Abstract
The Clinical Proteomic Tumor Analysis Consortium (CPTAC) has produced large proteomics datasets from the mass spectrometric interrogation of tumor samples previously analyzed by The Cancer Genome Atlas (TCGA) program. The availability of the genomic and proteomic data is enabling proteogenomic study for both reference (i.e., contained in major sequence databases) and non-reference markers of cancer. The CPTAC labs have focused on colon, breast, and ovarian tissues in the first round of analyses; spectra from these datasets were produced from 2D LC-MS/MS analyses and represent deep coverage. To reduce the variability introduced by disparate data analysis platforms (e.g., software packages, versions, parameters, sequence databases, etc.), the CPTAC Common Data Analysis Platform (CDAP) was created. The CDAP produces both peptide-spectrum-match (PSM) reports and gene-level reports. The pipeline processes raw mass spectrometry data according to the following: (1) Peak-picking and quantitative data extraction, (2) database searching, (3) gene-based protein parsimony, and (4) false discovery rate (FDR)-based filtering. The pipeline also produces localization scores for the phosphopeptide enrichment studies using the PhosphoRS program. Quantitative information for each of the datasets is specific to the sample processing, with PSM and protein reports containing the spectrum-level or gene-level (“rolled-up”) precursor peak areas and spectral counts for label-free or reporter ion log-ratios for 4plex iTRAQ™. The reports are available in simple tab-delimited formats and, for the PSM-reports, in mzIdentML. The goal of the CDAP is to provide standard, uniform reports for all of the CPTAC data, enabling comparisons between different samples and cancer types as well as across the major ‘omics fields.
Keywords: Proteomics data resource, bioinformatics, cancer, CPTAC, data analysis pipeline
Introduction
The National Cancer Institute formed a Clinical Proteomic Tumor Analysis Consortium (CPTAC) in 2011 to facilitate the discovery of cancer-specific protein biomarkers. The current program is a follow-on to the original CPTAC program (2006–2010), which focused on reproducibility. The current consortium has eight institutions as lead centers, with approximately thirty collaborating groups. NCI sought to leverage the results of their patient tumor sequencing program (The Cancer Genome Atlas (TCGA)) to inform the proteomics. Discovery proteomics provides evidence of protein sequence and abundance, including the identification and quantitation of post-translational modifications (PTMs) that may be critical to cell signaling pathways and networks. The proteins from 105 breast tumors were analyzed at the Broad Institute (MIT); from 95 colorectal tumor samples at Vanderbilt University1; and from 115 ovarian cancer tumors split between the Johns Hopkins School of Medicine (72); and the Pacific Northwest National Laboratory (75), with 32 samples in common between the two sites. The consortium members selected complementary methods of trypsin-digested, bottom-up, peptide analysis with state-of-the-art 2D LC-MS/MS methods using Orbitrap™ mass analyzers. The resulting 5,860 LC/MS/MS tumor runs required ~10,160 hours of instrument time and produced >91 million MS/MS spectra, occupying 3 terabytes of storage for the raw data files. Additionally, each laboratory used their same analytical procedures for human-in-mouse xenograft reference standard (“system suitability” or CompRef) tumor samples, run before and after 10 human tumor samples. These repeat analyses required 790 LC-MS/MS analytical runs requiring 1,122 hours of instrument time, and producing >14M tandem mass spectra. Together, this analytical data represents an important public resource for research in human cancer proteomics, which are accessible through an online portal: the CPTAC Data Portal2 managed by the CPTAC Data Coordinating Center (DCC) https://cptac-data-portal.georgetown.edu/cptacPublic/.
Following data acquisition, each laboratory used its preferred software tools to analyze its own data to extract maximum information relevant to tumor analysis and cancer biology for publication. In order to remove the multiple sources of variability that would otherwise result when comparing peptides and proteins inferred by each group using different software, the consortium agreed upon the need for a Common Data Analysis Pipeline (CDAP) to produce uniform report files for public release. Because all of the datasets are of interest to cancer researchers both inside and outside of the proteomics field, the uniformity of processing also eliminates differences due to the use of various reference proteome databases or varying software parameters, two well-known sources of variation in comparative proteomic analyses. This paper documents the Common Data Analysis Pipeline, an integral component of this multi-institutional research program in cancer proteomics.
Experimental Section
Selection of the component programs in the data analysis pipeline was based on the diversity of sample processing and instruments used in the studies. Since both label-free and iTRAQ™ 4plex strategies were used during early “system suitability” (xenograft analysis) evaluation studies, the pipeline was designed to handle both data types as well as data from phosphopeptide and glycopeptide enrichment studies. All laboratories used Thermo Fisher Orbitrap™-based high-resolution mass spectrometers for TCGA samples. In general, the design of the pipeline was based on group consensus by a steering committee of collaborators as well as the availability of tools for the NIST hardware and operating system infrastructure. Details of the pipeline are given below. Importantly, the database file and software versions were not changed throughout the processing of the “system suitability” and TCGA tumor analysis data files. (See Table 1 for a detailed list of software and parameters used.)
Table 1.
Software and options used in the Common Data Analysis Pipeline.
| Program | Versi on |
Source | Refere nce |
Purpos e |
Options Used | iTR AQ |
Phos pho |
|---|---|---|---|---|---|---|---|
|
ReAdW4Masc ot2.exe |
ftp://chemdata.nist.gov/download/peptide_library/software/current_releases/ReAdw4Mascot2/ | None | MS and MS/MS data extracti ons, precurs or m/z and charge state re- evaluati on |
-c -ChargeMgfOrbi - FixPepmass -MaxPI - metadata - MonoisoMgfOrbi - NoPeaks1 -PIvsRT - sep1 -sepZC -msfr - XmlOrbiMs1Profile - iTRAQ -TolPPM 20 |
- iTRA Q |
||
| MS-GF+ | v973 3 |
http://omics.pnl.gov/software/ms-gf | Kim et al.8 |
Sequen ce databas e search |
java –Xmx3500M –jar MSGFPlus.jar -d <file>.fasta -t 20ppm -e 1 -m (3 for QExactive, 1 for Orbitrap) -inst (1 for QExactive, 1 for Orbitrap) -ntt 1 -thread 2 -tda 1 -ti 0,0 -n 1 - maxLength 50 -mod <file>.txt |
- proto col 2 (3 for phos pho and iTRA Q) |
- protoc ol 1 |
| NIST-ProMS | NIST (developer communication) | MS1 data analysis |
in a file called proms.ini (<mzXML file>.raw.mzXML <search result file>.raw.FT.hcd.ch.M GF.mzid.tsv <output file>.raw.txt <search engine name: MS-GF+, MSPepSearch, SpectraST, OMSSA) <instrument: ORBI_HCD, ORBI, LTQ, QTOF> |
||||
| PhosphoRS | 1.0 | http://ms.imp.ac.at/?goto=phosphors | Taus et al.11 |
Phosph osite localiza tion |
ActivationTypes="HCD " MassTolerance Value="0.02" |
Data File Staging
To quality control the files received from the proteome characterization centers, Thermo raw data files (*.RAW) were downloaded from a private staging area (maintained by the DCC) to NIST servers for analysis. MD5 checksums were used to verify file integrity. If RAW files passed file quality control, they were entered into the processing queue and associated with a minimal set of metadata, including the following: site, labeling, enrichment status, and instrument make and model. All samples were digested with trypsin. More information on each of the sample processing conditions (e.g., alkylation, labeling, etc.) can be found in the metadata available with each dataset at the DCC or in the primary publications.
Raw Data Conversions
Raw files were processed by the NIST converter ReAdW4Mascot2.exe, a heavily modified version of the original ReAdW.exe (by Patrick Pedrioli) developed at The Institute for Systems Biology (ISB) for use in early versions of the Trans-Proteomic Pipeline (TPP)3,4. This converter produces peak lists of very similar content to those produced by msconvert5 (data not shown.) ReAdW4Mascot2.exe produces several output files for each raw file, including metadata (*.metadata) files with values such as date, instrument serial number, method and tune file names and parameters. File and instrument specific metadata were extracted directly from the raw files at conversion time. Most importantly, mzXML (not mzML) and MGF (Mascot generic file) files were produced by this converter. The software uses OCX calls directly to the XCalibur libraries, if available, or to those provided from an installation of MSFileReader (https://thermo.flexnetoperations.com). mzXML files are produced for legacy reasons and are used as input for MS1 intensity-based quantitation performed by NIST-ProMS in the next step of processing. MGF files follow the Matrix Science™ standard but provide substantial, additional information embedded in the TITLE lines. However, the major purpose of the MGF files is to provide MS2 peak lists for identification by the sequence search engine, MS-GF+ (described in a following section). MGF files are not distributed for public consumption but could be provided by request.
ReAdW4Mascot2.exe parameters can be found in Table 1. One important note regarding RAW file conversion is the setting “-FixPepMass.” This option forces the program to re-assess the accuracy of the monoisotopic precursor m/z by looking at both the previous and next MS1 scans to discern the accuracy of the monoisotopic peak assignment. If the assignment is inaccurate, the software will either change a precursor m/z, precursor charge or will attempt to assign a charge if one is missing. The frequency of these changes is heavily affected by instrument settings. “Exclusion of unassigned charge states” was not selected by one lab (PNNL) in the instrument method resulting in approximately 17% of all precursor or charge state values being modified at conversion-time, in comparison to approximately 3% for other contributing labs (data not shown). A second reason for choosing this option was to reduce false identifications of deamidations which are characterized by a precursor m/z difference of +1 (roughly equivalent to the mass of a single neutron) but lack fragment ion evidence. It is not uncommon for incorrect monoisotopic precursor m/z assignments to be incorrectly identified as deamidations6. To ensure accurate reporting of these conversion-time changes, the XCalibur-assigned precursor m/z and charge as well as the corrected values are given in the final PSM-level output files (*.psm and *.mzIdentML) for reference. One other note on precursor assignment: if ReAdW4Mascot2.exe was unable to assign either a precursor m/z or charge, the spectra were excluded from further processing. For at least PNNL, these spectra were included for lab-prepared data analysis for publication(s). Comparison indicated that a small but significant percentage of these spectra are identifiable (data not shown), representing one more area of possible variability.
iTRAQ Peak Processing and Reporting
Extraction of iTRAQ reporter ion peaks was added to ReAdW4Mascot2.exe for iTRAQ 4plex files. Along with the intensity values for the reporter ions (m/z’s 114, 115, 116, 117), a quality score is also computed. A value for variability of each iTRAQ channel is the dMZ/HWHM where dMZ = (measured peak m/z) − (exact m/z of iTRAQ reporter ion). A value >1 for dMZ/HWHM typically indicates reporter ion contamination. These values can be used to impose penalties on identified spectra with abundant impurities. ‘AbFract’ is also calculated; this is the fraction of the MS2 TIC (total ion current) accounted for by the reporter ions. All iTRAQ values are included in the PSM-level reports.
Along with the intensity of each channel and its quality, a column in the PSM-level reports called ‘iTRAQFlags’ is also created. For this field, ‘I’ is added if the geometric mean of the two ‘PrecursorPurity’ (isolation window purity calculations, also computed by ReAdW4Mascot2.exe) values is <90% (percentage of intensity in the isolation window (typically 2 m/z) attributable to the assigned precursor and its isotopes). Empirical evidence suggests that values >80% are usually reliable (i.e., lack significant fragmentation impurities (i.e., co-fragmentation) giving rise to so-called ratio compression7). It is also worth noting that filtering using ‘I’ as a flag would be strict for typical analyses since these flags are present on all spectra with purities <90%. Instead, it may be worthwhile for end users to include spectra with values <80%. This value will be used in the next release of the PSM files. A flag of ‘M’ is added if one or more iTRAQ channel intensities is a zero or a missing value, and ‘D’ is added if the quality score for one or more iTRAQ channels is > 1.
Search Engine
After a preliminary performance comparison of the major open-source or freeware search engines available to the community, MS-GF+8 was chosen for the CDAP. This search engine is under active develop at PNNL, which enabled rapid and effective communication between CDAP engineers and the developer (Dr. Sangtae Kim.) Search engine settings for MS-GF+ are given in Table 1. MS-GF+ uses a machine learning (i.e., training step) to improve its accuracy. Consequently, settings are based on a given instrument’s make and model. For example, a fragment ion tolerance setting is not necessary. Also, and somewhat atypical for MS/MS-search engines, MS-GF+ does not include a maximum setting for missed enzyme cleavages. Semi-specific tryptic in silico digestion was used for searching all data. This adds considerable overhead to the search space and may diminish some sensitivity but was an important parameter because some breast tumor samples contained semi-tryptic peptides at rates as high as 25%. This value is about 10× higher than what is typical for tryptic digestions, but plausibly reflective of post-mortem collection interval (ischemic time) and sample history.
The confidence of MS/MS assignments was determined by MS-GF+’s automatic target-decoy routines (search setting ‘-tda 1’). The engine calculates its own ‘SpecEvalues’ (as well as Evalues) from which QValues (q-values) are derived. Additionally, the setting ‘-ti 0,0’ requires that the monoisotopic precursor m/z value of a query spectrum match the database exactly (i.e., no “isotope wobble”). This setting was chosen because an attempt to fix incorrect monisotope selection is made at conversion-time. Prior to reporting, a q-value threshold of 0.01 (1% FDR on the PSM-level) was applied to all data files, and only Rank 1 hits were kept. Additionally, the decoy hits were removed unless scores tied with a target match. An in depth discussion of q-values on the PSM-, peptide- and gene-level, as well the consequences of removing the decoys, is given in the Results section below.
Sequence Database
The protein FASTA file used for system suitability analysis was concatenated RefSeq H. sapiens (build 37), M. musculus (build 37), and the sequence for S. scrofa (porcine) trypsinogen. The FASTA file used for analysis of the TCGA human samples lacks the M. musculus sequences. Decoy sequences were appended automatically by MS-GF+ by using the option ‘-tda 1’ as mentioned above.
Peptide Spectral Libraries
Peptide tandem mass spectral libraries were built from all CPTAC data at NIST. These were either used to build new libraries or add to existing public libraries and are suitable for MS/MS searching by MSPepSearch (NIST library search algorithm for batch identification of peptides http://chemdata.nist.gov/dokuwiki/doku.php?id=peptidew:mspepsearch) or SpectraST9 (library search algorithm integrated into the TPP). Importantly, these data have now contributed to the construction of very large iTRAQ libraries for human and mouse accessible to the public at http://chemdata.nist.gov/dokuwiki/doku.php?id=peptidew:cdownload. Libraries (human and mouse xenografts) were constructed from the MS-GF+ results and were separately compiled for ion trap (CID), beam-type collision cell (HCD), iTRAQ and label-free. HCD libraries are composed of best-replicate spectra at specific collision energies and ion trap entries are consensus spectra. Since different labs using HCD analysis operated instruments at different ‘NCE’ (normalized collision energy) values, best replicate spectra for small CE bins were used to represent each peptide ion. Consequently, each peptide ion may be represented by more than one spectrum in a given HCD library. A summary of the additional content can be found in Table 2.
Table 2.
NIST Peptide Mass Spectral Libraries created from CPTAC data
| Instrument/Mode | Species | Derivative/PTM | Number Spectra |
|---|---|---|---|
| Ion trap - HCD | Human | iTRAQ-4 part 1 | 581,416 |
| Ion trap - HCD | Human | iTRAQ-4 part2 | 620,216 |
| Ion trap - HCD | Human | iTRAQ-4/Phospho | 223,340 |
| Ion trap - HCD | Mouse | iTRAQ-4 | 17,851 |
| Ion trap - HCD | Mouse | iTRAQ-4/Phospho | 15,746 |
MS1 Data Analysis
MS1 data analysis was performed by the NIST-developed program, NIST-ProMS. This program was originally developed as part of the NISTMSQC metrics for calculating precursor areas from extracted ion chromatograms10. While this software provides many functions, it was used in the CDAP exclusively for calculating the intensity of precursor ions, applicable mainly to label-free analyses (i.e., not iTRAQ data, for which a precursor is the mixture of the isobaric, labeled forms). NIST-ProMS works by finding and then calculating the area of isotope groups. The program reads the output from MS-GF+ to annotate peak areas with peptide sequences when an MS/MS spectrum has been used to make a confident assignment. NIST-ProMS data can be found in the columns ‘PrecursorArea’ and ‘PrecursorRelAb’ in the PSM and mzIdentML files. ‘PrecursorArea’ is the total area for the precursor ion and ‘PrecursorRelAb’ is the ‘PrecursorArea’ normalized by the precursor area of the largest identified peptide ion in that file (i.e., fraction.)
QC Metrics
A subset of the NISTMSQC metrics was calculated on all of the RAW data files10. These reports were used to detect outliers and troubleshoot analytical problems within the program. QC reports are not publicly accessible but were used during internal communications between NIST, the DCC, and the collaborating labs. Briefly, QC calculations were designed to cover the full proteomics pipeline and highlight batch effects or other analytical problems. The QC metrics cover the following areas: chromatography, ESI, MS1, MS2, and data analysis.
Phosphosite localization
The software PhosphoRS11 was added to the pipeline for assignment of phosphosites after a preliminary performance comparison using a set of commonly used tools (not shown). Its purpose is to calculate site assignment probabilities which can be used to gauge the quality of a phosphosite assignment. This type of post-search analysis is needed as search engines often do not make explicit use of site-determining fragment ions when assigning a phosphosite in peptides containing n+1 potential phosphorylation sites, where n is the number of S, T or Y residues. This program was run according to the ReadMe.html file present with the download. Briefly, spectra in MGF format, for which non-trivial phospho identifications were identified below a QValue threshold of 0.01, were converted to the XML format described by the authors. Next the XML files were processed in batch by the software and the scores in the results were parsed into the report files. PhosphoRS scores are reported in the column ‘PhosphoRSPeptide’ in the PSM and mzIdentML files. Additionally, ‘nPhospho’ reports the number of phosphosites assigned by MS-GF+ and ‘FullyLocalized’ is set to Y (yes) if all phosphosites score >99.0 (a strict filter), otherwise this value is set to N (no).
Peptide-spectrum-match (PSM) Report Format
The PSM-level reports are the subject of extensive documentation, available from the CPTAC Data Portal under the “About the data” tab (https://cptac-data-portal.georgetown.edu/cptac/aboutData/show?scope=dataLevels), and have been partially described in a separate publication2. Briefly, * .psm files are the tab-delimited files produced by the CDAP at NIST from the search engine results. These files list PSMs as rows with data in columns. Data include MS-GF+ output as well as the precursor, iTRAQ, and phosphosite data when appropriate. ‘FileName’ and ‘ScanNum’ uniquely identify a single MS/MS spectrum. When two or more identifications score identically for the top-ranked position, the field ‘AmbiguousMatch’ is given a value of 1, and the row is repeated with the alternate identification(s). The mzIdentML files are produced by the DCC and are translations of the PSM files into the PSI standard12 from the CDAP PSM files. The informatics methods for the mzIdentML conversions are also available from the CPTAC Data Portal under the “About the data” tab.
Protein Reports (Gene-level)
The protein reports have been described elsewhere2 and in documentation available from the CPTAC Data Portal under the “About the data” tab. However, the concepts and thresholds are worth repeating, here. To generate the protein reports, the PSM-level data was aggregated to the peptide-level and peptides were parsimoniously assembled for gene inference. Peptides were associated with genes through their protein sequences and NCBI Gene and UniProt annotations. Assembling at the gene-level removes the potential problems associated with inappropriately assigning quantitative data to minor protein isoforms. Comparisons at the gene-level are also commonplace for genomics scientists, as many such users are interested in proteoegenomic comparisons. Following assembly, the gene-level FDR was assessed using the MAYU (not an acronym) technique to more accurately model inferences with both target and decoy hits13. The parsimony analysis required at least two unshared peptides per gene and two spectra per peptide. The PSM-level QValue was reduced until a MAYU-estimated gene-level FDR of 1%, for the entire assembly, was achieved. A summary of the QValue thresholds necessary to achieve this level of confidence can be found in Table 3.
Table 3.
A summary of q-value (FDR) thresholds used in the CDAP to filter major TCGA datasets.
| Dataset | Max Spec. FDR |
MAYU FDR |
Target Genes |
|---|---|---|---|
| TCGA_Colorectal_VU_Proteome | 0.115% | 1.00% | 5561 |
| TCGA_Breast_BI_Proteome | 0.077% | 1.00% | 10599 |
| TCGA_Breast_BI_Phosphoproteome | 0.185% | 0.99% | 7526 |
| TCGA_Ovarian_JHUZ_Proteome | 0.165% | 1.00% | 8588 |
| TCGA_Ovarian_JHUZ_Glycoproteome | 1.000% | 0.32% | 891 |
| TCGA_Ovarian_PNNL_Proteome | 0.271% | 0.99% | 7471 |
| TCGA_Ovarian_PNNL_Phosphoproteome | 1.000% | 0.46% | 5161 |
The protein reports are distributed across several files. For label-free quantitation (e.g., spectral counting as in Zhang et al.1), *.precursor_area.tsv and *.spectral_counts.tsv files list the sample-level precursor areas for total and unshared peptides or spectral counts along with metadata by gene. For iTRAQ data, gene-level iTRAQ log-ratios, with respect to the common POOL sample, are reported in the *.itraq.tsv files. For these files, the first 3 rows provide the mean, median and standard deviation of the sample log-ratios. Gene-level “roll-up” was performed in the following way, aggregated for all peptide ions or only the unshared peptide ions: (1) select the peptide ion spectrum with maximum total reporter intensity from each spectrum data file, (2) remove outliers using the “libra” technique implemented by the TransProteomicPipeline (TPP)3,4 and find the arithmetic mean of the retained log-ratios (sample : pooled control channel), (3) normalize each sample’s aggregate log-ratios using the sample median. Metadata, including organism, chromosome and locus are associated with each row in these files as columnar data.
Regardless of the quantitation workflow, protein and peptide identification summary reports are provided: *.peptide.tsv and *.summary.tsv. These files explicitly record the peptide-to-gene mapping information as well as the data summarized across the whole analysis, respectively.
Results
The goal of the CDAP is to produce uniform reports that can be used to compare across the participating proteomic centers, major TCGA sample types, and major -omics technologies (e.g., RNAseq vs. MS-based proteomics).
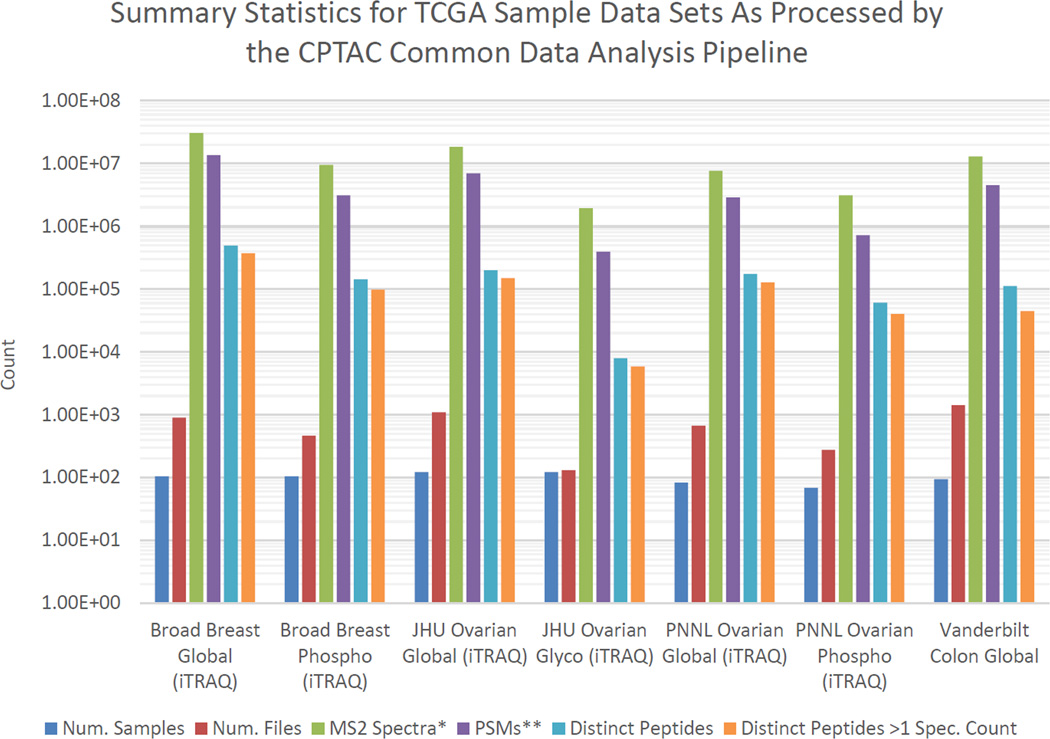
Logistically, it was straightforward to (1) create peak lists, (2) search them, and (3) filter the results to 1% PSM-level FDR using q-values provided by MS-GF+. Summary counts (number of identifications, peptide sequences, files, etc.) from each of the datasets are shown in Fig. 1. Note that these numbers were calculated by removing the fraction labeled ‘A’ from the breast cancer datasets and by ignoring PSMs which cannot be unambiguously assigned. Removing ambiguous peptides from the phospho datasets may be removing relatively more peptides than for the global sets because peptides with different localizations frequently have tie scores. As these peptides are enriched in phospho datasets, more peptides will not be counted. The purpose of Fig. 1 is simply to indicate the relative numbers of identifications and their magnitude for each dataset as well as the actual values in order to provide scale to the acquired data and processed data units.
Figure 1.
Summary of CDAP results for major CPTAC analysis of TCGA samples. * MS2 counts derived from MGF files at NIST. Identification results are listed at 1% PSM-level FDR. ** PSM counts exclude identifications marked as ambiguous (i.e., >1 equivalently-scoring peptide matches.)
Examining the PSM and gene-level FDRs
We also examined the gene-level FDR resulting from the gene-based parsimony analysis using the full 1% PSM-level FDR filtered TCGA datasets. As datasets of this size become increasingly available, it is important to note that a gene/protein-level FDR assessment is critical to determine and report. At the 1% PSM-level, the MAYU gene-FDR varied from 20–40% (not shown). Such identification-estimated FDRs as high as these are unacceptable by proteomics journal standards, but are an unavoidable consequence of processing very large datasets. The FDRs were refined slightly by applying the MAYU technique to account for the possibility that some of the potential false positive genes, whose number is estimated from inferred decoy genes, coincide with true positive target genes. The MAYU correction for the gene-FDR is more pronounced as the number of inferred target genes increases (not shown.) However, even with this correction, gene-level FDRs remain unacceptably high at 1% PSM-level FDR for most datasets.
To investigate features that may help discriminate true from false identifications, we ran a portion of the TCGA colon dataset through Percolator14. This analysis revealed SpecEValue (the score on which the discriminate score used for filtering, QValue, is based) to be the feature weighted most heavily (by absolute value) (Figure S1). This result is expected and served as a positive control. The second two most heavily-weighted features were the number of missed cleavages and the number of spectral counts for a given peptide sequence. This result is consistent with low spectral count peptides carrying higher decoy/target peptide rates (Figure 2). Not only do low spectral count peptides carry higher target/decoy rates, they also contribute a large number of potentially false peptide sequences (Figure 3).
Figure 2.
Decoy / Target identification rate over a range of spectra / peptide values. This figure show that peptides with fewer identifying spectra are more likely to be false or incorrect identifications.
Figure 3.
The number of decoy sequences contributed by identifications over a range of spectra / peptide values. This plot shows that the majority of decoy peptide sequences (which can disproportionately affect the protein-level FDR) are contributed by identifications with fewer spectra / peptide.
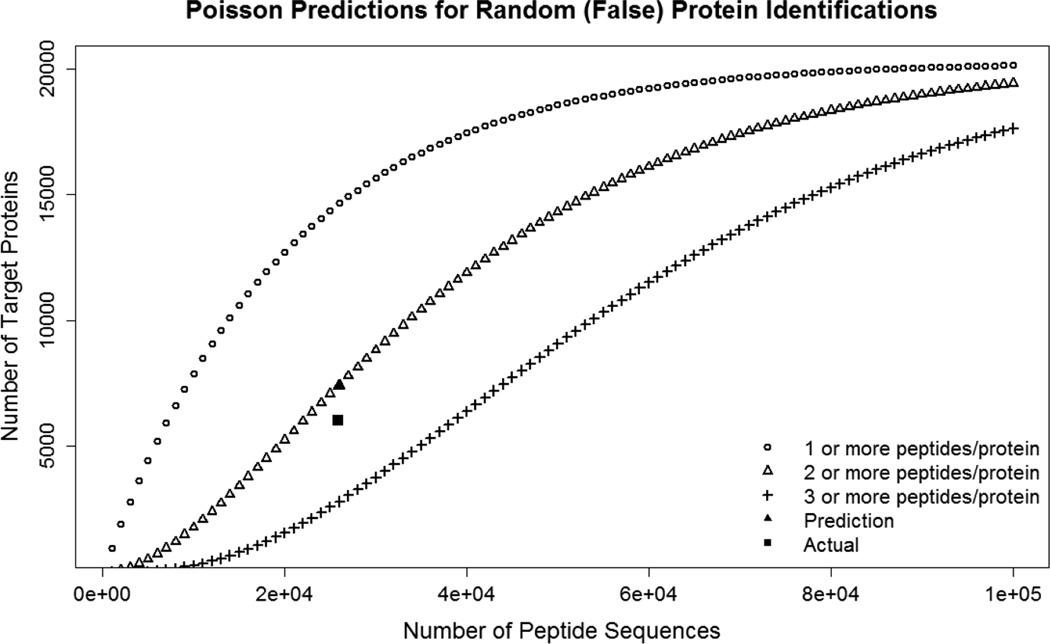
While Reiter et al.13 and others indirectly address the issue of disproportionately large protein-level FDRs in large datasets, this has not been thoroughly described in the literature. It can be approximated using Poisson statistics15. In Figure 4, we have plotted the relationship between false target peptide sequences (assumed to be a number equivalent to the number of decoy sequences) and false protein identifications at greater than or equal to one, two and three shared peptide sequences per protein. This plot shows that in order to maintain a <1% gene-level FDR before MAYU correction, the number of decoy peptide sequences must be restricted to less than 2,000 for a target gene identification set of ~10,000 genes at >=2 unshared peptide per protein.
Figure 4.
Poisson predictions for random target identification for a range of random (e.g., decoy) peptide sequence values. Green shows values for requiring >=1 peptide / protein for identification; red shows >=2 peptides / protein; purple >=3 peptides / protein. Red square shows Poisson prediction for the colorectal cancer dataset; red circle shows actual value. This figure shows that Poisson predictions using the number of decoy sequences can accurately predict the number of false protein identifications, and that requiring more peptides can greatly reduce the number of target protein identifications for the same number of random peptide assignments.
High numbers of false peptide sequences in large datasets are exacerbated due to the following: after multiple LC/MS/MS runs have been accumulated, a level of “saturation” is achieved. That is, fewer and fewer new target protein identifications are made by new peptides; the majority tend to re-identify (or confirm) existing peptide IDs, and it is not uncommon to see highly abundant true peptides represented by thousands of PSMs. On the contrary, false peptide sequences tend to randomly distribute over the entire proteome at an approximate probability of 1 / n where n is the number of protein sequences in the database. The consequence is that the ratio of false / true peptide sequences increases disproportionately to the number of PSMs, creating a situation where the gene-level FDR can be >>10× the PSM-level FDR, as was observed to be the case for all of the global TCGA datasets, prior to gene-level filtering.
Filtering the TCGA datasets to 1% gene-level FDR
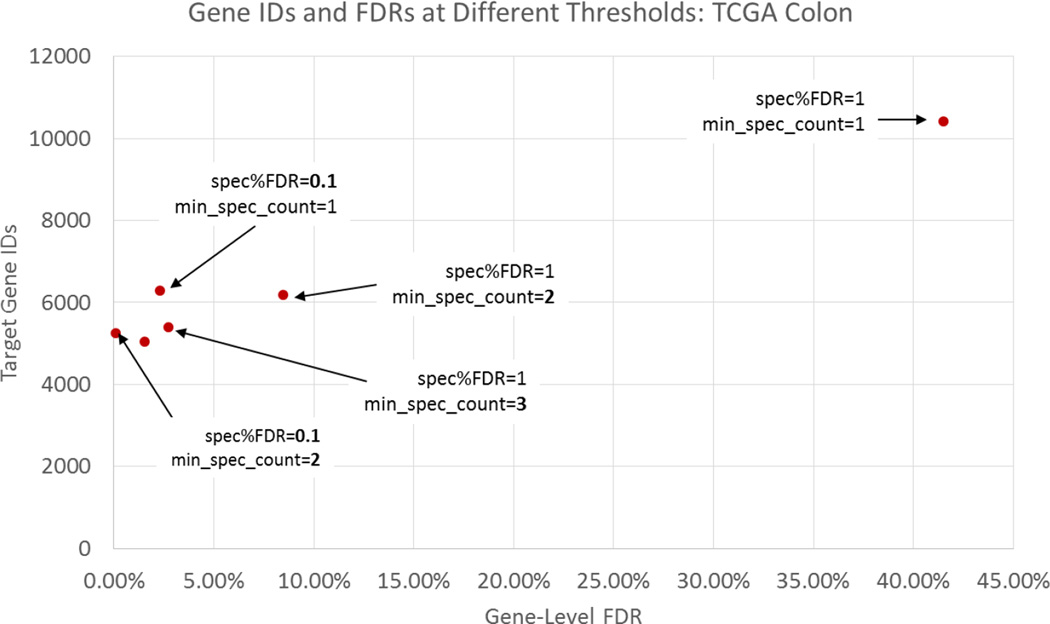
Guided by the Percolator results, we elected to impose a >1 spectrum / peptide filter. The effects of this and other filtering strategies can be observed in Figure 5 for the TCGA colon dataset. As this figure shows, an increase in the spectral count requirement by one reduced the gene-level FDR from 42% to 8% for the colorectal dataset. We chose to use this strategy in combination with a reduction in the allowed QValue threshold (per file) to reduce the gene-level FDR to 1%. Table 3 shows the QValue thresholds required for each of the datasets to achieve this level of confidence on the gene-level. These thresholds were determined empirically, and only in the case of two of the datasets was 1% PSM-level FDR able to sufficiently control the gene-level FDR.
Figure 5.
The number of target gene identifications and corresponding gene-level FDR for a number of threshold settings. Point furthest to the right shows the unacceptably high gene-level FDR resulting from only requiring a PSM-level FDR of 1% and one spectrum / peptide.
Comparing analyses from CDAP reports and Zhang et al.1
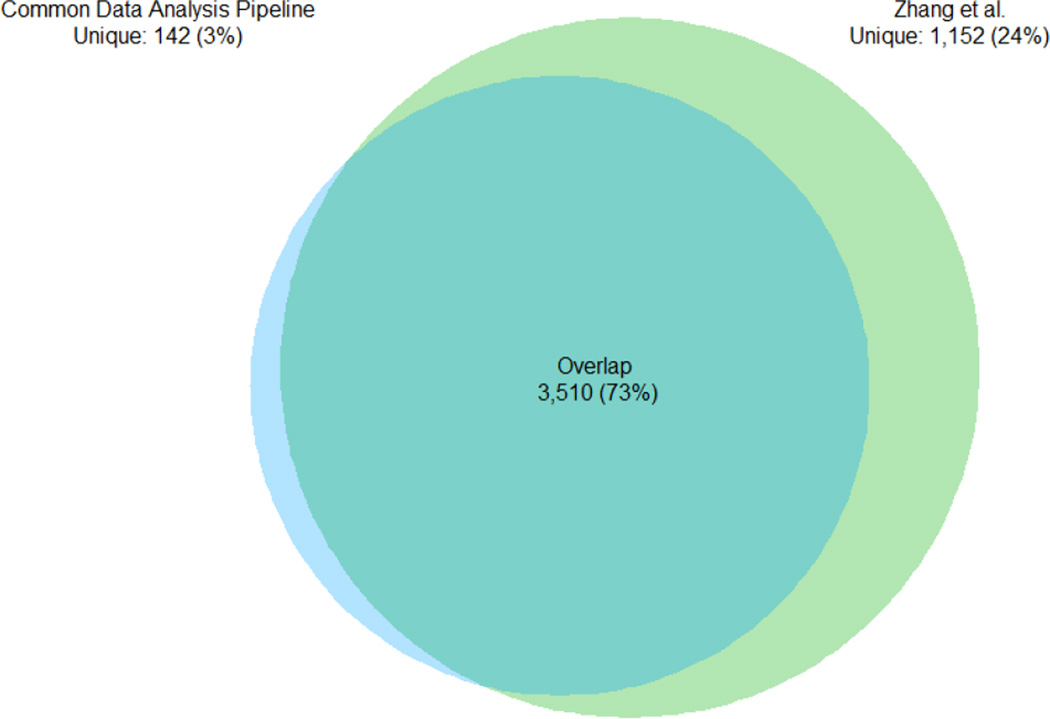
The goal of the CDAP is to provide common standardized analyses across all of the CPTAC datasets, while each contributing laboratory pursued independent analyses. The Vanderbilt University colorectal cancer study has been published, and consequently, we compared gene-level overlap and the similarity of spectral counts as assembled by the two separate pipelines. Gene-level summaries include all steps of the data processing: (1) spectrum identification, (2) protein assembly and (3) gene mapping. For one sample, TCGA-A6-3807-01A-22, the gene IDs and total spectral counts were extracted from both analyses. Figure 6 shows the overlap in gene identifications and Figure 7 shows the pairwise correlation of the natural logarithms of gene-level spectral counts. Figure 6 shows that about 1,100 genes were additionally identified in the Zhang et al. assembly with an overlap of about 73%. This is inevitably due to variations in the data analysis pipelines. In particular, the Zhang et al. assembly used 3 search engines (MS-GF+, MyriMatch16 and the spectral library search engine Pepitome17) and a 2.6% protein-level FDR for filtering. Moreover, the CDAP assembly excluded single spectrum peptides, Zhang et al. did not1. These factors also affected the correlation. On average, the Vanderbilt assembly identified more spectra for the lower abundance genes, consistent with the added sensitivity from combining multiple search engines. Other differences visible by comparing the two gene sets may be attributable to parsimony (e.g., handling of shared peptides) as well. However, considering the large number of data analytical differences, identification overlap and correlation is reasonably good. It is also worth noting that strict filtering also has the unwanted consequence of eliminating many good assignments. Here, were have tried to strike a reasonable balance and produce gene-level datasets with a low, estimated FDR.
Figure 6.
Gene identification overlap between CDAP analysis and Zhang et al1.
Figure 7.
Gene-level spectral count correlation between the CDAP analysis and Zhang et al1.
Discussion
The CDAP was designed to facilitate a fully-described common data analysis across all of the CPTAC datasets for public data access. CDAP was intended to perform conservatively. Hence we designed a filtering strategy to minimize (but not eliminate) most of the potential for incorrect gene identifications. In order to maximize performance for a given parameter set, it makes sense to find the settings that maximize the number of target gene identifications at a given (low) FDR. However, it should be noted that at a 1% gene-level FDR for a dataset containing 10,000 gene IDs, approximately 100 are estimated to be completely spurious. This should give caution to those expecting results free of errors.
Phosphopeptide datasets
Protein- or gene-assembly is more commonly performed on non-enriched data. Phosphorylation studies usually include a “roll-up” to the phosphosite-level, instead of the gene-level, for the purpose of analyzing phospho-signaling network biology. However, the CDAP assembly algorithms have not yet been applied to the phosphopeptide datasets for public release. Instead, we provide the peptide-spectrum-match data from which phospho-site data may be “rolled-up” by the end-user. When completed, CDAP gene-level summaries will be described in documentation to be posted to the DCC as soon as they are available. As such, no gene-level or site-level assemblies are available at the time of writing for the enriched datasets. Selecting and developing these methods is particularly challenging when assigning quantitative information to a site as sometimes peptides overlap or peptides with multiple phosphorylations are present which overlap, and it is unclear how best (or most appropriately) to “roll-up” the iTRAQ information. Additionally, since our references are on the gene-level, assigning phosphosites requires designating a representative protein sequence, which can be non-trivial.
Decoy peptides
With the initial release of the CDAP PSM files, decoy hits were removed for the sake of simplicity. While this makes for cleaner reports, it also prevents the use of 3rd party tools (e.g., Scaffold, IDPicker, TPP, etc.) for independent assembly and FDR-filtering. To remedy this, the next planned release of the PSM files will include decoy hits.
Non-reference peptides
The CDAP processed cancer tumor data should be useful for many researchers inside and outside of the field of proteomics for conducting pan-cancer analyses. However, since the sample-specific sequence databases (generated by the TCGA) were not used in the primary processing of the data files, non-reference peptides (those not occurring in RefSeq or other sequence databases) are not observable without adding them or adding a de novo search node to the pipeline. These results will be useful for labs seeking to detect and quantify protein products that correspond to splice variants, mutations, insertions, deletions, rearrangements, copy number aberrations, or epigenomic changes that were detected at the genome level. As work on the construction of protein FASTA files from individual patients continues within the program, future releases may include the addition of these sequences.
Supplementary Material
Acknowledgments
Funding Sources
NCI CPTAC 2
NIST/NCI IAA ACO13004 (“Proteomics Measurement Quality Assurance Program”) This project has been funded in whole or in part with Federal funds from National Cancer Institute, National Institutes of Health, under Contract No. HHSN261200800001E. The content of this publication does not necessarily reflect the views of policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
The authors thank all of the contributing CPTAC labs for their work generating these large datasets and for valuable feedback during the design and testing phases of the pipeline. In particular, the authors would like to thank Karl Clauser and Philip Mertins (Broad Institute), Sam Payne, Matt Monroe, and Tao Liu (PNNL), David Fenyo (NYU), Rob Slebos, Bing Zhang, and David Tabb (Vanderbilt University Medical Center), Bai Zhang (JHU), and Alexey Nesvizhskii (UMich). We are also grateful to Nuno Bandeira and June Snedecor for their help with assessing phosphosite assignment algorithms.
Acronyms and Abbreviations
- CDAP
Common data analysis pipeline
- CE
collision energy
- CID
collisional-induced dissociation
- CPTAC
Clinical Proteomic Tumor Analysis Consortium
- DCC
Data coordinating center
- dMZ
delta m/z
- ESI
electrospray ionization
- FASTA
sequence database standard
- FDR
false discovery rate
- HCD
higher-energy collisional dissociation
- HWHM
half width at half max
- ID
identification
- ISB
The Institute for Systems Biology
- iTRAQ
Isobaric tags for relative and absolute quantitation
- LC-MS/MS
liquid chromatography tandem mass spectrometry
- M
million
- MGF
Mascot generic format
- MS/MS
tandem mass spectrometry
- MS1
primary mass spectrometry signal
- MS2
tandem mass spectrometry signal
- MS-GF+
mass spectrometry generating function
- mzIdentML
HUPO PSI standard for peptide identification results
- mzML
HUPO PSI standard for mass spectrometry data
- mzXML
deprecated standard for mass spectrometry data developed by ISB
- NCE
normalized collision energy
- NIST
National Institute for Standards and Technology
- NISTMSQC
NIST mass spectrometry quality control
- OCX
object linking and embedding control developed by Microsoft
- PNNL
Pacific Northwest National Laboratories
- PSM
Peptide-spectrum match
- PTM
post-translational modification
- QValue
multiple hypothesis testing-correct p-value (false discovery statistic)
- RAW
Thermo-specific raw mass spectrometry data files
- TCGA
The Cancer Genome Atlas
- TIC
total ion current
- TPP
Trans-Proteomic Pipeline
Footnotes
DISCLAIMER
Certain commercial instruments are identified in this document. Such identification does not imply recommendation or endorsement by the National Institute of Standards and Technology, nor does it imply that the products identified are necessarily the best available for the purpose.
- Percolator Analysis
References
- 1.Zhang B, Wang J, Wang X, Zhu J, Liu Q, Shi Z, Chambers MC, Zimmerman LJ, Shaddox KF, Kim S, et al. Proteogenomic characterization of human colon and rectal cancer. Nature. 2014;513(7518):382–387. doi: 10.1038/nature13438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Edwards NJ, Oberti M, Thangudu RR, Cai S, McGarvey PB, Jacob S, Madhavan S, Ketchum KA. The CPTAC Data Portal: A Resource for Cancer Proteomics Research. J. Proteome Res. 2015 doi: 10.1021/pr501254j. [DOI] [PubMed] [Google Scholar]
- 3.Deutsch EW, Mendoza L, Shteynberg D, Slagel J, Sun Z, Moritz RL. Trans-Proteomic Pipeline, a standardized data processing pipeline for large-scale reproducible proteomics informatics. Proteomics Clin. Appl. 2015;9(7–8):745–754. doi: 10.1002/prca.201400164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Keller A, Eng J, Zhang N, Li X, Aebersold R. A uniform proteomics MS/MS analysis platform utilizing open XML file formats. Mol. Syst. Biol. 2005;1 doi: 10.1038/msb4100024. 2005.0017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chambers MC, Maclean B, Burke R, Amodei D, Ruderman DL, Neumann S, Gatto L, Fischer B, Pratt B, Egertson J, et al. A cross-platform toolkit for mass spectrometry and proteomics. Nat. Biotechnol. 2012;30(10):918–920. doi: 10.1038/nbt.2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nepomuceno AI, Gibson RJ, Randall SM, Muddiman DC. Accurate Identification of Deamidated Peptides in Global Proteomics Using a Quadrupole Orbitrap Mass Spectrometer. J. Proteome Res. 2014;13(2):777–785. doi: 10.1021/pr400848n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Karp NA, Huber W, Sadowski PG, Charles PD, Hester SV, Lilley KS. Addressing Accuracy and Precision Issues in iTRAQ Quantitation. Mol. Cell. Proteomics. 2010;9(9):1885–1897. doi: 10.1074/mcp.M900628-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim S, Mischerikow N, Bandeira N, Navarro JD, Wich L, Mohammed S, Heck AJR, Pevzner PA. The Generating Function of CID, ETD, and CID/ETD Pairs of Tandem Mass Spectra: Applications to Database Search. Mol. Cell. Proteomics. 2010;9(12):2840–2852. doi: 10.1074/mcp.M110.003731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lam H, Deutsch EW, Eddes JS, Eng JK, King N, Stein SE, Aebersold R. Development and validation of a spectral library searching method for peptide identification from MS/MS. Proteomics. 2007;7(5):655–667. doi: 10.1002/pmic.200600625. [DOI] [PubMed] [Google Scholar]
- 10.Rudnick PA, Clauser KR, Kilpatrick LE, Tchekhovskoi DV, Neta P, Blonder N, Billheimer DD, Blackman RK, Bunk DM, Cardasis HL, et al. Performance Metrics for Liquid Chromatography-Tandem Mass Spectrometry Systems in Proteomics Analyses. Mol. Cell. Proteomics. 2010;9(2):225–241. doi: 10.1074/mcp.M900223-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Taus T, Köcher T, Pichler P, Paschke C, Schmidt A, Henrich C, Mechtler K. Universal and Confident Phosphorylation Site Localization Using phosphoRS. J. Proteome Res. 2011;10(12):5354–5362. doi: 10.1021/pr200611n. [DOI] [PubMed] [Google Scholar]
- 12.Seymour SL, Farrah T, Binz P-A, Chalkley RJ, Cottrell JS, Searle BC, Tabb DL, Vizcaíno JA, Prieto G, Uszkoreit J, et al. A standardized framing for reporting protein identifications in mzIdentML 1.2. PROTEOMICS. 2014;14(21–22):2389–2399. doi: 10.1002/pmic.201400080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reiter L, Claassen M, Schrimpf SP, Jovanovic M, Schmidt A, Buhmann JM, Hengartner MO, Aebersold R. Protein identification false discovery rates for very large proteomics data sets generated by tandem mass spectrometry. Mol. Cell. Proteomics MCP. 2009;8(11):2405–2417. doi: 10.1074/mcp.M900317-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Käll L, Canterbury JD, Weston J, Noble WS, MacCoss MJ. Semi-supervised learning for peptide identification from shotgun proteomics datasets. Nat. Methods. 2007;4(11):923–925. doi: 10.1038/nmeth1113. [DOI] [PubMed] [Google Scholar]
- 15.Wiley: Exploring the Human Plasma Proteome - Gilbert S. Omenn. ISBN: 978-3-527-60942-0. Section 1.2.3. [Google Scholar]
- 16.Tabb DL, Fernando CG, Chambers MC. MyriMatch: highly accurate tandem mass spectral peptide identification by multivariate hypergeometric analysis. J. Proteome Res. 2007;6(2):654–661. doi: 10.1021/pr0604054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dasari S, Chambers MC, Martinez MA, Carpenter KL, Ham A-JL, Vega-Montoto LJ, Tabb DL. Pepitome: evaluating improved spectral library search for identification complementarity and quality assessment. J. Proteome Res. 2012;11(3):1686–1695. doi: 10.1021/pr200874e. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.