute DNA from spin columns with TE buffer and quantify using UV.
Ligate double digested pET28a(+) and PCR-product encoding DnaE IN. Use a 0.5 mL centrifuge tube, add ≈ 100 ng of Sal I, Not I-digested pET28a, ≈ 50 ng of Sal I, Not I-digested PCR-amplified DNA encoding DnaE IN, enough pure sterile H2O to make a final reaction volume of 20 μL, 2 μL of 10X T4 DNA ligase buffer, 1 μL of 10 mM ATP and 1 μL (400 units) T4 DNA ligase. Incubate at 16° C overnight.
Transform the ligation mixture into DH5α competent cells. ≈100 μL of chemical competent cells are thawed on ice and mixed with the ligation mixture (20 μL) for 30 min. Heat-shock the cells are heat-shocked at 42° C for 45 s and then keep on ice for an extra 10 min. Add 900 μL of SOC medium and incubate at 37° C for 1 h in an orbital shaker. Plate 100 μL on LB agar plate containing kanamycin (25 μg/mL) and incubate the plate at 37° C overnight.
Pick up several colonies (most of the times 5 colonies should be enough) and inoculate into 5 ml of LB medium containing kanamycin (25 μg/mL). Incubate tubes at 37° C overnight in an orbital shaker.
Pellet down cells and extract DNA using a miniprep kit following the manufacturer protocol and quantify plasmid using UV spectroscopy.
Verify the presence of DNA encoding DnaE IN in each colony using PCR and the same conditions described in step 1 of this section.
Screen colonies containing DNA encoding DnaE IN for protein expression (Steps 11 through 17).
Transform chemical competent BL21 (DE3) cells with plasmids containing the DNA encoding DnaE IN (Note 2). Transformed cells are plated on LB plate containing kanamycin (25 μg/mL) and incubated at 37° C overnight.
Resuspend the colonies from 1 plate in 1 mL of LB and inoculate 100 mL of LB containing kanamycin (25 μg/mL) in a 250 mL flask.
Grow cells in an orbital shaker incubator at 37° C for 2–3 h to reach mid-log phase (OD at 600 ≈ nm 0.5). Add IPTG to reach a final concentration of 1 mM and incubate cells for 3 h at 37° C.
Pellet 1 mL of cells by centrifugation at 6,000 × g for 15 min at 4° C.
Discard the supernatant and resuspend pellets in fresh SDS-PAGE sample buffer.
Heat samples at 94 °C for 5 min and separate the soluble cell lysate fraction by centrifugation at 15,000 × g for 20 min at 4° C
Analyze the soluble fraction by SDS-PAGE analysis to estimate the protein expression of the DnaE IN intein in each clone analyzed. The DnaE IN polypeptide should give a band around 15 kDa.
3.2 Cloning of DNA encoding YY1-IN construct into expression vector pET28a
Amplify the gene containing the DNA binding domain of YY1 by PCR using the cDNA for human YY1 (cDNA clone IMAGE: 5261384) as template using primers p5-YY1 and p3-YY1 (Table 2). The 5′-primer and 3′-primer introduce Nde I and BamH I restriction sites, respectively. Carry out the PCR reaction as follows: 40 μL sterile pure H2O, 1 μL of DNA template (≈10 ng/μL), 5 μL of 10× thermopol reaction buffer, 1.0 μL of dNTP solution (10 mM each), 1 μL of p5-YY1 primer solution (0.2 μM), 1 μL of p3-YY1 primer solution (0.2 μM), and 1 μL Vent DNA polymerase (2 units). Use the following PCR cycle conditions: initial denaturation at 94° C for 5 min followed by 30 cycles (94° C denaturation for 30 s, annealing at 52° C for 45 s, and extension at 72° C for 60 s) and final extension at 72° C for 10 min.
Purify the PCR amplified fragment encoding YY1 using a PCR purification kit following the manufacturer instructions and quantify by UV spectroscopy.
Digest plasmid pET28a encoding DnaE IN (pET-IN) (obtained in section 3.1) and PCR-amplified gene encoding YY1 construct with restriction enzymes Nde I and BamH I. Use a 0.5 mL centrifuge tube and add 5 μL of 10× restriction buffer (e.g. NEB buffer 3.1 from New England Biolabs), add enough pure sterile water to have a final volume reaction of 50 μL, add ≈10 μg of the corresponding dsDNA to be digested and finally add 1 μL (20 units) of restriction enzyme Nde I. Incubate at 37° C for 3 h. Then, add 1 μL (20 units) of restriction enzyme BamH I to the same tube and incubate at 37° C for 1 h.
Purify the double digested PCR-product and pET-IN plasmid by agarose (0.8% and 2% agarose gels for pET-IN and PCR product should be used, respectively) gel electrophoresis. Cut out the bands corresponding to the double digested DNA and purify them using a gel extraction kit. Elute DNA from the spin columns with TE buffer and quantify using UV spectroscopy.
Ligate double digested pET-IN and PCR-product encoding the DBD of YY1. Use a 0.5 mL centrifuge tube, add ≈100 ng of Nde I, BamH I-digested pET-IN, ≈50 ng of Nde I, BamH I-digested PCR-amplified DNA encoding the DBD of YY1, enough pure sterile H2O to make a final reaction volume of 20 μL, 2 μL of 10X T4 DNA ligase buffer, 1 μL of 10 mM ATP and 1 μL (400 units) T4 DNA ligase. Incubate at 16° C overnight.
Transform the ligation mixture into DH5α competent cells, plate them on a LB agar plate containing kanamycin (25 μg/mL), and incubate the plate at 37° C overnight as described previously (Section 3.1, steps 5 and 6).
Pick up several colonies (most of the times 5 colonies should be enough) and screen for the presence of DNA inserts encoding the DBD of YY1 by PCR as described previously (Section 3.1, steps 7 through 9).
Screen colonies containing DNA encoding the YY1-IN construct for protein expression (Steps 9 through 12).
Transform chemical competent Rosetta (DE3) cells with plasmids containing the DNA encoding YY1-IN (Note 3). Transformed cells are plated on LB plate containing kanamycin (25 μg/mL) and and chloramphenicol (34 μg/mL) incubated at 37° C.
Resuspend the colonies from 1 plate in 1 mL of LB and inoculate 100 mL of LB containing kanamycin (25 μg/mL) and chloramphenicol (34 μg/mL) in a 250 mL flask.
Grow cells in an orbital shaker incubator at 37° C for 2–3 h to reach mid-log phase (OD at 600 nm ≈ 0.5). Add IPTG to reach a final concentration of 1 mM and incubate cells for 3 h at 37° C.
Pellet, lyse cells and analyze protein expression level of YY1-IN construct by SDS-PAGE as described in sections 3.1.14 through 3.1.17. The YY1-IN construct should give a band around 30 kDa.
3.3 Bacterial expression of the YY1-IN construct
Transform chemical competent Rosetta (DE3) cells with plasmid containing the DNA encoding YY1-IN (plasmid pET-YY1-IN). Transformed cells are plated on LB plate containing kanamycin (25 μg/mL) and chloramphenicol (34 μg/mL) and incubated at 37° C overnight (Note 4).
Resuspend the colonies from 2 plates in 2 mL of LB and inoculate 1 L of LB containing kanamycin (25 μg/mL) and chloramphenicol (34 μg/mL) in a 2.5 L flask.
Grow cells in an orbital shaker incubator at 37° C for 2–3 h to reach mid-log phase (OD at 600 nm ≈ 0.5). Add IPTG to reach a final concentration of 1 mM and incubate cells for 3 h at 37° C.
Pellet cells by centrifugation at 6,000 × g for 15 min at 4° C. Discard the supernatant and process the pellet immediately (Note 5).
3.4 Purification of YY1-IN
Protein YY1-IN was purified from inclusion bodies. Resuspend cell pellet with 30 mL of lysis buffer containing 1 mM PMSF. Lyse cells by sonication on ice using 25 s bursts spaced 30 s each (Note 6). Repeat the cycle six times (Note 7).
Separate the soluble cell lysate fraction by centrifugation at 15,000 × g for 20 min at 4° C. Store the pellets at −80° C in case they need to be re-processed.
Resuspend the insoluble fraction with lysis buffer (30 mL) containing detergent (0.1% Triton X-100). Separate soluble fraction by centrifugation at 15,000 × g for 20 min at 4° C. Repeat this process 2 more times.
Wash the insoluble fraction with lysis buffer without detergent as described in step 3.
Extract YY1-IN with 10 mL of PBS containing 4 M urea (Note 8).
Characterize protein purity by SDS-PAGE analysis and determine concentration by UV spectroscopy (ε280 = 17,180 M−1 cm−1). Use immediately for in vitro trans-splicing experiments.
3.5 In vitro labeling of YY1 using protein trans-splicing
Pure polypeptide IC (Table 1) and YY1-IN fusion protein are combined in freshly prepared and degassed trans-splicing buffer to a concentration of ≈ 1 μM and 0.1 μM, respectively. Dissolve IC peptide in pure 30% acetonitrile in pure H2O containing 0.1% trifluoroacetic acid (TFA) to obtain a stock solution of ≈ 1 mg/mL (≈160 μM). Add 3.1 μL of IC stock solution and the appropriate amount of YY1-IN dissolved in lysis buffer containing 4 M urea (Section 3.4, step 6) to 500 μL of fresh trans-splicing buffer. Keep the reaction at room temperature with occasional gently shaking.
Take aliquots (50 μL) at different times (1, 3, 7, 10, 15, 30, and 45 min) and rapidly quench them by adding 15 μL of 4X SDS-PAGE sample buffer and heating them at 94° C for 2 min.
Monitor the progression of the trans-splicing reaction by using SDS-PAGE (Fig. 3). Load 25 μL of each time point onto an SDS-4–20% PAGE gel. Run the samples at 125 V for about 1 h and 30 min in 1X SDS running buffer. Remove SDS with pure water. Visualize trans-splice reaction by epifluorescence (e.g. Storm 860 Molecular Imager), and silver stain kit (e.g. Thermo scientific, USA) following manufacturer protocol (Fig. 3) (Note 9).
Figure 3.
SDS-PAGE analysis of the in vitro protein trans-splicing/labeling reaction between FRET-quenched DnaE IC (Table 1) and YY1-IN. Protein detection was performed by silver staining (top) and epifluorescence (bottom). TS = trans-splicing.
3.6 Cloning of DNA encoding YY1-IN construct into mammalian expression vector pcDNA4/TO/myc-His
-
1
Amplify the DNA encoding the DBD of YY1 fused to the N-terminus of the DnaE IN (YY1-IN) by PCR using plasmid pET-YY1-IN (Section 3.2) as template using primers p5-YY1-IN and p3-YY1-IN (Table 2). The 5′-primer and 3′-primer introduce Kpn I and Not I restriction sites, respectively. Carry out the PCR reaction as follows: 40 μL sterile pure H2O, 1 μL of DNA template (≈10 ng/μL), 5 μL of 10X thermopol reaction buffer, 1.0 μL of dNTP solution (10 mM each), 1 μL of p5-YY1-IN primer solution (0.2 μM), 1 μL of p3-YY1-IN primer solution (0.2 μM), and 1 μL Vent DNA polymerase (2 units). Use the following PCR cycle conditions: initial denaturation at 94° C for 5 min followed by 30 cycles (94° C denaturation for 30 s, annealing at 52° C for 45 s, and extension at 72° C for 60 s) and final extension at 72° C for 10 min.
-
2
Purify the PCR amplified fragment encoding the YY1-IN fusion protein using the a PCR purification kit following the manufacturer instructions and quantify it by UV-visible spectroscopy.
-
3
Digest plasmid pcDNA4/TO/myc-His (Invitrogen) and PCR-amplified gene encoding YY1-IN with restriction enzymes Not I and Kpn I. Use a 0.5 mL centrifuge tube and add 5 μL of 10× restriction buffer (e.g. NEB buffer 2.1 from New England Biolabs), add enough pure sterile water to have a final volume reaction of 50 μL, add ≈10 μg of the corresponding dsDNA to be digested, add 1 μL (20 units) of restriction enzyme Not I and 1 μL (20 units) of restriction enzyme Kpn I. Incubate at 37° C for 16 h.
-
4
Purify the double digested PCR-product and pcDNA4 plasmid by agarose (0.8% and 2% agarose gels for pCDNA4 and PCR product should be used, respectively) gel electrophoresis. Cut out the bands corresponding to the double digested DNA and purify using a gel extraction kit. Elute DNA from the spin columns with TE buffer and quantify using UV-visible spectroscopy.
-
5
Ligate double digested pcDNA4 and PCR-product encoding YY1-IN. Use a 0.5 mL centrifuge tube, add ≈ 100 ng of Kpn I, Not I-digested pcDNA4, ≈ 50 ng of Kpn I, Not I-digested PCR-amplified DNA encoding YY1-IN, enough pure sterile H2O to make a final reaction volume of 20 μL, 2 μL of 10X T4 DNA ligase buffer, 1 μL of 10 mM ATP and 1 μL (400 units) T4 DNA ligase. Incubate at 16° C overnight.
-
6
Transform the ligation mixture into DH5α competent cells. Thaw ≈ 100 μL of chemical competent cells on ice and mix with the ligation mixture (20 μL) for 30 min. Heat-shock the cells at 42° C for 45 s and then keep on ice for an extra 10 min. Add 900 μL of SOC medium and incubate at 37° C for 1 h in an orbital shaker. Plate 100 μL on LB agar plate containing ampicillin (100 μg/mL) and incubate the plate at 37° C overnight.
-
7
Pick up several colonies (most of the times 5 colonies should be enough) and inoculate into 5 mL of LB medium containing ampicillin (100 μg/mL). Incubate tubes at 37° C overnight in an orbital shaker.
-
6
Pellet down cells and extract DNA using a miniprep kit following the manufacturer protocol and quantify plasmid using UV-visible spectroscopy.
-
7
Verify the presence of DNA encoding YY1-IN construct in each colony using PCR. Carry out the PCR reaction out as follows: 40 μL sterile pure H2O, 1 μL of plasmid DNA (≈50 ng/μL), 5 μL of 10× TaqDNA polymerase buffer, 1.0 μL of dNTP solution (10 mM each), 1 μL primer p5-YY1-IN solution (0.2 μM), 1 μL of primer p3-YY1-IN (0.2 μM), and 1 μL Taq DNA polymerase (5 units).
-
8
Use the following PCR cycle conditions: initial denaturation at 94° C for 5 min followed by 30 cycles (94° C denaturation for 30 s, annealing at 56° C for 45 s, and extension at 72° C for 60 s) and final extension at 72° C for 10 min.
3.7 Expression of YY1-IN fusion protein in U2OS and HeLa cells
Grow U2OS and HeLa cells in DMEM containing 10% heat inactivated FBS, 1% L-glutamine and 1% penicillin-Streptomycin solution in a humidified incubator under 5% CO2 atmosphere at 37 °C.
The transfection conditions and expression conditions should be optimized for each protein and cell line. Grow U2OS and HeLa cells in 100 mm nonpyrogenic sterile polystrene plates to ≈60% confluency.
Transiently transfect the cells with plasmid pcDNA4-YY1-IN (1 μg DNA) using transfection reagent (e.g. Fugene-6) following the manual instructions. The transfected cells are then incubated for 24 h at 37° C.
Expression of YY1-IN protein is estimated by western-blot analysis (steps 5–14).
Wash cells twice with PBS. Discard PBS wash. Resuspend cells in 0.2 mL RIPA buffer in the presence of 0.1% SDS, 1 mM PMSF and complete protease cocktail. Use a cell scrapper to dislodge cells.
Transfer the cell suspension to 1.5 mL centrifuge tubes and incubate on ice for 10 min.
Separate the insoluble and soluble fractions by centrifugation at 14,000 rpm in a microcentrifuge at 4° C for 30 min.
Incubate 100 μL of cell lysate with 20 μL of 4X SDS-PAGE sample buffer containing 20% mercaptoethanol. Heat the sample at 94°C for 2 min.
Analyze the cell lysate samples using SDS-PAGE. Load samples (25 μL) onto an SDS-4–20% PAGE gel. Run the samples at 125 V for about 1 h and 30 min in 1X SDS running buffer.
Transfer proteins from PAGE gel to a PVDF membrane using standard protocol.
Block membrane with 5% skim milk in TBST buffer for 1 h at room temperature. Add primary anti-His antibody (e.g. murine anti-His IgG, Invitrogen) (Note 10) in TBST buffer containing 5% skim milk and incubate overnight at 4°C on an orbital shaker.
Wash the membrane 3 times with TBST buffer for 5 min. Add secondary antibody (e.g. horseradish peroxidase-cojugated anti-murine IgG, Vector Lab) in TBST buffer containing 5% skim, and incubate at room temperature for 1 h.
Wash membrane 3 times with TBST buffer for 5 min. Develop membrane with ECL mix (Life Technologies) following manufacturer instructions.
Visualize bands using a molecular imager system (e.g. Storm 860, Amersham Biosciences) or using an X-ray film (Fig. 4)
Figure 4.
Expression of protein YY1-IN in U2OS (and HeLa) cells. Cells were collected after 24 h post-transfection, lysed and the soluble fraction analyzed and quantified by western blot using an anti-His antibody.
3.8 In-cell labeling using protein trans-splicing
-
1
Grow U2OS and HeLa cells in either 35 mm glass bottom plates (when fluorescence microscopy is required) or 100 mm nonpyrogenic sterile polystrene plates to 40–50% confluency (see 3.7.1–3.7.3) and then transiently transfected with plasmid pcDNA4-YY1-IN (1 μg DNA) using a transfection reagent (e.g. Fugene-6) following the manual instructions. Incubate transfected cells for 24 h at 37° C.
-
2
Wash cells with PBS. Transfect peptide IC (50 nM) using the Chariot protein delivery reagent (Active Motif) for 1 h as described in the manufacturer manual (Note 11).
-
3
Wash transfected cells with full media 2 times and incubate for 1 h at 37° C.
-
4
Wash cells three times with serum free media and observe fluorescence-labeled proteins in live cells using a fluorescence microscope (Fig. 5A and 5B) (Note 12).
-
4
Lyse cells using RIPA buffer and analyze soluble cell lysate by SDS-PAGE as described previously (Section 3.7, Steps 5 through 9).
-
5
Visualize fluoresceine-labeled protein on PAGE-gel using a molecular imaging system (Fig. 5C).
-
5
Use the same PAGE-gel to analyze the efficiency of the labeling reaction by Western blotting as described previously (Section 3.7, steps 10 through 14) (Fig. 5C).
Figure 5.
In-cell site-specific labeling of YY1 DBD with a nuclear localization signal and concomitant fluorescence activation using protein trans-splicing. A. U2OS (and HeLa) cells were first transiently transfected with a plasmid encoding YY1-IN and with DnaE IC polypeptides as described. Cells were then extensively washed and examined by fluorescence microscopy. B. Magnification of cells after 18 h of incubation showing the migration of the labeled-YY1 to the nuclear compartment. C. Quantification of labeling yield for in-cell trans-splicing reaction. Identification of labeled YY1 DBD protein and quantification of in-cell trans-splicing yield was performed by western blot (right panel) and epifluorescence (left panel), respectively. Bar represents 25 μm in panels A and B. TS = trans-splicing.
Footnotes
The buffer should be degassed before adding the TCEP. The buffer should be prepared fresh every time and not stored as TCEP has a limited stability at pH 7.0 in the presence of air.
We recommend to screen at least 3–4 different colonies for protein expression in BL21(DE3) cells to make sure that the polypeptide encoding DnaE IN is expressed efficiently.
We recommend to screen at least 3–4 different colonies for protein expression in Rosetta(DE3) cells to make sure that the clone with highest expression yield is selected. Expression in Rosetta cells is recommended when the protein of interest (POI) contains rare codons in E. coli. Select the clone with the highest expression yield and proceed to the next section.
When plating the transformed cells with pET-YY1-IN, it is better to aim for plates containing 200–300 colonies.
Cell pellets can be stored at −80° C for no more than 2–3 weeks before being processed.
During sonication, be sure the temperature of the sample does not overheat.
A french press can be also used to lyse cells, depending on the availability.
Use the highest quality urea available. PBS containing urea should be prepared fresh and not stored for more than 1 day, as urea is not stable in buffers at pH above 7.
It is extremely import to check that the labeling/trans-splicing reaction works efficiently in vitro before trying it in live cells.
In this particular work the N-terminal of the DBD of YY1 contained a His-tag and therefore an anti-His IgG was used to detect the protein by Western blotting.
Peptide transfection should be optimized for any particular case. In our hands, the best peptide transfection efficiency was obtained for 50 nM IC using a molecular ratio IC: Pep-1 of 1:20. Pep-1 is the name of the peptide used in the Chariot transfection system and its sequence is Ac-KETWWETWWTEWSQPKKKRKV-cysteamine. This peptide can be also obtained from different commercial sources (e.g. Anaspec).
The use of a FRET-quenched IC polypeptide allows concomitant labeling of the protein with the corresponding fluorophore and activation of its fluorescence.
References
- 1.Xie XS, Yu J, Yang WY. Living cells as test tubes. Science. 2006;312:228–230. doi: 10.1126/science.1127566. [DOI] [PubMed] [Google Scholar]
- 2.Chen I, Ting AY. Site-specific labeling of proteins with small molecules in live cells. Curr Opin Biotechnol. 2005;16:35–40. doi: 10.1016/j.copbio.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 3.Miller LW, Cornish VW. Selective chemical labeling of proteins in living cells. Curr Opin Chem Biol. 2005;9:56–61. doi: 10.1016/j.cbpa.2004.12.007. [DOI] [PubMed] [Google Scholar]
- 4.Perler FB. Protein splicing mechanisms and applications. IUBMB Life. 2005;57:469–476. doi: 10.1080/15216540500163343. [DOI] [PubMed] [Google Scholar]
- 5.Saleh L, Perler FB. Protein splicing in cis and in trans. Chem Rec. 2006;6:183–193. doi: 10.1002/tcr.20082. [DOI] [PubMed] [Google Scholar]
- 6.Xu MQ, Evans TC., Jr Recent advances in protein splicing: manipulating proteins in vitro and in vivo. Curr Opin Biotechnol. 2005;16:440–446. doi: 10.1016/j.copbio.2005.06.012. [DOI] [PubMed] [Google Scholar]
- 7.Muir TW. Semisynthesis of proteins by expressed protein ligation. Annu Rev Biochem. 2003;72:249–289. doi: 10.1146/annurev.biochem.72.121801.161900. [DOI] [PubMed] [Google Scholar]
- 8.Shi J, Muir TW. Development of a tandem protein trans-splicing system based on native and engineered split inteins. J Am Chem Soc. 2005;127:6198–6206. doi: 10.1021/ja042287w. [DOI] [PubMed] [Google Scholar]
- 9.Kwon Y, Coleman MA, Camarero JA. Selective Immobilization of Proteins onto Solid Supports through Split-Intein-Mediated Protein Trans-Splicing. Angew Chem Int Ed. 2006;45:1726–1729. doi: 10.1002/anie.200503475. [DOI] [PubMed] [Google Scholar]
- 10.Kurpiers T, Mootz HD. Site-specific chemical modification of proteins with a prelabelled cysteine tag using the artificially split Mxe GyrA intein. Chembiochem. 2008;9:2317–2325. doi: 10.1002/cbic.200800319. [DOI] [PubMed] [Google Scholar]
- 11.Giriat I, Muir TW. Protein semi-synthesis in living cells. J Am Chem Soc. 2003;125:7180–7181. doi: 10.1021/ja034736i. [DOI] [PubMed] [Google Scholar]
- 12.Woo YH, Stubbs L, Camarero JA. In vivo protein labeling via protein trans-splicing. The 21st Annual Symposium of the Protein Society; San Diego. 2008. p. 32. [Google Scholar]
- 13.Borra R, Dong D, Elnagar AY, Woldemariam GA, Camarero JA. In-cell fluorescence activation and labeling of proteins mediated by FRET-quenched split inteins. J Am Chem Soc. 2012;134:6344–6353. doi: 10.1021/ja300209u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dassa B, Amitai G, Caspi J, Schueler-Furman O, Pietrokovski S. Trans protein splicing of cyanobacterial split inteins in endogenous and exogenous combinations. Biochemistry. 2007;46:322–330. doi: 10.1021/bi0611762. [DOI] [PubMed] [Google Scholar]
- 15.Zettler J, Schutz V, Mootz HD. The naturally split Npu DnaE intein exhibits an extraordinarily high rate in the protein trans-splicing reaction. FEBS Lett. 2009;583:909–914. doi: 10.1016/j.febslet.2009.02.003. [DOI] [PubMed] [Google Scholar]
- 16.Mootz HD, Blum ES, Tyszkiewicz AB, Muir TW. Conditional protein splicing: a new tool to control protein structure and function in vitro and in vivo. J Am Chem Soc. 2003;125:10561–10569. doi: 10.1021/ja0362813. [DOI] [PubMed] [Google Scholar]
- 17.Vila-Perello M, Hori Y, Ribo M, Muir TW. Activation of protein splicing by protease- or light-triggered O to N acyl migration. Angew Chem Int Ed. 2008;47:7764–7767. doi: 10.1002/anie.200802502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Berrade L, Kwon Y, Camarero JA. Photomodulation of protein trans-splicing through backbone photocaging of the DnaE split intein. Chembiochem. 2010;11:1368–1372. doi: 10.1002/cbic.201000157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Binschik J, Zettler J, Mootz HD. Photocontrol of protein activity mediated by the cleavage reaction of a split intein. Angew Chem Int Ed Engl. 2011;50:3249–3252. doi: 10.1002/anie.201007078. [DOI] [PubMed] [Google Scholar]
- 20.Wu H, Hu Z, Liu XQ. Protein trans-splicing by a split intein encoded in a split DnaE gene of Synechocystis sp. PCC6803. Proc Natl Acad Sci USA. 1998;95:9226–9231. doi: 10.1073/pnas.95.16.9226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wei XY, Sakr S, Li JH, Wang L, Chen WL, Zhang CC. Expression of split dnaE genes and trans-splicing of DnaE intein in the developmental cyanobacterium Anabaena sp. PCC 7120. Res Microbiol. 2006;157:227–234. doi: 10.1016/j.resmic.2005.08.004. [DOI] [PubMed] [Google Scholar]
- 22.Iwai H, Zuger S, Jin J, Tam PH. Highly efficient protein trans-splicing by a naturally split DnaE intein from Nostoc punctiforme. FEBS Lett. 2006;580:1853–1858. doi: 10.1016/j.febslet.2006.02.045. [DOI] [PubMed] [Google Scholar]
- 23.Wang CC, Chen JJ, Yang PC. Multifunctional transcription factor YY1: a therapeutic target in human cancer? Expert Opin Ther Targets. 2006;10:253–266. doi: 10.1517/14728222.10.2.253. [DOI] [PubMed] [Google Scholar]
- 24.Gronroos E, Terentiev AA, Punga T, Ericsson J. YY1 inhibits the activation of the p53 tumor suppressor in response to genotoxic stress. Proc Natl Acad Sci U S A. 2004;101:12165–12170. doi: 10.1073/pnas.0402283101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sui G, Affar el B, Shi Y, Brignone C, Wall NR, Yin P, Donohoe M, Luke MP, Calvo D, Grossman SR. Yin Yang 1 is a negative regulator of p53. Cell. 2004;117:859–872. doi: 10.1016/j.cell.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 26.Taki M, Sisido M. Leucyl/phenylalanyl(L/F)-tRNA-protein transferase-mediated aminoacyl transfer of a nonnatural amino acid to the N-terminus of peptides and proteins and subsequent functionalization by bioorthogonal reactions. Biopolymers. 2007;88:263–271. doi: 10.1002/bip.20678. [DOI] [PubMed] [Google Scholar]
- 27.Oeemig JS, Aranko AS, Djupsjobacka J, Heinamaki K, Iwai H. Solution structure of DnaE intein from Nostoc punctiforme: structural basis for the design of a new split intein suitable for site-specific chemical modification. FEBS Lett. 2009;583:1451–1456. doi: 10.1016/j.febslet.2009.03.058. [DOI] [PubMed] [Google Scholar]