Abstract
Pathogenesis-related proteins play multiple roles in plant development and biotic and abiotic stress tolerance. Here, we characterize a rice defense related gene named “jasmonic acid inducible pathogenesis-related class 10” (JIOsPR10) to gain an insight into its functional properties. Semi-quantitative RT-PCR analysis showed up-regulation of JIOsPR10 under salt and drought stress conditions. Constitutive over-expression JIOsPR10 in rice promoted shoot and root development in transgenic plants, however, their productivity was unaltered. Further experiments exhibited that the transgenic plants showed reduced susceptibility to rice blast fungus, and enhanced salt and drought stress tolerance as compared to the wild type. A comparative proteomic profiling of wild type and transgenic plants showed that overexpression of JIOsPR10 led to the differential modulation of several proteins mainly related with oxidative stresses, carbohydrate metabolism, and plant defense. Taken together, our findings suggest that JIOsPR10 plays important roles in biotic and abiotic stresses tolerance probably by activation of stress related proteins.
Keywords: abiotic stress, JIOsPR10, Magnaporthe oryzae, proteomics, rice
Plants are continuously exposed to various biotic and abiotic stresses that results in significant decrease in their productivity. During the course of evolution, plants have developed multiple pathways that allow them to sense and adapt these environmental stresses (Gupta et al., 2015). Plant’s responses to stress include activation of ion channels, production of reactive oxygen species (ROS) scavenging enzymes, accumulation of hormones, and expression of stress tolerance genes (Fraire-Velázquez et al., 2011). During multiple stresses, plants lead to a complex interaction and crosstalk with multiple signals, and decide the final output of stress-tolerance related genes (Rizhsky et al., 2004). Therefore, plenty of genes are likely to function together and play multiple roles against combined environmental stresses.
Among identified proteins, the pathogenesis-related (PR) family proteins are a highly conserved protein family which is involved in plant immune responses (Stintzi et al., 1993). Overexpression of PR proteins has ability to enhance plants tolerance to pathogen infections. For instance, overexpression of PR-5 protein in rice enhances tolerance to Rhizoctonia solani (Datta et al., 1999), while overexpression of pepper PR-1 in tobacco enhances host tolerance to Phytophthora nicotianae, Ralstonia solanacearum, and Pseudomonas syringae (Sarowar et al., 2005). Those PR proteins have also been reported to have multiple roles in adaption to abiotic stresses, like express pepper PR-1 protein in tobacco can enhance heavy metal tolerance (Sarowar et al., 2005). Moreover, transgenic tobacco overexpressing a Trichoderma harzianum induced endochitinase enhances the tolerance to both biotic (P. syringae), and abiotic (salt and heavy metals) stresses (de las Mercedes Dana et al., 2006). Therefore, study and utilization of PR proteins, which functions in both biotic and abiotic tolerance, may provide new challenges in agriculture.
The PR proteins have been classified into 17 classes based on their amino acid sequence, serological relationship, and biological activities (van Loon et al., 1994; van Loon and van Strien, 1999). Among them, the PR-10 family proteins are involved in multiple anti-pathogen processes, and are generally localized in the intracellular spaces (van Loon and van Strien, 1999). PR-10 family proteins have conserved N-terminal sequences, but very divergent in the C-terminus (Fernandes et al., 2013). The PR10 family proteins consist of three α-helices and seven antiparallel β-strands. Those structure elements enclose a large hydrophobic cavity which is most likely related with their functional relevance (Fernandes et al., 2013). Several studies have shown that PR-10 proteins contain RNase activities (Chen et al., 2007; Kim et al., 2008b; Krishnaswamy et al., 2011; Xie et al., 2010), and cysteine protease inhibitor activity (Andrade et al., 2010).
Previous studies have revealed that PR-10 family proteins are involved in anti-biotic stresses, such as antifungus (Chen et al., 2007; Xie et al., 2010), anti-bacteria (Flores et al., 2002; Xie et al., 2010), anti-virus (Park et al., 2004), and anti-nematode (Andrade et al., 2010). To date, four different PR10 family genes have been characterized in rice: PR10a (McGee et al., 2001), PR10b (McGee et al., 2001), jasmonic acid inducible pathogenesis-related class 10 (JIOsPR10) (Jwa et al., 2001), and RSOsPR10 (Hashimoto et al., 2004). All those PR10 family genes are induced by Magnaporthe oryzae infection and jasmonic acid treatment (Hashimoto et al., 2004; Jwa et al., 2001; McGee et al., 2001), suggesting that those PR10 family genes may be functional redundant in rice. Moreover, phytohormone salicylic acid could activate the transcriptional expression of leaf accumulated PR10a and JIOsPR10, but not the root specific RSOsPR10 (Hashimoto et al., 2004), indicating that PR10 gene regulation in root and shoot may have different mechanisms. Moreover, rice PR10 genes are also induced by various abiotic stresses, such as cold, salt and drought (Hashimoto et al., 2004; Kim et al., 2008b), suggesting that PR10 protein may have multiple function in tolerance to both biotic and abiotic stresses. In this study, transgenic rice constitutively overexpressing JIOsPR10 was constructed. The transgenic plants showed a promotion in root and shoot development, and enhance tolerance to rice blast fungus, drought and salt stresses. Proteomics analysis reveals that plant defense related proteins as well as ROS related proteins were also highly accumulated in overexpression plant. These results implied that JIOsPR10 play a role in multiple stress tolerance, which may important for future agricultural applicants.
Materials and Methods
Plant material, growth conditions, and stress treatment
Rice seeds (Oryzae sativa cv. Dongjin) were sterilized with 70% ethanol for 10 min, then in 3% sodium hypochlorite for 30 min, followed by washing with sterilized water for three times. Sterilized seeds were imbibed in water at 4°C in dark for 3 days, and allowed to germinate in standard rice soil (Monsanto, Seoul, Korea) in greenhouse or on Murashige & Skoog (MS) phytagel medium at 28°C growth chamber (light/dark time, 16/8 h). For drought and salt stress treatments, 10-day-old seedlings were transferred into container containing either 20% polyethylene glycol (PEG) 4000 or 150 mM NaCl solution. For drought stress to 5- to 6-leaf stages, pots were kept on dried papers to remove water from the pots, and plants were re-watered after 5 days. For salt stress to 4- to 5-leaf stage plants, rice in growth pots were transferred to a container filled with 150 mM NaCl. NaCl solution was changed each two days to maintain the concentration of salt stress.
Fungal spore preparation and infection
M. oryzae conidia (KJ301) was cultured on rice-bran agar medium (25 g/l rice bran, 1 g/l sucrose, and 20 g/l agar). Aerial mycelia were removed under fluorescent light to induce synchronous conidia as described previously (Kim et al., 2013). Conidial suspension (1 × 106 conidia/ml) was inoculated to rice leaves using an air sprayer. Inoculated plants were kept in a humidity chamber at 28°C. Leaf phenotypes were confirmed at 72-h post-inoculation, and the infection area were measured by Assess 2.0 software (The American Phytopathological Society Press, St. Paul, MN, USA).
Vector construction, plant transformation, and genotyping
Full length of JIOsPR10 coding sequence was cloned into pDONR201 vector and sequenced. Then, the JIOsPR10 was constructed into pMJ101 vector containing a rice cytochrome c promoter (OsCc1 pro) and basta selection marker (BAR) by using Gateway system. The construct was transformed into Agrobacterium LBA4404, and introduced into rice embryonic calli by selection on MS medium contain 50 μM phosphinothricin. Regenerated plants were transferred to soil pots and grown in greenhouse. Genotyping of these plants were carried out by PCR with specific primer sets (Supplementary Table 1).
Southern blot analysis
Genomic DNA was extracted from 10-day-old seedlings of wild type (WT) and JIOsPR10 transgenic lines. The genomic DNA was digested with EcoRI, electrophoresed on a 0.8% agarose gel and then blotted onto a nylon membrane (NEN Life Science, Boston, MA, USA). BAR gene in the binary vector was amplified by PCR and used as probe. BAR fragment was labeled with [α-32P]-dCTP by using Prime-A-Gene Labeling System (Promega, Madison, WI, USA), and hybridized with the blotted membrane. The membrane was exposed to X-ray film (Fuji Film, Tokyo, Japan) for 3 days at −80°C.
RNA extraction and RT-PCR
Total RNA was extracted from the leaves of WT and transgenic lines using TRIzol regent (ThermoFisher, Darmstadt, Germany). Extracted RNA was treated with DNase and precipitated with 2.5 vol of ethanol and 1/10 vol of 4 M sodium acetate. Five micrograms of RNA was used for cDNA synthesis by Superscript II reverse transcriptase (Invitrogen, Carlsbad, CA, USA). Gene specific primers were designed by Primer Designer 4 (Sci Ed Central, Cary, NC, USA), and summarized in Supplementary Table 1. The equal amount of cDNA was optimized by OsActin2 (Os10g36650) primer sets (He et al., 2009).
Measurement of water loss
The water loss from detached leaves of WT and transgenic lines was measured by monitoring the fresh weight loss at the indicated time points (Duan et al., 2012). For each time point, at least twenty independent leaves from each genotype were used for the measurement of fresh and dried weights.
Protein extraction, proteomics analysis, and protein identification
Total leaf proteins were extracted from three independent biological samples and subjected to PEG fractionation as described previously (Kim et al., 2001; Wang et al., 2014). Briefly, leaf powder was homogenized in the Mg/NP-40 extraction buffer (0.5 M Tris-HCl [pH 8.3], 2% [v/v] NP-40, 20 mM MgCl2, and 2% [v/v] β-mercaptoethanol) and centrifuged to obtain clear supernatant. Supernatant layer was then precipitated by 15% PEG solution and stand by on ice for 30 min. Supernatant was collected after centrifugation at 12,000 rpm for 15 min, and precipitated with 4 volume of methanol contain 0.1 M ammonium acetate. The 2-DE gels were performed as described previously (Kim et al., 2008a). Briefly, 250 μg of proteins of each samples were dissolved in the rehydration buffer 9 M (w/v) CO(NH2)2, 2% (w/v) 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate hydrate (CHAPS), 0.002% (w/v) bromophenol blue, 20 mM dithiothreitol (DTT), and 0.5% (v/v) pharmalyte (pH 5–8) and were loaded on 18 cm IPG strips (pH 4–7) by rehydration loading for 12 h. Second dimensional separation was carried out on 12% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDSPAGE) and the gels were stained with Coomassie brilliant blue G-250 (CBB). The stained gels were scanned using transmissive scanner (Powerlook III; UMAX, Fort Worth, TX, USA), and images were analyzed with ImageMaster 2D Platinum software 6.0 (Amersham Biosciences AB, Uppsala, Sweden). Protein spots were excised from CBB-stained gels, digested with trypsin, and extracted with method describes previously (Kim et al., 2008a). Extracted peptide mixture was re-dissolved in dissolve solution (distilled water:acetonitrile:trifluoacetic acid = 93:5:2), sonicated for 5 min, and spin down at 14,000 rpm for 2 min. The peptide mixture was dropped on MALDI plate in matrix solution α-cyano-4-hydroxycinnamic acid (Sigma, St. Louis, MO, USA) in acetone (40 mg/ml). The MALDI plate was then washed with 0.1% (v/v) TFA. The gel spots were analyzed using a Voyager-DE STR MALDI-TOF mass spectrometer (PerSeptiveBiosystems, Framingham, MA, USA). Database searches were performed using Mascot (http://www.matrixscience.com) and ProteinProspector (http://prospector.ucsf.edu/prospector/mshome.htm) as described previously (Kim et al., 2008a).
Western blot analysis
Equal amount of total leaf proteins were separated on 12% SDS-PAGE and transblotted to a PVDF membrane using semidry electrophoretic apparatus (Hoefer, Holliston, MA, USA). The transferred PVDF membranes were blocked for 4 h at room temperature in 1× TTBS buffer (50 mM Tris-HCl, pH 8.2, 0.1% v/v Tween 20, and 150 mM NaCl) containing 7% skim milk. Membranes were then incubated with primary antibodies of DUF26-like protein, Beta-1,3-glucanse and Cu/Zu-SOD, which were generated as described previously (Kim et al., 2014), at 1,000-fold dilution for 1 h at room temperature. Treated membranes were then incubated with a secondary antibody (anti-rabbit lgG conjugated with horseradish peroxidase; 5,000-fold dilution) for 2 h. Membranes were washed with 1× TTBS (Tris-buffered saline, 0.1% Tween 20) for 15 min, 3 times. Washed membranes were then subjected to protein detection by ECL (Perkin Elmer Life Sciences, Boston, MA, USA) using LAS4000 image system (Fujifilm, Tokyo, Japan).
Results
JIOsPR10 is induced under drought and salt stresses
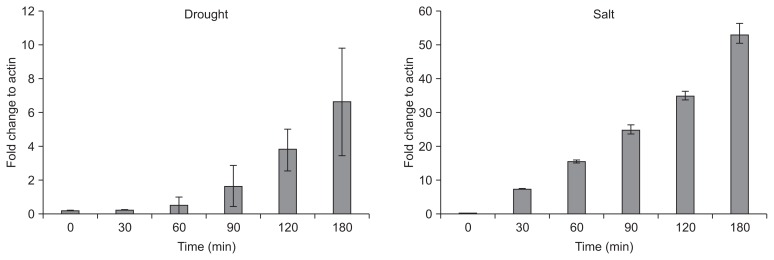
Previous report showed that JIOsPR10 protein was induced by drought and salt stress in rice (Kim et al., 2008b). To understand whether JIOsPR10 transcripts were induced in response to salt and drought stress, rice seedlings of WT were exposed to drought (20% PEG) and salt (150 mM NaCl) solutions and semi-quantitative RT-PCR analysis was carried out. JIOsPR10 transcripts were induced after 60 min and 30 min upon exposure to drought and salt stress, respectively, and increased in a time dependent manner (Fig. 1).
Fig. 1.
Transcriptional regulations of jasmonic inducible pathogenesis-related class 10 (JIOsPR10) under drought (A) and salt (B) stresses. Total RNAs were isolated from 10-day-old seedlings of wild type plant at indicated time points after treatment of 150 mM NaCl (A) or 20% polyethylene glycol (PEG) 4000 (B). Rice actin1 was set as internal control. Error bars represent standard error of three biological replicates of samples.
Molecular Characterization of transgenic rice overexpressing JIOsPR10
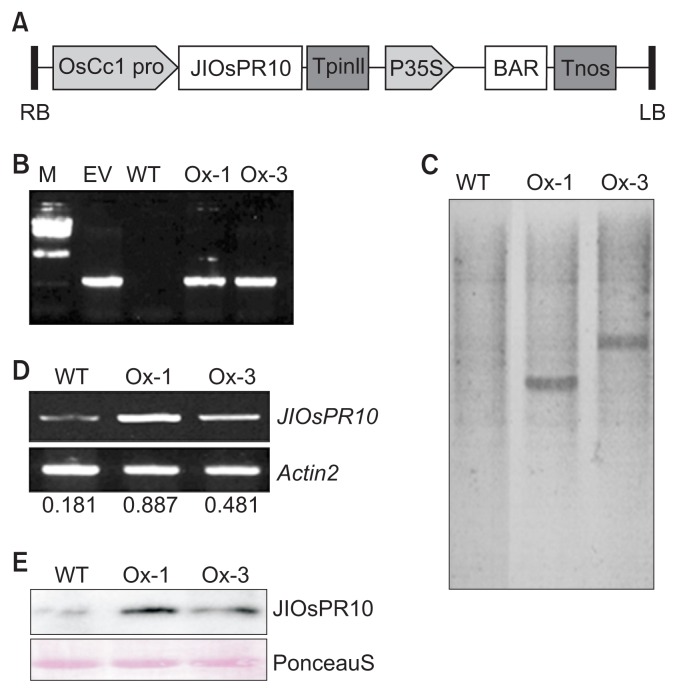
The role of JIOsPR10 in plant defense is well established (Kim et al., 2004). Our results also showed induction of JIOsPR10 transcripts by drought and salt stress, which prompted us to analyze the biological functions of this gene in both biotic and abiotic stress tolerance. The JIOsPR10 cDNA was constructed under constitutive promoter (Fig. 2A), and introduced into rice via Agrobacterium-mediated transformation. Two independent transgenic lines, Ox-1 and Ox-3, were produced and confirmed by PCR after Basta selection. A 500 bp PCR band was detected in transgenic plant, so as the constructed vector plasmid which set as positive control (Fig. 2B). Southern blot analysis indicated that both transgenic lines have single-copy T-DNA insertion (Fig. 2C). Transcriptional expression of JIOsPR10 in transgenic lines was then confirmed by semi-quantitative PCR, the results of which showed 4.9- and 2.6-fold increase of JIOsPR10 transcripts in Ox-1 and Ox-3 plant than WT plant (Fig. 2D). Subsequently, the protein level of JIOsPR10 was also detected by Western blot analysis which was 25 folds higher in each transgenic line, compared to the WT (Fig. 2E).
Fig. 2.
Characterization of jasmonic inducible pathogenesis-related class 10 (JIOsPR10) overexpression plants. (A) Schematic diagram of JIOsPR10 under rice cytochrome c promoter with basta selection marker was used for transformation. RB, right border; OsCc1 pro, rice cytochrome c promoter; TpinII, pinII terminator; BAR, basta resistance marker; Tnos, nos terminator; LB, left border. (B) Genotyping of overexpression plants by PCR with Basta selection marker specific primers. M, DNA marker; EV, empty vector plasmid set as positive control; WT; wild type. (C) Southern blot analysis of JIOsPR10 overexpression plants with BAR probe. (D) RT-PCR analysis of JIOsPR10 expression in WT and overexpression plants. Number indicate the relative expression level of JIOsPR10 compare to actin2. (E) Western blot analysis of JIOsPR10 in WT and overexpression plants with JIOsPR10 specific antibody. Equal loading of protein samples was detected by PonceauS staining.
Overexpression of JIOsPR10 promotes rice development
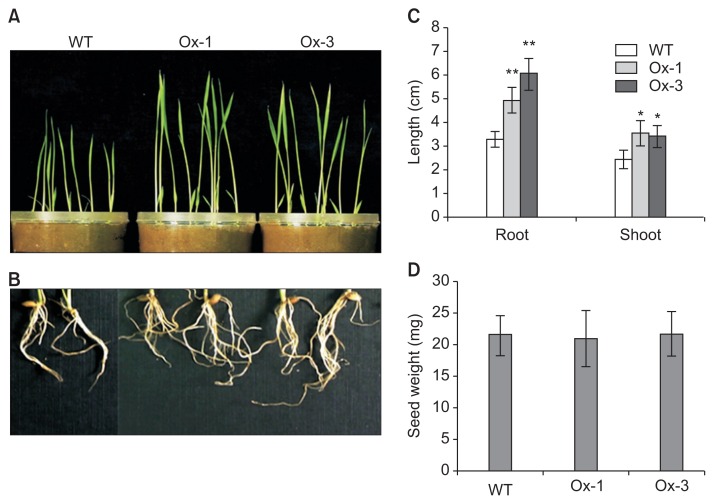
At first, the growth phenotype of transgenic plants was analyzed. Compared to the WT, transgenic lines showed a growth promotion at early developmental stages (Fig. 3A, B). Root and shoot lengths were 39% and 18% higher in Ox-1, and 34% and 29% higher in Ox-3 in comparison with WT at the 7-day-old seedling stage (Fig. 3C). In addition, seed weight and total productivity was also measured which did not show any significant differences in transgenic lines as compared to the WT (Fig. 3D).
Fig. 3.
Overexpression of jasmonic inducible pathogenesis-related class 10 (JIOsPR10) promotes root and shoot growth. Shoot (A) and root (B) phenotypes of 7-day-old seedlings. (C) Average shoot and root length of wild type (WT) and JIOsPR10 overexpression plants. (D) Seed weight of WT and JIOsPR10 overexpression plants. Error bars represent standard error of 20 individual plants. *P < 0.01, **P < 0.001.
JIOsPR10 overexpression lines increase drought and salt stress tolerance
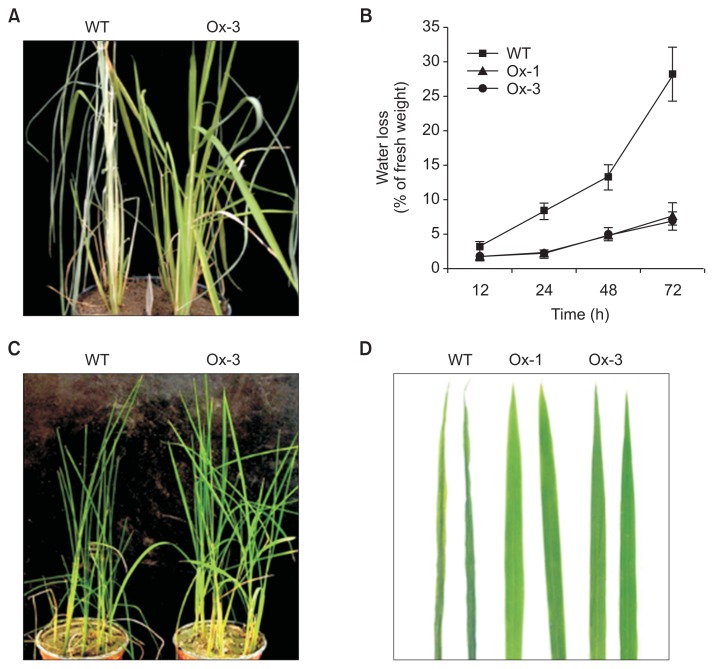
To analyze whether JIOsPR10 overexpression enhances abiotic stress tolerance in transgenic rice, the Ox-1 and Ox-3 lines were exposed to drought and salt stresses (Fig. 4). Over-expression lines (Ox-1 and Ox-3) have similar phenotypes in drought and salt stresses. For drought stress treatment, 4- to 5-week-old plants in growth pot were dried on paper in greenhouse, and rewatered after 5 days. OX-1 and Ox-3 plants showed successful growth recovery, but not WT (Fig. 4A). The leaf water loss was measured in WT, Ox-1, and Ox-3 lines at different treated time points. In WT, a water loss to nearly 30% of leaf fresh weight was detected under drought stress at 72-h post treatment (hpt) (Fig. 4B). Comparably, the water loss rate in Ox-1 and Ox-3 lines was less than 10% at 72 hpt (Fig. 4B). Salt stress treatment was also performed on JIOsPR10-overexpression plants. The growth pots were incubated in 150 mM NaCl solution and phenotype were measured after 24 h. Overexpression lines showed enhanced salt stress tolerance as compared with the WT (Fig. 4C), and a significant leaf curling was detected after 24 hpt in WT, but not in Ox-1 and Ox-3 plants (Fig. 4D).
Fig. 4.
Overexpression plant of jasmonic inducible pathogenesis-related class 10 (JIOsPR10) enhances drought and salt stress tolerance. (A) Phenotype of wild type (WT) and JIOsPR10 overexpression plants rewatered after 5 days under drought condition. (B) Leaf water loss of WT and overexpression plants. Error bars represent standard error of samples. (C) Phenotype of WT and JIOsPR10 overexpression under salt stress (150 mM NaCl). (D) Closeup view of leaf phenotype under salt stress. At least 20 independent plants were measured for each experiment.
Differential protein expression profile in WT and JIOsPR10 overexpression plant
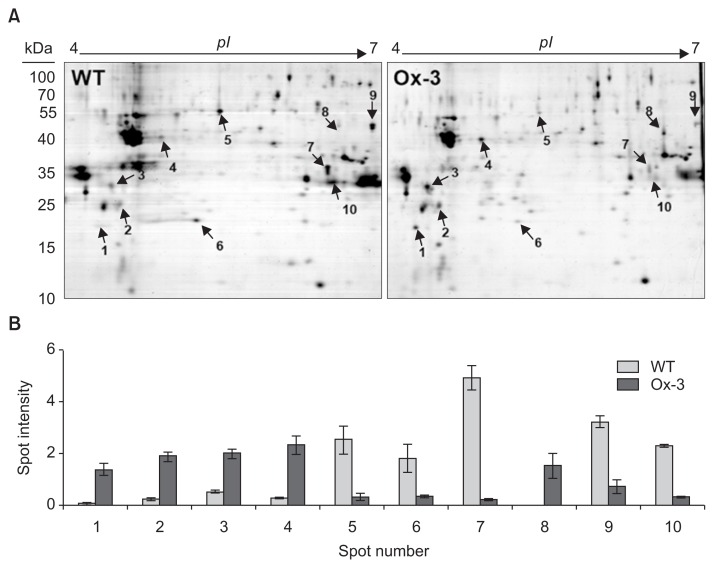
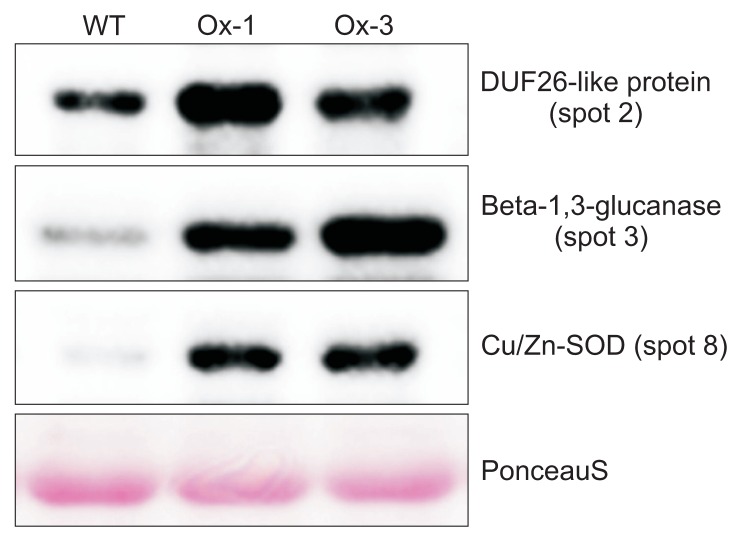
Our results also showed that both Ox-1 and Ox-3 lines showed very similar response to both drought and salt stresses, indicating the higher accumulation of JIOsPR10 in Ox-3 lines is sufficient for stress tolerance. Therefore, we selected the Ox-3 line for proteomic analysis. Total protein profiles of both WT and JIOsPR10 overexpression plants were analyzed by 2-DE analysis. The 2-DE gels were stained by CBB, and the differentially expressed protein spots were detected (Fig. 5A, indicated by arrows). Five protein spots (spot 1, 2, 3, 4, and 8) were up-regulated in JIOsPR10 overexpression plants (Fig. 5B), including thaumatin-like protein (TLP), DUF26-like protein, beta-1,3-glucanase, peroxidase, and Cu/Zn-SOD (Table 1). Five protein spots, which encoding enolase (spot 5), nucleoside diphosphate kinase (NDK, spot 6), formate dehydrogenase (spot 7), xylanse inhibitor protein 1 (spot 9), and cytoplasmic malate dehydrogenase (spot 10), were down-regulated in Ox-3 line than in WT (Fig. 5B, Table 1). Western blot analysis using gene specific antibody of DUF26-like protein, beta-1,3-glucanase, and Cu/Zn-SOD was employed to confirm the differential expression of identified proteins in Ox-1 and Ox-3 lines. The protein accumulation of all three proteins in Ox-1 and Ox-3 was strongly increased comparing with WT (Fig. 6), indicating a good correlation of proteomics data.
Fig. 5.
Proteomic analysis of jasmonic inducible pathogenesisrelated class 10 (JIOsPR10) overexpression plants showing a different protein expression profile as compared to the wild type (WT). (A) Gel map of 2-DE analysis. Same amount of total rice leaf proteins were separated on 2-DE gels, and stained with Coomassie brilliant blue. Arrows indicate the differentially expressed protein spots. (B) On-gel intensity of each protein spots in WT and overexpression plants. Error bars represent standard error of three biological replicates of experiments. pI, isoelectric point.
Table 1.
Identification of differentially modulated protein spots in wild type and JIOsPR10 overexpression plants using MALDI-TOF mass spectrometer
| Spot No. | Protein name | Accession No. | Score | Database | MP | SC (%) | Ex Mr (kDa) | Th Mr (kDa) | Ex pI | Th pI | Biological function |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Thaumatin-like protein | ACA50508 | 479 | Prospector | 3 | 21.0 | 18.0 | 18.1 | 4.4 | 6.5 | Defense |
| 2 | DUF26-like protein | AAW34053 | 475 | Prospector | 6 | 38.4 | 24.4 | 27.3 | 4.7 | 5.0 | Defense |
| 3 | Beta-1,3-glucanase | BAA77783 | 422 | Prospector | 4 | 15.9 | 32.2 | 34.2 | 4.6 | 4.6 | Carbohydrate metabolic process |
| 4 | Peroxidase | CAA46916 | 86 | Mascot | 7 | 29.0 | 37.1 | 32.9 | 5.2 | 5.8 | Response to oxidative stress |
| 5 | Enolase | ABB46862 | 374 | Prospector | 13 | 32.0 | 48.0 | 48.0 | 5.5 | 5.4 | Glycolytic process |
| 6 | Nucleoside diphosphate kinase | AAT08712 | 396 | Prospector | 4 | 27.0 | 18.5 | 17.7 | 5.4 | 9.1 | Energy |
| 7 | Formate dehydrogenase | NP_001057666 | 112 | Mascot | 8 | 32.0 | 36.6 | 41.2 | 6.4 | 6.9 | Response to oxidative stress |
| 8 | Cu/Zn-superoxide dismutase | BAB21760 | 228 | Prospector | 7 | 42.0 | 41.4 | 21.3 | 6.6 | 5.8 | Response to oxidative stress |
| 9 | Xylanase inhibitor protein 1 | NP_001057599 | 65 | Mascot | 5 | 24.0 | 44.6 | 27.4 | 6.9 | 6.4 | Carbohydrate metabolic process |
| 10 | Cytoplasmic malate dehydrogenase | AAG13573 | 372 | Prospector | 8 | 21.0 | 32.9 | 35.6 | 6.5 | 5.7 | Tricarboxylic acid cycle |
JIOsPR10, jasmonic inducible pathogenesis-related class 10; MP, matched peptides; SC, sequence coverage; Ex Mr, experimental molecular mass; Th Mr, theoretical molecular mass; Ex pI, experimental isoelectric point; Th pI, theoretical isoelectric point.
Fig. 6.
Overexpression of jasmonic inducible pathogenesis-related class 10 (JIOsPR10) induces highly accumulation of reactive oxygen species related protein. Same amount of protein was loaded on sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE). Antibody against DUF26-like protein, beta-1,3-glucanase, and Cu/Zn-SOD were used to detect the protein level expression in wild type (WT) and JIOsPR10 overexpression plant. Equal loading of protein samples was checked by PonceauS staining.
JIOsPR10 enhances tolerance to rice blast fungus
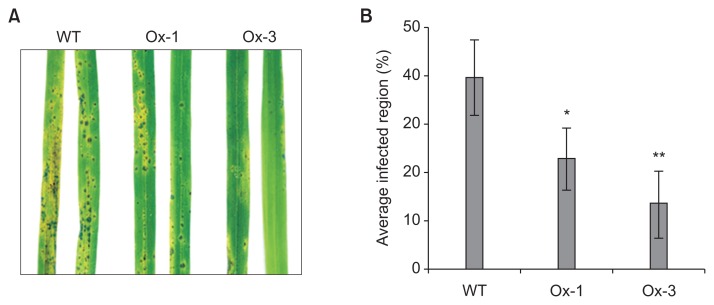
JIOsPR10 is a member of PR family genes, which provide biotic stress tolerance in rice. Therefore, the response of M. oryzae infection on transgenic lines was investigated. Spores of M. oryzae were sprayed on the leaves of WT and transgenic lines and the infection phenotype was analyzed after 72 h of spore inoculation. The number of infection lesions of M. oryzae was reduced in Ox-1 and Ox-3 lines in comparison with the WT (Fig. 7A). Average infected area on the leaves of Ox-1 and Ox-3 lines was reduced to 22.96% and 13.6%, respectively, from 39.72%, calculated in the WT (Fig. 7B).
Fig. 7.
Jasmonic inducible pathogenesis-related class 10 (JIOsPR10) overexpression plants showing increased tolerance to rice blast fungus. (A) Phenotype of wild type (WT) and JIOsPR10 overexpression plants infected with rice blast fungus at 3 days post inoculation (dpi). (B) Average percentage of infected area on WT, Ox-1, and Ox-3 leaves. Student’s t-test was applied on the data analysis. *P < 0.01; **P < 0.05.
Discussion
Crop productivity is strongly affected by the biotic and abiotic stresses in surrounding environments. Previous studies have shown that overexpression of stress responsive genes in plant may improve its adaption to various stresses and increase the yield (Li et al., 2014; Shi et al., 2014). Previously, JIOsPR10 was reported to be involved in the biotic stress tolerance, such as M. oryzae infections (Kim et al., 2004). In this study, we showed that JIOsPR10 was also induced by abiotic stresses, including drought and salt, suggesting JIOsPR10 has multiple functions in rice and might be important for agricultural usage (Fig. 1).
Previous studies have shown that PR10 family protein has ribonuclease (Bantignies et al., 2000; Zhou et al., 2002) and cysteine protease inhibition activities (Andrade et al., 2010). Rice JIOsPR10 was also reported to contain RNase activity, suggesting that biochemical activity might be essential for the role of JIOsPR10 (Kim et al., 2008b). Therefore, characterization of downstream regulated proteins contributes the understanding of molecular mechanism of JIOsPR10 induced stress tolerance. Proteomics, consists of protein separation and identification steps, is a widely used tool for large-scale identification of proteins. To investigate the mechanism of how JIOsPR10 promotes rice growth, and enhances stress tolerance, we therefore applied a 2-DE based comparative proteomic analysis of WT and JIOsPR10 overexpression plants. To increase the detection of low abundance proteins, a PEG mediated protein fractionation method, which depletes high-abundant proteins such as Rubisco, was used for protein extraction (Kim et al., 2001). The protein expression profile was observed by 2-DE gel analysis coupled with CBB staining. Differentially modulated protein spots from gel map of WT and transgenic plants were selected and identified by MALDI-TOF mass spectrometer, which revealed that proteins involved in metabolism, defense, and oxidative stress were predominant. In particular, several proteins related with glycolytic process (enolase, spot 5), tricarboxylic acid cycle (cytoplasmic malate dehydrogenase, spot 10), and energy (nucleoside diphosphate kinase, spot 6) were down regulated in transgenic plants, indicating a down regulation of metabolism inside the plant. The metabolic and energy related pathways generate ROS as by product in cells in response to environmental stresses (Andreyev et al., 2005). Previous studies have suggested that the early down regulation of metabolism is one of the consistent trends of plants in response to abiotic stress (Cramer et al., 2011). Therefore, overexpression of JIOsPR10 in rice may affect the balance between development and environmental stress adaption.
ROSs are key secondary signal molecules associated with both biotic and abiotic stresses in plants (Bailey-Serres and Mittler, 2006). Maintenance of ROS balance inside cell is essential for stress tolerance. Peroxidase has ability to generate oxidative burst and trigger immune responses in plants (Chittoor et al., 1997; Daudi et al., 2012; Sasaki et al., 2004). A peroxidase (spot 4) was highly expressed in Ox-3 lines as compared to the WT, suggesting overexpression of JIOsPR10 may lead to an induction of ROS. Meanwhile, Cu/Zn-SOD (spot 8) was highly accumulated in JIOsPR10 overexpression lines (Fig. 5). Cu/Zn-SOD has ability of catalyzing O2− into H2O2. Higher accumulation of Cu/Zn-SOD in transgenic plants might be related with the maintenance of in intracellular oxidative stress balances, therefore lead to a different phenotype in plant growth and stress resistance.
We also found that the expression of three proteins related with rice defense mechanism including TLP, DUF26-like protein, beta-1,3-glucanase was increased in the transgenic plants. TLP belongs to the PR-5 family based on the amino acid sequence and structural similarities (Edens et al., 1982). Overexpression of TLP enhances tolerance to different fungus (Acharya et al., 2013; Mackintosh et al., 2007; Munis et al., 2010), salt, drought, and oxidative stresses (Munis et al., 2010). The DUF26-like protein is also involved in multiples stress tolerance, such as rice blast fungus (Kim et al., 2004), salt stress (Zhang et al., 2009), and ozone stress (Wrzaczek et al., 2010). In plants, beta-1,3-glucanases are related with the pathogen infection, ozone, H2O2, wounding stresses, and plant development (Chandler and Robertson, 1994; Cheong et al., 2000; Hwang et al., 2007). The induction of those proteins may be triggered by the peroxidase induced oxidative stresses. Taken together, we suggest that enhancement of biotic and abiotic stresses of JIOsPR10 overexpression plant is probably because of its increased oxidative stress tolerance as compared to the WT.
Moreover, ROS were also emerged as important regulator of cell growth and plant development, and ROS are highly accumulated in plant during the germination and early development stages (Gapper and Dolan, 2006). In this study, we observed that overexpression of JIOsPR10 promotes the root and shoot development in early development stages (Fig. 3) which could be the result of enhanced ROS scavenging ability in transgenic plants. Similarly, overexpression of PR10 in Brassica napus enhanced seed germination under salinity conditions (Srivastava et al., 2004). Based on proteomics results, the up-regulated proteins such as TLP (Futamura et al., 2006), DUF26-like protein (Jiang et al., 2007), beta-1,3-glucanase (Cheong et al., 2000; Vögeli-Lange et al., 1994), and SOD (Kliebenstein et al., 1998) were also reported to be involved in plant development, indicating differential expression of those ROS and defense related proteins may be involved in plant growth promoting process. Furthermore, we also observed that the rice productivity was not changed in overexpression plants (Fig. 3D). However, these plants were grown in absence of any environmental stresses. In natural conditions, the overexpression plant may have high yield than WT due to its increase in tolerance to environmental stresses.
Here, we also confirmed that overexpression of JIOsPR10 in rice enhances M. oryzae tolerance. Meanwhile, transgenic plants also showed drought and salt stress tolerance phenotype. These results together indicate that JIOsPR10 is involved in the multiple processes of rice including early development, enhancement of host to fungal infection, and oxidative, drought and salt stress tolerance, demonstrating that JIOsPR10 is a good candidate gene for agricultural application to protect the crops against biotic and abiotic stresses. However, we also found that the transcriptional expression of JIOsPR10 in Ox-1 is higher than Ox-3 (Fig. 2), but Ox-3 rather than Ox-1 exhibits higher resistance against M. oryzae infection (Fig. 7). We also found that PR proteins such as DUF26-like protein and β-1,3-glucanase are differentially accumulated in Ox-1 and Ox-3 plant. Therefore, different correlation between transcriptional expression of JIOsPR10 and phenotype may cause by the differential accumulation of other PR in different transgenic lines due to its RNase enzyme activity or post-transcriptional modification.
Supplementary materials
Acknowledgments
This work was supported by a 2-Year Research Grant of Pusan National University.
References
- Acharya K, Pal AK, Gulati A, Kumar S, Singh AK, Ahuja PS. Overexpression of Camellia sinensis thaumatin-like protein, CsTLP in potato confers enhanced resistance to Macrophomina phaseolina and Phytophthora infestans infection. Mol Biotechnol. 2013;54:609–622. doi: 10.1007/s12033-012-9603-y. [DOI] [PubMed] [Google Scholar]
- Andrade LB, Oliveira AS, Ribeiro JK, Kiyota S, Vasconcelos IM, de Oliveira JT, de Sales MP. Effects of a novel pathogenesis-related class 10 (PR-10) protein from Crotalaria pallida roots with papain inhibitory activity against root-knot nematode Meloidogyne incognita. J Agric Food Chem. 2010;58:4145–4152. doi: 10.1021/jf9044556. [DOI] [PubMed] [Google Scholar]
- Andreyev AY, Kushnareva YE, Starkov AA. Mitochondrial metabolism of reactive oxygen species. Biochemistry (Mosc) 2005;70:200–214. doi: 10.1007/s10541-005-0102-7. [DOI] [PubMed] [Google Scholar]
- Bailey-Serres J, Mittler R. The roles of reactive oxygen species in plant cells. Plant Physiol. 2006;141:311. doi: 10.1104/pp.104.900191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bantignies B, Séguin J, Muzac I, Dédaldéchamp F, Gulick P, Ibrahim R. Direct evidence for ribonucleolytic activity of a PR-10-like protein from white lupin roots. Plant Mol Biol. 2000;42:871–881. doi: 10.1023/A:1006475303115. [DOI] [PubMed] [Google Scholar]
- Chandler PM, Robertson M. Gene expression regulated by abscisic acid and its relation to stress tolerance. Annu Rev Plant Physiol Plant Mol Biol. 1994;45:113–141. doi: 10.1146/annurev.pp.45.060194.000553. [DOI] [Google Scholar]
- Chen ZY, Brown RL, Damann KE, Cleveland TE. Identification of maize kernel endosperm proteins associated with resistance to aflatoxin contamination by Aspergillus flavus. Phytopathology. 2007;97:1094–1103. doi: 10.1094/PHYTO-97-9-1094. [DOI] [PubMed] [Google Scholar]
- Cheong YH, Kim CY, Chun HJ, Moon BC, Park HC, Kim JK, Lee S, Han C, Lee SY, Cho MJ. Molecular cloning of a soybean class III beta-1,3-glucanase gene that is regulated both developmentally and in response to pathogen infection. Plant Sci. 2000;154:71–81. doi: 10.1016/S0168-9452(00)00187-4. [DOI] [PubMed] [Google Scholar]
- Chittoor JM, Leach JE, White FF. Differential induction of a peroxidase gene family during infection of rice by Xanthomonas oryzae pv. oryzae. Mol Plant-Microbe Interact. 1997;10:861–871. doi: 10.1094/MPMI.1997.10.7.861. [DOI] [PubMed] [Google Scholar]
- Cramer GR, Urano K, Delrot S, Pezzotti M, Shinozaki K. Effects of abiotic stress on plants: a systems biology perspective. BMC Plant Biol. 2011;11:163. doi: 10.1186/1471-2229-11-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta K, Velazhahan R, Oliva N, Ona I, Mew T, Khush GS, Muthukrishnan S, Datta SK. Over-expression of the cloned rice thaumatin-like protein (PR-5) gene in transgenic rice plants enhances environmental friendly resistance to Rhizoctonia solani causing sheath blight disease. Theor Appl Genet. 1999;98:1138–1145. doi: 10.1007/s001220051178. [DOI] [Google Scholar]
- Daudi A, Cheng Z, O’Brien JA, Mammarella N, Khan S, Ausubel FM, Bolwell GP. The apoplastic oxidative burst peroxidase in Arabidopsis is a major component of pattern-triggered immunity. Plant Cell. 2012;24:275–287. doi: 10.1105/tpc.111.093039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de las Mercedes Dana M, Pintor-Toro JA, Cubero B. Transgenic tobacco plants overexpressing chitinases of fungal origin show enhanced resistance to biotic and abiotic stress agents. Plant Physiol. 2006;142:722–730. doi: 10.1104/pp.106.086140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan J, Zhang M, Zhang H, Xiong H, Liu P, Ali J, Li J, Li Z. OsMIOX, a myo-inositol oxygenase gene, improves drought tolerance through scavenging of reactive oxygen species in rice (Oryza sativa L.) Plant Sci. 2012;196:143–151. doi: 10.1016/j.plantsci.2012.08.003. [DOI] [PubMed] [Google Scholar]
- Edens L, Heslinga L, Klok R, Ledeboer AM, Maat J, Toonen MY, Visser C, Verrips CT. Cloning of cDNA encoding the sweet-tasting plant protein thaumatin and its expression in Escherichia coli. Gene. 1982;18:1–12. doi: 10.1016/0378-1119(82)90050-6. [DOI] [PubMed] [Google Scholar]
- Fernandes H, Michalska K, Sikorski M, Jaskolski M. Structural and functional aspects of PR-10 proteins. FEBS J. 2013;280:1169–1199. doi: 10.1111/febs.12114. [DOI] [PubMed] [Google Scholar]
- Flores T, Alape-Girón A, Flores-Díaz M, Flores HE. Ocatin. A novel tuber storage protein from the andean tuber crop oca with antibacterial and antifungal activities. Plant Physiol. 2002;128:1291–1302. doi: 10.1104/pp.010541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraire-Velázquez S, Rodríguez-Guerra R, Sánchez-Calderón L. Abiotic and biotic stress response crosstalk in plants. In: Shanker AK, Venkateswarlu B, editors. Abiotic stress response in plants: physiological, biochemical and genetic perspectives. InTech; Rijeka, Croatia: 2011. pp. 3–26. [Google Scholar]
- Futamura N, Tani N, Tsumura Y, Nakajima N, Sakaguchi M, Shinohara K. Characterization of genes for novel thaumatin-like proteins in Cryptomeria japonica. Tree Physiol. 2006;26:51–62. doi: 10.1093/treephys/26.1.51. [DOI] [PubMed] [Google Scholar]
- Gapper C, Dolan L. Control of plant development by reactive oxygen species. Plant Physiol. 2006;141:341–345. doi: 10.1104/pp.106.079079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta R, Lee SE, Agrawal GK, Rakwal R, Park S, Wang Y, Kim ST. Understanding the plantpathogen interactions in the context of proteomics-generated apoplastic proteins inventory. Front Plant Sci. 2015;6:352. doi: 10.3389/fpls.2015.00352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto M, Kisseleva L, Sawa S, Furukawa T, Komatsu S, Koshiba T. A novel rice PR10 protein, RSOsPR10, specifically induced in roots by biotic and abiotic stresses, possibly via the jasmonic acid signaling pathway. Plant Cell Physiol. 2004;45:550–559. doi: 10.1093/pcp/pch063. [DOI] [PubMed] [Google Scholar]
- He C, Lin Z, McElroy D, Wu R. Identification of a rice actin2 gene regulatory region for high-level expression of transgenes in monocots. Plant Biotechnol J. 2009;7:227–239. doi: 10.1111/j.1467-7652.2008.00393.x. [DOI] [PubMed] [Google Scholar]
- Hwang DH, Kim ST, Kim SG, Kang KY. Comprehensive analysis of the expression of twenty-seven beta-1, 3-glucanase genes in rice (Oryza sativa L.) Mol Cells. 2007;23:207–214. [PubMed] [Google Scholar]
- Jiang J, Li J, Xu Y, Han Y, Bai Y, Zhou G, Lou Y, Xu Z, Chong K. RNAi knockdown of Oryza sativa root meander curling gene led to altered root development and coiling which were mediated by jasmonic acid signalling in rice. Plant Cell Environ. 2007;30:690–699. doi: 10.1111/j.1365-3040.2007.01663.x. [DOI] [PubMed] [Google Scholar]
- Jwa NS, Agrawal GK, Rakwal R, Park CH, Agrawal VP. Molecular cloning and characterization of a novel Jasmonate inducible pathogenesis-related class 10 protein gene, JIOsPR10, from rice (Oryza sativa L.) seedling leaves. Biochem Biophys Res Commun. 2001;286:973–983. doi: 10.1006/bbrc.2001.5507. [DOI] [PubMed] [Google Scholar]
- Kim JY, Wu J, Kwon SJ, Oh H, Lee SE, Kim SG, Wang Y, Agrawal GK, Rakwal R, Kang KY, Ahn IP, Kim BG, Kim ST. Proteomics of rice and Cochliobolus miyabeanus fungal interaction: insight into proteins at intracellular and extracellular spaces. Proteomics. 2014;14:2307–2318. doi: 10.1002/pmic.201400066. [DOI] [PubMed] [Google Scholar]
- Kim SG, Wang Y, Lee KH, Park ZY, Park J, Wu J, Kwon SJ, Lee YH, Agrawal GK, Rakwal R, Kim ST, Kang KY. In-depth insight into in vivo apoplastic secretome of rice-Magnaporthe oryzae interaction. J Proteomics. 2013;78:58–71. doi: 10.1016/j.jprot.2012.10.029. [DOI] [PubMed] [Google Scholar]
- Kim ST, Cho KS, Jang YS, Kang KY. Twodimensional electrophoretic analysis of rice proteins by polyethylene glycol fractionation for protein arrays. Electrophoresis. 2001;22:2103–2109. doi: 10.1002/1522-2683(200106)22:10<2103::AID-ELPS2103>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- Kim ST, Kang SY, Wang Y, Kim SG, Hwang DH, Kang KY. Analysis of embryonic proteome modulation by GA and ABA from germinating rice seeds. Proteomics. 2008a;8:3577–3587. doi: 10.1002/pmic.200800183. [DOI] [PubMed] [Google Scholar]
- Kim ST, Kim SG, Hwang DH, Kang SY, Kim HJ, Lee BH, Lee JJ, Kang KY. Proteomic analysis of pathogen-responsive proteins from rice leaves induced by rice blast fungus, Magnaporthe grisea. Proteomics. 2004;4:3569–3578. doi: 10.1002/pmic.200400999. [DOI] [PubMed] [Google Scholar]
- Kim ST, Yu S, Kang YH, Kim SG, Kim JY, Kim SH, Kang KY. The rice pathogen-related protein 10 (JIOsPR10) is induced by abiotic and biotic stresses and exhibits ribonuclease activity. Plant Cell Rep. 2008b;27:593–603. doi: 10.1007/s00299-007-0485-6. [DOI] [PubMed] [Google Scholar]
- Kliebenstein DJ, Monde RA, Last RL. Superoxide dismutase in Arabidopsis: an eclectic enzyme family with disparate regulation and protein localization. Plant Physiol. 1998;118:637–650. doi: 10.1104/pp.118.2.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnaswamy S, Baral PK, James MN, Kav NN. Site-directed mutagenesis of histidine 69 and glutamic acid 148 alters the ribonuclease activity of pea ABR17 (PR10.4) Plant Physiol Biochem. 2011;49:958–962. doi: 10.1016/j.plaphy.2010.10.010. [DOI] [PubMed] [Google Scholar]
- Li JB, Luan YS, Yin YL. SpMYB overexpression in tobacco plants leads to altered abiotic and biotic stress responses. Gene. 2014;547:145–151. doi: 10.1016/j.gene.2014.06.049. [DOI] [PubMed] [Google Scholar]
- Mackintosh CA, Lewis J, Radmer LE, Shin S, Heinen SJ, Smith LA, Wyckoff MN, Dill-Macky R, Evans CK, Kravchenko S, Baldridge GD, Zeyen RJ, Muehlbauer GJ. Overexpression of defense response genes in transgenic wheat enhances resistance to Fusarium head blight. Plant Cell Rep. 2007;26:479–488. doi: 10.1007/s00299-006-0265-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGee JD, Hamer JE, Hodges TK. Characterization of a PR-10 pathogenesis-related gene family induced in rice during infection with Magnaporthe grisea. Mol Plant-Microbe Interact. 2001;14:877–886. doi: 10.1094/MPMI.2001.14.7.877. [DOI] [PubMed] [Google Scholar]
- Munis MF, Tu L, Deng F, Tan J, Xu L, Xu S, Long L, Zhang X. A thaumatin-like protein gene involved in cotton fiber secondary cell wall development enhances resistance against Verticillium dahliae and other stresses in transgenic tobacco. Biochem Biophys Res Commun. 2010;393:38–44. doi: 10.1016/j.bbrc.2010.01.069. [DOI] [PubMed] [Google Scholar]
- Park CJ, Kim KJ, Shin R, Park JM, Shin YC, Paek KH. Pathogenesis-related protein 10 isolated from hot pepper functions as a ribonuclease in an antiviral pathway. Plant J. 2004;37:186–198. doi: 10.1046/j.1365-313X.2003.01951.x. [DOI] [PubMed] [Google Scholar]
- Rizhsky L, Davletova S, Liang H, Mittler R. The zinc finger protein Zat12 is required for cytosolic ascorbate peroxidase 1 expression during oxidative stress in Arabidopsis. J Biol Chem. 2004;279:11736–11743. doi: 10.1074/jbc.M313350200. [DOI] [PubMed] [Google Scholar]
- Sarowar S, Kim YJ, Kim EN, Kim KD, Hwang BK, Islam R, Shin JS. Overexpression of a pepper basic pathogenesis-related protein 1 gene in tobacco plants enhances resistance to heavy metal and pathogen stresses. Plant Cell Rep. 2005;24:216–224. doi: 10.1007/s00299-005-0928-x. [DOI] [PubMed] [Google Scholar]
- Sasaki K, Iwai T, Hiraga S, Kuroda K, Seo S, Mitsuhara I, Miyasaka A, Iwano M, Ito H, Matsui H, Ohashi Y. Ten rice peroxidases redundantly respond to multiple stresses including infection with rice blast fungus. Plant Cell Physiol. 2004;45:1442–1452. doi: 10.1093/pcp/pch165. [DOI] [PubMed] [Google Scholar]
- Shi W, Hao L, Li J, Liu D, Guo X, Li H. The Gossypium hirsutum WRKY gene GhWRKY39-1 promotes pathogen infection defense responses and mediates salt stress tolerance in transgenic Nicotiana benthamiana. Plant Cell Rep. 2014;33:483–498. doi: 10.1007/s00299-013-1548-5. [DOI] [PubMed] [Google Scholar]
- Srivastava S, Fristensky B, Kav NN. Constitutive expression of a PR10 protein enhances the germination of Brassica napus under saline conditions. Plant Cell Physiol. 2004;45:1320–1324. doi: 10.1093/pcp/pch137. [DOI] [PubMed] [Google Scholar]
- Stintzi A, Heitz T, Prasad V, Wiedemann-Merdinoglu S, Kauffmann S, Geoffroy P, Legrand M, Fritig B. Plant ‘pathogenesis-related’ proteins and their role in defense against pathogens. Biochimie. 1993;75:687–706. doi: 10.1016/0300-9084(93)90100-7. [DOI] [PubMed] [Google Scholar]
- van Loon LC, Pierpoint WS, Boller T, Conejero V. Recommendations for naming plant pathogenesisrelated proteins. Plant Mol Biol Report. 1994;12:245–264. doi: 10.1007/BF02668748. [DOI] [Google Scholar]
- van Loon LC, van Strien EA. The families of pathogenesis-related proteins, their activities, and comparative analysis of PR-1 type proteins. Physiol Mol Plant Pathol. 1999;55:85–97. doi: 10.1006/pmpp.1999.0213. [DOI] [Google Scholar]
- Vögeli-Lange R, Fründt C, Hart CM, Nagy F, Meins F. Developmental, hormonal, and pathogenesis-related regulation of the tobacco class I β-1,3-glucanase B promoter. Plant Mol Biol. 1994;25:299–311. doi: 10.1007/BF00023245. [DOI] [PubMed] [Google Scholar]
- Wang Y, Kim SG, Wu J, Kim ST, Kang KY. Differential proteome and secretome analysis during rice-pathogen interaction. Methods Mol Biol. 2014;1072:563–572. doi: 10.1007/978-1-62703-631-3_38. [DOI] [PubMed] [Google Scholar]
- Wrzaczek M, Brosché M, Salojärvi J, Kangasjärvi S, Idänheimo N, Mersmann S, Robatzek S, Karpiński S, Karpińska B, Kangasjärvi J. Transcriptional regulation of the CRK/DUF26 group of receptor-like protein kinases by ozone and plant hormones in Arabidopsis. BMC Plant Biol. 2010;10:95. doi: 10.1186/1471-2229-10-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie YR, Chen ZY, Brown RL, Bhatnagar D. Expression and functional characterization of two pathogenesis-related protein 10 genes from Zea mays. J Plant Physiol. 2010;167:121–130. doi: 10.1016/j.jplph.2009.07.004. [DOI] [PubMed] [Google Scholar]
- Zhang L, Tian LH, Zhao JF, Song Y, Zhang CJ, Guo Y. Identification of an apoplastic protein involved in the initial phase of salt stress response in rice root by two-dimensional electrophoresis. Plant Physiol. 2009;149:916–928. doi: 10.1104/pp.108.131144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou XJ, Lu S, Xu YH, Wang JW, Chen XY. A cotton cDNA (GaPR-10) encoding a pathogenesisrelated 10 protein with in vitro ribonuclease activity. Plant Sci. 2002;162:629–636. doi: 10.1016/S0168-9452(02)00002-X. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.