Abstract
Background:
Soft-tissue sarcomas (STS) are a diverse group of malignancies that remain a diagnostic and therapeutic challenge. Relatively few reliable cell lines currently exist. Rapidly developing technology for genomic profiling with emerging insights into candidate functional (driver) aberrations raises the need for more models for in vitro functional validation of molecular targets.
Methods:
Primary cell culture was performed on STS tumours utilising a differential attachment approach. Cell lines were characterised by morphology, immunocytochemistry, proliferation assays, short tandem repeat (STR) and microarray-based genomic copy number profiling.
Results:
Of 47 STS cases of various subtypes, half formed adherent monolayers. Seven formed self-immortalised cell lines, including three undifferentiated pleomorphic sarcomas, two dedifferentiated liposarcomas (one of which had received radiotherapy), a leiomyosarcoma and a myxofibrosarcoma. Two morphologically distinct yet genetically identical variants were established in separate cultures for the latter two tumours. All cell lines demonstrated genomic and phenotypic features that not only confirm their malignant characteristics but also confirm retention of DNA copy number aberrations present in their parent tumours that likely include drivers.
Conclusions:
These primary cell lines are much-needed additions to the number of reliable cell lines of STS with complex genomics available for initial functional validation of candidate molecular targets.
Keywords: sarcoma, primary cell line, in vitro models, soft-tissue sarcoma, array CGH, copy number profiles, short tandem repeat
Soft-tissue sarcomas (STS) are a diverse group of malignant tumours that arise in mesenchymal tissues and represent ∼1% of adult human malignancies. Comprising over 50 clinicobiologic/molecular subtypes, the majority of STS subtypes remain a significant diagnostic and treatment challenge (Fletcher et al, 2013; Italiano et al, 2016). Only a small proportion (∼20%) have known specific diagnostic markers such as gene mutations or chromosomal translocations and fewer still possess identified molecular therapeutic targets (Taylor et al, 2011; Fletcher et al, 2013). The remainder are characterised by pervasive chromosomal instability evidenced by multiple seemingly random somatic DNA copy number aberrations. Elucidation of as yet unknown patterns among these complex genomic abnormalities is believed to be the key to accurate diagnosis and identification of molecular therapeutic targets in these heterogenic tumours (Barretina et al, 2010; Taylor et al, 2011).
Traditionally, the first step of therapeutic target validation for candidate driver genes or proteins utilises in vitro disease models (Taylor et al, 2011). Recent improvements in the technology for genomic aberration mapping such as high-resolution microarray-based comparative genomic hybridisation (array CGH) and next-generation sequencing have led to an increased rate of candidate identification and therefore a growing need to establish a wider range of STS cell lines for functional testing (Barretina et al, 2010; Taylor et al, 2011). In STS, however, there is a limited number of in vitro disease models (tumour cell lines) available for functional testing and target validation. Data from large-scale cancer cell line studies such as the Cancer Cell Line Encyclopaedia and Sanger Cancer Cell Line Project showed that <2% of the commercially available cell lines studied are derived from STS and the majority of these belong to the translocation-driven subgroup (Forbes et al, 2011; Barretina et al, 2012).
There has been increasing recognition of the limitations of commercial cell lines as a disease model stemming from reports of poor correlation of the response in these cell lines with in vivo tumour behaviour (Cree et al, 2010; Kamb, 2010). With cellular adaptation to artificial culture conditions, cell lines have been shown to grow more rapidly than parent tumour cells and acquire phenotypic changes that may alter their therapeutic response, such as dependence on growth factors in culture media or adherence to plastic (Kato et al, 2008; Pan et al, 2009). Furthermore, heterogeneity of tumour cell clones, which is characteristic of many cancers is lacking in cell lines in which single clones have been selected for and is widely believed to account for the poor correlation of preclinical and clinical data as the cell lines may not reflect resistant tumour cell clones or cancer ‘stem' cells that are believed to be responsible for tumour recurrence and late therapeutic failure (Kamb, 2010).
One widely accepted alternative to traditionally established (or commercially available) cell lines is the use of cells cultured directly from tumours (primary cell cultures) as in vitro disease models. This is however fraught with many problems (Luca et al, 2007). Fresh tumour tissue has to be donated by patients and obtained during surgery with the associated ethical and logistic constraints. When available, the behaviour of primary tumour cells in culture is generally unpredictable with a variable rate of successful establishment (Luca et al, 2007).
This paper describes our experience of primary tissue culture of STS cells and the establishment and molecular characterisation of self-immortalised primary cell lines, including morphologic variants derived from seven soft-tissue sarcomas.
Materials and methods
Ethics statement
National Research Ethics Committee approval was obtained for the collection and use of tumour tissue (Reference number 09/H1313/52). Written informed consent was obtained from all patients before the tumour tissue collection, and all tissues were stored according to the principles of the Declaration of Helsinki and were used in compliance with the Human Tissue Act 2004.
Tumour samples
Fresh tumour samples were obtained from 47 patients receiving surgical treatment for biopsy-confirmed STS at Sheffield Teaching Hospitals. Preoperative sarcoma diagnoses were confirmed by pathological assessment of the resected tumours, which were classified according to WHO diagnostic categories (Fletcher et al, 2013) and reported according to Royal College of Pathologists guidelines (Fisher, 2014). Within 30 min of resection, tumours were macroscopically sampled by specialised sarcoma pathologists and collected in sterile phosphate-buffered saline with additional tumour samples snap frozen and stored in liquid nitrogen or at −80 °C until DNA extraction. Normal tissue, where available typically as part of a wide resection specimen, was obtained from sites macroscopically distant from the tumour and snap frozen in liquid nitrogen.
Establishment of primary tumour cell cultures
Tumour cell cultures were set up under sterile conditions within 1 h of surgical resection by simple mechanical tissue dissociation using RPMI-1640 culture medium supplemented with penicillin (100 U ml−1), streptomycin (100 μg ml−1), amphotericin B (5 μg ml−1), foetal calf serum (20% v v−1) and D-glucose (0.4% v v−1). Briefly, a small piece (∼10 × 10 × 5 mm3) of fresh tumour was placed in a sterile Petri dish with a few drops prewarmed (37 °C) culture media and minced with a sterile scalpel until very fine. Minced tumour was then suspended in warm media and centrifuged before transferring from fresh warm media to sterile 25 mm2 tissue culture (T25) flasks and 5 mm2 flat-sided tubes (slopes) and placed in a 5% CO2 incubator at 37 °C in 95% humidified air. All cultures were inspected daily using a phase-contrast microscope and media were changed as required.
A modification of the differential attachment approach described by Nayak et al (2000) was used and washes were set up when it appeared that some viable cells in early cultures remained unattached to culture flasks. The media containing non-adherent cells were collected and replaced with fresh media, then centrifuged and the pellet resuspended in fresh prewarmed media and transferred into a new T25 flask for incubation. Adherent cell cultures were maintained by serial passage with gentle trypsinisation at confluence. In case they were undergoing a crisis period, cultures in which the majority of cells appeared senescent were maintained for at least a further 3 months with daily visual inspection and the media were changed as required.
Cultures were given STS laboratory designations based on the chronological order and year in which they were established and passage numbers were indicated using a ‘p' prefix. For example, STS 03/10 p2 refers to the second passage of cultures derived from the third tumour obtained in 2010. Cultures that were set up as washes were designated with a ‘w' prefix to the passage number. For example, cells from the second wash of an original T25 flask culture set-up and currently in their third passage were designated w2p3, whereas cells from a wash of an original slope set up at the same passage were designated wsp3. Cells from original culture set-up and washes were maintained separately with all relevant precautions to prevent cross-contamination. To make them more recognisable, the nomenclature of cultures that formed stable cell lines was revised to reflect the STS subtype they represent and their city of origin (see below).
Proliferation assay and doubling time
Cells were seeded at a density of 2 × 103 in individual wells of a 96-well plate and the MTT ((4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) proliferation assay was performed as described previously (Canovas et al, 2008). The ratio of absorbance in the test wells to control wells (containing 100 μl DMSO only) was calculated as relative MTT activity and used as a surrogate for the number of viable cells remaining in culture. Average relative MTT activity from four replicate wells was plotted against time and the exponential growth curve plotted and doubling time calculated using GraphPad Prism software v.6.0 (GraphPad software, LaJolla, CA, USA).
Short tandem repeat profiling
Short tandem repeat (STR) profiling to establish unique genomic identities and exclude cross-contamination of cell lines was performed using services provided by the University of Sheffield Core Genomic Facility. Alleles for 10 human loci including THO1, D21S11, D5S818, D13S317, D7S820, D16S539, CSFIPO, AMEL, vWA and TPOX were assessed and compared with large cell line STR profile databases using the standard match threshold of 80% (Capes-Davis et al, 2013).
Array CGH
Genomic DNA extraction and array CGH were performed using the Agilent 180K platform (Agilent, Cheshire, UK) as described previously (Salawu et al, 2012). Control DNA was extracted from the normal tissue obtained from the same patient where available. Otherwise, pooled sex-matched genomic DNA (Promega, Southampton, UK) was used. Agilent Genomic Workbench Software v.6.0 (Agilent Technologies, Santa Clara, CA, USA) was used for copy number data analysis and graphical representation on ideograms.
Immunocytochemistry
Cells were prepared for immunocytochemistry by culturing on sterile glass slides for 48 h before fixing with ice-cold acetone–methanol (1 : 1 mixture). Fixed cells were then pre-treated with Triton (0.1% v v−1) and H2O2 (3%) before immunostaining. Primary antibodies for cytokeratin (CK), vimentin, smooth muscle actin (SMA) and CD117 (c-KIT) were used (see Supplementary Table S1 for details). The Vectastain system appropriate for each primary antibody was then used according to the manufacturer's instructions for secondary antibody staining and detection. Confirmation of staining pattern was performed by light microscopy at appropriate magnification.
Results
Establishment of primary tumour cell cultures in various STS subtypes
Forty-seven STS cases comprising 16 different subtypes were set up in culture (see Table 1 for details). Failure to establish adherent cultures or early senescence within the first or second passage (p1–p2) was observed in 10 cases. Later senescence occurred in about a third of cases and was generally observed at two points. The first was around the fifth passage (p4–p7) and the second was after ∼10 passages (p9–p12). All cultures that continued beyond 12 passages went on to become stable self-immortalised cell lines that maintained reliably proliferative cultures for over 3 years with consistent and reliable proliferation rates. In all but one of these cases (a leiomyosarcoma) that formed stable cell lines, the cells adapted to proliferative tissue culture quite rapidly (within days) and no significant crisis period was seen (discussed later). Of note, long-term cultures leading to stable cell lines were derived not only from the original culture set-ups but also from washes in three out of seven tumours, and in two of these cases, separate washes led to the establishment of morphologically variant cultures of the same tumours, lending support to the differential attachment approach that was used.
Table 1. A summary of fresh soft-tissue sarcoma subtypes obtained and their primary cell culture outcomes.
| STS subtype | Obtained | Not established in culture | Early senescence | Senescent around p5 | Senescent around p10 | Long term culture |
|---|---|---|---|---|---|---|
|
Number of cases | ||||||
| Alveolar soft part sarcoma | 2 | – | – | 1 | 1 | – |
| Angiosarcoma | 6 | – | 3a | 2 | 1 | – |
| Dedifferentiated liposarcoma | 7 | 3 | 2 | – | – | 2b |
| Ewing's sarcoma | 1 | – | – | – | 1b | – |
| Extraskeletal myxoid chondrosarcoma | 1 | – | – | 1 | – | – |
| Leiomyosarcoma | 3 | 1 | – | – | 1 | 1 |
| Low-grade myofibroblastic sarcoma | 1 | – | – | 1 | – | – |
| Malignant peripheral nerve sheath tumour | 1 | – | – | 1 | – | – |
| Malignant solitary fibrous tumour | 1 | – | – | 1 | – | – |
| Myxofibrosarcoma | 5 | – | 3 | – | 1 | 1 |
| Myxoid liposarcoma | 1 | 1 | – | – | – | – |
| Pleomorphic liposarcoma | 2 | 1b | – | 1a | – | – |
| Pleomorphic rhabdomyosarcoma | 1 | – | – | – | 1b | – |
| Synovial sarcoma | 1 | – | – | 1 | – | – |
| Undifferentiated pleomorphic sarcoma | 7 | 2c | 1 | – | 1 | 3 |
| Well-differentiated liposarcoma | 7 | 6d | 1 | – | – | – |
| Total | 47 | 14 | 10 | 9 | 7 | 7 |
Abbreviation: STS=soft-tissue sarcoma.
One case was a metastatic tumour.
One case received neoadjuvant radiotherapy or chemotherapy.
Both cases received neoadjuvant radiotherapy or chemotherapy.
Three cases were recurrent tumours.
Neoadjuvant local or systemic treatment is expected to adversely impact tissue culture outcomes. It was interesting to note, however, that both STS cases in which the patient had received neoadjuvant chemotherapy were able to establish cells that were passaged up to 10 times before subsequent senescence and one out of four tumours that received neoadjuvant radiotherapy resulted in a stable cell line (Table 1). Undifferentiated pleomorphic sarcoma (UPS) was the STS subtype that had the most successful outcome with stable cell lines established in three out of seven cases. Of the UPS cultures that did not result in a stable cell line, two out of the four had received neoadjuvant treatment. Well-differentiated liposarcoma, on the other hand, was the STS subtype that had the poorest culture outcome overall with only one of the seven cases able to establish any cells in adherent culture and these subsequently underwent early senescence (Table 1).
To minimise the effects of genomic drift and cultural adaptation in future experiments, cultured cells were frozen at −80 °C and subsequently in liquid nitrogen every 3–4 passages, within the first 15 passages. Table 2 summarises the nomenclature, clinical and cultural phenotypic characteristics of the seven tumours that established long-term primary cell cultures and which are the focus of the rest of this manuscript.
Table 2. Characteristics of primary soft tissue sarcoma cell lines and their parent tumours.
| Cell line | Laboratory designation | Morphology | Doubling timea (h) | Age (years)/gender | STS subtype | Site | Size | TNM stage | Immunocytochemistry |
|---|---|---|---|---|---|---|---|---|---|
| Shef-UPS 01 | STS 14/10 | Spindle-shaped cells with distinct nuclei No pleomorphism No distinct colony formation | p70=40.55 | 53/F | Undifferentiated pleomorphic sarcoma | Lower limb | 230 mm | pT2b Stage III | Vimentin – positive CK, SMA, c-Kit – negative |
| Shef-UPS 02 | STS 06/11 | Pleomorphic mostly spindle-shaped cells No distinct colony formation | p91=35.38 | 76/M | Undifferentiated pleomorphic sarcoma | Lower limb | 170 mm | pT2b Stage III | Vimentin – positive CK, SMA, c-Kit – negative |
| Shef-UPS 03 | STS 09/11 | Long spindle-shaped cells No pleomorphism Loose colony formation | p35=63.97 | 66/F | Undifferentiated pleomorphic sarcoma | Lower limb | 115 mm | pT2b Stage III | Vimentin – positive CK, SMA, c-Kit – negative |
| Shef-DDLPS 01 | STS 09/10 | Mostly spindle-shaped cells Some pleomorphism No distinct colony formation | w2p35=49.5 | 68/F | Dedifferentiated liposarcoma | Retroperitoneum | 300 mm | pT2b Stage III | Vimentin – positive CK, SMA, c-Kit – negative |
| bShef-DDLPS 02 | STS 20/11 | Two distinct cell types in same culture (A) long spindle-shaped cells (B) Rounded, histiocyte-like cells with distinct nuclei Both form tight colonies | p23=58.22 | 70/F | Dedifferentiated liposarcoma | Lower limb | 170 mm | ypT2b Stage III | Vimentin, SMA – positive CK, c-Kit – negative |
| Shef-LMS 01 | STS 02/11 | ||||||||
| w1 | Long spindle-shaped cells No pleomorphism Tight colony formation | w1p54=27.44 | 62/F | Leiomyosarcoma | Pelvis | 135 mm | pT1b Stage IIA | Vimentin, SMA – positive CK, c-Kit – negative | |
| ws | Rounded, histiocyte-like cells with distinct nuclei No pleomorphism Tight colony formation | wsp63=44.62 | |||||||
| Shef-MFS 01 | STS 21/11 | ||||||||
| w1 | Rounded, histiocyte-like cells No pleomorphism Tight colony formation | w1p35=59.93 | 73/M | Myxofibrosarcoma | Upper limb | 50 mm | pT1b Stage IIA | Vimentin – positive CK, SMA, c-Kit – negative | |
| w2 | Polygonal cells No pleomorphism Loose colony formation | w2p31=56.30 |
Abbreviations: CK=cytokeratin; c-Kit=CD117; DDLPS=dedifferentiated liposarcoma; LMS=leiomyosarcoma; MFS=myxofibrosarcoma; SMA=smooth muscle actin; UPS=undifferentiated pleomorphic sarcoma.
Passage number at which proliferation assay was performed is indicated.
Patient received neoadjuvant radiotherapy.
Morphology and culture characteristics
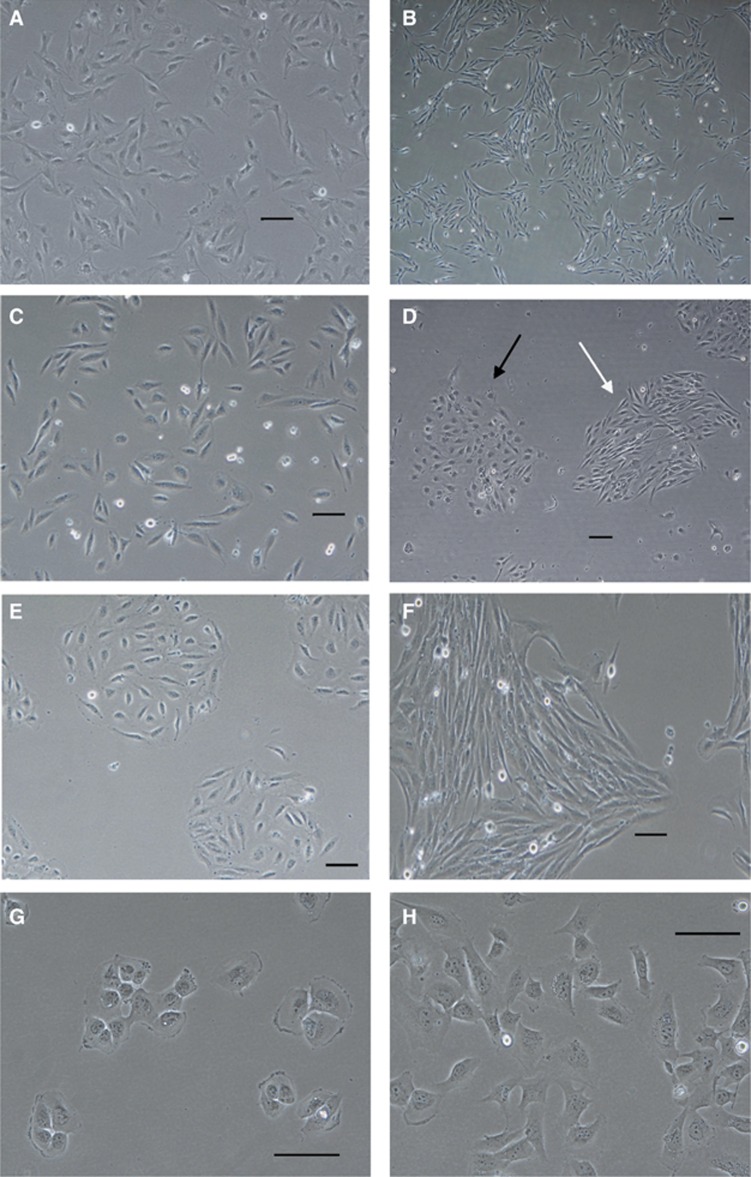
The majority of the STS primary cell lines were composed of cells that were morphologically homogeneous (Figures 1A and B). The exceptions were Shef-DDLPS 01, Shef-UPS 02 and Shef-DDLPS 02 (Table 2). Cells in the former two cultures were mostly spindle shaped but showed significant variation in their size and shape (Figure 1C). Neither showed distinct colony formation while in adherent culture. Shef-DDLPS 02 cultures on the other hand were dimorphic with one clone of long, spindle-shaped cells that were more numerous and a second population of rounded, histiocyte-like cells with distinct nuclei. Both cell populations tended to grow in tight colonies (Figure 1D). In common with Shef-DDLPS 02, Shef-LMS 01 and Shef-MFS 01 cultures were composed of two distinct morphologic cell populations (variants), but these were established as separate morphologically homogeneous cultures.
Figure 1.
Representative phase-contrast micrographs of STS primary cell lines.(A) Shef-UPS 01-spindle-shaped cells at passage 69 without distinct colony formation in culture. (B) Shef-UPS 03 cultures at passage 35 showing homogeneous cultures of long spindle-shaped cells growing in loose colonies. (C) Shef-DDLPS 01 cultures at passage 71 showing pleomorphic cells without distinct colony formation. (D) Shef-DDLPS 02 cultures at passage 22 composed of a combination of spindle-shaped cells (white arrows) and round, histiocyte-like cells (black arrows), both growing in distinct colonies. (E, F) Cells derived from Shef-LMS 01 growing in separate cultures designated as Ws (passage 69) and w1 (passage 56), respectively. (G, H) Morphologically distinct cells derived from Shef-MFS 01 growing in separate adherent cultures designated as w1 (passage 35) and w2 (passage 31), respectively. Scale bars=100 μm. DDLPS=dedifferentiated liposarcoma; LMS=leiomyosarcoma; MFS=myxofibrosarcoma.
The first Shef-LMS 01 clone was established as a wash from an original slope set-up following an ∼8-week period of crisis and was designated as ws variant. These cells had a rounded histiocyte-like morphology in adherent culture and grew in distinct, tight colonies (Figure 1E). A subsequent clone became established independently (after ∼12 weeks of crisis) in the first wash of an original T25 flask setup and was designated as w1 variant. They were slightly longer, spindle-shaped cells that also formed tight colonies (Figure 1F). Similarly, Shef-MFS 01 cells established in the initial and subsequent washes derived from the original T25 flask set-up, designated as w1 and w2, respectively, showed distinct appearance in culture. Although the former comprised cells with a rounded, histiocyte-like morphology that grew in distinct colonies (Figure 1G), the w2 variant cells had a more polygonal morphology and formed less distinct colonies (Figure 1H). Short tandem repeat profiling of the morphologic variants in both tumours showed identical profiles that reassuringly were significantly distinct from those of all the other cell lines in our laboratory and the examined databases (Table 3). Further details of the culture characteristics are shown in Supplementary Figures S1–S7.
Table 3. STR profiles of primary STS cultures.
| Cell line/passage | THO1 | D21S11 | D5S818 | D13S317 | D7S820 | D16S539 | CSFIPO | AMEL | vWA | TPOX |
|---|---|---|---|---|---|---|---|---|---|---|
| aShef-UPS 01 | ||||||||||
| p31 | 6,7 | 27,30 | 12,13 | 8,11 | 8 | 14 | 12 | X | 16 | 8 |
| p68 | 6,7 | 27,30 | 12,13 | 8,11 | 8 | 13,14 | 12 | X | 16 | 8 |
| aShef-UPS 02 | ||||||||||
| p41 | 6,9.3 | 29,31 | 9,13 | 14 | 8,11 | 11 | 10,11 | X, Y | 17,18 | 8 |
| p83 | 6,9.3 | 29,31 | 9,13 | 14 | 8,11 | 11 | 10,11 | X | 17,18 | 8 |
| Shef-UPS 03 | ||||||||||
| p34 | 9.3 | 28,31.2 | 9,12 | 11 | 10 | 9 | 10 | X | 17 | 8 |
| aShef-DDLPS 01 | ||||||||||
| w2p35 | 6 | 29,32 | 12,13 | 8,14 | 9,10 | 9,10 | 10,14 | X | 17,18 | 11 |
| w2p70 | 6 | 29,32 | 12,13 | 8,14 | 9, 10 | 9,10 | 10,14 | X | 17,18 | 11 |
| Shef-DDLPS 02 | ||||||||||
| p2 | 9 | 29 | 9 | 14 | 10,12 | 12 | 12 | X | 20 | 9,11 |
| aShef-LMS 01 | ||||||||||
| w1p16 | 9 | 27,30 | 11 | 14 | 10,11 | 11 | 10,12 | X | 16 | 8 |
| w1p54 | 9 | 27,30 | 11 | 14 | 10,11 | 11 | 10 | X | 16 | 8 |
| wsp27 | 9 | 27,30 | 11 | 14 | 10,11 | 11 | 10,12 | X | 16 | 8 |
| wsp63 | 9 | 27,30 | 11 | 14 | 10,11 | 11 | 10,12 | X | 16 | 8 |
| Shef-MFS 01 | ||||||||||
| w1p35 | 8 | 30,31.2 | 12,13 | 13 | 8,9 | 12 | 10 | X, Y | 14,16 | 11 |
| w2p31 | 8 | 30,31,2 | 12,13 | 13 | 8,9 | 12 | 10 | X, Y | 14,16 | 11 |
Abbreviations: DDLPS=dedifferentiated liposarcoma; LMS=leiomyosarcoma; MFS=myxofibrosarcoma; STR=short tandem repeat; STS=soft-tissue sarcoma; UPS=undifferentiated pleomorphic sarcoma.
Profiles comprise the alleles at 10 STR loci. No significant match (>80% relatedness) was found when compared with profiles in the COGcell database (http://strdb.cogcell.org/).
Profiles for morphologically distinct variants derived from two STS cases (shown in bold) are identical
STR profiling repeated after 1 year (30–40 subsequent passages) and disparities in allele matching is in italics.
Short tandem repeat profiling
Short tandem repeat profiling of primary STS cell lines using 10 loci was used to establish cell line identity and exclude cross-contamination. Analysis confirmed unique genomic identities for cell lines derived from all seven cases and matched the identity for those cases with separate morphologically variant cultures (Table 3). There was no evidence of significant relatedness with any other known cell lines or intralaboratory cross-contamination. Alleles for the AMEL locus were also concordant with the known gender of the patients from which all cell lines were derived, adding further confidence in the origin of these cell lines.
Profiling was repeated after a further year in culture (30–40 passages) in five cases. All retained overall unique profiles when checked against the cell line databases. Minimal evidence of genomic drift with prolonged in vitro culture was observed with only single-locus loss of heterozygosity (LOH) noted in three cell lines despite their characteristic genomic instability (Table 3).
Immunocytochemistry
Immunocytochemistry for vimentin was positive in all seven primary STS cell lines, whereas CK was consistently negative in keeping with their mesenchymal origin (Table 2). These results were also concordant with the immunophenotype at diagnosis of the corresponding tumour tissue of origin in all cases. The leiomyosarcoma cell line Shef-LMS 01 remained strongly positive for SMA and negative for c-KIT expression even after 61 passages, supporting its smooth muscle lineage of differentiation (not shown). None of the other cell lines stained positive for c-KIT or SMA, except Shef-DDLPS 02, a dedifferentiated liposarcoma that stained weakly positive for SMA (Table 2).
Ploidy
As expected in tumours known to possess complex karyotypes, metaphase chromosome spreads from all the STS cell lines examined showed significant aneuploidy with chromosome counts as high as 200 in some cases (see Supplementary Table S2). The highest chromosome counts were seen in Shef-LMS 01 and Shef-UPS 02, with a mean chromosome count of ∼120. It is interesting to note that these cell lines also had the shortest doubling times (Table 2). The lowest chromosome counts were seen in Shef-UPS 03, with a mean count of 58 chromosomes.
The majority of the cell lines, including those with very homogeneous cytomorphologic appearance on microscopy, showed significant intratumour heterogeneity with wide ranges of chromosome counts in each metaphase spread. The sole exception was Shef-UPS 01, which showed a fairly consistent near-triploid chromosome count (Supplementary Table S2). Further characterisation using conventional cytogenetics was not performed in favour of molecular cytogenetic profiling as described below.
DNA copy number profiling
Given the characteristic karyotypic complexity of these STS subtypes, our ability to demonstrate similarities in presumably random somatic copy number abnormalities (SCNAs) across the genome serves as an important method of establishing how well a cell line represents its parent tumour. Whole-genome copy number profile comparisons of paired DNA samples from each parent STS tumour and at least one of the corresponding primary tumour cell lines was therefore carried out using a high-resolution array CGH platform. Control DNA was obtained from normal viscera such as the kidney and uterus, which were resected as part of surgical excision in Shef-DDLPS 01 and Shef-LMS 01, respectively. In all other cases, pooled genomic DNA was used as control. Identical control DNA samples were used for all parent tumour and corresponding cell line pairs that were compared.
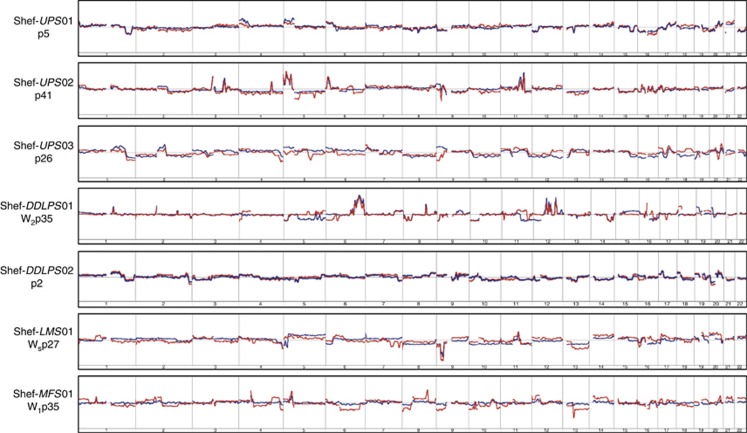
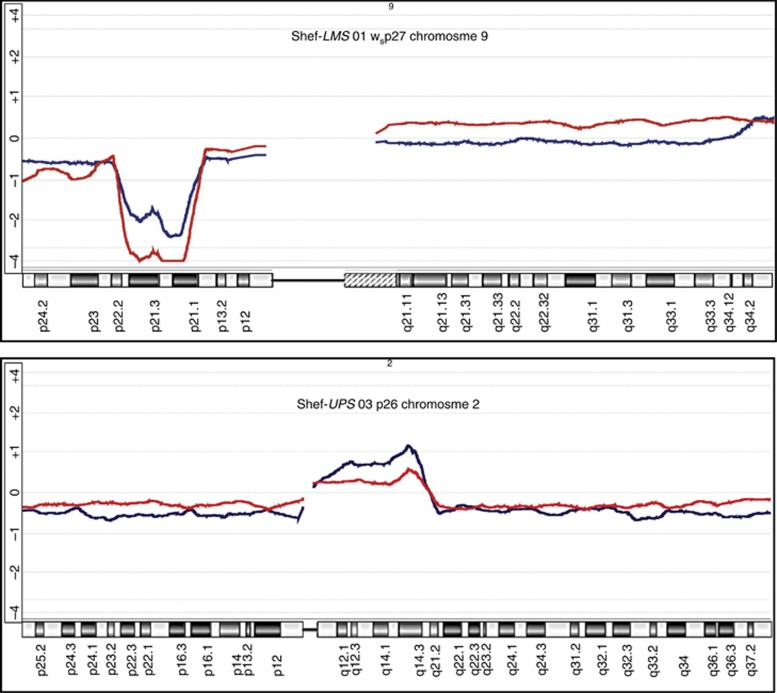
The analysis confirmed that all seven cell cultures analysed were related to their parent tumours. The greatest similarity across the whole genome was seen with Shef-DDLPS 02, Shef-UPS 01 and Shef-UPS 02 (Figure 2). Although this was expected in the former two, where the cultured cell DNA used for comparison was extracted within the first five passages, that from Shef-UPS 02, which was performed after over 40 passages, was rather interesting (Figure 2). When examined at higher resolution, even the cell lines that showed the greatest dissimilarity across the whole genome retained striking similarities at certain genomic loci with very similar moving average log2 ratio patterns and identical SCNA breakpoints. Examples as shown in Figure 3 include the deletion on the short arm of chromosome 9 seen in Shef-LMS 01, the proximal 2q amplicon in Shef-UPS 03 and the 5p amplification of Shef-MFS 01. Notably, two of these parent tumours were able to establish more than one morphologically distinct cell type in long-term culture, suggesting that the dissimilarity in the SCNA profile comparisons may be a reflection of the inherent heterogeneity of the parent tumour DNA when compared with the clonal homogeneity of the corresponding cell culture.
Figure 2.
Genomic copy number profile comparisons of seven soft-tissue sarcoma (STS) primary cell lines (shown on the left) with their parent tumours.Individual cell lines and passage number at which genomic DNA was extracted are shown to the left of the corresponding autosome ideograms. The overlaid red and blue lines represent the moving average of log2 ratios of the cultured cells and parent tumour tissue, respectively. Deviations above and below the horizontal baseline represent amplifications and deletions, respectively. Relative amplitude of deviation shows the log2 ratio and represents DNA copy number at the corresponding genomic locus. Note the close similarity and/or near-identical breakpoints in the moving average patterns in each case over the majority of the genome. Copy number analysis was performed on the Agilent 4 × 180K DNA microarray platform and data were analysed using Agilent Genomic Workbench Software v.6.0 (Agilent Technologies).
Figure 3.
Selected chromosome copy number profile comparisons of soft-tissue sarcoma (STS) primary cell lines with their parent tumours.Ideograms of specific chromosomes are as shown at the top of each panel with the corresponding regions at the bottom. The overlaid red and blue lines represent the moving average of log2 ratios (vs normal genomic DNA) of the cultured cells and parent tumour tissue, respectively. Deviations above and below the horizontal baseline represent amplifications and deletions, respectively. Amplitude of deviation shows the relative log2 ratio and represents relative DNA copy number. Note the close similarity and/or near-identical breakpoints in the moving average patterns in each case. Copy number analysis was performed on the Agilent 4 × 180K DNA microarray platform and data analysed using Agilent Genomic Workbench Software v.6.0 (Agilent Technologies).
Discussion
Commercial cell lines are a widely used in vitro disease model for initial functional validation studies of molecular candidate drivers in spite of their recognised limitations. This is because they are a readily available, endlessly replicating source of tumour material from which results obtained are usually reproducible (Cree et al, 2010). Many researchers, however, believe that successful bench to bedside translation of in vitro results is well worth the effort of obtaining primary cell cultures while others use results from primary cultures to augment those from the readily available commercial cell lines, instead of replacing them entirely (Cree et al, 2010).
In this study, establishment of primary cell cultures was attempted with all fresh STS tissue collected with a success rate of over 70% confluent cultures and more than half the adherent monolayers able to undergo four passages or more (Table 1). This rate is very comparable to previous studies, which reported success rates of between 5 and 33% when cultures were attempted from primary solid tumours and slightly higher success rates from tumour metastases (McBain et al, 1984; Gazdar et al, 1998; Nayak et al, 2000) or xenograft-derived cultures (Dangles-Marie et al, 2007; Kamiyama et al, 2013). Seven tumours (15% of cases) have established stable cell lines whose genomic and phenotypic characteristics were evaluated and compared with those of their corresponding parent tumours to confirm their suitability for functional validation studies. Cells were frozen down at intervals at early passages to minimise culture-related genomic drift when cells are used in experiments.
Among the 14 cases that failed to establish adherent cultures, eight were well-differentiated or myxoid liposarcomas that had high fat and/or myxoid components relative to cell number, which is believed to have reduced the likelihood of adherent culture using the manual mechanical tissue dissociation applied in this study. Simultaneous use of the explant method of culture establishment (Mitra et al, 2013) in some of these cases only yielded slow-growing, fibroblast-like cells that failed to reach confluence (data not shown).
Application of the principle of differential attachment as described by Nayak et al (2000) led to the establishment of long-term cultures in washes in three cases (Shef-DDLPS 01, Shef-LMS 01 and Shef-MFS 01) when cells adherent in the original culture set-ups became senescent. In the latter two cases, it also led to the establishment of separate cultures of two morphologically distinct variants each (Figure 1 and Table 2). Since both cell populations in these cultures were exposed to otherwise identical culture conditions, it is most likely that the variants represent separate clones present within the parent tumour rather than differential adaptation to culture conditions. Similar observations have been made in multiple cases of primary breast carcinoma tissue culture (McBain et al, 1984).
Short tandem repeat profiling, which is the current recommended standard for cell line identification (American Type Culture Collection Standards Development Organization Workgroup ASN, 2010), not only reliably confirmed identical profiles for the morphologic variants in Shef-LMS 01 and Shef-MFS 01, but established unique genomic identities for all the primary tumour cell lines reported in this study (Table 3). Loss of heterozygosity events were observed when STR profiling was repeated after ∼40 subsequent cell passages, which likely represent genomic drift that would be expected in cells undergoing progressive in vitro culture (Capes-Davis et al, 2013). The rate of genomic drift has been shown to be higher in cells that possess microsatellite instability (Masramon et al, 2006), a phenotype that has demonstrated in several studies of soft-tissue and bone sarcomas, where it is believed to be due to their overall genomic instability and not necessarily due to the classical defects in mismatch repair as seen in other cancers (Monument et al, 2012). While reassuring, the low frequency of LOH events seen in these cell lines after prolonged culture (Table 3) was therefore rather surprising.
All the cell lines remained proliferative in culture for at least 3 years and had each been passaged at least 60 times. Assessment of their doubling times showed proliferation rates that are comparable to those of well-known sarcoma cell lines such as SK-LMS1 and U2-OS as well as other tumour cell lines (McBain et al, 1984). Similarly, clonal-plating efficiency of the primary UPS and LMS subtypes of these cell lines (data not shown) ranged between 25 and 50% comparing favourably with ∼30% efficiency reported for the SK-LMS1 cell line in our lab and by other investigators (Kappler et al, 2004; Murphy et al, 2008). These results reflect the cancer hallmarks of increased proliferation and survival among these primary cell lines (Hanahan and Weinberg, 2011).
Characterisation of primary cells cultures for use as models in target validation studies is essential because of the potential for fibroblast overgrowth in early cultures (Mitra et al, 2013) and cross-contamination by other established cell lines in longer-term cultures (Gillet et al, 2013). Morphological characterisation may be unreliable because of the potential effects of an artificial in vitro microenvironment on tumour cell morphology. In most cancers, therefore, detection of diagnostic biomarker expression by immunochemistry or flow cytometry are commonly used for characterisation. The common biomarkers of tumour cell lineage such as CK, vimentin, SMA, S100 and CD34, however, have variable sensitivity and are not specific for many STS subtypes (Coindre, 2003; Fisher, 2011). Immunochemistry for panels of these markers, however, remains relevant to diagnostic practice because when interpreted with the appropriate expertise and as an adjunct to histology, they are helpful for the exclusion of benign and non-mesenchymal tumours as well as suggest differentiation lineages in certain STS subtypes. To this end, the immunochemistry results from this study provided support for a mesenchymal lineage for the established cell lines and were concordant with those of their parent tumours, suggesting that they represent those tumours at least to some extent (Table 2).
All the STS subtypes from which cell lines were developed in this study are known to possess genomic instability and complex karyotypes (Taylor et al, 2011; Fletcher et al, 2013). All seven cases showed highly abnormal chromosome numbers, and in some cases, significant changes in chromosome numbers with increased time in culture that reflects inherent genomic instability that is part of the cancer cell phenotype (Hanahan and Weinberg, 2011). Array CGH, a method that allows the mapping of the complex DNA copy number abnormalities across entire genomes and contributed significantly to the recent WHO classification of STS (Fletcher et al, 2013) was therefore used for definitive genomic characterisation of the tumour cell lines. Importantly, it also permits the matching of unique and presumably random genomic aberrations that the cell lines and their parent tumours have in common.
Genomic copy number profiles in all seven long-term cell cultures when compared with the corresponding parent tumours showed overall similarity in log ratio patterns (Figure 2). A number of regions with differences in their SCNA pattern were noted. In line with the clonal evolution model of cancer, the genomic instability that is inherent with these tumours combined with their rapid proliferation is expected to result in significant heterogeneity of tumour cell clones that may be under-represented in the cell line (Anderson et al, 2011; Greaves and Maley, 2012). This was likely the case in Shef-LMS 01 and Shef-MFS 01 where the genome profile of the parent tumour was compared with that of one of two confirmed clonal tumour cell variants in culture (Figure 2 and Supplementary Figures S3 and S7). Further, prolonged in vitro cell culture in addition to the aforementioned factors is expected to result in the accumulation of genomic copy number or structural karyotypic aberrations, most of which are functionally neutral, that is, ‘passengers' (Greaves and Maley, 2012; Gillet et al, 2013).
Overall, however, the genomic regions with dissimilar aberrations were few when compared with those regions that showed very similar SCNA patterns with near-identical breakpoints. This was seen even after 40 passages in culture in the case of Shef-UPS 02 (Figures 2 and 3) and suggests that these cell lines are a suitable in vitro disease model as even large-scale studies evaluating the relevance of established cell lines in various cancers have shown that ‘driver' genomic aberrations are nearly always retained in well-established commercial cell lines and vice versa despite evidence of genomic drift (Beroukhim et al, 2010; Gazdar et al, 2010; Barretina et al, 2012). Moreover, early passages of all the cell lines in this study that presumably bear a closer genomic and phenotypic resemblance to the parent tumour have been banked and can be used for target validation studies.
Among the seven tumours that established long-term cultures, three were undifferentiated pleomorphic sarcomas, representing half of the cases that were obtained of this characteristically aggressive STS subtype (Fletcher et al, 2013). The other STS subtypes that formed long-term cultures were also of a high grade. This suggests that overall, high-grade and aggressive clinical course in STS may correlate with amenability to in vitro growth, as was observed in primary cultures of breast (Gazdar et al, 1998) and colorectal cancer tissue (McBain et al, 1984). The three undifferentiated pleomorphic sarcoma cell lines notably demonstrate very different morphology, doubling time and chromosome numbers, which is not unexpected given that this diagnosis is largely one of exclusion (Fletcher et al, 2013) and likely includes a number of as yet undefined biologic tumour entities.
Shef-DDLPS 02 was derived from a patient who received neoadjuvant radiotherapy for dedifferentiated liposarcoma with partial response and the cell line probably represents a radioresistant clone of cells present within that tumour. It is notable that array CGH analysis of this cell line (and its parent tumour) did not show 12q amplification (typically involving the MDM2 and/or CDK4 genes) that is frequently seen well- and del-differentiated liposarcomas and was present in Shef-DDLPS 01 (Figure 2) and four other liposarcoma cases in this study (Table 1; array CGH data not shown). This is in keeping with data from large-scale genomic studies of STS such as the sarcoma genome project, which showed that up to 10% of DDLPS have neither CDK4 nor MDM2 amplification (Barretina et al, 2010). Review of the tumour histology confirmed a biphasic appearance with an area of atypical adipocytic differentiation and other areas of poorly differentiated sarcoma (Supplementary Figure S5) in keeping with the diagnostic criteria for DDLPS. Further, FISH analysis performed on the diagnostic tissue sample for MDM2 copy number (data not shown) was concordant with the array CGH results. It is therefore likely that this tumour belongs in this category of DDLPS with as yet undetermined specific genomic aberration.
Given their range of subtypes, clinical, genomic and in vitro cultural features, the primary cell lines in this study are potentially much-needed additions to the number of cell lines of STS with complex genomics available that could comprise a drug testing panel akin to the NCI-60 panel that includes only more prevalent cancers (Shoemaker, 2006; Taylor et al, 2011). Further, a radioresistant STS cell line may be used for the study of tumour response to radiation either alone or in combination with sensitising agents, as demonstrated with PARP inhibitors in Ewings' sarcoma cell lines (Garnett et al, 2012).
The results of primary tissue culture in this study show the potential for establishment of short- and long-term cell cultures from STS tissue. The need for validation studies in short-term cultures coupled with their finite nature and limited supply is, however, a practical limitation to their use by the wider research community. All the long-term cell lines in this study demonstrate genomic and phenotypic features that not only confirm their malignant characteristics but also confirm the retention of the majority of SCNA present in their parent tumours that likely include ‘driver' aberrations. When combined with their rapid proliferation and abundance, they represent an excellent model in vitro validation of genomic and transcriptomic targets in STS.
Further, the establishment of separate clones that have been genetically confirmed as belonging to the same tumour in two cases will permit the evaluation of differential responses to therapeutic agents. The combination of cryopreservation of early passages of these characterised cell lines as well as the potential to develop prospective short-term cultures from other tumours of the same STS subtypes will serve to mitigate some of the concerns that have been raised with existing commercially available cell lines. For further information on the availability of these cell lines, interested researchers should please contact the corresponding author.
Acknowledgments
Sarcoma Trust/Sarcoma UK and The Betty Waind Trust Weston Park Hospital Cancer Charity provided financial support for this work. The contributions of Sarcoma Pathologists David Hughes, Malee Fernando and John Goepel as well as general advice and technical support from Dave Hammond, Carmel Nichols and Marwa Jama are greatly appreciated. General clinical support for the research and patient identification for inclusion of samples was provided by the Sheffield Experimental Cancer Medical Centre.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on British Journal of Cancer website (http://www.nature.com/bjc)
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 4.0 Unported License.
Supplementary Material
References
- American Type Culture Collection Standards Development Organization Workgroup ASN (2010) Cell line misidentification: the beginning of the end. Nat Rev Cancer 10(6): 441–448. [DOI] [PubMed] [Google Scholar]
- Anderson K, Lutz C, van Delft FW, Bateman CM, Guo Y, Colman SM, Kempski H, Moorman AV, Titley I, Swansbury J, Kearney L, Enver T, Greaves M (2011) Genetic variegation of clonal architecture and propagating cells in leukaemia. Nature 469(7330): 356–361. [DOI] [PubMed] [Google Scholar]
- Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, Wilson CJ, Lehar J, Kryukov GV, Sonkin D, Reddy A, Liu M, Murray L, Berger MF, Monahan JE, Morais P, Meltzer J, Korejwa A, Jane-Valbuena J, Mapa FA, Thibault J, Bric-Furlong E, Raman P, Shipway A, Engels IH, Cheng J, Yu GK, Yu J, Aspesi P Jr, de Silva M, Jagtap K, Jones MD, Wang L, Hatton C, Palescandolo E, Gupta S, Mahan S, Sougnez C, Onofrio RC, Liefeld T, MacConaill L, Winckler W, Reich M, Li N, Mesirov JP, Gabriel SB, Getz G, Ardlie K, Chan V, Myer VE, Weber BL, Porter J, Warmuth M, Finan P, Harris JL, Meyerson M, Golub TR, Morrissey MP, Sellers WR, Schlegel R, Garraway LA (2012) The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 483(7391): 603–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barretina J, Taylor BS, Banerji S, Ramos AH, Lagos-Quintana M, Decarolis PL, Shah K, Socci ND, Weir BA, Ho A, Chiang DY, Reva B, Mermel CH, Getz G, Antipin Y, Beroukhim R, Major JE, Hatton C, Nicoletti R, Hanna M, Sharpe T, Fennell TJ, Cibulskis K, Onofrio RC, Saito T, Shukla N, Lau C, Nelander S, Silver SJ, Sougnez C, Viale A, Winckler W, Maki RG, Garraway LA, Lash A, Greulich H, Root DE, Sellers WR, Schwartz GK, Antonescu CR, Lander ES, Varmus HE, Ladanyi M, Sander C, Meyerson M, Singer S (2010) Subtype-specific genomic alterations define new targets for soft-tissue sarcoma therapy. Nat Genet 42(8): 715–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beroukhim R, Mermel CH, Porter D, Wei G, Raychaudhuri S, Donovan J, Barretina J, Boehm JS, Dobson J, Urashima M, Mc Henry KT, Pinchback RM, Ligon AH, Cho YJ, Haery L, Greulich H, Reich M, Winckler W, Lawrence MS, Weir BA, Tanaka KE, Chiang DY, Bass AJ, Loo A, Hoffman C, Prensner J, Liefeld T, Gao Q, Yecies D, Signoretti S, Maher E, Kaye FJ, Sasaki H, Tepper JE, Fletcher JA, Tabernero J, Baselga J, Tsao MS, Demichelis F, Rubin MA, Janne PA, Daly MJ, Nucera C, Levine RL, Ebert BL, Gabriel S, Rustgi AK, Antonescu CR, Ladanyi M, Letai A, Garraway LA, Loda M, Beer DG, True LD, Okamoto A, Pomeroy SL, Singer S, Golub TR, Lander ES, Getz G, Sellers WR, Meyerson M (2010) The landscape of somatic copy-number alteration across human cancers. Nature 463(7283): 899–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canovas D, Rennie IG, Nichols CE, Sisley K (2008) Local environmental influences on uveal melanoma. Cancer 112(8): 1787–1794. [DOI] [PubMed] [Google Scholar]
- Capes-Davis A, Reid YA, Kline MC, Storts DR, Strauss E, Dirks WG, Drexler HG, MacLeod RA, Sykes G, Kohara A, Nakamura Y, Elmore E, Nims RW, Alston-Roberts C, Barallon R, Los GV, Nardone RM, Price PJ, Steuer A, Thomson J, Masters JR, Kerrigan L (2013) Match criteria for human cell line authentication: where do we draw the line? Int J Cancer 132(11): 2510–2519. [DOI] [PubMed] [Google Scholar]
- Coindre JM (2003) Immunohistochemistry in the diagnosis of soft tissue tumours. Histopathology 43(1): 1–16. [DOI] [PubMed] [Google Scholar]
- Cree IA, Glaysher S, Harvey AL (2010) Efficacy of anti-cancer agents in cell lines versus human primary tumour tissue. Curr Opin Pharmacol 10(4): 375–379. [DOI] [PubMed] [Google Scholar]
- Dangles-Marie V, Pocard M, Richon S, Weiswald LB, Assayag F, Saulnier P, Judde JG, Janneau JL, Auger N, Validire P, Dutrillaux B, Praz F, Bellet D, Poupon MF (2007) Establishment of human colon cancer cell lines from fresh tumors versus xenografts: comparison of success rate and cell line features. Cancer Res 67(1): 398–407. [DOI] [PubMed] [Google Scholar]
- Fisher C (2011) Immunohistochemistry in diagnosis of soft tissue tumours. Histopathology 58(7): 1001–1012. [DOI] [PubMed] [Google Scholar]
- Fisher C (2014) Standards and datasets for reporting cancers Dataset for histopathology reporting of soft tissue sarcomas. The Royal College of Pathologists. Available at https://www.rcpath.org/asset/55DBFC41-F06A-4A71-BCC35A97821F8E8D/ (last accessed 11 July 2016).
- Fletcher CDM, Bridge JA, Hogendoorn PCW, Mertens F (2013) Classification of Tumours of Soft Tissue and Bone Classification of Tumours. World Health Organization: Geneva, Switzerland. [Google Scholar]
- Forbes SA, Bindal N, Bamford S, Cole C, Kok CY, Beare D, Jia M, Shepherd R, Leung K, Menzies A, Teague JW, Campbell PJ, Stratton MR, Futreal PA (2011) COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res 39(Database issue): D945–D950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garnett MJ, Edelman EJ, Heidorn SJ, Greenman CD, Dastur A, Lau KW, Greninger P, Thompson IR, Luo X, Soares J, Liu Q, Iorio F, Surdez D, Chen L, Milano RJ, Bignell GR, Tam AT, Davies H, Stevenson JA, Barthorpe S, Lutz SR, Kogera F, Lawrence K, McLaren-Douglas A, Mitropoulos X, Mironenko T, Thi H, Richardson L, Zhou W, Jewitt F, Zhang T, O'Brien P, Boisvert JL, Price S, Hur W, Yang W, Deng X, Butler A, Choi HG, Chang JW, Baselga J, Stamenkovic I, Engelman JA, Sharma SV, Delattre O, Saez-Rodriguez J, Gray NS, Settleman J, Futreal PA, Haber DA, Stratton MR, Ramaswamy S, McDermott U, Benes CH (2012) Systematic identification of genomic markers of drug sensitivity in cancer cells. Nature 483(7391): 570–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazdar AF, Gao B, Minna JD (2010) Lung cancer cell lines: useless artifacts or invaluable tools for medical science? Lung Cancer 68(3): 309–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazdar AF, Kurvari V, Virmani A, Gollahon L, Sakaguchi M, Westerfield M, Kodagoda D, Stasny V, Cunningham HT, Wistuba II, Tomlinson G, Tonk V, Ashfaq R, Leitch AM, Minna JD, Shay JW (1998) Characterization of paired tumor and non-tumor cell lines established from patients with breast cancer. Int J Cancer 78(6): 766–774. [DOI] [PubMed] [Google Scholar]
- Gillet JP, Varma S, Gottesman MM (2013) The clinical relevance of cancer cell lines. J Natl Cancer Inst 105(7): 452–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greaves M, Maley CC (2012) Clonal evolution in cancer. Nature 481(7381): 306–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA (2011) Hallmarks of cancer: the next generation. Cell 144(5): 646–674. [DOI] [PubMed] [Google Scholar]
- Italiano A, Di Mauro I, Rapp J, Pierron G, Auger N, Alberti L, Chibon F, Escande F, Voegeli A-C, Ghnassia J-P, Keslair F, Laé M, Ranchère-Vince D, Terrier P, Baffert S, Coindre J-M, Pedeutour F (2016) Clinical effect of molecular methods in sarcoma diagnosis (GENSARC): a prospective, multicentre, observational study. Lancet Oncol 17(4): 532–538. [DOI] [PubMed] [Google Scholar]
- Kamb A (2010) At a crossroads in oncology. Curr Opin Pharmacol 10(4): 356–361. [DOI] [PubMed] [Google Scholar]
- Kamiyama H, Rauenzahn S, Shim JS, Karikari CA, Feldmann G, Hua L, Kamiyama M, Schuler FW, Lin MT, Beaty RM, Karanam B, Liang H, Mullendore ME, Mo G, Hidalgo M, Jaffee E, Hruban RH, Jinnah HA, Roden RB, Jimeno A, Liu JO, Maitra A, Eshleman JR (2013) Personalized chemotherapy profiling using cancer cell lines from selectable mice. Clin Cancer Res 19(5): 1139–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kappler M, Bache M, Bartel F, Kotzsch M, Panian M, Wurl P, Blumke K, Schmidt H, Meye A, Taubert H (2004) Knockdown of survivin expression by small interfering RNA reduces the clonogenic survival of human sarcoma cell lines independently of p53. Cancer Gene Ther 11(3): 186–193. [DOI] [PubMed] [Google Scholar]
- Kato S, Espinoza N, Lange S, Villalón M, Cuello M, Owen GI (2008) Characterization and phenotypic variation with passage number of cultured human endometrial adenocarcinoma cells. Tissue Cell 40(2): 95–102. [DOI] [PubMed] [Google Scholar]
- Luca T, Privitera G, Lo Monaco M, Prezzavento C, Castorina S (2007) Validation study of a cell culture model of colorectal cancer. Ital J Anat Embryol 112(2): 81–92. [PubMed] [Google Scholar]
- Masramon L, Vendrell E, Tarafa G, Capellà G, Miró R, Ribas M, Peinado MA (2006) Genetic instability and divergence of clonal populations in colon cancer cells in vitro. J Cell Sci 119(Part 8): 1477–1482. [DOI] [PubMed] [Google Scholar]
- McBain JA, Weese JL, Meisner LF, Wolberg WH, Willson JK (1984) Establishment and characterization of human colorectal cancer cell lines. Cancer Res 44(12, Part 1): 5813–5821. [PubMed] [Google Scholar]
- Mitra A, Mishra L, Li S (2013) Technologies for deriving primary tumor cells for use in personalized cancer therapy. Trends Biotechnol 31(6): 347–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monument MJ, Lessnick SL, Schiffman JD, Randall RT (2012) Microsatellite instability in sarcoma: fact or fiction? ISRN Oncol 2012: 473146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy JD, Lucas DR, Somnay YR, Hamstra DA, Ray ME (2008) Gemcitabine-mediated radiosensitization of human soft tissue sarcoma. Transl Oncol 1(1): 50–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nayak SK, Kakati S, Harvey SR, Malone CC, Cornforth AN, Dillman RO (2000) Characterization of cancer cell lines established from two human metastatic breast cancers. In Vitro Cell Dev Biol Anim 36(3): 188–193. [DOI] [PubMed] [Google Scholar]
- Pan C, Kumar C, Bohl S, Klingmueller U, Mann M (2009) Comparative proteomic phenotyping of cell lines and primary cells to assess preservation of cell type-specific functions. Mol Cell Proteomics 8(3): 443–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salawu A, Ul-Hassan A, Hammond D, Fernando M, Reed M, Sisley K (2012) High quality genomic copy number data from archival formalin-fixed paraffin-embedded leiomyosarcoma: optimisation of universal linkage system labelling. PLoS One 7(11): e50415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker RH (2006) The NCI60 human tumour cell line anticancer drug screen. Nat Rev Cancer 6(10): 813–823. [DOI] [PubMed] [Google Scholar]
- Taylor BS, Barretina J, Maki RG, Antonescu CR, Singer S, Ladanyi M (2011) Advances in sarcoma genomics and new therapeutic targets. Nat Rev Cancer 11(8): 541–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.