SUMMARY
Heritability and genome stability are shaped by meiotic recombination, which is initiated via hundreds of DNA double-strand breaks (DSBs). The distribution of DSBs throughout the genome is not random, but mechanisms molding this landscape remain poorly understood. Here we exploit genome-wide maps of mouse DSBs at unprecedented nucleotide resolution to uncover previously invisible spatial features of recombination. At fine scale, we reveal a stereotyped hotspot structure—DSBs occur within narrow zones between methylated nucleosomes—and identify relationships between SPO11, chromatin, and the histone methyltransferase PRDM9. At large scale, DSB formation is suppressed on non-homologous portions of the sex chromosomes via the DSB-responsive kinase ATM, which also shapes the autosomal DSB landscape at multiple size scales. We also provide a genome-wide analysis of exonucleolytic DSB resection lengths and elucidate spatial relationships between DSBs and recombination products. Our results paint a comprehensive picture of features governing successive steps in mammalian meiotic recombination.
Graphical Abstract
INTRODUCTION
Meiotic recombination promotes pairing and segregation of homologous chromosomes and disrupts linkage relationships, thus ensuring faithful genome transmission and increasing genetic diversity (de Massy, 2013; Hunter, 2015). The DSBs that initiate recombination are distributed nonrandomly in most species (reviewed in Baudat et al., 2013). The shape of this “DSB landscape” governs inheritance and genome evolution but also influences the risk of mutations and genome rearrangements (Kim et al., 2016). The factors shaping this landscape remain poorly understood.
In budding yeast, DSB distributions are molded by many factors working in a hierarchical and combinatorial fashion over size scales ranging from single base pairs (bp) to whole chromosomes (Pan et al., 2011). For example, DSBs form preferentially in small (~150–200 bp) “hotspots”, which in this organism are principally in nucleosome-depleted regions (NDRs) in promoters. However, hotspots are just one organizational level among many: DSB frequencies also vary substantially between large chromosomal domains and between different chromosomes (Pan et al., 2011; Thacker et al., 2014).
Similar principles may operate in mammals, but most attention has focused on hotspots and how the histone methyltransferase PRDM9 specifies hotspot locations via its sequence-specific DNA binding (e.g., Brick et al., 2012; Smagulova et al., 2016; Davies et al., 2016). This hotspot-centric view leaves largely unexplored the possible hierarchies of factors working on different size scales. Also, current DSB maps lack spatial resolution needed to reveal fine-scale structure within hotspots. We leverage a key feature of how DSBs form to explore these understudied aspects.
SPO11 makes DSBs through a topoisomerase-like reaction linking a SPO11 molecule to each 5′ DNA end (Figure 1A). DNA nicks nearby release SPO11 covalently bound to short oligonucleotides (SPO11 oligos), and 5′→3′ exonucleolytic resection generates single-stranded DNA (ssDNA) that is bound by strand-exchange proteins DMC1 and RAD51 and engages in homology search (Hunter, 2015). DSB repair is templated from homologous DNA and is completed as a crossover (reciprocal exchange) or a noncrossover. Either outcome can be accompanied by gene conversion (non-reciprocal transfer of sequence polymorphisms).
Figure 1. Nucleotide-Resolution Map of Meiotic DSBs in Wild-Type Mice.
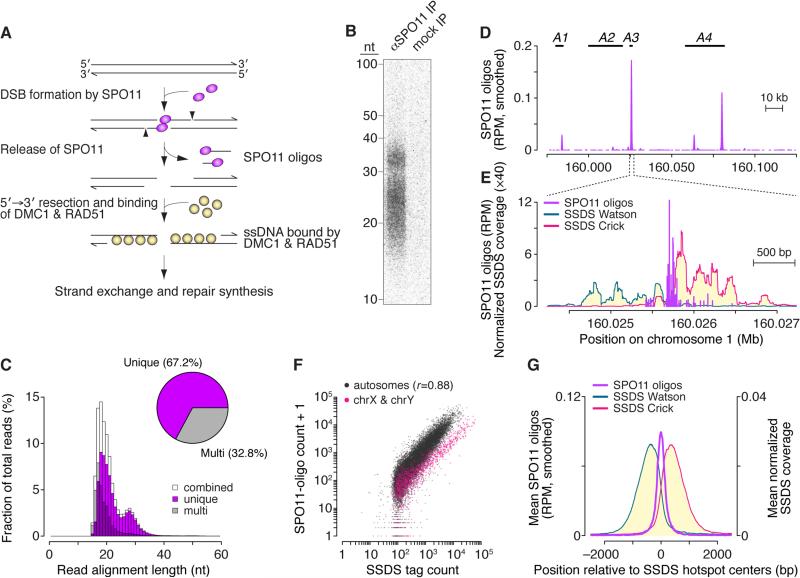
(A) Early steps in recombination and the protein–DNA complexes (SPO11 oligos and ssDNA bound by DMC1 and RAD51) used to generate genome-wide recombination initiation maps.
(B) SPO11 oligos immunoprecipitated (IP) from B6 mouse spermatocytes, deproteinized, 3′-end-labeled, and resolved in a denaturing 15% polyacrylamide gel. Anti-SPO11 antibody was omitted from the mock IP processed in parallel.
(C) Length distribution of SPO11 oligos that map uniquely or to multiple sites. Oligos appear longer on gels (panel B) because of nucleotides added for labeling and amino acid(s) left after SPO11 proteolysis.
(D) SPO11-oligo map (smoothed with a 1001-bp Hann filter) compared to positions of four known crossover hotspots (A1–A4) (Table S2A).
(E) SPO11 oligos and SSDS coverage (Brick et al., 2012) in a 3001-bp window around hotspot A3. SSDS coverage at each position was normalized to the total strand-specific coverage in the genome and multiplied by 106. See also Figure S1C.
(F) In SSDS hotspots (n=18,294), SPO11-oligo counts correlated strongly (Pearson's r) with SSDS tag counts. One SPO11-oligo read was added to permit plotting of hotspots with no oligos.
(G) Distribution of SPO11 oligos (51-bp Hann filter) and SSDS coverage around centers of SSDS hotspots.
See also Figure S1.
Meiotic DSBs were mapped genome-wide in men and male mice by deep-sequencing DMC1-bound ssDNA (Brick et al., 2012; Pratto et al., 2014). This ssDNA sequencing (SSDS) provides insight into the shape and evolution of the mammalian DSB landscape (e.g., Pratto et al., 2014; Smagulova et al., 2016; Davies et al., 2016). However, although SSDS can be used to estimate DSB hotspot midpoints, it does not directly report at high resolution where DSBs occur because resection spreads a kilobase (kb) or more on each side of a DSB. Moreover, midpoint positions alone shed little light on DSB distributions at sub-kb scales. To overcome these limitations, we sequenced mouse SPO11 oligos to provide nucleotide-resolution DSB maps with low background and high dynamic range. These maps uncover previously invisible relationships between DSB formation and PRDM9, identify factors on various size scales that influence DSB formation and repair, reveal spatial relationships between DSBs and downstream events, and elucidate the contribution of ATM.
RESULTS AND DISCUSSION
Nucleotide-Resolution Map of Meiotic DSBs
We purified SPO11 oligos from testes of C57BL/6J (B6) mice (Figure 1B). A mock-purified sample had little or no signal, indicating negligible nonspecific contamination. We ligated adaptors to the oligos, amplified them, then sequenced and mapped reads to the mouse genome (Figure S1A and Table S1). Read lengths matched the bimodal distribution of oligo sizes (Lange et al., 2011) (Figures 1B and 1C).
SPO11 oligos mapped in clusters as expected for DSB hotspots (Figures 1D and 1E), and these clusters matched known recombination hotspots. For example, four crossover hotspots mapped in pedigrees (Kelmenson et al., 2005) contained SPO11-oligo clusters (Figure 1D and Table S2A), and clusters accounted for 15 of 16 crossover hotspots defined by analysis of sperm DNA from hybrids of B6 with other strains (Figures 1E and S1B and Table S2B). (In the unaccounted crossover hotspot, recombination likely initiates on the non-B6 chromosome.) The SPO11-oligo map also agreed with SSDS hotspots (Brick et al., 2012): SPO11-oligo counts correlated well with SSDS counts genome wide (Figure 1F; sex chromosomes are discussed below) and SPO11 oligos were enriched near SSDS hotspot centers (Figures 1E, 1G and S1C). Thus, SPO11 oligos faithfully capture the genome-wide meiotic DSB distribution.
Of >69 million mapped reads, 67.2% mapped uniquely (Figure 1C). Multi-mapped reads were more prevalent in mice (32.8%; Figure 1C) than in S. cerevisiae (1.8%) (Pan et al., 2011). This is attributable to more repeated sequences in mouse and to the ~300-fold larger mouse genome increasing the probability that a read from an otherwise unique position fortuitously aligns to multiple places. Note that multi-mappers were especially abundant among reads <20 nucleotides (nt) (Figure 1C) and were enriched in hotspots (Figures S1D–S1F). Unless stated otherwise, analyses below focus on uniquely mapped reads, but the same conclusions were reached if multi-mapped reads were included.
Fine-Scale Anatomy of Mouse DSB Hotspots
Detailed understanding of mouse hotspots cannot be extrapolated from yeast. While most budding yeast hotspots share nucleosome depletion and relatively narrow width, individual hotspots often differ substantially from average, and patterns are not conserved in S. pombe (Pan et al., 2011; Fowler et al., 2014). Furthermore, the mechanism of hotspot specification via PRDM9 in mouse is unlike that in yeasts. Previous data lacked the resolution needed for a close-up view of mouse hotspot structure, but SPO11-oligo mapping surmounts this barrier.
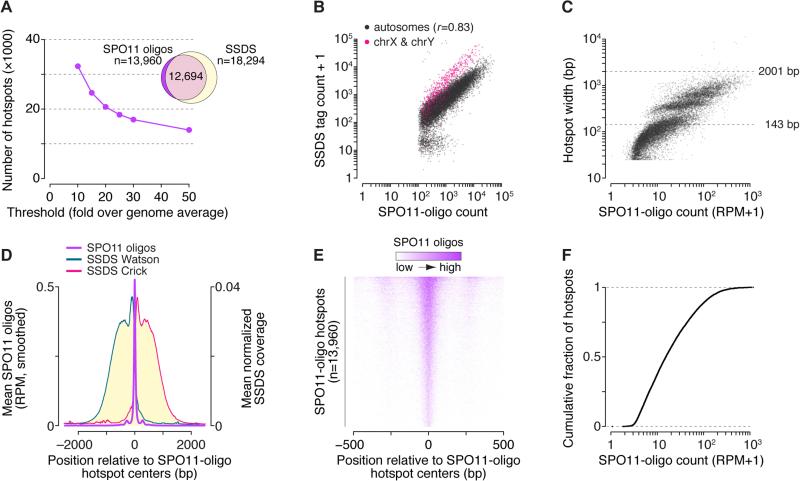
We defined SPO11-oligo hotspots using an established algorithm (Pan et al., 2011): regions exceeding an arbitrary threshold were identified in the smoothed SPO11-oligo map, then hotspot coordinates were defined in each region as the 5′-most and 3′-most positions with reads (see STAR Methods for details). We varied thresholds from 10× to 50× over the genome average of 0.000377 reads per million (RPM) per base pair (Figure 2A and Table S3), but for analyses below we focused on the most stringent definition (50×). For the 13,960 hotspots at this cutoff, 91% overlapped an SSDS hotspot (Figure 2A) and SPO11-oligo counts correlated well with SSDS coverage (Figure 2B). Of the minority of SSDS hotspots not in our list, many were weak sites below our threshold (Figures S2A and S2B). Some may overlap repetitive regions sampled better by the longer reads of SSDS. We conclude that SPO11-oligo hotspots are bona fide DSB hotspots.
Figure 2. DSB Hotspots Revealed by the SPO11-Oligo Map.
(A) Hotspot calling at various thresholds above genome average. Venn diagram shows overlap of SSDS hotspots with the most stringently defined SPO11-oligo hotspots.
(B) Good correlation (Pearson's r) between SPO11-oligo counts and SSDS tag counts in 2001-bp windows around SPO11-oligo hotspot centers.
(C) Read counts versus widths of SPO11-oligo hotspots.
(D) Distribution of SPO11 oligos (51-bp Hann filter) and SSDS coverage around SPO11-oligo hotspot centers. Note different y-axis scale for SPO11 oligos compared to Figure 1G. See also Figure S2E.
(E) Heat map of SPO11 oligos (5-bp bins) around hotspot centers, ordered by total read count. Most hotspots display a strong central cluster flanked by weaker peaks on one or both sides.
(F) Hotspot intensities vary over a smooth continuum.
See also Figure S2.
With the parameters used, median SPO11-oligo hotspot width was 143 bp, and the vast majority (99.8%) were under the 2001-bp width assigned to most SSDS hotspots (Brick et al., 2012) (Figure 2C). This difference derives from greater spatial precision when defining DSB positions using SPO11 oligos (Figures S2C and S2D). This greater precision uncovered spikes in the averaged SSDS coverage immediately adjacent to the bulk of SPO11 oligos (Figure 2D), likely from a systematic bias in SSDS (see section on DSB resection). SPO11 oligos clustered more narrowly when averaged around hotspot centers defined by our algorithm than around SSDS hotspot centers (Figure 1G and 2D).
The averaged SPO11-oligo profile displayed a narrow primary peak at hotspot centers that dwarfed secondary peaks 277 bp to either side (Figure 2D and S2E). The same pattern was seen if only multi-mappers were used (Figure S2F), so this substructure is not caused by mapping biases. Most hotspots are well represented by the average (Figure 2E), albeit with substantial diversity (Figure S1B). This structure explains the sub-groups when hotspots are arrayed by width as in Figure 2C: hotspot boundaries tend to occur at discrete distances from midpoints depending on whether there are two, one, or no secondary peaks strong enough to exceed the threshold. The stereotyped structure of mouse hotspots is thus a determinant of hotspot widths, but also contributes to the arbitrariness of hotspot definitions. The central cluster accounts for the vast majority of hotspot-associated oligos (Figure S2G), with 48.2% of uniquely mapped SPO11 oligos originating from 0.1% of the genome (≤95 bp from hotspot centers). Hotspot substructure differs markedly in mouse and S. cerevisiae, although median hotspot widths end up similar (189 bp in yeast (Pan et al., 2011)) because secondary peaks offset the more compact central cluster in mouse (Figure S2H). S. pombe hotspots are much wider (median 965 bp (Fowler et al., 2014)).
We conclude that mouse hotspots, to a remarkable degree, share structural features that shape the DSB distribution. As discussed next, chromatin is likely a key element. Importantly, however, 40.4% of uniquely mapped oligos fell outside clear hotspots. Furthermore, when rank-ordered by heat, hotspots formed a smooth continuum over three orders of magnitude (Figure 2F). Similarly smooth continua occur in yeasts (Pan et al., 2011; Fowler et al., 2014). The lack of a clear break in these distributions shows that the cutoff is arbitrary between sites that are or are not hotspots, illustrating limitations of a purely hotspot-centric view.
H3K4 Trimethylation and PRDM9 Binding
H3K4 trimethylation (H3K4me3) and DSBs are targeted near sites recognized by the polymorphic zinc-finger DNA binding domain of PRDM9 (de Massy, 2013). However, lack of direct, high-resolution information about DSB positions has left it unclear how PRDM9 binding and H3K4me3 relate spatially to SPO11 activity.
H3K4me3
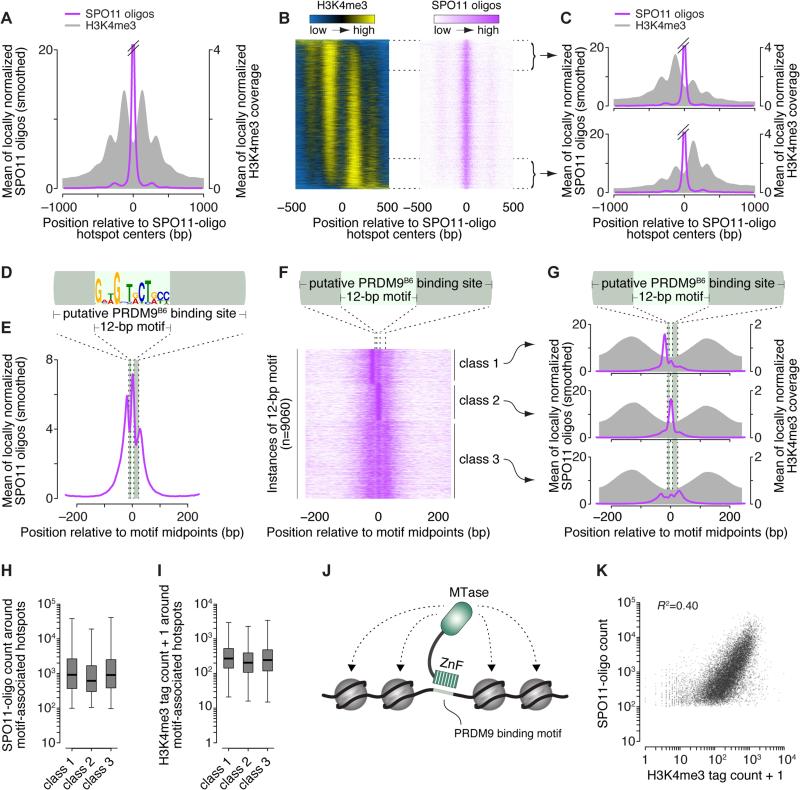
Conflicting proposals posit that DSB hotspot centers coincide with peaks in nucleosome occupancy (Smagulova et al., 2011) or are located instead between nucleosomes (Baker et al., 2014). To resolve this issue, we re-examined a map of H3K4me3 nucleosomes specific for PRDM9B6 (Baker et al., 2014). SPO11-oligo and H3K4me3 profiles were divided by the mean signal in a 5001-bp window around each hotspot's center, then the locally normalized profiles were averaged among hotspots. This normalization keeps the hottest sites from dominating the average and allows focus on spatial patterns separate from differences between hotspots in DSB frequency or total H3K4me3.
The primary SPO11-oligo peak precisely straddled a valley between oscillating H3K4me3 peaks, with secondary SPO11-oligo peaks nestled into neighboring valleys (Figures 3A and S3A). It was speculated that every hotspot has a symmetric array of methylated nucleosomes (Baker et al., 2014), but this is incorrect: Although all hotspots centers were depleted of H3K4me3, and methylated nucleosomes, when seen, lay in similar positions at most hotspots, the H3K4me3 signal ranged on a continuum of left–right asymmetry (Figure 3B). This asymmetry (not related to PRDM9 motif orientation; see below) predicted only weakly concordant asymmetry for SPO11 oligos (Figures 3C and S3B).
Figure 3. Spatial Relationships Between PRDM9 Binding, H3K4me3, and DSBs.
(A) SPO11 oligos map primarily between methylated nucleosomes. Data were locally normalized by dividing the signal at each base pair by the mean signal within each 2001-bp window, then were averaged across hotspots. The SPO11-oligo profile was smoothed with a 51-bp Hann filter. See also Figure S3A.
(B) H3K4me3 is often highly asymmetric around hotspots. Heat maps (data in 5-bp bins) were ordered according to H3K4me3 asymmetry. Data were locally normalized, so color-coding reflects the local spatial pattern, not relative signal strength between hotspots.
(C) Similar SPO11-oligo patterns between hotspots with opposite H3K4me3 asymmetry. Each panel shows mean of locally normalized profiles (51-bp Hann filter for SPO11-oligo data) across the 20% of hotspots with the most asymmetric H3K4me3 patterns (left > right in top panel; right > left in bottom panel). See also Figure S3B.
(D) The hotspot-enriched 12-bp motif and its disposition within the larger PRDM9B6 binding site.
(E) Asymmetric average profile of SPO11 oligos (15-bp Hann filter) around motif midpoints (n=9060).
(F) SPO11-oligo spatial classes from k-means clustering.
(G) H3K4me3 patterns are similar despite different SPO11-oligo patterns (15-bp Hann filter) between the motif classes from panel F.
(H, I) Similar SPO11-oligo counts (F) and H3K4me3 tag counts (G) for hotspots in each of the three PRDM9 motif classes. Counts are for 1001-bp windows around hotspot centers. Boxplots are as defined in Figure S2A legend. In panel I, a value of 1 was added to each hotspot to permit plotting of hotspots with no H3K4me3 tags.
(J) Schematic of modular PRDM9 DNA binding and histone methylation activities. ZnF, zinc-finger domain; MTase, methyltransferase domain.
(K) H3K4me3 is an imperfect predictor of DSB frequency. SPO11 oligos and H3K4me3 tag counts were summed in the 1001-bp around hotspot centers. One H3K4me3 tag was added to each hotspot to permit plotting of hotspots with no H3K4me3 tags. Eight outliers (H3K4me3 >104) are not shown.
See also Figure S3.
A simple interpretation is that most hotspots have a central NDR with positioned, methylated nucleosomes on one or both sides, and that SPO11 usually cleaves in the NDR but can also cut less frequently in flanking linkers. The preference of mouse SPO11 for nonnucleosomal DNA is similar to budding yeast but different from fission yeast (Pan et al., 2011; Fowler et al., 2014). Methylation apparently plays only a modest role in shaping DSB distributions within hotspots beyond the influence of nucleosome occupancy per se.
PRDM9
Independent approaches identified nearly identical motifs in SSDS hotspots and near PRDM9B6 H3K4me3 peaks (Brick et al., 2012; Baker et al., 2014; Baker et al., 2015), and we found the same 12-bp motif enriched precisely in SPO11-oligo hotspot centers (Figures 3D, S3C and S3D and Table S4). This motif matches part of the 36-bp predicted PRDM9B6 binding sequence (Brick et al., 2012; Baker et al., 2015) (Figure S3D).
Molecular events during DSB targeting by PRDM9 are unknown. A “hit-and-run” possibility is that PRDM9 binds DNA, methylates a nearby nucleosome(s), then departs before SPO11 cuts. Alternatively, PRDM9 may remain bound and occlude SPO11 access. These models make different predictions about DSB arrayal around PRDM9 sites, so we examined SPO11 oligos near PRDM9 motifs.
On average, SPO11 oligos formed three asymmetric peaks near the motifs (Figure 3E). K-means clustering divided motif instances into three classes: one with a SPO11-oligo peak to the left (class 1), one with a peak within the motif (class 2), and one with SPO11 oligos spread more evenly (Figures 3F and 3G). All three classes had similar average H3K4me3 (Figure 3G) and similar H3K4me3 asymmetry (Figure S3E), reinforcing the conclusion that histone methylation plays little role in shaping fine-scale DSB distributions in hotspots. On average, hotspots with class 2 instances of the motif had modestly fewer SPO11 oligos and less H3K4me3, but the values overlapped extensively with the other classes (Figures 3H and 3I). PRDM9 might have lower affinity for class 2 instances of the motif, but sequences were indistinguishable between the classes (Figure S3F) so the cause of a putative affinity difference is unclear.
Many SPO11 oligos mapped in the 36-bp PRDM9 binding site in all classes. We infer that PRDM9 often does not block SPO11 access to DNA, consistent with a hit-and-run model or with SPO11 displacing PRDM9 or cutting PRDM9-bound DNA. Because motif orientation can sometimes predict local DSB asymmetry without related H3K4me3 asymmetry, PRDM9 might still be DNA-bound and able to influence SPO11 position when DNA is cleaved. Different hotspots may be more or less prone to a particular mode of PRDM9–SPO11 interaction.
Further Implications
Our findings provide insight into roles of PRDM9 and chromatin structure. The lack of correlation between motif direction and H3K4me3 (a)symmetry implies that, while the PRDM9 zinc finger domain is tethered to DNA in an orientation dictated by the motif, the methyltransferase domain enjoys substantial freedom in accessing nucleosomes nearby (Figure 3J). This could arise via conformational flexibility between domains, PRDM9 multimerization (Baker et al., 2015), and/or DNA-binding dynamics. The source of frequent H3K4me3 asymmetry is unclear. It might reflect nucleosome availability and/or the influence of as yet unknown local factors that bias PRDM9 conformational states. A non-exclusive possibility is that asymmetry reflects H3K4me3 map biases, for example if methylated nucleosomes do not fully protect DNA from MNase or are difficult to liberate as mononucleosomes.
How does PRDM9 influence chromatin structure and DSBs? It was proposed that PRDM9 induces chromatin remodeling (Baker et al., 2014). The stereotyped position of H3K4me3 nucleosomes agrees with this hypothesis (Figure 3B), but is equally consistent with PRDM9 binding only the subset of chromosomes that already have a compatible nucleosome pattern. Note that the disposition of unmethylated nucleosomes is unknown: H3K4me3 ChIP is an affirmative signal of PRDM9 activity, but population-average measures of total nucleosomes merge signal from PRDM9-bound and -unbound chromosomes.
We favor that PRDM9 chromatin modification provides a landmark to attract SPO11 to locations at sub-kilobase scale (de Massy, 2013), but that other features including nucleosome occupancy (but not H3K4me3) and PRDM9 itself then shape the DSB distribution within the hotspot. Fine-scale relationships between PRDM9 binding and DSBs could only have been identified by nucleotide-resolution DSB maps.
PRDM9-dependent H3K4me3 is an imperfect predictor of hotspot heat, accounting for only ~40% of the variation in SPO11-oligo count (Figure 3K). Although this correlation is substantial—especially compared with budding yeast, where H3K4me3 levels do not predict hotspot heat (Tischfield and Keeney, 2012)—the data highlight the importance of other factors that may work combinatorially and on different size scales to determine whether a DSB will form at a site where PRDM9 has acted.
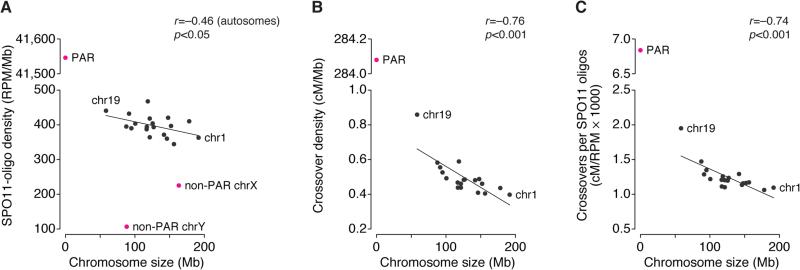
Chromosome-Scale Patterns
Smaller chromosomes tended to yield more SPO11 oligos per Mb, as SPO11-oligo density showed a weak negative correlation with chromosome size (Figure 4A and Table S5). A similar but stronger pattern is seen in budding yeast (Pan et al., 2011). There, differential DSB formation reflects chromosome size per se and is caused by negative feedback in which DSB formation is inhibited once chromosomes successfully engage their homologs (Thacker et al., 2014; Lam and Keeney, 2015). Smaller chromosomes may tend to take longer to engage their homologs and thus continue to make DSBs longer on average (Keeney et al., 2014). Mice display similar negative feedback driven by displacement of pro-DSB factors upon formation of synaptonemal complex (Wojtasz et al., 2009; Kauppi et al., 2013). Although the negative correlation between chromosome size and SPO11-oligo density was weak, it suggests that the mouse DSB landscape is shaped by this regulatory system.
Figure 4. Large-Scale Patterns of DSB Formation and Recombination.
(A,B) Per unit length, smaller autosomes incur more DSBs (A) and crossovers (B, centimorgans [cM]; data from Froenicke et al., 2002). SPO11-oligo density was exceptionally low in the non-PAR segments of the sex chromosomes, but very high in the PAR. The crossover rate in the PAR was set at 50 cM.
(C) The greater crossover density on smaller autosomes is explained only in part by the higher SPO11-oligo density.
Mouse chromosome size also correlates negatively with density of the crossover marker MLH1 (Froenicke et al., 2002) (Figure 4B and Table S5). This correlation had a steeper slope than the SPO11-oligo pattern, so the ratio of crossovers to SPO11 oligos was itself negatively correlated with chromosome size (Figure 4C). We infer that chromosome size-associated control of crossing over is asserted in at least two stages: smaller chromosomes tend to receive a higher density of DSBs, and a DSB is more likely to become a crossover on a smaller chromosome. The latter mechanism plays a larger role, unlike in budding yeast where DSB number control is the dominant mechanism (Pan et al., 2011).
Sex Chromosomes
The X and Y chromosomes share homology only in a small (<1 Mb) segment, the pseudoautosomal region (PAR). For accurate sex chromosome segregation in males, the PAR must form DSBs at higher frequency than typical autosome segments (Kauppi et al., 2011). Indeed, SPO11-oligo density in the PAR was ~110-fold higher than genome average (Figure 4A). Assuming ~200–300 DSBs per spermatocyte (e.g., Kauppi et al., 2013), SPO11-oligo density matches an average of 1.5–2.2 PAR DSBs. Consistent with PAR DSBs also being more likely to acquire a crossover fate (Kauppi et al., 2011), the ~570-fold higher crossover density in the PAR compared with genome average translates to a ~5-fold higher yield of crossovers per DSB (Figures 4B and 4C). The non-PAR parts of the sex chromosomes also experience DSBs, but at lower densities than autosomes (Figure 4A) (see section on ATM).
Sex-chromosome hotspots had a higher ratio of SSDS to SPO11 oligos than autosomes (Figures 1F, 2B, and S4A). The population-average amount of DMC1-bound ssDNA reflects DSB number and the life span of relevant recombination intermediates. SPO11 oligos have a long life span relative to prophase I, so whole-testis SPO11-oligo levels are proportional to DSB numbers and are insensitive to life span variation (Lange et al., 2011). Thus, elevated SSDS signal for sex-chromosome hotspots is readily explained if DSBs have a longer life span in non-PAR sex-chromosome regions because of delayed repair (Figure S4B), as proposed for human spermatocytes (Pratto et al., 2014). Unexpectedly, however, SSDS was disproportionately more elevated in stronger sex-chromosome hotspots than in weaker ones (Figures 1F and 2B). We propose that all DSBs on sex chromosomes have an extended life span because they persist until constraints against sister-chromatid recombination are relaxed in mid-to-late pachynema, but that DSB life span is even further extended in stronger hotspots because these tend to break earlier (Figure S4B). In this model, intrinsically weaker hotspots account for much of the prolonged DSB formation accompanying sex-chromosome asynapsis (Kauppi et al., 2013).
DSB Resection
Little is known about the exonucleolytic resection that prepares DSBs for recombination. In S. cerevisiae, comprehensive analysis of one side of an artificial DSB hotspot, HIS4LEU2, documented resection ranging ~350 to ~1550 nt with a mean of ~800 nt (Zakharyevich et al., 2010), and recent genome-wide analysis yielded similar measurements (Mimitou et al., 2016). There is no similar information in other organisms. We used SPO11-oligo and SSDS data to explore resection in mouse.
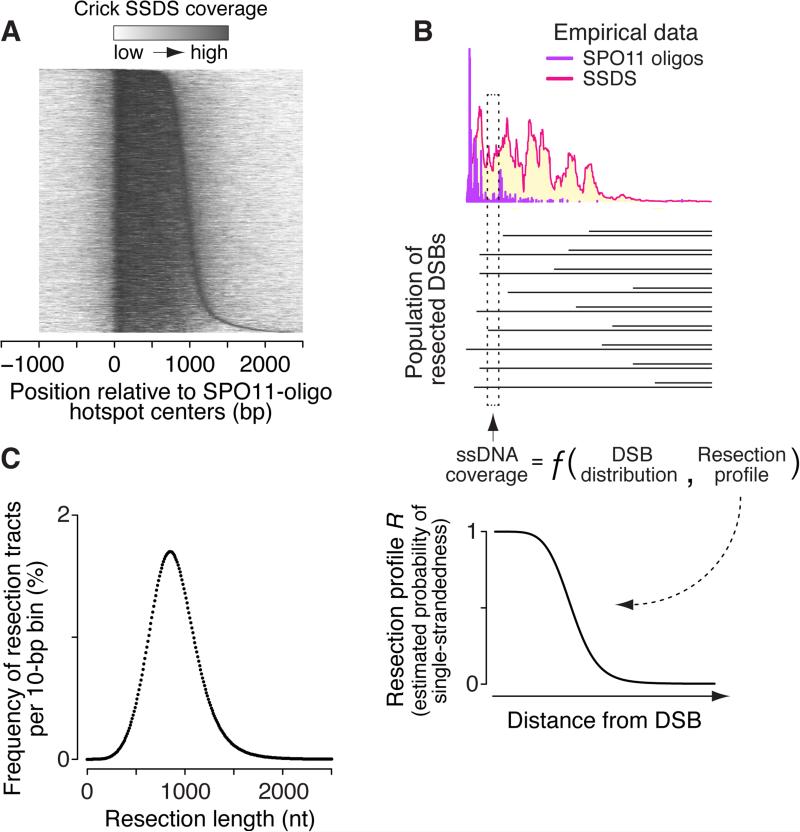
Strand-specific SSDS coverage around SPO11-oligo hotspots tapered off sharply <1 kb from hotspot centers, falling to undetectable levels typically by ~1.5–2 kb (Figures 2D and 5A). This indicates average resection of roughly 0.75–1 kb.
Figure 5. Analysis of DSB Resection.
(A) General resection trends. Heat map shows locally normalized Crick-strand SSDS coverage (10-bp bins) relative to centers of SPO11-oligo hotspots that overlap SSDS hotspots and that have no other hotspot within 5 kb. Hotspots were ordered by distance from 1005 bp left of the center to the point right of the center where cumulative SSDS signal was 90% of total.
(B) Combining SPO11-oligo and SSDS data to estimate the length distribution of resection tracts genome-wide. The ssDNA coverage at a given position is a function of the nearby DSB distribution and the per-DSB resection profile R.
(C) Estimated resection tract length distribution. See also Figure S5.
To obtain a more precise estimate, we combined SPO11-oligo and SSDS data to derive a resection profile R, which is the probability of single-strandedness a given distance from a DSB (Figure 5B; see STAR Methods and Figure S5 for further details). The amount of ssDNA at a genomic position is a function of how many DSBs are nearby, how far away they are, and the likelihood that resection reaches the position in question (Figure 5B). For the bottom strand to be single stranded, resection comes from DSBs on the left, and vice versa for the top strand (Figures S5A and S5B). Thus, we modeled strand-specific SSDS coverage at each genomic position as a multinomial equation combining the DSB distribution (SPO11 oligos) with the resection profile R (Figures 5B and S5B). We estimated R by solving these multinomial equations simultaneously for both strands at all positions around hotspots (Figure 5B, bottom). The spike in SSDS coverage adjacent to SPO11-oligos (Figure 2E) was disregarded because it probably reflects artifactual preference for sequencing of DSB-proximal ssDNA (Figures S5C–F). The resection length distribution is the first derivative of 1 – R (Figure 5C).
Resection lengths (per DSB end) ranged 300–1800 nt (mean = 894 nt, median = 870 nt; Figure 5C), remarkably similar in scale to yeast (Zakharyevich et al., 2010; Mimitou et al., 2016). This predicts a total of ~350–530 kb of ssDNA per meiosis assuming 200–300 DSBs. This analysis yields average resection behavior; individual hotspots may differ significantly, but exploring site-specific variation is precluded by bias in SSDS caused by a ssDNA enrichment step requiring foldback annealing (Figures 1E, 5B and S5D). Resection lengths are almost certainly underestimated because SSDS relies on repeats for foldback annealing and because DMC1 likely binds only a portion of ssDNA.
We note the following implications. First, the wide distribution of resection lengths (Figure 5C) means that there is substantial heterogeneity between individual DSBs—even at the same hotspot—in amount of ssDNA available for homology search and strand exchange. It is tempting to speculate that stochastic differences in resection length may influence recombination outcome or kinetics of DSB repair. Second, the resection machinery must traverse multiple nucleosomes given the frequent occurrence of positioned nucleosomes next to hotspots (Figures 3A and 3B). Chromatin thus has ample opportunity to shape resection (Mimitou et al., 2016). Third, total resection at each DSB (0.6–3.6 kb total for both sides; 1.8 kb on average) is almost always far longer than gene conversion tracts, unlike budding yeast despite similar resection length distribution (next section).
Spatial Relationship Between DSBs, Resection, and Recombination Products
Current models often depict gene conversion tracts centered on and including the DSB site (reviewed in Hunter, 2015) (Figure 6A). However, few direct spatial comparisons between DSBs and gene conversion tracts test these models. In yeast, studies often focused on a few sequence polymorphisms engineered at hotspots (e.g., Jessop et al., 2005). Low polymorphism density obscures the extent of gene conversion, and DSB locations were not always well known. Genome-wide studies of crosses of divergent strains mapped recombination at greater spatial precision because of higher polymorphism density (e.g., Mancera et al., 2008; Martini et al., 2011). However, DSB distributions are unknown, and limited numbers of meioses were analyzed so few recombination events were observed at any locus. In mouse and human, spatial distributions of crossovers and noncrossover gene conversions are known at numerous hotspots from typing of sperm or oocyte DNA (e.g., Cole et al., 2014), but until now precise locations of DSBs were not known. SPO11 oligos, along with SSDS and published sperm typing data, afforded an unprecedented look at spatial relationships between successive steps in recombination.
Figure 6. Spatial Relationships Between Sequential Steps in Meiotic Recombination.
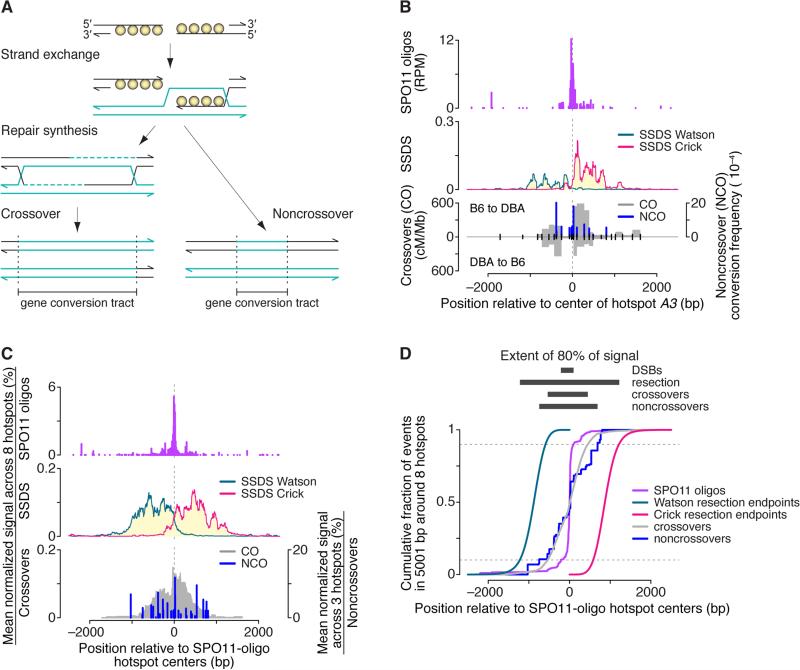
(A) Standard model for meiotic recombination, depicting strand exchange, repair synthesis, and completion of recombination as either a crossover or noncrossover. Note that gene conversion tracts overlap the location of the DSB.
(B) Spatial relationships between DSBs (SPO11 oligos), resection (SSDS, normalized coverage; data from Brick et al., 2012), and recombination products at hotspot A3 (Cole et al., 2010). Gray filled areas show density of crossover breakpoints (cM/Mb) detected by allele-specific PCR on sperm DNA using forward primers specific for the B6 haplotype and reverse primers for the DBA haplotype (“B6 to DBA”) or vice versa (“DBA to B6”). Blue bars show noncrossover gene conversion frequencies for individual polymorphisms, from the B6 allele to either DBA or A/J. Noncrossovers usually involve conversion of only a single polymorphism. In the bottom graph, ticks denote tested polymorphisms. See Figure S6 for other hotspots. (C,D) Multi-hotspot composite. Data were locally normalized by dividing by the mean of values in the 5001-bp window encompassing each hotspot, then were averaged across eight hotspots (three for noncrossovers). See STAR Methods for details about hotspots chosen for the composite. In panel D, resection endpoints were based on the estimated genome-wide average. See also Figure S6.
Figures 6B and S6 compare data for fifteen crossover hotspots (Table S2B). We also computed average profiles (Figure 6C) using eight particularly informative loci chosen on the basis of adequate polymorphism density, presence of a single SPO11-oligo hotspot, and availability of recombination data from mice with the same Prdm9 allele as B6. The average captures well the features of individual hotspots.
Several patterns emerge. First, 80% of crossover breakpoints fell within a zone less than 1 kb wide centered on the DSB hotspots’ centers (Figures 6C and 6D). This extends results of mouse tetrad analysis, in which crossover-associated gene conversions at the A3 hotspot averaged 626 bp and often traversed the hotspot center (Cole et al., 2014).
Second, noncrossovers did not cluster close to DSB positions but instead spread across much of the resection zone (Figures 6C and 6D) (Cole et al., 2014). Noncrossover gene conversions are typically very short, often just a single polymorphism (e.g., Cole et al., 2014). Thus, we infer that there is often a stretch of non-converted polymorphisms between the DSB and the position of a gene conversion, and that most noncrossovers are asymmetrically disposed relative to the DSB, as in budding yeast (Jessop et al., 2005). Displacement of noncrossover gene conversion tracts from DSB sites is consistent with re-synthesis of resected DNA proceeding initially via invasion and copying of the sister chromatid, followed by template switch to the homolog and further synthesis (Oh et al., 2007; Martini et al., 2011; Cole et al., 2014). Alternatively, spatial bias in mismatch repair could preferentially restore heteroduplex DNA near the DSB to the original sequence but convert heteroduplex DNA further away to the allelic sequence (Radford et al., 2007; Martini et al., 2011; Cole et al., 2014), although some studies in S. cerevisiae argue against this model (Kirkpatrick et al., 1998).
Third, nearly all recombination events were entirely within the extent of SSDS coverage (Figures 6C and 6D). Thus, despite similar resection lengths in yeast and mouse, crossover and noncrossover gene conversion tract lengths are much shorter in mice: ~500 bp (crossovers) and <100 bp (noncrossovers) for mice (Cole et al., 2014, and references therein), versus medians of 2 kb (crossovers) and 1.8 kb (noncrossovers) in yeast (Mancera et al., 2008). This suggests species-specific differences in meiotic recombination pathways.
ATM Shapes the DSB Landscape at Multiple Levels
ATM kinase is activated by DNA damage to trigger checkpoint signaling and promote DSB repair. Spermatocytes lacking ATM produce >10-fold more SPO11 oligos because of a conserved negative feedback circuit in which DSB-activated ATM inhibits further cleavage by SPO11 (Lange et al., 2011; Garcia et al., 2015). How does this feedback affect the DSB landscape? One possibility is a ‘rising tide raises all boats’ scenario, in which additional DSBs have the same distribution as in normal meiosis. However, because ATM is activated near DSBs, we proposed that this feedback is spatially patterned, in turn predicting that the DSB distribution is altered in Atm−/− meiocytes (Lange et al., 2011; Keeney et al., 2014). Indeed, yeast Tel1 (ATM ortholog) suppresses formation of clustered DSBs (Garcia et al., 2015). To distinguish between these models, we generated SPO11-oligo maps from Atm−/− mice (“Atm null”). Controls were ATM-proficient animals (Atm+/+ [“Atm wt”] and Atm+/− [“Atm het”]) from the same breeding colony, which is a mixed background of 129/Sv (129) and B6. These strains carry the same Prdm9 allele.
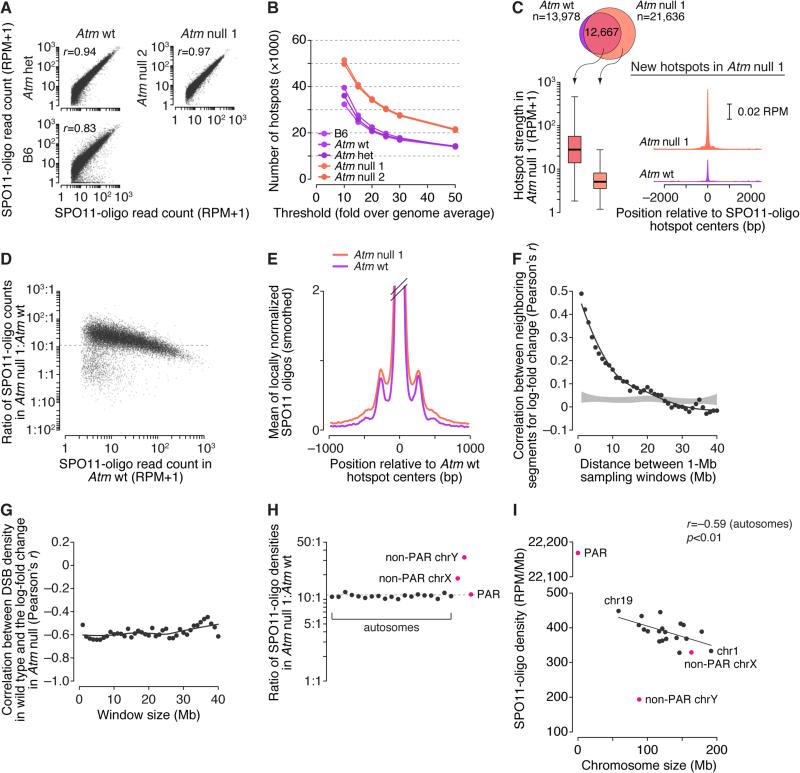
Replicate maps from Atm null mice agreed well and the Atm wt and Atm het maps also matched well, affirming reproducibility (Figure 7A). The Atm wt map also agreed with the B6 map, but less well than with Atm het (Figure 7A). Sequence differences between the strains likely affect strengths of subsets of PRDM9 binding sites (Figure S7A) (Smagulova et al., 2016). SPO11 oligos are longer on average in Atm null spermatocytes (Lange et al., 2011), also reflected in read lengths (Figure S7B).
Figure 7. DSB Patterns in the Absence of ATM.
(A) Reproducibility of SPO11-oligo maps. SPO11-oligo read counts were summed in 1001-bp windows.
(B) Atm null spermatocytes display more hotspots than ATM-proficient spermatocytes. Data for B6 are reproduced from Figure 2A.
(C) New hotspots in Atm null are weak hotspots that also yield small numbers of DSBs in wild type. Boxplot is as defined Figure S2A legend; SPO11-oligo profiles were smoothed with a 51-bp Hann filter.
(D) In ATM-deficient spermatocytes, weaker hotspots increase more than stronger hotspots. Each point represents a 1001-bp window around a hotspot called in the Atm wt map (one outlier is not shown). The dashed horizontal line marks the 11.3-fold increase in whole-testis SPO11-oligo levels in Atm null mice (Lange et al., 2011).
(E) Wider average SPO11-oligo distribution around hotspots in the absence of ATM. SPO11-oligo profiles were smoothed with a 51-bp Hann filter.
(F) Local domains of correlated behavior. Each point compares the log-fold change in SPO11-oligo density in 1-Mb segments on autosomes to the log-fold change in neighboring segments the indicated distance away. Shaded area denotes estimated 95% confidence intervals for data randomized within-chromosome.
(G) Domains that are relatively DSB-poor in wild type tend to be more strongly suppressed by ATM. Each point compares the log-fold change in SPO11-oligo density in Atm null to the SPO11-oligo density in Atm wt when autosomes are segmented into non-overlapping windows of the indicated size.
(H) In the absence of ATM, DSB densities increase more on the non-PAR segments of the sex chromosomes than on autosomes or the PAR.
(I) In Atm null spermatocytes, SPO11-oligo density remains negatively correlated with chromosome size, and the X chromosome more closely matches expectation from its size.
See also Figure S7.
More hotspots were identified in ATM-deficient samples at all thresholds tested, whereas ATM-proficient samples yielded similar numbers of hotspots that overlapped extensively between maps (Figures 7B and S7C and Table S3). Hotspots called only in the Atm null maps were much weaker on average than hotspots also seen in ATM-proficient mice (Figure 7C). Moreover, sites of “new” hotspots tended to have weak SPO11-oligo clusters in Atm wt (Figure 7C) and were enriched for a similar 15-bp motif (Figure S3D and Table S4). Thus, new hotspots are not a qualitatively distinct class, but instead are simply weak SPO11 targets that rise above the arbitrary threshold when ATM is absent. Of note, new Spo11-oligo hotspots did not emerge in S. cerevisiae in the absence of the ATM ortholog Tel1 (Mohibullah and Keeney, 2016). This difference may indicate qualitatively different functions for Tel1/ATM in yeast versus mice, or may simply reflect that ATM is quantitatively a more important controller of DSB activity in mice than in yeast, where Tel1 absence increases Spo11-oligo complexes only two fold.
Because hotspots were called in RPM-normalized data relative to each sample's genome average, the selective increase in numbers of weaker hotspots implies that strong and weak hotspots respond differently to ATM deficiency. Indeed, if we account for increased SPO11 oligos from Atm null spermatocytes by scaling the Atm null map by 11.3 (the average increase in SPO11-oligo complexes (Lange et al., 2011)), nearly all hotspots experienced more DSBs in the absence of ATM, but weaker hotspots increased more (Figure 7D). These patterns are not an artifact of better mapping efficiency of longer SPO11 oligos: When we truncated Atm null reads in silico to match the length distribution in Atm wt and regenerated the SPO11-oligo map (Figure S7D), Atm null still yielded more hotspots because of preferential strengthening of weak hotspots (Figures S7E–S7G and Table S6).
We further asked how SPO11-oligo distributions were altered at different size scales. Hotspots in Atm null had a similar average profile as wild type, with a narrow primary peak and weaker secondary peaks, but hotspots tended to be wider and the secondary peaks accounted for a greater fraction of SPO11 oligos (Figure 7E). ATM deficiency may alter local chromatin structure before DSBs form, making SPO11 less constrained toward hotspot centers. Or ATM may prevent SPO11 from breaking the same chromatid more than once, as in yeast (Garcia et al., 2015); if so, our results suggest that extra DSBs sometimes spill into a wider zone around the PRDM9 binding site. Population average SPO11-oligo measurement cannot distinguish between these possibilities, but we favor the multiple-DSB model given the yeast precedent.
To test if ATM also shapes the DSB landscape at larger size scales, we asked if neighboring regions on the same chromosome respond to ATM absence in a correlated manner. We calculated log-fold change in SPO11-oligo density in 1-Mb segments, then assessed the correlation of all segments with their neighboring segments located 1–40 Mb away. The change in SPO11-oligo density within a given genomic region was strongly correlated with the change in regions close by; correlation strength decayed with distance and reached background levels (no correlation) by ~15–20 Mb (Figure 7F). We further found a strong negative correlation between change in Atm null and local density of SPO11 oligos in wild type, for window sizes up to at least 40 Mb (Figure 7G). Thus, megabase-scale domains that are normally DSB-poor experience a disproportionately large increase in DSB formation in the absence of ATM. This finding explains the inverse relationship between the SPO11-oligo increase in Atm null and hotspot strength when individual hotspots were considered on a genome-wide scale (Figure 7D). More importantly, the results demonstrate a major role for ATM in shaping large-scale features of the DSB landscape in wild type. We speculate that differential sensitivity to ATM-mediated DSB control may reflect underlying variation in density of ATM phospho-targets, e.g., HORMAD1 (Wojtasz et al., 2009).
All autosomes responded similarly to the absence of ATM (Figure 7H) so SPO11-oligo density remained negatively correlated with chromosome size (Figure 7I). This finding is consistent with the hypothesis that negative feedback control of DSB formation via ATM and homolog engagement are distinct pathways (Keeney et al., 2014).
The PAR behaved similarly to autosomes, but the non-PAR sex chromosome portions increased disproportionately (~1.6-fold more than genome average on X and ~2.9-fold on Y; Figure 7H). As a result, SPO11-oligo density on the X chromosome more closely matched expectation from chromosome size (Figure 7I). One possibility is that DNA can be insulated from the DSB machinery by the sex body, a heterochromatic domain encompassing the non-PAR sex chromosomes and that requires ATM for its formation (Bellani et al., 2005). Alternatively, the same ATM-dependent negative feedback operating on autosomes may be stronger on sex chromosomes. Regardless of mechanism, these findings reveal an unexpected role for ATM in suppressing DSB formation on the non-PAR portions of the X and Y chromosomes.
Conclusions
SPO11-oligo maps provide a comprehensive look at the meiotic DSB landscape in male mice. We find common threads running through DSB patterning processes in different species, but also striking differences. One difference is that fine-scale hotspot structure is more homogeneous in mouse than in either budding or fission yeast, reflecting in part a more consistent chromatin structure for mouse hotspots caused by PRDM9-provoked chromatin remodeling and/or by PRDM9 selecting from a pre-existing state. A consequence of this stereotyped structure is that many DSBs arise from what are essentially point sources, somewhat more so than in budding yeast (Pan et al., 2011) and much more so than in fission yeast (Fowler et al., 2014). A practical implication is that constrained DSB locations facilitate comparisons to downstream events. Another implication is that hotspots are a more prominent component of the DSB landscape in mice.
Nonetheless, hotspots are just one organizational level among many. More than a third of DSBs apparently occur outside of hotspots, and even in hotspots H3K4me3 signal (a proxy for PRDM9 activity) explains only 40% of the variation in DSB frequency. Beyond the hotspot scale, we find differences between sub-chromosomal domains, variation between autosomes correlated with chromosome length, and exceptionally high PAR DSB activity. Because hotspots with similar H3K4me3 show a different likelihood of DSB formation in different larger domains, we infer that features defining the domains are more apical than hotspot-level determinants in the hierarchy of landscape architects. These findings and others (Davies et al., 2016) thus support the hypothesis that a hierarchical combination of factors shapes the DSB landscape in mouse. This is an important common thread between species.
Another common thread is the role of regulatory mechanisms. DSB landscape architects can be divided into two broad categories: intrinsic (or proactive) factors such as chromosome structures, the DSB machinery, or histone modifications; and extrinsic (or reactive) factors layered on top, such as feedback circuits described above (Keeney et al., 2014; Cooper et al., 2016). Our findings support this hypothesis by demonstrating that ATM not only controls DSB numbers but also molds the DSB landscape. Furthermore, DSB regulators interact in complex, combinatorial ways. Specifically, DSB formation is actively suppressed on non-homologous parts of the X and Y chromosomes, and we show an unexpected role (whether direct or indirect) for ATM in this process. But DSBs also continue to form longer on unsynapsed regions including on the X and Y chromosomes (Kauppi et al., 2013), thus the final number and distribution of DSBs on the sex chromosomes are shaped by a balance between antagonistic regulatory systems. Understanding complex interplay between the factors shaping the DSB terrain is an important challenge, and SPO11-oligo mapping will be a valuable resource.
STAR METHODS
KEY RESOURCES TABLE
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for reagents may be directed to, and will be fulfilled by, the corresponding author Scott Keeney at Memorial Sloan Kettering Cancer Center (skeeney@ski.mskcc.org).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice
C57BL/6J (B6) mice at 2–3 months of age were purchased from The Jackson Laboratory and were dissected upon delivery. The Atm mutation was described previously (Barlow et al., 1996) and was bred onto a mixed 129/B6 background. A single breeding pair of Atm+/− mice with this mixed background was used to establish the colony. Offspring of Atm+/− × Atm+/− breeding pairs were maintained on regular rodent chow with continuous access to food and water until dissection at 2–3 months of age. Experiments conformed to regulatory standards and were approved by the Memorial Sloan Kettering Cancer Center Institutional Animal Care and Use Committee.
METHOD DETAILS
Purification of Mouse SPO11 Oligos
We purified SPO11 oligos from adult mouse testes by adapting previously described methods in which we isolate SPO11 oligos by immunoprecipitation then deproteinize the oligos (Pan et al., 2011; Lange et al., 2011), as detailed here. Mouse testes were dissected at 2–3 months of age, decapsulated and stored at −80°C until use. We prep ared testis lysates and immunoprecipitated SPO11-oligo complexes using reagent volumes indicated in the table below, with minor modifications by sample. Because Atm−/− testes produce >10-fold more SPO11 oligos than ATM-proficient testes (Lange et al., 2011), fewer mice and smaller reagent volumes were required to generate the Atm null maps.
For each experiment, testes were Dounce-homogenized in lysis buffer (1% Triton X-100, 400 mM NaCl, 25 mM HEPES-NaOH at pH 7.4, 5 mM EDTA) containing EDTA-free protease inhibitors, then extracts were cleared by centrifugation at >300,000 g for 1 h. The supernatant was incubated with protein A–agarose beads for mock IP1 (5 h at 4°C with mixing by end-over-end rotation). The supernatant was transferred to a fresh tube and mock IP1 beads were stored at 4°C. The supernatant was incubated with anti-mSP O11 monoclonal antibody Spo11-180 at 4°C for 1 h with mixing by end-over-end rotation, t hen protein A–agarose beads were added for a further 3 h of mixing. The supernatant was transferred to a fresh tube and SPO11 IP1 beads were stored at 4°C. To maximally extract SPO11-olig o complexes, the supernatant was reimmunopreciptated (SPO11 re-IP1) with anti-mSPO11 antibody at 4°C for 1 h followed by addition of protein A–agarose beads and mixing overnight. SPO11 IP1 and re-IP1 beads were consolidated during washing steps in which the mock IP1 beads and the combined SPO11 IP1 beads were washed five times with IP buffer (1% Triton X-100, 150 mM NaCl, 15 mM Tris-HCl at pH 8.0). Immunoprecipitates were eluted with Laemmli sample buffer and diluted 6-fold in IP buffer. Eluates were subjected to a second round of immunoprecipitation (SPO11 IP2 and mock IP2) with or without anti-mSPO11 antibody at 4°C fo r 1 h, followed by the addition of protein A– agarose beads and end-over-end mixing overnight.
| ATM-proficient samples |
ATM-deficient samples |
||||
|---|---|---|---|---|---|
| B6 | Atm wt | Atm het | Atm null 1 | Atm null 2 | |
| number of mice | 200 | 100 | 100 | 15 | 15 |
| volume of lysis buffer (mL) | 80 | 24 | 60 | 4 | 4 |
| mock IP1 | |||||
| volume of beads (μL) | 1000 | 500 | 1000 | 150 | 150 |
| SPO11 IP1 | |||||
| amount of antibody (μg) | 600 | 300 | 600 | 100 | 100 |
| volume of beads (μL) | 800 | 400 | 800 | 100 | 100 |
| SPO11 re-IP1 | |||||
| amount of antibody (μg) | 300 | 300 | 300 | 50 | 50 |
| volume of beads (μL) | 500 | 250 | 500 | 50 | 100 |
| mock IP2 | |||||
| volume of beads (μL) | 300 | 300 | 300 | 150 | 150 |
| SPO11 IP2 | |||||
| amount of antibody (μg) | 400 | 200 | 400 | 100 | 100 |
| volume of beads (μL) | 300 | 300 | 300 | 150 | 150 |
SPO11 IP2 and mock IP2 beads were washed five times with IP buffer and once with Proteinase K buffer lacking SDS (100 mM Tris-HCl, pH 7.4, 1 mM EDTA, 1 mM CaCl2). Beads were then resuspended with 100 μg purified Proteinase K in Proteinase K buffer (100 mM Tris-HCl, pH 7.4, 1 mM EDTA, 0.5% SDS, 1 mM CaCl2) and incubated overnight at 50°C on a ThermoMixer (Eppendorf) at 300 rpm. The supernatant was collected using a Spin-X filter (Corning) and ethanol precipitated with 0.3 volume of 9 M ammonium acetate, 10 mg of DNA-free glycogen and 2.5 volumes of 100% ethanol. SPO11 oligos were quantified by radiolabeling with terminal deoxynucleotidyl transferase and [α-32P] GTP and comparing to a known quantity of a similarly labeled 25-nt synthetic oligo (Pan et al., 2011).
Library Preparation, Sequencing and Mapping
HiSeq libraries were prepared from one third to one half (≥100 fmol) of purified SPO11 oligos, essentially as described (Thacker et al., 2014). Briefly, purified SPO11 oligos were tailed on their 3′ ends with GTP and terminal deoxynucleotidyl transferase, then tailed oligos were ligated to a dsDNA adaptor with a 3′-CCCC overhang using T4 RNA ligase 2. The reverse complement of each SPO11 oligo was synthesized by Klenow DNA polymerase, then purified on denaturing PAGE, tailed with GTP, ligated to a second dsDNA adaptor, amplified by low numbers of PCR cycles using Taq DNA polymerase, and sequenced (Illumina HiSeq). Sequencing was performed using Illumina HiSeq (2×75-bp paired-end reads) in the Integrated Genomics Operation at Memorial Sloan Kettering Cancer Center.
Adaptor sequences were removed in silico and SPO11-oligo reads were mapped to mouse genome assembly GRCm38/mm10 by the Bioinformatics Core Facility at Memorial Sloan Kettering Cancer Center. Previously described pipelines for mapping S. cerevisiae and S. pombe oligos (Thacker et al., 2014; Fowler et al., 2014) were customized for the mouse genome. A full copy of the source code is available online at https://github.com/soccin/Spo11_v3. In brief, adaptor sequences were removed from both the 5′ and 3′ ends, then reads were mapped to the mouse genome using gmapper-ls (2_1_1b) from the SHRiMP mapping package (Rumble et al., 2009). The specific mapping parameters used were -N 18 -U -g -1000 -q -1000 -m 10 -i -20 -h 100 -r 50% -n 1 -o 100001 -Q -E --sam-unaligned --strata. After mapping, the reads were separated into unique and multiple mapping sets. Multi-mapped reads were distributed in two ways:
-
1)
Imputation: We used the CSEM method (Chung et al., 2011; Fowler et al., 2014), which imputes the likely fractional occupancy of multi-mapped reads using an Expectation-Maximization method that uses the local density of uniquely mapping reads at each multi mapped position. Version 2.4 of CSEM was used and called as follows: csem-2.4/run-csem --bam -p 12 --no-extending-reads --upper-bound 1000.
-
2)
Normalization: Each multi-mapped read was fractionally divided equally among its mapped positions.
Genome Size
The mappable genome size of 2,652,767,201 bp was calculated by subtracting the 78,088,274-bp sum of gaps from the total size of 2,730,855,475 bp (http://genome.ucsc.edu). The PAR has been estimated to be ~700 kb in length (Perry et al., 2001). Our PAR analyses (Figure 4) are based on the combined mappable size of 176,007 bp (161,532 bp for the X chromosome and 14,475 for the Y chromosome). PAR boundaries were set at position 169,969,767 on the X chromosome and position 90,830,223 on the Y chromosome. Oligos mapping to either the X- or Y-assigned PAR sequences were summed to estimate PAR totals.
SSDS and H3K4me3 Datasets
We combined SSDS data from GEO accession numbers GSM869781 and GSM869782 (anti-DMC1 B6 sample 1 and anti-DMC1 B6 sample 2 from GSE35498), kindly remapped to mm10 by Kevin Brick and R. Daniel Camerini-Otero. SSDS reads were converted to strand-specific coverage maps. For each strand, the coverage at each position was normalized to the total strand-specific coverage in the genome and multiplied by 106.
For comparison with our SPO11-oligo map, we converted the sequence coordinates for published SSDS hotspots (Brick et al., 2012) to mm10 using the UCSC Genome Browser LiftOver tool (http://genome.ucsc.edu/cgi-bin/hgLiftOver). (All but 10 out of 18,313 hotspots were successfully converted.) Additionally, we removed nine hotspots that localized to chrM, chr4_GL456216_random (an unplaced contig on chr4) or chrUn_GL456370 (an unassigned contig) because the remapped SSDS reads did not include these elements. We defined SSDS hotspot centers as the midpoints between start and end coordinates. We determined the SSDS hotspot tag count for the remapped datasets using the coverageBed algorithm in the BEDTools suite version 2.25.0.
H3K4me3 data (Baker et al., 2014) from GEO accession numbers GSM1273023 and GSM1273024 (B6_H3K4me3_ChIP_sample_1 and B6_H3K4me3_ChIP_sample_2 from GSE52628) were remapped to mm10 using the BWA package (Li and Durbin, 2009). Data were filtered to retain only uniquely mapping reads, which were then converted to coverage maps. Coverage maps for the two datasets were subsequently combined.
Analysis of Hotspots
We defined DSB hotspots as SPO11-oligo clusters that met cutoffs for six thresholds for oligo density over the genome average of 0.000377 RPM/bp, which was determined using the mappable genome size of 2,652,767,201 bp. Hotspot calls were made based on a previously described algorithm (Pan et al., 2011). For each chromosome, the raw SPO11-oligo map was smoothed with a 201-bp Hann window, and regions where the smoothed profile met or exceeded a given threshold were identified. Within each such region, hotspot coordinates (“Start” and “End” in Tables S3 and S6) were defined as the 5′-most and 3′-most positions with mapped reads. For each hotspot, the SPO11-oligo read count (as RPM) was summed from Start to End positions (“Hits” in Tables S3 and S6). The hotspot center was defined as the position of the smoothed peak in the SPO11-oligo density between Start and End; and the hotspot median was defined as the position of the weighted mean in SPO11-oligo density from Start to End. Hotspots with widths of less than 25 bp or with a strand bias greater than 100:1 were discarded. Hotspots separated by an edge-to-edge distance less than 1000 bp were merged and hotspot data were recalculated.
Prior to comparing SPO11-oligo read counts and SSDS tag counts around SPO11-oligo hotspots, we censored 17 SPO11-oligo hotspots: two neighboring hotspots on chromosome 2 because of a substantial spike in SSDS signal likely due to a mapping artifact, and 15 hotspots on unplaced or unassigned contigs. Thus, the comparisons shown in Figures 2 and S2 are for the remaining 13,943 SPO11-oligo hotspots. SSDS tag counts for these SPO11-oligo hotspots were determined for the center position ± 1000 bp using the coverageBed algorithm in the BEDTools suite. To compare H3K4me3 signal and SPO11 oligos around all 13,960 SPO11-oligo hotspots, we determined H3K4me3 tag counts (using the coverageBed algorithm in the BEDTools suite) and SPO11-oligo read counts in a 1001-bp window around each hotspot center.
S. cerevisiae Spo11-oligo data (Mohibullah and Keeney, 2016) were used for comparing average profiles around hotspots.
PRDM9 Motif Analysis
We searched for enrichment of DNA sequence motifs within hotspots using the MEME suite of tools version 4.11.0 (http://meme-suite.org). We supplied MEME-ChIP (Machanick and Bailey, 2011) with the genome sequences ± 1000 bp from the center positions of all 13,960 hotspots. Default parameters for MEME-ChIP were used, except that we required a minimum of 11 bp and a maximum of 15 bp. The most significant motifs included some or all of the 12-bp motif that we denoted as the primary motif, which matched part of a putative 36-bp PRDM9B6 binding site identified by independent approaches (Figure S3D) (Brick et al., 2012; Baker et al., 2014; Baker et al., 2015; Davies et al., 2016). Similar motifs were identified when we limited the searched genome space to 1) Start to End of all hotspots, or 2) the center position ± 100 bp of all hotspots (data not shown).
Because the 12-bp primary motif is highly enriched immediately around hotspot centers (Figure S3C), we limited the search space for calling instances of the motif to hotspot centers ± 100 bp. We queried these 13,960 201-bp sequences using MAST (Bailey and Gribskov, 1998) with the “-hit_list” parameter, which identified 9865 instances of the motif distributed among 9060 hotspots (Table S4A).
When we searched by MEME-ChIP the remaining 4900 hotpots for enrichment of other motifs, we identified a 15-bp sequence that substantially overlaps the primary motif (Figure S3D). Querying the central 201 bp of these 4900 hotspots using MAST identified 1622 instances of this secondary motif distributed among 1534 hotspots (Table S4B). We observed essentially identical SPO11-oligo patterns around the secondary motif as around the primary motif (data not shown).
When we searched for enrichment of DNA sequence motifs in the 8969 new hotspots arising in the Atm null 1 map, we identified a 15-bp sequence that substantially overlaps the primary motif (Figure S3D). In querying the central 201 bp of these hotspots using MAST, we found 5763 instances of this motif distributed among 5218 hotspots (Table S4C) and essentially identical SPO11-oligo patterns as around the primary motif (data not shown).
We used k-means clustering to group hotspot-associated instances of the motif by local SPO11-oligo pattern. The SPO11-oligo map in the 501 bp surrounding each motif midpoint was smoothed with a 15-bp Hann window, the smoothed pattern was normalized across the window, and the normalized signals were clustered. When a queried region (hotspot centers ± 100 bp) contained more than one motif, the central-most instance was selected for plotting (Figures 3F and 3G). Highly similar results were obtained when we analyzed all instances of the motif or only motifs from regions containing a single instance of the motif, as well as the secondary motif and motifs in new Atm null 1 hotspots (data not shown).
Chromosome-Scale Patterns
Chromosome-scale analyses were performed after combining uniquely mapping and imputed multi-mapped reads. SPO11-oligo and crossover densities in the PAR were calculated with the combined PAR size of 176,007 bp (see above). When we set the PAR size to 700 kb (Perry et al., 2001), densities were ~28-fold and ~143-fold higher than their respective genome averages. This translated to a similar ~5-fold higher yield of crossovers per DSB. When plotting ratios of SSDS tag counts to SPO11-oligo read counts in SSDS hotspots, all sex-chromosome hotspots were within non-PAR segments of the X and Y chromosomes.
Analysis of DSB Resection Tract Lengths
Estimating a Global Average Resection Profile by Combining SSDS Coverage with SPO11-Oligo Data
SPO11 oligos are a measure of the genome-wide distribution of DSBs, i.e., resection start sites. SSDS measures DMC1-bound ssDNA, encompassing some or all of the ssDNA revealed by resection. Both measures are population averages, so they do not directly reveal the lengths of individual resection tracts. Note, however, that the amount of ssDNA at a given genomic position is a function of how often a DSB forms near that position, how far such DSBs are away, and the likelihood that resection of each of these DSBs will reach the position in question (Figures 5B, S5A, and S5B). The steady-state amount of ssDNA as a function of distance from the DSB will also be influenced by the speed of resection if this is slow compared to the lifespan of resected DSBs, because in such a scenario partially resected DSBs would contribute to the population average. However, DSBs in yeast are resected extremely rapidly, such that they appear to be fully resected as soon as they are detectable (Zakharyevich et al., 2010). Here we assume the same is true for mouse meiosis, in which case we can safely ignore resection speed as a contributing factor. We sought to estimate a genome-wide average profile of resection lengths by combining the strand-specific SSDS coverage at each genomic position with information about the DSB distribution (SPO11 oligos), using the following approach.
Let S be the SPO11-oligo distribution in 10-bp bins: S = {s1, s2, ..., si}. Thus, si is the empirical estimate of the DSB frequency within genomic bin i. (We used binned data to make the analysis computationally tractable.) Similarly, let C be the binned SSDS coverage on the Crick (bottom) strand, C = {c1, c2, ..., ci}, and let W be the binned SSDS coverage on the Watson (top) strand, W = {w1, w2, ..., wi}. Thus, ci and wi are empirical estimates of how much ssDNA is present within bin i. Let E be an error function encompassing all sources of error in measuring true DSB and ssDNA distributions for each strand, E = {εc1, εc2, ..., εci, εw1, εw2, ..., εwi}.
Let R be the distribution model for the genome-average resection profile, R = {r0, r1, ..., r250}, where rk is the probability that resection will extend >10k nucleotides from the DSB. We empirically chose 2.5 kb (i.e., max k = 250) as the resection limit based on examination of the spread of SSDS signal around SPO11-oligo hotspots (Figures 5A). R can also be viewed as the probability of single-strandedness, i.e., rk is also the probability that the DNA ≤10k nt from a DSB will be single stranded.
We wished to estimate R from the SPO11-oligo and SSDS data. The ssDNA present at a given base in the genome is the sum of the resection tracts that overlap that base. For the Crick strand to be single stranded, resection must come from DSBs located to the left (Figures S5A and S5B), thus C can be described as a series of linear equations combining S, R, and E:
Conversely, for the Watson strand to be single stranded, resection must come from DSBs located to the right, so the equations describing W take the form:
We therefore have two equations (one for each strand) for every 10-bp bin at position i across the genome. To simplify further calculations and to ensure that the analysis focused on informative loci, we selected a subset of SPO11-oligo hotspots (11,804; 84% of hotspots) by the following criteria: 1) The SPO11-oligo hotspot must overlap an SSDS hotspot, to exclude uninformative loci (e.g., hotspots with SPO11-oligo signal but no SSDS signal). 2) The SPO11-oligo hotspot must not have a neighboring SPO11-oligo or SSDS hotspot within 5 kb of its peak position, to correctly attribute SSDS signal to the specified hotspot. We set values of si, ci, and wi as 0 if the position i was >2.5 kb away from the peak of one of the selected hotspots. Values of si, ci, and wi were locally normalized across each window so that SPO11-oligo and SSDS data are equally comparable at each hotspot, and each hotspot contributes equally to the SPO11-oligo and SSDS spatial patterns without regard to hotspot heat.
The number of non-trivial equations (i.e., not of the form 0 = 0) is >1.2 × 107 (11,804 hotspots considered × 501 10-bp bins centered on each hotspot × 2 strands). We computed a best fit solution (minimizing the sum of absolute errors) to this system of equations for R using the Moore–Penrose pseudoinverse. The initial solution (R′) is shown in the left graph in Figure S5C. Because the empirical data are measured in different types of units (coverage for SSDS, tag counts for SPO11 oligos), the calculated value of R′ has somewhat arbitrary units. To convert R′ to the desired probability distribution (R), we set the maximum to 1 and the minimum to 0, after removing the end bias introduced by the SSDS mapping method (see the next section for details) (Figure S5C, middle graph). We then smoothed R using logistic regression and determined the underlying distribution of resection tract lengths as the first derivative of 1 – R (Figure S5C, right graph, and Figure 5C).
Accommodating Elements of Bias in the SSDS Map
Individual hotspots show a general trend where SSDS coverage extends up to somewhat over 1 kb from hotspot centers (Figure 5A). However, we note two distinct elements of bias in the SSDS profiles. First, individual hotspots displayed a strong peaks-and-valleys appearance for SSDS coverage that was not as expected if reads were derived randomly from ssDNA emanating out from the DSB sites (see Figure 1E as an example). To further illustrate, we plotted a heat map of the locally normalized Crick-strand SSDS coverage around SPO11-oligo hotspots ordered by the distance to the point with maximal signal (Figure S5D). This analysis reveals a tendency for individual hotspots to display a significant peak in SSDS coverage whose distance from the site of most DSBs varies continuously out to nearly 1 kb. We infer from this pattern that some ssDNA segments are preferentially sequenced over others, most likely in a sequence-dependent manner. The SSDS method relies on foldback annealing to provide specificity for ssDNA (Khil et al., 2012; Brick et al., 2012). It is likely that this requirement for appropriately spaced sites of microcomplementarity imposes much of the observed non-randomness in the SSDS coverage. It is also possible that non-random binding by DMC1 and/or other technical biases in PCR amplification and deep sequencing contribute. We accommodate this type of bias by averaging genome-wide, because the spatial distribution of sequence-dependent bias is different for different hotspots and thus largely cancels out when averaging across the genome (Figure S5D). Note, however, that this issue largely precludes detailed analysis of individual loci.
Second, when SSDS signal was averaged across all hotspots, we observed a strong spike in coverage immediately adjacent to SPO11-oligo hotspot centers (Figures 2E and S5D). At least some degree of hotspot-proximal enrichment was seen even in cases where the highest peak of SSDS coverage lay further away from the hotspot center (see quantiles 3 and 4 in the graphs of Figure S5Dii). It is likely that this “end effect” is an artifact caused by nonrandom ssDNA fragmentation. In SSDS, sequenced ssDNA ranged from 50 to 200 nt (Khil et al., 2012; Brick et al., 2012). At an early step of the protocol, immunoprecipitated DMC1-bound ssDNA is sheared by sonication. Note that a ssDNA fragment of sequenceable length can be recovered from the 3′ end of the resected strand with just a single break from sonication, but sequenceable fragments from segments further from the DSB require two breaks from sonication (Figure S5E). This feature creates a bias in favor of DSB-proximal segments of ssDNA at DSBs.
To evaluate whether such an effect would be sufficient to account for the observed enrichment of SSDS coverage near hotspots, we simulated the isolation and sequencing of ssDNA in the length range targeted by the SSDS protocol. To do so, we randomly subdivided one million hypothetical ssDNAs with breaks spaced on average 100 nt apart. This results in a population of “ssDNA fragments” that span either from the original ssDNA end (i.e., the DSB site) to a “sonication” site, or from one sonication site to the next. Because fragments were size selected during SSDS (long fragments appear to have been excluded) (Khil et al., 2012; Brick et al., 2012), we sampled with replacement from the population of ssDNA fragments to match the length distribution of published SSDS reads. We then evaluated the coverage provided by this sample relative to the DSB ends of the original ssDNAs. The results show strong enrichment for the 100–150 nt adjacent to the DSB site (Figure S5F), indicating that this recovery bias is sufficient to account for the elevated SSDS coverage we observe.
To accommodate the bias from this end effect, we made the straightforward assumption that ssDNA coverage should be maximal within a plateau extending for some distance from DSB sites, with the length of the plateau representing the minimal resection tract length. In this scenario, the sonication end effect yields extra coverage above this plateau, and thus artificially inflates values of R (the resection profile) close to the DSB (Figure S5C, left graph), so we manually removed the end-effect spike by determining the maximum SSDS coverage to the right of the peak, then applying that value to all positions back to the DSB site (Figure S5C, middle graph).
In principle, an alternative explanation for the DSB-proximal enrichment of SSDS coverage could be that there are two distinct types of DSBs: one that gives rise to short resection tracts and another that is resected to longer lengths. There is no evidence for such distinct resection classes in yeast (Zakharyevich et al., 2010), and our simulations indicate that sonication bias alone is sufficient to explain the observed patterns, so we consider this explanation unlikely.
Crossover and Noncrossover Data
To compare the SPO11-oligo maps with recombination maps, we determined the mm10 coordinates for all DNA sequence polymorphisms used to define crossovers and noncrossovers (Kauppi et al., 2007; Bois, 2007; Wu et al., 2010; Getun et al., 2010; Cole et al., 2010; Getun et al., 2012; de Boer et al., 2015). Data underlying published B6×DBA crossover maps were kindly provided by Irina Getun and Philippe Bois.
To examine average patterns of recombination around hotspots analyzed by sperm typing, we narrowed down the 16 recombination hotspots using three criteria. First, to focus our analysis on recombination hotspots that had been defined with adequate resolution, we included only those with a substantial number of polymorphisms between B6 and DBA or between B6 and A/J. Second, to exclude recombination data that could have arisen from nearby neighboring hotspots, we required recombination hotspots to be well separated from adjacent hotspots. Third, to eliminate recombination hotspots in which most DSBs were formed on the non-B6 allele, we included only those loci that were also called as hotspots by SPO11-oligo mapping. These criteria narrowed the 16 recombination hotspots down to eight informative loci: three on chromosome 1 (A3, central and distal), which have been analyzed for both crossovers and noncrossovers (Cole et al., 2010; de Boer et al., 2015) and five on chromosome 19 (H22, HS23.9, HS59.5, HS61.1 and HS61.2), which have been assessed only for crossovers (Bois, 2007; Wu et al., 2010; Getun et al., 2012).
Shortening of Read Lengths in Atm Null Maps
Because longer reads are more likely to map uniquely, we tested whether better mapping efficiency in Atm null could account for the observed differences with wild type. For each of the two Atm null datasets, we randomly selected 5% of the reads, shortened their 3′ ends to generate an overall length distribution matching that of randomly selected 5% of the Atm wt reads, and remapped the transformed reads (Figure S7D). All previous observations for Atm null were recapitulated. Chromosome-scale analyses were conducted with uniquely mapping reads only.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical analyses were performed using R versions 3.2.0 to 3.3.0 (http://www.r-project.org). Statistical parameters and tests are reported in the Figures and corresponding Figure Legends. In cases where outliers were removed for plotting purposes, none of the data points were removed from the statistical analysis.
DATA AND SOFTWARE AVAILABILITY
Data Resources
Raw and processed data files for the five SPO11-oligo maps have been deposited in the NCBI Gene Expression Omnibus (GEO) under accession number GSE84689. A copy of source code used for mapping SPO11 oligos is available at https://github.com/soccin/Spo11_v3.
Supplementary Material
Article highlights.
Hierarchical combinations of factors shape DSB landscapes at small and large scales
Local chromatin structure strongly shapes DSB distributions
Exonucleolytic DSB resection averages 900 base pairs with wide variation
ATM plays a dominant role in molding genome-wide DSB distributions
ACKNOWLEDGMENTS
We thank A. Viale (Integrated Genomics Operation, MSKCC) for sequencing; M. Thelen (Lawrence Livermore National Laboratory) for the anti-mSPO11 hybridoma cell line; K. Brick and R. D. Camerini-Otero (NIH) for SSDS data remapped to mm10; I. Getun and P. Bois (Scripps) for data underlying published crossover maps; and members of the Keeney and Jasin laboratories for comments and discussions. This work was supported by NIH grants GM105421 (M.J. and S. Keeney) and HD053855 (S. Keeney and M.J.). J.L. was supported in part by American Cancer Society fellowship PF-12-157-01-DMC. J.P. and S. Kim were supported in part by Leukemia and Lymphoma Society fellowships. N.D.S. was supported in part by NIH/NCI Cancer Center Support Grant P30 CA008748.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
STAR METHODS
- KEY RESOURCES TABLE
- CONTACT FOR REAGENT AND RESOURCE SHARING
- EXPERIMENTAL MODEL AND SUBJECT DETAILS
- Mice
- METHOD DETAILS
- Purification of Mouse SPO11 Oligos
- Library Preparation, Sequencing and Mapping
- Genome Size
- SSDS and H3K4me3 Datasets
- Analysis of Hotspots
- PRDM9 Motif Analysis
- Chromosome-Scale Patterns
- Analysis of DSB Resection Tract Lengths
- Crossover and Noncrossover Data
- Shortening of Read Lengths in Atm Null Maps
- QUANTIFICATION AND STATISTICAL ANALYSIS
- DATA AND SOFTWARE AVAILABILITY
- Data Resources
SUPPLEMENTAL INFORMATION
Supplemental Information includes seven figures and six tables and can be found with this article online at http://.
AUTHOR CONTRIBUTIONS
Conceptualization, J.L., M.J., and S. Keeney; Methodology, J.L., J.P., and S. Keeney; Software, N.D.S.; Formal Analysis, J.L., S.Y., S.E.T., S. Kim, S. Keeney; Investigation, J.L., X.Z.; Data Curation, N.D.S.; Writing – Original Draft, J.L. and S. Keeney; Supervision, M.J. and S. Keeney; Funding Acquisition, M.J. and S. Keeney.
REFERENCES
- Bailey TL, Gribskov M. Combining evidence using p-values: application to sequence homology searches. Bioinformatics. 1998;14:48–54. doi: 10.1093/bioinformatics/14.1.48. [DOI] [PubMed] [Google Scholar]
- Baker CL, Kajita S, Walker M, Saxl RL, Raghupathy N, Choi K, Petkov PM, Paigen K. PRDM9 drives evolutionary erosion of hotspots in Mus musculus through haplotype-specific initiation of meiotic recombination. PLoS Genet. 2015;11:e1004916. doi: 10.1371/journal.pgen.1004916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker CL, Walker M, Kajita S, Petkov PM, Paigen K. PRDM9 binding organizes hotspot nucleosomes and limits Holliday junction migration. Genome Res. 2014;24:724–732. doi: 10.1101/gr.170167.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlow C, Hirotsune S, Paylor R, Liyanage M, Eckhaus M, Collins F, Shiloh Y, Crawley JN, Ried T, Tagle D, et al. Atm-deficient mice: a paradigm of ataxia telangiectasia. Cell. 1996;86:159–171. doi: 10.1016/s0092-8674(00)80086-0. [DOI] [PubMed] [Google Scholar]
- Baudat F, Imai Y, de Massy B. Meiotic recombination in mammals: localization and regulation. Nat Rev Genet. 2013;14:794–806. doi: 10.1038/nrg3573. [DOI] [PubMed] [Google Scholar]
- Bellani MA, Romanienko PJ, Cairatti DA, Camerini-Otero RD. SPO11 is required for sex-body formation, and Spo11 heterozygosity rescues the prophase arrest of Atm−/− spermatocytes. J Cell Sci. 2005;118:3233–3245. doi: 10.1242/jcs.02466. [DOI] [PubMed] [Google Scholar]
- Bois PR. A highly polymorphic meiotic recombination mouse hot spot exhibits incomplete repair. Mol Cell Biol. 2007;27:7053–7062. doi: 10.1128/MCB.00874-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brick K, Smagulova F, Khil P, Camerini-Otero RD, Petukhova GV. Genetic recombination is directed away from functional genomic elements in mice. Nature. 2012;485:642–645. doi: 10.1038/nature11089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung D, Kuan PF, Li B, Sanalkumar R, Liang K, Bresnick EH, Dewey C, Keles S. Discovering transcription factor binding sites in highly repetitive regions of genomes with multi-read analysis of ChIP-Seq data. PLoS Comput Biol. 2011;7:e1002111. doi: 10.1371/journal.pcbi.1002111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole F, Baudat F, Grey C, Keeney S, de Massy B, Jasin M. Mouse tetrad analysis provides insights into recombination mechanisms and hotspot evolutionary dynamics. Nat Genet. 2014;46:1072–1080. doi: 10.1038/ng.3068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole F, Keeney S, Jasin M. Comprehensive, fine-scale dissection of homologous recombination outcomes at a hot spot in mouse meiosis. Mol Cell. 2010;39:700–710. doi: 10.1016/j.molcel.2010.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper TJ, Garcia V, Neale MJ. Meiotic DSB patterning: A multifaceted process. Cell Cycle. 2016;15:13–21. doi: 10.1080/15384101.2015.1093709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies B, Hatton E, Altemose N, Hussin JG, Pratto F, Zhang G, Hinch AG, Moralli D, Biggs D, Diaz R, et al. Re-engineering the zinc fingers of PRDM9 reverses hybrid sterility in mice. Nature. 2016;530:171–176. doi: 10.1038/nature16931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Boer E, Jasin M, Keeney S. Local and sex-specific biases in crossover vs. noncrossover outcomes at meiotic recombination hot spots in mice. Genes Dev. 2015;29:1721–1733. doi: 10.1101/gad.265561.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Massy B. Initiation of meiotic recombination: how and where? Conservation and specificities among eukaryotes. Annu Rev Genet. 2013;47:563–599. doi: 10.1146/annurev-genet-110711-155423. [DOI] [PubMed] [Google Scholar]
- Fowler KR, Sasaki M, Milman N, Keeney S, Smith GR. Evolutionarily diverse determinants of meiotic DNA break and recombination landscapes across the genome. Genome Res. 2014;24:1650–1664. doi: 10.1101/gr.172122.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Froenicke L, Anderson LK, Wienberg J, Ashley T. Male mouse recombination maps for each autosome identified by chromosome painting. Am J Hum Genet. 2002;71:1353–1368. doi: 10.1086/344714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia V, Gray S, Allison RM, Cooper TJ, Neale MJ. Tel1(ATM)-mediated interference suppresses clustered meiotic double-strand-break formation. Nature. 2015;520:114–118. doi: 10.1038/nature13993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Getun IV, Wu ZK, Bois PR. Organization and roles of nucleosomes at mouse meiotic recombination hotspots. Nucleus. 2012;3:244–250. doi: 10.4161/nucl.20325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Getun IV, Wu ZK, Khalil AM, Bois PR. Nucleosome occupancy landscape and dynamics at mouse recombination hotspots. EMBO Rep. 2010;11:555–560. doi: 10.1038/embor.2010.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter N. Meiotic recombination: the essence of heredity. Cold Spring Harb Perspect Biol. 2015;7 doi: 10.1101/cshperspect.a016618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessop L, Allers T, Lichten M. Infrequent co-conversion of markers flanking a meiotic recombination initiation site in Saccharomyces cerevisiae. Genetics. 2005;169:1353–1367. doi: 10.1534/genetics.104.036509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauppi L, Barchi M, Baudat F, Romanienko PJ, Keeney S, Jasin M. Distinct properties of the XY pseudoautosomal region crucial for male meiosis. Science. 2011;331:916–920. doi: 10.1126/science.1195774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauppi L, Barchi M, Lange J, Baudat F, Jasin M, Keeney S. Numerical constraints and feedback control of double-strand breaks in mouse meiosis. Genes Dev. 2013;27:873–886. doi: 10.1101/gad.213652.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauppi L, Jasin M, Keeney S. Meiotic crossover hotspots contained in haplotype block boundaries of the mouse genome. Proc Natl Acad Sci U S A. 2007;104:13396–13401. doi: 10.1073/pnas.0701965104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeney S, Lange J, Mohibullah N. Self-organization of meiotic recombination initiation: general principles and molecular pathways. Annu Rev Genet. 2014;48:187–214. doi: 10.1146/annurev-genet-120213-092304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelmenson PM, Petkov P, Wang X, Higgins DC, Paigen BJ, Paigen K. A torrid zone on mouse chromosome 1 containing a cluster of recombinational hotspots. Genetics. 2005;169:833–841. doi: 10.1534/genetics.104.035063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khil PP, Smagulova F, Brick KM, Camerini-Otero RD, Petukhova GV. Sensitive mapping of recombination hotspots using sequencing-based detection of ssDNA. Genome Res. 2012;22:957–965. doi: 10.1101/gr.130583.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Peterson SE, Jasin M, Keeney S. Mechanisms of germ line genome instability. Semin Cell Dev Biol. 2016;54:177–187. doi: 10.1016/j.semcdb.2016.02.019. [DOI] [PubMed] [Google Scholar]
- Kirkpatrick DT, Dominska M, Petes TD. Conversion-type and restoration-type repair of DNA mismatches formed during meiotic recombination in Saccharomyces cerevisiae. Genetics. 1998;149:1693–1705. doi: 10.1093/genetics/149.4.1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam I, Keeney S. Nonparadoxical evolutionary stability of the recombination initiation landscape in yeast. Science. 2015;350:932–937. doi: 10.1126/science.aad0814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange J, Pan J, Cole F, Thelen MP, Jasin M, Keeney S. ATM controls meiotic double-strand-break formation. Nature. 2011;479:237–240. doi: 10.1038/nature10508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machanick P, Bailey TL. MEME-ChIP: motif analysis of large DNA datasets. Bioinformatics. 2011;27:1696–1697. doi: 10.1093/bioinformatics/btr189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancera E, Bourgon R, Brozzi A, Huber W, Steinmetz LM. High-resolution mapping of meiotic crossovers and non-crossovers in yeast. Nature. 2008;454:479–485. doi: 10.1038/nature07135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martini E, Borde V, Legendre M, Audic S, Regnault B, Soubigou G, Dujon B, Llorente B. Genome-wide analysis of heteroduplex DNA in mismatch repair-deficient yeast cells reveals novel properties of meiotic recombination pathways. PLoS Genet. 2011;7:e1002305. doi: 10.1371/journal.pgen.1002305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimitou EP, Yamada S, Keeney S. A global view of meiotic double-strand break end resection. bioRxiv. 2016 doi: 10.1126/science.aak9704. doi: 10.1101/067199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moens PB, Chen DJ, Shen Z, Kolas N, Tarsounas M, Heng HH, Spyropoulos B. Rad51 immunocytology in rat and mouse spermatocytes and oocytes. Chromosoma. 1997;106:207–215. doi: 10.1007/s004120050241. [DOI] [PubMed] [Google Scholar]
- Mohibullah N, Keeney S. Numerical and Spatial Patterning of Yeast Meiotic DNA Breaks by Tel1. bioRxiv. 2016 doi: 10.1101/gr.213587.116. doi: 10.1101/059022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh SD, Lao JP, Hwang PY, Taylor AF, Smith GR, Hunter N. BLM ortholog, Sgs1, prevents aberrant crossing-over by suppressing formation of multichromatid joint molecules. Cell. 2007;130:259–272. doi: 10.1016/j.cell.2007.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan J, Sasaki M, Kniewel R, Murakami H, Blitzblau HG, Tischfield SE, Zhu X, Neale MJ, Jasin M, Socci ND, et al. A hierarchical combination of factors shapes the genome-wide topography of yeast meiotic recombination initiation. Cell. 2011;144:719–731. doi: 10.1016/j.cell.2011.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry J, Palmer S, Gabriel A, Ashworth A. A short pseudoautosomal region in laboratory mice. Genome Res. 2001;11:1826–1832. doi: 10.1101/gr.203001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratto F, Brick K, Khil P, Smagulova F, Petukhova GV, Camerini-Otero RD. Recombination initiation maps of individual human genomes. Science. 2014;346:1256442. doi: 10.1126/science.1256442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radford SJ, Sabourin MM, McMahan S, Sekelsky J. Meiotic recombination in Drosophila Msh6 mutants yields discontinuous gene conversion tracts. Genetics. 2007;176:53–62. doi: 10.1534/genetics.107.070367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rumble SM, Lacroute P, Dalca AV, Fiume M, Sidow A, Brudno M. SHRiMP: accurate mapping of short color-space reads. PLoS Comput Biol. 2009;5:e1000386. doi: 10.1371/journal.pcbi.1000386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smagulova F, Brick K, Pu Y, Camerini-Otero RD, Petukhova GV. The evolutionary turnover of recombination hot spots contributes to speciation in mice. Genes Dev. 2016;30:266–280. doi: 10.1101/gad.270009.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smagulova F, Gregoretti IV, Brick K, Khil P, Camerini-Otero RD, Petukhova GV. Genome-wide analysis reveals novel molecular features of mouse recombination hotspots. Nature. 2011;472:375–378. doi: 10.1038/nature09869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thacker D, Mohibullah N, Zhu X, Keeney S. Homologue engagement controls meiotic DNA break number and distribution. Nature. 2014;510:241–246. doi: 10.1038/nature13120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tischfield SE, Keeney S. Scale matters: the spatial correlation of yeast meiotic DNA breaks with histone H3 trimethylation is driven largely by independent colocalization at promoters. Cell Cycle. 2012;11:1496–1503. doi: 10.4161/cc.19733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojtasz L, Daniel K, Roig I, Bolcun-Filas E, Xu H, Boonsanay V, Eckmann CR, Cooke HJ, Jasin M, Keeney S, et al. Mouse HORMAD1 and HORMAD2, two conserved meiotic chromosomal proteins, are depleted from synapsed chromosome axes with the help of TRIP13 AAA-ATPase. PLoS Genet. 2009;5:e1000702. doi: 10.1371/journal.pgen.1000702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu ZK, Getun IV, Bois PR. Anatomy of mouse recombination hot spots. Nucleic Acids Res. 2010;38:2346–2354. doi: 10.1093/nar/gkp1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakharyevich K, Ma Y, Tang S, Hwang PY, Boiteux S, Hunter N. Temporally and biochemically distinct activities of Exo1 during meiosis: double-strand break resection and resolution of double Holliday junctions. Mol Cell. 2010;40:1001–1015. doi: 10.1016/j.molcel.2010.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.