Abstract
MicroRNAs (miRs) are a class of small noncoding RNAs that negatively regulate the gene expression by directly binding to the 3′ untranslated region of their target mRNA, thus resulting in mRNA degradation or translational repression. miR-9 has recently been demonstrated to play a role in the development and progression of malignant melanoma (MM), but the regulatory mechanism of miR-9 in the malignant phenotypes of MM still remains largely unknown. In this study, a total of 73 pairs of MM tissues and adjacent normal tissues were collected. Real-time reverse transcription polymerase chain reaction and Western blot were used to detect the mRNA and protein expression of miR-9. MTT assay, wound healing assay, and transwell assay were conducted to determine the cell proliferation, migration, and invasion. Luciferase reporter assay was used to determine the targeting relationship between miR-9 and NRP1. Our data demonstrated that miR-9 expression was significantly downregulated in MM tissues compared with that in adjacent normal tissues. The decreased miR-9 level was significantly associated with the tumor stage and metastasis of MM. We also found that the expression level of miR-9 was decreased in MM cell lines (G361, B16, A375, and HME1) compared with normal skin HACAT cells. Ectopic expression of miR-9 led to a significant decrease in the ability of proliferation, migration, and invasion in A375 cells. NRP1 was further identified as a direct target gene of miR-9, and the protein expression of NRP1 was negatively regulated by miR-9 in A375 cells. Furthermore, overexpression of NRP1 reversed the suppressive effects of miR-9 on the malignant phenotypes of A375 cells. In vivo study revealed that miR-9 overexpression decreased the tumor growth, while overexpression of NRP1 increased MM growth. In summary, our findings suggest that the miR-9/NRP1 axis may serve as a potential target for the treatment of MM.
Keywords: malignant melanoma, microRNA, neuropilin1, proliferation, migration, invasion
Introduction
Malignant melanoma (MM) is a malignant tumor originating from melanocytes and is a dangerous disease entity that is ranked as the seventh most common malignancy in females and the fifth most common cancer in males globally.1,2 It is characterized by a rapid progression and lymph node metastasis.3 Furthermore, the recurrence of MM is common if not detected in the early stage, and thus has become the deadliest skin cancer.3 Recently, the incidence of MM is estimated to have increased by 3.1% in 1 year, and thus has become a continuing threat to health.2 Recent studies have identified some oncogenes and tumor suppressors involved in MM, which show promise to serve as therapeutic targets for the treatment of MM.4–6
MicroRNAs (miRs) are a class of small noncoding RNAs that negatively regulate the gene expression via directly binding to the 3′ untranslational region (UTR) of their target mRNA, thus resulting in mRNA degradation or translational repression.7 Through regulation of gene expression, miRs are involved in a variety of biological processes including development and differentiation, cell survival and proliferation, cell cycle progression, apoptosis and autophagy, migration and invasion, angiogenesis, as well as tumorigenesis.8–10 Moreover, aberrant upregulation or downregulation of miRs has been observed in MM, some of which have been confirmed to play key roles in the development and progression of this disease.11,12 At the invasive front of primary cutaneous melanoma, the decreased levels of miR-200 and miR-203 are significantly associated with the disease progression.13 miR-340 acts as a tumor suppressor in MM through suppressing the activity of RAS–RAF–MAPK signaling.14 Our previous study has also demonstrated that miR-203 inhibits MM cell migration via inhibition of versican expression.15
miR-9 has been implicated in various human cancers, and it functions as either an oncogene or a tumor suppressor due to the tumor specificity.16 For instance, miR-9 expression was recently reported to be significantly upregulated in non-small-cell lung cancer (NSCLC) tissues, which further promoted the growth of NSCLC cells and decreased the growth inhibitory effect of erlo-tinib.17 In contrast, miR-9 played a suppressive role by targeting TAZ in hepatocellular carcinoma cells.18 Recently, several studies revealed that miR-9 was involved in the pathogenesis of MM. It suppressed the migration and invasion of MM cells through inhibiting the NF-κB1 signaling and Snail1, while increasing the expression of E-cadherin.19,20 Moreover, Zhao et al21 reported that YY1 inhibited the tumorigenesis of MM through upregulating miR-9, which subsequently downregulated its target RYBP. However, the detailed mechanism of the role of miR-9 in the regulation of the malignant phenotypes of MM cells still remains to be fully investigated.
In this study, we aimed to investigate the expression of miR-9 in MM and the regulatory mechanism of miR-9 in the malignant phenotypes of MM cells.
Materials and methods
Ethics statement
This study was approved by the Ethics Committee of Third Xiangya Hospital, Central South University, Changsha, People’s Republic of China. All animal experiments complied with the Animal Care Guideline of Central South University, Changsha, People’s Republic of China.
Tissue collection
A total of 73 pairs of MM tissues and adjacent normal tissues were collected from Third Xiangya Hospital. All specimens were confirmed as MM by pathological diagnosis. All patients involved in this study received no radiation therapy or chemotherapy before surgical resection. Written informed consents were obtained from the MM patients involved in this study. The study included 35 females and 30 males, ranging in age from 41 to 74 years with a mean age of 56.5 years. The clinicopathological information is summarized in Table 1. Tissues were snap frozen in liquid nitrogen, and then stored at −80°C before use.
Table 1.
Relationship between miR-9 expression and clinicopathologic features of malignant melanoma patients
| Feature | Cases (n) |
miR-9 expression
|
χ2 | P-value | |
|---|---|---|---|---|---|
| Low (n=45) |
High (n=28) |
||||
| Sex | 0.252 | 0.615 | |||
| Male | 39 | 23 | 16 | ||
| Female | 34 | 22 | 12 | ||
| Age (years) | 0.027 | 0.868 | |||
| ≤56 | 33 | 20 | 13 | ||
| >56 | 40 | 25 | 15 | ||
| Tumor thickness | 0.847 | 0.357 | |||
| ≤1 mm | 31 | 21 | 10 | ||
| >1 mm | 42 | 24 | 18 | ||
| TNM stag e | 4.447 | 0.035 | |||
| I/II | 28 | 13 | 15 | ||
| III | 45 | 32 | 13 | ||
| Lymph node metastasis | 5.726 | 0.017 | |||
| No | 34 | 16 | 18 | ||
| Yes | 39 | 29 | 10 | ||
Abbreviations: miR-9, microRNA-9; TNM, tumor node metastasis.
Cell culture
Human normal skin HACAT cells and MM cell lines (G361, B16, A375, and HME1) were purchased from the Cell Bank of Central South University. All cell lines were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM) (Thermo Fisher Scientific, Waltham, MA, USA) added with 10% fetal bovine serum (FBS; Thermo Fisher Scientific) in a 37°C humidified atmosphere with 5% CO2.
Real-time reverse transcription PCR assay
Total RNA was extracted using Trizol Reagent (Thermo Fisher Scientific) according to the manufacturer’s instructions. Total RNA (1,000 ng) was reverse transcribed into cDNA using Reverse Transcription Kit (Thermo Fisher Scientific), according to the manufacturer’s instruction. For mRNA detection, a Q-PCR Detection Kit (Thermo Fisher Scientific) was used to conduct real-time polymerase chain reaction (PCR) on ABI 7500 thermocycler (Thermo Fisher Scientific). GAPDH was used as internal reference. The PCR steps were: 95°C for 5 minutes, 45 cycles of denaturation at 95°C for 15 seconds, and annealing/elongation step at 60°C for 30 seconds. For miR-9 detection, an MiRNA Q-PCR Detection Kit (GeneCopoeia, Rockville, MD, USA) was used to conduct real-time PCR on ABI 7500 thermocycler. U6 gene was used as internal reference. The PCR steps were 95°C for 10 minutes, 40 cycles of denaturation at 95°C for 15 seconds, and annealing/elongation step at 60°C for 60 seconds. The relative expression was analyzed by the 2−ΔΔCt method.
Western blot
Cells were lysed with ice-cold lysis buffer (50 mM Tris–HCl, pH 6.8, 100 mM 2-mercaptoethanol, 2%w/v sodium dodecyl sulfate, 10% glycerol). Proteins were quantified and separated with 10% sodium dodecyl sulfate–polyacrylamide electrophoresis and then transferred onto a polyvinylidene difluoride (PVDF) membrane (Thermo Fisher Scientific), followed by incubation with phosphate buffered saline (PBS) containing 5% milk (Mengniu, Beijing, People’s Republic of China) at room temperature for 3 hours. After washing with PBS containing Tween 20 (PBST) three times, the PVDF membrane was incubated with mouse anti-NRP1 polyclonal antibody (1:1,000, Abcam, Cambridge, UK) and mouse anti-GAPDH polyclonal antibody (Abcam) at room temperature for 3 hours, respectively. After washing with PBST for 15 minutes, it was then incubated with goat anti-mouse IgG (1:3,000, Abcam) at room temperature for 40 minutes. After washing with PBST for 15 minutes, and PBS for 5 minutes, Super Signal West Pico Chemiluminescent Substrate Kit (Pierce Biotechnology, Waltham, MA, USA) was used to detect signals. The relative protein expression was analyzed by Image-Pro plus software 6.0 (Media Cybernetics, Rockville, MD, USA). GAPDH was used as internal reference.
miR mimic and inhibitor, and plasmid
The miR-9 mimic, scramble miR mimic (miR-NC), miR-9 inhibitor, negative control (NC) inhibitor, pcDNA3.1-NRP1 plasmid, pLVTH-miR-9 lentiviral plasmid, and pLVTH-NRP1 lentiviral plasmid were generated from Amspring (Changsha, People’s Republic of China).
MTT assay
MTT assay was used to examine the cell proliferating capacity. A375 cells were seeded at a density of 5,000 cells per well in a 96-well plate. After culturing for 0, 12, 24, 48, and 72 hours, cells were incubated with MTT at a final concentration of 0.5 mg/mL at 37°C for 4 hours. Then, the medium containing MTT was removed, and dimethyl sulfoxide solution (150 μL) was added to dissolve the formazan crystals. The absorbance was read at 570 nm using a UV2600 spectrophotometer reader (Shimadzu, Kyoto, Japan).
Cell migration assay
Wound healing assay was used to examine the cell migration. A375 cells in each group were cultured to full confluence. Approximately, 1 mm width of wound was created by using a plastic scriber. Cells were washed with PBS and incubated in DMEM medium without FBS for 24 hours. After that, cells were incubated in the DMEM medium added with 10% FBS for 48 hours. Then, cells were observed under a CX31microscope (Olympus, Tokyo, Japan).
Cell invasion assay
Cell invasion assay was performed using transwell chambers precoated with Matrigel (BD, Franklin Lakes, NJ, USA). DMEM with 10% FBS was added into the lower chamber. A375 cell suspension (5×105 cells/mL) was prepared in DMEM, 300 μL of which was added into the upper chamber. After culturing at 37°C for 24 hours, a cotton-tipped swab was used to wipe out the cells that did not invade through the membrane on the upper chamber. The filters were fixed in absolute alcohol, stained by crystal violet (Sigma-Aldrich Co., St Louis, MO, USA), and observed under a CX31 microscope.
Bioinformatics predication and luciferase reporter assay
Bioinformatics predication was conducted to analyze the putative target genes of miR-9 using the TargetScan software (www.targetscan.org), and NRP1 was indicated as a potential target. The wild type (WT) of NRP1 UTR was amplified and cloned into the downstream of the firefly luciferase coding region of pMIR-GLOTM Luciferase vector (Promega Corporation, Fitchburg, WI, USA). Mutations of miR-9 binding sites were introduced using Easy Mutagenesis System Kit (Promega Corporation), in accordance with the manufacturer’s protocol, which was also cloned into the pMIR-GLOTM Luciferase vector, generating the mutant type (MT) of NRP1 3′UTR reporter plasmid. A375 cells cotransfected with WT-NRP1-3′UTR or MUT-NRP1-3′UTR vector and miR-9 mimic or miR-NC were seeded into 24-well plates. After culturing at 37°C for 48 hours, cells were lysed using the lysis buffer (Promega Corporation), and Dual-Luciferase Reporter Assay System (Promega Corporation) was used to detect the luciferase activity according to the manufacturer’s instruction.
Tumor growth in vivo
Male BALB/C-nu/nu nude mice (8 weeks) were maintained under pathogen-free conditions at the Animal Center of Central South University. A375 cells were stably transfected with the pLVTH-miR-9 lentiviral plasmid or pLVTH-NRP1 lentiviral plasmid. In the control group, A375 cells were stably transfected with the blank pLVTH vector. Nude mice (n=6 for each group) were injected subcutaneously in the dorsal flank with 1×107 A375 cells of each group. Survival time was recorded in each group. At 60 days after tumor implantation, all mice were sacrificed if not already dead. Tumor weight was also recorded. Tumor volume was measured every 5 days, which was calculated by using the formula V (mm3) =0.5× a × b2 (a is the maximum length to diameter and b is the maximum transverse diameter).
Statistical analysis
SPSS 17.0 software (SPSS Inc., Chicago, IL, USA) was used for statistical analysis. Data were expressed as mean ± standard deviation. Data were analyzed by using a Student’s t-test for two-group comparison and one-way analysis of variance for multiple-group comparison. P-value less than 0.05 was considered statistically significant.
Results
miR-9 is downregulated in MM and associated with its malignant progression
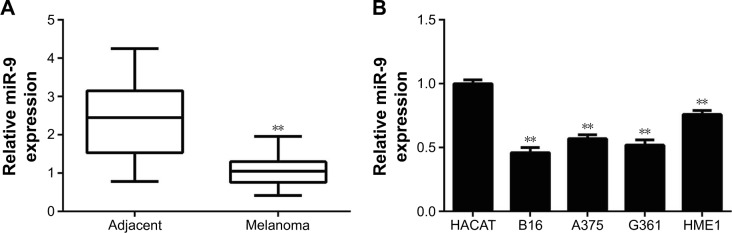
In the present study, we first examined the miR-9 expression in a total of 73 pairs of MM tissues and adjacent normal tissues by using real-time reverse transcription PCR. Our data showed that miR-9 expression was significantly downregulated in MM tissues compared with adjacent normal tissues (Figure 1A). According to the mean value of the miR-9 expression as the cutoff point, we divided MM patients into two groups, high miR-9 level group (n=28) and low miR-9 level (n=45) group. We analyzed the correlation between the miR-9 level and the clinicopathological features of MM. Although no significant association was observed in the miR-9 expression with respect to age, sex, and tumor thickness in MM patients, the decreased level of miR-9 was significantly associated with the advanced tumor stage and lymph node metastasis of MM (Table 1). These findings suggest that downregulation of miR-9 is involved in the malignant progression of MM.
Figure 1.
(A) Real-time PCR data indicated that miR-9 was significantly downregulated in melanoma tissues compared to their matched adjacent normal tissues. **P<0.01 vs adjacent. (B) Real-time PCR data indicated that miR-9 was significantly downregulated in human melanoma cell lines (G361, B16, A375, and HME1) compared to human normal skin HACAT cells. **P<0.01 vs HACAT.
Abbreviations: PCR, polymerase chain reaction; miR, microRNA.
In addition, we further analyzed the expression of miR-9 in several MM cell lines including G361, B16, A375, and HME1, and human normal skin HACAT cells. Real-time PCR data demonstrated that miR-9 was also markedly downregulated in MM cell lines compared with HACAT cells (Figure 1B). Taken together, miR-9 was significantly downregulated in MM tissues and cell lines.
Ectopic expression of miR-9 reduces the proliferation, migration, and invasion of MM cells
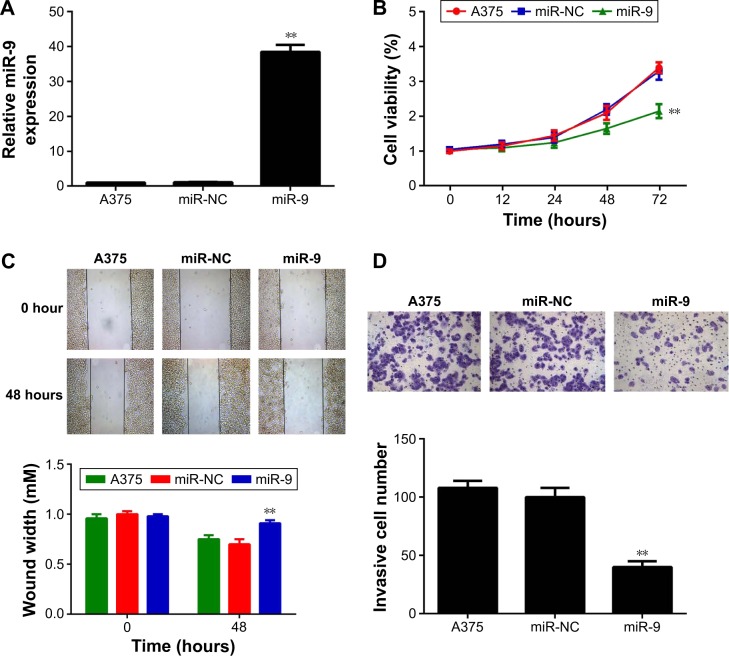
A375 cells originating from human melanoma were then used to investigate the regulatory role of miR-9 in MM in vitro. miR-9 mimic was transfected into A375 cells to upregulate miR-9 expression level. Real-time PCR data showed that the expression level of miR-9 was significantly increased after transfection with miR-9 mimic when compared with the control group; however, transfection with miR-NC showed no effect on the miR-9 level in A375 cells (Figure 2A).
Figure 2.
(A) Real-time PCR was used to examine the miR-9 level in A375 cells transfected with scramble miR mimic (miR-NC) or miR-9 mimic. Nontransfected A375 cells were used as control group. (B) MTT assay was used to measure cell viability in A375 cells transfected with miR-NC or miR-9 mimic, or the untreated cells. (C) Wound healing assay was used to measure the ability of cell migration in A375 cells transfected with miR-NC or miR-9 mimic, or the untreated cells (40×). (D) Transwell assay was used to measure the ability of cell invasion in A375 cells transfected with miR-NC or miR-9 mimic, or the untreated cells (200×). **P<0.01 vs negative control (miR-NC).
Abbreviations: PCR, polymerase chain reaction; miR, microRNA.
To reveal the role of miR-9 in MM cell proliferation, MTT assay was conducted in MM cells in each group. As demonstrated in Figure 2B, the ectopic expression of miR-9 significantly reduced the proliferation of A375 cells compared with the control group (Figure 2B), suggesting that miR-9 has an inhibitory effect on MM growth. Cell migration and invasion are important for metastasis. Therefore, we further conducted wound healing assay and transwell assay to examine the cell migration and invasion of A375 cells with or without ectopic expression of miR-9. Our data demonstrated that ectopic expression of miR-9 led to a significantly decreased migration and invasion of A375 cells when compared with the control group (Figure 2C and D, respectively). Therefore, miR-9 also plays a suppressive role in MM metastasis.
NRP1 is identified as a direct target gene of miR-9
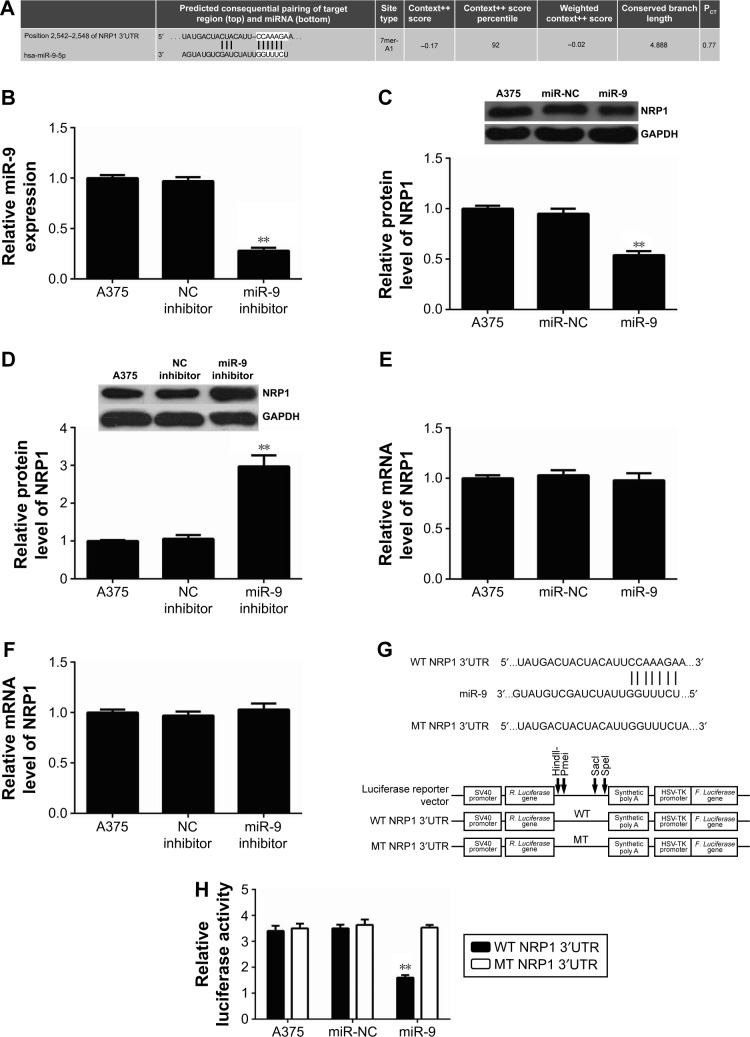
As miRs functions were achieved through negatively regulating the expression of their target genes, we further used the computational algorithm, TargetScan (Whitehead Institute for Biomedical Research, Cambridge, MA, USA), to analyze the putative targets of miR-9. As indicated in Figure 3A, perfect base pairing was observed between the 3′UTR of NRP1 mRNA and the seed sequence of miR-9. To verify whether NRP1 was indeed a target gene of miR-9, we firstly examined the effect of miR-9 overexpression or downregulation on the NRP1 expression in A375 cells. Real-time PCR data showed that the miR-9 level was significantly reduced after transfection with miR-9 inhibitor, while transfection with NC inhibitor did not affect the miR-9 level in A375 cells (Figure 3B). Our data also showed that overexpression of miR-9 caused a decreased protein level of NRP1, while downregulation of miR-9 enhanced the protein expression of NRP1 (Figure 3C and D), indicating that miR-9 negatively mediates the protein expression of NRP1 in A375 cells. In addition, neither miR-9 overexpression nor miR-9 knockdown affected the mRNA expression of NRP1 in A375 cells (Figure 3E and F). These data indicate that miR-9 indeed negatively regulates the NRP1 expression at a posttranscriptional level.
Figure 3.
(A) TargetScan software showed a perfect base pairing between the 3′UTR of NRP1 mRNA and the seed sequence of miR-9. (B) Real-time PCR was used to examine the miR-9 level in A375 cells transfected with negative control (NC) inhibitor or miR-9 inhibitor. Nontransfected A375 cells were used as control group. (C–F) Western blot assay and real-time PCR were used to examine the protein and mRNA level of NRP1 in A375 cells transfected with scramble miR mimic (miR-NC), miR-9 mimic, NC inhibitor, or miR-9 inhibitor. Nontransfected A375 cells were used as control group. (G) The wild type (WT) or mutant type (MT) of NRP1 3′UTR was subcloned into the luciferase reporter vector. (H) Luciferase reporter assay data indicated that cotransfection with WT-NRP1 3′UTR reporter plasmid and miR-9 mimic caused a significant decrease in the luciferase activity; however, cotransfection with MT-NRP1 3′UTR reporter plasmid and miR-9 mimic did not affect the luciferase activity when compared to the control group. **P<0.01 vs A375.
Abbreviations: UTR, untranslated region; PCR, polymerase chain reaction; miR, microRNA.
To further confirm the targeting relationship between miR-9 and NRP1, the WT or MT of NRP1 3′UTR was sub-cloned into the luciferase reporter vector (Figure 3G), and luciferase reporter assay was further conducted. As indicated in Figure 3H, cotransfection with WT-NRP1 3′UTR reporter plasmid and miR-9 mimic caused a significant decrease in the luciferase activity; however, cotransfection with MT-NRP1 3′UTR reporter plasmid and miR-9 mimic did not affect the luciferase activity when compared to the control group. These data indicate that miR-9 indeed directly binds to the 3′UTR of NRP1. Taken together these aforementioned findings, we demonstrate that miR-9 negatively regulates the protein expression of NRP1 via directly binding to its 3′UTR.
Overexpression of NRP1 reverses the inhibitory effects of miR-9 on the proliferation, migration, and invasion of MM cells
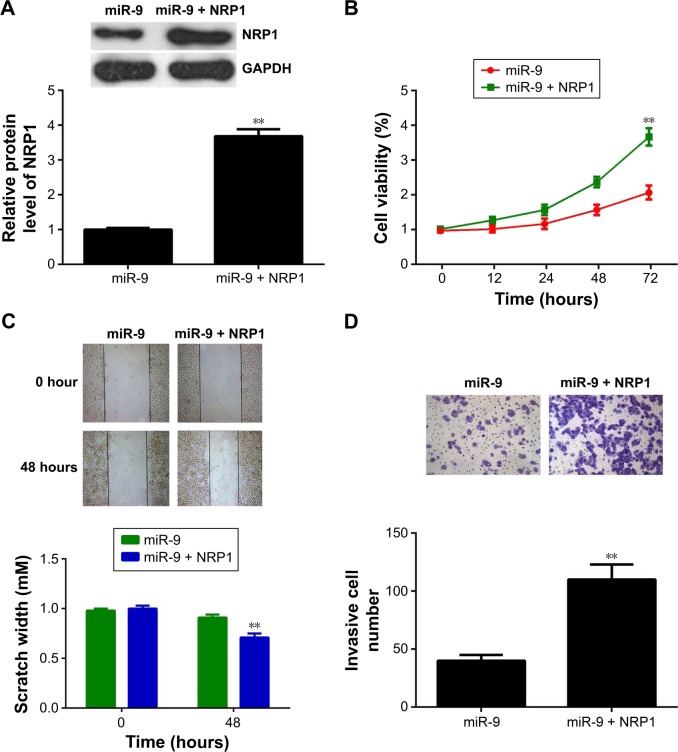
As upregulation of miR-9 reduces the proliferation, migration, and invasion of MM cells, and NRP1 is a target gene of miR-9, we speculated that NRP1 might act as a downstream effector of miR-9 in MM cells. Therefore, to clarify this speculation, we then investigated whether NRP1 was involved in the miR-9-mediated malignant phenotypes of MM cells. The pcDNA3.1-NRP1 ORF plasmid was used to transfect miR-9-overexpressing A375 cells. Our data indicated that after transfection with pcDNA3.1-NRP1 ORF plasmid, the NRP1 level was significantly increased when compared with that in miR-9-overexpressing A375 cells (Figure 4A). We then examined the cell proliferation, migration, and invasion of MM cells in each group. MTT assay data showed that the proliferating capacity was higher in A375 cells cotransfected with miR-9 mimic and NRP1 plasmid than in A375 cells only transfected with miR-9 mimic (Figure 4B), suggesting that overexpression of NRP1 effectively reversed the suppressive effects of miR-9 on MM cell proliferation. Similarly, we also found that the migratory and invading capacities of A375 cells were also higher in A375 cells cotransfected with miR-9 mimic and NRP1 plasmid than in those transfected with only miR-9 mimic (Figure 4C and D). Therefore, our findings demonstrate that overexpression of NRP1 reverses the inhibitory effects of miR-9, that is, the proliferation, migration, and invasion of A375 cells.
Figure 4.
(A) Western blot was conducted to determine the NRP1 protein levels in A375 cells transfected with miR-9 alone or cotransfected with miR-9 mimic and pcDNA3.1-NRP1 plasmid. (B) MTT assay was used to measure cell viability in A375 cells transfected with miR-9 alone or cotransfected with miR-9 mimic and pcDNA3.1-NRP1 plasmid. (C) Wound healing assay was used to measure the ability of cell migration in A375 cells transfected with miR-9 alone or cotransfected with miR-9 mimic and pcDNA3.1-NRP1 plasmid. (D) Transwell assay was used to measure the ability of cell invasion in A375 cells transfected with miR-9 alone or cotransfected with miR-9 mimic and pcDNA3.1-NRP1 plasmid. **P<0.01 vs miR-9.
Abbreviation: miR, microRNA.
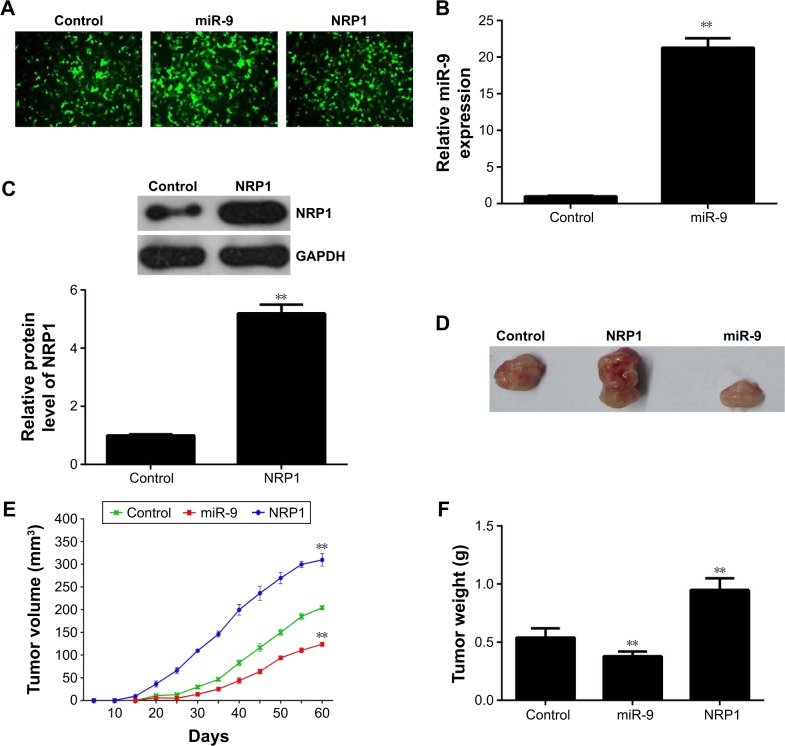
Effects of miR-9 and NRP1 on the tumor growth of MM cells in vivo
To further study the effect of the miR-9/NRP1 axis on the tumor growth of MM cells, pLVTH-miR-9 lentiviral plasmid and pLVTH-NRP1 lentiviral plasmid were stably transfected separately into A375 cells (Figure 5A). In the control group, A375 cells were stably transfected with the blank pLVTH vector. The miR-9 levels and NRP1 protein levels were significantly increased in A375 cells transfected with pLVTH-miR-9 lentiviral plasmid and pLVTH-NRP1 lentiviral plasmid compared with their control group (Figure 5B and C). After that, nude mice were subcutaneously implanted with the cells of each group. In the control group, four mice died within 60 days after implantation; however, only one mouse died in the miR-9 overexpression group. In contrast, all six mice died in the NRP1 overexpression group. All remaining mice were sacrificed 60 days after implantation. The xenograft was obtained and photographed (Figure 5D). Our data indicated that the tumor volume and weight were significantly reduced in the miR-9 overexpression group, but increased in the NRP1 overexpression group when compared with the control group (Figure 5E and F). Accordingly, we demonstrate that the inhibitory effect of miR-9 on the tumor growth of MM cells in vivo may be due to the direct inhibition of the protein expression of NRP1.
Figure 5.
(A) pLVTH-miR-9 lentiviral plasmid and pLVTH-NRP1 lentiviral plasmid were stably transfected into A375 cells. A375 cells stably transfected with the blank pLVTH vector were used as control (200×). (B) Real-time PCR was performed to examine the miR-9 level in A375 cells transfected with pLVTH-miR-9 lentiviral plasmid or blank pLVTH vector. (C) Western blots were used to examine NRP1 protein level in A375 cells transfected with pLVTH-NRP1 lentiviral plasmid or blank pLVTH vector. After that, nude mice were subcutaneously implanted with the cells of each group. All mice were sacrificed at 60 days after implantation. (D) The xenografted tumors were obtained and photographed. (E) The tumor volume was significantly reduced in the miR-9 overexpression group, but increased in the NRP1 overexpression group when compared with that in the control group. (F) The tumor weight was significantly reduced in the miR-9 overexpression group, but increased in the NRP1 overexpression group when compared with that in the control group. **P<0.01 vs control.
Abbreviations: PCR, polymerase chain reaction; miR, microRNA.
Discussion
Here, we investigated the exact role and regulatory mechanism of miR-9 in MM. Our data showed that miR-9 expression was significantly downregulated in MM tissues compared with adjacent normal tissues. The decreased miR-9 level was further found to be significantly associated with the advanced tumor stage and metastasis of MM. Besides, miR-9 was also downregulated in four MM cell lines compared with normal skin HACAT cells. Ectopic expression of miR-9 caused reduced cell proliferation, migration, and invasion in A375 cells, as well as the decreased protein expression of NRP1, while overexpression of NRP1 reversed the inhibitory effects of miR-9 on the malignant phenotypes of A375 cells. Furthermore, the in vivo study showed that overexpression of miR-9 inhibits the tumor growth, which is enhanced by NRP1 overexpression, suggesting that miR-9 inhibits MM in vitro and in vivo, at least partly via directly targeting NRP1.
A variety of miRs have recently been reported to play critical roles in the tumorigenesis and malignant progression of MM.22 The expression levels of miR-200c, miR-205, and miR-211 were found to be significantly decreased in MM, and overexpression of miR-200c and miR-205 inhibited anchorage-independent colony formation, while miR-211 overexpression suppressed both anchorage-independent colony formation and invasion.23 In addition, the miR let-7b and miR-199a were reported to inhibit the proliferation of MM B16F10 cells.24 Recently, several studies reported that deregulation of miR-9 was involved in the pathogenesis of MM.25 Shiiyama et al25 evaluated the serum levels of six miRs including miR-9, miR-145, miR-150, miR-155, miR-203, and miR-205 in eleven patients with metastatic MM and in 16 patients without MM, and found that they were differentially expressed between metastatic MM patients and healthy controls. In addition, the combination of miR-9, miR-145, miR-150, miR-155, and miR-205 was more sensitive than when each miR was used individually to distinguish metastatic MM patients from healthy controls.25 Liu et al20 found that miR-9 was downregulated in metastatic MM compared to primary MM. Here, we showed that miR-9 was significantly downregulated in MM tissues compared to adjacent normal tissues. Moreover, the miR-9 level was also decreased in MM cell lines, including G361, B16, A375, and HME1, than in the normal skin HACAT cells. In addition, the decreased miR-9 level was associated with higher tumor stage and metastasis, suggesting that the downregulation of miR-9 plays a role during the malignant progression of MM.
To further study the exact role of miR-9 in MM, miR-9 mimics were transfected into A375 cells to increase their level. Our data demonstrated that ectopic expression of miR-9 resulted in a significant decrease in the proliferation, migration, and invasion of A375 cells, suggesting that miR-9 has suppressive effects on the malignant phenotypes of MM cells. In fact, another two studies also reported similar findings. For instance, Liu et al19 showed that miR-was significantly downregulated in highly invasive uveal MM cell lines, and its upregulation suppressed the migration and invasion of highly invasive cells. Zhao et al21 reported that the upregulation of miR-9 reduced the proliferation, cell cycle progression, migration, and invasion of MM cells. These findings were consistent with ours. Next, we further investigated the role of miR-9 in MM growth in vivo and found that overexpression of miR-9 significantly inhibited the tumor growth and increased tumor-induced deaths in nude mice. Therefore, our study demonstrated that miR-9 acts as a tumor suppressor in MM.
miR functions are achieved mainly through negatively regulating the expression of their target genes at posttranscriptional levels. Therefore, we further investigated the potential target of miR-9 by using bioinformatics analysis and luciferase reporter assay. NRP1 is identified as a direct target gene of miR-9, and its protein expression is negatively regulated by miR-9 in A375 cells. NRP1 contains a large N-terminal extracellular domain, a short membrane-spanning domain, and a small cytoplasmic domain.26 NRP1 can bind many ligands and various types of coreceptors, such as vascular endothelial growth factor and semaphorin family members.27,28 Through the mediation of several different signaling pathways, NRP1 is involved in the regulation of cell survival and motility.29,30 Recently, Lu et al31 reported that NRP1 was significantly upregulated in primary and metastatic MM compared with common nevi and dysplas-tic nevi. Moreover, higher NRP1 levels were significantly associated with worse overall and disease-specific survival of MM patients, and NRP1 was found to be an independent prognostic marker for MM,31 indicating that NRP1 may be an oncogene in MM. However, the regulatory mechanism of NRP1 in MM remains largely unclear. Here, we show that NRP1 is negatively regulated by miR-9 in A375 cells, and overexpression of NRP1 reverses the inhibitory effect of miR-9 overexpression on the malignant phenotypes of A375 cells. Moreover, overexpression of NRP1 promotes the tumor growth of A375 cells in vivo. These data indicate that NRP1 may act as a downstream effector in the miR-9-mediated malignant phenotypes of MM cells. In addition, miR-365 was also found to inhibit the growth, invasion, and metastasis of MM by targeting NRP1 expression.32 Therefore, our study expands the importance of the miRs/NRP1 axis in MM.
To our knowledge, our study is the first to demonstrate that miR-9 suppresses the growth, migration, and invasion of MM cells, partly at least, via directly inhibiting the protein expression of NRP1. Therefore, our study suggests that the miR-9/NRP1 axis may serve as a potential therapeutic target for the treatment of MM.
Footnotes
Disclosure
The authors report no conflicts of interest in this study.
References
- 1.Trotter SC, Sroa N, Winkelmann RR, Olencki T, Bechtel M. A global review of melanoma follow-up guidelines. J Clin Aesthet Dermatol. 2013;6(9):18–26. [PMC free article] [PubMed] [Google Scholar]
- 2.Linos E, Swetter SM, Cockburn MG, Colditz GA, Clarke CA. Increasing burden of melanoma in the United States. J Invest Dermatol. 2009;129(7):1666–1674. doi: 10.1038/jid.2008.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rastrelli M, Tropea S, Rossi CR, Alaibac M. Melanoma: epidemiology, risk factors, pathogenesis, diagnosis and classification. In Vivo. 2014;28(6):1005–1011. [PubMed] [Google Scholar]
- 4.Kozubek J, Ma Z, Fleming E, et al. In-depth characterization of microRNA transcriptome in melanoma. PLoS One. 2013;8(9):e72699. doi: 10.1371/journal.pone.0072699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Poliseno L, Haimovic A, Segura MF, et al. Histology-specific microRNA alterations in melanoma. J Invest Dermatol. 2012;132(7):1860–1868. doi: 10.1038/jid.2011.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Populo H, Soares P, Lopes JM. Insights into melanoma: targeting the mTOR pathway for therapeutics. Expert Opin Ther Targets. 2012;16(7):689–705. doi: 10.1517/14728222.2012.691472. [DOI] [PubMed] [Google Scholar]
- 7.Ambros V. The functions of animal microRNAs. Nature. 2004;431(7006):350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 8.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 9.Moss EG. MicroRNAs: hidden in the genome. Curr Biol. 2002;12(4):R138–R140. doi: 10.1016/s0960-9822(02)00708-x. [DOI] [PubMed] [Google Scholar]
- 10.Ambros V. microRNAs: tiny regulators with great potential. Cell. 2001;107(7):823–826. doi: 10.1016/s0092-8674(01)00616-x. [DOI] [PubMed] [Google Scholar]
- 11.Zhou J, Liu R, Wang Y, et al. miR-199a-5p regulates the expression of metastasis-associated genes in B16F10 melanoma cells. Int J Clin Exp Pathol. 2014;7(10):7182–7190. [PMC free article] [PubMed] [Google Scholar]
- 12.Liu SM, Lu J, Lee HC, Chung FH, Ma N. miR-524-5p suppresses the growth of oncogenic BRAF melanoma by targeting BRAF and ERK2. Oncotarget. 2014;5(19):9444–9459. doi: 10.18632/oncotarget.2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van Kempen LC, van den Hurk K, Lazar V, et al. Loss of microRNA-200a and c, and microRNA-203 expression at the invasive front of primary cutaneous melanoma is associated with increased thickness and disease progression. Virchows Arch. 2012;461(4):441–448. doi: 10.1007/s00428-012-1309-9. [DOI] [PubMed] [Google Scholar]
- 14.Poenitzsch SA, Setaluri V, Spiegelman VS. MicroRNA-340 as a modulator of RAS-RAF-MAPK signaling in melanoma. Arch Biochem Biophys. 2014;563:118–124. doi: 10.1016/j.abb.2014.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bu P, Yang P. MicroRNA-203 inhibits malignant melanoma cell migration by targeting versican. Exp Ther Med. 2014;8(1):309–315. doi: 10.3892/etm.2014.1708. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 16.Yuva-Aydemir Y, Simkin A, Gascon E, Gao FB. MicroRNA-9: functional evolution of a conserved small regulatory RNA. RNA Biol. 2011;8(4):557–564. doi: 10.4161/rna.8.4.16019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen X, Zhu L, Ma Z, et al. Oncogenic miR-9 is a target of erlotinib in NSCLCs. Sci Rep. 2015;5:17031. doi: 10.1038/srep17031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Higashi T, Hayashi H, Ishimoto T, et al. miR-9-3p plays a tumour-suppressor role by targeting TAZ (WWTR1) in hepatocellular carcinoma cells. Br J Cancer. 2015;113(2):252–258. doi: 10.1038/bjc.2015.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu N, Sun Q, Chen J, et al. MicroRNA-9 suppresses uveal melanoma cell migration and invasion through the NF-kappaB1 pathway. Oncol Rep. 2012;28(3):961–968. doi: 10.3892/or.2012.1905. [DOI] [PubMed] [Google Scholar]
- 20.Liu S, Kumar SM, Lu H, et al. MicroRNA-9 up-regulates E-cadherin through inhibition of NF-kappaB1-Snail1 pathway in melanoma. J Pathol. 2012;226(1):61–72. doi: 10.1002/path.2964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao G, Li Q, Wang A, Jiao J. YY1 regulates melanoma tumorigenesis through a miR-9 ~ RYBP axis. J Exp Clin Cancer Res. 2015;34:66. doi: 10.1186/s13046-015-0177-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mannavola F, Tucci M, Felici C, Stucci S, Silvestris F. miRNAs in melanoma: a defined role in tumor progression and metastasis. Expert Rev Clin Immunol. 2016;12(1):79–89. doi: 10.1586/1744666X.2016.1100965. [DOI] [PubMed] [Google Scholar]
- 23.Xu Y, Brenn T, Brown ER, Doherty V, Melton DW. Differential expression of microRNAs during melanoma progression: miR-200c, miR-205 and miR-211 are downregulated in melanoma and act as tumour suppressors. Br J Cancer. 2012;106(3):553–561. doi: 10.1038/bjc.2011.568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu D, Tan J, Zhou M, et al. Let-7b and microRNA-199a inhibit the proliferation of B16F10 melanoma cells. Oncol Lett. 2012;4(5):941–946. doi: 10.3892/ol.2012.878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shiiyama R, Fukushima S, Jinnin M, et al. Sensitive detection of melanoma metastasis using circulating microRNA expression profiles. Melanoma Res. 2013;23(5):366–372. doi: 10.1097/CMR.0b013e328363e485. [DOI] [PubMed] [Google Scholar]
- 26.Zachary I. Neuropilins: role in signalling, angiogenesis and disease. Chem Immunol Allergy. 2014;99:37–70. doi: 10.1159/000354169. [DOI] [PubMed] [Google Scholar]
- 27.Yoshida A, Shimizu A, Asano H, et al. VEGF-A/NRP1 stimulates GIPC1 and Syx complex formation to promote RhoA activation and proliferation in skin cancer cells. Biol Open. 2015;4(9):1063–1076. doi: 10.1242/bio.010918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Watanabe Y, Sakuma C, Yaginuma H. NRP1-mediated Sema3A signals coordinate laminar formation in the developing chick optic tectum. Development. 2014;141(18):3572–3582. doi: 10.1242/dev.110205. [DOI] [PubMed] [Google Scholar]
- 29.Dong JC, Gao H, Zuo SY, et al. Neuropilin 1 expression correlates with the radio-resistance of human non-small-cell lung cancer cells. J Cell Mol Med. 2015;19(9):2286–2295. doi: 10.1111/jcmm.12623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cariboni A, Davidson K, Dozio E, et al. VEGF signalling controls GnRH neuron survival via NRP1 independently of KDR and blood vessels. Development. 2011;138(17):3723–3733. doi: 10.1242/dev.063362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lu J, Cheng Y, Zhang G, et al. Increased expression of neuropilin 1 in melanoma progression and its prognostic significance in patients with melanoma. Mol Med Rep. 2015;12(2):2668–2676. doi: 10.3892/mmr.2015.3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bai J, Zhang Z, Li X, Liu H. MicroRNA-365 inhibits growth, invasion and metastasis of malignant melanoma by targeting NRP1 expression. Int J Clin Exp Pathol. 2015;8(5):4913–4922. [PMC free article] [PubMed] [Google Scholar]