Abstract
The endoplasmic reticulum (ER) is the largest organelle in the cell, and its functions have been studied for decades. The past several years have provided novel insights into the existence of distinct domains between the ER and other organelles, known as membrane contact sites (MCSs). At these contact sites, organelle membranes are closely apposed and tethered, but do not fuse. Here, various protein complexes can work in concert to perform specialized functions such as binding, sensing and transferring molecules, as well as engaging in organelle biogenesis and dynamics. This Review describes the structure and functions of MCSs, primarily focusing on contacts of the ER with mitochondria and endosomes.
The endoplasmic reticulum (ER) is the largest membrane-bound organelle in eukaryotic cells and performs a variety of essential cellular functions, including protein synthesis and processing, lipid synthesis, and calcium (Ca2+) storage and release. It consists of multiple structural domains that are interconnected and contiguous (FIG. 1a). The largest domain of the ER flattens around the cell nucleus to form the double membrane bilayer barrier, termed the nuclear envelope. Branching out of the outer nuclear membrane is the peripheral ER, which consists of two structural domains: flat membrane cisternae (also known as sheets) and tubules. ER sheets are covered with ribosomes for the synthesis, translocation and folding of membrane, luminal and secreted proteins. ER tubules are branched and spread throughout the cytosol. They associate with significantly fewer ribosomes and are therefore considered ‘smooth’ ER1–6. The tubular ER network forms abundant membrane contact sites (MCSs) with other organelles and with the plasma membrane (FIG. 1a). This Review describes and compares the structure, factors and functions found at ER MCSs with two very different cytosolic organelles: mitochondria and endosomes (TABLE 1). ER MCSs with the Golgi, peroxisomes and lipid droplets will only be discussed briefly, because these MCSs have additional complexities stemming from the fact that their biogenesis begins on the ER membrane itself, and thus they are not entirely autonomous organelles (see FIG. 1a and TABLE 1).
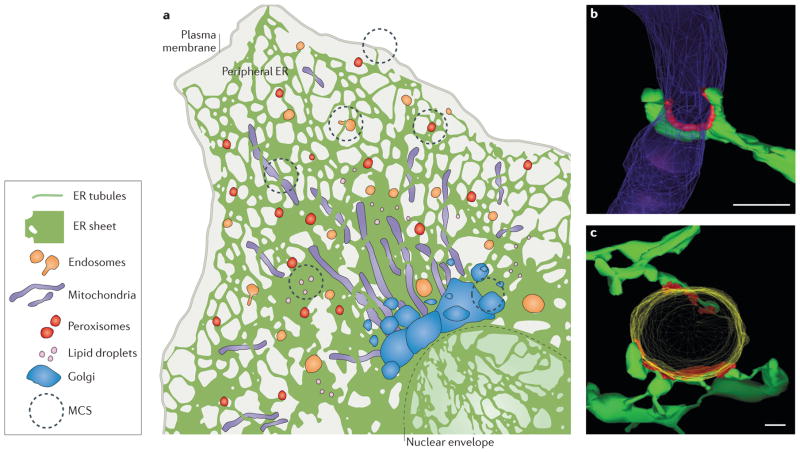
Figure 1. Structure of endoplasmic reticulum (ER) membrane-contact sites (MCSs).
a | The ER consists of the nuclear envelope (outlined with a dashed line) and the peripheral ER, which spreads into the cytosol as a network of sheets and tubules. The peripheral ER forms MCSs with the plasma membrane, mitochondria, endosomes, peroxisomes, lipid droplets and the Golgi. b, c | Electron tomography reveals the three-dimensional structure of MCSs (coloured red) between ER tubules (green) and mitochondria (purple) in a yeast cell b) or an endosome (yellow) in an animal cell (c). Scale bars represent 200 nm in parts b–c. Image in parts b reproduced with permission from REF. 9, AAAS. Image in part c republished with permission of the American Society for Cell Biology, from Endoplasmic reticulum-endosome contact increases as endosomes traffic and mature. Friedman, J. R., Dibenedetto, J. R., West, M., Rowland, A. A. & Voeltz, G. K. Mol. Biol. Cell 24, 1030–1040 (2013); permission conveyed through Copyright Clearance Center, Inc.
Table 1.
Location, characteristics and proposed functions of mammalian and yeast ER–organelle MCS proteins
| MCS proteins | Characteristics | Proposed function | Refs |
|---|---|---|---|
| Mitochondria–ER | |||
| MFN2–MFN2 or MFN1–MFN2 |
|
Calcium (Ca2+) transfer at ER–mitochondrial contact sites | 15 |
| VDAC–GRP75–Ins(1,4,5)P3R |
|
Ins(1,4,5)P3R is the Ca2+-release channel on the ER, when stimulated Ca2+ can be transferred to mitochondria at MCSs and taken up into the mitochondria through VDACs. The GRP75 chaperone couples the Ins(1,4,5)P3R and the VDAC | 73, 81, 84 |
| PTPIP51–VAPs |
|
Lipid transfer between ER and mitochondria may be facilitated by the PtdInsP-transfer domain of PTPIP51 | 114 |
| FIS1–BAP31 |
|
FIS1–BAP31 interaction allows for transmission of apoptotic signals from the mitochondria to the ER | 115 |
| Mmr1* | Localizes to yeast cortical ER–mitochondria contact sites | Important for mitochondrial inheritance into yeast bud | 12 |
| ERMES complex* | Contains both ER- and mitochondria-localized proteins | In yeast, tethers ER and mitochondria. ERMES components contain SMP domains that are potentially capable of transferring lipids | 14,44, 46,47, 50 |
| Endosome–ER | |||
| ORP1L–VAP-A |
|
Senses sterol levels and regulates endosome positioning. Under low cholesterol concentrations, ORP1L negatively regulates late endosome association with dynein. Dynein no longer translocates late endosomes to the cell centre | 28 |
| STARD3–VAP-A |
|
Possible role in sterol sensing and endosome positioning | 7, 59 |
| STARD3NL–VAP-A |
|
Possible role in sterol sensing and endosome tabulation | 7 |
| NPC1–ORP5 |
|
Proposed mechanism for cholesterol transfer through late endosome–ER MCSs by the ORD domain of ORP5 | 58 |
|
|
Regulates endosome positioning. Protrudin transfers kinesin-1 from the ER to late endosomes. Kinesin facilitates late endosome translocation to the cell periphery | 34 |
| EGFR–PTP1B |
|
PTP1B dephosphorylates receptors to regulate EGFR signalling | 11 |
| G-CSFR–PTP1B | G-CSFR, receptor protein in the endosome membrane PTP1B, ER-localized phosphatase | PTP1B dephosphorylates receptors to regulate G-CSFR signalling | 116 |
| Golgi–ER | |||
| OSBP–VAP | OSBP associates with the Golgi membrane through PtdIns4P binding and contains a FFAT domain capable of interacting with ER VAPs | OSBP regulates PtdIns(4)P levels in the Golgi by transferring PtdIns(4)P from Golgi to the ER. OSBP transfers sterol in the opposite direction, from ER to Golgi | 60, 64 |
| CERT–VAP | CERT associates with the Golgi membrane through PtdIns(4)P binding and contains a FFAT domain capable of interacting with ER VAPs | CERT has a role in ceramide transfer at ER–Golgi MCSs | 60, 61 |
| FAPP2–VAP | FAPP2 associates with the Golgi membrane through PtdIns(4)P binding and contains a FFAT domain capable of interacting with ER VAPs | FAPP2 has a role in glucosylceramide transfer at ER–Golgi MCSs | 62 |
| NIR2–VAP | NIR2 associates with the Golgi membrane and contains a FFAT motif capable of interacting with ER VAPs | NIR2 plays a part in maintaining diacylglycerol levels in the Golgi | 60, 63, 60 |
| Lipid droplet–ER | |||
| DGAT2–FATP1 | DGAT2 localizes to lipid droplets FATP1 localizes to the ER | DGAT2 and FATP1 coordinate lipid droplet expansion at lipid droplet–ER MCSs | 117 |
| Peroxisome–ER | |||
| Pex3–Inp1–Pex3* | Pex3, integral membrane protein localized to both peroxisomes and ER Inp1 cytosolic factor | In yeast, Inp3 binds to Pex3 and regulates tethering of peroxisomes to ER | 118 |
CERT, ceramide-transfer protein; DGAT2, diacylglycerol O-acyltransferase 2; EGFR, epidermal growth factor receptor; ER, endoplasmic reticulum; ERMES, ER–mitochondrial encounter structure; FAPP2, four-phosphate adaptor protein 2; FATP1, fatty acid transport protein 1; FFAT, diphenylalanine in an acidic tract; FM, fluorescence microscopy; G-CSFR, granulocyte–macrophage colony-stimulating receptor; GRP75, glucose-regulated protein 75; Inp, inheritance of peroxisomes; Ins(1,4,5)P3R, inositol-1,4,5-trisphosphate receptor; LE, late endosome; MCS, membrane contact site; MFN, mitofusin; Mmr1, mitochondrial Myo2p receptor-related 1; NIR2, PYK2 N-terminal domain-interacting receptor 2; NPC1, Niemann–Pick C1 protein; ORD, oxysterol-binding-related domain; ORP, oxysterol-binding-related protein; Pex3, peroxin 3; PtdInsP, phosphatidylinositol phosphate; PTP1B, protein-Tyr phosphatase 1B; STARD3, START domain-containing protein 3; STARD3NL, STARD3 N-terminal-like protein; VAP, VAMP-associated protein; VDAC, voltage-dependent anion channel.
indicates yeast proteins, all other proteins are of mammalian origin.
The structure of MCSs
The combined efforts of electron microscopy and live-cell fluorescence microscopy have revealed the structures of ER MCSs with mitochondria and endosomes. Studies using electron microscopy and tomography have captured features of these contact sites at nanometre resolution (FIG. 1b,c). One defining feature of all MCSs is that ribosomes are excluded from the ER membrane at the interface with the partner organelle membrane6–11. The absence of ribosomes from these locations implies that there are specialized ER proteins that maintain the structure of contact sites and thereby prevent the ribosome-bound translocation machinery from diffusing into these regions.
Electron micrographs have also been used to measure the distance between the ER and the apposing organelles. The gap distances are quite similar: 3–15 nm for ER–endosome7 and 6–15 nm for ER–mitochondria8,9. Such short tethering distances allow channelling of smooth ER materials, such as lipids and Ca2+ (discussed below). Electron microscopy and tomography also revealed the frequency of membrane contact. Typically, the ER network will interact at multiple small and discrete positions with an individual organelle9,10,12–14 (FIG. 1b,c). When these contacts are cumulatively analysed, mammalian ER MCSs cover about 2–5% of the surface area of an average mitochondrion15,16 and 3–5% of the surface of an endosome7,10,11. These multiple discrete contact sites could be functionally redundant, or they may each mediate different activities.
MCSs regulate organelle dynamics
Multi-colour live-cell fluorescence microscopy has been used to visualize how organelle trafficking affects the integrity of inter-organelle MCSs (FIG. 2a). Strikingly, once bound, the endosomes and mitochondria appear to be tightly tethered to the ER, but do not fuse with it. Tethering is maintained when the organelles traffic, even over very long distances. As a result of this tethering, the moving organelles will ‘drag’ ER tubules with them10,17 (FIG. 2a,b). At any given time, dozens of endosomes and mitochondria bound to ER tubules traffic within the cell. Therefore, it can be expected that the overall dynamics and structure of the ER are influenced by these events.
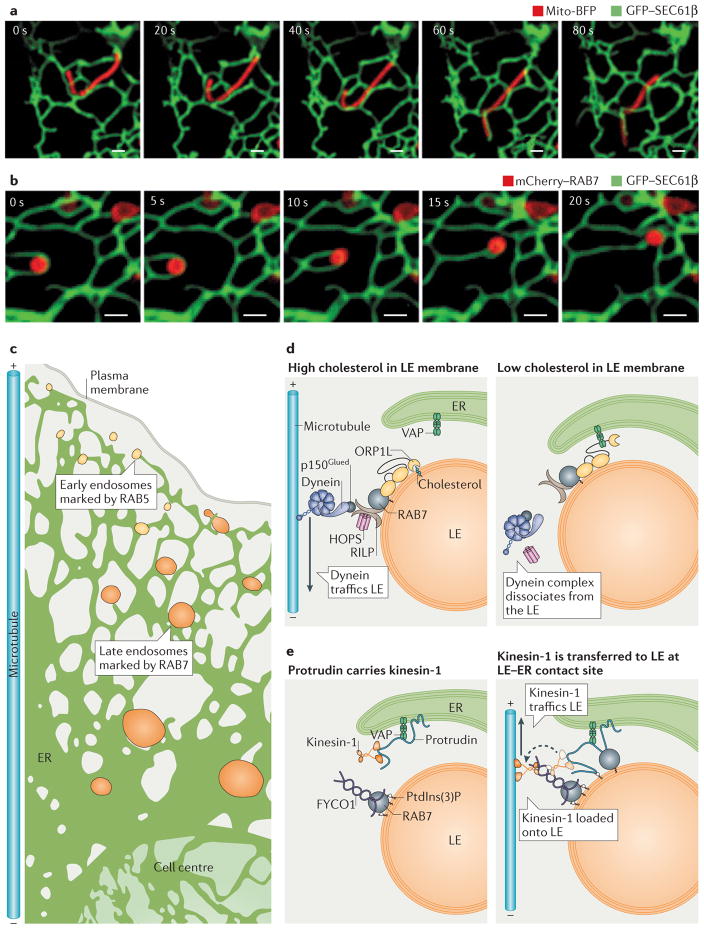
Figure 2. Dynamics of endoplasmic reticulum (ER) membrane contact sites (MCSs).
a,b | Endosomes and mitochondria are tightly tethered to ER tubules even as they traffic. Time-lapse fluorescence images of ER and organelle dynamics in live Cos-7 cells expressing GFP–SEC61β (labelling ER in green) and (a) mito-BFP (labelling mitochondria in red) or (b) mCherry–RAB7 (labelling late endosomes (LEs) in red). Note how the contact sites are maintained as the apposing organelles move. Scale bars represent 1 μm. c | Endosomes mature as they traffic from the cell periphery along microtubules to the cell centre. ER–endosome contact increases as endosomes mature, with 53% of early endosomes (EEs; marked by RAB5) and 99% of LEs (marked by RAB7) remaining in contact with the ER during trafficking. d | Model of how cholesterol levels regulate the composition of ER–LEs MCSs and LE trafficking. When the LE contains high cholesterol levels (left panel), oxysterol-binding-related protein 1L (ORP1L) can bind to cholesterol on the LE membrane and does not associate with ER VAPs (VAMP-associated proteins). In addition, ORP1L interacts with RAB7 GTPase, which stimulates minus-end-directed LE trafficking through a complex that includes RILP (RAB-interacting lysosomal protein), the HOPS (homotypic fusion and vacuole protein sorting) complex, dynactin (p150Glued) and the motor protein dynein. At low cholesterol levels (right panel), ORP1L is not bound to cholesterol and instead interacts with ER VAPs. The ORP1L–VAP interaction displaces dynein from the endosome. e | Protrudin is an ER integral membrane protein that interacts with VAP and kinesin-1 (left panel). Protrudin binds to RAB7 and phosphatidylinositol-3-phosphate (PtdIns(3)P) on the LE membrane. Protrudin can bind to and transfer kinesin-1 to the LE protein FYCO1 (FYVE and coiled-coil domain-containing protein 1), and this promotes plus-end-directed microtubule trafficking of LEs (right panel).
MCS-bound organelles in motion
The tubular ER network is very dynamic and constantly rearranges its structure along the microtubule cytoskeleton. In animal cells, these rearrangements, referred to as ER sliding, occur in both directions and are mediated by the molecular motors dynein and kinesin18. Mitochondria and endosomes are also very dynamic organelles that traffic along microtubules. As the ER, endosomes and mitochondria are all dynamic organelles, it is unclear how stable MCSs are maintained as the organelles move. One explanation could be that endosomes and mitochondria traffic using the same motors as the ER — kinesin-1 and dynein18–22 — so that the joined organelles can travel together. In addition, a few proteins have been identified that may contribute to maintaining ER MCS stability during organelle trafficking. Miro, for example, is an outer mitochondrial membrane component and one of the proteins enriched at ER–mitochondrial contact sites23. It is linked to dynein through the cytosolic factor Milton21,24 and additionally contains multiple Ca2+-binding domains that could be regulated by Ca2+ fluxes at the ER–mitochondria interface23,25 (see below). Importantly, the yeast orthologue of Miro, Gem1, is also enriched at ER–mitochondrial contact sites23, and its deletion has been shown to regulate the assembly and disassembly of the yeast ER–mitochondrial encounter structure (ERMES) complex, which maintains ER–mitochondria MCSs (also discussed below)16.
Regulation of organelle trafficking by MCSs
The formation of the MCSs does not only affect the structure of the ER itself but has also been implicated in regulating the trafficking and localization of both endosomes and mitochondria. Following budding from the plasma membrane, endosomes traffic along microtubules towards the microtubule organizing centre (MTOC) and mature on the way, transitioning from early endosomes to late endosomes, to finally fuse with the lysosome26. A large percentage of endosomes maintain contact with the ER as they traffic (FIG. 2b,c). In fact, approximately 53% of all early endosomes, and a staggering 99% of late endosomes, remain in contact with the ER during trafficking. Thus, contacts between the endosomes and the ER are very pervasive and increase as endosome maturation progresses10. Notably, it has been shown that the composition of the ER–late endosome MCS is not constant, and various proteins that are implicated in regulating the trafficking of late endosomes can be recruited to these sites. Consequently, the observed abundance of MCSs between late endosomes and ER can be involved in regulating localization of late endosomes within the cell.
Cholesterol levels affect the composition of ER–late endosome MCSs, and this affects late endosome trafficking and localization (FIG. 2d). Late endosomes accumulate near the microtubule plus ends (at the cell periphery) when cholesterol levels are low, or at the minus ends (at the cell centre) when levels of cholesterol are high. As endosomes mature, they travel to the centre of the cell to fuse with the lysosome and degrade cargo; disrupting late endosome trafficking can result in lysosomal storage disorders27,28. This mechanism is mediated by a cholesterol-sensing protein, oxysterol-binding-related protein 1L (ORP1L), which is a member of the highly conserved and ubiquitous oxysterol-binding-related protein family. ORP1L contains an oxysterol-binding-related domain (ORD) that has been shown to be capable of binding sterols in vitro29 and localizes to the surface of late endosomes. The current model of how cholesterol influences localization of late endosomes indicates that when the ORP1L ORD domain senses cholesterol in the late endosome membrane, ORP1L acquires a conformation that allows its interaction with a complex that includes RAB7 GTPase (a marker of late endosomes associated with their membranes), RILP (RAB-interacting lysosomal protein), the HOPS (homotypic fusion and vacuole protein sorting) late endosome tethering complex, and the dynactin (p150Glued)–dynein motor complex, resulting in minus-end-directed late endosome trafficking28,30 (FIG. 2d, left panel). Conversely, at low cholesterol levels, the ORP1L FFAT (diphenylalanine in an acidic tract) domain is free to bind to the ER-resident protein VAP (VAMP-associated protein) instead of the RAB7–RILP–p150 complex, which reduces dynein-facilitated trafficking towards microtubule minus ends28 (FIG. 2d, right panel).
START domain-containing protein 3 (STARD3; also known as MLN64) is another candidate that could sense sterol at ER–late endosome MCSs and provide regulation of late endosome trafficking by means of altering the protein composition and function of the MCSs. Similar to ORP1L, STARD3 is capable of binding to cholesterol in vitro through its conserved START protein domain (see further discussion of START family members in lipid section below)31 and contains an FFAT motif that is capable of interacting with ER VAP proteins7. Overexpression of STARD3 results in accumulation of late endosomes at the perinuclear region and enrichment of actin patches on late endosomes, which may play a part in budding domain formation or in late endosome positioning32. Conversely, STARD3 knockdown results in late endosome scattering to the cell periphery and loss of actin patches on late endosomes32. The specific cytoskeletal machinery that interacts with STARD3 and regulates late endosome positioning is yet to be discovered, but it seems that STARD3 functions by influencing late endosome association with the actin cytoskeleton.
Yet another protein that functions at ER–late endosome contact sites to regulate late endosome trafficking is protrudin33,34. It has been demonstrated that by localizing to MCSs, protrudin promotes plus-end-directed trafficking of late endosomes in neurite outgrowths34. Protrudin is an ER transmembrane protein that interacts with VAP. It binds to RAB7 on the late endosome and also contains a FYVE domain, which allows it to bind to phosphatidylinositol-3-phosphate (PtdIns(3)P), a lipid that is enriched on the endosomal membrane34. Overexpression of protrudin and RAB7 causes ER to wrap around late endosomes, and these late endosomes accumulate in the cell periphery. Conversely, protrudin depletion results in perinuclear accumulation of late endosomes34. Protrudin functions in this regulation of late endosome positioning via recruitment of kinesin-1 to contact sites, followed by transfer of this motor onto FYCO1 (FYVE and coiled-coil domain-containing protein). FYCO1 localizes on the late endosome membrane (via RAB7 and PtdIns(3)P) and functions as an adaptor to link kinesin-1 to late endosomes. Kinesin-1 association with late endosomes promotes plus-end-directed late endosome trafficking34–36 (FIG. 2e). An interesting possibility is that protrudin–RAB7 and VAP–ORP1L could localize together to ER–late endosome contact sites and drive anterograde trafficking (that is, towards plus ends of microtubules) of late endosomes by coordinating the recruitment of kinesin-1 and the release of dynein, respectively34.
MCSs can also be involved in regulating mitochondrial trafficking. One notable example has been described for budding yeast, in which ER–mitochondria MCSs are important for proper inheritance of mitochondria by a daughter cell upon division. This mechanism is mediated by mitochondrial Myo2p receptor- related 1 (Mmr1), a putative tether that localizes to MCSs between mitochondria and cortical ER12. More specifically, Mmr1 is found at both the ER and mitochondrial surfaces of the MCSs in the bud tip. Deletion of this tether perturbs the proper anchorage of mitochondria in the forming bud, leading to their slippage out of the bud and, consequently, defects in mitochondrial (but not ER) inheritance by the daughter cell12.
In sum, it appears that MCSs formed between ER and endosomes as well as mitochondria can influence the dynamics, intracellular trafficking and localization of these organelles. This can be mediated by the regulated recruitment of various proteins and protein complexes, which alter the molecular composition and functions of these MCSs.
MSCs in lipid biosynthesis and exchange
Phospholipids, sterols and the precursors for sphingo-lipids and cardiolipin are largely synthesized at the ER and need to be transported to other membrane-bound cellular compartments. Lipids can be transferred from the ER to other organelles by vesicular transport. However, mounting evidence suggests that rapid transport also occurs by non-vesicular transport mechanisms at positions where the ER membrane is closely apposed to other membrane- bound compartments37–39. The short (<30 nm) distance between the ER and other membranes at MCSs provides an excellent hub for non-vesicular transfer. Non-vesicular transport of lipids at MCSs requires machinery that can extract a lipid molecule out of the outer leaflet of the originating membrane, shield it in a hydrophobic pocket from the cytosolic aqueous environment, bridge the cytosolic gap between the apposing membranes to allow the transfer of the molecule between membranes and finally insert it into the outer leaflet of the target membrane.
It is likely that multiple factors work together to coordinate lipid transfer. In the 1960s, potential lipid- transfer proteins (LTPs) were first detected in vitro as soluble factors that accelerated the transfer of lipids between mitochondria and ER-derived microsomes38. However, it took many more years to actually identify the proteins involved. Now, LTPs are much better understood, and in this Review, three major protein families will be discussed: the oxysterol-binding protein-related protein family40; the START (steroidogenic acute regulatory protein-related lipid transfer domain) protein family31; and the synaptotagmin-like mitochondrial protein (SMP)/tubular lipid-binding protein (TULIP) super-family41. Members of these protein families contain a hydrophobic pocket that is possibly capable of binding a lipid monomer, and, importantly, many contain domains that can recruit them to ER MCSs. Below, we discuss the role of tight coupling between particular organelles and the ER in the process of lipid biosynthesis and exchange, thus emphasizing the importance of the formation of MCSs as well as the role of LTPs and their recruitment to MCSs in this fundamental pathway.
ER–mitochondria
Biosynthetic enzymes that coordinate synthesis of major cellular phospholipids are localized to both the ER membrane and the mitochondrial matrix. As an example, phosphatidylserine synthesized at the ER can be altered by mitochondria-localized phospholipid synthase to generate phosphatidylethanolamine, which can be converted to phosphatidylcholine by ER-localized enzymes42,43 (FIG. 3a). In addition, the precursor for mitochondrial-specific cardiolipin, phosphatidic acid, is synthesized at the ER and must be transferred to the mitochondria for modification42,43. Directional transfer of these biosynthetic precursors between the ER and the outer and inner mitochondrial membranes would be facilitated by rapid non-vesicular lipid transfer at the ER–mitochondria interface39. Ideally, this machinery would also monitor levels of precursors at each organelle to balance the synthesis and transfer process, thereby regulating the proper composition for each individual organelle.
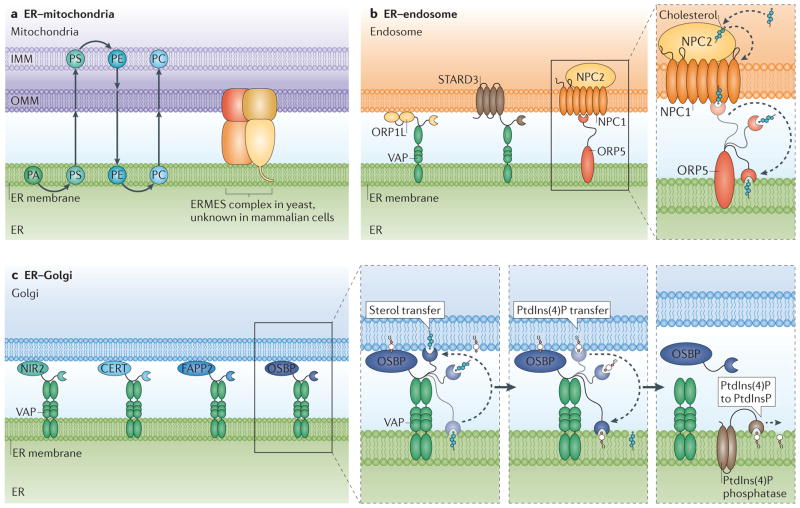
Figure 3. Endoplasmic reticulum (ER) membrane contact sites (MCSs) function in lipid biosynthesis and exchange.
a | ER– mitochondria MCSs are rich in lipids and lipid-synthesis enzymes. Lipids are transferred between organelles at MCSs. In one pathway, phosphatidic acid (PA) is converted to phosphatidylserine (PS) at the ER. PS is transferred to the inner mitochondria membrane (IMM) where it is converted to phosphatidylethanolamine (PE). PE is transferred back to the ER, where it is converted to phosphatidylcholine (PC). PC is likely to also be transported back to mitochondria. ER–mitochondria membrane-tethering proteins (such as the ER–mitochondrial encounter structure (ERMES) in yeast) may aid this process; however, the exact mechanism of their action is currently elusive, and a mammalian counterpart has not been identified. b | Various complexes sense, modify or potentially transfer lipids at ER–endosome MCSs. Oxysterol-binding-related protein 1L (ORP1L) and START domain-containing protein 3 (STARD3) on the endosome have both been shown to interact with ER-resident VAMP-associated proteins (VAPs), but how they aid the exchange of lipids at ER–endosome MCSs is unclear. Niemann–Pick type C2 protein (NPC2) resides in the endosome lumen and interacts with endosome membrane protein NPC1. NPC1 interacts with the ER protein ORP5. The interactions between NPC2, NPC1 and ORP5 provide a potential mechanism for cholesterol transfer between the endosome lumen and the ER (blow-up). In this model, NPC2 transfers cholesterol from the endosome lumen to NPC1 on the endosome membrane. ER-resident ORP5 then accepts cholesterol from NPC1 and may transfer the cholesterol to the ER for redistribution. c | Multiple potential lipid-transfer proteins localize to the Golgi membrane and interact with ER VAPs. These include the phosphatidylinositol-transfer protein NIR2 (PYK2 N-terminal domain-interacting receptor 2), the ceramide-transfer protein (CERT), the glycosylceramide-transfer protein Golgi-associated four-phosphate adaptor protein (FAPP2) and the cholesterol and phosphatidylinositol-4-phosphate (PtdIns(4)P)-transfer protein oxysterol-binding protein (OSBP). Studies specifically on OSBP (right panels) show that it associates with the Golgi membrane through PtdIns(4)P binding. The OSBP oxysterol-binding-related domain (ORD domain) can bind and transfer sterol from the ER to the Golgi and PtdIns(4)P from the Golgi to the ER. When PtdIns(4)P levels are depleted at the Golgi, OSBP dissociates from the Golgi membranes. PtdIns(4)P at the ER is converted back to PtdInsP by ER-associated PtdIns(4)P phosphatase. OMM, outer mitochondria membrane.
In yeast, ERMES is the primary candidate for tethering non-vesicular transfer of lipids between ER and mitochondria23 (FIG. 3a). Fluorescence microscopy has revealed that all ERMES complex components localize to punctate structures at contact sites between the ER and mitochondria14,16,23. Three of the four components of the yeast ERMES complex contain SMP domains and thus are members of the SMP/TULIP family of LTPs14,41,44. Structural analysis of other SMP/TULIP protein family members suggests that SMP domains are capable of binding to lipids to shield them from an aqueous environment41,45. It remains to be tested whether ERMES directly binds to and transfers lipids itself, or whether it functions as a structural tether to facilitate transfer of lipids by other proteins. In addition, there have been conflicting reports as to whether ERMES complex depletion in yeast alters the lipid transfer rates between the ER and mitochondria14,46–50. ERMES may not be the only machinery facilitating lipid transfer from ER to mitochondria. In fact, recent evidence has shown that the MCSs between the yeast vacuole and mitochondria can function as a circuitous route for lipid transfer, which occurs via a vCLAMP (vacuole and mitochondria patch) complex51,52. Thus, even in the absence of ERMES, phospholipids could traffic through an alternative route from the ER via the vacuole to the mitochondria. Interestingly, when ERMES is depleted, the contact between mitochondria and vacuole at vCLAMP expands51,52. Elimination of both vCLAMP and ERMES contact sites leads to significant defects in phospholipid transfer to mitochondria51,52. These findings suggest that lipid trafficking at ER–mitochondria MCSs can be rescued by vacuole–mitochondria MCSs.
ER–endosome
Endosomes store cholesterol and can redistribute this cholesterol from the endosomal membrane to the ER. Different protein pairs at the ER–endosome interface may be capable of sensing and regulating cholesterol at this MCS (FIG. 3b). Cholesterol enters the cell in low-density lipoprotein (LDL) particles. LDL particles bind to cell surface receptors and accumulate in late endosomes and lysosomes53. Studies monitoring the localization of cholesterol indicate that approximately 30% of cholesterol in late endosomes and lysosomes is directly transported to the ER54. This LDL-bound cholesterol (LDL-C) can then be distributed to other parts of the cell, providing membrane rigidity, fluidity and permeability. Improper export of LDL from late endosomes and lysosomes has been linked to Niemann–Pick Type C disease, which is an autosomal recessive neurodegenerative disorder characterized by accumulation of LDL-derived cholesterol in late endosomes and lysosomes55. Niemann–Pick C2 protein (NPC2) is a soluble LTP in the late endosome or lysosome lumen that binds to LDL-C and transfers it to NPC1, a late endosome integral membrane protein, which facilitates LDL-C export56,57. It is not fully understood how NPC1 exports cholesterol out of the late endosome or lysosome. ORP5 is an ER transmembrane protein and is a potential acceptor that could receive LDL-C from NPC1 (FIG. 3b). ORP5 is capable of binding sterol in a conserved pocket29 and co-immunoprecipitates with NPC1. This has led to a model in which the two proteins function in the same complex to transfer sterol from late endosomes to the ER membrane58. In support of this model, depletion of NPC1 or ORP5 will prevent sterol transfer from the late endosome to the ER, and this results in sterol accumulation on the late endosome membrane58.
ORP1L and STARD3 are other sterol-binding proteins that are present on late endosomes, but their role in sterol transfer at ER–endosome MCSs remains unclear. ORP1L and STARD3 can both interact with ER-localized VAP and form ER–endosome MCSs (see discussion above). Interestingly ORP1L is found on the same population of endosomes as ORP5, but STARD3 is found on a less mature population of late endosomes containing a different sterol-transfer protein, ATP-binding cassette transporter 3 (ABCA3)59. The purpose of this separate population of STARD3 and ABCA3 double-positive late endosomes and the exact function of ABCA3 are both unknown. One idea is that ABCA3 facilitates the recycling of newly hydrolysed cholesterol back to the plasma membrane, whereas the hydrolysed cholesterol in more mature late endosome populations is transferred to the ER by NPC1–ORP5 interaction59.
Overall, multiple complexes that bind to lipids are present at the ER–late endosome interface, and these could have cooperative or opposing roles in the regulation of sterol flux at MCSs. Although overexpression of many of these pairs will increase ER–endosome contact, no one pair is essential for MCS formation and maintenance, because their depletion does not prevent contact site formation. Furthermore, contact is already observed with early endosomes10,17, and the above-mentioned proteins are all recruited to late endosomes. Thus, it seems that it is not the formation of the ER–late endosome MCSs per se but the regulation of their composition that affects important cellular processes such as lipid biogenesis.
ER–Golgi
The past several years have revealed that MCSs exist between the ER and the Golgi, and that they are able to regulate direct lipid transfer. A variety of LTPs have been localized to ER–Golgi MCSs (TABLE 1). All of these proteins have the ability to bind to lipids to facilitate ER–Golgi lipid transfer60–64. These include the ceramide-transfer protein (CERT)61, the glycosylceramide-transfer protein Golgi-associated four-phosphate adaptor protein 2 (FAPP2; also known as PLEKHA8)62, the phosphatidylinositol-transfer protein NIR2 (PYK2 N-terminal domain-interacting receptor 2)63, and the cholesterol- and PtdIns(4)P-transfer protein oxysterol-binding protein (OSBP)64,65 (FIG. 3c). All four proteins contain a pleckstrin homology (PH) domain that allows them to bind to PtdIns(4)P on the Golgi, and an FFAT motif that is capable of interacting with the ER-localized VAPs39,66. CERT and FAPP2 regulate ceramide and glucosylceramide transfer, respectively, at ER–Golgi MCSs61,62,67. NIR2 plays a part in maintaining diacylglycerol levels in the Golgi60,63,68.
Since all of these LTPs are recruited to the Golgi by PtdIns(4)P binding, it is especially important to understand what regulates Golgi PtdIns(4)P levels. A recent elegant study has shown that OSBP regulates both PtdIns(4)P and sterol transfer at ER–Golgi MCSs. OSBP can bridge ER–Golgi MCS, because it contains a PH domain that binds to PtdIns(4)P in the Golgi and an FFAT domain that interacts with ER VAPs64. In vitro and in vivo data demonstrate that the OSBP ORD domain can bind to and transfer both sterol and PtdIns(4)P64. In the overall model (FIG. 3c), OSBP promotes anterograde sterol transfer from the ER to the Golgi and retrograde PtdIns(4)P transfer from the Golgi back to the ER64. This process is regulated by a feedback mechanism: OSBP dissociates from the Golgi when Golgi PtdIns(4)P levels are low, because it is no longer recruited64. This also disrupts direct transfer of sterol from the Golgi to the ER64.
Collectively, proper lipid synthesis and intracellular lipid distribution seem to be tightly coupled to the existence of MCSs between various membranous organelles within the cell. These tight membrane contacts have been implicated in supporting the non-vesicular exchange of lipids between the organelles through specialized lipid-binding and lipid-transferring proteins, the LTPs, which can specifically associate with various MCSs. Exactly how these proteins function at MCSs and how their localization is regulated are still poorly understood, and both of these questions open up exciting new avenues for future studies.
MCSs in Ca2+ exchange
Ca2+ must be transferred across membrane interfaces to propagate signals throughout the cell. The intracellular Ca2+ concentration is controlled by regulated opening of Ca2+-permeable channels on the plasma membrane and ER. Multiple mechanisms regulate Ca2+ concentration to maintain extremely low Ca2+ levels in the cytosol, and low micromolar Ca2+ levels in endosomes and mitochondria (FIG. 4a). The ER lumen houses the major Ca2+ store in mammalian cells (FIG. 4a), and this store is released through inositol-1,4,5-trisphosphate receptor (Ins(1,4,5)P3R) channels that are found throughout the ER membrane69. External stimuli activate receptors on the plasma membrane that subsequently activate phospholipase C (PLC). PLC cleaves phosphatidylinositol-4,5- bisphosphate (PtdIns(4,5)P2) within the plasma membrane, releasing cytosolic Ins(1,4,5)P3 that binds to and stimulates ER Ins(1,4,5)P3Rs70,71 and thus Ca2+ release from ER stores. This cascade enables the cell to convey external signals to intracellular organelles through Ca2+ signalling. These ER stores can release Ca2+ into the cytosol or onto its neighbouring organelles when higher levels are needed.
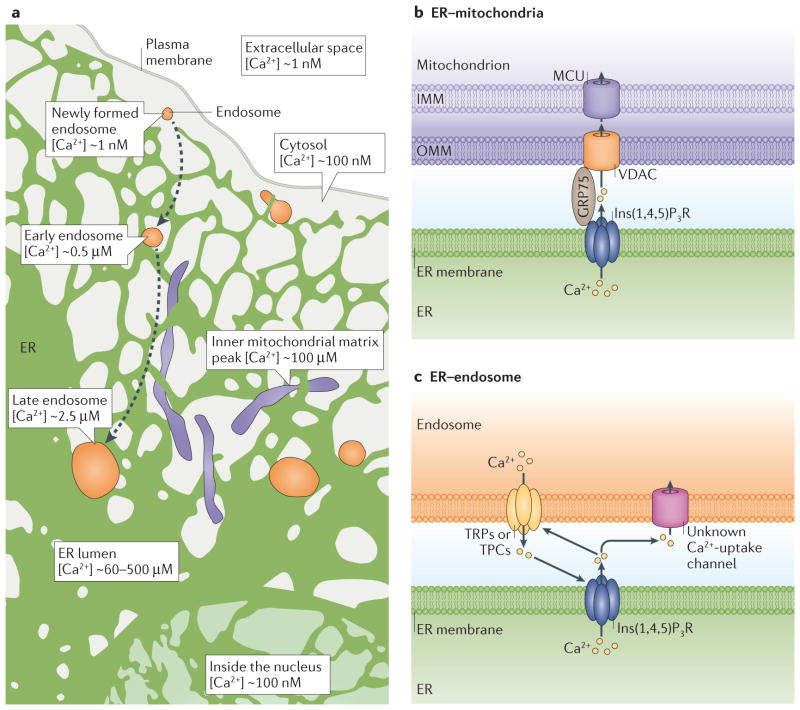
Figure 4. Calcium (Ca2+) exchange at endoplasmic reticulum (ER) membrane contact sites (MCSs).
a | The ER lumen is the major Ca2+ store in the cell, with a Ca2+ concentration ([Ca2+]) of ~60–500 μM). In the extracellular space, [Ca2+] is high (~1mM) compared to the intracellular cytosol (~100nM). Newly formed endosomes have taken up Ca2+ from the extracellular space, so the luminal [Ca2+] is close to the same as that of the extracellular space (~1 mM). Luminal Ca2+ is then released so that early endosomes have [Ca2+] ~0.5 μM and late endosomes have [Ca2+] ~2.5 μM. The ER–endosome MCS is a site of dynamic Ca2+ crosstalk. Endosomes may be able to sequester Ca2+ released from the ER. The ER transfers Ca2+ to mitochondria, with peak mitochondrial Ca2+ concentrations reaching 100 μM. b | ER Ca2+ released from the ER through inositol-1,4,5- trisphosphate receptors (Ins(1,4,5)P3Rs) provides a concentrated Ca2+ spike that can be taken up through the outer mitochondrial membrane (OMM) by VDACs (voltage dependent anion channels) and then through the inner mitochondrial membrane (IMM) by the mitochondrial Ca2+ uniporter (MCU) ion transporter into the mitochondrial matrix. The 75 kDa glucose-regulated protein (GRP75) functions as a chaperone, coupling Ins(1,4,5)P3R to the VDACs. c | Endosomes are capable of releasing Ca2+ though transient receptor potential channels (TRPs) or two-pore channels (TPCs). ER Ca2+ released from ER via Ins(1,4,5)P3Rs could be taken up into endosomes through unknown endosome Ca2+-uptake channels. Ca2+ release from endosomes can also stimulate Ca2+ release from the ER through Ins(1,4,5)P3Rs and vice versa.
MCSs have been found to be involved in regulating this important signalling pathway by means of concentrating and directing Ca2+ transfer. Opening of Ins(1,4,5)P3R Ca2+ channels on the ER leads to an increase in local Ca2+ concentration, but this spike in Ca2+ diffuses, substantially decreasing approximately 100 nm away from the channels72. Thus, signalling crosstalk through the release of Ca2+ from the ER to other organelles can be expected to be much more efficient at tight interfaces73,74.
ER–mitochondria
Localized Ca2+ spikes released from ER by Ins(1,4,5)P3Rs stimulate mitochondrial Ca2+ uptake. Ca2+ passes through voltage-dependent anion channels (VDACs) on the outer mitochondrial membrane and mitochondrial Ca2+ uniporters (MCUs) on the inner mitochondrial membrane75,76. The low-affinity MCU requires a large, localized Ca2+ concentration to facilitate Ca2+ transfer to the mitochondrial matrix75 (FIG. 4b). Uptake of Ca2+ into the mitochondrial matrix alters mitochondrial activity in several ways. For example, Ca2+ stimulates dehydrogenases in the tricarboxylic acid cycle, resulting in more energy production for the cell77. In addition, fluctuations in mitochondrial Ca2+ levels regulate cell death programmes77, and the Ca2+ released from the ER stimulates apoptosis by opening the mitochondrial transition pore78,79.
It has been known for some time that Ca2+ released from the ER can be sequestered by mitochondria80. Further research using a sensor targeted to the outside of mitochondria showed that upon Ins(1,4,5)P3-induced Ca2+ mobilization, mitochondria are exposed to higher Ca2+ concentrations than the bulk cytosol73,81. Ca2+ transfer occurs specifically at ER–mitochondria MCSs, and it is abrogated by increasing the gap distance between the organelles8,81. Evidence suggests that the mitochondrial dynamin-related family member mitofusin 2 (MFN2) localizes to both ER and mitochondria and regulates inter-organelle linkage at Ca2+-transfer sites82. However, owing to conflicting results15,83, further research in this field is currently striving to fully elucidate the role of MFN2 at ER–mitochondria MCSs. For example, there is an open question as to how MFN2 targeting to two different organelles is regulated.
Recent studies have identified additional regulators of Ca2+ transfer at ER–mitochondrial MCSs. The 75 kDa glucose-regulated protein (GRP75) is required for coupling the VDAC channel to Ins(1,4,5)P3R channels84 (FIG. 4b). However, as overexpression of GRP75 does not result in increased ER–mitochondria contact, it is likely that GRP75 functions at established contacts to regulate mitochondrial Ca2+ uptake. Promyelocytic leukaemia tumour suppressor (PML) also regulates Ins(1,4,5) P3R activity to control the amount of Ca2+ at the ER– mitochondria membrane interface and facilitates mitochondrial Ca2+ uptake85. PML was recently found within mitochondria-associated membrane fractions in a complex with AKT and protein phosphatase 2A (PP2A)85. Active, phosphorylated AKT phosphorylates Ins(1,4,5) P3R and inhibits Ins(1,4,5) P3R Ca2+ release, protecting mitochondria from raising a Ca2+-mediated apoptotic response, whereas PP2A phosphatase activity is capable of deactivating AKT by means of dephosphorylation86,87. The amount of phosphorylated AKT was found to be increased in PML-knockout mouse embryonic fibroblasts (MEFs)85. Additionally, in these knockout MEFs, mitochondrial and cytosolic Ca2+ did not respond as dramatically to apoptotic stimuli, but a normal response to stimuli was recovered by introducing an ER-localized PML85. Therefore, PML may recruit PP2A, which inactivates AKT, and regulate Ins(1,4,5)P3R-mediated Ca2+ release in response to apoptotic stimuli. These findings show that ER–mitochondria MCSs are tightly regulated interfaces that easily respond to various cues, including cell stress stimuli, thereby regulating various mitochondrial functions.
ER–endosome
Current research aims to better understand the purpose of endosome Ca2+ stores and how this storage may be regulated by MCSs. Newly formed endocytic vesicles contain material from the Ca2+-rich extracellular space, so, inadvertently, Ca2+ concentration within the endosomal lumen is similar to that in the extracellular space. Ca2+ is quickly released from endosomes, suggesting that Ca2+ release may be required for early steps in endocytic maturation or that it may be coupled to the acidification of endosomes88–90 (FIG. 4a). Specifically, early endosomes marked by RAB5 GTPase have a luminal Ca2+ concentration of around 0.5 μM, and late endosomes marked by RAB7 GTPase have a luminal Ca2+ concentration of around 2.5 μM. Late endosomes and lysosomes contain Ca2+ levels that are close to ER Ca2+ levels91,92 (FIG. 4a). Notably, in late endosomes Ca2+ levels are known to fluctuate, and these fluctuations may be a result of the existence of abundant MCSs between late endosomes and the Ca2+-rich ER92,93.
Several lines of evidence suggest that the ER–endosome interface is a dynamic site for Ca2+ crosstalk between these organelles, with Ca2+ being released from both endosomes and the ER. Studies indicate that endosomes can release Ca2+ stores through both transient receptor potential channels (TRPs) and two-pore channels (TPCs), which have homology to TRP channels94–96 (FIG. 4c). Interestingly, stimulating Ca2+ release from acidic endocytic vesicles can stimulate ER Ca2+ mobilization97 and vice versa, release of Ca2+ from the ER, induced by either Ins(1,4,5)P3 or cyclic ADP-ribose (cADPR) can activate Ca2+ release from acidic vesicles74. Additional evidence indicates that ER Ca2+ release can stimulate increases in fluorescence of a calcium indicator in lysosomes, suggesting that ER Ca2+ could be sequestered into endosomes and/or lysosomes through unknown Ca2+-uptake channels98 (FIG. 4c). Together, these data reveal that substantial Ca2+ crosstalk occurs between the ER and the endo-lysosomal system. However, further studies are needed to determine whether Ca2+ transfer from the ER lumen to the endosome lumen occurs specifically at ER–endosome MCSs, and how this exchange would be regulated.
Organelle biogenesis
Mitochondria and endosomes are dynamic organelles that are constantly undergoing fission and fusion, which is important for maintaining cellular homeostasis26,99. Surprisingly, live-cell imaging has revealed that ER contact sites define the position of fission on mitochondria and endosomes9,13 (FIG. 5). In addition, ER MCSs regulate the sorting and degradation of at least one endocytic cargo, the epidermal growth factor receptor (EGFR)11.
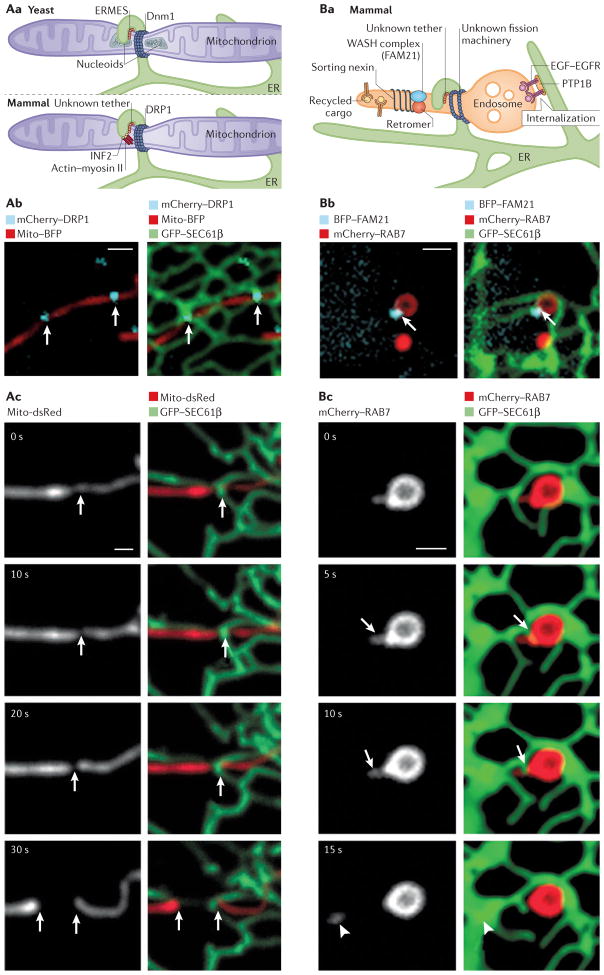
Figure 5. Endoplasmic reticulum (ER) membrane contact sites (MCSs) define the timing and position of both mitochondrial and endosome fission.
Aa. In yeast (top panel), mitochondrial ER-marked constriction and fission sites contain the ER–mitochondrial tethering complex (ERMES), mitochondrial nucleoid DNA and the fission-machinery protein dynamin-related protein 1 (Dnm1). In mammalian cells (bottom panel), an ER-localized inverted formin (INF2), actin and myosin II are candidates for driving ER-associated constriction of mitochondria. Then, the fission-machinery protein dynamin-related protein 1 (DRP1) is recruited by adaptor proteins to ER-marked constrictions, where it drives fission. Ab | Live confocal fluorescence microscopy images of a Cos7 cell expressing mito-BFP (mitochondria in red) and mCherry–DRP1 (in cyan), merged with GFP–SEC61 β (ER in green) in the right panel. ER tubules contact two mitochondrial constrictions labelled with DRP1, as marked by the white arrows. Ac. Live fluorescence microscopy, as in Ab, of a cell expressing mito-dsRed (mitochondria in grey in left panels, red in right panels) and GFP-–SEC61β (ER in green). Note that the ER tubule circumscribes the position of constriction and fission (white arrows) (t=30s). Ba. In ER-associated endosome fission in animal cells, cargo is sorted into tubules marked by the retromer, sorting nexins and WASH complex protein FAM21. ER tubules are recruited to these sorting domains by an unidentified tether, and fission is rapid following ER recruitment. Note that another ER–endosome MCS regulates dephosphorylation and internalization of epidermal growth factor receptor (EGFR) by ER-localized protein-Tyr phosphatase 1B (PTP1B). Bb. Live confocal fluorescence microscopy images of a Cos7 cell expressing mCherry–RAB7 (late endosome in red) and BFP–FAM21 (late endosome cargo-sorting domain in cyan), merged in the right panel with GFP–SEC61β (ER in green). The arrow marks a MCS between the tip of an ER tubule and the FAM21-labelled sorting domain on the late endosome. Bc. Time-lapsed images of a cell expressing mCherry– RAB7 (late endosome shown in grey in the left panels and red in the right panels) and GFP–SEC61β (ER in green) show ER tubule recruitment to the neck of the late endosome bud (t=5 s, arrow at the constriction), followed by fission (arrow, between t=10 s and t=15 s; bud marked by arrowhead, t=15 s). Scale bars in Ab, Ac and Bb, Bc represent 1 μm. Images in Ab courtesy of Jason Lee, University of Colorado Boulder, USA. Images in part Ac were adapted with permission from REF. 9, AAAS. Images in parts Bb and Bc were adapted with permission from REF. 13, Elsevier.
ER–mitochondria
The central player in mitochondrial fission is the dynamin-related protein DRP1 (Dnm1 in yeast)100–102. These proteins oligomerize into spirals that circumscribe mitochondria and mediate their fisson103,104. In both yeast and mammalian cells, ER MCSs define the position at which this fission machinery will assemble and, consequently, where mitochondrial fission will occur9,16 (FIG. 5a). Notably, the mean diameter of mitochondria is at least twofold larger than the diameter of the DRP1 (Dnm1) spiral assembly103,105,106. However, it has been observed that at points at which ER and mitochondria are in contact, mitochondrial membranes are constricted, and this is where DRP1 spirals preferentially assemble on mitochondria9 (FIG. 5a). These data suggest that ER contact sites play a direct part in mitochondrial membrane constriction and facilitate the recruitment of the machinery that drives mitochondrial fission.
An important question is how this site of constriction and recruitment of fission machinery is defined. In yeast, the majority of ER-marked mitochondrial fission sites colocalize with the position of the nucleoid16 (FIG. 5a). Synergizing nucleoid position, ER contact and fission machinery would help to ensure that upon fission, both daughter mitochondria can inherit nucleoids and ER contact sites. Several layers of tethering complexes would be required to coordinate the position of nucleoids, the inner and outer mitochondrial membranes and the apposing ER, as well as the fission machinery. In yeast, the ER–mitochondrial tethering complex (and potential lipid transfer complex) ERMES also colocalizes with nucleoids at ER-marked constriction and fission sites, suggesting that it may have a role in this process16 (FIG. 5a). In animal cells, the ER–mitochondrial tether responsible for regulating contact at constriction and fission sites has not yet been discovered.
ER–endosome
ER contact sites also mark the positions at which early and late endosomes undergo fission during cargo sorting13 (FIG. 5b). Live-cell imaging of multiple fluorescently tagged components has revealed the order of events during endosome sorting and fission. First, endosomal cargoes are sorted between vacuolar and budding domains. Then, the ER–endosome MCS is assembled at the base of the budding domain, where it colocalizes with puncta of FAM21 (REF. 13) (FIG. 5b). FAM21 is a component of the actin nucleation-promoting WASH complex and interacts with VPS35, a subunit of the retromer cargo-sorting complex, which could potentially recruit the ER to endosome fission sites. Within seconds of ER recruitment, an ER tubule rearranges around the base of the bud, and this rearrangement is accompanied by bud fission (FIG. 5b). When fission is inhibited, ER tubules form stable contacts with stalled constrictions on tubular endosomes13. Thus, the ER seems to have a similar role in constriction and fission of both endosomes and mitochondria.
How do ER contact sites regulate the constriction and fission of two very different organelles? First, MCSs may provide a general platform for the recruitment of cytoskeletal proteins, which then mediate constriction. In mammalian cells, actin–myosin complexes are recruited to ER–mitochondria contact sites by an ER-localized protein, inverted formin 2 (INF2)107,108. Mitochondria are elongated in cells depleted of INF2 and are shorter in the presence of dominant-active INF2 (REF. 107). Consequently, it has been proposed that the assembly of actin–myosin complexes mediated by INF2 drives the initial constriction of the mitochondrial membrane at ER MCSs107,108 (FIG. 5a). ER MCSs could also provide a platform for the recruitment of lipid-modifying enzymes that would work with LTPs to transfer lipids, promoting the acquisition of high membrane curvature at the constriction site. Once formed, such ER-associated constriction sites would recruit the fission machinery, which could be facilitated by the specific association of additional adaptor proteins at these constricted sites. For instance, in mitochondria, mitochondrial fission factor (MFF), which is an adaptor protein for DRP1, colocalizes with the ER-associated constrictions9 and is required for DRP1 recruitment9,109–113. However, what targets such adaptor proteins to the ER MCSs is so far elusive. Furthermore, ER Ca2+ release could have an additional, regulatory role in triggering the completion of the fission process. Currently, these are only speculations, and further studies are needed to better understand the factors and the mechanism that regulate the assembly of ER-marked constriction and fission sites on mitochondria and endosomes.
As well as being involved in fission, ER–endosome MCSs are implicated in regulating sorting and degradation of the EGFR. Endocytosis of ligand-bound EGFR targets it for degradation by the lysosome and thus regulates EGF signalling. Following endocytosis, EGFR is internalized into intra-luminal vesicles (ILVs) that will be degraded upon fusion with the lysosome. EGFR is dephosphorylated on the cytosolic surface of the endosome by the ER-localized protein-Tyr phosphatase 1B (PTP1B)11, and this dephosphorylation event is required for EGFR internalization into ILVs11. Immuno-electron microscopy shows that PTP1B and EGFR colocalize at ER–endosome MCSs11 and, on the basis of the co-immunoprecipitation experiments, it can be suggested that the two proteins may in fact interact directly to bridge the MCS (although they are not required to maintain contact)11. Interestingly, when visualized with the use of fluorescently labelled EGF, EGFR–EGF can be observed to localize to punctate structures along the endosomal membrane, and these puncta colocalize with the positions of ER–endosome contacts13. These MCSs only partially overlap with the sites at which endosome fission events take place, reinforcing the initially postulated idea that the ER is able to form multiple discrete contacts with other organelles and that these discrete contacts are probably not all functionally redundant.
Conclusions
The expansive ER network extends throughout the cell interior to make stable contacts with multiple organelles that are ensnared like flies in a spider’s web. The continuity of the ER and the extensive contacts that it makes with other organelles indicate that this is probably a mechanism that allows various signals to be propagated throughout the ER network, thereby rapidly reaching several contacting organelles and subsequently coordinating a widespread cellular response to a particular cue. In this way, a signal originating from the extracellular space and passing through ER–plasma membrane contact sites could be delivered to contacting organelles and routed back to the plasma membrane in rapid succession, promoting efficient intra- or intercellular signalling. The mitochondrial field has already begun to elucidate the role of cellular signalling throughout the ER with respect to coordinating the apoptotic signalling cascade through the timely release of Ca2+. Unveiling the importance of ER MCSs for other signalling pathways within the cell will provide crucial insight into how the cell coordinates signalling events that require a network response spanning the entire cell. Future work will be aimed at identifying the composition of the MCS machinery, as well as the functional impact of each of the MCSs. This will provide important insight into the role of these complex multimembrane interactions in maintaining cellular health and homeostasis.
Acknowledgments
The authors thank Matt West, Jonathan Friedman, Jason Lee, Ashley Rowland and Patrick Chitwood for images used here, and Laura Westrate for comments on the manuscript. This work was supported by grants from the American Cancer Society and from the U.S. National Institutes of Health (NIH) (GM083977) to G.K.V.. M.J.P. was supported by a U.S. National Science Foundation (NSF) Graduate Research Fellowship (DGE 1144083) and by a pre-doctoral training grant from the NIH (T32 GM08759).
Glossary
- Peripheral ER
The ER network that spans from the perinuclear region of the cell to the cell periphery
- ER sliding
ER tubules attach to a motor protein on a stable microtubule. The motor protein then pulls the ER tubule along the microtubule
- Early endosomes
Endosomes that have been recently internalized into cells and labelled with RAB5 GTPase, have a relatively low pH, and have not further internalized cargo, such as signalling receptors, from the plasma membrane into intraluminal vesicles
- Late endosomes
Mature endosomes that have not yet fused with the lysosome. These endosomes are labelled with RAB7 GTPase, have a relatively high pH, and have abundant intraluminal vesicles internalized into the lumen for easier degradation of cargo when the late endosome fuses with the lysosome
- Cortical ER
Peripheral ER that is found directly underneath and tethered to the plasma membrane
- Microsomes
ER vesicles resulting from the breakage of the ER network as the ER is isolated from cells
- Nucleoid
Mitochondrial DNA associated with proteins that compact into one region of the mitochondrion
Footnotes
Competing interests statement
The authors declare no competing interests.
References
- 1.Fawcett DW. The Cell. W. B. Saunders; 1981. [Google Scholar]
- 2.Ogata T, Yamasaki Y. Ultra-high-resolution scanning electron microscopy of mitochondria and sarcoplasmic reticulum arrangement in human red, white, and intermediate muscle fibers. Anat Rec. 1997;248:214–223. doi: 10.1002/(SICI)1097-0185(199706)248:2<214::AID-AR8>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 3.Rolls MM, Hall DH, Victor M, Stelzer EHK, Rapoport TA. Targeting of rough endoplasmic reticulum membrane proteins and ribosomes in invertebrate neurons. Mol Biol Cell. 2002;13:1778–1791. doi: 10.1091/mbc.01-10-0514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shibata Y, Voeltz GK, Rapoport TA. Rough sheets and smooth tubules. Cell. 2006;126:435–439. doi: 10.1016/j.cell.2006.07.019. [DOI] [PubMed] [Google Scholar]
- 5.Shibata Y, et al. Mechanisms determining the morphology of the peripheral ER. Cell. 2010;143:774–788. doi: 10.1016/j.cell.2010.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.West M, Zurek N, Hoenger A, Voeltz GK. A 3D analysis of yeast ER structure reveals how ER domains are organized by membrane curvature. J Cell Biol. 2011;193:333–346. doi: 10.1083/jcb.201011039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alpy F, et al. STARD3 or STARD3NL and VAP form a novel molecular tether between late endosomes and the ER. J Cell Sci. 2013;126:5500–5512. doi: 10.1242/jcs.139295. Measured ER–late endosome contact site distance using electron microscopy. Showed that the STARD3 and STARD3NL FFAT domain can interact with ER VAP proteins. Overexpression of STARD3 resulted in expansion of ER–endosome contact sites. [DOI] [PubMed] [Google Scholar]
- 8.Csordás G, et al. Structural and functional features and significance of the physical linkage between ER and mitochondria. J Cell Biol. 2006;174:915–921. doi: 10.1083/jcb.200604016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Friedman JR, et al. ER tubules mark sites of mitochondrial division. Science. 2011;334:358–362. doi: 10.1126/science.1207385. Demonstrated that ER tubules mark the site of mitochondrial division and that ER contact occurs prior to recruitment of the mammalian division machinery DRP1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Friedman JR, Dibenedetto JR, West M, Rowland AA, Voeltz GK. Endoplasmic reticulum– endosome contact increases as endosomes traffic and mature. Mol Biol Cell. 2013;24:1030–1040. doi: 10.1091/mbc.E12-10-0733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eden ER, White IJ, Tsapara A, Futter CE. Membrane contacts between endosomes and ER provide sites for PTP1B–epidermal growth factor receptor interaction. Nat Cell Biol. 2010;12:267–272. doi: 10.1038/ncb2026. [DOI] [PubMed] [Google Scholar]
- 12.Swayne TC, et al. Role for cER and Mmr1p in anchorage of mitochondria at sites of polarized surface growth in budding yeast. Curr Biol. 2011;21:1994–1999. doi: 10.1016/j.cub.2011.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rowland AA, Chitwood PJ, Phillips MJ, Voeltz GK. ER contact sites define the position and timing of endosome fission. Cell. 2014;159:1027–1041. doi: 10.1016/j.cell.2014.10.023. Demonstrated that ER tubules are recruited to pre-established endosome sorting domains that undergo fission to sort cargo, and that ER dynamics are required for endosome fission. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kornmann B, et al. An ER–mitochondria tethering complex revealed by a synthetic biology screen. Science. 2009;325:477–481. doi: 10.1126/science.1175088. Performed a yeast screen for mutants that could be rescued by an artificial ER–mitochondria tether. Identified a role for the ERMES complex in ER– mitochondria tethering in yeast. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cosson P, Marchetti A, Ravazzola M, Orci L. Mitofusin-2 independent juxtaposition of endoplasmic reticulum and mitochondria: an ultrastructural study. PLoS ONE. 2012;7:e46293. doi: 10.1371/journal.pone.0046293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Murley A, et al. ER-associated mitochondrial division links the distribution of mitochondria and mitochondrial DNA in yeast. eLife. 2013;2:e00422. doi: 10.7554/eLife.00422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zajac AL, Goldman YE, Holzbaur ELF, Ostap EM. Local cytoskeletal and organelle interactions impact molecular-motor-driven early endosomal trafficking. Curr Biol. 2013;23:1173–1180. doi: 10.1016/j.cub.2013.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.WoŸniak MJ, et al. Role of kinesin-1 and cytoplasmic dynein in endoplasmic reticulum movement in VERO cells. J Cell Sci. 2009;122:1979–1989. doi: 10.1242/jcs.041962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoepfner S, et al. Modulation of receptor recycling and degradation by the endosomal kinesin KIF16B. Cell. 2005;121:437–450. doi: 10.1016/j.cell.2005.02.017. [DOI] [PubMed] [Google Scholar]
- 20.Hurd DD, Saxton WM. Kinesin mutations cause motor neuron disease phenotypes by disrupting fast axonal transport in Drosophila. Genetics. 1996;144:1075–1085. doi: 10.1093/genetics/144.3.1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Glater EE, Megeath LJ, Stowers RS, Schwarz TL. Axonal transport of mitochondria requires milton to recruit kinesin heavy chain and is light chain independent. J Cell Biol. 2006;173:545–557. doi: 10.1083/jcb.200601067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tanaka Y, et al. Targeted disruption of mouse conventional kinesin heavy chain, kif5B, results in abnormal perinuclear clustering of mitochondria. Cell. 1998;93:1147–1158. doi: 10.1016/s0092-8674(00)81459-2. [DOI] [PubMed] [Google Scholar]
- 23.Kornmann B, Osman C, Walter P. The conserved GTPase Gem1 regulates endoplasmic reticulum– mitochondria connections. Proc Natl Acad Sci USA. 2011;108:14151–14156. doi: 10.1073/pnas.1111314108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stowers RS, Megeath LJ, Górska-Andrzejak J, Meinertzhagen IA, Schwarz TL. Axonal transport of mitochondria to synapses depends on Milton, a novel Drosophila protein. Neuron. 2002;36:1063–1077. doi: 10.1016/s0896-6273(02)01094-2. [DOI] [PubMed] [Google Scholar]
- 25.Saotome M, et al. Bidirectional Ca2+-dependent control of mitochondrial dynamics by the Miro GTPase. Proc Natl Acad Sci USA. 2008;105:20728–20733. doi: 10.1073/pnas.0808953105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huotari J, Helenius A. Endosome maturation. EMBO J. 2011;30:3481–3500. doi: 10.1038/emboj.2011.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vihervaara T, et al. Sterol binding by OSBP-related protein 1L regulates late endosome motility and function. Cell Mol Life Sci. 2011;68:537–551. doi: 10.1007/s00018-010-0470-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rocha N, et al. Cholesterol sensor ORP1L contacts the ER protein VAP to control Rab7–RILP–p150Glued and late endosome positioning. J Cell Biol. 2009;185:1209–1225. doi: 10.1083/jcb.200811005. Discovered that late-endosome-localized ORP1L interacts with ER membrane protein VAP when cholesterol levels are low in the late endosome membrane. ORP1L–VAP interaction inhibits dynein-directed positioning of late endosomes to the cell centre, resulting in late endosomes in the cell periphery. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Suchanek M, et al. The mammalian oxysterol-binding protein-related proteins (ORPs) bind 25-hydroxycholesterol in an evolutionarily conserved pocket. Biochem J. 2007;405:473–480. doi: 10.1042/BJ20070176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johansson M, et al. Activation of endosomal dynein motors by stepwise assembly of Rab7–RILP–p150Glued, ORP1L, and the receptor βIII spectrin. J Cell Biol. 2007;176:459–471. doi: 10.1083/jcb.200606077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tsujishita Y, Hurley JH. Structure and lipid transport mechanism of a StAR-related domain. Nat Struct Biol. 2000;7:408–414. doi: 10.1038/75192. [DOI] [PubMed] [Google Scholar]
- 32.Hölttä-Vuori M, et al. MLN64 is involved in actin-mediated dynamics of late endocytic organelles. Mol Biol Cell. 2005;16:3873–3886. doi: 10.1091/mbc.E04-12-1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chang J, Lee S, Blackstone C. Protrudin binds atlastins and endoplasmic reticulum-shaping proteins and regulates network formation. Proc Natl Acad Sci USA. 2013;110:14954–14959. doi: 10.1073/pnas.1307391110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Raiborg C, et al. Repeated ER–endosome contacts promote endosome translocation and neurite outgrowth. Nature. 2015;520:234–238. doi: 10.1038/nature14359. Analysis of protrudin domains showed that protrudin interacts with the late endosome through PtdIns(3)P and RAB7, creating an ER–late endosome MCS. When the ER–late endosome MCS is formed, protrudin delivers kinesin-1 to FYCO1, which links the kinesin-1 to the late endosome RAB7. This promotes trafficking of late endosomes to the cell exterior. [DOI] [PubMed] [Google Scholar]
- 35.Pankiv S, et al. FYCO1 is a Rab7 effector that binds to LC3 and PI3P to mediate microtubule plus end-directed vesicle transport. J Cell Biol. 2010;188:253–269. doi: 10.1083/jcb.200907015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Matsuzaki F, Shirane M, Matsumoto M, Nakayama KI. Protrudin serves as an adaptor molecule that connects KIF5 and its cargoes in vesicular transport during process formation. Mol Biol Cell. 2011;22:4602–4620. doi: 10.1091/mbc.E11-01-0068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vance JE, Aasman EJ, Szarka R. Brefeldin A does not inhibit the movement of phosphatidylethanolamine from its sites for synthesis to the cell surface. J Biol Chem. 1991;266:8241–8247. [PubMed] [Google Scholar]
- 38.Wirtz KW, Zilversmit DB. Exchange of phospholipids between liver mitochondria and microsomes in vitro. J Biol Chem. 1968;243:3596–3602. [PubMed] [Google Scholar]
- 39.Lev S. Non-vesicular lipid transport by lipid-transfer proteins and beyond. Nat Rev Mol Cell Biol. 2010;11:739–750. doi: 10.1038/nrm2971. [DOI] [PubMed] [Google Scholar]
- 40.Im YJ, Raychaudhuri S, Prinz WA, Hurley JH. Structural mechanism for sterol sensing and transport by OSBP-related proteins. Nature. 2005;437:154–158. doi: 10.1038/nature03923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kopec KO, Alva V, Lupas AN. Bioinformatics of the TULIP domain superfamily. Biochem Soc Trans. 2011;39:1033–1038. doi: 10.1042/BST0391033. [DOI] [PubMed] [Google Scholar]
- 42.Dennis EA, Kennedy EP. Intracellular sites of lipid synthesis and the biogenesis of mitochondria. J Lipid Res. 1972;13:263–267. [PubMed] [Google Scholar]
- 43.Osman C, Voelker DR, Langer T. Making heads or tails of phospholipids in mitochondria. J Cell Biol. 2011;192:7–16. doi: 10.1083/jcb.201006159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Toulmay A, Prinz WA. A conserved membrane-binding domain targets proteins to organelle contact sites. J Cell Sci. 2012;125:49–58. doi: 10.1242/jcs.085118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schauder CM, et al. Structure of a lipid-bound extended synaptotagmin indicates a role in lipid transfer. Nature. 2014;510:552–555. doi: 10.1038/nature13269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Osman C, et al. The genetic interactome of prohibitins: coordinated control of cardiolipin and phosphatidylethanolamine by conserved regulators in mitochondria. J Cell Biol. 2009;184:583–596. doi: 10.1083/jcb.200810189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tamura Y, et al. Role for two conserved intermembrane space proteins, Ups1p and Ups2p, [corrected] in intra-mitochondrial phospholipid trafficking. J Biol Chem. 2012;287:15205–15218. doi: 10.1074/jbc.M111.338665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tan T, Ozbalci C, Brügger B, Rapaport D, Dimmer KS. Mcp1 and Mcp2, two novel proteins involved in mitochondrial lipid homeostasis. J Cell Sci. 2013;126:3563–3574. doi: 10.1242/jcs.121244. [DOI] [PubMed] [Google Scholar]
- 49.Nguyen TT, et al. Gem1 and ERMES do not directly affect phosphatidylserine transport from ER to mitochondria or mitochondrial inheritance. Traffic. 2012;13:880–890. doi: 10.1111/j.1600-0854.2012.01352.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Voss C, Lahiri S, Young BP, Loewen CJ, Prinz WA. ER-shaping proteins facilitate lipid exchange between the ER and mitochondria in S. cerevisiae. J Cell Sci. 2012;125:4791–4799. doi: 10.1242/jcs.105635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Elbaz-Alon Y, et al. A dynamic interface between vacuoles and mitochondria in yeast. Dev Cell. 2014;30:95–102. doi: 10.1016/j.devcel.2014.06.007. [DOI] [PubMed] [Google Scholar]
- 52.Hönscher C, et al. Cellular metabolism regulates contact sites between vacuoles and mitochondria. Dev Cell. 2014;30:86–94. doi: 10.1016/j.devcel.2014.06.006. [DOI] [PubMed] [Google Scholar]
- 53.Möbius W, et al. Recycling compartments and the internal vesicles of multivesicular bodies harbor most of the cholesterol found in the endocytic pathway. Traffic. 2003;4:222–231. doi: 10.1034/j.1600-0854.2003.00072.x. [DOI] [PubMed] [Google Scholar]
- 54.Neufeld EB, et al. Intracellular trafficking of cholesterol monitored with a cyclodextrin. J Biol Chem. 1996;271:21604–21613. doi: 10.1074/jbc.271.35.21604. [DOI] [PubMed] [Google Scholar]
- 55.Liscum L, Ruggiero RM, Faust JR. The intracellular transport of low density lipoprotein-derived cholesterol is defective in Niemann-Pick type C fibroblasts. J Cell Biol. 1989;108:1625–1636. doi: 10.1083/jcb.108.5.1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Infante RE, et al. NPC2 facilitates bidirectional transfer of cholesterol between NPC1 and lipid bilayers, a step in cholesterol egress from lysosomes. Proc Natl Acad Sci USA. 2008;105:15287–15292. doi: 10.1073/pnas.0807328105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kwon HJ, et al. Structure of N-terminal domain of NPC1 reveals distinct subdomains for binding and transfer of cholesterol. Cell. 2009;137:1213–1224. doi: 10.1016/j.cell.2009.03.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Du X, et al. A role for oxysterol-binding protein-related protein 5 in endosomal cholesterol trafficking. J Cell Biol. 2011;192:121–135. doi: 10.1083/jcb.201004142. Demonstrated that depletion of tail-anchored ER protein ORP5 resulted in cholesterol accumulation in the external membranes of late endosomes, leading to the model in which ORP5 accepts cholesterol from late endosome NPC1 and transfers it to the ER. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Van der Kant R, Zondervan I, Janssen L, Neefjes J. Cholesterol-binding molecules MLN64 and ORP1L mark distinct late endosomes with transporters ABCA3 and NPC1. J Lipid Res. 2013;54:2153–2165. doi: 10.1194/jlr.M037325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Peretti D, Dahan N, Shimoni E, Hirschberg K, Lev S. Coordinated lipid transfer between the endoplasmic reticulum and the Golgi complex requires the VAP proteins and is essential for Golgi-mediated transport. Mol Biol Cell. 2008;19:3871–3884. doi: 10.1091/mbc.E08-05-0498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hanada K, et al. Molecular machinery for non-vesicular trafficking of ceramide. Nature. 2003;426:803–809. doi: 10.1038/nature02188. [DOI] [PubMed] [Google Scholar]
- 62.D’Angelo G, et al. Glycosphingolipid synthesis requires FAPP2 transfer of glucosylceramide. Nature. 2007;449:62–67. doi: 10.1038/nature06097. [DOI] [PubMed] [Google Scholar]
- 63.Litvak V, Dahan N, Ramachandran S, Sabanay H, Lev S. Maintenance of the diacylglycerol level in the Golgi apparatus by the Nir2 protein is critical for Golgi secretory function. Nat Cell Biol. 2005;7:225–234. doi: 10.1038/ncb1221. [DOI] [PubMed] [Google Scholar]
- 64.Mesmin B, et al. A four-step cycle driven by PI(4)P hydrolysis directs sterol/PI(4)P exchange by the ER–Golgi tether OSBP. Cell. 2013;155:830–843. doi: 10.1016/j.cell.2013.09.056. Demonstrated that OSBP binding to PtdIns(4)P localizes OSBP to the Golgi. OSBP moves sterol from the ER to the Golgi. OSBP moves PtdIns(4)P to the ER, where it is hydrolysed. Depletion of PtdIns(4)P from the Golgi membrane results in OSBP dissociation from the Golgi membrane. [DOI] [PubMed] [Google Scholar]
- 65.Perry RJ, Ridgway ND. Oxysterol-binding protein and vesicle-associated membrane protein-associated protein are required for sterol-dependent activation of the ceramide transport protein. Mol Biol Cell. 2006;17:2604–2616. doi: 10.1091/mbc.E06-01-0060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Loewen CJR, Roy A, Levine TP. A conserved ER targeting motif in three families of lipid binding proteins and in Opi1p binds VAP. EMBO J. 2003;22:2025–2035. doi: 10.1093/emboj/cdg201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lev S, Ben Halevy D, Peretti D, Dahan N. The VAP protein family: from cellular functions to motor neuron disease. Trends Cell Biol. 2008;18:282–290. doi: 10.1016/j.tcb.2008.03.006. [DOI] [PubMed] [Google Scholar]
- 68.Amarilio R, Ramachandran S, Sabanay H, Lev S. Differential regulation of endoplasmic reticulum structure through VAP–Nir protein interaction. J Biol Chem. 2005;280:5934–5944. doi: 10.1074/jbc.M409566200. [DOI] [PubMed] [Google Scholar]
- 69.Foskett JK, White C, Cheung K, Mak DD. Inositol trisphosphate receptor Ca2+ release channels. Physiol Rev. 2007;87:593–658. doi: 10.1152/physrev.00035.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tovey SC, Dedos SG, Taylor EJA, Church JE, Taylor CW. Selective coupling of type 6 adenylyl cyclase with type 2 IP3 receptors mediates direct sensitization of IP3 receptors by cAMP. J Cell Biol. 2008;183:297–311. doi: 10.1083/jcb.200803172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Taylor CW, Tovey SC. IP3 receptors: toward understanding their activation. Cold Spring Harb Perspect Biol. 2010;2:a004010. doi: 10.1101/cshperspect.a004010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shuai J, Parker I. Optical single-channel recording by imaging Ca2+ flux through individual ion channels: theoretical considerations and limits to resolution. Cell Calcium. 2005;37:283–299. doi: 10.1016/j.ceca.2004.10.008. [DOI] [PubMed] [Google Scholar]
- 73.Rizzuto R, et al. Close contacts with the endoplasmic reticulum as determinants of mitochondrial Ca2+ responses. Science. 1998;280:1763–1766. doi: 10.1126/science.280.5370.1763. [DOI] [PubMed] [Google Scholar]
- 74.Morgan AJ, et al. Bidirectional Ca2+ signaling occurs between the endoplasmic reticulum and acidic organelles. J Cell Biol. 2013;200:789–805. doi: 10.1083/jcb.201204078. Demonstrated that stimulated ER Ca2+ release can activate NAADP-regulated channels on the lysosome and result in Ca2+ release from lysosomes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.De Stefani D, Raffaello A, Teardo E, Szabò I, Rizzuto R. A forty-kilodalton protein of the inner membrane is the mitochondrial calcium uniporter. Nature. 2011;476:336–340. doi: 10.1038/nature10230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Baughman JM, et al. Integrative genomics identifies MCU as an essential component of the mitochondrial calcium uniporter. Nature. 2011;476:341–345. doi: 10.1038/nature10234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rizzuto R, De Stefani D, Raffaello A, Mammucari C. Mitochondria as sensors and regulators of calcium signalling. Nat Rev Mol Cell Biol. 2012;13:566–578. doi: 10.1038/nrm3412. [DOI] [PubMed] [Google Scholar]
- 78.Scorrano L, et al. BAX and BAK regulation of endoplasmic reticulum Ca2+: a control point for apoptosis. Science. 2003;300:135–139. doi: 10.1126/science.1081208. [DOI] [PubMed] [Google Scholar]
- 79.Zong WX, et al. Bax and Bak can localize to the endoplasmic reticulum to initiate apoptosis. J Cell Biol. 2003;162:59–69. doi: 10.1083/jcb.200302084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rizzuto R, Brini M, Murgia M, Pozzan T. Microdomains with high Ca2+ close to IP3-sensitive channels that are sensed by neighboring mitochondria. Science. 1993;262:744–747. doi: 10.1126/science.8235595. [DOI] [PubMed] [Google Scholar]
- 81.Csordás G, et al. Imaging interorganelle contacts and local calcium dynamics at the ER–mitochondrial interface. Mol Cell. 2010;39:121–132. doi: 10.1016/j.molcel.2010.06.029. Adjusted ER–mitochondria contact site distance using artificial tethers and showed that distance between ER and mitochondria affects Ca2+ transfer at the ER–mitochondria MCS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.De Brito OM, Scorrano L. Mitofusin 2 tethers endoplasmic reticulum to mitochondria. Nature. 2008;456:605–610. doi: 10.1038/nature07534. [DOI] [PubMed] [Google Scholar]
- 83.Filadi R, et al. Mitofusin 2 ablation increases endoplasmic reticulum–mitochondria coupling. Proc Natl Acad Sci USA. 2015;112:E2174–E2181. doi: 10.1073/pnas.1504880112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Szabadkai G, et al. Chaperone-mediated coupling of endoplasmic reticulum and mitochondrial Ca2+ channels. J Cell Biol. 2006;175:901–911. doi: 10.1083/jcb.200608073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Giorgi C, et al. PML regulates apoptosis at endoplasmic reticulum by modulating calcium release. Science. 2010;330:1247–1251. doi: 10.1126/science.1189157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Marchi S, et al. Akt kinase reducing endoplasmic reticulum Ca2+ release protects cells from Ca2+-dependent apoptotic stimuli. Biochem Biophys Res Commun. 2008;375:501–505. doi: 10.1016/j.bbrc.2008.07.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Marchi S, et al. Selective modulation of subtype III IP3R by Akt regulates ER Ca2+ release and apoptosis. Cell Death Dis. 2012;3:e304. doi: 10.1038/cddis.2012.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gerasimenko JV, Tepikin aV, Petersen OH, Gerasimenko OV. Calcium uptake via endocytosis with rapid release from acidifying endosomes. Curr Biol. 1998;8:1335–1338. doi: 10.1016/s0960-9822(07)00565-9. [DOI] [PubMed] [Google Scholar]
- 89.Pryor PR, Mullock BM, Bright NA, Gray SR, Luzio JP. The role of intraorganellar Ca2+ in late endosome–lysosome heterotypic fusion and in the reformation of lysosomes from hybrid organelles. J Cell Biol. 2000;149:1053–1062. doi: 10.1083/jcb.149.5.1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Morgan AJ, Platt FM, Lloyd-Evans E, Galione A. Molecular mechanisms of endolysosomal Ca2+ signalling in health and disease. Biochem J. 2011;439:349–374. doi: 10.1042/BJ20110949. [DOI] [PubMed] [Google Scholar]
- 91.Albrecht T, Zhao Y, Nguyen TH, Campbell RE, Johnson JD. Fluorescent biosensors illuminate calcium levels within defined beta-cell endosome subpopulations. Cell Calcium. 2015;57:263–274. doi: 10.1016/j.ceca.2015.01.008. [DOI] [PubMed] [Google Scholar]
- 92.Christensen KA, Myers JT, Swanson J. A pH-dependent regulation of lysosomal calcium in macrophages. 2002;115:599–607. doi: 10.1242/jcs.115.3.599. [DOI] [PubMed] [Google Scholar]
- 93.Lloyd-Evans E, et al. Niemann-Pick disease type C1 is a sphingosine storage disease that causes deregulation of lysosomal calcium. Nat Med. 2008;14:1247–1255. doi: 10.1038/nm.1876. [DOI] [PubMed] [Google Scholar]
- 94.Abe K, Puertollano R. Role of TRP channels in the regulation of the endosomal pathway. Physiology. 2011;26:14–22. doi: 10.1152/physiol.00048.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lelouvier B, Puertollano R. Mucolipin-3 regulates luminal calcium, acidification, and membrane fusion in the endosomal pathway. J Biol Chem. 2011;286:9826–9832. doi: 10.1074/jbc.M110.169185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ruas M, et al. Purified TPC isoforms form NAADP receptors with distinct roles for Ca2+ signaling and endolysosomal trafficking. Curr Biol. 2010;20:703–709. doi: 10.1016/j.cub.2010.02.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kilpatrick BS, Eden ER, Schapira AH, Futter CE, Patel S. Direct mobilisation of lysosomal Ca2+ triggers complex Ca2+ signals. J Cell Sci. 2013;126:60–66. doi: 10.1242/jcs.118836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.López-Sanjurjo CI, Tovey SC, Prole DL, Taylor CW. Lysosomes shape Ins(1,4,5)P3-evoked Ca2+ signals by selectively sequestering Ca2+ released from the endoplasmic reticulum. J Cell Sci. 2013;126:289–300. doi: 10.1242/jcs.116103. Measured pH-adjusted Ca2+ levels in the lysosome and demonstrated that Ca2+ levels increase in the lysosome upon ER Ins(1,4,5)P3R stimulation, leading to the idea that lysosomes can sequester ER Ca2+ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Nunnari J, Suomalainen A. Mitochondria: in sickness and in health. Cell. 2012;148:1145–1159. doi: 10.1016/j.cell.2012.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Otsuga D, et al. The dynamin-related GTPase, Dnm1p, controls mitochondrial morphology in yeast. J Cell Biol. 1998;143:333–349. doi: 10.1083/jcb.143.2.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Bleazard W, et al. The dynamin-related GTPase Dnm1 regulates mitochondrial fission in yeast. Nat Cell Biol. 1999;1:298–304. doi: 10.1038/13014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Smirnova E, Griparic L, Shurland DL, van der Bliek AM. Dynamin-related protein Drp1 is required for mitochondrial division in mammalian cells. Mol Biol Cell. 2001;12:2245–2256. doi: 10.1091/mbc.12.8.2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Mears JA, et al. Conformational changes in Dnm1 support a contractile mechanism for mitochondrial fission. Nat Struct Mol Biol. 2011;18:20–26. doi: 10.1038/nsmb.1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ingerman E, et al. Dnm1 forms spirals that are structurally tailored to fit mitochondria. J Cell Biol. 2005;170:1021–1027. doi: 10.1083/jcb.200506078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Yoon Y, Pitts KR, McNiven MA. Mammalian dynamin-like protein DLP1 tubulates membranes. Mol Biol Cell. 2001;12:2894–2905. doi: 10.1091/mbc.12.9.2894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Legesse-Miller A, Massol RH, Kirchhausen T. Constriction and Dnm1p recruitment are distinct processes in mitochondrial fission. Mol Biol Cell. 2003;14:1953–1963. doi: 10.1091/mbc.E02-10-0657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Korobova F, Ramabhadran V, Higgs HN. An actin-dependent step in mitochondrial fission mediated by the ER-associated formin INF2. Science. 2013;339:464–467. doi: 10.1126/science.1228360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Korobova F, Gauvin TJ, Higgs HN. A role for myosin II in mammalian mitochondrial fission. Curr Biol. 2014;24:409–414. doi: 10.1016/j.cub.2013.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Losón OC, Song Z, Chen H, Chan DC. Fis1, Mff, MiD49, and MiD51 mediate Drp1 recruitment in mitochondrial fission. Mol Biol Cell. 2013;24:659–667. doi: 10.1091/mbc.E12-10-0721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Tieu Q, Nunnari J. Mdv1p is a WD repeat protein that interacts with the dynamin-related GTPase, Dnm1p, to trigger mitochondrial division. J Cell Biol. 2000;151:353–366. doi: 10.1083/jcb.151.2.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Gandre-Babbe S, van der Bliek AM. The novel tail-anchored membrane protein Mff controls mitochondrial and peroxisomal fission in mammalian cells. Mol Biol Cell. 2008;19:2402–2412. doi: 10.1091/mbc.E07-12-1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Palmer CS, et al. MiD49 and MiD51, new components of the mitochondrial fission machinery. EMBO Rep. 2011;12:565–573. doi: 10.1038/embor.2011.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Mozdy AD, McCaffery JM, Shaw JM. Dnm1p GTPase-mediated mitochondrial fission is a multi-step process requiring the novel integral membrane component Fis1p. J Cell Biol. 2000;151:367–380. doi: 10.1083/jcb.151.2.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.De Vos KJ, et al. VAPB interacts with the mitochondrial protein PTPIP51 to regulate calcium homeostasis. Hum Mol Genet. 2012;21:1299–1311. doi: 10.1093/hmg/ddr559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Iwasawa R, Mahul-Mellier AL, Datler C, Pazarentzos E, Grimm S. Fis1 and Bap31 bridge the mitochondria–ER interface to establish a platform for apoptosis induction. EMBO J. 2011;30:556–568. doi: 10.1038/emboj.2010.346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Palande K, et al. Peroxiredoxin-controlled G-CSF signalling at the endoplasmic reticulum–early endosome interface. J Cell Sci. 2011;124:3695–3705. doi: 10.1242/jcs.089656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Xu N, et al. The FATP1–DGAT2 complex facilitates lipid droplet expansion at the ER–lipid droplet interface. J Cell Biol. 2012;198:895–911. doi: 10.1083/jcb.201201139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Knoblach B, et al. An ER–peroxisome tether exerts peroxisome population control in yeast. EMBO J. 2013;32:2439–2453. doi: 10.1038/emboj.2013.170. [DOI] [PMC free article] [PubMed] [Google Scholar]