Abstract
Quantum dots (QDs) have been intensively investigated for bioimaging, drug delivery, and labeling probes because of their unique optical properties. In this study, CdSe/ZnS QDs-based nonviral vectors with the dual functions of delivering miR-26a plasmid and bioimaging were formulated by capping the surface of CdSe/ZnS QDs with polyethyleneimine (PEI). The PEI-coated QDs were capable of condensing miR-26a expression vector into nanocomplexes that can emit strong red luminescence when loaded with CdSe/ZnS QDs. Further results showed that PEI-modified nanoparticles (NPs) could transfect miR-26a plasmid into HepG2 cells in vitro. Meanwhile, imaging of living cells could be achieved based on the CdSe/ZnS QDs. Further study suggested that miR-26a transfection up-regulated miR-26a expression, induced cycle arrest, and triggered proliferation inhibition in HepG2 cells. The results indicated that PEI-coated QD NPs possess the capability of bioimaging and gene delivery and could be a promising vehicle with the engineering of QD NPs for gene therapy in the future.
Keywords: miR-26a, PEI/QDs, HepG2, gene delivery, bioimaging
Introduction
With the significant advances in the development of diverse functional nanomaterials, there have been increasing interests in fabricating a single nano-object with multiple functionalities. The development of new diagnostic and treatment methods based on these nanomaterials represents a major opportunity for basic and applied biomedical research.1–3 The nanostructural properties of these materials have a strong influence on the microenvironment of the biological system in which they interact. Especially, internalization of these particles through endocytosis by the cell membrane is highly influenced by its particle size and surface charge.4–6 Among them, quantum dots (QDs) have attracted increasing number of researchers to explore for bioimaging and labeling probes because of their considerable advantages, including high quantum yield, wide band excitation, narrow emission, photostability, and size-tunable multicolor imaging.7,8 To explore the imaging capability with the therapy potential, much work has been focused on the surface modification of QDs with biocompatible ligands, including biomacromolecules, peptides, antibodies, and so on.3,9–11 Recently, Yang et al12 transfected VEGF genes into endothelial progenitor cells and realized in vivo imaging using QDs in an ischemia hind limb model. Park et al13 reported a novel QD–amphiphilic polyethyleneimine (amPEI) nanocomplex that has been shown to successfully deliver silencing RNA (siRNA) into cancer cells, with real-time imaging of siRNA delivery and ratiometric oxygen sensing in live cells. Meanwhile, our previous works demonstrated that PEI-coupled nanoparticle (NP) is an effective transfection carrier for gene delivery.14–16 It was found that poly(l-lactic-co-glycolic acid) NPs coated with PEI can be delivered into HepG2 cells, enabling the endosomal escape of the NPs and the release of the particles into the cytoplasm, followed by cell cycle arrest and apoptosis.
MicroRNAs (miRNAs), a class of single-stranded non-coding small RNAs, play pivotal roles in gene expression regulation. Although miRNAs have been found to be aberrantly over-expressed, they are generally down-regulated in many cancer types.17–22 Therefore, the therapeutic applications of miRNA have been developed using various vectors, including adenovirus, adeno-associated virus (AAV), liposomes, polycationic polymers, and organic/inorganic NPs.23–28 Recently, a study demonstrated that when miR-26a, a down-regulated miRNA in liver cancer, was delivered into liver cancer cells mediated by AAV in vitro, it induced cell cycle arrest, associated with direct targeting of cyclins D2 and E2, by inhibiting cancer cell proliferation and inducing tumor-specific apoptosis in the mouse hepatocellular carcinoma model.29 Ibrahim et al30 applied sole PEI-mediated delivery of unmodified miR-145 and miR-33a into a mouse model of colon carcinoma, by which the PEI/miRNA complexes were delivered into mouse xenograft tumors, where they caused profound antitumor effects.
Inspired by previous research, herein we explored the use of PEI to modify QDs into larger complex structures to apply as multifunctional imaging vehicle for gene delivery visualization. CdSe/ZnS QDs were selected as labeling probes for living cells since the gradient alloy CdSe/ZnS QDs possess excellent opticophysical properties and are more compatible with a biological environment by shielding the Cd2+-containing core from the ZnS shell structure.31,32 The recombinant plasmid expression vector of miR-26a, which is capable of inducing green fluorescence and cell cycle arrest in HepG2 cells,29 was used for the evaluation of gene delivery of PEI/QD NPs in HepG2 cells. CdSe/ZnS QDs were fabricated with PEI coating using a simple method of electrostatic attraction. The size and morphology of CdTe QDs were characterized by dynamic light scattering (DLS) and transmission electron microscopy (TEM). The effects of PEI/QD NPs on transfected HepG2 cells were investigated by polymerase chain reaction (PCR), Western blot, and confocal laser scanning microscopy (CLSM) analyses.
Materials and methods
Materials
PEI (25 kDa) was purchased from Sigma-Aldrich Co. (St Louis, MO, USA). CdSe/ZnS QDs with emission maxima at 640 nm were obtained from Mesolight Nanotechnology Co. Ltd. (Suzhou, Jiangsu, People’s Republic of China). To ensure its stability in an aqueous solution, the CdSe/ZnS QDs were modified with 3-mercaptopropionic acid, resulting in a net negative charge on the surface for facilitating interaction with cationic PEI. Dulbecco’s Modified Eagle’s Medium (DMEM) and 0.25% trypsin were purchased from Corning Life Sciences (Mabassas, VA, USA). Fetal bovine serum (FBS) was purchased from Thermo Fisher Scientific. Agarose, glycine, N,N,N′,N′-tetramethylethylenediamine, dimethyl sulfoxide, and chloroform were purchased from Sangon Biotech Co. Ltd. (Shanghai, People’s Republic of China). The primary and secondary antibodies of cyclin-D2 (CCND2), cyclin-E2 (CCNE2), and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA). Primer was designed and synthesized by Sangon Biotech Co. Ltd. Other reagents were of analytical grade and used without further purification.
Preparation and characterization of PEI/QD NPs
Aqueous solutions of monodisperse QDs were prepared according to the manufacturer’s protocol. Briefly, PEI (2 mg) was dissolved in 10 mL of phosphate-buffered saline (PBS)-buffered saline for 1 h at 65°C. Then, the PEI solutions were filtered through a 0.22 μm syringe filter (Pall Corporation, New York, NY, USA). QDs (4 nmol/mL) and PEI solutions were mixed with deionized water (1 mL) to produce the PEI/QD NP suspension. The as-formed PEI/QD NP suspension was then added to 1 mg/mL of solution containing miR-26a expression vectors at various PEI/QD concentrations (0–4 nmol/mL) and vortexed gently to obtain PEI/QD nanocomplexes.
The average diameter of the PEI/QD NPs was measured by DLS (Zetasizer Nano ZS; Malvern Instruments, Malvern, UK). In brief, the PEI/QD nanocomplexes were suspended in deionized water at a concentration of 2 nmol/mL. The mean hydrodynamic diameter was determined by cumulative analysis. The zeta potential (surface charge) of the polymers and polyplexes was determined at 25°C using a scattering angle of 90° with potential measurement analyzer (Zetasizer Nano ZS; Malvern Instruments). Samples were prepared in PBS and diluted 1:10 with deionized water to ensure that the measurements were performed under conditions of low ionic strength. The surface morphologies of the multiple QD-bundled NPs were determined by TEM (JEM-2100; JEOL, Tokyo, Japan). The samples were prepared by dropping PEI/QDs suspension onto a copper grid, followed by air drying at 37°C.
Cytotoxic assessment and cell proliferation assay
HepG2 cells were seeded in 96-well plates at an initial density of 1×104 cells per well in 0.5 mL of growth medium and incubated for 24 h prior to the addition of different concentrations of PEI/QD NPs. Untreated cells were taken as control with 100% viability. Cell Counting Kit-8 (CCK-8; Dojindo Molecular Technologies, Kumamoto, Japan) was used for assessing the cytotoxicity of PEI/QD. Details of the process are provided in manufacturer’s instruction. Cell proliferation was determined in a separate set of experiments using CCK-8 to detect changes in cell viability after being treated with PEI/QD/miRNA. HepG2 cells were seeded in 24-well plates at an initial density of 2×104 cells per well in 1 mL of growth medium and incubated for 24 h prior to the addition of PEI/QD nanocomplexes. Untreated cells were taken as control with 100% viability. Triton X-100 1% was used as positive control.
Construction of miR-26a expression vectors and cell cultures
MiR-26a expression vector with the cytomegalo virus promoter that drives the expression of green fluorescent protein (GFP) and its negative control (miR-NC) with GFP were constructed as described previously,33 and verified by DNA sequencing. The cell line (HepG2 cells) used in this study was purchased from Shanghai Institute of Cell Bank (Shanghai, People’s Republic of China). HepG2 cells were cultured in DMEM medium (Invitrogen, Carlsbad, CA, USA) supplemented with 10% FBS (Invitrogen). Cells were maintained at 37°C and 5% CO2 during all experiments.
Preparation of QD/pDNA complexes and affinity analysis
First, PEI/QD and miR-26a plasmid vector were separately dissolved in 0.5 mL of PBS (pH 7.4) and the PEI/QD suspensions were filtered with 0.22 mm sterile filters. The different concentrations of PEI/QD suspension diluted with PBS were added to 1 μg of miR-26a vector solution, and then vortexed immediately at room temperature. Finally, the solution was allowed to stand for 30 min to enable the formation of PEI/QD/pDNA nanocomplexes. In this study, the complexing ratio was expressed as the nitrogen to phosphate (N/P) ratio of PEI/QD/pDNA. For affinity analysis of the PEI/QDs and miR-26a vector complex, 10 μL of the nanocomplex suspension was subjected to electrophoresis in 1% tris-borate-ethylene diamine tetraacetic acid agarose gel at 100 V for 40 min. Images were acquired using a Tanon gel imaging system (Tanon Science & Technology, Shanghai, People’s Republic of China).
Gene transfer and miR-26a expression assay
HepG2 cells seeded in a 6-well plate at a density of 2×105 cells per well were transfected with the miRNA expression vectors described earlier using the PEI/QDs nanocomplex. Cells were viewed by fluorescence microscopy at different time points after transfection (24, 48, and 72 h), and imaged based on fluorescein isothiocyanate (FITC) channels. Finally, the cells were harvested after being transfected for 72 h, and the total RNA was extracted with Trizol according to the manufacturer’s instruction. miR-26a reverse-transcript primers (CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGAGCCTATC) and reverse transcriptase were used to convert mature miRNA to cDNA.34 The reaction system was heated to 80°C for 5 min to denature the RNA, and it was cooled to room temperature quickly, then the remaining reagents (5× buffer, dNTPs, dithiothreitol, RNase inhibitor, prime script RTase) were added as specified according to the manufacturer’s protocol. The reaction proceeded for 45 min at 42°C followed by 5 min incubation at 85°C to inactivate the reverse transcriptase. cDNA may be stored indefinitely at −20°C or −80°C.
Then quantitative real-time PCR (qRT-PCR) was performed to evaluate miR-26a expression in HepG2 cells post 3 days of transfection using standard protocols on an Applied Biosystems 7500 Sequence Detection System. Briefly, 1 μL of cDNA was added to 10 μL of the 2× SYBR green PCR master mix (TaKaRa Biotechnology, Dalian, People’s Republic of China), then 1 μL of 10 μM of each primer (forward: ACACTCCAGCTGGGTTCAAGTAATC CAGGA; reverse : TGGTGTCGTGGAGTC) was added and the volume was made up to 20 μL with water. The reaction system was amplified for 15 s at 95°C and 1 min at 60°C for 40 cycles. Reactions are typically run in triplicate. The cycle number at which the reaction crossed an arbitrarily placed threshold was determined for each gene and the relative amount of each miRNA was calculated. Target gene expression was normalized to the housekeeping gene U6 for each sample. Data were analyzed using the 2−ΔΔCT method.35
Confocal laser scanning microscopy analysis
For the distribution of PEI/QDs/miRNA in HepG2 cells, different treated cells were washed 3 times with PBS to remove excess nanocomplexes and visualized using a CLSM (Nikon Eclipse 80i; Nikon, Tokyo, Japan). The fluorescence was monitored in the FITC and Cy5 channels.
Cell cycle assay
Cell cycle profiles of PEI/QD NPs transfected HepG2 cells were determined using FACSCalibur flow cytometer and CellQuest™ software (BD Biosciences, Mountain View, CA, USA). HepG2 cells were seeded into 12-well plates with 5×104 cells per well. Cells were treated with different formulations at a concentration of 1 μg of miRNA expression vector in serum-containing medium at 37°C for 72 h. Cells were washed once with PBS and then fixed in 75% ethanol. After fixation, the cells were stained with propidium iodide (PI) according to manufacturer’s protocol.
Western blot assay
For Western blot analysis, the HepG2 cells were transfected as previously described for 48 h. After transfection, the cell total protein was extracted with lysis buffer (Boster Biological Technology, Wuhan, People’s Republic of China). The protein was quantified by the bicinchoninic acid protein assay kit (CWBio, Beijing, People’s Republic of China). An equal amount of protein was separated on the sodium dodecyl sulfate polyacrylamide gel electrophoresis, and then transferred onto polyvinylidene difluoride membrane. The membrane was then blocked and incubated overnight with monoclonal antibodies against CCND2 (1:500), CCNE2 (1:500), and GAPDH (1:600). After washing, the membrane was incubated with horseradish peroxidase-conjugated secondary antibody (1:5,000) for 2 h at room temperature. The bands were visualized using the enhanced chemiluminescence kit (CWBio) and the expression of CCND2 and CCNE2 was normalized with GAPDH housekeeping gene expression.
Statistical analysis
Data were expressed as mean ± standard deviation and statistical significance was calculated by either two-tailed unpaired Student’s t-test or analysis of variance (ANOVA) as appropriate. P-values <0.05 were considered statistically significant.
Results and discussion
Characterization of PEI/QD NPs
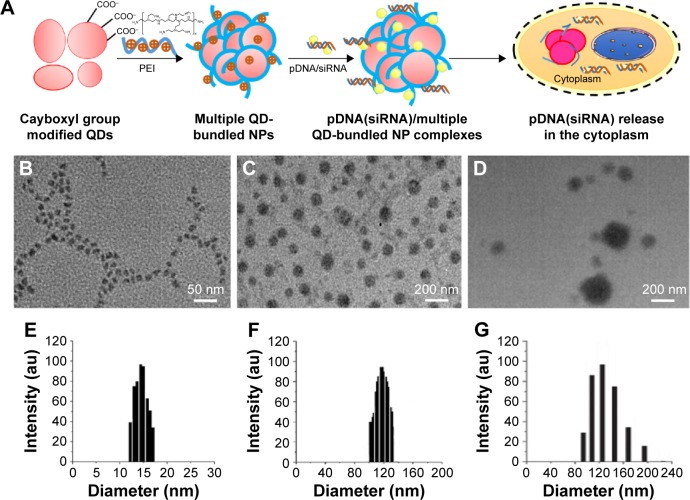
The formulated PEI/QD NP delivery vehicle is demonstrated in Figure 1A. Generally, the size and zeta potential of NP play vital roles in the cellular uptake process. Appropriately sized (100–200 nm) NPs can be more easily internalized by target cells.36 In this study, the prepared QD was monodispersed with an average diameter of 9 nm (Figure 1B). With the addition of PEI solution, the single QDs gathered together with PEI to form a larger NP, and the size of the PEI/QDs increased according to the PEI concentration. When 5 μg/mL of PEI, the maximum concentration which does not cause obvious cytotoxicity when used for gene transfection, was introduced, the multiple QDs coupled with PEI resulting in three-dimensional spherical structures with the diameter of about 100 nm (Figure 1C). When miRNA expression vector was added to the PEI/QDs suspension, the sizes of complexed PEI/QD NPs containing pDNA increased to about 120 nm, or even larger (Figure 1D). The particle size from DLS data (Figure 1E–G) is slightly larger than that of TEM, the possible reason lies in the hydrous membrane which was coated on the surface of NPs in the process of detection, therefore increasing the particle size. Overall, these results of the DLS observations are consistent with those of TEM, and indicate that the size of PEI/QDs/miRNA nanocomplexes could be increased with PEI or polyplexed PEI/pDNA coating.
Figure 1.
Schematic representations and TEM images of PEI/QDs/miRNA nanoparticles.
Notes: (A) Schematic representations of the preparing route for PEI/QD nanoparticles; TEM micrographs of QDs (B), PEI/QDs (C), and PEI/QD/miRNA (D) nanoparticles. (E, F, and G) DLS data corresponding to (B, C, and D), respectively.
Abbreviations: DLS, dynamic light scattering; miRNA, microRNA; NP, nanoparticle; PEI, polyethyleneimine; QD, quantum dot; siRNA, silencing RNA; TEM, transmission electron microscopy.
Next, we investigated the surface charge (zeta potential) of the formulated NPs. The single QDs had a negative charge of about −24 mV due to the carboxylic groups located on the surface, because the PEI-containing primary and secondary amine groups had a mean positive charge of +28 mV, and the miR-26a expression vector had a negative surface charge owing to anionic phosphate groups on their structure. The positive charge of the PEI coating on the negatively charged QD NPs resulted in an average zeta potential of +22 mV. While the miR-26a expression vector was introduced to the PEI/QD NPs by electrostatic interaction, surface charge of the nanocomplexes reduced to +16 mV due to the introduction of miR-26a vector with negative charge.
Cytotoxicity assessment and gel retardation assay of the PEI/QD NPs
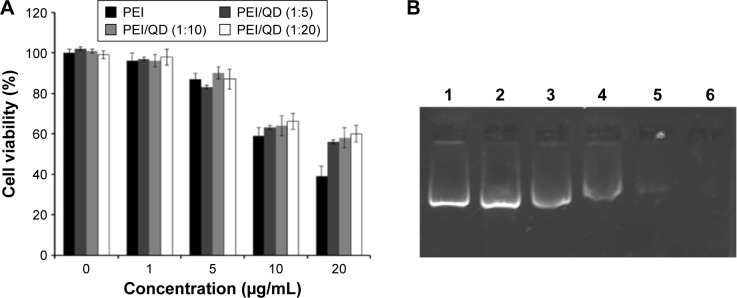
The cytotoxicity of the PEI/QD NPs on HepG2 cells was determined by the CCK-8 assay. As seen in Figure 2A, with the concentration of the PEI exceeding 5 μg/mL, the cytotoxicity increased obviously. When the concentration was more than 20 μg/mL, the viability of HepG2 cells dropped to about 40%. However, QDs combined with NPs (PEI/QD, 1:5, 1:10, 1:20) showed relatively good biocompatibility. The reason may be that the negatively charged QDs offset the excess positive charge on the PEI surface, avoiding damage to the cell membrane. Considering the impact of the surface charge on the delivery of miRNA, the PEI concentration of 5 μg/mL and PEI/QDs in the 1:10 ratio was focused for the further study.
Figure 2.
Cell viability and gel electrophoresis assays of different nanocomplexes.
Notes: (A) Cell viability of HepG2 cells treated with different formulated PEI/QD nanoparticles. (B) Gel electrophoresis assays of PEI/QD/pDNA nanocomplexes: lane 1, miR-26a vector alone; lanes 2 to 6, PEI/QD/pDNA complexes with N/P ratio 0.5, 1, 2, 4, and 6, respectively.
Abbreviations: PEI, polyethyleneimine; QD, quantum dot; N/P, nitrogen to phosphate.
To explore complex formation, PEI/QD NPs were mixed with miR-26a plasmid vector in ultrapure water at different N/P ratios, and affinity formation was assessed by a gel retardation assay. As shown in Figure 2B, the NPs were able to fully retard the mobility of DNA in agarose gel when the N/P ratio was 6:1 or higher. When the N/P ratio was lower than 6:1, the retardation was not complete, and DNA bands were visible in N/P complexes in lanes 2 to 5, indicating the presence of free pDNA in the nanocomplexes of 0.5, 1, 2, and 4 N/P ratios, which were comparable with the band observed in lane 1 containing only free miR-26a vector. Weaker DNA bands were observed in the subsequent lanes of N/P ratio 6, indicating nearly complete complex formation of miR-26a vector. The results indicated that the nanocomplexes with miR-26a vector can be easily prepared by simply mixing cationic polymer and miR-26a solution, and the results also showed that N/P ratio of 6:1 is optimal for the preparation, resulting in PEI/QDs/miRNA NPs with a diameter of 120 nm and a surface charge of 16 mV.
Gene transfection and fluorescence monitoring
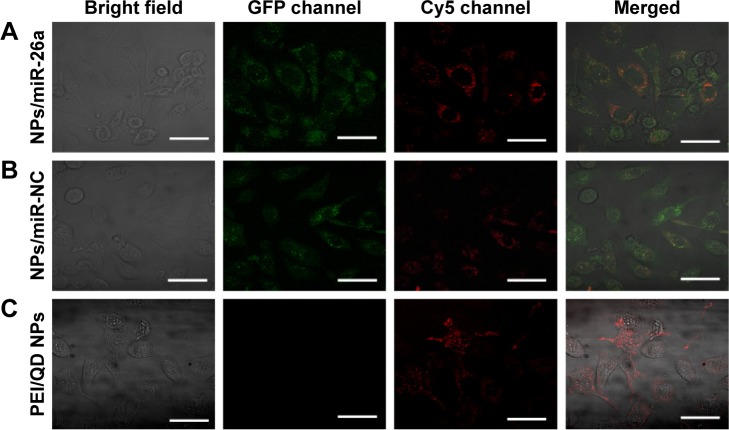
Gene transfection was visualized by confocal laser scanning microscopy 48 h after the addition of PEI/QDs/miRNA nanocomplex. Green fluorescence in Figure 3 showed that internalization of miR-26a vector was translated into proteins from GFP pDNA complexed with PEI/QDs in HepG2 cells. Meanwhile, the QDs were observed to produce red fluorescence with Cy5 channel. The merged images of the HepG2 cells transfected with PEI/QDs nanocomplexes containing miR-26a vector show that PEI/QD colocalizated with GFP in cytosol. Interestingly, the red fluorescence of QDs was mainly distributed around the nucleus of the HepG2 cells. The merged colors present in the cytosol suggested that the PEI/QDs nanocomplex entered into the cells and that they primarily existed in the cytosol of HepG2 cells.
Figure 3.
CLSM analysis of PEI/QD nanocomplex transfected HepG2 cells.
Notes: Cells transfected with PEI/QD/miR-26a vector (A); cells transfected with PEI/QD/miR-NC (B); cells transfected with negative control (PEI/QD/miR-NC vector) (C). Scale bar =50 μm.
Abbreviations: CLSM, confocal laser scanning microscopy; PEI, polyethyleneimine; QD, quantum dot; NC, negative control; NP, nanoparticle; GFP, green fluorescent protein.
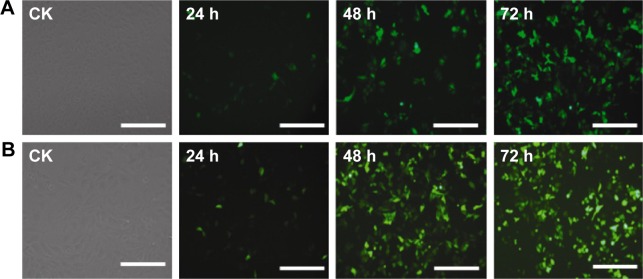
In addition, in order to assess the transfection efficiency of the PEI/QD NPs, HepG2 cells transfected with PEI/QDs and commercial Lipo2000 were monitored for GFP expression with inverted fluorescence microscopy (Nikon Ti, Tokyo, Japan) at different time points after transfection. After 3 days of continuous monitoring, the results suggested that transfection efficiency of PEI/QDs was close to Lipo2000 in HepG2 cells. Fluorescence intensity of GFP was obviously enhanced with the increase of transfection time in PEI/QDs or Lipo2000 groups (Figure 4), and the GFP expression was highest at 72 h post-transfection. The results also indicated that PEI/QD NPs and Lipo2000 show maximum internalization after 72 h in transfected HepG2 cells. These results are coincident with the internalization of PEI/QDs into HepG2 cells observed by confocal laser microscopy (data not shown). Taken together, these results suggested the successful integration of gene delivery and bioimaging functions in the PEI/QD NPs.
Figure 4.
GFP expression in transfected HepG2 cells.
Notes: CK: untransfected cells; 24, 48, and 72 h represent different time points post-transfection. Cells transfected GFP with PEI/QDs (A); Cells transfected GFP with Lipo2000 (B). Scale bar =100 μm.
Abbreviations: GFP, green fluorescent protein; PEI, polyethyleneimine; QD, quantum dot.
Up-regulation of miR-26a mediated by nanocomplex
To investigate miR-26a expression mediated by PEI/QD NPs in transfected HepG2 cells, which were harvested after 72 h of transfection, total RNA was extracted for monitoring miR-26a expression by qRT-PCR. The results showed that the miR-26a level in HepG2 cells transfected with recombination miR-26a vector nanocomplex (PEI/QDs/miR-26a) was higher than in the untransfected and negative control (PEI/QDs/miR-NC) transfected cells (Figure 5). In contrast, the expression level of miR-26a showed no obvious change in cells transfected with miR-NC (P>0.05).
Figure 5.
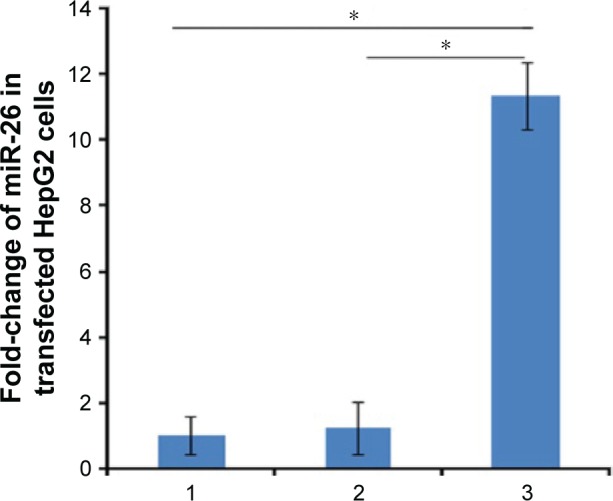
Relative expression change of miR-26a level.
Notes: 1: untransfected group; 2: negative control group (PEI/QD/miR-NC); 3: miR-26a vector transfected group. *P<0.05 compared with groups 3 and 1 (n=3).
Abbreviations: PEI, polyethyleneimine; QD, quantum dot; NC, negative control.
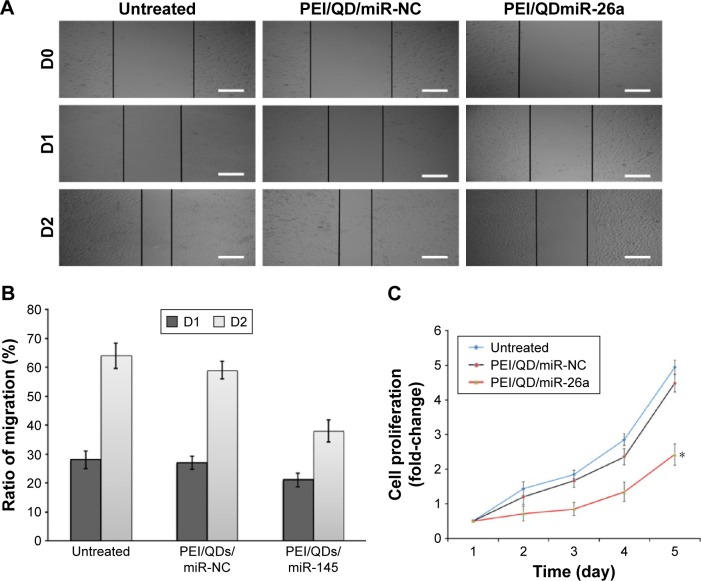
Inhibition of HepG2 cell migration and proliferation
To determine tumor inhibition activities of the PEI/QDs nanocomplex containing miR-26a vector in vitro, we assessed cell migration and proliferation with a 5-day continuous monitoring protocol using the wound healing assay and CCK-8, respectively. After culturing HepG2 cells for 2 h with a negative control (PEI/QDs/miR-NC) and miR-26a, cell monolayers were scratched, washed with PBS, and cultured for another 24 or 48 h. Microscopy was used to measure the migration distance. Figure 6A and B showed that the wound-like gaps healed in 48 h in untreated or PEI/QDs/miR-NCtreated cells, indicating a significant cell migration in untreated or PEI/QDs/miR-NCtreated HepG2 cells. However, PEI/QDs/miR-26a treated cells had wounds persisting at 24 and 48 h. Furthermore, the results of cell proliferation showed that the proliferation of HepG2 cells transfected with miR-26a vector was remarkably inhibited, especially on the fifth day, compared with cells that were not transfected or transfected with control vector (Figure 6C). It should be noted that due to slight toxicity caused by QD coated with PEI, cells transfected with PEI/QD containing the control vector showed a lower viability than the untransfected cells. Moreover, the results from the quantitative analysis showed a statistically significant reduction in the wound closures in the cells treated with miR-26a vector compared with the negative control and the untreated cells. The results showed that the miR-26a-included nanocomplexes could effectively inhibit HepG2 cell migration and proliferation.
Figure 6.
Inhibition of HepG2 cell migration and proliferation after transfection.
Notes: (A) Cell migration after treatment with different nanoparticles; D0, D1, and D2 represent the first, second, and third day when cell monolayers were scratched; Scale bar =100 μm. (B) Ratio of migration of HepG2 cells corresponding to A. (C) Cells proliferation after treatment with different nanoparticles. *P<.0.05.
Abbreviations: PEI, polyethyleneimine; QD, quantum dot; NC, negative control.
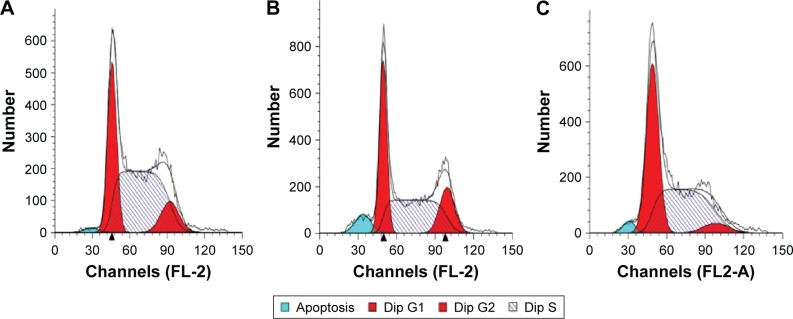
Inhibition of cell cycle induction
As shown in Figure 7, the miR-26a-containing PEI/QD nanocomplexes significantly inhibit the cell cycle progression by induction of G1 phase arrest in the transfected HepG2 cells, whereas the effect in control groups (untreated and PEI/QDs/miR-NC) was not obvious. Table 1 also indicates that HepG2 cells with enforced miR-26a expression were characterized by significantly increased number of cells arrested in G1 phase, which was more than that of tumor cells treated with miR-NC containing PEI/QD nanocomplex or untreated control. This result is consistent with that reported by previous studies,37 and up-regulation of miR-26a expression results in the inhibition of cell proliferation, induction of tumor-specific apoptosis, and consequent enhancement of antitumor activity.29
Figure 7.
Cell cycle profiles of PEI/QDs transfected HepG2 cells.
Notes: (A) untreated group, (B) miR-NC contained nanoparticles transfected HepG2 cells, (C) miR-26a vector contained nanoparticles transfected HepG2 cells.
Abbreviations: PEI, polyethyleneimine; QD, quantum dot; NC, negative control.
Table 1.
Cell cycle profiles of HepG2 cells treated with different nanoparticles (n=3)
| Phase | Untreated | NPs/miR-NC | NPs/miR-26a |
|---|---|---|---|
| G1 | 30.61±2.3 | 34.63±2.6 | 48.81±3.4 |
| G2 | 13.32±1.5 | 17.56±1.2 | 9.43±1.5 |
| S | 56.07±2.1 | 47.81±1.4 | 41.76±2.2 |
Note: Data presented as mean ± standard deviation.
Abbreviations: NP, nanoparticle; NC, negative control.
Western blot analysis of protein expression
The appealing property of miRNAs as therapeutic agents lies in their capability to regulate multiple genes by efficiently regulating definite cell processes.38 To investigate the further mechanism as to how miR-26a inhibits cell proliferation, we examined the expression of CCND2, CCNE2, predicted targets of miR-26a, which play important roles in the G1/S phases of the cell cycle.29 As shown in Figure 8A and B, the untreated and NPs/miR-NC groups did not down-regulate the protein levels of CCND2 and CCNE2. In contrast, the nanocomplex containing miR-26a vector substantially down-regulated the protein levels of CCND2 and CCNE2. The phenomena suggested that the miR-26a expression level plays an important role in the regulation of the CCND2 and CCNE2 expression in HepG2 cells. These results demonstrated that the PEI/QD NPs coupled with miR-26a vector exhibited excellent anti-proliferative activity and efficient gene silencing in HepG2 cells.
Figure 8.
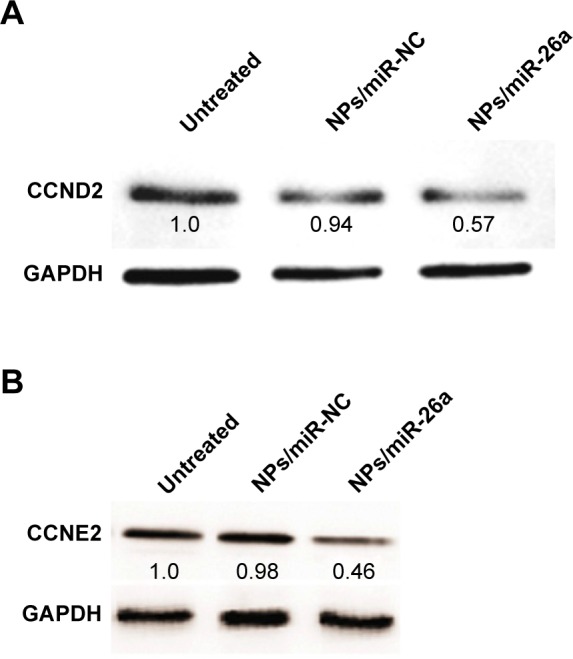
Western blot analysis of protein expression in HepG2 cells.
Notes: The protein expression of CCND2 (A), CCNE2 (B) after treatment with different nanoparticles. Untreated, NPs/miR-NC, NPs/miR-26a: HepG2 cells treated with no nanoparticles, PEI/QD/miR-NC, PEI/QD/miR-26a nanocomplex, respectively.
Abbreviations: PEI, polyethyleneimine; QD, quantum dot; NC, negative control; NP, nanoparticle.
Conclusion
In this study, PEI-coated QDs were fabricated to investigate the miRNA delivery into HepG2 cells. Cationic PEI can capture anionic QDs via electrostatic interaction, which ultimately form QD-bundled NPs through inter-particular aggregation with each other. The QDs with cationic PEI coating are capable of binding pDNA molecules for gene delivery and bioimaging. It was also demonstrated that miR-26a-loaded QD nanocomplexes can up-regulate miR-26a expression, arrest cell cycle, and suppress the protein levels of CCND2 and CCNE2, thus resulting in the inhibition of cancer cell proliferation in HepG2 cells. This finding perhaps will lead to improvements in the design of nano-materials for various biomedical applications, including gene therapy, molecular diagnostics, and bioimaging.
Acknowledgments
This work was financially supported by the National Natural Science Foundation of China (U1404824), Science & Technology Agency of Henan province (142107000023), Young Backbone Teacher Project of Henan Provincial Universities (2015GGJS-049), and Doctor’s Research Staring Foundation of Henan University of Science and Technology (09001635).
Footnotes
Disclosure
The authors report no conflicts of interests in this work.
References
- 1.Wu C, Han D, Chen T, et al. Building a multifunctional aptamer-based DNA nanoassembly for targeted cancer therapy. J Am Chem Soc. 2013;135(49):18644–18650. doi: 10.1021/ja4094617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huang P, Rong PF, Jin A, et al. Dye-loaded ferritin nanocages for multimodal imaging and photothermal therapy. Adv Mater. 2014;26(37):6401–6408. doi: 10.1002/adma.201400914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen F, Huang P, Qi C, et al. Multifunctional biodegradable mesoporous microspheres of Eu3+-doped amorphous calcium phosphate: microwave-assisted preparation, pH-sensitive drug release, and bioimaging application. J Mater Chem B. 2014;2(41):7132–7140. doi: 10.1039/c4tb01193g. [DOI] [PubMed] [Google Scholar]
- 4.Yu D, Zhang Y, Zhou X, Mao Z, Gao C. Influence of surface coating of PLGA particles on the internalization and functions of human endothelial cells. Biomacromolecules. 2012;13(10):3272–3282. doi: 10.1021/bm3010484. [DOI] [PubMed] [Google Scholar]
- 5.Zhang W, Liu J, Tabata Y, Meng J, Xu H. The effect of serum in culture on RNAi efficacy through modulation of polyplexes size. Biomaterials. 2014;35(1):567–577. doi: 10.1016/j.biomaterials.2013.09.102. [DOI] [PubMed] [Google Scholar]
- 6.Shao X-R, Wei X-Q, Song X, et al. Independent effect of polymeric nanoparticle zeta potential/surface charge, on their cytotoxicity and affinity to cells. Cell Prolif. 2015;48(4):465–474. doi: 10.1111/cpr.12192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zibik EA, Grange T, Carpenter BA, et al. Long lifetimes of quantum-dot intersublevel transitions in the terahertz range. Nature Mater. 2009;8(10):803–807. doi: 10.1038/nmat2511. [DOI] [PubMed] [Google Scholar]
- 8.Zhao BX, Huang P, Rong PF, et al. Facile synthesis of ternary CdMnS QD-based hollow nanospheres as fluorescent/magnetic probes for bioimaging. J Mater Chem B. 2016;4(7):1208–1212. doi: 10.1039/c5tb01963j. [DOI] [PubMed] [Google Scholar]
- 9.Choi Y, Kim K, Hong S, Kim H, Kwon Y-J, Song R. Intracellular protein target detection by quantum dots optimized for live cell imaging. Bioconjug Chem. 2011;22(8):1576–1586. doi: 10.1021/bc200126k. [DOI] [PubMed] [Google Scholar]
- 10.Mansur HS, Mansur AAP, Soriano-Araujo A, Lobato ZIP, de Carvalho SM, Leite MDF. Water-soluble nanoconjugates of quantum dot-chitosan-antibody for in vitro detection of cancer cells based on “enzyme-free” fluoroimmunoassay. Mater Sci Eng C Mater Biol Appl. 2015;52:61–71. doi: 10.1016/j.msec.2015.03.022. [DOI] [PubMed] [Google Scholar]
- 11.Shao D, Li J, Guan FY, et al. Selective inhibition of liver cancer growth realized by the intrinsic toxicity of a quantum dot-lipid complex. Int J Nanomed. 2014;9:5753–5769. doi: 10.2147/IJN.S73185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang HN, Park JS, Woo DG, Jeon SY, Park KH. Transfection of VEGF(165) genes into endothelial progenitor cells and in vivo imaging using quantum dots in an ischemia hind limb model. Biomaterials. 2012;33(33):8670–8684. doi: 10.1016/j.biomaterials.2012.08.012. [DOI] [PubMed] [Google Scholar]
- 13.Park J, Lee J, Kwag J, et al. Quantum dots in an amphiphilic polyethyleneimine derivative platform for cellular labeling, targeting, gene delivery, and ratiometric oxygen sensing. ACS Nano. 2015;9(6):6511–6521. doi: 10.1021/acsnano.5b02357. [DOI] [PubMed] [Google Scholar]
- 14.Liang GF, Zhu YL, Jing A, et al. Cationic microRNA-delivering nanocarriers for efficient treatment of colon carcinoma in xenograft model. Gene Ther. 2016 Sep 1; doi: 10.1038/gt.2016.60. Epub. [DOI] [PubMed] [Google Scholar]
- 15.Liang G, Li P, Lei W. Delivery of miRNA using Fe2O3 nanoparticles capped polyethyleneimine as a nonviral carrier. Zhang CL, Zhang LC, editors. Materials Science and Nanotechnology I. 2013;531–532:543–546. [Google Scholar]
- 16.Zhu Y, Liang G, Sun B, Tian T, Hu F, Xiao Z. A novel type of self-assembled nanoparticles as targeted gene carriers: an application for plasmid DNA and antimicroRNA oligonucleotide delivery. Int J Nanomed. 2016;11:399–411. doi: 10.2147/IJN.S84927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang J-X, Song W, Chen Z-H, et al. Prognostic and predictive value of a microRNA signature in stage II colon cancer: a microRNA expression analysis. Lancet Oncol. 2013;14(13):1295–1306. doi: 10.1016/S1470-2045(13)70491-1. [DOI] [PubMed] [Google Scholar]
- 18.Soares RJ, Cagnin S, Chemello F, et al. Involvement of microRNAs in the regulation of muscle wasting during catabolic conditions. J Biol Chem. 2014;289(32):21909–21925. doi: 10.1074/jbc.M114.561845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vogel B, Keller A, Frese KS, et al. Multivariate miRNA signatures as biomarkers for non-ischaemic systolic heart failure. Eur Heart J. 2013;34(36):2812–2822. doi: 10.1093/eurheartj/eht256. [DOI] [PubMed] [Google Scholar]
- 20.Jiang W, Zhang Y, Meng F, et al. Identification of active transcription factor and miRNA regulatory pathways in Alzheimer’s disease. Bioinformatics. 2013;29(20):2596–2602. doi: 10.1093/bioinformatics/btt423. [DOI] [PubMed] [Google Scholar]
- 21.Farazi TA, Horlings HM, ten Hoeve JJ, et al. MicroRNA sequence and expression analysis in breast tumors by deep sequencing. Cancer Res. 2011;71(13):4443–4453. doi: 10.1158/0008-5472.CAN-11-0608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liang G, Li J, Sun B, et al. Deep sequencing reveals complex mechanisms of microRNA deregulation in colorectal cancer. Int J Oncol. 2014;45(2):603–610. doi: 10.3892/ijo.2014.2474. [DOI] [PubMed] [Google Scholar]
- 23.Kortylewski M, Nechaev S. How to train your dragon: targeted delivery of microRNA to cancer cells in vivo. Mol Ther. 2014;22(6):1070–1071. doi: 10.1038/mt.2014.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rincon MY, VandenDriessche T, Chuah MK. Gene therapy for cardiovascular disease: advances in vector development, targeting, and delivery for clinical translation. Cardiovasc Res. 2015;108(1):4–20. doi: 10.1093/cvr/cvv205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hatakeyama H, Murata M, Sato Y, et al. The systemic administration of an anti-miRNA oligonucleotide encapsulated pH-sensitive liposome results in reduced level of hepatic microRNA-122 in mice. J Controlled Release. 2014;173:43–50. [PubMed] [Google Scholar]
- 26.Scomparin A, Polyak D, Krivitsky A, Satchi-Fainaro R. Achieving successful delivery of oligonucleotides – From physico-chemical characterization to in vivo evaluation. Biotechnol Adv. 2015;33(6):1294–1309. doi: 10.1016/j.biotechadv.2015.04.008. [DOI] [PubMed] [Google Scholar]
- 27.Kheirolomoom A, Kim CW, Seo JW, et al. Multifunctional nanoparticles facilitate molecular targeting and miRNA delivery to inhibit atherosclerosis in ApoE(−/−) mice. ACS Nano. 2015;9(9):8885–8897. doi: 10.1021/acsnano.5b02611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen F, Huang P, Zhu YJ, Wu J, Zhang CL, Cui DX. The photoluminescence, drug delivery and imaging properties of multifunctional Eu3+/Gd3+ dual-doped hydroxyapatite nanorods. Biomaterials. 2011;32(34):9031–9039. doi: 10.1016/j.biomaterials.2011.08.032. [DOI] [PubMed] [Google Scholar]
- 29.Kota J, Chivukula RR, O’Donnell KA, et al. Therapeutic microRNA delivery suppresses tumorigenesis in a murine liver cancer model. Cell. 2009;137(6):1005–1017. doi: 10.1016/j.cell.2009.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ibrahim AF, Weirauch U, Thomas M, Gruenweller A, Hartmann RK, Aigner A. MicroRNA replacement therapy for miR-145 and miR-33a is efficacious in a model of colon carcinoma. Cancer Res. 2011;71(15):5214–5224. doi: 10.1158/0008-5472.CAN-10-4645. [DOI] [PubMed] [Google Scholar]
- 31.Maikov GI, Vaxenburg R, Sashchiuk A, Lifshitz E. Composition-tunable optical properties of colloidal IV–VI quantum dots, composed of core/shell heterostructures with alloy components. ACS Nano. 2010;4(11):6547–6556. doi: 10.1021/nn101760t. [DOI] [PubMed] [Google Scholar]
- 32.Soenen SJ, Manshian BB, Himmelreich U, Demeester J, Braeckmans K, De Smedt SC. The performance of gradient alloy quantum dots in cell labeling. Biomaterials. 2014;35(26):7249–7258. doi: 10.1016/j.biomaterials.2014.05.023. [DOI] [PubMed] [Google Scholar]
- 33.Liang G, Chen S, Zhu Y, Xiao Z. In: Construction of MiRNA Eukaryotic Expression Vector and its Stable Expression in Human Liver Cancer Cells. Dan Y, editor. Amsterdam: Elsevier; 2011. [Google Scholar]
- 34.Chen CF, Ridzon DA, Broomer AJ, et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005;33(20):e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(T)(−Delta Delta C) method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 36.Yang HN, Park JS, Jeon SY, Park W, Na K, Park K-H. The effect of quantum dot size and poly(ethylenimine) coating on the efficiency of gene delivery into human mesenchymal stem cells. Biomaterials. 2014;35(29):8439–8449. doi: 10.1016/j.biomaterials.2014.06.024. [DOI] [PubMed] [Google Scholar]
- 37.Chen L, Zheng J, Zhang Y, et al. Tumor-specific expression of microRNA-26a suppresses human hepatocellular carcinoma growth via cyclin-dependent and -independent pathways. Mol Ther. 2011;19(8):1521–1528. doi: 10.1038/mt.2011.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gandhi NS, Tekade RK, Chougule MB. Nanocarrier mediated delivery of siRNA/miRNA in combination with chemotherapeutic agents for cancer therapy: current progress and advances. J Control Release. 2014;194:238–256. doi: 10.1016/j.jconrel.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]