Abstract
Quaternary glaciations and mostly last glacial maximum have shaped the contemporary distribution of many species in the Alps. However, in the Maritime and Ligurian Alps a more complex picture is suggested by the presence of many Tertiary paleoendemisms and by the divergence time between lineages in one endemic species predating the Late Pleistocene glaciation. The low number of endemic species studied limits the understanding of the processes that took place within this region. We used species distribution models and phylogeographical methods to infer glacial refugia and to reconstruct the phylogeographical pattern of Silene cordifolia All. and Viola argenteria Moraldo & Forneris. The predicted suitable area for last glacial maximum roughly fitted current known distribution. Our results suggest that separation of the major clades predates the last glacial maximum and the following repeated glacial and interglacial periods probably drove differentiations. The complex phylogeographical pattern observed in the study species suggests that both populations and genotypes extinction was minimal during the last glacial maximum, probably due to the low impact of glaciations and to topographic complexity in this area. This study underlines the importance of cumulative effect of previous glacial cycles in shaping the genetic structure of plant species in Maritime and Ligurian Alps, as expected for a Mediterranean mountain region more than for an Alpine region.
Introduction
Quaternary glaciations and mostly late Pleistocene glaciation (0.120–0.018 Ma) have shaped the contemporary distribution of many species [1]. In general, species survived cold adverse periods in so-called glacial refugia and later recolonised areas newly available during warmer post-glacial periods [2, 3]. Populations that survived glaciations in refugia could have accumulated large amounts of genetic diversity, whereas those now found in post-glacially recolonised regions may have less diversity as a result of bottlenecks during their recent expansions [3, 4].
In the last few years several phylogeographical studies have supported this general view concerning the importance of the last glacial maximum (LGM; about 0.020 Ma) in shaping species distribution [5] and contemporary intraspecific genetic diversity [6, 4]. However, it has also been recently demonstrated that Mediterranean refugia represent climatically stable areas for which it is necessary to consider the cumulative effects of changes, rather than just the changes that occurred during the last glacial period [7]. The high genetic diversity detected in the Mediterranean refugia is due to the preservation of genotypes that went extinct in other places and to the intensity and accumulation of a number of processes in a patchy landscape across a varied topography [8]. A detailed understanding of the effects of past climate changes on the distribution and genetic pattern of organisms may help us to better predict the effects of ongoing climate change [9, 10]. It has been recently demonstrated that the combination of paleo-distribution modelling with phylogeographical approaches may led to new interpretations of population genetic patterns and to new hypotheses about glacial survival and postglacial colonization [11, 12, 13].
Situated at the crossroads of the Mediterranean Basin and the Alps, the Maritime and Ligurian Alps are a hotspot for plant biodiversity [14, 15] and well-known peripheral refugia [16]. Molecular investigations on the endemic plants of this region have underlined the role of vicariance events in modelling the genetic pattern of species during the last glaciation [17, 18, 19]. Nevertheless, Casazza et al. [13] recently demonstrated that the narrow endemic Primula allionii Loisel. was separated into two geographically disjoined groups during the Early/Mid Pleistocene border (0.781 Ma) and the region probably contains other paleoendemic species representing living examples of an ancient Tertiary flora present prior to the onset of the Mediterranean climate regime and subsequent Quaternary glaciations. However, the low number of endemic species that have been studied in this region limits our capacity to understand the processes that took place within the Maritime and Ligurian Alps [13, 17, 18, 19, 20].
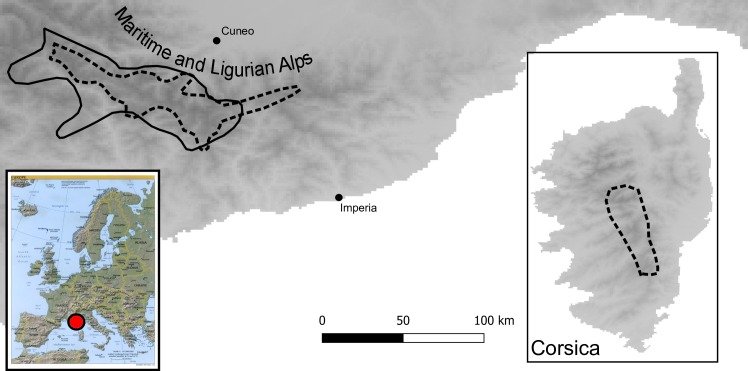
In the present study we used species distribution models (SDMs) and phylogeographical methods to infer glacial refugia and to reconstruct the phylogeographical pattern of two endemic species of the area, Silene cordifolia All. and Viola argenteria Moraldo & Forneris (Fig 1). Silene cordifolia is a strict endemic species restricted to the Maritime Alps that represents an old Tertiary lineage [21] and currently occurs on siliceous cliffs and erratic rocks between 700 and 3000 m of altitude. Viola argenteria is a disjoint endemic species (sensu [22]) that has a small number of populations on Corsica (Fig 1). This species grows on scree slopes from 2000 to 3000 m altitude and may have originated more recently, as suggested by morphological, cytogenetic and molecular evidences [23]. More specifically, we asked the following questions: (1) Is there any evidence of haplotype divergence within S. cordifolia and V. argenteria before the last Pleistocene glaciation? (2) Is there any evidence that populations of S. cordifolia and V. argenteria were influenced by Pleistocene climate fluctuations, and, if so, how?
Fig 1. Area of distribution of S. cordifolia (continuous black line) and V. argenteria dotted black line).
The names of major geographical areas are reported.
Materials and Methods
Species distribution modelling
We collected all occurrence data from the SILENE database (www.silene.eu) of the Conservatoire Botanique National Méditerranéen de Porquerolles (CBNMed), from Florence, Turin and Genoa Herbaria (FI, TO, GE) and from our own field surveys. We thus gathered a total of 369 points (266 from SILENE, 49 from Herbaria and 54 from field surveys) for S. cordifolia and 224 points (121 from SILENE, 54 from Herbaria and 49 from field surveys) for V. argenteria.
At the local scale, species distribution models provide excellent spatial projections by additionally using non-climatic predictors [24]. For this reason, a layer reporting the presence/absence of suitable substrate was elaborated, starting from the global lithological map dataset, GLiM [25]. The bioclimatic layers for the current (30 arcsec resolution) and LGM (2.5 arcmin resolution) conditions, data were downloaded from the WorldClim 1.4 database website (http://www.worldclim.org). We used data from the Community Climate System Model (CCSM), the Model for Interdisciplinary Research on Climate (MIROC) and from the Max Planck Institute for Meteorology (MPI) to hindcast LGM (http://www.worldclim.org/paleo-climate1#com). These data were statistically downscaled from the original resolution to 30 arcsec with the Delta method [26]. This downscaling method is based on the interpolation of anomalies (differences between LGM and current conditions) at 30 arcsec resolution through the thin-plate smoothing spline algorithm. The interpolated anomalies were finally added to the layers of current condition. To reduce the multicollinearity between predictors and to minimize model overfitting, after normalizing the 19 bioclimatic predictors we performed a pairwise Pearson correlation between them (S1 Table) using R v. 3.1.1 [27]. We used the predictors that are considered physiologically important for plants [28] and that were not strongly correlated with each other (Pearson correlation coefficient, r2 <|0.80|). To model species distributions, we used the following six variables: BIO3, isothermality; BIO4, temperature seasonality; BIO14, precipitation of driest month; BIO19, precipitation of coldest quarter; the presence/absence of suitable substrate. We removed duplicates using ENMTools [29]. Finally, 360 for S. cordifolia and 223 presence records for V. argenteria were applied in SDM analysis.
To account for model-based uncertainties, we applied six SDM techniques: multivariate adaptive regression splines (MARS–[30]), generalized linear models (GLM–[31]), classification tree analysis (CTA–[32]), flexible discriminant analysis (FDA–[33]), random forest (RF–[34]), and Maximum entropy modelling (MaxEnt–[35]). These techniques belong to three different categories of models (i.e. regression methods–MARS and GLM; classification methods–CTA and FDA; and machine learning algorithms–RF and MaxEnt).
Analyses were implemented with BIOMOD2 v. 1.3.5 package [36] for R v. 3.1.1 [27]. The number of replicate sets of pseudo-absences may influence the predictive accuracy of the models. For this reason, we chose the best strategy in pseudo-absences selection according to Barbet-Massin et al. [37] tips (S2 Table). The predictive performance of the models was evaluated for each pseudo-absence run using a random subset (70%) of the initial dataset each time to calibrate the models, while the remaining 30% were used to evaluate the models. The predictive performance of the models was evaluated based on the true skill statistic (TSS), which takes into account both omission and commission errors and it is not affected by prevalence [38]. To transform the inferred continuous probability values to binary presence-absence form we used the TSS. Consensus projection under current conditions was created summing binary predictions of the six modelling techniques. We also created consensus projection under LGM conditions summing the binary predictions of the six modelling techniques for the three LGM climates, thus outlining those areas predicted by most modelling techniques under the different climates.
DNA sampling and sequencing
A total of 94 individuals from 19 populations for S. cordifolia and a total of 80 individuals from 21 populations for V. argenteria were sampled, covering the entire geographical range of species (Table 1). Permissions to collect samples in field were obtained from Parc National du Mercantour and form Parco Naturale Alpi Marittime. Total DNA was extracted from leaf tissue using the DNeasy Plant Mini Kit (Qiagen AG, Hombrechtikon, Switzerland), following the manufacturer’s instructions. Seven cpDNA regions in S. cordifolia (rps16 gene, trnQ-rps16, trnG2G-trnG, trnH-psbA, trnC-ycf6R, trnTa-trnLb, trnL-trnF) and seven cpDNA regions in V. argenteria (atpF-atpH, rpoC1 gene, rps12-rpL20, trnG2G-trnG, trnH-psbA, trnL-trnF, trnTa-trnLb) were chosen (S3 Table). PCR reactions were performed using Eppendorf thermocycler, with 20 μL total volume reactions containing 5–30 ng DNA, 10 μL GoTaq Green Master Mix (Promega, Milan, Italy), 10 pmol of each primer and purified water. We used the following PCR conditions: 2 min pre-treatment at 94°C, followed by 35 cycles of 120 s denaturation (94°C), 45 s annealing (48–63°C), 75–120 s extension (72°C). After the last cycle the temperature was kept at 72°C for the last 7 min of extension and then lowered to 4°C (S3 Table). The PCR products were checked on 2% agarose gels in biotium staining. The amplification products were then purified using the commercial kit QIAquick PCR Purification Kit (Qiagen, USA). All DNA sequencing was performed by Macrogen company (Korea). Sequences were aligned using MAFFT v. 6.903 [39] and adjusted by hand. The cpDNA regions were concatenated in a single matrix. We considered indels as character because previous studies exploring the utility of cpDNA indels have concluded that they are informative and that they might increase resolution between geographical regions of high diversity [40]. All sequences have been deposited in GenBank database (S4 Table).
Table 1. Information about the populations of S. cordifolia and V. argenteria sampled for the analysis.
Population code (C), Latitude N (Lat), Longitude E (Long) and elevation (Elev.) as metres above sea level, are reported. Populations from Corsica were sampled in Florence Herbarium (FI) and the coordinates are approximate.
| Taxon | Population | C | Lat | Long | Elev. |
|---|---|---|---|---|---|
| S. cordifolia | Vallon de Rabuons, Vallèe du Tinée, FR | 1 | 44.2704 | 6.96283 | 2400 |
| L'illions, Vallée du Var, FR | 2 | 44.0490 | 6.95815 | 1638 | |
| Vallone di Sant'Anna, Valle Stura, IT | 3 | 44.2735 | 7.13497 | 1341 | |
| S. Anna di Vinadio, Valle Stura, IT | 4 | 44.2264 | 7.10274 | 2154 | |
| Isola 2000, Vallèe du Tinée, FR | 5 | 44.2069 | 7.11142 | 1715 | |
| La Barre-Roure, Vallèe du Tinèe, FR | 6 | 44.1015 | 7.08885 | 1378 | |
| Mt. Pépoiri, Vallèe du Tinèe, FR | 7 | 44.1141 | 7.18859 | 2315 | |
| Vallone del Valasco, Val Gesso, IT | 8 | 44.2143 | 7.23606 | 2440 | |
| Vallone della Meris, Val Gesso, IT | 9 | 44.2439 | 7.22745 | 2300 | |
| L. di Fremamorta, Val Gesso, IT | 10 | 44.1745 | 7.25362 | 2107 | |
| M. Ray, Val Gesso, IT | 11 | 44.2262 | 7.36828 | 1971 | |
| Bacino del Chiotas, Val Gesso, IT | 12 | 44.1700 | 7.33278 | 2095 | |
| Rifugio Soria-Ellena, Val Gesso, IT | 13 | 44.1401 | 7.36359 | 1884 | |
| Col de Fenestre, Vallée du Vésubie, FR | 14 | 44.1082 | 7.35565 | 2314 | |
| V. de la Madone de Fenestre, V. Vésubie, FR | 15 | 44.0841 | 7.28705 | 1314 | |
| St. Grat de Gordolasque, V. Vésubie, FR | 16 | 44.0727 | 7.39797 | 1775 | |
| Lago Veil del Buc, Val Gesso, IT | 17 | 44.1491 | 7.42930 | 2444 | |
| Mt. Peirafica, Vallée de la Roya, FR | 18 | 44.1297 | 7.51889 | 1990 | |
| Ref. Merveilles, Vallée de la Roya, FR | 19 | 44.0563 | 7.46058 | 2097 | |
| V. argenteria | Ref. Lausa, Vallèe du Tinèe, FR | 1 | 44.2978 | 6.96232 | 2544 |
| Vallon de Rabuons, Vallèe du Tinèe, FR | 2 | 44.2607 | 6.98320 | 2678 | |
| Collalunga, Valle Stura, IT | 3 | 44.2235 | 7.03190 | 2445 | |
| Tesina/Sabulé, Valle Stura, IT | 4 | 44.2271 | 7.08179 | 2466 | |
| Mt Saint Sauveur, Vallèe du Tinèe, FR | 5 | 44.1700 | 7.13032 | 2414 | |
| Bassa del Druos, | 6 | 44.1902 | 7.19138 | 2610 | |
| Col de Barn-Mt. Pepoiri, Vallèe du Tinèe, FR | 7 | 44.1140 | 7.20159 | 2493 | |
| Alta Valle Meris, Val Gesso, IT | 8 | 44.2319 | 7.21665 | 2620 | |
| Lago inferiore di Valrossa, Val Gesso, IT | 9 | 44.2161 | 7.22214 | 2507 | |
| Laghi Colle di Fremamorta, Val Gesso, IT | 10 | 44.1601 | 7.25121 | 2433 | |
| Altopiano del Baus, Val Gesso, IT | 11 | 44.1741 | 7.31499 | 2644 | |
| Valle del Brocan, Val Gesso, IT | 12 | 44.1526 | 7.31649 | 2504 | |
| Passo delle Finestrelle, Val Gesso, IT | 13 | 44.1572 | 7.34966 | 2433 | |
| Col de Fenestre, Vallée du Vésubie, FR | 14 | 44.1130 | 7.36051 | 2384 | |
| C. Vallette de Prals, Vallée du Vésubie, FR | 15 | 44.0639 | 7.35972 | 2385 | |
| Rifugio Pagari, ValGesso, IT | 16 | 44.1226 | 7.40698 | 2597 | |
| Monte Carboné, Val Gesso, IT | 17 | 44.1520 | 7.43214 | 2654 | |
| P. dell'Arpette, Vallée de la Roya, FR | 18 | 44.0599 | 7.42640 | 2528 | |
| Monte Rotondo, Corse, FR* | 19 | 42.2200 | 9.07000 | 2400 | |
| Monte d'Oro, Corse, FR** | 20 | 42.1400 | 9.10000 | 2100 | |
| Monte Renoso, Corse, FR*** | 21 | 42.0600 | 9.13000 | 2500 |
* Monte Rotondo, 7.1907, Martelli (FI)
** Monte d’Oro, 25.7.1907, Martelli (FI)
*** Monte Renoso, 2500 m, Levriere (FI).
Genetic diversity and differentiation
Haplotype number (H), haplotype diversity (Hd), number of segregating sites (S) and nucleotide diversity (π) [41] were calculated with DNASP 4.10 [42]. The total gene diversity (HT), population differentiation (GST) and an estimation of population subdivision for phylogenetically ordered alleles (NST) were calculated using the program PERMUT [43]. We further tested the presence of phylogeographical structure (sensu [43]), comparing NST to GST using 1000 permutations to obtain statistical significance with the program PERMUT. To evaluate the genetic structure, we conducted an analysis of molecular variance (AMOVA) in ARLEQUIN 3.1 [44], partitioning the genetic diversity into the diversity among groups, the diversity among populations within groups, and the diversity within populations. The correlation between the genetic (FST) and geographic pairwise population distance matrices was evaluated using a Mantel test [45] with 10,000 permutations using ARLEQUIN 3.1. The Mantel test was calculated on all populations analysed both in S. cordifolia and V. argenteria and on the Maritime and Ligurian Alps’ populations only in V. argenteria.
Analysis of haplotypes
To determine the genetic relationship among populations, we followed the method implemented by Muñoz-Pajares [46] in the SIDIER package. The method gathers the evolutionary information contained in insertions and deletions (indels) and combines such information with the p-distance matrix inferred from substitutions. This method improves the resolution of phylogenetic inference of low-divergence organisms, yielding results compatible with other evolutionary procedures [46]. Based on recommendations by Simmons et al. [47], indels in the concatenated dataset were coded as single mutational event using a variation of modified complex indel coding (MCIC—sensu [48]).
Spatial analysis of molecular variance (SAMOVA—[49], http://web.unife.it/progetti/genetica/Isabelle/samova.html) was used to identify groups of populations that are geographically homogeneous and maximally differentiated from each other without assuming any prior association between populations. The algorithm identifies the optimal number of groups of populations (k) by maximizing FCT (the proportion of total genetic variance due to differences among groups of populations) and minimizing FSC (genetic differentiation among populations within groups). SAMOVA was run for 10,000 iterations from each of 100 random initial conditions, and testing the predefined number of groups (k) from 2 to 10.
Best-fit models of nucleotide substitution were determined to be a TIM1 + I model for all S. cordifolia sequences and F81 model for V. argenteria, by the Akaike Information Criterion using JMODELTEST 2.1.1 [50]. Divergence time among haplotypes was estimated using BEAST v.1.8.0 [51]. We used a strict molecular clock and a coalescent Gaussian Markov random field (GMRF) Bayesian skyride tree prior. We sampled all parameters once every 2000 steps from 1 x 109 MCMC steps after a burn-in of 2.5 x 108 steps. The program TRACER [52] was used to examine convergence of chains to the stationary distribution [effective sample size (ESS) > 200]. In this study, we used normal distribution priors with a mean of 2 x 10−9 and a standard deviation of 5 x 10−10 substitutions per site per year to cover the cpDNA substitution rates for angiosperm (1–3 x 10−9 substitutions per site per year; [53, 54, 55]).
Results
Species distribution models
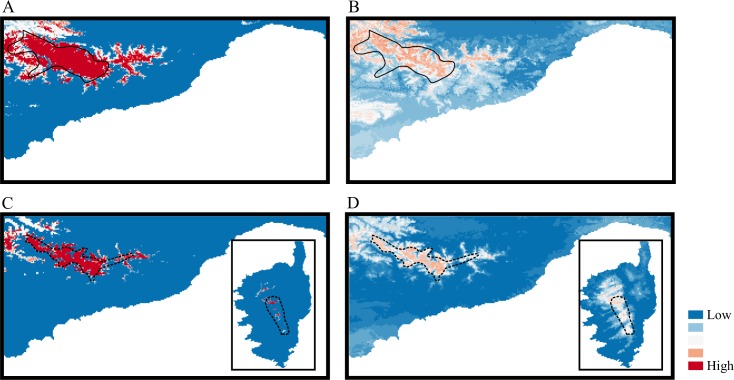
Under current climate conditions, following Swets’ scale modified by Araújo et al. [56], TSS (Table 2) indicated excellent model performance for both species. The SDMs projections for current conditions (Fig 2A and 2C) roughly fitted the current species distribution. The predicted potential distribution during the LGM was different among climates and algorithms (S1 Fig). In both species, LGM models based on CCSM and MIROC climate identified regions similar to present models, but reduced in range. On the contrary, MPI climate predicted a range wider than present in S. cordifolia while in V. argenteria it predicted a range wider than present in Corsica and narrower in the Maritime and Ligurian Alps. Despite this variation, the ensemble maps of all climates and algorithms suggested the presence of suitable areas in the current distributional range (Fig 2B and 2D).
Table 2. Evaluation of individual modelling techniques for S. cordifolia and V. argenteria.
Statistics given are the mean values and the associated standard deviations (in brackets) for the true skill statistic (TSS). Values given are. Accuracy classification for TSS following Swets’ scale modified by Araújo et al. [56]: 1> excellent>0.8>good>0.6>fair>0.4>poor>0.2>fail. MARS Multiple Additive Regression Spline; GLM, Generalized Linear Models; CTA, Classification Tree Analysis; FDA, Flexible Discriminant Analysis; RF, Random Forest; MAX, Maximum Entropy.
| S. cordifolia | V. argenteria | |
|---|---|---|
| TSS | TSS | |
| MARS | 0.887 (0.053) | 0.932 (0.077) |
| GLM | 0.893 (0.008) | 0.931 (0.021) |
| CTA | 0.938 (0.014) | 0.940 (0.033) |
| FDA | 0.929 (0.050) | 0.963 (0.033) |
| RF | 0.975 (0.013) | 0.972 (0.019) |
| MaxEnt | 0.887 (0.013) | 0.941 (0.016) |
Fig 2.
Maps showing the consensus among the 6 binary maps of the current distribution (A and C) and of the 18 binary maps of the Last Glacial Maximum distribution (B and D; LGM ~ 21 kya) for S. cordifolia (A and B) and V. argenteria (C and D). Models were obtained using six algorithms (MARS, GLM, CTA, FDA, RF and MAX). LGM models are obtained using three palaeoclimate data (MIROC, CCSM and MPI). The inferred continuous probability values were converted to binary using TSS threshold. Red indicates greatest consensus among models and blue indicates lowest consensus among models. Current known distribution area of S. cordifolia (continuous black line) and V. argenteria (dotted black line) is reported.
Sequence data
The total combined length of the aligned sequences of the seven cpDNA was 4461 bp in S. cordifolia and 3730 bp in V. argenteria. We detected variations in four out of seven cpDNA regions both in S. cordifolia and in V. argenteria (Table 3). In particular, we recorded 12 variable and 9 parsimony-informative sites in S. cordifolia and 15 variable and parsimony-informative sites in V. argenteria. Alignments of the sequences of S. cordifolia and V. argenteria showed 1 and 17 insertion/deletion sites (indels) respectively. In all, there were 13 variable sites among the sampled individuals of S. cordifolia and 65 in V. argenteria (S5 Table). We recovered 12 haplotypes within S. cordifolia and 16 haplotypes within V. argenteria (Table 3).
Table 3. Chloroplast genetic diversity within S. cordifolia and within V. argenteria.
Number of sampled individuals (NI), number of haplotypes (h), number of private haplotypes (ph), haplotype diversity (Hd) and nucleotide diversity (π) are indicated for populations from each species.
| Taxon/population | NI | h | ph | Hd | π |
|---|---|---|---|---|---|
| S. cordifolia | |||||
| 1 | 5 | 1 | 0 | 0 | 0 |
| 2 | 5 | 2 | 1 | 0.4 | 0.00027 |
| 3 | 5 | 1 | 0 | 0 | 0 |
| 4 | 5 | 2 | 1 | 0.4 | 0.00018 |
| 5 | 5 | 2 | 0 | 0.7 | 0.00031 |
| 6 | 5 | 3 | 0 | 0.7 | 0.00027 |
| 7 | 5 | 1 | 0 | 0 | 0 |
| 8 | 5 | 1 | 0 | 0 | 0 |
| 9 | 4 | 4 | 1 | 1 | 0.00086 |
| 10 | 4 | 4 | 1 | 1 | 0.00101 |
| 11 | 5 | 2 | 0 | 0.6 | 0.00013 |
| 12 | 6 | 2 | 0 | 0.6 | 0.00013 |
| 13 | 5 | 2 | 0 | 0.6 | 0.00013 |
| 14 | 5 | 2 | 0 | 0.6 | 0.00081 |
| 15 | 5 | 1 | 0 | 0 | 0 |
| 16 | 5 | 2 | 1 | 0.4 | 0.00018 |
| 17 | 5 | 1 | 0 | 0 | 0 |
| 18 | 5 | 1 | 0 | 0 | 0 |
| 19 | 5 | 1 | 0 | 0 | 0 |
| 94 | 12 | 0.824 | 0.00089 | ||
| V. argenteria | |||||
| 1 | 4 | 1 | 0 | 0 | 0 |
| 2 | 4 | 1 | 0 | 0 | 0 |
| 3 | 5 | 1 | 0 | 0 | 0 |
| 4 | 5 | 2 | 1 | 0.6 | 0.00049 |
| 5 | 6 | 1 | 0 | 0 | 0 |
| 6 | 5 | 1 | 0 | 0 | 0 |
| 7 | 5 | 3 | 2 | 0.7 | 0 |
| 8 | 3 | 1 | 0 | 0 | 0 |
| 9 | 2 | 1 | 0 | 0 | 0 |
| 10 | 3 | 1 | 0 | 0 | 0 |
| 11 | 5 | 2 | 1 | 0.6 | 0.00016 |
| 12 | 4 | 4 | 2 | 1 | 0.00036 |
| 13 | 5 | 2 | 0 | 0.4 | 0.00022 |
| 14 | 3 | 1 | 0 | 0 | 0 |
| 15 | 5 | 1 | 0 | 0 | 0 |
| 16 | 3 | 2 | 1 | 0.7 | 0 |
| 17 | 4 | 2 | 1 | 0.5 | 0 |
| 18 | 3 | 1 | 1 | 0 | 0 |
| 19 | 2 | 1 | 1 | 0 | 0 |
| 20 | 2 | 1 | 1 | 0 | 0 |
| 21 | 2 | 1 | 1 | 0 | 0 |
| 80 | 16 | 0.834 | 0.00081 |
Phylogeographical relationships and divergence times of haplotypes
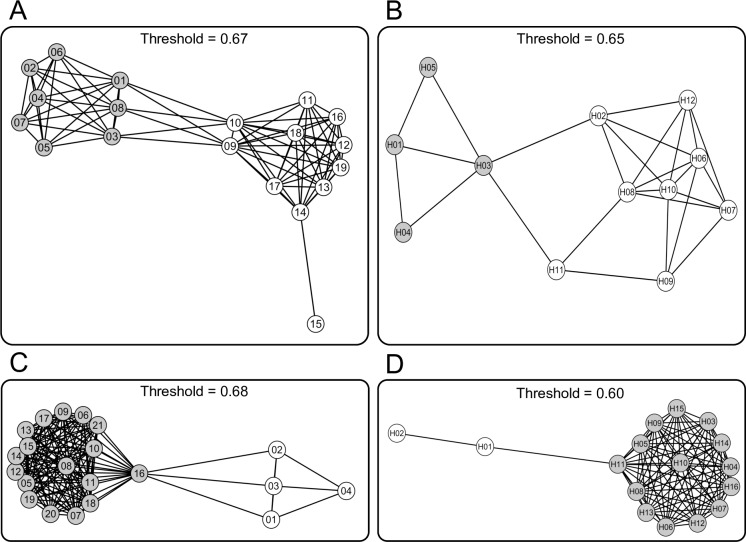
The dendrogram shows a clear divergence between two groups of haplotypes in both species. In S. cordifolia the first group (grey haplotypes in Fig 3A) is present in the north-western populations while the second group (white haplotypes in Fig 3C) is present throughout the entire range of distribution. In V. argenteria the first group (grey haplotypes in Fig 3B) is present in the north-western populations while the second group (white haplotypes in Fig 3D) is only present in the south-eastern populations.
Fig 3. Percolation network obtained combining distance matrices of indels and substitutions.
Relationships among populations (A and B) and haplotypes (C and D) are showed for S. cordifolia (A and C) and V. argenteria (B and D). The network was built connecting distances lower than the estimated percolation threshold (reported in the figure). Groups (i.e., subsets of nodes conforming densely connected subgraphs) are represented in different grey tones.
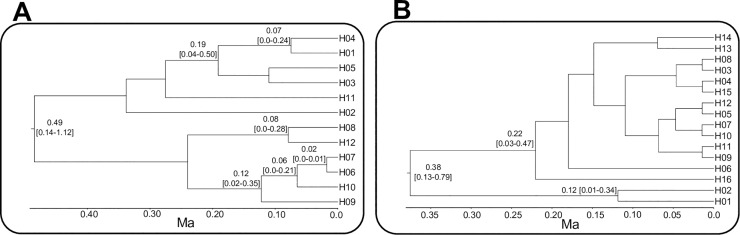
Diversification of S. cordifolia and V. argenteria haplotype lineages occurred during the Pleistocene, with the main lineages diverging during the middle Pleistocene. Phylogenetic analysis of cpDNA haplotypes showed that S. cordifolia differentiated into two main lineages at 0.49 (0.14–1.12) Ma. The first (H01-H05, H11) is mainly present in south-eastern populations (H01-H04) but also in some north-western populations (H05 and H11) while the second lineage (H06-H10, H12) is mainly present in north-western populations (H06-H07, H09-H10, H12) but also in some south-eastern populations (H08). Similarly, V. argenteria differentiated into two main lineages at 0.37 (0.12–0.79) Ma. The first lineage (H01 and H02) is exclusive of the north-western populations while the second lineage (H03-H16) is exclusive of the south-eastern and Corsican populations. However, the node age estimates should be interpreted with caution given the width of the 95% confidence intervals for each node (Fig 4).
Fig 4.
BEAST derived chronogram for haplotypes in S. cordifolia (A) and V. argenteria (B) based on cpDNA regions. Mean divergence time and 95% Highest Posterior Density (HPD) intervals are reported for nodes with posterior probability higher than 50%. Haplotype codes correspond to those in Table 1.
Genetic diversity and differentiation of populations
In S. cordifolia eight populations contained just one haplotype, seven populations contained two haplotypes, two populations contained three haplotypes and the remaining two populations contained four haplotypes. Five populations distributed throughout the range of the species showed a private haplotype (2, 4, 9, 10 and 16). The populations showing the higher values of haplotype diversity were located in the central-northern part of distribution range. The total gene diversity was high (HT = 0.847, SE = 0.0251) while the within-population diversity was low (HS = 0.368, SE = 0.0824). The differentiation among populations based on cpDNA variation (GST = 0.565, SE = 0.0960) indicates a moderate degree of population genetic structure within S. cordifolia. Allelic differentiation (NST = 0.749, SE = 0.0835) is greater than but not significantly different from GST, indicating a weak genetic differentiation among populations. In V. argenteria 14 populations contained just one haplotype, five populations contained two haplotypes and the remnant two populations contained three and four haplotypes, respectively. Ten populations, mainly located in the southern part of Maritime Alps and in Corsica, showed private haplotype (4, 7, 11, 12, 16, 17, 18, 19, 20 and 21). The populations showing the higher values of haplotype diversity were located in the central-southern part of distribution range. The total gene diversity was high (HT = 0.885, SE = 0.0417) while the within-population diversity was low (HS = 0.213, SE = 0.0710). The differentiation among populations based on cpDNA variation (GST = 0.760 SE = 0.0785) indicates a large degree of population genetic structure within V. argenteria. Allelic differentiation (NST = 0.921 SE = 0.0486) is greater than and significantly different from GST, indicating a strong genetic differentiation among populations. AMOVA (Table 4) showed that a large proportion (S. cordifolia: 77.32%; V. argenteria: 87.70%) of the chloroplast variation was partitioned among the population groups identified by SAMOVA. In contrast, the component of among-populations variation within groups was the lowest (S. cordifolia: 7.09%; V. argenteria: 4.74%). The proportion of variation residing within populations (S. cordifolia: 15.59%; V. argenteria: 7.56%) was consistent with the relatively low value of within-population diversity. Mantel test revealed a significant isolation-by-distance pattern in both species (S. cordifolia: r = 0.417, P < 0.0001; V. argenteria all populations: r = 0.296, P < 0.0001; V. argenteria Maritime and Ligurian Alps populations: r = 0.586, P < 0.0001).
Table 4. Results of AMOVA of cpDNA data from 19 populations of S. cordifolia and 21 populations of V. argenteria.
df: degree of freedom; SS: sum of square; variation %: percentage of total variance; ** P<0.001.
| Species | Source of variation | df | SS | Variation% | F statistic |
|---|---|---|---|---|---|
| S. cordifolia | |||||
| Among population | 18 | 170.409 | 77.96** | FST = 0.77957 | |
| Within population | 75 | 38.400 | 22.04 | ||
| Among SIDIER groups | 1 | 117.707 | 70.65** | FCT = 0.70655 | |
| Among population within groups | 17 | 52.702 | 14.84** | FSC = 0.50558 | |
| Within population | 75 | 38.400 | 14.51** | FST = 0.85491 | |
| Among SAMOVA groups | 2 | 143.813 | 77.32** | FCT = 0.77320 | |
| Among population within groups | 16 | 26.595 | 7.09** | FSC = 0.31259 | |
| Within population | 75 | 38.400 | 15.59** | FST = 0.84409 | |
| V. argenteria | |||||
| Among population | 20 | 768.596 | 90.20** | FST = 0.90196 | |
| Within population | 59 | 63.217 | 9.80 | ||
| Among SIDIER groups | 1 | 399.431 | 69.37** | FCT = 0.69369 | |
| Among population within groups | 19 | 369.165 | 25.12** | FSC = 0.82023 | |
| Within population | 59 | 63.217 | 5.51** | FST = 0.94493 | |
| Among SAMOVA groups | 707.960 | 85.18** | FCT = 0.85175 | ||
| Among population within groups | 60.636 | 7.69** | FSC = 0.51905 | ||
| Within population | 63.217 | 7.13** | FST = 0.92870 |
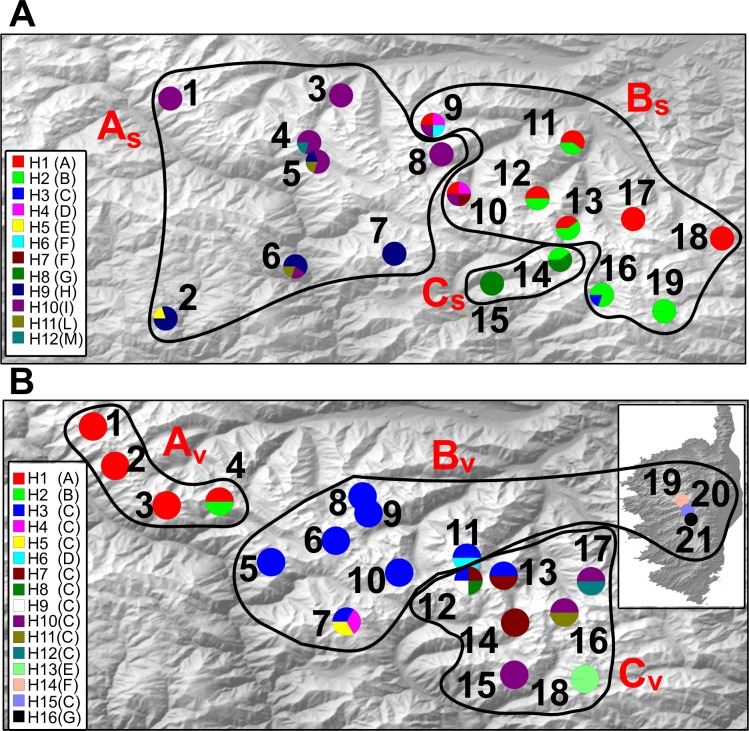
SAMOVA analysis detected 3 groups in S. cordifolia (AS, BS and CS in Fig 5) and V. argenteria (AV, BV and CV in Fig 5). Although the FCT continued to increase slightly and the FSC continued to decrease slightly as the number of groups was increased for each species, the FCT largely plateaued and the FSC markedly decreased at these respective groupings (S2 Fig). Basing on population distance, SIDIER analysis clearly separates populations into two discrete clusters: north-western and south-eastern populations (Fig 3A and 3B) in both species. In particular, in S. cordifolia the north-western cluster corresponds to SAMOVA group AS while south-eastern cluster corresponds to BS and CS groups. Similarly, in V. argenteria the north-western cluster corresponds to SAMOVA group AV while south-eastern cluster corresponds to BV and CV groups of SAMOVA.
Fig 5.
Geographical distribution of haplotypes across sampled populations of S. cordifolia (A) and V. argenteria (B). Pie charts represent haplotype proportions. Haplotype codes correspond to those in Table 1. Population groups identified by SAMOVA are delimited by continuous black lines and named by capital red letters for S. cordifolia (AS-CS) and V. argenteria (AV-DV).
Discussion
Palaeoclimatic models and haplotype divergence
In general, Alpine species are thought to have persisted LGM (0.12–0.01 Ma–[16, 57]) in glacial refugia and later to have recolonised newly available areas [4, 6]. As a consequence, the LGM had a strong influence on the genetic structure, distribution and evolution of plant species [2, 6, 58]. In fact, the LGM is recognized as a primary factor causing distributional patterns of endemic plants in Alps [5] while, the current ranges of many Alpine plants may also have been shaped by delayed Holocene recolonisation of suitable sites from refugial areas [59].
The palaeo-distribution models inferred in this work together with the low and patchy ice sheet coverage of LGM in the Maritime and Ligurian Alps [60, 61] suggest that the two study species, S. cordifolia and V. argenteria, probably survived the LGM in the sites where they were already present (in situ survival sensu [57]). The weak impact of the LGM in Maritime and Ligurian Alps may have been due to the climatic mitigation effect of the nearby Mediterranean Sea and to the relatively steep relief of this sector which was probably unfavourable to the formation of major glacier basins [61]. Recently, Patsiou et al. [62] showed that Saxifraga florulenta Moretti, endemic to Maritime Alps, survived the last glaciation in several microrefugia close to sites of current occurrence. Our finding supports the idea of a weak impact of the LGM on the distribution pattern of endemic species in the Maritime and Ligurian Alps, in marked contrast to its probable effect in other parts of the Alps [2, 4]. In line with the possibility of in situ survival during the LGM detected by SDM and previously debated, our analyses (Fig 4) suggest a divergence time between lineages predating the Late Pleistocene glaciation (0.115–0.018 Ma) both in S. cordifolia (0.490 Ma; 95% HPD:0.140–1.120 Ma) and V. argenteria (0.370 Ma; 95% HPD: 0.120–0.790 Ma). We thus suggest that these two species illustrate the persistence of an ancient genetic structure through several Pleistocene climatic cycles. A similar time frame has been previously suggested for other animal and plant species from other biogeographical regions [63, 64].
Our results are also congruent with the palaeo-climatic reconstructions. In fact, the most extensive glaciation in Mediterranean mountains occurred during the Middle Pleistocene (0.780–0.120 Ma), in particular a major phase probably occurred during Mindel glaciation (0.470–0.420 Ma–[65, 66]). Moreover, pollen-inferred historical climate indicates that maxima temperatures for interglacial stages in the Middle Pleistocene (0.780–0.120 Ma) were similar to the lowest values observed during the Late Pleistocene glacial period (0.115–0.018 Ma–[67]). In contrast, in the central Alps there is evidence of extensive glaciation during Riss (0.190–0.115 Ma–[68]) and Würm glaciation (0.115–0.018 Ma–[60]). Although the differentiation between main lineages is most likely to have begun in the Middle Pleistocene, haplotype divergence within main lineages probably occurred successively (Fig 4). This is congruent with the observation that in some Alpine animal species the separation of the major clades occurred long before the LGM and the following repeated glacial and interglacial periods may have been the motor for marked differentiation that is currently observed [69, 70]. A similar time frame was detected in another endemic species in the calcareous cliffs of the Maritime and Ligurian Alps, P. allionii where the separation into two groups dates back to the Middle Pleistocene [13]. In contrast, in S. florulenta the differentiation of two ancestral gene pools seems to have been influenced by range contractions that were more recent than the glacial oscillations [20], an interpretation that is supported by SDMs [62]. Nevertheless, the difference in differentiation time for S. florulenta compared to those of our study species agrees with the observation that, although recurrent patterns exist, some species may exhibit distinct patterns reflecting the unique, rather than the shared, aspects of species’ histories [71, 72].
In general, our results for S. cordifolia and V. argenteria point out the importance of not just the LGM but also previous glacial cycles in shaping the contemporary genetic structure of plant species in the Maritime and Ligurian Alps. The results thus corroborate the notes inferring the necessity to go beyond interpretations that relate only to the role of a small number of major historical events, such as the LGM and the Messinian Salinity Crisis [7].
The distinctiveness of current populations
In general, current patterns of genetic variation are explained by contraction/expansion model in association with climatic oscillations. In this model refuge populations are associated with multiple genetic lineages and higher genetic diversity compared to more recently established populations [3]. Due to the recurrent founder effect affecting colonizing populations, the genetic diversity is supposed to decrease by increasing the distance from refugia, producing a genetic signature [73]. Nevertheless, this pattern can be obscured by secondary contact of vicariant lineages which results in admixing populations with high heterozygosity and genetic diversity [74].
The total gene diversity and haplotype diversity (Table 2) detected in S. cordifolia (HT = 0.847; Hd = 0.824) and V. argenteria (HT = 0.885; Hd = 0.834) are similar and the total gene diversity is higher than the mean value (HT = 0.670; [64]) detected in 170 of the plant species studied by Petit et al. [74]. Comparably, high total gene diversity and haplotype diversity have been detected in several other endemic species (e.g., Phyllodoce nipponica Makino: HT = 0.852, [75]; Rhodiola dumulosa (Franchet) S.H. Fu: HT = 0.981, [76]; 2010; Arenaria provincialis Chater & Halliday: Hd = 0.94, [77]). Although both species show relatively high genetic differentiation among populations, the within-population values of haplotype and nucleotide diversity are lower and the number of private haplotypes higher in V. argenteria than in S. cordifolia (Table 3), resulting in a higher level of genetic differentiation between populations and in a more marked phylogeographical structure (S. cordifolia: NST = 0.749; GST = 0.565; V. argenteria: NST = 0.921; GST = 0.760, see also Table 4). Low within population diversity we observed suggests bottleneck or founder effects during glacial population contraction and post-glacial expansions, resulting in few haplotype being fixed in most populations. This result, together with the weak phylogeographical structure, also suggests that a repeated pattern of contraction/expansion, throughout the glacial cycles, may have shaped the contemporary genetic structure of the studied species. This result is in line with our SDM and molecular dating findings. The level of genetic differentiation detected among populations of both species even indicates low levels of recurrent gene flow among populations, as also confirmed by IBD results. The high haplotype and gene diversity, taken together with the patchy distribution and the numerous private haplotype, may be explained by a long history of differentiation and in situ glacial survival. Two factors may contribute to the higher cpDNA-based population subdivision in V. argenteria compared to S. cordifolia. First, vicariance events due to glaciation cycles may have had a stronger impact on V. argenteria, growing at higher altitude. Second, a more efficient seed dispersal mechanism in S. cordifolia may contribute to the lack of correspondence between population groups and haplotype clades in this species. This last explanation seems unlikely because V. argenteria may be dispersed by bird (see discussion below). However, the IBD result suggests a similar immigration by seed in Maritime and Ligurian Alps populations of both species (S. cordifolia: r = 0.417, P < 0.0001; V. argenteria: r = 0.586, P < 0.0001). In addition, the high proportion of variation residing among the population groups identified by SAMOVA indicates that past climatic oscillations would have stimulated a stronger intraspecific diversification and population genetic differentiation in V. argenteria. According to this result, the higher within-population diversity detected in S. cordifolia may be due to its larger altitudinal range. This may have facilitated short-distance altitudinal migrations likely decreasing the impact of founder effects and the genetic signature of glaciations. A similar pattern of short-distance altitudinal shift and recurrent re-colonizations of neighbouring populations was reported in Armeria genus growing at high altitude [78]. This is congruent with the higher haplotype diversity observed in central populations for S. cordifolia and in southern populations for V. argenteria, and with the stronger effect of glaciation on the latest. V. argenteria growing only in the alpine belt probably survived glaciation in a southern peripheral refugium while S. cordifolia may have survived to last glaciation in several refugia at lower altitudes, showing so its higher diversity in the contact zone.
The recent origin of haplotypes detected in Corsica suggests that the disjoint Corsican populations of V. argenteria may have originated by a rare long-distance dispersal of seed by birds rather than by vicariance due to Corsica-Sardinia microplate disjunction (15 Ma). In fact, seeds of violets have been shown to be capable of bird-mediated endozoochorous dispersal [79]. Moreover, Corsica is located along one of the main present-day migration routes of birds from European continental regions to the African areas [80]. This random dispersion is congruent with the weak IBD detected among all populations of V. argenteria (r = 0.296, P < 0.0001).
The complex phylogeographical pattern observed in the studied species would indicates that, even if some species-specific differences are detected, both population and genotype extinction was minimal during the LGM, probably due to the low impact of glaciations and to topographic complexity in this area [81]. This result is in line with evidences from SDM and divergence time among haplotypes. In the south western Alps, survival in peripheral refuges and nunataks during the LGM has recently been demonstrated in Primula spp. using both SDM and phylogeographical approaches [12]; however, our results are congruent with previous findings in the Maritime and Ligurian Alps where glaciations seem to have had a low influence on plant distribution and their effect seems to be weakened by high level of postglacial migrations [81]. These findings are more similar to that expected for a Mediterranean mountain region [7, 8] rather than for an Alpine region [16, 58], where patterns were simplified to a great extent by major losses of diversity during glacial periods.
Supporting Information
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Acknowledgments
We are grateful to the CBNMed for allowing us to use their occurrence points. We also thank John Thompson for insightful discussion and English revision and Nils Moosdorf for the global lithological map dataset.
Data Availability
DNA sequences are available at GenBank data base. Accession numbers are reported in S3 Table. Climatic data are available from WorldClim website (http://www.worldclim.org). Data from a third party are available upon request directly to the proprietary. In particular, occurrence data from the SILENE and FLORE databases are available upon request from Conservatoire Botanique National Méditerranéen de Porquerolles (http://www.cbnmed.fr/pres/index.php). The global lithological map dataset (GLiM) is available upon request via email from Dr. Nils Moosdorf (Institute for Biogeochemistry and Marine Chemistry, Universität Hamburg, Germany; http://www.zmt-bremen.de).
Funding Statement
Interreg Alcotra programme 2007-2013, project n. 192 BIODIVAM.
References
- 1.Davis MB, Shaw RG. Range shifts and adaptive responses to quaternary climate change. Science. 2001; 292: 673–679. 10.1126/science.292.5517.673 [DOI] [PubMed] [Google Scholar]
- 2.Hewitt GM. Post-glacial re-colonization of European biota. Biol J Linn Soc. 1999; 68: 87–112. [Google Scholar]
- 3.Hewitt GM. The genetic legacy of quaternary ice ages. Nature. 2000; 405: 907–913. 10.1038/35016000 [DOI] [PubMed] [Google Scholar]
- 4.Hewitt GM. Genetic consequences of climatic oscillations in the Quaternary. Philos T R Soc B. 2004; 359: 183–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tribsch A. Areas of endemism of vascular plants in the Eastern Alps in relation to Pleistocene glaciation. J Biogeogr. 2004; 31: 747–760. [Google Scholar]
- 6.Taberlet P, Fumagalli L, Wust-Saucy AG, Cosson J-F. Comparative phylogeography and postglacial colonization routes in Europe. Mol Ecol. 1998; 7: 453–464. [DOI] [PubMed] [Google Scholar]
- 7.Médail F, Diadema K. Glacial refugia influence plant diversity patterns in the Mediterranean Basin. J Biogeogr. 2009; 36: 1333–1345. [Google Scholar]
- 8.Nieto Feliner G. Southern European glacial refugia: a tale of tales. Taxon. 2011; 60: 365–372. [Google Scholar]
- 9.Parmesan C. Ecological and evolutionary responses to recent climate change. Annu Rev Ecol Evol S. 2006; 37: 637–669. [Google Scholar]
- 10.Pauls SU, Nowak C, Bálint M, Pfenninger M. The impact of global climate change on genetic diversity within populations and species. Mol Ecol. 2013; 22: 925–946. 10.1111/mec.12152 [DOI] [PubMed] [Google Scholar]
- 11.Carstens BC, Richards CL. Integrating coalescent and ecological niche modeling in comparative phylogeography. Evolution. 2007; 61: 1439–1454. 10.1111/j.1558-5646.2007.00117.x [DOI] [PubMed] [Google Scholar]
- 12.Schorr G, Holstein N, Pearman PB, Guisan A, Kadereit JW. Integrating species distribution models (SDMs) and phylogeography for two species of Alpine Primula. Ecol Evol. 2012; 2: 1260–1277. 10.1002/ece3.100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Casazza G, Grassi F, Zecca G, Mariotti MG, Guerrina M, Minuto L. Phylogeography of Primula allionii (Primulaceae), a narrow endemic of the Maritime Alps. Bot J Linn Soc. 2013; 173: 637–653. [Google Scholar]
- 14.Médail F, Verlaque R. Ecological characteristics and rarity of endemic plants from southeast France and Corsica: implications for biodiversity conservation. Biol Conserv. 1997; 80: 269–281. [Google Scholar]
- 15.Casazza G, Barberis G, Minuto L. Ecological characteristics and rarity of endemic plants of the Italian Maritime Alps. Biol Conserv. 2005; 123: 361–371. [Google Scholar]
- 16.Schönswetter P, Stehlik I, Holderegger R, Tribsch A. Molecular evidence for glacial refugia of mountain plants in the European Alps. Mol Ecol. 2005: 14: 3547–3555. 10.1111/j.1365-294X.2005.02683.x [DOI] [PubMed] [Google Scholar]
- 17.Diadema K, Bretagnolle F, Affre L, Yuan YM, Médail F. Geographic structure of molecular variation of Gentiana ligustica (Gentianaceae) in the Maritime and Ligurian regional hotspot, inferred from ITS sequences. Taxon. 2005; 54: 887–894. [Google Scholar]
- 18.Minuto L, Grassi F, Casazza G. Ecogeographic and genetic evaluation of endemic species in the Maritime Alps: the case of Moehringia lebrunii and M. sedoides (Caryophyllaceae). Pl Biosyst. 2006; 140: 146–155. [Google Scholar]
- 19.Grassi F, Labra M, Minuto L, Casazza G, Sala F. Haplotype richness in refugial area: phylogeographical structure of Saxifraga callosa. J Plant Res. 2009; 122: 377–387. 10.1007/s10265-009-0230-z [DOI] [PubMed] [Google Scholar]
- 20.Szövényi P, Arroyo K, Guggisberg A, Conti E. Effects of Pleistocene glaciations on the genetic structure of Saxifraga florulenta (Saxifragaceae), a rare endemic of the Maritime Alps. Taxon. 2009; 58: 532–543. [Google Scholar]
- 21.Sloan DB, Oxelman B, Rautenberg A, Taylor DR. Phylogenetic analysis of mitochondrial substitution rate variation in the angiosperm tribe Sileneae. BMC Evol Biol. 2009; 9: 260–275. 10.1186/1471-2148-9-260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thompson JD. Plant evolution in the Mediterranean Oxford, UK: Oxford University Press; 2005. [Google Scholar]
- 23.Yockteng R, Ballard HE Jr, Mansion G, Dajoz I, Nadot S. Relationships among pansies (Viola section Melanium) investigated using ITS and ISSR markers. Plant Syst Evol. 2003; 241: 153–170. [Google Scholar]
- 24.Lomba A, Pellissier L, Randin C, Vicente J, Moreira F, Honrado J, Guisan A. Overcoming the rare species modelling paradox: a novel hierarchical framework applied to an Iberian endemic plant. Biol Conserv. 2010; 143: 2647–2657. [Google Scholar]
- 25.Hartmann J, Moosdorf N. The new global lithological map database (GLiM): A representation of rock properties at the Earth surface. Geochem Geophy Geosy. 2012; 12: Q12004. [Google Scholar]
- 26.Ramirez-Villegas J, Jarvis A. Downscaling Global Circulation Model Outputs: The Delta Method Decision and Policy Analysis Working Paper No.1. Cali, Colombia: International Center for Tropical Agriculture, CIAT; 2010.
- 27.R Development Core Team. R: A language and environment for statistical computing R Foundation for Statistical Computing; Vienna, Austria; 2005. [Google Scholar]
- 28.Körner C. Alpine plant life 1st edn. Berlin, Germany: Springer; 1999. [Google Scholar]
- 29.Warren DL, Glor RE, Turelli M. ENMtools: a toolbox for comparative studies of environmental niche models. Ecography. 2010; 33: 607–611. [Google Scholar]
- 30.Friedman J. Multivariate adaptive regression splines. Ann. Stat. 1991; 19: 1–141. [Google Scholar]
- 31.McCullagh P, Nelder JA. Generalized Linear Models (2nd edn). London, UK: Chapman & Hall; 1989. [Google Scholar]
- 32.Breiman L, Friedman JH, Olshen RA, Stone CJ. Classification and regression trees New York, USA: Chapman and Hall; 1984. [Google Scholar]
- 33.Hastie T, Tibshirani R, Buja A. Flexible discriminant analysis by optimal scoring. J. Am. Stat. Assoc. 1994; 89: 1255–1270. [Google Scholar]
- 34.Breiman L. Statistical modeling: the two cultures. Stat. Sci. 2001; 16: 199–215. [Google Scholar]
- 35.Phillips SJ, Anderson RP, Schapire RE. Maximum entropy modeling of species geographic distributions. Ecological Modelling 2006; 190: 231–259. [Google Scholar]
- 36.Thuiller W, Lafourcade B, Engler R, Araujo MB. BIOMOD–a platform for ensemble forecasting of species distributions. Ecography. 2009; 32: 369–373. [Google Scholar]
- 37.Barbet-Massin M, Jiguet F, Albert CH, Thuiller W. Selecting pseudo-absences for species distribution models: how, where and how many? Methods Ecology Evol. 2012; 3: 327–338. [Google Scholar]
- 38.Allouche O, Tsoar A, Kadmon R. Assessing the accuracy of species distribution models: prevalence, kappa and the true skill statistic (TSS). Journal of Applied Ecology 2006; 43: 1223–1232. [Google Scholar]
- 39.Katoh K, Toh H. Recent developments in the MAFFT multiple sequence alignment program. Brief Bioinform. 2008; 9: 286–298. 10.1093/bib/bbn013 [DOI] [PubMed] [Google Scholar]
- 40.Saltonstall K, Lambertini C. The value of repetitive sequences in chloroplast DNA for phylogeographic inference: a comment on Vachon and Freeland 2011. Mol Ecol Resour. 2012; 12: 581–585. 10.1111/j.1755-0998.2012.03146.x [DOI] [PubMed] [Google Scholar]
- 41.Nei M, Li WH. Mathematical model for studying genetic variation in terms of restriction endonucleases. P Natl Acad Sci USA. 1979; 76: 5269–5273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rozas J, Sánchez-Del Barrio JC, Messeguer X, Rozas R. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics. 2003; 19: 2496–2497. [DOI] [PubMed] [Google Scholar]
- 43.Pons O, Petit RJ. Measuring and testing genetic differentiation with ordered versus unordered alleles. Genetics. 1996; 144: 1237–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Excoffier L, Laval G, Schneider S. Arlequin version 3.01: an integrated software package for population genetics data analysis. Evol Bioinform. 2005; 1: 47–60. [PMC free article] [PubMed] [Google Scholar]
- 45.Mantel NA. The detection of disease clustering and a generalized regression approach. Cancer Res. 1967; 27: 209–220. [PubMed] [Google Scholar]
- 46.Muñoz-Pajares AJ. SIDIER: substitution and indel distances to infer evolutionary relationships. Methods Ecology Evol. 2013; 4: 1195–1200. [Google Scholar]
- 47.Simmons MP, Müller K, Norton AP. The relative performance of indel-coding methods in simulations. Mol Phylogenet Evol. 2007; 44: 724–740. 10.1016/j.ympev.2007.04.001 [DOI] [PubMed] [Google Scholar]
- 48.Muller K. Incorporating information from length-mutational events into phylogenetic analysis. Methods Ecology Evol. 2006; 38: 667–676. [DOI] [PubMed] [Google Scholar]
- 49.Dupanloup I, Schneider S, Excoffier L. A simulated annealing approach to define the genetic structure of populations. Mol Ecol. 2002; 11: 2571–2581. [DOI] [PubMed] [Google Scholar]
- 50.Darriba D, Taboada GL, Doallo R, Posada D. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 2012; 9: 772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Drummond AJ, Rambaut A. Beast: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007; 7: 214 10.1186/1471-2148-7-214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rambaut A, Drummond AJ. Tracer v1.5; 2007 [cited 2015 Aug 1]. Available at: http://beast.bio.ed.ac.uk/Tracer.
- 53.Wolfe KH, Li W-H, Sharp PM. Rates of nucleotide substitution vary greatly among plant mitochondrial, chloroplast, and nuclear DNAs. P Natl Acad Sci USA. 1987; 84: 9054–9058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Graur D, Li WH. Fundamentals of molecular evolution 2nd edition Sunderland, Massachussets: Sinauer Associates Inc.; 2000. [Google Scholar]
- 55.Wang LY, Ikeda H, Liu TL, Wang YJ, Liu JQ. Repeated range expansion and glacial endurance of Potentilla glabra (Rosaceae) in the Qinghai-Tibetan Plateau. J Integr Plant Biol. 2009; 51: 698–706. 10.1111/j.1744-7909.2009.00818.x [DOI] [PubMed] [Google Scholar]
- 56.Araújo MB, Pearson RG, Thuiller W, Erhard M. Validation of species-climate impact models under climate change. Global Change Biol. 2005; 11: 1504–1513. [Google Scholar]
- 57.Holderegger R. Thiel-Egenter C. A discussion of different types of glacial refugia used in mountain biogeography and phylogeography. J Biogeogr. 2009; 36: 476–480. [Google Scholar]
- 58.Tribsch A, Schönswetter P. Patterns of endemism and comparative phylogeography confirm palaeoenvironmental evidence for Pleistocene refugia in the Eastern Alps. Taxon. 2003; 52: 477–497. [Google Scholar]
- 59.Dullinger S, Willner W, Plutzar C, Englisch T, Schratt-Ehrendorfer L, Moser D, et al. Post-glacial migration lag restricts range filling of plants in the European Alps. Global Ecol Biogeogr. 2012; 21: 829–840. [Google Scholar]
- 60.Ehlers J, Gibbard PL. Quaternary glaciations–extent and chronology part I: Europe. Developments in Quaternary Science. Amsterdam, The Netherlands: Elsevier; 2004. Vol. 2a. [Google Scholar]
- 61.Federici PR, Granger DE, Ribolini A, Spagnolo M, Pappalardo M, Cyr AJ. Last Glacial Maximum and the Gschnitz stadial in the Maritime Alps according to 10Be cosmogenic dating. Borea. 2011; 41: 277–291. [Google Scholar]
- 62.Patsiou TS, Conti E, Zimmermann NE, Theodoridis S, Randin CF. Topo-climatic microrefugia explain the persistence of a rare endemic plant in the Alps during the last 21 Millennia. Global Change Biol. 2014; 20: 2286–2300. [DOI] [PubMed] [Google Scholar]
- 63.Fernández-Mazuecos M, Vargas P. Congruence between distribution modelling and phylogeographical analyses reveals Quaternary survival of a toadflax species (Linaria elegans) in oceanic climate areas of a mountain ring range. New Phytol. 2013; 198: 1274–1289. 10.1111/nph.12220 [DOI] [PubMed] [Google Scholar]
- 64.Sun Y, Moore MJ, Yue L, Feng T, Chu H, Chen S, et al. Chloroplast phylogeography of the East Asian Arcto-Tertiary relict Tetracentron sinense (Trochodendraceae). J Biogeogr. 2014; 41: 1721–1732. [Google Scholar]
- 65.Hughes PD, Woodward JC, van Calsteren PC, Thomas LE. The glacial history of the Dinaric Alps, Montenegro. Quaternary Sci Rev. 2011; 30: 3393–3412. [Google Scholar]
- 66.Hughes PD, Braithwaite RJ. Application of a degree-day model to reconstruct Pleistocene glacial climates. Quaternary Res. 2008; 69: 11–116. [Google Scholar]
- 67.Combourieu-Nebout N, Bertini A, Russo-Ermolli E, Peyrone O, Klotz S, Montade V, et al. Climate changes in the central Mediterranean and Italian vegetation dynamics since the Pliocene. Rev Palaeobot Palyno. 2015; 218: 127–147. [Google Scholar]
- 68.Dehnert A, Preusser F, Kramers JD, Akçar N, Kubik PW, Reber R, Schlüchter C. A multi-dating approach applied to proglacial sediments attributed to the most extensive glaciation of the Swiss Alps. Boreas. 2010; 39: 620–632. [Google Scholar]
- 69.Pfenninger M, Posada D. Phylogeographic history of the land snail Candidula unifasciata (Helicellinae, Stylommatophora): fragmentation, corridor migration, and secondary contact. Evolution. 2002; 56: 1776–1788. [DOI] [PubMed] [Google Scholar]
- 70.Borer M, Alvarez N, Buerki S, Margraf N, Rahier M, Naisbit RE. The phylogeography of an alpine leaf beetle: divergence within Oreina elongata spans several ice ages. Mol Phylogenet Evol. 2010; 57: 703–709. 10.1016/j.ympev.2010.08.017 [DOI] [PubMed] [Google Scholar]
- 71.Soltis DE, Morris AB, McLachlan JS, Manos PS, Soltis PS. Comparative phylogeography of unglaciated eastern North America. Mol Ecol. 2006; 15: 4261–4293. 10.1111/j.1365-294X.2006.03061.x [DOI] [PubMed] [Google Scholar]
- 72.Poelchau MF, Hamrick JL. Comparative phylogeography of three common Neotropical tree species. J Biogeogr. 2013; 40: 618–631. [Google Scholar]
- 73.Hewitt GM, Ibrahim KM. Inferring glacial refugia and historical migrations with molecular phylogenies In: Silvertown J, Antonovics J, eds. Integrating ecology and evolution in a spatial context. BES symposium volume. Oxford, UK: Blackwells; 2001. 271–294 p. [Google Scholar]
- 74.Petit RJ, Duminil J, Fineschi S, Hampe A, Salvini D, Vendramin GG. Comparative organization of chloroplast, mitochondrial and nuclear diversity in plant populations. Mol Ecol. 2005; 14: 689–701. 10.1111/j.1365-294X.2004.02410.x [DOI] [PubMed] [Google Scholar]
- 75.Ikeda H, Setoguchi H. Phylogeography and refugia of the Japanese endemic alpine plant, Phyllodoce nipponica Makino (Ericaceae). J Biogeogr. 2007; 34: 169–176. [Google Scholar]
- 76.Hou Y, Lou A. Phylogeographical patterns of an alpine plant, Rhodiola dumulosa (Crassulaceae), inferred from chloroplast DNA sequences. J Hered. 2014; 105: 101–110. 10.1093/jhered/est072 [DOI] [PubMed] [Google Scholar]
- 77.Pouget M, Youssef S, Migliore J, Juin M, Médail F, Baumel A. Phylogeography sheds light on the central–marginal hypothesis in a Mediterranean narrow endemic plant. Ann Bot-London. 2013; 112: 1409–1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gutiérrez Larena B, Fuertes Aguilar J, Nieto Feliner G. Glacial-induced altitudinal migrations in Armeria (Plumbaginaceae) inferred from patterns of chloroplast DNA haplotype sharing. Mol Ecol. 2002; 11: 1965–1974. [DOI] [PubMed] [Google Scholar]
- 79.Ballard HE, Sytsma KJ. Evolution and biogeography of the woody Hawaiian violets (Viola, Violaceae): Arctic origins, herbaceous ancestry and bird dispersal. Evolution. 2000; 54: 1521–1532. [DOI] [PubMed] [Google Scholar]
- 80.Spina F, Volponi S. Atlante delle Migrazioni degli Uccelli in Italia Ministero dell’Ambiente e della Tutela del Territorio e del Mare. Roma, Italy: Tipografia SCR; 2008. [Google Scholar]
- 81.Casazza G, Zappa E, Mariotti MG, Medáil F, Minuto L. Ecological and historical factors affecting distribution pattern and richness of endemic plant species: the case of Maritime and Ligurian Alps hotspot. Divers Distrib. 2008; 14: 47–58. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Data Availability Statement
DNA sequences are available at GenBank data base. Accession numbers are reported in S3 Table. Climatic data are available from WorldClim website (http://www.worldclim.org). Data from a third party are available upon request directly to the proprietary. In particular, occurrence data from the SILENE and FLORE databases are available upon request from Conservatoire Botanique National Méditerranéen de Porquerolles (http://www.cbnmed.fr/pres/index.php). The global lithological map dataset (GLiM) is available upon request via email from Dr. Nils Moosdorf (Institute for Biogeochemistry and Marine Chemistry, Universität Hamburg, Germany; http://www.zmt-bremen.de).