Abstract
Background
Melanoma skin cancer remains the leading cause of skin cancer related deaths. Spitz lesions represent a subset of melanocytic skin lesions characterized by epitheloid or spindled melanocytes organized in nests. These lesions occupy a spectrum ranging from benign Spitz and atypical Spitz lesions all the way to malignant Spitz tumors. Appropriate management is reliant on accurate diagnostic classification, yet this effort remains challenging utilizing current light microscopic techniques. The discovery of novel biomarkers such as microRNAs (miR) may ultimately be a useful diagnostic adjunct for the evaluation of Spitz lesions. miR expression profiles have been suggested for non-Spitz melanomas but have yet to be ascribed to Spitz lesions. We hypothesized that distinct miR expression profiles would be associated with different lesions along the Spitz spectrum.
Materials and Methods
RNA extracted from paraffin embedded, formalin fixed tissues of 11 resected skin lesions including benign nevi (n=2), benign Spitz lesions (n=3), atypical Spitz lesions (n=3), and malignant Spitz tumors (n=3) were analyzed by the NanoString platform for simultaneous evaluation of over 800 miRs in each patient sample.
Results
Benign Spitz lesions had increased expression of miR-21-5p and miR-363-3p compared to benign nevi. Malignant Spitz lesions exhibited over-expression of miR-21-5p, miR-155-5p, and miR-1283 relative to both benign nevi and benign Spitz tumors. Notably, atypical Spitz tumors had increased expression of miR-451a and decreased expression of miR-155-5p expression relative to malignant Spitz lesions. Conversely, atypical Spitz lesions had increased expression of miR-21-5p, miR-34a-5p, miR-451a, miR-1283, and miR-1260a relative to benign Spitz tumors.
Conclusions
Overall, distinct miR profiles are suggested among Spitz lesions of varying malignant potential with some similarities to non-Spitz melanoma tumors. This work demonstrates the feasibility of this analytic method and forms the basis for further validation studies.
Keywords: Spitz, Melanoma, MicroRNA
1. Introduction
Spitz melanocytic lesions were first described by Dr. Sophie Spitz in 1948 (1). These lesions are characterized by epithelioid or spindled shaped melanocytes arranged in nests (1). A spectrum exists, ranging from benign Spitz lesions to malignant Spitz melanomas (2). Herein, we refer to an intermediate category as atypical Spitz tumors although the terms indeterminate Spitz and melanocytic tumors of uncertain malignant potential (MELTUMP) have also been used interchangeably. There has been some difficulty surrounding the accurate diagnosis of these lesions based upon standard histopathologic analysis. Byrne et al. found discordance in the diagnosis of 36% of Spitz lesions when evaluated by different pathologists (3). Barnhill et al. suggested the rate may be higher than anticipated as their group reported dermatopathologists were unable to achieve diagnostic consensus in 16 of 17 Spitz lesions (4). The clinical ramifications of an inaccurate diagnosis can be immense. Improper pathologic designation of a benign Spitz tumor as an atypical Spitz lesion may promote a transition in clinical management from a non-operative approach to one involving surgical resection. Thereby, patients may unnecessarily be subjected to the morbidity of surgery. While the exact incidence of complete lymph node dissections for Spitz lesions in the United States is difficult to discern, a systematic review of 541 patients with atypical Spitz lesions revealed that sentinel lymph node biopsy was performed in 303 (56%) patients (5). 119 of these patients (39%) had a positive sentinel lymph node and the majority (97/119 patients) went on to receive a completion lymph node dissection (5). Importantly, 99% of patients with a positive sentinel lymph node were alive at a median follow-up of 59 months. This suggests that while sentinel lymph node biopsy in the setting of an atypical Spitz tumor is more commonly involved with microscopic disease than conventional cutaneous malignant melanoma (10-16%), sentinel lymph node involvement does not appear to be prognostic for atypical Spitz lesions (5). Roatan et al. found a 57% complication rate in melanocytic skin lesions requiring completion lymph node dissections, most commonly from lymphedema and infections (6). Conversely, misdiagnosis of an atypical lesion as a benign Spitz tumor may allow for disease progression through omission of surgical intervention. Ultimately, diagnostic adjuncts using novel biomarkers may be a valuable tool for clinicians faced with diagnostic uncertainty.
MicroRNAs (miR) are 19-22 nucleotide, non-coding RNA molecules that inhibit protein translation. Over 800 different microRNAs are present within human cells and the expression profile of these miRs may vary across neoplastic lesions at different stages of malignancy (7). Our group has previously shown malignant, (non-Spitz) melanomas have over-expression of miR-21 and miR-155 compared to benign nevi (8). Similarly, Jiang et al. discovered a sequential increase in miR-21 expression between benign nevi to primary melanoma and from primary melanoma to metastatic melanoma (P<0.05) (9). Exploitation of differential miR expression patterns has been suggested as a diagnostic adjunct for other malignancies including gastric cancer, colon cancer, and hepatocellular carcinoma (10-12).
The miR profile of Spitz lesions remains unknown. We hypothesize distinct miR expression profiles are present within lesions along the Spitz spectrum. Discovery of a differential expression profile between benign Spitz, atypical Spitz, and malignant Spitz may serve as a diagnostic aid to standard histopathology. Important clinical benefits may result from the establishment of an accurate diagnosis. In this study, the first use of NanoString nCounter miR expression profiling for global analysis of Spitz lesions is described.
2. Materials and Methods
2.1 Tissue Samples
Between 2006-2012, 11 adults with Spitz lesions or benign nevi that underwent biopsy or resection of primary lesions at The Ohio State University Wexner Medical Center were selected for analysis. 2-3 sections with sufficient paraffin embedded tumor were selected at random for further analysis. The diagnosis was confirmed using histopathologic evaluation by co-author SBP. Paraffin embedded tissue samples of lesions were obtained for further analysis. This work was conducted under the auspices of The Ohio State University IRB protocol 2007C0015 .
2.2 RNA Isolation and NanoString microRNA Profiling
RNA isolation from paraffin embedded samples was performed using RecoverAll Total Nucleic Acid Isolation Kit® (Ambion, Foster City, CA). 100 ng of isolated RNA was loaded onto a NanoString nCounter (NanoString Technologies, Seattle, WA) platform and miR quantification was carried out as previously described (13). Briefly, miRs were ligated with DNA tags to normalize melting temperatures and provide unique identification of each miR. Excess tags were removed and a panel of capture and reporter probes containing unique fluorescent signals corresponding to individual miRs were hybridized at 64°C for 18 hours. Immobilization of hybridized probes onto a streptavidin coated cartridge was performed using nCounter Prep Station (NanoString Technologies, Seattle, WA). The fluorescence of each hybridized miR was measured by an nCounter Digital analyzer (NanoString Technologies, Seattle, WA) with a high density scan containing 600 fields of view. The investigation included 5 negative and 5 positive synthetic controls containing sequences not homologous to any known organism as well as 5 housekeeping genes supplied by NanoString Technologies (Seattle, WA).
2.3 Technical normalization of NanoString nCounter microRNA expression
Six positive controls at known concentrations supplied by NanoString Technologies (Seattle, WA) were included in each nCounter microRNA expression assay to calculate a Normalization Factor and control for inter-lane variation. For each lane, the counts for all 6 positive controls were summed to generate a value representing the “Sum of Positives.” A Normalization Reference was calculated by taking the average of all Sum of Positives values. In order to generate a Normalization Factor for each lane, each Sum of Positives value was divided by the Normalization Reference. Normalized endogenous miR targets were obtained by multiplying each target count value by the Normalization Factor for that lane. The Normalization Factor is recommended to be between 0.3 and 3 (NanoString Technologies, Seattle, WA). One benign nevi sample that was outside of this range was removed from further analysis to yield 11 samples with reliable measurements.
2.4 Unsupervised Hierarchical Clustering of miRs
Broad Institute GENE-E software was used to conduct unsupervised hierarchical clustering across the four sample types using all miR expression features: http://www.broadinstitute.org/cancer/software/GENE-E/. Euclidean distance metric and the average linkage method was implemented for dendrogram construction.
2.5 Differential miR expression analysis and Statistics
edgeR software was used for differential expression in technically normalized miR expression data between sample groups at False Discovery Rate (FDR) <0.05. Implementation of edgeR analysis was conducted via the EDDA platform: http://edda.gis.a-star.edu.sg/dad/ (14).
2.6 Pathway enrichment analysis of miR target genes
Differentially expressed miRs discovered via edgeR analysis were mapped to their experimentally validated target genes contained within the miRTarBase, and the union of the enriched KEGG pathways were identified using a hypergeometric test and FDR-correction for p-values via the DIANA miRPath program (15-17). Dendrogram branches were generated to visualize clustered miRs and pathways.
3. Results
3.1 Clinical Results
10 females and 1 male were included in the sample and the average age of patients was 23.2 years old (Table 1). There were no local recurrences or deaths. Patient 7 had a right upper extremity lesion and was the only individual to undergo a sentinel lymph node biopsy. 0/6 nodes were positive for microscopic disease. She has been followed for 75 months during which she developed metachronous cutaneous lesions at different sites containing atypical foci of: vulva, left thigh, left arm, left arm and chest at 12, 54, 57, 68 and 70 months, respectively. Additionally, she developed malignant melanoma (T1b) of the left arm at 59 months and underwent a sentinel lymph node biopsy that revealed 0/2 nodes were positive for microscopic disease. She currently has no evidence of disease 16 months following resection of her malignant melanoma.
Table 1.
Clinical characteristics of patients.
| Patient | Age at Diagnosis | Sex | Primary Tumor Site | Sentinel Lymph Node Biopsy* | Local Recurrence | Survival | Follow-up (months) |
|---|---|---|---|---|---|---|---|
| Benign nevi | |||||||
| 1 | 18 | F | head | No | - | - | - |
| 2 | 36 | F | arm | No | none | alive | 26 |
| Benign Spitz | |||||||
| 3 | 18 | F | buttock | No | none | alive | 75 |
| 4 | 19 | M | back | No | - | - | - |
| 5 | 17 | F | face | No | - | - | - |
| Atypical Spitz | |||||||
| 6 | 20 | F | chest | No | - | - | - |
| 7 | 35 | F | arm | Yes- 0/6 positive | none | alive | 68 |
| 8 | 16 | F | leg | No | - | - | - |
| Spitzoid Melanoma | |||||||
| 9 | 26 | F | arm | No | none | alive | 92 |
| 10 | 20 | F | axilla | No | - | - | - |
| 11 | 30 | F | face | No | none | alive | 50 |
Clinical parameters of patients containing benign nevi, benign Spitz, atypical Spitz, and Spitzoid melanomas are shown.
Total number of sentinel lymph nodes containing tumor to total number of lymph nodes biopsied.
(-) Indicates no data available.
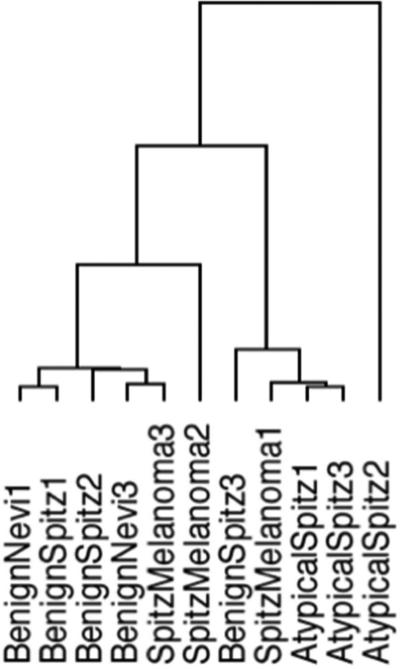
3.2. Unsupervised hierarchical clustering analysis of microRNA expression
In order to investigate how global patterns of miR expression could influence groupings of Spitz tumors and benign nevi, we performed unsupervised hierarchical clustering using all miR features across all samples (n=11) in this study. The resultant dendrogram from this analysis depicts the relative closeness of distinct patient samples (Figure 1). 4 out of 5 benign nevi and benign Spitz tumors are clustered together. Additionally, 2 of 3 atypical Spitz tumors are tightly clustered. Conversely, Spitz melanoma tumors do not appear to cluster together according to their global miR expression patterns, perhaps to due heterogeneous tumor biology among these select lesions.
Figure 1. Unsupervised hierarchical clustering of Spitz tumors and benign nevi across 805 miRNAs.
Unsupervised hierarchical clustering was performed on microRNA expression data of all samples using the Euclidean distance metric and average linkage method.
3.3 MicroRNA pattern distinguishes benign Spitz lesions
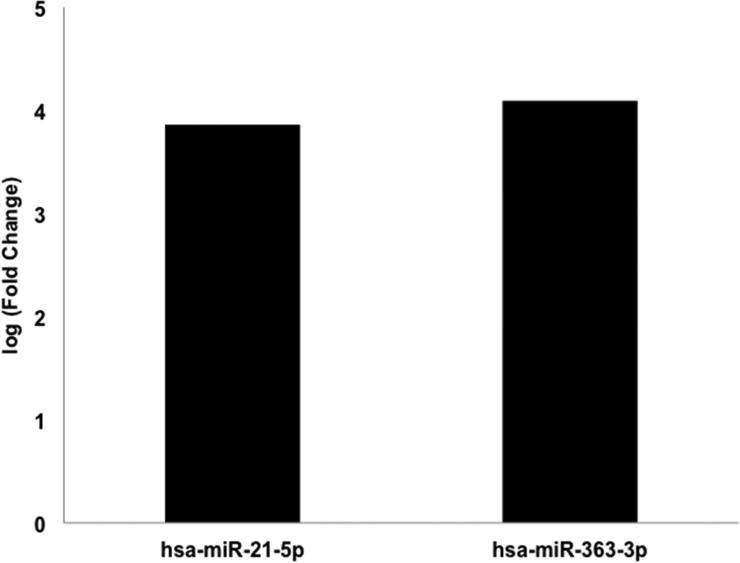
To identify differentially expressed miRs between tumor samples, miR expression within patient samples (n=11) was quantified using the NanoString platform and analyzed using the edgeR program. Differentially expressed miRs were considered significant at an FDR < 0.05 (Supplemental Tables S1-S6). To determine the miR expression profile suggestive of a Spitz lesion, benign Spitz tumors (n=3) were compared to benign nevi (n=2). Benign Spitz lesions exhibited an up-regulation of miR-21-5p and miR-363-3p relative to benign nevi (Figure 2) (FDR<0.05).
Figure 2. Differentially expressed microRNAs between benign Spitz and benign Nevi.
microRNA expression of benign Spitz (n=3) compared to benign nevi (n=2) was analyzed for 805 microRNAs by NanoString nCounter. Differential expression of microRNAs with False Discovery Rate (FDR) <0.05 are shown.
3.4 MicroRNA pattern distinguishing malignant Spitz lesions
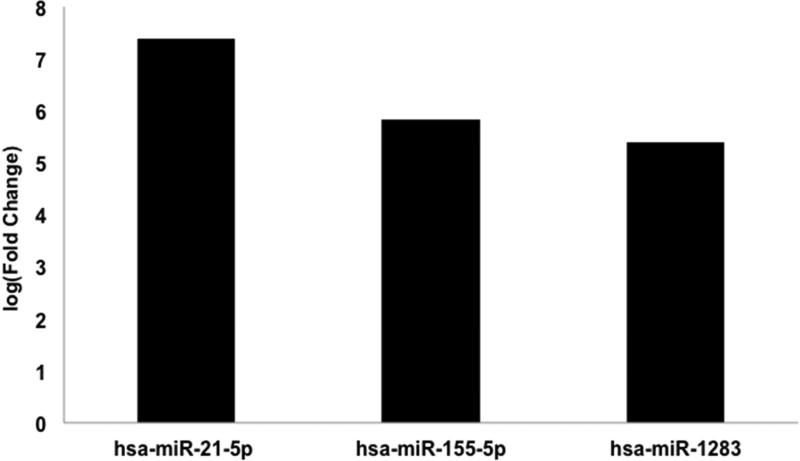
To characterize the miR profile of a malignant Spitz lesion, malignant Spitz tumors (n=3) were first compared to benign nevi (n=2). Similar to benign Spitz lesions, malignant Spitz tumors were also distinguished from benign nevi by up-regulation of miR-21-5p (Figure 3) (FDR<0.05). Additionally, malignant Spitz tumors had up-regulation of miR-155-5p and miR-1283 relative to benign nevi by (FDR<0.05). To confirm these findings, malignant Spitz melanomas (n=3) were compared to benign Spitz tumors (n=3). This comparison also revealed an increase in miR-21-5p, miR-155-5p, and miR-1283 expression associated with malignant Spitz tumors (Figure 4) (FDR<0.05).
Figure 3. Differentially expressed microRNAs between Spitz melanoma and benign Nevi.
microRNA expression of Spitz melanoma (n=3) compared to benign nevi (n=2) was analyzed for 805 microRNAs by NanoString nCounter. Differential expression of microRNAs with False Discovery Rate (FDR) <0.05 are shown.
Figure 4. Differentially expressed microRNAs between Spitz melanoma and benign Spitz.
microRNA expression of Spitz melanoma (n=3) compared to benign Spitz (n=3) was analyzed for 805 microRNAs by NanoString nCounter. Differential expression of microRNAs with False Discovery Rate (FDR) <0.05 are shown.
3.5 MicroRNA pattern distinguishing Atypical Spitz Lesions
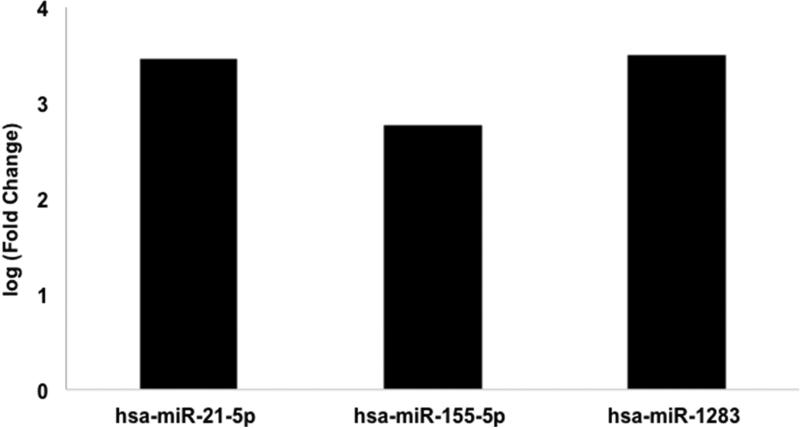
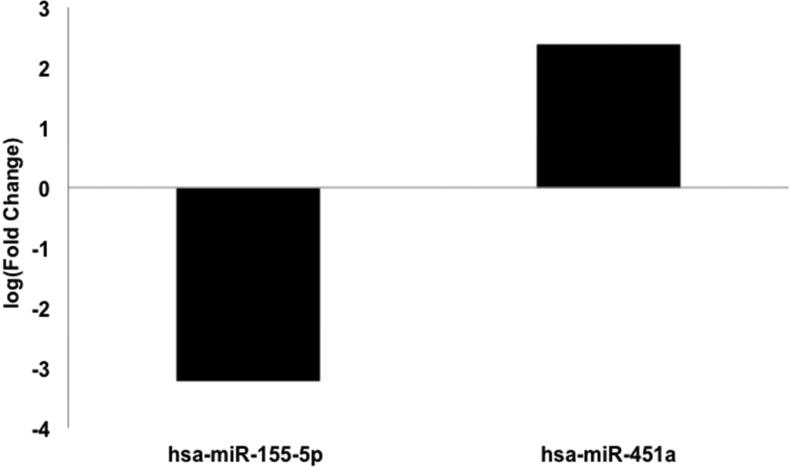
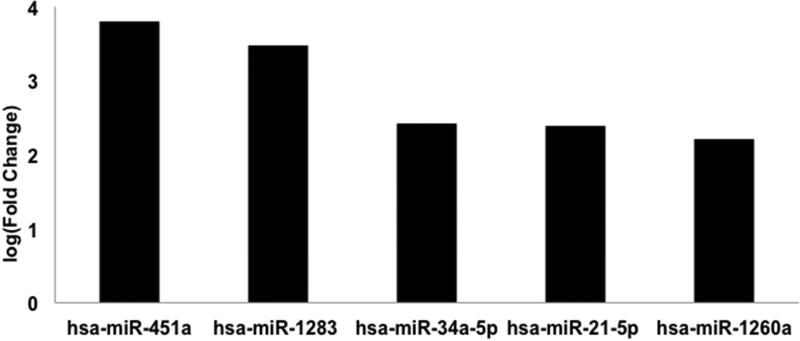
We next explored potential differences in the miR pattern between atypical Spitz lesions and other categories of Spitz lesions. Atypical Spitz lesions were compared separately to benign and malignant Spitz lesions. A comparison of atypical Spitz lesions to malignant Spitz lesions revealed increased miR-451a and decreased miR-155-5p expression associated with atypical Spitz lesions (Figure 5). In contrast, a comparison of atypical Spitz lesions (n=3) to benign Spitz lesions (n=3) revealed increased expression of miR-21-5p, miR-34a-5p, miR-451a, miR-283, and miR-1260a associated with atypical lesions. These results suggest a potential incremental increase in miR-21 expression from a benign nevi towards a malignant Spitz lesion (Figure 6).
Figure 5. Differentially expressed microRNAs between atypical Spitz and malignant Spitz.
microRNA expression of atypical Spitz lesions (n=3) compared to malignant Spitz (n=3) was analyzed for 805 microRNAs by NanoString nCounter. Differential expression of microRNAs with False Discovery Rate (FDR) <0.05 are shown.
Figure 6. Differentially expressed microRNAs between atypical Spitz and benign Spitz.
microRNA expression of atypical Spitz lesions (n=3) compared to benign Spitz (n=3) was analyzed for 805 microRNAs by NanoString nCounter. Differential expression of microRNAs with False Discovery Rate (FDR) <0.05 are shown.
3.6 Downstream targets of dysregulated microRNAs
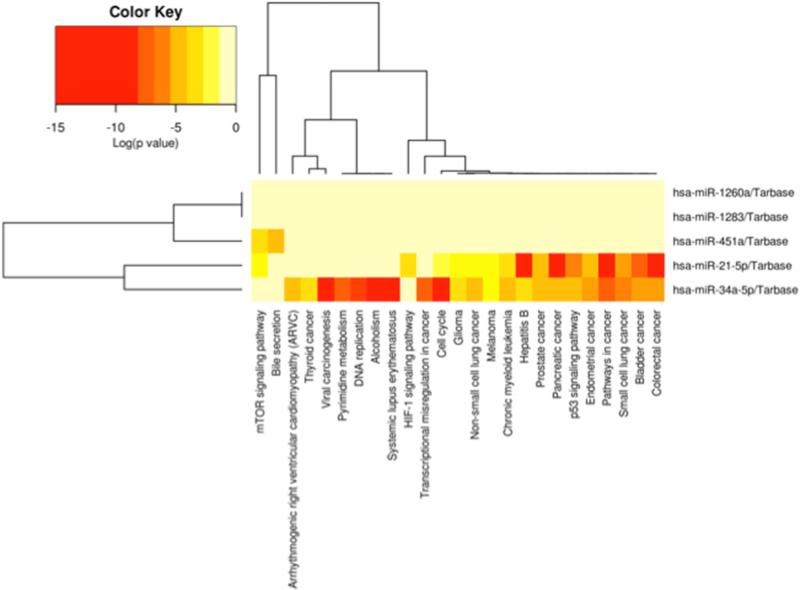
In order to understand the functional impact of differentially expressed miRs on the various Spitz lesions, the downstream mRNA targets of differentially expressed miRs and their involvement within biological pathways were explored. It is widely recognized that individual miRs can control dozens of genes, and many other miRs have been shown to synchronize their activity in targeting a diverse array of cellular and molecular processes (18). In order to decipher the biological pathways affected the complex cross-talk among differentially expressed miRs, we used a computational approach to determine significantly enriched pathways. Differentially expressed miRs were mapped to their experimentally validated target genes via miRTarBase, and enriched biological pathways were identified using the DIANA miRPath program (hypergeometric distribution). Clustered miRs and pathways were visualized in a heatmap in order to demonstrate sets of significantly enriched pathways. As an example, the results for 5 differentially expressed miRs (miR-21-5p, miR-34a-5p, miR-451a, miR-1283, and miR-1260a) derived from comparing benign Spitz vs. atypical Spitz tumors are shown in Figure 7. This comparison yielded 25 significantly enriched target pathways (FDR<0.05, Supplemental Table S8). miR-34a-5p and miR-21-5p showed the highest degree of enriched biological pathways in Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways (http://www.genome.jp/kegg/). Interestingly, the pattern of significantly enriched pathways appeared to be mutually exclusive between miR-34a-5p and miR-21-5p, suggesting these miRs may regulate specific biological functions in a non-redundant manner. For example, miR-34a-5p appears to be enriched for a set of pathways involved in DNA processes, including DNA replication, pyrimidine metabolism, transcriptional misregulation in cancer and cell cycle. Highly significantly enriched pathways for miR-21-5p target genes appear to be disease-specific, including colorectal cancer, bladder cancer, pancreatic cancer, hepatitis B and pathways in cancer. The full list of enriched target pathways are provided in Supplemental Tables S7-12. The results of these biological pathway enrichment studies may be used to inform future functional studies that seek to confirm the roles of specific miRs on the development of Spitz lesions.
Figure 7. Heatmap of significantly enriched biological pathways targeted by miRs from atypical Spitz vs. benign Spitz comparison.
Differentially expressed miRs discovered via edgeR analysis of atypical Spitz vs. benign Spitz were mapped to their experimentally validated target genes contained within the miRTarBase, and the union of the enriched KEGG pathways were identified using a hypergeometric test and FDR-correction for p-values via the DIANA miRPath program. Dendrogram branches are shown for clustered miRs and pathways. The corresponding pathways and their significance values are listed in Supplemental Table S8.
4. Discussion
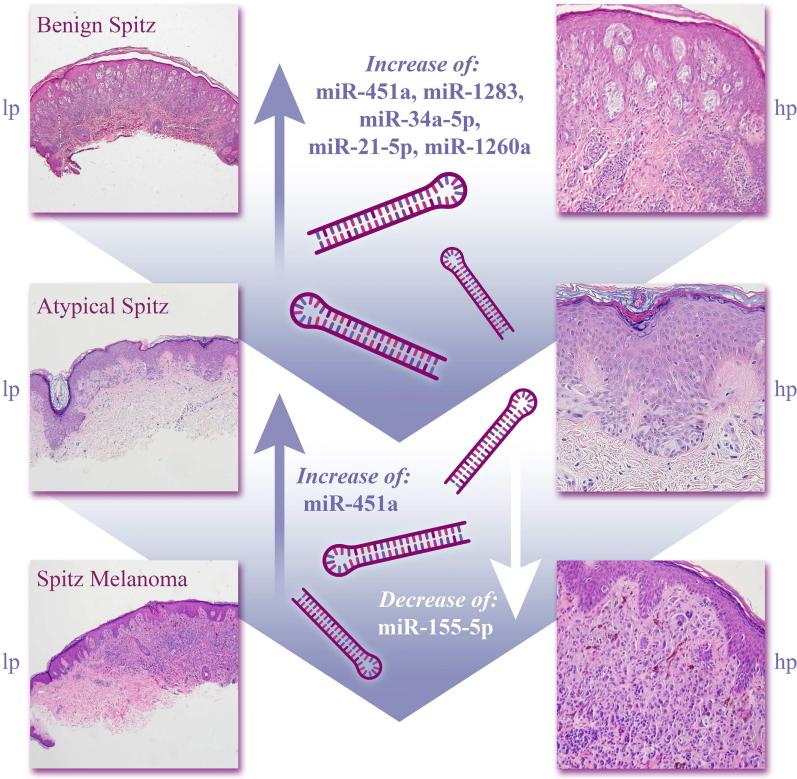
The current report describes the first use of the NanoString platform for characterization of the miR transcriptome in melanocytic Spitz lesions. Specifically, benign Spitz lesions were characterized by an up-regulation of miR-21-5p and miR-363-3p relative to benign nevi, whereas malignant Spitz lesions are differentiated from both benign nevi and benign Spitz tumors by up-regulation of miR-21-5p, miR-155-5p, and miR-1283. Atypical Spitz tumors have increased miR-451a and decreased miR-155-5p expression relative to malignant Spitz lesions, while increased expression of miR-21-5p, miR-34a-5p, miR-451a, miR-1283, and miR-1260a was present in atypical Spitz lesions relative to benign Spitz tumors. A summary of key results is shown in Figure 8. Together, these results suggest distinct miR expression profiles may be present across Spitz lesions of varying malignant potential and could serve as a basis for further validation studies.
Figure 8. Summary of select differentially expressed miRs.
Increased expression of miR-451a, miR-1283, miR-34a-5p, miR-21-5p and miR-1260a distinguishes atypical Spitz relative to benign Spitz. Increased expression of miR-451a and decreased expression of miR-155-5p distinguishes Spitz melanoma relative to atypical Spitz.
Atypical Spitz lesions remain a diagnostic challenge (4). As such, new biomarkers have been explored in an attempt to augment diagnostic light microscopy. Investigations into BRAF mutations reveal the presence of V600E mutations in benign Spitz, atypical Spitz, and malignant Spitz thereby, precluding its usefulness as a means for determining malignant potential (19). Da Forno et al. made similar findings when employing NRAS and BRAF mutational analysis across the Spitz spectrum (20). Immunohistochemical methods focused on Ki-67, S-100, and HMB-45 levels have also been investigated as a means to distinguish Spitz lesions however, these efforts have been only partially useful for diagnostic differentiation (21). Fluorescent in situ hybridization (FISH) and comparative genomic hybridization (CHG) of DNA copy changes across Spitz lesions have been explored. Gaiser et al. found no clinical usefulness for FISH in a series of 22 melanocytic lesions containing probes against chromosome 6p25, centromere 6, 6q23, and 11q13 (22). The present study provides a rationale for an alternative approach using miRs for diagnostic evaluation of Spitz lesions given the ongoing development of the aforementioned methodologies.
The use of miRs as a diagnostic aid for distinguishing Spitz lesions of varying malignant potentials has not been previously reported. Similar approaches have been performed in non-Spitz melanomas with some overlapping miR patterns being identified. Our group has reported miR-21 up-regulation in primary melanomas relative to benign and dysplastic nevi (8, 9, 23). miR-21 up-regulation has also been found in borderline primary lesions that contain atypical cellular features (8). Similar to trends identified in non-Spitz melanoma by Jiang et al., this study suggests a sequential increase in miR-21 expression from benign nevi to benign Spitz and benign Spitz compared to atypical Spitz and malignant Spitz lesions (9). miR-21 up-regulation in melanoma may promote oncogenesis through a variety of mechanisms, including alteration of cellular pathways involved with proliferation, migration, invasion and apoptosis (9, 24, 25). The mechanism of miR-21 mediated oncogenesis may involve inhibition of metalloprotinease-3 (TIMP3), BTG family member 2 (BTG2), programmed cell death 4 (PDCD4), or phosphate and tensin homologue (PTEN) (24, 25). Specifically, miR-21 over-expression in melanoma cells in vitro has been shown by our group to inhibit TIMP-3 expression and can promote increased cellular invasion (24).
These results also suggest miR-155-5p up-regulation in malignant Spitz lesions relative to benign and atypical Spitz lesions. miR-155 expression has previously been observed in malignant melanoma relative to benign nevi (8). Likewise, Segura et al. also found elevated miR-155 in primary and metastatic melanoma samples compared to benign nevi (P<0.02) (26). The mechanism of miR-155 oncogenesis in melanoma is incompletely understood but may involve negative regulation of v-ski avian sarcoma viral oncogene homolog (SKI) gene, which is implicated in cell growth and invasion (27).
Herein, miR-34a-5p, miR-451a, miR-1283, and miR-1260a up-regulation was present in atypical Spitz lesions relative to benign Spitz tumors. miR-1260 up-regulation has previously been described in cutaneous malignant melanoma (28). Contrary to our findings, miR-34a down-regulation has been reported in melanoma cell lines and leads to inhibition of cellular proliferation, migration, invasion, and motility which may be mediated, in part, through inhibition of c-Met (29-32). Advantages of a global approach to analysis of miR transcriptome across Spitz lesions includes the potential to explore unfamiliar miR in melanocytic lesions. Notably, this is the first study implicating miR-363, miR-451a, and miR-1283 as potential players in melanocytic skin tumors including Spitz lesions as well as cutaneous melanomas.
While our current efforts were geared toward diagnostic differentiation, miRs have the capacity to potentially address other pertinent challenges related to Spitz lesions. In particular, the determination of prognosis for Spitz lesions remains difficult and sentinel lymph biopsies are performed to assist in predictions and guidance of treatment (33, 34). Indeed, miRs have been used in prognostication of other neoplastic settings with promising results. Tian et al. found miR-206 downregulation in malignant melanoma correlated with poorer 5 year survival and disease free survival (35). Investigation into the relationship of miRs for prognostication of Spitz lesions may be of interest and could be explored given current pitfalls of management.
The drawback of this study includes the limited number of patients analyzed. NanoString platforms have the capacity to hold up to 12 samples per analysis. Thus, the potential for Type II error is present when comparing multiple groups given inherent structural limitations. The female predominance of individuals within the sample may limit the generalizability of these findings. Additionally, validation of NanoString findings using PCR is necessary to control for potential Type I errors. In spite of these setbacks, the ability to analyze miR transcriptome without the need for amplification and reverse transcription has made NanoString an attractive platform (36). Furthermore, as in this investigation, minute quantities of RNA input are necessary while the capability of single miR discrimination per cell is retained (36, 37). In a recent study using fresh, frozen and formalin-fixed paraffin-embedded (FFPE) clinical samples from a wide array of tissue types, the NanoString platform was shown to be highly sensitive and reproducible across replicates (38). NanoString remains advantageous relative to other high throughput modalities given lower relative costs and less complex analysis relative to RNA-seq while providing greater sensitivity and the ability to overcome saturation issues seen with microarray techniques (39).
Validation of these miR findings in additional samples is the next step in evaluating the role of miRs Spitz lesions. Future NanoString studies should include multiple assays with balanced numbers of group samples (n=24 per group; power 0.5, alpha 0.05) to account for inter-assay variability. In particular, it will be important to evaluate the miR features individually and combinations thereof with the other histopathological features in order to determine the sensitivity, specificity and area under the curve (AUC) values for the potential diagnostic classifiers, as has been done for miR biomarkers in other cancers (39, 40). More definitive studies will be needed to evaluate these potential diagnostic miR biomarkers for additional prognostic and predictive value with regards to surgical management (41, 42).
We contend analyses utilizing Spitz lesions with unfavorable biology that progress to distant metastasis would be of great interest however the feasibility of this approach might be challenged by the rarity of such events. For instance, Ludgate et al. identified 67 patients with atypical Spitz lesions at the University of Michigan over a 13 year period and only 1 individual within this series developed recurrent disease and distant metastasis (33). To overcome these limitations, utilization of institutions with high volume of experience with melanocytic lesions and/or multi-institutional collaborations would be necessary to identify individuals with poor biology that could be used for robust comparison.
In conclusion, the present study has suggested a distinct miR transcriptome for different Spitz lesions. This effort should form the framework for further investigation of miRs in Spitz lesions.
Supplementary Material
Acknowledgements
NIH Grants P01 CA095426 (to M.A. Caligiuri), P30 CA16058 (to M.A. Caligiuri), T15 LM011270 (to K. Regan) T32 CA090223 (to W.E. Carson), K24 CA093670 (to W.E. Carson).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author contributions: NL, KER, JHA, MAR, SBP, KKN, SPA, PF, XZ, AG and PROP contributed to acquisition and analysis of data. KER, PROP and XZ provided statistical analysis and interpretation. NL, KER and WEC drafted the manuscript. NL, KER, JHH, LPS, REP, WEC were responsible for critical revision. WEC was the senior author who provided supervision of this work and was responsible for the study design and conceptualization.
Author Disclosure Statement: The authors report no proprietary or commercial interest in any product mentioned or concept discussed in this article. Conflicts of interest: none.
References
- 1.SPITZ S. Melanomas of childhood. Am J Pathol. 1948;24:591–609. [PMC free article] [PubMed] [Google Scholar]
- 2.Joyce DP, Prichard RS, Gulmann C, Hill AD. The surgical management of Spitz naevi and atypical spitzoid neoplasms: A review of the literature. The surgeon : journal of the Royal Colleges of Surgeons of Edinburgh and Ireland. 2013;11:205–209. doi: 10.1016/j.surge.2013.01.001. [DOI] [PubMed] [Google Scholar]
- 3.Byrne M, Regan PJ, Kelly JL, Hussey A. Analysis of features associated with equivocal diagnosis and diagnostic discordance of Spitz naevi over an 18-year period. Eur J Dermatol. 2015;25:162–168. doi: 10.1684/ejd.2014.2497. [DOI] [PubMed] [Google Scholar]
- 4.Barnhill RL, Argenyi ZB, From L, Glass LF, Maize JC, et al. Atypical Spitz nevi/tumors: lack of consensus for diagnosis, discrimination from melanoma, and prediction of outcome. Hum Pathol. 1999;30:513–520. doi: 10.1016/s0046-8177(99)90193-4. [DOI] [PubMed] [Google Scholar]
- 5.Lallas A, Kyrgidis A, Ferrara G, Kittler H, Apalla Z, et al. Atypical Spitz tumours and sentinel lymph node biopsy: a systematic review. Lancet Oncol. 2014;15:e178–183. doi: 10.1016/S1470-2045(13)70608-9. [DOI] [PubMed] [Google Scholar]
- 6.Roaten JB, Partrick DA, Pearlman N, Gonzalez RJ, Gonzalez R, et al. Sentinel lymph node biopsy for melanoma and other melanocytic tumors in adolescents. J Pediatr Surg. 2005;40:232–235. doi: 10.1016/j.jpedsurg.2004.09.022. [DOI] [PubMed] [Google Scholar]
- 7.Gowrishankar B, Ibragimova I, Zhou Y, Slifker MJ, Devarajan K, et al. MicroRNA expression signatures of stage, grade, and progression in clear cell RCC. Cancer Biol Ther. 2014;15:329–341. doi: 10.4161/cbt.27314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grignol V, Fairchild ET, Zimmerer JM, Lesinski GB, Walker MJ, et al. miR-21 and miR-155 are associated with mitotic activity and lesion depth of borderline melanocytic lesions. Br J Cancer. 2011;105:1023–1029. doi: 10.1038/bjc.2011.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jiang L, Lv X, Li J, Li X, Li W, et al. The status of microRNA-21 expression and its clinical significance in human cutaneous malignant melanoma. Acta Histochem. 2012;114:582–588. doi: 10.1016/j.acthis.2011.11.001. [DOI] [PubMed] [Google Scholar]
- 10.Guo J, Miao Y, Xiao B, Huan R, Jiang Z, et al. Differential expression of microRNA species in human gastric cancer versus non-tumorous tissues. J Gastroenterol Hepatol. 2009;24:652–657. doi: 10.1111/j.1440-1746.2008.05666.x. [DOI] [PubMed] [Google Scholar]
- 11.Thomas J, Ohtsuka M, Pichler M, Ling H. MicroRNAs: Clinical Relevance in Colorectal Cancer. Int J Mol Sci. 2015;16:28063–28076. doi: 10.3390/ijms161226080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang YC, Xu Z, Zhang TF, Wang YL. Circulating microRNAs as diagnostic and prognostic tools for hepatocellular carcinoma. World J Gastroenterol. 2015;21:9853–9862. doi: 10.3748/wjg.v21.i34.9853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alder H, Taccioli C, Chen H, Jiang Y, Smalley KJ, et al. Dysregulation of miR-31 and miR-21 induced by zinc deficiency promotes esophageal cancer. Carcinogenesis. 2012;33:1736–1744. doi: 10.1093/carcin/bgs204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Luo EC, Chang YC, Sher YP, Huang WY, Chuang LL, et al. MicroRNA-769-3p down-regulates NDRG1 and enhances apoptosis in MCF-7 cells during reoxygenation. Sci Rep. 2014;4:5908. doi: 10.1038/srep05908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hsu SD, Tseng YT, Shrestha S, Lin YL, Khaleel A, et al. miRTarBase update 2014: an information resource for experimentally validated miRNA-target interactions. Nucleic acids research. 201442:D78–85. doi: 10.1093/nar/gkt1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vlachos IS, Kostoulas N, Vergoulis T, Georgakilas G, Reczko M, et al. DIANA miRPath v.2.0: investigating the combinatorial effect of microRNAs in pathways. Nucleic acids research. 2012;40:W498–504. doi: 10.1093/nar/gks494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vlachos IS, Zagganas K, Paraskevopoulou MD, Georgakilas G, Karagkouni D, et al. DIANA- miRPath v3.0: deciphering microRNA function with experimental support. Nucleic acids research. 2015;43:W460–466. doi: 10.1093/nar/gkv403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peter ME. Targeting of mRNAs by multiple miRNAs: the next step. Oncogene. 2010;29:2161–2164. doi: 10.1038/onc.2010.59. [DOI] [PubMed] [Google Scholar]
- 19.Fullen DR, Poynter JN, Lowe L, Su LD, Elder JT, et al. BRAF and NRAS mutations in spitzoid melanocytic lesions. Mod Pathol. 2006;19:1324–1332. doi: 10.1038/modpathol.3800653. [DOI] [PubMed] [Google Scholar]
- 20.Da Forno PD, Pringle JH, Fletcher A, Bamford M, Su L, et al. BRAF, NRAS and HRAS mutations in spitzoid tumours and their possible pathogenetic significance. Br J Dermatol. 2009;161:364–372. doi: 10.1111/j.1365-2133.2009.09181.x. [DOI] [PubMed] [Google Scholar]
- 21.Zedek DC, McCalmont TH. Spitz nevi, atypical spitzoid neoplasms, and spitzoid melanoma. Clin Lab Med. 2011;31:311–320. doi: 10.1016/j.cll.2011.03.008. [DOI] [PubMed] [Google Scholar]
- 22.Gaiser T, Kutzner H, Palmedo G, Siegelin MD, Wiesner T, et al. Classifying ambiguous melanocytic lesions with FISH and correlation with clinical long-term follow up. Mod Pathol. 2010;23:413–419. doi: 10.1038/modpathol.2009.177. [DOI] [PubMed] [Google Scholar]
- 23.Satzger I, Mattern A, Kuettler U, Weinspach D, Niebuhr M, et al. microRNA-21 is upregulated in malignant melanoma and influences apoptosis of melanocytic cells. Exp Dermatol. 2012;21:509–514. doi: 10.1111/j.1600-0625.2012.01510.x. [DOI] [PubMed] [Google Scholar]
- 24.Martin del Campo SE, Latchana N, Levine KM, Grignol VP, Fairchild ET, et al. MiR-21 enhances melanoma invasiveness via inhibition of tissue inhibitor of metalloproteinases 3 expression: in vivo effects of MiR-21 inhibitor. PloS one. 2015;10:e0115919. doi: 10.1371/journal.pone.0115919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang CH, Yue J, Pfeffer SR, Handorf CR, Pfeffer LM. MicroRNA miR-21 regulates the metastatic behavior of B16 melanoma cells. J Biol Chem. 2011;286:39172–39178. doi: 10.1074/jbc.M111.285098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Segura MF, Belitskaya-Lévy I, Rose AE, Zakrzewski J, Gaziel A, et al. Melanoma MicroRNA signature predicts post-recurrence survival. Clin Cancer Res. 2010;16:1577–1586. doi: 10.1158/1078-0432.CCR-09-2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Levati L, Pagani E, Romani S, Castiglia D, Piccinni E, et al. MicroRNA-155 targets the SKI gene in human melanoma cell lines. Pigment Cell Melanoma Res. 2011;24:538–550. doi: 10.1111/j.1755-148X.2011.00857.x. [DOI] [PubMed] [Google Scholar]
- 28.Sand M, Skrygan M, Sand D, Georgas D, Gambichler T, et al. Comparative microarray analysis of microRNA expression profiles in primary cutaneous malignant melanoma, cutaneous malignant melanoma metastases, and benign melanocytic nevi. Cell and tissue research. 2013;351:85–98. doi: 10.1007/s00441-012-1514-5. [DOI] [PubMed] [Google Scholar]
- 29.Yamazaki H, Chijiwa T, Inoue Y, Abe Y, Suemizu H, et al. Overexpression of the miR-34 family suppresses invasive growth of malignant melanoma with the wild-type p53 gene. Exp Ther Med. 2012;3:793–796. doi: 10.3892/etm.2012.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yan D, Zhou X, Chen X, Hu DN, Dong XD, et al. MicroRNA-34a inhibits uveal melanoma cell proliferation and migration through downregulation of c-Met. Invest Ophthalmol Vis Sci. 2009;50:1559–1565. doi: 10.1167/iovs.08-2681. [DOI] [PubMed] [Google Scholar]
- 31.Mazar J, Khaitan D, DeBlasio D, Zhong C, Govindarajan SS, et al. Epigenetic regulation of microRNA genes and the role of miR-34b in cell invasion and motility in human melanoma. PLoS One. 2011;6:e24922. doi: 10.1371/journal.pone.0024922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dong F, Lou D. MicroRNA-34b/c suppresses uveal melanoma cell proliferation and migration through multiple targets. Mol Vis. 2012;18:537–546. [PMC free article] [PubMed] [Google Scholar]
- 33.Ludgate MW, Fullen DR, Lee J, Lowe L, Bradford C, et al. The atypical Spitz tumor of uncertain biologic potential: a series of 67 patients from a single institution. Cancer. 2009;115:631–641. doi: 10.1002/cncr.24047. [DOI] [PubMed] [Google Scholar]
- 34.Duncan LM. Atypical Spitz tumours and sentinel lymph nodes. Lancet Oncol. 2014;15:377–378. doi: 10.1016/S1470-2045(13)70397-8. [DOI] [PubMed] [Google Scholar]
- 35.Tian R, Liu T, Qiao L, Gao M, Li J. Decreased serum microRNA-206 level predicts unfavorable prognosis in patients with melanoma. Int J Clin Exp Pathol. 2015;8:3097–3103. [PMC free article] [PubMed] [Google Scholar]
- 36.Moreno-Moya JM, Vilella F, Simón C. MicroRNA: key gene expression regulators. Fertil Steril. 2014;101:1516–1523. doi: 10.1016/j.fertnstert.2013.10.042. [DOI] [PubMed] [Google Scholar]
- 37.Brumbaugh CD, Kim HJ, Giovacchini M, Pourmand N. NanoStriDE: normalization and differential expression analysis of NanoString nCounter data. BMC Bioinformatics. 2011;12:479. doi: 10.1186/1471-2105-12-479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Veldman-Jones MH, Brant R, Rooney C, Geh C, Emery H, et al. Evaluating Robustness and Sensitivity of the NanoString Technologies nCounter Platform to Enable Multiplexed Gene Expression Analysis of Clinical Samples. Cancer research. 2015;75:2587–2593. doi: 10.1158/0008-5472.CAN-15-0262. [DOI] [PubMed] [Google Scholar]
- 39.McDermott AM, Miller N, Wall D, Martyn LM, Ball G, et al. Identification and validation of oncologic miRNA biomarkers for luminal A-like breast cancer. PloS one. 2014;9:e87032. doi: 10.1371/journal.pone.0087032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schultz NA, Dehlendorff C, Jensen BV, Bjerregaard JK, Nielsen KR, et al. MicroRNA biomarkers in whole blood for detection of pancreatic cancer. Jama. 2014;311:392–404. doi: 10.1001/jama.2013.284664. [DOI] [PubMed] [Google Scholar]
- 41.Mandrekar SJ, Sargent DJ. Clinical trial designs for predictive biomarker validation: one size does not fit all. Journal of biopharmaceutical statistics. 2009;19:530–542. doi: 10.1080/10543400902802458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kulasingam V, Diamandis EP. Strategies for discovering novel cancer biomarkers through utilization of emerging technologies. Nature clinical practice. Oncology. 2008;5:588–599. doi: 10.1038/ncponc1187. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.