Abstract
OBJECTIVES
Lung cancer is one of the most lethal cancers. Currently, there are no biomarkers for early detection, monitoring treatment response, and detecting recurrent lung cancer. We undertook this study to determine if 1H magnetic resonance spectroscopy (MRS) of sputum and exhaled breath condensate (EBC), as a noninvasive tool, can identify metabolic biomarkers of lung cancer.
MATERIALS AND METHODS
Sputum and EBC samples were collected from 20 patients, comprising patients with pathologically confirmed non-small cell lung cancer (n = 10) and patients with benign respiratory conditions (n = 10). Both sputum and EBC samples were collected from 18 patients; 2 patients provided EBC samples only. 1H MR spectra were obtained on a Bruker Avance 400 MHz nuclear magnetic resonance (NMR) spectrometer. Sputum samples were further confirmed cytologically to distinguish between true sputum and saliva.
RESULTS
In the EBC samples, median concentrations of propionate, ethanol, acetate, and acetone were higher in lung cancer patients compared to the patients with benign conditions. Median concentration of methanol was lower in lung cancer patients (0.028 mM) than in patients with benign conditions (0.067 mM; P = 0.028). In the combined sputum and saliva and the cytologically confirmed sputum samples, median concentrations of N-acetyl sugars, glycoprotein, propionate, lysine, acetate, and formate were lower in the lung cancer patients than in patients with benign conditions. Glucose was found to be consistently absent in the combined sputum and saliva samples (88%) as well as in the cytologically confirmed sputum samples (86%) of lung cancer patients.
CONCLUSION
Absence of glucose in sputum and lower concentrations of methanol in EBC of lung cancer patients discerned by 1H MRS may serve as metabolic biomarkers of lung cancer for early detection, monitoring treatment response, and detecting recurrence.
Keywords: magnetic resonance spectroscopy, sputum, breath condensate, lung cancer
Introduction
Global lung cancer mortality is alarming. The primary reasons for the high mortality in lung cancer patients are as follows: lack of an established screening tool to detect early-stage lung cancer, advanced stage at presentation, and absence of biomarkers to monitor treatment response and to detect disease recurrence contributing to treatment failure and poor prognosis.2,3 Currently, implementing screening programs to detect early-stage lung cancer remains a challenge. In the National Lung cancer Screening Trial (NLST), more than 50,000 participants underwent low-dose CT (LDCT) with 20% reduction in mortality with LDCT as compared to chest X-ray. However, LDCT triggered invasive diagnostic procedures in 7% of the patients who did not have lung cancer. Considered as a low radiation dose procedure, the dose received by the participants who underwent LDCT was approximately 8 mSv per participant over three years. However, this translates into a risk of one death for screening every 2500 persons.4 Hence, there is an urgent need to explore noninvasive, pragmatic, and cost-effective tools to screen for early-stage lung cancer, which can complement and perhaps replace LDCT. It has been further suggested that analysis of biofluids (sputum, blood, and urine) might help identify subjects who should undergo LDCT.5–7
Metabolomics research has opened new biological platforms to explore the disease processes, complementing our existing knowledge of genomic, proteomic, and transcriptomic changes in a living cell.8 In the process of carcinogenesis, it has been postulated that cancer cells develop new biochemical adaptations with quantitative changes in endogenous metabolites.9 There has been a surge of investigators exploring magnetic resonance spectroscopy (MRS) and mass spectrometry (MS) techniques as noninvasive means to identify metabolomic signatures of cancer in body fluids and tissues.10–13 MRS can provide structural details of small organic molecules and can detect variations in the concentration of endogenous metabolites. It requires very little sample preparation and has several analytical advantages such as being nondestructive, quantitative, and reproducible.12 It has been used to analyze body fluids in both malignant and benign conditions, including lung cancer, cystic fibrosis (CF), and chronic obstructive pulmonary disease (COPD).14–16
We undertook the current pilot study to determine whether 1H MRS can identify lung cancer-specific metabolic biomarkers in the sputum and exhaled breath of patients with known lung cancer. The hypothesis in question, in this study, is based on our previous work where we demonstrated that there was a relative “absence of glucose” in the sputum of lung cancer patients compared to those with benign conditions.17
Materials and Methods
Patients
Informed consent was obtained from each subject, and the study protocol conformed to the ethical guidelines of the Declaration of Helsinki as reflected in a priori approval by the Institution’s Human Research Ethics Board. Sputum and exhaled breath condensate (EBC) samples were prospectively collected according to a preapproved protocol from 22 patients. Characteristics of the patients are summarized in Table 1. Patients with pathologically confirmed non-small cell lung cancer (NSCLC, n = 12), stage III–IV, before any oncologic treatment, and patients with respiratory conditions other than lung cancer (controls, n = 10) were enrolled at the respiratory clinic, University of Manitoba. The patients in the control group were confirmed not to have any clinical, radiological, and/or pathological evidence of lung cancer or any other malignant conditions. One patient with undetermined pathology and one with B-cell lymphoma were excluded from the analysis. Both sputum and EBC samples were collected from 18 patients; two patients provided EBC samples only. All specimens were collected by a dedicated respiratory research nurse and a respiratory technician in the institution’s pulmonary function laboratory. Specimens were stored at −80°C immediately after collection and later transported (on dry ice) to the University of Winnipeg for MRS analysis.
Table 1.
Clinical characteristics of the patients enrolled in the study.
| CHARACTERISTICS | PATIENTS WITH NSCLC % (n) | PATIENTS WITH BENIGN CONDITIONS % (n) |
|---|---|---|
| Gender (male:female) | 40:60 | 80:20 |
| Age mean (STD) | 68.3 (10.0) | 63.0 (13.3) |
| COPD | 40 (4) | 70 (7) |
| Associated respiratory infection | 30 (3) | 29 (2) |
| Diabetes | 0 | 10 (1) |
| Previous cancer | 40 (4) | 22 (2) |
| Steroid user | 10 (1) | 56 (5) |
| Current smoker | 0 | 22 (2) |
| Asbestosis | 0 | 11 (1) |
| Histopathology | Adenocarcinoma: 50 (5) | |
| Squamous cell carcinoma: 50 (5) | ||
| Stage | III: 40 (4), IV: 50 (5), unknown: 10 (1) | |
| Tumor location | Central: 60 (6); peripheral: 40 (4) | |
| Other conditions | 0 | |
| Churg strauss vasculitis | 0 | 10 (1) |
| Granulamatous inflammation | 0 | 10 (1) |
| Sarcoidosis | 0 | 10 (1) |
| Neurofibromatosis | 0 | 10 (1) |
| Exudative pleural effusion | 0 | 10 (1) |
Specimen collection
Induced sputum samples (2–5 mL) were obtained from patients by inhalation of hypertonic saline solution. EBC was collected noninvasively by having the patient exhale into a cooled collecting container and breathing comfortably through a mouthpiece with a nose clip in place (to prevent loss of exhaled volume through the nose). Approximately 2–3 mL of EBC was collected from each patient over a period of 10–20 minutes. A custom-built condenser with a metal element surrounding a tightly fit inner collection tube was used for this purpose (Fig. 1). Both our collecting device and the technique followed the methodological recommendations set by the American Thoracic Society and European Respiratory Society Task Force published in the European Respiratory Journal.18
Figure 1.

Exhaled breath condensate (EBC) collection device with a nasal clip.
Chemicals
Deuterium oxide (D2O), sodium chloride (NaCl), PBS buffer tablets, and 3-(trimethylsilyl) propionic-2,2,3,3-d4 acid sodium salt (TSP) were purchased from Sigma-Aldrich.
MRS data collection and analysis
MRS staff were blinded to the clinical characteristics and the pathological diagnoses of the patients.
For sputum analysis, samples were thawed for 10–15 minutes, and ~300 µL 2M NaCl solution (buffered with PBS/D2O, pH = 7.4) was added to each sample in 1:1 ratio (v/v), mixed by vortex to get homogenous clear suspensions. Frozen EBC samples were thawed, and neat samples were used for analysis. All MRS experiments were performed on a Bruker Avance 400 MHz NMR spectrometer at the University of Winnipeg. A volume of 500 µL of the processed sputum or EBC sample was transferred into a 5-mm NMR tube along with separate reusable coaxial capillary tubes containing a standard solution of TSP prepared in D2O (TSP concentration = 1.538 and 1.763 mM for sputum and EBC samples, respectively). TSP served as both chemical shift reference for aligning the spectra (0.0 ppm) and external concentration reference, whereas D2O was used as a “deuterium lock”. One-dimensional (1D) 1H MRS experiments with single pulse or excitation sculpting (ES) pulse sequence were performed on all samples with no spinning at 25°C. The following acquisition parameters were employed in all 1D experiments: number of scans = 32, a 45° pulse (single pulse: zgpr) and a 90° pulse (for ES: zgesgp), number of points in time domain = 32 k, inter-pulse delay = 5 seconds, spectral width = 5556 Hz, acquisition time = 2.95 seconds, and line broadening for exponential window function = 0.3 Hz. When using excitation sculpting sequence, the power of the shaped pulse (sp1) was optimized to get a better suppression of water signal. For more details on the acquisition parameters, see the Appendices 1 (zgpr) and 2 (zgesgp).
Various metabolite peaks were confirmed by performing 1H-1H COSY or TOCSY experiments on representative samples. Moreover, we have also compared 1H chemical shifts of metabolites with those from NMR databases such as the human metabolome database (HMDB).19 The peak areas of the signals from metabolites of interest (Tables 2 and 3) were measured by manual integration using TSP as an external reference. Concentrations of various metabolites were calculated as previously shown by Ijare et al.20
Table 2.
Metabolites identified in EBC samples.
| METABOLITE | LUNG CANCER | CONTROL GROUP (BENIGN CONDITIONS) | P |
|---|---|---|---|
| Propionate | 0.022 (0–0.060) | 0.012 (0–0.077) | 0.94 |
| Ethanol | 0.24 (0.14–0.43) | 0.19 (0.14–0.36) | 0.53 |
| Acetate | 0.16 (0.09–0.22) | 0.11 (0.08–0.20) | 0.53 |
| Acetone | 0.026 (0.013–0.039) | 0.023 (0–0.049) | 0.79 |
| Methanol | 0.028 (0.023–0.057) | 0.067 (0.043–0.09) | 0.028 |
Notes: Values are presented as median (interquartile) concentrations (mM). Samples (continuous data) are compared using the Wilcoxon two-sample test.
Table 3.
Metabolites in sputum samples without cytological confirmation (sputum and saliva) and with cytological confirmation (true sputum).
| METABOLITES IN SPUTUM | WITHOUT CYTOLOGICAL CONFIRMATION | P | WITH CYTOLOGICAL CONFIRMATION | ||
|---|---|---|---|---|---|
| LUNG CANCER | CONTROL GROUP (BENIGN CONDITIONS) | LUNG CANCER | CONTROL GROUP (BENIGN CONDITIONS) | ||
| Glycoprotein | 1.58 (1.17–2.62) | 2.28 (1.74–3.43) | 0.14 | 1.76 | 3.43 |
| Propionate | 0.46 (0.42–0.92) | 0.59 (0.29–0.88) | 0.90 | 0.46 | 0.58 |
| Lactate | 0.52 (0.38–0.83) | 0.50 (0–0.75) | 0.69 | 0.51 | 0.64 |
| Acetate | 2.40 (1.24–5.40) | 2.69 (1.71–4.17) | 0.9 | 2.21 | 2.53 |
| N-acetyl sugars | 3.46 (3.08–4.74) | 5.09 (4.35–8.39) | 0.1 | 3.58 | 8.39 |
| Lysine | 1.23 (0.48–1.48) | 1.46 (0.69–1.89) | 0.19 | 1.35 | 1.89 |
| Choline | 0 (0–0.10) | 0.03 (0–0.16) | 0.77 | 0 | 0.16 |
| Formate | 0.20 (0.04–0.33) | 0.22 (0.12–0.28) | 0.63 | 0.13 | 0.19 |
| % of subjects with absent glucose | 88% | 30% | 0.59 | 86% | 40% |
Notes: Values are presented as median (interquartile) concentrations (mM). Samples (continuous data) are compared using the Wilcoxon two-sample test except for glucose where data were compared with Fisher’s exact test since the data were dichotomous.
Cytological confirmation of the sputum samples
An independent dedicated lung pathologist analyzed sputum samples for cytological confirmation of sputum. Presence or absence of alveolar macrophages differentiated between sputum and saliva. The pathologist was blinded to the clinical characteristics and the observed metabolic profiles of the collected specimens.
Statistical analysis
Dichotomous outcomes were compared using Fisher’s exact test. Continuous data were analyzed with the Wilcoxon two-sample test. SAS9.4© software was used for the statistical analyses.
Results
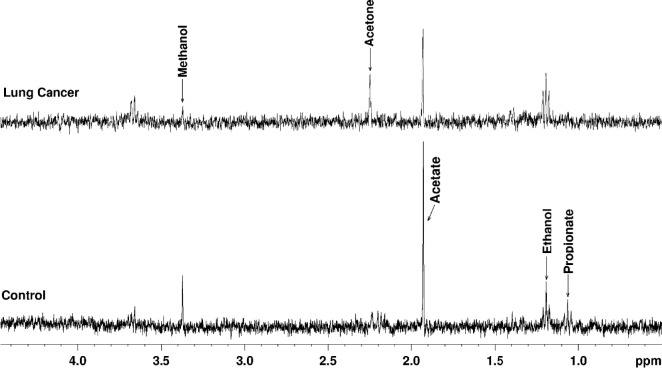
1H MRS of EBC samples (Fig. 2) showed the presence of metabolites such as propionate, ethanol, acetate, acetone, and methanol. Propionate, ethanol, acetate, and acetone were elevated in lung cancer patients compared to the patients with benign conditions. The median concentration of methanol was found to be lower in lung cancer patients (0.028 mM) than in patients with benign conditions (0.067 mM; P = 0.028). However, the difference in the median concentrations of other metabolites between the two groups was not statistically significant (Table 2). Furthermore, subset analysis based on the tumor location in cancer group revealed that the median concentration of methanol from tumors in the central location (6/10) was lower compared to the peripheral location (4/10), but the difference was not statistically significant (0.028 vs. 0.032 mM; P = 0.76).
Figure 2.
1H MR spectra (ES sequence) of exhaled breath condensate (EBC) samples from control and lung cancer patients (adenocarcinoma) showing relative levels of metabolites.
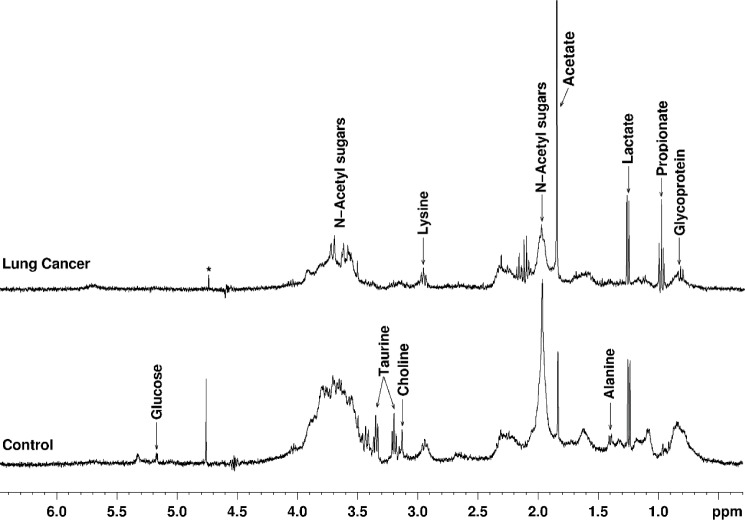
Cytology of the sputum samples revealed that 7 of 8 lung cancer patients and 5 of 10 patients with benign diseases had cytologically confirmed sputum. 1H MRS of sputum samples (Fig. 3) showed the presence of N-acetyl sugars, glycoprotein, propionate, lysine, acetate, and lactate. Median concentration of these metabolites was lower in the lung cancer patients compared to patients with benign conditions. Glucose was absent in six of seven (86%) lung cancer patients and two of five (40%) patients with benign diseases (Table 3). Median concentration of metabolites in samples without cytological confirmation of sputum (sputum plus saliva) was also obtained from both groups of patients (Table 3).
Figure 3.
1H MR spectra (ES sequence) of sputum samples from a control subject and a lung cancer patient (adenocarcinoma) showing relative levels of metabolites including the absence of glucose in lung cancer patient (*residual water signal).
Discussion
The process of carcinogenesis alters the metabolism of the cells due to genetic aberrations in their normal signaling pathways.21 Newly developed malignant cells undergo a metabolic transformation by generating ATP, macromolecules, and altering the impact of reactive oxygen species to support their proliferation. As a consequence, tumor microenvironment manifests hypoxia, alteration in its pH, and nutritional status.21,22 MRS or MS has been used to identify altered metabolic environment of cancer cells in different body fluids.12 In this study, using a home-built EBC-collecting device, it was feasible and convenient to obtain EBC samples from all the patients. 1H MRS identified specific compounds (propionate, ethanol, acetone, and acetate) with higher concentration in cancer patients; however, the median concentration of methanol was 41% lower in EBC samples of cancer patients compared to the control group (P = 0.028). In a recent review, based on the studies carried out during 1985–2015, the most frequent volatile organic compounds (VOCs) identified as biomarkers in the breath of lung cancer patients were 2-butanone, 1-propanol, isoprene, ethylbenzene, styrene, and hexanal. These bio-markers were identified mostly by using gas chromatography in combination with mass spectrometry (GC-MS).23 However, in this review, the largest series included 220 lung cancer patients and 441 healthy volunteers using proton transfer reaction mass spectrometry (PTR-MS). This study reported lower median concentration of methanol in the breath samples of lung cancer patients than healthy controls; 118.5 vs. 142.0 ppb; (P = 0.011).24 Interestingly, methanol is present in very small amounts in the breath of healthy adults with a concentration in the range of 32–2319 partsper-billion by volume. It is primarily produced in the gut by the interaction of bacteria with the unabsorbed carbohydrates and can be detected in body fluids including blood, urine, and breath condensate. Ingested fruits, vegetables, and aspartame can effect the endogenous production of methanol and hence its level of concentration in body fluids.25–28 So far, there has not been any previous report explaining the altered metabolism of methanol in malignant cells. Significantly reduced levels of methanol in the breath of lung cancer patients observed in this study and in the study by Bajtarevic et al24 indicated a possible alteration in its metabolic pathway triggered by the malignant transformation of the normal cells and warrants further investigation. Moreover, it is also worth noting in our study that patients with central lesions either in close proximity or involving the tracheobronchial tree with or without a lymph node mass had lower methanol concentration as compared to peripheral lesions. The difference, however, is not statistically significant possibly due to the small sample size. Future studies with larger sample size may be able to differentiate the metabolic profile of tumors based on their location and tumor burden.
Sputum analysis with and without cytological confirmation has consistently demonstrated absence of glucose in most of the pathologically confirmed lung cancer patients enrolled in our study. This is a novel finding only reported by our group previously.17 Interestingly, some of the other metabolites (specifically glycoproteins and N-acetyl sugars) in both sets of samples—combined sputum and saliva and cytologically confirmed true sputum samples—also consistently revealed lower median concentrations in the lung cancer group (Table 3). However, since only 50% of the samples from the benign diseases group had cytologically confirmed sputum, we remain uncertain about the observed metabolic profile in this group. It is also worthwhile to note that there were two non-cancer patients who had cytologically confirmed sputum samples but whose spectra did not show the presence of glucose signal. One of them was a diabetic patient and currently on metformin. The other patient had increased glucose level in the blood around the period of sample collection.
The authors acknowledge that sputum could not be obtained from all the patients. However in spite of the difficulties in obtaining induced sputum samples, sputum, as a surrogate of altered metabolism of the malignant process, is emerging as an important noninvasive tool to diagnose benign diseases and lung cancer. Guzmán et al have demonstrated methylation of tumor suppressor genes in sputum to diagnose lung cancer and COPD.29 Another group recently has discovered a set of metabolites in the sputum of lung cancer patients with flow infusion electro-spray ion MS.30 Such metabolic phenotype may be an outcome of proliferating cancer cells that can reprogram their own metabolic activity or enzymatic metabolic pathways through activated oncogenes and inactivated tumor suppressor genes at mitochondrial level.31–33 Cancer cells initiate anaerobic pathway for their survival even in the presence of oxygen and generate energy through glycolysis, previously described as Warburg effect.11,34 With metabolic transformation of the cells, mitochondrial oxidative phosphorylation is suppressed and replaced by glycolysis and accumulation of lactate, providing a more efficient form of ATP production for the rapidly proliferating tumor cells.35 However, compromised mitochondrial activity and enhanced glycolysis in proliferating cancer cells may not be the entire story. The two sources of energy; oxidative phosphorylation in mitochondria and glycolysis may operate preferentially and under different conditions of the tumor microenvironment including hypoxia and rate of tumor growth. Oncogenes and associated proteins such as HIF-1α, RAS, C-MYC, SRC, and p53 can also influence the glycolytic pathway and the process of oxidative phosphorylation.36 Gao et al37 have demonstrated the role of C-MYC regulating the glutamine metabolism at mitochondrial level providing ATP for the proliferating lymphoma and prostate cancer cells. Reduced availability of glucose in the sputum of lung cancer patients as demonstrated in our study is intriguing and novel. It may provide a new dimension of the altered metabolism specifically in the context of lung cancer.
Limitations of Our Study
This was a pilot study with a small sample size. It was not possible to correlate several clinical variables including smoking, COPD, diabetes, age of the patient, stage of the cancer, drug profile location of the disease, and molecular markers (EGFR, ALK) with the identified metabolites. We also acknowledge that there were fewer samples of cytologically confirmed sputum in the group with benign diseases that makes statistical evaluation difficult. We need to find novel techniques to improve our ability to obtain induced sputum more reliably.
Conclusion
Lower levels of methanol in EBC samples and the absence of glucose in sputum of lung cancer patients detected by MRS is novel and significant, warranting further investigation in a larger clinical study.
Supplementary Materials
Appendix 1. “ased” output of pulse sequence “zgpr” used for collecting 1H NMR data on sputum and EBC samples.
Appendix 2. “ased” output of pulse sequence “zgesgp” used for collecting 1H NMR data on sputum and EBC samples.
Acknowledgments
We also thank Ann Szabo and Garth Rodgers for their technical assistance.
Footnotes
ACADEMIC EDITOR: Sendhil Velan, Editor in Chief
PEER REVIEW: Four peer reviewers contributed to the peer review report. Reviewers’ reports totaled 1932 words, excluding any confidential comments to the academic editor.
FUNDING: We sincerely thank Department of Respirology, Health Sciences Center, Winnipeg, for the funding support. The authors confirm that the funder had no influence over the study design, content of the article, or selection of this journal.
COMPETING INTERESTS: Authors disclose no potential conflicts of interest.
NOTE: This work was in part presented at the 24th Annual Meeting of the International Society of Magnetic Resonance in Medicine, Singapore (May 7–13, 2016).1
Paper subject to independent expert single-blind peer review. All editorial decisions made by independent academic editor. Upon submission manuscript was subject to anti-plagiarism scanning. Prior to publication all authors have given signed confirmation of agreement to article publication and compliance with all applicable ethical and legal requirements, including the accuracy of author and contributor information, disclosure of competing interests and funding sources, compliance with ethical requirements relating to human and animal study participants, and compliance with any copyright requirements of third parties. This journal is a member of the Committee on Publication Ethics (COPE).
Author Contributions
Conceived and designed the experiments: NA, TB, OI, RM, SB, and ZB. Analyzed the data: ZN, NA, OI, and RA. Wrote the first draft of the manuscript: NA, OI, and TB. Contributed to the writing of the manuscript: JK and GQ. Agree with manuscript results and conclusions: NA, TB, OI, RM, RA, MA, ZB, SB, ZN, JK, and GQ. Jointly developed the structure and arguments for the paper: NA and OI. Made critical revisions and approved final version: NA, OI, TB, and MA. All authors reviewed and approved of the final manuscript.
REFERENCES
- 1.Ahmed N, Bezabeh T, Myers R, et al. Proton magnetic resonance spectroscopy (1-H MRS) of sputum and exhaled breath condensate: a non-invasive tool for lung cancer screening; 24th Scientific Meeting and Exhibition of the International Society for Magnetic Resonance in Medicine; Singapore: ISMRM; May, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61(2):69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 3.Morgensztern D, Ng SH, Gao F, Govindan R. Trends in stage distribution for patients with non-small cell lung cancer: a National Cancer Database survey. J Thorac Oncol. 2010;5(1):29–33. doi: 10.1097/JTO.0b013e3181c5920c. [DOI] [PubMed] [Google Scholar]
- 4.Bach PB, Mirkin JN, Oliver TK, et al. Benefits and harms of CT screening for lung cancer: a systematic review. JAMA. 2012;307(22):2418–2429. doi: 10.1001/jama.2012.5521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.National Lung, Screening Trial, Research Team, Aberle DR, Adams AM, et al. Reduced lung-cancer mortality with low-dose computed tomographic screening. N Engl J Med. 2011;365(5):395–409. doi: 10.1056/NEJMoa1102873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kramer BS, Berg CD, Aberle DR, Prorok PC. Lung cancer screening with low-dose helical CT: results from the National Lung Screening Trial (NLST) J Med Screen. 2011;18(3):109–111. doi: 10.1258/jms.2011.011055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tanoue LT, Tanner NT, Gould MK, Silvestri GA. Lung cancer screening. American Journal of Respiratory and Critical Care Medicine. 2015;191(1):19–33. doi: 10.1164/rccm.201410-1777CI. [DOI] [PubMed] [Google Scholar]
- 8.Gadian DG. Nuclear Magnetic Resonance and Its Applications to Living Systems. Oxford, UK: Clarendon Press; Oxford University Press; 1982. [Google Scholar]
- 9.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324(5930):1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hao D, Sarfaraz MO, Farshidfar F, et al. Temporal characterization of serum metabolite signatures in lung cancer patients undergoing treatment. Metabolomics. 2016;12:58. doi: 10.1007/s11306-016-0961-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Louis E, Adriaensens P, Guedens W, et al. Detection of lung cancer through metabolic changes measured in blood plasma. J Thorac Oncol. 2016;11(4):516–523. doi: 10.1016/j.jtho.2016.01.011. [DOI] [PubMed] [Google Scholar]
- 12.Lenz EM, Wilson ID. Analytical strategies in metabonomics. J Proteome Res. 2007;6(2):443–458. doi: 10.1021/pr0605217. [DOI] [PubMed] [Google Scholar]
- 13.Patel S, Ahmed S. Emerging field of metabolomics: big promise for cancer biomarker identification and drug discovery. J Pharm Biomed Anal. 2015;107:63–74. doi: 10.1016/j.jpba.2014.12.020. [DOI] [PubMed] [Google Scholar]
- 14.Wang L, Tang Y, Liu S, et al. Metabonomic profiling of serum and urine by (1)H NMR-based spectroscopy discriminates patients with chronic obstructive pulmonary disease and healthy individuals. PLoS One. 2013;8(6):e65675. doi: 10.1371/journal.pone.0065675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sofia M, Maniscalco M, de Laurentiis G, Paris D, Melck D, Motta A. Exploring airway diseases by NMR-based metabonomics: a review of application to exhaled breath condensate. J Biomed Biotechnol. 2011;2011:7. doi: 10.1155/2011/403260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Montuschi P, Paris D, Melck D, et al. NMR spectroscopy metabolomic profiling of exhaled breath condensate in patients with stable and unstable cystic fibrosis. Thorax. 2012;67(3):222–228. doi: 10.1136/thoraxjnl-2011-200072. [DOI] [PubMed] [Google Scholar]
- 17.Bezabeh T, Ijare OB, Marginean EC, Nicholas G. Proton magnetic resonance spectroscopy of sputum for the non-invasive diagnosis of lung cancer: preliminary findings. J Anal Oncol. 2012;1(1):14–18. [Google Scholar]
- 18.Horvath I, Hunt J, Barnes PJ, et al. Exhaled breath condensate: methodological recommendations and unresolved questions. Eur Respir J. 2005;26(3):523–548. doi: 10.1183/09031936.05.00029705. [DOI] [PubMed] [Google Scholar]
- 19.Wishart DS, Jewison T, Guo AC, et al. HMDB 3.0—the human metabolome database in 2013. Nucleic Acids Res. 2013;41(Database issue):D801–D807. doi: 10.1093/nar/gks1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ijare OB, Bezabeh T, Albiin N, et al. Simultaneous quantification of glycine-and taurine-conjugated bile acids, total bile acids, and choline-containing phospholipids in human bile using 1 H NMR spectroscopy. J Pharm Biomed Anal. 2010;53(3):667–673. doi: 10.1016/j.jpba.2010.05.028. [DOI] [PubMed] [Google Scholar]
- 21.Cairns R, Harris I, McCracken S, Mak T. Cold Spring Harbor symposia on quantitative biology. Vol. 76. Cold Spring Harbor Laboratory Press; 2011. Cancer cell metabolism; pp. 299–311. [DOI] [PubMed] [Google Scholar]
- 22.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 23.Saalberg Y, Wolff M. VOC breath biomarkers in lung cancer. Clin Chim Acta. 2016;459:5–9. doi: 10.1016/j.cca.2016.05.013. [DOI] [PubMed] [Google Scholar]
- 24.Bajtarevic A, Ager C, Pienz M, et al. Noninvasive detection of lung cancer by analysis of exhaled breath. BMC Cancer. 2009;9:348. doi: 10.1186/1471-2407-9-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Komarova TV, Petrunia IV, Shindyapina AV, et al. Endogenous methanol regulates mammalian gene activity. PLoS One. 2014;9(2):e90239. doi: 10.1371/journal.pone.0090239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smith D, Spanel P. Pitfalls in the analysis of volatile breath biomarkers: suggested solutions and SIFT–MS quantification of single metabolites. J Breath Res. 2015;9(2):022001. doi: 10.1088/1752-7155/9/2/022001. [DOI] [PubMed] [Google Scholar]
- 27.Lee HJJ, Pahl MV, Vaziri ND, Blake DR. Effect of hemodialysis and diet on the exhaled breath methanol concentration in patients with ESRD. J Ren Nutr. 2012;22(3):357–364. doi: 10.1053/j.jrn.2011.07.003. [DOI] [PubMed] [Google Scholar]
- 28.Španěl P, Dryahina K, Vicherková P, Smith D. Increase of methanol in exhaled breath quantified by SIFT-MS following aspartame ingestion. J Breath Res. 2015;9(4):047104. doi: 10.1088/1752-7155/9/4/047104. [DOI] [PubMed] [Google Scholar]
- 29.Guzmán L, Depix MS, Salinas AM, et al. Analysis of aberrant methylation on promoter sequences of tumor suppressor genes and total DNA in sputum samples: a promising tool for early detection of COPD and lung cancer in smokers. Diagn Pathol. 2012;7(1):1. doi: 10.1186/1746-1596-7-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.O’Shea K, Cameron SJ, Lewis KE, Lu C, Mur LA. Metabolomic-based biomarker discovery for non-invasive lung cancer screening: a case study. Biochim Biophys Acta. 2016;1860(11 pt B):2682–2687. doi: 10.1016/j.bbagen.2016.07.007. [DOI] [PubMed] [Google Scholar]
- 31.Cantor JR, Sabatini DM. Cancer cell metabolism: one hallmark, many faces. Cancer Discov. 2012;2(10):881–898. doi: 10.1158/2159-8290.CD-12-0345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Munoz-Pinedo C, El Mjiyad N, Ricci J. Cancer metabolism: current perspectives and future directions. Cell Death Dis. 2012;3(1):e248. doi: 10.1038/cddis.2011.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sciacovelli M, Gaude E, Hilvo M, Frezza C. The metabolic alterations of cancer cells. Methods Enzymol. 2014;542:1–23. doi: 10.1016/B978-0-12-416618-9.00001-7. [DOI] [PubMed] [Google Scholar]
- 34.Upadhyay M, Samal J, Kandpal M, Singh OV, Vivekanandan P. The Warburg effect: insights from the past decade. Pharmacol Ther. 2013;137(3):318–330. doi: 10.1016/j.pharmthera.2012.11.003. [DOI] [PubMed] [Google Scholar]
- 35.Gillies RJ, Robey I, Gatenby RA. Causes and consequences of increased glucose metabolism of cancers. J Nucl Med. 2008;49(suppl 2):24S–42S. doi: 10.2967/jnumed.107.047258. [DOI] [PubMed] [Google Scholar]
- 36.Jose C, Bellance N, Rossignol R. Choosing between glycolysis and oxidative phosphorylation: a tumor’s dilemma? Biochim Biophys Acta. 2011;1807(6):552–561. doi: 10.1016/j.bbabio.2010.10.012. [DOI] [PubMed] [Google Scholar]
- 37.Gao P, Tchernyshyov I, Chang T-C, et al. c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature. 2009;458(7239):762–765. doi: 10.1038/nature07823. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 1. “ased” output of pulse sequence “zgpr” used for collecting 1H NMR data on sputum and EBC samples.
Appendix 2. “ased” output of pulse sequence “zgesgp” used for collecting 1H NMR data on sputum and EBC samples.