Abstract
Cellular signaling is often propagated by multivalent interactions. Multivalency creates avidity, allowing stable biophysical recognition. Multivalency is an attractive strategy for achieving potent binding to protein targets, as the affinity of bivalent ligands is often greater than the sum of monovalent affinities. The BET family of transcriptional coactivators features tandem bromodomains, through which BET proteins naturally bind acetylated histones and transcription factors. All reported BRD4 antagonists bind in a monovalent fashion. Here, we report the first bivalent BET bromodomain inhibitor, MT1 that has unprecedented potency. Biophysical and biochemical studies suggest MT1 is an intramolecular bivalent BRD4 binder that is over 100-fold more potent in cellular assays compared to the corresponding monovalent antagonist, JQ1. MT1 significantly delayed leukemia progression in mice (Mus musculus) compared to JQ1. These data qualify a powerful chemical probe for BET bromodomains and extensible rationale for further development of multidomain epigenetic reader protein inhibitors.
INTRODUCTION
Cell–cell interactions and signal transduction often depend on multivalent interactions between receptors and their corresponding ligands1. As is often the case in binding of carbohydrates (glycoproteins, glycolipids, polysaccharides or proteoglycans) to lectins that have several binding sites, individual weak interactions can be enhanced more than 1,000-fold through multivalent interactions, a phenomenon known as the avidity effect2. Multivalent ligands that have either homo- or hetero-binding motifs show avidity by several mechanisms, such as interactions with oligomeric receptors, oligomerization of monomeric receptors, or increasing effective molarity of binding ligands3. Further, multivalent ligands can exhibit a prolonged residence time4,5. These historical observations from the natural world establish a strong rationale for multivalent ligand discovery4–8.
Molecular recognition of chromatin by transcriptional or epigenetic complexes is often mediated by proteins with single or multiple “reader” domains, which bind histone proteins, DNA, or transcription factors in specific post-translational modification states. In the context of transcriptional activation, recruitment of histone acetyltransferases leads to N-acetylation (Kac) of lysine residues on histone proteins and transcription factors. Local hyperacetylation leads to subsequent recruitment of co-activator proteins with acetyl-lysine recognition domains, or bromodomains. A bromodomain is an antiparallel bundle of alpha helices that recognizes mono- or di-acetylated peptides via a hydrophobic pocket with an adjacent, conserved asparagine residue9. The BET (bromodomain and extra-terminal domain) family of human bromodomains are transcriptional co-activators involved in cell cycle progression, transcriptional activation, and elongation10,11. BET bromodomains (BRD2, BRD3, BRD4 and BRDT) are critical mediators of chromatin-dependent signal transduction from master regulatory transcription factors to RNA Polymerase II. BRD4, in particular, has emerged as a therapeutic target in cancer, as a co-activator protein for the prevalent oncoprotein, MYC12,13. Further, BRD4 facilitates transcriptional elongation via recruitment or activation of the positive transcription elongation factor (P-TEFb) and displacement of negative regulators (HEXIM1 and 7SK snRNA)14,15.
In 2010, our laboratory collaboratively reported the first direct-acting bromodomain antagonist, JQ1 (Fig. 1a)16. JQ1 is a potent and BET-selective thieno-1,4-diazepine which binds the critical asparagine via a methyl-triazolo moiety. JQ1 has proven a valuable chemical probe for mechanistic and translational research, providing pharmacologic target validation in predictive models of solid tumors and hematological malignancies. Already, more than eight BRD4 antagonists have advanced to human clinical investigation as cancer therapy17. To date, all validated BRD4 antagonists are monovalent inhibitors that interact with either bromodomain 1 (BD1) or bromodomain 2 (BD2) in a selective or non-selective fashion. The significance of the tandem BET bromodomains is not mechanistically resolved18. Chromatin binding is principally influenced by BD119, but genetic and chemical genetic studies identify a role for both domains in transcriptional activation20.
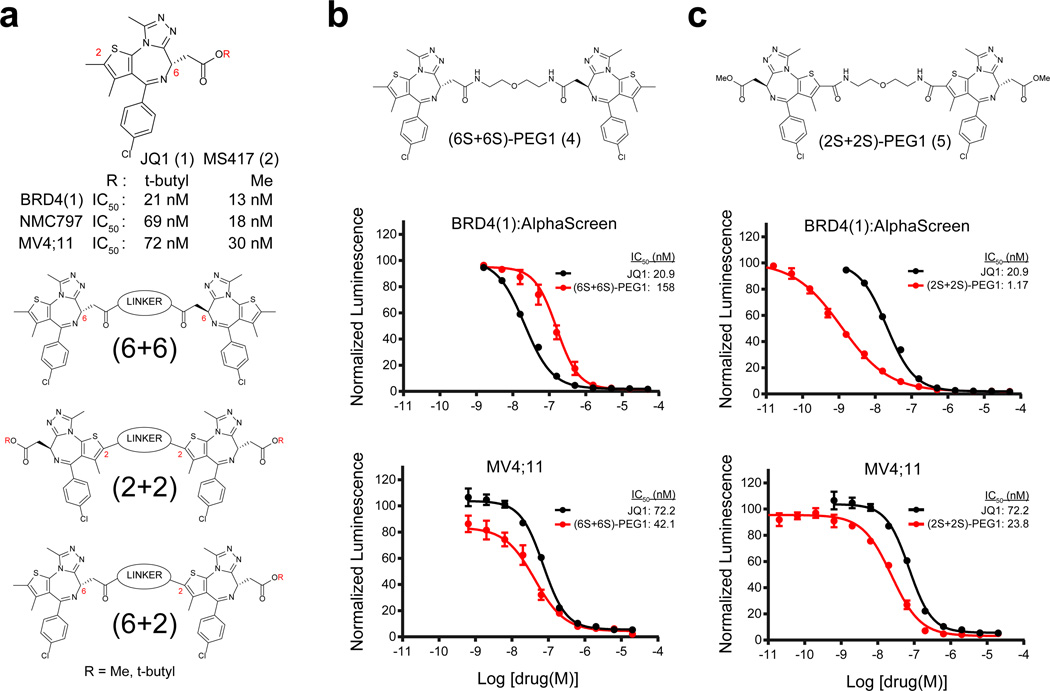
Figure 1. Rational design of bivalent BET family bromodomain inhibitors.
(a) Structure and activity of the prototype BET family bromodomain inhibitor JQ1 and its close analog MS417. General chemical structure and design of dimeric JQ1-like molecules are also shown. Linkage sites at the 6 position of the diazepine and the 2 position of the thiophene are indicated. (b,c) BRD4(1) biochemical and MV4;11 cellular activity of the (b) (6+6) homo-dimer and the (c) (2+2) homo-dimer. Biochemical and cellular data represent the mean of at least 2 or 4 replicates, respectively ± SD.
Motivated by the tandem bromodomain primary structure of BRD4, and guided by our structural data explaining the mode of molecular recognition of BD1 and BD2 by JQ1, we pursued bivalent inhibitors of BET bromodomains. Here, we present structure–function principles, biochemical, and biological data supporting bivalent inhibition of BET bromodomains as a facile approach to highly potent inhibitors of epigenetic reader proteins. Through iterative optimization of attachment and linker chemistry, we identified a series of BET inhibitors with unprecedented potency, among which is MT1. While these compounds maintained the relative selectivity profile of JQ1 against a panel of bromodomains and off-target receptors, they induced stronger growth arrest in leukemia cells associated with MYC downregulation and HEXIM1 upregulation. Biophysical and crystallographic experiments revealed that these compounds bind to bromodomains in a bivalent fashion. Half an equivalent of MT1 induced dimerization of tandem BRD4 constructs when there was a point mutation at either key conserved asparagine, but failed to dimerize a wild type construct, suggesting an intramolecular bivalent binding mode. Remarkably, MT1 possessed not only the desirable properties of a chemical probe, but also tolerable pharmacokinetic (PK) profiles for animal studies. MT1 reduced tumor burden in two experimental mouse leukemia models, and showed greater efficacy compared to JQ1.
RESULTS
Potent and selective bivalent BET bromodomain inhibitors
To design bivalent inhibitors of BRD4, we selected our prototype bromodomain antagonist JQ1 (1) as a lead scaffold to leverage our knowledge both in chemistry and pharmacology (Fig. 1a). Our prior high-resolution structure of JQ1 bound to BD1 and BD216, and internal structure–activity relationship (SAR) guidance, supports chemical substitution of the t-butyl ester on the diazepine ring and substitution of the methyl moiety at C2 on the thiophene ring, as both are positioned toward solvent (Supplementary Results, Supplementary Fig. 1a). As the BD1 and BD2 bromodomains are separated by a 280 residue linker region (Supplementary Fig. 1b), and as binding may be intramolecular or intermolecular, we considered the chemical spacer between monomeric ligands as a variable for focused library synthesis. As limited biochemical data are available on full-length BRD4, owing to challenges in active protein preparation, we further considered the mode of linker attachment to monomeric ligands in index library design. Three series of dimeric JQ1 molecules were first prepared, defined by conjugation of a variably spaced polyethylene glycol (PEG) linker to either the C6 or C2 positions, hereafter referred to as (6+6), (2+2), and (6+2) (Fig. 1a).
We first synthesized (6+6) and (2+2) homodimers with a minimal PEG spacer (Fig. 1b and 1c), and compared these compounds to control ligands in biochemical and cell-based assays of direct BRD4 inhibition. For the synthesis of (2+2) homodimers, the methyl ester analog of JQ1 (MS417, 2)21 was adopted as an alternative scaffold due to synthetic compatibility (Fig. 1a). To assess competitive binding with JQ1 to the BET bromodomains, we adapted a luminescence homogeneous, nanomaterial-based proximity assay (AlphaScreen™) for BD1 of BRD4 (hereafter referred to as BRD4(1)) as the primary screening method22,23. In this assay system, the active (S)-enantiomer of JQ1 exhibited an IC50 of 21 nM (Supplementary Fig. 2a and 2b). In contrast, the IC50 for the inactive (R)-enantiomer (3) was >5 µM. Growth effects of BRD4 inhibition were evaluated using BRD4–rearranged carcinoma cells (NUT midline carcinoma; NMC797) and acute myeloid leukemia (AML) cell lines (MV4;11). (S)-JQ1 attenuates proliferation of each cell line with IC50 values of 69 and 72 nM, respectively. MS417 showed similar levels of inhibition in both AlphaScreen™ and in cells (Fig. 1a).
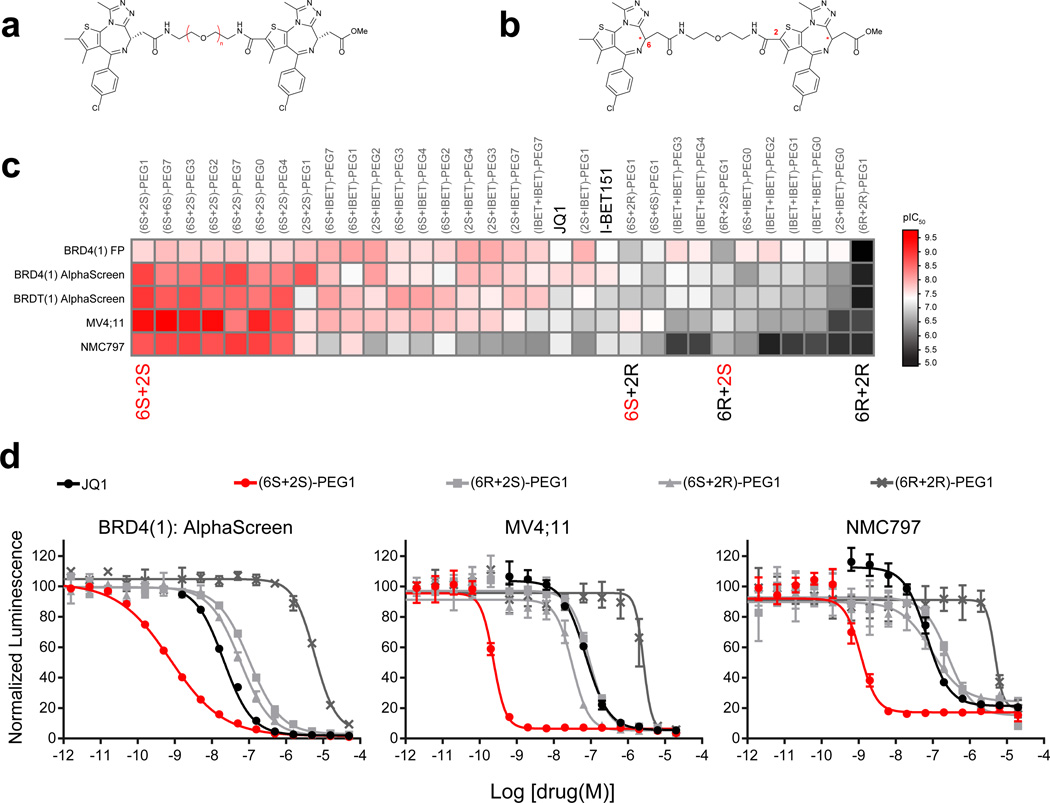
The (6+6) homo-dimer, which has a PEG1 linker (hereafter referred to as (6S+6S)-PEG1, 4), did not show improved activity in either the biochemical or cellular assays (Fig. 1b). Interestingly, the (2+2) homo-dimer, (2S+2S)-PEG1, 5) had a 20-fold increase in biochemical potency (IC50 = 1.17 nM) (Fig. 1c). However, this effect was not properly reflected in cellular activity, perhaps because of decreased permeability or metabolic liability arising from two appending ester moieties. In order to overcome these challenges, we turned our focus to (6+2) hetero-dimers with various PEG linker lengths. These molecules feature a single C6 position ester (Fig. 2a). Notably, all of the (6+2) heterodimers we synthesized (6–11) showed profound improvements in potency (single-digit to sub-nanomolar IC50 values) in biochemical assays (Supplementary Fig. 3a and 3b). Importantly, the increased biochemical potency was maintained in cellular assays (IC50s = 0.22–2.6 nM). To rule out non-specific assay interference from the compounds (e.g., aggregation)24, we synthesized all of the (6+2)-PEG1 diastereomers (7 and 12–14, Fig. 2b–d). Only the homo-combination of active (S)-enantiomers of JQ1 ((6S+2S)-PEG1) retained single-digit to sub-nanomolar activity in both the biochemical and cellular assays, while the homodimer of inactive (R)-JQ1 ((6R+2R)-PEG1) showed much weaker activity (IC50 = 5,956 nM by AlphaScreen™). The hetero-combinations of (S)- and (R)-JQ1 ((6S+2R)-PEG1 and (6R+2S)-PEG1) retained almost the same level of activity as JQ1 in biochemical assays (IC50 = 59 and 103 nM, respectively), suggesting a capacity for linker conjugation at either the C2 or C6 positions. Similar results were obtained for another BET family protein BRDT(1) (Fig. 2c). We also synthesized a single warhead analog with a PEG1 linker (2S-PEG1, 15, Supplementary Fig. 3c). This compound and JQ1 demonstrated a similar level of activity (IC50 = 115 nM by AlphaScreen™). Together, these data support dramatic enhancement of BRD4 inhibition by bivalent molecules in both biochemical and cellular assays.
Figure 2. Heterodimeric bivalent inhibitors and activity optimization.
(a, b) The general chemical structure of heterodimeric bivalent inhibitors with changes in both (a) linker length and (b) stereochemistry. (c) Heat-map of dimeric compound activity (pIC50s) in biochemical (BRD4(1) FP, BRD4(1) AlphaScreen, BRDT(1) AlphaScreen) and cellular (MV4;11, NMC797) assays compared to the prototypical BRD4 inhibitor JQ1 or I-BET151 (in black). All biochemical and cellular data represent the mean of at least 2 or 4 replicates respectively. (d) Biochemical and cellular activity of the different (6+2)-PEG1 diastereomers were measured by BRD4(1) AlphaScreen and ATPLite. Biochemical and cellular data points represent the mean of at least 2 or 4 replicates, respectively ± SD.
To further profile bivalent inhibitors, we utilized fluorescent polarization (FP) and phage-based, multiplexed bromodomain displacement assays (BROMOscan™, DiscoverX). All analogs showed a similar tendency in the FP assay (Fig. 2c), with somewhat increased sensitivity of the BROMOscan™ assay to JQ1 compared to AlphaScreen™ (BROMOscan™, Kd = 8.0 nM; AlphaScreen™, IC50 = 20.9 nM for BRD4(1)). This tendency was more pronounced for (6S+2S)-PEG1 (Supplementary Fig. 4). The compound exhibited picomolar displacement of phage expressing BRD4(1), BRD4(2), a tandem bromodomain construct (BRD4(1,2)), and full length protein (BRD4(full)). These dramatic increases in activities were confirmed to be selective for the BET family using a panel of 40 phage-displayed bromodomains (Supplementary Table 1). The (6S+2S)-PEG1 compound did not show significant binding to non-BET family bromodomains at higher concentrations (Kd = 2.5 µM for CBP, 5.0 µM for EP300, 9.9 µM for WDR9(2), >10 µM for others). The relative selectivity of the JQ1 scaffold is reflected in this JQ1 dimer.
To validate bivalent BET inhibition using an orthogonal chemical scaffold, we adapted the more recently reported isoxazole BET inhibitor, I-BET151 (16, Supplementary Fig. 2a)25, to a homodimeric strategy. I-BET151 exhibits comparable performance to JQ1 in biochemical and cellular assays (Supplementary Fig. 2b). Guided by the reported cocrystal structure of -BET151 bound to BRD4(1) (PDB: 3ZYU), the isoxazole ring binds to the conserved asparagine, while an amino moiety within the cyclic urea is faced toward a solvent exposed region. We therefore synthesized hetero- and homo-dimeric combinations of I-BET151 and JQ1 again using a series of PEG linkers (17–34, Supplementary Fig. 5). Though the increase in potency was not as dramatic as for JQ1 dimers, we observed a 5-fold increase in biochemical inhibition for some combinations (Fig. 2c, Supplementary Fig. 5). Differences in geometry or substitution off of the nitrogen atom of the cyclic urea may hinder binding or ability to effectively dimerize. Although further optimization for the I-BET151 scaffold is surely required, this result suggests that the dual warhead strategy may be versatile and thus applied to other molecular recognition modules.
Bivalent inhibitors simultaneously bind two bromodomains
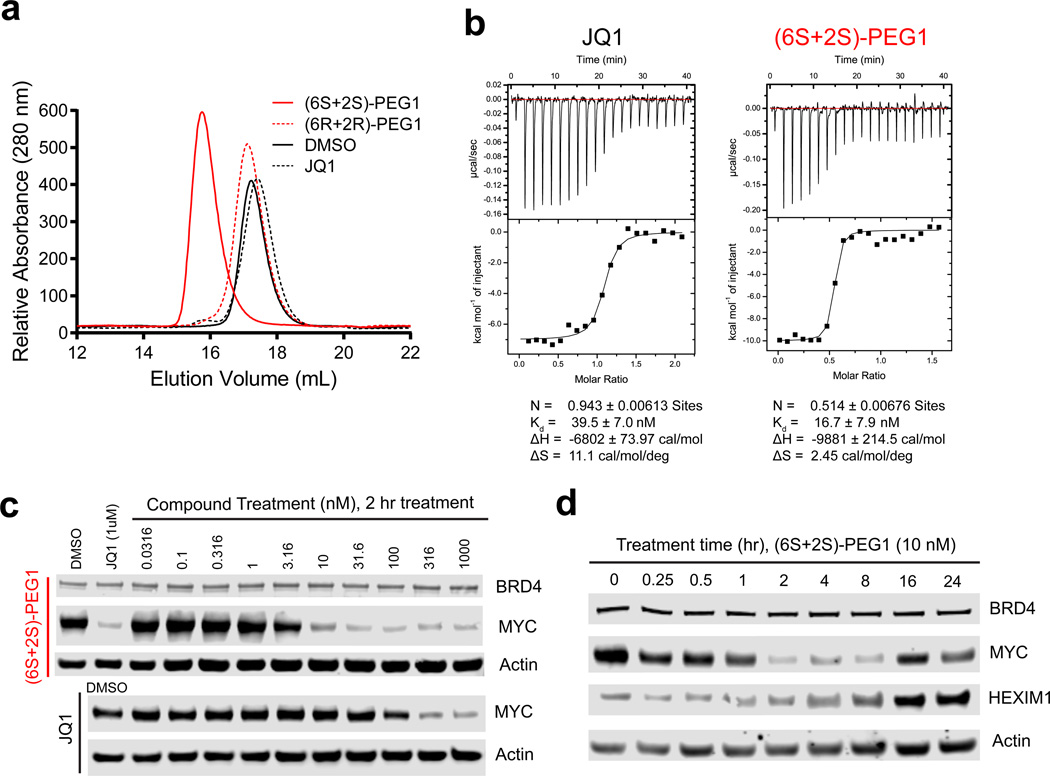
To determine whether the observed increase in potency is mediated by bivalent interaction with two discrete bromodomains, we employed size-exclusion chromatography (SEC) and isothermal titration calorimetry (ITC). SEC showed that the active (6S+2S)-PEG1 molecule completely shifted the BRD4(1) monomeric peak to a more slowly migrating dimeric peak when added in a 1:2 ratio to the protein, whereas excess JQ1 and the inactive (6R+2R)-PEG1 molecule failed to shift the monomer peak (Fig. 3a). Experimentally, overt signs of protein aggregation were not observed. ITC experiments revealed that JQ1 and (6S+2S)-PEG1 bound with a similar range of Kd values (40 and 17 nM, respectively) to isolated BRD4(1), as expected. However, the stoichiometry of binding was divergent. JQ1 bound to BRD4(1) in a 1:1 ratio, whereas (6S+2S)-PEG1 bound to BRD4(1) in a 1:2 ratio (Fig. 3b). Together, these findings support the possibility of an avidity effect via dimerization of individual BET bromodomains.
Figure 3. Biochemical and biophysical properties of bivalent inhibitors.
(a) (6S+2S)-PEG1, (6R+2R)-PEG1 or JQ1 were incubated with BRD4(1) and complex formation was assayed by size exclusion chromatography. (b) Stoichiometry of (6S+2S)-PEG1 or JQ1 binding to BRD4(1) as analyzed by Isothermal titration calorimetry. (c) Immunoblot for BRD4, MYC, and actin after treatment of MV4;11 cells with (6S+2S)-PEG1 or JQ1 at the indicated concentrations for 2 hours. (d) Immunoblot for BRD4, MYC, HEXIM1, and actin after treatment of MV4;11 cells with (6S+2S)-PEG1 at 10 nM at the indicated time points. Full blots can be found in Supplementary Figure 15.
Bivalent inhibitors induce growth arrest in AML cells
We next assessed the cellular consequences of bivalent BET inhibitors in a series of cancer cell assays, previously credentialed for BRD4-specific biology using JQ1. To assess if cell growth inhibition by (6S+2S)-PEG1 is mediated through downregulation of MYC transcription as observed with JQ1 we measured expression levels of MYC and HEXIM1 after compound treatment by immunoblot. Within two hours of treatment with (6S+2S)-PEG1 in MV4;11 cells, MYC was downregulated in a concentration-dependent manner at concentrations as low as 10 nM (Fig. 3c). This effect was time-dependent and reversible (Fig. 3d), with a maximum effect achieved at 2 hours. Consistent with an inhibitory effect on transcriptional elongation, upregulation of the compensatory negative elongation factor HEXIM1 was observed at later time points (4–24 hours). Together, these data support on-target BRD4 activity in cells26,27. Recently, we and others reported highly potent compounds capable of degrading BET bromodomain proteins, via chemical conjugation of E3 ligase-recruiting moieties28–30. To rule out enhanced potency via BET degradation, we performed immunoblots following compound treatment. No effect on protein stability was observed, suggesting that this effect is not likely due to oligomerization and subsequent degradation of BRD4 in cells. (Fig. 3c and 3d) These findings support that the cell growth inhibition by (6S+2S)-PEG1 was associated with specific BRD4 engagement.
MT1: a bivalent chemical probe of BET bromodomains
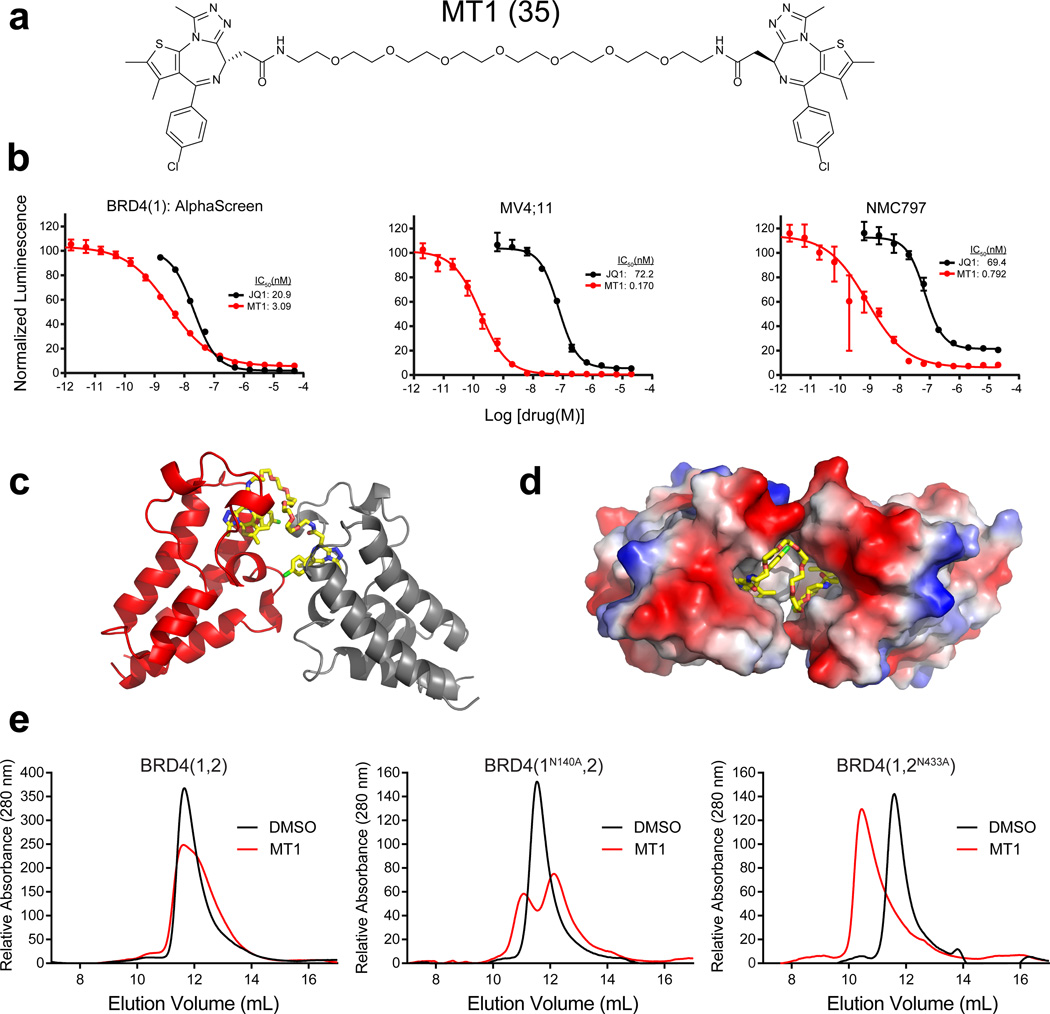
Although JQ1 has acceptable PK properties for animal studies, the in vivo plasma half-life of the hetero-dimer (6S+2S) was extremely short, perhaps suggesting extensive metabolism of the newly introduced linker section or the ester (Supplementary Fig. 6a and 6b)31. We therefore sought a biostable derivative to support use as a chemical probe in vivo. During linker optimization, we learned that the homo-dimer (6S+6S) derivative with a long PEG7 linker (hereafter referred to as MT1, 35) has comparable activity (IC50 = 3.09 nM for BRD4(1)) to the hetero-dimer molecules explored earlier, but improved PK properties (Fig. 4a and Supplementary Fig. 7a). MT1 does not possess ester moieties, which are prone to metabolism. We also note that the hetero-dimer (6S+2S) with a short “PEG0” (ethylene diamine) linker ((6S+2S)-PEG0) has comparable PK properties to MT1 (Supplementary Fig. 7b). The biochemical activity of MT1 translated well in cellular activities and the compound was confirmed to be selective for the BET family (Fig. 4b and Supplementary Table 1). We have also found MT1 to exhibit few off-target effects on cellular receptors and ion channels, among more than 50 tested. As with the parent scaffold JQ1, only partial inhibition of binding of a [Nleu-10]-NKA radioligand agonist for the neurokinin NK2 receptor was observed (Supplementary Fig. 8)16. These data encouraged us to further profile the molecule.
Figure 4. MT1: an intramolecular bivalent chemical probe of BET bromodomains.
(a) Chemical structure of MT1. (b) BRD4(1) biochemical and MV4;11 and NMC797 cellular activity of MT1. Biochemical and cellular data points represent the mean of at least 2 or 4 replicates respectively ± SD. (c) Cocrystal structure of MT1 bound to two BRD4(2) monomers (PDB: 5JWM).(d) Cocrystal structure illustrating the hydrophobic pocket where MT1 binds between two bromodomain monomers. (e) Size-exclusion chromatography of wild-type and mutant (N140A or N433A) tandem bromodomain constructs with MT1.
To assess whether MT1 dimerizes monomeric BET bromodomains, we adapted an AlphaScreen™ assay to estimate ligand-induced proximity of two differentially tagged BRD4(1) constructs. Luminescence transfer was observed between GSH-donor beads and nickel-acceptor beads when GST-BRD4(1) and His-BRD4(1) constructs were incubated with MT1. Increase in signal over baseline levels was observed in the micromolar range, similar to the previously described molecules above (Supplementary Fig. 9). The luminescent signal deteriorated at higher concentrations, characteristic of the Hook effect seen with multivalent inhibitors32.
To establish the binding mode of MT1, we attempted to solve cocrystal structures of the ligand in complex with purified, recombinant BRD4(1) and BRD4(2). We successfully obtained a cocrystal structure of BRD4(2) when excess MT1 was added (for data collection and refinement statistics see Supplementary Table 2) (Fig. 4c, 4d, and Supplementary Fig. 10). A similarly high concentration of MT1 was not compatible with BRD4(1) crystallization. The determined high-resolution structure clearly revealed one molecule of MT1 simultaneously recognizing two bromodomains of BRD4(2) (Fig. 4c). MT1 binding established a newly created hydrophobic pocket between the two bromodomain monomers (Fig. 4d). Overall, the binding mode of each warhead is very close to that of JQ1 in BRD4(1) (Supplementary Fig. 10)16. Each triazole ring formed a hydrogen bond with the evolutionarily conserved asparagine and exhibited great shape complementarity with the Kac binding site, occupying the entire binding pocket. Again, these findings support a bromodomain dimerization-based avidity effect for MT1 efficacy.
To address whether MT1 binds to BRD4 in an intramolecular or intermolecular fashion we once again employed SEC, this time using tandem bromodomain constructs that are either wild-type or have a point mutation at one of the key conserved asparagine residues in each bromodomain binding pocket (N140A in BRD4(1) and N433A in BRD4(2)), rendering them unable to bind JQ19. Complete dimerization was observed for the second bromodomain mutant (BRD4(1,2N433A)) when half an equivalent of MT1 was added in contrast to the absence of dimerization when the wild-type BRD4(1,2) protein was used. Likewise partial dimerization was observed for the BRD4(1N140A,2) construct (Fig. 4e). These results support a model whereby MT1 binds to tandem bromodomains in an intramolecular fashion and that both BRD4(1) and BRD4(2) are directly involved in this binding event.
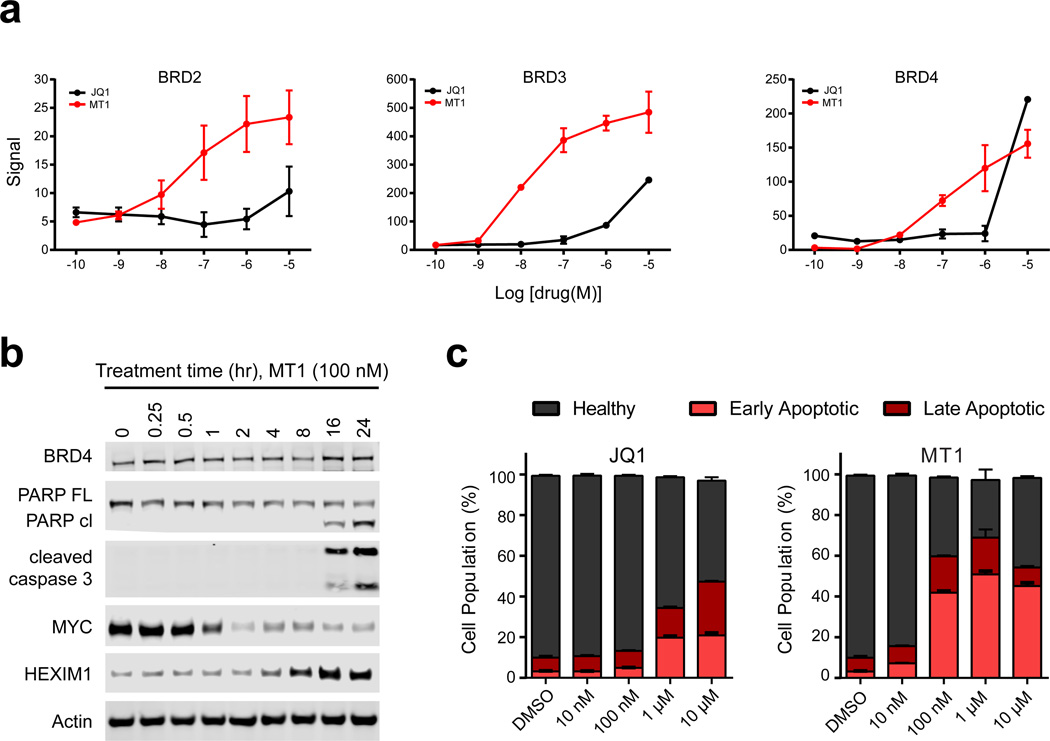
To establish target engagement of MT1 in a cellular context we performed a cellular thermal shift assay (CETSA)33. In the CETSA assay cells are treated with vehicle or drug, heated to denature and precipitate proteins, and lysed. Cellular debris and aggregates are separated from the soluble protein fraction by centrifugation. Whereas unbound proteins denature and precipitate at elevated temperatures, ligand-bound proteins remain in solution. Stabilized protein in the supernatant was measured by quantitative immunoblotting. Both JQ1 and MT1 stabilized BRD2, 3 and 4 in a concentration dependent manner although MT1 was able to bind and stabilize BET proteins at lower concentrations (Fig. 5a and Supplementary Fig. 11). These results effectively demonstrate MT1 cellular target engagement of BET family proteins.
Figure 5. Kinetic and anti-tumor advantage of dual-BET bromodomain inhibition.
(a) Cellular thermal shift assay (CETSA) of MT1 and JQ1 in MV4;11 cells; technical triplicates, mean ± SEM. Blots can be found in Supplementary Figure 11. (b) Immunoblot for BRD4, PARP-cleavage, cleaved caspase 3, MYC, HEXIM1 and actin from MV4;11 cells treated with 100 nM of MT1 for the indicated times. Full blots can be found in Supplementary Figure 15. (c) Percent early and late apoptotic and healthy MV4;11 cells after treatment with either MT1, JQ1, or DMSO for 24 hours as analyzed by Annexin-V and propidium iodide staining and flow cytometric analysis. Error bars represent the mean ± SD of three biological replicates.
Next, we assessed the effect of MT1 on cancer cell viability. Significant apoptosis was observed by caspase-3 and PARP cleavage after treatment with MT1 (Fig. 5b). These cellular events followed after HEXIM1 upregulation and MYC downregulation. Early and late apoptosis were assessed with annexin-V and propidium iodide staining to compare apoptosis induction between MT1 and JQ1 (Fig. 5c and Supplementary Fig. 12). Importantly, MT1 induced a greater degree of apoptosis at 10-fold lower concentrations than JQ1.
MT1 possesses all desirable qualities of a chemical probe34, such as high target potency in homogeneous and cellular assays, a well-characterized profile of selectivity, and synthetic accessibility (for synthetic schemes of MT1 see the Supplementary Note).
Antitumor efficacy of MT1 in leukemia xenograft models
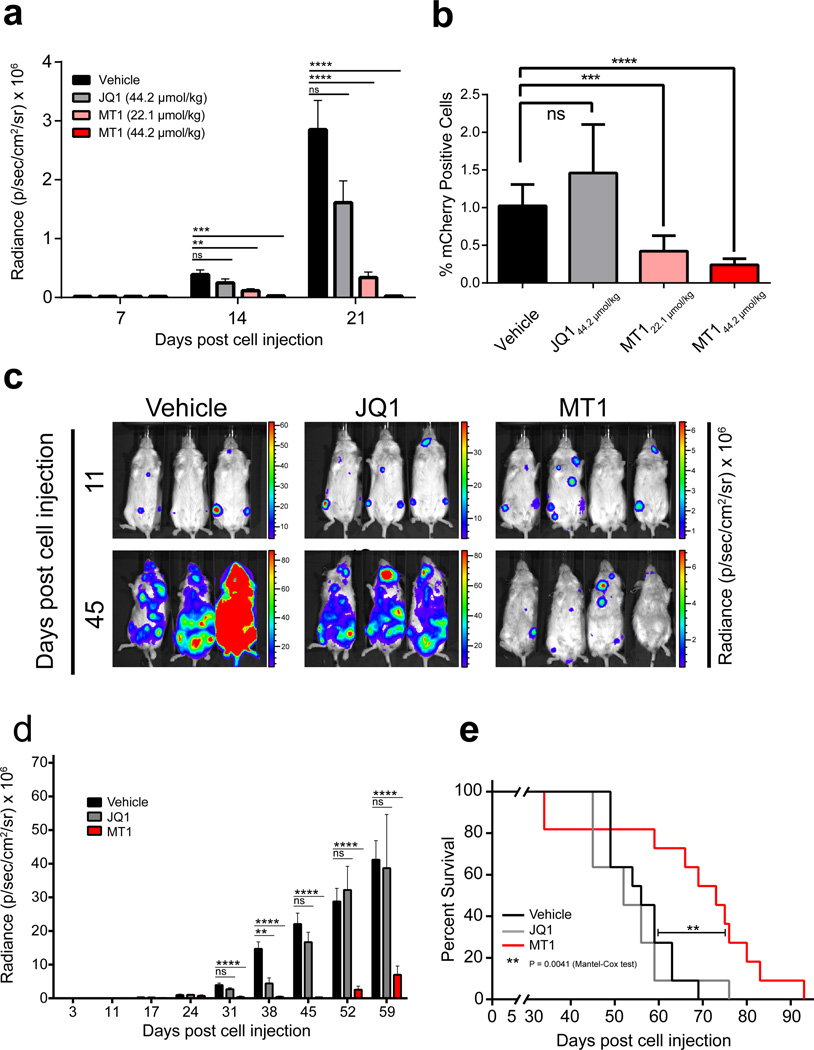
To determine whether MT1 could attenuate the growth of BRD4-dependent leukemia as a single agent in vivo, we selected an aggressive disseminated leukemia model (mCherry+, Luciferase+, MV4;11) and treated animals with established disease using equimolar (44.2 µmol/kg) and half an equivalent (22.1 µmol/kg) of MT1 compared to JQ1 for 14 days. During the study, leukemic burden was monitored by non-invasive bioluminescence imaging. Even half an equivalent of MT1 significantly reduced leukemic burden over the course of the study compared to either vehicle or JQ1 (Fig. 6a). Post-mortem analysis of leukemic burden in bone marrow by FACS also revealed significantly decreased mCherry+ disease with MT1 administration at 22.1 µg/kg (Fig. 6b).
Figure 6. Efficacy of dual-BET bromodomain inhibition in vivo.
(a) Luminal radiance (mean ± SEM) of mice treated with either MT1 at 44.2 µmol/kg (n=8), 22.1 µmol/kg (n=9), JQ1 at 44.2 µmol/kg (n=8) or vehicle (n=8) from day 7 to day 21 after initial leukemic cell injection of MV4;11 cells in a disseminated xenograft model. (b) Percentage of mCherry+ leukemic cells (means ± SEM) in flushed bone marrow from mice in (a). (c) Images of luminal radiance of three representative cages before (day 11) and after (day 45) treatment. MT1 (n = 11), JQ1 (n = 11), or vehicle (n = 11) was administered once daily at 44.2 µmol/kg from days 17–28 and days 31–35 after initial leukemia cell injection of MV4;11 cells in a disseminated xenograft model. (d) Luminescence (mean ± SEM) of MT1, JQ1 or vehicle treated mice from (c). (e) Kaplan–Meier survival summary plot for MT1, JQ1 and vehicle-treated mice from (c). ns = not significant, *P = 0.01 to 0.05, **P = 0.001 to 0.01, ***P = 0.0001 to 0.001, ****P <0.0001. (Kaplan-Meier: Mantel-Cox test. All other comparisons: unpaired two-tailed Welch’s t test, no correction for multiple comparisons).
We performed a second study to assess the survival advantage endowed by MT1 over JQ1 or vehicle using a similar xenograft model where mice were treated with equimolar concentrations of JQ1 and MT1 for 12 days (44.2 µmol/kg). After a two-day drug holiday we continued treatment for another 5 days. At this high dose of drug, two mice (of 11) in the MT1 group needed to be sacrificed due to emaciation from drug toxicity. Drug administration was stopped after this point and survival of the remaining mice was monitored. It is notable that those mice that had lost weight on MT1 recovered body weight after drug was withdrawn (Supplementary Fig. 13). As previously, disease monitored by bioluminescence was significantly reduced with MT1 treatment compared to JQ1 (Fig. 6c and 6d). Importantly MT1 significantly increased overall survival compared to vehicle or JQ1 treated mice (Fig. 6e).
DISCUSSION
In this study, we tested a structural hypothesis regarding multivalent recognition of BET family bromodomain proteins by bivalent organic ligands. Encouraged by the fact that both intramolecular (e.g. PSD-95 PDZ domains) and intermolecular (e.g. CBFβ-SMMHC) bivalent inhibitions are successfully accomplished by linking two monomeric ligands via PEG linkers4,5, we adopted PEG linkers to the monovalent BET antagonist JQ1 substituted off two separate sites. Initial homodimeric derivatives failed to achieve increased biochemical or cellular potency due to improper linker length, low permeability, or inability to dimerize target protein. However, hetero-dimeric (6S+2S) derivatives showed greatly improved activity both biochemically and in cells with little linker length dependence. Inversion of either stereocenter reverted heterodimeric compounds to JQ1-like potency, and the enantiomer (6R+2R) analog resulted in almost total loss of activity, suggesting a target specific effect. A single warhead analog with a PEG1 linker (2S-PEG1) did not increase activity, excluding the possibility that the newly introduced linker strongly interacts with the protein. The efficacy of (6S+2S)-PEG1 extended to both BRD4(1) and BRD4(2) as well as full length constructs. The combination between I-BET151 and JQ1 also increased potency, although to a lesser extent, revealing feasibility of the dual warhead strategy.
Improved activity of dimeric compounds is likely due to their ability to dimerize bromodomains as evidenced by SEC, ITC, and nanomaterial-based proximity assays. In particular, SEC with bromodomain mutants supports binding of bivalent compounds avidly via simultaneous intramolecular recognition of tandem bromodomains. Despite JQ1 and a bivalent ligand having similar dissociation constants for a single bromodomain, bivalent compounds had profoundly greater effects in cells (CETSA and cell proliferation), further supporting the conclusion that there is a functional avidity effect in play in cellular contexts. These bivalent inhibitors have pronounced efficacy and rapid kinetic downregulation of MYC in cultivated human cancer cells, establishing a rationale for drug-like derivatives to be advanced to human clinical investigation. Indeed, our best optimized bivalent inhibitor MT1 exhibits a 400-fold improvement in activity in AML compared to JQ1, and highly prolonged exposure in vivo. It is noteworthy that improvements of metabolic stability and PK properties are accomplished by varying the linker length and the attachment points to create a functional in vivo probe, despite MT1’s rather large size in comparison to canonical small molecule inhibitors (molecular weight 1134 Da). Significantly, although 44.2 µmol administration of JQ1 did not decrease leukemic burden in the mouse model, a half equivalence of MT1 (22.1 µmol) profoundly reduced leukemic burden, supporting functional avidity even in an in vivo setting.
A pressing need exists for the development of qualified probes of transcriptional and epigenomic proteins. Among these compelling targets are epigenetic reader proteins, which function through protein-protein interactions with post-translationally modified chromatin and transcription factors. Inhibitors of individual protein-protein interactions are historically difficult to realize, but epigenetic reader proteins commonly possess multivalent recognition modules. Among the 46 bromodomain-containing proteins in the human proteome, some of them outside the BET family (e.g. BRWD3, TAF1, and PB1) contain more than one bromodomain35,36. Importantly, our work shows that only the potencies for the on-target proteins of the parent monovalent scaffold were profoundly increased by bivalent inhibition, while the activities for off-targets did not appreciably increase. It may be possible to apply our strategy to promiscuous monovalent bromodomain inhibitors, such as bromosporine37 to selectively improve the activities that are associated with only the muti-bromodomain-containing proteins that it targets. We therefore anticipate the strategy taken herein for bivalent BET inhibition to guide efforts in epigenetic drug discovery more broadly.
ONLINE METHODS
Cell culture
MV4;11 cells were purchased from ATCC (CRL-9591), and NMC 797 cells were a kind gift from Dr. Christopher French (Brigham and Women’s Hospital), described previously38. Cells matched their expected cell type morphology. Cells were cultured at 37°C with 5% CO2 in either DMEM (NMC797) or RPMI1640 (MV4;11) (both Gibco) supplemented with 10 % FBS (Sigma), 100 U/mL penicillin, 100 µg/mL streptomycin, and 2 mM glutamine (Gibco). Cells tested negative for mycoplasma using the MycoAlert™ kit (Lonza). None of the utilized cell lines are among those that are commonly misidentified as listed by ICLAC.
BRD4(1) and BRDT(1) AlphaScreen™
Assays were performed as previously described with the following modifications: final concentrations of His-BRD4(1) or His-BRDT(1) (see protein expression section) were 20 nM39.
BRD4(1) dimerization AlphaScreen™
Assays were performed as previously described with the following modifications: final concentrations of His-BRD4(1) (see protein expression) and GST-BRD4(1) (see protein expression) were 500 nM and were added in 5 µL to 384-well plates (AlphaPlate-384 ProxiPlate, PerkinElmer)39. Following compound transfer, plates were allowed to incubate for 30 minutes before addition of Ni-coated Acceptor and GSH-coated Donor Beads. Data was analyzed using GraphPad PRISM.
BRD4(1) fluorescence polarization assay
In 384-well black plates (Nunc), 250 nM His-BRD4(1) (see protein expression section) and 20 nM JQ1-FITC were diluted in 20 µL assay buffer (50 mM HEPES, 150 mM NaCl, 0.01% w/v Tween20, pH 7.5) containing competitor compound or DMSO. Following 30 min incubation at RT, fluorescence polarization (mP) was measured using Envision 2104 Multilabel Reader (FP FITC dual optical module; Excitation: 480 nm, Emission: 535 nm for both S- and P-channels). Normalized mP values were calculated by setting the average background (no enzyme wells) to 0% the average DMSO wells to 100% activity. Standard deviations were determined from four replicate measurements for each compound concentration. Data were analyzed and plotted using GraphPad PRISM v6 and IC50 values were determined using the ‘log(inhibitor) vs normalized response –variable slope’ analysis module.
Analysis of cell viability by ATPLite
Cells were plated at 1000 cells/well in 50 µL/well of media in 384 well white culture plates (Thermo). NMC797 cells were allowed to adhere overnight before adding 100 nL of compound in DMSO from compound stock plates using a Janus Workstation pin tool (PerkinElmer). After addition of compound, plates were incubated for 72 hours at 37°C. Cell viability was read out using the ATPlite kit (PerkinElmer). Plates were brought to room temperature prior to reagent addition. Lyophilized powder was resuspended in lysis buffer and diluted 1:2 with DI water. 20 µL of this solution was added to each well and plates were incubated for 15 min at room temperature before signal was read on an Envision 2104 plate reader (Perkin Elmer).
Cellular Thermal Shift Assay (CETSA)
Compound or DMSO was incubated with 4x106 MV411 cells for 1 hr at 37°C. Cells were put on ice and washed with PBS and transferred to PCR tubes. Supernatant was aspirated to leave ∼5 µl in each tube and then cells were heat shocked in a thermocycler at 48.5°C for 3 min to denature proteins. Cells were then allowed to cool at RT for 3 min and then resuspended in 35 µl lysis buffer (50 mM Tris-HCl pH 7.5, 5% glycerol, 100 mM NaCl, 1.5 mM MgCl, 0.2% NP-40 + halt protease inhibitor cocktail (Thermo)) and freeze-thawed 3 times with liquid nitrogen to lyse cells. Lysates were then spun at 20,000xg for 20 min to clarify and pellet aggregated protein. Supernatant was boiled with LDS and split into three samples for immunoblotting. Bands were quantified using Image Studio™ software and plotted using GraphPad PRISM v6 as triplicate means ± SEM.
Xenograft experiments
MV4;11 human leukemia cells (mCherry+ and Luciferase+) were injected via tail-vein into immunocompromised (NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ, NSG) 8 week old female mice (2x10e6 cells/animal) purchased from The Jackson Laboratory (005557). Cells tested negative for mycoplasma and rodent infectious agents at Charles River Laboratories (Mouse Comprehensive Panel). Luminescence was utilized to monitor engraftment (evident 7 days after injection), at which point mice were randomly assigned into four cohorts that received MT1 at 44.2 µmol/kg (n=8), MT1 at 22.1 µmol/kg (n=9), JQ1 at 44.2 µmol/kg (n=8) or vehicle (n =8) formulated in a 10% captisol, 10% DMSO solution in water. Mice that failed to engraft were excluded from the study. Each group was dosed once daily for 14 days (day 7–21) and bioluminescence was monitored weekly at 7, 14 and 21 days post injection. At day 21, mice were sacrificed, bone marrow was extracted from both femurs of each mouse and leukemic burden was determined by measuring %mcherry+ cells with an LSRFortessa X-20 flow cytometer (Fig. 6b).
A second experiment was performed as above using 12 week old female mice (NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ, NSG) with engraftment evident at 17 days after injection, at which point mice were randomly assigned into three cohorts that received MT1 (n=11), JQ1 (n=11) or vehicle (n=11) treatment once daily for 12 subsequent days. A drug holiday was given for 2 days (day 29–30), Treatment resumed for 5 more days (31–35) and then ended. Mice were monitored and sacrificed when hind-limb paralysis was evident or body weight dropped by 15%. Both agents were dosed at equimolar concentrations of 44.2 µmol/kg (50 mg/kg and 20.2 mg/kg for MT1 and JQ1, respectively).
No statistical methods were used to predetermine sample size. The experiments were not randomized, and the investigators were not blinded to allocation during experiments and outcome assessment. Animal experiments were conducted following protocol 13–053 approved by the Dana-Farber Cancer Institute Animal Care and Use Committee and adherent to DFCI institutional standards.
Flow cytometry
For analysis of apoptotic cells, cells were washed with Annexin V binding buffer (140 mM NaCl, 10 mM HEPES, 2.5 mM CaCl2, pH 7.4) and then stained with 250 ng/mL FITC-Annexin V and 500 ng/mL propidium iodide (Thermo, P1304MP) in Annexin V binding buffer40. All centrifugation steps were performed at 400xg at 4°C for 5 minutes. Flow cytometry analyses were performed on an LSRFortessa X-20 flow cytometer (BD Biosciences) and all data analyzed with FlowJo software (v10, Tree Star).
Immunoblotting
Cells were lysed with RIPA buffer (Boston BioProducts) supplemented with halt protease inhibitor cocktail (Thermo) and 0.1% benzonase (Novagen) on ice for 20 minutes. The lysates were spun at 20,000xg for 15 minutes at 4°C. A BCA assay (Pierce) was used to quantify protein concentration. The following antibodies were used in this study: BRD4 (Bethyl labs, A301–985A, 1:5000 dilution), BRD3 (abcam, ab56342, 1:1000), BRD2 (Bethyl labs, A302–582A, 1:1000), c-MYC (Santa Cruz, sc-764, 1:1000), actin (Santa Cruz, sc-8432, 1:1000), HEXIM1 (Cell Signaling, 9064S, 1:1000), PARP (Cell Signaling, 9542S, 1:1000) and cleaved caspase 3 (Cell Signaling, 9579S, 1:1000). Blots were imaged after incubating with fluorescence-labeled secondary antibodies anti-mouse-680 (LI-COR, 926–68070, 1:7000) or anti-rabbit-800 (LI-COR, 926–32211, 1:7000) on the OdysseyCLxImager (LI-COR).
Protein expression and purification
Human BRD4 covering residues 44–168 (His-BRD4(1)) or 333–460 (His-BRD4(2)) in the pNIC28Bsa4 vector (Addgene) were overexpressed in E. coli BL21 (DE3) as previously described28. For His-BRD4(1) cell pellets were sonicated in buffer A (50mM HEPES pH 7.4, 400 nM NaCl, 1mM BME, 10 mM imidazole) and for His-BRD4(2) cell pellets were sonicated in buffer B (50 mM HEPES pH 7.5, 300 mM NaCl, 10% glycerol, 10 mM imidazole, 3 mM BME) and the resulting lysate was centrifuged at 30,000 xg for 30 min. Ni-NTA beads (Qiagen) were mixed with lysate supernatant for 30 min and washed with buffer A or B. Beads were transferred to an FPLC-compatible column and the bound protein was washed with 15% buffer C (50mM hepes pH 7.4, 400 nM NaCl, 1mM BME, 500 mM imidazole) or buffer D (50 mM HEPES pH 7.5, 300 mM NaCl, 10% glycerol, 300 mM imidazole, and 3 mM BME) and eluted with 100% buffer C or D. His-BRD4(1) was dialyzed against 20 mM HEPES pH 7.5, 150 mM NaCl, 1 mM BME and frozen at –80°C for use in AlphaSceen™ and FP assays. For crystallography studies of BRD4(2) TEV was added to the eluted protein and incubated at 4°C overnight. The sample was then passed through a desalting column (26/10 column) pre-equilibrated with buffer B without imidazole, and the eluted protein was subjected to a second Ni-NTA step to remove the His-tag and TEV site. The eluent was concentrated and passed through a Superdex 200 10/300 column (GE healthcare) in a buffer containing 20 mM HEPES 7.5, 150 mM NaCl, and 1 mM DTT. Fractions were pooled, concentrated to 14 mg/ml (BD1) or 48 mg/ml (BD2), and frozen at –80°C. For tandem domain SEC assays a construct of human BRD4 covering residues 1–463 (His-BRD4(1,2)) or similar constructs with N140A (His-BRD4(1N140A,2)) and N433A (His-BRD4(1,2N433A)) mutations in the pDEST17 vector (Invitrogen) was overexpressed in E. coli BL21 (DE3) in LB medium in the presence of 50 mg/ml of carbenicillin. Cells were grown at 37°C to an OD of 0.8, cooled to 17°C, induced with 200 µM isopropyl-1-thio-D-galactopyranoside (IPTG), incubated overnight at 17°C, collected by centrifugation, and stored at –80°C. Cell pellets were resuspended in buffer E (50mM HEPES pH 8.0, 300 mM NaCl, 10% glycerol) and then lysozyme was added to a concentration of 0.33 mg/mL, and the pellet incubated at RT for 30 minutes and then sonicated. The resulting lysate was centrifuged at 30,000 xg for 30 min. 4 mL of HIS-Select HF nickel affinity beads (Sigma, H0537) were applied to a 14 x 1.5 cm Econo-Pac Column (Bio-Rad, 9704652) and washed with buffer E + 20 mM imidazole. Lysate supernatant was gravity filtered through the column at 4°C and washed with buffer A + 20 mM imidazole. His-tagged proteins were eluted with 4mL of buffer A + 500 mM imidazole. Proteins were further purified on a HiLoad 16/600 Superdex 200 (GE healthcare), and eluted with buffer E. Fractions were pooled, concentrated to 2.4 mg/ml and frozen at –80°C.
For BRD4(1) dimerization AlphaSceen™ assays a construct of human BRD4 covering residues 2–170 (GST-BRD4(1)) in a gateway compatible pgex-6p-1 vector (Amersham) was overexpressed in E. coli BL21 (DE3) in LB medium in the presence of 50 mg/ml of carbenicillin. Cells were grown at 37°C to an OD of 0.6, induced with 500 µM isopropyl-1-thio-D-galactopyranoside (IPTG), incubated for 4 hr at 37°C, collected by centrifugation, and stored at –80°C. Cell pellets were resuspended in buffer E (50mM HEPES pH 8.0, 300 mM NaCl, 10% glycerol) and then lysozyme was added to a concentration of 0.33 mg/mL, and the pellet incubated at RT for 30 minutes and then sonicated. The resulting lysate was centrifuged at 30,000 xg for 30 min. Supernatant was added to 2 mL of packed Glutathione Sepharose® 4B (GE Healthcare, 17-0756-01) beads and incubated overnight at 4°C and then purified in batch mode with buffer E + 16 mM glutathione. Batches were pooled and then dialyzed against buffer E to get rid of contaminating glutathione and frozen at –80°C at a concentration of 3.23 mg/ml.
His-BRDT(1) was obtained as described previously16. An SDS page gel showing the purity of each recombinant protein can be found in Supplementary Figure 14.
Crystallization, data collection and structure determination
A half equivalence of MT1 (10 mM in DMSO) was mixed with 500 µM protein and crystallized by sitting-drop vapor diffusion at 20 °C in the following crystallization buffer: 2 M NH4SO4 and 0.1 M BisTris pH 5.5. Crystals were transferred briefly into crystallization buffer containing 25% glycerol prior to flash-freezing in liquid nitrogen. Diffraction data from complex crystals were collected at beamline 24ID-E of the NE-CAT at the Advanced Photon Source (Argonne National Laboratory). Data sets were integrated and scaled using XDS41. Structures were solved by molecular replacement using the program Phaser and the search model PDB entry 3MXF42. The ligand was positioned and preliminarily refined using Buster and Rhofit43. Iterative manual model building and refinement using Phenix and Coot led to a model with excellent statistics44,45. The solved structure has been submitted to the PDB as 5JWM.
Size-Exclusion Chromatography
The oligomeric state of the BRD4(1) in solution was analyzed by gel filtration in a buffer containing 20 mM Hepes pH 7.5, 150 mM NaCl using a Superdex 200 10/300GL column (GE Healthcare) calibrated with globular proteins of known molecular weight (GE Healthcare, 28-4038-41/42). Protein (50 µM) and ligands (6S+2S)-PEG1 (25 µM), (6R+2R)-PEG1 (100 µM), JQ1 (100 µM) or DMSO were mixed and incubated at 20°C for 20 min before injection. Eluting peaks were monitored using ultraviolet absorbance at 280 nm. BRD4(1,2) and domain mutants BRD4(1N140A,2) and BRD4(1,2N433A) corresponding to the N140A and N433A mutants respectively were analyzed using the same method but 42 µM of protein was used with a half equivalence of MT1.
Isothermal Titration Calorimetry
Experiments were carried out on an Auto-ITC200 titration microcalorimeter (Malvern Instruments). All experiments were carried out at 25 °C while stirring at 1000 rpm, in ITC buffer (50 mM HEPES pH 7.4 at 25°C, 150 mM NaCl). The microsyringe was loaded with a solution of the ligand sample. All titrations were conducted using an initial injection of 0.4 µl followed by 19 identical injections of 2.0 µl with a duration of 4 sec (per injection) and a spacing of 120 sec between injections. The heat of dilution was determined by independent titrations (ligand into buffer) and was subtracted from the experimental data. The collected data were analyzed in the MicroCal™ Origin software supplied with the instrument to yield enthalpies of binding (ΔH) and binding constants (KB) as previously described46. Thermodynamic parameters were calculated (ΔG = ΔH – TΔS = –RTlnKB, where ΔG, ΔH and ΔS are the changes in free energy, enthalpy and entropy of binding respectively). In all cases a single binding site model was employed.
Receptor Profiling Studies
Selectivity profiling (ExpresSProfile) was performed on MT1 at 1 µM against 55 ligand receptors, ion channels, and transport proteins by CEREP (Eurofins) using manufacturer’s protocols.
Statistical Methods
No statistical methods were used to predetermine sample size. The experiments were not randomized, and the investigators were not blinded to allocation during experiments and outcome assessment. For all experiments, number of replicates (n), center values, error bars, and p-value cutoffs are described in the respective figure legends. Error bars are shown for all data points with replicates as a measure of variation within each data group. All t-tests performed were Welch’s t-tests that allows for unequal variance and distributions assumed to follow a Student’s t distribution. These assumptions are not contradicted by the data. All t-tests were two-sided.
Supplementary Material
Acknowledgments
We thank J. A. Perry for critical reading of the manuscript and discussions. Crystallographic work was based upon research conducted at the Advanced Photon Source on the Northeastern Collaborative Access Team beamlines. We thank non-research support of M.T. by Mitsubishi Tanabe Pharma Corporation. This research was supported by the William Lawrence & Blanche Hughes Foundation (J.E.B.), The Leukemia & Lymphoma Society (J.E.B.) and the National Institutes of Health (CA066996-01A1, J.E.B.).
Footnotes
ACCESSION CODES
Atomic coordinates and structure factors for the reported cocrystal structures have been deposited with the Protein Data Bank under accession code 5JWM (BRD4(2)–MT1).
AUTHOR CONTRIBUTIONS
M.T. and J.M.R. designed and led the study under the supervision of J.E.B. M.T. designed bivalent molecules and performed organic chemistry and medicinal chemistry experiments. J.M.R. designed and performed biochemical, cellular, and in vivo experiments. J.P. performed BRD4(1) FP experiments. S.D., H.S., and S.L.D. performed crystallographic studies. H.S. performed SEC and ITC experiments. A.S. and T.G.S. performed in vivo studies. M.T., J.M.R., and J.E.B. wrote the manuscript with input from all authors.
COMPETING FINANCIAL INTERESTS
The authors claim the following competing financial interests: M.T. is a visiting scientist from Mitsubishi Tanabe Pharma Corporation and supported by the company for non-research funds. J.E.B. is a founder of Tensha Therapeutics, a biotechnology company that develops drug-like derivatives of JQ1 as investigational cancer therapies. Dana-Farber Cancer Institute has filed patent applications (62/259,797) that include MT1 and its analogs described in this manuscript. J.E.B. is currently an employee of the Novartis Institutes of BioMedical Research, effective January 1, 2016.
REFERENCES
- 1.Mammen M, Choi S-K, Whitesides GM. Polyvalent Interactions in Biological Systems: Implications for Design and Use of Multivalent Ligands and Inhibitors. Angewandte Chemie International Edition. 1998;37:2754–2794. doi: 10.1002/(SICI)1521-3773(19981102)37:20<2754::AID-ANIE2754>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 2.Monsigny M, Mayer R, Roche AC. Sugar-lectin interactions: sugar clusters, lectin multivalency and avidity. Carbohydrate letters. 2000;4:35–52. [PubMed] [Google Scholar]
- 3.Kiessling LL, Gestwicki JE, Strong LE. Synthetic multivalent ligands in the exploration of cell-surface interactions. Current opinion in chemical biology. 2000;4:696–703. doi: 10.1016/s1367-5931(00)00153-8. [DOI] [PubMed] [Google Scholar]
- 4.Bach A, et al. Design and Synthesis of Highly Potent and Plasma-Stable Dimeric Inhibitors of the PSD-95-NMDA Receptor Interaction. Angewandte Chemie International Edition. 2009;48:9685–9689. doi: 10.1002/anie.200904741. [DOI] [PubMed] [Google Scholar]
- 5.Illendula A, et al. Chemical biology. A small-molecule inhibitor of the aberrant transcription factor CBFbeta-SMMHC delays leukemia in mice. Science. 2015;347:779–784. doi: 10.1126/science.aaa0314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Profit AA, Lee TR, Lawrence DS. Bivalent Inhibitors of Protein Tyrosine Kinases. Journal of the American Chemical Society. 1999;121:280–283. [Google Scholar]
- 7.Sun H, et al. Design, synthesis, and characterization of a potent, nonpeptide, cell-permeable, bivalent Smac mimetic that concurrently targets both the BIR2 and BIR3 domains in XIAP. J Am Chem Soc. 2007;129:15279–15294. doi: 10.1021/ja074725f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bach A, et al. A high-affinity, dimeric inhibitor of PSD-95 bivalently interacts with PDZ1-2 and protects against ischemic brain damage. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:3317–3322. doi: 10.1073/pnas.1113761109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Filippakopoulos P, Knapp S. Targeting bromodomains: epigenetic readers of lysine acetylation. Nature reviews. Drug discovery. 2014;13:337–356. doi: 10.1038/nrd4286. [DOI] [PubMed] [Google Scholar]
- 10.Zeng L, Zhou MM. Bromodomain: an acetyl-lysine binding domain. FEBS letters. 2002;513:124–128. doi: 10.1016/s0014-5793(01)03309-9. [DOI] [PubMed] [Google Scholar]
- 11.Smith SG, Zhou MM. The Bromodomain: A New Target in Emerging Epigenetic Medicine. ACS chemical biology. 2016;11:598–608. doi: 10.1021/acschembio.5b00831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zuber J, et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature. 2011;478:524–528. doi: 10.1038/nature10334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Delmore JE, et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146:904–917. doi: 10.1016/j.cell.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang Z, et al. Recruitment of P-TEFb for stimulation of transcriptional elongation by the bromodomain protein Brd4. Molecular cell. 2005;19:535–545. doi: 10.1016/j.molcel.2005.06.029. [DOI] [PubMed] [Google Scholar]
- 15.Krueger BJ, Varzavand K, Cooper JJ, Price DH. The mechanism of release of P-TEFb and HEXIM1 from the 7SK snRNP by viral and cellular activators includes a conformational change in 7SK. PloS one. 2010;5:e12335. doi: 10.1371/journal.pone.0012335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Filippakopoulos P, et al. Selective inhibition of BET bromodomains. Nature. 2010;468:1067–1073. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tanaka M, Roberts JM, Qi J, Bradner JE. Inhibitors of emerging epigenetic targets for cancer therapy: a patent review (2010–2014) Pharmaceutical patent analyst. 2015;4:261–284. doi: 10.4155/ppa.15.16. [DOI] [PubMed] [Google Scholar]
- 18.Schroder S, et al. Two-pronged binding with bromodomain-containing protein 4 liberates positive transcription elongation factor b from inactive ribonucleoprotein complexes. The Journal of biological chemistry. 2012;287:1090–1099. doi: 10.1074/jbc.M111.282855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baud MG, et al. Chemical biology. A bump-and-hole approach to engineer controlled selectivity of BET bromodomain chemical probes. Science. 2014;346:638–641. doi: 10.1126/science.1249830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Picaud S, et al. RVX-208, an inhibitor of BET transcriptional regulators with selectivity for the second bromodomain. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:19754–19759. doi: 10.1073/pnas.1310658110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang G, et al. Down-regulation of NF-kappaB transcriptional activity in HIV-associated kidney disease by BRD4 inhibition. The Journal of biological chemistry. 2012;287:28840–28851. doi: 10.1074/jbc.M112.359505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McKeown MR, et al. Biased multicomponent reactions to develop novel bromodomain inhibitors. Journal of medicinal chemistry. 2014;57:9019–9027. doi: 10.1021/jm501120z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roberts JM, Bradner JE. A Bead-Based Proximity Assay for BRD4 Ligand Discovery. Current protocols in chemical biology. 2015;7:263–278. doi: 10.1002/9780470559277.ch150024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dahlin JL, et al. PAINS in the assay: chemical mechanisms of assay interference and promiscuous enzymatic inhibition observed during a sulfhydryl-scavenging HTS. Journal of medicinal chemistry. 2015;58:2091–2113. doi: 10.1021/jm5019093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dawson MA, et al. Inhibition of BET recruitment to chromatin as an effective treatment for MLL-fusion leukaemia. Nature. 2011;478:529–533. doi: 10.1038/nature10509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chaidos A, et al. Potent antimyeloma activity of the novel bromodomain inhibitors I-BET151 and I-BET762. Blood. 2014;123:697–705. doi: 10.1182/blood-2013-01-478420. [DOI] [PubMed] [Google Scholar]
- 27.Bartholomeeusen K, Xiang Y, Fujinaga K, Peterlin BM. Bromodomain and extra-terminal (BET) bromodomain inhibition activate transcription via transient release of positive transcription elongation factor b (P-TEFb) from 7SK small nuclear ribonucleoprotein. The Journal of biological chemistry. 2012;287:36609–36616. doi: 10.1074/jbc.M112.410746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Winter GE, et al. DRUG DEVELOPMENT. Phthalimide conjugation as a strategy for in vivo target protein degradation. Science. 2015;348:1376–1381. doi: 10.1126/science.aab1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lu J, et al. Hijacking the E3 Ubiquitin Ligase Cereblon to Efficiently Target BRD4. Chemistry & biology. 2015;22:755–763. doi: 10.1016/j.chembiol.2015.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zengerle M, Chan KH, Ciulli A. Selective Small Molecule Induced Degradation of the BET Bromodomain Protein BRD4. ACS chemical biology. 2015;10:1770–1777. doi: 10.1021/acschembio.5b00216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matzuk MM, et al. Small-molecule inhibition of BRDT for male contraception. Cell. 2012;150:673–684. doi: 10.1016/j.cell.2012.06.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dutta Roy R, Stefan MI. Cooperative Binding Mitigates the High-Dose Hook Effect. bioRxiv. 2015 doi: 10.1186/s12918-017-0447-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martinez Molina D, et al. Monitoring drug target engagement in cells and tissues using the cellular thermal shift assay. Science. 2013;341:84–87. doi: 10.1126/science.1233606. [DOI] [PubMed] [Google Scholar]
- 34.Arrowsmith CH, et al. The promise and peril of chemical probes. Nature chemical biology. 2015;11:536–541. doi: 10.1038/nchembio.1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schultz J, Copley RR, Doerks T, Ponting CP, Bork P. SMART: a web-based tool for the study of genetically mobile domains. Nucleic acids research. 2000;28:231–234. doi: 10.1093/nar/28.1.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sanchez R, Meslamani J, Zhou MM. The bromodomain: from epigenome reader to druggable target. Biochimica et biophysica acta. 2014;1839:676–685. doi: 10.1016/j.bbagrm.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Structural Genomics Consortium website. [accessed June 19, 2016]; http://www.thesgc.org/chemical-probes/Bromosporine.
ONLINE METHODS REFERENCES
- 38.French CA, et al. Midline carcinoma of children and young adults with NUT rearrangement. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2004;22:4135–4139. doi: 10.1200/JCO.2004.02.107. [DOI] [PubMed] [Google Scholar]
- 39.Anders L, et al. Genome-wide localization of small molecules. Nature biotechnology. 2014;32:92–96. doi: 10.1038/nbt.2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brumatti G, Sheridan C, Martin SJ. Expression and purification of recombinant annexin V for the detection of membrane alterations on apoptotic cells. Methods (San Diego, Calif.) 2008;44:235–240. doi: 10.1016/j.ymeth.2007.11.010. [DOI] [PubMed] [Google Scholar]
- 41.Kabsch W. Integration, scaling, space-group assignment and post-refinement. Acta crystallographica. Section D, Biological crystallography. 2010;66:133–144. doi: 10.1107/S0907444909047374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McCoy AJ, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smart OS, et al. Exploiting structure similarity in refinement: automated NCS and target-structure restraints in BUSTER. Acta crystallographica. Section D, Biological crystallography. 2012;68:368–380. doi: 10.1107/S0907444911056058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta crystallographica. Section D, Biological crystallography. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta crystallographica. Section D, Biological crystallography. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 46.Wiseman T, Williston S, Brandts JF, Lin LN. Rapid measurement of binding constants and heats of binding using a new titration calorimeter. Analytical biochemistry. 1989;179:131–137. doi: 10.1016/0003-2697(89)90213-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.