Abstract
The emergence of whole-genome annotation approaches is paving the way for the comprehensive annotation of the human genome across diverse cell and tissue-types exposed to various environmental conditions. This has already unmasked the positions of thousands of functional cis-regulatory elements integral to transcriptional regulation, such as enhancers, promoters and anchors of chromatin interactions that populate the noncoding genome. Recent studies showed that cis-regulatory elements are commonly the targets of genetic and epigenetic alterations associated with aberrant gene expression in cancer. Here we review these findings to showcase the contribution of the noncoding genome and its alteration in the development and progression of cancer. We also highlight the opportunities to translate the biological characterization of genetic and epigenetic alterations in the noncoding cancer genome into novel approaches to treat or monitor disease.
Keywords: noncoding DNA, functional elements, cancer, genetics, epigenetics, chromatin, somatic mutations, single nucleotide variants, histone modification, DNA methylation
The identification and annotation of cis-regulatory elements in the human genome
The human genome is more than a collection of genes
The diploid human genome consists of over 6 billion bases of DNA that provide the genetic basis for our phenotypic individuality (1,2). Approximately 20,000 genes are encoded in the human genome and are transcribed into ~80,000 transcripts that are subsequently translated into various proteins. Despite the importance of proteins in diverse cellular processes, protein coding sequences account for under 2% of the human genome (less than 120 million bases of DNA in the diploid genome). The role for the remaining noncoding bases (~98%) is a source of investigation and debate (1,3). Nearly half of the noncoding genome consists of repetitive elements including interspersed satellites, short interspersed nuclear elements, long interspersed nuclear elements, ribosomal DNA, DNA transposons and retrotransposons that impact various biological functions (4). Additionally, the noncoding genome harbors non-repetitive elements, including cis-regulatory elements such as promoters, enhancers and anchors of chromatin interactions (5,6). These cis-regulatory elements are directly involved in modulating gene expression and noncoding RNA transcription through long-range chromatin interactions (Figure 1A–D) (7,8). Identifying and characterizing functional noncoding elements within the genome hold great promise to improve our understanding of the human genome in health and disease. In this review, we focus on the progress in noncoding functional element annotation and recent advances demonstrating the central role of genetic and epigenetic alterations affecting noncoding cis-regulatory elements of relevance to cancer initiation and progression.
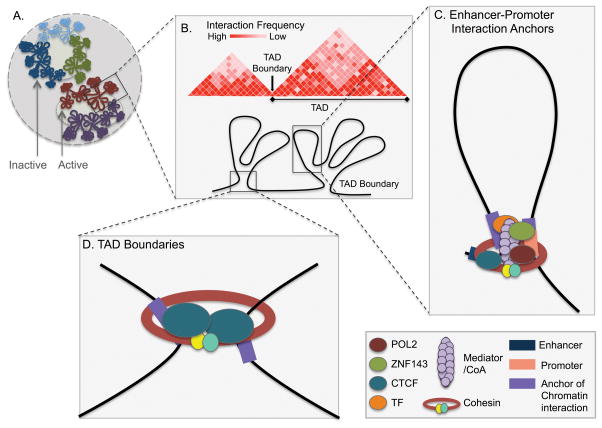
Figure 1. The genome is organized through a hierarchy of long-range interactions.
A) Large chromosomal neighborhoods associate with each other in the nuclear space. Euchromatic regions that are associated with high transcriptional activity tend to cluster in the center of the nucleus. In contrast, heterochromatic regions associated with transcriptional repression tend to cluster at the nuclear periphery. B) Heat map representing virtual genome-wide chromatin interaction maps. Mega-base scale chromatin interaction partitions the genome into domains of interactions known as Topologically Associated Domains (TADs). TAD boundaries preclude interactions between neighboring TADs, therefore restricting most interactions to within their borders. C) Enhancer-promoter chromatin interactions are mediated by the chromatin-interaction factors ZNF143 and CTCF, in concert with several accessory/co-binding proteins. These factors act in concert with several co-binding/accessory/associated proteins to influence genome organization via enhancer-promoter interactions. Enhancer-promoter interactions are at the kilobase (Kb) scale and are highly cell-type specific. D) Anchors of chromatin interactions that define TAD boundaries are enriched for CTCF and cohesin binding. TADs are up to a megabase (Mb) in scale and are highly conserved across cell types.
Identification and annotation of noncoding functional elements across the genome
The noncoding genome has historically been overlooked because of technical limitations hindering the characterization of its genetic and epigenetic nature. Recent advances in whole- genome annotation, inclusive of next-generation sequencing technologies, now offer the means to effectively delineate functional noncoding regions of the human genome. This annotation takes into account multiple definitions of functionality to incorporate the evolutionary, genetic and molecular biology perspectives.
From an evolutionary perspective, comparative analyses are commonly used to identify conservation of DNA sequences across related species (9). Genetic elements that are retained across species are generally considered biologically important and are thus considered functional. In a recent study, about 8% of the human genome has been reported to be under evolutionary constraint (10). Taking into account protein-coding sequences, this implies that functionality can be ascribed to approximately 6% of the noncoding genome (10,11). Identifying conserved DNA sequences through sequence alignment along a linear genome however disregards the three-dimensionality of the genome in which the sequence identity of cis- regulatory elements regulating the same gene, for instance, may be conserved across species despite localizing to neighboring yet distinct positions along the linear genome of different species. This is suggested by a comparative study of cis-regulatory elements between the human and mouse genomes revealing conservation at the level of transcription factor networks between these two species (12). Similar multi-species comparative analyses based on the chromatin binding profiles for multiple transcription factors with different DNA binding motifs exhibit conserved binding DNA sequence preferences but with limited binding event alignment across species (13–15). This supports the integration of epigenomics and comparative genomic to assist in the identification of functional elements in the context of the evolutionary perspective.
From a genetic perspective, functional elements of the genome are defined by the ability of a variation in their DNA sequence, either a structural alteration or a Single Nucleotide Variant (SNV), to cause quantifiable phenotypic changes, inclusive of differential gene expression. This is exemplified by the mutations reported in the telomerase reverse transcriptase (hTERT) gene promoter in melanoma patients that increases hTERT gene expression (16,17). Until recently, the genetic approach was hampered by low-to-modest throughput methodologies. The recent development of high-throughput in vitro assays however, including the Massively Parallel Reporter Assay (MPRA), Massively Parallel Functional Dissection (MPFD) assay, Self-Transcribing Active Regulatory Region sequencing (STARR-seq) and Protein Binding Microarrays (PBMs) are now allowing to measure biochemical differences across cis-regulatory elements and genetically modified variants (18–21). These assays are further complemented with newly designed in silico approaches such as IntraGenomic Replicates (IGR) and Function Based Prioritization of Sequence Variations (FunSeq) that predict changes in DNA-protein interaction induced by genetic alterations (22–26). Together, these technologies enable researchers to assess the impact of thousands of genetic alterations found across the genome to quantifiably change phenotypic traits, thereby accelerating the identification of functional noncoding elements based on a genetic perspective.
From a molecular biology perspective, functional noncoding elements are identified based on biochemical measurements. Following-up on the work from independent laboratories characterizing biochemical activity across the noncoding genome, the Encyclopedia of DNA Elements (ENCODE) project launched in 2003 significantly accelerated the identification of biochemically active noncoding elements of the human genome. This was made possible using a series of high-throughput assays including Chromatin Immunoprecipitation-sequencing (ChIP-seq), RNA-sequencing and DNase I hypersensitive site sequencing (DNase-seq) across a collection of normal and cancer cell lines from different tissues of origin (27–29). Overall, this led the ENCODE project to predict biochemical activity across approximately 80% of the human genome (27). Other initiatives, including The Functional Annotation of the Mammalian Genome (FANTOM) project and the International Human Epigenome Consortium (IHEC), inclusive of the Roadmap Epigenomics project, Blueprint, DEEP, Canadian Epigenetics, Environment and Health Research Consortium (CEEHRC) and Core Research for Evolutional Science and Technology (CREST), are further contributing to the biochemical characterization of the human genome to identify functional elements (30–32). Overall, these efforts led to the annotation of a diverse set of functional elements, inclusive of cis-regulatory elements, populating the noncoding genome to establish transcriptional programs in a lineage-specific manner across many cell and tissue-types.
In accordance to the aberrant changes in gene expression profiles promoting cellular dedifferentiation and pluripotency during cancer development, cis-regulatory elements integral to transcriptional programs are garnering attention in the field of cancer genetics.
Cis-regulatory elements are targets of genetic and epigenetic alterations in cancer
Genetic and epigenetic alterations target promoters in cancer
Promoters correspond to the basic unit of regulation required for the expression of any transcript located upstream of the target gene transcription start site (33). The initiation of gene transcription involves the recruitment of coactivator proteins to assist in a series of steps, culminating in the assembly of the transcription pre-initiation complex consisting of general transcription factors (TFIIA, TFIIB, TFIID, TFIIE, TFIIF and TFIIH) and RNA polymerase II. The activation of the RNA Polymerase II results in transcriptional elongation(34,35).
Genetic alterations populate promoters in cancer
Given the fundamental role of promoters in transcription, genetic alterations targeting their underlying sequences can directly contribute to aberrant gene expression (Figure 2A). This is exemplified by the recurrent somatic mutations identified in the hTERT gene promoter in multiple human cancers including melanoma, glioma, glioblastoma, medulloblastoma, lung adenocarcinoma, thyroid cancer, bladder cancer and hepatocellular carcinoma (16,17,36–40). This same promoter is also affected by genetic predisposition, specifically by the rs2853669 Single Nucleotide Polymorphism (SNP) located 246 base-pairs upstream from the start codon (17). These genetic alterations create DNA recognition motifs for members of the E26 Transformation-Specific (ETS) transcription factor family, leading to the increased binding of ETS factors, promoting an increase in hTERT expression (16,17,41). The continued hTERT gene expression normally suppressed in somatic cells can result in the aberrant lengthening of telomeres to favor cellular immortalization and oncogenesis (42). In addition to the hTERT gene promoter, recent whole-genome analyses of various cancer types have also reported a significant burden of mutations at the WDR74, MED16, SDHD and TFPI2 gene promoters (41,43). Furthermore, genetic alterations in the SDHD and hTERT promoters have also been shown to discriminate patient outcome in a collection of cancer types, including melanoma, glioma, glioblastoma, medulloblastoma, thyroid, liver and bladder cancer, supporting their usefulness as biomarker for patient stratification (36–38,41,43–46).
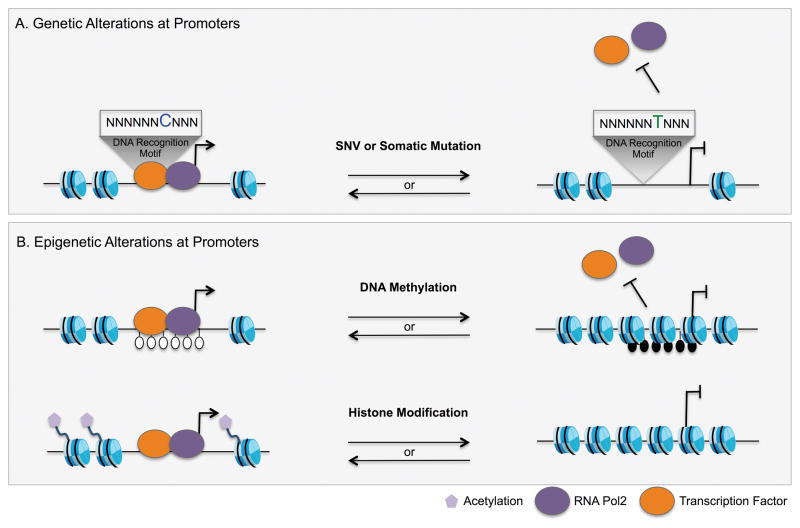
Figure 2. Genetic and epigenetic alterations are observed at gene promoters in cancer.
A) Alterations in the sequences of promoters can modulate transcription factor binding affinity for the DNA to change the expression of the associated gene. This can arise through somatic mutations or inherited Single Nucleotide Variants (SNVs). B) Changes in the epigenetic identity, either based on changes in the DNA methylation or histone modifications was reported in cancer initiation and progression that influence promoter activity and results in altered gene expression in cancer.
Structural variations involving promoters can also contribute to oncogenesis. A prototypical example consists of a fusion between the ETS factor ETS-Related Gene (ERG) proto-oncogene and the promoter region of the Transmembrane Protease, Serine 2 (TMPRSS2) gene through an intronic deletion on chromosome 22q22.2-3 (47). The TMPRSS2:ERG (T2E) fusion, reported in approximately 50% of prostate cancer patients, places ERG gene expression under the control of the androgen-regulated TMPRSS2 promoter, resulting in an oncogenic increase in ERG transcript and protein levels (47,48). The increase in ERG protein resulting from the T2E fusion can upregulate the expression of target genes that favor prostate cancer cell migration and invasion, including CXCR4 and ADAMTS-1 (47,49), and has been associated with higher grade prostate cancer (50). Fusion of the TMPRSS2 promoter with other ETS family members, such as ETS Variant 1, 4 and 5 (ETV1, ETV4, ETV5), have also been reported in another 5–10% of prostate cancer patients (48). More recently, structural variations including deletions, duplications, inversions and translocations associated with breakpoints at chromosome 9p24 that cluster within the 3′-Untranslated Region (UTR) of the PD-L1 gene have been reported in multiple cancers, including adult T-cell leukemia, large B-cell lymphoma and stomach adenocarcinoma (51). These aberrant structural alterations targeting the 3′-UTR of PD-L1 elevate the stability of PD-L1 transcripts and expression, suggested to aid cancer cells in escaping anti-tumor immunity (51). Together, these studies showcase that promoters can be targeted by either inherited or acquired genetic alterations, inclusive of both SNVs and structural variants, that contribute to oncogenesis.
Epigenetic alterations accumulate at promoters in cancer
The activity of cis-regulatory elements is greatly dependent on their chromatin accessibility. Promoters found in compacted chromatin, known as heterochromatin, are inactive while those found in accessible chromatin, known as euchromatin, are actively engaged in transcriptional regulation. Epigenetic modifications, inclusive of DNA methylation, histone modifications or variants, readily influence chromatin accessibility by affecting the density of nucleosomes (Figure 2B) (29,52,53). Changes in chromatin accessibility, either increasing or decreasing its compaction, through epigenetic alterations can directly impact cancer development.
This is highlighted by changes in DNA methylation at CpG dinucleotides commonly reported at promoters of target gene or noncoding transcribed regions across different types of cancer (Figure 2B) (54,55). For example, the promoters of numerous tumor suppressor genes such as RASSF1A, BRCA1, APC, MLH1 and p16 are hypermethylated in osteosarcoma, endometrial carcinoma, glioblastoma, pancreatic, breast, colorectal, ovarian and non-small cell lung cancer (56–63). The hypermethylation of these promoters correlates with the reduced expression of their associated gene (58–60,64–66). Similar results were reported for noncoding transcribed regions, inclusive of micro (miR) and long noncoding RNAs (lncRNA). For instance, the hypermethylation of the miR-124a promoter is associated with reduced miR-124a expression in leukemia, lymphoma and colon, breast and lung cancer (67). Similarly, DNA hypermethylation of the bidirectional miR-34b/c promoter relates to miR-34b/c silencing in colorectal cancer cells, favorable to colony formation (68). Finally, DNA hypermethylation of the MEG3 lncRNA promoter is linked with the reduced MEG3 expression in multiple cancers (69–71) and associates with poor prognosis in gastric cancer patients (72). In summary, aberrant DNA hypermethylation can affect both coding and noncoding gene promoters to impact oncogenesis. Noteworthy, differential methylation status at promoters can inform on clinical outcome. For instance, DNA methylation of the GSTP1 promoter on chromosome 11q13 can discriminate malignant from normal prostate tissue (73,74). Methylation status of the HOXD3 promoter on chromosome 2q31 also segregates low-grade prostate cancer from intermediate and high-grade prostate cancer (75). Moreover, the methylation status of the MGMT promoter in glioma patients on chromosome 10q26 has been suggested to be a predictor of treatment response and post-treatment survival to temozolomide and alkylating agents (76,77). These studies support that DNA methylation patterns at promoters can also potentially serve as biomarkers to stratify cancer patients for treatment response and distinct clinical outcomes.
Conversely, promoters can undergo DNA demethylation as cancer develops (78), and the loss of DNA methylation at promoters associates with the overexpression of the associated gene. Demethylation at the ELMO3 gene promoter for instance is associated with its overexpression in human lung cancer (79). In accordance with the proposed role of ELMO3 in cellular migration, the overexpression of ELMO3 has been documented in metastatic lung cancer (79,80). Promoter demethylation driving the aberrant expression of uPA, involved in tumor progression and metastasis is similarly reported in invasive prostate cancer (81,82). The treatment of invasive prostate cancer PC-3 cells with S-adenosylmethionine, previously shown to favor hypermethylation, inhibits uPA gene expression and cell invasion in vitro, suggesting that the inhibition of promoter demethylation may be a potential therapeutic strategy against aberrantly activated tumor-promoting genes (81).
Promoters can also be epigenetically marked by specific histone modifications (Figure 2B), and aberrant fluctuation of these modifications has been linked to oncogenesis. The co-occupancy of lysine 4 and 27 trimethylation on histone H3 (H3K4me3 and H3K27me3, associated with activation and repression of transcription, respectively) defines a “bivalent” state found at the promoters of genes poised for expression (83–85). These bivalent promoters can transit into either active (H3K4me3-positive and H3K27me3-negative) or silent (H3K4me3-negative and H3K27me3-positive) states during cell differentiation (85). During colorectal cancer initiation, gains and losses of the H3K4me3 modification at promoters are associated with differential gene expression (86). The loss of the H3K27me3 modification has also been linked to aberrant activation of oncogenic gene transcription, including MKI67 and CD133, a proliferation marker and a cancer stem cell marker, respectively (87). Moreover, the loss of both H3K4me3 and H3K27me3 modifications is associated with aberrant gains in promoter methylation in colorectal cancer (87). Apparent gains and losses of the H3K27me3 modification at promoters also discriminates androgen deprivation-resistant versus sensitive prostate cancer cells, suggesting a role for epigenetic alterations at promoters during cancer progression (88). Unfortunately, these observations do not delineate the causal role of changes in either H3K4me3 or H3K27me3 at promoter in cancer. Future work relying on the recent development of epigenetic editing technologies, such as Transcription Activator-Like Effector (TALE) or deactivated-Cas9 (dCas9) fused with epigenetic writer or eraser proteins (e.g. TALE-TET1, TALE-LSD1, dCas9-p300)(89–93) will provide an opportunity to directly assess the role for the changes in histone modifications targeting promoters in oncogenesis. In support, increased IL1RN gene expression was achieved in cells expressing the dCas9-p300 fusion protein targeted to the IL1RN promoter, which allowed for lysine 27 acetylation of histone H3 (H3K27ac) (91).
Genetic and epigenetic alterations target enhancers in cancer
Enhancers are cis-regulatory elements found tens to thousands of base-pairs away from their target gene promoter that can modulate gene expression independently of their orientation compared that of their target gene. They serve to modulate the activation of promoters and fine-tune transcription in a cell-type specific manner, a property that renders enhancer activity modulation ideal for genetic and epigenetic alterations to impact cell identity, abrogate cellular differentiation and promote oncogenesis (94).
Genetic alterations at enhancers affect gene expression in cancer
Genetic predispositions for various traits and diseases identified through genome-wide association studies preferentially map to noncoding cis-regulatory elements, particularly enhancers, in a disease- and tissue-specific manner (Figure 3A) (95). Risk-SNPs found in enhancers can change the DNA recognition motifs of transcription factors to alter their binding to the chromatin, directly impacting the transactivation potential of enhancers, and modulate target gene expression (95).
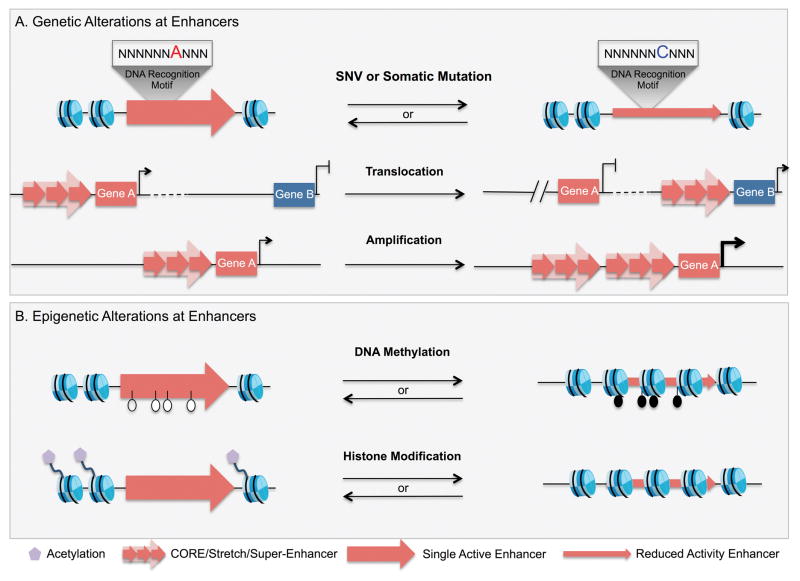
Figure 3. Genetic and epigenetic alterations are observed at enhancers in cancer.
A) Single Nucleotide Variants (SNVs) and structural variations can alter enhancer activity. SNVs and somatic mutations observed in enhancers can modulate the activity of these regulatory elements by changing their affinity for transcription factors. Translocation of a region that acts as an enhancer that places it in proximity of an oncogene can drive its aberrant expression. Similarly, amplification of an active enhancer element that is associated with an oncogene can drive its over-expression and subsequently contribute to oncogenesis. These genetic alterations to enhancers ultimately serve to modulate expression of oncogenes or tumor-suppressor genes. B) Changes in the epigenetic identity have been reported at enhancers in cancer. Hyper- or hypomethylation of CpGs at enhancers affects the accessibility of the DNA to transcription factors. Changes in the composition of post-translational modifications to histone in enhancers are thought to impact transcription factor binding to the chromatin. Increased histone acetylation increases chromatin accessibility to favor transcription factor binding, whereas loss of acetylation decreases chromatin accessibility thereby modulating the activity of enhancer.
This is exemplified by the colorectal cancer risk-associated SNP rs6983267 identified on chromosome 8q24 (96,97) that maps to an enhancer containing a consensus TCF4 recognition motif upstream of the MYC gene (98,99). The variant risk-allele of this SNP increases the binding of TCF4 to the enhancer compared to the reference allele, driving the aberrant overexpression of MYC (98,99). In agreement with the 8q24 region physically interacting with the MYC promoter in other cancer types (100), the rs6983267 locus is associated with the risk of developing other cancers including liver, lung and prostate cancer (101,102). More recently, multiple lymphoma risk-associated SNPs (rs2445610, rs13255292, rs7826019, rs59602790) were shown to map to subtype-specific lymphoma cis-regulatory elements within the chromosome 8q24 risk-locus (103). Separately, the risk-associated SNP rs1859961 maps to a prostate cancer-specific enhancer in the chromosome 17q24.3 prostate cancer risk locus that regulates SOX9 gene expression (104). Aberrant SOX9 gene expression is associated with increased risk for prostate cancer and is involved in prostate oncogenesis in mice (105). The rs4784227 SNP at the chromosome 16q12.1 breast cancer risk-locus provides further evidence of genetic alterations targeting enhancers in cancer. The variant risk-allele of the rs4784227 SNP changes the sequence of a Forkhead DNA recognition motif within an enhancer that regulates the transcription of the TOX3 gene, favoring the binding of the FOXA1 transcription factor (106). The increased binding of FOXA1 represses the transactivation ability of the enhancer through the recruitment of the transcriptional repressor Groucho/TLE, resulting in the decreased expression of the TOX3 tumor suppressor gene (106).
Finally, SNPs can also target units of enhancers referred to as Clusters Of Regulatory Elements (COREs), such as super-enhancers or stretch-enhancers, which correspond to multiple enhancers in close proximity to each other (Figure 3A) (107,108). This is showcased by the rs2168101 SNP mapping to a tissue-specific super-enhancer near the LMO1 neuroblastoma oncogene on chromosome 11p15 (109). The variant allele of this SNP disrupts the binding of GATA3 to lower the expression of LMO1, reducing the risk of developing neuroblastoma (109,110).
While these examples consist of single functional SNPs changing the activity of enhancers, recent work demonstrated that multiple SNPs within a risk-locus can impact distinct enhancers, classifying these as Multiple Enhancer Variants (MEVs) risk-loci (111). These MEVs have been reported to contribute to disease onset, including cancer. For instance, three SNPs (rs12352658, rs7847449 and rs10759944) in linkage disequilibrium with each other within the chromosome 9q22 thyroid cancer risk-locus can change the transactivation potential of two different enhancers that physically interact with the promoter of the FOXE1 and PTCSC2 genes (112). This likely account for the reduced expression of the FOXE1 and PTCSC2 genes associated with the 9q22 risk-locus in normal thyroid tissue from cancer patients (113). Overall, the functional interpretation of risk-loci identified through genome-wide association studies showcases the contribution of genetic alteration in enhancers to promote cancer development.
Similar to inherited genetic variants, acquired somatic mutations can alter enhancer activity and contribute to oncogenesis (Figure 3A). While enhancers are typified by a reduced mutational density compared to the genome found in heterochromatin, argued to result from active DNA repair in these elements (114), mutations do preferentially accumulate in enhancers present in the tissue from which the tumor originates (115). For example, a putative enhancer region of the PAX5 gene essential for the commitment of lymphoid progenitors into the B-cell lineage located on chromosome 9p13 is found to be recurrently mutated in chronic lymphocytic leukemia tumors (116,117). These mutations mapping to this enhancer are significantly correlated with altered PAX5 gene expression. Moreover, the CRISPR/Cas9-mediated deletion and introduction of mutations at this enhancer region in B-cells reduced PAX5 gene expression, suggesting these mutations directly alter the activity of the enhancer to disrupt PAX5 expression (116). Moreover, heterozygous 2–18 basepair indel mutations map to an intergenic site 7.5 kilobases upstream of the TAL1 transcription start site reported in 8 of 146 T-cell acute lymphoblastic leukemia samples, all changing the enhancer landscape by creating binding motifs for the MYB transcription factor (118). This allows MYB binding to the chromatin followed by the recruitment of its binding partner CBP, a lysine acetyltransferase, leading to the formation of a super-enhancer upstream of the TAL1 oncogene to drive its overexpression (118).
In addition to point mutations, enhancer activity is also affected by structural variants in cancer, such as inversions, translocations and copy number alterations (Figure 3A). In medulloblastoma, the GFI1 and GFI1B loci are translocated from transcriptionally silent chromatin regions into the proximity of active super-enhancers, presenting them as novel oncogenic drivers (119). Similarly, the repositioning of an enhancer near the GATA2 gene on chromosome 3q21 to an ectopic region near the EVI1 gene through inversions and translocations has been reported in acute myeloid leukemia (120). This leads to the formation of a super-enhancer that physically interacts with the EVI1 promoter through chromatin interactions, reducing the expression of GATA2 while simultaneously increasing the expression of the EVI1 proto-oncogene (120). In glioblastoma, the aberrant expression of the hTERT gene is also suggested to be affected by the rearrangement of a super-enhancer normally found on chromosome 10q22 to the hTERT gene promoter located on chromosome 5p15 (121). Likewise, the overexpression of MYC reported in multiple myeloma is mediated through a translocation of a 3′ IgH super-enhancer adjacent to the MYC oncogene (107). Moreover, as a result from the fusion between MYB and QKI (MYB-QKI) in angiocentric gliomas, active enhancers including two super-enhancers demarcated by H2K27ac are translocated from the QKI gene locus located on chromosome 6q26 near the MYB gene located on chromosome 6q23 to support aberrant MYB expression (122). The MYB gene is also targeted by translocations in adenoid cystic carcinoma with the NFIB and TGFBR3 loci (123). The translocations juxtapose enhancers, including super-enhancers, to the MYB locus, giving rise to a positive feedback loop regulating the aberrant expression of this potent oncogene mediated by the binding of the MYB protein to the translocated super-enhancer (123). Finally, copy number alterations in regions that harbor super-enhancers can also contribute to aberrant gene expression (Figure 3A). For instance, two focal amplification events of regions harboring super-enhancers were identified and associated with the aberrant expression of the MYC oncogene in uterine corpus endometrial carcinoma and lung adenocarcinoma (124). The CRISPR/Cas9-mediated deletion of a 1.7 kb enhancer, part of the super-enhancer region driving MYC overexpression in lung adenocarcinoma NCI-H2009 cells led to a significant reduction in MYC expression and impaired clonogenic growth, suggesting that super-enhancer amplification can prompt aberrant gene expression (124). Additional amplification events of regions inclusive of super-enhancers associated with an increase in gene expression are also observed near the KLF5, USP12, and PARD6B genes in head and neck squamous cell carcinoma, colorectal cancer and liver hepatocellular carcinoma, respectively (124).
In summary, various types of genetic alterations targeting enhancers can adversely modulate their activity to impact normal transcription and gene expression, and contribute to cancer development.
Epigenetic alterations accumulate at enhancers in cancer
Enhancer activity is also subject to epigenetic regulation (Figure 3B). Aberrant DNA methylation observed at enhancers in cancer was suggested to be more closely relate to changes in gene expression than at promoters (125). This may partly be due to differential transcription factor binding. Transcription factors are suggested to bind DNA hypomethylated enhancers more readily than DNA methylated enhancers as exemplified by the enrichment of FOXQ1 binding within DNA hypomethylated enhancers previously implicated in colorectal cancer oncogenesis (125–127). DNA hypomethylated enhancers responsive to ESR1 binding in breast cancer are also suggested to be critical for the development of ESR1-positive breast cancer (128). Moreover, aberrant enhancer DNA hypomethylation during oncogenesis is suggested to associate with the upregulation of cancer-related gene expression, whereas DNA hypermethylation at enhancers correlates with reduced target gene expression (128,129). In support of this, a putative DNA hypomethylated enhancer is associated with the increased expression of its target genes including the MYC and RNF43 oncogenes, and DNA hypermethylation at enhancers are associated with the reduced expression of DAXX and GET4 in breast cancer (126,128). These studies suggest a linkage between aberrant DNA methylation at enhancers and its potential role in altering transcription factor binding and gene expression in cancer development.
Enhancers permissive to transcription factor binding are commonly flanked by nucleosomes mono- and dimethylated on lysine 4 of histone H3 (H3K4me1 and H3K4me2) (130–133). Moreover, active enhancers are further discriminated from poised enhancers by being flanked with H3K27ac nucleosomes (Figure 3B) (134). Genome-wide profiling for H3K4me1 in both normal colon epithelia and colorectal cancer cells revealed thousands of enhancers, termed Variant Enhancer Loci (VEL) that are either lost or gained in colorectal cancer cells compared to normal colon crypts, suggestive of ectopic enhancer activity in the process of cancer initiation (86). These VEL associate with differential expression of their putative target gene in normal versus colon cancer cells (86). Specifically, enhancers active in normal colon but inactive in colorectal cancer cells, are found near genes that are part of the normal colon gene expression profile and vice versa (86). VEL also characterize cancer progression. For instance, thousands of enhancers active in endocrine therapy-sensitive breast cancer cells are no longer active in endocrine therapy-resistant cells (135). This change in enhancer usage reflects differences in the transcriptional machinery that inform on alternative therapeutic strategies (135). Moreover, cells resistant to gamma-secretase inhibitor (GSI) in T- cell acute lymphoblastic leukemia appear to be epigenetically labile as they can readily re- activate a transcriptional program typical of GSI sensitive cells upon GSI withdrawal. Furthermore, these cells are sensitive to BRD4 inhibition (136), known to antagonize the activity of super-enhancers (137).
The cause of epigenetic alterations at enhancers is still under investigation but whole exome sequencing of tumor samples supports a role for genetic alterations in chromatin remodeling factors (138). This is exemplified by the mutational load in EP300 (p300), ARID1A, CREBBP (CBP), MLL3/4 and LDB1 genes reported in bladder cancer, hepatocellular carcinoma, non-Hodgkin lymphoma, medulloblastoma, breast cancer and colon cancer (139– 145). Mutations in MLL3/4 are proposed to destabilize the MLL3/4 protein, reduce its binding to transcription factors or to inactivate its catalytic domain that can impact the methylation of nucleosomes at enhancers (138,146). Likewise, evidence suggests that mutations in the tumor suppressor ARID1A gene (147) can impinge upon the activity of this SWI/SNF chromatin remodeling complex subunit to favor oncogenesis (148). Overall, this warrants further characterization of mutations in chromatin factors to delineate their impact on cis-regulatory element activation.
Anchors of chromatin interaction are targets of genetic and epigenetic alterations in cancer
Chromosomes are organized into a hierarchy of chromatin interactions that coordinate the interplay between enhancers and promoters to regulate the expression of their target transcripts (Figure 1). Chromatin interactions, also referred to as chromatin loops, mediate the communication between diverse types of cis-regulatory elements separated by large genomic distances at the kilobases scale by bringing them into close physical proximity. Megabase-scale chromatin interactions define topologically associated domains (TADs) separated by boundaries that are broadly conserved across cell- and tissue-types and demarcate active from inactive chromatin domains (Figure 1A–B) (149–151). Smaller range chromatin interactions anchored at promoters facilitate the interactions with enhancers in a cell type-specific manner and relate to cell type-specific gene expression profiles (Figure 1C) (152–155). These promoter-enhancer chromatin interactions are constrained within TAD boundaries because these limit their formation across adjoining TADs to insulate target gene promoters from aberrant enhancer activity (Figure 1D) (156). Chromatin interactions are mediated by factors that recognize the DNA sequence at loop anchors in conjunction with intermediary proteins. The anchors that define TAD boundaries are occupied by the CCCTC-binding protein (CTCF), which recognizes a specific 12 basepair consensus motif, and the cohesin complex consisting of RAD21, STAG1, SMC1a and SMC3 (Figure 1D) (157).
Although the vast majority of TAD boundaries harbor CTCF/cohesin binding sites, co-binding of these factors does not necessarily create these boundaries. In fact, several studies have shown that the majority of CTCF/cohesin binding sites do not block physical long-range chromatin interactions, and are therefore considered to be located outside of TAD boundaries (152,155,158). A subset of CTCF and cohesin co-bound sites are implicated in interactions involving anchors within a few hundred kilobases from each other, such as promoter-enhancer or enhancer-enhancer interactions (152,159,160). While CTCF binding appears to be directed to distal cis-regulatory elements as opposed to promoters (149,161,162), the chromatin-interaction factor ZNF143 directly occupies promoters (153,154,163,164) to anchor chromatin interactions (Figure 1C) (154). Studies that have examined genome-wide interaction maps in conjunction with transcription factor binding profiles have identified additional factors that preferentially occupy anchors of chromatin interactions (150,153,154). Some of these additional proteins found at anchors, such as the Mediator complex, assist in the formation of chromatin interactions (161). However, the role for most of the factors present at anchors of chromatin interactions remains to be determined.
Genetic alterations target anchors of chromatin interaction
Maintaining the genetic identity of anchors of chromatin interaction ensures appropriate chromatin folding to guide the regulation of transcriptional programs in normal cells. Chromatin interaction frequencies can be affected by genetic alterations targeting anchors of chromatin interaction (Figure 4A), as showcased by the rs12913832 human pigmentation-associated risk-SNP mapping to the HERC2 enhancer modulating the loop interaction with the oculocutaneous albinism II (OCA2) promoter (165). Similarly, the ZC3HAV1 gene-locus harbors a functional SNP rs13228237 capable of altering the interaction frequency between the ZC3HAV1 gene promoter and a distal enhancer located 200 kilobases away by imposing an allele-specific bias in the binding of the chromatin interaction factor ZNF143 (154). Furthermore, an analysis of mutations reported in the International Cancer Genome Consortium (ICGC) pan-cancer database revealed that the DNA recognition motifs for CTCF and ZNF143 are among the motifs with the highest average number of cancer-associated mutations (166). Moreover, in a study of 213 colorectal tumors, mutations were reported to accumulate in the DNA recognition motif for CTCF and its flanking sequences (167). Variation in the sequences flanking core transcription factor binding sites has a significant impact on binding. Indeed, these variations can explain why factors from the same family, which often recognize nearly identical core recognition motifs, have distinct genome-wide binding profiles and serve different biological functions in vivo (168–172).
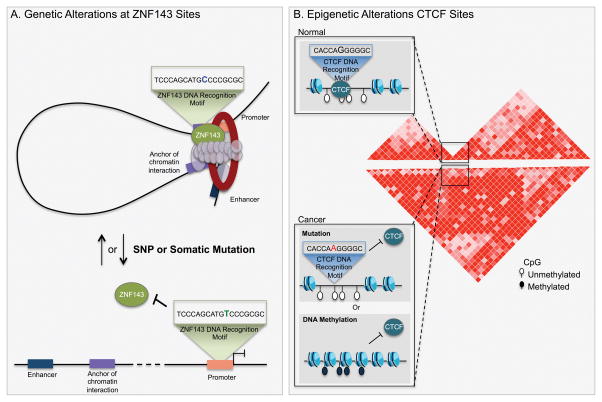
Figure 4. Genetic and epigenetic alterations targeting anchors of chromatin interactions.
A) ZNF143 recognizes a DNA binding motif that is enriched at promoters. Genetic alteration in the consensus motif of ZNF143 can deter ZNF143 binding and result in the impaired chromatin interactions between promoters and enhancers and impact the expression of target genes. B) Anchors of chromatin interactions that define topologically associated domains (TADs) are bound by CTCF. Disruption of CTCF binding at these anchors can abrogate the formation of chromatin interactions to ultimately disrupt the three-dimensional organization of the genome in cancer. CTCF recognizes a 12 base-pair (bp) consensus motif mutated in various cancer types. The binding of CTCF to the DNA can also be compromised by DNA methylation, as reported in glioblastoma.
While these studies did not distinguish between mutations affecting TAD boundaries versus inner-TAD promoter-enhancer or enhancer-enhancer interactions, several recent reports have focused on the role of genetic alterations at TAD boundaries. For instance, CTCF binding sites that define TAD boundaries show a striking enrichment for mutations compared to non-boundary CTCF binding-sites in liver and esophageal carcinomas (173). Moreover, the CRISPR/Cas9-mediated deletion of two CTCF/cohesin binding sites commonly mutated in T cell acute lymphoblastic leukemia (T-ALL) results in the loss of TAD boundaries and leads to a significant increase in LMO2 and TAL1 genes expression, two proto-oncogenes involved in hematopoiesis (173). Hence, genetic alterations to the anchors of chromatin interaction can disrupt the activity of noncoding regulatory elements and impact downstream target gene expression.
Aberrant epigenetic modifications target anchors of chromatin interactions in cancer
A distinctive feature of CTCF binding sites is the absence of DNA methylation (174–176). Genome-wide CTCF binding in multiple cell types negatively correlates with DNA methylation (176,177). Changes to the DNA methylation profile at anchors of chromatin interactions can compromise CTCF binding and its activity (Figure 4B). This was recently reported in human IDH mutant gliomas that exhibit a CpG Island Methylator Phenotype (CIMP), characterized by genome-wide hypermethylation at CTCF/cohesin binding sites (178). Moreover, IDH1 mutants were shown to be sufficient in driving the CIMP phenotype in gliomas resulting in aberrant gene expression programs (179). Hypermethylation CTCF/cohesin binding sites interferes with CTCF binding to the chromatin, which results in altered chromatin interactions and aberrant expression of the PDGFRA oncogene (178). The CIMP phenotype is known in several other cancer types including colorectal, breast, endometrial cancer (180). Although the effect of genome-wide methylation on the binding of chromatin-interaction factors in these cancer types is yet to be assessed, results in gliomas combined with the well-established mutual-exclusivity between CTCF binding and DNA methylation on the chromatin suggests that epigenetic alterations impacting chromatin interactions might be common across many cancer types.
Clinical implications for the functional noncoding genome
Identifying therapeutic opportunities and biomarkers within the functional noncoding cancer genome
Specific factors are recruited to cis-regulatory elements including BRD4, a chromatin reader featuring two N-terminal bromodomains that bind to acetylated histones to subsequently recruit transcriptional activators (137,181). BRD4 inactivation with bromodomain inhibitors such as JQ1 and iBET can inhibit the cis-regulatory element activity as reported for super-enhancers that can drive oncogene overexpression. This is showcased in the repression of aberrant MYC expression in various malignancies, including medulloblastoma, B-cell acute lymphoblastic leukemia, acute myeloid leukemia, Merkel cell carcinoma and resistance in T-cell acute lymphoblastic leukemia that halt proliferation (136,181–186). Moreover, BRD4 inhibition induces differentiation and growth arrest in patient-derived NUT midline carcinoma cells (186). This example suggests that chemical modulation of cis-regulatory element activity can be of benefit to treat cancer.
The identification of genetic and epigenetic alterations in functional noncoding cis-regulatory elements also provides a source of biomarkers to monitor cancer development (187–190). For instance, genetic alterations mapping to the hTERT promoter are associated with advanced cancer staging and poor patient survival in glioma, bladder and thyroid cancer patients (40,45,191–193). Mutations in the hTERT promoter in glioma patients particularly have been suggested to confer radioresistance and resistance to temozolomide treatment (191,194). Moreover, aberrant hypermethylation of the hTERT promoter can also serve as a predictive biomarker for poor survival in ependymoma patients (195). Similarly, the CIMP phenotype has shown promise as a discriminator for patient stratification and outcome in ependymoma, glioma, glioblastoma, colorectal cancer and hepatocellular carcinoma (180,196–201). These studies collectively suggest that genetic and epigenetic alterations targeting noncoding elements offer new opportunities for biomarker discovery in cancer to guide treatment and disease monitoring.
Conclusion
In summary, evidence supports the critical role of the noncoding genome in maintaining normal transcriptional programs and cell identity. Genetic and epigenetic alterations targeting functional noncoding cis-regulatory elements reported in cancer can alter these transcriptional programs and promote oncogenesis. These alterations inform on tumor biology and also reveal new biomarkers for patient stratification associated with distinct outcome. Hence, the comprehensive characterization of the noncoding cancer genome offers a promising avenue to delineate new therapeutic opportunities, identify biomarkers for disease monitoring and ultimately improve patient care.
Statement of Significance.
The majority of genetic and epigenetic alterations accumulate in the noncoding genome throughout oncogenesis. Discriminating driver from passenger events is a challenge that holds great promise to improve our understanding of the etiology of different cancer types. Advancing our understanding of the noncoding cancer genome may thus identify new therapeutic opportunities and accelerate our capacity to find improved biomarkers to monitor various stages of cancer development.
Acknowledgments
Grant Support
The National Cancer Institute (NCI) at the National Institute of Health (NIH) (R01CA155004 to M.L.), the Princess Margaret Cancer Foundation (M.L.), The Canadian Cancer Society (CCSRI702922 to M.L.) supported this work. M.L. holds an investigator award from the Ontario Institute for Cancer Research (OICR), a new investigator salary award from the Canadian Institute of Health Research (CIHR) and a Movember Rising Star award from Prostate Cancer Canada (PCC) (RS2014-04).
The authors apologize to colleagues whose work was not cited in this review due to space limitation. The authors thank Drs. Swneke D. Bailey and Paul Guilhamon (Princess Margaret Cancer Research Centre), Jérôme Eeckoute (Université de Lille) as well as Michael D. Wilson (Department of Molecular Genetics, University of Toronto) for their valuable comments towards this review.
Footnotes
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest to disclose.
References
- 1.Kellis M, Wold B, Snyder MP, Bernstein BE, Kundaje A, Marinov GK, et al. Defining functional DNA elements in the human genome. Proc Natl Acad Sci U S A. 2014;111:6131–8. doi: 10.1073/pnas.1318948111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.1000 Genomes Project Consortium. Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, et al. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Graur D, Zheng Y, Price N, Azevedo RBR, Zufall RA, Elhaik E. On the Immortality of Television Sets: “Function” in the Human Genome According to the Evolution-Free Gospel of ENCODE. Genome Biol Evol. 2013;5:578–90. doi: 10.1093/gbe/evt028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Treangen TJ, Salzberg SL. Repetitive DNA and next-generation sequencing: computational challenges and solutions. Nat Rev Genet. 2012;13:36–46. doi: 10.1038/nrg3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wittkopp PJ, Kalay G. Cis-regulatory elements: molecular mechanisms and evolutionary processes underlying divergence. Nat Rev Genet. 2012;13:59–69. doi: 10.1038/nrg3095. [DOI] [PubMed] [Google Scholar]
- 6.Bouwman BAM, de Laat W. Getting the genome in shape: the formation of loops, domains and compartments. Genome Biol. 2015;16:154. doi: 10.1186/s13059-015-0730-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maston GA, Evans SK, Green MR. Transcriptional regulatory elements in the human genome. Annu Rev Genomics Hum Genet. 2006;7:29–59. doi: 10.1146/annurev.genom.7.080505.115623. [DOI] [PubMed] [Google Scholar]
- 8.Cech TR, Steitz JA. The noncoding RNA revolution-trashing old rules to forge new ones. Cell. 2014;157:77–94. doi: 10.1016/j.cell.2014.03.008. [DOI] [PubMed] [Google Scholar]
- 9.Alföldi J, Lindblad-Toh K. Comparative genomics as a tool to understand evolution and disease. Genome Res. 2013;23:1063–8. doi: 10.1101/gr.157503.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rands CM, Meader S, Ponting CP, Lunter G. 8. 2% of the Human genome is constrained: variation in rates of turnover across functional element classes in the human lineage. PLoS Genet. 2014;10:e1004525. doi: 10.1371/journal.pgen.1004525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Siepel A, Bejerano G, Pedersen JS, Hinrichs AS, Hou M, Rosenbloom K, et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–50. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yue F, Cheng Y, Breschi A, Vierstra J, Wu W, Ryba T, et al. A comparative encyclopedia of DNA elements in the mouse genome. Nature. 2014;515:355–64. doi: 10.1038/nature13992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schmidt D, Wilson MD, Ballester B, Schwalie PC, Brown GD, Marshall A, et al. Five-vertebrate ChIP-seq reveals the evolutionary dynamics of transcription factor binding. Science. 2010;328:1036–40. doi: 10.1126/science.1186176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ballester B, Medina-Rivera A, Schmidt D, Gonzàlez-Porta M, Carlucci M, Chen X, et al. Multi-species, multi-transcription factor binding highlights conserved control of tissue-specific biological pathways. Elife. 2014;3:e02626. doi: 10.7554/eLife.02626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stefflova K, Thybert D, Wilson MD, Streeter I, Aleksic J, Karagianni P, et al. Cooperativity and rapid evolution of cobound transcription factors in closely related mammals. Cell. 2013;154:530–40. doi: 10.1016/j.cell.2013.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang FW, Hodis E, Xu MJ, Kryukov GV, Chin L, Garraway LA. Highly recurrent TERT promoter mutations in human melanoma. Science. 2013;339:957–9. doi: 10.1126/science.1229259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Horn S, Figl A, Rachakonda PS, Fischer C, Sucker A, Gast A, et al. TERT promoter mutations in familial and sporadic melanoma. Science. 2013;339:959–61. doi: 10.1126/science.1230062. [DOI] [PubMed] [Google Scholar]
- 18.Melnikov A, Murugan A, Zhang X, Tesileanu T, Wang L, Rogov P, et al. Systematic dissection and optimization of inducible enhancers in human cells using a massively parallel reporter assay. Nat Biotechnol. 2012;30:271–7. doi: 10.1038/nbt.2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arnold CD, Gerlach D, Stelzer C, Boryń ŁM, Rath M, Stark A. Genome-wide quantitative enhancer activity maps identified by STARR-seq. Science. 2013;339:1074–7. doi: 10.1126/science.1232542. [DOI] [PubMed] [Google Scholar]
- 20.Mukherjee S, Berger MF, Jona G, Wang XS, Muzzey D, Snyder M, et al. Rapid analysis of the DNA-binding specificities of transcription factors with DNA microarrays. Nat Genet. 2004;36:1331–9. doi: 10.1038/ng1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Patwardhan RP, Hiatt JB, Witten DM, Kim MJ, Smith RP, May D, et al. Massively parallel functional dissection of mammalian enhancers in vivo. Nat Biotechnol. 2012;30:265–70. doi: 10.1038/nbt.2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gonzalez-Perez A, Abel G-P, Ville M, Boris R, Ritchie GRS, Pau C, et al. Computational approaches to identify functional genetic variants in cancer genomes. Nat Methods. 2013;10:723–9. doi: 10.1038/nmeth.2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ghoussaini M, Edwards SL, Michailidou K, Nord S, Cowper-Sal Lari R, Desai K, et al. Evidence that breast cancer risk at the 2q35 locus is mediated through IGFBP5 regulation. Nat Commun. 2014;4:4999. doi: 10.1038/ncomms5999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang D, Ovcharenko I. Identifying causal regulatory SNPs in ChIP-seq enhancers. Nucleic Acids Res. 2015;43:225–36. doi: 10.1093/nar/gku1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li S, Ovcharenko I. Human Enhancers Are Fragile and Prone to Deactivating Mutations. Mol Biol Evol. 2015;32:2161–80. doi: 10.1093/molbev/msv118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Khurana E, Fu Y, Colonna V, Mu XJ, Kang HM, Lappalainen T, et al. Integrative annotation of variants from 1092 humans: application to cancer genomics. Science. 2013;342:1235587. doi: 10.1126/science.1235587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dunham I, Kundaje A, Aldred SF, Collins PJ, Davis CA, Doyle F, et al. Nature. Vol. 489. Nature Publishing Group; 2012. An integrated encyclopedia of DNA elements in the human genome; pp. 57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.ENCODE Project Consortium. The ENCODE (ENCyclopedia Of DNA Elements) Project. Science. 2004;306:636–40. doi: 10.1126/science.1105136. [DOI] [PubMed] [Google Scholar]
- 29.Thurman RE, Rynes E, Humbert R, Vierstra J, Maurano MT, Haugen E, et al. The accessible chromatin landscape of the human genome. Nature. 2012;489:75–82. doi: 10.1038/nature11232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kawai J, Shinagawa A, Shibata K, Yoshino M, Itoh M, Ishii Y, et al. Functional annotation of a full-length mouse cDNA collection. Nature. 2001;409:685–90. doi: 10.1038/35055500. [DOI] [PubMed] [Google Scholar]
- 31.Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–63. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 32.Bernstein BE, Stamatoyannopoulos JA, Costello JF, Ren B, Milosavljevic A, Meissner A, et al. The NIH Roadmap Epigenomics Mapping Consortium. Nat Biotechnol. 2010;28:1045–8. doi: 10.1038/nbt1010-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shandilya J, Roberts SGE. The transcription cycle in eukaryotes: from productive initiation to RNA polymerase II recycling. Biochim Biophys Acta. 2012;1819:391–400. doi: 10.1016/j.bbagrm.2012.01.010. [DOI] [PubMed] [Google Scholar]
- 34.Jonkers I, Lis JT. Getting up to speed with transcription elongation by RNA polymerase II. Nat Rev Mol Cell Biol. 2015;16:167–77. doi: 10.1038/nrm3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kwak H, Lis JT. Control of transcriptional elongation. Annu Rev Genet. 2013;47:483–508. doi: 10.1146/annurev-genet-110711-155440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mosrati MA, Malmström A, Lysiak M, Krysztofiak A, Hallbeck M, Milos P, et al. TERT promoter mutations and polymorphisms as prognostic factors in primary glioblastoma. Oncotarget. 2015;6:16663–73. doi: 10.18632/oncotarget.4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Remke M, Ramaswamy V, Peacock J, Shih DJH, Koelsche C, Northcott PA, et al. TERT promoter mutations are highly recurrent in SHH subgroup medulloblastoma. Acta Neuropathol. 2013;126:917–29. doi: 10.1007/s00401-013-1198-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Eckel-Passow JE, Lachance DH, Molinaro AM, Walsh KM, Decker PA, Sicotte H, et al. Glioma Groups Based on 1p/19q, IDH, and TERT Promoter Mutations in Tumors. N Engl J Med. 2015;372:2499–508. doi: 10.1056/NEJMoa1407279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vinagre J, Almeida A, Pópulo H, Batista R, Lyra J, Pinto V, et al. Frequency of TERT promoter mutations in human cancers. Nat Commun. 2013;4:2185. doi: 10.1038/ncomms3185. [DOI] [PubMed] [Google Scholar]
- 40.Killela PJ, Reitman ZJ, Jiao Y, Bettegowda C, Agrawal N, Diaz LA, Jr, et al. TERT promoter mutations occur frequently in gliomas and a subset of tumors derived from cells with low rates of self-renewal. Proc Natl Acad Sci U S A. 2013;110:6021–6. doi: 10.1073/pnas.1303607110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weinhold N, Jacobsen A, Schultz N, Sander C, Lee W. Genome-wide analysis of noncoding regulatory mutations in cancer. Nat Genet. 2014;46:1160–5. doi: 10.1038/ng.3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chiba K, Johnson JZ, Vogan JM, Wagner T, Boyle JM, Hockemeyer D. Cancer-associated TERT promoter mutations abrogate telomerase silencing. eLife. 2015;4:e07918. doi: 10.7554/eLife.07918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fujimoto A, Furuta M, Totoki Y, Tsunoda T, Kato M, Shiraishi Y, et al. Whole-genome mutational landscape and characterization of noncoding and structural mutations in liver cancer. Nat Genet. 2016;48:500–9. doi: 10.1038/ng.3547. [DOI] [PubMed] [Google Scholar]
- 44.Muzza M, Colombo C, Rossi S, Tosi D, Cirello V, Perrino M, et al. Telomerase in differentiated thyroid cancer: promoter mutations, expression and localization. Mol Cell Endocrinol. 2015;399:288–95. doi: 10.1016/j.mce.2014.10.019. [DOI] [PubMed] [Google Scholar]
- 45.Rachakonda PS, Hosen I, de Verdier PJ, Fallah M, Heidenreich B, Ryk C, et al. TERT promoter mutations in bladder cancer affect patient survival and disease recurrence through modification by a common polymorphism. Proc Natl Acad Sci U S A. 2013;110:17426–31. doi: 10.1073/pnas.1310522110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nagore E, Heidenreich B, Rachakonda S, Garcia-Casado Z, Requena C, Soriano V, et al. TERT promoter mutations in melanoma survival. Int J Cancer. 2016;139:75–84. doi: 10.1002/ijc.30042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tomlins SA, Rhodes DR, Perner S, Dhanasekaran SM, Mehra R, Sun X-W, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–8. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 48.Cancer Genome Atlas Research Network. The Molecular Taxonomy of Primary Prostate Cancer. Cell. 2015;163:1011–25. doi: 10.1016/j.cell.2015.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Carver BS, Jennifer T, Anuradha G, Zhenbang C, Safa S, Arkaitz C, et al. Aberrant ERG expression cooperates with loss of PTEN to promote cancer progression in the prostate. Nat Genet. 2009;41:619–24. doi: 10.1038/ng.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Demichelis F, Rubin MA. TMPRSS2-ETS fusion prostate cancer: biological and clinical implications. J Clin Pathol. 2007;60:1185–6. doi: 10.1136/jcp.2007.046557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kataoka K, Shiraishi Y, Takeda Y, Sakata S, Matsumoto M, Nagano S, et al. Aberrant PD-L1 expression through 3′-UTR disruption in multiple cancers. Nature. 2016;534:402–6. doi: 10.1038/nature18294. [DOI] [PubMed] [Google Scholar]
- 52.Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–19. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 53.Voss TC, Hager GL. Dynamic regulation of transcriptional states by chromatin and transcription factors. Nat Rev Genet. 2014;15:69–81. doi: 10.1038/nrg3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kulis M, Esteller M. DNA methylation and cancer. Adv Genet. 2010;70:27–56. doi: 10.1016/B978-0-12-380866-0.60002-2. [DOI] [PubMed] [Google Scholar]
- 55.Baylin SB, Jones PA. A decade of exploring the cancer epigenome — biological and translational implications. Nat Rev Cancer. 2011;11:726–34. doi: 10.1038/nrc3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xing X-B, Cai W-B, Luo L, Liu L-S, Shi H-J, Chen M-H. The Prognostic Value of p16 Hypermethylation in Cancer: A Meta-Analysis. PLoS One. 2013;8:e66587. doi: 10.1371/journal.pone.0066587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dammann R, Schagdarsurengin U, Liu L, Otto N, Gimm O, Dralle H, et al. Frequent RASSF1A promoter hypermethylation and K-ras mutations in pancreatic carcinoma. Oncogene. 2003;22:3806–12. doi: 10.1038/sj.onc.1206582. [DOI] [PubMed] [Google Scholar]
- 58.Esteller M, Sparks A, Toyota M, Sanchez-Cespedes M, Capella G, Peinado MA, et al. Analysis of adenomatous polyposis coli promoter hypermethylation in human cancer. Cancer Res. 2000;60:4366–71. [PubMed] [Google Scholar]
- 59.Esteller M, Silva JM, Dominguez G, Bonilla F, Matias-Guiu X, Lerma E, et al. Promoter hypermethylation and BRCA1 inactivation in sporadic breast and ovarian tumors. J Natl Cancer Inst. 2000;92:564–9. doi: 10.1093/jnci/92.7.564. [DOI] [PubMed] [Google Scholar]
- 60.Jin Z, Tamura G, Tsuchiya T, Sakata K, Kashiwaba M, Osakabe M, et al. Adenomatous polyposis coli (APC) gene promoter hypermethylation in primary breast cancers. Br J Cancer. 2001;85:69–73. doi: 10.1054/bjoc.2001.1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hou P, Ji M, Yang B, Chen Z, Qiu J, Shi X, et al. Quantitative analysis of promoter hypermethylation in multiple genes in osteosarcoma. Cancer. 2006;106:1602–9. doi: 10.1002/cncr.21762. [DOI] [PubMed] [Google Scholar]
- 62.Herman JG, Umar A, Polyak K, Graff JR, Ahuja N, Issa JP, et al. Incidence and functional consequences of hMLH1 promoter hypermethylation in colorectal carcinoma. Proc Natl Acad Sci U S A. 1998;95:6870–5. doi: 10.1073/pnas.95.12.6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Esteller M, Levine R, Baylin SB, Ellenson LH, Herman JG. MLH1 promoter hypermethylation is associated with the microsatellite instability phenotype in sporadic endometrial carcinomas. Oncogene. 1998;17:2413–7. doi: 10.1038/sj.onc.1202178. [DOI] [PubMed] [Google Scholar]
- 64.Lahtz C, Pfeifer GP. Epigenetic changes of DNA repair genes in cancer. J Mol Cell Biol. 2011;3:51–8. doi: 10.1093/jmcb/mjq053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ruscito I, Dimitrova D, Vasconcelos I, Gellhaus K, Schwachula T, Bellati F, et al. BRCA1 gene promoter methylation status in high-grade serous ovarian cancer patients--a study of the tumour Bank ovarian cancer (TOC) and ovarian cancer diagnosis consortium (OVCAD) Eur J Cancer. 2014;50:2090–8. doi: 10.1016/j.ejca.2014.05.001. [DOI] [PubMed] [Google Scholar]
- 66.Baylin SB, Herman JG. DNA hypermethylation in tumorigenesis: epigenetics joins genetics. Trends Genet. 2000;16:168–74. doi: 10.1016/s0168-9525(99)01971-x. [DOI] [PubMed] [Google Scholar]
- 67.Lujambio A, Ropero S, Ballestar E, Fraga MF, Cerrato C, Setién F, et al. Genetic unmasking of an epigenetically silenced microRNA in human cancer cells. Cancer Res. 2007;67:1424–9. doi: 10.1158/0008-5472.CAN-06-4218. [DOI] [PubMed] [Google Scholar]
- 68.Toyota M, Suzuki H, Sasaki Y, Maruyama R, Imai K, Shinomura Y, et al. Epigenetic silencing of microRNA-34b/c and B-cell translocation gene 4 is associated with CpG island methylation in colorectal cancer. Cancer Res. 2008;68:4123–32. doi: 10.1158/0008-5472.CAN-08-0325. [DOI] [PubMed] [Google Scholar]
- 69.Zhao J, Dahle D, Zhou Y, Zhang X, Klibanski A. Hypermethylation of the promoter region is associated with the loss of MEG3 gene expression in human pituitary tumors. J Clin Endocrinol Metab. 2005;90:2179–86. doi: 10.1210/jc.2004-1848. [DOI] [PubMed] [Google Scholar]
- 70.Benetatos L, Dasoula A, Hatzimichael E, Georgiou I, Syrrou M, Bourantas KL. Promoter hypermethylation of the MEG3 (DLK1/MEG3) imprinted gene in multiple myeloma. Clin Lymphoma Myeloma. 2008;8:171–5. doi: 10.3816/CLM.2008.n.021. [DOI] [PubMed] [Google Scholar]
- 71.Zhang X, Gejman R, Mahta A, Zhong Y, Rice KA, Zhou Y, et al. Maternally expressed gene 3, an imprinted noncoding RNA gene, is associated with meningioma pathogenesis and progression. Cancer Res. 2010;70:2350–8. doi: 10.1158/0008-5472.CAN-09-3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sun M, Xia R, Jin F, Xu T, Liu Z, De W, et al. Downregulated long noncoding RNA MEG3 is associated with poor prognosis and promotes cell proliferation in gastric cancer. Tumour Biol. 2014;35:1065–73. doi: 10.1007/s13277-013-1142-z. [DOI] [PubMed] [Google Scholar]
- 73.Gonzalgo ML, Pavlovich CP, Lee SM, Nelson WG. Prostate cancer detection by GSTP1 methylation analysis of postbiopsy urine specimens. Clin Cancer Res. 2003;9:2673–7. [PubMed] [Google Scholar]
- 74.Cairns P, Esteller M, Herman JG, Schoenberg M, Jeronimo C, Sanchez-Cespedes M, et al. Molecular detection of prostate cancer in urine by GSTP1 hypermethylation. Clin Cancer Res. 2001;7:2727–30. [PubMed] [Google Scholar]
- 75.Kron KJ, Liu L, Pethe VV, Demetrashvili N, Nesbitt ME, Trachtenberg J, et al. DNA methylation of HOXD3 as a marker of prostate cancer progression. Lab Invest. 2010;90:1060–7. doi: 10.1038/labinvest.2010.57. [DOI] [PubMed] [Google Scholar]
- 76.Hegi ME, Diserens A-C, Gorlia T, Hamou M-F, de Tribolet N, Weller M, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352:997–1003. doi: 10.1056/NEJMoa043331. [DOI] [PubMed] [Google Scholar]
- 77.Esteller M, Garcia-Foncillas J, Andion E, Goodman SN, Hidalgo OF, Vanaclocha V, et al. Inactivation of the DNA-repair gene MGMT and the clinical response of gliomas to alkylating agents. N Engl J Med. 2000;343:1350–4. doi: 10.1056/NEJM200011093431901. [DOI] [PubMed] [Google Scholar]
- 78.Ehrlich M. DNA methylation in cancer: too much, but also too little. Oncogene. 2002;21:5400–13. doi: 10.1038/sj.onc.1205651. [DOI] [PubMed] [Google Scholar]
- 79.Søes S, Daugaard IL, Sørensen BS, Carus A, Mattheisen M, Alsner J, et al. Hypomethylation and increased expression of the putative oncogene ELMO3 are associated with lung cancer development and metastases formation. Oncoscience. 2014;1:367–74. doi: 10.18632/oncoscience.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gumienny TL, Brugnera E, Tosello-Trampont AC, Kinchen JM, Haney LB, Nishiwaki K, et al. CED-12/ELMO, a novel member of the CrkII/Dock180/Rac pathway, is required for phagocytosis and cell migration. Cell. 2001;107:27–41. doi: 10.1016/s0092-8674(01)00520-7. [DOI] [PubMed] [Google Scholar]
- 81.Shukeir N, Pakneshan P, Chen G, Szyf M, Rabbani SA. Alteration of the methylation status of tumor-promoting genes decreases prostate cancer cell invasiveness and tumorigenesis in vitro and in vivo. Cancer Res. 2006;66:9202–10. doi: 10.1158/0008-5472.CAN-06-1954. [DOI] [PubMed] [Google Scholar]
- 82.Pakneshan P, Xing RH, Rabbani SA. Methylation status of uPA promoter as a molecular mechanism regulating prostate cancer invasion and growth in vitro and in vivo. FASEB J. 2003;17:1081–8. doi: 10.1096/fj.02-0973com. [DOI] [PubMed] [Google Scholar]
- 83.Kim TH, Barrera LO, Zheng M, Qu C, Singer MA, Richmond TA, et al. A high-resolution map of active promoters in the human genome. Nature. 2005;436:876–80. doi: 10.1038/nature03877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Barrera LO, Li Z, Smith AD, Arden KC, Cavenee WK, Zhang MQ, et al. Genome-wide mapping and analysis of active promoters in mouse embryonic stem cells and adult organs. Genome Res. 2008;18:46–59. doi: 10.1101/gr.6654808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–26. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 86.Akhtar-Zaidi B, Cowper-Sal middle dot lari R, Corradin O, Saiakhova A, Bartels CF, Balasubramanian D, et al. Epigenomic Enhancer Profiling Defines a Signature of Colon Cancer. Science. 2012;336:736–9. doi: 10.1126/science.1217277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hahn MA, Li AX, Wu X, Yang R, Drew DA, Rosenberg DW, et al. Loss of the polycomb mark from bivalent promoters leads to activation of cancer-promoting genes in colorectal tumors. Cancer Res. 2014;74:3617–29. doi: 10.1158/0008-5472.CAN-13-3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Xu K, Wu ZJ, Groner AC, He HH, Cai C, Lis RT, et al. EZH2 oncogenic activity in castration-resistant prostate cancer cells is Polycomb-independent. Science. 2012;338:1465–9. doi: 10.1126/science.1227604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Maeder ML, Angstman JF, Richardson ME, Linder SJ, Cascio VM, Tsai SQ, et al. Targeted DNA demethylation and activation of endogenous genes using programmable TALE-TET1 fusion proteins. Nat Biotechnol. 2013;31:1137–42. doi: 10.1038/nbt.2726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Mendenhall EM, Williamson KE, Reyon D, Zou JY, Ram O, Joung JK, et al. Locus-specific editing of histone modifications at endogenous enhancers. Nat Biotechnol. 2013;31:1133–6. doi: 10.1038/nbt.2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hilton IB, D’Ippolito AM, Vockley CM, Thakore PI, Crawford GE, Reddy TE, et al. Epigenome editing by a CRISPR-Cas9-based acetyltransferase activates genes from promoters and enhancers. Nat Biotechnol. 2015;33:510–7. doi: 10.1038/nbt.3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kearns NA, Genga RMJ, Enuameh MS, Garber M, Wolfe SA, Maehr R. Cas9 effector-mediated regulation of transcription and differentiation in human pluripotent stem cells. Development. 2014;141:219–23. doi: 10.1242/dev.103341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Shalem O, Sanjana NE, Zhang F. High-throughput functional genomics using CRISPR-Cas9. Nat Rev Genet. 2015;16:299–311. doi: 10.1038/nrg3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kron KJ, Bailey SD, Lupien M. Enhancer alterations in cancer: a source for a cell identity crisis. Genome Med. 2014;6:77. doi: 10.1186/s13073-014-0077-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zhang X, Bailey SD, Lupien M. Laying a solid foundation for Manhattan – “setting the functional basis for the post-GWAS era. Trends Genet. 2014;30:140–9. doi: 10.1016/j.tig.2014.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Pomerantz MM, Ahmadiyeh N, Jia L, Herman P, Verzi MP, Doddapaneni H, et al. The 8q24 cancer risk variant rs6983267 shows long-range interaction with MYC in colorectal cancer. Nat Genet. 2009;41:882–4. doi: 10.1038/ng.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Nan H, Morikawa T, Suuriniemi M, Imamura Y, Werner L, Kuchiba A, et al. Aspirin use, 8q24 single nucleotide polymorphism rs6983267, and colorectal cancer according to CTNNB1 alterations. J Natl Cancer Inst. 2013;105:1852–61. doi: 10.1093/jnci/djt331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wright JB, Brown SJ, Cole MD. Upregulation of c-MYC in cis through a large chromatin loop linked to a cancer risk-associated single-nucleotide polymorphism in colorectal cancer cells. Mol Cell Biol. 2010;30:1411–20. doi: 10.1128/MCB.01384-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Tuupanen S, Turunen M, Lehtonen R, Hallikas O, Vanharanta S, Kivioja T, et al. The common colorectal cancer predisposition SNP rs6983267 at chromosome 8q24 confers potential to enhanced Wnt signaling. Nat Genet. 2009;41:885–90. doi: 10.1038/ng.406. [DOI] [PubMed] [Google Scholar]
- 100.Ahmadiyeh N, Pomerantz MM, Grisanzio C, Herman P, Jia L, Almendro V, et al. 8q24 prostate, breast, and colon cancer risk loci show tissue-specific long-range interaction with MYC. Proc Natl Acad Sci U S A. 2010;107:9742–6. doi: 10.1073/pnas.0910668107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Yeager M, Orr N, Hayes RB, Jacobs KB, Kraft P, Wacholder S, et al. Genome-wide association study of prostate cancer identifies a second risk locus at 8q24. Nat Genet. 2007;39:645–9. doi: 10.1038/ng2022. [DOI] [PubMed] [Google Scholar]
- 102.Park SL, Chang S-C, Cai L, Cordon-Cardo C, Ding B-G, Greenland S, et al. Associations between variants of the 8q24 chromosome and nine smoking-related cancer sites. Cancer Epidemiol Biomarkers Prev. 2008;17:3193–202. doi: 10.1158/1055-9965.EPI-08-0523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ryan RJH, Drier Y, Whitton H, Cotton MJ, Kaur J, Issner R, et al. Detection of Enhancer-Associated Rearrangements Reveals Mechanisms of Oncogene Dysregulation in B-cell Lymphoma. Cancer Discov. 2015;5:1058–71. doi: 10.1158/2159-8290.CD-15-0370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zhang X, Cowper-Sal lari R, Bailey SD, Moore JH, Lupien M. Integrative functional genomics identifies an enhancer looping to the SOX9 gene disrupted by the 17q24. 3 prostate cancer risk locus. Genome Res. 2012;22:1437–46. doi: 10.1101/gr.135665.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Huang Z, Hurley PJ, Simons BW, Marchionni L, Berman DM, Ross AE, et al. Sox9 is required for prostate development and prostate cancer initiation. Oncotarget. 2012;3:651–63. doi: 10.18632/oncotarget.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Cowper-Sal lari R, Zhang X, Wright JB, Bailey SD, Cole MD, Eeckhoute J, et al. Breast cancer risk-associated SNPs modulate the affinity of chromatin for FOXA1 and alter gene expression. Nat Genet. 2012;44:1191–8. doi: 10.1038/ng.2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Hnisz D, Abraham BJ, Lee TI, Lau A, Saint-André V, Sigova AA, et al. Super-enhancers in the control of cell identity and disease. Cell. 2013;155:934–47. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Parker SCJ, Stitzel ML, Taylor DL, Orozco JM, Erdos MR, Akiyama JA, et al. Chromatin stretch enhancer states drive cell-specific gene regulation and harbor human disease risk variants. Proc Natl Acad Sci U S A. 2013;110:17921–6. doi: 10.1073/pnas.1317023110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Oldridge DA, Wood AC, Weichert-Leahey N, Crimmins I, Sussman R, Winter C, et al. Genetic predisposition to neuroblastoma mediated by a LMO1 super-enhancer polymorphism. Nature. 2015;528:418–21. doi: 10.1038/nature15540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wang K, Diskin SJ, Zhang H, Attiyeh EF, Winter C, Hou C, et al. Integrative genomics identifies LMO1 as a neuroblastoma oncogene. Nature. 2011;469:216–20. doi: 10.1038/nature09609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Corradin O, Saiakhova A, Akhtar-Zaidi B, Myeroff L, Willis J, Cowper-Sal-lari R, et al. Combinatorial effects of multiple enhancer variants in linkage disequilibrium dictate levels of gene expression to confer susceptibility to common traits. Genome Res. 2014;24:1–13. doi: 10.1101/gr.164079.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.He H, Li W, Liyanarachchi S, Srinivas M, Wang Y, Akagi K, et al. Multiple functional variants in long-range enhancer elements contribute to the risk of SNP rs965513 in thyroid cancer. Proc Natl Acad Sci U S A. 2015;112:6128–33. doi: 10.1073/pnas.1506255112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.He H, Li W, Liyanarachchi S, Jendrzejewski J, Srinivas M, Davuluri RV, et al. Genetic Predisposition to Papillary Thyroid Carcinoma: Involvement of FOXE1, TSHR, and a Novel lincRNA Gene, PTCSC2. J Clin Endocrinol Metab. 2015;100:E164–72. doi: 10.1210/jc.2014-2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Polak P, Lawrence MS, Haugen E, Stoletzki N, Stojanov P, Thurman RE, et al. Reduced local mutation density in regulatory DNA of cancer genomes is linked to DNA repair. Nat Biotechnol. 2014;32:71–5. doi: 10.1038/nbt.2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Polak P, Karlić R, Koren A, Thurman R, Sandstrom R, Lawrence MS, et al. Cell-of-origin chromatin organization shapes the mutational landscape of cancer. Nature. 2015;518:360–4. doi: 10.1038/nature14221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Puente XS, Silvia B, Rafael V-M, Neus V, Jesús G-A, Martín-Subero JI, et al. Non-coding recurrent mutations in chronic lymphocytic leukaemia. Nature. 2015;526:519–24. doi: 10.1038/nature14666. [DOI] [PubMed] [Google Scholar]
- 117.Cobaleda C, Schebesta A, Delogu A, Busslinger M. Pax5: the guardian of B cell identity and function. Nat Immunol. 2007;8:463–70. doi: 10.1038/ni1454. [DOI] [PubMed] [Google Scholar]
- 118.Mansour MR, Abraham BJ, Anders L, Berezovskaya A, Gutierrez A, Durbin AD, et al. Oncogene regulation. An oncogenic super-enhancer formed through somatic mutation of a noncoding intergenic element. Science. 2014;346:1373–7. doi: 10.1126/science.1259037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Northcott PA, Lee C, Zichner T, Stütz AM, Erkek S, Kawauchi D, et al. Enhancer hijacking activates GFI1 family oncogenes in medulloblastoma. Nature. 2014;511:428–34. doi: 10.1038/nature13379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Gröschel S, Sanders MA, Hoogenboezem R, de Wit E, Bouwman BAM, Erpelinck C, et al. A single oncogenic enhancer rearrangement causes concomitant EVI1 and GATA2 deregulation in leukemia. Cell. 2014;157:369–81. doi: 10.1016/j.cell.2014.02.019. [DOI] [PubMed] [Google Scholar]
- 121.Francis JM, Zhang C-Z, Maire CL, Jung J, Manzo VE, Adalsteinsson VA, et al. EGFR variant heterogeneity in glioblastoma resolved through single-nucleus sequencing. Cancer Discov. 2014;4:956–71. doi: 10.1158/2159-8290.CD-13-0879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Bandopadhayay P, Ramkissoon LA, Jain P, Bergthold G, Wala J, Zeid R, et al. MYB-QKI rearrangements in angiocentric glioma drive tumorigenicity through a tripartite mechanism. Nat Genet. 2016;48:273–82. doi: 10.1038/ng.3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Drier Y, Cotton MJ, Williamson KE, Gillespie SM, Ryan RJH, Kluk MJ, et al. An oncogenic MYB feedback loop drives alternate cell fates in adenoid cystic carcinoma. Nat Genet. 2016;48:265–72. doi: 10.1038/ng.3502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Zhang X, Choi PS, Francis JM, Imielinski M, Watanabe H, Cherniack AD, et al. Identification of focally amplified lineage-specific super-enhancers in human epithelial cancers. Nat Genet. 2016;48:176–82. doi: 10.1038/ng.3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Aran D, Sabato S, Hellman A. DNA methylation of distal regulatory sites characterizes dysregulation of cancer genes. Genome Biol. 2013;14:R21. doi: 10.1186/gb-2013-14-3-r21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Heyn H, Vidal E, Ferreira HJ, Vizoso M, Sayols S, Gomez A, et al. Epigenomic analysis detects aberrant super-enhancer DNA methylation in human cancer. Genome Biol. 2016;17:11. doi: 10.1186/s13059-016-0879-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Kaneda H, Arao T, Tanaka K, Tamura D, Aomatsu K, Kudo K, et al. FOXQ1 is overexpressed in colorectal cancer and enhances tumorigenicity and tumor growth. Cancer Res. 2010;70:2053–63. doi: 10.1158/0008-5472.CAN-09-2161. [DOI] [PubMed] [Google Scholar]
- 128.Stone A, Zotenko E, Locke WJ, Korbie D, Millar EKA, Pidsley R, et al. DNA methylation of oestrogen-regulated enhancers defines endocrine sensitivity in breast cancer. Nat Commun. 2015;6:7758. doi: 10.1038/ncomms8758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Bell RE, Golan T, Sheinboim D, Malcov H, Amar D, Salamon A, et al. Enhancer methylation dynamics contribute to cancer plasticity and patient mortality. Genome Res. 2016;26:601–11. doi: 10.1101/gr.197194.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Lupien M, Eeckhoute J, Meyer CA, Wang Q, Zhang Y, Li W, et al. FoxA1 translates epigenetic signatures into enhancer-driven lineage-specific transcription. Cell. 2008;132:958–70. doi: 10.1016/j.cell.2008.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–12. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39:311–8. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 133.He HH, Meyer CA, Shin H, Bailey ST, Wei G, Wang Q, et al. Nucleosome dynamics define transcriptional enhancers. Nat Genet. 2010;42:343–7. doi: 10.1038/ng.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci U S A. 2010;107:21931–6. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Magnani L, Stoeck A, Zhang X, Lánczky A, Mirabella AC, Wang T-L, et al. Genome-wide reprogramming of the chromatin landscape underlies endocrine therapy resistance in breast cancer. Proc Natl Acad Sci U S A. 2013;110:E1490–9. doi: 10.1073/pnas.1219992110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Knoechel B, Birgit K, Roderick JE, Williamson KE, Jiang Z, Lohr JG, et al. An epigenetic mechanism of resistance to targeted therapy in T cell acute lymphoblastic leukemia. Nat Genet. 2014;46:364–70. doi: 10.1038/ng.2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Lovén J, Hoke HA, Lin CY, Lau A, Orlando DA, Vakoc CR, et al. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell. 2013;153:320–34. doi: 10.1016/j.cell.2013.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Herz H-M, Hu D, Shilatifard A. Enhancer malfunction in cancer. Mol Cell. 2014;53:859–66. doi: 10.1016/j.molcel.2014.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Cleary SP, Jeck WR, Zhao X, Chen K, Selitsky SR, Savich GL, et al. Identification of driver genes in hepatocellular carcinoma by exome sequencing. Hepatology. 2013;58:1693–702. doi: 10.1002/hep.26540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Morin RD, Mendez-Lago M, Mungall AJ, Goya R, Mungall KL, Corbett RD, et al. Frequent mutation of histone-modifying genes in non-Hodgkin lymphoma. Nature. 2011;476:298–303. doi: 10.1038/nature10351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Gui Y, Guo G, Huang Y, Hu X, Tang A, Gao S, et al. Frequent mutations of chromatin remodeling genes in transitional cell carcinoma of the bladder. Nat Genet. 2011;43:875–8. doi: 10.1038/ng.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Parsons DW, Li M, Zhang X, Jones S, Leary RJ, Lin JC-H, et al. The genetic landscape of the childhood cancer medulloblastoma. Science. 2011;331:435–9. doi: 10.1126/science.1198056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Pugh TJ, Weeraratne SD, Archer TC, Pomeranz Krummel DA, Auclair D, Bochicchio J, et al. Medulloblastoma exome sequencing uncovers subtype-specific somatic mutations. Nature. 2012;488:106–10. doi: 10.1038/nature11329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Pasqualucci L, Dominguez-Sola D, Chiarenza A, Fabbri G, Grunn A, Trifonov V, et al. Inactivating mutations of acetyltransferase genes in B-cell lymphoma. Nature. 2011;471:189–95. doi: 10.1038/nature09730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Bitler BG, Aird KM, Garipov A, Li H, Amatangelo M, Kossenkov AV, et al. Synthetic lethality by targeting EZH2 methyltransferase activity in ARID1A-mutated cancers. Nat Med. 2015;21:231–8. doi: 10.1038/nm.3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Hu D, Gao X, Morgan MA, Herz H-M, Smith ER, Shilatifard A. The MLL3/MLL4 branches of the COMPASS family function as major histone H3K4 monomethylases at enhancers. Mol Cell Biol. 2013;33:4745–54. doi: 10.1128/MCB.01181-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Huang J, Jian H, Qing D, Qun W, Kun-Yu L, Ji-Hong D, et al. Exome sequencing of hepatitis B virus–associated hepatocellular carcinoma. Nat Genet. 2012;44:1117–21. doi: 10.1038/ng.2391. [DOI] [PubMed] [Google Scholar]
- 148.Wu JN, Roberts CWM. ARID1A mutations in cancer: another epigenetic tumor suppressor? Cancer Discov. 2013;3:35–43. doi: 10.1158/2159-8290.CD-12-0361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Phillips-Cremins JE, Sauria MEG, Sanyal A, Gerasimova TI, Lajoie BR, Bell JSK, et al. Architectural protein subclasses shape 3D organization of genomes during lineage commitment. Cell. 2013;153:1281–95. doi: 10.1016/j.cell.2013.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Rao SSP, Huntley MH, Durand NC, Stamenova EK, Bochkov ID, Robinson JT, et al. A 3D Map of the Human Genome at Kilobase Resolution Reveals Principles of Chromatin Looping. Cell. 2014;159:1665–80. doi: 10.1016/j.cell.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Dowen JM, Fan ZP, Hnisz D, Ren G, Abraham BJ, Zhang LN, et al. Control of cell identity genes occurs in insulated neighborhoods in mammalian chromosomes. Cell. 2014;159:374–87. doi: 10.1016/j.cell.2014.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Sanyal A, Lajoie BR, Jain G, Dekker J. The long-range interaction landscape of gene promoters. Nature. 2012;489:109–13. doi: 10.1038/nature11279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Heidari N, Phanstiel DH, He C, Grubert F, Jahanbani F, Kasowski M, et al. Genome-wide map of regulatory interactions in the human genome. Genome Res. 2014;24:1905–17. doi: 10.1101/gr.176586.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Bailey SD, Zhang X, Desai K, Aid M, Corradin O, Cowper-Sal Lari R, et al. ZNF143 provides sequence specificity to secure chromatin interactions at gene promoters. Nat Commun. 2015;2:6186. doi: 10.1038/ncomms7186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Schoenfelder S, Furlan-Magaril M, Mifsud B, Tavares-Cadete F, Sugar R, Javierre B-M, et al. The pluripotent regulatory circuitry connecting promoters to their long-range interacting elements. Genome Res. 2015;25:582–97. doi: 10.1101/gr.185272.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Pope BD, Tyrone R, Vishnu D, Feng Y, Weisheng W, Olgert D, et al. Topologically associating domains are stable units of replication-timing regulation. Nature. 2014;515:402–5. doi: 10.1038/nature13986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Vietri Rudan M, Barrington C, Henderson S, Ernst C, Odom DT, Tanay A, et al. Comparative Hi-C reveals that CTCF underlies evolution of chromosomal domain architecture. Cell Rep. 2015;10:1297–309. doi: 10.1016/j.celrep.2015.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Nora EP, Lajoie BR, Schulz EG, Giorgetti L, Okamoto I, Servant N, et al. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature. 2012;485:381–5. doi: 10.1038/nature11049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Shen Y, Yue F, McCleary DF, Ye Z, Edsall L, Kuan S, et al. A map of the cis-regulatory sequences in the mouse genome. Nature. 2012;488:116–20. doi: 10.1038/nature11243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Liu Z, Scannell DR, Eisen MB, Tjian R. Control of embryonic stem cell lineage commitment by core promoter factor, TAF3. Cell. 2011;146:720–31. doi: 10.1016/j.cell.2011.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL, et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010;467:430–5. doi: 10.1038/nature09380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Faure AJ, Schmidt D, Watt S, Schwalie PC, Wilson MD, Xu H, et al. Cohesin regulates tissue-specific expression by stabilizing highly occupied cis-regulatory modules. Genome Res. 2012;22:2163–75. doi: 10.1101/gr.136507.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Myslinski E, M-AG, Krol A, Carbon P. A Genome Scale Location Analysis of Human Staf/ZNF143-binding Sites Suggests a Widespread Role for Human Staf/ZNF143 in Mammalian Promoters. J Biol Chem. 2006;281:39953–62. doi: 10.1074/jbc.M608507200. [DOI] [PubMed] [Google Scholar]
- 164.Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–93. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Visser M, Kayser M, Palstra RJ. HERC2 rs12913832 modulates human pigmentation by attenuating chromatin-loop formation between a long-range enhancer and the OCA2 promoter. Genome Res. 2012;22:446–55. doi: 10.1101/gr.128652.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Ji X, Dadon DB, Powell BE, Fan ZP, Borges-Rivera D, Shachar S, et al. 3D Chromosome Regulatory Landscape of Human Pluripotent Cells. Cell Stem Cell. 2016;18:262–75. doi: 10.1016/j.stem.2015.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Katainen R, Dave K, Pitkänen E, Palin K, Kivioja T, Välimäki N, et al. CTCF/cohesin-binding sites are frequently mutated in cancer. Nat Genet. 2015;47:818–21. doi: 10.1038/ng.3335. [DOI] [PubMed] [Google Scholar]
- 168.Levo M, Zalckvar E, Sharon E, Dantas Machado AC, Kalma Y, Lotam-Pompan M, et al. Unraveling determinants of transcription factor binding outside the core binding site. Genome Res. 2015;25:1018–29. doi: 10.1101/gr.185033.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Maerkl SJ, Quake SR. A systems approach to measuring the binding energy landscapes of transcription factors. Science. 2007;315:233–7. doi: 10.1126/science.1131007. [DOI] [PubMed] [Google Scholar]
- 170.Sharon E, Kalma Y, Sharp A, Raveh-Sadka T, Levo M, Zeevi D, et al. Inferring gene regulatory logic from high-throughput measurements of thousands of systematically designed promoters. Nat Biotechnol. 2012;30:521–30. doi: 10.1038/nbt.2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Jolma A, Arttu J, Jian Y, Thomas W, Jarkko T, Nitta KR, et al. DNA-Binding Specificities of Human Transcription Factors. Cell. 2013;152:327–39. doi: 10.1016/j.cell.2012.12.009. [DOI] [PubMed] [Google Scholar]
- 172.Rajkumar AS, Dénervaud N, Maerkl SJ. Mapping the fine structure of a eukaryotic promoter input-output function. Nat Genet. 2013;45:1207–15. doi: 10.1038/ng.2729. [DOI] [PubMed] [Google Scholar]
- 173.Hnisz D, Weintraub AS, Day DS, Valton A-L, Bak RO, Li CH, et al. Activation of proto-oncogenes by disruption of chromosome neighborhoods. Science. 2016;351:1454–8. doi: 10.1126/science.aad9024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Ong C-T, Corces VG. CTCF: an architectural protein bridging genome topology and function. Nat Rev Genet. 2014;15:234–46. doi: 10.1038/nrg3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Stadler MB, Rabih M, Lukas B, Robert I, Florian L, Anne S, et al. DNA-binding factors shape the mouse methylome at distal regulatory regions. Nature. 2011;480:490–5. doi: 10.1038/nature10716. [DOI] [PubMed] [Google Scholar]
- 176.Wang H, Maurano MT, Qu H, Varley KE, Gertz J, Pauli F, et al. Widespread plasticity in CTCF occupancy linked to DNA methylation. Genome Res. 2012;22:1680–8. doi: 10.1101/gr.136101.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Dubois-Chevalier J, Oger F, Dehondt H, Firmin FF, Gheeraert C, Staels B, et al. A dynamic CTCF chromatin binding landscape promotes DNA hydroxymethylation and transcriptional induction of adipocyte differentiation. Nucleic Acids Res. 2014;42:10943–59. doi: 10.1093/nar/gku780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Flavahan WA, Drier Y, Liau BB, Gillespie SM, Venteicher AS, Stemmer-Rachamimov AO, et al. Insulator dysfunction and oncogene activation in IDH mutant gliomas. Nature. 2016;529:110–4. doi: 10.1038/nature16490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Turcan S, Rohle D, Goenka A, Walsh LA, Fang F, Yilmaz E, et al. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature. 2012;483:479–83. doi: 10.1038/nature10866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Hughes LAE, Melotte V, de Schrijver J, de Maat M, Smit VTHBM, Bovée JVMG, et al. The CpG island methylator phenotype: what’s in a name? Cancer Res. 2013;73:5858–68. doi: 10.1158/0008-5472.CAN-12-4306. [DOI] [PubMed] [Google Scholar]
- 181.Delmore JE, Issa GC, Lemieux ME, Rahl PB, Shi J, Jacobs HM, et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146:904–17. doi: 10.1016/j.cell.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Ott CJ, Kopp N, Bird L, Paranal RM, Qi J, Bowman T, et al. BET bromodomain inhibition targets both c-Myc and IL7R in high-risk acute lymphoblastic leukemia. Blood. 2012;120:2843–52. doi: 10.1182/blood-2012-02-413021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Zuber J, Shi J, Wang E, Rappaport AR, Herrmann H, Sison EA, et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature. 2011;478:524–8. doi: 10.1038/nature10334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Sengupta D, Kannan A, Kern M, Moreno MA, Vural E, Stack B, Jr, et al. Disruption of BRD4 at H3K27Ac-enriched enhancer region correlates with decreased c-Myc expression in Merkel cell carcinoma. Epigenetics. 2015;10:460–6. doi: 10.1080/15592294.2015.1034416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Bandopadhayay P, Bergthold G, Nguyen B, Schubert S, Gholamin S, Tang Y, et al. BET bromodomain inhibition of MYC-amplified medulloblastoma. Clin Cancer Res. 2014;20:912–25. doi: 10.1158/1078-0432.CCR-13-2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, et al. Selective inhibition of BET bromodomains. Nature. 2010;468:1067–73. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Costa-Pinheiro P, Montezuma D, Henrique R, Jerónimo C. Diagnostic and prognostic epigenetic biomarkers in cancer. Epigenomics. 2015;7:1003–15. doi: 10.2217/epi.15.56. [DOI] [PubMed] [Google Scholar]
- 188.Rodríguez-Paredes M, Esteller M. Cancer epigenetics reaches mainstream oncology. Nat Med. 2011;17:330–9. doi: 10.1038/nm.2305. [DOI] [PubMed] [Google Scholar]
- 189.Herceg Z, Hainaut P. Genetic and epigenetic alterations as biomarkers for cancer detection, diagnosis and prognosis. Mol Oncol. 2007;1:26–41. doi: 10.1016/j.molonc.2007.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Issa J-P. CpG island methylator phenotype in cancer. Nat Rev Cancer. 2004;4:988–93. doi: 10.1038/nrc1507. [DOI] [PubMed] [Google Scholar]
- 191.Gao K, Li G, Qu Y, Wang M, Cui B, Ji M, et al. TERT promoter mutations and long telomere length predict poor survival and radiotherapy resistance in gliomas. Oncotarget. 2016;7:8712–25. doi: 10.18632/oncotarget.6007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Landa I, Ganly I, Chan TA, Mitsutake N, Matsuse M, Ibrahimpasic T, et al. Frequent somatic TERT promoter mutations in thyroid cancer: higher prevalence in advanced forms of the disease. J Clin Endocrinol Metab. 2013;98:E1562–6. doi: 10.1210/jc.2013-2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Liu X, Bishop J, Shan Y, Pai S, Liu D, Murugan AK, et al. Highly prevalent TERT promoter mutations in aggressive thyroid cancers. Endocr Relat Cancer. 2013;20:603–10. doi: 10.1530/ERC-13-0210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Chen C, Han S, Meng L, Li Z, Zhang X, Wu A. TERT promoter mutations lead to high transcriptional activity under hypoxia and temozolomide treatment and predict poor prognosis in gliomas. PLoS One. 2014;9:e100297. doi: 10.1371/journal.pone.0100297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Castelo-Branco P, Choufani S, Mack S, Gallagher D, Zhang C, Lipman T, et al. Methylation of the TERT promoter and risk stratification of childhood brain tumours: an integrative genomic and molecular study. Lancet Oncol. 2013;14:534–42. doi: 10.1016/S1470-2045(13)70110-4. [DOI] [PubMed] [Google Scholar]
- 196.Mack SC, Witt H, Piro RM, Gu L, Zuyderduyn S, Stütz AM, et al. Epigenomic alterations define lethal CIMP-positive ependymomas of infancy. Nature. 2014;506:445–50. doi: 10.1038/nature13108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Roessler J, Ammerpohl O, Gutwein J, Steinemann D, Schlegelberger B, Weyer V, et al. The CpG island methylator phenotype in breast cancer is associated with the lobular subtype. Epigenomics. 2015;7:187–99. doi: 10.2217/epi.14.74. [DOI] [PubMed] [Google Scholar]
- 198.Cohen SA, Wu C, Yu M, Gourgioti G, Wirtz R, Raptou G, et al. Evaluation of CpG Island Methylator Phenotype as a Biomarker in Colorectal Cancer Treated With Adjuvant Oxaliplatin. Clin Colorectal Cancer. 2016;15:164–9. doi: 10.1016/j.clcc.2015.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Mah W-C, Lee CG. DNA methylation: potential biomarker in Hepatocellular Carcinoma. Biomark Res. 2014;2:5. doi: 10.1186/2050-7771-2-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Mur P, Rodríguez de Lope Á, Díaz-Crespo FJ, Hernández-Iglesias T, Ribalta T, Fiaño C, et al. Impact on prognosis of the regional distribution of MGMT methylation with respect to the CpG island methylator phenotype and age in glioma patients. J Neurooncol. 2015;122:441–50. doi: 10.1007/s11060-015-1738-9. [DOI] [PubMed] [Google Scholar]
- 201.Amary MF, Bacsi K, Maggiani F, Damato S, Halai D, Berisha F, et al. IDH1 and IDH2 mutations are frequent events in central chondrosarcoma and central and periosteal chondromas but not in other mesenchymal tumours. J Pathol. 2011;224:334–43. doi: 10.1002/path.2913. [DOI] [PubMed] [Google Scholar]